SUMMARY
In eukaryotes, the differentiation of cellular extensions such as cilia or neuronal axons depends on the partitioning of proteins to distinct plasma membrane domains by specialized diffusion barriers. However, examples of this compartmentalization strategy are still missing for prokaryotes, although complex cellular architectures are widespread among this group of organisms. This study reveals the existence of a protein-mediated membrane diffusion barrier in the stalked bacterium Caulobacter crescentus. We show that the Caulobacter cell envelope is compartmentalized by macromolecular complexes that prevent the exchange of both membrane and soluble proteins between the polar stalk extension and the cell body. The barrier structures span the cross-sectional area of the stalk and comprise at least four proteins that assemble in a cell cycle-dependent manner. Their presence is critical for cellular fitness, as they minimize the effective cell volume, allowing faster adaptation to environmental changes that require de novo synthesis of envelope proteins.
INTRODUCTION
Proper spatiotemporal regulation of protein localization and mobility is crucial for cellular organization and development. In eukaryotes, proteins are commonly sorted to subcellular compartments such as the endoplasmatic reticulum or the Golgi apparatus, where they are separated from other cellular regions by a membrane bilayer. In addition, membrane systems can themselves be compartmentalized into functionally distinct domains by protein-mediated diffusion barriers, a compartmentalization strategy that is critically involved in the differentiation of cellular extensions such as buds, axons, dendritic spines or primary cilia (Caudron and Barral, 2009). In most cases, the precise composition of the diffusion barriers and their mechanisms of function are still unclear. Similar to eukaroytes, prokaryotic cells have evolved strategies to compartmentalize proteins within the cell. These include the formation of various kinds of intracytoplasmic membrane vesicles or so-called microcompartments, highly specialized reaction chambers that encapsulate a defined set of metabolic enzymes in a protein shell (Murat et al., 2010). However, protein-mediated diffusion barriers with a role in membrane organization have not been identified in prokaryotes so far, although cellular extensions are also widespread among this group of organisms.
The Gram-negative bacterium Caulobacter crescentus (henceforth Caulobacter) develops a polar stalk that is formed by local extension of the cell body. It largely consists of cell envelope (i.e. outer membrane, peptidoglycan and inner membrane) surrounding a thin cytoplasmic core devoid of DNA, ribosomes and most cytoplasmic proteins (Ireland et al., 2002; Poindexter and Cohen-Bazire, 1964; Wagner et al., 2006). At its tip, the stalk carries an adhesive organelle (holdfast) mediating permanent surface attachment (Curtis and Brun, 2010). Moreover, it is segmented at irregular intervals by so-called crossbands (Poindexter and Cohen-Bazire, 1964), disk-like structures that traverse the entire width of the stalk perpendicular to the long-axis of the cell. Crossbands are observed in a variety of prosthecate species and were hypothesized to have an architectural or stabilizing function (Jones and Schmidt, 1973; Poindexter and Cohen-Bazire, 1964; Schmidt, 1973; Schmidt and Swafford, 1975). However, their precise role and molecular composition has remained unclear, because, the lack of mutants has so far prevented the elucidation of crossband biogenesis and function.
Previous studies have established that crossband formation is coupled to cell cycle progression (Poindexter and Staley, 1996). Early in the Caulobacter life cycle, the polar flagellum is substituted for a stalk, marking the developmental reprogramming of a motile, DNA replication-arrested swarmer cell into a sessile, replication-competent stalked cell. After transition into S-phase, the stalked cell elongates, assembles a new flagellum at the pole opposite the stalk, and finally divides asymmetrically to produce a new swarmer cell and a stalked cell. During the late stages of cell division, a new crossband is added at the stalk base (Poindexter and Staley, 1996). It is then gradually displaced as the stalk elongates by insertion of new cell wall material at the junction between the stalk and the cell body (Schmidt and Stanier, 1966; Seitz and Brun, 1998; Smit and Agabian, 1982). Additionally, stalk extension is significantly stimulated in response to phosphate starvation (Gonin et al., 2000). Based on this observation, current models suggest that the stalk promotes phosphate uptake by increasing the surface area of the cell. Since the ABC transporter complex that translocates phosphate across the inner membrane (PstCAB) is restricted to the cell body, phosphate was proposed to be shuttled from the stalk to the cell body by the periplasmic phosphate-binding protein PstS (Wagner et al., 2006).
Here, we demonstrate that crossbands represent multi-protein complexes that act as diffusion barriers separating the Caulobacter stalk and cell body into functionally independent domains. While eukaryotic diffusion barriers are mainly involved in organizing lipids or membrane proteins, crossbands restrict the diffusion of both membrane-associated and soluble proteins. They provide cells with a significant fitness advantage by retaining newly synthesized membrane and periplasmic proteins in the cell body. This compartmentalization strategy minimizes the physiologically active part of the cell envelope, reducing the energy cost for protein synthesis and allowing faster adaptation of the cell envelope proteome to changing environmental conditions.
RESULTS
The Caulobacter cell is compartmentalized by protein diffusion barriers
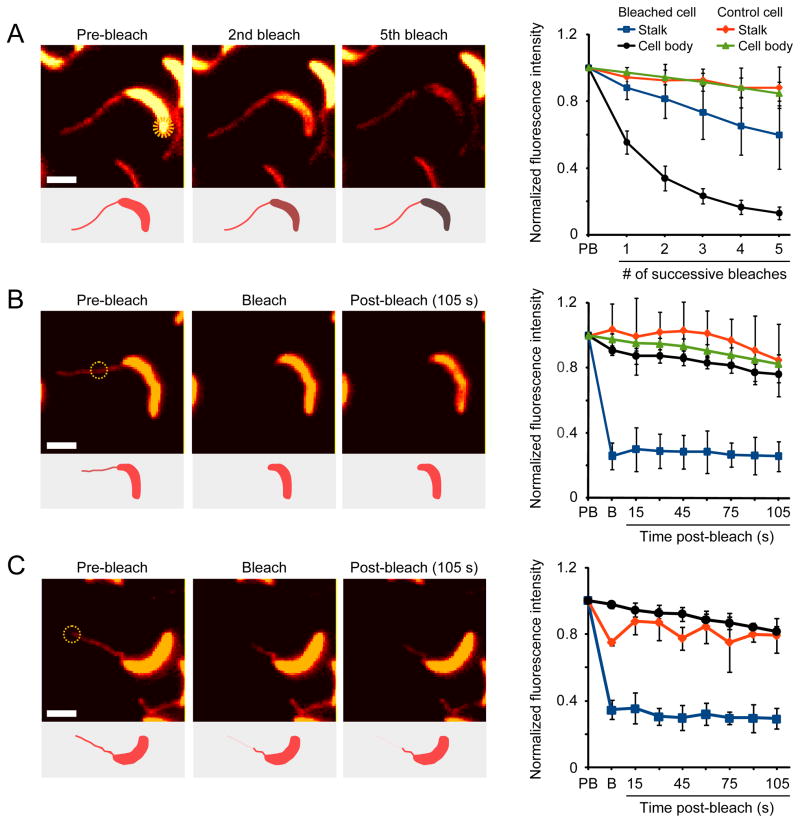
When grown in phosphate-limiting conditions, Caulobacter cells display highly elongated stalks (Gonin et al., 2000). The resulting increase in the cellular surface area-to-volume ratio was proposed to facilitate phosphate scavenging, mediated through the shuttling of phosphate from the stalk to the cell body by the periplasmic phosphate-binding protein PstS (Wagner et al., 2006). To assay PstS mobility, we performed both FLIP (fluorescence loss in photobleaching) and FRAP (fluorescence recovery after photobleaching) studies of cells expressing a functional PstS-mCherry fluorescent protein fusion (Figures S1A and S1B). When a laser pulse was applied to the stalk-distal cell pole, fluorescence was lost throughout the cell body but not within the stalk (Figure 1A). Control experiments with fixed cells verified that the FLIP/FRAP setup used can bleach a small subregion of the cell and that protein diffusion is required for the total loss of fluorescence observed (Figures S1C and S1D). Thus, PstS-mCherry molecules can readily diffuse within the cell body periplasm but not across the stalk-cell body boundary, challenging the model of PstS-mediated phosphate shuttling. To test whether the observed diffusion barrier was bidirectional, we photobleached PstS-mCherry molecules in the stalk and, consistently, detected no recovery of stalk fluorescence (Figure 1B). Furthermore, when a laser pulse was applied to the tip of the stalk, PstS-mCherry fluorescence decreased only in a region close to the tip (Figure 1C), suggesting the existence of additional intra-stalk compartmentalization.
Figure 1. A diffusion barrier compartmentalizes the Caulobacter periplasm.
(A–C) Diffusion of a xylose-inducible PstS-mCherry fusion assayed using FLIP (A and C) and FRAP (B). Cells (EK363) were bleached with seven ~4 nsec pulses in the region indicated by yellow circles. Bleaching was performed either multiple times in succession (A) or once followed by a 105 sec recovery (B–C). Insets show schematic representations of the results, and graphs show the quantification of fluorescence in multiple cells (panel A: n = 6, p < 0.0002; panel B: n = 7, p < 2×10−7; panel C: n = 3, p < 0.002; error bars = SD). Fluorescence intensities were measured in the stalk (blue) and cell body (black) of the bleached cell or the stalk (red) and cell body (green) of a nearby control cell. For intra-stalk bleaching, the bleached (blue) and unbleached (red) portions of the stalk as well as the cell body (black) fluorescence were quantified. The color-maps of the fluorescent images were scaled for easier visualization. However, all quantifications were performed using raw image data. The fluorescence intensity of each region of interest was normalized to its pre-bleach intensity. Abbreviations: PB, pre-bleach; B, bleach. Scale bars: 2 μm. See also Figure S1.
Identification of novel stalk proteins that localize in a crossband-like pattern
Assuming a potential link between formation of the diffusion barrier and stalk biogenesis, we sought to identify the constituents of the barrier structure by focusing on uncharacterized open reading frames that were transcriptionally upregulated at the onset of stalk formation (McGrath et al., 2007). Candidate genes were fused to mcherry and ectopically expressed from a xylose-inducible promoter (Pxyl). Microscopic analysis of the resulting fluorescent protein fusions turned our attention to two conserved hypothetical proteins, CCNA_02562 and CCNA_02561 (Marks et al., 2010), that produced distinct foci distributed at irregular intervals along the length of the stalk. The two proteins, which are encoded in a putative operon, were designated StpA and StpB (Stalk protein A and B), respectively (Figure 2A).
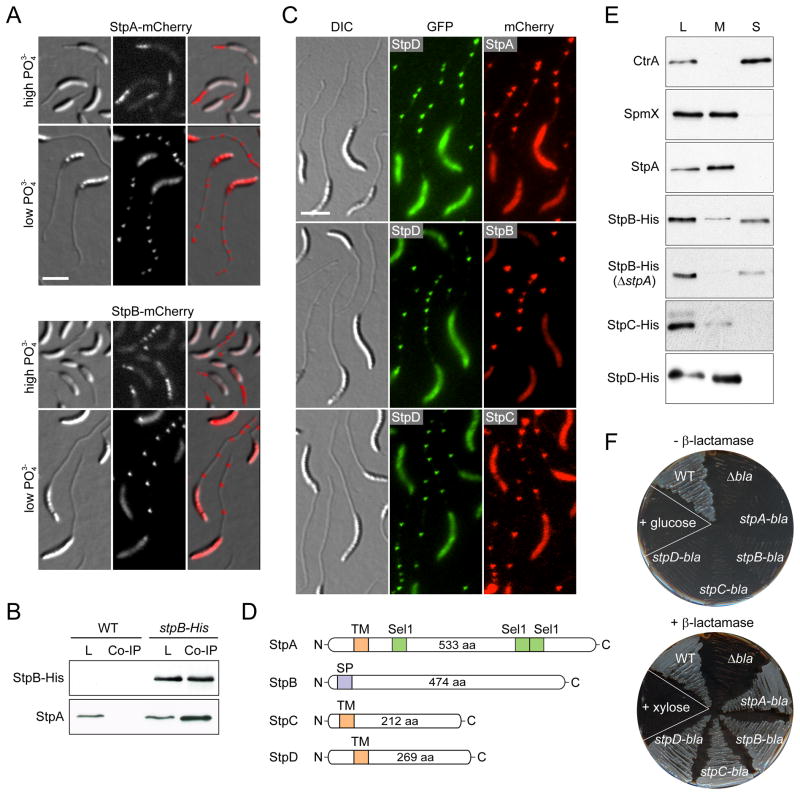
Figure 2. Identification and subcellular localization of novel stalk proteins.
(A) Stalk localization of StpA-mCherry (SW33) and StpB-mCherry (SW30) produced from the xylose-inducible Pxyl promoter after 24 h of growth in phosphate-rich (M2G, high PO43−) or phosphate-poor medium (M2G−P, low PO43−) containing 0.3% xylose.
(B) Co-immunoprecipitation analysis of stpB-His (SS233) and wild-type cells reveals an interaction between StpA and StpB. Whole-cell lysates (L) and eluates from co-immunoprecipitation experiments (Co-IP) were subjected to immunoblot analysis using anti-His and anti-StpA antibodies.
(C) StpA,B,C,D localization in stalks is reminiscent of the distribution of crossbands. Cells of strain SS243 (stpD::stpD-gfp Pxyl::Pxyl-stpA-mcherry), SS88 (stpB::stpB-mcherry stpD::stpD-gfp) and SS389 (stpC::stpC-mcherry stpD::stpD-gfp) were grown in M2G−P for 24 h. Synthesis of StpA-mCherrry was induced with 0.3% xylose for 24 h.
(D) Schematic depicting the domain organization of the stalk proteins with the predicted transmembrane domains (orange), the signal peptide (purple) and the Sel1 motifs (green).
(E) Cell fractionation analysis reveals that StpA, StpC, and StpD are membrane-bound proteins, whereas StpB is soluble. Whole-cell lysates (L) and the corresponding membrane (M) and soluble (S) fractions of cells producing His-tagged Stp proteins (SS233, SS220, SS244 and SS247) were subjected to Western blot analysis using an anti-His antibody. Fractionation efficiency was verified by probing the same fractions with anti-CtrA and anti-SpmX antibodies. Note, the absence of stpA and stpAB does not reduce the levels of StpB and StpC, respectively (Figure S2C).
(F) The Stp proteins are targeted to the periplasm. The TEM-1 β-lactamase gene (bla) was fused to the 3’ end of stpA, stpB, stpC and stpD, respectively. The gene fusions were placed under the Pxyl promoter in a β-lactam-sensitive reporter strain. Cells (SS165, SS172, SS273, SS274) were patched on PYE agar containing ampicillin and either 0.2% glucose or 0.3% xylose. Scale bars: 3 μm. See also Figure S2.
Prompted by the similar localization patterns of StpA and StpB, we tested whether the two proteins could interact with each other. Co-immunoprecipitation analysis followed by immunodetection showed that StpA co-sedimented with hexahistidine-tagged StpB (Stp-His) (Figure 2B), indicating that the two proteins formed a complex. Further analysis of the immunoprecipitates by mass spectrometry led to the identification of two additional stalk proteins, StpC (CCNA_02560) and StpD (CCNA_02271). Interestingly, stpC is likely to be co-transcribed with stpA and stpB, supporting the idea that the three gene products functionally interact. Fluorescently tagged StpC and StpD displayed the characteristic multi-focus localization pattern observed for StpA and StpB and co-localized with StpA and StpB in the stalk (Figure 2C). Of note, the bodies of cells producing fluorescently tagged Stp proteins displayed higher fluorescence when grown in low-phosphate medium. This effect was not due to instability of the fluorescent protein fusions (Figures S2A and S2B) but may result from starvation-induced changes in gene expression and growth dynamics. Collectively, the biochemical interaction and colocalization data suggest that the four Stp proteins assemble into a multi-subunit complex.
Bioinformatic analysis showed that StpB, StpC, and StpD lack known functional domains. StpA, by contrast, harbors three Sel1-like motifs, mediating protein-protein interactions (Blatch and Lassle, 1999). In addition, StpA, StpC, and StpD are each predicted to possess a single transmembrane helix close to the N-terminus, while StpB is predicted to be a soluble periplasmic protein (Figure 2D). Protein fractionation experiments confirmed that StpA, StpC, and StpD exclusively co-sedimented with the cell membranes, whereas StpB was detected in both the membrane and the soluble protein fractions (Figure 2E). Notably, StpB was completely soluble in StpA-deficient cells, suggesting that StpA functions to tether StpB to the inner membrane. To further clarify the subcellular localization and membrane topology, we engineered C-terminal fusions of StpA, StpB, StpC, and StpD to a TEM-1 β-lactamase reporter, which needs to be translocated to the periplasm in order to confer resistance to β-lactam antibiotics. Expression of each of the four fusion proteins restored resistance to a β-lactam-sensitive reporter strain, demonstrating that StpB and the C-terminal portions of StpA, StpC and StpD are positioned in the periplasmic space (Figure 2F).
The StpABCD complex forms static crossband structures
The Stp proteins show the same localization pattern as crossbands and are, at least in part, conserved in other stalked bacteria that synthesize crossbands, such as Asticcacaulis and Brevundimonas species (Figure S2D). To investigate whether they play a role in crossband formation, we examined stalk ultrastructure in a ΔstpAB mutant by ECT. In tomograms of wild-type cells, crossbands appear as distinct densities that transect the inner membrane, peptidoglycan and outer membrane layers as well as the cytoplasmic core of the stalk (Figure 3A and Movie S1). Intriguingly, in StpAB-deficient cells, crossbands were undetectable, while stalk length and morphology remained unperturbed, indicating that crossband formation is not required for stalk biogenesis per se (Figure 3A). Strains lacking stpC or stpD, by contrast, still exhibited clearly discernible crossbands (Figure S3A).
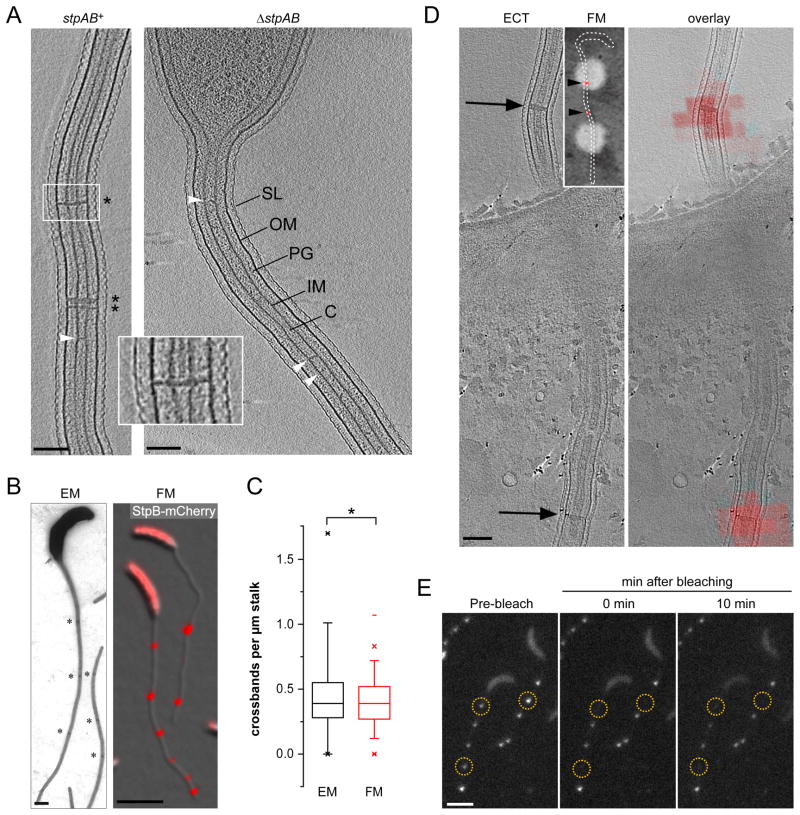
Figure 3. Crossbands are static multi-protein complexes.
(A) StpAB-deficient cells consistently lack crossbands. Cells with and without stpAB (SW51, n = 8) were grown in PYE and imaged by ECT. The images show a longitudinal section of the stalk. Asterisks denote crossbands. Arrowheads point at unidentified structures spanning the stalk core. Scale bars: 100 nm.
(B and C) The distribution of StpB-mCherry foci reflects the distribution of crossbands in stalks. Cells of strains CB15N (WT) and SS160 (stpB-mcherry) were grown in M2G−P and imaged either by electron (EM) or fluorescence (FM) microscopy, respectively. Electron micrographs were acquired of negatively stained wild-type cells. From the respective images, the number of crossbands (n = 68 cells) and StpB-mCherry foci per μm stalk (n = 316 cells) was quantified (*p > 0.2, t-test; error bars = SEM). Asterisks denote crossbands. Scale bars: 500 nm (EM) and 3 μm (FM).
(D) StpB spatially overlaps with crossbands. Strain SW30 (Pxyl::Pxyl-stpB-mcherry) was grown in M2G−P with 0.3% xylose. Cells were fixed on EM grids and imaged first by low-magnification phase contrast/fluorescence microscopy (inset; arrowheads indicate StpB-mCherry foci) and then by ECT. Shown is an ECT slice of a stalk with arrows pointing to crossband structures (left panel) and the respective correlated image showing the ECT slice overlayed with a fluorescence micrograph of the same region (right panel). Scale bar: 100 nm.
(E) FRAP analysis reveals that crossbands are static protein complexes. Cells of strain SS160 (stpB-mcherry) were cultured in M2G−P and imaged by fluorescence microscopy to identify StpB-mCherry localization. A laser pulse was applied to selected regions (yellow circles), and StpB-mCherry signals were bleached. Cells were imaged immediately and 10 min after the laser pulse. Scale bar: 3 μm. See also Figure S3.
Next, we compared the frequency of StpB-mCherry foci and crossbands in phosphate-starved wild-type cells as visualized by fluorescence and transmission electron microscopy, respectively (Figures 3B and 3C). The data revealed that Stp complexes showed the same average spacing (2.5 to 3 μm) as crossbands. To determine whether the Stp complex colocalized with crossbands, phosphate-starved cells producing StpB-mCherry (n = 11) were analyzed by correlated light microscopy and ECT. Alignment of ECT slices with fluorescence micrographs of the same region verified that the StpABCD complex invariantly assembles at the sites of crossband formation (Figures 3D and S3B).
The Stp proteins lack domains with enzymatic activity and may therefore have a structural role. Indeed, when StpB-mCherry foci were photobleached, the fluorescence signal did not recover over time, and the intensity of neighboring StpB-mCherry foci remained unchanged (Figure 3E). Identical results were obtained for fluorescently tagged StpA, StpC and StpD (data not shown). The four Stp proteins thus assemble into static multi-protein scaffolds that localize to the same subcellular sites as crossbands. Previously, crossbands were proposed to consist of peptidoglycan (Schmidt, 1973), with their synthesis depending on the essential cell division protein FtsZ (Divakaruni et al., 2007). However, we found that stalks of a conditional ftsZ mutant still displayed crossbands (Figure S4A) and continued to accumulate StpB-mCherry foci with the typical crossband-like distribution pattern (Figure S4B), supporting the idea that crossbands represent macromolecular assemblies of StpABCD rather than cell wall material.
StpA is the central regulator of cell cycle-dependent crossband formation
To characterize the molecular mechanism of crossband assembly, we determined the abundance of the Stp proteins over the course of the Caulobacter cell cycle. Each of the proteins was only barely detectable in swarmer cells but started to accumulate gradually after the swarmer-to-stalked-cell transition, with a distinct peak at the late pre-divisional stage (Figure 4A). This pattern correlates well with the cell cycle expression patterns of stpA, stpB and stpD determined previously by global transcriptome analysis (McGrath et al., 2007), suggesting that synthesis of the Stp complex may largely be regulated at the transcriptional level. Consistent with this notion, continuous expression of stpA from an inducible promoter in a ΔstpA background abolished the observed fluctuations in StpA protein levels (data not shown). To investigate whether the cell cycle-dependent accumulation of the four Stp proteins in fact correlated with the appearance of crossbands, we monitored synchronously growing cells producing a StpB-mCherry fusion from the native stpB promoter (Figure 4B). In line with the observed abundance patterns, no fluorescence was observed in swarmer cells. However, polar StpB-mCherry foci became detectable once the cells had progressed halfway through the cell cycle. Furthermore, stalked cells consistently displayed a second StpB-mCherry focus after passing through an additional cell cycle (data not shown), indicating that the Stp proteins assemble in a cell cycle-dependent manner. Notably, the intensity of the fluorescent foci increased over time, suggesting a gradual maturation of the complexes.
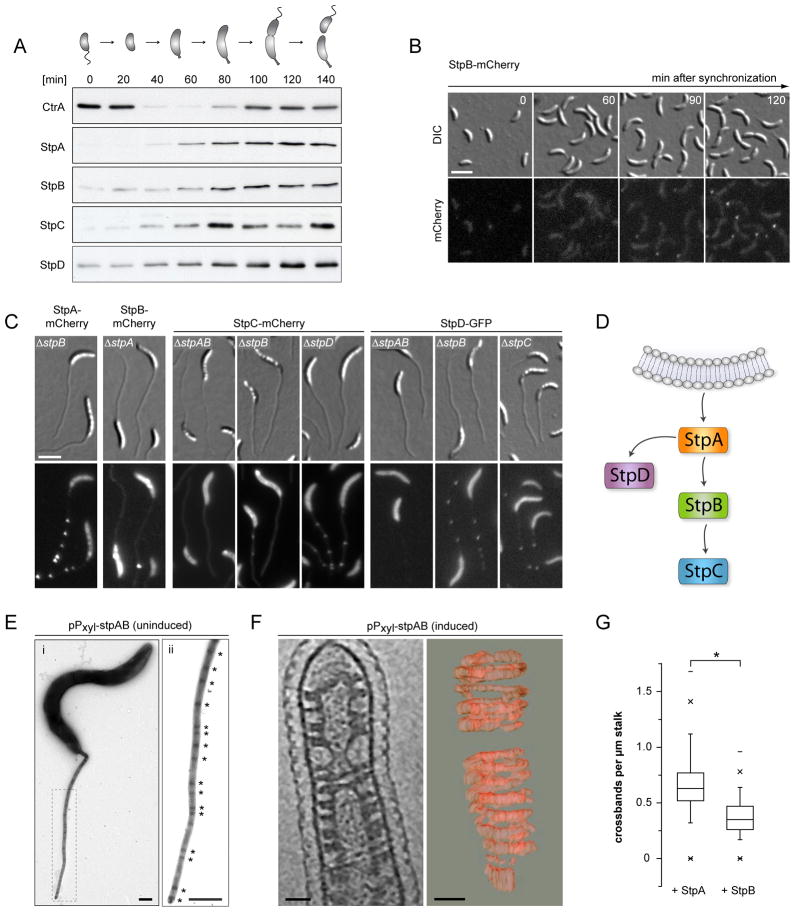
Figure 4. Crossband synthesis is cell cycle-dependent and relies on hierarchal self-assembly of the Stp proteins.
(A) Western blot analysis of Stp protein levels during the cell cycle. Swarmer cells of SS233 (stpB::stpB-His), SS247 (stpC::stpC-His) and SS244 (stpD::stpD-His) were grown in M2G. Samples were taken from the culture in 20 min intervals and probed with anti-CtrA, anti-StpA and anti-His antiserum. The schematic illustrates the different morphological stages of the cell cycle.
(B) Timecourse microscopy of StpB-mCherry localization, starting with isolated swarmer cells of SS160 (stpB-mcherry). Cells were grown in M2G. Scale bar: 3 μm.
(C) Localization hierarchy of the Stp proteins. Xylose-inducible fluorescent protein fusions to StpA, StpB, StpC, and StpD were analyzed in the indicated strain backgrounds (SS141, SS142, SS234, SS236, SS240, SS263, SS264, SS265). Cells were grown in M2G−P and induced for 24 h with 0.3% xylose. Scale bar: 3 μm.
(D) Schematic illustrating the order of StpABCD complex assembly.
(E) Stalk ultrastructure of cells carrying an inducible copy of stpAB on a multi-copy plasmid (SS214). Cells were cultivated in M2G−P in the absence inducer, negatively contrasted with uranyl acetate and imaged by transmission electron microscopy. The dashed rectangle in (i) indicates the region magnified in (ii). Asterisks indicate crossbands. Scale bars: 500 nm. A strain carrying the empty plasmid (SS258) showed the wild-type frequency of crossbands (data not shown).
(F) Visualization and 3D-reconstruction of helical StpAB assemblies. Cells carrying a plasmid-encoded copy of stpAB under the control of Pxyl (SS214) were pre-cultured in PYE, grown in M2G−P containing 0.3% xylose, and analyzed by ECT. Shown is a section through a representative tomogram of a stalk (left panel) and a 3D reconstruction of the helical assemblies induced by StpAB overproduction (right panel). Scale bars: 50 nm.
(G) Constitutive production of StpA increases the frequency of crossbands. Cells of strain SW33 (Pxyl ::Pxyl-stpA-mcherry) and SW30 (Pxyl::Pxyl-stpB-mcherry) were grown in M2G−P with 0.3% xylose for 24 h and imaged by fluorescence microscopy. The number of fluorescent foci per μm stalk was determined for cells of SW30 (n = 120) and SW33 (n = 194) (*p < 0.05, t-test; error bars = SEM). See also Figure S4.
Next, we examined the order of Stp complex assembly. To this end, we engineered xylose-inducible fluorescent protein fusions to StpA-D and examined the localization pattern of each fusion in cells lacking either single or multiple Stp proteins (Figure 4C). StpB, StpC and StpD were all mislocalized in a ΔstpA background, demonstrating that StpA provides a scaffold necessary for Stp complex formation. StpC localization further depended on the presence of StpB. Occasionally, we observed single StpB-mCherry or StpC-mCherry foci in ΔstpA or ΔstpB cells, respectively. However, these cells also showed occasional fluorescent dots within the cell body, suggesting that these foci likely represent protein aggregates that were accidentally inserted into the growing stalk. The localization hierarchy deduced from these analyses (Figure 4D) was corroborated by time-lapse microscopy of cells that co-produced StpD-GFP and mCherry-labeled derivatives of StpA, StpB or StpC (Movies S2, S3 and S4). StpD-GFP consistently localized to the stalked pole significantly later than StpA-mCherry but earlier than StpC-mCherry. The temporal order of StpB and StpD recruitment could not be unambiguously resolved, suggesting that StpA independently and concurrently recruits both StpB and StpD to the nascent Stp complex.
A key role of StpA in crossband formation is also supported by overexpression experiments. We noticed that cells carrying a plasmid with an additional copy of stpAB under the control of the Pxyl promoter displayed considerably more crossbands, even in the absence of inducer (Figure 4E). This effect likely resulted from the elevated production of StpA and StpB due to leaky expression of the plasmid-borne genes (Figure S4C). Interestingly, induction of stpAB overexpression dramatically reduced cellular fitness (Figure S4D), accompanied by the formation of slender, elongated cells with short or misshapen stalks (data not shown). ECT analysis of StpAB-overproducing cells revealed that the stalks contained massive helical densities lining the periplasmic face of the inner membrane, supporting the idea that the Stp proteins assemble spontaneously into high-molecular weight structures (Figures 4F and S4E and Movie S5). These helical arrays did not extend into the cytoplasm or the outer membrane, consistent with the finding that StpB and the C-terminal region of StpA form a plasma membrane-associated periplasmic complex (Figure 2). The reason for the observed growth disadvantage is unclear. However, in many tomograms, the cytoplasmic membrane at the stalked pole was covered by an extensive layer of electron-dense material (Figure 4E). These structures may be related to the accumulation of StpAB and interfere with the function of polar protein complexes involved in the regulation of Caulobacter development.
To dissect the mechanism of Stp complex formation, we individually expressed stpA-mCherry and stpB-mCherry fusions integrated at the chromosomal Pxyl locus in an otherwise wild-type background and compared the number of fluorescent foci per μm stalk in cells after growth in low-phosphate medium. Importantly, induction of stpA-mCherry, but not stpB-mCherry, was sufficient to significantly increase the frequency of Stp complexes (Figure 4G). Crossband assembly thus appears to be stimulated by StpA in a concentration-dependent, nucleation-like process.
The central importance of StpA in Stp complex assembly raises the question of how StpA itself is recruited to the stalked pole. Although the underlying mechanism still needs to be determined, we can exclude the involvement of several known polar proteins, including the cell polarity determinant DivJ, the pole-organizing protein PopZ, the stalk-specific protein StpX, the penicillin-binding protein PpbC and the scaffolding proteins BacA and BacB (Curtis and Brun, 2010; Hughes et al., 2010; Kühn et al., 2010) (Figure S4F).
Crossbands are non-specific barriers to protein diffusion
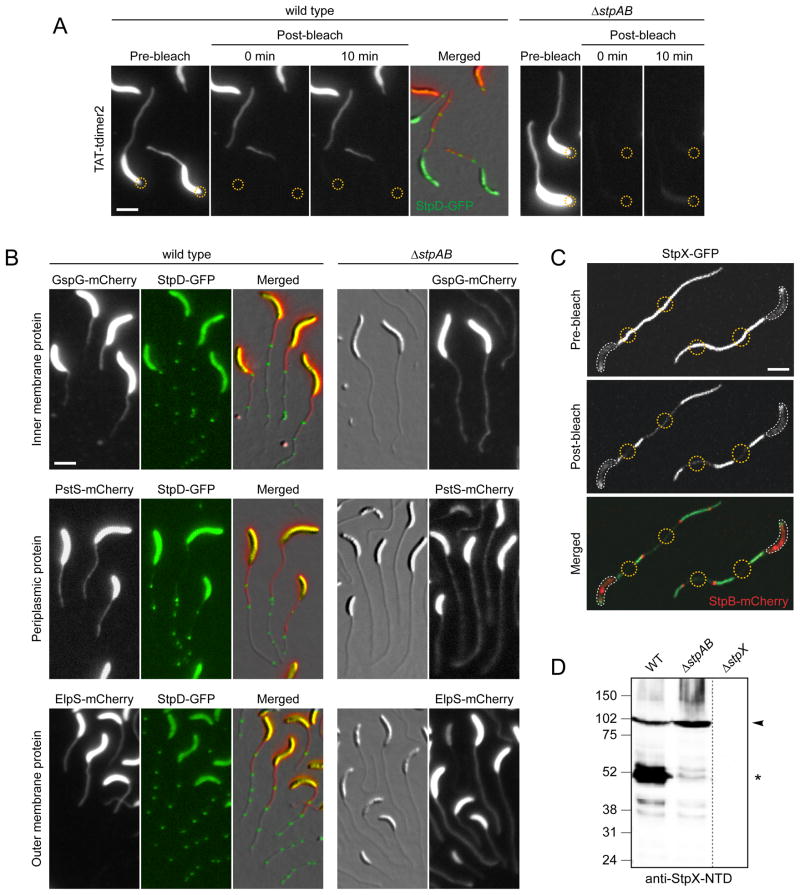
To test whether crossbands were responsible for the observed compartmentalization of Caulobacter cells, we compared the mobility of a soluble periplasmic red fluorescent protein (TAT-tdimer2) in the wild type and a StpAB-deficient mutant using FLIP analysis (Figure 5A). In both strains, diffuse red fluorescence was detected throughout the stalk and the cell body periplasm prior to photobleaching. When wild-type cells were exposed to a laser pulse focused onto the stalk-distal pole, TAT-tdimer2 fluorescence was completely bleached within the cell body. The bleached region extended to the crossband closest to the stalk base and did not recover any fluorescence within a 10-min interval. In about 20% of the cells (n = 20), TAT-tdimer2 fluorescence decreased up to the second crossband, indicating that crossband assembly may still have been in progress at the time of the bleaching event (data not shown). In contrast, when the same experiment was performed on StpAB-deficient cells, TAT-tdimer2 was completely bleached throughout the cell, including the stalk. Thus, periplasmic protein diffusion is no longer restricted in the absence of crossbands. Notably, in a ΔstpCD mutant, which still forms electron-dense crossband structures (Figure S3A), TAT-tdimer2 was only partly compartmentalized. Approximately 50% of the cells (n = 122) lost fluorescence in the unbleached region during a 10-min recovery period (Figure S5A), indicating that StpC and StpD are critical for tightening the diffusion barrier.
Figure 5. Crossbands serve as protein diffusion barriers.
(A) Analysis of the compartmentalization of soluble and periplasmic red fluorescent protein (tdimer2) using FLIP. Cells of strain SS269 (stpD::stpD-gfp pPxyl-TAT-tdimer2) and SS216 (ΔstpAB pPxyl-TAT-tdimer2) were cultured in M2G−P containing 0.3% xylose for 24 h. Cells were mounted on an agarose pad and exposed to a laser pulse in the regions indicated by a yellow circle. Scale bar: 3 μm.
(B) Crossbands compartmentalize periplasmic, inner- and outer membrane proteins. Cells of strains SS277 (stpD::stpD-gfp Pxyl-gspG-mcherry), SS272 (ΔstpAB Pxyl::Pxyl-gspG-mcherry), SS299 (stpD::stpD-gfp Pxyl::Pxyl-pstS-mcherry), SS302 (ΔstpAB Pxyl::Pxyl-pstS-mcherry), SS283 (stpD::stpD-gfp Pxyl::Pxyl-elpS-mcherry), and SS284 (ΔstpAB Pxyl::Pxyl-elspS-mcherry) were first grown in M2G−P for 36 h and then induced with 0.3% xylose for 11–13 h. Scale bar: 3 μm.
(C) StpX-GFP mobility requires compartmentalization of the stalk from the cell body by the newest crossband. Cells of strain YB5058 (stpX::stpX-gfp Pxyl::Pxyl-stpB-mcherry) were grown in HIGG-30 μM phosphate containing 0.3% xylose and mounted on an agarose pad. First, StpB-mCherry fluorescence was imaged to identify regions of interest (yellow circles). Then, these regions were simultaneously bleached for 50 sec, followed by the acquisition of a post-bleach image. White lines outline the cell bodies. Scale bar: 3 μm.
(D) Crossbands affect the processing of the stalk-specific protein StpX. Wild-type, ΔstpAB (SW51) and ΔstpX (YB5231) cells were grown to stationary phase in HIGG-30 μM phosphate and subjected to immunoblot analysis using an antibody raised against the N-terminal domain of StpX (anti-StpX-NTD). Arrowheads denote the full-length version of StpX, asterisks the dominant short fragment. See also Figure S5.
We then determined whether crossbands also compartmentalize the inner and outer membrane. For this purpose, FLIP analysis was performed on wild-type and ΔstpAB mutant cells producing mCherry fusions to the inner-membrane type II secretion protein GspG, the outer-membrane TonB-dependent receptor MalA (Neugebauer et al., 2005) and the outer-membrane lipoprotein ElpS (Le Blastier et al., 2010) (Figure S5E). In the wild-type background, fluorescence was only bleached in a defined sub-region of the cell and did not show any recovery within a 10-min period following the bleaching event (n > 30 per strain). The ΔstpAB mutant, by contrast, showed no sign of compartmentalization for any of the proteins investigated (n > 79 per strain). Crossbands thus act as general diffusion barriers that restrict the mobility of proteins in all layers of the cell envelope.
Next, we investigated how proteins can enter the stalk despite the presence of diffusion barriers. Our analyses showed that fluorescently labeled envelope proteins are distributed throughout the stalk when induced concomitantly with the onset of stalk growth (Figures 5A and S5E). Since crossbands are inserted at intervals, diffusible proteins are likely to be trapped randomly in intra-stalk compartments during stalk elongation. To test this hypothesis, cells synthesizing a xylose-inducible PstS-mCherry, GspG-mCherry or ElpS-mCherry fusion were first grown in low-phosphate medium lacking inducer to stimulate stalk elongation. Subsequently, synthesis of the fusion proteins was induced, and images were taken after an additional growth period (Figure 5B). In the wild-type background, the newly produced proteins only entered the stalk up to the newest crossband. Moreover, they were occasionally trapped in the compartment formed by the two proximal crossbands whenever a new Stp complex had assembled during the time of induction. In crossband-deficient cells, by contrast, the fusions were able to diffuse freely throughout the entire cell envelope including the stalk. These results indicate that the timing of synthesis determines whether an envelope protein localizes to the stalk, with diffusion barriers helping to retain newly produced proteins in the cell body.
ECT analysis suggested that crossbands might also compartmentalize the stalk cytoplasm (Figure 3A and Movie S1). However, consistent with the reported absence of cytoplasmic proteins in the stalk (Ireland et al., 2002; Wagner et al., 2006), we observed that the fluorescent protein YFP remained excluded from the stalk even in a ΔstpAB background (data not shown). To probe for stalk core compartmentalization, we therefore took advantage of the stalk-specific bitopic inner membrane protein StpX. Previously, StpX was found to be immobile near the stalk base but mobile in stalk regions distal to the cell body (Hughes et al., 2010). We found that the spatial boundary between the immobile and mobile subpopulations consistently (100%; n > 30) coincided with the newest crossband (Figure 5C). In contrast, StpX-GFP was largely immobile in barrier-deficient cells (Figure S5B). The mobile fraction was proposed to result from post-translational processing of StpX (Hughes et al., 2010). Indeed, while StpX accumulated as a dominant short fragment in wild-type cells, this fragment was undetectable in StpAB-deficient cells (Figure 5D). The processed form presumably results from cleavage within the cytoplasmic domain of StpX (Figures 5D, S5C and S5D). Since the cleaved C-terminal domain is soluble (Hughes et al., 2010), the apparent mobility of StpX-GFP in wild-type cells is likely explained by the release of a GFP-containing fragment into the cytoplasm of intra-stalk compartments, where its diffusion is constrained by crossbands. Although the function of StpX and the nature of its processing are still unclear, these data demonstrate that diffusion barriers create intra-stalk compartments that differ from the cell body in protein composition and/or activity.
Protein diffusion barriers are critical for bacterial fitness
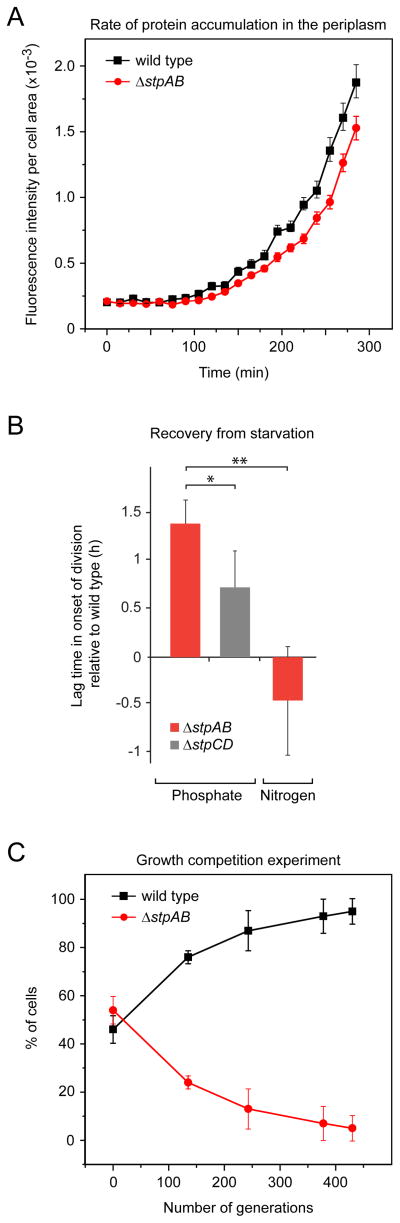
Upon prolonged phosphate starvation, the stalk can account for the majority of the periplasmic volume and of the inner and outer membrane areas. Since crossbands retain newly synthesized envelope proteins in the cell body (Figures 5A and 5B), they should allow faster protein accumulation during periods of protein upregulation. To test this hypothesis, we monitored the accumulation of a xylose-inducible PstS-mCherry fusion in the cell body periplasm of wild-type and StpAB-deficient cells after addition of the inducer (Figure 6A). Cells deficient in diffusion barriers showed a delay in PstS-mCherry accumulation of 22.4 ± 0.8 min compared to compartmentalized cells. Thus, a lack of diffusion barriers increases the effective periplasmic volume, necessitating a higher production of periplasmic proteins to reach final steady-state levels.
Figure 6. Diffusion barriers are crucial for bacterial fitness.
(A) Rate of periplasmic accumulation of an inducible PstS-mCherry protein fusion in the wild-type (EK363) and ΔstpAB (EK389) background. Cells were grown in HIGG-30 μM phosphate with 0.3% glucose to induce long stalks while repressing the synthesis of PstS-mCherry. The cells were seeded on pads with 0.3% xylose, and PstS-mCherry accumulation was monitored by timelapse microscopy. Mean fluorescence/cell body area was measured for ~ 300 cells per strain. Error bars = SD. Fitting the data to an exponential function and solving the equations in the exponential region (t = 165 to 255 min) for equal fluorescence intensities yielded a time difference in the accumulation of PstS-mCherry of 22.4 (± 0.8) min.
(B) Competitive growth of wild-type and diffusion barrier-deficient cells. To analyze the effect of diffusion barriers on the rate of recovery from nutrient starvation, wild-type, ΔstpAB and ΔstpCD cells were differentially labeled with the fluorescent proteins YFP (EK392, EK417, EK486) or mCherry (EK416, EK393, EK487) and starved for either phosphate or nitrogen. Mutant and wild-type cells were combined at equal ratios, transferred to nutrient-replete medium and grown to late-exponential phase. More than 1000 cells were analyzed by fluorescence microscopy before and after recovery to determine shifts in the relative composition of the cultures. The differentials were then used to calculate the lag in the onset of cell division (see Figure S6B). Values represent the average of four experiments, including two in which the fluorescent marker was switched (error bars = SD; * p < 0.02; ** p < 0.002).
(C) Wild-type cells outcompete a diffusion barrier-deficient mutant. Wild-type and ΔstpAB cells (SW51) were grown in PYE and mixed at equal optical densities. Mixed cultures (n = 5) were repeatedly diluted into fresh PYE and cultured to stationary phase. At the indicated timepoints, cells were withdrawn and spread on PYE agar. The ratio of wild-type and barrier-deficient cells was determined by colony PCR. Error bars show SD (n ~ 450). See also Figure S6.
To test for a possible selective advantage of diffusion barriers, we performed competitive growth experiments. Wild-type and ΔstpAB mutant cells, labeled with distinct fluorescent proteins, were first grown individually in low-phosphate medium and then mixed at equal ratios. After transfer of the mixed culture to phosphate-rich medium and cultivation to late-exponential phase, the fraction of wild-type cells was determined by fluorescence microscopy (n > 1000). We consistently found that wild-type cells outcompeted barrier-deficient cells in recovery from phosphate starvation. Since the growth rates of the two strains are equal in phosphate-rich medium (Figure S6A), we reasoned that the competitive advantage of wild-type cells stems from a delay in the onset of cell division in the non-compartmentalized ΔstpAB cells. The delay calculated for ΔstpAB cells relative to the wild type was 1.39 ± 0.25 h (Figures 6B, S6B and S6C). We additionally performed growth competition experiments using ΔstpCD mutant cells, which display leaky diffusion barriers and are thus only partially compartmentalized (see Figure S5A). While cells deficient in StpCD were still outcompeted by wild-type cells, the calculated lag in restarting cell division was significantly shorter (0.72 ± 0.38 h, p < 0.02) (Figure 6B). These data support the hypothesis that diffusion barriers provide a fitness advantage by reducing the effective envelope area and periplasmic volume of the cell and thus accelerating the rate of new protein accumulation.
To provide additional evidence that the effective cell envelope volume is critical for fitness, we asked whether the observed competition advantage of wild-type cells was directly linked to stalk length. Given the lack of true stalk-less mutants, we took advantage of the fact that nitrogen-limiting growth conditions do not stimulate stalk elongation (Figure S6D), yielding relatively similar envelope volumes for wild-type and diffusion barrier-deficient (ΔstpAB) cells. As expected, we found that wild-type cells had no measurable growth advantage over the mutant strain after transfer from nitrogen-limited to nitrogen-rich medium (Figure 6B), consistent with a positive effect of volume reduction on cellular fitness.
The Stp complex is synthesized in both starvation and nutrient-rich conditions (see Figure 4), suggesting that subcellular compartmentalization is critical even when only short stalks are produced. Since short-term competition assays may not be sensitive enough to detect small fitness differences, we carried out long-term experiments with untagged cells that were fully (wild-type), partially (ΔstpCD) or not (ΔstpAB) compartmentalized. Mixed cultures initially containing an equal ratio of wild-type and ΔstpAB or ΔstpCD cells were repeatedly grown to stationary phase and then re-diluted into fresh rich medium for approximately 450 generations. Monitoring changes in the relative abundance of the respective strains over time, we observed that wild-type cells efficiently outcompeted a mutant with defective diffusion barriers (Figures 6C and S6E). The selection coefficients (Lang et al., 2009) calculated for the ΔstpAB and ΔstpCD strains were −0.7% and −0.4%, respectively. The value obtained for StpAB-deficient cells corresponds to a difference in doubling times of only 4.6%. This delay is too small to be resolved by short-term growth analyses but provides a critical advantage over longer time scales. Collectively, our results thus show that diffusion barriers make an important general contribution to cellular fitness.
DISCUSSION
Intracellular compartmentalization by protein-mediated diffusion barriers has previously been thought to be a characteristic of eukaryotic cells. In this study, we show that general protein diffusion barriers, analogous to those reported for cilia or neuronal axons (Chih et al., 2012; Nakada et al., 2003), also exist in prokaryotes. These structures are functionally and structurally different from bacterial microcompart-ments (Murat et al., 2010) as they do not only encapsulate a distinct set of functionally related enzymes but rather divide the whole cell into physiologically distinct domains. Unlike in eukaryotic cells, these diffusion barriers not only laterally compartmentalize cellular membranes but also limit the free diffusion of soluble proteins, thereby providing a significant fitness advantage. Diffusion barrier formation in Caulobacter therefore represents a thus far unrecognized mechanism to optimize the growth of a prokaryote by restricting protein mobility within the cell.
Timing and assembly of the StpABCD diffusion barrier complex
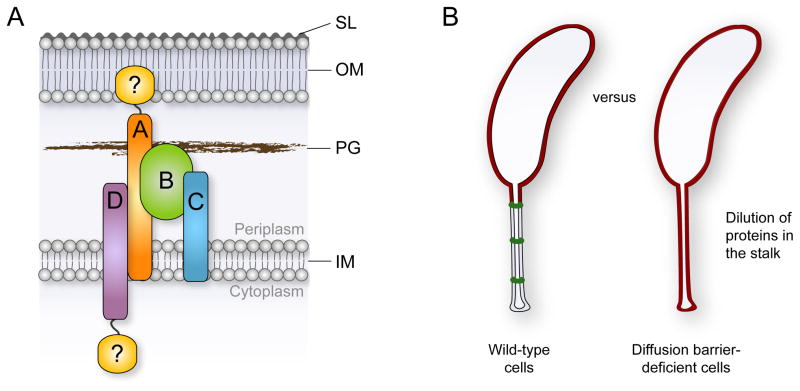
The formation of the StpABCD diffusion barrier complex is coordinated with the developmental program and occurs in the second half of the cell cycle, consistent with earlier reports on the temporal appearance of crossbands as visualized by electron microscopy (Poindexter and Staley, 1996; Staley and Jordan, 1973). Upon synthesis, the Stp proteins are targeted to the stalked pole in a hierarchal order, with StpA taking the top position in the localization hierarchy. The cellular abundance of StpA appears to be the rate-limiting factor in de novo diffusion barrier formation because the frequency of crossbands scales with the expression level of the stpA gene. Thus, diffusion barrier assembly may be triggered as soon as a critical number of StpA molecules have accumulated in the cell. After the recruitment of StpB, which is also essential for the formation of microscopically discernible crossband structures, StpC and StpD are incorporated to seal the diffusion barrier complex (Figure 7A). Notably, overproduction of StpAB leads to the formation of large helical assemblies in the stalk periplasm (Figure 4F and Movie S5). The two proteins thus self-assemble to form a membrane-associated macromolecular complex (Figure 2) that provides the basic scaffold for diffusion barrier formation.
Figure 7. Model for diffusion barrier formation and function.
(A) Diffusion barrier assembly can be envisioned as a nucleation-like process in which StpA (orange) and StpB (green) form the basic scaffold. StpC (blue) and StpD (purple) are accessory components that are required to seal the diffusion barrier. Potential unidentified components of the complex that may establish a connection to the outer membrane or assemble at the cytoplasmic face of the inner membrane are depicted in yellow.
(B) The synthesis of diffusion barriers minimizes the effective volume of the periplasmic space and reduces the physiologically active membrane surface area. In the absence of diffusion barriers, newly synthesized proteins that are targeted to the cell envelope are constantly diluted due to diffusion into the stalk extension.
The establishment of diffusion barriers is uncoupled from the cell cycle when cells are grown in low-phosphate medium, a condition that causes a general arrest of cell cycle progression but leads to constitutive elongation of the stalk (Gonin et al., 2000). Despite the lack of developmental cues, such as DNA replication and cell division, the growing stalk continues to be segmented by crossbands, albeit at somewhat irregular intervals. The precise mechanism that controls the timing of Stp complex formation in these conditions remains unclear. However, it is conceivable that nucleation of a crossband leads to a transient drop in the cellular concentration of StpA, which prevents formation of another complex until StpA levels rise again above a critical threshold level.
If StpA is required for recruiting the remaining Stp proteins to the stalk base, what targets StpA to the stalked pole in the first place? Thus far, we have been unable to identify a localization factor upstream of StpA. However, formation of the stalk creates a region of positive membrane curvature at the stalk base, which could be specifically recognized by StpA, reminiscent of the mechanism described for the bacterial proteins SpoVM and DivIVA.
Our data strongly suggest that crossbands are high-molecular weight protein complexes rather than discs made of peptidoglycan, as proposed previously (Jones and Schmidt, 1973; Schmidt, 1973). In support of this hypothesis and consistent with the observation that FtsZ does not localize to the stalked pole (Thanbichler and Shapiro, 2006), crossband formation was found to be independent of FtsZ-mediated peptidoglycan synthesis. Moreover, while the peptidoglycan biosynthetic apparatus is localized to the periplasmic space, crossbands extend on both sides of the cytoplasmic membrane, reaching from the outer membrane to the cytoplasmic core (Figure 3A). Most importantly, however, crossbands are not detectable in purified murein sacculi, although they are significantly thicker than the cellular peptidoglycan layer (Gan et al., 2008; A. Briegel and G. J. Jensen, unpublished). The previous observation that fewer crossbands are detectable upon treatment of stalks with lysozyme is thus likely explained by an indirect, stabilizing effect of the cell wall on StpABCD complex assembly.
Physiological significance of protein diffusion barriers in stalked bacteria
Caulobacter thrives in oligotrophic aquatic environments, where inorganic phosphorus commonly represents the limiting nutrient (Paerl, 1982). The Caulobacter stalk elongates in response to phosphate starvation, leading to the hypothesis that it acts as a “nutrient scavenging antenna” (Gonin et al., 2000; Schmidt and Stanier, 1966; Wagner et al., 2006). The presence of diffusion barriers, however, challenges the currently held model that phosphate-bound PstS diffuses through the stalk to deliver its cargo to the PstCAB inner-membrane transporter in the cell body (Wagner et al., 2006). However, it is possible that previous experiments have simply failed to detect the PstCAB complex in the stalk, opening the possibility that PstS-bound phosphate is targeted immediately to the stalk cytoplasm. Although the diffusion of proteins is restricted in the stalk, small molecules such as phosphate may be able to pass crossbands and then use the stalk core to travel to the cell body.
Regardless of a possible role in phosphate uptake, the stalk may predominantly function to spatially separate the cell body from the point of holdfast attachment. Elevation of the cell body may provide various selective advantages such as the ability to rise above an existing biofilm or to expose the immobilized cell to bulk nutrients (Wagner and Brun, 2007). Since the stalk is a true extension of the cell envelope, lengthening of this polar structure leads to an increase in the membrane surface area and periplasmic volume. While under optimal growth conditions the stalk periplasm only accounts for ~ 10% of the total periplasmic volume, this value can increase considerably in response to phosphate starvation. We have demonstrated that periplasmic, inner and outer membrane proteins can diffuse freely throughout the cell in the absence of diffusion barriers, so that newly synthesized proteins are constantly diluted in the stalk extension (Figure 7B). Notably, membrane proteins make up approximately 20% to 30% of the total protein in a bacterial cell (Wallin and von Heijne, 1998). Diffusion barrier formation is therefore an efficient mechanism to minimize the effective area/volume of the cell envelope and, thus, reduce the energetic cost of establishing or maintaining a pool of physiologically active envelope proteins. Moreover, physiological compartmentalization could facilitate faster adaptation to sudden environmental or developmental cues that require the induction and accumulation of a different set of envelope proteins to ensure overall fitness. The benefit of such diffusion barriers may be relatively widespread as crossbands have been identified in a range of stalked bacteria. Notably, the StpB homologue of the prosthecate species Asticcacaulis excentricus displays a similar stalk localization pattern as Caulobacter StpB (Figure S2B), indicating that crossband formation is also mediated by the Stp complex in these organisms.
Collectively, we have identified and functionally characterized a previously unrecognized protein compartmentalization mechanism that relies on the assembly of at least four proteins into a diffusion barrier complex. Structural analyses may provide insights into the precise mechanism underlying the self-assembly and subcellular localization of this structure. Our findings open the possibility that diffusion barriers could exist in a wider range of prokaryotes, providing a stimulating framework for future studies.
EXPERIMENTAL PROCEDURES
Bacterial strains, plasmids and growth conditions
The strains and plasmids used in this study are described in Tables S1, S2 and S3. Caulobacter wild-type strain CB15N and its derivatives were grown at 28°C in peptone-yeast-extract (PYE) medium (Poindexter, 1964) or M2-glucose (M2G) minimal medium. To achieve stalk elongation in response to phosphate starvation, stationary cells were diluted 1:20 in M2G−P (Kühn et al., 2010) containing 3.9 mM KCl and cultured for additional 24 h. Alternatively, cells were directly grown in Hutner imidazole-buffered glucose-glutamate (HIGG) medium containing 30 μM phosphate (Poindexter, 1978). Cell synchronization and growth competition experiments are detailed in the Extended Supplemental Procedures.
Immunoblot analysis
Immunoblot analysis was performed as described previously (Thanbichler and Shapiro, 2006). Details on the antibodies used are given in the Extended Supplemental Procedures.
Cell fractionation and co-immunoprecipitation analysis
Biochemical fractionation of cells was performed as described previously (Möll et al., 2010). For co-immunoprecipitation analysis, StpB-His was isolated from dodecyl maltoside-treated cell extracts using anti-His affinity beads and analyzed for interactors using immunoblot analysis or mass spectrometry. Experimental details are given in the Extended Supplemental Procedures.
Fluorescence microscopy
For light microscopy, cells were spotted onto pads made of 1% agarose (for still images) or 1% agarose in M2G medium (for time-lapse analyses). Details on the fluorescence microscopy and FRAP/FLIP setups are given in the Extended Supplemental Procedures.
Electron microscopy
The analysis of negatively stained Caulobacter cells by transmission electron microscopy is detailed in the Extended Supplemental Procedures.
Electron cryo-tomography was performed as described (Möll et al., 2010). Correlated fluorescence light microscopy (FLM) combined with ECT was essentially carried out as reported previously (Ingerson-Mahar et al., 2010), with the exception that cells were immobilized on C flat™ 2/2 London finder copper TEM grids with a ~40 nm thick holey carbon coat (Protochips Inc., Raleigh, NC, USA), which were treated with 5 μl of 1 mg/ml sterile-filtered poly-L-lysine (Sigma, P1524) before use. The correlative analysis was performed manually using Photoshop software (Adobe Systems). The prominent holes in the carbon foil together with the cell body and stalk densities were sufficient to unambiguously overlay the FLM images and the ECT slices. Details on the tools used for image analysis are given in the Extended Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Caulobacter cells are compartmentalized by protein diffusion barriers.
The diffusion barriers are mediated by membrane-spanning multiprotein complexes.
Barrier complex formation minimizes the effective volume of the cell.
The presence of diffusion barriers is critical for Caulobacter fitness.
Acknowledgments
We thank Stephanie Wick for excellent technical assistance, Nikolay Ouzounov for helpful discussions, Juliane Kühn, Patrick Viollier and Grant Bowman for providing plasmids and strains, and Sarah Cheng for help with image segmentation. This work was supported by funds from the Max Planck Society to M.T., a Young Investigator Grant (RGY0069/2008-C) from the Human Frontier Science Program to Z.G. and M.T., a National Institutes of Health Director’s New Innovator Award (DP2OD004389) to Z.G., an NRSA postdoctoral fellowship (F32GM906842) from the National Institute of General Medical Sciences to E.A.K., and grant GM51986 from the National Institutes of Health to Y.V.B.
Footnotes
Supplemental material, including extended experimental procedures, six figures, four tables and five movies, is available online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Caudron F, Barral Y. Septins and the lateral compartmentalization of eukaryotic membranes. Dev Cell. 2009;16:493–506. doi: 10.1016/j.devcel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Chih B, Liu P, Chinn Y, Chalouni C, Komuves LG, Hass PE, Sandoval W, Peterson AS. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat Cell Biol. 2012;14:61–72. doi: 10.1038/ncb2410. [DOI] [PubMed] [Google Scholar]
- Curtis PD, Brun YV. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol Mol Biol Rev. 2010;74:13–41. doi: 10.1128/MMBR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakaruni AV, Baida C, White CL, Gober JW. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol Microbiol. 2007;66:174–188. doi: 10.1111/j.1365-2958.2007.05910.x. [DOI] [PubMed] [Google Scholar]
- Gan L, Chen S, Jensen GJ. Molecular organization of Gram-negative peptidoglycan. Proc Natl Acad Sci U S A. 2008;105:18953–18957. doi: 10.1073/pnas.0808035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonin M, Quardokus EM, O’Donnol D, Maddock J, Brun YV. Regulation of stalk elongation by phosphate in Caulobacter crescentus. J Bacteriol. 2000;182:337–347. doi: 10.1128/jb.182.2.337-347.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes HV, Huitema E, Pritchard S, Keiler KC, Brun YV, Viollier PH. Protein localization and dynamics within a bacterial organelle. Proc Natl Acad Sci U S A. 2010;107:5599–5604. doi: 10.1073/pnas.0909119107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat Cell Biol. 2010;12:739–746. doi: 10.1038/ncb2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland MM, Karty JA, Quardokus EM, Reilly JP, Brun YV. Proteomic analysis of the Caulobacter crescentus stalk indicates competence for nutrient uptake. Mol Microbiol. 2002;45:1029–1041. doi: 10.1046/j.1365-2958.2002.03071.x. [DOI] [PubMed] [Google Scholar]
- Jones HC, Schmidt JM. Ultrastructural study of crossbands occurring in the stalks of Caulobacter crescentus. J Bacteriol. 1973;116:466–470. doi: 10.1128/jb.116.1.466-470.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn J, Briegel A, Mörschel E, Kahnt J, Leser K, Wick S, Jensen GJ, Thanbichler M. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. EMBO J. 2010;29:327–339. doi: 10.1038/emboj.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Murray AW, Botstein D. The cost of gene expression underlies a fitness tradeoff in yeast. Proc Natl Acad Sci U S A. 2009;106:5755–5760. doi: 10.1073/pnas.0901620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Blastier S, Hamels A, Cabeen M, Schille L, Tilquin F, Dieu M, Raes M, Matroule JY. Phosphate starvation triggers production and secretion of an extracellular lipoprotein in Caulobacter crescentus. PLoS One. 2010;5:e14198. doi: 10.1371/journal.pone.0014198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks ME, Castro-Rojas CM, Teiling C, Du L, Kapatral V, Walunas TL, Crosson S. The genetic basis of laboratory adaptation in Caulobacter crescentus. J Bacteriol. 2010;192:3678–3688. doi: 10.1128/JB.00255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Lee H, Zhang L, Iniesta AA, Hottes AK, Tan MH, Hillson NJ, Hu P, Shapiro L, McAdams HH. High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nat Biotechnol. 2007;25:584–592. doi: 10.1038/nbt1294. [DOI] [PubMed] [Google Scholar]
- Möll A, Schlimpert S, Briegel A, Jensen GJ, Thanbichler M. DipM, a new factor required for peptidoglycan remodelling during cell division in Caulobacter crescentus. Mol Microbiol. 2010;77:90–107. doi: 10.1111/j.1365-2958.2010.07224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat D, Byrne M, Komeili A. Cell biology of prokaryotic organelles. Cold Spring Harb Perspect Biol. 2010;2:a000422. doi: 10.1101/cshperspect.a000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada C, Ritchie K, Oba Y, Nakamura M, Hotta Y, Iino R, Kasai RS, Yamaguchi K, Fujiwara T, Kusumi A. Accumulation of anchored proteins forms membrane diffusion barriers during neuronal polarization. Nat Cell Biol. 2003;5:626–632. doi: 10.1038/ncb1009. [DOI] [PubMed] [Google Scholar]
- Neugebauer H, Herrmann C, Kammer W, Schwarz G, Nordheim A, Braun V. ExbBD-dependent transport of maltodextrins through the novel MalA protein across the outer membrane of Caulobacter crescentus. J Bacteriol. 2005;187:8300–8311. doi: 10.1128/JB.187.24.8300-8311.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paerl HW. Factors limiting productivity of freshwater ecosystems. In: Marhal KC, editor. Advances in microbial ecology. 1982. pp. 75–110. [Google Scholar]
- Poindexter JLS, Cohen-Bazire G. Fine structure of stalked bacteria belonging to family Caulobacteraceae. J Cell Biol. 1964;23:587. doi: 10.1083/jcb.23.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poindexter JS. Biological Properties and Classification of the Caulobacter Group. Bacteriol Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poindexter JS. Selection for nonbuoyant morphological mutants of Caulobacter crescentus. J Bacteriol. 1978;135:1141–1145. doi: 10.1128/jb.135.3.1141-1145.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poindexter JS, Staley JT. Caulobacter and Asticcacaulis stalk bands as indicators of stalk age. J Bacteriol. 1996;178:3939–3948. doi: 10.1128/jb.178.13.3939-3948.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Lecuyer S, Stone HA, Losick R. Geometric cue for protein localization in a bacterium. Science. 2009;323:1354–1357. doi: 10.1126/science.1169218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JM. Effect of lysozyme on crossbands in stalks of Caulobacter crescentus. Arch Mikrobiol. 1973;89:33–40. [Google Scholar]
- Schmidt JM, Stanier RY. The development of cellular stalks in bacteria. J Cell Biol. 1966;28:423–436. doi: 10.1083/jcb.28.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JM, Swafford JR. Ultrastructure of crossbands in prosthecae of Asticcacaulis species. J Bacteriol. 1975;124:1601–1603. doi: 10.1128/jb.124.3.1601-1603.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz LC, Brun YV. Genetic analysis of mecillinam-resistant mutants of Caulobacter crescentus deficient in stalk biosynthesis. J Bacteriol. 1998;180:5235–5239. doi: 10.1128/jb.180.19.5235-5239.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit J, Agabian N. Cell surface patterning and morphogenesis: biogenesis of a periodic surface array during Caulobacter development. J Cell Biol. 1982;95:41–49. doi: 10.1083/jcb.95.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JT, Jordan TL. Crossbands of Caulobacter crescentus stalks serve as indicators of cell age. Nature. 1973;246:155–156. doi: 10.1038/246155a0. [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Wagner JK, Brun YV. Out on a limb: how the Caulobacter stalk can boost the study of bacterial cell shape. Mol Microbiol. 2007;64:28–33. doi: 10.1111/j.1365-2958.2007.05633.x. [DOI] [PubMed] [Google Scholar]
- Wagner JK, Setayeshgar S, Sharon LA, Reilly JP, Brun YV. A nutrient uptake role for bacterial cell envelope extensions. Proc Natl Acad Sci U S A. 2006;103:11772–11777. doi: 10.1073/pnas.0602047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner JN, Chen EY, Guberman JM, Zippilli AR, Irgon JJ, Gitai Z. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc Natl Acad Sci U S A. 2009;106:7858–7863. doi: 10.1073/pnas.0901781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.