Abstract
The clostridial neurotoxins (CNTs) are among the most potent protein toxins for humans and are responsible for botulism, a flaccid paralysis elicited by the botulinum toxins (BoNT), and spastic paralysis elicited by tetanus toxin (TeNT). Seven serotypes of Botulinum neurotoxins (A–G) and tetanus toxin showed different toxicities and cleave their substrate with different efficiencies. However the molecular basis of their different catalytic activities to their substrates is not clear. BoNT/B light chain (LC/B) and TeNT light chain (LC/T) cleave Vesicle-Associated Membrane Protein 2 (VAMP2) at the same scissile bond, but possess different catalytic activities and substrate requirements, which make them the best candidates to study the mechanisms of their different catalytic activities. The recognition of five major P sites of VAMP2 (P7, P6, P1, P1′ and P2′) and fine alignment of P2, P3 and P2, P4 sites by LC/B and LC/T respectively, contributed to their substrate recognition and catalysis. Significantly, we found that the S1 pocket mutation LC/T(K168E) increased the rate of native VAMP2 cleavage to approach the rate of LC/B, which explains the molecular basis for the lower kcat that LC/T possesses for VAMP2 cleavage relative to LC/B. This analysis explains the molecular basis underlining the VAMP2 recognition and cleavage by LC/B and LC/T and provides insight that may extend the pharmacologic utility of these neurological reagents.
Keywords: BoNT/B, TeNT, VAMP-2, substrate recognition, structure function
INTRODUCTION
The clostridial neurotoxins (CNTs) are among the most potent protein toxins for humans and are responsible for botulism, a flaccid paralysis elicited by the botulinum toxins (BoNT), and spastic paralysis elicited by tetanus toxin (TeNT). CNTs are 150 kDa dichain proteins with typical A-B structure-function properties, where the B (binding) domain binds to surface components on the mammalian cell and translocates the A (active) domain to an intracellular location (1). CNTs are organized into three functional domains: an N-terminal catalytic domain (light chain, LC), an internal translocation domain (heavy chain, HCT), and a C-terminal receptor binding domain (heavy chain, HCR) (2). The CNTs are zinc metalloprotease that cleave SNARE (Soluble NSF Attachment REceptor) proteins which interferes with synaptic vesicle fusion to the plasma membrane and ultimately blocks neurotransmitter release in nerve cells (1, 3). Mammalian neuronal exocytosis is driven by the formation of protein complexes between the vesicle SNARE protein, VAMP2, and the plasma membrane SNAREs, SNAP25 and syntaxin 1a (4). There are seven BoNT serotypes (termed A–G) that cleave specific residues on one of three SNARE proteins: BoNT serotypes B, D, F, G, and TeNT cleave VAMP2, BoNT serotypes A and E cleave SNAP25, and BoNT serotype C cleaves SNAP25 and syntaxin 1a (3, 5–7).
BoNTs are considered as potential biological warfare and have been classified as category A agent by the CDC (Center for Disease Control and Prevention) in the U.S.A. [6,7]. To date, there is no effective treatment for BoNT intoxication and the development of anti-botulism drugs is of upmost importance. Effective small molecular antitoxin is the most efficient way for the treatment of human botulism, and the prerequisite to develop these inhibitors is to understand the molecular mechanism of the action of CNTs. In addition, BoNT/B has been approved by USFDA to treat cervical dystonia, while the relative low activity of BoNT/B requires higher dose of toxin for effective treatment, which results in the development of immunoresistance in some patients(8–10). The understanding of the mechanisms of substrate recognition of BoNT/B may help to develop novel therapies with better pharmacological properties.
Unlike other zinc proteases, BoNTs and TeNT recognize an extended region of the SNARE proteins for substrate cleavage (16–18). Recent studies using protein crystallography, protein modelling and biochemical characterization of BoNTs revealed the mechanisms of substrate recognition by BoNT/A, E and F (11–14). While all these three BoNTs recognize their substrates through important exosites and share a common theme, the substrate recognition is unique for each of the BoNTs (11, 13, 14), which requires that the mechanisms of substrate recognition and specificity be thoroughly investigated for each serotype. LC/B and LC/T cleave VAMP2, at the same scissile bond, but differ in catalytic activity, substrate requirement, and sensitivity to inhibitors (12, 15). Previous studies showed that LC/B and LC/T cleave VAMP2 with different Km and kcat (12, 16). Alanine-scanning mutagenesis and kinetic analysis identified three regions within VAMP2 that were recognized by LC/B and LC/T: residues adjacent to the site of scissile bond cleavage (cleavage region) and residues located within N-terminal region and C-terminal region relative to the cleavage region (12). Mutations at the P7, P4, P2, and P1′ residues of VAMP2 had the greatest inhibition of LC/B cleavage (>32-fold), while mutations at P7, P4, P1′, and P2′ residues of VAMP2 had the greatest inhibition of LC/T cleavage (>64-fold) (12) The different Km of LC/B and LC/T for VAMP2 may be attributed to the different compositions of binding sites N- and C- terminal to the LC active sites, while different kcat for VAMP2 may be due to different substrate recognition within the LC active site. This study addresses the molecular basis for the different recognition and cleavage of VAMP2 by LC/B and LC/T and may provide insights for the engineering of novel neurotoxin derivatives with improved therapeutic properties.
Experimental Procedures
Plasmid construction for protein expression
Plasmids for the expression of BoNT LC/B(1-430), LC/T(1-436) and VAMP2(1-97) and subsequent protein expression and purification were performed as previously described (11, 13, 17). Site directed mutagenesis of pLC/B, pLC/T and pVAMP2 were performed using QuickChange (Stratagene) protocols as previously described (11, 13). Plasmids were sequenced to confirm the mutation and that additional mutations were not present within the ORFs. Mutated proteins were produced and purified as described above (11–13, 17).
Linear velocity and kinetic constant determinations for VAMP2 cleavage by LC/B and LC/T
Linear velocity reactions (10μl) were performed as previously described (11–13). VAMP2 proteins (5 μM) were incubated with varying concentrations of LC/B, LC/T or LC derivatives in 10 mM Tris-HCl (pH 7.6) with 20 mM NaCl at 37°C for 10 min. Reactions were stopped by adding SDS-PAGE buffer, and VAMP2 and cleavage product were resolved by SDS-PAGE. The amount of VAMP2 cleaved was determined by densitometry. Km and kcat determinations were performed with the same assay where VAMP2 concentrations were adjusted between 1 and 300 μM to achieve ~ 10% cleavage by LC/B and LC/T. Reaction velocity versus substrate concentration was fit to the Michaelis-Menten equation and kinetic constants were derived using the GraphPad Program (San Diego, CA).
Compensatory assay
Effect of compensatory mutations within LC/B and LC/T on the cleavage of VAMP2 and mutated forms of VAMP2 was performed as previously described with modification (13). Briefly, 5μM VAMP2 or VAMP2 derivatives were incubated with LC/B, LC/T or LC derivatives at 37°C for 20min. The reactions were stopped by adding SDS-PAGE sample buffer and uncleaved and cleave VAMP2 were resolved by SDS-PAGE. The amount of wild type LC/B, LC/T or LC derivatives in the reaction were plotted verses % cleavage and the amount of LC required to cleave 50% of VAMP2 or VAMP2 derivative were calculated.
Molecular modeling
Complex structures of LC/B-VAMP2 and LC/T-VAMP2 were modeled using SWISS-MODEL and refined with PyMol (www.pymol.com) as described previously (22). PDB coordinates used in this analysis were 1f82 for LC/B, 1z7h for LC/T and 1xtg for LC/A–SNAP25.
RESULTS
Molecular modeling was used to predict physical contacts between LC/B-VAMP2 and LC/T-VAMP2 to initiate assessment of interactions that contribute to productive substrate cleavage (Supplementary Fig 1). VAMP2 recognition within the active pockets of LC/B and LC/T shared common contacts, and also possessed unique associations that included a variation of the overall shape of the LCs active site. Additional structure based alignment of LC/B and LC/T showed that the amino acid composition of potential substrate recognition pockets differed at several of the pockets that contacted the VAMP2 residues that have been implicated in LC recognition. This may contribute to the different kcat of LC/B and LC/T for VAMP2. Biochemical approaches were used to define the different substrate recognition pockets so that the molecular basis of the differential catalytic activity of LC/B and LC/T may be addressed. Trypsin sensitivity analysis indicated that point mutations generated within LC/B and LC/T did not affect the rate of trypsin cleavage or the tryptic peptide generated indicating that the overall conformations of the mutated LCs were similar to the wild type LCs (Supplementary Fig 2).
VAMP2 recognition by LC/B and LC/T
Modeling predicted the direct interactions for the S7, S6, S1, S1′ and S2′ substrate recognition pockets within the active sites of LC/B and LC/T with the respective P sites of VAMP2. This study focused on the characterization of the S7, S6, S1, S1′ and S2′ pockets within LC/B and LC/T along with the respective P sites of VAMP2.
S7 pocket recognition
In LC/B and LC/T, the S7 pocket comprised F26 and Y26, respectively, which contacted L70, the P7 residue of VAMP2 (Fig 1a). LC/B and LC/T cleavage of VAMP2(L70A) was ~195-and ~230-fold less efficient than cleavage of wild type VAMP2, respectively (Table 1). The LC/B(F26A) and LC/T(Y26A) mutations had an ~5-fold increase in Km, and ~10-fold decrease in kcat for VAMP2 cleavage (Table 2), while the mutation to Asp had a greater effect on kinetic values. The fact that the LC/B(F26Y) and LC/T(Y26F) mutations did not affect hydrolysis of VAMP2, indicated that L70 of VAMP2 and the aromatic S7 pocket residue of LC/B and LC/T interact through a similar hydrophobic interaction. Thus, although the physical interaction is unique to the analogous ionic S5-P5 contact between LC/A and SNAP25, the molecular outcome of the S7-P7 interactions are similar, affecting both the affinity and rate of VAMP2 cleavage (12).
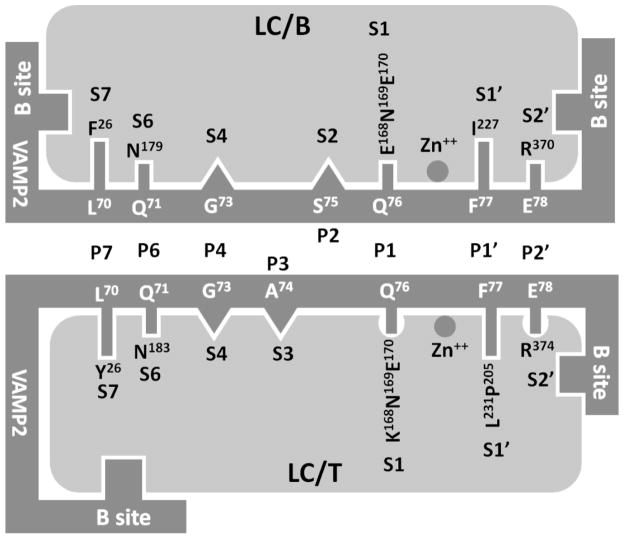
Fig 1. Recognition of VAMP2 P sites by LC/B and LC/T substrate recognition pockets.
Computational and experimental data from the current study provides a model for the interaction of LC/B (left panel) and LC/T (right panels) with VAMP2. (a) Recognition of P7 and P6 sites by S7 and S6 pockets of LC/B and LC/T. The S7 pockets of LC/B was formed by F26, which recognized P7, L70 and the S6 pocket of LC/B was formed by N179, which recognized P6, Q71. The S7 of LC/T was formed by Y26, which recognized P7, L70 and the S6 pocket of LC/B was formed by N183, which specifically recognize P6, Q71. (b) Recognition of P1, P1′ and P2′ sites of VAMP2 by S1, S1′ and S2′ pockets of LC/B and LC/T. The S1 pocket of LC/B and LC/T was formed by residues E168N169E 170 and K168N169E 170 respectively, recognized P1, Q76 of VAMP2. The S1′ pocket of LC/B and LC/T was formed by I227 and L231, P205, respectively, which recognized F77 of VAMP2. The S2′ of LC/B and LC/T was formed by R370 and R374, respectively, which recognized P2′, E78 of VAMP2. (c) Fine alignment of P4, P3 and P2 sites of VAMP2 into S4, S3, and S2 pockets of LC/B and LC/T. The large side chain of S4 pocket residue, R, allows the alignment of smaller side chain residue, G. The different shapes of S2 and S3 pockets of LC/B and LC/T, while not the residue composition of the pockets enable S2 and S3 to tolerate different P2 and P3 site residues.
Table 1.
Kinetic constants of LC/B and LC/T on VAMP2 point mutations
| VAMP P sites | Wt | P7, L70 | P6, Q71 | P1,Q76 | P1′, F77 | P2′, E78 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Point Mutation | No | A | A | A | Y | F | R | E | A | A | D | R | Y |
| Km (μM) | 1.7± 0.3 | 6.5± 0.8 | 1.9± 0.2 | 1.8± 0.3 | 1.7± 0.3 | 1.8± 0.4 | 1.7± 0.4 | 1.8± 0.4 | 1.6± 0.2 | 2.4± 0.4 | 2.6± 0.2 | 2.2± 0.2 | 1.8± 0.2 |
| LC/B | |||||||||||||
| kcat (min−1) | 66.0 | 1.2 | 4.4 | 9.6 | 33.2 | 11.0 | 5.6 | 1.3 | 0.2 | 12.2 | 67.3 | 1.2 | 1.3 |
| kcat/Km (min−1 • μM−1) | 39.0 | 0.2 | 2.3 | 5.3 | 19.5 | 6.1 | 3.3 | 0.7 | 0.1 | 5.1 | 25.9 | 0.6 | 0.7 |
| Km (μM) | 4.1±0.2 | 20.8±0.8 | 4.1±1.2 | 4.0±0.6 | 4.1±0.2 | 4.1±0.3 | 4.0±0.2 | 4.1±0.6 | 3.8±0.4 | 4.3±0.4 | 4.1±0.4 | 3.9±0.3 | N/D |
| LC/T | |||||||||||||
| kcat (min−1) | 9.6 | 0.1 | 1.6 | 1.3 | 9.4 | 9.4 | 1.5 | 1.9 | 0.02 | 0.05 | 9.7 | 0.2 | N/D |
| kcat/Km (min−1 • μM−1) | 2.3 | 0.01 | 0.4 | 0.3 | 2.3 | 2.3 | 0.4 | 0.5 | 0.005 | 0.01 | 2.4 | 0.05 | N/D |
Table 2.
Kinetic constants of LC/B, LC/T and LC derivatives
| LCs | VAMP2 (Residue) | LC Pockets | LC derivative | Km (μM) | kcat (min−1) | kcat/Km (min−1 • μM−1) |
|---|---|---|---|---|---|---|
| LC/B | Wt-LC/B | 1.7±0.3 | 66 | 39 | ||
|
|
||||||
| P7(L70) | S7 | F26A | 9.1±0.8 | 5.6 | 0.6 | |
| F26D | 35.6±2.4 | 0.4 | 0.01 | |||
| F26Y | 1.8±0.3 | 68 | 39 | |||
|
|
||||||
| P6 (Q71) | S6 | N179L | 1.7±0.4 | 16 | 9.4 | |
|
|
||||||
| P1 (Q76) | S1 | E168A | 1.7±0.4 | 3.2 | 1.9 | |
| E168K | 1.6±0.2 | 0.4 | 0.3 | |||
| N169A | 1.8±0.3 | 6.5 | 3.6 | |||
| E170A | 1.8±0.4 | 6.6 | 3.7 | |||
|
|
||||||
| P1′(F77) | S1′ | I227A | 1.6±0.2 | 0.8 | 0.5 | |
| I227E | 1.8±0.3 | 0.08 | 0.05 | |||
| I227L | 1.8±0.4 | 0.1 | 0.07 | |||
|
|
||||||
| P2′ (E78) | S2′ | R370A | 1.8±0.4 | 0.6 | 0.4 | |
| R370E | 1.8±0.2 | 0.2 | 0.08 | |||
|
| ||||||
| LC/T | Wt-LC/T | 4.1±0.2 | 9.6 | 2.3 | ||
|
|
||||||
| P7(L70) | S7 | Y26A | 20.8±0.8 | 0.9 | 0.05 | |
| Y26D | N/D | N/D | N/D | |||
| Y26F | 4.2±0.2 | 9.9 | 2.4 | |||
|
|
||||||
| P6 (Q71) | S6 | N183L | 4.2±0.4 | 4.5 | 1.1 | |
|
|
||||||
| P1 (Q76) | S1 | K168A | 4.2±0.2 | 10.6 | 2.5 | |
| K168E | 4.1±0.2 | 79.8 | 19.5 | |||
| N169A | 4.0±0.2 | 2.0 | 0.5 | |||
| E170A | 4.1±0.6 | 2.6 | 0.63 | |||
|
|
||||||
| P1′(F77) | S1′ | L231A | 4.2±0.2 | 0.3 | 0.8 | |
| L231K | N/D* | N/D | N/D | |||
| P205A | 4.0±0.3 | 0.3 | 0.8 | |||
| P205K | N/D | N/D | N/D | |||
|
|
||||||
| P2′ (E78) | S2′ | R374A | N/D | N/D | N/D | |
| R374E | N/D | N/D | N/D | |||
N/D, Not determined, the mutant is too inactive to determine the kinetic constants
S6 pocket recognition
In LC/B and LC/T, the S6 pocket comprised N179 and N183, respectively, which contacted Q71, the P6 residue of VAMP2 (Fig 1a). LC/B and LC/T cleaved VAMP2(Q71A) ~15 and 6-fold lower kcat than wild type VAMP2, respectively (Table 1). The N1791L and N183L mutation in LC/B and LC/T had ~4- and ~2-fold reduction in kcat, respectively, without affecting the Km for VAMP2 cleavage (Table 2). The compensatory mutation sets, LC/B(N179L) and LC/T(N183L) cleaved VAMP2(Q71A) as efficiently as LC/B cleaved wild type VAMP2 (Table 3). The positive correlation of the compensatory mutations, supported a direct interaction (possibly via hydrogen bond) between the S6 pocket N179 or N183 of the LCs and Q71 of VAMP2.
Table 3.
Compensatory mutational assay for LC/B and LC/T interactions with VAMP2
| LCs | 50% Cleavage of VAMP2 (nM) | Ratio of activity (LC/LC derivative) | 50% Cleavage of VAMP2 Q71A (nM) | Ratio of activity (LC/LC derivative) |
|---|---|---|---|---|
| LC/B | 6 | 4 | 96 | 1/16 |
| LC/B(N179L) | 24 | 6 | ||
| LC/T | 120 | 2 | 720 | 1/6 |
| LC/T(N183L) | 240 | 120 | ||
|
| ||||
| 50% Cleavage of VAMP2 G73A (nM) | ||||
|
| ||||
| LC/B | 6 | 1 | 192 | 1/32 |
| LC/B(R184M) | 6 | 6 | ||
| LC/T | 120 | 1 | 7680 | 1/64 |
| LC/T(R188M) | 20 | 60 | ||
|
| ||||
| 50% Cleavage of VAMP2 E78R (nM) | ||||
|
| ||||
| LC/B | 6 | 400 | 300 | 1/50 |
| LC/B(R370E) | 2400 | 6 | ||
| LC/T | 120 | 300 | >36000 | ND* |
| LC/T(R374E) | 36000 | >36000 | ||
ND*, Not determined
S1 pocket recognition
LC/B cleaved the VAMP2(Q76A) with an ~7-fold lower kcat than VAMP2 (Table 1). Mutations within the P1 site of VAMP2 (Q76Y, Q76F, Q76R and Q76E) resulted in ~2-, ~6-, ~12-, and ~50-fold reductions in LC/B kcat, respectively, without an effect on Km, showing that hydrophobic residues were favored at this site and that the P1 pocket could accommodate an aromatic ring (Table 1). The S1 pocket of LC/B was formed by E168, N169, and E 170 (Table 2, Fig 1b). None of these residues showed a dominant interaction with the P1 residue Q76 (Fig 1b), since the LC/B mutations E168A, N169A and E170A showed reductions in kcat by ~20, ~11 and ~11 folds, respectively. The charge reversal mutation, LC/B(E170K), had an ~ 130-fold slower kcat (Table 2), suggesting that the negatively charged S1 pocket of LC/B favored recognition of the polar residue Q76.
LC/T cleaved VAMP2(Q76A) ~8-fold slower than VAMP2 (Table 1). In contrast to LC/B, mutations at P1 site of VAMP2 (Q76Y, Q76F, Q76R and Q76E) showed no, no, ~6-, and ~5-fold reduction on the cleavage activity of LC/T, suggesting that the S1pocket residues may interact with main chain components of VAMP2 (Table 1). The S1 pocket of LC/T was formed by K168, N169, and E170 (Fig 1b). Mutations K168A, N169A or E170A to LC/T had no, ~5-, and ~4-fold reduction to the kcat, while, unexpectedly, LC/T(K168E) cleaved VAMP2 with an ~8-fold increased kcat. The increased activity of LC/T(K168E) suggested that an acidic S1 pocket is optimal for Q76 recognition. Thus, the S1-P1 interaction may have structural and charge tolerance, implicating the S1 pocket of LC/T and LC/B as a candidate region to engineer and expand the substrate potential to non-neuronal VAMP derivatives with therapeutic potential.
S1′ pocket recognition
In LC/B, the S1′ pocket residue I227 contacted the hydrophobic P1′ site residue F77 (Fig 1b). The VAMP2 P1′ mutation (F77A) decreased the ability of LC/B to hydrolyze by ~320 fold (Table 1). LC/B(I227A) did not affect Km, but showed an ~80-fold reduction of kcat, while LC/B(I227E) also did not affect Km,, but showed an ~800-fold reduction of kcat (Table 2). The mutation of other LC/B residues adjacent to I227, including, F195A, V200A, L226A and S201A, did not affect LC/B cleavage of VAMP2 (data not shown), supporting a direct interaction between LC/B-I227 and VAMP2 F77. The conservative mutation of I227L did not affect Km,, but had an ~550-fold reduction of kcat, supporting the importance of the R-group (side chain of the amino acid residue) orientation for optimal VAMP2 cleavage.
In LC/T, the S1′ pocket residues, L231 and P205, contacted the hydrophobic P1′ site residue, F77 (Fig 1b). The VAMP2 P1′ mutation (F77A) decreased the ability of LC/T to hydrolyze VAMP2 by ~460-fold (Table 1). The L231A or P205A mutations did not affect Km, but reduced the kcat ~ 30-fold, while L231K became too inactive to determine its kinetic constants (Table 2). Mutation of other residues adjacent to S2 pocket residues, L231 and P205 including, L232, F199A, V204A, and L230A, did not affect LC/T cleavage of VAMP2 (data not shown).
S2′ pocket recognition
LC/B cleaved the P2′ residue mutation VAMP2(E78A) slower than VAMP2 (Table 1). The (E78R) or (E78Y) mutations did not affect Km, but reduced the kcat ~ 50-fold, while the E78D mutation did not affect LC/B cleavage. This suggested that negatively charged residues were favored at P2′ site (Table 1). In LC/B, S2′ pocket comprised R370 (Fig 1b). LC/B(R370A) cleaved VAMP2 with an ~ 100-fold lower kcat, while LC/B(R370E) had an ~ 500-fold lower kcat (Table 2). This was unexpected, since the corresponding Arg, R369, within LC/A has been implicated in the transition state coordination of P1 and P1′ site of SNAP25 (18). The direct contribution of R370 to P2′ site recognition was examined with compensatory mutation sets. This analysis showed that LC/B cleaved VAMP2(E78R) ~50-fold slower than VAMP2 and LC/B(R370E) cleaved VAMP2 ~400-fold slower than LC/B, while LC/B(R370E) has a similar rate of VAMP2(E78R) cleavage as LC/B cleavage of VAMP2 (Table 3). These data support a role for a salt-bridge between R370 of LC/B and E78 VAMP2, providing evidence for a direct interaction between P2′ residue E78 and the S2′ pocket residue, R370, of LC/B.
LC/T cleaved VAMP2(E78A), a P2′ site mutation, ~230-fold slower that VAMP2 (Table 1). Like LC/B, LC/T had an ~50- fold lower kcat for VAMP2(E78R) and the kinetic constants could not be determined for VAMP2(E78Y) due to the extreme low activity of LC/T on this VAMP mutation, while the E78D mutation did not affect catalysis (Table 1). Also like LC/B, S2′ pocket of LC/T contains an Arg, R374 (Fig 1b). LC/T(R374A) and LC/T(R374E) became too inactive to determine their kinetic constants (Table 2). However, in contrast to LC/B, analysis of compensatory mutation sets showed that LC/T(R374E) did not cleave VAMP2(E78R) (Table 3). These data suggest that R374 of LC/T and E78 of VAMP2 do not directly interact or that the interaction is due to non-ionic interactions between the two residues. This indicates a unique role for the S2′ pocket Arg of LC/B and LC/T which could be to positional differences within the respective S2′ pockets (Fig 1b).
VAMP2-LC interactions at P site residues not predicted by computational predictions
While protein modeling predicted direct interactions between the S pocket residues of LC/B and LC/T and VAMP2, other LC S pockets including S4, S3, and S2, were not predicted to have direct interactions with their respective R-group of the P site residues. However, directed mutagenesis showed that G73 (P4) and S75 (P2) of VAMP2 were required for optimal LC/B cleavage and A72 (P5), G73 (P4) and A74 (P3) of VAMP2 were required for optimal LC/T cleavage (12). Thus, while modeled complexes of LC/B-VAMP2 and LC/T-VAMP2 did not predicted direct R-group interactions by these small R-group amino acids (A72G73A74S75) these residues appear to facilitate alignment of VAMP2 into the active sites of LC/B and LC/T. Data support the former since the 3 amino acid-mutated VAMP2(A72G,A74G,S75G) was not cleaved by LC/B or LC/T (data not shown). Together, this data implies a role for the S4, S3, and S2 pockets in LC cleavage of VAMP2.
S4 pocket
In LC/B, the S4 pocket that specifically recognized the P4 site of VAMP-2 G73 comprised one positively charged residue, R184 (Fig 1c). The mutation of LC/B R184M that retained its positive charged property, while with smaller side chain, showed similar hydrolytic activity as the wild type LC/B (Table 3). However, LC/B R184M was able to cleave VAMP G73A as efficient as wild type LC/B to cleave wild type VAMP-2 (Table 3). These data suggested that the 32-fold decreased hydrolytic activity of LC/B on VAMP-2 G73A was due to the space constraint caused by adding a methyl group to P4 Glycine and the replacement of R184 with a small side chain M complemented the larger side chain Ala at P4 position.
In LC/T, the S4 pocket that specifically recognized the P4 of VAMP-2 G73, was formed by R188 (Fig 3). The mutation of LC/TeNT R188M that remained its positive charged property, while with smaller side chain, showed ~5 fold higher activity on VAMP-2 than wild type LC/B (Table 3). Furthermore, LC/T R188M was able to cleave VAMP G73A slightly more efficient than wild type LC/T to cleave wild type VAMP-2. These data also suggested that the 64-fold decreased hydrolytic activity of LC/T on VAMP-2 G73A was due to the space constraint caused by the larger side chain residue Ala and the smaller side chain mutation of R183M can complemented the larger side chain Ala at the P4 position.
S3 pocket recognition
While LC/B cleaved VAMP2(A74S), a P3 mutation, as efficiently as VAMP2, LC/T cleaved VAMP2(A74S) less efficiently than VAMP2 (12). The S3 pocket residues comprise T172 in LC/B and V176 in LC/T (Fig 1c). LC/B(T172V) and LC/T(V176T) cleaved VAMP2 as efficiently as the wild type LCs (data not shown), suggesting the modifications in the polarity (-OH) at the S3 site was tolerated by both LCs. Structural comparison of S3 pockets indicated that the S3 pocket of LC/B was smaller than LC/T. This suggests that LC/T may be able to tolerate an increase in size of P3 site residues, which may be coordinated by an adjacent β-sheet that is not conserved between LC/B and LC/T (Fig 1c).
S2 pocket recognition
While LC/T cleaved VAMP2(S75A), a P2 mutation, as efficiently as VAMP2, LC/B cleaved VAMP2(S75A) less efficiently than VAMP2 (12). The S2 pocket was formed by Q265 in LC/B and S269 in LC/T (Fig 1c). LC/B(Q265S) and LC/T(S269Q) were as efficient as wild type LCs for VAMP2 cleavage (data not shown). This suggests that the differential efficiency of LC/B and LC/T cleavage of VAMP2(A74S) was not due to composition of the corresponding substrate recognition pockets. Comparison of S2 pockets of LC/B and LC/T showed that the S2 pocket of LC/B was wider than LC/T, which could enabled LC/B to tolerate the addition of the –OH of Ser (Fig 1c).
DISCUSSION
Earlier studies showed that LC/B and LC/T recognized three regions in VAMP2 that contribute to substrate recognition: a cleavage region (residues 70~78) that immediately surround the scissile bond and two regions that contribute to high affinity binding to VAMP2 which are located N-terminal and C-terminal to the cleavage region (12, 16). Together with the current study, the generation of a detailed model for how LC/B and LC/T recognize VAMP2 and a molecular basis to explain the similarities and differences in substrate cleavage are now possible. By analogy with LC/A-SNAP25 interactions (13, 19), upon N- terminal and C-terminal VAMP2 binding, the S7 pocket residue (F26 in LC/B and Y26 in LC/T) associate with L70 in the P7 site through hydrophobic interactions and the S2′ pocket residue (R370 for LC/B and R374 for LC/T) associates through an ionic interaction with E78 the P2′ residue. These interactions facilitate P1 and P1′ (Q76, F77) association to the LCs to align the scissile bond for substrate cleavage. In addition, interactions of the internal S pocket residues of LC/B and LC/T (S5, S4, S3 and S2) align VAMP2 through physical interactions that allow physical orientations for the effective cleavage of a coiled substrate.
The similarity of LC/B and LC/T substrate recognition includes two binding sites that are unique in Clostridium neurotoxins and the specific recognition of several P sites by active site pockets of the LCs, where the P7-S7 and P1′-S1′ interaction are the most important anchoring points, while other P-S interactions contribute to the tighter and more specific substrate recognition. The different substrate recognition between these two neurotoxins includes their different binding sites that may contribute to their different Km on substrate VAMP2 and their different composition of the S pockets, which may contribute to their different kcat on VAMP2. The active site substrate recognition for LC/B includes the direct recognition of P7, P6, P1, P1′, and P2′ sites of VAMP2 by the corresponding S pockets in LC/B and the fine alignment of P4 and P3 sites to the S4 and S3 pockets, while the substrate recognition for LC/T includes the direct recognition of P7, P6, P1, P1′, and P2′ sites of VAMP2 by the corresponding S pockets in LC/T and the fine alignment of P4 and P2 sites to the S4 and S2 pockets. The less optimal composition of S1 pocket of and the more complex interaction between P2′-S2′ interactions in LC/T may contribute to the lower kcat of LC/T on VAMP2 (Fig 2).
Fig 2. Similarity and difference of substrate recognition by LC/B and LC/T.
The active site substrate recognition for LC/B includes the direct recognition of P7, P6, P1, P1′, and P2′ sites of VAMP2 by the corresponding S pockets in LC/B and the fine alignment of P4 and P3 sites to the S4 and S3 pockets, while the substrate recognition for LC/T includes the direct recognition of P7, P6, P1, P1′, and P2′ sites of VAMP2 by the corresponding S pockets in LC/T and the fine alignment of P4 and P2 sites to the S4 and S2 pockets. The less optimal composition of S1 pocket of and the more complex interaction between P2′-S2′ interaction in LC/T may contribute to the lower kcat of LC/T on VAMP2.
The residues within the cleavage regions vary among the BoNT serotypes. Comparing the cleavage regions that are recognized by the LCs: the P2′ and P2 sites of LC/E and LC/F, the P4′ and P5 sites of LC/A, and the P2′ and P7 sites of LC/B and LC/T contribute to P1′-P1 scissile bond stabilization. The different arrangements of P sites that are recognized by LCs to stabilize the scissile bond correlates with LCs catalytic potential. LC/E and LC/F have high kcat and compact cleavage regions while LC/B and LC/T have low kcat and extended cleavage regions. The larger cleavage region may result in weak stabilization of scissile bond cleavage (11–13, 16, 19, 20).
Relative to LC/B, the low kcat for LC/T appears to be attributed to less than optimal interactions between the P1 residue with the S1 pocket residues and possibly the P2′ residue with the S2′ pocket. The observation that the introduction of an ionic bond between the P1 residue and the S1 pocket residue enhances the kcat provided a basis for the lower kcat of LC/T relative to LC/B. This suggests that the fastest rate of substrate cleavage may not be optimal in a biological setting. This also indicates that there is the potential for additional optimization to modulate BoNTs with higher activity, which may be a potential solution to the immune-resistance issue of BoNT-based therapies.
Although the precise mechanism for peptide bond cleavage by the BoNTs remains to be resolved, cleavage of the scissile peptide bond appears to follow a general base-catalyzed mechanism (18, 21, 22). Arg362 and Tyr365 interact with the carbonyl oxygen of the P1 and P1′-residues of SNAP25, respectively, and stabilization of the oxyanion in the transition state. Peptide bond cleavage is initiated by a water molecule that is polarized by the Glu within the zinc binding motif (HEXXH) and Zn2+, which causes a nucleophilic attack on the carbonyl carbon of the scissile bond to form an oxyanion. Peptide bond cleavage is likely achieved by a proton transfer from the attacking water mediated by the carboxyl group of the downstream Glu to form a protonated amine. The crystal structure of LC/E bound to the C terminus of LC/E-cleaved SNAP25 product showed that P1′ oxygen interacts with the side chain NH1 and NH2 groups of the conserved residue Arg347 and Tyr350 OH interacts with P2′: Met182 oxygen and helps in stabilizing its main chain, which confirms that Arg347 and Tyr350 play a crucial role in transition state stabilization by allowing proper docking of the main chain of P1, P1′, and P2′ residues at the active site. The current study shows a different (additional) role for R370 of LC/B. Complementation assays indicated that R370 of LC/B is directly involved in substrate recognition and specifically recognized P2, E78 of VAMP2. In contrast, although the S2′ pockets in LC/B and LC/T were very similar, while R368 of LC/T and P2, E78 did not show direct interaction. The role of R374 in LC/T is not clear and may be involved in the coordination of substrate catalysis. The role of this analogous Arg in other serotypes of BoNTs will be needed for further confirmation.
In conclusion, the comparative characterization addressed the molecular mechanisms of VAMP2 recognition and cleavage by LC/B and LC/T and described the molecular basis of their similarity and difference. The novel information regarding BoNT substrate recognition presented may facilitate the engineering of novel BoNTs to extend BoNT based therapies.
Supplementary Material
Acknowledgments
We thank the members of the Sheng’s laboratory for helpful discussion. This work was sponsored by the RGC/Hong Kong PolyU Competitive Research Grants A-PK05 and G-YJ15 for SC, and the Great Lakes Regional Center of Excellence U54 AI057153 for JTB.
Abbreviations
- BoNT/B
Botulinum neurotoxin serotype B
- TeNT
Tetanus neurotoxin
- LC
light chain
- VAMP2
vesicle associated membrane protein-2
- SNARE
soluble NSF attachment receptor
Footnotes
This work was sponsored by the RGC/Hong Kong PolyU Competitive Research Grants A-PK05 and G-YJ15 for SC, and the Great Lakes Regional Center of Excellence U54 AI057153 for JTB.
Two figures reporting the “recognition of VAMP2 P sites residues within the active sites regions of LC/B and LC/T.” and “trypsin digestion profiles of LC/B, LC/T and their derivatives”. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Montecucco C, Schiavo G. Mechanism of action of tetanus and botulinum neurotoxins. Mol Microbiol. 1994;13:1–8. doi: 10.1111/j.1365-2958.1994.tb00396.x. [DOI] [PubMed] [Google Scholar]
- 2.Davletov B, Bajohrs M, Binz T. Beyond BOTOX: advantages and limitations of individual botulinum neurotoxins. Trends Neurosci. 2005;28:446–452. doi: 10.1016/j.tins.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Montecucco C, Schiavo G. Tetanus and botulism neurotoxins: a new group of zinc proteases. Trends Biochem Sci. 1993;18:324–327. doi: 10.1016/0968-0004(93)90065-u. [DOI] [PubMed] [Google Scholar]
- 4.Brunger AT. Structure and function of SNARE and SNARE-interacting proteins. Q Rev Biophys. 2005;38:1–47. doi: 10.1017/S0033583505004051. [DOI] [PubMed] [Google Scholar]
- 5.Schiavo G, Benfenati F, Poulain B, Rossetto O, Polverino de Laureto P, DasGupta BR, Montecucco C. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature. 1992;359:832–835. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- 6.Schiavo G, Malizio C, Trimble WS, Polverino de Laureto P, Milan G, Sugiyama H, Johnson EA, Montecucco C. Botulinum G neurotoxin cleaves VAMP/synaptobrevin at a single Ala-Ala peptide bond. J Biol Chem. 1994;269:20213–20216. [PubMed] [Google Scholar]
- 7.Schiavo G, Rossetto O, Benfenati F, Poulain B, Montecucco C. Tetanus and botulinum neurotoxins are zinc proteases specific for components of the neuroexocytosis apparatus. Ann N Y Acad Sci. 1994;710:65–75. doi: 10.1111/j.1749-6632.1994.tb26614.x. [DOI] [PubMed] [Google Scholar]
- 8.Brashear A, Lew MF, Dykstra DD, Comella CL, Factor SA, Rodnitzky RL, Trosch R, Singer C, Brin MF, Murray JJ, Wallace JD, Willmer-Hulme A, Koller M. Safety and efficacy of NeuroBloc (botulinum toxin type B) in type A-responsive cervical dystonia. Neurology. 1999;53:1439–1446. doi: 10.1212/wnl.53.7.1439. [DOI] [PubMed] [Google Scholar]
- 9.Atassi MZ, Jankovic J, Steward LE, Aoki KR, Dolimbek BZ. Molecular immune recognition of botulinum neurotoxin B. The light chain regions that bind human blocking antibodies from toxin-treated cervical dystonia patients. Antigenic structure of the entire BoNT/B molecule. Immunobiology. 2012;217:17–27. doi: 10.1016/j.imbio.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Atassi MZ, Dolimbek BZ, Jankovic J, Steward LE, Aoki KR. Molecular recognition of botulinum neurotoxin B heavy chain by human antibodies from cervical dystonia patients that develop immunoresistance to toxin treatment. Mol Immunol. 2008;45:3878–3888. doi: 10.1016/j.molimm.2008.06.031. [DOI] [PubMed] [Google Scholar]
- 11.Chen S, Barbieri JT. Multiple pocket recognition of SNAP25 by botulinum neurotoxin serotype E. J Biol Chem. 2007;282:25540–25547. doi: 10.1074/jbc.M701922200. [DOI] [PubMed] [Google Scholar]
- 12.Chen S, Hall C, Barbieri JT. Substrate recognition of VAMP-2 by botulinum neurotoxin B and tetanus neurotoxin. J Biol Chem. 2008;283:21153–21159. doi: 10.1074/jbc.M800611200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S, Kim JJ, Barbieri JT. Mechanism of substrate recognition by botulinum neurotoxin serotype A. J Biol Chem. 2007;282:9621–9627. doi: 10.1074/jbc.M611211200. [DOI] [PubMed] [Google Scholar]
- 14.Chen S, Wan HY. Molecular mechanisms of substrate recognition and specificity of botulinum neurotoxin serotype F. Biochem J. 2011;433:277–284. doi: 10.1042/BJ20101310. [DOI] [PubMed] [Google Scholar]
- 15.Foran P, Shone CC, Dolly JO. Differences in the protease activities of tetanus and botulinum B toxins revealed by the cleavage of vesicle-associated membrane protein and various sized fragments. Biochemistry. 1994;33:15365–15374. doi: 10.1021/bi00255a017. [DOI] [PubMed] [Google Scholar]
- 16.Sikorra S, Henke T, Galli T, Binz T. Substrate recognition mechanism of VAMP/synaptobrevin-cleaving clostridial neurotoxins. J Biol Chem. 2008;283:21145–21152. doi: 10.1074/jbc.M800610200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen S, Wan HY. Molecular mechanisms of substrate recognition and specificity of botulinum neurotoxin serotype F. Biochem J. 433:277–284. doi: 10.1042/BJ20101310. [DOI] [PubMed] [Google Scholar]
- 18.Binz T, Bade S, Rummel A, Kollewe A, Alves J. Arg(362) and Tyr(365) of the botulinum neurotoxin type a light chain are involved in transition state stabilization. Biochemistry. 2002;41:1717–1723. doi: 10.1021/bi0157969. [DOI] [PubMed] [Google Scholar]
- 19.Chen S, Barbieri JT. Unique substrate recognition by botulinum neurotoxins serotypes A and E. J Biol Chem. 2006;281:10906–10911. doi: 10.1074/jbc.M513032200. [DOI] [PubMed] [Google Scholar]
- 20.Sikorra S, Henke T, Swaminathan S, Galli T, Binz T. Identification of the amino acid residues rendering TI-VAMP insensitive toward botulinum neurotoxin B. J Mol Biol. 2006;357:574–582. doi: 10.1016/j.jmb.2005.12.075. [DOI] [PubMed] [Google Scholar]
- 21.Agarwal R, Schmidt JJ, Stafford RG, Swaminathan S. Mode of VAMP substrate recognition and inhibition of Clostridium botulinum neurotoxin F. Nat Struct Mol Biol. 2009;16:789–794. doi: 10.1038/nsmb.1626. [DOI] [PubMed] [Google Scholar]
- 22.Agarwal R, Eswaramoorthy S, Kumaran D, Binz T, Swaminathan S. Structural analysis of botulinum neurotoxin type E catalytic domain and its mutant Glu212-->Gln reveals the pivotal role of the Glu212 carboxylate in the catalytic pathway. Biochemistry. 2004;43:6637–6644. doi: 10.1021/bi036278w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.