Abstract
Rfa2 is a ssDNA (single-stranded DNA)-binding protein that plays an important role in DNA replication, recombination and repair. Rfa2 is regulated by phosphorylation, which alters its protein–protein interaction and protein–DNA interaction. In the present study, we found that the Pph3–Psy2 phosphatase complex is responsible for Rfa2 dephosphorylation both during normal G1-phase and under DNA replication stress in Candida albicans. Phosphorylated Rfa2 extracted from pph3Δ or psy2Δ G1 cells exhibited diminished binding affinity to dsDNA (double-stranded DNA) but not to ssDNA. We also discovered that Cdc28 (cell division cycle 28) and Mec1 are responsible for Rfa2 phosphorylation in G1-phase and under DNA replication stress respectively. Moreover, MS revealed that the domain of Rfa2 that was phosphorylated in G1-phase differed from that phosphorylated under the stress conditions. The results of the present study imply that differential phosphorylation plays a crucial role in RPA (replication protein A) regulation.
Keywords: dephosphorylation, DNA replication, phosphatase, protein–DNA interaction, replication protein A (RPA)
Abbreviations: AP, alkaline phosphatase; ATM, ataxia telangiectasia mutated; Cdc, cell division cycle; CDK, cyclin-dependent kinase; co-IP, co-immunoprecipitation; CTD, C-terminal domain; DBD, DNA-binding domain; DNA-PK, DNA-dependent protein kinase; DSB, double-strand break; dsDNA, double-stranded DNA; EMSA, electrophoretic mobility-shift assay; FL, full-length; GFP, green fluorescent protein; GMM, glucose minimal medium; HF, hyphal form; HU, hydroxyurea; λPPase, lambda phosphatase; NTD, N-terminal domain; PI3K, phosphoinositide 3-kinase; RPA, replication protein A; ssDNA, single-stranded DNA; TBST, Tris-buffered saline containing 0.1% Tween 20; TGE, Tris-glycine-EDTA; UTR, untranslated region; WT, wild-type; YF, yeast form
INTRODUCTION
RPA (replication protein A) is a heterotrimeric complex that functions in DNA replication, repair and recombination pathways in eukaryotes [1–4]. It consists of three subunits of 70, 32 and 14 kDa respectively, namely RPA1 or RPA70, RPA2 or RPA32, and RPA3 or RPA14 (known as Rfa1, Rfa2 and Rfa3 respectively in Saccharomyces cerevisiae) [1,3–6]. RPA1 consists of four OB (oligonucleotide-binding) fold DBDs (DNA-binding domains), named DBD-F, DBD-A, DBD-B and DBD-C [3,4]. The N-terminal DBD-F and DBD-A domains of RPA1 have been implicated in the recruitment of other factors during DNA replication, repair or recombination [7–9], whereas the C-terminal DBD-C domain is essential for RPA trimerization [3,4,10,11]. RPA2 contains a flexible NTD (N-terminal domain) that is highly phosphorylated, a DBD, DBD-D, and a CTD (C-terminal domain) that mediates protein–protein interactions [3,12–15]. RPA3 contains a single DBD, DBD-E, which interacts with DBD-D of RPA2 and DBD-C of RPA1 to form a stable RPA trimer.
The RPA heterotrimeric complex has a high affinity for ssDNA (single-stranded DNA) in a 5′→3′ polar manner [16–22]. It binds to ssDNA ranging from 8 to 30 nt [11], seemingly with low sequence specificity. DBD-A and DBD-B of RPA1 have the highest DNA-binding affinities [3,21]. Various binding modes requiring different DBDs in the trimeric RPA have been presented [3,22–25]. However, distinct from RPA1, DBD-D of RPA2 also interacts with dsDNA (double-stranded DNA) and plays an important role in DNA priming during early DNA replication [4]. Moreover, Dickson et al. [26] showed that an RPA2 DBD-D mutant with compromised DNA-binding activity has little effect on cell viability.
RPA is phosphorylated in a cell-cycle-dependent manner [27–29]. RPA2 is preferentially phosphorylated, especially at its NTD [3–5]. RPA2 is phosphorylated at Ser23 in S-phase, and simultaneously at Ser23 and Ser29 in M-phase [30]. Phosphorylated RPA2 extracted from mitotic HeLa cells resulted in lower dsDNA-binding affinity, but had no effect on ssDNA binding [31]. RPA2 phosphorylation also abolished its binding to DNA replication and repair proteins such as ATM (ataxia telangiectasia mutated), DNA-PK (DNA-dependent protein kinase) and DNA polymerase α [31]. Furthermore, phosphorylated RPA also showed weaker RPA1–RPA2 interactions [32]. In addition, DNA damage is another key factor triggering RPA hyperphosphorylation, which is, however, distinct from the cell-cycle-dependent phosphorylation. Ionizing radiation led to RPA2 localization to DNA damage loci and RPA2 hyperphosphoryation which involves ATM and DNA-PK [30]. Bleomycin-treated cells undergoing mitosis led to RPA2 phosphorylation at Ser4/Ser8 and Thr21 which are primed by phosphorylation at Ser23 and Ser29 [33,34]. Ser4 and Ser8 was demonstrated to be phosphorylated during DSB (double-strand break) of DNA leading to stalled DNA replication [13,35–37]. Furthermore, RPA2 was shown to be phosphorylated at Ser11, Ser12, Ser13, Thr21, Ser23, Ser29 and Ser33 by ATM and DNA-PK in vitro, many of which coincide with those phosphorylated in vivo in response to DNA damage [13,36,38–41]. Hence, phosphorylation plays a pivotal role in regulating RPA activity [39,42]. Currently, studies of the phosphatases involved in the phosphoregulation of RPA2 are limited. The PP2A-like phosphatases PP2AC and PP4C have been implicated in dephosphorylating RPA2 in the DNA-damage response [43,44]. However, the phosphatase(s) responsible for RPA2 dephosphorylation during G1-phase remains elusive [43,44].
In the present study, we found that the Pph3–Psy2 phosphatase complex is responsible for Rfa2 dephosphorylation both during normal G1-phase and under DNA replication stress in Candida albicans. Moreover, the results of the present study showed that the domain of Rfa2 phosphorylated during G1-phase differed from that phosphorylated in response to DNA replication stress, indicating that differential phosphorylation plays a crucial role in RPA regulation in C. albicans.
MATERIALS AND METHODS
Strains and culture conditions
All C. albicans strains used in the present study are listed in Supplementary Table S1 (at http://www.BiochemJ.org/bj/449/bj4490673add.htm). Except where noted, C. albicans were routinely grown at 30°C in YPD medium (1% yeast extract, 2% peptone and 2% glucose), in GMM (glucose minimal medium; 2% glucose and 6.79 g/l yeast nitrogen base without amino acids) or in GMM supplemented with the required nutrients for auxotrophic mutants. Solid media contained 2% agar.
Preparation of G1 cells, induction of hyphal growth by serum and HU (hydroxyurea) treatment of cells
G1 cells were obtained by growing yeast cultures at 30°C for 72 h until >90% of cells were found in G1-phase under the microscope. Then the cells were released into fresh YPD medium as described previously [45].
For hyphal growth, bovine serum was added to C. albicans yeast cells in YPD medium to a final concentration of 20% and the cells were incubated at 37°C for 4 h before harvesting the cells for analysis.
To cause DNA replication stress, HU was added to C. albicans cultures in YPD medium to a final concentration of 20 mM, and the cells were incubated at 30°C for a specified time. Recovery from the DNA replication stress was achieved by shifting the cells to fresh HU-free YPD medium and incubation at 30°C for 4–6 h before harvesting cells for analysis.
Construction of C. albicans mutant strains
C. albicans homologues of S. cerevisiae genes were identified by sequence alignment in the C. albicans genome database (http://www.candidagenome.org). C. albicans deletion mutants were constructed by sequentially deleting the two copies of the target gene with two deletion cassettes from the WT (wild-type) strain of BWP17. The deletion cassettes were constructed by flanking a selectable marker gene (ARG4 or HIS1) with the AB and CD DNA fragments (~400 bp each), which correspond to the 5′ and 3′ UTRs (untranslated regions) of the target gene respectively [45]. Homozygous deletion mutants were verified by PCR.
For rescue experiments, the entire ORF (open reading frame) of the target gene, together with its promoter (~1000 bp), was cloned into the CIp10-based URA3-marked plasmid at KpnI and ClaI sites, followed by the GAL4 3′ UTR. The construct was linearized with StuI, whose site exists in the RP10 sequence of the plasmid CIp10, and finally introduced into the gene deletion strains [45].
Construction of C. albicans strains expressing C-terminal Myc-tagged Rfa2 or truncated Rfa2 fragments was carried out as described previously [46]. C-terminal GFP (green fluorescent protein)-tagged Rfa2 was constructed in the WT C. albicans strain as described above. For affinity purification of Rfa2, C-terminal His-tagged full-length Rfa2 was constructed in the WT strain, and pph3Δ and psy2Δ mutants in a similar fashion.
Protein extraction, Western blotting, protein dephosphorylation and co-IP (co-immunoprecipitation)
To extract proteins, cells were harvested by centrifugation (4000 g for 5 min at 4°C), and ~100 mg of cell pellet was resuspended in 300 μl of ice-cold RIPA buffer [50 mM Tris/HCl (pH 7.4), 150 mM NaCl, 1% Nonidet P40, 0.5% sodium deoxycholate and 0.1% SDS]. After adding an equal volume of acid-washed glass beads (Sigma–Aldrich), the cells were lysed by four rounds of 45 s of beating at 5000 rev./min in a MicroSmash MS-100 bead beater (Tomy Medico) with 2 min of cooling on ice between rounds. The supernatant was collected after centrifugation of the cell lysate at 16000 g for 20 min at 4°C. The protein concentration of the lysate was determined using the bicinchoninic acid protein assay (Galen).
For Western blot analysis, 30 μg of total protein was separated by SDS/PAGE (10% or 12% gels) and transferred on to a PVDF membrane (Millipore). The membrane was immersed in TBST [TBS (Tris-buffered saline, pH 7.4) containing 0.1% Tween 20] and 5% non-fat dried skimmed milk for 1 h at room temperature (25°C), followed by primary antibody and secondary antibody conjugated to hydrogen peroxidase or AP (alkaline phosphatase) consecutively for 1 h each, both in TBST containing 1% non-fat dried skimmed milk. The target protein was visualized by using the ECL (enhanced chemiluminescence) system or AP system. Anti-Myc and anti-Cdc28 (cell division cycle 28) (PSTAIRE) antibodies were purchased from Santa Cruz Biotechnology.
Protein dephosphorylation was carried out as described previously [45]. λPPase (lambda phosphatase) was purchased from New England BioLabs (catalogue number P07535).
A co-IP assay was performed by using an anti-Myc antibody to first pull down the Myc-tagged protein. Then co-immunoprecipitated proteins were detected by Western blot analysis with appropriate antibodies as described above.
Protein purification and EMSA (electrophoretic mobility-shift assay)
WT C. albicans and pph3Δ and psy2Δ mutant cells expressing C-terminal His-tagged full-length Rfa2 were grown in YPD medium at 30°C for 3 days. Cells were harvested, lysed by cell disruption (Avestin) and centrifuged at 20000 g at 4°C for 30 min. The supernatant was purified on gravity-flow Ni-NTA (Ni2+-nitrilotriacetate) columns (Qiagen) followed by anion-exchange chromatography (SourceQ, GE Healthcare) on an Äkta purifier (GE Healthcare). Purified Rfa2 was concentrated by using centrifugal concentration tubes (Amicon, Millipore) until a final concentration of 0.8–1.0 mg/ml was reached.
An EMSA was performed with ssDNA of the sequence 5′-FITCCCCCTCTCCTTCTTGGCCTCTTCCTTCCCC-3′ or dsDNA annealed from complementary strands of the sequences 5′-FITCCCCCTCTCCTTCTTGGCCTCTTCCTTCCCC-3′ and 5′-GGGGAAGGAAGAGGCCAAGAAGGAGAGGGG-3′ using standard protocols. Purified Rfa2 protein was mixed at ratios of 0, 3.12, 6.25 and 12.5 pmol to a constant amount of DNA of 400 pmol in a total reaction mixture of 20 μl and incubated at room temperature for 30 min. All reaction mixtures were loaded on to an 8% TGE (Tris-glycine-EDTA) gel and run at 100 V at 4°C for 1 h.
RESULTS
The Pph3–Psy2 complex is responsible for Rfa2 dephosphorylation during G1-phase in C. albicans
Human RPA2 was found to be phosphorylated during mitosis and became hyperphosphorylated on DNA damage [3–5,31,33]. PP2A and PP4 were found to dephosphorylate RPA2 in humans after DNA damage [43,44]. However, the phosphatase responsible for RPA2 dephosphorylation during G1-phase remained elusive. In the present study, we found that Pph3, the yeast homologue of PP4, dephosphorylates Rfa2 during G1-phase in vivo and in vitro in C. albicans.
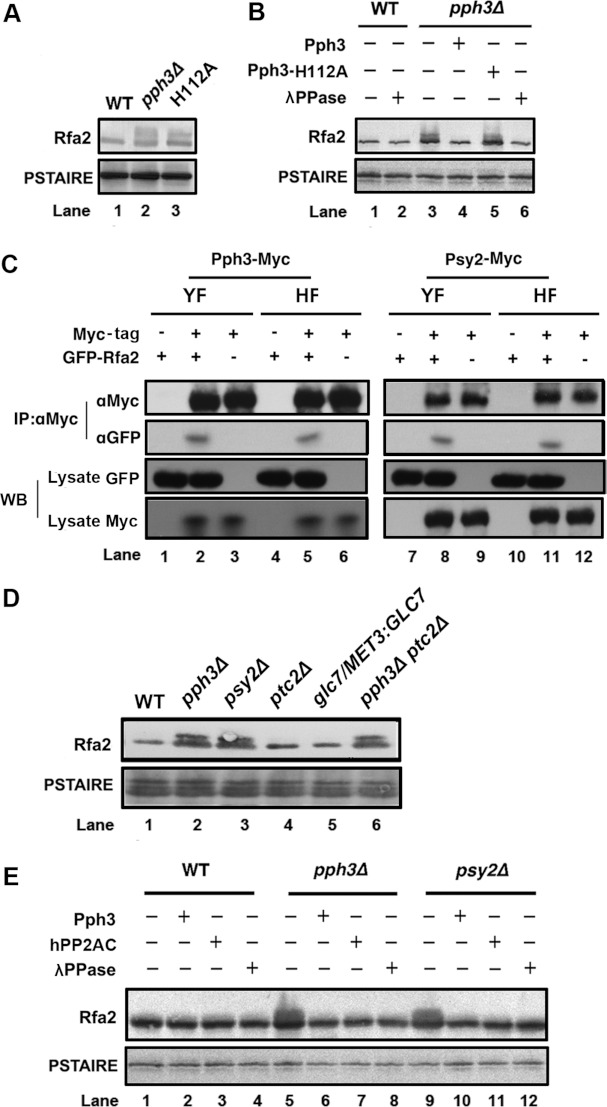
Western blot analysis of Rfa2 extracted from WT C. albicans G1 cells exhibited a dephosphorylated form (Figure 1A). The deletion of PPH3 or replacement of WT PPH3 with the catalytic mutant allele pph3-H112A led to accumulation of phosphorylated Rfa2 in G1 cells (Figure 1A). Furthermore, addition of antibody-purified Pph3 dephosphorylated Rfa2 in pph3Δ cell lysate (Supplementary Figure S1 at http://www.BiochemJ.org/bj/449/bj4490673add.htm), whereas addition of Pph3-H112A had no such effect (Figure 1B). Similarly, Rfa2 was dephosphorylated by the broad-specificity λPPase in the positive control (Figure 1B). Hence, the results strongly suggest that Pph3 dephosphorylates Rfa2 during G1-phase in C. albicans.
Figure 1. Pph3 is responsible for Rfa2 dephosphorylation during G1-phase.
(A) Western blot analysis of Rfa2 extracted from WT cells (HT1), pph3Δ cells (HT2) and pph3Δ cells expressing Pph3-H112A (HT29.1) in G1-phase. G1 cells were obtained by growing cells at 30°C for 3 days until >90% of the cells were in G1-phase. (B) Western blot analysis of Rfa2 extracted from WT (HT1) and pph3Δ (HT2) G1 cells. Pph3 affinity-purified from strain HT28, Pph3-H112A from HT29 or λPPase was used to treat immune-purified Rfa2 in vitro. (C) Co-IP analysis of Pph3 and Psy2 with Rfa2. C. albicans cells co-expressing Pph3–Myc and GFP–Rfa2 (HT26), and those co-expressing Psy2–Myc and GFP–Rfa2 (HT27) were grown as yeast (YF) or hyphae (HF). Cell lysates were prepared for co-IP. IP was performed using an anti-Myc antibody followed by Western blot (WB) analysis with an anti-Myc or anti-GFP antibody. (D) Western blot analysis of Rfa2 extracted from WT (HT1), pph3Δ (HT2), psy2Δ (HT3), ptc2Δ (HT4), glc7/MET3:GLC7 (HT25) and pph3Δ ptc2Δ (HT5) cells during G1 arrest. To shut down GLC7 expression in HT25, the medium contained 0.5 mM each of methionine and cysteine. (E) Western blot analysis of Rfa2 extracted from WT (HT1), pph3Δ (HT2) and psy2Δ (HT3) G1 cells. C. albicans Pph3, human PP2Ac or λPPase was used to treat purified Rfa2 in vitro.
In addition, we found that the regulatory subunit Psy2 (corresponding to human PP4R3) of Pph3 is involved in the interaction between Pph3 and Rfa2. Co-IP assays revealed that Rfa2 could be pulled down with Pph3 and Psy2 from both yeast and hyphal cells of C. albicans (Figure 1C). Whether both subunits directly interact with Rfa2 has not been investigated. Moreover, Rfa2 displayed a phosphorylated form in both the pph3Δ and the psy2Δ mutant (Figure 1D). This indicates that Psy2 is required for proper Rfa2 dephosphorylation during G1-phase. Further investigations with deletion mutants of ptc2 and glc7, the yeast homologues of PP2C and PP1, demonstrated that the pph3Δ ptc2Δ mutant resulted in Rfa2 phosphorylation during G1-phase, whereas Rfa2 remained dephosphorylated in ptc2Δ and glc7Δ cells, similar to WT cells (Figure 1D). In addition, in vitro addition of Pph3, human PP2AC or λPPase dephosphorylated Rfa2 immunopurified from pph3Δ and psy2Δ cell lysates (Figure 1E). Hence, we concluded that Rfa2 is specifically dephosphorylated by the Pph3–Psy2 complex during G1-phase in C. albicans.
Pph3 is responsible for Rfa2 dephosphorylation under DNA replication stress in C. albicans
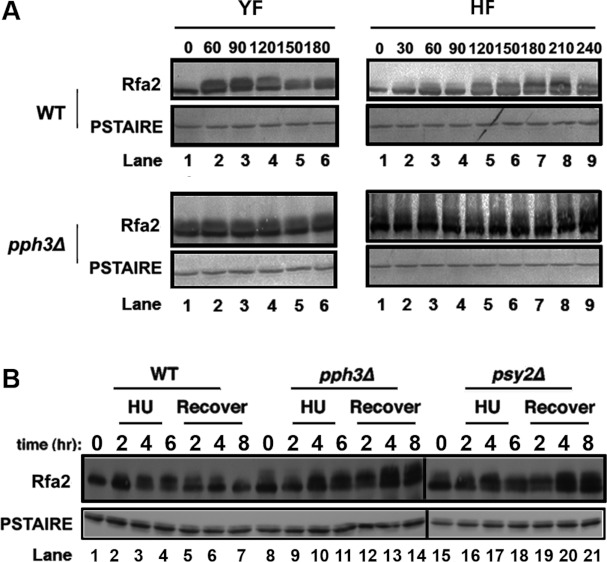
In humans, DNA damage is known to lead to RPA2 hyperphosphorylation in a manner distinct from its phosphorylation during mitosis [3,4,33,34]. We next asked whether Pph3 is involved in Rfa2 dephosphorylation under DNA replication stress in C. albicans. Before addressing this question, we first wanted to check whether cell-cycle phases affect Rfa2 phosphorylation, as observed for RPA2 in human cells. We monitored Rfa2 phosphorylation at timed intervals after releasing G1 yeast cells into both yeast and hyphal growth conditions. In WT YF (yeast form) cells, Rfa2 was not phosphorylated at time zero, but became phosphorylated between 60 and 120 min, and the phosphorylation started to diminish at 150 min (Figure 2A, top left-hand panel). The results indicated that Rfa2 undergoes cell-cycle-dependent phosphorylation. In HF (hyphal form) WT cells induced by serum at 37°C, Rfa2 phosphorylation was observed after 120 min and started to diminish at 240 min after hyphal induction (Figure 2A, top right-hand panel). In sharp contrast, Rfa2 was persistently phosphorylated in pph3Δ mutant cells under both yeast and hyphal growth conditions (Figure 2A, bottom panels).
Figure 2. Pph3 is responsible for Rfa2 dephosphorylation during normal growth and under DNA replication stress.
(A) WT (HT1) and pph3Δ (HT2) G1 cells were released into conditions for yeast (YF) or hyphal (HF) growth. Aliquots of cells were harvested at the indicated times for Western blot analysis of Rfa2. (B) WT (HT1), pph3Δ (HT2) and psy2Δ (HT3) G1 cells were first treated with 20 mM HU and then allowed to recover by transferring cells to HU-free medium for further growth. Aliquots of cells were harvested at the times indicated during both HU treatment and the recovery period for Western blot analysis of Rfa2.
We next tested the effect of the DNA replication inhibitor HU on Rfa phosphorylation. We observed that growing WT yeast cells in the presence of HU resulted in Rfa2 hyperphosphorylation, and subsequent shifting of the cells to HU-free media caused its gradual dephosphorylation (Figure 2B). In contrast, while Rfa2 in pph3Δ and psy3Δ cells became hyperphosphorylated in response to HU treatment, the level of Rfa2 phosphorylation continued to increase after HU removal (Figure 2B).
Hence, we concluded that Pph3 is responsible for Rfa2 dephosphorylation both during the cell cycle and under HU treatment.
Phosphorylated Rfa2 exhibited diminished binding activity to dsDNA, but not to ssDNA
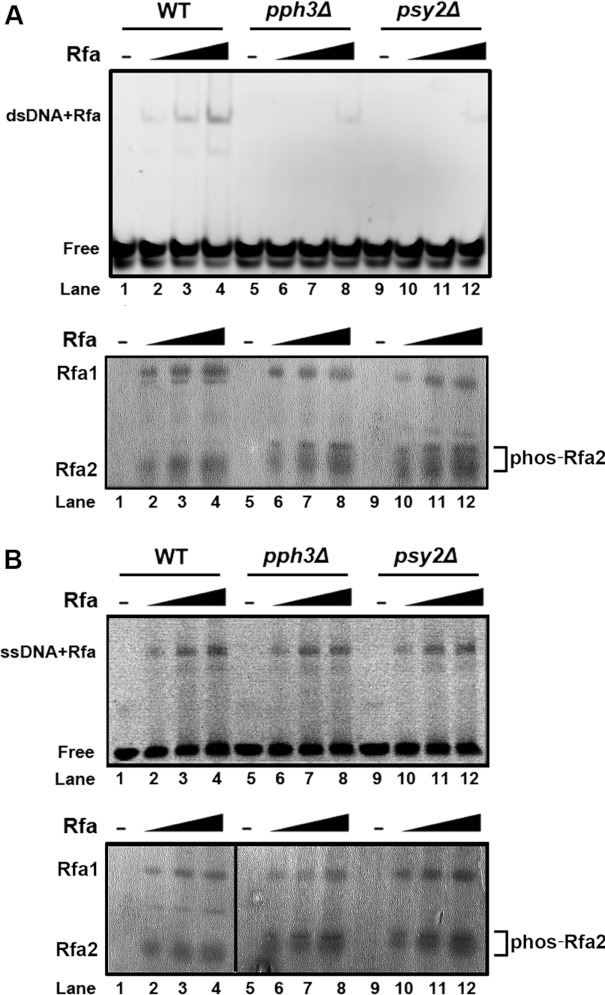
Oakley et al. [31] showed previously that phosphorylated RPA2 extracted from mitotic human cells had little dsDNA-binding activity, whereas unphosphorylated recombinant RPA2 purified from bacteria exhibited dsDNA-binding activity. In the present study, we discovered that Rfa2 extracted from pph3Δ or psy2Δ cells also exhibited diminished binding to dsDNA compared with Rfa2 from WT cells in an EMSA (Figure 3A). Consistent with previous studies [31], ssDNA binding of Rfa2 purified from pph3Δ or psy2Δ cells was unaffected (Figure 3B). Therefore the results of the present study are consistent with the concept that phosphorylation of Rfa2 abolishes its binding to dsDNA, but not ssDNA [3,4,31].
Figure 3. EMSA of Rfa2 with dsDNA and ssDNA.
(A) Top panel: Rfa2 affinity-purified from WT (HT41), pph3Δ (HT42) and psy2Δ (HT43) cells was incubated with FITC-labelled dsDNA at molar ratios of DNA/protein of 0, 8, 16 and 32 at room temperature for 30 min. The protein/DNA mixture was then resolved by TGE/PAGE (8% gel) and detected by fluorescence at an emission wavelength of 320 nm. Bottom panel: the same amounts of purified Rfa2 as above were resolved by SDS/PAGE (12% gel) as a protein loading control. (B) The experiment described above was repeated using ssDNA instead of dsDNA.
Different domains of Rfa2 are (de)phosphorylated under different conditions in C. albicans
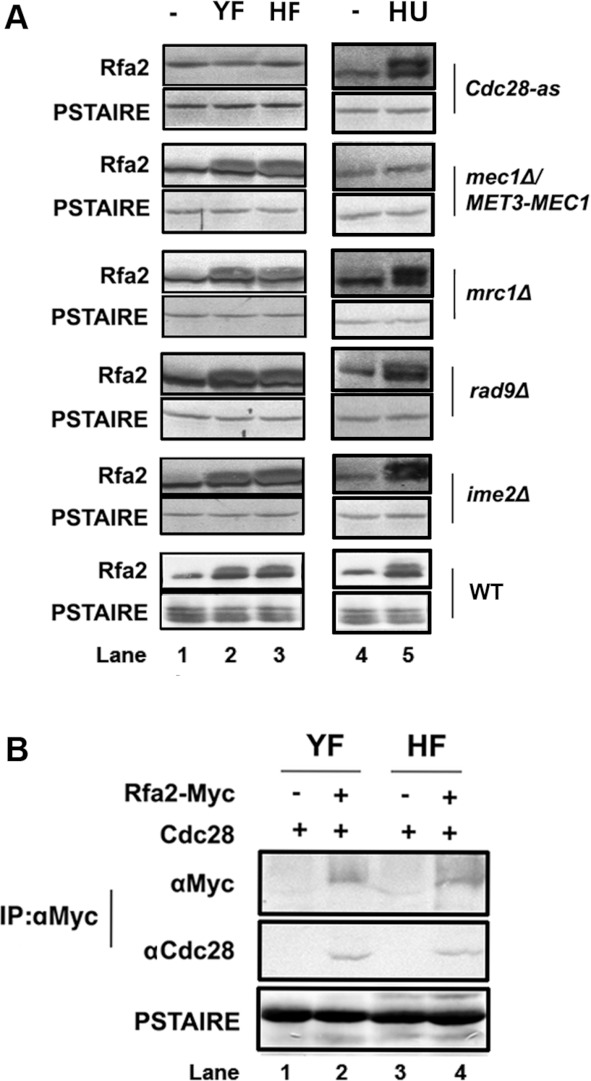
Thr43 in DBD-D is the only consensus CDK (cyclin-dependent kinase) phosphorylation site present on C. albicans Rfa2. Next, we tried to decipher the sites of phosphorylation on Rfa2 under normal growth conditions and DNA replication inhibition. We examined Rfa2 phosphorylation by Western blot analysis in different kinase-deficient C. albicans mutants. We first used a strain expressing the mutant Cdc28-as kinase which can be inhibited by the ATP analogue 1NM-PP1. We found that Rfa2 phosphorylation was not detectable in either YF or HF cells when 1NM-PP1 was added to the cultures. In comparison, Rfa2 phosphorylation was detected to be normal in the mec1Δ/MET3-MEC1 (MEC1 was shown to decrease by adding methionine and cysteine to the medium), mrc1Δ, rad9Δ and ime2Δ mutants grown under normal growth conditions (Figure 4A, left-hand panel). However, Rfa2 remained dephosphorylated only in the mec1Δ/MET3-MEC1 mutant after HU treatment (Figure 4A, right-hand panel). Moreover, co-IP showed that Cdc28 directly interacted with Rfa2 in both yeast and hyphal cells under normal growth conditions (Figure 4C). However, in spite of repeated efforts, we failed to observe an interaction between Rfa2 and Mec1 in co-IP experiments (results not shown). We cannot rule out the possibility of transient or weak interaction between the proteins. Nevertheless, the results of the present study support a proposal that Cdc28 and Mec1 are responsible for Rfa2 phosphorylation during a normal cell cycle and under HU stress respectively.
Figure 4. Role of Cdc28 and Mec1 in Rfa2 phosphorylation during normal growth and under HU treatment.
(A) Western blot analysis of Rfa2 extracted from WT (HT1), Cdc28-as (HT40), mec1Δ/MET3-MEC1 (HT37), mrc1Δ (HT34), rad9Δ (HT35) and ime2Δ (HT39) cells in G1-phase, yeast growth (YF) and hyphal growth (HF) (left-hand panel) or after HU treatment for 4 h (right-hand panel). The anti-PSTAIRE antibody was used to show that equal amounts of protein were used. (B) Co-IP analysis of Cdc28 with Rfa2. Cells expressing Rfa2–Myc (HT1) were grown as yeast (YF) or hyphae (HF) for co-IP analysis. Rfa2 was pulled down with an anti-Myc antibody, followed by Western blot analysis with anti-Myc or anti-PSTAIRE antibodies.
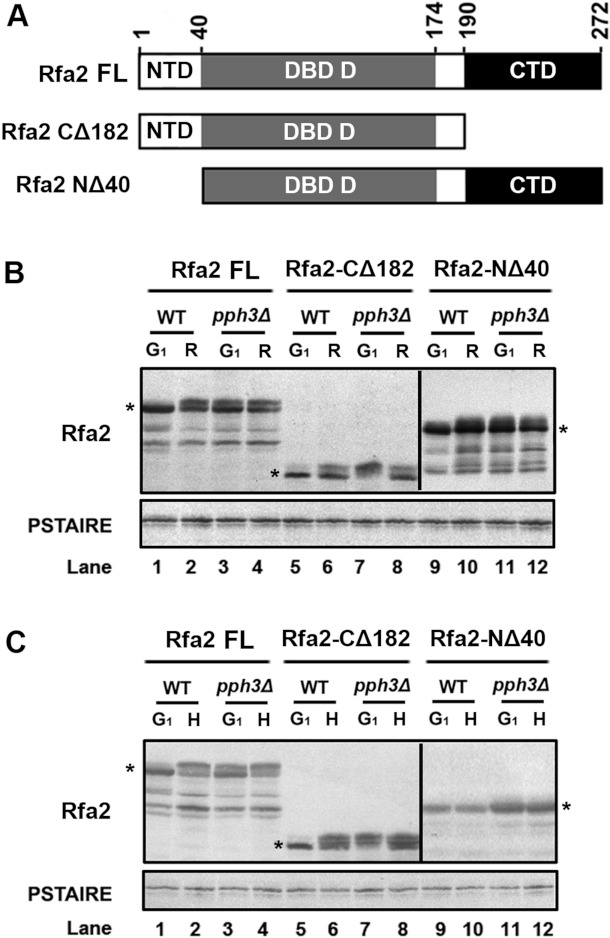
Next, we wanted to determine whether the phosphorylation sites on Rfa2 that occurred during normal growth differ from those induced by HU treatment. First, we constructed two truncated forms of Rfa2: Rfa2-NΔ40 containing residues 41–272 and Rfa2-CΔ182 containing residues 1–190 (Figure 5A). We then expressed the two truncated versions of Rfa2 in WT and pph3Δ cells under normal growth conditions and HU treatment. The phosphorylation status of the two truncated Rfa2 proteins was examined by Western blot analysis in comparison with the FL (full-length) WT Rfa2 (Rfa2-FL). The results showed that in WT cells both truncated forms of Rfa2 were dephosphorylated during G1-phase (Figure 5B, lanes 1, 5 and 9) and became phosphorylated 90 min after release (Figure 5B, lanes 2, 6 and 10). In contrast, Rfa2-FL, Rfa2-NΔ40 and Rfa2-CΔ182 expressed in the pph3Δ mutant cells were phosphorylated during G1 arrest (Figure 5B, lanes 3, 7 and 11), and the phosphorylation persisted at 90 min (Figure 5B, lanes 4, 8 and 12). Therefore these results demonstrated that: (i) unlike the predominant phosphorylation of RPA2 in the NTD reported in previous studies [13,36,38–41], the NTD of Rfa2 is not the only domain phosphorylated during normal growth in C. albicans; and (ii) Pph3 is responsible for dephosphorylating Rfa2 during G1-phase.
Figure 5. Different domains on Rfa2 are (de)phosphorylated under different circumstances.
(A) Schematic description of FL and truncated versions of Rfa2. Rfa2-FL (HT1 and HT2), Rfa2-CΔ182 (HT31 and HT33) containing residues 1–190 and Rfa2-NΔ40 (HT30 and HT32) containing residues 41–272 were expressed in WT or pph3Δ (SJL2) mutant cells. (B) Western blot analysis of Rfa2 extracted from WT (HT1, HT30 and HT31) and pph3Δ (HT2, HT32 and HT33) cells expressing different versions of Rfa2 in G1-phase and after release. An anti-PSTAIRE antibody was used to detect Cdc28 as a loading control. (C) Western blot analysis of Rfa2 extracted from the same set of cells as above during G1-phase and after HU treatment.
Next, Western blot analysis of HU-treated WT and pph3Δ cells expressing Rfa2-FL, Rfa2-NΔ40 and Rfa2-CΔ182 showed that Rfa2-FL and Rfa2-CΔ182, but not Rfa2-NΔ40, became phosphorylated in WT cells (Figure 5C, lanes 2, 6 and 8), whereas all three proteins were phosphorylated in pph3Δ cells (Figure 5C, lanes 4, 8 and 12). Therefore we observed that Rfa2-NΔ40 containing the DBD-D and CTDs existed in dephosphorylated form in WT, but not pph3Δ, cells after HU treatment in C. albicans. We deduced that the persistent phosphorylation of Rfa2-NΔ40 in pph3Δ cells before and after HU treatment (Figure 5C, lanes 11 and 12) might reflect the background level of phosphorylation as seen in normal cells (Figure 5B, lanes 11 and 12). Hence, this result suggests that the NTD is involved in response to DNA replication stress.
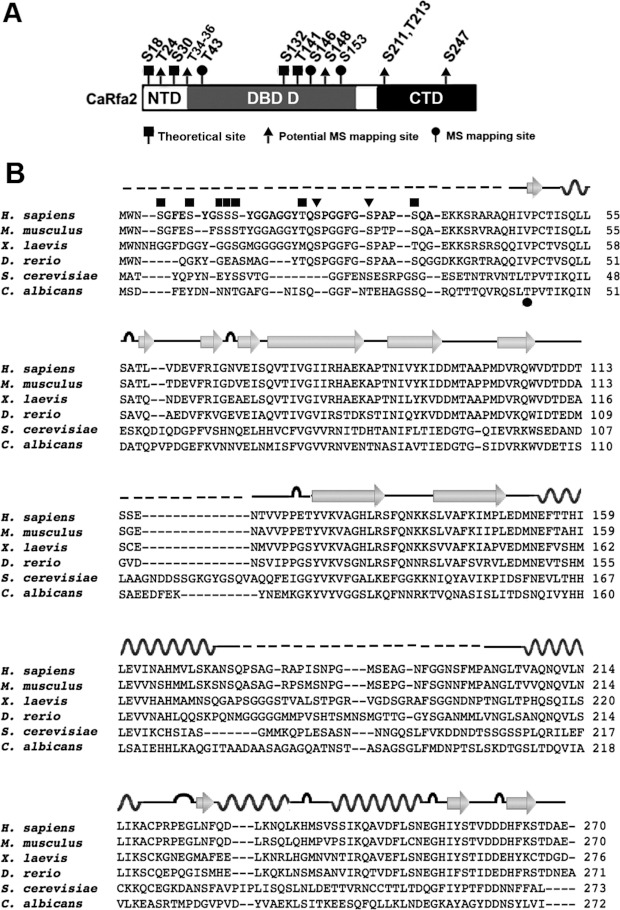
Next, we tried to map phosphorylation sites on Rfa2 extracted from pph3Δ mutant cells under normal growth conditions by MS. Results showed that the only consensus CDK phosphorylation site, Thr43, was phosphorylated (Figure 6A). Additionally, Ser146 and Ser153 were two other phosphorylation sites with the highest probabilities (Figure 6A). Eight other potential phosphorylation sites, Thr24, Thr34/Thr35/Thr36, Ser148, Ser211, Thr213 and Ser247, were also identified (Table 1). Sequence alignment of these potential phosphorylation sites of C. albicans Rfa2 with RPA2 in human, mouse, frog and zebrafish revealed little homology. However, Thr43 of C. albicans Rfa2 is conserved with Thr40 in S. cerevisiae Rfa2 (Figure 6B).
Figure 6. Potential dephosphoryation sites on Rfa2 mediated by Pph3 during normal growth.
(A) Domain structure of Rfa2 depicting potential dephosphorylation sites mediated by Pph3. Rfa2 extracted from pph3Δ (HT2) cells was subject to MS analysis to map potential dephosphorylation sites mediated by Pph3 during normal growth phase. A black square denotes theoretical phosphorylation sites listed on the Uniprot database; a black triangle denotes phosphorylated sites mapped by MS with medium-high probability; and a black circle denotes phosphorylated sites mapped by MS with high probability. (B) Sequence alignment of Rfa2 and homologues in Homo sapiens, Mus musculus, Xenopus laevis, Danio rerio, S. cerevisiae and C. albicans. The secondary structure of human RPA2 (PDB codes 2PQA and 1Z1D) is illustrated above the sequence, where wave lines denotes α-helix, arrows denote β-strand, loops denote turns and broken lines denote unstructured regions. Black squares denote empirical DNA-PK and PI3K phosphorylation sites on human RPA2; black triangles denote CDK phosphorylation sites on human RPA2; and the black circle denotes the phospho-threonine mapped on C. albicans Rfa2 in the present study.
Table 1. MS mapping of phosphorylation sites in Rfa2.
Phosphorylation site mapping by tandem MS. Asterisks indicate phosphoserine or phosphothreonine residues. Consensus CDK phosphorylation sites are underlined.
| Sequence of identified peptide | Position of phospho-residues |
|---|---|
| SQGGFNT*EHAGSSQRQ | Thr24 |
| RQT*T*TQVRQ | Thr34, Thr35 |
| RQSLT*PVTIKQ | Thr43 |
| RQSLT*PVT*IKQ | Thr43, Thr46 |
| RQS*LT*PVTIKQ | Ser41,Thr43 |
| TTQVRQSLT*PVTIKQ | Thr43 |
| RQTTT*QVRQSLT*PVTIKQ | Thr36, Thr43 |
| RQTTT*QVRQSLT*PVT*IKQ | Thr36, Thr43, Thr46 |
| KTVQNASIS*LIT | Ser148 |
| RKTVQNASISLITDS*NQI | Ser153 |
| KT*VQNAS*ISLITDSNQIVYHHLSAIEHHLKA | Thr141, Ser146 |
| KEES*QFQLLKL | Ser247 |
| RTMPDGVPVDYVAEKLSITKEES*QFQLLKL | Ser247 |
| KDTGS*LT*DQVIAVLKE | Ser211, Thr213 |
| KAYAGYDDNS*YLVIEQKL | Ser268 |
| KYNEMKGKYVYVGGS*LKQFNNRK | Ser132 |
| KAQGITAADAASAGAGQATNSTAS*AGS* | Ser192, Ser195, Thr203, Ser204, Ser206 |
| GLFMDNPT*S*LS*KD |
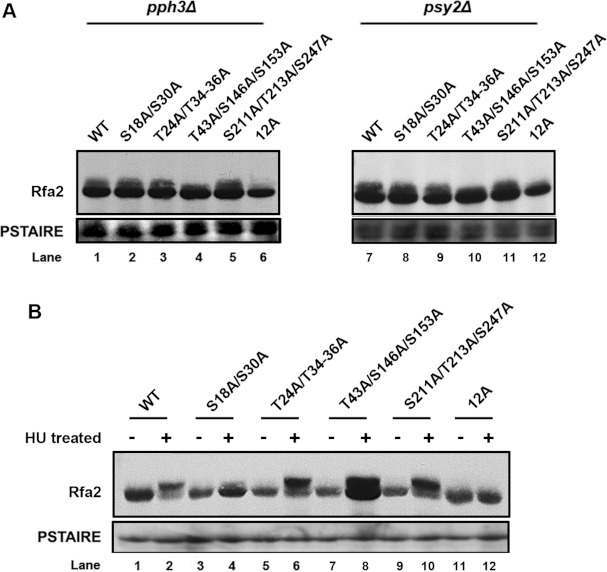
Finally, we performed alanine mutagenesis on the most probable phosphorylation sites occurring in different domains of Rfa2 identified by MS (Table 1) and investigated the phosphorylation state of these mutant proteins in G1 cells and cells treated with HU. Western blot analysis showed that the Rfa2-T43A/S146A/S153A mutant remained unphosphorylated in G1 cells (Figure 7A), but not in HU-treated cells (Figure 7B). This observation is consistent with our finding that domains other than the NTD is phosphorylated in G1-phase (Figure 5B), and suggests that Thr43, Ser146 and Ser153 in the DBD may play a critical role in a normal cell cycle. Ser18 and Ser30 in the NTD were predicted to be DNA-PK and PI3K (phosphoinositide 3-kinase) phosphorylation sites. We found that the Rfa2-S18A/S30A mutant showed significantly reduced phosphorylation on HU treatment (Figure 7B), confirming that the NTD, but not other domains, is essentially involved in Rfa2 phosphorylation under HU treatment (Figure 5C). The observation that the level of Rfa2-S18A/S30A phosphorylation was comparable with WT Rfa2 in G1-phase (Figure 7A) further supports our finding that domains other than the NTD were phosphorylated during G1-phase (Figure 5B). Furthermore, the Rfa2-S211A/T213A/S247A mutant showed a similar level of phosphorylation to WT Rfa2 both in G1-phase (Figure 7A) and under HU treatment (Figure 7B), suggesting that the CTD plays a minor role in Rfa2 phosphoregulation. Other mutants manifested similar phosphorylation patterns as WT Rfa2. It is to be noted that the mutant Rfa2 (Rfa2-12A) with all 12 selected sites of Ser18, Ser30, Thr24, Thr34, Thr35, Thr36, Thr43, Ser146, Ser153, Ser211, Thr213 and Ser247 mutated to alanine, remained unphosphorylated in G1-phase in both pph3Δ and psy2Δ mutants (Figure 7A) and on HU treatment in WT cells (Figure 7B), indicating that these sites are determinant for Rfa2 phosphorylation collectively. Taken together, these results support the idea that domains of Rfa2 that are (de)phosphorylated during a normal cell cycle are different from those phosphorylated in response to DNA replication stress in C. albicans.
Figure 7. Identification of Rfa2 phosphorylation sites under different circumstances.
(A) Western blot analysis of Rfa2 in pph3Δ (HT2, HT45, HT48, HT51, HT54 and HT57) and psy2Δ (HT3, HT46, HT49, HT52, HT55 and HT58) cells expressing different serine/threonine mutant alleles of RFA2 in G1 cells. An anti-PSTAIRE antibody was used to probe Cdc28 as a loading control. (B) Western blot analysis of Rfa2 in WT (HT1, HT44, HT47, HT50, HT53 and HT56) cells expressing different serine/threonine mutant alleles of RFA2 before and after HU treatment.
DISCUSSION
Pph3 dephosphorylates Rfa2 during G1-phase and HU treatment
In the present study, we found that the Pph3–Psy2 complex is responsible for Rfa2 dephosphorylation during G1-phase and on HU treatment. However, it was reported that PP4R2, but not PP4R3 (corresponding to Psy4 and Psy2 respectively in yeast) mediated RPA2 dephosphorylation in humans [44]. Although we could not rule out a role for Psy4 in Rfa2 dephosphorylation in C. albicans, the results of the present study clearly demonstrated that, distinct from humans, Psy2 is required for Rfa2 dephosphorylation. Further studies on other regulatory subunits of Pph3 need to be carried out to determine whether other Pph3 complexes have a role in the regulation of Rfa2 dephosphorylation.
We also showed that Rfa2 extracted from pph3Δ cells exhibited diminished binding to dsDNA, but not ssDNA, a result consistent with reports by Oakley et al. [31]. However, Patrick et al. [47] showed that phosphorylated RPA2 binds more weakly to short ssDNA (8–11 nt), but not long ssDNA. In the present study, the length of ssDNA used was 30 nt and, therefore, our EMSA result is not contradictory to previous observations.
Rfa2 regulation involves differential phosphorylation of different domains
Different kinases have been inferred in Rfa2 phosphorylation. For instance, in human cells, the CDK Cdc2 was shown to phosphorylate RPA2 during S-phase [29], and Mec1 was implicated in RPA2 hyperphosphorylation during HU treatment [48]. In S. cerevisiae, the meiosis-specific kinase Ime2 was found to be an Rfa2 kinase [49] which was shown to phosphorylate Ser27 of Rfa2 in a distinct manner from the mitotic Cln2–Cdk1 complex [50]. In addition to phosphorylation, previous studies have also shown that the phosphatase PP2A dephosphorylates Thr21 and Ser33 of RPA2 to trigger DNA-repair pathways in response to HU treatment in humans [43], and that PP4 was responsible for RPA2 dephosphorylation triggered by DNA DSBs induced by camptothecin [44]. Thus differential phosphorylation is prevalent in RPA2 under different conditions.
Human RPA2 is phosphorylated during mitosis [31,33]. Ser23 and Ser29 in human RPA2 are two consensus CDK phosphorylation sites (SP/TP) which are conserved from zebrafish to humans (Figure 6B, black triangle); however, these two SP/TP sites are not conserved in plants such as Arabidopsis thaliana or yeast such as S. cerevisiae or C. albicans. We noticed that Thr43 in C. albicans Rfa2 (corresponding to Thr40 in S. cerevisiae) is the sole predicted CDK site that aligns to a conserved valine–proline site in RPA2 of higher eukaryotes (Figure 6B, black circle). In the present study, MS indicated that Thr43 was phosphorylated in pph3Δ mutant cells, which replicated slightly faster than WT cells under unperturbed conditions (Supplementary Figure S2 at http://www.BiochemJ.org/bj/449/bj4490673add.htm), but lost viability more rapidly than WT cells in response to genotoxic stress [46]. In support of this observation, the Rfa2-T43A/S146A/S153A triple mutant remained unphosphorylated in G1-phase in both pph3Δ and psy2Δ cells (Figure 7A). Although the results of the present study suggest that mutating Thr43, Ser146 and Ser153 is sufficient to block Rfa2 phosphorylation during G1-phase, single-residue mutation is necessary to delineate the role of each residue. Nevertheless, the results of the present study support the notion that the DBD which harbours these residues is phosphorylated during G1-phase (Figures 5B, 6A and 7A). Sequence identities of C. albicans Rfa2 to human, mouse, Xenopus and zebrafish RPA2 are 22.7, 23.1, 23.7 and 20.4% respectively, compared with a slightly higher identity of 29.4% to S. cerevisiae Rfa2, where homology mainly exists in their DBDs. In this context, it is hard to predict whether phosphorylation in regions other than the NTD of RPA2 occurs in other eukaryotes. Currently, reports on RPA2 are limited in other organisms. Hence, we do not know whether this phenomenon is unique in C. albicans.
Under certain stress conditions, many sites were shown to be phosphosphorylated in human RPA2 [13,35–41] (Figure 6B, black square), and the majority of these sites were found to be located in the NTD. Among these sites, Ser18 and Ser30 in C. albicans Rfa2 (corresponding to Thr21 and Ser33 in human RPA2) are conserved. The results of the present study show that, on HU treatment, Rfa2-FL and Rfa2-CΔ182 became phosphorylated, whereas Rfa2-NΔ40 remained dephosphorylated in WT cells, indicating possible phosphorylation in the NTD in C. albicans Rfa2 after HU treatment. Mutagenesis on Ser18 and Ser30 demonstrated that these sites partially contribute to Rfa2 phosphorylation during HU treatment (Figure 7B). Therefore we speculate that regulation of Rfa2 phosphorylation in C. albicans upon HU treatment also occurs in its NTD, similar to humans.
In summary, the present study has shown that: (i) the Pph3–Psy2 complex is responsible for Rfa2 dephosphorylation both during G1-phase and under DNA replication stress in C. albicans; and (ii) different domains of Rfa2 are (de)phosphorylated under different circumstances.
Online data
AUTHOR CONTRIBUTION
Haitao Wang, Ada Wong, Jianli Sang and Yue Wang conceived and designed the experiments. Haitao Wang, Jiaxin Gao and Kangdi Hu performed the experiments. Haitao Wang, Ada Wong, Jianli Sang, Wanjie Li and Yue Wang analysed the data. Ada Wong, Yue Wang and Haitao Wang wrote the paper.
ACKNOWLEDGEMENTS
We thank members of the Sang laboratory for comments and suggestions on the paper before submission.
FUNDING
This work was supported by the National Basic Research Program of China [grant number 2007CB914401], the National Natural Science Foundation of China [grant number 31270113] and the Fundamental Research Funds for the Central Universities [grant number 105566GK].
References
- 1.Brill S. J., Stillman B. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 1991;5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- 2.Santocanale C., Neecke H., Longhese M. P., Lucchini G., Plevani P. Mutations in the gene encoding the 34 kDa subunit of yeast replication protein A cause defective S phase progression. J. Mol. Biol. 1995;254:595–607. doi: 10.1006/jmbi.1995.0641. [DOI] [PubMed] [Google Scholar]
- 3.Fanning E., Klimovich V., Nager A. R. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broderick S., Rehmet K., Concannon C., Nasheuer H. P. Eukaryotic single-stranded DNA binding proteins: central factors in genome stability. Subcell. Biochem. 2010;50:143–163. doi: 10.1007/978-90-481-3471-7_8. [DOI] [PubMed] [Google Scholar]
- 5.Binz S. K., Lao Y., Lowry D. F., Wold M. S. The phosphorylation domain of the 32-kDa subunit of replication protein A (RPA) modulates RPA–DNA interactions. Evidence for an intersubunit interaction. J. Biol. Chem. 2003;278:35584–35591. doi: 10.1074/jbc.M305388200. [DOI] [PubMed] [Google Scholar]
- 6.Henricksen L. A., Umbricht C. B., Wold M. S. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 7.He Z., Henricksen L. A., Wold M. S., Ingles C. J. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature. 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- 8.Weisshart K., Pestryakov P., Smith R. W., Hartmann H., Kremmer E., Lavrik O., Nasheuer H. P. Coordinated regulation of replication protein A activities by its subunits p14 and p32. J. Biol. Chem. 2004;279:35368–35376. doi: 10.1074/jbc.M403825200. [DOI] [PubMed] [Google Scholar]
- 9.Sugiyama T., Kowalczykowski S. C. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 2002;277:31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- 10.Gomes X. V., Wold M. S. Structural analysis of human replication protein A. Mapping functional domains of the 70-kDa subunit. J. Biol. Chem. 1995;270:4534–4543. doi: 10.1074/jbc.270.9.4534. [DOI] [PubMed] [Google Scholar]
- 11.Wold M. S. Replication protein A: a heterotrimeric, single-stranded DNAbinding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 12.Oakley G. G., Loberg L. I., Yao J., Risinger M. A., Yunker R. L., Zernik-Kobak M., Khanna K. K., Lavin M. F., Carty M. P., Dixon K. UV-induced hyperphosphorylation of replication protein a depends on DNA replication and expression of ATM protein. Mol. Biol. Cell. 2001;12:1199–1213. doi: 10.1091/mbc.12.5.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nuss J. E., Patrick S. M., Oakley G. G., Alter G. M., Robison J. G., Dixon K., Turchi J. J. DNA damage induced hyperphosphorylation of replication protein A. 1. Identification of novel sites of phosphorylation in response to DNA damage. Biochemistry. 2005;44:8428–8437. doi: 10.1021/bi0480584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mer G., Bochkarev A., Gupta R., Bochkareva E., Frappier L., Ingles C. J., Edwards A. M., Chazin W. J. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- 15.Arunkumar A. I., Klimovich V., Jiang X., Ott R. D., Mizoue L., Fanning E., Chazin W. J. Insights into hRPA32 C-terminal domain-mediated assembly of the simian virus 40 replisome. Nat. Struct. Mol. Biol. 2005;12:332–339. doi: 10.1038/nsmbXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Laat W. L., Appeldoorn E., Sugasawa K., Weterings E., Jaspers N. G., Hoeijmakers J. H. DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev. 1998;12:2598–2609. doi: 10.1101/gad.12.16.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bochkareva E., Korolev S., Lees-Miller S. P., Bochkarev A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002;21:1855–1863. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bochkareva E., Belegu V., Korolev S., Bochkarev A. Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J. 2001;20:612–618. doi: 10.1093/emboj/20.3.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bastin-Shanower S. A., Brill S. J. Functional analysis of the four DNA binding domains of replication protein A. The role of RPA2 in ssDNA binding. J. Biol. Chem. 2001;276:36446–36453. doi: 10.1074/jbc.M104386200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wyka I. M., Dhar K., Binz S. K., Wold M. S. Replication protein A interactions with DNA: differential binding of the core domains and analysis of the DNA interaction surface. Biochemistry. 2003;42:12909–12918. doi: 10.1021/bi034930h. [DOI] [PubMed] [Google Scholar]
- 21.Arunkumar A. I., Stauffer M. E., Bochkareva E., Bochkarev A., Chazin W. J. Independent and coordinated functions of replication protein A tandem high affinity single-stranded DNA binding domains. J. Biol. Chem. 2003;278:41077–41082. doi: 10.1074/jbc.M305871200. [DOI] [PubMed] [Google Scholar]
- 22.Blackwell L. J., Borowiec J. A., Mastrangelo I. A. Single-stranded-DNA binding alters human replication protein A structure and facilitates interaction with DNA-dependent protein kinase. Mol. Cell. Biol. 1996;16:4798–4807. doi: 10.1128/mcb.16.9.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bochkarev A., Pfuetzner R. A., Edwards A. M., Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 24.Pestryakov P. E., Khlimankov D. Y., Bochkareva E., Bochkarev A., Lavrik O. I. Human replication protein A (RPA) binds a primer-template junction in the absence of its major ssDNA-binding domains. Nucleic Acids Res. 2004;32:1894–1903. doi: 10.1093/nar/gkh346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pestryakov P. E., Weisshart K., Schlott B., Khodyreva S. N., Kremmer E., Grosse F., Lavrik O. I., Nasheuer H. P. Human replication protein A. The C-terminal RPA70 and the central RPA32 domains are involved in the interactions with the 3′-end of a primer-template DNA. J. Biol. Chem. 2003;278:17515–17524. doi: 10.1074/jbc.M301265200. [DOI] [PubMed] [Google Scholar]
- 26.Dickson A. M., Krasikova Y., Pestryakov P., Lavrik O., Wold M. S. Essential functions of the 32 kDa subunit of yeast replication protein A. Nucleic Acids Res. 2009;37:2313–2326. doi: 10.1093/nar/gkp090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henricksen L. A., Wold M. S. Replication protein A mutants lacking phosphorylation sites for p34cdc2 kinase support DNA replication. J. Biol. Chem. 1994;269:24203–24208. [PubMed] [Google Scholar]
- 28.Dutta A., Stillman B. cdc2 family kinases phosphorylate a human cell DNA replication factor, RPA, and activate DNA replication. EMBO J. 1992;11:2189–2199. doi: 10.1002/j.1460-2075.1992.tb05278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang F., Newport J. W. Distinct roles of cdk2 and cdc2 in RP-A phosphorylation during the cell cycle. J. Cell Sci. 1993;106:983–994. doi: 10.1242/jcs.106.3.983. [DOI] [PubMed] [Google Scholar]
- 30.Stephan H., Concannon C., Kremmer E., Carty M. P., Nasheuer H. P. Ionizing radiation-dependent and independent phosphorylation of the 32-kDa subunit of replication protein A during mitosis. Nucleic Acids Res. 2009;37:6028–6041. doi: 10.1093/nar/gkp605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oakley G. G., Patrick S. M., Yao J., Carty M. P., Turchi J. J., Dixon K. RPA phosphorylation in mitosis alters DNA binding and protein–protein interactions. Biochemistry. 2003;42:3255–3264. doi: 10.1021/bi026377u. [DOI] [PubMed] [Google Scholar]
- 32.Binz S. K., Wold M. S. Regulatory functions of the N-terminal domain of the 70-kDa subunit of replication protein A (RPA) J. Biol. Chem. 2008;283:21559–21570. doi: 10.1074/jbc.M802450200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anantha R. W., Sokolova E., Borowiec J. A. RPA phosphorylation facilitates mitotic exit in response to mitotic DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2008;105:12903–12908. doi: 10.1073/pnas.0803001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anantha R. W., Borowiec J. A. Mitotic crisis: the unmasking of a novel role for RPA. Cell Cycle. 2009;8:357–361. doi: 10.4161/cc.8.3.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liaw H., Lee D., Myung K. DNA-PK-dependent RPA2 hyperphosphorylation facilitates DNA repair and suppresses sister chromatid exchange. PLoS ONE. 2011;6:e21424. doi: 10.1371/journal.pone.0021424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zernik-Kobak M., Vasunia K., Connelly M., Anderson C. W., Dixon K. Sites of UV-induced phosphorylation of the p34 subunit of replication protein A from HeLa cells. J. Biol. Chem. 1997;272:23896–23904. doi: 10.1074/jbc.272.38.23896. [DOI] [PubMed] [Google Scholar]
- 37.Niu H., Erdjument-Bromage H., Pan Z. Q., Lee S. H., Tempst P., Hurwitz J. Mapping of amino acid residues in the p34 subunit of human single-stranded DNA-binding protein phosphorylated by DNA-dependent protein kinase and Cdc2 kinase in vitro. J. Biol. Chem. 1997;272:12634–12641. doi: 10.1074/jbc.272.19.12634. [DOI] [PubMed] [Google Scholar]
- 38.Carty M. P., Zernik-Kobak M., McGrath S., Dixon K. UV light-induced DNA synthesis arrest in HeLa cells is associated with changes in phosphorylation of human single-stranded DNA-binding protein. EMBO J. 1994;13:2114–2123. doi: 10.1002/j.1460-2075.1994.tb06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olson E., Nievera C. J., Klimovich V., Fanning E., Wu X. RPA2 is a direct downstream target for ATR to regulate the S-phase checkpoint. J. Biol. Chem. 2006;281:39517–39533. doi: 10.1074/jbc.M605121200. [DOI] [PubMed] [Google Scholar]
- 40.Shao R. G., Cao C. X., Zhang H., Kohn K. W., Wold M. S., Pommier Y. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 1999;18:1397–1406. doi: 10.1093/emboj/18.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morgan S. E., Kastan M. B. Dissociation of radiation-induced phosphorylation of replication protein A from the S-phase checkpoint. Cancer Res. 1997;57:3386–3389. [PubMed] [Google Scholar]
- 42.Vassin V. M., Wold M. S., Borowiec J. A. Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol. Cell. Biol. 2004;24:1930–1943. doi: 10.1128/MCB.24.5.1930-1943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng J., Wakeman T., Yong S., Wu X., Kornbluth S., Wang X. F. Protein phosphatase 2A-dependent dephosphorylation of replication protein A is required for the repair of DNA breaks induced by replication stress. Mol. Cell. Biol. 2009;29:5696–5709. doi: 10.1128/MCB.00191-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee D. H., Pan Y., Kanner S., Sung P., Borowiec J. A., Chowdhury D. A PP4 phosphatase complex dephosphorylates RPA2 to facilitate DNA repair via homologous recombination. Nat. Struct. Mol. Biol. 2010;17:365–372. doi: 10.1038/nsmb.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi Q. M., Wang Y. M., Zheng X. D., Lee R. T., Wang Y. Critical role of DNA checkpoints in mediating genotoxic-stress-induced filamentous growth in Candida albicans. Mol. Biol. Cell. 2007;18:815–826. doi: 10.1091/mbc.E06-05-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang H., Gao J., Li W., Wong A. H., Hu K., Chen K., Wang Y., Sang J. Pph3 dephosphorylation of Rad53 is required for cell recovery from MMS-induced DNA damage in Candida albicans. PLoS ONE. 2012;7:e37246. doi: 10.1371/journal.pone.0037246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patrick S. M., Oakley G. G., Dixon K., Turchi J. J. DNA damage induced hyperphosphorylation of replication protein A.2. Characterization of DNA binding activity, protein interactions, and activity in DNA replication and repair. Biochemistry. 2005;44:8438–8448. doi: 10.1021/bi048057b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brush G. S., Morrow D. M., Hieter P., Kelly T. J. The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc. Natl. Acad. Sci. U.S.A. 1996;93:15075–15080. doi: 10.1073/pnas.93.26.15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clifford D. M., Marinco S. M., Brush G. S. The meiosis-specific protein kinase Ime2 directs phosphorylation of replication protein A. J. Biol. Chem. 2004;279:6163–6170. doi: 10.1074/jbc.M306943200. [DOI] [PubMed] [Google Scholar]
- 50.Sawarynski K. E., Kaplun A., Tzivion G., Brush G. S. Distinct activities of the related protein kinases Cdk1 and Ime2. Biochim. Biophys. Acta. 2007;1773:450–456. doi: 10.1016/j.bbamcr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.