Background: GATA-type transcription factor Ams2 required for core histone transcription is degraded by the SCF during G2 and M phases.
Results: Ams2 is also a substrate for the APC/C ubiquitin system, and its deregulation alters progression through S phase.
Conclusion: Ams2 is regulated by both the APC/C and SCF ubiquitin ligases.
Significance: Discovering ubiquitylation and proteolysis events by the APC/C is crucial for understanding cell division cycle control.
Keywords: Cell Cycle, Histone Modification, Transcription Regulation, Ubiquitin Ligase, Ubiquitination, APC/C, Ams2, Fission Yeast, Proteolysis
Abstract
Histone transcription and deposition are tightly regulated with the DNA replication cycle to maintain genetic integrity. Ams2 is a GATA-containing transcription factor responsible for core histone gene expression and for CENP-A loading at centromeres in fission yeast. Ams2 levels are cell cycle-regulated, and after the S phase Ams2 is degraded by the SCFpof3 ubiquitin ligase; however, the regulation of Ams2 in G1 or meiosis is poorly understood. Here we show that another ubiquitin ligase, the anaphase-promoting complex/cyclosome (APC/C) targets Ams2 for destruction in G1. Ubiquitylation and destruction of Ams2 is dependent upon a coactivator Cdh1/Ste9 and the KEN box in the C terminus of Ams2. We also find that stabilization of Ams2 sensitizes cells to the anti-microtubule drug thiabendazole and the histone deacetylase inhibitor tricostatin A when a histone deacetylase gene hst4 is deleted, suggesting that histone acetylation together with Ams2 stability ensures the coupling of mitosis to DNA replication. Furthermore, in meiosis, the failure of the APC/C-mediated destruction of Ams2 is deleterious, and pre-meiotic DNA replication is barely completed. These data suggest that Ams2 destruction via both the APC/C and the SCF ubiquitin ligases underlies the coordination of histone expression and DNA replication.
Introduction
The ubiquitin system is an ATP-dependent tagging process, by which ubiquitin is attached to acceptor lysine residue(s) on a substrate via an enzymatic cascade consisting of a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3) (1, 2). Depending upon the number of ubiquitins or type of ubiquitin chain formed, the signal may differ from destruction of the substrate by the 26 S proteasome to a change in substrate function or localization. Two highly conserved cell cycle-regulated E3 ubiquitin ligases are the anaphase-promoting complex/cyclosome (APC/C)2 and the SCF (Skp1-Cullin 1-F-box) protein complexes (3, 4). APC/C E3 catalytic activity is tightly regulated during the cell cycle and is active from metaphase to the end of G1. This is mainly achieved by phosphorylation of APC/C subunits and binding to one of two cell cycle-specific coactivator proteins, Cdc20/Slp1 and Cdh1/Ste9. In contrast, SCF E3 catalytic activity can be present throughout the cell cycle, and the timing of substrate destruction is regulated by the phosphorylation status of those substrates. Therefore, the two E3 ligases can be active at different times in the cell cycle and thus complement each other.
The APC/C controls cell cycle transition by catalyzing the ubiquitylation of cell cycle regulators. Securin and cyclin B are two major substrates whose destruction is required for sister chromatid separation and mitotic exit, respectively (5–7). In addition, the APC/C controls other cell cycle events such as spindle dynamics, spindle checkpoint, cytokinesis, and DNA replication. However, it is believed that more APC/C substrates are still awaiting discovery, and thus systematic genome-wide screening for APC/C substrates is required. In addition to its well studied role in the mitotic cycle, the APC/C is essential for meiosis, a specialized form of cell division that gives rise to haploid gametes from diploid germ cells (8). Meiosis occurs from one round of DNA replication followed by two consecutive rounds of chromosome segregation, known as meiosis I (MI) and meiosis II (MII) (9). Progression through meiosis in fission yeast has been shown to require Mes1, which acts as a competitive inhibitor of the APC/C in MI, ensuring sufficient levels of cyclin B/Cdk1 remain to initiate MII (10, 11). In addition, the APC/C regulates proteins that function in a variety of meiotic processes. The destruction of a transcriptional repressor Ume6 by the APC/C-Cdc20 is required for meiotic gene transcription, as well as meiotic progression (12). We have recently shown that destruction of Rhp54 (Rad54 homolog in fission yeast) in G1 prior to entry into meiosis is required for faithful meiotic recombination and thus the generation of genetic variation (13).
In eukaryotic cells, histone gene expression is regulated in a cell cycle-dependent manner. Histone proteins are essential for nucleosome formation. Nucleosomes consist of ∼150 base pairs of DNA wrapped around a histone octamer containing two sets of each of the histone H2A/H2B and H3/H4 dimers (14). New nucleosomes are assembled as DNA replication occurs, and thus histone expression and DNA replication need to be coupled, so histone proteins are regulated in a delicate balance transcriptionally, post-transcriptionally, and post-translationally. Ams2 in fission yeast was originally identified as a multicopy suppressor of the temperature-sensitive cnp1-1 mutant, defective in the centromere-specific histone H3 variant CENP-A (15). Ams2 promotes the loading of CENP-A and centromere nucleosome formation; thus the loss or overproduction of Ams2 interferes with the core centromere structure (15, 16). Ams2 is a member of the GATA-type transcription factor family (15) and regulates the transcription of all core histone genes during the S phase, and expression levels oscillate periodically during the cell cycle, peaking at the S phase when core histones are expressed. Transcriptional activation of ams2+ from the G1 to S phases is partly involved in this spike expression. In addition, the destruction of Ams2 in the post-S phase by the SCF (Skp1-Cullin-F box) E3 ligase containing Pof3 as an F-box helps to prevent unnecessary histone gene expression during G2 and M phase (16). Because ectopic expression of core histone genes has a deleterious effect on chromosome transmission fidelity (17), the regulation of histone gene expression is crucial. Furthermore, histone modification, such as acetylation, plays a role in nucleosome assembly as well as DNA damage response control. In budding yeast, acetylation of histone H3 at lysine 56 (H3-K56Ac) has been shown to occur during the S phase and can be stimulated when DNA is damaged (18–21); however, how the acetylation of histones and expression of core histones are coordinated to preserve genome integrity remains to be understood.
Using a cell-free APC/C-dependent destruction assay, we have set out a genome-wide search of APC/C substrates in Schizosaccharomyces pombe and identified Ams2 as a new substrate. We demonstrate that Ams2 is ubiquitylated by APC/CCdh1, but not APC/CCdc20. We also show that Ams2 is degraded in vivo, and its destruction in G1 is a prerequisite for timely progression during DNA replication, in particular in meiosis. Ams2 is regulated by both SCF and APC/C ubiquitin ligases, underscoring a role of Ams2 proteolysis in the maintenance of histone homeostasis and genome integrity, in conjunction with the control of histone acetylation.
EXPERIMENTAL PROCEDURES
Xenopus Egg Extracts and Destruction Assays
Xenopus cytostatic factor-arrested egg extracts (CSF extracts) were prepared as described previously (22). A cell-free Cdh1-APC/C-dependent destruction assay was preformed as previously described (13).
Yeast General Methods
Methods of handling S. pombe were described previously (23). Thiamine (2 μm) was added to the medium to repress the nmt1 promoter. The strains used in this study are shown in supplemental Table S1.
Plasmid Construction and Mutagenesis
The coding region of ams2+ was amplified from an S. pombe cDNA library and subcloned using the Invitrogen gateway system. Ams2 constructs with mutations were generated by PCR-based mutagenesis. All of the constructs were confirmed by DNA sequencing (Cogenics and University College London in-house).
Synchronous Cultures
To induce synchronous meiosis, homozygous diploid (h−/h−) cells containing the pat1-114 mutation were grown in Edinburgh minimal medium 2 (EMM2) to mid-log phase (Asyn), washed into EMM2-nitrogen at 25 °C for 15 h (G1), and shifted to 34 °C to inactivate Pat1 kinase and induce meiosis. Cells were collected every 20 min and analyzed by microscopy and immunoblotting.
Flow Cytometry
CyAn ADP high performance flow cytometry was used to analyze samples, and FlowJo software and the Watson Pragmatic were used to analyze the percentage of cells in the S and G1 phases.
RNA Analysis
RNA samples were prepared, and Northern Blotting was carried out as previously described (24). The probes were prepared as described in Ref. 25. The one-step Mesa Green qRT-PCR MasterMix for SYBR assay (Eurogentec, Southampton, U.K.) was used for quantitative RT-PCR experiments. The data were analyzed using MJ Opticon monitor analysis software 3.0.
Ubiquitylation Assay
Ubiquitylation assays were essentially performed as described (26). Xenopus APC/C was immunoprecipitated from 15 μl of interphase extract using anti-Apc3 mAb (AF3.1) immobilized on Dynabeads protein A (Invitrogen). Reactions were performed at 23 °C in 10 μl of buffer (20 mm Tris-HCl, pH 7.5, 100 mm KCl, 2.5 mm MgCl2, 2 mm ATP, 0.3 mm DTT) containing 0.05 mg/ml E1, 0.025 mg/ml UbcX, 0.75 mg/ml ubiquitin, 1 μm ubiquitin-aldehyde, 150 μm MG132, 0.01 mg/ml purified His-Cdh1 protein, and 1 μl of 35S-labeled substrates. The reactions were stopped at the indicated time points with SDS sample buffer and resolved by SDS-PAGE followed by autoradiography.
Antibodies
Antibodies were used as follows: anti-Pk (AbD Serotec, 1:200), anti-Myc 4A6 (Millipore 05–724, 1:300), anti-Cdc2 (mAb Y100, 1:2,000), anti-Cdc13 (RbAb HY1, 1:1,000), anti-Cig2 (mAb 3A11, 1:1,000), anti-Cut2 (RbAb HY19, 1:100), anti-Cdc2 pTpY (mAb CP3.2, a gift from Dr. J. Gannon, 1:10), and anti-histone H3 (Abcam ab1791, 1:500).
RESULTS
Ams2 Is a New APC/C Substrate
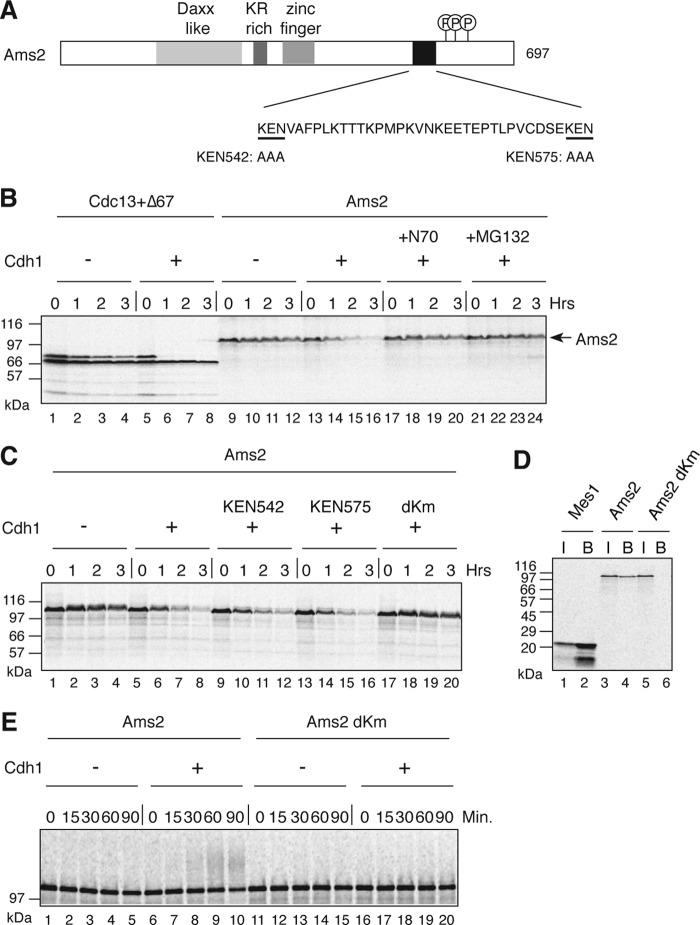
In a genome-wide screen for a KENXP motif (where X is any amino acid) containing proteins in the fission yeast S. pombe (13), Ams2 was shown to be a substrate of the APC/C in a cell-free APC/C-dependent destruction assay reconstituted in Xenopus egg extract. Ams2 was radiolabeled by coupled transcription and translation in the presence of [35S]methionine and added into a Cdh1-driven destruction assay. We found that Ams2 destruction was dependent upon the addition of recombinant Xenopus Cdh1 (Fig. 1B), but not Cdc20 (supplemental Fig. S1). In addition, Ams2 destruction was blocked by the presence of the N-terminal 70 amino acids of cyclin B, a known competitive inhibitor of the APC/C (27), as well as the 26 S proteasome inhibitor, MG132. Sequence inspection of Ams2 revealed two KEN boxes within its C terminus region, KEN 542 and KEN 575 (Fig. 1A). A single KEN box mutation did not stabilize Ams2, whereas when both of these KEN boxes were mutated (dKm), Ams2 became stable (Fig. 1C), suggesting that recognition of either KEN box by Xenopus APC/CCdh1 is sufficient to target Ams2 for destruction. We also confirmed that S. pombe Cdh1, Ste9, was able to bind Ams2 in a KEN box-dependent manner (Fig. 1D). Finally, to determine whether the APC/CCdh1 complex was directly capable of ubiquitylating Ams2, we employed an in vitro APC/C ubiquitin ligase assay. In this assay, APC/C purified from interphase egg extracts and recombinant Xenopus Cdh1 were used together with 35S-labeled substrate, Ams2. Wild type Ams2 was ubiquitylated only when Cdh1 was added, but mutation of both KEN boxes (dKm) abolished its ubiquitylation (Fig. 1E). These results indicate that Ams2 is an APC/C-Cdh1/Ste9 substrate.
FIGURE 1.
Ams2 is a substrate of the APC/C. A, schematic diagram of Ams2 showing the Daxx homology domain, a basic amino region rich in arginine and lysine residues, putative zinc finger domain, KEN box sequences, and the Hsk1 phosphorylation region. B, a cell-free destruction assay reconstituted in Xenopus egg interphase extracts in the presence/absence of Cdh1. 35S-Labeled in vitro translated Cdc13 together with a version lacking the N-terminal 67 residues (Δ67, stable control) (lanes 1–8) or Ams2 (lanes 9–24) was used. The N-terminal 70 amino acids of Cdc13 (lanes 17–20) or MG132 (lanes 21–24) were added to the reaction prior to the addition of Ams2. C, Ams2 destruction is dependent upon two KEN boxes; same as B, but 35S-labeled Ams2 (lanes 1–8), KEN542/AAA (lanes 9–12), KEN575/AAA (lanes 13–16), or dKm (lanes 17–20) were used as substrates. D, Ams2 binds to S. pombe Cdh1, Ste9. HA-Ste9 was incubated with 35S-labeled Ams2 (lanes 3 and 4) or dKm (lanes 5 and 6) and pulled down with anti-HA antibody beads. The input (lanes I) and bound (lanes B) values are shown. 35S-Labeled Mes1 was used as a control (lanes 1 and 2). E, Ams2 is ubiquitylated in Cdh1 and KEN box-dependent manner. 35S-Labeled Ams2 (lanes 1–10) or dKm (lanes 11–20) were subjected to an in vitro ubiquitylation assay using purified Xenopus APC/C in the presence or absence of Cdh1.
Ams2 Is Regulated in a Cell Cycle-dependent Manner
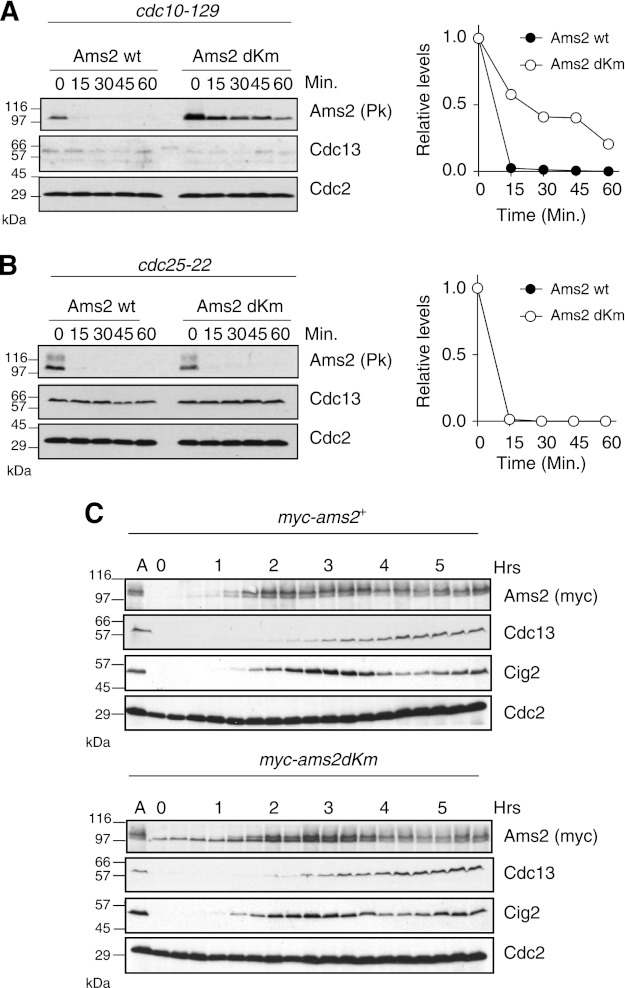
Next, we investigated the stability of Ams2 in S. pombe by measuring the half-life of Ams2 (WT and dKm). Pk-tagged versions of Ams2 were expressed from an inducible nmt81 promoter in cells blocked in G1 or G2 using a temperature-sensitive allele of cdc10 or cdc25, respectively, and then cycloheximide and thiamine were added to block de novo translation and transcription. In G1, Ams2 WT was quickly destroyed with a half-life of ∼8 min, whereas the half-life of dKm was much longer (>20 min) (Fig. 2A). In contrast, in G2, WT and dKm of Ams2 were both unstable (Fig. 2B), indicating that Ams2 destruction in G1 is dependent upon the two KEN boxes, but destruction in G2 is not. These results are in agreement with recent evidence that Ams2 is a substrate of the other E3 ubiquitin ligase SCF in late S/G2 (16). Overexpression of Ams2 in G2 causes deregulation of histone deposition into centromeres and genetic instability (16). Here, to study the impact of Ams2 stabilization in G1 without overexpression, we created yeast strains carrying a single Myc-tagged ams2+ and ams2-dKm under a native promoter and examined Ams2 levels during the normal cell cycle (Fig. 2C). Nitrogen starvation arrests cells at G1 and subsequent nitrogen addition releases the G1 block, achieving a synchronous cell culture. Ams2 WT was absent in G1 (0–1 h) but appeared just before cells enter the S phase, as judged by the appearance of the S phase cyclin Cig2 (28). As cells progressed through the S phase, Ams2 became a doublet. This doublet has already been published and is due to phosphorylation by Hsk1/Cdc7 (16), but CDK also seems to phosphorylate Ams2.3 In striking contrast to WT, Ams2-dKm was present during G1 (even from time 0) as a single form, which became a doublet (phosphorylated) with similar timing to wild type in S phase. Following the S phase, both Ams2 WT and dKm were reduced and oscillated similarly. We checked the expression of histone to assess the function of Ams2 in these cells (supplemental Fig. S2). In the ams2-dKm strain, histone H2A transcript was expressed at a higher level and earlier in G1 than WT strain. Furthermore, to investigate whether endogenous Ams2 levels fluctuate in a cell cycle-dependent manner, a synchronous culture was prepared by centrifugal elutriation and analyzed (supplemental Fig. S3). Because wild type S. pombe cells have a very short G1, the wee1 mutation was used to extend G1 phase. At time 0 (G2), both Ams2 WT and Ams2-dKm proteins were absent because of SCF-dependent proteolysis (16). As the cell cycle progressed, both proteins were still being degraded in G2 and M, but Ams2 WT started to accumulate in the next S phase, whereas Ams2-dKm, which is refractory to APC/C-dependent proteolysis, started to accumulate in G1. These results indicate that Ams2 is degraded in G1 in an APC/C-dependent manner in vivo.
FIGURE 2.
Ams2 is destroyed in G1 in a KEN box-dependent manner in vivo. A and B, Ams2 stability in G1 and G2 phase was measured. Ams2 was expressed from the nmt1 (rep81) promoter in cdc10-129 (HYY96) or cdc25-22 (HYY8) cells that had been incubated at the restrictive temperature to arrest cells at G1 or G2, respectively. Then cycloheximide (a protein synthesis inhibitor) and thiamine (a repressor of the nmt1 promoter) were added at time 0. Samples were taken at the indicated times and analyzed by immunoblotting using indicated antibodies. As control, endogenous Cdc13 and Cdc2 were used. The graph represents relative levels of Ams2 and Ams2-dKm. C, cells expressing Myc-Ams2 (HYY908) and Myc-Ams2-dKm (HYY909) from the endogenous ams2+ promoter were grown in EMM2 (asynchronous, lane A), washed with EMM2-N and then cultured for 15 h at 25 °C. Upon addition of NH4Cl2 (nitrogen source), the cells were released from the G1 block into the cell cycle. The samples were taken every 20 min and analyzed by immunoblotting.
Ams2 and HDAC Act in Concert to Control Cell Cycle Progression
In most eukaryotes, newly synthesized histones during S phase are acetylated and deposited into nucleosomes. We hypothesized that histone gene expression by Ams2 might be coupled with histone acetylation. To explore this model, we created double mutant strains containing ams2-dKm with HDACs and investigated whether the presence of Ams2 in G1 had an impact on their cellular sensitivity toward genotoxic stress or anti-mitotic agents. We studied HDACs from the “classical” family, which has two classes: class I Hda1/Hos2 and Clr6 and class II Clr3, as well as the class III SIR2 family of NAD+-dependent HDACs: Sir2, Hst2, and Hst4 (29). Clr6 is an essential gene; thus a temperature-sensitive strain clr6-1 was used (30). The growth and sensitivity of these double mutants to a variety of drugs were investigated (Fig. 3 and supplemental Fig. S4). Stabilization of Ams2 in a clr3Δ, hos2Δ, sir2Δ, hst2Δ, or clr6-1 background did not show any clear synergistic effects, whereas stabilization of Ams2 in a hst4Δ background (hst4Δams2-dKm) significantly elevated sensitivity to thiabendazole (a microtubule inhibitor) and tricostatin A (an inhibitor of classical HDACs), suggesting that Ams2 levels and/or histone acetylation may be involved in monitoring DNA replication and mitosis. The generation times of these mutant cells also supports this notion, 3 h in hst4Δ and 4 h in hst4Δams2-dKm, compared with 2.25 h for both WT and ams2-dKm. We investigated the phenotype of hst4Δams2-dKm in the presence of CBZ (carbendazim a microtubule inhibitor), compared with hst4Δ. Intriguingly, hst4Δams2-dKm resulted in a significant increase in mitotic defective phenotypes such as “cut” (cell untimely torn) or “multiple septa” in the presence of CBZ, compared with hst4Δ (Fig. 3, B and C), implying that the presence of Ams2 in G1 may uncouple DNA replication and mitotic events. It is also possible that this is due to Ams2-dKm-dependent premature CENP-A loading and/or Hst4, which may contribute to mitosis indirectly.
FIGURE 3.
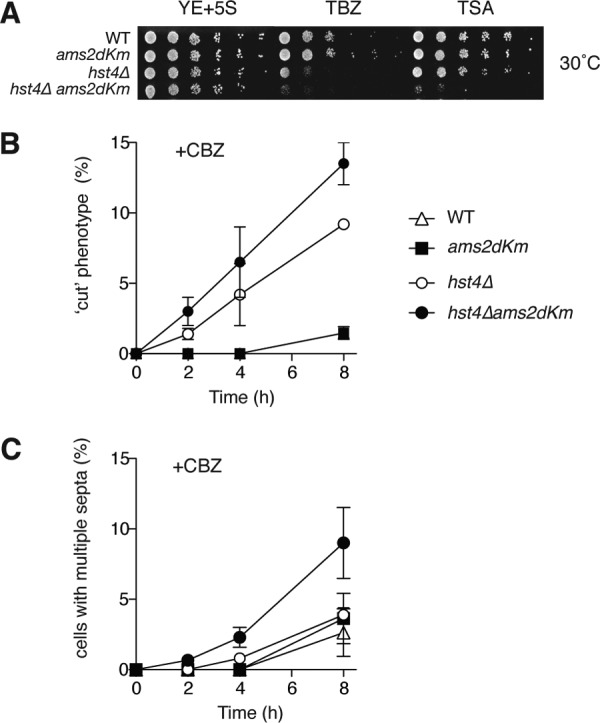
Genetic interaction between ams2-dKm and histone deacetylases. A, Ams2 shows a genetic interaction with Hst4. Asynchronous wild type (HYY908), ams2-dKm (HYY909), hst4Δ (HYY1032), and hst4Δams2-dKm (HYY1051) cells were spotted with decreasing cell numbers onto rich medium containing 12.5 μg/ml thiabendazole (TBZ) or 12.5 μg/ml trichostatin A (TSA), and incubated at 30 °C for 4 days. B and C, WT (HYY908), ams2-dKm (HYY909), hst4Δ (HYY1032), and hst4Δams2-dKm (HYY1051) cells were incubated in the presence of 50 μg/ml CBZ and “cut” phenotype together with cells with multiple septa were counted at the indicated time points. Error bars, S.E. from three independent experiments.
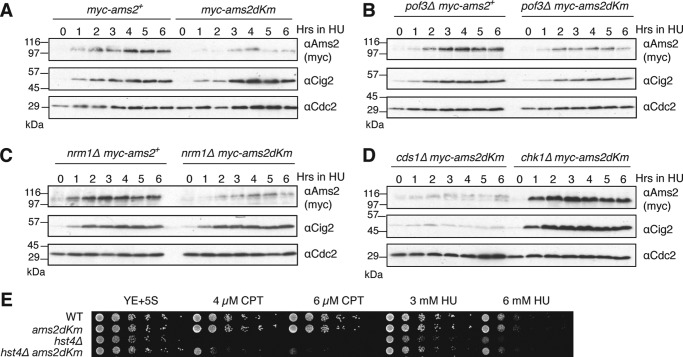
In G2, Ams2 stability is regulated by Pof3, the F-box protein responsible for Ams2 destruction via the SCF (16). To investigate the relationship between Ams2 abundance and drug sensitivity, we created double mutants with pof3Δ. The spot test analysis revealed pof3Δams2-dKm to be more sensitive to camptothecin (CPT), hydroxyurea (HU), and methyl methane sulfonate (MMS) than the single pof3Δ or ams2-dKm mutant, illustrating the additive effect of Ams2 stability from both the APC/C and the SCF ubiquitin ligases (supplemental Fig. S5).
Destruction of Ams2 in G1 Is Required for Proper Meiotic Progression
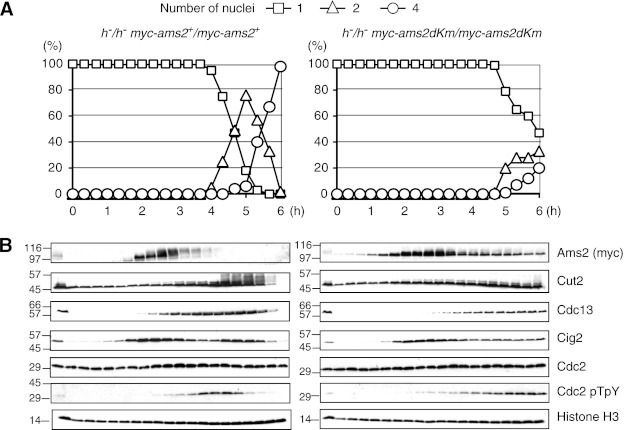
Meiosis is a specialized form of cell division generating haploid gametes and is normally induced through G1. Because the KEN box mutation stabilizes Ams2 in G1, we next investigated what effect ams2-dKm might have upon entry into and progression through meiosis. To this end, we constructed strains where either Myc-tagged wild type ams2+ or ams2-dkm had been integrated into the ams2 locus and expressed from the native promoter. Meiosis was zygotically induced in these cells, and meiotic progression was monitored by counting the number of nuclei. Meiosis was synchronously induced by thermal inactivation of the Pat1 kinase in prestarved homozygous (h−/h−) diploid cells. As shown in Fig. 4A, the presence of Ams2 (ams2-dkm) in G1 significantly delayed meiotic progression, meiosis I not starting until 4.5 h after Pat1 inactivation. In contrast, WT (ams2+) entered meiosis I around 4 h, completing it by 5 h. As cells progressed through the pre-meiotic S phase, WT Ams2 appeared and became phosphorylated. Ams2 was then destroyed prior to MI as judged by nuclei counting and was absent from the rest of the time course (Fig. 4B). The destruction of Ams2 around 3 h (after pre-meiotic S phase) is likely to be via the SCF as seen in mitosis. On the other hand, Ams2-dKm was present in G1 cells as a single form (Fig. 4B), which became phosphorylated at a similar time to WT Ams2 and persisted throughout the experiment, albeit at lower levels after 3.5 h. Unlike WT cells, which entered and passed through S phase by 3 h, the ams2-dKm cells did not appear to complete S phase, as seen by the presence of a persistent 1C peak by flow cytometry (data not shown). This observation was further supported by examination of protein levels of APC/C substrates such as Cut2, Cig2, and Cdc13. Cut2/securin, a known substrate of the APC/C, became phosphorylated as cells entered meiotic divisions (MI and MII) and was destroyed at the end of MII. However, in ams2-dKm cells, Cut2 did not become phosphorylated because cells never completely entered MI. Cig2 had two peaks during premeiotic S phase and MI, as previously reported (31). It should be noted that ams2-dKm cells are not sterile; thus they do complete meiosis eventually, but the meiotic progression is severely delayed if Ams2 is present in G1 before meiosis is initiated.
FIGURE 4.
Roles of Ams2 destruction in meiosis. Meiosis was induced by thermal inactivation of Pat1 (pat1-114), and meiotic progression was monitored in WT myc-ams2+ (HYY952) and myc-ams2-dKm (HYY953) diploid cells. The samples were examined microscopically for the number of nuclei in a cell to monitor progression through meiosis (A) and analyzed by immunoblotting using specific antibodies every 20 min (B).
Transcriptional Control of Histone Genes Is Deregulated by Ams2-dKm in Meiosis
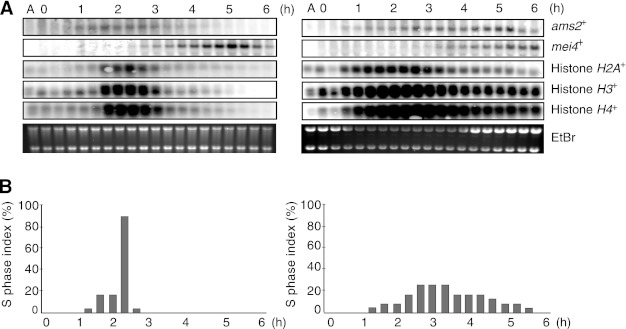
Because Ams2 has been shown to be a transcriptional activator of core histones during mitosis (25), we analyzed the mRNA levels of the core histones (H2A, H3, and H4) during meiotic progression (Fig. 5A), as well as ams2+ and mei4+ (a key regulator of several middle phase meiotic genes). In WT, the ams2+ transcript appeared before those of the core histones and then disappeared as the cells enter into meiosis. The core histones had a narrow window of transcription appearing around 1.5 h and vanishing by 3 h, coinciding with DNA replication (Fig. 5, A and B). In contrast, in ams2-dKm mutant, the ams2+ transcripts and those of the core histones appeared earlier than WT and persisted until 6 h (during the whole experiment). Although levels were reduced at later time points as cells attempted to cross the MI boundary, overall levels of histone mRNAs were significantly enhanced in the ams2-dKm cells compared with WT. Because the S phase index was evident for over 5 h in ams2-dKm cells, it is likely that ams2-dKm cells are impaired for S phase entry and/or progression and ams2-dKm transcription stays on, which in turn keeps core histone transcription switched on. Expression of the meiosis-specific transcription factor, mei4+ was also monitored. Mei4 is not required for pre-meiotic S phase but is essential for MI and sporulation (32), which is achieved by regulating cdc25+ expression (33). In WT cells, a peak of mei4+ expression was seen around 5 h (at the end of MI), whereas in ams2-dKm cells mei4+ expression never peaked even after 6 h, which is consistent with the failure of the mutant to complete MI (Fig. 5A). To investigate whether this S phase delay is dependent on the activation of checkpoint proteins Cds1 or Chk1, we have made cds1Δams2-dKm and chk1Δams2-dKm homozygous (h−/h−) diploid cells, However, neither cds1Δ nor chk1Δ mutation could rescue the ams2-dKm phenotype in meiosis, indicating that this delay is not due to Cds1 or Chk1 (see supplemental Fig. S6). Please note that elevated levels of the histone transcripts did not affect the global level of histone proteins (Fig. 4B). This result is consistent with previously published data in an ams2+ shut off strain where no ams2+ or histone transcripts are observed (25).
FIGURE 5.
Ams2-dKm stimulates histone gene regulation in meiosis. A, Northern blot analysis of pat1-114-induced synchronized meiosis in WT myc-ams2+ (HYY952) and myc-ams2-dKm (HYY953) diploid cells. Total RNAs were isolated from samples in Fig. 4, and RNA levels of ams2+, mei4+, histone H2A+, H3+, and H4+ were examined. EtBr acts as a loading control. B, percentage of cells in the S phase (above) was estimated using FlowJo software and the Watson Pragmatic.
Feedback Control of Ams2 Expression
Ams2-dKm is more stable than wild type Ams2 in G1 (Fig. 2A); however, we noticed that Ams2-dKm levels are less abundant than wild type Ams2 in the presence of HU, a competitive inhibitor of ribonucleotide reductase that depletes the deoxyribonucleotide pool and stalls DNA replication (Fig. 6A). The addition of HU into ams2-dkm cells released from G1 arrest also gave a similar result (supplemental Fig. S7). This unexpected result suggests the existence of a cellular mechanism by which Ams2 protein levels are monitored in G1 and then down-regulated, presumably to avoid uncoordinated histone expression with DNA replication. First, we examined whether this is due to SCFpof3, which is responsible for Ams2 destruction after the S phase and in the G2 phase. However, deletion of pof3 (pof3Δ) did not rescue the low abundance of Ams2-dKm (Fig. 6B), so Pof3 is unlikely to be involved in this regulation. Next, we examined whether this is due to repression of transcriptional activation. Ams2 is a Mlu I cell cycle box-binding factor (MBF)-regulated G1/S transcript. It has been shown that Nrm1 is an inhibitor of MBF, and in the presence of HU, Cds1 phosphorylates and inactivates Nrm1 to maintain MBF-dependent transcripts (34). We observed an increase of RNA levels of both ams2+ and ams2-dKm after HU addition (supplemental Fig. S8), so it is unlikely that low abundancy of Ams2-dKm is because of transcriptional repression. In agreement with this, deletion of Nrm1 repressor (nrm1Δ) was unable to rescue low levels of Ams2-dKm in the presence of HU (Fig. 6C). In addition, we investigated whether checkpoint proteins Cds1 or Chk1 might be involved in this negative feedback. As expected, Ams2 is poorly expressed if cds1+ is deleted (cds1Δ), because Nrm1 stably inhibits MBF. Intriguingly, the absence of Chk1 mostly rescued the phenotype and showed early accumulation of Ams2-dKm (Fig. 6D). Expression of ams2-dKm mRNA in chk1Δams2-dkm was very similar to that in ams2-dkm (supplemental Fig. S8), so deletion of chk1 (chk1Δ) does not regulate ams2-dKm transcription in the presence of HU. Thus, Chk1 presumably regulates Ams2-dKm levels via proteolysis or translation, but we do not know the precise mechanism(s) at the moment. We examined whether this feedback mechanism would reflect the sensitivity of cells to S phase DNA damage. Because hst4Δ and hst4Δams2-dKm were both sensitive to higher doses of S phase DNA-damaging agents such as CPT, HU, or MMS (supplemental Fig. S4), lower doses of CPT and HU were used (Fig. 6E). Although hst4Δ cells were still sensitive to low doses of these agents, ams2-dKm partly rescued the drug sensitivity of hst4Δ.
FIGURE 6.
A feedback mechanism of Ams2 levels in G1. A, WT myc-ams2+ (HYY908) and myc-ams2-dKm (HYY909) cells were grown to mid-log phase in YE+5S medium, and hydroxyurea was added to 20 mm final concentration. The samples were taken every hour for analysis by immunoblotting with indicated antibodies. B–D, same as A, but pof3Δmyc-ams2+ (HYY1094) and pof3Δmyc-ams2-dKm (HYY1067) (B), nrm1Δmyc-ams2+ (HYY1117) and nrm1Δmyc-ams2-dKm (HYY1119) (C), or cds1Δ myc-ams2-dKm (HYY1083) and chk1Δ myc-ams2-dKm (HYY1080) (D) cells were analyzed. E, asynchronous WT ams2+ (HYY908), ams2-dKm (HYY909), hst4Δ (HYY1032), and hst4Δams2-dKm (HYY1051) cells were spotted with decreasing cell number onto rich medium containing indicated DNA-damaging agents and incubated for 4 days.
DISCUSSION
Histone expression and deposition has to be coordinated with DNA replication. The results presented here demonstrate how Ams2, a GATA-containing transcription factor responsible for core histone gene expression, is controlled during G1 in fission yeast. Using a cell-free system reconstituted in Xenopus egg extracts, we searched for new APC/C substrates in fission yeast and identified Ams2 as a target of Cdh1/Ste9-APC/C. Ams2 has two KEN boxes, and when both were mutated, it was no longer ubiquitylated or degraded in G1. To see the physiological effect of Ams2 destruction in G1, cells expressing Ams2-dKm under a native promoter (ams2-dKm) were carefully investigated rather than using overexpression based experiments. We showed a collaborative role for both the APC/C and SCF ubiquitin ligases in regulating Ams2. In addition, our data highlight the important role of Ams2 in histone homeostasis in both the mitotic and meiotic cell cycles. The absence of Ams2 in G1 apparently plays a role in the transcription repression of the core histone genes in this phase of the cell cycle.
The regulation of acetylation of lysine 56 of histone H3 (H3K56-Ac) is important for the deposition of newly synthesized histones into nucleosomes, as well as for the maintenance of DNA damage responses (18–20, 35, 36). H3K56-Ac is carried out by Rtt109 in fungi (37, 38), whereas Hst4 is responsible for deacetylation during G2/M (39, 40). The genetic interaction observed between ams2-dKm and hst4Δ (Fig. 3) may suggest a relationship between the G1 stability of Ams2 and hyperacetylation of histone H3. Histone acetylation alongside histone expression seems to be another important element coupling DNA replication and mitosis (41–44). Ams2 might directly or indirectly regulate the status of H3K56-Ac using histone acetyltransferases. Because we have been unable to detect an interaction between Ams2 and histone H3, it is possible that the Ams2 effect we are seeing involves histone chaperones Caf1 and Rtt106, which promote histone H3 acetylation on lysine 56 (20). It should be noted that members of the GATA transcription factor family are known to be substrates of acetylation themselves; both human GATA-1 and GATA-4 are acetylated by p300 stimulating GATA-dependent transcription (45, 46).
When cells are treated by genotoxic stress such as HU, it appears that Ams2 levels are significantly reduced if Ams2 is present in G1, suggesting the existence of a feedback system that might monitor Ams2 and/or histone levels in G1. In this paper, we have observed, in pof3Δ cells where Ams2 can only be destroyed in G1, a normal accumulation of Ams2 in the presence of HU. Less Ams2 could result in a slower synthesis/incorporation of histone H3 into nucleosomes and thus rescue from DNA-damaging agents such as CPT, HU, and MMS in hst4Δams2-dKm might be observed (Fig. 6E), compared with hst4Δ where restoring Ams2 levels could result in more or faster incorporation of H3, which compromises genomic integrity (16). It is also possible that double KEN box mutations (ams2-dkm) might result in a loss or gain of function of Ams2, and thus phenotypes of hst4Δ such as DNA damage sensitivity are indirectly rescued in hst4Δams2-dKm. It seems that Chk1 is involved in this feedback control, although the mechanism remains unclear. In budding yeast, transcriptional repression via Chk1 following DNA damage has already been elucidated. Under normal conditions, Chk1 acts through histone H3 phosphorylation to recruit GCN5 to the promoters of relevant genes such as Cdk1 and cyclin B. Upon DNA damage, however, Chk1 dissociates from chromatin reducing transcription levels. The transcription of some 200 genes and the transcriptional elongation/3′ processing machinery have been found to be effected by Chk1 (47, 48).
We surmise that through the destruction of Ams2 in G1, the APC/C plays a role in histone regulation in meiosis by repressing the early expression of histone transcripts, as seen for Ume6 (Figs. 4 and 5) (12). The pre-meiotic S phase is prolonged in ams2-dkm cells. However, this delay is not due to a histone surveillance mechanism or DNA checkpoint Cds1/Rad53 or Chk1 (supplemental Fig. S6) (49, 50). Because failure of Ams2 destruction in G1 has more evident effects on meiotic progression than mitosis, it is tempting to speculate that additional layers of histone regulation might be present in mitosis, which are absent in meiosis. Indeed, S. pombe histone transcription is regulated by the HIRA-like protein Hip1, which represses transcription outside of the S phase (24). In ams2-dKm, histone H2A, H3, and H4 transcripts appear early in G1, during a period when Hip1 silences histone transcription, suggesting that it is the elevated Ams2 levels that override the repression system and that this is important as cells progress into meiosis. In higher metazoans, the mechanism is slightly different, but HBP/SLBP (histone hairpin binding protein/stem-loop-binding protein) binds to the 3′-end of histone mRNAs and regulates histone gene expression during the S phase in mitosis (51).
Transcription factors can be both targets of the APC/C and enhancers of APC/C activity. The forkhead transcription factor M1 (FoxMI), which is required for the expression of the mitotic regulators cyclin B, Aurora B, and Plk1, is a target of APC/C-Cdh1 (52) and is critical for entry into the S phase (53). In contrast, CBP/p300 (54) and Atf1 (55) have been found to interact with APC/C subunits and stimulate APC/C activity. Through interaction with APC/C, CBP-p300 acetyltransferase activity is also activated and thus potentiates CBP-p300-dependent transcription (54). Although Ams2 could bind to Cdh1/Ste9 (Fig. 1D), we could not detect it binding to the core APC/C (data not shown); thus we believe the meiotic defect to be due to the inability of Ams2 to be targeted by the APC/C rather than an inability to enhance APC/C activity. The consequences of these elevated histone mRNAs are unclear at the moment. Overexpressing Ams2 and overriding the SCF pathway in mitotic cells produces constitutive expression of the canonical histones, resulting in the inclusion of histone H3 at centromeric chromatin at the exclusion of the centromeric histone H3 variant CENP-A. This in turn leads to chromosome instability and a slow growth phenotype (16). These traits were not observed in our ams2-dKm cells where Ams2-dKm is expressed under a native promoter, because centromere structure and chromosome stability all seem to be normal (data not shown).
It might be noteworthy that Ams2 regulation is reminiscent of Cig2, S phase cyclin, which is targeted by the APC/C in anaphase/G1 and the SCF in G2/M to produce a peak of Cig2 activity in S phase (28, 56). Cdc18/Cdc6, an essential regulator of DNA replication, is also regulated by both APC/C and SCF ubiquitin ligases (57, 58).3 DNA replication is essential and fundamental for genome duplication, but it must be restricted to occur only once per cell cycle. Although these key proteins are regulated at multiple levels including transcriptionally and translationally, dual regulation of protein turnover by the APC/C and the SCF plays a pivotal role in allowing protein expression to peak in G1/S when both ubiquitin ligases are inactive. Moreover, as demonstrated in the relationship with HDACs, it is conceivable that post-translational modification such as phosphorylation and acetylation of regulators, as well as histones, are also involved in fine-tuning during the processes of DNA synthesis and chromatin assembly. Further study is clearly required for understanding the mechanism involved.
Supplementary Material
Acknowledgments
We thank members of the Yamano laboratory for helpful discussions and critical reading of the manuscript. We also thank Drs J. Gannon for anti-Cdc2 pTpY antibody; A. Carr, A. Klar, T. Toda, and M. Yanagida for strains; Y. Takayama for helpful advice on Northern blotting; R. de Bruin for strains and performing qRT-PCR; Tim Hunt and Hiro Mahbubani for access to the Cancer Research UK Clare Hall Laboratories Xenopus colony in the early stages of this project; and the staff at the University College London Biological Services Unit for taking care of the Xenopus colony in University College London.
This work was supported by Marie Curie Cancer Care, the Association for International Cancer Research and Cancer Research UK.

This article contains supplemental Table S1 and Figs. S1–S8.
M. Trickey and H. Yamano, unpublished data.
- APC/C
- anaphase-promoting complex/cyclosome
- HDAC
- histone deacetylase
- SCF
- Skp1-Cullin 1-F-box
- MI
- meiosis I
- MII
- meiosis II
- EMM2
- Edinburgh minimal medium 2
- HU
- hydroxyurea
- CPT
- camptothecin
- MMS
- methyl methane sulfonate
- MBF
- Mlu I cell cycle box-binding factor.
REFERENCES
- 1. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2. Hochstrasser M. (1996) Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30, 405–439 [DOI] [PubMed] [Google Scholar]
- 3. Deshaies R. J. (1999) SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435–467 [DOI] [PubMed] [Google Scholar]
- 4. Peters J. M. (1998) SCF and APC. The Yin and Yang of cell cycle regulated proteolysis. Curr. Opin. Cell Biol. 10, 759–768 [DOI] [PubMed] [Google Scholar]
- 5. Harper J. W., Burton J. L., Solomon M. J. (2002) The anaphase-promoting complex. It's not just for mitosis any more. Genes Dev. 16, 2179–2206 [DOI] [PubMed] [Google Scholar]
- 6. Peters J. M. (2006) The anaphase promoting complex/cyclosome. A machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7, 644–656 [DOI] [PubMed] [Google Scholar]
- 7. Thornton B. R., Toczyski D. P. (2006) Precise destruction. An emerging picture of the APC. Genes Dev. 20, 3069–3078 [DOI] [PubMed] [Google Scholar]
- 8. Pesin J. A., Orr-Weaver T. L. (2008) Regulation of APC/C activators in mitosis and meiosis. Annu. Rev. Cell Dev. Biol. 24, 475–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Petronczki M., Siomos M. F., Nasmyth K. (2003) Un menage a quatre. The molecular biology of chromosome segregation in meiosis. Cell 112, 423–440 [DOI] [PubMed] [Google Scholar]
- 10. Izawa D., Goto M., Yamashita A., Yamano H., Yamamoto M. (2005) Fission yeast Mes1p ensures the onset of meiosis II by blocking degradation of cyclin Cdc13p. Nature 434, 529–533 [DOI] [PubMed] [Google Scholar]
- 11. Kimata Y., Trickey M., Izawa D., Gannon J., Yamamoto M., Yamano H. (2008) A mutual inhibition between APC/C and its substrate Mes1 required for meiotic progression in fission yeast. Dev. Cell 14, 446–454 [DOI] [PubMed] [Google Scholar]
- 12. Mallory M. J., Cooper K. F., Strich R. (2007) Meiosis-specific destruction of the Ume6p repressor by the Cdc20-directed APC/C. Mol. Cell 27, 951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Trickey M., Grimaldi M., Yamano H. (2008) The anaphase-promoting complex/cyclosome controls repair and recombination by ubiquitylating Rhp54 in fission yeast. Mol. Cell Biol. 28, 3905–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 15. Chen E. S., Saitoh S., Yanagida M., Takahashi K. (2003) A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol. Cell 11, 175–187 [DOI] [PubMed] [Google Scholar]
- 16. Takayama Y., Mamnun Y. M., Trickey M., Dhut S., Masuda F., Yamano H., Toda T., Saitoh S. (2010) Hsk1- and SCF(Pof3)-dependent proteolysis of S. pombe Ams2 ensures histone homeostasis and centromere function. Dev. Cell 18, 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meeks-Wagner D., Hartwell L. H. (1986) Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell 44, 43–52 [DOI] [PubMed] [Google Scholar]
- 18. Li Q., Zhou H., Wurtele H., Davies B., Horazdovsky B., Verreault A., Zhang Z. (2008) Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134, 244–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Masumoto H., Hawke D., Kobayashi R., Verreault A. (2005) A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436, 294–298 [DOI] [PubMed] [Google Scholar]
- 20. Chen C. C., Carson J. J., Feser J., Tamburini B., Zabaronick S., Linger J., Tyler J. K. (2008) Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 134, 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu F., Zhang K., Grunstein M. (2005) Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121, 375–385 [DOI] [PubMed] [Google Scholar]
- 22. Murray A. W. (1989) Cyclin synthesis and degradation and the embryonic cell cycle. J. Cell Sci. Suppl. 12, 65–76 [DOI] [PubMed] [Google Scholar]
- 23. Moreno S., Klar A., Nurse P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823 [DOI] [PubMed] [Google Scholar]
- 24. Blackwell C., Martin K. A., Greenall A., Pidoux A., Allshire R. C., Whitehall S. K. (2004) The Schizosaccharomyces pombe HIRA-like protein Hip1 is required for the periodic expression of histone genes and contributes to the function of complex centromeres. Mol. Cell Biol. 24, 4309–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Takayama Y., Takahashi K. (2007) Differential regulation of repeated histone genes during the fission yeast cell cycle. Nucleic Acids Res. 35, 3223–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kimata Y., Baxter J. E., Fry A. M., Yamano H. (2008) A role for the Fizzy/Cdc20 family of proteins in activation of the APC/C distinct from substrate recruitment. Mol. Cell 32, 576–583 [DOI] [PubMed] [Google Scholar]
- 27. Yamano H., Tsurumi C., Gannon J., Hunt T. (1998) The role of the destruction box and its neighbouring lysine residues in cyclin B for anaphase ubiquitin-dependent proteolysis in fission yeast. Defining the D-box receptor. EMBO J. 17, 5670–5678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamano H., Kitamura K., Kominami K., Lehmann A., Katayama S., Hunt T., Toda T. (2000) The spike of S phase cyclin Cig2 expression at the G1-S border in fission yeast requires both APC and SCF ubiquitin ligases. Mol. Cell 6, 1377–1387 [DOI] [PubMed] [Google Scholar]
- 29. Bjerling P., Silverstein R. A., Thon G., Caudy A., Grewal S., Ekwall K. (2002) Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol. Cell Biol. 22, 2170–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grewal S. I., Bonaduce M. J., Klar A. J. (1998) Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics 150, 563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Borgne A., Murakami H., Ayté J., Nurse P. (2002) The G1/S cyclin Cig2p during meiosis in fission yeast. Mol. Biol. Cell 13, 2080–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Horie S., Watanabe Y., Tanaka K., Nishiwaki S., Fujioka H., Abe H., Yamamoto M., Shimoda C. (1998) The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol. Cell Biol. 18, 2118–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murakami-Tonami Y., Yamada-Namikawa C., Tochigi A., Hasegawa N., Kojima H., Kunimatsu M., Nakanishi M., Murakami H. (2007) Mei4p coordinates the onset of meiosis I by regulating cdc25+ in fission yeast. Proc. Natl. Acad. Sci. U.S.A. 104, 14688–14693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Bruin R. A., Wittenberg C. (2009) All eukaryotes. Before turning off G1-S transcription, please check your DNA. Cell Cycle 8, 214–217 [DOI] [PubMed] [Google Scholar]
- 35. Wurtele H., Kaiser G. S., Bacal J., St-Hilaire E., Lee E. H., Tsao S., Dorn J., Maddox P., Lisby M., Pasero P., Verreault A. (2012) Histone H3 lysine 56 acetylation and the response to DNA replication fork damage. Mol. Cell Biol. 32, 154–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fillingham J., Kainth P., Lambert J. P., van Bakel H., Tsui K., Peña-Castillo L., Nislow C., Figeys D., Hughes T. R., Greenblatt J., Andrews B. J. (2009) Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol. Cell 35, 340–351 [DOI] [PubMed] [Google Scholar]
- 37. Han J., Zhou H., Horazdovsky B., Zhang K., Xu R. M., Zhang Z. (2007) Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315, 653–655 [DOI] [PubMed] [Google Scholar]
- 38. Driscoll R., Hudson A., Jackson S. P. (2007) Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315, 649–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Haldar D., Kamakaka R. T. (2008) Schizosaccharomyces pombe Hst4 functions in DNA damage response by regulating histone H3 K56 acetylation. Eukaryot. Cell 7, 800–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xhemalce B., Miller K. M., Driscoll R., Masumoto H., Jackson S. P., Kouzarides T., Verreault A., Arcangioli B. (2007) Regulation of histone H3 lysine 56 acetylation in Schizosaccharomyces pombe. J. Biol. Chem. 282, 15040–15047 [DOI] [PubMed] [Google Scholar]
- 41. Brachmann C. B., Sherman J. M., Devine S. E., Cameron E. E., Pillus L., Boeke J. D. (1995) The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 9, 2888–2902 [DOI] [PubMed] [Google Scholar]
- 42. Celic I., Masumoto H., Griffith W. P., Meluh P., Cotter R. J., Boeke J. D., Verreault A. (2006) The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr. Biol. 16, 1280–1289 [DOI] [PubMed] [Google Scholar]
- 43. Maas N. L., Miller K. M., DeFazio L. G., Toczyski D. P. (2006) Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol. Cell 23, 109–119 [DOI] [PubMed] [Google Scholar]
- 44. Thaminy S., Newcomb B., Kim J., Gatbonton T., Foss E., Simon J., Bedalov A. (2007) Hst3 is regulated by Mec1-dependent proteolysis and controls the S phase checkpoint and sister chromatid cohesion by deacetylating histone H3 at lysine 56. J. Biol. Chem. 282, 37805–37814 [DOI] [PubMed] [Google Scholar]
- 45. Boyes J., Byfield P., Nakatani Y., Ogryzko V. (1998) Regulation of activity of the transcription factor GATA-1 by acetylation. Nature 396, 594–598 [DOI] [PubMed] [Google Scholar]
- 46. Kawamura T., Ono K., Morimoto T., Wada H., Hirai M., Hidaka K., Morisaki T., Heike T., Nakahata T., Kita T., Hasegawa K. (2005) Acetylation of GATA-4 is involved in the differentiation of embryonic stem cells into cardiac myocytes. J. Biol. Chem. 280, 19682–19688 [DOI] [PubMed] [Google Scholar]
- 47. Beckerman R., Donner A. J., Mattia M., Peart M. J., Manley J. L., Espinosa J. M., Prives C. (2009) A role for Chk1 in blocking transcriptional elongation of p21 RNA during the S-phase checkpoint. Genes Dev. 23, 1364–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shimada M., Niida H., Zineldeen D. H., Tagami H., Tanaka M., Saito H., Nakanishi M. (2008) Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell 132, 221–232 [DOI] [PubMed] [Google Scholar]
- 49. Gunjan A., Verreault A. (2003) A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115, 537–549 [DOI] [PubMed] [Google Scholar]
- 50. Murakami H., Nurse P. (1999) Meiotic DNA replication checkpoint control in fission yeast. Genes Dev. 13, 2581–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Koseoglu M. M., Dong J., Marzluff W. F. (2010) Coordinate regulation of histone mRNA metabolism and DNA replication. Cyclin A/cdk1 is involved in inactivation of histone mRNA metabolism and DNA replication at the end of S phase. Cell Cycle 9, 3857–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Laoukili J., Alvarez-Fernandez M., Stahl M., Medema R. H. (2008) FoxM1 is degraded at mitotic exit in a Cdh1-dependent manner. Cell Cycle 7, 2720–2726 [DOI] [PubMed] [Google Scholar]
- 53. Park H. J., Costa R. H., Lau L. F., Tyner A. L., Raychaudhuri P. (2008) Anaphase-promoting complex/cyclosome-CDH1-mediated proteolysis of the forkhead box M1 transcription factor is critical for regulated entry into S phase. Mol. Cell Biol. 28, 5162–5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Turnell A. S., Stewart G. S., Grand R. J., Rookes S. M., Martin A., Yamano H., Elledge S. J., Gallimore P. H. (2005) The APC/C and CBP/p300 cooperate to regulate transcription and cell-cycle progression. Nature 438, 690–695 [DOI] [PubMed] [Google Scholar]
- 55. Ors A., Grimaldi M., Kimata Y., Wilkinson C. R., Jones N., Yamano H. (2009) The transcription factor Atf1 binds and activates the APC/C ubiquitin ligase in fission yeast. J. Biol. Chem. 284, 23989–23994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yamano H., Kominami K., Harrison C., Kitamura K., Katayama S., Dhut S., Hunt T., Toda T. (2004) Requirement of the SCFPop1/Pop2 ubiquitin ligase for degradation of the fission yeast S phase cyclin Cig2. J. Biol. Chem. 279, 18974–18980 [DOI] [PubMed] [Google Scholar]
- 57. Perkins G., Drury L. S., Diffley J. F. (2001) Separate SCF(CDC4) recognition elements target Cdc6 for proteolysis in S phase and mitosis. EMBO J. 20, 4836–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Petersen B. O., Wagener C., Marinoni F., Kramer E. R., Melixetian M., Lazzerini Denchi E., Gieffers C., Matteucci C., Peters J. M., Helin K. (2000) Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 14, 2330–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.