Background: Prion proteins adopt different conformations, known as variants, each with a distinct phenotype.
Results: Deletion of specific chaperone genes (HSC82, AHA1, CPR6, CPR7, SBA1, TAH1, SSE1) alters established [PIN+] variants in S. cerevisiae.
Conclusion: Chaperone proteins have a role in determining prion variants.
Significance: Chaperone activity helps to regulate cell prion phenotype.
Keywords: Amyloid, Epigenetics, Hsp90, Molecular Chaperone, Prions, Yeast, Rnq1, Sup35, Variant, Cyclophilin
Abstract
Prions are proteins that can adopt different infectious conformations known as “strains” or “variants,” each with a distinct, epigenetically inheritable phenotype. Mechanisms by which prion variants are determined remain unclear. Here we use the Saccharomyces cerevisiae prion Rnq1p/[PIN+] as a model to investigate the effects of chaperone proteins upon prion variant determination. We show that deletion of specific chaperone genes alters [PIN+] variant phenotypes, including [PSI+] induction efficiency, Rnq1p aggregate morphology/size and variant dominance. Mating assays demonstrate that gene deletion-induced phenotypic changes are stably inherited in a non-Mendelian manner even after restoration of the deleted gene, confirming that they are due to a bona fide change in the [PIN+] variant. Together, our results demonstrate a role for chaperones in regulating the prion variant complement of a cell.
Introduction
The mammalian prion protein, PrP, was originally identified as the causative agent of a group of neurodegenerative disorders collectively known as the transmissible spongiform encephalopathies (1). When the prion-determining domain (PrD) of PrP misfolds, PrP can switch from a non-infectious conformation (PrPC) to a conformation prone to forming self-propagating, β-sheet-rich, amyloid polymers (PrPSc) (1, 2). These amyloids spread by catalyzing the transformation of PrPC into PrPSc, eventually forming large aggregates (for review, see Ref. 3).
Although genetic polymorphisms of the PrP gene do affect its disease pathology (4, 5), distinct sets of symptoms arising from genetically identical PrPSc, called strains, have been described (6, 7). Protease treatment of different strains of PrPSc aggregates revealed that they differed by the size and composition of their amyloid core region, suggesting that it is specific conformations of the amyloid that give rise to PrPSc strains (8).
Prions have also been characterized in non-mammalian model organisms, including bakers' yeast, Saccharomyces cerevisiae, where they have been demonstrated to occur frequently in the wild (9–13). Yeast prions can also form different strains, called variants (14–16). Studies have shown that different prion conformations distinguish each prion variant (17–19).
The prion protein Rnq1p can adopt different variants, each with a distinct phenotype. Although the function of its non-prion conformation remains unknown, the prion conformation of Rnq1p, [PIN+], acts to help another yeast prion, Sup35p/[PSI+], adopt its own prion state (20, 21). Rnq1p has a prion-determining domain between amino acids 153 and 405, with a non-prion domain N terminus (11). Some [PIN+] variants include [PIN+]low, [PIN+]medium, and [PIN+]high, each named for its respective [PSI+] induction efficiency (16). Another [PIN+] variant-linked phenotype is observed when GFP-tagged Rnq1p is overproduced in vivo (22). [PIN+]high strains form multiple Rnq1-GFP foci per cell, whereas [PIN+]medium and [PIN+]low strains generally form only a single Rnq1-GFP focus. Another phenotypic difference between [PIN+] variants is the size and stability of their amyloid aggregates. [PIN+]high contains less stable aggregates that break down into smaller subparticles when heated, as opposed to [PIN+]low or [PIN+]medium aggregates, which remain stable when heated (16, 23, 24). When two [PIN+] variants are introduced into the same cell, either through mating or cytoduction, the diploid and all haploid progeny adopt the phenotype of the dominant variant. [PIN+]high is dominant over [PIN+]medium, which is in turn dominant over [PIN+]low (16). These multiple, distinct phenotypes make [PIN+] an ideal model system with which to study the etiology of prion variants.
Mutations in a prion protein can affect its amyloid structure and, through that, the type of variant it adopts (25, 26). Still, variants can arise from prion proteins with identical sequences (14–16, 27), suggesting that other cellular factors may influence the variant conformation that a given prion will adopt. Chaperone proteins are strong candidates for such variant regulating factors. Chaperones are known to affect the conformation of a wide array of client proteins (for review, see Ref. 28) and have been implicated in other aspects of prion biology, including the de novo formation, propagation, and curing of prions (for review, see Ref. 29–31). Additionally, changes to chaperone activity and levels have been shown to affect prion variants. Yeast strains expressing N- and C-terminal truncations of the primary stress response transcriptional regulator, Hsf1p, which were shown to increase Hsp104p and decrease Hsp90 levels, respectively, preferentially formed specific [PSI+] variants upon de novo induction (32). Likewise, strains over- or underexpressing SSE1, which is important to Hsp70 activity, gave rise to specific [PSI+] variants when induced (33). To date, only one genetic mutation has been reported to lead to a change in a pre-existing variant. Sondheimer et al. (27) demonstrated that deletion of the Sis1p G/F domain altered Rnq1-GFP aggregation pattern in a manner stably propagated even after reintroduction of wild-type Sis1p.
Here, we report the findings of our investigations into the actions of chaperone proteins upon already established [PIN+] variants. We found that disruption of several chaperone genes gives rise to shifts in [PIN+] variant-linked phenotypes. Genetic analysis showed that the phenotypic shifts are inherited in a non-Mendelian manner, confirming that a bona fide change in [PIN+] variant was achieved. Our findings provide evidence that chaperones can affect established prion variants and highlight a potential role for chaperones in regulating prion-linked phenotypes through their modulation of prion variants.
EXPERIMENTAL PROCEDURES
Yeast Strains, Culture, and Genetic Manipulation
S. cerevisiae strains used in this study are listed in supplemental Table S1. All yeast strains were cultured at 30 °C with the exception of diploids undergoing sporulation, which were cultured at 25 °C. Media were as follows: YEPD (1% yeast extract, 2% peptone, 2% glucose); CSM (0.67% yeast nitrogen base without amino acids, 2% glucose, 1 × Complete supplement mixture (Bio 101, Vista, CA)); CSM auxotrophic marker growth medium (same as CSM but containing 1 × CSM minus the appropriate auxotrophic selection (−ADE, −HIS, −LEU, −URA, −LYS-URA, −TRP-URA)); CSM auxotrophic marker induction medium (same as CSM but with 2% galactose in place of 2% glucose). For solid media, the same recipes were used with 2% agar added. Deletions were made using a HIS3 cassette as described (34) and confirmed by PCR. Isogenic MATα strains were made using YCpGAL::HO and mating-type switching (35). Mating, diploid sporulation, and dissection of haploids were performed as previously described with modified sporulation medium (0.3% potassium acetate, 0.02% raffinose) (36). Haploid progeny of dissections were tested for the presence of a gene deletion by culture on CSM-HIS plates.
Yeast Plasmids and Cloning
Plasmids used in this study were constructed as follows; pYES2.0-SUP35NM-GFP was constructed by ligating the sequence encoding enhanced GFP (37) in-frame with the 3′-end of base pairs 1–762 of the SUP35 gene into pYES2.0 (Invitrogen). pYES2.0-GFP was similarly constructed with only the GFP-encoding sequence. pYES2.0-Rnq1-GFP was constructed by amplifying sequence coding for Rnq1p C-terminally tagged with GFP from genomic DNA of a commercially available GFP-tagged library strain (Invitrogen) and ligating it into pYES2.0. pGREG535-Rnq1 was constructed by amplifying RNQ1 and inserting it into pGREG535 according to the Drag & Drop protocol (38). Plasmids were verified by sequencing. Escherichia coli and yeast were transformed with plasmid using standard chemical transformation protocols (34, 39).
Assay for Nonsense Suppression
To quantify [PSI+] induction efficiency based upon nonsense suppression, Sup35NM-GFP or GFP alone was overproduced by culturing strains harboring pYES2.0-SUP35NM-GFP or pYES2.0-GFP, respectively, in liquid CSM medium for 24 h followed by subculturing in liquid CSM induction medium for 48 h. When testing primary deletions and diploids, cell cultures were normalized to an A600 of 1.0, and when testing tetrads, cultures were normalized to an A600 of 0.25. After normalization, 10-fold serial dilutions were made. 5 μl of each dilution were spotted onto both CSM and CSM−ADE plates, and colony-forming units (cfu) were counted after 2 and 4 days, respectively. Percent [PSI+] induction efficiency was calculated as cfu (CSM−ADE)/cfu (CSM) × 100. Four to six independent experiments were performed for each strain.
Assay for Plasmid Retention
To determine if a given strain retained pYES2.0-SUP35NM-GFP, induced strains were plated for single colonies on both CSM and CSM−ADE, and colonies from both platings were then replica-plated onto CSM and CSM-URA media. Percent plasmid retention was calculated as cfu (CSM-URA)/cfu (CSM) × 100. Between 100 and 200 colonies were compared for each strain.
Assay for Prion Curing
To test induced [PSI+] strains for curability, colonies growing on CSM−ADE medium plates were streaked onto curing plates (YEPD + 3 mm guanidine HCl) and allowed to grow at 30 °C for 3 days. Putatively “cured” colonies were then selected based on their pigmentation, restreaked onto CSM and CSM−ADE plates, and allowed to grow at 30 °C for 2 and 4 days, respectively. At least four colonies were tested for each strain.
Characterization of Rnq1-GFP Aggregates in Vivo
Strains carrying pYES2.0-Rnq1-GFP were cultured for 24 h in liquid CSM growth medium then subcultured in liquid CSM induction medium for 24 h. The number of Rnq1-GFP foci per cell was then quantified by acquiring random wide-field micrographs of cells using an Olympus IX-80 fluorescence microscope. The frequency of multiple foci was expressed as a percentage of all cells containing multiple Rnq1-GFP foci. 800–1000 cells were counted and categorized over the course of 4 independent experiments.
Protein Analysis
Strains carrying pGREG535-Rnq1 were cultured for 24 h in liquid CSM growth medium, then subcultured in liquid CSM induction medium for 8 h. Protein samples were prepared and pelleted as described (24). The pellet fraction, enriched for insoluble HA-Rnq1p, was heated briefly at 55 °C and analyzed by semi-denaturing detergent-agarose gel electrophoresis (SDD-AGE)3 (24, 40). Blots were probed with anti-HA antibody (F-7 Sc7392, Santa Cruz Biotechnology, Santa Cruz, CA) and detected with HRP-conjugated anti-mouse IgG (GE Healthcare).
Yeast Two-hybrid Analysis
Yeast two-hybrid analysis was performed using the yeast strain HF7c and the plasmids pGAD424 (prey) and pGBT9 (bait) (Clontech, Mountain View, CA) following standard protocols (41). Genes encoding Rnq1p or chaperone proteins were ligated in-frame into pGAD424 and pGBT9 for expression. Interactions were scored by growth of cells on CSM-HIS medium after 4 days of incubation relative to growth on YEPD. At least three independent experiments were done for each tested pair of proteins.
RESULTS
Deletion of Chaperone Genes Affects [PSI+] Induction Efficiency in a [PIN+] Variant-dependent Manner
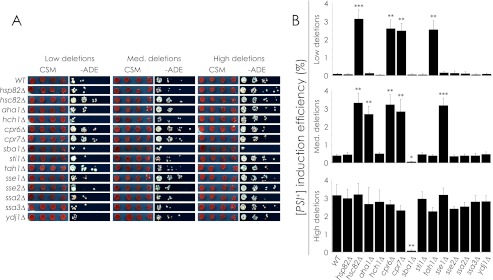
To investigate the role of chaperones in [PIN+] variant determination, we made deletions of several chaperone-encoding genes in strains carrying characterized [PIN+] variants and measured changes in [PSI+] induction efficiency. Each strain was [psi−] and carried a nonsense-suppression reporter gene (ADE1–14 UGA) that allowed growth on CSM−ADE medium only when [PSI+] was induced (Fig. 1A). We measured the relative [PSI+] induction efficiencies of deletion strains using a nonsense suppression assay (Fig. 1B) as described under “Experimental Procedures.” In brief, we overproduced Sup35NM-GFP using the galactose-driven expression vector pYES2.0-SUP35NM-GFP and quantified cfu on CSM−ADE medium plates relative to cfu on CSM medium plates. The strength of the induced [PSI+] variants was not factored into our calculations of [PSI+] induction efficiency; only the number of colonies and not their size or pigmentation was considered.
FIGURE 1.
Effect of chaperone gene deletion on [PSI+] induction efficiency in relation to [PIN+] variants. A, shown is a nonsense suppression assay. Chaperone gene deletions were made in [psi−] [PIN+]low/med/high strains that were induced to become [PSI+] by overproduction of Sup35NM-GFP. Normal growth is shown on CSM medium, and [PSI+]-dependent growth is shown on −ADE medium. B, [PSI+] induction efficiency is shown. The efficiency of [PSI+] induction was determined by quantification of the nonsense suppression assay, expressing average −ADE cfu as a percentage of average CSM cfu. Error bars represent S.E. Student's t tests were done to compare the [PSI+] induction efficiency of deletion strains with that of their parental wild-type strain. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
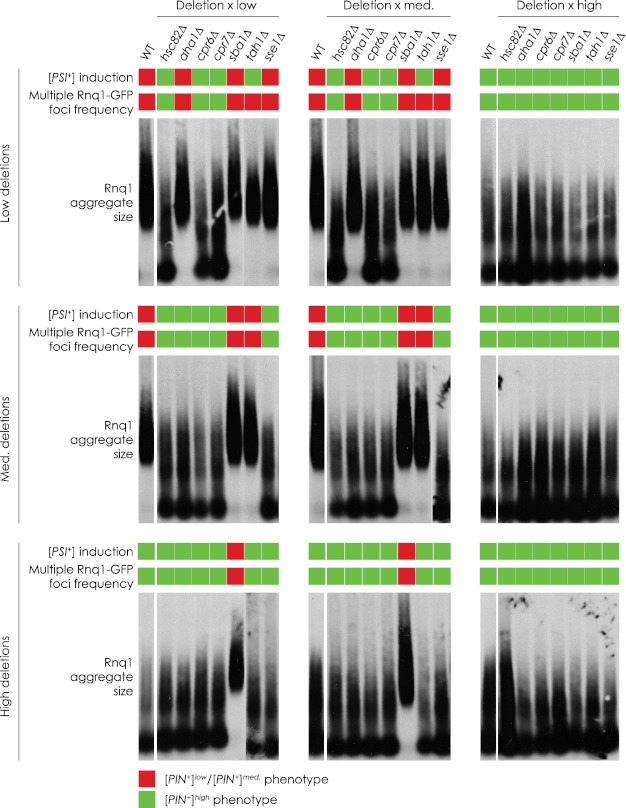
Deletion of HSC82, CPR6, CPR7, or TAH1 in the [PIN+]low strain as well as HSC82, AHA1, CPR6, CPR7, or SSE1 in the [PIN+]medium strain increased strain growth on CSM−ADE medium to levels significantly higher than those of wild-type [PIN+]low and wild-type [PIN+]medium strains and similar to those of the wild-type [PIN+]high strain. Deletion of these same genes in the [PIN+]high strain did not affect [PSI+] induction efficiency. Conversely, deletion of SBA1 in the [PIN+]medium and [PIN+]high backgrounds decreased the efficiency of [PSI+] induction to levels matching those of the wild-type [PIN+]low strain but did not significantly decrease the efficiency of [PSI+] induction when deleted in the [PIN+]low strain. Nearly all strains gave rise to colonies with variable levels of pigment ranging from pink to almost white (Fig. 1A). The only exception was the [PIN+]low strain deleted for TAH1 in which colonies were almost entirely white after induction, suggesting that the induced [PSI+] colonies all carry a strong [PSI+] variant.
ADE-competent colonies were confirmed to be prion-linked because they lost ADE competence after treatment with guanidine HCl (supplemental Fig. S1). Also, the extent of [PSI+] induction in a strain being dependent on the levels of plasmid retention by that strain was excluded as a possibility, as both wild-type and deletion strains retained plasmid at levels between 90 and 95% whether or not the cells were ADE-competent (data not shown).
Deletion of Chaperone Genes Affects the Aggregation of Rnq1p in a [PIN+] Variant-dependent Manner
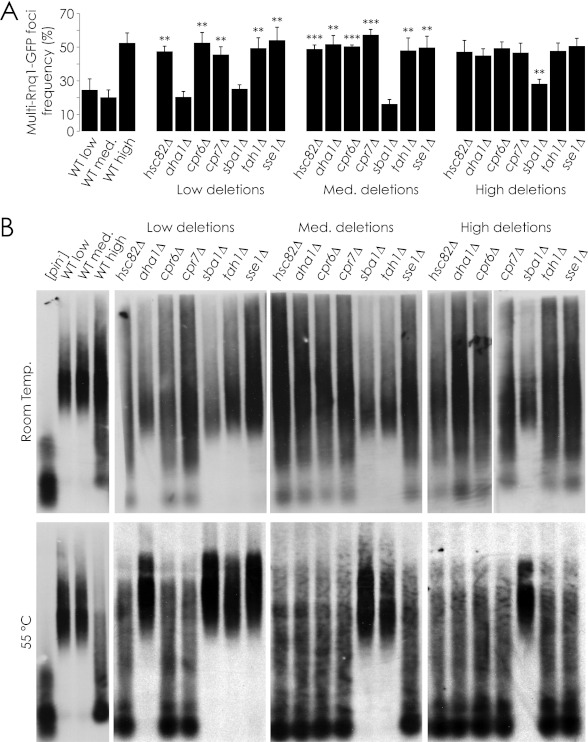
Our results showing that the effects of gene deletion were [PIN+] variant-specific and resulted in shifts in [PSI+] induction efficiency from levels found in one wild-type variant strain, e.g. [PIN+]low, to a different wild-type variant strain, e.g. [PIN+]high, suggested that the chaperone gene deletions may have given rise to a change in the [PIN+] variant carried by the cell. If the [PIN+] variant of a strain had been altered by deletion of a chaperone gene, then other variant-specific phenotypes should also have been affected. Accordingly, we characterized the effects of chaperone gene deletion upon the phenotype of Rnq1p aggregates.
We first quantified the localization of Rnq1-GFP in all the deletion strains affected in [PSI+] induction efficiency (Fig. 2A). When GFP-tagged Rnq1p was overproduced from the galactose-driven plasmid pYES2.0-Rnq1-GFP, ∼75% of focus-containing wild-type [PIN+]low and wild-type [PIN+]medium cells contained a single Rnq1-GFP focus, whereas ∼25% contained more than one focus. ∼50% of wild-type [PIN+]high focus-containing cells contained multiple foci, in agreement with previous findings (22). [PIN+]low and [PIN+]medium strains deleted for the genes HSC82, CPR6, CPR7, TAH1, or SSE1 that gave rise to increased [PSI+] induction also displayed multiple Rnq1-GFP foci at the frequency displayed by the wild-type [PIN+]high strain. Deletion of AHA1 in the [PIN+]medium strain but not in the [PIN+]low strain led to increased [PSI+] induction (Fig. 1B). Likewise, deletion of AHA1 in the [PIN+]medium strain but not in the [PIN+]low strain led to an increase in multiple Rnq1-GFP foci to the frequency exhibited by the wild-type [PIN+]high strain. In contrast, deletion of the SBA1 gene from the [PIN+]high strain led to fewer cells with multiple Rnq1-GFP foci, consistent with the frequencies exhibited by wild-type [PIN+]low and wild-type [PIN+]medium strains. Interestingly, increases in the number of multiple Rnq1-GFP foci were also observed when TAH1 and SSE1 were deleted in [PIN+]medium and [PIN+]low strains, respectively, even though there was no observed increase in [PSI+] induction efficiency in these deletion strains.
FIGURE 2.
Effect of chaperone gene deletion on [PIN+] variant-dependent phenotypes. A, Rnq1-GFP was overproduced in deletion strains of interest, and the frequency of multiple Rnq1-GFP foci in foci-containing cells was calculated as a percentage of total cells. Error bars represent S.E. Student's t tests were performed to compare the frequency of multiple Rnq1-GFP foci in cells containing foci with that of their parental wild-type strain. **, p < 0.01; ***, p < 0.001. B, the Rnq1p aggregate size in deletion strains of interest was compared by SDD-AGE. Samples were incubated at room temperature (top) or 55 °C (bottom) before loading.
We next characterized the sizes of Rnq1p amyloid subparticles in the deletion strains. The sizes of Rnq1p amyloid subparticles have been shown to vary depending on whether the [PIN+] variant displays a single Rnq1-GFP focus per cell, as with the wild-type [PIN+]low and wild-type [PIN+]medium strains, or multiple foci per cell as observed in the wild-type [PIN+]high strain (23). Strains with a single focus display a relatively narrow range of large subparticles, whereas strains with multiple foci display subparticles ranging in size from monomer to as large as, or larger than, those found in strains with a single focus. Accordingly, we produced HA-tagged Rnq1p from the plasmid pGREG535-Rnq1 in the wild-type [PIN+]low, [PIN+]medium, and [PIN+]high strains and in these strains harboring a gene deletion. We then prepared samples enriched for proteins in an insoluble prion state and incubated them at room temperature and 55 °C, as Rnq1p aggregates purified from strains with multiple Rnq1p foci, specifically [PIN+]high, have been shown to be more sensitive to changes in temperature and to degrade partially at moderately elevated temperatures, whereas other Rnq1p variant aggregates do not (24). Finally, we compared the sizes of the purified HA-Rnq1p amyloid aggregates using SDD-AGE (Fig. 2B). We found that deleting SBA1 in the wild-type [PIN+]high strain eliminated small HA-Rnq1p amyloid aggregates, leaving a narrower range of larger aggregates, consistent with the patterns observed in the wild-type [PIN+]low and wild-type [PIN+]medium strains. Conversely, we observed that hsc82Δ, aha1Δ, cpr6Δ, cpr7Δ, and sse1Δ strains, which increased the efficiency of [PSI+] induction in [PIN+]low and/or [PIN+]medium backgrounds to levels observed in the wild-type [PIN+]high strain, exhibited amyloid aggregates with a broader range of sizes, consistent with aggregates from the wild-type [PIN+]high strain. These aggregates, like wild-type [PIN+]high aggregates, also proved to be sensitive to temperature, degrading in part to a monomer at 55 °C (Fig. 2B). It is important to note that the pattern of protein migration displayed by these partly degraded protein aggregates is easily distinguished from that produced by protein aggregates isolated from a [pin−] strain. [pin−] aggregates were monomeric and of low molecular weight, whereas partially degraded aggregates from [PIN+] strains still displayed species of high molecular weight. Our results suggest that deletion of specific chaperone genes leads to changes in the prion amyloid physical properties, thereby affecting its heat sensitivity.
Our results show a correlation between chaperone gene deletion strains displaying altered [PSI+] induction efficiency and those displaying changes in other [PIN+]-linked phenotypes. The tah1Δ strains were exceptions. They were unaffected in the sizes of their HA-Rnq1p aggregates vis à vis the sizes of the aggregates in their corresponding wild-type [PIN+] variant background. Also of note is that although the sse1Δ strain in the [PIN+]low background showed multiple Rnq1-GFP foci per cell, its aggregates were limited to a narrow range of large sizes.
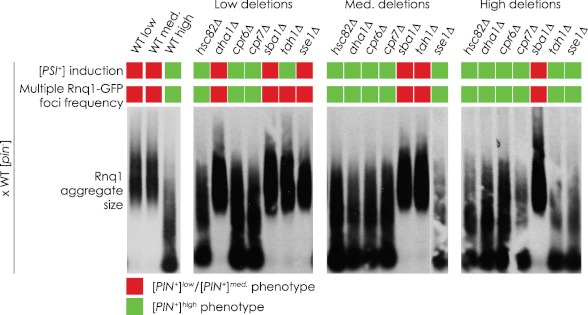
Deletion-induced Phenotypes Are Inherited in a Non-Mendelian Manner
The [PIN+] variant-linked phenotypic changes we observed in strains deleted for chaperone genes could be due directly to the effects of gene deletion or could be due simply to changes in the cell chaperone complement and to the overproduction of tagged protein resulting from our methodology. For example, loss of a chaperone could impair the ability of the cell to deal with the overexpression of tagged Rnq1p, leading to formation of denatured, non-prion aggregates that are detectable in our assays as multiple Rnq1-GFP foci and/or low molecular weight aggregates in SDD-AGE. To eliminate this trivial explanation and to confirm that it is indeed deletion of the chaperone genes that gives rise to the observed [PIN+] variant shifts, we reintroduced a wild-type copy of a deleted chaperone gene by crossing a deletion strain with an isogenic wild-type strain. This wild-type strain was [pin−] so as to avoid any convolution related to introducing dominant [PIN+] variants (16). If the effects of deletions were specific to the deletion of the gene, then restoring the chaperone gene would be expected to restore the original [PIN+] variant phenotype. If on the other hand a permanent change in the [PIN+] variant had occurred in the deletion strain, then the phenotypic changes should persist after reintroduction of the chaperone gene. Fig. 3 shows that, for the most part, [PSI+] induction efficiencies, the frequency of Rnq1-GFP foci, and the sizes of Rnq1p aggregates of diploid strains did not revert to the phenotypes of the original wild-type strains from which the deletion strains were derived. Instead, these diploid strains maintained the phenotypes of the deletion strains. The only exceptions were diploids arising from matings with sse1Δ in the [PIN+]low background and with tah1Δ in both the [PIN+]low and [PIN+]medium backgrounds. The increased frequency of Rnq1-GFP foci in the original haploid strains deleted for SSE1 or TAH1 was eliminated in the diploid strains, suggesting that these changes were [PIN+]-independent. It should be noted that under our experimental conditions we were unable to statistically distinguish [PIN+]low from [PIN+]medium based upon [PSI+] induction efficiencies (supplemental Fig. S2). As such, we could categorize our results only as being consistent with either [PIN+]low/medium or [PIN+]high levels.
FIGURE 3.
Maintenance of chaperone gene deletion-induced changes in [PIN+] variant-dependent phenotypes. Chaperone deletion strains were mated with a wild-type [psi−][pin−] strain, and the resulting diploids were analyzed for [PSI+] induction efficiency, frequency of multiple Rnq1-GFP foci in cells containing foci, and Rnq1p aggregate size. These phenotypes were then categorized as either [PIN+]low/medium or [PIN+]high. Quantifications of [PSI+] induction efficiency and frequency of multiple Rnq1-GFP foci in foci-containing cells can be found in supplemental Fig. S2.
There was the remote possibility that spurious mutations had been introduced into our deletion strains and had produced dominant effects mimicking an apparent variant switch. If this were the case, it would be expected that this trait would be co-inherited in a Mendelian manner as opposed to the non-Mendelian manner of a true change in variant. To eliminate the possibility of introduced spurious mutations, we sporulated and dissected our diploid strains, yielding wild-type and deletion-carrying haploid progeny. The presence of the chaperone gene deletion cassette (HIS3) was detected by growth on CSM-HIS medium and showed a 2:2 ratio in the progeny as expected (data not shown). We then measured [PSI+] induction efficiency of the dissected tetrads by nonsense suppression assay (Fig. 4). The specificity of this assay was demonstrated by mating wild-type strains carrying different [PIN+] variants with a strain deleted for HSP104, which is required for both [PSI+] and [PIN+] maintenance (20, 42). The wild-type haploid progeny were inducible at levels similar to their parental strains, whereas the hsp104Δ haploids were unable to be induced. When the chaperone gene deletion strains were mated with the wild-type [pin−] strain, we found that the haploid progeny of 24 tetrads all maintained the same [PSI+] induction efficiency as the parental deletion strain regardless of the presence or absence of the chaperone gene of interest. These levels were consistent with either [PIN+]low/medium or [PIN+]high levels. Haploid progeny were found to retain pYES2.0-Sup35NM-GFP at a rate consistent with the parental strains, with no strain losing or retaining the plasmid at a markedly higher level than any other strain. Also, putative [PSI+] colonies that arose after [PSI+] induction were shown to be curable by growth on medium containing guanidine HCl (supplemental Fig. S3).
FIGURE 4.
Non-Mendelian inheritance of chaperone gene deletion-induced changes in [PSI+] induction efficiency. Diploids generated by mating chaperone deletion stains with a wild-type [psi−][pin−] strain were sporulated and dissected. The resulting tetrads were analyzed for [PSI+] induction efficiency by nonsense suppression assay on −ADE medium. Wild-type strains were mated with hsp104Δ strains to demonstrate genetic specificity for this assay. Wild-type (+) and deletion-carrying (−) haploid progeny are presented. Twenty-four tetrads were analyzed per diploid.
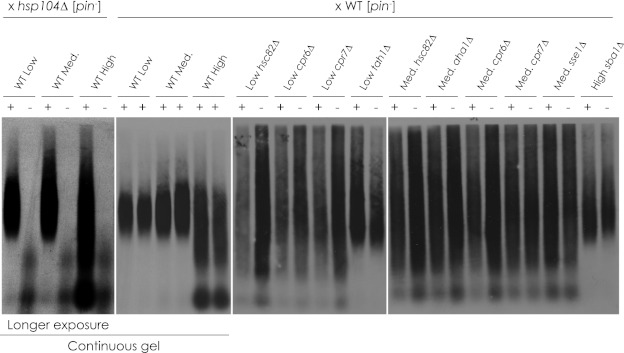
The Rnq1p aggregates of wild-type and mutant haploid progeny derived from the mating of deletion strains that gave rise to [PIN+] variant-related phenotypic changes were characterized using SDD-AGE (Fig. 5). We found that the deletion-induced changes in aggregate size were stable even after tetrad dissection. Deletion strains that exhibited no change in aggregate size were found to be consistent with the wild-type strain in which they were made (data not shown). Taken altogether, our results show that deletion of chaperone genes gives rise to bona fide changes in [PIN+] variant.
FIGURE 5.
Non-Mendelian inheritance of chaperone gene deletion-induced changes in HA-Rnq1p aggregate size. Diploids generated by mating chaperone deletion stains with a wild-type [psi−][pin−] strain were sporulated and dissected. The resulting tetrads were analyzed for HA-Rnq1p aggregate size by SDD-AGE. Wild-type strains were mated with hsp104Δ strains to demonstrate genetic specificity for this assay. Wild-type (+) and deletion-carrying (−) haploid progeny are presented. The hsp104Δ samples and wild-type controls were run on a continuous gel and exposed for a longer period. Twenty-four tetrads were analyzed per diploid.
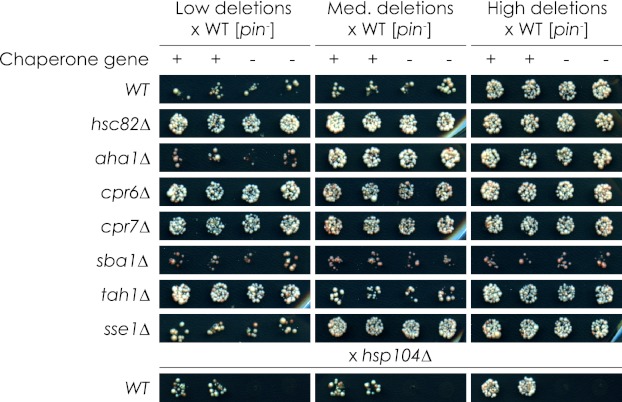
The Hierarchy of Inheritance of Induced Variants Is Consistent with Characterized [PIN+] Variants
Another characteristic of prion variants is that one is often dominant over another. In the case of [PIN+] variants, [PIN+]high is dominant over [PIN+]medium, which in turn is dominant over [PIN+]low (16). In most cases, deletion-induced variants display phenotypes comparable with [PIN+]high, with the exception of sba1Δ, which gives rise to a variant similar to [PIN+]low/medium. Still, it remained unclear if these variants were actually [PIN+]high, [PIN+]low/medium, or novel, previously uncharacterized types of variants. If these variants were not novel, then they would be expected to follow the same hierarchy of variant dominance in relation to other [PIN+] variants. To determine the hierarchy of the induced variants, we crossed deletion strains to isogenic wild-type strains carrying [PIN+]low, [PIN+]medium, or [PIN+]high. We then documented the [PIN+] variant-related phenotypes of the resulting diploid strains (Fig. 6). As before, we were unable to distinguish statistically [PIN+]low from [PIN+]medium based on their [PSI+] induction efficiencies (supplemental Fig. S4).
FIGURE 6.
Dominance of chaperone gene deletion-induced variants. Diploids were generated by mating chaperone deletion stains with a wild-type [psi−] strain carrying the [PIN+]low or [PIN+]medium or [PIN+]high variant. These diploids were analyzed for [PSI+] induction efficiency, frequency of multiple Rnq1-GFP foci in foci-containing cells, and Rnq1p aggregate size. These phenotypes were then categorized as either [PIN+]low/medium or [PIN+]high. Quantification of [PSI+] induction efficiency and frequency of multiple Rnq1-GFP foci in foci-containing cells can be found in supplemental Fig. S4.
We found that diploids generated by mating putative [PIN+]high strains with any wild-type strain exhibited phenotypes consistent with wild-type [PIN+]high. Also, the presumed [PIN+]low/medium variant induced by the deletion of SBA1 was shown to be eliminated by the introduction of the more dominant [PIN+]high. In the case of tah1Δ generated in [PIN+]low, its [PIN+]high-like [PSI+] induction efficiency was dominant over [PIN+]low/medium wild-type phenotypes, whereas wild-type [PIN+]high was dominant over its other [PIN+]-linked phenotypes. Finally, the frequency of [PIN+]high-like Rnq1-GFP foci observed when SSE1 was deleted in a [PIN+]low strain reverted to a [PIN+]low/med-like phenotype when mated with a wild-type [PIN+]low strain or a wild-type [PIN+]medium strain. All other traits followed wild-type patterns of inheritance.
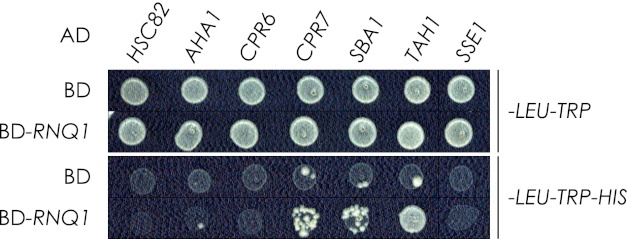
Rnq1p Interacts Physically with Chaperone Proteins
We performed a yeast two-hybrid analysis to investigate possible physical interactions between our chaperones of interest and Rnq1p (Fig. 7). With growth on CSM-HIS medium as a reporter of interaction, we detected a previously documented interaction between Rnq1p and Tah1p (43). We also identified two novel interactions: Cpr7p and Sba1p with Rnq1p. No interaction was detected between the remaining chaperones and Rnq1p. When we characterized the [PIN+] variant of our yeast two-hybrid strain (HF7c) by SDD-AGE and localization of Rnq1-GFP (supplemental Fig. S5), we found HF7c to be [pin−], suggesting that the interactions we detected occur when Rnq1p is in its non-prion conformation.
FIGURE 7.
Yeast two-hybrid analysis. Cpr7p, Sba1p, and Tah1p were found to interact with Rnq1p based on growth of yeast on selective CSM-HIS medium. Total growth (top) and growth arising from protein interaction (bottom) are shown. The pattern presented is representative of four independent experiments. AD, activation domain; BD, binding domain.
DISCUSSION
Although recombinant S. cerevisiae prion proteins do not require cofactors to misfold into multiple infectious variants in vitro, they need the activity of different chaperone proteins to propagate stably in vivo. Most prominent among these chaperones are Hsp104p, Sis1p, and members of the Ssa subfamily (for review, see Refs. 29–31). Together these chaperones facilitate the fragmentation of growing amyloid fibrils, thereby exposing more fibril growing ends and generating more infectious prion seeds. It has been proposed that conformational differences between variants alter their susceptibility to fragmentation and their rate of fibril growth and that equilibrium between these processes determines the stability of prion propagation and the strength of prion phenotype (18, 44, 45). For example, compared with [PSI+]weak, [PSI+]strong has a smaller amyloid core that fragments more easily, allowing for the generation of more growing ends and a higher rate of Sup35p incorporation into a greater number of prion seeds (18). These seeds in turn lead to increased nonsense suppression and more stable propagation of [PSI+]. Chaperones have also been implicated in variant determination, with alteration of Hsf1p or Sse1p activity affecting the de novo induction of the [PSI+] variant and truncation of Sis1p leading to stable changes in the established [PIN+] variant (27, 32, 33).
Our study provides further evidence that chaperones are important for the selection of prion variants. Deletion of AHA1 in a [PIN+]medium background or deletion of HSC82, CPR6, or CPR7 in either a [PIN+]low or a [PIN+]medium background increases the efficiency of [PSI+] induction to levels comparable to those seen for the wild-type [PIN+]high strain. Conversely, deletion of SBA1 in the [PIN+]high background decreases the efficiency of [PSI+] induction to wild-type [PIN+]low/medium levels.
Could the effects of deleting these chaperone-encoding genes on [PSI+] induction be independent of [PIN+]? For example, the [PIN+] variant-specific effects on [PSI+] induction could be explained by different chaperone requirements for the de novo formation of [PSI+] in the presence of different [PIN+] variants. This scenario is not without precedent, as a [PSI+] variant with exceptionally large aggregates was shown to require increased Hsp104p levels to propagate stably (46). However, such a scenario is unlikely in our case, because the changes we observed in [PSI+] induction in our chaperone gene deletion strains were also accompanied by changes in the size and localization pattern of Rnq1p, consistent with a shift in [PIN+] variant. Additionally, these phenotypic shifts persist after sporulation of diploids into wild-type and deletion-carrying haploid progeny upon reintroduction of the deleted chaperone gene. Together, our results demonstrate that the chaperone gene deletion-induced phenotypes we observed are due to stable shifts in the [PIN+] variant.
Like other chaperone gene deletions, deletion of TAH1 or SSE1 in the [PIN+]low or [PIN+]medium backgrounds gave rise to changes in [PIN+] variant-dependent phenotypes. However, in contrast to what was observed for the other chaperone gene deletion strains, not all of the tested phenotypes were altered in TAH1 or SSE1 gene deletion strains, and of those phenotypes that were altered, not all were maintained after the wild-type gene was restored. This suggests that at least some of the changes observed in the tah1Δ and sse1Δ strains are not the result of shifts in the [PIN+] variant and that any putative variant shift that may have occurred in these deletion strains does not correspond to a previously characterized variant (16, 22). In contrast, variants arising from deletion of HSC82, AHA1, CPR6, or CPR7 are consistent with [PIN+]high, whereas variants in sba1Δ strains match [PIN+]low or [PIN+]medium. These deletion-induced variants may eventually be shown to be distinct from classical [PIN+] variants but for the purpose of discussion are referred to here as [PIN+]high and [PIN+]low/medium, respectively.
[PIN+]high phenotypes were readily identified, but [PIN+]low and [PIN+]medium were difficult to distinguish from each other. Their [PSI+] induction efficiencies did not vary greatly, and the properties of their aggregates were indistinguishable. As such, it is interesting that deletion of AHA1, TAH1, or SSE1 had specific effects depending upon whether they were deleted in [PIN+]low or [PIN+]medium backgrounds. This finding reinforces that [PIN+]low and [PIN+]medium are distinct variants and that these chaperone genes could serve as a genetic fingerprint for experimentally differentiating between the two variants in future studies.
By what mechanism might the variant changes we observed occur? Hsc82p, Aha1p, Cpr6p, Cpr7p, and Sba1p are known to act in the Hsp90 cycle (for review, see Ref. 47); Hsc82p is the constitutively expressed isomer of Hsp90; Aha1p, Cpr6p, and Cpr7p have been shown to increase Hsp90 ATP hydrolysis; Sba1p, the yeast homologue of p23, stabilizes the Hsp90-client complex, thereby decreasing Hsp90 ATPase activity (48–51). Our finding that deleting genes linked to increased Hsp90 ATPase activity led to a [PIN+]high phenotype whereas deleting SBA1 led to a [PIN+]low/medium phenotype suggests that Hsp90 activity is important for variant determination, with Rnq1p a possible Hsp90 client. This possibility is supported by the finding that a strain expressing a C-terminal truncation of Hsf1p and predisposed to form unstable [PSI+] also had markedly decreased Hsp90 levels (32).
We detected physical interactions of Rnq1p with Cpr7p, Sba1p, and Tah1p but not with the other chaperones. This suggests that some of these chaperones may act upon Rnq1p independently of Hsp90. Cpr7p, for example, has intrinsic proline isomerase activity (52, 53). Rnq1p contains three proline residues: one in the N-terminal region, a region shown to affect prion propagation (54), and two in a putative loop region between the β-sheets of the amyloid core (55). Isomerization of these prolines could potentially affect Rnq1p conformation.
Additionally, as numerous physical interactions between these chaperones and other chaperone complexes have been documented, changes in the levels of these chaperones may affect how Hsp40s, Hsp70s, and/or Hsp104p interact with Rnq1p, resulting in variant change. Cpr7p, for example, has been shown to interact with Hsp104p in a manner that is not essential for its thermotolerance activity (56, 57). Additionally, inhibition of Hsp90 ATPase activity has been shown to increase the levels of both Hsp104p and Hsp70 (58). The effect of SSE1 deletion that we report here also implicates Hsp70s in variant change, as SSE1 encodes an important Hsp70 nucleotide exchange factor (59). Fan et al. found that manipulation of Sse1p levels affected the [PSI+] variant (33), although our sse1Δ strain did not display the same disposition toward an unstable weak [PSI+] variant. The difference between our and their results could be due to [PIN+] variant-specific effects, as Fan et al. (33) did not characterize the [PIN+] variant of their strain. It is also interesting that it has been reported that mutations in Sis1p give rise to an increase in Rnq1-GFP foci (27), similar to what we observed. It may be that our gene deletions indirectly impaired the activity of Sis1p and/or its association with Rnq1p.
How the loss of specific chaperones alters the levels and activities of other chaperones, in addition to their interaction with Rnq1p, will be an important avenue of future investigation to clarify the mechanisms underlying the variant changes that we observed. Chaperones could mediate [PIN+] variant changes by altering the conformation of Rnq1p. For example, chaperones could regulate the folding of monomeric Rnq1p in such a way as to predispose it to adopt a specific variant upon de novo formation or upon encountering prion seeds. More likely, because we observed changes to established variants, chaperones could work together to affect the conformation of existing amyloid polymers. For this to be effective, only the growing ends of polymers need be remodeled. Alternatively, if multiple or unstable variants are present at the same time in the cell, as has been shown to occur for [PSI+] (60), changes in the chaperone environment could alter the rates of amyloid polymer fragmentation and/or prion seed generation in a variant-specific manner. In this way, one variant could be selected over others.
In closing, we have demonstrated that altering the chaperone complement of a cell can alter existing prion variants without the introduction of exogenous prion material. By modulating existing prion variants, the cell can maintain prion seeds within the cell while mitigating potential negative effects of stronger prion phenotypes. Also, when environmental pressures demand, the existing prion variant could be quickly altered to provide a more advantageous phenotype to the cell. In light of recent findings reporting the prevalence of prions in wild strains of yeast as well as the apparent survival advantages that they bestow (13), prion variant regulation represents a powerful mechanism for modulating a cell response and adaptability to changing environmental conditions.
Supplementary Material
Acknowledgments
We thank Susan Liebman (University of Illinois) for wild-type [PIN+] variant strains and Troy Locke (Microarray and Proteomics Facility, University of Alberta) for help in screening haploid progeny of genetic crosses. We also thank Richard Poirier, Hanna Kroliczak, Dwayne Weber, and Elena Savidov for technical assistance and Christopher Power, Paul Melançon, Paul LaPointe, Andrei Fagarasanu, Barbara Knoblach, Jinlan Chang, Robert Tower, Fred Mast, and Erin Brown for helpful discussion.

This article contains supplemental Table S1 and Figs. S1–S5.
- SDD-AGE
- semi-denaturing detergent-agarose gel electrophoresis.
REFERENCES
- 1. Prusiner S. (1982) Novel proteinaceous infectious particles cause scrapie. Science 216, 136–144 [DOI] [PubMed] [Google Scholar]
- 2. Pan K. M., Baldwin M., Nguyen J., Gasset M., Serban A., Groth D., Mehlhorn I., Huang Z., Fletterick R. J., Cohen F. E. (1993) Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. U.S.A. 90, 10962–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Colby D. W., Prusiner S. B. (2011) Prions. Cold Spring Harb. Perspect. Biol. 3, a006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown K., Mastrianni J. A. (2010) The prion diseases. J. Geriatr. Psychiatry Neurol. 23, 277–298 [DOI] [PubMed] [Google Scholar]
- 5. Ohhashi Y., Ito K., Toyama B. H., Weissman J. S., Tanaka M. (2010) Differences in prion strain conformations result from non-native interactions in a nucleus. Nat. Chem. Biol. 6, 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fraser H., Dickinson A. G. (1968) The sequential development of the brain lesion of scrapie in three strains of mice. J. Comp. Pathol. 78, 301–311 [DOI] [PubMed] [Google Scholar]
- 7. Bruce M. E. (1993) Scrapie strain variation and mutation. Br. Med. Bull. 49, 822–838 [DOI] [PubMed] [Google Scholar]
- 8. Bessen R. A., Marsh R. F. (1994) Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J. Virol. 68, 7859–7868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wickner R. B. (1994) [URE3] as an altered URE2 protein. Evidence for a prion analog in Saccharomyces cerevisiae. Science 264, 566–569 [DOI] [PubMed] [Google Scholar]
- 10. Paushkin S. V., Kushnirov V. V., Smirnov V. N., Ter-Avanesyan M. D. (1996) Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 15, 3127–3134 [PMC free article] [PubMed] [Google Scholar]
- 11. Sondheimer N., Lindquist S. (2000) Rnq1. An epigenetic modifier of protein function in yeast. Mol. Cell 5, 163–172 [DOI] [PubMed] [Google Scholar]
- 12. Alberti S., Halfmann R., King O., Kapila A., Lindquist S. (2009) A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137, 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Halfmann R., Jarosz D. F., Jones S. K., Chang A., Lancaster A. K., Lindquist S. (2012) Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature 482, 363–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Derkatch I. L., Chernoff Y. O., Kushnirov V. V., Inge-Vechtomov S. G., Liebman S. W. (1996) Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144, 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schlumpberger M., Prusiner S. B., Herskowitz I. (2001) Induction of distinct [URE3] yeast prion strains. Mol. Cell. Biol. 21, 7035–7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bradley M. E., Edskes H. K., Hong J. Y., Wickner R. B., Liebman S. W. (2002) Interactions among prions and prion “strains” in yeast. Proc. Natl. Acad. Sci. U.S.A. 99, 16392–16399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tanaka M., Chien P., Naber N., Cooke R., Weissman J. S. (2004) Conformational variations in an infectious protein determine prion strain differences. Nature 428, 323–328 [DOI] [PubMed] [Google Scholar]
- 18. Tanaka M., Collins S. R., Toyama B. H., Weissman J. S. (2006) The physical basis of how prion conformations determine strain phenotypes. Nature 442, 585–589 [DOI] [PubMed] [Google Scholar]
- 19. Toyama B. H., Kelly M. J., Gross J. D., Weissman J. S. (2007) The structural basis of yeast prion strain variants. Nature 449, 233–237 [DOI] [PubMed] [Google Scholar]
- 20. Derkatch I. L., Bradley M. E., Zhou P., Chernoff Y. O., Liebman S. W. (1997) Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 147, 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Derkatch I. L., Bradley M. E., Hong J. Y., Liebman S. W. (2001) Prions affect the appearance of other prions. The story of [PIN+]. Cell 106, 171–182 [DOI] [PubMed] [Google Scholar]
- 22. Bradley M. E., Liebman S. W. (2003) Destabilizing interactions among [PSI+] and [PIN+] yeast prion variants. Genetics 165, 1675–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bagriantsev S., Liebman S. W. (2004) Specificity of prion assembly in vivo. [PSI+] and [PIN+] form separate structures in yeast. J. Biol. Chem. 279, 51042–51048 [DOI] [PubMed] [Google Scholar]
- 24. Liebman S. W., Bagriantsev S. N., Derkatch I. L. (2006) Biochemical and genetic methods for characterization of [PIN+] prions in yeast. Methods 39, 23–34 [DOI] [PubMed] [Google Scholar]
- 25. Chien P., Weissman J. S. (2001) Conformational diversity in a yeast prion dictates its seeding specificity. Nature 410, 223–227 [DOI] [PubMed] [Google Scholar]
- 26. Kushnirov V. V., Kryndushkin D. S., Boguta M., Smirnov V. N., Ter-Avanesyan M. D. (2000) Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr. Biol. 10, 1443–1446 [DOI] [PubMed] [Google Scholar]
- 27. Sondheimer N., Lopez N., Craig E. A., Lindquist S. (2001) The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 20, 2435–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morano K. A., Grant C. M., Moye-Rowley W. S. (2012) The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 190, 1157–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Masison D. C., Kirkland P. A., Sharma D. (2009) Influence of Hsp70s and their regulators on yeast prion propagation. Prion 3, 65–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Romanova N. V., Chernoff Y. O. (2009) Hsp104 and prion propagation. Protein Pept. Lett. 16, 598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Summers D. W., Douglas P. M., Cyr D. M. (2009) Prion propagation by Hsp40 molecular chaperones. Prion 3, 59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Park K.-W., Hahn J.-S., Fan Q., Thiele D. J., Li L. (2006) De novo appearance and “strain” formation of yeast prion [PSI+] are regulated by the heat-shock transcription factor. Genetics 173, 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fan Q., Park K.-W., Du Z., Morano K. A., Li L. (2007) The role of Sse1 in the de novo formation and variant determination of the [PSI+] prion. Genetics 177, 1583–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gietz R. D., Woods R. A. (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96 [DOI] [PubMed] [Google Scholar]
- 35. Herskowitz I., Jensen R. E. (1991) Putting the HO gene to work. Practical uses for mating-type switching. Methods Enzymol. 194, 132–146 [DOI] [PubMed] [Google Scholar]
- 36. Rose M. D., Winston F. M., Heiter P. (1990) Methods in Yeast Genetics: A Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37. Scholz O., Thiel A., Hillen W., Niederweis M. (2000) Quantitative analysis of gene expression with an improved green fluorescent protein. Eur. J. Biochem. 267, 1565–1570 [DOI] [PubMed] [Google Scholar]
- 38. Jansen G., Wu C., Schade B., Thomas D. Y., Whiteway M. (2005) Drag&Drop cloning in yeast. Gene 344, 43–51 [DOI] [PubMed] [Google Scholar]
- 39. Hanahan D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580 [DOI] [PubMed] [Google Scholar]
- 40. Kryndushkin D. S., Alexandrov I. M., Ter-Avanesyan M. D., Kushnirov V. V. (2003) Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 278, 49636–49643 [DOI] [PubMed] [Google Scholar]
- 41. Feilotter H. E., Hannon G. J., Ruddell C. J., Beach D. (1994) Construction of an improved host strain for two hybrid screening. Nucleic Acids Res. 22, 1502–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chernoff Y. O., Lindquist S. L., Ono B., Inge-Vechtomov S. G., Liebman S. W. (1995) Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268, 880–884 [DOI] [PubMed] [Google Scholar]
- 43. Yu H., Braun P., Yildirim M. A., Lemmens I., Venkatesan K., Sahalie J., Hirozane-Kishikawa T., Gebreab F., Li N., Simonis N., Hao T., Rual J.-F., Dricot A., Vazquez A., Murray R. R., Simon C., Tardivo L., Tam S., Svrzikapa N., Fan C., de Smet A.-S., Motyl A., Hudson M. E., Park J., Xin X., Cusick M. E., Moore T., Boone C., Snyder M., Roth F. P., Barabási A.-L., Tavernier J., Hill D. E., Vidal M. (2008) High-quality binary protein interaction map of the yeast interactome network. Science 322, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sindi S. S., Serio T. R. (2009) Prion dynamics and the quest for the genetic determinant in protein-only inheritance. Curr. Opin. Microbiol. 12, 623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Derdowski A., Sindi S. S., Klaips C. L., DiSalvo S., Serio T. R. (2010) A size threshold limits prion transmission and establishes phenotypic diversity. Science 330, 680–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Borchsenius A. S., Müller S., Newnam G. P., Inge-Vechtomov S. G., Chernoff Y. O. (2006) Prion variant maintained only at high levels of the Hsp104 disaggregase. Curr. Genet. 49, 21–29 [DOI] [PubMed] [Google Scholar]
- 47. Taipale M., Jarosz D. F., Lindquist S. (2010) HSP90 at the hub of protein homeostasis. Emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 11, 515–528 [DOI] [PubMed] [Google Scholar]
- 48. Borkovich K. A., Farrelly F. W., Finkelstein D. B., Taulien J., Lindquist S. (1989) hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 9, 3919–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Prodromou C., Siligardi G., O'Brien R., Woolfson D. N., Regan L., Panaretou B., Ladbury J. E., Piper P. W., Pearl L. H. (1999) Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 18, 754–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Panaretou B., Siligardi G., Meyer P., Maloney A., Sullivan J. K., Singh S., Millson S. H., Clarke P. A., Naaby-Hansen S., Stein R., Cramer R., Mollapour M., Workman P., Piper P. W., Pearl L. H., Prodromou C. (2002) Activation of the ATPase activity of Hsp90 by the stress-regulated cochaperone Aha1. Mol. Cell 10, 1307–1318 [DOI] [PubMed] [Google Scholar]
- 51. McLaughlin S. H., Sobott F., Yao Z.-P., Zhang W., Nielsen P. R., Grossmann J. G., Laue E. D., Robinson C. V., Jackson S. E. (2006) The co-chaperone p23 arrests the Hsp90 ATPase cycle to trap client proteins. J. Mol. Biol. 356, 746–758 [DOI] [PubMed] [Google Scholar]
- 52. Duina A. A., Chang H. C., Marsh J. A., Lindquist S., Gaber R. F. (1996) A cyclophilin function in Hsp90-dependent signal transduction. Science 274, 1713–1715 [DOI] [PubMed] [Google Scholar]
- 53. Mayr C., Richter K., Lilie H., Buchner J. (2000) Cpr6 and Cpr7, two closely related Hsp90-associated immunophilins from Saccharomyces cerevisiae, differ in their functional properties. J. Biol. Chem. 275, 34140–34146 [DOI] [PubMed] [Google Scholar]
- 54. Kurahashi H., Ishiwata M., Shibata S., Nakamura Y. (2008) A regulatory role of the Rnq1 nonprion domain for prion propagation and polyglutamine aggregates. Mol. Cell. Biol. 28, 3313–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wickner R. B., Dyda F., Tycko R. (2008) Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register β-sheet structure. Proc. Natl. Acad. Sci. U.S.A. 105, 2403–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Abbas-Terki T., Donzé O., Briand P. A., Picard D. (2001) Hsp104 interacts with Hsp90 cochaperones in respiring yeast. Mol. Cell. Biol. 21, 7569–7575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mackay R. G., Helsen C. W., Tkach J. M., Glover J. R. (2008) The C-terminal extension of Saccharomyces cerevisiae Hsp104 plays a role in oligomer assembly. Biochemistry 47, 1918–1927 [DOI] [PubMed] [Google Scholar]
- 58. Reidy M., Masison D. C. (2010) Sti1 regulation of Hsp70 and Hsp90 is critical for curing of Saccharomyces cerevisiae [PSI+] prions by Hsp104. Mol. Cell. Biol. 30, 3542–3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shaner L., Sousa R., Morano K. A. (2006) Characterization of Hsp70 binding and nucleotide exchange by the yeast Hsp110 chaperone Sse1. Biochemistry 45, 15075–15084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sharma J., Liebman S. W. (2012) [PSI+] prion variant establishment in yeast. Mol. Microbiol. 4, 866–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.