Abstract
RNA editing is a post-transcriptional process that can act upon transcripts from mitochondrial, nuclear, and chloroplast genomes. In chloroplasts, single-nucleotide conversions in mRNAs via RNA editing occur at different frequencies across the plant kingdom. These range from several hundred edited sites in some mosses and ferns to lower frequencies in seed plants and the complete lack of RNA editing in the liverwort Marchantia polymorpha. Here, we report the sequence and edited sites of the chloroplast genome from the liverwort Pellia endiviifolia. The type and frequency of chloroplast RNA editing display a pattern highly similar to that in seed plants. Analyses of the C to U conversions and the genomic context in which the editing sites are embedded provide evidence in favor of the hypothesis that chloroplast RNA editing evolved to compensate mutations in the first land plants.
Keywords: liverwort, plastid, RNA editing, evolution, land plants
Introduction
Chloroplast genomes descend from the genome of a free-living, cyanobacterial-like ancestor, which was engulfed by a eukaryotic cell and reduced to a chloroplast (Martin et al. 2002; Hempel et al. 2007; Bolte et al. 2009). Several genomic characteristics of cyanobacteria, such as operon structures, are still maintained in chloroplast genomes. However, other attributes such as various transcript maturation processes or the phage-type RNA polymerase are lacking in cyanobacteria and hence can be thought to have arisen during plastid evolution (Maier et al. 2008; Stern et al. 2010; Tillich and Krause 2010). This is arguably true for RNA editing in plastids as well, that is, the post-transcriptional alteration of one to a few bases in maturing mRNAs (Tillich et al. 2006).
RNA editing was observed in some minicircle-encoded plastid genes in dinoflagellates (Zauner et al. 2004; Dang and Green 2009) but was never observed in plastid-encoded RNAs in other algae groups so far. Nevertheless, it is common in land plants where it occurs at frequencies that vary across lineages. Seed plant plastids typically exhibit 26–54 editing sites (Wakasugi et al. 1996; Tillich et al. 2006; Jiang et al. 2012). By contrast, the fern Adiantum capillus-veneris (Wolf et al. 2004) and the hornwort Anthoceros formosae (Kugita et al. 2003) harbor hundreds of sites that are post-transcriptionally modified by editing. At the high end of the spectrum, the lycophyte Isoetes engelmannii appears to have more than 1,500 edited sites in total (Grewe et al. 2011). At the low end, the moss Physcomitrella patens maintains chloroplast RNA-editing machinery for two sites only (Miyata and Sugita 2004; Rüdinger et al. 2009), whereas in the liverwort Marchantia polymorpha, no detectable RNA editing occurs at all (Steinhauser et al. 1999).
What underlies this diversity in RNA-editing frequency among plastids? Several hypotheses have been put forth to explain the evolution of chloroplast RNA editing in land plants. Covello and Gray (1993) proposed a three-step model for the evolution of RNA editing, in which first an RNA-editing activity appears in an enzyme, followed by mutation at editable positions and genetic drift, and finally fixation of the edited sites and their corresponding editing machinery; the general underlying principle of evolving complicated processes, like editing, without advantage was later dubbed constructive neutral evolution (Stoltzfus 1999; Gray et al. 2010). Jobson and Qiu (2008) took a more selectionist stance on the issue, arguing that RNA editing arose in land plant chloroplasts through natural selection imposed by functional constraints on the encoded proteins. Our own suggestion is that chloroplasts encountered a high load of mutations during the transition from an aquatic to a terrestrial life style and that RNA-editing mechanisms suppressed these mutations, thereby eliminating nonsynonymous amino acid changes in coding regions and compensating for their potentially deleterious effects in the chloroplast genome (Tillich et al. 2006; Maier et al. 2008). In addition, the incorporation of a phage-type RNA polymerase into plastid genome expression could have helped to overcome mutations in promoter regions, which are not transcribed (Maier et al. 2008).
Regardless of the causes behind the origin of chloroplast editing, edited sites can be lost by back mutation (or remutation) of the edited site into the “correct” (ancestral) base. Lineage-specific variation in this parameter could account for the different frequencies of editing across the chloroplast of different groups. However, in that case, the bottleneck for the elimination of editing sites would entail mutational dynamic that vary across sequence contexts and species, and indeed genomic contexts with a low remutation rate (Tillich et al. 2006) were found for flowering plants to be especially TCA trinucleotides (Morton 1995, 2003; Morton et al. 1997). In chloroplast genomes with a high editing load, no strict consensus with respect to flanking nucleotides can be defined, whereas in seed plants with a low editing frequency, a biased RNA-editing pattern is present in which the editing site C is preferentially located within a TCA genomic context (Tillich et al. 2006).
Marchantia polymorpha figures prominently in this issue, because no RNA-editing sites exist in the chloroplast genome (Freyer et al. 1997; Steinhauser et al. 1999), indicating that chloroplast functions are independent of RNA editing and that regulatory mechanisms in other lineages that involve editing might have evolved secondarily. Thus, M. polymorpha and the whole marchantiid subclade might never have had the “load” of RNA editing or might have lost it secondarily. Analysis of jungermanniid liverwort plastid genomes could be instructive. As chloroplast and mitochondrial RNA editing generally coincides (Freyer et al. 1997; Steinhauser et al. 1999; Tillich et al. 2006), the detection of mitochondrial RNA-editing sites in some jungermanniid liverworts are possibly a proxy for chloroplast RNA editing in this group (Groth-Malonek et al. 2007; Rüdinger et al. 2008). In line with that view, RNA editing was identified in the chloroplast of the liverworts Bazzania trilobata and Pellia epiphylla by analyzing ndhB and rbcL transcripts (Freyer et al. 1997), and extensive RNA editing was observed in the liverwort Haplomitrium mnioides (Groth-Malonek et al. 2005, 2007). Haplomitrium (Haplomitriopsida) was shown to be, to some extent, an isolated group in the clade of liverworts, which most probably diverged before the separation of Jungermanniopsida and Marchantiopsida (Groth-Malonek et al. 2007). Taken together, the data suggest that M. polymorpha and the whole marchantiid subclade might have lost RNA editing in organelles secondarily.
To test this “secondary lost” prediction and to illuminate the distribution and evolution of RNA-editing sites in the clade of liverworts and plants in general, we sequenced the chloroplast genome of the liverwort P. endiviifolia (Dicks.) Dumort and determined the frequency and the genomic context of chloroplast RNA editing. Our data support the view of a secondary loss of RNA editing in M. polymorpha and indicate that chloroplast RNA editing was present in early land plants. In addition, we show that patterns in the RNA-editing sites in the chloroplast of a liverwort are comparable to those in seed plants, indicating retention of RNA-editing sites in a genomic context known to have a low remutation rate (Morton 1995, 2003; Morton et al. 1997; Tillich et al. 2006).
Materials and Methods
Culture Conditions
Pellia endiviifolia (Dicks.) Dumort was obtained from the Botanical Garden of the Philipps University of Marburg. Plants were cultured under natural light/dark cycle conditions at 20°C on soil.
DNA and RNA Isolation
Total DNA was isolated by the CTAB method (Doyle and Doyle 1990). For RNA isolation, 100 mg of plant material was collected, frozen in liquid nitrogen, and ground to a fine powder with mortar and pestle. One milliliter of Trizol (Life Technologies, Darmstadt, Germany) was added, and grinding was continued for 2 min. The mixture was transferred to a microcentrifuge tube and spun for 10 min at 12,000 × g and 4°C. Supernatant was transferred into a new tube and incubated at room temperature for 5 min, after which 200 µl chloroform was added, mixed, and incubated at room temperature again for 5 min. After centrifugation at 12,000 × g for 10 min at room temperature, the aqueous phase was collected and transferred into a new tube. An equal volume of isopropanol was added, mixed, and spun for 20 min at 12,000 × g and 4°C. The pellet was washed once with 70% ethanol, dried, and dissolved in 50 µl H2O. Before cDNA synthesis, the RNA was treated with DNAseI (Roche, Munich, Germany or Fermentas, St. Leon-Roth, Germany) and reverse transcribed using Omniscript Reverse Transciptase, OneStep RT-PCR Kit (both Qiagen, Hilden, Germany), or MuLV Reverse Transcriptase (Fermentas, St. Leon-Rot, Germany).
PCR and Sequencing
For genome sequencing, Long PCR Enzyme Mix (Fermentas, St. Leon-Rot, Germany) was used. Normal PCR was made with Biotools DNA polymerase (Biotools, Madrid, Spain) or Phusion® High-Fidelity polymerase (Finnzymes, Vantaa, Finland). PCR products were purified with NucleoSpin Gel and PCR Clean-up Kit (Macherey-Nagel, Düren, Germany) and directly sequenced using the DYEnamic ET Terminator Cycle Sequencing Kit (GE-Healthcare, Munich, Germany) on an ABI PRISM® 377 DNA Sequencer (Applied Biosystems, Darmstadt, Germany). Oligonucleotides used for PCR and sequencing were purchased from MWG (Ebersberg, Germany) or Sigma-Aldrich (Munich, Germany). Coverage of sequencing was at least 2-fold based on independent isolations. In some cases, we had to clone PCR products in pJet Cloning Vector (Fermentas, St. Leon-Rot, Germany). In this case, sequencing coverage was at least 6-fold.
Data Assembly and Annotation
Data were assembled and edited using the Sequencher 4.7-5.0 (Gene Codes Corporation, Ann Arbor, MI). Open reading frames or tRNAs were identified using tRNAscan SE (Schattner et al. 2005) and ORF Finder and Blast tools from National Center for Biotechnology Information. DOGMA (Wyman et al. 2004) and OGDraw (Lohse et al. 2007) were used for annotation and map drawing, respectively.
Results and Discussion
Pellia endiviifolia cpDNA Gene Content and Structure
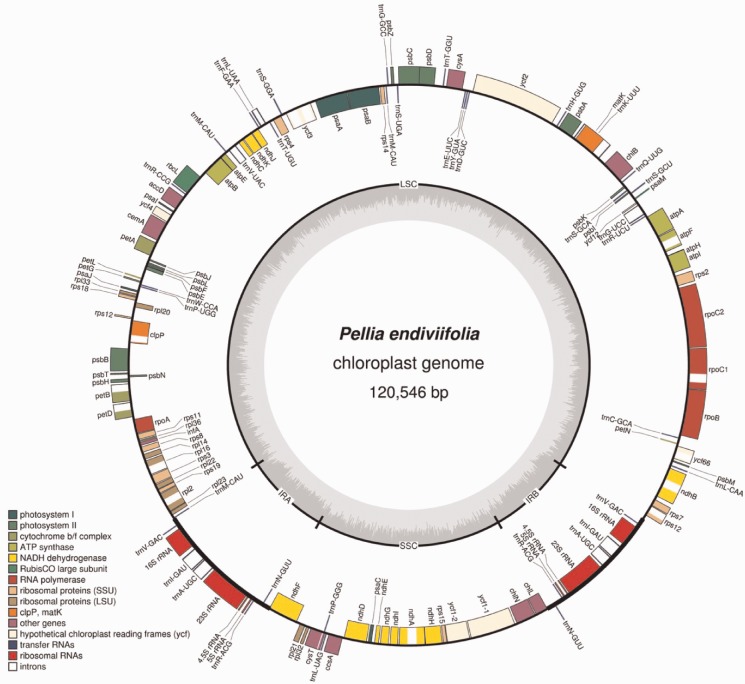
The chloroplast genome of P. endiviifolia was amplified from genomic DNA and sequenced yielding a contig of 120,546 bp (fig. 1 and table 1). The sequence was nonpolymorphic except for some differing sequences in the 3′-region of the cysT gene. That polymorphic stretch might indicate a chromosomal region involved in a switch from circular to linear chromosomes as described by others (Bendich 2004; Oldenburg and Bendich 2004). We detected 123 genes including YCFs and genes for hypothetical proteins, all known from orthologs from other chloroplast genomes (fig. 1). In comparison to the chloroplast genome of M. polymorpha, the first sequenced chloroplast genome of a liverwort, we observed strong conservation in arrangement, gene order, and gene content, and the general borders between IR, SSC, and LSC are conserved as well. The chloroplast genome of P. endiviifolia (120,546 bp) is slightly smaller than that of M. polymorpha (121,025 bp), whereas the overall A/T content differs clearly (M. polymorpha 71.19% and P. endiviifolia 64.11%). Size difference in comparison to M. polymorpha is due to various small deletions and insertions. Inverted repeat regions are more compact in P. endiviifolia, whereas at the border between IRb and rps12-3’, P. endiviifolia has a 301-bp-long noncoding stretch. An obvious difference is observable in the coding region of ycf2, which carries ∼550 bp deletion in comparison to M. polymorpha. Smaller deletions are detectable in ycf1-1 and ycf1-2 (orf464 and orf1068 in M. polymorpha). In general, ycf’s are not well conserved between these two species. We further identified 21 introns in the chloroplast genome of P. endiviifolia, most of which are known to be present in other nonparasitic liverwort chloroplast genomes. Differences in existing introns in comparison to M. polymorpha comprised an additional intron in the ycf3 gene in P. endiviifolia and an intron close to the end of the coding region of cysT.
Fig. 1.—
The chloroplast genome of Pellia endiviifolia. The plastome of the liverwort is displayed in a circular pattern. LSC, large single copy region; SSC, small single region; IRA/B, inverted repeat A/B. Color code see legend on the left. Drawing was made with OGDraw (Lohse et al. 2007).
Table 1.
Characteristics of the Chloroplast Genome of Pellia endiviifolia
| Region | Begin–End | Length (bp) | A/T Content (%) |
|---|---|---|---|
| LSC | 1–82,508 | 82,508 | 66.05 |
| SSC | 91,601–111,454 | 19,854 | 66.97 |
| IRA | 82,509–91,600 | 9,092 | 52.14 |
| IRB | 111,455–120,546 | 9,092 | 52.14 |
| Complete genome | 120,546 | 64.11 | |
| Coding regions | |||
| Protein | 73,257 | 64.33 | |
| t-/rRNAs | 11,922 | 46.19 | |
| Complete coding regions | 85,179 | 61.79 | |
| Ptilidium pulcherrimuma | 119,007 | 66.77 | |
| Marchantia polymorphab | 121,025 | 71.19 | |
Recently, the chloroplast genome of the jungermanniid liverwort Ptilidium pulcherrimum was published (Forrest et al. 2011). By comparing their results with the then available data, the authors proposed a stasis in chloroplast genome structure of liverworts. Our data from P. endiviifolia support that view because the plastid genome of P. pulcherrimum is nearly identical with respect to the described characteristics.
All members of the “core set” of chloroplast encoded genes indicating the minimal set of genes encoded by all chloroplast genomes of photosynthetic active organisms (Martin et al. 1998; Martin 2003) are present in the P. endiviifolia chloroplast genome.
RNA-Editing Pattern
In contrast to M. polymorpha, which shows no organellar RNA editing, we identified 54 editing sites in the chloroplast of P. endiviifolia by comparing mRNAs with genomic sequences. Editing events were observed in 26 of 87 protein-coding genes (table 2). For ycf66, we have no indications of transcriptional activity, therefore no cDNA sequences could be determined. The exact function of the protein product of ycf66 is still unknown, but recently a loss of this gene was described for ferns where it was lost in at least four independent cases. It therefore appears that ycf66 is, at least in ferns, not essential (Gao et al. 2011). Nevertheless, the ycf66 reading frame in P. endiviifolia is intact, indicating functional constraints.
Table 2.
All Identified RNA-Editing Sites in the Chloroplast Genome of Pellia endiviifolia
| Gene | Editing Sites | Nt Position | AA Position | Codon | Context | Code | Remarks | |
|---|---|---|---|---|---|---|---|---|
| atpB | 1 | 1097 | 366 | tCg | S → L | atpBeU1097SL | ||
| atpF | 1 | 113 | 38 | cCa | P → L | atpFeU113PL | ||
| atpH | 2 | 188 | 63 | tCa | S → L | atpHeU188SL | ||
| 224 | 75 | cCg | P → L | atpHeU224PL | ||||
| atpI | 4 | 292 | 98 | Ctt | tCtt | L → F | atpIeU292LF | |
| 431 | 144 | cCt | P → L | atpIeU431PL | ||||
| 632 | 211 | cCa | P → L | atpIeU632PL | ||||
| 704 | 235 | cCa | P → L | atpIeU704PL | ||||
| ccsA | 3 | 2 | 1 | aCg | T → M | ccsAeU2TM | Start codon | |
| 403 | 135 | Cgg | cCgg | R → W | ccsAeU403RW | |||
| 482 | 161 | cCt | P → L | ccsAeU482PL | ||||
| clpP | 3 | 81 | 27 | ctC | ctCt | L | clpPeU81LL | Silent |
| 320 | 107 | tCa | S → L | clpPeU320SL | ||||
| 533 | 178 | tCt | S → F | clpPeU533SF | ||||
| ndhB | 1 | 1427 | 476 | tCt | S → F | ndhBeU1427SF | ||
| ndhD | 5 | 536 | 179 | tCa | S → L | ndhDeU536SL | ||
| 548 | 183 | tCa | S → L | ndhDeU548SL | ||||
| 824 | 275 | cCa | P → L | ndhDeU824PL | ||||
| 1037 | 346 | tCt | S → F | ndhDe1037SF | ||||
| 1324 | 442 | Cat | tCat | H → Y | ndhDeU1324HY | |||
| ndhE | 1 | 242 | 81 | tCa | S → L | ndhEeU242SL | ||
| ndhF | 5 | 566 | 187 | tCa | S–L | ndhFeU566SL | Partial | |
| 581 | 194 | tCa | S–L | ndhFeU581SL | ||||
| 1067 | 356 | tCa | S → L | ndhFeU1067SL | ||||
| 2072 | 691 | tCt | S → F | ndhFeU2072SF | ||||
| 2078 | 693 | cCa | P → L | ndhFeU2078PL | ||||
| ndhG | 1 | 133 | 45 | Cgt | tCgt | R → C | ndhGeU133RC | |
| ndhK | 1 | 56 | 19 | tCa | S → L | ndhKeU56SL | ||
| petA | 3 | 308 | 103 | cCa | P → L | petAeU308PL | Partial | |
| 311 | 104 | tCa | S → L | petAeU311SL | ||||
| 748 | 250 | Ccg | tCcg | P → L | petAeU748PL | |||
| petB | 3 | 167 | 56 | tCc | S → F | petBeU167SF | ||
| 506 | 169 | tCa | S → L | petBeU506SL | ||||
| 593 | 198 | tCt | S → F | petBeU593SF | ||||
| petG | 1 | 80 | 27 | tCg | S → L | petGeU80SL | ||
| petL | 2 | 23 | 8 | tCt | S → F | petLeU23SF | ||
| 74 | 25 | tCa | S → L | petLeU74SL | ||||
| petN | 1 | 86 | 29 | tCg | S → L | petNeU86SL | ||
| psaB | 3 | 764 | 255 | cCa | P → L | psaBeU764PL | ||
| 782 | 261 | tCt | S → F | psaBeU782SF | ||||
| 1595 | 532 | cCa | P → L | psaBeU1595PL | ||||
| psbB | 2 | 448 | 150 | Cgt | tCgt | R → C | psbBeU448RC | |
| 740 | 247 | tCc | S → F | psbBeU740SF | ||||
| psbC | 1 | 859 | 287 | Cgt | tCgt | S → L | psbCeU859RC | |
| psbJ | 1 | 60 | 20 | ctC | ctCg | L | psbJeU60LL | Silent, partial |
| psbM | 2 | 41 | 14 | tCt | S → F | psbMeU41SF | ||
| 47 | 16 | tCa | S → L | psbMeU47SL | ||||
| rpl23 | 2 | 2 | 1 | aCg | T → M | rpl23eU2TM | Start codon | |
| 127 | 43 | Ctt | tCtt | L → F | rpl23eU127LF | |||
| rpoB | 3 | 863 | 288 | tCa | S → L | rpoBeU863SL | ||
| 1414 | 472 | Cat | tCat | H → Y | rpoBeU1414HY | |||
| 1940 | 647 | tCa | S → L | rpoBeU1940SL | ||||
| rpoC2 | 1 | 290 | 97 | cCt | P → L | rpoC2eU290PL | ||
| ycf3 | 1 | 65 | 22 | cCg | P → L | ycf3eU65PL | ||
| 54 | ||||||||
For all editing sites, we observed only C to U conversions. Two editing events restore a start methionine (ccsAeU2TM and rpl23eU2TM), and three were found to be only partially edited. Additionally, we detected two silent editing sites with no impact on the encoded amino acid. One of latter is edited partially (psbJeU60LL) (table 2). Most affected by RNA editing is the ndh-gene family with 14 editing events, followed by pet genes (10), genes encoding subunits of the photosystem (psa/psb) with 9 editing sites and atp with 8 editing events.
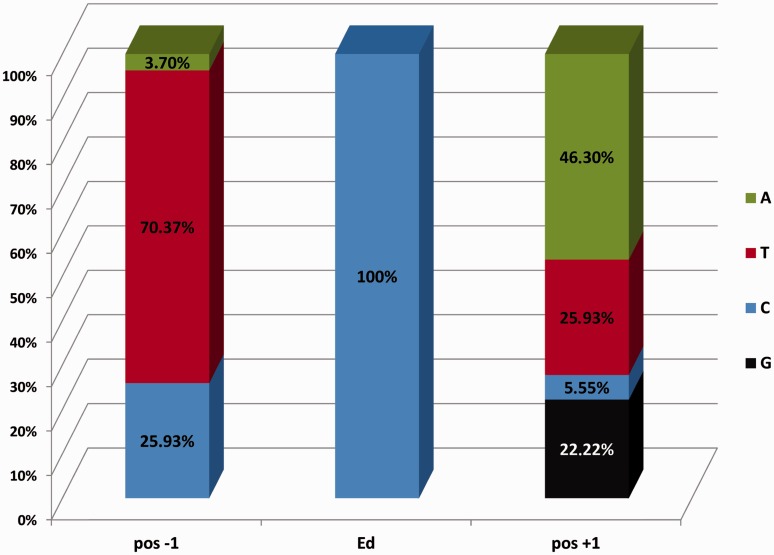
We examined the genomic context in which RNA-editing sites occur (table 2). In seed plants, the editing sites converting C→U were shown to have a bias toward 5’-T_A-3’ flanking bases (Tillich et al. 2006), a genomic context known to have a lower substitution rate in plastids (Morton 2003). Analysis of the flanking bases of the editing sites (fig. 2) reveals that 17 show a tCa bias (31.5%), whereas in 29 cases (53.7%), we could observe a variation of this motif (tCN or NCa). Overall, we observed a 5′-pyrimidine at 96% of all edited sites (thymine in 70.4% and cytosine in 25.9%). The occurrence of a 5′-pyrimidine was reported to result in lower substitution rate (Morton et al. 1997). Thus, 96% of the editing sites in P. endiviifolia cpDNA are embedded in a “low re-mutation biased” genomic context.
Fig. 2.—
Nucleotide context of editing sites in Pellia endiviifolia. The diagram shows the direct upstream (pos −1) and downstream (pos +1) nucleotide context of the identified editing sites (Ed) independent of position within the codon. Editing sites are embedded in a context of known low mutation rate in plastid DNA with predominantly 5′-pyrimidine (T or C) and 3′-purine bases (A or G) (see text for a detailed description).
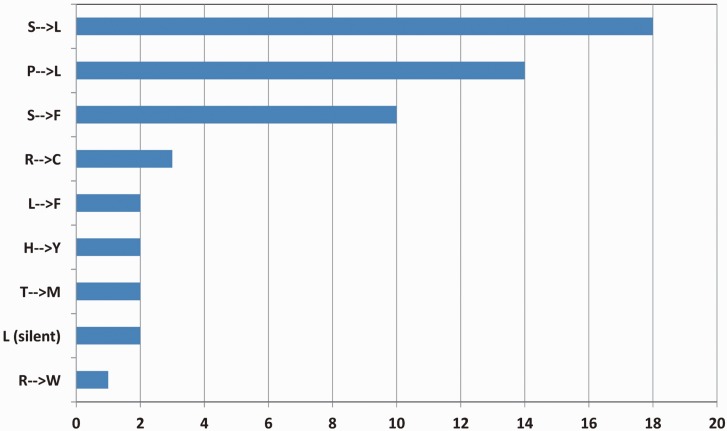
The editing events in the chloroplast genome of P. endiviifolia lead to many nonsynonymous changes (fig. 3). Serine to leucine or serine to phenylalanine changes are most frequently observed, followed by proline to leucine changes. This is in line with previously observed predominant amino acid exchanges as a consequence of RNA editing (Tillich et al. 2006). The genomic context of flanking bases for the observed editing sites clearly illustrates why we frequently observe the amino acid exchanges S → L, S → F, and P → L (fig. 3). Serine is encoded by the tCn codon. This codon is affected in 28 of 54 editing events (51.9%) in P. endiviifolia (table 2) and is known, as described, for a low remutation rate (Morton et al. 1997; Morton 2003). Proline is encoded by cCn, but 8 of 13 editing events affecting this codon are editing events on cCa and thereby a sequence context of lower remutation rate as well (Morton et al. 1997; Morton 2003). These data indicate that the frequently observed amino acid exchanges, as a result of plastid RNA editing, are predominantly a consequence of a sequence context of known low remutation rate in chloroplast DNA. Other mutations of the plastid DNA might have occurred, but these resulted either in no exchange of the encoded amino acid or a conservative (functionally similar) amino acid exchange likely without negative effects on protein function.
Fig. 3.—
Amino acid exchanges resulting from plastid RNA editing. The types (y axis) and quantities (x axis) of all amino acid exchanges as a result of plastid RNA editing in Pellia endiviifolia are shown. Editing leads to amino acid exchanges, which should have an impact on protein function because affected amino acids have differing biochemical properties or restore a start codon.
Evolutionary Implications
The evolution of RNA editing is incompletely understood, and in particular, the question “why editing” is still open. A general answer that would hold for all cases and kinds of editing might not be possible due to various manifestations and functions of RNA editing in different compartments and organisms. Here, we have focused on RNA editing in chloroplasts and the hypothesis proposed by Tillich et al. (2006) which, in the main, posits the following: chloroplast RNA editing originated in the first land plants whose plastids experienced, during the transition to land, a high mutation load. Mutations effecting coding regions were possibly compensated via RNA editing, whereas mutations in important regulatory, nontranscribed cis-acting sequences, the promoter regions, possibly necessitate recruitment of a chloroplast-directed phage-type RNA polymerase (Maier et al. 2008). Although RNA-editing sites could be eliminated via genomic remutations of the editing site into the “correct” (ancestral) base with no need for editing anymore, gains of editing sites might contribute to the different rate of RNA editing in different species as well. However, recent studies indicate that gains are rare (Hayes and Hanson 2008; Tillich et al. 2009).
If the “Tillich” hypothesis is correct, one would predict to observe the following in newly analyzed land plant chloroplasts.
-
i)
RNA editing should not be detectable, caused by secondary loss of the need for RNA editing. The mechanism of loss should be remutations, which restore the ancetral (pre-editing) base.
-
ii)
If RNA editing is observed, then either
-
iia)
RNA editing should present at high frequencies (approximately >100 editing sites), whereby the prediction is that no strong genomic context of the editing sites (i.e., the 5′- and 3′-flanking base) should be found and edits other than C to U transitions might be detectable.
Alternatively:
-
iib)
in cases where approximately <100 editing sites per chloroplast genome (∼30 in seed plants) occur, a strong tendency for the location of the editing site within TCA trinucleotides (with C being the edited base) and/or a generally increasing number of 5′-pyrimidines should be observed, because these genomic backgrounds were shown to be the genomic context with the lowest remutation rate (Morton et al. 1997; Morton 2003). This additionally implies that only C to U transitions might be present as seen in the seed plant examples (Tillich et al. 2006).
Because every cytosine edited in P. endiviifolia is a thymine in M. polymorpha, we can conclude that these positions remutated and led to a secondary loss of RNA editing in M. polymorpha. However, although closely related, not every C–T difference between M. polymorpha and P. endiviifolia coding regions reflects an actual editing site because only sites that are important for protein function seem to undergo editing. Editing frequently results in radical amino acid changes, an exchange of amino acids with clearly different physicochemical properties, which could impact protein function (Maier et al. 1996; Jiang et al. 2012) (fig. 3). Such sites might require editing or remutations. Every C that is edited to U in P. endiviifolia mRNA is a T in M. polymorpha chloroplast DNA. Additionally, the flanking bases, as we see them today, are the same in many cases or even have a 5′-thymine in M. polymorpha (data not shown). Thus, chloroplast genomes of marchantiid liverworts might have overcome the low mutation bottleneck in the respective genomic context and now encode the “right” base. Therefore, it is likely that the chloroplast genome of the marchantiid liverwort progenitor underwent a period of accelerated evolution with an increased substitution rate. Accelerated evolution and a corresponding low RNA-editing frequency have been shown for Geraniaceae (Parkinson et al. 2005). Such a “progressed evolution” might additionally be seen in the comparatively high A/T content of the chloroplast genome of M. polymorpha (table 1). Vice versa, a high editing frequency in correlation with a high G/C content was shown for the chloroplast genome of the lycophyte Selaginella (Smith 2009), congruent with our postulates.
Another possibility is that RNA editing evolved after the diversification of liverworts. Available data for RNA editing in the mitochondrion genome of other liverworts and from two identified chloroplast RNA-editing sites in B. trilobata and P. epiphylla (Freyer et al. 1997) suggest that liverworts inherited an RNA-editing load but secondarily lost it in the case of marchantiopsida (Groth-Malonek et al. 2007; Rüdinger et al. 2008). This view is supported by our finding of “only” 54 editing sites in the chloroplast of the liverwort P. endiviifolia. Thus, it is possible that all groups of land plants might need RNA editing in the chloroplast, and only rarely they lose it secondarily.
In comparison to the hornwort A. formosae or the fern A. capillus-veneris, a small number of editing sites is present in the chloroplast genome of P. endiviifolia, which is comparable to the situation in seed plants. The genomic context of the 54 editing sites in P. endiviifolia (fig. 2) show that 17 positions (31.5%) have the edited C embedded in a TCA context. In addition, 21 editing sites show the edited C in a genomic context of TCN (38.9%) and 8 in NCA (14.8%). Only eight editing sites (14.8%) do not match TCN or NCA. Of the latter, two editing sites cure a start codon. Together with the finding that in P. endiviifolia chloroplast RNA editing involves only C to U transitions, this supports the idea that RNA editing evolved in the first land plants (Maier et al. 2008).
The chloroplast genomes of M. polymorpha (Ohyama et al. 1988), P. pulcherrimum (Forrest et al. 2011), and P. endiviifolia are highly conserved in their structure and gene order. Forrest et al. (2011) suggested “evolutionary stasis” for chloroplast genome structure in liverworts. However, this might be a secondary effect, because the low editing rates or even the loss of RNA editing in Jungermanniopsida and Marchantiopsida, respectively, imply the contrary, namely rapid evolutionary rates in liverwort chloroplast genomes after splitting of the Haplomitrium subclade.
The present data are consistent with the hypothesis that M. polymorpha and the marchantiid liverworts secondarily lost the need for RNA editing in the chloroplast. This implies that such “primitive” land plants show a highly derived expression machinery in the chloroplast, one in which the mutational burden incurred during the transition from water to land was suppressed. In addition, the type of RNA editing (C to U), the genomic context of the editing site (TCA), and the amount of RNA-editing sites detected in the chloroplast genome of P. endiviifolia are consistent with the view that RNA editing initially evolved to eliminate deleterious mutations introduced into the first land plant plastid genomes. Subsequent gains of edited sites might have also contributed to the species-specific distribution of chloroplast RNA editing, but at present this contribution seems to be minor.
Acknowledgments
C.G. and S.Z. thank Judith Köbis for skillful laboratory assistance. This work was supported by the Deutsche Forschungsgemeinschaft through the Collaborative Research Centre/Transregio1 and the DFG Research Training Group 1216 Intra- and Intercellular Transport and Communication.
Literature Cited
- Bendich AJ. Circular chloroplast chromosomes: the grand illusion. Plant Cell. 2004;16:1661–1666. doi: 10.1105/tpc.160771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte K, et al. Protein targeting into secondary plastids. J Eukaryot Microbiol. 2009;56:9–15. doi: 10.1111/j.1550-7408.2008.00370.x. [DOI] [PubMed] [Google Scholar]
- Covello PS, Gray MW. On the evolution of RNA editing. Trends Genet. 1993;9:265–268. doi: 10.1016/0168-9525(93)90011-6. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Dang Y, Green BR. Substitutional editing of Heterocapsa triquetra chloroplast transcripts and a folding model for its divergent chloroplast 16S rRNA. Gene. 2009;442:73–80. doi: 10.1016/j.gene.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Forrest LL, Wickett NJ, Cox CJ, Goffinet B. Deep sequencing of Ptilidium (Ptilidiaceae) suggests evolutionary stasis in liverwort plastid genome structure. Plant Ecol Evol. 2011;144:29–43. [Google Scholar]
- Freyer R, Kiefer-Meyer MC, Kossel H. Occurrence of plastid RNA editing in all major lineages of land plants. Proc Natl Acad Sci U S A. 1997;94:6285–6290. doi: 10.1073/pnas.94.12.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Zhou Y, Wang ZW, Su YJ, Wang T. Evolution of the rpoB-psbZ region in fern plastid genomes: notable structural rearrangements and highly variable intergenic spacers. BMC Plant Biol. 2011;11:64. doi: 10.1186/1471-2229-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Lukes J, Archibald JM, Keeling PJ, Doolittle WF. Cell biology. Irremediable complexity? Science. 2010;330:920–921. doi: 10.1126/science.1198594. [DOI] [PubMed] [Google Scholar]
- Grewe F, et al. A unique transcriptome: 1782 positions of RNA editing alter 1406 codon identities in mitochondrial mRNAs of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 2011;39:2890–2902. doi: 10.1093/nar/gkq1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth-Malonek M, Pruchner D, Grewe F, Knoop V. Ancestors of trans-splicing mitochondrial introns support serial sister group relationships of hornworts and mosses with vascular plants. Mol Biol Evol. 2005;22:117–125. doi: 10.1093/molbev/msh259. [DOI] [PubMed] [Google Scholar]
- Groth-Malonek M, Wahrmund U, Polsakiewicz M, Knoop V. Evolution of a pseudogene: exclusive survival of a functional mitochondrial nad7 gene supports Haplomitrium as the earliest liverwort lineage and proposes a secondary loss of RNA editing in marchantiidae. Mol Biol Evol. 2007;24:1068–1074. doi: 10.1093/molbev/msm026. [DOI] [PubMed] [Google Scholar]
- Hayes ML, Hanson MR. High conservation of a 5' element required for RNA editing of a C target in chloroplast psbE transcripts. J Mol Evol. 2008;67:233–245. doi: 10.1007/s00239-008-9101-9. [DOI] [PubMed] [Google Scholar]
- Hempel F, et al. Transport of nuclear-encoded proteins into secondarily evolved plastids. Biol Chem. 2007;388:899–906. doi: 10.1515/BC.2007.119. [DOI] [PubMed] [Google Scholar]
- Jiang Y, et al. Identification of RNA editing sites in cotton (Gossypium hirsutum) chloroplasts and editing events that affect secondary and three-dimensional protein structures. Genet Mol Res. 2012;11:987–1001. doi: 10.4238/2012.April.19.4. [DOI] [PubMed] [Google Scholar]
- Jobson RW, Qiu Y-L. Did RNA editing in plant organellar genomes originate under natural selection or through genetic drift? Biol Direct. 2008;3:43. doi: 10.1186/1745-6150-3-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugita M, et al. The complete nucleotide sequence of the hornwort (Anthoceros formosae) chloroplast genome: insight into the earliest land plants. Nucleic Acids Res. 2003;31:716–721. doi: 10.1093/nar/gkg155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Bock R. Organellargenomedraw (OGdraw): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- Maier RM, et al. RNA editing in plant mitochondria and chloroplasts. Plant Mol Biol. 1996;32:343–365. doi: 10.1007/BF00039390. [DOI] [PubMed] [Google Scholar]
- Maier UG, et al. Complex chloroplast RNA metabolism: just debugging the genetic programme? BMC Biol. 2008;6:36–36. doi: 10.1186/1741-7007-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W. Gene transfer from organelles to the nucleus: frequent and in big chunks. Proc Natl Acad Sci U S A. 2003;100:8612–8614. doi: 10.1073/pnas.1633606100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, et al. Gene transfer to the nucleus and the evolution of chloroplasts. Nature. 1998;393:162–165. doi: 10.1038/30234. [DOI] [PubMed] [Google Scholar]
- Martin W, et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci U S A. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata Y, Sugita M. Tissue- and stage-specific RNA editing of rps14 transcripts in moss (Physcomitrella patens) chloroplasts. J Plant Physiol. 2004;161:113–115. doi: 10.1078/0176-1617-01220. [DOI] [PubMed] [Google Scholar]
- Morton BR. Neighboring base composition and transversion/transition bias in a comparison of rice and maize chloroplast noncoding regions. Proc Natl Acad Sci U S A. 1995;92:9717–9721. doi: 10.1073/pnas.92.21.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton BR. The role of context-dependent mutations in generating compositional and codon usage bias in grass chloroplast DNA. J Mol Evol. 2003;56:616–629. doi: 10.1007/s00239-002-2430-1. [DOI] [PubMed] [Google Scholar]
- Morton BR, Oberholzer VM, Clegg MT. The influence of specific neighboring bases on substitution bias in noncoding regions of the plant chloroplast genome. J Mol Evol. 1997;45:227–231. doi: 10.1007/pl00006224. [DOI] [PubMed] [Google Scholar]
- Ohyama K, et al. Structure and organization of Marchantia polymorpha chloroplast genome. I. Cloning and gene identification. J Mol Biol. 1988;203:281–298. doi: 10.1016/0022-2836(88)90001-0. [DOI] [PubMed] [Google Scholar]
- Oldenburg DJ, Bendich AJ. Most chloroplast DNA of maize seedlings in linear molecules with defined ends and branched forms. J Mol Biol. 2004;335:953–970. doi: 10.1016/j.jmb.2003.11.020. [DOI] [PubMed] [Google Scholar]
- Parkinson CL, et al. Multiple major increases and decreases in mitochondrial substitution rates in the plant family Geraniaceae. BMC Evol Biol. 2005;5:73. doi: 10.1186/1471-2148-5-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüdinger M, Funk HT, Rensing SA, Maier UG, Knoop V. RNA editing: only eleven sites are present in the Physcomitrella patens mitochondrial transcriptome and a universal nomenclature proposal. Mol Genet Genomics. 2009;281:473–481. doi: 10.1007/s00438-009-0424-z. [DOI] [PubMed] [Google Scholar]
- Rüdinger M, Polsakiewicz M, Knoop V. Organellar RNA editing and plant-specific extensions of pentatricopeptide repeat proteins in jungermanniid but not in marchantiid liverworts. Mol Biol Evol. 2008;25:1405–1414. doi: 10.1093/molbev/msn084. [DOI] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS Web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR. Unparalleled GC content in the plastid DNA of Selaginella. Plant Mol Biol. 2009;71:627–639. doi: 10.1007/s11103-009-9545-3. [DOI] [PubMed] [Google Scholar]
- Steinhauser S, Beckert S, Capesius I, Malek O, Knoop V. Plant mitochondrial RNA editing. J Mol Evol. 1999;48:303–312. doi: 10.1007/pl00006473. [DOI] [PubMed] [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR. Chloroplast RNA metabolism. Annu Rev Plant Biol. 2010;61:125–155. doi: 10.1146/annurev-arplant-042809-112242. [DOI] [PubMed] [Google Scholar]
- Stoltzfus A. On the possibility of constructive neutral evolution. J Mol Evol. 1999;49:169–181. doi: 10.1007/pl00006540. [DOI] [PubMed] [Google Scholar]
- Tillich M, Krause K. The ins and outs of editing and splicing of plastid RNAs: lessons from parasitic plants. N Biotechnol. 2010;27:256–266. doi: 10.1016/j.nbt.2010.02.020. [DOI] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Morton BR, Maier UG. The evolution of chloroplast RNA editing. Mol Biol Evol. 2006;23:1912–1921. doi: 10.1093/molbev/msl054. [DOI] [PubMed] [Google Scholar]
- Tillich M, et al. Loss of matK RNA editing in seed plant chloroplasts. BMC Evol Biol. 2009;9:201–201. doi: 10.1186/1471-2148-9-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi T, et al. Creation of a novel protein-coding region at the RNA level in black pine chloroplasts: The pattern of RNA editing in the gymnosperm chloroplast is different from that in angiosperms. Proc Natl Acad Sci U S A. 1996;93:8766–8770. doi: 10.1073/pnas.93.16.8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf PG, Rowe CA, Hasebe M. High levels of RNA editing in a vascular plant chloroplast genome: analysis of transcripts from the fern Adiantum capillus-veneris. Gene. 2004;339:89–97. doi: 10.1016/j.gene.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- Zauner S, Greilinger D, Laatsch T, Kowallik KV, Maier UG. Substitutional editing of transcripts from genes of cyanobacterial origin in the dinoflagellate Ceratium horridum. FEBS Lett. 2004;577:535–538. doi: 10.1016/j.febslet.2004.10.060. [DOI] [PubMed] [Google Scholar]