Abstract
Plakophilin 2, a member of the arm-repeat protein family, is a dual location protein that occurs both in the cytoplasmic plaques of desmosomes as an architectural component and in an extractable form in the nucleoplasm. Here we report the existence of two nuclear particles containing plakophilin 2 and the largest subunit of RNA polymerase (pol) III (RPC155), both of which colocalize and are coimmunoselected with other pol III subunits and with the transcription factor TFIIIB. We also show that plakophilin 2 is present in the pol III holoenzyme, but not the core complex, and that it binds specifically to RPC155 in vitro. We propose the existence of diverse nuclear particles in which proteins known as plaque proteins of intercellular junctions are complexed with specific nuclear proteins.
In recent years, several components of the plaques of intercellular junctions surprisingly have also been identified in cell nuclei, suggesting their involvement in nuclear functions and regulatory interactions between the cell periphery and the nucleus. Such proteins include members of the arm-repeat family, which are characterized by variable numbers of a 42-amino acid motif (1). Among these, the plakophilins (PKPs) are characteristic of desmosomes and also occur constitutively in the nucleoplasm of a wide range of cells (2–11). These positively charged proteins (isoelectric points ranging from 9.3 to 10.1) represent the products of three genes, PKP1, PKP2, and PKP3, and at least PKP1 and PKP2 occur in two splice variants a and b (3–12). Both variants can also be detected in the nuclei of various cells devoid of desmosomes and after cDNA transfection have also been shown to accumulate in the nuclei (9).
The observation of plakophilins in the nucleus, however, has been possible only with the use of appropriate immunocytochemical protocols that minimize the extraction of soluble nucleoplasmic proteins and particles (e.g., see refs. 6, 9, and 13–15). Moreover, in a wide range of different kinds of cells lacking desmosomes including fibroblasts, lymphocytes, and chicken erythrocytes, plakophilins have been detected exclusively in the nucleus (6, 7, 10, 11). This finding suggests that plakophilins are constitutive nuclear proteins that, in certain pathways of cell differentiation, are recruited to a second specific location in desmosomal plaques (6–9).
Plakophilins 2a (92.75 kDa) and 2b (97.41 kDa) are of special importance, because they display the most widespread occurrence in desmosomes of all simple and complex epithelia, in lower layers of many stratified epithelia, in all carcinomas, in heart muscle, in reticulum cells of lymph node follicles, and in early embryos (6–8). Therefore, we have decided to characterize the plakophilin 2-containing structures of nuclei. Here we show plakophilin 2 in complexes with the largest subunit of RNA polymerase (pol) III and other pol III polypeptides, and we describe nucleoplasmic particles combining plakophilin 2 with pol III proteins.
Materials and Methods
Cell Cultures.
The following human cell culture lines were used: HaCaT keratinocytes (16), vulvar squamous carcinoma-derived A-431, hepatocellular carcinoma PLC, mammary gland carcinoma MCF-7, cervical adenocarcinoma HeLa, HEK (embryonic kidney), acute myeloid leukemia KG1a, and promyelocytic leukemia HL60 (all from the American Type Culture Collection). In addition, several cells and tissues of human and other vertebrate origins were examined including chicken erythrocytes and Xenopus laevis ovary (7). Several of these cell lines, notably HaCaT cells, were also used for the isolation of nuclei according to the method of Lee and Green (17)
Antibodies.
Immunoselections were performed with guinea pig antibodies specific for plakophilin 2 (6) and rabbit antisera against one of the following components of human pol III: hRPC155 (CSH499), hRPC39, hRPC82, hTFIIIB90, and hTFIIIC63 (18–21). Guinea pig antibodies specific for RPC155 were generated by immunization with the synthetic carboxyl-terminal peptide PKRPLIFDTNEFHIPLVT (residues 1374–1391) coupled to keyhole limpet hemocyanin and affinity-purified. Immunoblotting for plakophilin 2 was performed with the monoclonal antibodies PP2–150, PP2–86 (6–8), or plakophilin 2a (Becton Dickinson and Transduction Laboratories, Lexington, KY).
Sucrose Gradient Centrifugation and Gel Filtration Chromatography.
For sucrose gradient centrifugation, cells grown on 10-cm plates were lysed with either 0.6 ml of ice-cold 5:1 buffer [83 mM KCl, 17 mM NaCl, 10 mM Tris-HCl, pH 7.4, 5 mM EDTA, 3 mM DTT, 1 mM PMSF, and 1× Complete protease inhibitors (Roche Molecular Biochemicals)] or physiological salt buffer (140 mM NaCl, 1.5 mM MgCl2, 15 mM Hepes, pH 7.4, and 1× Complete), both containing 0.1% Triton X-100. Cells were scraped off with a rubber policeman, resuspended by pipetting, and transferred to a 1.5-ml tube. To analyze the influence of nucleases, extracts were incubated with either 16 μg/ml RNase A (Roche) or 416 μg/ml DNase for 15 min on ice. Digestion with DNase was performed with cell extracts in 2 mM MgCl2 instead of 5 mM EDTA. After centrifugation at 15,000 × g and 4°C for 10 min, the supernatant was centrifuged for 1 h at 100,000 × g. Then, 0.5-ml aliquots were layered on linear 10–40% (in 10 mM Tris-HCl, pH 7.5) or 10–60% (in 5:1 buffer without detergent) sucrose gradients and subjected to centrifugation for 16 h at 23,000 rpm (10–40%) or for 18 h at 35,000 rpm (10–60%) in an SW40 rotor (Beckman Instruments, Munich). Fractions of 0.4 ml (from 10–40% gradients) or 0.8 ml (from 10–60% gradients) were collected from top to bottom, and proteins were analyzed by SDS/PAGE directly or after immunoselection. In parallel gradients, BSA (Sigma), catalase, thyroglobulin (Amersham Pharmacia), or ribosomal subunits from X. laevis ovaries were used as reference proteins or particles.
For gel filtration, cell extracts prepared by lysing cells from three confluent 10-cm plates in a final volume of 0.6 ml of 5:1 buffer containing 0.1% Triton X-100 and 5 mM β-mercaptoethanol instead of DTT were centrifuged at 15,000 × g for 10 min, followed by 100,000 × g for 1 h. Then, 0.2 ml of the supernatant was loaded on a Superose 12-h 10/30 column (Amersham Pharmacia), and proteins were eluted with 5:1 buffer at a flow rate of 0.2 ml/min. Proteins from 0.4-ml fractions were precipitated with 3 volumes of absolute methanol and analyzed by SDS/PAGE.
Protein Fractions Containing Pol III.
HeLa cells stably transfected with flag epitope-tagged RPC53 (line BN51) were used. Pol III was affinity-purified from S100 extracts by using BC buffer (20 mM Hepes, pH 7.9, 20% glycerol, 0.5 mM EDTA, 1 mM DTT, and 0.5 mM PMSF) containing 300 mM KCl and 0.1% Nonidet P-40 (20). Pol III holoenzyme was enriched from nuclear extracts and immunopurified in BC buffer containing 100 mM KCl and 0.05% Nonidet P-40 (19).
Immunoisolation and Matrix-Assisted Laser Desorption Ionization Analysis.
Confluent A-431 cells of 200 Petri dishes (10-cm diameter) were lysed (0.6 ml per dish) in ice-cold modified RIPA buffer (20 mM Hepes, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM DTT, 1 mM PMSF, and 1× Complete protease inhibitors) and homogenized in a Dounce-type glass homogenizer (all procedures were performed at 4°C). After centrifugation for 1 h at 100,000 × g, the supernatant was incubated for 2 h with 20 mg of protein A-Sepharose beads (Amersham Pharmacia) and cleared by centrifugation. The lysate was incubated overnight with the beads bound with 200 μl of plakophilin 2 antibodies, and the selected protein complexes were washed four times in RIPA buffer and analyzed by SDS/PAGE. Bands seen after Coomassie blue staining were excised and digested in the gel for peptide fingerprinting by matrix-assisted laser desorption ionization mass spectrometry (22). For sequence comparisons, the programs Peptide Search (European Molecular Biology Laboratory), MS-Fit (University of California, San Francisco), and ProFound (Rockefeller University, New York) were used.
Immunoselections.
Immunoselected complexes from sucrose gradient fractions were obtained with material from five parallel gradients (10–60%). The fractions were pooled, adjusted by adding the double volume of 2× RIPA buffer, and obtained after preclearing with protein A-Sepharose for 2 h. Samples were incubated overnight with protein A-Sepharose beads coated with plakophilin 2 antibodies. Before analysis by SDS/PAGE, the immunoselected material was washed four times with RIPA buffer.
Extracts from lysed MCF-7 cells were prepared by immunoselection buffer (20 mM Hepes, pH 7.9, 10% glycerol, 2 mM MgCl2, 100 mM KCl, 0.1% Triton X-100, 1 mM DTT, 1 mM PMSF, and 1× Complete protease inhibitors). After centrifugation at 15,000 × g for 20 min, the supernatants were preincubated with Sepharose beads and subjected to immunoselection as described by using rabbit antibodies against one of the pol III components. In all washes, the buffer was the same as that used for lysis.
Blot Binding Assays.
A pol III-enriched fraction was prepared from HeLa S100 extracts (18–20), separated by SDS/PAGE, and transferred to nitrocellulose membrane sheets. [35S]methionine-labeled plakophilin 2 was obtained by coupled in vitro transcription-translation of plakophilin 2 cDNA or a subclone encoding the amino-terminal domain only (residues 1–367; ref. 6) using the coupled transcription and translation reticulocyte lysate system (Promega). Binding assays were as described (7).
Immunofluorescence Microscopy.
For immunofluorescence microscopy, cultured cells grown on coverslips were briefly rinsed in phosphate-buffered saline prewarmed to 37°C. Cells were fixed at −20°C in methanol (for 5 min) and acetone (for 30 sec) and air-dried. For double-staining experiments, guinea pig antibodies against plakophilin 2 and rabbit antisera specific for RPC155 were applied for 30 min followed by three phosphate-buffered saline washes for 3 min each. Secondary antibodies coupled to Texas red or fluorescein isothiocyanate (Dianova, Hamburg, Germany) were applied for 30 min. After washing with phosphate-buffered saline, the specimens were rinsed in water, briefly dipped in ethanol, air-dried, and mounted with Fluoromount (Biozol, Eching, Germany). Confocal laser scanning immunofluorescence images were acquired with a Zeiss LSM 410 microscope (Zeiss; ref. 6–8).
Results
Plakophilin 2-Containing Complexes.
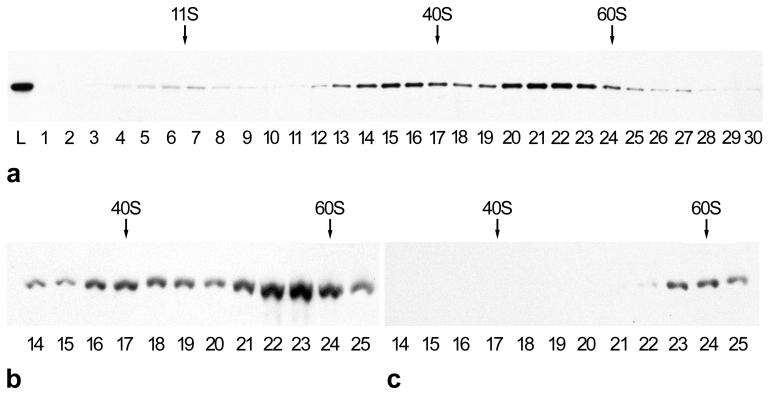
To study the native state of nuclear plakophilin 2, rapid particle extracts from cultured cells in Triton X-100 and EDTA were subjected to sucrose density gradient centrifugation, and the fractions obtained were analyzed by SDS/PAGE and immunoblotting (Fig. 1a). Plakophilin 2 was recovered in three peak fractions. The majority of the protein sedimented in large complexes of 30–35 S (fractions 14–17) and 50–55 S (fractions 20–23), whereas a minor proportion appeared at ≈11 S (fractions 6 and 7). Corresponding sedimentation profiles were obtained with extracts from various human cell lines such as HaCaT, A-431, or PLC, although the relative recovery in the different peak fractions varied somewhat.
Figure 1.
Sucrose gradient sedimentation of plakophilin 2-containing particles extracted with 5:1 buffer (a) or physiological salt buffer (b) from human keratinocyte cultures (HaCaT) or from HaCaT nuclei (c) in a linear 10–40% sucrose density gradient. Fractions were collected from top to bottom (1–30) and analyzed by SDS/PAGE and immunoblotting for plakophilin 2, which is recovered in fractions corresponding to S values of ≈11, 30–35, and 50–55 S (a), 40 and 55 S (b), and 55–60 S (c). L, loading sample. Size references: catalase, 11 S; X. laevis ribosomal subunits, 40 and 60 S.
Because plakophilin 2 is rather susceptible to proteolytic breakdown, most of the experiments were performed with rapidly isolated complexes. Under more protective conditions (physiological salt buffer) without EDTA, the two major peaks appeared at about 40 and 55 S (Fig. 1b). By contrast, when isolated nuclei were washed and subjected to particle extractions, only a single 55–60 S peak was predominant (Fig. 1c).
Plakophilin 2 in Complexes with the Largest Subunit of Pol III.
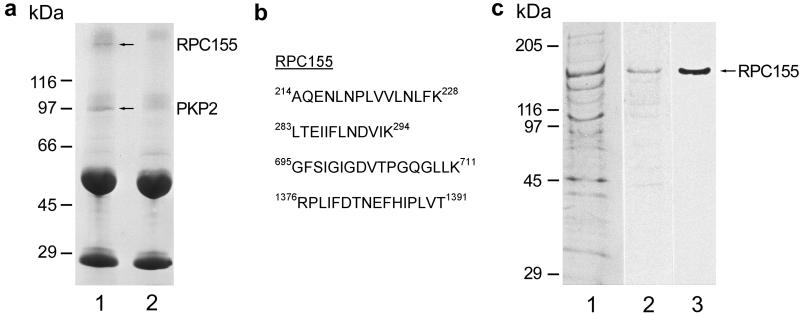
When extracts from A-431 cells were subjected to immunoselection with antibodies against plakophilin 2, a 155-kDa polypeptide was specifically coselected (Fig. 2a, arrow in lane 1; see also ref. 7). On analysis by mass spectrometry and peptide fingerprinting, this polypeptide was identified as the largest subunit of pol III (Fig. 2b, RPC155; cf. ref. 18–20).
Figure 2.
Plakophilin 2 interacts with RPC155. (a) Coimmunoselection of RPC155 with plakophilin 2. Extracts from cultured A-431 cells were subjected to immunoselection with plakophilin 2 antiserum (lane 1; lane 2, control showing the proteins of the same antiserum bound to protein A-Sepharose), and the proteins selected were analyzed by SDS/PAGE and stained with Coomassie brilliant blue (the sizes of reference proteins are indicated). The 155-kDa polypeptide (upper arrow) coselected with plakophilin 2 (lower arrow) was excised from the gel and identified by matrix-assisted laser desorption ionization analysis as the largest subunit of RNA polymerase III (RPC155). (b) Peptide sequences determined in the 155-kDa binding partner of plakophilin 2 by matrix-assisted laser desorption ionization analysis. In addition, myosin heavy chain and major vault protein (42) were identified as apparently nonspecifically bound coadsorbed proteins (cf. ref. 7). (c) Blot-overlay assay of polypeptides of a chromatographic fraction enriched in pol III, separated by SDS/PAGE, and visualized with Coomassie brilliant blue (lane 1) or transferred to nitrocellulose membranes (lanes 2 and 3). After incubation with in vitro translated 35S-labeled plakophilin 2, the binding partners of plakophilin 2 within the pol III fraction were detected by autoradiography (lane 2). A 155-kDa protein, which was identified subsequently as the largest subunit of pol III by immunoblot analysis using RPC155-specific antibodies (lane 3), binds plakophilin 2 selectively. The position of reference proteins is indicated on the left.
In Vitro Binding of Plakophilin 2.
The association between plakophilin 2 and RPC155 was demonstrable also in vitro. When fractions of nuclear proteins enriched in pol III were subjected to SDS/PAGE (Fig. 2c, lane 1), blotted onto nitrocellulose membranes, and reacted with radioactively labeled plakophilin 2, specific and intense binding to the band containing RPC155 was noted (Fig. 2c, lanes 2 and 3).
Biochemical Characterizations.
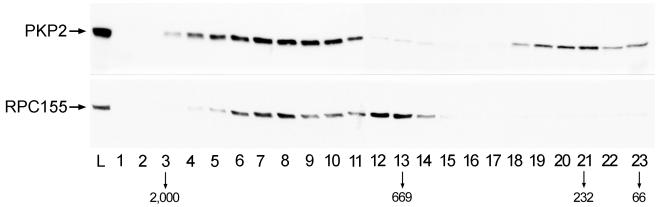
To examine the distribution of particles containing plakophilin 2 and RPC155, extracts from various cell cultures were subjected to gel filtration on a Superose 12 column, and the fractions obtained were analyzed by immunoblotting (Fig. 3). The majority of plakophilin 2 eluted in a rather broad peak (fractions 4–11) briefly after the void volume. In agreement with the sedimentation analysis (Fig. 1), smaller amounts coeluted with the reference protein catalase (fraction 21). RPC155 copurified with plakophilin 2 in peak fractions 6–9, corresponding to a mean molecular mass of ≈1.5 × 106. In addition, some RPC155 eluted as a distinct complex with a relative weight of about 700,000, which corresponds to the weight of the native catalytically active 16-subunit pol III complex (23).
Figure 3.
Gel filtration of plakophilin 2 and RPC155. Triton X-100-soluble extracts from human PLC cell cultures were chromatographed on a Superose 12 column, and individual fractions were analyzed by immunoblotting with antibodies against plakophilin 2 and RPC155. The elution peaks of the size markers (kDa) Dextran blue 2000 (2,000), thyroglobulin (669), catalase (232), and BSA (66) are indicated by arrows. Plakophilin 2 and RPC155 co-elute in a position corresponding to ≈1,500 kDa (fractions 6–8). L, loading sample.
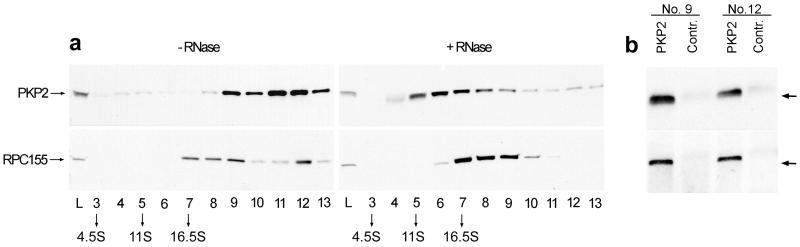
To characterize particles containing plakophilin 2 and RPC155 further, they were subjected to centrifugation on 10–60% sucrose gradients. Both RPC155 and plakophilin 2 were detected in fractions 9 and 12, corresponding to 30–35- and 50–55-S complexes, respectively (Fig. 4a, Left). Moreover, we noted that treatment with RNase A had an effect on the sedimentation behavior of the plakophilin 2-containing particles, which shifted to positions corresponding to smaller sizes (Fig. 4a, Right). By contrast, DNase treatments did not alter the sedimentation profiles (data not shown; ref. 7). These results may suggest that plakophilin 2 and RPC155 are components of particles that also contain some RNA of an as yet unknown nature.
Figure 4.
Analysis of plakophilin 2: RPC155 complexes by ribonuclease treatment and immunoselection. (a) Sedimentation of plakophilin 2 and RPC155 in sucrose density gradients (10–60%) from A-431 cell extracts without and with RNase A treatment. After centrifugation, proteins from individual fractions were precipitated with absolute methanol, subjected to SDS/PAGE, and analyzed by immunoblotting with antibodies against plakophilin 2 and RPC155. Size references: BSA, 4.5 S; catalase, 11 S; and thyroglobulin, 16.5 S. (b) For coimmunoselection of plakophilin 2 and RPC155 from sucrose density gradient fractions (no RNase treatment), fractions 9 and 12 (a) were subjected to immunoselection by using either antibodies specific for plakophilin 2 or unrelated antibodies (Contr.). Fractions were analyzed by immunoblotting with antibodies against plakophilin 2 and RPC155. RPC155 coimmunoselects with plakophilin from both fractions tested, whereas no signal is found in the negative controls.
To characterize the particles containing plakophilin 2 and RPC155 further, plakophilin 2 was immunoselected from the peak sucrose gradient fractions (Fig. 4b presents fractions 9 and 12, for example). Immunoblot analysis demonstrated that RPC155 was associated physically with plakophilin 2 (Fig. 4b), indicating that both proteins are part of the same complex.
We also scored the sucrose gradient fractions for other known desmosomal components. Desmogleins, desmoplakin, and plakoglobin appeared in fractions showing no cosedimentation with plakophilin 2 (data not shown; ref. 7). Plakophilins 1 and 3 were recovered in similarly sized particle fractions, which however did not contain pol III.
The Pol III Holoenzyme Complex.
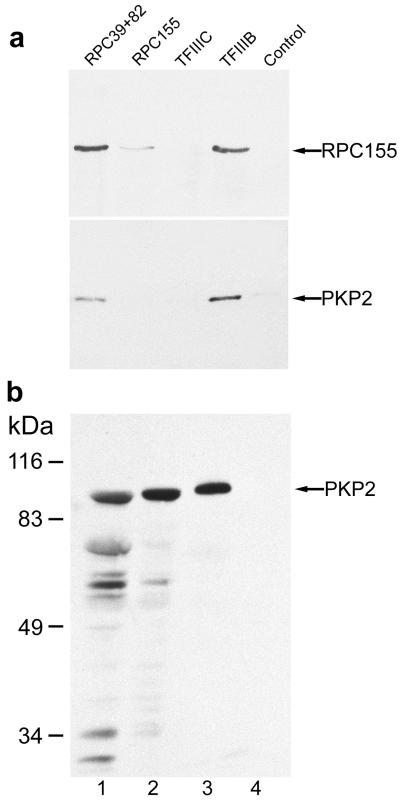
The size of the complex containing plakophilin 2 and RPC155 corresponds to that of the pol III holoenzyme, which consists of pol III and all basal factors essential for transcription (23). To examine whether plakophilin 2 was also associated with other components of the pol III holoenzyme, extracts prepared from MCF-7 cells were immunoselected with antibodies against other pol III components and analyzed for the presence of RPC155 and plakophilin 2 by immunoblotting (Fig. 5a). RPC155 and plakophilin 2 both appeared together with the 39-kDa subunit (RPC39) in combination with the 82-kDa pol III subunit (RPC82) and were associated also with transcription initiation factor TFIIIB. No binding was found to the primary DNA-binding factor TFIIIC (Fig. 5a).
Figure 5.
Plakophilin 2 is a component of the pol III holoenzyme complex. (a) For immunoselection, proteins extracted from human MCF-7 carcinoma cells were reacted with antibodies against the pol III subunits RPC39, RPC82, and RPC155 as well as with antibodies specific for the transcription factors TFIIIB (90-kDa subunit) and TFIIIC (63-kDa subunit), respectively. For control, preimmune serum was used. The immunoselected fractions were analyzed by immunoblotting using antibodies against RPC155 and plakophilin 2. The result shows the presence of plakophilin 2 in particles specifically selected with antibodies to different pol III components (RPC39, RPC82, RPC155, and TFIIIB) but not to TFIIIC (note that here the specific enrichment of RPC155 is too low to show plakophilin 2). (b) Immunoblot detection of plakophilin 2 in the purified pol III holoenzyme. Lane 1, S100 extract from human embryonic kidney (HEK) cells; lane 2, nuclear extract from HEK cells; lane 3, 5 μl of the FLAG-eluted pol III holoenzyme; lane 4, 5 μl of the FLAG-eluted pol III core enzyme.
Affinity-purification studies using nuclear extracts from HeLa cells stably transfected with a FLAG-tagged form of the RPC53 subunit resulted in the recovery of a functional pol III-containing complex, the holoenzyme, capable of transcription initiation and consisting of a multitude of distinct factors (cf. refs. 18–21). Among them one was identified as a ≈100-kDa polypeptide corresponding in size to plakophilin 2 (6–8). Indeed, immunoblot analysis (Fig. 5b) revealed a strong plakophilin 2 signal in fractions containing the affinity-purified pol III holoenzyme (lane 3). By contrast, no plakophilin 2 reaction was observed in fractions containing exclusively the 16-subunit pol III, the so-called core enzyme (lane 4), indicating that plakophilin 2 was associated with the pol III holoenzyme but not the pol III core enzyme complex.
Immunolocalization Microscopy of the Nuclear Particles Containing Plakophilin 2 and Pol III.
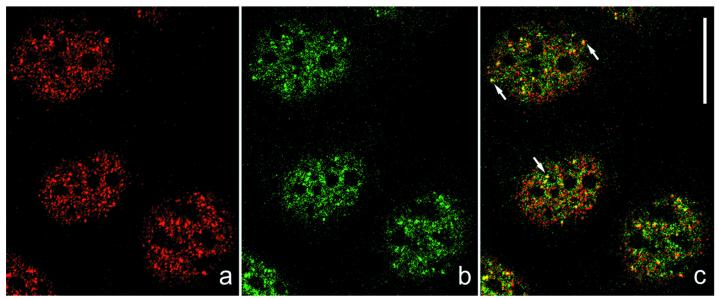
The nuclear particles containing plakophilin 2 and RPC155 were visualized directly by double-label immunofluorescence microscopy in several cell culture lines. For example, the reaction on PLC cells is shown in Fig. 6. Plakophilin 2 and RPC155 displayed a granular nucleoplasmic immunostaining and colocalized in distinct nuclear structures (Fig. 6c, yellow dots). However, in addition to these particles containing both proteins, each of them also occurred in separate granules as indicated by the non-yellow dots. This finding suggests that only a portion of nuclear plakophilin 2 is associated with complexes containing RPC155 and that both proteins also exist in other forms specific for either protein. Moreover, the plakophilin 2-positive dots were different from those immunostained for plakophilins 1 and 3 (data not shown).
Figure 6.
Colocalization of plakophilin 2 and RPC155 in nuclear particles. Cultured PLC cells were double-immunolabeled with antibodies against plakophilin 2 (a, red) and RPC155 (b, green) and observed by confocal laser scanning microscopy. Both proteins exhibit a finely punctate nucleoplasmic immunostaining, sparing the nucleoli. They colocalize in many nuclear particles as indicated (yellow) in the merged image (c, arrows). These structures are different from various other nucleoplasmic granules examined in parallel (for antibodies used see refs. 43 and 44). In addition, one sees some dots mutually exclusive for pol III (green) or plakophilin 2 (red). (Scale bar, 20 μm.)
Discussion
We have begun to characterize the nucleoplasmic forms of plakophilin 2, a protein also known as a major component of desmosomes. In particle fractionation and immunoselection experiments as well as in in vitro binding assays, plakophilin 2 has been detected in specific complexes with the largest subunit of RNA polymerase III (RPC155), the pol III subunit of 39 kDa and transcription factor TFIIIB and also in distinct nucleoplasmic granules. Both plakophilin 2 and RPC155 are also part of two large multisubunit complexes that appear, depending on the specific extraction conditions, between 30–35 and 50–60 S. Moreover, we have identified plakophilin 2 as one of the polypeptides enriched in affinity-purified preparations of the pol III holoenzyme. This interaction between plakophilin 2 and pol III components may provide a clue to one of the hitherto unknown nuclear functions of a junctional plaque protein.
Of the three nuclear RNA polymerases (I–III), pol III transcribes genes encoding ribosomal 5S RNA, the tRNAs, signal recognition particle RNA, U6 small nuclear RNA, and several small viral RNAs (for review see ref. 23). The inclusion of plakophilin 2 in nucleoplasmic assembly forms of the pol III transcription machinery would be compatible with the widespread and constitutive occurrence of plakophilin 2 in nuclei (6–8), a situation that is fundamentally different from the transient signal-induced nuclear translocation reported for other proteins of the arm-repeat family such as β-catenin and plakoglobin (24–31) and suggests that it serves some general nuclear function.
The human pol III of ≈700 kDa representing the core complex contains 16 subunits that remain tightly associated with each other even under partially denaturing conditions (e.g., see refs. 18–21). Its largest subunit (RPC155), shown here in complexes with plakophilin 2, has been highly conserved throughout evolution (18, 23, 32), as have amino acid sequence motifs in other pol III subunits (23, 33, 34). Accurate transcription by pol III requires the initiation factors TFIIIB and TFIIIC (35) and an ordered sequence of steps for their assembly into preinitiation complexes. On the other hand, affinity purification of pol III from nuclear extracts has revealed a preassembled nucleoplasmic particle (holoenzyme) containing the 16-subunit pol III and all basal factors essential for transcription (19, 23, 35, 36). Our results indicate that plakophilin 2 may be added to the list of components that can be present in this pol III holoenzyme complex.
The pol III complexes with plakophilin 2 seem to be located free in interchromatin spaces of the nucleoplasm, although their specific distribution in relation to transcriptionally engaged chromatin sites (37) remains to be established. We think that the particles described are not engaged in transcription but represent preinitiation assembly or storage forms, from which complexes active in transcription or RNA processing might be recruited (19, 23, 27, 38). A special kind of nucleoplasmic assembly granules has been described in amphibian oocytes and shown to harbor not only pol III but also other kinds of polymerases (transcriptosomes) and to aggregate to very large sizes up to 20 μm in diameter (Cajal bodies; refs. 39–41). Clearly, future studies will have to reveal the functional roles of both the Cajal bodies and the much smaller plakophilin 2-containing granules of somatic nuclei.
Acknowledgments
We thank Drs. H. Spring (German Cancer Research Center), N. Hernandez (Cold Spring Harbor Laboratory), and R. G. Roeder (Rockefeller University) for valuable discussions and experimental help as well as the German Research Foundation (Deutsche Forschungsgemeinschaft) for financial support.
Abbreviations
- PKP
plakophilin
- pol
RNA polymerase
References
- 1.Peifer M, Berg S, Reynolds A B. Cell. 1994;76:789–791. doi: 10.1016/0092-8674(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 2.Kapprell H-P, Owaribe K, Franke W W. J Cell Biol. 1988;106:1679–1691. doi: 10.1083/jcb.106.5.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hatzfeld M, Kristjansson G I, Plessmann U, Weber K. J Cell Sci. 1994;107:2259–2270. doi: 10.1242/jcs.107.8.2259. [DOI] [PubMed] [Google Scholar]
- 4.Heid H W, Schmidt A, Zimbelmann R, Schäfer S, Winter-Simanowski S, Stumpp S, Keith M, Figge U, Schnölzer M, Franke W W. Differentiation (Berlin) 1994;58:113–131. doi: 10.1046/j.1432-0436.1995.5820113.x. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt A, Heid H W, Schäfer S, Nuber U A, Zimbelmann R, Franke W W. Eur J Cell Biol. 1994;65:229–245. [PubMed] [Google Scholar]
- 6.Mertens C, Kuhn C, Franke W W. J Cell Biol. 1996;135:1009–1025. doi: 10.1083/jcb.135.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mertens C. Ph.D. thesis. Heidelberg: University of Heidelberg; 1999. [Google Scholar]
- 8.Mertens C, Kuhn C, Moll R, Schwetlick I, Franke W W. Differentiation (Berlin) 1999;64:277–290. doi: 10.1046/j.1432-0436.1999.6450277.x. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt A, Langbein L, Rode M, Prätzel S, Zimbelmann R, Franke W W. Cell Tissue Res. 1997;290:481–499. doi: 10.1007/s004410050956. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt A, Langbein L, Prätzel S, Rode M, Rackwitz H-R, Franke W W. Differentiation (Berlin) 1999;64:291–306. doi: 10.1046/j.1432-0436.1999.6450291.x. [DOI] [PubMed] [Google Scholar]
- 11.Bonné S, van Hengel J, Nollet F, Kools P, van Roy F. J Cell Sci. 1999;112:265–276. doi: 10.1242/jcs.112.14.2265. [DOI] [PubMed] [Google Scholar]
- 12.Franke W W, Mueller H, Mittnacht S, Kapprell H-P, Jorcano J L. EMBO J. 1983;2:2211–2215. doi: 10.1002/j.1460-2075.1983.tb01725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krohne G, Franke W W. Exp Cell Res. 1980;129:167–189. doi: 10.1016/0014-4827(80)90341-9. [DOI] [PubMed] [Google Scholar]
- 14.Krohne G, Franke W W. Proc Natl Acad Sci USA. 1980;77:1034–1038. doi: 10.1073/pnas.77.2.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ankenbauer T, Kleinschmidt J A, Walsh M J, Weiner O H, Franke W W. Nature (London) 1989;342:822–825. doi: 10.1038/342822a0. [DOI] [PubMed] [Google Scholar]
- 16.Boukamp P, Petrussevska R T, Breitkreutz D, Hornung J, Markham A, Fusenig N E. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee K A W, Green M R. Methods Enzymol. 1990;181:20–30. doi: 10.1016/0076-6879(90)81108-7. [DOI] [PubMed] [Google Scholar]
- 18.Sepehri S, Hernandez N. Genome Res. 1997;7:1006–1019. doi: 10.1101/gr.7.10.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Luo T, Roeder R G. Genes Dev. 1997;11:2371–2382. doi: 10.1101/gad.11.18.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Roeder R G. Genes Dev. 1997;11:1315–1326. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- 21.Hsieh Y-J, Wang Z, Kovelman R, Roeder R G. Mol Cell Biol. 1999;19:4944–4952. doi: 10.1128/mcb.19.7.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmelz M, Way D L, Borgs P, Peitsch W K, Schmidt H, Witte M H, Witte C L, Franke W W, Moll R. Cell Tissue Res. 1998;294:11–25. doi: 10.1007/s004410051152. [DOI] [PubMed] [Google Scholar]
- 23.Roeder R G. Methods Enzymol. 1996;273:165–171. doi: 10.1016/s0076-6879(96)73016-1. [DOI] [PubMed] [Google Scholar]
- 24.Funayama N, Fagotto F, McCrea P, Gumbiner B M. J Cell Biol. 1995;128:959–968. doi: 10.1083/jcb.128.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karnovsky A, Klymkowsky M W. Proc Natl Acad Sci USA. 1995;92:4522–4526. doi: 10.1073/pnas.92.10.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Behrens J, von Kries J P, Kühl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Nature (London) 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 27.Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann B G, Kemler R. Mech Dev. 1996;59:3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 28.Miller J R, Moon R T. Genes Dev. 1996;10:2527–2539. doi: 10.1101/gad.10.20.2527. [DOI] [PubMed] [Google Scholar]
- 29.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destrée O, Clevers H. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 30.Schneider S, Steinbeisser H, Warga R M, Hausen P. Mech Dev. 1996;57:191–198. doi: 10.1016/0925-4773(96)00546-1. [DOI] [PubMed] [Google Scholar]
- 31.Yost C, Torres M, Miller J R, Huang E, Kimelman D, Moon R T. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 32.Dieci G, Hermann-Le Denmat S, Lukhtanov E, Thuriaux P, Werner M, Sentenac A. EMBO J. 1995;14:3766–3776. doi: 10.1002/j.1460-2075.1995.tb00046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gabrielsen O S, Sentenac A. Trends Biochem Sci. 1991;16:412–416. doi: 10.1016/0968-0004(91)90166-s. [DOI] [PubMed] [Google Scholar]
- 34.Chiannikulchai N, Stalder R, Riva M, Carles C, Werner M, Sentenac A. Mol Cell Biol. 1992;12:4433–4440. doi: 10.1128/mcb.12.10.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roeder R G. Cold Spring Harbor Symp Quant Biol. 1998;63:201–208. doi: 10.1101/sqb.1998.63.201. [DOI] [PubMed] [Google Scholar]
- 36.Ferri M-L, Peyroche G, Siaut M, Lefebvre O, Carles C, Conesa C, Sentenac A. Mol Cell Biol. 2000;20:488–495. doi: 10.1128/mcb.20.2.488-495.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pombo A, Jackson D A, Hollinshead M, Wang Z, Roeder R G, Cook P R. EMBO J. 1999;18:2241–2253. doi: 10.1093/emboj/18.8.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flores A, Briand J-F, Gadal O, Andrau J-C, Rubbi L, Van Mullem V, Boschiero C, Goussot M, Marck C, Carles C, Thuriaux P, Sentenac A, Werner M. Proc Natl Acad Sci USA. 1999;96:7815–7820. doi: 10.1073/pnas.96.14.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gall J G, Bellini M, Wu Z, Murphy C. Mol Biol Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morgan G T, Doyle O, Murphy C, Gall J G. J Struct Biol. 2000;129:258–268. doi: 10.1006/jsbi.2000.4231. [DOI] [PubMed] [Google Scholar]
- 41.Bauer D W, Gall J G. Mol Biol Cell. 1997;8:73–82. doi: 10.1091/mbc.8.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kickhoefer A A, Vasu S K, Rome L H. Trends Cell Biol. 1996;6:174–178. doi: 10.1016/0962-8924(96)10014-3. [DOI] [PubMed] [Google Scholar]
- 43.Brandner J M, Reidenbach S, Kuhn C, Franke W W. Eur J Cell Biol. 1998;75:295–308. doi: 10.1016/S0171-9335(98)80063-0. [DOI] [PubMed] [Google Scholar]
- 44.Eilbracht J, Schmidt-Zachmann M S. Proc Natl Acad Sci USA. 2001;98:3849–3854. doi: 10.1073/pnas.071042298. [DOI] [PMC free article] [PubMed] [Google Scholar]