Abstract
Directed motility, or chemotaxis, is required for Helicobacter pylori to establish infection in the stomach, although the full repertoire of this bacterium’s chemotactic responses is not yet known. Here we report that H. pylori responds to zinc as an attractant and nickel as a repellent. To reach this conclusion, we employed both a temporal chemotaxis assay based on bacterial reversals and a supplemented soft agar spatial assay. We refined the temporal assay using a previously described chemorepellent, acid, and found that H. pylori requires rich media with serum to maintain optimal swimming motility. Surprisingly, we found that some strains respond to acid as an attractant, and that the TlpC chemoreceptor correlated with whether acid was sensed as an attractant or repellent. Using this same assay, we detected weak repellent responses to nickel and copper, and a varied response to zinc. We thus developed an alternative spatial chemotactic assay called the supplemented soft agar assay, which utilizes soft agar medium supplemented with the test compound. With Escherichia coli, the attractant serine slowed overall bacterial migration, while the repellent nickel increased the speed of overall migration. In H. pylori we detected slowed migration with doubled tryptone media, as well as zinc, consistent with an attractant response. In contrast, nickel increased migration, consistent with repulsion.
Introduction
Helicobacter pylori is a Gram-negative bacterium that requires directed motility, or chemotaxis, to colonize its gastric niche (Foynes et al., 2000; Lowenthal et al., 2009; Terry et al., 2005). H. pylori infection can cause ulcers and gastric cancers (Yamada et al., 1994; Parsonnet et al., 1991; Uemura et al., 2001). One well-established H. pylori chemotactic signal is pH (Croxen et al., 2006; Schreiber et al., 2004). Acid chemotaxis fits well with H. pylori’s gastric mucosal habitat, which is much less acidic than the lumen (Bahari et al., 1982; Baumgartner & Montrose, 2004; Chu et al., 1999; Ross et al., 1982; Schreiber & Scheid, 1997; Williams & Turnberg, 1982). There is a gap in our knowledge, however, of the full set of H. pylori chemotactic signals.
Bacterial chemotaxis derives from regulated interspersal of forward swimming with direction reorientations that arise from changes in flagellar rotation (reviewed by Minamino et al., 2008; Wadhams & Armitage, 2004). In the well-studied bacterium Escherichia coli, counterclockwise (CCW) flagellar rotation causes forward swimming, while clockwise (CW) flagellar rotation causes the bacterial body to twitch or tumble, and reorient. The frequency of forward swimming and reorientations is altered by beneficial attractant molecules and harmful repellent molecules. Specifically, attractant molecules cause bacteria to swim forward with few reorientations, while repellents increase reorientations.
Flagellar rotational direction switches are controlled by the chemotaxis response regulator CheY. When CheY is phosphorylated (CheY-P), it binds to the flagellar motor and causes reorientations. Non-phosphorylated CheY does not bind to the motor, leaving the motor to rotate in its default CCW direction that confers forward swimming. CheY-P levels are controlled by a chemotaxis signal transduction pathway that begins when chemoreceptors detect specific environmental characteristics and transmit this information to the CheA kinase. CheA in turn phosphorylates CheY to create CheY-P. This system has been most extensively studied in E. coli, but appears to operate similarly in H. pylori (Beier et al., 1997; Foynes et al., 2000; Jiménez-Pearson et al., 2005; Lertsethtakarn & Ottemann, 2010; Lertsethtakarn et al., 2011; Lowenthal et al., 2009).
Although chemotaxis provides an advantage to H. pylori in stomach colonization, we still know relatively little about specific attractant and repellent molecules. H. pylori produces four chemoreceptor proteins: TlpA, TlpB, TlpC and TlpD (Croxen et al., 2006; Draper et al., 2011; Lertsethtakarn et al., 2011; Tomb et al., 1997; Williams et al., 2007). These proteins share significant homology with other chemoreceptors in the signal transduction region, but share no homology in the ligand-sensing portion. There are a handful of reported H. pylori chemotaxis-active compounds. Urea is one such chemoattractant (Cerda et al., 2003; Mizote et al., 1997; Worku et al., 2004), although H. pylori mucosal localization is not affected in vivo when the urea/ammonium gradient is altered (Schreiber et al., 2004). Other reported attractants include bicarbonate (Cerda et al., 2003; Mizote et al., 1997), NaCl (Mizote et al., 1997), cholesterol (Wunder et al., 2006), and the amino acids aspartate, serine and arginine (Cerda et al., 2003). H. pylori is repelled by NiCl2 (Cerda et al., 2003), low pH (Croxen et al., 2006; Howitt et al., 2011; Rader et al., 2011) and autoinducer-2 (Rader et al., 2011), the latter two via the TlpB chemoreceptor. Alterations in proton motive force trigger both attractant and repellent responses (Schweinitzer et al., 2008).
Chemotactic responses can be assessed by a variety of methods (reviewed by Miller et al., 2009), some of which have been used for H. pylori (Lertsethtakarn et al., 2011). Chemotaxis assays are either spatial, in which bacteria migrate to particular areas in response to a gradient of chemoeffectors, or temporal, in which flagellar switching is monitored. A widely used assay for H. pylori and other bacteria in general is the spatial soft agar migration assay. This assay analyses movement of the bacterial cells over time in low-percentage-agar media (Hazelbauer et al., 1969; Miller et al., 2009; Wolfe & Berg, 1989). In this assay, bacteria create gradients by nutrient consumption and waste product excretion, and migrate to fresh regions. DeLoney-Marino et al. (2003) employed a variation on the soft agar assay by adding particular compounds to the bacterial colony edge to elevate their concentration. Chemoattractants slowed bacterial migration, presumably due to a decreased rate of gradient formation. Another common assay is microscopic filming of bacteria in liquid media. This assay has been used for many bacteria including H. pylori (Berg & Brown, 1972; Croxen et al., 2006; Howitt et al., 2011; Rader et al., 2011; Schweinitzer et al., 2008; Smith & Doetsch, 1969). This method avoids complications of chemical diffusion and metabolism, but analysis of bacterial cell behaviour under the microscope can be complex. Certain behaviours are predicted to indicate chemotaxis based on observations of E. coli, but H. pylori exhibits divergent swimming behaviours that are not clearly categorized. Previous analysis of H. pylori swimming behaviour has utilized reversal or stop frequency (Rader et al., 2011; Schweinitzer et al., 2008; Terry et al., 2006), fixed-time diffusion (Lowenthal et al., 2009), straight-line and curvilinear velocity (Foynes et al., 2000; Karim et al., 1998), and various measurements of bacterial path linearity (Foynes et al., 2000; Karim et al., 1998).
Here, we examine which of these swimming behaviour parameters best capture H. pylori chemotactic response to a well-accepted chemoeffector, acid, and then use reversal frequency combined with a spatial assay to analyse the response to three biologically relevant metals.
Methods
Bacterial strains, media and chemicals.
All strains are listed in Table 1. H. pylori mG27 was poorly motile in liquid medium, so we isolated a more motile variant of this strain, called mG27m, by inoculating mG27 into Brucella Broth soft agar plates and collecting cells from the outer ring of the colony.
Table 1. Strains used in this study.
| Strain | Relevant characteristics | Ottemann strain | Reference or source |
| E. coli strains | |||
| RP437 | E. coli K-12 derivative, chemotactic wild-type | KO40 | Parkinson (1978) |
| RP8611 | E. coli K-12 derivative Δtsr, Δtar-tap, Δtrg | KO186 | J. S. Parkinson* (Surette & Stock, 1996) |
| H. pylori strains | |||
| G27 | Wild-type | N. Salama† (Censini et al., 1996) | |
| mG27 | Mouse-adapted isolate of G27 | KO625 | Castillo et al. (2008) |
| mG27m | Soft agar-selected mG27 isolate | KO1253 | This study |
| mG27m cheY | mG27m ΔcheY : : cat; CmR | KO1257 | This study (cheY allele in Terry et al., 2005) |
| mG27m tlpD | mG27m ΔtlpD : : cat; CmR | KO1259 | This study (tlpD allele in Williams et al., 2007) |
| SS1 | Wild-type | KO441 | J. O’Rourke‡ (Lee et al., 1997) |
| SS1 cheA | SS1 ΔcheA : : cat; CmR | KO855 | Terry et al. (2005) |
| SS1 tlpA | SS1 ΔtlpA : : cat; CmR | KO661 | Andermann et al. (2002) |
| SS1 tlpC | SS1 ΔtlpC : : cat; CmR | KO565 | Andermann et al. (2002) |
| SS1 tlpD | SS1 ΔtlpD : : cat; CmR | KO914 | Williams et al. (2007) |
| J99 | Wild-type | KO479 | N. Salama† (Alm et al., 1999) |
| CYP3401 | Wild-type | KO637 | D. Berg§ (Mizote et al., 1997) |
| M6 | Wild-type | KO613 | E. Joyce|| (Eaton et al., 2002) |
| ATCC43504 | Wild-type | KO669 | ATCC. Also known as NCTC 11637 (Joyce et al., 2000) |
The University of Utah.
Fred Hutchinson Cancer Research Center.
University of New South Wales.
Washington University, St. Louis and University of California, San Diego.
Tufts University School of Medicine.
H. pylori was grown at 37 °C in 5 % O2, 10 % CO2 and 85 % N2. Solid media consisted of Columbia Agar (Remel or BD) with 5 % defibrinated horse blood (Hemostat Laboratories), 0.2 % (w/v) β-cyclodextrin, plus H. pylori-selective antibiotics: 50 µg cycloheximide ml−1, 10 µg vancomycin ml−1, 5 µg cefsulodin ml−1, 2.5 units polymyxin B ml−1 (CHBA). For liquid growth we used 90 % (v/v) Brucella Broth (BD) and 10 % (v/v) fetal bovine serum (FBS; Gibco) (BB10) or 90 % (v/v) HAMS-F12 (Invitrogen) plus 10 % FBS (HAMS10). H. pylori was stored at −80 °C in 10 % (v/v) filter-sterilized FBS, 25 % (v/v) glycerol, 5 % (v/v) DMSO with 200 mg β-cyclodextran ml−1, and 60 % brain-heart infusion broth. All chemicals were from Sigma Aldrich or Fisher Scientific unless otherwise mentioned.
Isogenic mutants of mG27m were constructed by transforming mG27m with genomic DNA (Wizard kit, Promega) from mG27 ΔtlpD : : cat-D1 (originally called ΔhylB : : cat-D1; Williams et al., 2007) or SS1 ΔcheY : : cat102 (Terry et al., 2005) to chloramphenicol resistance (15 µg ml−1). Potential transformants were confirmed by PCR using primers homologous to upstream and downstream flanking regions for each gene: primers cheY3 (5′-GGAAGCTGCAGGTTTATCGTCAAACG-3′) and cheY4 (GCTCATTGAACGCTCCATTTAGC-3′) for cheY, and hylB3 and hylB4 for tlpD (Williams et al., 2007).
Determination of media metal content.
Media metal content was determined by inductively coupled-MS (ICP-MS) on a Thermo ELEMENT XR (magnetic sector inductively coupled plasma mass spectrometer) on samples diluted 1 : 100 with 1 % (v/v) HNO3. Measurements were averaged over two technical replicates, recorded as counts per second and normalized to scandium, which was used to spike the samples. The counts were then normalized to a standard (MS-2, personal communication, Rob Franks, UCSC Keck Isotope Lab), converted to parts per billion (p.p.b.), and corrected against the water blank to obtain the p.p.b. of each metal in solution.
Microscopic analysis.
For bacterial swimming analysis, H. pylori was grown with shaking in BB10 at 37 °C for 15–17 h (mid-exponential growth phase). Motility was confirmed microscopically, and then cultures were diluted to a final OD600 of 0.12, into prewarmed test solutions prepared immediately prior to use. Acidified solutions were created by mixing HCl with BB10 to a final concentration of 17 mM (pH 4.7), 34 mM (pH 3.5) or 68 mM HCl (pH 2.5). Metal test solutions were created with a final concentration of 10 µM–10 mM ZnCl2, CuSO4 or NiCl2. Concentrations below 10 mM did not alter the pH of BB10 from 7 (our unpublished observations). Ten-second phase-contrast microscopic films of glass slide-mounted cultures were recorded with Simple PCI software (version 5.2.1.1609, Compix 2003) using a Hamamatsu digital camera mounted on a Nikon Eclipse E600 microscope at ×400 magnification. Videos were recorded at maximum acquisition speed (~16 frames s−1), exported into .avi format and relabelled to blind the analyser to the condition. To assess motility in phosphate buffers, cells were isolated from 6-h-old CHBA cultures, and placed in PBS (14 mM NaCl, 0.02 % KCl, 0.14 % Na2HPO4, 0.024 % KH2PO4). Cells were then centrifuged at 250 g for 2 min, resuspended gently in PBS, mixed with 10-fold PBS, BB10 or phosphate buffer (10 mM potassium or sodium phosphate, pH 7).
Reversal frequency analysis.
Individual bacterial swimming paths were analysed by tracing a randomly selected cell visible for at least 5 s, using transparency paper affixed to the computer monitor. Each clear direction change was counted. For all strains and conditions, we utilized two separate overnight cultures to accumulate at least 20 bacteria per treatment. For each strain on each day, we determined the reversal frequency of a sample without supplemented metals, and made all comparisons with that sample. We removed any samples from analysis that were deemed outliers by Grubb’s outlier test (99 % confidence interval).
Analysis of velocities and track linear percentage (TL%).
Velocity and TL% analysis were calculated using the Compix Simple PCI Motion Tracking Analysis program. A Simple PCI workfile was created to identify bacteria using object enhancement. Bacterial cell positions were tracked for cells that were motile (straight-line velocity >0.29, and straight-line distance >9.9) and whose paths were at least 2 s long (32 frames). Straight-line velocity (VSL) is defined as the velocity of the cell if it were to go directly from the start to the end point. Curvilinear velocity (VCL) is the velocity of the cell along its actual track. TL% is VSL/VCL×100 %. Data from Simple PCI were further processed by OpenOffice.org Calc to calculate the means, sems and Student’s t-test significance (P) values.
Soft agar migration chemotaxis assay.
H. pylori soft agar plates were composed of 0.35 % (w/v) Bacto agar (BD), 2.5 % v/v heat-inactivated FBS, H. pylori-selective antibiotics plus 5 µg trimethoprim ml−1 and 8 µg amphotericin B ml−1, and either 2.8 % (w/v) Brucella Broth mix (1×) or tryptone (BD) at 1 % (1×), 2 % (2×) or 3 % (3×) (w/v). E. coli soft agar plates were composed of 0.35 % Bacto agar, 1 % tryptone and 0.5 % NaCl (Wolfe & Berg, 1989). Soft agar plates were modified by adding metal stock solutions [sterile-filtered NiSO4 (Alfa Aesar) or unfiltered FeCl3, CuSO4, NiCl2 and ZnCl2] or sterile-filtered serine after autoclaving the medium. H. pylori was inoculated using a pipette tip to stab a small amount from a plate culture into the soft agar, and migration diameter was measured after 5 days. E. coli inoculations were from liquid cultures and migration diameters were measured after 20 h. Statistical significance was calculated by OpenOffice.org Calc Student’s t test, and results were significant for P<0.05.
Results
H. pylori temporal swimming assays work optimally with FBS and analysis of direction reversals
We aimed to assess the chemotactic response of H. pylori to metals. In other H. pylori work, chemotactic responses have been studied using temporal assays that monitored bacterial swimming behaviour for number of stops (Rader et al., 2011; Schweinitzer et al., 2008), number of reversals (Terry et al., 2006) or ability to promote diffusion (Lowenthal et al., 2009), or compared various velocity measurements (Foynes et al., 2000; Karim et al., 1998). We thus first sought to compare these various measurements to ascertain which would be the most sensitive for detecting chemotaxis responses. For our comparison, we employed a previously shown chemoeffector, acidic pH (Croxen et al., 2006), and attempted our first studies with bacteria suspended in potassium phosphate buffer or PBS, as has been used in other H. pylori and E. coli work (Mizote et al., 1997). Unlike many other motile bacteria, all H. pylori strains tested (G27, SS1, J99, CYP3401, M6 and 43504) were non-motile in the simple buffers PBS and phosphate buffer, even when supplemented with polyvinylpyrrolidone, a solution employed in some studies (Cerda et al., 2003). This observation suggests that H. pylori does not have an adequate internal energy store, as E. coli does, to power the flagella. Brucella broth supported some motility, but a large number of cells were observed to swim only when FBS was present. FBS is known to promote robust H. pylori growth (Testerman et al., 2001), but our data show that it also promotes motility. For this reason, we used FBS-containing medium (BB10) for all further analyses.
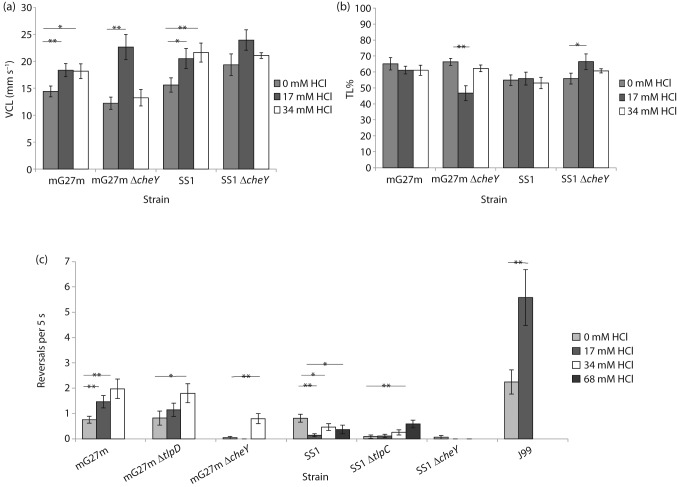
With these basal conditions in hand, we filmed H. pylori swimming in BB10 and BB10+acid, and compared reversal frequency, VCL and TL%. VCL measures velocity along the bacterial path, as opposed to VSL, which is the speed of the bacterium in a straight line from the beginning to the end of its path. TL% is the ratio of VSL to VCL multiplied by 100. Acid increased the VCL of H. pylori strains mG27m and SS1, as reported before for H. pylori (Merrell et al., 2003; Sidebotham et al., 2003) (Fig. 1a). The VCL of non-chemotactic cheY mutants of mG27m and SS1 strains also increased (Fig. 1a), suggesting that acid acts both on the flagellar motor to affect speed and on the chemotaxis pathway to control flagellar reversals (Croxen et al., 2006).
Fig. 1.
pH influences H. pylori speed and reversal frequency. (a) Low pH increases VCL in a chemotaxis-independent manner. (b) pH affects TL% to a modest extent. (c) pH has a significant influence on reversal frequency. All behaviour was measured in BB10 supplemented to the final concentration of HCl indicated. Each measurement is shown as mean±sem. (a, b) Each measurement derives from five bacteria that were tracked from three to four independently grown cultures. (c) Each measurement derives from at least 20 bacteria from at least two independently grown cultures, and is given as the mean per bacterium. *Statistically significant response (P<0.05); **highly statistically significant response (P<0.01) using Student’s t test.
We next compared TL% and reversal frequency. Repellents such as acid and autoinducer 2 cause a less linear path, due to reversals and stopping behaviour (Rader et al., 2011), although the reported behaviour has also been described as arc-shaped (Croxen et al., 2006). A repellent would thus increase the VCL relative to the VSL, as the cell deviates from a straight path and in turn decreases the TL%. An attractant, on the other hand, would cause the VCL to approximate the VSL and increase the TL%. Acid caused minimal detectable TL% decreases in both wild-type and cheY mutant strains (Fig. 1b), suggesting that TL% is not a sensitive or specific indicator of chemotactic response.
We next evaluated reversal frequency as a measure of chemotactic response. H. pylori strains mG27m and J99 both showed significant increases in the number of reversals upon acid exposure, while strain SS1 responded with fewer reversals (Fig. 1c). It was surprising that strain SS1 responded to acid as an attractant in this assay; other assay systems have found that SS1 is repelled by acid, suggesting that this may be a condition-specific response (Howitt et al., 2011). One difference between SS1 and mG27m is that the former expresses all four predicted chemoreceptors, while G27-derived strains do not express tlpC due to a frameshift (Rader et al., 2011; Williams et al., 2007) (K. M. Ottemann, unpublished observations). We thus examined whether loss of tlpC affected the acid response. H. pylori SS1 ΔtlpC responded to acid as a repellent (Fig. 1c). This strain was very strongly straight swimming-biased, but at the highest acid exposure changed its behaviour to have more reversals (Fig. 1c). TlpB has been shown to drive the acid-repellent response in that loss of tlpB in G27 creates strains that no longer increase reversal frequency in response to acid (Rader et al., 2011). Our results suggest, however, that tlpC can modulate the TlpB-mediated acid behaviour by an as-yet-unknown mechanism.
We compared the TL% and reversal frequency of non-chemotactic mutants, which lacked cheY, to test whether reversal frequency also detected chemotaxis-independent acid responses. We found that cheY mutants of strain SS1 displayed no TL% or reversal frequency changes, while strain mG27m cheY displayed a weak response to high acid exposure (Fig. 1b, c). Taken together, these data suggest that reversal frequency is a sensitive measure of the chemotaxis response to acid, with modest non-chemotaxis effects.
Metal supplementation changes H. pylori swimming reversal frequency
Our next goal was to assess chemotactic response to specific metals, focusing on three that are needed for H. pylori metabolism: copper, nickel and zinc (Whitmire et al., 2007). We first determined the concentration of biologically relevant metals in our medium. The concentrations of copper and nickel were very low (<0.5 µM) in Brucella broth and another common H. pylori medium, HAMS-F12 (Table 2). Zinc and iron, on the other hand, were the most abundant metals (Table 2). Our values approximated manufacturer-published numbers for HAMS-F12. FBS contained <0.5 µM of each of the tested metals, except copper, iron and zinc, which were found at 1, 16 and 16 µM concentrations, respectively (Table 2). From these measurements, we calculated the concentrations of zinc, copper and nickel in BB10 (Table 2), and used concentrations in excess of those amounts for our tests.
Table 2. Analysis of media metal content.
Brucella broth (BB) and HAMS F-12 ICP-MS metal measurements derived from two technical replicates are shown. Complete BB10 and HAMS10 metal contents were calculated from Brucella broth or a 0.8× stock solution of HAMS F-12 plus 10 % FBS. −, Readings <1 µM.
| Metal | FBS (μM) | BB (μM) | BB10 calculated (μM) | HAMS (μM) | HAMS10 calculated (μM) |
| Cr-52 | 0.011 | 0.913 | 0.823 | 0.001 | 0.002 |
| Fe-56 | 16.397 | 8.954 | 9.698 | 2.387 | 3.788 |
| Co-56 | 0.001 | 0.059 | 0.053 | 0.379 | 0.341 |
| Ni-60 | − | − | − | − | − |
| Cu-63 | 1.084 | 0.151 | 0.244 | − | 0.043 |
| Zn-66 | 15.855 | 5.173 | 6.240 | 2.747 | 4.058 |
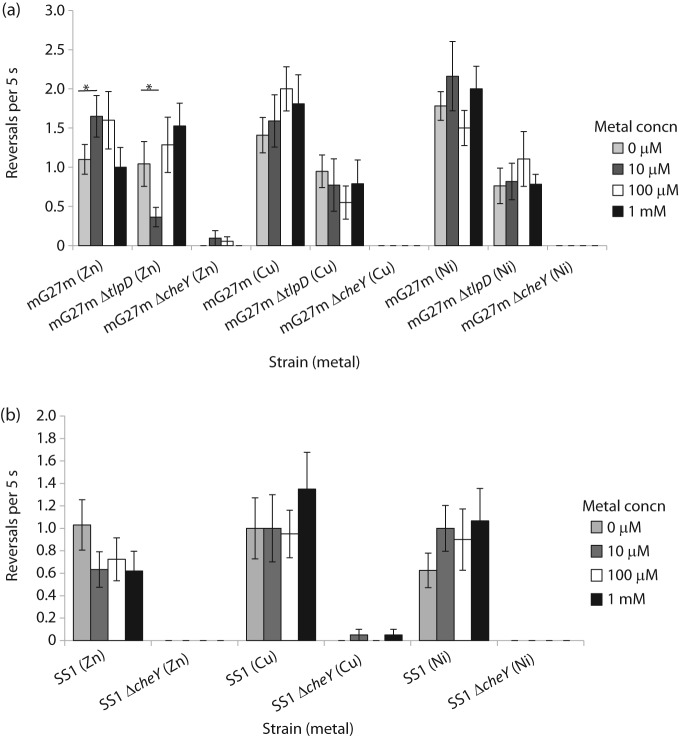
We next analysed the bacterial swimming behaviour response to copper, nickel and zinc in strain mG27m. Zinc treatment caused a small but significant increase in reversal frequency (Fig. 2a), consistent with repellent behaviour, at 10 and 100 µm. Both copper and nickel triggered a repellent-type response (Fig. 2a), but neither of these reached statistical significance. Nickel repellent responses are common (Seymour & Doetsch, 1973; Tso & Adler, 1974), including in H. pylori (Cerda et al., 2003), and our findings are consistent with these observations. These directional changes were all chemotaxis-dependent, as shown by the lack of response of a cheY mutant (Fig. 2a).
Fig. 2.
H. pylori reversal response to zinc, copper and nickel. All behaviour was measured in BB10 supplemented to the final concentration of metal indicated in the legend. (a) Results with strain mG27m and its isogenic chemotaxis mutants. (b) Results with strain SS1 and its isogenic chemotaxis mutants. Each measurement derives from at least 20 bacteria from two or more independently grown cultures, and is given as the mean per bacterium. *Statistically significant response (P<0.05) using Student’s t test.
H. pylori strain SS1 was also tested for metal chemotaxis using reversal frequency analysis. Interestingly, strain SS1 had a different response to zinc from that of H. pylori mG27m: zinc triggered fewer reversals, as if it were an attractant (Fig. 2b). Not all chemotaxis responses were inverted between SS1 and mG27m, as SS1 responded to nickel and copper as repellents, like mG27m, although these responses were not significant (Fig. 2b). All responses were chemotaxis-dependent, as the mutant lacking cheY did not respond to any metals (Fig. 2b).
As a first step toward determining how these metals are sensed, we examined the behaviour of mG27m lacking the tlpD chemoreceptor, because this receptor has recently been found to bear a zinc-containing domain termed CZB (Draper et al., 2011). The mutant strain mG27m tlpD displayed a muted response to all metals, suggesting that tlpD is important for the normal response to these metals (Fig. 2a). These findings were somewhat difficult to interpret, as the tlpD mutant displayed behaviour that was biased toward straight smooth swimming as compared with the wild-type (Fig. 2a). This bias makes it difficult to ascertain whether TlpD is needed to sense the metals directly, or whether it is needed to set the flagellar switch bias properly to allow a normal response. In addition, the tlpD mutant strain did have a modest but altered behaviour upon zinc addition (Fig. 2a), suggesting that TlpD may contribute to but not play an exclusive role in zinc sensing.
A supplemented soft agar assay can be used to determine attractants and repellents
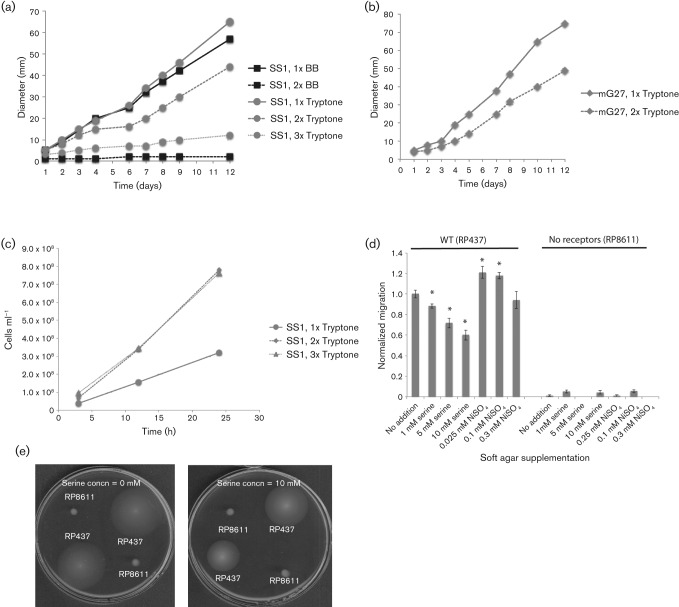
The above analysis of H. pylori reversal behaviour suggested that this microbe might respond chemotactically to metals, although the results were difficult to interpret because of strain variability, the small magnitude of the responses and the skewed basal reversal frequencies of tlpC and tlpD mutant H. pylori strains. We thus sought a different type of analysis to help characterize the chemotactic response to zinc and nickel. The Brucella broth soft agar migration assay has worked well for H. pylori (Beier et al., 1997; McGee et al., 2005), and so we embarked on adapting this assay to monitor specific chemoeffectors. In the standard version of this assay, bacteria migrate outwards from an inoculation point in response to gradients created either by consuming nutrients or producing waste products. Others have analysed specific chemicals in this assay by spotting high concentrations of attractants at the colony edge, and monitoring whether this addition deformed the migrating chemotaxis colonies (DeLoney-Marino et al., 2003); presumably the high attractant concentration takes additional time to metabolize, and the ensuing slowed migration is easily seen. We tried spotting high concentrations of reported chemoeffectors such as arginine (Cerda et al., 2003) on the edge of an H. pylori soft agar colony, but did not find any colony deformation (data not shown). We presumed that this lack of response was due to the relatively slow migration of H. pylori of ~5 mm day−1 (Andermann et al., 2002), as compared with E. coli migration of 5 mm h−1 (Weis & Koshland, 1990), such that small chemical gradients dissipate before they influence migration. We thus decided upon a different approach in which potential chemoeffectors were added homogeneously throughout the soft agar. In this case, we predicted a colony-wide slow down in response to the need to deplete the chemoeffector before migrating outward. Indeed, when we used 2× H. pylori media, the overall migration rate slowed (Fig. 3a, b), while growth was enhanced (Fig. 3c). This response suggests that elevated levels of chemotactic compounds have a detectable output in this assay.
Fig. 3.
Homogeneous attractants slow and repellents increase bacterial soft agar migration. (a, b) Colony expansion of H. pylori SS1 (a) or G27m (b) in soft agar media composed of 1× tryptone, 2× tryptone, 3× tryptone, 1× Brucella broth (BB) or 2× BB, all with additional 2.5 % FBS. Data shown are representative of three or more replicates. (c) H. pylori grows more robustly in 2× or 3× tryptone/FBS media than in 1× tryptone. (d) E. coli in tryptone soft agar supplemented with 1–10 mM serine or 0.025–0.3 mM NiSO4. Migration distances are the means of two to four biological replicates, which included seven to eight technical replicates each. Error bars, sem. Distances and errors were normalized to the wild-type RP437 no-addition sample (2.2 cm); RP8611 yielded 0.01 cm at the same time point. *P<0.05 using Student’s t test on the non-normalized data. (e) Appearance of E. coli soft agar plates supplemented with 10 mM serine as compared with no addition.
To further validate the supplemented soft agar assay, we employed the well-developed E. coli system. The attractant serine showed a dose-dependent decrease in colony expansion rate (Fig. 3d), presumably because E. coli created gradients after consuming serine, and this process took longer if there was more serine. Nickel, on the other hand, promoted an enhanced colony expansion rate at the two lowest concentrations used (Fig. 3d). Both of these responses were dependent on chemoreceptors, as a strain lacking these proteins showed little response to the elevated serine or nickel (Fig. 3d, e).
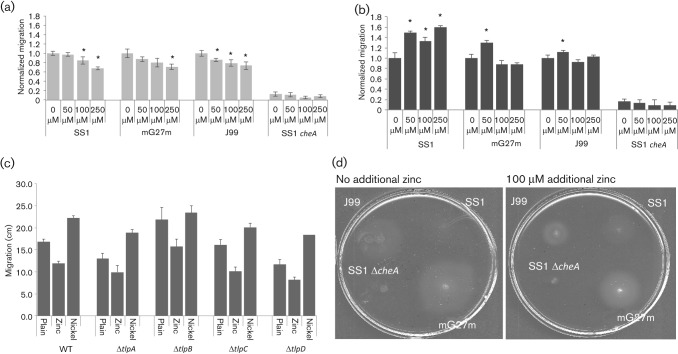
A soft agar migration assay confirms zinc as an attractant and nickel as a repellent
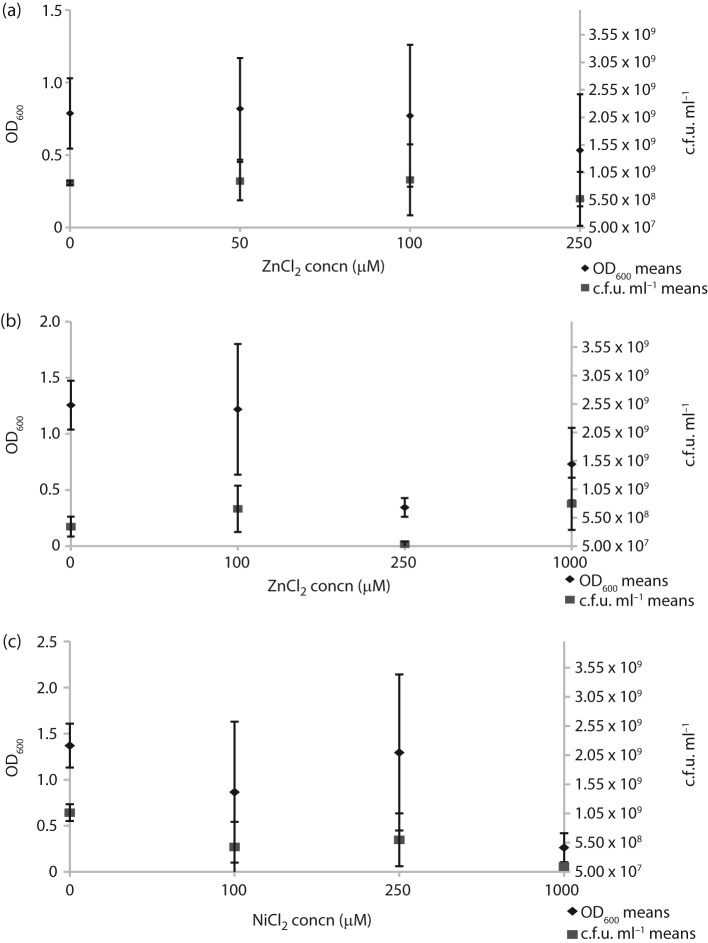
We then tested how H. pylori would respond to elevated zinc and nickel. We first determined the highest concentration of each metal that would not impact growth in liquid media (Fig. 4). Although there was some variability between H. pylori strains, neither mG27m nor SS1 was affected by 100 µM ZnCl2 or 250 µM NiCl2 (Fig. 4). We then evaluated how these non-toxic concentrations of ZnCl2 or NiCl2 would impact soft agar migration. We found that ZnCl2 uniformly acted to decrease colony migration of wild-type strains mG27m, SS1 and J99, but did not impact a non-chemotactic variant that lacked cheA (Fig. 5a). This behaviour suggests that zinc acts as a chemoattractant for H. pylori in this assay. NiCl2 supplementation, on the other hand, resulted in enhanced colony migration (Fig. 5b), consistent with this metal acting as a repellent. We also repeated these experiments with H. pylori SS1 mutants missing each known chemoreceptor individually, but did not find one that affected the response (Fig. 5c). Thus it appears that no single chemoreceptor is responsible for the nickel or zinc chemotaxis response.
Fig. 4.
ZnCl2 and NiCl2 toxicity in H. pylori. Cultures of each strain were grown overnight and diluted to OD600 0.1 with metal added to the indicated concentrations. The cultures were then incubated for 20–21 h, followed by determination of OD600 and c.f.u. ml−1. (a) H. pylori SS1 exposed to ZnCl2 (two biological replicates); (b) H. pylori mG27m exposed to ZnCl2 (three biological replicates); (c) H. pylori mG27m exposed to NiCl2 (two biological replicates).
Fig. 5.
H. pylori responds to zinc as an attractant and nickel as a repellent. (a) H. pylori in Brucella broth/2.5 % FBS soft agar supplemented with 50–250 µM ZnCl2. (b) Same as (a), but with NiCl2 supplementation. (c) Same as (b), but comparing H. pylori SS1 wild-type with isogenic single chemoreceptor mutants. Migration distances are the means of two to four biological replicates, which included seven to eight technical replicates each. Error bars, sem. Both distances and errors were normalized to the wild-type 0 µM sample by dividing by the distance for that sample. *P<0.05 using Student’s t test on the non-normalized data. (d) Image of H. pylori colonies on unsupplemented and zinc-supplemented soft agar plates.
Discussion
In this manuscript, we describe that H. pylori senses zinc as a chemoattractant and nickel as a chemorepellent. H. pylori requires zinc for growth in vitro (Testerman et al., 2001, 2006). We do not know whether zinc is limiting in the stomach, but studies have found that H. pylori infection lowers mucosal zinc concentrations (Sempértegui et al., 2007), possibly due to the presence of neutrophils, which produce the zinc-chelating agent calprotectin (Kehl-Fie & Skaar, 2010). Thus, perhaps the infiltrating neutrophils create a zinc-poor environment in which H. pylori benefits by moving toward zinc. Our results did have some variability in that zinc was always an attractant in the supplemented soft agar assay, but was a repellent for mG27m in the reversal assay. While we do not yet know the basis for these differences, we believe the supplemented soft agar assay is more accurate due to the lower variability.
H. pylori does not require additional nickel for growth (Testerman et al., 2001, 2006), although it requires nickel as a cofactor for two critical enzymes: urease, which mediates pH homeostasis, and hydrogenase, which is used for hydrogen metabolism (Maier et al., 2007). The basal level of nickel in the medium is apparently sufficient for these enzymes, but it is not yet known whether in vivo nickel is limiting or toxic. Our data support the notion that nickel is encountered at toxic levels that require a repellent chemotaxis response, and agree with mechanisms identified in H. pylori to efflux and sequester nickel (Seshadri et al., 2007; Stähler et al., 2006).
Metal chemotaxis is a relatively understudied area of chemotactic responses, despite it being reported in early bacterial chemotaxis papers (Seymour & Doetsch, 1973; Tso & Adler, 1974). Both laboratories reported a chemorepellent response to nickel, but otherwise differed in their findings. Seymour & Doetsch (1973) detected repellent responses to aluminium, copper, iron, lanthanum and nickel amongst 10 diverse bacterial species. Interpreting these results, however, was complicated by the fact that all of the metals lowered the pH at the concentrations used. We avoided this complication by using lower concentrations. For example, for NiCl2, Seymour and Doetsch used 10 % of a saturated solution (~300 mM final concentration), while we used 10–100 µM final concentration. The study of Tso and Adler, in contrast, focused only on E. coli and utilized metal concentrations near 10 µM (Tso & Adler, 1974). These authors reported that E. coli is repelled by both nickel and cobalt, and does not respond chemotactically to copper, magnesium, chromium or sodium. We now know that both cobalt and nickel are sensed by the Tar chemoreceptor, although the exact mechanism is unknown (Englert et al., 2010). These early experiments did not test zinc, nor did they find any metals that were attractants.
Metal chemotaxis has also been studied in the context of movement toward optimal terminal electron acceptors. Environmental microbes such as Shewanella and Geobacter spp. are attracted to redox-active compounds that are used as terminal electron acceptors, including iron, manganese and nitrate (Bencharit & Ward, 2005). This response is categorized as energy taxis, because it depends on the electron transport chain. A set of chemoreceptors has been linked to this response (Baraquet et al., 2009). Shewanella oneidensis, additionally, has a chemotactic response to non-redox metals, including attraction to zinc and repulsion from nickel (Bencharit & Ward, 2005). This finding suggests that S. oneidensis may perform true chemotaxis to metals, although the basis for this response is not yet known.
Our studies clarify that monitoring bacterial reversals is a more powerful mechanism for detecting chemotactic responses than are other measures of bacterial paths, including TL%. Our temporal assays, however, were variable and challenging to interpret, especially with regard to the zinc response. The initial report of H. pylori pH taxis described an acid response that consisted of increased arcing swimming as opposed to straight, non-arcing paths (Croxen et al., 2006). The arcing behaviour might be explained by observations that E. coli swims in circles near the glass surface, due to bacterial body rotation that traps it near the surface (Lauga et al., 2006). The diameter of the circular bacterial paths is affected by the ionic strength of the medium (Vigeant & Ford, 1997): a parameter that pH would be predicted to influence. Subsequent work has employed different measures that corroborate H. pylori’s acid-repellent response (Rader et al., 2011; Howitt et al., 2011). Our work also suggests, surprisingly, that acid can act as a chemoattractant for strain SS1. Others have reported that acid is an SS1 repellent (Howitt et al., 2011); while we do not yet know the basis for these different responses, there are substantial condition and time-frame differences between these studies that may underlie the distinct responses. Therefore it might by plausible that H. pylori modulates its chemotactic response to acid.
Finally, we describe a simple chemotaxis assay that works well for H. pylori. This assay involves examining the effect of putative attractants or repellents on soft agar migration. Others have observed that attractants slow E. coli and Vibrio fischeri migration rates, presumably because it takes time to deplete the attractant and create a gradient (DeLoney-Marino et al., 2003; Wolfe & Berg, 1989). The effect of repellents has been studied less, and is harder to predict. We hypothesize that the nickel-induced faster migration rate is due to a sustained increase in reversal frequency. We have previously reported that an H. pylori cheV3 mutant has increased reversal frequency and similarly migrates more rapidly in soft agar than does the wild-type (Lowenthal et al., 2009).
In sum, we show here that H. pylori responds to zinc as an attractant and nickel as a repellent. These studies thus increase the known H. pylori chemotactome, such that it now includes zinc and nickel, as well as several amino acids, pH, autoinducer 2, energy and cholesterol (Lertsethtakarn et al., 2011; Rader et al., 2011). Nickel is toxic, and chemotaxis away from this compound appears to benefit diverse bacteria in dissimilar niches (Bencharit & Ward, 2005; Englert et al., 2010; Seymour & Doetsch, 1973; Tso & Adler, 1974). Attraction to zinc is much less common, with the only reported bacterial example being in S. oneidensis (Bencharit & Ward, 2005). Most attractants are bacterial nutrients, as is zinc for H. pylori, suggesting that H. pylori uses chemotaxis to acquire this nutrient.
Acknowledgements
We are grateful to several members of the Ottemann lab who helped figure out that FBS promotes H. pylori swimming and who worked on earlier iterations of the supplemented soft agar assay: Alvin Go, Yu-Ting Chen, Andrew Lowenthal and Amber Fair, as well as to Jennifer Chang for assisting with the supplemented soft agar assays. The described project was supported by the National Institutes of Allergy and Infectious Disease (NIAID) at the National Institutes of Health via grant number AI050000 (to K. M. O.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Abbreviations:
- CCW
counterclockwise
- ICP-MS
inductively coupled-MS
- TL%
track linear percentage
- VCL
curvilinear velocity
- VSL
straight-line velocity
References
- Alm R. A., Ling L. S., Moir D. T., King B. L., Brown E. D., Doig P. C., Smith D. R., Noonan B., Guild B. C. & other authors (1999). Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397, 176–180. 10.1038/16495 [DOI] [PubMed] [Google Scholar]
- Andermann T. M., Chen Y.-T., Ottemann K. M. (2002). Two predicted chemoreceptors of Helicobacter pylori promote stomach infection. Infect Immun 70, 5877–5881. 10.1128/IAI.70.10.5877-5881.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahari H. M., Ross I. N., Turnberg L. A. (1982). Demonstration of a pH gradient across the mucus layer on the surface of human gastric mucosa in vitro. Gut 23, 513–516. 10.1136/gut.23.6.513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraquet C., Théraulaz L., Iobbi-Nivol C., Méjean V., Jourlin-Castelli C. (2009). Unexpected chemoreceptors mediate energy taxis towards electron acceptors in Shewanella oneidensis. Mol Microbiol 73, 278–290. 10.1111/j.1365-2958.2009.06770.x [DOI] [PubMed] [Google Scholar]
- Baumgartner H. K., Montrose M. H. (2004). Regulated alkali secretion acts in tandem with unstirred layers to regulate mouse gastric surface pH. Gastroenterology 126, 774–783. 10.1053/j.gastro.2003.11.059 [DOI] [PubMed] [Google Scholar]
- Beier D., Spohn G., Rappuoli R., Scarlato V. (1997). Identification and characterization of an operon of Helicobacter pylori that is involved in motility and stress adaptation. J Bacteriol 179, 4676–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencharit S., Ward M. J. (2005). Chemotactic responses to metals and anaerobic electron acceptors in Shewanella oneidensis MR-1. J Bacteriol 187, 5049–5053. 10.1128/JB.187.14.5049-5053.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg H. C., Brown D. A. (1972). Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature 239, 500–504. 10.1038/239500a0 [DOI] [PubMed] [Google Scholar]
- Castillo A. R., Woodruff A. J., Connolly L. E., Sause W. E., Ottemann K. M. (2008). Recombination-based in vivo expression technology identifies Helicobacter pylori genes important for host colonization. Infect Immun 76, 5632–5644. 10.1128/IAI.00627-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Censini S., Lange C., Xiang Z., Crabtree J. E., Ghiara P., Borodovsky M., Rappuoli R., Covacci A. (1996). cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci U S A 93, 14648–14653. 10.1073/pnas.93.25.14648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda O., Rivas A., Toledo H. (2003). Helicobacter pylori strain ATCC700392 encodes a methyl-accepting chemotaxis receptor protein (MCP) for arginine and sodium bicarbonate. FEMS Microbiol Lett 224, 175–181. 10.1016/S0378-1097(03)00423-3 [DOI] [PubMed] [Google Scholar]
- Chu S., Tanaka S., Kaunitz J. D., Montrose M. H. (1999). Dynamic regulation of gastric surface pH by luminal pH. J Clin Invest 103, 605–612. 10.1172/JCI5217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxen M. A., Sisson G., Melano R., Hoffman P. S. (2006). The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J Bacteriol 188, 2656–2665. 10.1128/JB.188.7.2656-2665.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLoney-Marino C. R., Wolfe A. J., Visick K. L. (2003). Chemoattraction of Vibrio fischeri to serine, nucleosides, and N-acetylneuraminic acid, a component of squid light-organ mucus. Appl Environ Microbiol 69, 7527–7530. 10.1128/AEM.69.12.7527-7530.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper J., Karplus K., Ottemann K. M. (2011). Identification of a chemoreceptor zinc-binding domain common to cytoplasmic bacterial chemoreceptors. J Bacteriol 193, 4338–4345. 10.1128/JB.05140-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton K. A., Gilbert J. V., Joyce E. A., Wanken A. E., Thevenot T., Baker P., Plaut A., Wright A. (2002). In vivo complementation of ureB restores the ability of Helicobacter pylori to colonize. Infect Immun 70, 771–778. 10.1128/IAI.70.2.771-778.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert D. L., Adase C. A., Jayaraman A., Manson M. D. (2010). Repellent taxis in response to nickel ion requires neither Ni2+ transport nor the periplasmic NikA binding protein. J Bacteriol 192, 2633–2637. 10.1128/JB.00854-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foynes S., Dorrell N., Ward S. J., Stabler R. A., McColm A. A., Rycroft A. N., Wren B. W. (2000). Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect Immun 68, 2016–2023. 10.1128/IAI.68.4.2016-2023.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer G. L., Mesibov R. E., Adler J. (1969). Escherichia coli mutants defective in chemotaxis toward specific chemicals. Proc Natl Acad Sci U S A 64, 1300–1307. 10.1073/pnas.64.4.1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt M. R., Lee J. Y., Lertsethtakarn P., Vogelmann R., Joubert L. M., Ottemann K. M., Amieva M. R. (2011). ChePep controls Helicobacter pylori infection of the gastric glands and chemotaxis in the Epsilonproteobacteria. MBio 2, e00098–e00011. 10.1128/mBio.00098-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Pearson M. A., Delany I., Scarlato V., Beier D. (2005). Phosphate flow in the chemotactic response system of Helicobacter pylori. Microbiology 151, 3299–3311. 10.1099/mic.0.28217-0 [DOI] [PubMed] [Google Scholar]
- Joyce E. A., Bassler B. L., Wright A. (2000). Evidence for a signaling system in Helicobacter pylori: detection of a luxS-encoded autoinducer. J Bacteriol 182, 3638–3643. 10.1128/JB.182.13.3638-3643.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim Q. N., Logan R. P., Puels J., Karnholz A., Worku M. L. (1998). Measurement of motility of Helicobacter pylori, Campylobacter jejuni, and Escherichia coli by real time computer tracking using the Hobson BacTracker. J Clin Pathol 51, 623–628. 10.1136/jcp.51.8.623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehl-Fie T. E., Skaar E. P. (2010). Nutritional immunity beyond iron: a role for manganese and zinc. Curr Opin Chem Biol 14, 218–224. 10.1016/j.cbpa.2009.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauga E., DiLuzio W. R., Whitesides G. M., Stone H. A. (2006). Swimming in circles: motion of bacteria near solid boundaries. Biophys J 90, 400–412. 10.1529/biophysj.105.069401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A., O’Rourke J., De Ungria M. C., Robertson B., Daskalopoulos G., Dixon M. F. (1997). A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112, 1386–1397. 10.1016/S0016-5085(97)70155-0 [DOI] [PubMed] [Google Scholar]
- Lertsethtakarn P., Ottemann K. M. (2010). A remote CheZ orthologue retains phosphatase function. Mol Microbiol 77, 225–235. 10.1111/j.1365-2958.2010.07200.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lertsethtakarn P., Ottemann K. M., Hendrixson D. R. (2011). Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol 65, 389–410. 10.1146/annurev-micro-090110-102908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenthal A. C., Simon C., Fair A. S., Mehmood K., Terry K., Anastasia S., Ottemann K. M. (2009). A fixed-time diffusion analysis method determines that the three cheV genes of Helicobacter pylori differentially affect motility. Microbiology 155, 1181–1191. 10.1099/mic.0.021857-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier R. J., Benoit S. L., Seshadri S. (2007). Nickel-binding and accessory proteins facilitating Ni-enzyme maturation in Helicobacter pylori. Biometals 20, 655–664. 10.1007/s10534-006-9061-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee D. J., Langford M. L., Watson E. L., Carter J. E., Chen Y.-T., Ottemann K. M. (2005). Colonization and inflammation deficiencies in Mongolian gerbils infected by Helicobacter pylori chemotaxis mutants. Infect Immun 73, 1820–1827. 10.1128/IAI.73.3.1820-1827.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell D. S., Goodrich M. L., Otto G., Tompkins L. S., Falkow S. (2003). pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infect Immun 71, 3529–3539. 10.1128/IAI.71.6.3529-3539.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. D., Russell M. H., Alexandre G. (2009). Diversity in bacterial chemotactic responses and niche adaptation. Adv Appl Microbiol 66, 53–75. 10.1016/S0065-2164(08)00803-4 [DOI] [PubMed] [Google Scholar]
- Minamino T., Imada K., Namba K. (2008). Molecular motors of the bacterial flagella. Curr Opin Struct Biol 18, 693–701. 10.1016/j.sbi.2008.09.006 [DOI] [PubMed] [Google Scholar]
- Mizote T., Yoshiyama H., Nakazawa T. (1997). Urease-independent chemotactic responses of Helicobacter pylori to urea, urease inhibitors, and sodium bicarbonate. Infect Immun 65, 1519–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. (1978). Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J Bacteriol 135, 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonnet J., Friedman G. D., Vandersteen D. P., Chang Y., Vogelman J. H., Orentreich N., Sibley R. K. (1991). Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med 325, 1127–1131. 10.1056/NEJM199110173251603 [DOI] [PubMed] [Google Scholar]
- Rader B. A., Wreden C., Hicks K. G., Sweeney E. G., Ottemann K. M., Guillemin K. (2011). Helicobacter pylori perceives the quorum-sensing molecule AI-2 as a chemorepellent via the chemoreceptor TlpB. Microbiology 157, 2445–2455. 10.1099/mic.0.049353-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross I. N., Bahari H. M., Turnberg L. A. (1982). Studies of the pH gradient across the mucus on rat gastric mucosa in vivo and across mucus on human gastric mucosa in vitro. Adv Exp Med Biol 144, 189–191. 10.1007/978-1-4615-9254-9_29 [DOI] [PubMed] [Google Scholar]
- Schreiber S., Scheid P. (1997). Gastric mucus of the guinea pig: proton carrier and diffusion barrier. Am J Physiol 272, G63–G70. [DOI] [PubMed] [Google Scholar]
- Schreiber S., Konradt M., Groll C., Scheid P., Hanauer G., Werling H. O., Josenhans C., Suerbaum S. (2004). The spatial orientation of Helicobacter pylori in the gastric mucus. Proc Natl Acad Sci U S A 101, 5024–5029. 10.1073/pnas.0308386101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinitzer T., Mizote T., Ishikawa N., Dudnik A., Inatsu S., Schreiber S., Suerbaum S., Aizawa S. I., Josenhans C. (2008). Functional characterization and mutagenesis of the proposed behavioral sensor TlpD of Helicobacter pylori. J Bacteriol 190, 3244–3255. 10.1128/JB.01940-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempértegui F., Díaz M., Mejía R., Rodríguez-Mora O. G., Rentería E., Guarderas C., Estrella B., Recalde R., Hamer D. H., Reeves P. G. (2007). Low concentrations of zinc in gastric mucosa are associated with increased severity of Helicobacter pylori-induced inflammation. Helicobacter 12, 43–48. 10.1111/j.1523-5378.2007.00476.x [DOI] [PubMed] [Google Scholar]
- Seshadri S., Benoit S. L., Maier R. J. (2007). Roles of His-rich Hpn and Hpn-like proteins in Helicobacter pylori nickel physiology. J Bacteriol 189, 4120–4126. 10.1128/JB.01245-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour F. W. K., Doetsch R. N. (1973). Chemotactic responses by motile bacteria. J Gen Microbiol 78, 287–296. 10.1099/00221287-78-2-287 [DOI] [PubMed] [Google Scholar]
- Sidebotham R. L., Worku M. L., Karim Q. N., Dhir N. K., Baron J. H. (2003). How Helicobacter pylori urease may affect external pH and influence growth and motility in the mucus environment: evidence from in-vitro studies. Eur J Gastroenterol Hepatol 15, 395–401. 10.1097/00042737-200304000-00010 [DOI] [PubMed] [Google Scholar]
- Smith J. L., Doetsch R. N. (1969). Studies on negative chemotaxis and the survival value of motility in Pseudomonas fluorescens. J Gen Microbiol 55, 379–391. 10.1099/00221287-55-3-379 [DOI] [PubMed] [Google Scholar]
- Stähler F. N., Odenbreit S., Haas R., Wilrich J., Van Vliet A. H., Kusters J. G., Kist M., Bereswill S. (2006). The novel Helicobacter pylori CznABC metal efflux pump is required for cadmium, zinc, and nickel resistance, urease modulation, and gastric colonization. Infect Immun 74, 3845–3852. 10.1128/IAI.02025-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette M. G., Stock J. B. (1996). Role of α-helical coiled-coil interactions in receptor dimerization, signaling, and adaptation during bacterial chemotaxis. J Biol Chem 271, 17966–17973. 10.1074/jbc.271.30.17966 [DOI] [PubMed] [Google Scholar]
- Terry K., Williams S. M., Connolly L., Ottemann K. M. (2005). Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect Immun 73, 803–811. 10.1128/IAI.73.2.803-811.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry K., Go A. C., Ottemann K. M. (2006). Proteomic mapping of a suppressor of non-chemotactic cheW mutants reveals that Helicobacter pylori contains a new chemotaxis protein. Mol Microbiol 61, 871–882. 10.1111/j.1365-2958.2006.05283.x [DOI] [PubMed] [Google Scholar]
- Testerman T. L., McGee D. J., Mobley H. L. (2001). Helicobacter pylori growth and urease detection in the chemically defined medium Ham’s F-12 nutrient mixture. J Clin Microbiol 39, 3842–3850. 10.1128/JCM.39.11.3842-3850.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testerman T. L., Conn P. B., Mobley H. L., McGee D. J. (2006). Nutritional requirements and antibiotic resistance patterns of Helicobacter species in chemically defined media. J Clin Microbiol 44, 1650–1658. 10.1128/JCM.44.5.1650-1658.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomb J.-F., White O., Kerlavage A. R., Clayton R. A., Sutton G. G., Fleischmann R. D., Ketchum K. A., Klenk H. P., Gill S. & other authors (1997). The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388, 539–547. 10.1038/41483 [DOI] [PubMed] [Google Scholar]
- Tso W. W., Adler J. (1974). Negative chemotaxis in Escherichia coli. J Bacteriol 118, 560–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura N., Okamoto S., Yamamoto S., Matsumura N., Yamaguchi S., Yamakido M., Taniyama K., Sasaki N., Schlemper R. J. (2001). Helicobacter pylori infection and the development of gastric cancer. N Engl J Med 345, 784–789. 10.1056/NEJMoa001999 [DOI] [PubMed] [Google Scholar]
- Vigeant M. A., Ford R. M. (1997). Interactions between motile Escherichia coli and glass in media with various ionic strengths, as observed with a three-dimensional-tracking microscope. Appl Environ Microbiol 63, 3474–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhams G. H., Armitage J. P. (2004). Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol 5, 1024–1037. 10.1038/nrm1524 [DOI] [PubMed] [Google Scholar]
- Weis R. M., Koshland D. E., Jr (1990). Chemotaxis in Escherichia coli proceeds efficiently from different initial tumble frequencies. J Bacteriol 172, 1099–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmire J. M., Gancz H., Merrell D. S. (2007). Balancing the double-edged sword: metal ion homeostasis and the ulcer bug. Curr Med Chem 14, 469–478. 10.2174/092986707779941069 [DOI] [PubMed] [Google Scholar]
- Williams S. E., Turnberg L. A. (1982). Studies of the protective properties of gastric mucus. Adv Exp Med Biol 144, 187–188. 10.1007/978-1-4615-9254-9_28 [DOI] [PubMed] [Google Scholar]
- Williams S. M., Chen Y. T., Andermann T. M., Carter J. E., McGee D. J., Ottemann K. M. (2007). Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect Immun 75, 3747–3757. 10.1128/IAI.00082-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe A. J., Berg H. C. (1989). Migration of bacteria in semisolid agar. Proc Natl Acad Sci U S A 86, 6973–6977. 10.1073/pnas.86.18.6973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worku M. L., Karim Q. N., Spencer J., Sidebotham R. L. (2004). Chemotactic response of Helicobacter pylori to human plasma and bile. J Med Microbiol 53, 807–811. 10.1099/jmm.0.45636-0 [DOI] [PubMed] [Google Scholar]
- Wunder C., Churin Y., Winau F., Warnecke D., Vieth M., Lindner B., Zähringer U., Mollenkopf H. J., Heinz E., Meyer T. F. (2006). Cholesterol glucosylation promotes immune evasion by Helicobacter pylori. Nat Med 12, 1030–1038. 10.1038/nm1480 [DOI] [PubMed] [Google Scholar]
- Yamada T., Searle J. G., Ahnen D., Aipers D. H., Greenberg H. B., Gray M., Joscelyn K. B., Kauffman G., Podolsky D. K. (1994). NIH Consensus Conference. Helicobacter pylori in peptic ulcer disease. NIH Consensus Development Panel on Helicobacter pylori in peptic ulcer. JAMA 272, 65–69. 10.1001/jama.272.1.65 [DOI] [PubMed] [Google Scholar]