Abstract
The PI3K pathway is one of the most commonly misregulated signaling pathways in human cancers, but its impact on the tumor microenvironment has not been considered as deeply as its autonomous impact on tumor cells. In this study we demonstrate that NF-κB is activated by the two most common PI3K mutations, PIK3CA E545K and H1047R. We found that markers of NF-κB are most strongly upregulated under conditions of growth factor deprivation. Gene expression analysis performed on cells deprived of growth factors identified the repertoire of genes altered by oncogenic PI3K mutations following growth factor deprivation. This gene set most closely correlated with gene signatures from claudin-low and basal-like breast tumors, subtypes frequently exhibiting constitutive PI3K/Akt activity. An NF-κB-dependent subset of genes driven by oncogenic PI3K mutations was also identified that encoded primarily secreted proteins, suggesting a paracrine role for this gene set. Interestingly, while NF-κB activated by oncogenes such as Ras and EGFR leads to cell-autonomous effects, abrogating NF-κB in PI3K-transformed cells did not decrease proliferation or induce apoptosis. However, conditioned media from PI3K mutant-expressing cells led to increased STAT3 activation in recipient THP-1 monocytes or normal epithelial cells in a NF-κB and IL-6-dependent manner. Together, our findings describe a PI3K-driven, NF-κB-dependent transcriptional profile which may play a critical role in promoting a microenvironment amenable to tumor progression. These data also indicate that NF-κB plays diverse roles downstream from different oncogenic signaling pathways.
Introduction
Phosphatidylinositol 3-kinase (PI3K) is an essential mediator of cellular processes critical to tumorigenesis such as growth, survival, and cell proliferation (1, 2). Consistent with these roles, the PI3K pathway is one of the most commonly misregulated signaling pathways in human cancers and the PI3K catalytic subunit (p110α, PIK3CA) itself is frequently amplified or mutated in cancer (2–5). In fact, about 30% of breast cancer samples harbor mutations in PIK3CA (3, 6), of which the most common are E545K and H1047R. These sites lie in mutational hotspots that account for >85% of oncogenic PIK3CA mutations (3, 6). Tissue-specific expression of the H1047R mutation in transgenic mice leads to both breast and lung tumors (7–9). In addition, both the E545K and H1047R mutations induce cell transformation characterized by growth factor independent AKT signaling and proliferation, as well as colony growth in soft agar, when expressed in a variety of cell types, including MCF10A cells (10–12).
The IKK/NF-κB pathway is a critical signaling axis which regulates diverse cellular functions such as inflammation, cell survival, proliferation, and senescence (13–15). Canonical NF-κB activation requires activation of the IκB kinase (IKK) complex, which phosphorylates the NF-κB inhibitory protein IκB, leading to IκB proteasomal degradation and nuclear accumulation of activated NF-κB heterodimers (14). NF-κB DNA binding is greatly enhanced by phosphorylation of the NF-κB subunit p65. NF-κB is frequently activated in a variety of solid tumors including melanoma and breast, prostate, and liver tumors (16–18). In addition, a number of oncogenic signaling pathways, such as Ras, EGFR, and HER2, depend on NF-κB for full transformation potential (19–23). NF-κB activities involved in oncogenesis are hypothesized to result from transcriptional upregulation of a large number of well-characterized anti-apoptotic and pro-proliferation NF-κB target genes. However, no comprehensive oncogenic NF-κB-dependent transcriptional profile has been identified.
IL-6 is a well-established NF-κB target gene and a critical inflammatory mediator, as its secretion potently activates T cells and macrophages (24). However, IL-6 is also associated with increased cancer cell proliferation and tumor progression (24, 25). In fact, expression of IL-6 in the serum or tissue of cancer patients correlates with poor prognosis (24, 25). Binding of IL-6 to its receptor activates JAK tyrosine kinases, which promote phosphorylation and nuclear translocation of STAT3 transcription factors. Many tumor types such as breast, colon, lung, and prostate, are characterized by increased STAT3 activation (26–28).
Here, we use MCF10A cells expressing PIK3CA E545K or H1047R to show that NF-κB is activated by these oncogenic mutations. Interestingly, while modest NF-κB activation was identified under growth conditions, markers of NF-κB activation were most dramatically increased under conditions of growth factor (GF) deprivation. While GF-independent signaling and proliferation are well-established hallmarks of cancer (29), few studies of cancer cells are performed under conditions of GF deprivation, and little is known regarding GF-independent gene expression changes occurring downstream from oncogenes. We therefore identified both a comprehensive transcriptional profile and an NF-κB-dependent gene set activated by the E545K and H1047R mutations. NF-κB activity downstream from EGFR and Ras plays an important role in cancer cell survival and proliferation (19, 20, 23). In contrast, we show that NF-κB activity driven by oncogenic PIK3CA mutations does not increase proliferation, survival, or anchorage-independent growth of the transformed cells. Rather, the primary function of PI3K-driven NF-κB activation is to promote expression and secretion of cytokines and chemokines, especially IL-6, which act in a paracrine and autocrine manner to activate STAT3 in nearby monocytes and normal epithelial cells. Together, these data support a model in which PI3K-driven NF-κB activation leads to increased activation of nearby stromal cells, helping to generate a pro-tumor microenvironment and facilitate tumor progression. In addition, these data suggest the exciting possibility that the role of NF-κB may vary dramatically downstream from different oncogenes and under different growth/growth factor conditions.
Materials and Methods
Antibodies, Plasmids, and Reagents
pJP1520-HA-GFP, pJP1520-HA-PIK3CA WT, pJP1520-HA-PIK3CA E545K, and p-JP1520-HA-PIK3CA H1047R, were a generous gift from Dr. Lewis Cantley (10). Anti-Phospho-AKT, anti-phospho-ERK, anti-phospho-p65, anti-phospho-IKK, anti-phospho-IκBα, anti-phospho-STAT3, anti-AKT, anti-p65, anti-IκBα, and anti-STAT3 were from Cell Signaling Technology. Anti-HA was from Covance. Anti-Actin was from Abcam. TaqMan real-time RT-PCR gene expression assays were from Applied Biosystems. IL-6 ELISA assays were from BD Biosciences. CXCL1 ELISA assays and the IL-6 receptor monoclonal antibody were from R&D Systems. LY294002 and UO126 were obtained from Cell Signaling Technology. BAY-65-1942 was from Bayer Pharmaceuticals.
Microarray Analysis
Total RNA was purified, reverse-transcribed, labeled, and hybridized to a custom Agilent 4X44K whole human DNA microarray as described previously (30). Microarrays were scanned using an Agilent DNA microarray scanner and features were extracted using Agilent Feature Extraction software version 10.7.3.1. Data was uploaded to the University of North Carolina Microarray Database (UMD) and to the Gene Expression Omnibus (GEO) under accession number GEO:GSE33403. Gene expression data was extracted from the UMD for each sample as log2 Cy5/Cy3 ratios, filtering for probes with Lowess normalized intensity values greater than 10 in both channels and for probes with data on greater than 70% of the microarrays. Hierarchical clustering was performed using Gene Cluster 3.0 (31) and data was viewed using Java Treeview version 1.1.5r2 (32). Expression changes were determined using a 1-Class significance analysis of microarray (SAM) analysis for each experimental set of conditions using the maximum number of permutations (33). In each case, on a single microarray, GFP control was compared to PIK3CA WT, PIK3CA E545K, or PIK3CA H1047R. Probes with a false discovery rate (FDR) of 0% were considered statistically significant. Categorical enrichment of the top 300 PIK3CA mutant induced genes (based on average fold-change) was determined using EASE version 2.0 with all genes found in the final dataset used as the background population list (34). Categories with a Bonferonni corrected p-value ≤ 0.05 were considered statistically significant.
Cell Culture and Western Blotting
MCF10A cells and THP-1 cells were purchased from ATCC and used for no longer than 6 months before being replaced. MCF10A cells expressing GFP, PIK3CA WT and mutants were generated and grown as described previously (10, 35). For starvation experiments, cells were grown for 24h in complete media lacking EGF and insulin, as described previously (10). For all Western blots, cells were lysed in RIPA buffer with protease and phosphatase inhibitors.
Soft Agar Assays
MCF10A cells expressing PIK3CA WT, E545K, or H1047R were infected with adenovirus expressing either GFP or IκBα superrepressor at 10 MOI overnight. Media was removed, and cells were allowed to recover in growth media for 24h. Infected cells were then plated in 0.6% Bacto Agar in the absence of growth factors. Media was replaced every 4 days. Colonies were counted after 25 days.
Results
NF-κB is activated in PI3K-transformed cells following growth factor deprivation
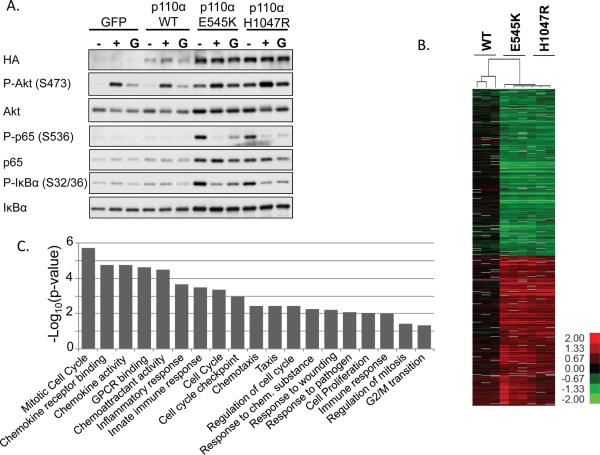
To determine whether the IKK/NF-κB signaling pathway is activated downstream from PIK3CA mutations, MCF10A cells stably expressing GFP (control), HA-PIK3CA WT, HA-PIK3CA E545K, or HA-PIK3CA H1047R were propagated in normal growth media (G), starvation media lacking EGF and insulin for 24h (−), or starvation media for 24h followed by 10 minutes of EGF and insulin stimulation (+) (Figure 1A and quantified in Figure S1). PIK3CA E545K and H1047R expression was higher than WT PIK3CA expression, consistent with previous studies demonstrating that these mutations confer resistance to proteasome-mediated degradation (36). In addition, total levels of AKT, p65, and IκBα were slightly increased in cells expressing E545K or H1047R. Under growth conditions cells expressing the E545K or H1047R mutations exhibited slightly increased phosphorylation of AKT and the NF-κB markers p65 and IκBα, when compared to cells expressing GFP or WT PIK3CA. It is well-established that WT MCF10A cells require exogenous EGF and insulin in order to proliferate, even in the presence of serum, while transformed MCF10A cells can undergo growth-factor independent proliferation (10). Consistent with this, following 24h of growth factor deprivation, cells expressing the oncogenic mutations exhibited dramatically increased AKT phosphorylation compared to cells expressing GFP or WT PIK3CA. Interestingly, under conditions of GF deprivation cells expressing the oncogenic mutations also demonstrated dramatically increased phosphorylation of p65 and IκBα (Figure 1A). However, surprisingly, while stimulating these GF-deprived cells with EGF and insulin led to increased AKT phosphorylation, p65 and IκBα phosphorylation rapidly and dramatically decreased (Figure 1A). While it is not clear how NF-κB is being so acutely downregulated following growth factor stimulation, these data suggest that NF-κB is not being regulated via a direct signaling pathway downstream from AKT.
Figure 1. IKK/NF-κB is activated by oncogenic PI3K mutations.
MCF10A cells stably expressing HA-GFP, HA-PIK3CA WT, HA-PIK3CA E545K, or HA-PIK3CA H1047R were generated by retroviral transduction. A) Cells were grown in growth medium (G), starvation medium lacking EGF and insulin (−) for 24h, or were starved for 24h and stimulated for 10 minutes with 20ng/mL EGF and 10μg/mL insulin (+). Lysates were evaluated by immunoblot. B) Cells expressing PIK3CA WT, E545K, or H1047R were GF-deprived for 24h. RNAs were evaluated by microarray and compared to expression in GFP control samples. Genes which are statistically significant by SAM analysis and altered more than 2-fold are shown in the heat map. Color key is for log2 ratio. C) A categorical enrichment analysis was performed on the top 300 genes upregulated by the E545K or H1047R mutation. Statistically enriched categories are shown (P < 0.05).
A comprehensive profile of genes upregulated by PIK3CA mutations has not been described. Therefore, we utilized microarrays to identify both global and NF-κB-dependent gene expression changes that occur in PI3K-transformed cells following GF deprivation. Supervised gene expression analyses were performed to find genes changed by the ectopic expression of mutant and WT PIK3CA. As expected, expression of WT PIK3CA led to few gene changes when compared to cells expressing GFP control (Figure 1B). However, expression of PIK3CA E545K or H1047R led to a statistically significant change in expression (based on SAM analysis) for 5513 genes, of which 1290 changed more than 2-fold (Figure 1B and Supplemental Table 1), suggesting that a large number of transcriptional programs are dramatically altered downstream from oncogenic PI3K mutations. While both the E545K and H1047R mutations increase the lipid kinase activity of PI3K, they act via different mechanisms (10, 11). However, our transcriptional analysis of these two mutations revealed that no statistically significant changes exist between the E545K and H1047R mutations when transcription of individual genes is examined (Figure 1B). For this reason, in later analyses we combine the gene expression data for the E545K and H1047R mutations and perform common analyses. Interestingly, a categorical enrichment analysis revealed that many of the gene categories statistically enriched by oncogenic PI3K mutations are known to be regulated by NF-κB, such as chemokine, inflammatory, and immune signaling pathways (Figure 1C). These data are consistent with Figure 1A, and suggest a critical role for inflammation in promoting growth factor-independent survival and tumorigenicity of PI3K-driven cancers.
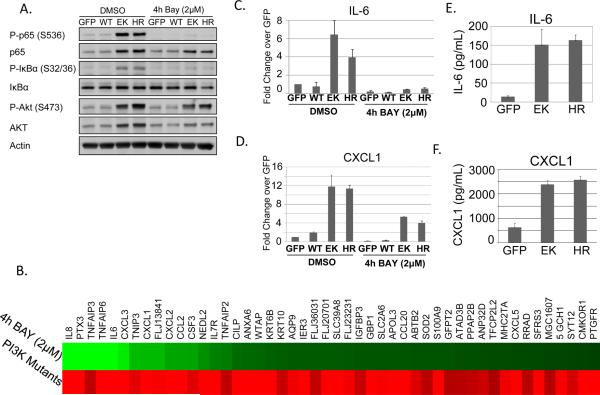
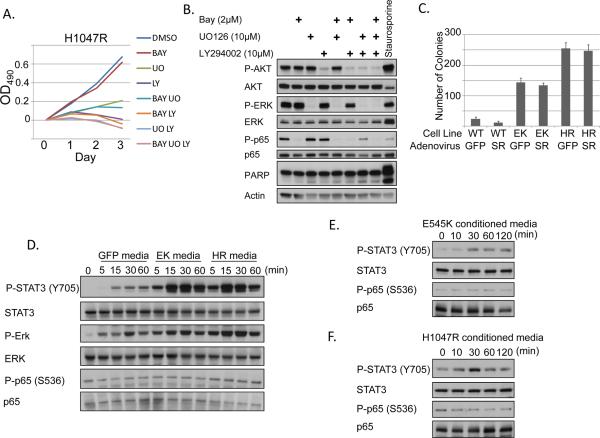
We next sought to determine what subset of genes upregulated by oncogenic PI3K is dependent on IKK/NF-κB signaling. Cells expressing the E545K or H1047R mutation were treated with DMSO or the well-established IKKβ-specific inhibitor BAY-65-1942 (BAY) for 4 hours (37). Phosphorylation of p65 and IκBα was completely abrogated following treatment with BAY-65-1942 (Figure 2A). Interestingly, phosphorylation of AKT and protein levels of both p65 and AKT were also slightly decreased, suggesting that expression and/or stability of these proteins may be partially NF-κB-dependent. Microarrays revealed that 48 genes are both upregulated in the presence of PI3K mutation and downregulated following 4 hours of IKKβ inhibition (Figure 2B and Supplemental Table 1). Many of these are established NF-κB target genes, while several are putative NF-κB targets which were previously unknown. Expression of several of these genes was validated by real-time RT-PCR (Figures 2C–D and Figure S2) and ELISA (Figures 2E–F, Figure S3). Expression of the PI3K-dependent, but NF-κB-independent, genes ENPP2 and S100A8 was also examined (Figure S2). Together, the data in Figures 1 and 2 show that both IKK signaling and NF-κB target gene expression are dramatically upregulated in cells expressing oncogenic PI3K mutations.
Figure 2. NF-κB target genes are upregulated by oncogenic PI3K mutations.
MCF10A cells stably expressing GFP, PIK3CA WT, E545K, or H1047R were starved for 24h and treated with DMSO or 2μM BAY-65-1942 for 4h. A) Cells were lysed and evaluated by immunoblot. B) Microarray analysis was performed. Genes which exhibit both increased expression in PIK3CA-transformed cells and decreased expression following IKK inhibition are shown. C–D) Real-time RT-PCR was used to verify expression changes for IL-6 and CXCL1. E–F) ELISA analysis shows increased secretion of IL-6 and CXCL1 by cells expressing the oncogenic PI3K mutations following 24h GF-deprivation.
Sustained activation of signaling pathways in PI3K-transformed cells following PI3K inhibition
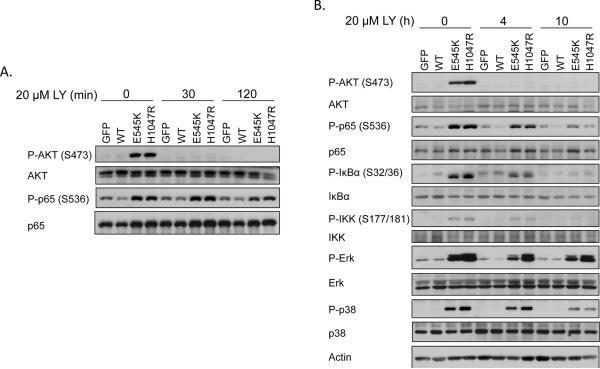
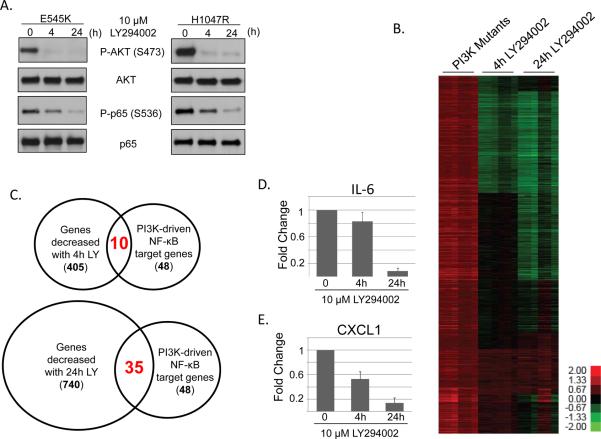
In order to determine whether inhibition of PI3K with LY294002 can disrupt IKK signaling in PI3K-transformed cells, MCF10A cells expressing the E545K or H1047R mutations were GF-deprived for 24 hours and treated with LY294002 for 30 or 120 minutes (Figure 3A). Interestingly, while AKT phosphorylation was completely abrogated following 30 minutes of LY294002 treatment, these short treatments with the PI3K inhibitor did not decrease phosphorylation of p65. In contrast, long (10h or 24h) periods of PI3K inhibition led to dramatically decreased phosphorylation of p65, IκBα, and IKK (Figure 3B and 4A). Further examination suggested that other stress-responsive signaling pathways, such as ERK and p38, are activated in the presence of the PI3K mutants following GF deprivation and showed sustained activation following PI3K inhibition (Figure 3B). These observations led us to perform further microarray analyses to identify gene expression changes following short (4h) or long (24h) treatment with LY294002 (Figure 4A). Consistent with our signaling data, of the 2856 genes upregulated by PI3K mutation, 405 were decreased following 4h of PI3K inhibition, and 740 genes were decreased following 24h of PI3K inhibition (Figure 4B). An examination of the NF-κB-dependent subset of genes showed that of the 48 NF-κB target genes upregulated by PIK3CA mutations, 10 were decreased following 4h PI3K inhibition, while 35 were decreased following 24h PI3K inhibition (Figure 4C). Expression of several of these genes, including IL-6 and CXCL1, was verified by real-time RT-PCR (Figure 4D–E). These results demonstrate that, following GF deprivation, oncogenic PI3K mutations activate a large number of long-lived signaling pathways and transcriptional complexes which are not appropriately feedback-inhibited.
Figure 3. Sustained PI3K inhibition decreases NF-κB activation by oncogenic PI3K mutations.
A) MCF10A cells stably expressing GFP, PIK3CA WT, E545K, or H1047R were starved for 24h, treated with 20μM LY294002 for 0–120 minutes and lysates were evaluated by immunoblot. B) MCF10A cells stably expressing GFP, PIK3CA WT, E545K, or H1047R were starved for 24h, treated with 20μM LY294002 for 0–10h and lysates were evaluated by immunoblot for the indicated signaling molecules.
Figure 4. Sustained PI3K inhibition decreases expression of NF-κB target genes in cells expressing oncogenic PI3K mutations.
A) MCF10A cells expressing PIK3CA H1047R were treated for 4h or 24h with 10μM LY294002 and analyzed by immunoblot. B) RNA samples from cells treated as in A) were evaluated by microarray. C) Venn diagram depicting changes in NF-κB-dependent gene expression following 4h or 24h of LY294002 treatment. C–D) MCF10A cells expressing PIK3CA E545K were treated for 4h or 24h with 10μM LY294002 and analyzed by real-time RT-PCR to evaluate expression of C) IL-6 or D) CXCL1.
NF-κB is not required for proliferation, survival, or anchorage-independent growth of PI3K-transformed cells
NF-κB target genes are critical for a wide range of cellular processes known to be required for tumorigenesis, such as proliferation, resistance to apoptosis, and response to cellular stress such as hypoxia (15, 16). Surprisingly, IKK inhibition did not decrease proliferation or metabolic activity of growth-factor deprived transformed MCF10A cells as assessed by MTT assay (Figures 5A and S4A). In addition, the IKK inhibitor did not synergize with a PI3K and/or MEK inhibitor to further decrease cell growth or increase PARP cleavage in PI3K-transformed cells (Figures 5A–B and S4A–B). An unphosphorylatable IκBα isoform (S32A/S36A) acts as an NF-κB “superrepressor” which prevents nuclear translocation of NF-κB. Expression of this NF-κB superrepressor in PI3K-transformed MCF10A cells failed to decrease colony formation in soft agar under GF-independent conditions (Figure 5C). Together, these data indicate that the primary function of NF-κB downstream from oncogenic PI3K mutations is not cell-autonomous. Rather, we observed that a large number of the NF-κB dependent genes upregulated by PI3K mutation are cytokines, chemokines, and other secreted proteins (Figure 2B). Therefore, we hypothesized that the role of NF-κB in PI3K-driven cell transformation may be to activate surrounding stromal and inflammatory cells in a paracrine (and autocrine) manner.
Figure 5. Secreted factors from cells expressing oncogenic PI3K mutations activate STAT3.
A) GF-deprived MCF10A cells expressing PIK3CA H1047R were treated for 3 days with the indicated inhibitors. Media and inhibitors were replaced daily. BAY = 2μM BAY-65-1942. LY = 10μM LY294002. UO = 10μM UO126 and cells were evaluated by MTT assay. B) Growth factor-starved MCF10A cells expressing H1047R were treated for 2 days with the indicated inhibitors. Media and inhibitors were replaced daily. Lysates were evaluated for PARP cleavage. 1μM staurosporine (positive control) was added 6h prior to lysis. C) MCF10A cells expressing PIK3CA WT, E545K, or H1047R were infected with adenovirus expressing either GFP or IκBα superrepressor (SR) and plated in 0.6% Bacto agar in MCF10A media lacking EGF and insulin. Colonies were counted after 25 days. D) MCF10A cells expressing GFP, PIK3CA E545K, or PIK3CA H1047R were starved for 24h. Conditioned media from these cells was used to treat THP-1 monocytes for 0–60 minutes. Lysates were evaluated by immunoblot. E–F) MCF10A cells stably expressing PIK3CA E545K (E) or H1047R (F) were starved for 24h. Conditioned media from these cells was then used to stimulate starved parental MCF10A cells for 0–120 minutes. Lysates were evaluated by immunoblot.
Secreted NF-κB gene products from PI3K-transformed cells activate STAT3 in monocytes and epithelial cells
Many advanced tumors are characterized by infiltration of macrophages and other immune cells to the site of the tumor. In order to determine whether factors secreted from PI3K-transformed cells can activate immune cells, conditioned media experiments were performed. MCF10A cells expressing GFP, PIK3CA E545K, or PIK3CA H1047R were deprived of growth factors for 24h. THP-1 monocytes were then resuspended in conditioned media from these cells to allow factors present in the media to activate the THP-1 cells. Interestingly, no phosphorylation of p65 was observed in recipient THP-1 cells, suggesting that no factors secreted by the PI3K-transformed cells activate NF-κB (Figure 5D). However, THP-1 cells treated with the media from E545K or H1047R-expressing cells showed robust activation of STAT3 and ERK in as little as 5 minutes. In addition, conditioned media from growth factor-deprived MCF10A cells expressing the E545K or H1047R mutations dramatically increased STAT3 phosphorylation in parental MCF10A cells within 30 minutes, suggesting that a factor(s) secreted by PI3K-transformed cells can also activate STAT3 signaling in normal epithelial cells (Figures 5E–F). Pretreating donor PI3K-transformed cells with an IKK inhibitor for 24h prior to stimulating either THP-1 cells or MCF10A cells with conditioned media led to a dramatic decrease in STAT3 phosphorylation, showing that the cytokine(s) activating STAT3 are being expressed in an NF-κB -dependent manner (Figure 6A–B and S5A–B).
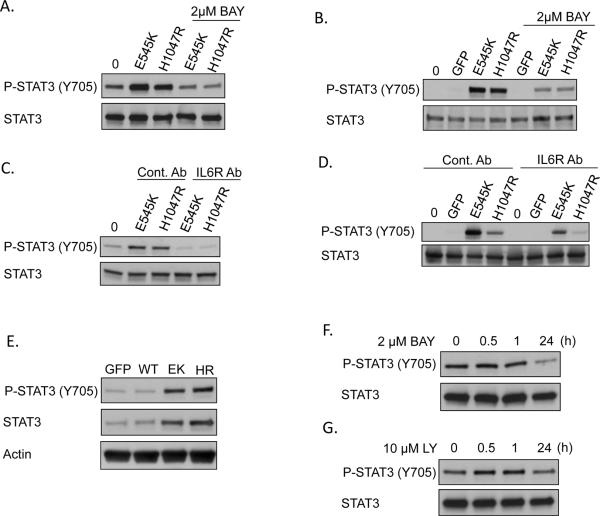
Figure 6. IL-6 contributes to STAT3 activation by E545K or H1047R conditioned media.
A) MCF10A cells stably expressing PIK3CA E545K or H1047R were starved for 24h in the presence or absence of 2μM BAY-65-1942. Conditioned media was used to stimulate parental MCF10A cells for 30 min and lysates were evaluated by immunoblot. B) MCF10A cells expressing GFP, PIK3CA E545K, or PIK3CA H1047R were starved for 24h in the presence or absence of 2μM BAY-65-1942. Conditioned media was used to stimulate THP-1 cells for 15 minutes and lysates were evaluated by immunoblot. C) Donor MCF10A cells expressing PIK3CA E545K or H1047R were starved for 24h. Both recipient parental MCF10A cells and donor cells were pretreated for 2h with 500 ng/mL IL-6 receptor antagonizing antibody or conrol antibody (anti-Flag mouse monoclonal). Conditioned media from donor cells was used to stimulate parental MCF10A cells for 30 minutes and lysates were evaluated by immunoblot. D) Donor MCF10A cells expressing GFP, PIK3CA E545K, or PIK3CA H1047R were starved for 24h. Recipient THP-1 cells and donor cells were pretreated for 2h with 500 ng/mL IL-6 receptor antagonizing antibody or control antibody (anti-Flag mouse monoclonal). Conditioned media from donor cells was used to stimulate THP-1 cells for 15 minutes and lysates were evaluated by immunoblot. E) MCF-10A cells expressing GFP, or PIK3CA WT, E545K, or H1047R were GF-deprived for 24h and immunoblotted with the indicated antibodies. F–G) MCF10A cells expressing PIK3CA H1047R were GR-deprived for 24h and treated for the indicated times with F) 2μM BAY-65-1942 or G) 10μM LY294002 and immunoblotted with the indicated antibodies.
IL-6 is required for E545K and H1047R-driven paracrine effects
IL-6 is a well-established pro-tumorigenic cytokine that activates STAT3 and ERK, the two signaling pathways which were activated in THP-1 cells by E545K or H1047R conditioned media. IL-6 is also one of the NF-κB target genes most dramatically upregulated in PI3K-transformed cells (Figures 2C and 2E). Therefore, we hypothesized that IL-6 may be the primary cytokine involved in these paracrine activities. Consistent with this, pretreating recipient MCF10A or THP-1 cells with an IL-6 receptor antagonizing monoclonal antibody for 2h prior to adding conditioned media from GF-deprived transformed MCF10A cells dramatically decreased STAT3 phosphorylation compared to cells pretreated with a control antibody (Figure 6C–D and S5C–D). Together, data in Figures 5 and 6 indicate that one of the primary consequences of NF-κB activation in PI3K-transformed cells is increased expression of IL-6, which can then activate STAT3 in both normal epithelial cells and immune cells in a paracrine manner.
The ability to stimulate parental MCF10A cells in a paracrine manner suggests that IL-6 may also activate STAT3 in an autocrine manner. Indeed, Figure 6E shows that STAT3 phosphorylation is increased in cells expressing the oncogenic PI3K mutations. Interestingly, total STAT3 protein levels are also increased. In addition, treating PI3K-transformed cells with an IKK inhibitor or PI3K inhibitor for short periods of time (<1h) does not affect phosphorylation of STAT3, consistent with an autocrine loop rather than direct intracellular signaling (Figures 6F–G, S5E–F, and data not shown). However, longer (24h) periods of IKK or PI3K inhibition do lead to decreased STAT3 phosphorylation, consistent with our microarray and real-time RT-PCR data showing that IL-6 is decreased at this timepoint. Treating PI3K-transformed MCF-10A cells with an IL-6R antagonizing antibody leads to decreased STAT3 phosphorylation, though this is not sufficient to decrease cell proliferation. These data are consistent with a model in which the primary role of PI3K-activated NF-κB is to induce paracrine activation of surrounding stromal cells (Figures S6A–B).
The PIK3CA mutant gene signature correlates with claudin-low and basal breast cancer subtypes
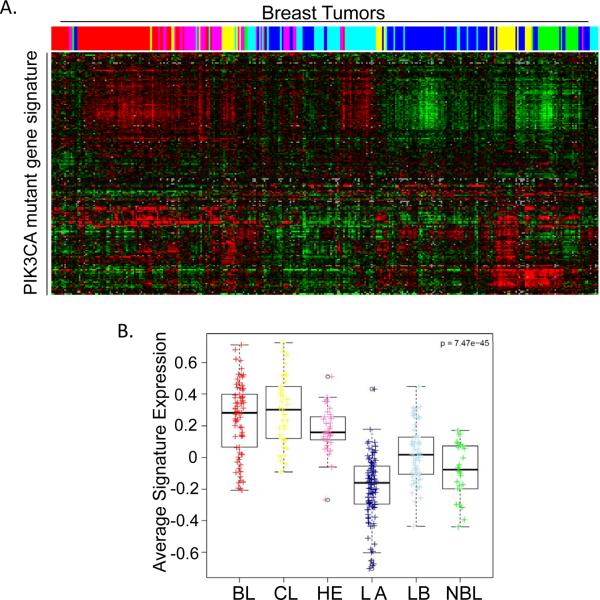
It has become clear over the last decade that breast cancer is not a single disease, but, rather, a set of diseases which can be classified into a variety of subtypes identified using protein or gene expression patterns (38, 39). The clinical relevance of these subtypes has been well-established, as different breast cancer subtypes display differences in patient survival and response to therapies (38, 39). It is therefore of great interest to determine which breast cancer subtype our PIK3CA-driven transformed cells resemble. As MCF10A cells are hormone receptor-negative, we hypothesized that transformed MCF10A cells might most closely resemble triple-negative, basal-like breast cancer (35, 40). Triple-negative breast cancers frequently display increased activation of the PI3K pathway, usually due to loss of either the PTEN or INPP4B tumor suppressors, and sometimes due to PIK3CA mutation (5, 41–45). We first identified the top 300 genes which are upregulated by PI3K mutation in MCF10A cells, which we refer to as the PIK3CA mutant gene signature. Of these, 192 probes correspond to those present in the publicly available UNC337 dataset which provides comprehensive gene expression and subtype data for 337 breast tumors. Gene expression data for these 192 probes was then evaluated within the UNC337 dataset using hierarchical clustering analysis (Figure 7A). Consistent with our hypothesis, the PIK3CA mutant gene signature most closely correlated with the triple-negative claudin-low and basal-like subtypes (Figure 7B). Interestingly, claudin-low tumors are also characterized by enrichment of immune response genes, including CCL2, CXCL2, and IL-6, which were also identified in our PIK3CA mutant gene signature (30, 39). MCF10A cells transformed by oncogenic PI3K mutations therefore provide an in vitro model that properly recapitulates both the signaling and gene expression profiles which occur in vivo in triple-negative tumors, many of which have constitutive PI3K activation.
Figure 7. The PIK3CA mutant gene signature correlates with claudin low and basal breast cancer subtypes.
A) 192 of the top 300 genes upregulated by PIK3CA mutation were identified in the UNC337 dataset. Expression of these genes is shown for each of the breast tumor samples present in the UNC337 dataset. Color-coding denotes tumor subtype: basal-like (red); claudin-low (yellow); HER2-enriched (pink); luminal A (dark blue); luminal B (light blue); normal breast-like (green). B) The expression value for each of the 192 PIK3CA upregulated genes was plotted by subtype and average signature value was calculated. Colored dots or boxes denote tumor subtype. BL (red), basal-like; CL (yellow), claudin-low H2 (pink), HER2-enriched; LA (dark blue), luminal A; LB (light blue), luminal B; NBL (green), normal breast-like.
Discussion
Cancer cells are characterized by the ability to proliferate in the absence of growth factors, but it is unclear what effects GF deprivation has on gene transcription downstream from oncoproteins. Here, we show that NF-κB signaling and gene expression are strongly upregulated under conditions of growth factor deprivation in PI3K-transformed cells, and categorical enrichment analysis showed that a number of cellular processes dependent on the transcription factor NF-κB are dramatically increased by these mutations (Figures 1C). Interestingly, we also observed that while stimulating these GF-deprived cells with EGF and insulin led to the expected increase in AKT phosphorylation, p65 and IκBα phosphorylation rapidly and dramatically decreased (Figure 1A). While it is not clear how NF-κB is being so acutely regulated under conditions of growth factor deprivation or stimulation, this disconnect between AKT phosphorylation and p65 phosphorylation suggests that NF-κB is not being activated via a direct signaling pathway downstream from AKT. Rather, we hypothesize that in the setting of a tumor, NF-κB is activated as part of a stress response to the aberrant cancer cell proliferation in the absence of growth factors. Once the stress is relieved by stimulation with growth factors, NF-κB is no longer required and is rapidly deactivated.
NF-κB can regulate any of hundreds of target genes depending on the type of stimulus, duration of stimulus, and the type of cell being activated (46). Interestingly, microarray analysis revealed that of the genes upregulated by oncogenic PI3K mutations, 48 were then downregulated following 4h treatment with an IKK inhibitor, providing a comprehensive NF-κB-dependent oncogenic transcriptional profile. Previous studies demonstrating a role for NF-κB downstream from oncogenes such as KRAS or EGFR have shown that the effects of NF-κB are largely cell-autonomous. In contrast, we find that inhibiting NF-κB in PI3K-transformed cells transformed does not lead to decreased cell proliferation, increased susceptibility to apoptosis, or decreased colony growth in soft agar (Figures 5A–C and S4). These data suggest the interesting possibility that even though numerous oncogenes may activate NF-κB, the set of NF-κB target genes that are transcribed may vary enormously between different cancers and under different growth/growth factor conditions.
Virtually all of the PI3K-driven NF-κB target genes identified by the microarray analyses are cytokines, chemokines, or other secreted proteins. Of these genes, IL-6 was of particular interest as it is highly expressed in PI3K-transformed cells in an NF-κB-dependent manner, and because its importance in tumorigenesis is well-established. Indeed, we find that conditioned media from cells expressing the E545K or H1047R mutations can dramatically upregulate STAT3 phosphorylation in both THP-1 monocytes and normal MCF10A breast epithelial cells in an NF-κB and IL-6 dependent manner (Figures 5–6). While the effect of this increased STAT3 activation will need to be confirmed in an in vivo model of PI3K-driven tumorigenesis, it is clear that increased macrophage infiltration and stromal inflammation correlates with poor prognosis in a number of tumor types. In addition, inhibition or genetic loss of STAT3 decreases progression of epithelial tumors in several animal models (26–28). We therefore propose that addition of an IL-6 receptor inhibitor, such as the recently FDA-approved tociluzimab, may dramatically increase the effectiveness of treatment regimens for PI3K-driven tumors by decreasing immune cell infiltration to the site of the tumor.
For virtually all solid tumors, the tumor microenvironment plays a critical role in the survival and progression of the tumor by influencing invasion, metastasis, angiogenesis, and recruitment of tumor-supporting macrophages (15, 47). While many signaling pathways regulate these stromal effects, the NF-κB pathway is well-positioned to be a critical regulator of virtually all of these processes as many of the proteins which regulate these diverse pathways are known NF-κB target genes (15, 16). Several recent reports have demonstrated that cells transformed by oncogenes such as Ras, EGFR, or HER2 lead to activation of NF-κB (19–23). These groups also demonstrated that inhibition of NF-κB in these transformed cells decreases proliferation and/or increases the susceptibility of cells to apoptosis in a largely cell-autonomous manner. However, non-cell autonomous roles for NF-κB in manipulating the tumor microenvironment have been less well-studied in these cancer models. Interestingly, a recent study by Ying, et al. showed that while KRAS mutation is sufficient to induce pancreatic ductal adenocarcinoma, additional loss of PTEN drives increased cytokine production and immune cell infiltration to the site of the tumor (48). However, this group also demonstrated a cell autonomous role for NF-κB (48). As RAS mutation alone has been shown to increase cell proliferation in an NF-κB-dependent manner (19, 21, 23), it is possible that KRAS mutation leads to NF-κB-driven expression of genes which promote proliferation, while additional activation of PI3K leads to a different NF-κB-driven cytokine and chemokine profile. Alternatively, as NF-κB can activate unique gene sets when in a complex with other transcription factors it is possible that KRAS-mediated ERK signaling leads to activation of transcription factors, such as AP-1, which cooperate with NF-κB to activate a different gene set than that observed in the presence of KRAS alone. It is therefore clear that while numerous oncogenes may all converge on NF-κB, it will be critical to define the gene sets regulated by NF-κB downstream from these various oncogenes in order to determine the effect of this NF-κB activity and to predict how interplay between NF-κB and other oncogenic signaling pathways may promote tumor growth.
Supplementary Material
Acknowledgements
We thank members of the Baldwin and Perou labs for helpful discussions. This work was supported by NIH grants CA75080 (A.S.B.), AI35098 (A.S.B.), CA73756 (A.S.B.), CA138255 (C.M.P.), CA148761 (C.M.P.), and the NCI Breast SPORE program (P50-CA58223). Additional support was provided by Department of Defense CDMRP BCRP grant W81XWH-10-1-0342 (J.E.H.) and the Samuel Waxman Cancer Research Foundation (A.S.B.). J.E.H. is a Lilly Research Labs fellow of the Life Sciences Research Foundation.
Warrell RP Jr, Frankel SR, Miller WH Jr, Scheinberg DA, Itri LM, Hittelman WN, et al. Differentiation therapy of acute promyelocytic 584 leukemia with tretinoin (all-trans-retinoic acid). N Engl J Med 1991;324:1385-93.
Footnotes
We have no conflicts of interest to disclose.
References
- 1.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296(5573):1655–7. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 2.Luo J, Manning BD, Cantley LC. Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer cell. 2003;4(4):257–62. doi: 10.1016/s1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- 3.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7(8):606–19. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 4.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nature reviews Cancer. 2002;2(7):489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 5.Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer research. 2009;69(10):4116–24. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27(41):5486–96. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu P, Cheng H, Santiago S, Raeder M, Zhang F, Isabella A, et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat Med. 2011;17(9):1116–20. doi: 10.1038/nm.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams JR, Xu K, Liu JC, Agamez NM, Loch AJ, Wong RG, et al. Cooperation between Pik3ca and p53 mutations in mouse mammary tumor formation. Cancer research. 2011;71(7):2706–17. doi: 10.1158/0008-5472.CAN-10-0738. [DOI] [PubMed] [Google Scholar]
- 9.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14(12):1351–6. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer research. 2005;65(23):10992–1000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 11.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(3):802–7. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikenoue T, Kanai F, Hikiba Y, Obata T, Tanaka Y, Imamura J, et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer research. 2005;65(11):4562–7. doi: 10.1158/0008-5472.CAN-04-4114. [DOI] [PubMed] [Google Scholar]
- 13.Chien Y, Scuoppo C, Wang X, Fang X, Balgley B, Bolden JE, et al. Control of the senescence-associated secretory phenotype by NF-kappaB promotes senescence and enhances chemosensitivity. Genes & development. 2011;25(20):2125–36. doi: 10.1101/gad.17276711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes & development. 2004;18(18):2195–224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 15.Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nature immunology. 2011;12(8):715–23. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- 16.Basseres DS, Baldwin AS. Nuclear factor-kappaB and inhibitor of kappaB kinase pathways in oncogenic initiation and progression. Oncogene. 2006;25(51):6817–30. doi: 10.1038/sj.onc.1209942. [DOI] [PubMed] [Google Scholar]
- 17.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nature reviews. 2005;5(10):749–59. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 18.Kim HJ, Hawke N, Baldwin AS. NF-kappaB and IKK as therapeutic targets in cancer. Cell death and differentiation. 2006;13(5):738–47. doi: 10.1038/sj.cdd.4401877. [DOI] [PubMed] [Google Scholar]
- 19.Basseres DS, Ebbs A, Levantini E, Baldwin AS. Requirement of the NF-kappaB subunit p65/RelA for K-Ras-induced lung tumorigenesis. Cancer research. 2010;70(9):3537–46. doi: 10.1158/0008-5472.CAN-09-4290. Epub 2010/04/22. doi: 10.1158/0008-5472.CAN-09-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bivona TG, Hieronymus H, Parker J, Chang K, Taron M, Rosell R, et al. FAS and NF-kappaB signalling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471(7339):523–6. doi: 10.1038/nature09870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finco TS, Westwick JK, Norris JL, Beg AA, Der CJ, Baldwin AS., Jr. Oncogenic Ha-Ras-induced signaling activates NF-kappaB transcriptional activity, which is required for cellular transformation. The Journal of biological chemistry. 1997;272(39):24113–6. doi: 10.1074/jbc.272.39.24113. [DOI] [PubMed] [Google Scholar]
- 22.Galang CK, Garcia-Ramirez J, Solski PA, Westwick JK, Der CJ, Neznanov NN, et al. Oncogenic Neu/ErbB-2 increases ets, AP-1, and NF-kappaB-dependent gene expression, and inhibiting ets activation blocks Neu-mediated cellular transformation. The Journal of biological chemistry. 1996;271(14):7992–8. doi: 10.1074/jbc.271.14.7992. [DOI] [PubMed] [Google Scholar]
- 23.Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, et al. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462(7269):104–7. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naugler WE, Karin M. The wolf in sheep's clothing: the role of interleukin-6 in immunity, inflammation and cancer. Trends in molecular medicine. 2008;14(3):109–19. doi: 10.1016/j.molmed.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 25.Grivennikov SI, Karin M. Inflammatory cytokines in cancer: tumour necrosis factor and interleukin 6 take the stage. Annals of the rheumatic diseases. 2011;70(Suppl 1):i104–8. doi: 10.1136/ard.2010.140145. [DOI] [PubMed] [Google Scholar]
- 26.Bollrath J, Phesse TJ, von Burstin VA, Putoczki T, Bennecke M, Bateman T, et al. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer cell. 2009;15(2):91–102. doi: 10.1016/j.ccr.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer cell. 2009;15(2):103–13. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nature reviews Cancer. 2009;9(11):798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 30.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast cancer research : BCR. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20(9):1453–4. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 32.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 33.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(9):5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hosack DA, Dennis G, Jr., Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome biology. 2003;4(10):R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30(3):256–68. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 36.Yuan TL, Wulf G, Burga L, Cantley LC. Cell-to-cell variability in PI3K protein level regulates PI3K-AKT pathway activity in cell populations. Current biology : CB. 2011;21(3):173–83. doi: 10.1016/j.cub.2010.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ziegelbauer K, Gantner F, Lukacs NW, Berlin A, Fuchikami K, Niki T, et al. A selective novel low-molecular-weight inhibitor of IkappaB kinase-beta (IKK-beta) prevents pulmonary inflammation and shows broad anti-inflammatory activity. British journal of pharmacology. 2005;145(2):178–92. doi: 10.1038/sj.bjp.0706176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 39.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Molecular oncology. 2011;5(1):5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soule HD, Maloney TM, Wolman SR, Peterson WD, Jr., Brenz R, McGrath CM, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer research. 1990;50(18):6075–86. [PubMed] [Google Scholar]
- 41.Aleskandarany MA, Rakha EA, Ahmed MA, Powe DG, Paish EC, Macmillan RD, et al. PIK3CA expression in invasive breast cancer: a biomarker of poor prognosis. Breast cancer research and treatment. 2010;122(1):45–53. doi: 10.1007/s10549-009-0508-9. [DOI] [PubMed] [Google Scholar]
- 42.Marty B, Maire V, Gravier E, Rigaill G, Vincent-Salomon A, Kappler M, et al. Frequent PTEN genomic alterations and activated phosphatidylinositol 3-kinase pathway in basal-like breast cancer cells. Breast cancer research : BCR. 2008;10(6):R101. doi: 10.1186/bcr2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gewinner C, Wang ZC, Richardson A, Teruya-Feldstein J, Etemadmoghadam D, Bowtell D, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer cell. 2009;16(2):115–25. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fedele CG, Ooms LM, Ho M, Vieusseux J, O'Toole SA, Millar EK, et al. Inositol polyphosphate 4-phosphatase II regulates PI3K/Akt signaling and is lost in human basal-like breast cancers. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(51):22231–6. doi: 10.1073/pnas.1015245107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saal LH, Johansson P, Holm K, Gruvberger-Saal SK, She QB, Maurer M, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(18):7564–9. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sen R, Smale ST. Selectivity of the NF-{kappa}B response. Cold Spring Harbor perspectives in biology. 2010;2(4):a000257. doi: 10.1101/cshperspect.a000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ying H, Elpek KG, Vinjamoori A, Zimmerman SM, Chu GC, Yan H, et al. Pten is a major tumor suppressor in pancreatic ductal adenocarcinoma and regulates an NF-kappaB-cytokine network. Cancer discovery. 2011;1(2):158–69. doi: 10.1158/2159-8290.CD-11-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.