Abstract
Autophagy is a tightly controlled degradation process involved in various developmental aspects of eukaryotes. However, its involvement in developmental processes of multicellular filamentous ascomycetes is largely unknown. Here, we analyzed the impact of the autophagic proteins SmATG8 and SmATG4 on the sexual and vegetative development of the filamentous ascomycete Sordaria macrospora. A Saccharomyces cerevisiae complementation assay demonstrated that the S. macrospora Smatg8 and Smatg4 genes can functionally replace the yeast homologs. By generating homokaryotic deletion mutants, we showed that the S. macrospora SmATG8 and SmATG4 orthologs were associated with autophagy-dependent processes. Smatg8 and Smatg4 deletions abolished fruiting-body formation and impaired vegetative growth and ascospore germination, but not hyphal fusion. We demonstrated that SmATG4 was capable of processing the SmATG8 precursor. SmATG8 was localized to autophagosomes, whereas SmATG4 was distributed throughout the cytoplasm of S. macrospora. Furthermore, we could show that Smatg8 and Smatg4 are not only required for nonselective macroautophagy, but for selective macropexophagy as well. Taken together, our results suggest that in S. macrospora, autophagy seems to be an essential and constitutively active process to sustain high energy levels for filamentous growth and multicellular development even under nonstarvation conditions.
Keywords: autophagy, Atg8, Atg4, filamentous ascomycete, fruiting-body development, Sordaria macrospora
Introduction
In all eukaryotic cells, from yeast to plants to man, autophagy (which literately translates to “self-eating”) is a conserved intracellular recycling process to overcome nutrient depletion.1 Mechanistically, autophagy is divided into three basic types: macroautophagy, microautophagy and chaperone-mediated autophagy.2 Macroautophagy (hereafter, autophagy) describes the formation of a double-membrane vesicle, the autophagosome, which encloses a portion of cytoplasm containing excessive or defective proteins and/or organelles.3-5
Selective and nonselective pathways have been described in autophagy. The selective degradation of surplus organelles such as peroxisomes, mitochondria, ribosomes and nuclei are termed pexo-, mito-, ribo- and nucleophagy, respectively.6-9 In yeast, but not in filamentous fungi, a specific form of autophagy has been described,2,10 the cytoplasm-to-vacuole targeting (Cvt) pathway, which transports the hydrolytic enzymes aminopeptidase I (Ape1) and α-mannosidase (Ams1) to the vacuole.
The molecular dissection of autophagy has been mostly performed in the budding yeast Saccharomyces cerevisiae and has led to the identification of autophagy-related, or ATG, genes.10 The high degree of conservation of these genes simplified the identification of orthologs in other organisms including filamentous ascomycetes.11
In S. cerevisiae, two ubiquitin-like conjugation systems are involved in formation of the initial sequestering compartment, the phagophore, and its expansion into an autophagosome.12-14 In one of these conjugation systems, the ubiquitin-like protein Atg8 is first C-terminally processed to a glycine-exposed form by the cysteine protease Atg4. The processed Atg8 is then activated by Atg7 and transferred to the E2-like enzyme Atg3. Finally, a conjugate of Atg8 and the lipid phosphatidylethanolamine (PE) is formed, which is a structural component of the autophagosome membrane.13-15 In addition to processing newly synthesized Atg8, Atg4 acts as a deconjugating enzyme and facilitates the recycling of Atg8 from the membrane.15
In filamentous ascomycetes, the process of autophagy has been thoroughly investigated in Podospora anserina, and the plant pathogens Magnaporthe grisea, Fusarium graminearum, Colletotrichum orbiculare and Ustilago maydis.16 It was demonstrated that autophagy is induced during heterokaryon incompatibility, aerial hyphae and fruiting-body formation as well as in appressorium formation, conidiation and penicillin production.17-27 So far, it is not exactly known how autophagy participates in fungal differentiation processes, but autophagy might be involved in the reconstitution of intracellular components during development of different cell types. Hyphae that are not in contact with the medium may acquire nutrients through recycling of intracellular components by autophagy.9,28
The haploid filamentous ascomycete Sordaria macrospora is an excellent fungal model organism to study multicellular fruiting-body development.29 S. macrospora is a coprophilic and homothallic (self-fertile) fungus that naturally lives on herbivore dung. It lacks an asexual cycle, but every strain is able to complete the sexual cycle without a mating partner. Thus, in contrast to heterothallic (self-sterile) ascomycetes, recessive mutations can directly be tested for defects in fruiting-body development. Under laboratory conditions, the S. macrospora life cycle is completed within 7 d. After ascosporic germination, S. macrospora grows as a two-dimensional mycelium accompanied by hyphal branching and fusion. At day 3 of development, the sexual cycle starts with the formation of ascogonial coils followed by prefruiting-body (protoperithecia) formation and ends after a self-fertilization event, karyogamy and meiosis with the mature perithecia, which contain asci with sexual ascospores.30-34
To analyze whether nonselective autophagy is necessary for vegetative growth and fruiting-body development in S. macrospora, we previously characterized Smatg7, encoding the common E1-like enzyme of the two conjugation systems. However, we were not able to generate a homokaryotic knockout mutant in S. macrospora, suggesting that Smatg7 is required for viability. Interestingly, a heterokaryotic ΔSmatg7/Smatg7 strain and transformants generated by RNA interference showed considerable morphological phenotypes during fruiting-body development.31 In this study, we elucidated the impact of Smatg8 and Smatg4 on sexual and vegetative development in S. macrospora by phenotypically characterizing deletion mutants. By means of microscopic analysis of enhanced green fluorescent protein (EGFP)-tagged versions of SmATG8 and SmATG4, the proteins were localized in vivo and the processing of the SmATG8 precursor by SmATG4 was visualized.
Results
Isolation of S. macrospora Smatg8 and Smatg4 genes
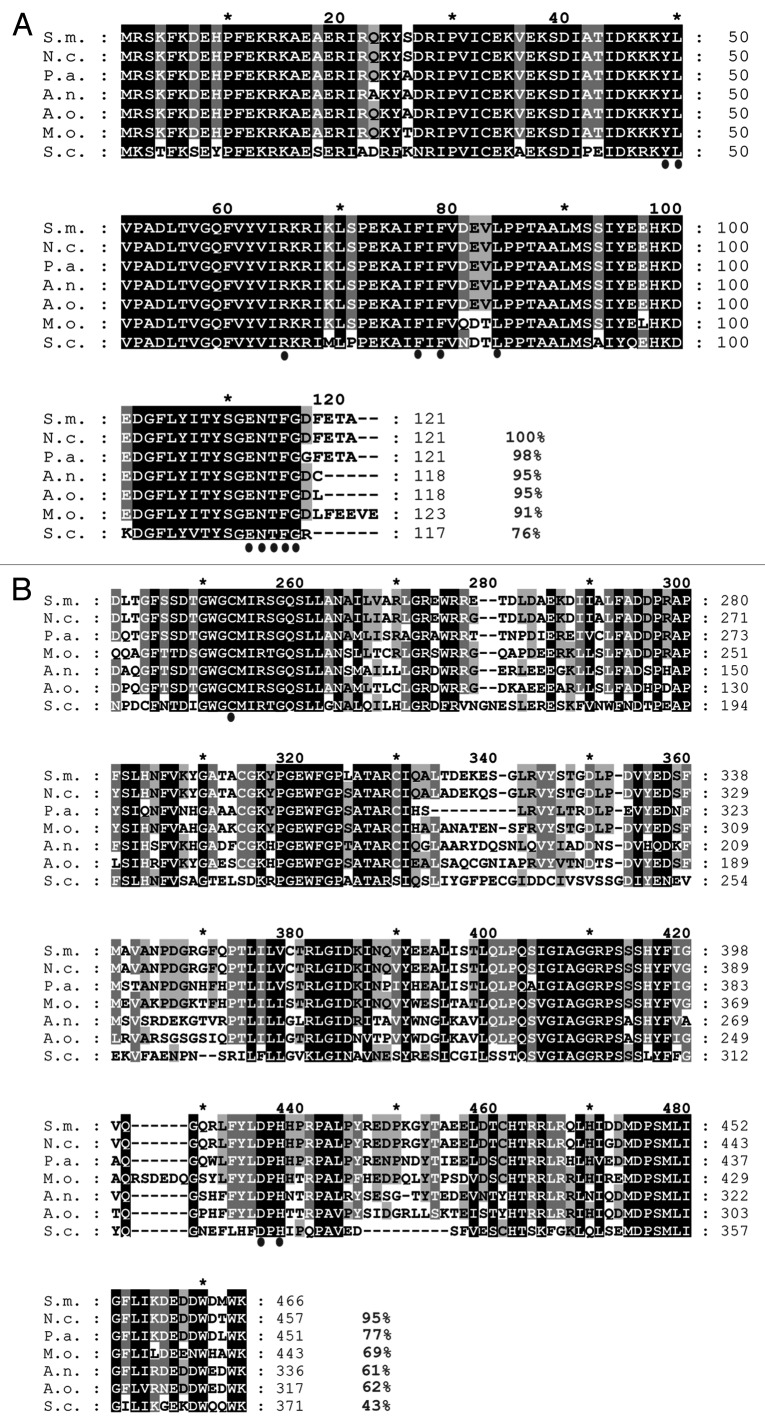
To isolate a gene encoding a homolog of the autophagosomal protein ATG8 from S. macrospora, a TBLASTN search was performed on the S. macrospora genome35 with the S. cerevisiae Atg8 (YBL078C) protein as a query sequence. We identified the 503 bp ORF SMAC_02305 (CCC07296.1) encoding a putative homolog of S. cerevisiae Atg8. Splicing of two predicted introns (69 and 68 bp) was confirmed by sequencing of the cDNA. SMAC_02305 encodes a protein of 121 amino acids (aa) with a predicted mass of 14.1 kDa and a theoretical isoelectric point (pI) of 6.15. Because the encoded protein shares 76% amino acid identity to the S. cerevisiae Atg8 protein, the gene was named Smatg8. Amino acid alignment of SmATG8 with homologs of other members of the Ascomycota phylum showed that this protein is highly conserved. Only the last five C-terminal aa (DEFTA) after Gly116, which is predicted to be C-terminally exposed after processing by Atg4, display a low level of identity (Fig. 1A). The alignment revealed that SmATG8 shares 100, 98, 95, 95 and 91% identity with orthologs of Neurospora crassa (Q8WZY7), P. anserina (Q8J282), Aspergillus nidulans (Q5B2U9), A. oryzae (Q2UBH5) and Magnaporthe oryzae (Q51MW4), respectively. To isolate an ortholog of the gene encoding the cysteine protease Atg4, which is required for Atg8 processing and recycling, we used S. cerevisiae Atg4 (YNL223W) for a TBLASTN search and identified the 1789 bp SMAC_08321 (CCC09424.1) sequence, encoding a protein of 515 amino acids with a predicted mass of 56.16 kDa and a theoretical pI of 4.88. The ORF is predicted to contain three introns of 83, 90 and 68 bp. Intron splicing was confirmed by cDNA sequencing. The encoded protein shows 43% identity in 254 aa overlap when compared with the S. cerevisiae homolog. However, an alignment of the predicted C54 peptidase domain (pfam03416) of the protein demonstrated that this domain is rather conserved among Atg4 orthologs of different ascomycetes. This domain includes an active cysteine residue that was previously identified in the Atg4 protease of M. oryzae (Fig. 1B).17 Due to this high degree of sequence identity, the isolated gene was named Smatg4. Quantitative real-time experiments revealed that Smatg8 and Smatg4 are constitutively expressed under both vegetative and sexual growth conditions (data not shown).
Figure 1. Multiple sequence alignment of the Atg8 and Atg4 proteins from members of the Ascomycota. (A) Amino acid alignment of Atg8 proteins. The ClustalX alignment was created using the following sequences: S.m., Sordaria macrospora, F7VP68; N.c., Neurospora crassa, Q8WZY7; P.a., Podospora anserina, Q8J282; A.n., Aspergillus nidulans, Q5B2U9; A.o., Aspergillus oryzeae, Q2UBH5; M.o., Magnaporthe oryzae, Q51MW4; S.c. Saccharomyces cerevisiae, P38182. Conserved residues important for the interaction with Atg4 and Atg7 in other organisms are indicated by dots below the alignment. (B) ClustalX alignment of catalytic C54 domain of Atg4 starting with Asp223 to Lys466 of the S. macrospora ATG4. Following sequences were utilized in the alignment: S.m., F7VV83; N.c., Q7S3X7; P.a., Q86ZL5; M.o., Q523C3; A.n., Q5B7L0; A.o., Q2U5B0; S.c., P53867. The conserved Cys, Asp and His residues of the catalytic triad of the C54 domain are marked with dots under the alignment. Identical amino acids, which are conserved in all proteins, are shaded in black; residues conserved in at least 6 of 7 sequences are shaded in dark gray and residues conserved in at least four sequences are shaded in light gray. Amino acid identity in % is given at the right margin.
Functional characterization of Smatg8 and Smatg4 in S. cerevisiae autophagy mutants
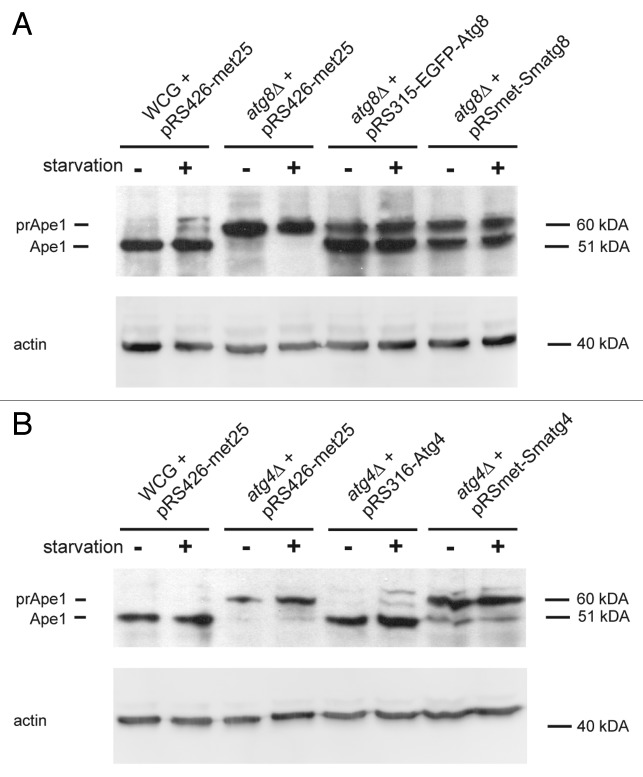
In order to confirm functional conservation of Smatg8 and Smatg4 and their yeast counterparts, we performed a complementation experiment using Smatg8 and Smatg4 cDNA under control of a MET25 promoter in S. cerevisiae to rescue atg8Δ and atg4Δ deletion strains, respectively. In S. cerevisiae, maturation of the precursor form of aminopetidase I (prApe1) to the mature enzyme (Ape1) depends on autophagy proteins, including Atg8 and Atg4.36 Rescue of autophagy of the S. cerevisiae mutants was thus monitored using a prApe1 maturation assay.37 In S. cerevisiae prApe1 is also localized to the vacuole by the Cvt pathway, which again depends on the autophagy proteins. Precursor Ape1 is synthesized as a 61- kDa soluble cytosolic protein and, upon delivery to the vacuole, the N-terminal propeptide is removed to yield the 50-kDa Ape1. We generated the yeast strains WCG + pRS426-met25, atg8∆ + pRS315-EGFP-Atg8, atg4∆ + pRS316-Atg4 as positive controls and atg8∆ + pRS426-met25 as well as atg4∆ + pRS426-met25 as negative controls (Table 1). Yeast strains atg8∆ + pRS-met-Smatg8 and atg4∆ + pRS-met-Smatg4 were used to assess whether the S. macrospora genes could rescue prApe1 processing in the yeast deletion strains. Yeast strains were grown overnight in synthetic defined (SD) minimal medium and adjusted to OD600 = 1. One set of cells was used for protein extraction, while another set was grown for 4 h in SD minimal medium lacking nitrogen (SD-N) that induces amino acid-starvation and, in turn, autophagy in S. cerevisiae (Fig. 2). The effect of starvation was not as drastic as expected, which might have been due to the fact that the cells were already in stationary phase at the start of the experiment and thus nutrient deprived. Figure 2A shows that prApe1 maturation occurred to the same extent when S. cerevisiae ATG8 or Smatg8 was expressed in atg8Δ, which indicated the capability of Smatg8 to complement the yeast atg8∆ strain. However, Smatg4 apparently only partially complemented the yeast atg4∆ strain. In the negative control (atg4∆ + pRS426-met25), in addition to the Ape1 precursor form, a very faint band of Ape1 was visible under starvation conditions, indicating inefficient prApe1 processing activity in the yeast atg4∆ mutant. Extracts from cells expressing the heterologous Smatg4 showed a clearly visible Ape1 signal, but when compared with the positive control, the signal of the strain expressing the endogenous ATG4 gene (atg4∆ + pRS316-Atg4), the signal was weaker (Fig. 2B).
Table 1.Saccharomyces cerevisiae and Sordaria macrospora strains used in this study.
| Strain | Characteristics | Source |
|---|---|---|
| Sc PJ69-4A |
MATa; trp1-901; leu2-3,112; ura3-52; his3-200; ga14D; ga18OD; LYS2::GAL1-HIS3; GAL2-ADE2; met2::GAL7-lacZ |
James et al. (1996)38 |
| Sc WCG4a |
MATα; his3-11,15; leu2-3,112; ura3-52 |
M. Thumm, Göttingena |
| Sc atg8Δ |
MATα; his3-11,15; leu2-3,112; ura3-52; atg8Δ::KAN |
M. Thumm, Göttingena |
| Sc atg4∆ |
MATα; his3-11,15; leu2-3,112; ura3-52; atg4Δ::HIS5 |
M. Thumm, Göttingena |
| Sc Y187 |
MATα; ura3-52; his3-200; ade2-101; trp1-901; leu2-3, 112; gal4∆; met∆; gal80∆; MEL1; URA3::GAL1UAS-GAL1TATA-lacZ |
Clontech |
| Sc AH109 |
MATa; trp1-901; leu2-3, 112; ura3-52; his3-200; ade2-101; gal4∆, gal80∆; LYS2::GAL1UAS-GAL1TATA-HIS3; GAL2UASGAL2TATAAde2; URA3::MEL1UAS-MEL1TATA-lacZ; MEL1 |
Clontech |
|
S48977 |
Wild type |
U. Kück, Bochum,b |
|
S66001 |
∆ku70::natR |
Pöggeler and Kück (2006)67 |
|
S23442 |
mutation in fus gene, brownish ascospores |
Nowrousian et al. (2012)39 |
| Sc WCG4a + pRS426-met25 |
MATα; his3-11,15; leu2-3,112; ura3-52 + pRS426-met25 |
This study |
| Sc atg8Δ + pRS426-met25 |
MATα his3-11,15; leu2-3,112; ura3; atg8Δ::KAN; + pRS426-met25 |
This study |
| Sc atg8Δ + pRS315-EGFP-Atg8 |
MATα; his3-11,15; leu2-3,112; ura3-52; atg8Δ::KAN; egfp-atg8 under control of endogenous promoter |
This study |
| Sc atg8Δ + pRS-met-Smatg8 |
MATα; his3-11,15; leu2-3,112; ura3-52; atg8Δ::KAN; Smatg8 under control of met25 promoter |
This study |
| Sc atg4∆ + pRS426-met25 |
MATα; his3-11,15; leu2-3,112; ura3-52; atg4Δ::HIS5; + pRS426-met25 |
This study |
| Sc atg4∆ + pRS316-Atg4 |
MATα; his3-11,15; leu2-3,112; ura3-52; atg4Δ::HIS5; ATG4 under control of endogenous promoter |
This study |
| Sc atg4∆ + pRS-met-Smatg4 |
MATα; his3-11,15; leu2-3,112; ura3-52; atg4Δ::HIS5; Smatg4 under control of met25 promoter |
This study |
| S ∆Smatg8 |
∆Smatg8::hygR, ssi |
This study |
| S ∆Smatg8::Smatg8ect |
∆Smatg8::hygR, Smatg8ect, natR, ssi |
This study |
| S ∆Smatg8::egfp-Smatg8ect |
∆Smatg8::hygR, egfp-Smatg8ect, natR, ssi |
This study |
| S ∆Smatg8::egfp-Smatg8-DsRed-SKLect |
∆Smatg8::hygR, egfp-Smatg8-DsRed-SKLect, natR, ssi |
This study |
| S ∆Smatg8::DsRed-SKLect |
∆Smatg8::hygR, DsRed-SKLect, natR |
This study |
| S ∆Smatg4 |
∆Smatg4::hygR, ssi |
This study |
| S ∆Smatg4::Smatg4ect |
∆Smatg4::hygR, Smatg4ect, natR, ssi |
This study |
| S ∆Smatg4::egfp-Smatg8ect |
∆Smatg4::hygR, egfp-Smatg8ect, natR |
This study |
| S ∆Smatg4::Smatg8-egfpect |
∆Smatg4::hygR, Smatg8-egfpect, natR |
This study |
| S ∆Smatg4::egfp-Smatg8G116ect |
∆Smatg4::hygR, egfp-Smatg8G116ect, natR |
This study |
| S ∆Smatg4::Smatg4-egfpect |
∆Smatg4::hygR, Smatg4-egfpect, natR, ssi |
This study |
| S ∆Smatg4::DsRed-SKLect |
∆Smatg4::hygR, DsRed-SKLect, natR |
This study |
| S wt::Smatg8-egfpect |
Smatg8-egfpect, natR, ssi |
This study |
| S wt::egfp-Smatg8mutect | egfp-Smatg8mutect, natR | This study |
Sc, Saccharomyces cerevisiae; S, Sordaria macrospora; hygR, hygromycin resistant; natR, nourseothricin resistant; ssi, single spore isolate. aDepartment of Biochemistry II, UMG, Georg-August University Göttingen, Germany bDepartment of General and Molecular Botany, Ruhr-University Bochum, Germany
Figure 2. Complementation of the S. cerevisiae autophagy mutants with S. macrospora autophagy genes. Complementation of respective yeast strains with Smatg8 and Smatg4 was analyzed using a precursor Ape1 maturation assay.37 In S. cerevisiae the cytosolic 61-kDa precursor of Ape1 (prApe1) is delivered to the vacuole via the Cvt pathway, which involves intact Atg4 and Atg8 proteins. Upon delivery to the vacuole prApe1 is processed to the mature Ape1 enzyme (Ape1). Identical amounts of yeast cell extracts (0.2 OD600 equivalents of cells) isolated before (-) and after starvation on SD-N medium for 4 h (+) were separated on 15% SDS-PAGE gels. After blotting, PVDF membranes were probed with an anti-Ape1 antibody as stated in the Materials and Methods. To verify equal protein concentrations after stripping of the membrane anti-actin antibody was used as loading control. Numbers at the right indicate the molecular mass. prApe1, precursor Ape1, Ape1, mature Ape1. (A) Complementation of S. cerevisiae atg8∆ with Smatg8 under control of the yeast MET25 promoter (pRSmet-Smatg8). As a positive control the S. cerevisiae wild-type strain WCG was transformed either with the empty plasmid pRS426-met25, or S. cerevisiae atg8∆ was transformed with the yeast egfp-ATG8 gene (pRS315-EGFP-Atg8). S. cerevisiae atg8∆ transformed with the empty vector pRS426-met25 served as a negative control. (B) Complementation of S. cerevisiae atg4∆ with Smatg4 under control of the yeast MET25 promoter (pRSmet-Smatg4). As positive control, the S. cerevisiae wt WCG was transformed with the empty plasmid pRS426-met25 or S. cerevisiae atg4∆ was transformed with the yeast ATG4 gene (pRS316-Atg4). S. cerevisiae atg4∆ transformed with the empty vector pRS426-met25 served as a negative control.
SmATG8 interacts with SmATG4 in the yeast two-hybrid system
A direct protein-protein interaction of Atg8 and Atg4 has been described in S. cerevisiae15 and was recently demonstrated in M. oryzae.17 To confirm that SmATG8 and SmATG4 interact with each other, a yeast two-hybrid analysis was performed. For this purpose, Smatg8 and Smatg4 cDNA was cloned into the two-hybrid vectors pGBKT7 (bait) and pGADT7 (prey), respectively. The resulting bait plasmids were transformed into yeast strain Y187, and prey plasmids into the strain AH109. Transactivation of pBD-Smatg8 and pBD-Smatg4 was tested by mating them with strain AH109 carrying the pGADT7 vector. Mating of Y187 carrying pGBKT7 with AH109 containing either pAD-Smatg8 or pAD-Smatg4 served as a negative control. As a positive control and to test the appropriate expression of the proteins encoded by the bait plasmids, mating of bait plasmids carrying strains with an AH109 strain containing pAD-RanBPM was conducted.40 The two-hybrid experiment clearly demonstrated an interaction of SmATG8 and SmATG4 (Fig. 3).
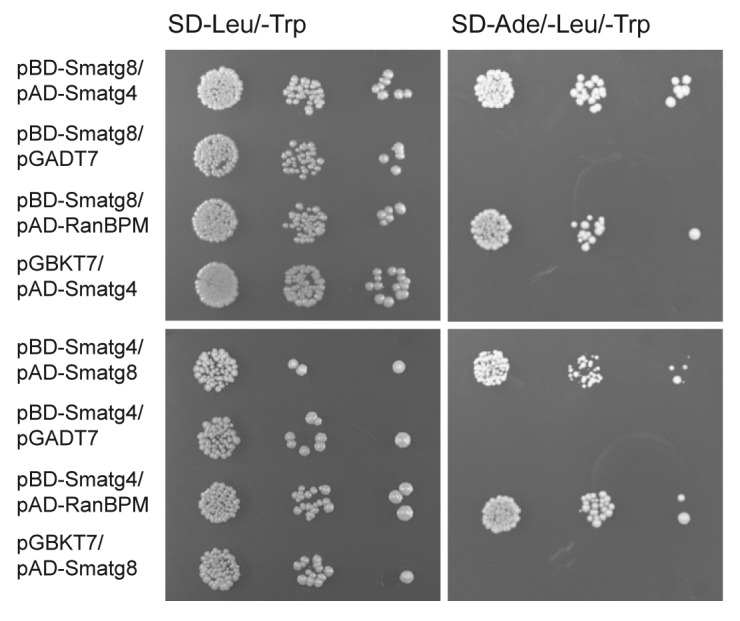
Figure 3. Yeast two-hybrid interaction of SmATG8 and SmATG4. Full-length cDNAs of Smatg8 and Smatg4 were used to generate Gal4 DNA binding domain (BD) and activation domain (AD) plasmids. Yeast strains Y187 and AH109 were transformed either with bait plasmid pGBKT7, pBD-Smatg8 and pBD-Smatg4 or with prey plasmids pGADT7, pAD-RanBPM, pAD-Smatg8 and pAD-Smatg4, respectively. After mating of both strains in all possible combinations, diploid cells were selected on SD-Leu/-Trp media. Strains carrying empty plasmids pGADT7 and pGBKT7 served as negative controls. To determine whether the bait proteins are expressed appropriately for an interaction to be detected, we used as a positive control a test, based on interaction with the Gal4 BD with protein RanBPM.40 Serial dilutions were made either on SD-Leu/-Trp to select for the presence of both plasmids (left) or on selection medium lacking adenine (SD-Ade/-Leu/-Trp) to verify the interaction of SmATG8 with SmATG4 (right).
Smatg8 and Smatg4 deletion strains are sterile and impaired in vegetative growth and ascospore germination
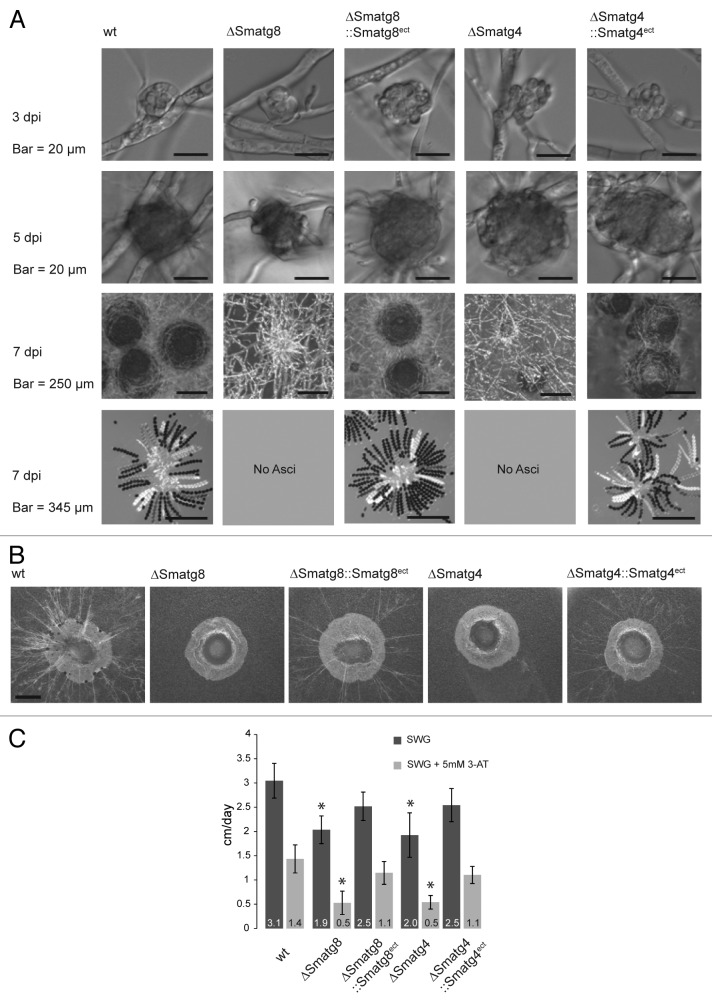
In order to analyze the effect of Smatg8 and Smatg4 on the viability and sexual reproduction of S. macrospora, we generated ΔSmatg8 and ΔSmatg4 deletion strains for phenotypic characterization (Figs. S1 and S2). Microscopy analysis of the knockout strains revealed that both were blocked during fruiting-body development. They were able to form the first stages of sexual development (ascogonia and protoperithecia) but failed to produce a mature fruiting body (perithecia) or ascospores. Complementation by inserting an ectopic copy of Smatg8 and Smatg4 into the ΔSmatg8 and ΔSmatg4 mutant, respectively, fully restored the wild-type (wt) phenotype (Fig. 4A). Furthermore, we were interested in determining whether autophagy impairment also led to an impairment of the foraging capability, as was recently shown for a F. graminearum Δatg8 mutant.41 Here, we use the term “foraging” to describe the growth of a filamentous fungus over a non-nutritious surface to reach nutrient-rich regions. For this type of growth, the required nutrients are thought to be provided by basal hyphae.28 To assess the foraging capability of the mutants, wt and the complemented strains, agar plugs were transferred into an empty Petri dish and incubated for 5 d. While the wt and complemented strains displayed extensive mycelial growth, ∆Smatg8 and ∆Smatg4 knockout strains were unable to grow on an inert plastic surface (Fig. 4B). In addition to this phenotype, both mutants showed a significant decrease in vegetative growth velocity [1.9 (± 0.46) and 2.0 (± 0.29) cm/d compared with 3.1 (± 0.36) cm/d of the wt strain]. The vegetative growth defect was partially rescued in the complemented strains (Fig. 4C). When histidine starvation was imposed by the drug 3-amino-1,2,4-triazole (3-AT), reduction of the growth velocity was more severe in the mutant strain than in the wt (Fig. 4C).
Figure 4. Phenotypic analysis of Smatg8 and Smatg4 deletion and complementation strains. (A) Microscopy investigation of sexual development in ΔSmatg8 and ΔSmatg4 compared with wt. Wt strains form ascogonia at day 3 which develop into protoperithecia at day 5 and mature perithecia after 7 d. Ascus rosettes are visible when cracking the perithecia. ΔSmatg8 and ΔSmatg4 generate only ascogonia and protoperithecia. Ectopically integrated copies of the Smatg8 and Smatg4 complement the mutant phenotype. (B) Deletion strains are unable to undergo foraging but wt and complemented strains exhibit the ability to grow on an inert plastic surface. Agar plugs of 5-mm diameter were transferred into empty Petri dishes and incubated for 5 d in a damp chamber. Scale bar: 2 mm. (C) Growth velocity of deletion strains and complemented strains. In comparison to the wt the vegetative growth velocity of ΔSmatg8 and ΔSmatg4 was reduced by 39% and 36%, respectively. On medium containing 5 mM 3-AT this effect was increased. Growth-rate analysis was conducted in 30-cm race tubes. Growth rates shown are averages from nine independent measurements of three independent experiments (n = 27), standard deviations are indicated by error bars. Values of the deletion mutants with asterisks differ significantly from wt and the complemented mutant according to Student’s t-test (p < 0.0000001).
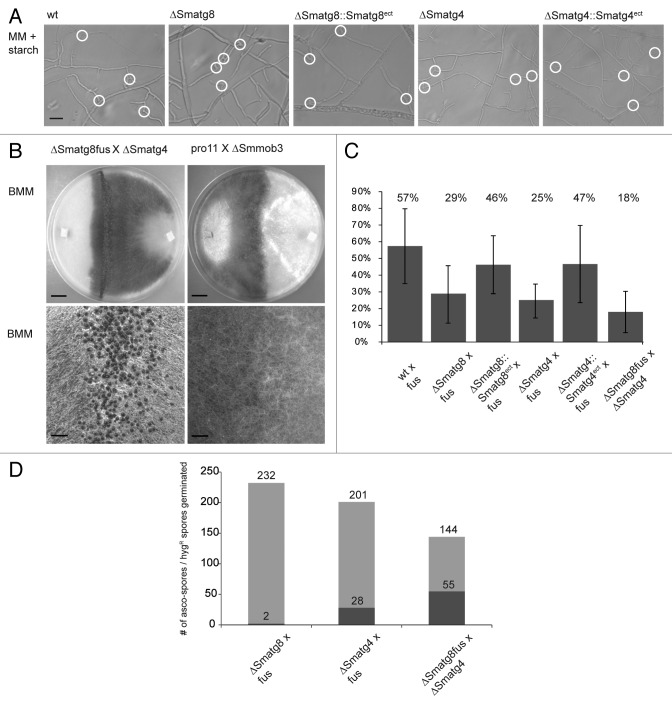
Previous work in S. macrospora has shown that sterile mutants, whose perithecia development ceases at a stage of protoperithecia formation similar to that of the ΔSmatg8 and ΔSmatg4 mutants, are often unable to perform hyphal fusion or form an interconnected mycelial network.42-44 Furthermore, it was recently reported for N. crassa that atg8 is required for intracolonial hyphal cell fusion.45 To test if ΔSmatg8 and ΔSmatg4 mutants were capable of undergoing hyphal fusion, we microscopically analyzed ΔSmatg8 and ΔSmatg4 for the presence of intracolonial hyphal cell fusion and monitored cytoplasmic flow at putative hyphal fusion sites. Figure 5A shows that both mutants exhibited no hyphal fusion defect. In comparison to the wt and the complemented strains, both mutant strains displayed a similar number of hyphal fusion events.
Figure 5. ∆Smatg8 and ∆Smatg4 strains display no defect in hyphal fusion. (A) Microscopy investigation of vegetative hyphal fusion. White circles indicate hyphal fusion events. Hyphal fusions were verified by monitoring cytoplasmic flow. Microscopy pictures were taken at subperiphal regions 5- to 10-mm from the growth front. MM + starch = 0.1% starch-containing minimal medium. Scale bar: 20 µm. (B) After crossing of sterile ∆Smatg8 and ∆Smatg4 strains, fertile perithecia were formed, whereas no fruiting-body development occurred when the sterile hyphal fusion mutants pro11 and ΔSmmob3 were crossed. Lower panel displays close up of the crossing zone. Strains were grown on BMM medium for 10 d. Scale bars: 1 cm and 1 mm (close-up). (C) Determination of germination efficiency of ascospores. As described in the Materials and Methods, sterile deletion strains, complemented and wt strains were crossed with the spore color-mutant fus or a fus isolate of ΔSmatg8 (ΔSmatg8fus) was crossed with ΔSmatg4. 400 spores of each color were isolated and inoculated on BMM supplemented with 0.5% sodium acetate. The percentage of germinated spores was determined for each strain. (D) Determination of the genotype (hygR resistance) of ascospores lines. Colonies grown from germinated ascospores were transferred to hygromycin-containing BMM medium. Light gray = spores germinated; dark gray = spores germinated with deletion background.
Another method to examine hyphal fusion in S. macrospora is to cross two sterile strains. When two sterile strains are used in a cross, complementation of genetic defects results in the formation of fertile perithecia only in the contact zone of two mutant mycelia, all of which are hybrid perithecia. As can be seen in Figure 5B, crossing of the sterile mutant ∆Smatg8 and ∆Smatg4 resulted in the formation of hybrid perithecia in the crossing zone. In contrast, crossing of two previously described hyphal fusion mutants, pro11 and ΔSmmob3 did not result in the formation of hybrid perithecia.42,43
To determine the role of SmATG8 and SmATG4 during ascospore germination, we investigated the ascospore-germination rate of ΔSmatg8 and ΔSmatg4 in comparison to wt and complemented strains by crossing them against a fertile fus strain, which has a mutation in the fus gene that leads to production of brown ascospores (Fig. 5C).39 To compare germination rates of all crosses, spores were only isolated from hybrid perithecia. S. macrospora is a homothallic fungus that produces self-fertile perithecia. The discrimination between self-fertile and hybrid perithecia in crosses of wt strains is difficult. To circumvent this problem, spore-color mutants can be used in crosses. Crosses between wt and spore-color mutants result in hybrid perithecia in the contact zone, with asci containing four black wt spores and four colored spores. In comparison to wt, the germination efficiency of ΔSmatg8 and ΔSmatg4 decreased by about 50% when both mutants were crossed against the fertile fus mutant. The ascospore germination rate dropped even more when the two mutant strains were crossed against each other. To determine the percentage of germinated ascospores carrying the ΔSmatg8 and ΔSmatg4 deletion background, the mycelia of the spore isolates were transferred to BMM medium containing hygromycin. In the case of ΔSmatg8 × fus, only 2 of the 232 ascospore isolates were hygromycin resistant and contained the ∆Smatg8 background, whereas in the ΔSmatg4 × fus cross, 28 of 201 spores displayed the ∆Smatg4 genotype. Crossing of ΔSmatg8fus × ∆Smatg4 resulted in 55 out of 144 hygromycin-resistant ascospore isolates (Fig. 5D). Thus, the autophagy mutants of S. macrospora are capable of hyphal fusion, but due to the impaired germination efficiency of the ascospores, the majority of ascospore isolates carry the wt background. From these experiments we concluded that autophagy as a nutrient-providing process was necessary for fruiting body-development, foraging, vegetative growth and ascospore germination, but not for hyphal fusion.
EGFP-SmATG8 is processed by SmATG4
In S. cerevisiae, it was clearly demonstrated that the C-terminal arginine residue of Atg8 is removed by the cysteine protease Atg4 resulting in a processed Atg8 with a C-terminally exposed glycine residue.15 The alignment of SmATG8 with Atg8 homologs from S. cerevisiae and other fungi (Fig. 1A) showed a high degree of amino acid identity throughout the protein, except for the C-terminal amino acids after the Gly116 residue. To verify that SmATG8 is cleaved by SmATG4, different versions of SmATG8 were either C- (SmATG8-EGFP) or N-terminally (EGFP-SmATG8) fused with EGFP. Protein extracts isolated from wt, ΔSmatg8 or ΔSmatg4 expressing the EGFP-tagged versions of SmATG8 were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and examined by western blot analysis using an anti-EGFP antibody. A wt strain expressing an ectopically integrated egfp gene and an untransformed wt were used as a control (Fig. 6). Signals of 40 kDa, representing the EGFP-SmATG8 fusion protein and of the free 26 kDa EGFP, were visible when proteins were extracted from ∆Smatg8::egfp-Smatg8ect. In the wt strain expressing an ectopic Smatg8-egfp fusion gene (wt::Smatg8-egfpect) a band that presumably represented the DFETA-EGFP apparently ran slightly slower than free EGFP (Fig. 6). Only the unprocessed fusion protein was visible when a mutated version of SmATG8 was produced in the wt (wt::Smatg8mut-egfpect). In this construct, amino acids 115–118 (including Gly116) were exchanged for Ala. Similarly, only the 40-kDa band was detected when N- or C-terminally EGFP-tagged versions of SmATG8 were produced in ΔSmatg4 (∆Smatg4::egfp-Smatg8ect, ∆Smatg4::Smatg8-egfpect). Based on these results, we concluded that SmATG4 is an essential protease for SmATG8 processing.
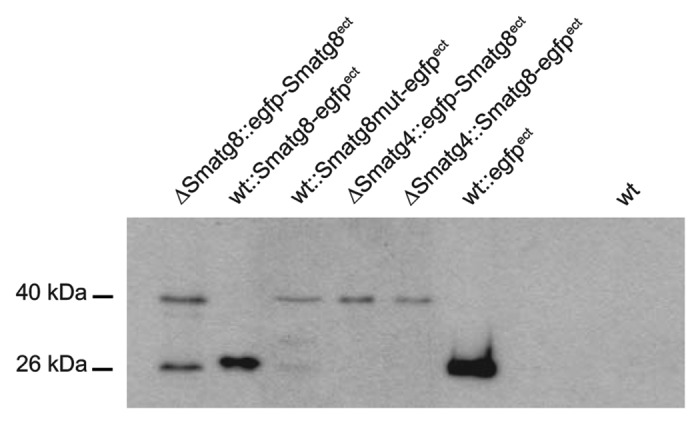
Figure 6. SmATG8 is C-terminally processed by SmATG4. Cleavage of EGFP-SmATG8 by SmATG4 was verified by western blotting using an anti-EGFP antibody. Protein crude extract of S. macrospora wt and mutant strains expressing EGFP, EGFP-SmATG8, SmATG8-EGFP or SmATG8mut-EGFP were separated on a 15% SDS-PAGE gel. The protein crude extract of the untransformed wt was used as a negative control. Cleaved or free EGFP is indicated by a 26 -kDa band, and fusion proteins of SmATG8 and EGFP are 40 kDa in size.
EGFP-SmATG8 localizes to autophagosomes and vacuoles, whereas SmATG4-EGFP is distributed within the cytoplasm
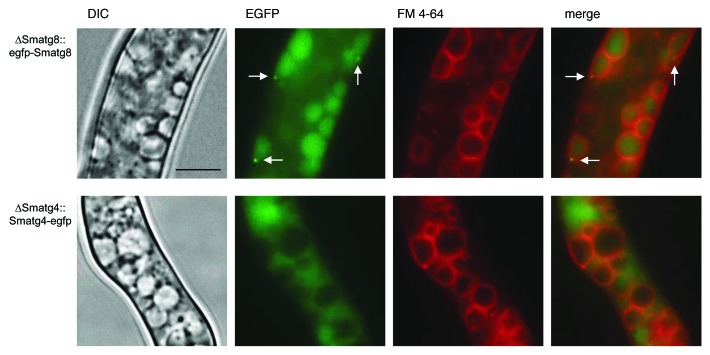
To analyze the cellular localization of SmATG8 and SmATG4 in S. macrospora, we performed fluorescence microscopy. We transformed plasmids pRS-Smatg4-egfp and pRS-egfp-Smatg8 carrying egfp fusion genes under control of their endogenous promoters into the corresponding S. macrospora deletion mutant. When compared with the wt, the obtained strains ∆Smatg8::egfp-Smatg8ect and ∆Smatg4::Smatg4-egfpect showed no obvious difference in vegetative growth and sexual development, indicating that EGFP-SmATG8 and SmATG4-EGFP fusion proteins were functional (data not shown). Fluorescence microscopy revealed that EGFP-SmATG8 was localized to punctate autophagosome-like structures in the cytoplasm and to the lumen of vacuoles, while SmATG4-EGFP was distributed diffusely within the cytoplasm (Fig. 7).
Figure 7. EGFP-SmATG8 localizes to autophagosomes and is degraded in vacuoles while SmATG4-EGFP is distributed within the cytoplasm. Fluorescence microscopy analysis of the S. macrospora ΔSmatg8 and ΔSmatg4 strain carrying plasmid pRS-egfp-Smatg8 and pRS-Smatg4-egfp, respectively. The upper panel shows that EGFP-SmATG8 localizes to autophagosomes and vacuoles. Autophagosomes are indicated by an arrow. SmATG4-EGFP expression in ΔSmatg4 leads to a diffuse cytoplasmic signal which is excluded from the vacuole. For the visualization of the vacuolar contours, vacuolar membranes were stained with FM 4-64 as described in the Materials and Methods. Strains were grown on SWG layered slides or cellophane. Scale bar: 10 µm.
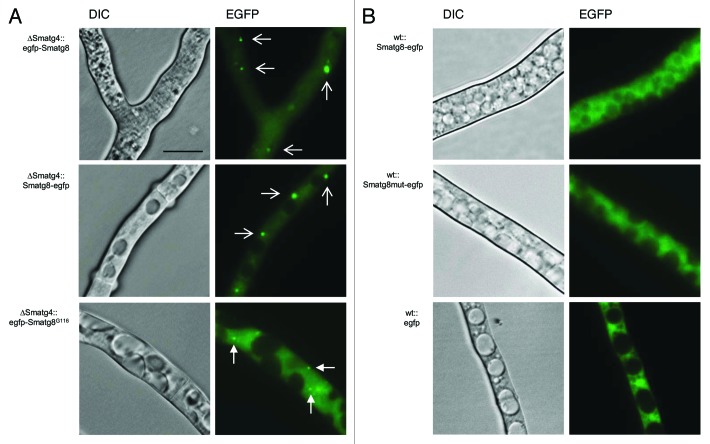
In the mutant ∆Smatg4, EGFP-SmATG8 and the C-terminally tagged version SmATG8-EGFP were observed in large aggregates that were excluded from the vacuole. Thus, in both strains, an accumulation of EGFP was observed, but fluorescence of small autophagosomes and vacuoles was absent (Fig. 8A). In contrast, the C-terminally tagged SmATG8-EGFP in the wt seemed to be cleaved, and displayed a similar distribution as the wt transformant expressing egfp under the control of the strong constitutive gpd promoter of A. nidulans (Fig. 8B). The same localization can be observed when the SmATG8-EGFP version with the mutated processing site was expressed in the wt (wt::Smatg8mut-egfpect) (Fig. 8B). To verify if a processed version of SmATG8 can complement the ΔSmatg4 mutant, we constructed a C-terminally truncated EGFP-SmATG8 version ending with Gly116 and the strain ∆Smatg4::egfp-Smatg8G116ect was generated. In this strain, the EGFP fluorescence localized to small autophagosome-like structures similar to those observed in ∆Smatg8::egfp-Smatg8ect (Fig. 7; Fig. 8A). However, expression of the SmATG8G116 processed version did not complement sterility or the slow growth phenotype of ΔSmatg4 (data not shown). Taken together, these experiments indicated that SmATG4 was able to C-terminally process SmATG8 most probably by cutting the C-terminal motif DFETA, but it seemed to be also required for SmATG8 recycling.
Figure 8. Localization of EGFP-tagged versions of SmATG8 in ΔSmatg4 and wt. (A) In the ∆Smatg4 mutant, EGFP-SmATG8 and SmATG8-EGFP localize to large aggregates (wide arrows) whereas a processed EGFP-SmATG8G116 version localizes to autophagosomes (narrow arrows). (B) In the wt, SmATG8-EGFP and its mutagenized version SmATG8mut-EGFP are mainly visible in the cytoplasm, similar to EGFP. Strains were grown on SWG layered slides or cellophane. Scale bar: 10 µm.
SmATG8 and SmATG4 are involved in pexophagy
Recently, it was demonstrated in A. oryzae that macroautophagy mediates degradation of nuclei, mitochondria and peroxisomes in basal cells of the mycelium to support the growth of tip cells.9 In order to analyze the involvement of SmATG8 and SmATG4 in pexophagy, we constructed plasmid pRS-egfp-Smatg8-DsRed-SKL encoding EGFP-SmATG8 together with DsRed fused to a C-terminal serine-lysine-leucine (SKL), which represents the peroxisomal targeting sequence 1 (PTS1) signal for peroxisomal matrix import. Previously, we demonstrated that in S. macrospora, expression of the DsRed-SKL fusion gene under the control of the A. nidulans gpd-promoter led to protein import of DsRed into peroxisomes.46
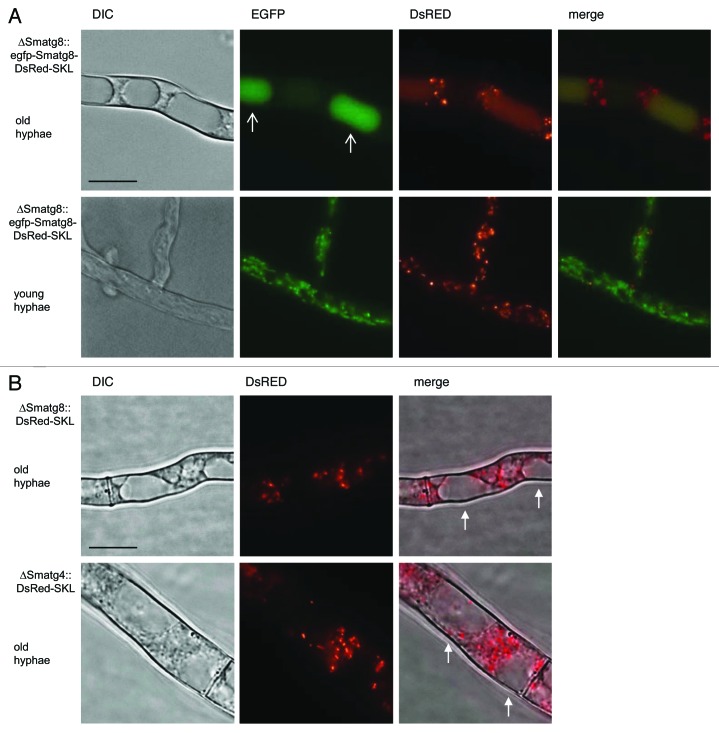
Plasmid pRS-egfp-Smatg8-DsRed-SKL was transformed into ΔSmatg8. DsRed and EFGP fluorescence was analyzed by fluorescence microscopy in the obtained strain ∆Smatg8::egfp-Smatg8-DsRed-SKLect. In the basal hyphae, EGFP signals were observed in the lumen of vacuoles, while the DsRed signal also localized to the lumen of vacuoles and to punctate peroxisomal structures (Fig. 9A, upper panel). The merged picture showed a yellow staining of the vacuolar lumen, indicating colocalization of DsRed and EGFP and the transport of these proteins into the vacuole. In the tip cells, EGFP-SmATG8 and DsRed-SKL localization differed from that in basal hyphae. The EGFP signal was mainly localized to autophagosomes and some small vacuoles, but excluded from larger vacuoles, while DsRed signals displayed only punctate fluorescent patterns (Fig. 9A, lower panel). An overlay showed no colocalization of EGFP and DsRed signals, indicating that pexophagy occurred in basal hyphae rather than in apical cells.
Figure 9.Smatg8 and Smatg4 are involved in pexophagy. (A) Localization of EGFP-SmATG8 and DsRed-SKL in basal hyphae (upper panel) and in apical hyphae (lower panel) of ∆Smatg8. Arrows indicate vacuoles. In basal hyphae DsRed-SKL and EGFP-SmATG8 are localized to the lumen of vacuoles. In apical hyphae peroxisomes are not degraded in the vacuoles. (B) DsRed-SKL is excluded from the vacuole in basal hyphae of ∆Smatg8 and ∆Smatg4. Arrows indicate vacuoles free of the DsRed-SKL fluorescence signals or cellophane. Scale bar: 10 µm.
To determine the impact of SmATG8 and SmATG4 on pexophagy, plasmid pDsRed-SKL was transformed into the ΔSmatg8 and ΔSmatg4 strains. Consistent with reports in A. oryzae,9 in the absence of Smatg8 and Smatg4, DsRed fluorescence was only visible in small spots, but no DsRed signals were observed in the vacuoles of basal hyphae (Fig. 9B). Therefore, we concluded that pexophagy was arrested in the Smatg8 and Smatg4 deletion mutants.
Discussion
Autophagy proteins SmATG8 and SmATG4 are highly conserved and can rescue S. cerevisiae mutants
The ubiquitin-like S. macrospora protein SmATG8 reveals a high degree of sequence identity when compared with the S. cerevisiae Atg8 and other fungal Atg8 orthologs. This includes the conserved core hydrophobic side chains Phe77, Phe79, as well as Leu84 and Arg65, which in S. cerevisiae and the human LC3 (an ATG8 homolog), were shown to be part of the recognition site for the cysteine protease Atg4 and the E1-enzyme Atg7. In addition, the latter enzyme interacts with conserved residues Glu112, Asn113 and Thr114 of the Atg8 tail region via a salt bridge and a network of hydrogen bonds. Furthermore, Tyr49 and Leu50, which were demonstrated to be essential for Atg8 function, and Phe115 and Gly116, which are substantial for the Atg4 dependent-processing, are also perfectly conserved in the Atg8 homologs of filamentous ascomycetes.47-49 However, the last C-terminal amino acids after the conserved Gly116 residues displayed a high variability (Fig. 1A). In contrast to Atg8, the overall amino acid identity of Atg4 orthologs were less conserved. Only the predicted peptidase C-54 domain at the C terminus was conserved among fungal Atg4 orthologs (Fig. 1B). Recently, studies on M. oryzae have shown that Cys206 of the C-54 domain is the catalytic center of MoAtg4, which is responsible for processing of the M. oryzae MoAtg8.17 This cysteine residue (Cys236) as well as two other residues of the catalytic triad of cysteine proteases (Asp348 and His350) were conserved in SmATG4 and in other fungal Atg4 orthologs (Fig. 1B).50
The functional conservation of the Smatg8 and Smatg4 orthologs was confirmed by rescue of S. cerevisiae atg8∆ and atg4∆ deletion strains. Complementation of the deletion strains was monitored using a prApe1 maturation assay.37 The Smatg8 cDNA was able to complement the prApe1 maturation defect of the atg8∆ strain to the same extent as the endogenous S. cerevisiae EGFP-ATG8 (Fig. 2A). This result was not surprising, considering the high degree of sequence identity between SmATG8 and the budding yeast Atg8 (76%), and the conservation of residues essential for interaction with Atg7 and Atg4. In a previous study, we demonstrated that Smatg7 was capable of partially complementing ascospore-formation deficiency in a S. cerevisiae atg7∆ strain.31 Similarly, complementation of the prApe1 maturation defect in the S. cerevisiae atg4∆ mutant was only partially rescued by expression of the Smatg4 cDNA (Fig. 2B). Recently, it was demonstrated that the M. oryzae MoATG4 cDNA rescued starvation sensitivity of the S. cerevisiae atg4∆ mutant. However, it was unclear whether this was a partial or a full complementation, since the determination of the prApe1 maturation efficiency seems to be more precise than the rescue of starvation sensitivity.17 Expression of the Arabidopsis thaliana AtATG8a and AtATG8d orthologs only partially rescues prApe1 maturation of a S. cerevisiae atg8∆ strain, whereas AtATG4b almost completely complements an atg4∆ strain.51
In accordance with studies performed using ATG8 and ATG4 orthologs of M. oryzae and A. thaliana, we demonstrated, by means of a yeast two-hybrid study, that SmATG8 and SmATG4 interact with each other, as do their yeast orthologs.17,51 Recently, Liu et al.17 tried to confirm in vivo interaction of M. oryzae MoAtg4 and MoAtg8 by means of a bimolecular fluorescence complementation assay, but detected an interaction only in vegetative hyphae grown under nitrogen starvation conditions. Furthermore, we demonstrated that SmATG8 is C-terminally processed by SmATG4. Western blot experiments revealed that the SmATG8-EGFP fusion protein can be processed in the wt, but not in the ΔSmatg4 deletion strain or when the putative processing site was mutated (Fig. 6). These results corroborate that SmATG8 and SmATG4 might also interact in vivo.
Smatg8 and Smatg4 are involved in vegetative growth, fruiting-body development and ascospore germination
In contrast to Smatg7, we succeeded in deleting Smatg8 and Smatg4 to generate homokaryotic ΔSmatg8 and ΔSmatg4 knockout strains from ascospore isolates of the primary transformants. Previously, we suggested that SmATG7-mediated autophagy is at least essential for ascospore germination and the establishment of mycelial growth during the regeneration of protoplasts.31 However, here we showed that germination of ascospores is not per se abolished in autophagy mutants, although germination efficiency of the ΔSmatg8 and ΔSmatg4 deletion strains was significantly decreased (Fig. 5C). Thus, autophagy seems to be important for a proper nutrient supply in the germinating ascospore. Previously, it was demonstrated that in A. oryzae and the plant pathogen M. grisea, autophagy-related proteins ATG8 and ATG4 were involved in early stages of conidial germination.21,25,27,28,52,53 In F. graminearum, FgATG8 deletion resulted only in the reduction of conidiospore production.41 However, despite the fact that ascospores carrying the ΔSmatg8 or ΔSmatg4 background can germinate, the growth velocity of the received mycelium was significantly reduced under normal conditions, but was restricted even more under amino acid starvation conditions or on an inert plastic surface when compared with wt (Fig. 4B and C). This was similar to reports on other filamentous fungi, which showed that deletion of either ATG8 or ATG4 also leads to a decrease in vegetative growth velocity, whereas idi7 deletion, the ortholog of ATG8 in P. anserina, shows no influence on the linear growth rate, but only on the hyphal density.17,23,25,27,41,53 In U. maydis, atg8 deletion affects survival during carbon starvation and pathogenic growth.19 Thus, autophagy is required for vegetative growth of filamentous fungi in general, but seems to be indispensable for growth under nutrient-limiting conditions. Recently, Shoji and Craven28 found that autophagy-mediated degradation of basal cell components, including nuclei and other organelles, were closely connected to the tip growth of filamentous fungi.
In addition to the reduced growth velocity, mycelial fungi deficient in ATG4 and ATG8 display a reduced density of the aerial hyphae and an absent or decreased conidiation.17,23,25,27,54 An altered morphology of aerial hyphae in the S. macrospora ΔSmatg8 and ΔSmatg4 mutants was not observed and the effect on conidiation cannot be analyzed because S. macrospora produces no asexual spores. However, as in P. anserina, M. oryzae and F. graminearum, deletion of Smatg8 and Smatg4 affected sexual development.17, 23, 39 ∆Smatg8 and ∆Smatg4 mutants are incapable of perithecia and ascospore formation and generated only the precursors of fruiting bodies, protoperithecia, in low numbers (Fig. 4A).
In S. macrospora, many genes involved in fruiting-body development have been characterized and several have shown a phenotype similar to ΔSmatg8 and ΔSmatg4. They have been described as pro-mutants, whose sexual development ceases at the point of protoperithecia formation.30 Another characteristic shared by most pro-mutants is their incapability of hyphal fusion, which is thought to be essential for the formation of multicellular sexual structures in ascomycetes.42-44 Surprisingly, hyphal fusion was not affected and hyphal fusion events occurred in equal numbers in ΔSmatg8 and ΔSmatg4 when compared with wt and complemented strains (Fig. 5A). In contrast to crossing of the hyphal fusion mutants pro11 and ∆Smmob3, crossing of sterile ∆Smatg8 and ∆Smatg4 strains resulted in mature hybrid perithecia at the contact zone of both mycelia (Fig. 5B), once again implying that autophagy affects sexual development, but not hyphal fusion. For N. crassa, it has recently been reported that deletion of Ncatg8 (NCU01545) prevents intracolonial hyphal cell fusion, but an effect on fruiting-body development was not analyzed in that study.45
EGFP-SmATG8 is localized to autophagosomes, while SmATG4-EGFP is distributed in the cytoplasm
Cellular localization studies using functionally expressed EGFP-SmATG8 fusion proteins revealed that SmATG8 was found to be localized in small dots, assumingly representing autophagosomes, and to vacuoles, indicating degradation of EGFP-SmATG8 delivered to vacuoles as structural components of autophagosomes (Fig. 7, upper panel). As previously described in yeast, EGFP-Atg8 fusion proteins assemble to autophagosomes and are visible as fluorescent dots that are also observed in other filamentous fungi, including P. anserina, M. oryzae and A. oryzae and even in mammals.21,22,27,55 In contrast to EGFP-SmATG8, SmATG4-EGFP was not focused in dot-like structures, but rather dispersed throughout the cytoplasm (Fig. 7, lower panel). The same findings have been reported for Atg4 localization experiments in M. oryzae and A. oryzae, whereas in S. cerevisiae, Atg4-EGFP is dispersed in the cytoplasm and localized to nuclei.17,21,25,56 Nuclear localization of SmATG4-EGFP could not be detected in our study. SmATG4 seems to be necessary for proper processing and autophagosome formation, since expression of EGFP-SmATG8 and SmATG8-EGFP in the ∆Smatg4 mutant led to the formation of larger aggregates instead of small punctate autophagosomes. Similar to our results, an accumulation of EGFP-Atg8 into aggregates has been observed in M. oryzae and A. oryzae ATG4 deletion strains.21,25 It remains unclear whether accumulation of unprocessed EGFP-SmATG8 is formed by aggregation of EGFP or SmATG8. However, when the wt expressed SmATG8-EGFP or a mutated version, SmATG8mut-EGFP, the fluorescence was dispersed in the cytoplasm and did not accumulate in aggregates (Fig. 8).
Interestingly, the expression of the putatively processed form EGFP-SmATG8Gly116 in ∆Smatg4 led to the formation of small punctate autophagosome-like structures confirming our assumption that Gly116 of SmATG8 is C-terminally exposed by the catalytic activity of SmATG4 und can undergo lipidation. Similar observations have been reported in S. cerevisiae, where introduction of EGFP-ATG8G116 in an atg4∆ mutant also resulted in formation of wt-like autophagosomes. Expression of the glycine-exposed Atg8G116 in the yeast atg4Δ mutant results in significant autophagy defects.15,56 Just like in yeast, we showed that expression of the putatively processed form of SmATG8 did not complement the ΔSmatg4 phenotype, suggesting that delipidation and recycling of SmATG8 is also an important function of SmATG4.
Deletion of Smatg8 and Smatg4 prevents pexophagy
In order to investigate the involvement of SmATG8 and SmATG4 in selective macroautophagy, we examined the extent of pexophagy in young and old hyphae of deletion mutants. Transformation of pRS-egfp-Smatg8-DsRed-SKL into ∆Smatg8 complemented the phenotype of the mutant. Thus, this construct allowed us to simultaneously follow fluorescence signals of DsRed-SKL-labeled peroxisomes and EGFP-labeled autophagosomes. Only in basal hyphae, but not in tip cells, both fluorescence signals were colocalized to the vacuole, indicating that peroxisomes were delivered to the vacuole as cargo of autophagosomes and were degraded together (Fig. 9A). As shown for A. oryzae and P. chrysogenum, our result suggested that autophagic degradation of peroxisomes preferentially takes place in old basal hyphae, but not in the growing hyphal tips.9,26 In Smatg8 and Smatg4 deletion mutants, pexophagy was abolished (Fig. 9B). Similarly, it was recently shown that pexophagy is not observed in a Pichia pastoris PpATG8 deletion mutant.6 In P. chrysogenum, deletion of atg1 prevents pexophagy and, furthermore, results in a significantly increased number of peroxisomes in subapical hyphae.26 However, in S. macrospora, an increased number of peroxisomes was not observed in ΔSmatg8 and ΔSmatg4 mutants.
In conclusion, our study demonstrated that in S. macrospora even under nonstarvation conditions, autophagy seems to be an essential and constitutively active process for filamentous growth, the development of multicellular fruiting bodies and the production of a sexual progeny. Our future studies will address the impact of filamentous fungus-specific genes on autophagy-mediated vegetative and sexual development processes.
Materials and Methods
Strains and culture conditions
Cloning and amplification of recombinant plasmids was achieved in an Escherichia coli Mach1 strain under standard culture conditions.57 All plasmids used in this study are listed in Table S1. All fungal strains used and generated during this work are listed in Table 1. Depending on the experimental setup S. macrospora was cultivated on Biomalt Maiz Medium (BMM), complete medium containing 10.8% saccharose (CMS) and fruiting-body development-inducing SWG medium.46,58 S. cerevisiae was inoculated on YEPD or SD minimal medium.59 S. macrospora was grown at 27°C in liquid medium for genomic DNA, RNA and protein extraction.60 Determination of growth velocity of the S. macrospora strains was performed as described by Nolting and Pöggeler.61 For microscopy analysis, S. macrospora was inoculated on objective slides coated with 1–2 ml SWG medium or on a cellophane layer on solid SWG medium and incubated at 27°C.42 For examination of the foraging abilities of S. macrospora strains, a plug test was performed according to Josefsen et al.41: an agar plug with a diameter of 5 mm was put into an empty Petri dish and incubated for 5 d in a damp chamber at 27°C. Mycelial growth was visualized using a Digital Microscope VHX-500F (Keyence).
To determine germination efficiency of ascospores from hybrid perithecia, deletion and complementation strains were crossed with a fus S23442 (spore-color mutant) strain. A total of 400 spores of each color were isolated from hybrid perithecia and inoculated on BMM supplemented with 0.5% sodium acetate. Germinated spores were counted after 1, 2 and 3 d. Obtained colonies were then transferred to hygromycin selective media to elucidate whether the ascospore isolates carried a wt or knockout phenotype.
Transformation techniques
Transformation of chemically competent E. coli was achieved by standard transformation protocols.57 S. cerevisiae transformations were performed by electroporation using an Eppendorf Electroporator 2510 (Eppendorf) at 1.5 kV.62 Transformation of S. macrospora was conducted as described previously.58,63 S. macrospora transformants were selected on media containing nourseothricin-dihydrogen sulfate (50 µg/ml) (WernerBioAgents, 5004000) or hygromycin B (110 U/ml) (Merck, 400051-10MU).
Preparation of nucleic acids and PCR
S. macrospora genomic gDNA isolation was performed as previously specified.64 PCR amplification of S. macrospora gDNA and cDNA was performed with HotStarTaq Master Mix Kit (Qiagen, 203443), Phusion High-Fidelity DNA polymerase (New England Biolabs, M0530S), Molzyme MolTaq polymerase (Molzym GmbH & Co. KG, P-010-1000) or Pfu polymerase (Promega GmbH, M7741) according to the manufacturer’s recommendations. Primers used in this study were obtained from MWG Biotech (Eurofins MWG GmbH) and are listed in Table S2.
Generation of the S. macrospora Smatg8 and Smatg4 deletion and complementation strains
For the isolation of the S. macrospora autophagy genes Smatg8 (SMAC_02305) and Smatg4 (SMAC_08321) a TBLASTN search of the S. macrospora genomic sequence35 was performed using the amino acid sequence of S. cerevisiae ATG8 (YBL078C) and ATG4 (YNL223W), respectively.
The generation of the deletion and complementation constructs was conducted using the homologous recombination mechanism of S. cerevisiae.65 For deletion cassette generation 5′- and 3′-regions of Smatg8 and Smatg4 were amplified from wt gDNA using the primer pair atg8-5f/atg8-5r and atg8-3f/atg8-3r for Smatg8 and atg4-5f/atg4-5r and atg4-3f/atg4-3r for Smatg4. During the PCR amplification, specific 29-bp overhangs either homologous to the S. cerevisiae shuttle vector pRS42666 or to the hygromycin-resistance cassette, were added to the 5′- and 3′-flank. The hph cassette was amplified using the primer pair hph-f/hph-r and plasmid pCB1003 as template. The three PCR amplicons obtained and the EcoRI/XhoI linearized vector pRS426, were co-transformed into S. cerevisiae strain PJ69-4A,38 where the fragments were fused by homologous recombination. The resulting plasmids pRS-∆atg8 and pRS-∆atg4 (Table S1) were isolated and used as a template to amplify deletion cassettes with primer pairs atg8-5f/atg8-3r and atg4-5f/atg4-3r, respectively. Subsequently, 3400-bp Smatg8 and Smatg4 deletion cassettes were transformed into the S. macrospora ∆ku70 strain which is enhanced in homologous recombination events.67 As S. macrospora transformants are often heterokaryotic and carry both transformed and nontransformed nuclei, single spore isolates were generated from the primary transformants. To eliminate the Δku70::natR background nourseothricin- and hygromycin- resistant single spore isolates were afterwards crossed with the S. macrospora spore color mutant fus (S23442) and hygromycin-resistant, respectively, nourseothricin-sensitive spores were isolated. The confirmation of the desired gene deletion was achieved by PCR and Southern blot analysis (Figs. S1 and S2). To verify the integration of the deletion cassette at the targeted Smatg8 locus, primer pairs atg8-5D1/tC1 and atg8-3D1 /h3-o were used whereas primer pair atg8-ver-f/atg8-ver-r was used to confirm the presence of Smatg8 in the S. macrospora wt and complemented strain and the absence of Smatg8 in the homokaryotic ΔSmatg8 deletion strain (Fig. S1). The verification of the Smatg4 deletion and complementation was performed in the same manner using the primer pairs atg4-5D1/tC1-o, atg4-3D1/h3-o and atg4-ver-f/atg4-ver-r (Fig. S2).
To complement the phenotype of ΔSmatg8 and ΔSmatg4 mutants, plasmids pRS-Smatg8-comp and pRS-Smatg4-comp (see Table S1) were constructed by amplifying the 5′- and 3′- regions together with the entire coding regions of Smtatg8 and Smatg4 with primer pairs atg8-5f/atg8-3r and atg4-5f/atg4-3r from gDNA of the wt, respectively. Amplified fragments were integrated into the EcoRI linearized vector pRSnat68 by homologous recombination in S. cerevisiae. Plasmid pRS-Smatg8-comp and pRS-Smatg4-comp was transformed into deletion strains ΔSmatg8 and ΔSmatg4, respectively. Complemented transformants were selected on medium containing nourseothricin and inspected for perithecia development. The reintegration of the ectopic copy of the desired wild-type gene was confirmed by PCR using the primer pair atg8-ver-f/atg8-ver-r for the ∆Smatg8::Smatg8ect and atg4-ver-f/atg4-ver-r for ∆Smatg4::Smatg4ect strains, respectively (Figs. S1 and S2).
In addition, deletion and complementation events were confirmed by Southern hybridization (Figs. S1 and S2). Southern blotting was done according to standard techniques.57 Hybridization was done with DIG High Prime DNA Labeling and Detection Starter Kit II (Roche Diagnostics GmbH, 1585614). DNA probes were obtained by PCR with primers hph-f and hph-r (see Table S2), and labeling and detection was done according to the manufacturer’s protocol.
Light and fluorescence microscopy
Egfp-fusion constructs were integrated ectopically into the genome of wt and deletion strains. To examine the localization of SmATG8 and EGFP-SmATG8 fusion proteins under their native promoters and terminators was constructed. The 5′- untranslated region (UTR) of Smatg8 was amplified using wt gDNA as template and the primer pair atg8-5f/atg8-egfp-5r (Table S2). The forward primer atg8–5f contains a 29-bp overhang to the pRSnat vector and the reverse primer atg8-egfp-5r an overhang to the egfp gene. The egfp gene without stop codon was amplified using the primer pair egfp-f/egfp-r and p178369 as template. Primer pair atg8-egfp-3f/atg8-egfp-3r was used to amplify the coding region of Smatg8 without the start codon and its 3′-UTR. Both primers have an overhang homologous to the 3′-end of egfp and to the pRSnat vector, respectively. The fragments were fused in S. cerevisiae via homologous recombination and plasmid pRS-egfp-Smatg8 was obtained. Generation of a C-terminal SmATG8-EGFP fusion construct was achieved using the primer pair atg8-gf/atg8-gr; here the forward primer generated a BglII overhang and the reverse primer a HindIII overhang. Applying these primers, the stop codon was removed from the Smatg8 ORF. Subsequently, the fragment was cloned into the BglII/HindIII linearized pDS23-eGFP (M. Nowrousian, unpublished), a pRS426 derivative with egfp under control of a gpd promoter and trpC terminator of A. nidulans (Table S1). The resulting plasmid was termed pRS-Smatg8-egfp. The construction of a C-terminal SmATG4-EGFP fusion encoded by plasmid pRS-Smatg4-egfp was conducted in the same manner with the primer pair atg4-gf/atg4-gr.
For the generation of an EGFP-SmATG8mut fusion protein encoded by plasmid pRS-egfp-Smatg8mut, we mutagenized the putative SmATG4 processing site of SmATG8. To amplify the mutated version of Smatg8 we used the primer pair atg8-gf/atg8-gr2 to substitute amino acid residues 115–118 of SmATG8 to Ala. This fragment was cloned also into pDS23-eGFP. Plasmid pRS-egfp-Smatg8G116 encodes the putatively processed form of SmATG8 that lacks amino acids 117–121 and exposes a C-terminal Gly116. It was generated using primer pair atg8-5f/atg8p5r and atg8p-3f/atg8-3r, respectively with pRS-egfp-Smatg8 as a template. Fragments consisting of Smatg8 5′-UTR, egfp and a truncated Smatg8 as well as the 3′-region of Smatg8 were amplified. The obtained fragments were cloned into pRSnat via homologous recombination in S. cerevisiae. To study the involvement of Smatg8 and Smatg4 in pexophagy we generated the plasmid pRS-egfp-Smatg8-DsRed-SKL encoding both an EGFP-SmATG8 and a DsRed-SKL fusion protein. Primer pair atg8-5f3/atg8-org-5r amplified a fragment consisting of Smatg8 5′-region, egfp, Smatg8 and its 3′-UTR using pRS-egfp-Smatg8 as template. A fragment containing a gpd promoter, DsRed-SKL and a trpC terminator was amplified using the primer pair atg8-org-3f/MiPe-r2 and pDsRed-SKL46 as a template. Both fragments were cloned into pRSnat by homologous recombination in S. cerevisiae.
For the visualization of hyphae, ascogonia and protoperithecia an AxioImager M1 microscope (Zeiss, purchased by Visitron Systems GmbH) was utilized. A Photometrics CoolSNAP2HQ camera (Roper Scientific, purchased by Visitron Systems GmbH) was used to obtain the images. EGFP fluorescence was visualized with the filter combination chroma filter set 49002 (exciter ET470/40×, emitter ET525/50 m and beamsplitter T495LP) and detection of DsRed and FM 4-64 dye (Invitrogen, F34653) was achieved with a chroma filter set 49005 (excitation/emission filter ET545/30/ET620/60, beam splitter T570lp) and an X-cite 120 PC lamp (EXFO). Image editing was performed with MetaMorph (Visitron Systems GmbH), Adobe Photoshop CS2 and Illustrator CS2. Staining with FM 4-64 was achieved by applying 20–40 µl of a FM 4-64 solution (1 µg/ml in dH2O) directly on the mycelium.
Immunoblotting
For detection of EGFP-SmATG8, SmATG8-EGFP, SmATG8mut-EGFP and EGFP-SmATG8G116 fusion proteins S. macrospora protein extracts were separated by SDS-PAGE using a 15% gel and blotted to a PVDF membrane.42,70,71 Immunodetection of the fusion proteins was achieved with monoclonal mouse anti-EGFP antibody (Santa Cruz Biotechnology, sc-9996, 1:5000). As secondary antibody a goat anti-mouse horseradish peroxidase (HRP)-linked antibody (Dianova, 115-035-003, 1:5000) was used. Detection of signals was achieved by the enhanced chemiluminescent reaction (Agilent Technologies, 204525). The prApe1 maturation assay in S. cerevisiae was performed as described previously37 with the polyclonal rabbit anti-Ape1 antibody (M. Thumm, 1:2500) and secondary HRP-linked anti-rabbit antibody (Invitrogen, G21234, 1:2500). After stripping of nitrocellulose membrane by incubating it for 1 h at RT in a solution of 0.2% Ponceau S dissolved in 3% TCA, and subsequent blocking, the calibration of the western blot was done by immunodetection of actin with a monoclonal mouse anti-actin (Novus, NB100-74340, 1:2500) and secondary HRP-linked goat anti-mouse antibody (Dianova, 115-035-003, 1:2500).
Yeast two-hybrid interaction studies
Bait plasmids pBD-Smatg8 and pBD-Smatg4 were constructed by cloning an amplified cDNA fragment using primer pair atg8-Hf/atg8-Hr and atg4-Hf3/atg4-Hr, respectively, into vector pGBKT7 (Clontech, 630443). To generate the prey vector pAD-Smatg8 the Smatg8 amplicon was cloned into the vector pGADT7 (Clontech, 630442). For cloning of the pAD-Smatg4 prey plasmid primers atg4-Hf3/atg4-Hr3 were utilized. Bait plasmids were transformed into the S. cerevisiae strain Y187 and prey plasmids into S. cerevisiae strain AH109, by a lithium acetate transformation protocol and selected on SD-Trp and SD-Leu, respectively.72 Mating of transformants was performed in YPDA liquid medium overnight at 30°C and mated cells were selected on SD-Trp/Leu solid medium. Interaction was verified on SD-Ade/Leu/Trp solid medium. To determine whether the bait proteins are expressed appropriately for an interaction to be detected we used a test, based on interaction with the Gal4 binding domains with protein RanBPM.40
Sequence analysis
Primers were synthesized at MWG Biotech AG (Ebersberg). DNA sequencing was performed by the G2L-sequencing service of the “Göttinger Genom Labor” (Georg-August University of Göttingen). Molecular weights and isoelectric points of proteins were calculated with programs from the ExPASy Proteomics Server (www.expasy.org). Protein sequence alignments were performed using the ClustalX program73 and visualized using GeneDoc.74 Protein and nucleotide sequence data of Atg8/ATG8 and Atg4/ATG4 from other organisms were obtained from the public databases at NCBI (www.ncbi.nlm.nih.gov/entrez/) or by BLAST searches of the complete sequenced genomes at the Broad Institute (www.broad.mit.edu/annotation/fungi/fgi/).
Supplementary Material
Acknowledgments
We thank Gertrud Stahlhut for excellent technical assistance, Regina Ricke and Nicole Nolting for help with some experiments and Britta Herzog for critically reading the manuscript. We are very grateful to Prof. Dr. Michael Thumm (Department of Biochemistry II, UMG, Georg-August University Göttingen) for kindly providing yeast ATG8 and ATG4 deletion strains and the Ape1 antibody, Peter Rube (Department of Biochemistry II, UMG, Georg-August University Göttingen) for his help with the yeast complementation studies and the prApe1 maturation assay, Dr. Minou Nowrousian and Prof. Dr. Ulrich Kück (Department of General and Molecular Botany, Ruhr-University Bochum, Germany) for providing plasmid pDS23-eGFP and S. macrospora strains, respectively.
Glossary
Abbreviations:
- aa
amino acids
- BLAST
Basic Local Alignment Search Tool
- BMM
Biomalt Maize Medium
- bp
base pairs
- CM
complete medium
- CMS
complete medium with saccharose
- DIC
differential interference contrast
- dpi
days past inoculation
- EGFP
enhanced green fluorescent protein
- kb
kilobase pairs
- kDa
kilodalton
- ORF
open reading frame
- PCR
polymerase chain reaction
- PE
phosphatidylethanolamine
- RT
room temperature
- RT-PCR
reverse transcriptase PCR
- SD
selective dropout
- SWG
Sordaria Westergaards medium
- wt
wild type
- YEPD
yeast extract, peptone, dextrose
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/22398
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/22398
References
- 1.Reggiori F, Klionsky DJ. Autophagy in the eukaryotic cell. Eukaryot Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uttenweiler A, Mayer A. Microautophagy in the yeast Saccharomyces cerevisiae. Methods Mol Biol. 2008;445:245–59. doi: 10.1007/978-1-59745-157-4_16. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–81. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–88. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farré JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14:365–76. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beau I, Esclatine A, Codogno P. Lost to translation: when autophagy targets mature ribosomes. Trends Cell Biol. 2008;18:311–4. doi: 10.1016/j.tcb.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Shoji JY, Kikuma T, Arioka M, Kitamoto K. Macroautophagy-mediated degradation of whole nuclei in the filamentous fungus Aspergillus oryzae. PLoS One. 2010;5:e15650. doi: 10.1371/journal.pone.0015650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klionsky DJ, Cregg JM, Dunn WA, Jr., Emr SD, Sakai Y, Sandoval IV, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–45. doi: 10.1016/S1534-5807(03)00296-X. [DOI] [PubMed] [Google Scholar]
- 11.Meijer WH, van der Klei IJ, Veenhuis M, Kiel JAKW. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:106–16. doi: 10.4161/auto.3595. [DOI] [PubMed] [Google Scholar]
- 12.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–8. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 13.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–6. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 14.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–92. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 15.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–76. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pollack JK, Harris SD, Marten MR. Autophagy in filamentous fungi. Fungal Genet Biol. 2009;46:1–8. doi: 10.1016/j.fgb.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 17.Liu TB, Liu XH, Lu JP, Zhang L, Min H, Lin FC. The cysteine protease MoAtg4 interacts with MoAtg8 and is required for differentiation and pathogenesis in Magnaporthe oryzae. Autophagy. 2010;6:74–85. doi: 10.4161/auto.6.1.10438. [DOI] [PubMed] [Google Scholar]
- 18.Pinan-Lucarré B, Clavé C. Monitoring autophagy in the filamentous fungus Podospora anserina. Methods Enzymol. 2008;451:251–70. doi: 10.1016/S0076-6879(08)03218-7. [DOI] [PubMed] [Google Scholar]
- 19.Nadal M, Gold SE. The autophagy genes ATG8 and ATG1 affect morphogenesis and pathogenicity in Ustilago maydis. Mol Plant Pathol. 2010;11:463–78. doi: 10.1111/j.1364-3703.2010.00620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asakura M, Ninomiya S, Sugimoto M, Oku M, Yamashita S, Okuno T, et al. Atg26-mediated pexophagy is required for host invasion by the plant pathogenic fungus Colletotrichum orbiculare. Plant Cell. 2009;21:1291–304. doi: 10.1105/tpc.108.060996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kershaw MJ, Talbot NJ. Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc Natl Acad Sci U S A. 2009;106:15967–72. doi: 10.1073/pnas.0901477106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinan-Lucarré B, Balguerie A, Clavé C. Accelerated cell death in Podospora autophagy mutants. Eukaryot Cell. 2005;4:1765–74. doi: 10.1128/EC.4.11.1765-1774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinan-Lucarré B, Paoletti M, Dementhon K, Coulary-Salin B, Clavé C. Autophagy is induced during cell death by incompatibility and is essential for differentiation in the filamentous fungus Podospora anserina. Mol Microbiol. 2003;47:321–33. doi: 10.1046/j.1365-2958.2003.03208.x. [DOI] [PubMed] [Google Scholar]
- 24.Richie DL, Fuller KK, Fortwendel J, Miley MD, McCarthy JW, Feldmesser M, et al. Unexpected link between metal ion deficiency and autophagy in Aspergillus fumigatus. Eukaryot Cell. 2007;6:2437–47. doi: 10.1128/EC.00224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kikuma T, Kitamoto K. Analysis of autophagy in Aspergillus oryzae by disruption of Aoatg13, Aoatg4, and Aoatg15 genes. FEMS Microbiol Lett. 2011;316:61–9. doi: 10.1111/j.1574-6968.2010.02192.x. [DOI] [PubMed] [Google Scholar]
- 26.Bartoszewska M, Kiel JA, Bovenberg RA, Veenhuis M, van der Klei IJ. Autophagy deficiency promotes beta-lactam production in Penicillium chrysogenum. Appl Environ Microbiol. 2011;77:1413–22. doi: 10.1128/AEM.01531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kikuma T, Ohneda M, Arioka M, Kitamoto K. Functional analysis of the ATG8 homologue Aoatg8 and role of autophagy in differentiation and germination in Aspergillus oryzae. Eukaryot Cell. 2006;5:1328–36. doi: 10.1128/EC.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shoji JY, Craven KD. Autophagy in basal hyphal compartments: A green strategy of great recyclers. Fungal Biol Rev. 2011;25:79–83. doi: 10.1016/j.fbr.2011.04.001. [DOI] [Google Scholar]
- 29.Kück U, Pöggeler S, Nowrousian M, Nolting N, Engh I. Sordaria macrospora, a model system for fungal development. In: Anke T, Weber D, eds. THE MYCOTA XV, Physiology and Genetics: Selected Basic and Applied Aspects. Heidelberg: Springer Verlag, 2009:17-39. [Google Scholar]
- 30.Engh I, Nowrousian M, Kück U. Sordaria macrospora, a model organism to study fungal cellular development. Eur J Cell Biol. 2010;89:864–72. doi: 10.1016/j.ejcb.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Nolting N, Bernhards Y, Pöggeler S. SmATG7 is required for viability in the homothallic ascomycete Sordaria macrospora. Fungal Genet Biol. 2009;46:531–42. doi: 10.1016/j.fgb.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 32.Read ND. A scanning electron microscopic study of the external features of perithecium development in Sordaria humana. Can J Bot. 1983;61:3217–29. doi: 10.1139/b83-359. [DOI] [Google Scholar]
- 33.Read ND, Beckett A. The anatomy of the mature perithecium in Sordaria humana and its significance for fungal multicellular development. Can J Bot. 1985;63:281–96. doi: 10.1139/b85-033. [DOI] [Google Scholar]
- 34.Lord KM, Read ND. Perithecium morphogenesis in Sordaria macrospora. Fungal Genet Biol. 2011;48:388–99. doi: 10.1016/j.fgb.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 35.Nowrousian M, Stajich JE, Chu M, Engh I, Espagne E, Halliday K, et al. De novo assembly of a 40 Mb eukaryotic genome from short sequence reads: Sordaria macrospora, a model organism for fungal morphogenesis. PLoS Genet. 2010;6:e1000891. doi: 10.1371/journal.pgen.1000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J Biol Chem. 1996;271:17621–4. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- 37.Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–36. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nowrousian M, Teichert I, Masloff S, Kück U. Whole-Genome Sequencing of Sordaria macrospora Mutants Identifies Developmental Genes. G3 (Bethesda) 2012;2:261–70. doi: 10.1534/g3.111.001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tucker CL, Peteya LA, Pittman AM, Zhong J. A genetic test for yeast two-hybrid bait competency using RanBPM. Genetics. 2009;182:1377–9. doi: 10.1534/genetics.109.103069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Josefsen L, Droce A, Sondergaard TE, Sørensen JL, Bormann J, Schäfer W, et al. Autophagy provides nutrients for nonassimilating fungal structures and is necessary for plant colonization but not for infection in the necrotrophic plant pathogen Fusarium graminearum. Autophagy. 2012;8:326–37. doi: 10.4161/auto.18705. [DOI] [PubMed] [Google Scholar]
- 42.Bloemendal S, Bernhards Y, Bartho K, Dettmann A, Voigt O, Teichert I, et al. A homologue of the human STRIPAK complex controls sexual development in fungi. Mol Microbiol. 2012;84:310–23. doi: 10.1111/j.1365-2958.2012.08024.x. [DOI] [PubMed] [Google Scholar]
- 43.Bernhards Y, Pöggeler S. The phocein homologue SmMOB3 is essential for vegetative cell fusion and sexual development in the filamentous ascomycete Sordaria macrospora. Curr Genet. 2011;57:133–49. doi: 10.1007/s00294-010-0333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bloemendal S, Lord KM, Rech C, Hoff B, Engh I, Read ND, et al. A mutant defective in sexual development produces aseptate ascogonia. Eukaryot Cell. 2010;9:1856–66. doi: 10.1128/EC.00186-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu C, Iyer P, Herkal A, Abdullah J, Stout A, Free SJ. Identification and characterization of genes required for cell-to-cell fusion in Neurospora crassa. Eukaryot Cell. 2011;10:1100–9. doi: 10.1128/EC.05003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elleuche S, Pöggeler S. Visualization of peroxisomes via SKL-tagged DsRed protein in Sordaria macrospora. Fungal Genet Reports. 2008;55:9–12. [Google Scholar]
- 47.Amar N, Lustig G, Ichimura Y, Ohsumi Y, Elazar Z. Two newly identified sites in the ubiquitin-like protein Atg8 are essential for autophagy. EMBO Rep. 2006;7:635–42. doi: 10.1038/sj.embor.7400698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noda NN, Satoo K, Fujioka Y, Kumeta H, Ogura K, Nakatogawa H, et al. Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol Cell. 2011;44:462–75. doi: 10.1016/j.molcel.2011.08.035. [DOI] [PubMed] [Google Scholar]
- 49.Satoo K, Noda NN, Kumeta H, Fujioka Y, Mizushima N, Ohsumi Y, et al. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28:1341–50. doi: 10.1038/emboj.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sugawara K, Suzuki NN, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F. Structural basis for the specificity and catalysis of human Atg4B responsible for mammalian autophagy. J Biol Chem. 2005;280:40058–65. doi: 10.1074/jbc.M509158200. [DOI] [PubMed] [Google Scholar]
- 51.Ketelaar T, Voss C, Dimmock SA, Thumm M, Hussey PJ. Arabidopsis homologues of the autophagy protein Atg8 are a novel family of microtubule binding proteins. FEBS Lett. 2004;567:302–6. doi: 10.1016/j.febslet.2004.04.088. [DOI] [PubMed] [Google Scholar]
- 52.Liu XH, Lu JP, Zhang L, Dong B, Min H, Lin FC. Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryot Cell. 2007;6:997–1005. doi: 10.1128/EC.00011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science. 2006;312:580–3. doi: 10.1126/science.1124550. [DOI] [PubMed] [Google Scholar]
- 54.Veneault-Fourrey C, Talbot NJ. Autophagic cell death and its importance for fungal developmental biology and pathogenesis. Autophagy. 2007;3:126–7. doi: 10.4161/auto.3529. [DOI] [PubMed] [Google Scholar]
- 55.Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 56.Nakatogawa H, Ishii J, Asai E, Ohsumi Y. Atg4 recycles inappropriately lipidated Atg8 to promote autophagosome biogenesis. Autophagy. 2012;8:177–86. doi: 10.4161/auto.8.2.18373. [DOI] [PubMed] [Google Scholar]
- 57.Sambrook J, Russell DW. Molecular cloning: A laboratory manual / Joseph Sambrook, David W. Russell. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory, 2001. [Google Scholar]
- 58.Nowrousian M, Masloff S, Pöggeler S, Kück U. Cell differentiation during sexual development of the fungus Sordaria macrospora requires ATP citrate lyase activity. Mol Cell Biol. 1999;19:450–60. doi: 10.1128/mcb.19.1.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amberg D, Burke D, Strathern J. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, 2005. [Google Scholar]
- 60.Nowrousian M, Cebula P. The gene for a lectin-like protein is transcriptionally activated during sexual development, but is not essential for fruiting body formation in the filamentous fungus Sordaria macrospora. BMC Microbiol. 2005;5:64. doi: 10.1186/1471-2180-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nolting N, Pöggeler S. A MADS box protein interacts with a mating-type protein and is required for fruiting body development in the homothallic ascomycete Sordaria macrospora. Eukaryot Cell. 2006;5:1043–56. doi: 10.1128/EC.00086-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Becker DM, Lundblad V. Introduction of DNA into yeast cells. Curr Protoc Mol Biol 2001; Chapter 13:Unit13 7. [DOI] [PubMed] [Google Scholar]
- 63.Pöggeler S, Nowrousian M, Jacobsen S, Kück U. An efficient procedure to isolate fungal genes from an indexed cosmid library. J Microbiol Methods. 1997;29:49–61. doi: 10.1016/S0167-7012(97)00018-3. [DOI] [Google Scholar]
- 64.Pöggeler S, Risch S, Kück U, Osiewacz HD. Mating-type genes from the homothallic fungus Sordaria macrospora are functionally expressed in a heterothallic ascomycete. Genetics. 1997;147:567–80. doi: 10.1093/genetics/147.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A. 2006;103:10352–7. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–22. doi: 10.1016/0378-1119(92)90454-W. [DOI] [PubMed] [Google Scholar]
- 67.Pöggeler S, Kück U. Highly efficient generation of signal transduction knockout mutants using a fungal strain deficient in the mammalian ku70 ortholog. Gene. 2006;378:1–10. doi: 10.1016/j.gene.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 68.Klix V, Nowrousian M, Ringelberg C, Loros JJ, Dunlap JC, Pöggeler S. Functional characterization of MAT1-1-specific mating-type genes in the homothallic ascomycete Sordaria macrospora provides new insights into essential and nonessential sexual regulators. Eukaryot Cell. 2010;9:894–905. doi: 10.1128/EC.00019-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pöggeler S, Masloff S, Hoff B, Mayrhofer S, Kück U. Versatile EGFP reporter plasmids for cellular localization of recombinant gene products in filamentous fungi. Curr Genet. 2003;43:54–61. doi: 10.1007/s00294-003-0370-y. [DOI] [PubMed] [Google Scholar]
- 70.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 71.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76:4350–4. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nicholas KB, Nicholas HB, Jr., Deerfield DW., II GeneDoc: Analysis and visualization of genetic variation. EMBNETnews. 1997;4:1–4. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.