Abstract
Epigenetic mechanisms regulate expression of the genome to generate various cell types during development or orchestrate cellular responses to external stimuli. Recent studies highlight that bacteria can affect the chromatin structure and transcriptional program of host cells by influencing diverse epigenetic factors (i.e., histone modifications, DNA methylation, chromatin-associated complexes, noncoding RNAs, and RNA splicing factors). In this article, we first review the molecular bases of the epigenetic language and then describe the current state of research regarding how bacteria can alter epigenetic marks and machineries. Bacterial-induced epigenetic deregulations may affect host cell function either to promote host defense or to allow pathogen persistence. Thus, pathogenic bacteria can be considered as potential epimutagens able to reshape the epigenome. Their effects might generate specific, long-lasting imprints on host cells, leading to a memory of infection that influences immunity and might be at the origin of unexplained diseases.
Bacteria can alter the epigenetic marks of host cells, either by activating signaling cascades or by acting directly in the nucleus. Some of these modifications may be long-lasting and heritable.
Upon a microbial attack, host cells undergo massive changes in their transcriptional program, mobilizing genes involved in key processes (e.g., immunity, cell death/survival, and adhesion/motility) to trigger an appropriate response (Jenner and Young 2005). It is thus not surprising that successful pathogens have developed specific mechanisms to deregulate the expression levels and/or kinetics of these defense genes. Host transcription factors are first obvious targets to reprogram the genome and bacteria use diverse tricks to alter their function. For instance, bacterial factors can hijack cellular signaling pathways that activate or sequester transcription factors (e.g., NF-κB, IRF/STATs, or AP-1) in the cytosol of targeted cells, or manipulate their half-lives via posttranslational modifications (Bhavsar et al. 2007; Ribet and Cossart 2010; Perrett et al. 2011). Some bacteria, such as the phytopathogen Xanthomonas, even produce transcriptional activators that function as eukaryotic transcription factors (Kay et al. 2007).
However, selective activation or silencing of specific genes not only depends on transcription factors, but also on their cross talk with epigenetic modulators, which regulate DNA accessibility by controlling the chromatin structure. Epigenetic modifications of chromatin during development and in response to distinct environmental factors contribute to adult phenotypic variability and susceptibility to a number of diseases, including cancers and metabolic and autoimmune disorders (van Vliet et al. 2007; Liu et al. 2008; Wilson 2008; Portela and Esteller 2010). This article uncovers the most recent data highlighting that epigenetic changes can also contribute to and/or result from bacterial infectious diseases. After an overview of the complex mechanisms governing chromatin dynamics, we will describe how they may contribute to host response to infection and/or are hijacked by bacteria to impose a transcriptional program beneficial for infection. Only animal pathogenic bacteria will be addressed here; for chromatin targeting by plant pathogenic bacteria, we refer to recent reviews (Ma et al. 2011b; Rivas 2011; Bierne and Cossart 2012).
CHROMATIN REGULATION OF GENE EXPRESSION AND THE EPIGENETIC MEMORY
In eukaryotic cells, DNA is packaged with histones into chromatin, allowing its confinement into the tight space of the nucleus. The state of compaction of chromatin not only organizes the genome but also plays a major role in nuclear processes requiring access to DNA, i.e., transcription, replication, recombination, and DNA repair. The structure of chromatin has several levels of organization (Woodcock and Dimitrov 2001). On the lowest level, the structure is based on repeating units, the nucleosomes, which consist of octamers of histone proteins (two copies of each of the core histones H2A, H2B, H3, and H4) around which ∼147 base pairs of DNA is wound. Nucleosomes line up along the DNA in nucleosomal arrays and associate with linker histone H1 (or its isoforms), nonhistone proteins, and RNAs, forming higher-order chromatin structures, such as the loosely packaged and transcriptionally active euchromatin, or the highly condensed and transcriptionally silent heterochromatin. Chromatin-remodeling and modifying mechanisms function together to dynamically control the state of chromatin compaction and open or close regions of the genome for appropriate outcomes.
ATP-Dependent Remodeling of Nucleosomes
Sequence-dependent physical properties of DNA, thermal motion, and binding of transcription factors to DNA influence nucleosome mobility. Yet, the movement of histone octamers relative to DNA is mainly catalyzed by ATP-dependent remodeling enzymes that use energy from ATP hydrolysis to move, destabilize, evict, or reassemble nucleosomes (Hota et al. 2011). These ATPases are encoded by ∼27 genes in humans and are usually associated with several other proteins within multiprotein complexes, grouped into four families: SWI/SNF, ISWI, CHD, INO80/SWR (all abbreviations are listed in Table 1) (Gangaraju and Bartholomew 2007; Hargreaves and Crabtree 2011). These complexes contribute either to activation or repression of transcription or both, depending on their interaction with histones and other chromatin-binding proteins.
Table 1.
Abbreviations and acronyms
| Name | Full name | Function |
|---|---|---|
| 5mC/5mU | 5-Methylcytosine/5-methyluracyl | Modified DNA base |
| 5hmC/5hmU | 5-Hydroxymethylcytosine/5′-hydroxymethyluracyl | Modified DNA base |
| 5fC | 5-Formylcytosine | Modified DNA base |
| 5caC | 5-Carboxylcytosine | Modified DNA base |
| AID/APOBEC | Activation-induced cytidine deaminase/apolipoprotein B mRNA-editing enzyme catalytic polypeptide | Nucleic acid mutator |
| BER/NER | Base excision repair /nucleotide excision repair | DNA repair |
| BAHD1 | Bromo adjacent homology domain-containing 1 | Chromatin-repressor complex subunit |
| CHD3/4 | Chromodomain-helicase-DNA-binding protein 3/4 (Mi-2α/Mi-2β) | Nucleosome remodeler |
| CIITA | Class II, major histocompatibility complex, transactivator | Transcription factor |
| DNMT | DNA methyltransferase | DNA writer |
| G9a (EHMT2) | Euchromatic histone-lysine N-methyltransferase 2 (EHMT2) | Histone writer |
| H2AK119ub | Histone H2A ubiquitylated at lysine 119 | Modified histone |
| H3K9me 2/3 | Histone H3 di/trimethylated at lysine 9 | Modified histone |
| H3K27me3 | Histone H3 trimethylated at lysine 27 | Modified histone |
| H3K9ac | Histone H3 acetylated at lysine 9 | Modified histone |
| H3K14ac | Histone H3 acetylated at lysine 14 | Modified histone |
| H3K23ac | Histone H3 acetylated at lysine 23 | Modified histone |
| H3T3 | Histone H3 phosphorylated at threonine 3 | Modified histone |
| H3K9ac | Histone H3 acetylated at lysine 9 | Modified histone |
| H3S10p | Histone H3 phosphorylated at serine 10 | Modified histone |
| H4K9me | Histone H4 methylated at lysine 9 | Modified histone |
| HMT | Histone methylatransferase | Modified histone |
| HDM | Histone demethylase | Histone writer |
| HDAC | Histone deacetylase | Histone writer |
| HMT | Histone methyltransferase | Histone writer |
| HP1 | Heterochromatin protein 1 | Chromatin-silencing factor reader |
| IFN-γ | Interferon γ | Cytokine |
| INO80 | INOsitol requiring protein 80 | Nucleosome remodeler |
| IKKα | IκB kinase α | Kinase |
| ISG | Interferon-stimulated gene | Gene |
| ISWI | Imitation switch (ISWI) | Nucleosome remodeler |
| JNK | c-Jun amino-terminal kinases | Kinase |
| LPS | Lipopolysaccharide | Major component of the outer membrane of gram-negative bacteria |
| LSD1 | Lysine (K)-specific demethylase 1A (KDM1A) | Histone writer |
| MAMPS | Microbe-associated molecular patterns | Bacterial molecules |
| MAPK | Mitogen-activated protein (MAP) kinases | Kinase |
| MBD1 | Methyl-CpG-binding protein 1 | Chromatin-silencing factor; 5mC reader |
| MSK1/2 | Mitogen- and stress-activated kinase 1/2 | Kinase-histone writer |
| MTA1/2/3 | Metastasis-associated gene 1/2/3 | Chromatin-remodeling complex subunit |
| NF-κB | Nuclear factor κ-light-chain enhancer of activated B cells | Transcription factor |
| NuRD | Nucleosome remodeling and histone deacetylase | Chromatin-remodeling complex |
| p300/CBP | E1A binding protein p300/CREB-binding protein | Histone writer, reader |
| PTB | Splicing factor | |
| PRC1/PRC2 | Polycomb repressive complex 1/2 | Chromatin-repressor complex |
| SETDB1 | SET domain, bifurcated 1 | Histone writer |
| SIN3A | SIN3 transcription regulator homolog A; histone deacetylase complex subunit Sin3a | Chromatin-repressor complex subunit |
| SWI/SNF | Switch/Sucrose Nonfermentable | Nucleosome remodeler |
| TET | Ten-eleven translocation | Enzyme converting 5-meC to 5hmC |
| TNF-α | Tumor necrosis factor-α | Cytokine |
| TLR | Toll-like receptor | Receptor for MAMPS |
| TNF-R | Tumor necrosis factor-receptor | Cytokine receptor |
| TSIX | TSIX transcript, XIST antisense RNA | Noncoding RNA |
| VRK1 | Vaccinia-related kinase 1 | Kinase |
| XIST | X-inactive specific transcript | Noncoding RNA |
CREB, cyclic AMP response element binding protein; PTB, phosphotyrosine binding; MAMPS, microbe-associated molecular patterns.
Posttranslational Modifications of Histones and DNA Methylation: The Epigenetic Code
Nucleosome stability is also deeply influenced by changes occurring in the nucleosome itself. The histone octamer can be modified by exchange of histone variants and by a plethora of posttranslational modifications (PTMs), such as acetylation, methylation, phosphorylation, ubiquitylation, sumoylation, ADP ribosylation, and others, mainly in the histone tails that protrude from the nucleosome (Kouzarides 2007; Suganuma and Workman 2011), but also, as shown more recently, in globular domains (Tropberger and Schneider 2011). DNA can also be modified by methylation of the 5 position of the pyrimidine ring of cytosine (5mC), mainly in the context of CpG dinucleotides in somatic mammalian cells (Klose and Bird 2006; Chen and Riggs 2011).
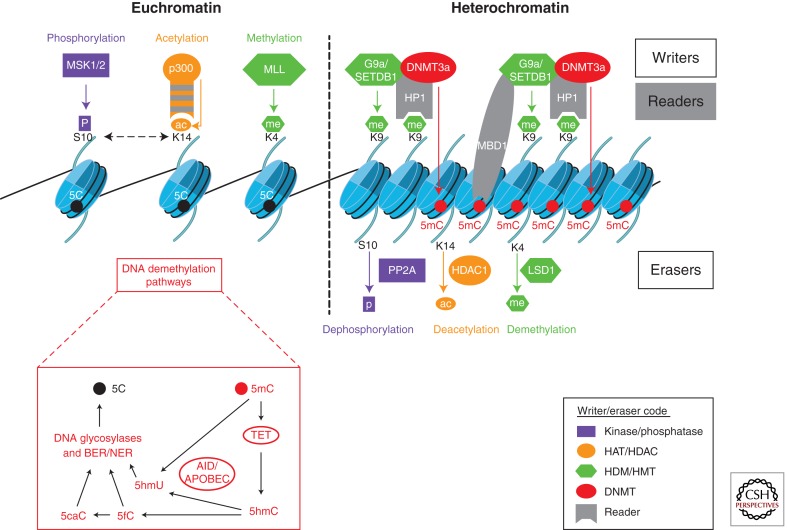
These covalent modifications of chromatin, the “chromatin marks,” are added or removed by a wide range of enzymes termed as “writers,” such as histone acetyltransferases (HATs), methyltransferases (HMTs), and kinases, and “erasers,” such as histone deacetylases (HDACs), demethylases (HDMs), and phosphatases (Fig. 1) (Zhou et al. 2011). In the case of DNA methylation, writers are DNA methyltransferases (DNMTs), either involved in establishing methylation (i.e., the “de novo” methyltransferases DNMT3a and DNMT3b) or copying methylation patterns to the newly synthesized DNA strand during replication (i.e., the “maintenance” methyltransferase DNMT1). 5mC is a relatively stable mark and its erasure is more complex than that of histone PTMs. Intense recent research has nevertheless started to identify enzymes capable of modifying preexisting methylation patterns. DNA methylation is reversible either by a passive way, when 5mC is not copied during DNA replication, or by active mechanisms involving intermediate chemical modifications of 5mC, followed by passive demethylation or DNA repair, as detailed in Figure 1 (Bhutani et al. 2011; Chen and Riggs 2011; Wu and Zhang 2011).
Figure 1.
Chromatin modifications and regulators. Chromatin modifications open or close the chromatin structure, thereby activating or repressing gene expression. Serine phosphorylation on lysine 10 of histone H3 (S10p), acetylation on lysine 14 of H3 (K14ac), and methylation on lysine 4 of H4 (K4me) are examples of activating marks. Conversely, dephosphorylation, deacetylation, and demethylation of the same residues are associated with repression. Methylation of lysine 9 of H3 (K9me) and 5-cytosine methylation of DNA (5mC) are also repressive marks. These modifications are catalyzed or reversed by different enzymes known as “writers” or “erasers,” respectively. An example of writer and eraser for each modification is shown in the color of the modification. Erasure of DNA methylation involves intermediate chemical modifications of 5mC, followed by passive demethylation or DNA repair. 5mC can be hydroxylated by TET dioxygenases (Tahiliani et al. 2009) to form 5hmC, and further oxidized to produce 5fC and 5caC (Wu and Zhang 2011). 5hmC is poorly recognized by DNMT1 and thus can lead to passive demethylation. In addition, 5mC and 5hmC can both be deaminated by AID/APOBEC deaminases to form modified uracyls, 5mU or 5hmU. Excision of modified bases (5mU, 5hmU, 5fC, and 5caC) by DNA glycosylases followed by repair via BER (base excision repair)/NER (nucleotide excision repair) is proposed to regenerate the unmethylated cytosine (Bhutani et al. 2011; Wu and Zhang 2011). Modified residues are recognized and interpreted by different protein modules, known as “readers.” For instance, the bromodomain of HAT p300 binds H3K14ac, the chromodomain of HP1 binds H3K9me, and the methyl-CpG-binding domain (MBD) of MBD1 binds 5meC. Examples of epigenetic cross talks are shown: H3S10p is often associated with H3K14ac and H3K9ac (not shown). H3K9me is often associated with 5mC, on cross-interactions between the H3K9 methyltransferase (G9a/SETDB1), DNMT, HP1, and MBD1. Overall, the chromatin compaction, loss of histone activation marks, and removal of transcription factors accompany gene silencing. HAT, histone acetyltransferase; HDAC, histone deacetylase; HMT, histone methyltransferase; HDM, histone demethyltransferase; DNMT, DNA methyltransferases. Abbreviations are listed in Table 1.
DNA methylation, especially at promoters and enhancers, is mainly coupled with transcriptional silencing, whereas histone modifications are involved in both positive and negative regulation of transcription. For instance, H3 methylated at lysine 4 (H3K4me), phosphorylated at serine 10 (H3S10p), and acetylated at lysine 14 (H3K14ac) are linked to transcriptional activation, whereas deacetylated histones and H3 methylated at lysine 9 (H3K9me) (Fig. 1) and lysine 27 (H3K27me) are associated with repression. Some genes can have both repressive and active histone marks, and constitutively bind RNA polymerase II at their proximal promoters, thus being prepared for efficient future transcription while remaining silent (Mikkelsen et al. 2007). Such so-called “poised” genes can operate in cells in which a rapid change in expression levels is required, enabling cell plasticity (Cuddapah et al. 2010).
Chromatin modifications affect gene expression in two nonexclusive ways. First, they can directly alter the nucleosome structure. In particular, acetylation and phosphorylation of histones are proposed to attenuate the electrostatic interactions between basic histones and negatively charged DNA, thereby increasing nucleosome mobility and access of transcription factors to DNA (Fenley et al. 2010). Second, histone PTMs and 5mC serve as “signaling platforms” that recruit chromatin-modifying and/or regulatory proteins, termed “readers,” which themselves interact or stabilize other chromatin components (Fig. 1). Readers are docked to histone PTMs or methylated DNA via chromatin-binding modules, such as methyl-CpG binding domains of MBD proteins that interact with methylated DNA (Sasai and Defossez 2009), or bromodomains, chromodomain, 14-3-3, Tudor, plant homeodomains (PHDs), etc., which interact with specific histone PTMs (Taverna et al. 2007). For instance, the chromodomain of heterochromatin protein 1 (HP1) binds H3K9me2/3, which initiates heterochromatin formation and gene silencing (Bannister et al. 2001; Lachner et al. 2001). Conversely, the bromodomain of the HAT p300/CBP binds acetylated-lysine histone residues, and is important for the enzyme activity and transcriptional activation (Fig. 1) (Chen et al. 2010).
A Ballet of Chromatin-Modifying and -Remodeling Complexes: The Epigenetic Language
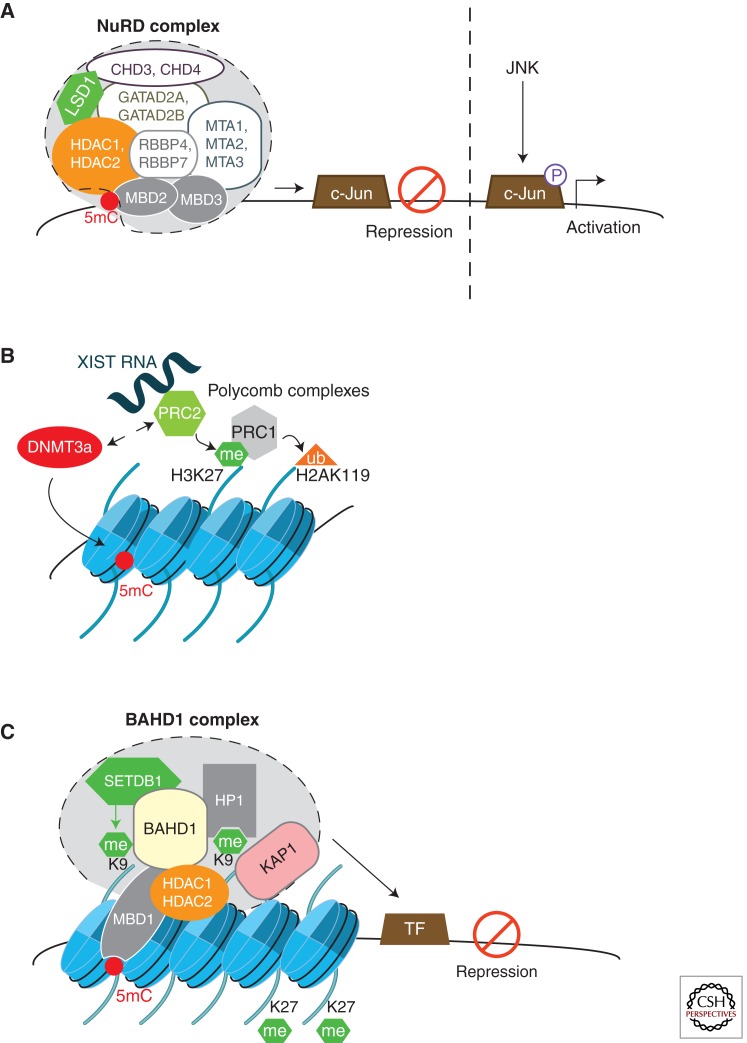
Writers, readers, and erasers are often modular proteins displaying several properties. This is well exemplified by p300/CBP, which is a writer (via its HAT module), a reader (via its bromodomain), and an adaptor (via other modules, such as transcription factor and steroid receptor interaction domains), thus functioning as a transcriptional integrator. In addition, chromatin-modifying enzymes usually assemble into multisubunit complexes, together with nonenzymatic subunits acting as readers and/or as linkers for sequence-specific DNA-binding factors. For instance, the NuRD complex (Fig. 2A) contains subunits involved in nucleosome remodeling (CHD3 and CHD4), histone deacetylation (HDAC1 and HDAC2), histone demethylation (LSD1), and in binding to other subunits and histones (RBBP4, RBBP7, GATAD2A, and GATAD2B), to methylated DNA (MBD2) and to transcription factors (MTA1, MTA2, MTA3, and MBD3) (Hayakawa and Nakayama 2011; Lai and Wade 2011). Combinatorial assembly of these subunits determines the function of NuRD in genomic targeting and mediating cell type-specific transcriptional regulations, such as transcriptional repression of tumor-suppressor genes.
Figure 2.
Three examples of chromatin-modifying complexes and their mode of action. (A) The nucleosome remodeling and histone deacetylase NuRD (or Mi-2) complex contains different subunits involved in nucleosome remodeling (CHD3/CHD4), histone deacetylation (HDAC1/HDAC2), histone demethylation (LSD1), and binding to transcription factors and histones. Additionally, MBD2 recruits NuRD to methylated DNA, whereas MBD3 (whose MBD region is mutated and does not bind methylated DNA) interacts with transcription factors. The function and targeting of the complex to specific loci depends on the combinatorial assembly of the different subunit isoforms. Most often, recruitment of the NuRD complex by a tissue-specific transcription factor to gene promoters mediates transcriptional repression. In the case of the transcription factor c-Jun of the AP1 family, recent studies suggest that its recruitment to the NuRD complex involves MBD3 interaction with unphosphorylated c-Jun. On activation of the JNK pathway, c-Jun phosphorylation mediates MBD3/c-Jun dissociation, resulting in derepression of transcription of target genes (e.g., cell-cycle and cell-differentiation genes) (Aguilera et al. 2011). Abbreviations are listed in Table 1. (B) Mammalian PcG proteins form two multiprotein complexes, polycomb repressive complexes 1 and 2 (PRC1 and PRC2). PRC2 contains a histone-methyltransferase subunit (EZH2, not shown) that generates H3K27me3. PRC1 binds H3K27me3 and catalyzes ubiquitinylation of H2AK119. An early event in chromosome X inactivation in mammalian females is the recruitment of PRC2 by the XIST long coding RNA, inducing H3K27me3 and H2AK119ub along the inactive X chromosome. PRC2 has the ability to recruit DNMTs to some of its target genes, thereby stimulating de novo DNA methylation. (C) The BAHD1-associated chromatin-silencing complex has been discovered by the study of the bacterial protein LntA from L. monocytogenes. Two-hybrid screen, coimmunoprecipitation, and colocalization experiments (Bierne et al. 2009), as well as tandem-affinity purification (Lebreton et al. 2011), identified components associated to BAHD1, i.e., HP1, MBD1, KAP1, histone methyltransferase (SETDB1), and deacetylases (HDAC1/2). Our data suggest that BAHD1 acts as a silencer by tethering chromatin regulators and modifying enzymes to sequence-specific transcription factors (TF), enabling local chromatin compaction. BAHD1-associated heterochromatin sites are enriched in H3K27me3, but the relationship with PRC2 is unknown. Like for NuRD, the function and targeting of the BAHD1 complex to specific genes likely depend on the combinatorial assembly of the different subunits, in response to signals to which cells are submitted. On Listeria infection, the BAHD1 complex assembles at promoters of a set of interferon-stimulated genes (ISGs), as shown in Figure 4.
These macromolecular complexes may function alone or with others to orchestrate, in time and space, a specific chromatin state. The vast array and different combinations of histone PTMs coordinate the sequential recruitment of complexes, a stepwise regulation that reinforces or reverses existing histone PTMs (Latham and Dent 2007; Lee et al. 2010; Suganuma and Workman 2011). For instance, H3S10p is required for acetylation at lysine 14 on the same histone by the HAT Gcn5. Another example is the sequential recruitment of the polycomb repressive complexes. PRC2 “writes” H3K27me3, while PRC1 “reads” it and induces monoubiquitylation of histone H2A, ultimately leading to chromatin compaction at target genes (Fig. 2B) (Margueron and Reinberg 2011). Such cross talk also takes place between histone PTMs and DNA methylation. Interaction of H3K9 HMT (e.g., SETDB1, G9a) or H3K27 HMT (PRC2) with DNMT3A/B is proposed to promote DNA methylation at specific regions (Figs. 1 and 2B) (Li et al. 2006; Vire et al. 2006). Once formed, 5mC sites recruit the methyl-CpG binding proteins, such as MBD1, which can interact with both H3K9 HMTs and HP1 (Fujita et al. 2003; Ichimura et al. 2005). Such sequence of events may explain the spreading of repressive marks at targeted loci.
The recruitment, regulation, and interactions of the complexes implementing chromatin marks trigger dynamic combinatorial patterns that are proposed to form a language (Lee et al. 2010). The “grammar” of this language also involves modification of nonhistone substrates by chromatin-modifying enzymes (Xu et al. 2007; Rathert et al. 2008), as well as a wide range of PTMs induced by cell signaling pathways that alter interaction or stability of subunits of chromatin complexes and transcription factors. This translates signal transduction information to the chromatin structure in response to different environmental signals (Mohammad and Baylin 2010). Phosphorylation and sumoylation are important modifications in the function of a variety of repressor complexes (Garcia-Dominguez and Reyes 2009; Baek 2011) and, for instance, play a central role in the targeting of HP1α to chromatin (Hiragami-Hamada et al. 2011; Maison et al. 2011). Phosphorylation of the transcription factor c-Jun by JNK kinase impairs the binding of c-Jun to the MBD3 subunit of the NuRD complex, thereby relieving repression of target genes (Fig. 2A) (Aguilera et al. 2011). JNK, MSK1/2, IKKα, and several other kinases of signal transduction pathways can also directly phosphorylate histones in the nucleus (Yamamoto et al. 2003; Baek 2011; Tiwari et al. 2011). Thus, signaling molecules activated in cells in response to external stimuli have key effects on the chromatin syntax.
Other Epigenetic Regulators: Noncoding RNAs and RNA Splicing Factors
Additional complexity was recently added to the epigenetic landscape by the discovery of a crucial role of RNAs and RNA-binding proteins in dynamic changes of the chromatin structure and spatial organization of the genome inside the nucleus (for reviews, see Bernstein and Allis 2005; Faghihi and Wahlestedt 2009; Chen and Carmichael 2010; Grewal 2010; Turner and Morris 2010; Kaikkonen et al. 2011).
Eukaryotes transcribe most of their genomic DNA, albeit only 1%–2% of all cellular transcripts encode proteins. The vast majority of RNAs are thus noncoding RNAs (ncRNA) of diverse size (from small 17–30 bp to long >200 bp ncRNAs), which can be generated from intergenic regions, exons, introns, or enhancers, in sense or antisense orientation (Mattick and Makunin 2005; Tisseur et al. 2011). Most reported data link ncRNAs to gene silencing. Well-known examples are two long noncoding transcripts, XIST and TSIX, which regulate inactivation of one of the two X chromosomes in female mammals (Nora and Heard 2010; Morey and Avner 2011). XIST transcripts coat the inactive X chromosome in cis and recruit chromatin-remodeling complexes, such as PRC2 and PRC1, that trigger repressive marks and heterochomatin formation (Fig. 2B) (Pontier and Gribnau 2011; Wutz 2011). The action of XIST is antagonized by TSIX, a noncoding antisense RNA to the Xist gene that represses Xist transcription through chromatin modifications. The mechanism at play might involve XIST/TSIX RNA duplexes processed by the RNAi machinery and subsequent recruitment of chromatin-remodeling complexes by small interfering RNAs (siRNAs; see below) (Ogawa et al. 2008). Numerous cases of sense–antisense transcript pairs have now been discovered in mammalian genomes and increasing evidence suggests that antisense RNAs might play a general role in epigenetic changes by recruiting DNA methyltransferase and/or histone-modifying enzymes (Faghihi and Wahlestedt 2009; Kaikkonen et al. 2011). Additionally, long ncRNAs were recently shown to act as scaffolds, localizing genes to specific subnuclear regions for either repressive or activating effects (Yang et al. 2011).
Mammalian cells also produce small ncRNAs that were originally characterized for their role in posttranscriptional gene silencing in the cytoplasm, by targeting specific mRNAs for degradation or translational inhibition. However, recent data indicate that small ncRNAs also play an important role in chromatin dynamics in the nucleus. These RNAs are generated by cleavage of double-stranded RNAs (dsRNAs) by dsRNA-specific RNase III-type enzymes: microRNAs (miRNAs) derive from unique hairpin-shaped precursors in long transcripts, whereas endogenous small interfering RNAs (endo-siRNAs) are formed by cleavage of long dsRNA generated at repeated sequences (e.g., centromere, telomere, and transposons) or by antisense transcription (Turner and Morris 2010; Rother and Meister 2011). Current models suggest that the regulatory functions of nuclear small ncRNAs and associated factors in gene expression are to serve as guides for chromatin-modifying complexes and target them at specific sites, in cis or trans, possibly through base pairing or formation of an RNA/DNA triple helix (Schmitz et al. 2010).
Last, but not least, mounting evidence suggests that chromatin modifications also contribute to pre-mRNA alternative splicing, which in eukaryotes allows for one gene to encode functionally distinct proteins (Allemand et al. 2008; Hnilicova and Stanek 2011). The spliceosome, a complex of small nuclear RNA (snRNA) and protein subunits (snRNPs, small nuclear ribonucleoprotein particles) that removes introns, seems to be influenced by chromatin modifications. For instance, HDAC inhibition changes alternative splicing of ∼700 genes in human cells (Hnilicova et al. 2011). In addition, splicing regulators, such as PTB or hnRNPs, have been shown to interact with chromatin regulators, such as MRG15 (Luco et al. 2010) or HP1, respectively (Piacentini and Pimpinelli 2010). Interestingly H3K9me2/3 marks are enriched in introns, in contrast to exons, and a reader of such marks, HP1γ, was recently shown to modulate alternative splicing (Saint-Andre et al. 2011). Furthermore, siRNA-mediated heterochromatin formation seems to contribute to splicing events (Allo et al. 2009). Splicing decisions are thus likely assisted by chromatin factors and ncRNAs.
The Epigenetic Memory
Chromatin modifications can promote mitotically heritable changes in gene expression without altering the DNA sequence, especially during cell differentiation. This property to persist through cell division, despite the fact that the initiating signal is no longer present, and thus allowing a cell to “remember” its transcriptional profile and identity, defines epigenetic regulation (Russo et al. 1996). Chromatin modifications are thus commonly referred to as “epigenetic” modifications. However, many marks induced by cell signaling, DNA repair, or cell-cycle transitions, are short-lived and do not give rise to long-term memory storage. To account for both transient and stable modifications of the epigenetic language, we thus prefer another definition of epigenetic events: “the structural adaptation of chromosomal regions so as to register, signal, or perpetuate altered activity states,” recently proposed by Adrian Bird (Bird 2007). When inherited, chromatin modifications generate an epigenetic memory.
BACTERIAL CYTOSOLIC SIGNALING TO CHROMATIN
Mechanisms controlling gene expression at the chromatin level show high levels of complexity. Various bacterial products can affect them in many ways (Table 2), through activation of signaling cascades or directly in the nucleus, as we will review now.
Table 2.
Bacterial species, factors, and effects
| Bacterial species | Bacterial factor | Effect |
|---|---|---|
| Anaplasma phagocytophilum | ? | Histone deacetylation and silencing of host defense genes |
| AnkA | Binding to chromatin at AT-rich sequences; silencing of CYBB | |
| Bacillus anthracis | LT | Inhibition of MAPK and of H3S10p and down-regulation of IL-8 and KC genes. |
| Campylobacter rectus | ? | Hypermethylation in the promoter region P0 of the IGF2 gene in the murine-infected placenta |
| Chlamydia trachomatis | NUE | Methylation of mammalian histones |
| Escherichia coli (uropathogenic) | ? | DNA methylation and down-regulation of CDKN2A |
| Ehrlichia chaffeensis | Binding to chromatin at Alu-Sx elements | |
| Helicobacter pylori | Induction of DNA methylation in gastric mucosa | |
| Induction of H3 modifications | ||
| Induction of miRNA expression | ||
| Induction of RNA splicing factors | ||
| Legionella pneumophila | Flagellin | Histone acetylation in infected lung epithelial cells |
| Listeria | ? | Acetylation of H4 and phosphorylation/acetylation of H3, in particular at the IL-8 promoter and decreased binding of HDAC1 at the IL8 promoter in Listeria-infected endothelial cells |
| LLO | Histone modifications and modulation of host defense genes by a signaling pathway involving K+ efflux | |
| ? | Down-regulation of ISG by BAHD1-associated complex in epithelial cells | |
| LntA | Up-regulation of ISG by LntA-mediated inhibition of BAHD1-associated complex | |
| ? | Change in expression of a subset of host miRNAs | |
| Moraxella catarrhalis | Reduction in the global expression and activity of HDAC1/2 in airway epithelial cells | |
| Mycobacterium tuberculosis | Control of chromatin complex at ISG downstream from IFN-γ | |
| Porphyromonas gingivalis | Reactivation of latent viruses via chromatin modification induced by butyric acid | |
| Salmonella | Change in expression of a subset of host miRNAs | |
| Shigella flexneri | OspF | Down-regulation of MAP kinase in the nucleus by eliminylation; inhibition of phosphorylation of H3S10; down-regulation of immune response genes inhibition of NF-κB chromatin access leading to the inactivation of IL-8 and other genes essential for innate immune responses |
| OspB | Binding to Rb | |
| IpaH9.3 | Ubiquitin ligase, which targets a splicing factor for degradation and impairs splicing | |
| Bacterial product | LPS | Inducer of innate immunity via activation of TLR4-mediated responses and the production of proinflammatory cytokines; induces immunosuppression by chromatin modifications on repeated challenge |
| Butyrate | Inhibition of HDAC activity |
LT, lethal toxin; LLO, listeriolysin O; LPS, lipopolysaccharide.
Histone Modifications
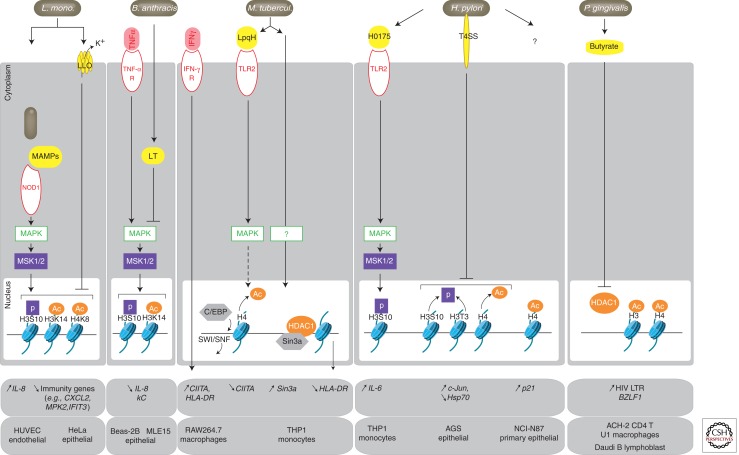
So far, most of the reported chromatin modifications induced by bacteria are histone acetylation/deacetylation and phosphorylation/dephosphorylation events generated through activation of host cell signaling cascades by bacterial components (e.g., microbe-associated molecular patterns, metabolites, and virulence factors) (Fig. 3). The effects are complex, because they differ according to the bacterial agonist, cell type, and kinetics parameters. Among the host signaling pathways that a number of bacteria activate, MAPKs (e.g., ERK and p38), NF-κB, and PI3K pathways are known to activate the kinases that phosphorylate H3S10 in the nucleus (i.e., MSK1/2, IKKα, and AKT, respectively) (Yamamoto et al. 2003; Baek 2011). Any bacterial stimulus activating these pathways has therefore the potential to induce H3S10p and associated acetylated histones. Examples of these are listed below.
Figure 3.
Bacterial signaling to histones and downstream effects. Schematic representation of Listeria monocytogenes-, Bacillus anthracis-, Mycobacterium tuberculosis-, Helicobacter pylori-, or Porphyromonas gingivalis-induced signaling pathways leading to histone modifications, as detailed in the text. Bacterial products inducing host cellular signaling are in yellow. Membrane (TNF-α-R, IFN-γ, TLR2, TLR4) or cytosolic (NOD1) receptors are indicated by a red oval. Effects on target genes are indicated by arrows (up for activation and down for repression). Cell types in which studies have been performed are listed below. Abbreviations are listed in Table 1.
In endothelial HUVEC cells, the causative agent of listeriosis, Listeria monocytogenes, rapidly increases phosphorylation and acetylation of histones (H3S10p, H3K14ac, and H4K8ac), and specifically at the promoter of the proinflammatory gene IL-8 (Schmeck et al. 2005; Opitz et al. 2006). This effect involves NOD1-mediated sensing of cytosolic bacteria and activation of MAP kinases (MAPK), followed by enhanced binding of HAT p300/CBP and decreased binding of HDAC1 to the IL-8 promoter (Fig. 3). Flagellin of Legionella pneumophila, the causative agent of Legionnaire disease and bacterial lipopolysaccharide (LPS), has similar effects on IL-8 in lung epithelial cells and dendritic cells, respectively (Saccani et al. 2002; Schmeck et al. 2008). Not only pathogenic but also commensal bacteria, such as Moraxella catarrhalis, a saprophytic bacterium of the respiratory tract, or Bacteroides vulgatus, a commensal of the intestinal flora, can induce phosphorylation/acetylation of H3 via induction of an inflammatory signaling cascade (Haller et al. 2003; Slevogt et al. 2006).
However, with such an important role on inflammation, MAPK signaling is often counteracted by bacterial-induced mechanisms. In the case of B. vulgatus, this is performed by bacterial induction of the TGF-β1 anti-inflammatory pathway, which in turn induces H3 deacetylation via HDAC recruitment at proinflammatory gene promoters (Haller et al. 2003). This prevents B. vulgatus from eliciting an inflammatory response in the gut and contributes to its tolerance by the host. Bacterial toxins also dampen the host innate immune responses by inhibiting H3 phosphorylation/acetylation. Once endocytosed into the host cell, lethal toxin (LT) from Bacillus anthracis, the agent of anthrax, cleaves and inactivates MAPKKs leading to disruption in MAPK signaling (Bardwell et al. 2004). In lung epithelial cells activated by TNF-α, LT-mediated MAPK inhibition promotes a decrease in the levels of H3S10p and H3K14ac at the promoter of IL-8 and KC genes (Fig. 3) (Raymond et al. 2009). Pore-forming toxins that “intoxicate” cells by disruption of membrane permeability use another signaling pathway to change histone marks. In epithelial HeLa cells, L. monocytogenes listeriolysin O (LLO) promotes deacetylation and dephosphorylation of histones, in particular at a subset of immune genes, such as CXCL2, MKP2, or IFIT3, which become repressed (Fig. 3) (Hamon et al. 2007). The mechanism at play involves a K+ efflux through pore formation at the membrane (Hamon and Cossart 2011). Other pore-forming toxins, such as PFO of Clostridium perfringens, PLY of Streptococcus pneumonia, and Aerolysin from Aeromonas hydrophila have similar effects, suggesting that all of these toxins activate an intracellular K+ sensor leading to a pathway that modifies histones and subsequent gene expression (Hamon and Cossart 2011).
Of note, MAP kinases have several downstream targets other than MSK1/2, some of which are involved in histone deacetylation and transcriptional repression at later time points (Yang et al. 2001). This might explain why bacterial-induced MAPK signaling does not always lead to transcriptional activation of immune genes, as exemplified by Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis. Control of Mtb infection requires IFN-γ, but some IFN-γ-induced genes are repressed in macrophages infected with Mtb or exposed to Mtb components, such as the lipoprotein LpqH (Wang et al. 2005; Pennini et al. 2006). These genes include CIITA, coding for the master regulator of MHC class II genes, as well as some of its targets (e.g., HLA-DR). Activation of the TLR2/MAPK-dependent pathway on Mtb infection stimulates recruitment of the transcriptional repressor C/EBP and histone deacetylation at the promoter of CIITA, antagonizing the nucleosome-remodeling activity of the SWI/SNF complex and down-regulating CIITA expression (Fig. 3) (Pathak et al. 2006; Pennini et al. 2007). Additionally, mycobacterial infection up-regulates expression of Sin3A encoding a corepressor that acts in concert with HDACs to repress a number of genes, including MHC class II genes (Wang et al. 2005). Thus, to counteract IFN-γ-induced pathways, Mtb not only silences CIITA, but also CIITA-regulated genes, such as HLA-DR, on increased recruitment of Sin3A-HDACs to their promoters (Fig. 3).
The carcinogenic bacterium Helicobacter pylori is also capable of modifying histones by diverse means, including the modulation of the MAPK pathway. The peptidyl prolyl cis-, trans-isomerase HP0175 secreted by H. pylori binds the innate immunity receptor TLR4, which in turn activates MAPKs and the downstream MSK1-dependent phosphorylation of H3, in particular, at the IL-6 promoter in THP-1 monocytes (Fig. 3) (Pathak et al. 2006). In contrast, in gastric epithelial cells H. pylori has an opposite effect by inducing dephosphorylation of H3S10 and H3T3, as well as deacetylation of H3K23 (Fehri et al. 2009; Ding et al. 2010a). H. pylori-mediated chromatin modifications are in this case dependent on a functional type IV secretion system, suggesting involvement of an injected bacterial product. H. pylori-induced dephosphorylation of H3S10 is transient and impacts both the cell cycle (Fehri et al. 2009) and transcription of oncogene c-Jun (positively) and heat shock gene hsp70 (negatively) (Ding et al. 2010a). H. pylori-mediated premitotic arrest involves dephosphorylation of H3S10 upon deregulation of the mitotic histone kinase VRK1, followed by rephosphorylation of H3S10 by an IKKα-dependent pathway. Another study showed that exposure of H. pylori to gastric epithelial cells promotes release of HDAC1 from the promoter of the cell-cycle regulator gene p21WAF, hyperacetylation of H4, and increased expression of p21WAF. Chromatin alterations might contribute to the effects of H. pylori on cell-cycle progression, cellular proliferation, and cell death (Xia et al. 2008).
Changes in the levels of chromatin-modifying components are another possible outcome of bacterial infections. As discussed above, Mtb infection up-regulates expression of Sin3A (Wang et al. 2005), whereas the tick-transmitted pathogen Anaplasma phagocytophilum up-regulates the expression of HDACs in infected cells (Garcia-Garcia et al. 2009a), an effect that contributes to silencing of host defense genes by deacetylation of histones. In contrast, the levels of HDAC transcripts are decreased in gingival epithelial cells treated with oral pathogen Porphyromonas gingivalis and nonpathogenic Fusobacterium nucleatum (Yin and Chung 2011).
Bacteria can also produce metabolites acting as inhibitors of chromatin-modifying enzymes. One such product is butyric acid, a short-chain fatty acid acting as a potent inhibitor of HDACs (Riggs et al. 1977). Interestingly, the adverse effect of Porphyromanas gingivalis in reactivating latent viruses, such as human immunodeficiency virus (HIV) and Epstein–Barr syndrome (EBV), seems to result from production of butyrate by this bacterium (Imai et al. 2009; Imai et al. 2011). It is proposed that viral genes maintained silent by HDAC-containing complexes are reactivated following inhibition of HDACs by butyric acid (Fig. 3). Thus, P. gingivalis infection might be a risk factor for viral diseases, such as AIDS or Herpes. Butyric acid also exerts beneficial anti-inflammatory effects on the host, at least in animal models, possibly via epigenetic up-regulation of anti-inflammatory genes. Such observations open up the interesting possibility to use butyrate-producing probiotic bacteria as immunosuppressors (Licciardi et al. 2010).
DNA Methylation
The importance of DNA methylation events associated with bacterial infections is also becoming increasingly appreciated. The best documented example is H. pylori infection that induces aberrant DNA methylation in the human gastric mucosa, strikingly at promoters of genes found methylated in gastric cancer cells (Maekita et al. 2006; Ding et al. 2010b; Ushijima and Hattori 2012). H. pylori-associated hypermethylation occurs, for instance, at the E-cadherin gene CDH1 (Chan et al. 2003), tumor-suppressor genes (e.g., USF1/2 and WWOX [Bussiere et al. 2010; Yan et al. 2011]), DNA repair genes (e.g., MLH1 [Yao et al. 2006]), as well as to CpG islands of miRNA genes (Ando et al. 2009). The ability of H. pylori to induce DNA methylation in gastric mucosa was confirmed in the gerbil animal model and, interestingly, this effect was diminished on treatment with the immunosuppressor cyclosporin A (Niwa et al. 2010). Indeed, in contrast to ethanol or NaCl stimuli that induce neutrophil infiltration in the stomach, H. pylori-mediated inflammation triggered lymphocyte and macrophage infiltration, which appears to have a key role in induction of methylation (Hur et al. 2011). Thus, although the mechanisms by which H. pylori induces DNA hypermethylation are still unclear, the infection-associated inflammatory response is a tempting explanation (Ding et al. 2010b; Ushijima and Hattori 2012). Among signals resulting from chronic inflammation, elevated levels of IL1β and nitric oxide (NO) are proposed to contribute to influence the recruitment of DNMTs at specific loci.
In fact, evidence for an epigenetic control of the inflammatory response is increasingly recognized (Medzhitov and Horng 2009; Bayarsaihan 2011). In the intestine, some PRC2 target genes are subject to aberrant DNA methylation following chronic inflammation (Hahn et al. 2008). In a noninflammatory context, the LPS-sensing receptor encoding gene TLR4 is down-regulated in intestinal epithelial cells and there is evidence for a role of commensal bacteria in TLR4 methylation (Takahashi et al. 2011). This is proposed to maintain intestinal homeostasis by preventing an excessive inflammatory reaction toward the gut microbiota. In the oral cavity, similar mechanisms may dampen uncontrolled inflammatory response triggered by bacterial-induced chronic infection. In particular, periodontally inflamed gingival biopsies showed a significant increase in promoter methylation of the gene encoding the proinflammatory enzyme COX-2, when compared with noninflamed biopsy samples (Zhang et al. 2010). This would allow a chronic inflammatory stimulus to be tolerated, preventing unrestricted tissue destruction. Whether this is a bacteria-triggered phenomenon is unknown, but it is of note that resident bacteria, such as P. gingivalis, can induce hypermethylation of specific genes in gingival epithelial cells (Yin and Chung 2011).
Bacteria-induced DNA methylation can also affect genes involved in cell proliferation, as illustrated by uropathogenic Escherichia coli (UPEC). In human uroepithelial cells, infection with UPEC results in the up-regulation of DNA methyltransferase (DNMT) activity and DNMT1 expression and induces CpG methylation and down-regulation of CDKN2A, a G1-cell-cycle inhibitor regulator (Tolg et al. 2011). This may increase uroepithelial cell proliferation and pathogen persistence, by counteracting infection-stimulated host cell apoptosis. Other organs can be targeted by bacteria-mediated epigenetic changes, including placenta. Indeed, maternal infection with Campylobacter rectus induces hypermethylation of the imprinted IGF2 gene promoter in murine placental tissue (Bobetsis et al. 2007). This result suggests that bacterial infections during pregnancy might epigenetically affect genes that play an important role in fetal development.
ncRNA and RNA Splicing
It is now well established that miRNAs are important regulators of immune responses (O’Connell et al. 2011). Not surprisingly, several of them are induced in response to pathogenic bacteria, such as H. pylori (Fassi Fehri et al. 2010; Matsushima et al. 2011), Salmonella typhimurium (Schulte et al. 2011), L. monocytogenes, and Mycobacterium bovis BCG (Ma et al. 2011a; Izar et al. 2012), and even by microbiota (Dalmasso et al. 2011). However, in all of the reported studies, miRNAs function in posttranscriptional repression of mRNAs in the cytosol. Whether a bacterial stimulus induces expression of miRNA or endogenous siRNA acts at the chromatin level in the nucleus is unknown.
Likewise, whereas numerous immunologically relevant genes undergo alternative splicing (Lynch 2004), it remains to be explored whether bacteria could affect alternative splicing by changes in chromatin to deregulate the function of the immune system. A recent study analyzing changes in the phosphoproteome of gastric cells on H. pylori infection described an enrichment of RNA processing and splicing factors in infected cells (Holland et al. 2011). Besides this study and the reported targeting of a splicing factor by the Shigella flexneri protein IpaH9.8 (see below) (Okuda et al. 2005), information of the link between splicing and bacterial infection is lacking.
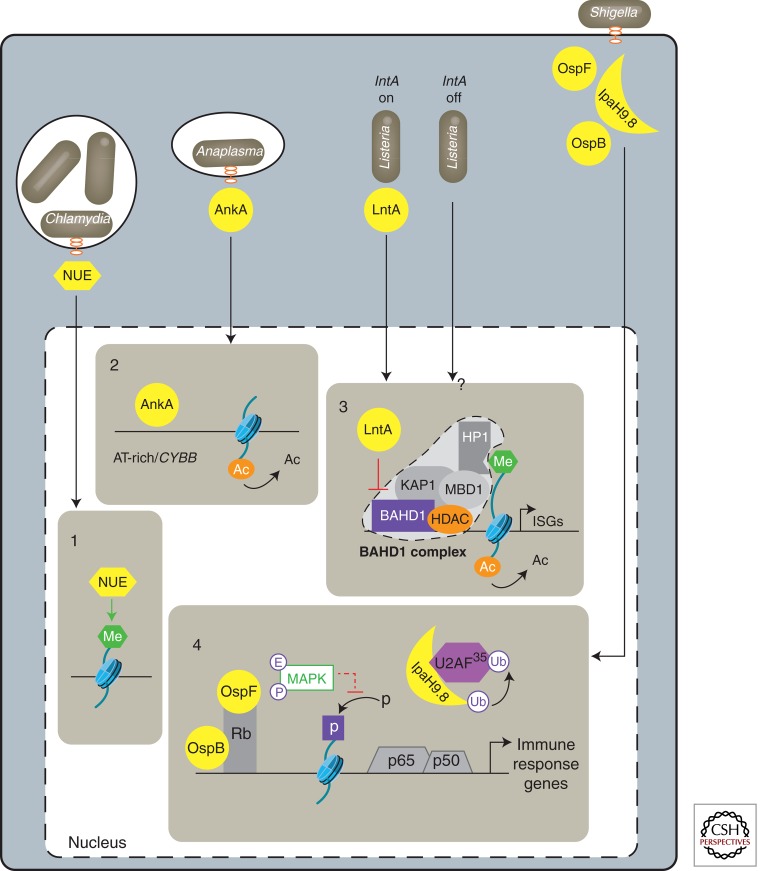
As we have seen, bacteria-mediated signaling pathways affect epigenetic processes in many ways. However, it is not always easy to distinguish what is the contribution of the host response to infection and what is a specific bacteria-targeted mechanism’s on the chromatin changes. The existence of bacterial factors that act directly in the nucleus, which we recently referred to as the “nucleomodulins” (Bierne and Cossart 2012), highlights that bacteria have evolved mechanisms to actively manipulate the chromatin-regulatory machinery in the nucleus and reprogram host gene expression to their advantage (Fig. 4).
Figure 4.
Bacterial nucleomodulins targeting chromatin. Schematic representation of Chlamydia, Anaplasma, Listeria, and Shigella secreted factors involved in the control of gene expression in the nucleus of host cells, as detailed in the text. The bacterial nucleomodulins are in yellow. 1. Chlamydia histone-methyltransferase NUE methylate’s mammalian histones. However, target genes are unknown. 2. Binding to AT-rich sequences and silencing of CYBB expression by Anaplasma Ank effector AnkA. 3. Inhibition of the BAHD1-associated heterochromatic complex and induction of interferon-stimulated genes by Listeria LntA. On Listeria infection, an unknown signaling pathway drives the BAHD1-associated chromatin complex (see Fig. 2C) to repress interferon-stimulated genes. When Listeria produces and secretes LntA, this factor enters the nucleus and interacts with BAHD1, destabilizes the silencing complex, restores histone acetylation (Ac), and enhances the expression of ISGs. 4. Control of a set of NF-κB (p65-p50) regulated genes by Shigella posttranslational modifiers OspF, which eliminylates MAP kinases preventing phosphorylation of histone H3, and IpaH9.8, which ubiquitinylates and promotes degradation of the splicing factor U2AF35. OspF and another effector, OspB, bind the retinoblastoma protein (Rb), which potentially recruits several chromatin-remodeling enzymes (not shown). Ac, Acetylation; Me, methylation; P, phosphorylation; E, eliminylation; UB, ubiquitinylation.
BACTERIAL PROTEINS MIMICKING OR CONTROLLING HOST CHROMATIN-REGULATORY FACTORS
Histone-Modifying Bacterial Enzymes
Several components of the eukaryotic protein methylation system might have their origins in bacteria. In particular, SET domain-containing HMT (e.g., G9a, SETDB1) may have first emerged in prokaryotes from the SAF superfamily of carbohydrate-binding domains (Aravind et al. 2011). Conversely, a subset of the bacterial SET domain-containing protein might have evolved the chromatin-related role of their eukaryotic counterparts. Evidence for this hypothesis emerged from studies of chlamydial SET proteins. A SET domain protein (cpnSET) from Chlamydophila pneumoniae, which causes acute respiratory diseases in humans, was first shown to methylate the chlamydial histone-like proteins HC1/HC2 (Murata et al. 2007). Its homolog nuclear effector (NUE) from Chlamydia trachomatis was subsequently identified as a T3SS effector that enters the nucleus of infected cells, where it associates with chromatin (Fig. 4) (Pennini et al. 2010). Importantly, NUE can methylate mammalian histones H2B, H3, and H4 in vitro, confirming its histone methyltransferase activity. Additionally, nuclear effector (NUE) can methylate itself, a property shared by eukaryotic G9a. However, experimental evidence for a direct relationship between NUE activity in the nucleus and host gene expression is still lacking. Nonetheless, with regard to the number of SET domain proteins present in many bacterial species that interact with eukaryotes, it is tempting to speculate that several bacteria might use this strategy.
Ankyrin Repeat (Ank)-Containing Bacterial Proteins
Several effectors of human intracellular bacterial pathogens, such as Anaplasma, Ehrlichia, Ricketssia, Orientia, Coxiella, and Legionella species contain proteins with Ank eukaryotic motifs. These motifs mediate protein–protein interactions in a multitude of host processes, including transcriptional regulation (Mosavi et al. 2004). Ank-containing proteins, AnkA and p200, from the related pathogens Anaplasma phagocytophilum and Ehrlichia chaffeensis, respectively, bind chromatin in the nucleus of infected cells (Fig. 4) (Park et al. 2004; Zhu et al. 2009). AnkA preferentially targets AT-rich chromatin sites and decreases expression of a set of genes. In particular, AnkA represses CYBB that encodes Cytochrome b-245, a component of the phagocyte oxidase that influences A. phagocytophilum survival (Garcia-Garcia et al. 2009b). This is concomitant to deacetylation of H3 at the CYBB locus. E. chaffeensis p200 binds to chromatin at Alu-Sx elements located in promoters and introns of various human genes (Zhu et al. 2009). Several p200 target genes are strongly up-regulated during infection, suggesting that p200 may affect gene transcription globally through mechanisms associated with Alu element gene regulation.
Bacterial Proteins Targeting or Modifying Chromatin-Remodeling Complexes
Our recent studies identified a bacterial protein that targets a subunit of a chromatin-repressive complex, highlighting that bacteria can directly hijack the heterochromatin machinery. The nuclear-targeted factor LntA from L. monocytogenes interacts with BAHD1, a previously unknown protein that we characterized as a novel heterochromatinization factor in vertebrates. BAHD1 acts in complex with other chromatin factors (e.g., HP1, MBD1, SETDB1, HDACs, and KAP1) to silence gene expression (Fig. 2C) (Bierne et al. 2009; Lebreton et al. 2011). The set of genes repressed by the BAHD1-associated complex depends on the signal to which cells are submitted and on the cell type. In particular, BAHD1 represses interferon-stimulated genes (ISG) in epithelial cells infected with L. monocytogenes (Lebreton et al. 2011). Upon an unknown signal, L. monocytogenes switches on expression of the lntA gene. The secreted LntA factor enters the nucleus and alleviates BAHD1 binding to ISG promoters, thereby up-regulating their expression. By preventing recruitment of BAHD1 and HDAC1/2 at ISG promoters, LntA is thought to trigger chromatin unwinding (Fig. 4). Consistent with this model, acetylation of histone H3 increases at ISG promoters in cells infected with LntA-producing bacteria. We propose that the LntA–BAHD1 interplay spatiotemporally modulates the interferon response to control bacterial colonization of the host (Lebreton et al. 2012). However, questions remain regarding how BAHD1 is specifically recruited to a subset of infection-dependent targets and what signal determines the switch between a lntA nonexpressing and expressing state of Listeria during the infection process.
Another strategy used by bacteria to target chromatin is to modify locally and specifically the activity of key chromatin/transcriptional regulatory factors. This is used by several T3SS effectors from gram-negative pathogens, such as OspF of S. flexneri. OspF is a phosphothreonine lyase that irreversibly modifies host MAPKs by a reaction called eliminylation (Li et al. 2007; Brennan and Barford 2009). This enzymatic reaction converts a phosphothreonine residue into a dehydrobutyrine residue that can no longer be phosphorylated, and hence locks the substrate in an inactive form. Inhibition of MAPK signaling by OspF abrogates phosphorylation of histone H3 at a set of NF-κB-regulated promoters and blocks the activation of a pool of proinflammatory genes (Arbibe et al. 2007). In addition, OspF and another nuclear-targeted effector, OspB, interact with the human retinoblastoma protein Rb, which is known to bind several chromatin-remodeling factors (Zurawski et al. 2009). Shigella likely uses OspF–OspB synergy to down-regulate host innate immunity via alteration of the chromatin structure at specific genes (Fig. 4).
S. flexneri secretes another effector, IpaH9.8, which also contributes to Shigella pathogenesis by impairing host inflammatory responses. IpaH9.8 is an E3 ubiquitin ligase that binds several host proteins, likely by its LRR domain, and targets them for proteasome-dependent degradation (Rohde et al. 2007). IpaH9.8 affects the NF-κB-dependent pathway in the cytosol (Ashida et al. 2010), as well as the activity of an mRNA splicing factor, U2AF35, in the nucleus (Fig. 4), thus interfering with U2AF35-dependent splicing (Toyotome 2001; Okuda et al. 2005; Seyedarabi et al. 2011). This Shigella IpaH9.8 effector is a member of a novel family of bacterial E3 ubiquitin ligases found in several animal and plant pathogens (Rohde et al. 2007; Singer et al. 2008; Zhu et al. 2008). Orthologs of IpaH9.8, such as SspH1 of Salmonella enterica and YopM of Yersinia pestis, have also been shown to migrate to the host-cell nucleus (Haraga and Miller 2003; Benabdillah et al. 2004) but their effect on chromatin regulation has not yet been examined.
THE EPIGENETIC MEMORY OF INFECTION: THE BACTERIAL IMPRINTS
As chromatin modifications may be transmitted to daughter cells during cell division, leading to heritable changes in gene function, it is possible that a bacterial infection could generate heritable marks after pathogen eradication. Deciphering whether histone modification and/or DNA methylation fingerprints imposed by bacterial components are maintained over time is thus a key issue. The following two examples support this idea.
The Epigenetic Component of Severe Systemic Inflammation and LPS Tolerance
Aberrant inflammatory reactions in response to sustained exposure to microbes and microbial products, such as LPS, lead to tissue damage, multiorgan dysfunction, septic shock, and death. To compensate these adverse effects, the immune system has developed postseptic immunosuppression (PSI) mechanisms that enable hematopoietic cells to become temporary hyporesponsive. This compensatory anti-inflammatory response counteracts the harmful effects of sepsis, but leaves individuals more susceptible to opportunistic infections for extended periods of time (weeks to years). Although PSI is a complex multifactorial process, the contribution of epigenetic regulations is increasingly recognized, as reviewed recently (McCall et al. 2010; Carson et al. 2011). One of the facets of PSI is LPS tolerance, in which LPS-elicited TLR4 responses are reprogrammed toward silencing of proinflammatory cytokine genes and expression of anti-inflammatory or antimicrobial mediators. LPS activation of TLR4 first elicits transcription of poised proinflammatory genes, which are rapidly derepressed and then returned to basal state within hours. As mentioned previously, opening the chromatin at target genes during this acute phase involves histone phosphorylation and acetylation. However, sustained exposure to LPS or subsequent LPS challenge activates a pathway leading to permanent gene repression, as characterized for TNF-α and IL1-β (El Gazzar et al. 2008; Chen et al. 2009). This relies on a change in the composition of NF-κB transcription factor at the proximal promoters of TNF-α and IL1-β, with a shift from activating p65-p50 to repressive RelB-p50, owing to the up-regulation of RelB expression by p65-p50. RelB interacts with H3K9 HMT G9a, leading to H3K9me2 and subsequent recruitment of HP1. The repressive complex formed by G9a and HP1 recruits DNMT3A/B, which induces de novo CpG methylation (McCall et al. 2010). This leads to assembly of silent, facultative heterochromatin in a similar cross talk as depicted in Figure 1.
LPS tolerance can last for weeks in humans, but whether this memory is passed through cell division is not yet proven. Additionally, even if imprinted hematopoietic cells divide, why new cells from progenitors in the bone marrow do not restore an efficient immune system is an open question. A tempting hypothesis would be that epigenetic imprinting also occurs at the level of stem cells. This hypothesis needs to be investigated by analyzing the epigenome of stem cells isolated from animal models of sepsis. The reversal of heterochromatin to euchromatin at genes targeted for LPS-mediated repression is also a key issue to understand how “imprinted” immune cells return to homeostasis.
Bacterial Infection-Associated Cancers
As mentioned above, H. pylori is an important acquired risk factor for gastric cancer. Besides H. pylori-mediated effects on cell proliferation and DNA integrity, H. pylori-induced aberrant DNA methylation emerges as an important mechanism in stomach carcinogenesis (Ding et al. 2010b; Ushijima and Hattori 2012). Remarkably, it has been shown that eradication of H. pylori infection in human patients leads to a decrease but not to full disappearance in methylation of promoter CpG islands of genes closely correlated with the risk of gastric cancer development (Nakajima et al. 2010). This is a strong indication that a bacterial infection may leave epigenetic imprints in a tissue enabling permanent changes in gene expression. Given the fact that cancer must arise from a cell that has the potential to divide, this bacterial reprogramming might be induced in long-living cells, such as stem cells or progenitors, thereby being propagated to daughter cells. It may also target mature epithelial cells for dedifferentiation by silencing components of the stem cell signaling network, leading to increased proliferation and survival (Katoh 2007).
The “Helicobacter paradigm” is transposable to bacteria targeting other tissues. It is speculated that E. coli infection may be linked with bladder carcinoma risk (Tolg et al. 2011) and intestinal bacteria might predispose to colon cancer (Sun 2010). More generally, deregulation of tumor-suppressor and/or stem cell-associated pathways (e.g., Wnt, JAK-STAT, JNK, and Notch) on genetic alteration and epigenetic reprogramming induced by bacteria is a possible cause of cancer development in epithelial niches.
CONCLUDING REMARKS
There is growing evidence for a role of bacterial infection in modulating the epigenetic information of host cells by diverse mechanisms. This is particularly well illustrated with the pathogen L. monocytogenes: on the one hand, this pathogen induces signaling events in host cells leading to activation (via activation of MAP kinases) or repression (via activation of the BAHD1 silencing complex) of immunity genes; on the other hand, this bacterium produces at least two proteins, LLO toxin and LntA nucleomodulin, to counteract these pathways. How these events are spatiotemporally coordinated, in particular, how expression and secretion of bacterial factors is controlled by Listeria during the infectious process, and how chromatin writers and erasers become specifically localized at specific regions of the genome, at a given time and in a specific cell, are important questions to be answered. In this regard, deciphering how chromatin modifications are spread throughout the genome (the “pathoepigenome”) is likely to provide important clues. Technological advances now make human genome-wide mapping of DNA methylation and histone modification profiles feasible. Not only Listeria, but all bacteria presented here, as well as others eliciting chromatin modification in mammalian cells, will undoubtedly benefit from these new technologies, highlighting genes epigenetically deregulated on bacterial infection. Epigenetics has been discussed here for its role in regulating transcription; however, it has additional roles in DNA repair, DNA replication, and cell division. Loss of epigenetic control of these other DNA-based processes may also become relevant in bacterial infectious diseases. Interestingly, studying bacterial modulation of epigenetics processes can help in understanding the fundamental principles of their occurrence and regulation, as we have shown with the discovery of the BAHD1-chromatin-silencing complex (Fig. 2C).
Understanding the role of chromatin modifications and their regulators in the physiopathology of infectious diseases is another challenging perspective. Investigations will have to be performed at the tissue and cell levels, with the objective of analyzing cells with long lifespans, to determine the “chromatin signature” of infection, not only during infection but also following recovery. Results of these investigations will probably depend on the cell type and host genetic background. With the better understanding of the connections between bacterial infectious diseases and the epigenome, opportunities will arise for therapeutic applications, especially as epigenetic processes can be reverted. Rapid elimination of microbe-induced pathoepigenetic changes may prevent chronic or latent infections, some cancers, or autoimmune diseases. This opens avenues for future research in the field of bacterial pathogenesis and chromatin-based regulation of defense genes.
ACKNOWLEDGMENTS
Studies in our laboratory are funded by Institut Pasteur, INRA, INSERM, French Ligue Nationale Contre le Cancer (LNCC RS10/75-76 Bierne), European Research Council (ERC Advanced Grant 233348), French National Research Agency (ANR, grant EPILIS), and Fondation Louis Jeantet, Fondation Le Roch les Mousquetaires.
Footnotes
Editors: Pascale Cossart and Stanley Maloy
Additional Perspectives on Bacterial Pathogenesis available at www.perspectivesinmedicine.org
REFERENCES
- Aguilera C, Nakagawa K, Sancho R, Chakraborty A, Hendrich B, Behrens A 2011. c-Jun N-terminal phosphorylation antagonises recruitment of the Mbd3/NuRD repressor complex. Nature 469: 231–235 [DOI] [PubMed] [Google Scholar]
- Allemand E, Batsche E, Muchardt C 2008. Splicing, transcription, and chromatin: A menage a trois. Curr Opin Genet Dev 18: 145–151 [DOI] [PubMed] [Google Scholar]
- Allo M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, et al. 2009. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol 16: 717–724 [DOI] [PubMed] [Google Scholar]
- Ando T, Yoshida T, Enomoto S, Asada K, Tatematsu M, Ichinose M, Sugiyama T, Ushijima T 2009. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: Its possible involvement in the formation of epigenetic field defect. Int J Cancer 124: 2367–2374 [DOI] [PubMed] [Google Scholar]
- Aravind L, Abhiman S, Iyer LM 2011. Natural history of the eukaryotic chromatin protein methylation system. Prog Mol Biol Transl Sci 101: 105–176 [DOI] [PubMed] [Google Scholar]
- Arbibe L, Kim DW, Batsche E, Pedron T, Mateescu B, Muchardt C, Parsot C, Sansonetti PJ 2007. An injected bacterial effector targets chromatin access for transcription factor NF-κB to alter transcription of host genes involved in immune responses. Nat Immunol 8: 47–56 [DOI] [PubMed] [Google Scholar]
- Ashida H, Kim M, Schmidt-Supprian M, Ma A, Ogawa M, Sasakawa C 2010. A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKγ to dampen the host NF-κB-mediated inflammatory response. Nat Cell Biol 12: 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek SH 2011. When signaling kinases meet histones and histone modifiers in the nucleus. Mol Cell 42: 274–284 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 [DOI] [PubMed] [Google Scholar]
- Bardwell AJ, Abdollahi M, Bardwell L 2004. Anthrax lethal factor-cleavage products of MAPK (mitogen-activated protein kinase) kinases exhibit reduced binding to their cognate MAPKs. Biochem J 378: 569–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayarsaihan D 2011. Epigenetic mechanisms in inflammation. J Dent Res 90: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benabdillah R, Jaime Mota Ls, Lützelschwab S, Demoinet E, Cornelis GR 2004. Identification of a nuclear targeting signal in YopM from Yersinia spp. Microbial Pathogenesis 36: 247–261 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Allis CD 2005. RNA meets chromatin. Genes Dev 19: 1635–1655 [DOI] [PubMed] [Google Scholar]
- Bhavsar AP, Guttman JA, Finlay BB 2007. Manipulation of host-cell pathways by bacterial pathogens. Nature 449: 827–834 [DOI] [PubMed] [Google Scholar]
- Bhutani N, Burns DM, Blau HM 2011. DNA demethylation dynamics. Cell 146: 866–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H, Cossart P 2012. When bacteria target the nucleus: The emerging family of nucleomodulins. Cell Microbiol 14: 622–633 [DOI] [PubMed] [Google Scholar]
- Bierne H, Tham TN, Batsche E, Dumay A, Leguillou M, Kerneis-Golsteyn S, Regnault B, Seeler JS, Muchardt C, Feunteun J, et al. 2009. Human BAHD1 promotes heterochromatic gene silencing. Proc Natl Acad Sci 106: 13826–13831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A 2007. Perceptions of epigenetics. Nature 447: 396–398 [DOI] [PubMed] [Google Scholar]
- Bobetsis YA, Barros SP, Lin DM, Weidman JR, Dolinoy DC, Jirtle RL, Boggess KA, Beck JD, Offenbacher S 2007. Bacterial infection promotes DNA hypermethylation. J Dent Res 86: 169–174 [DOI] [PubMed] [Google Scholar]
- Brennan DF, Barford D 2009. Eliminylation: A post-translational modification catalyzed by phosphothreonine lyases. Trends Biochem Sci 34: 108–114 [DOI] [PubMed] [Google Scholar]
- Bussiere FI, Michel V, Memet S, Ave P, Vivas JR, Huerre M, Touati E 2010. H. pylori-induced promoter hypermethylation downregulates USF1 and USF2 transcription factor gene expression. Cell Microbiol 12: 1124–1133 [DOI] [PubMed] [Google Scholar]
- Carson WF, Cavassani KA, Dou Y, Kunkel SL 2011. Epigenetic regulation of immune cell functions during post-septic immunosuppression. Epigenetics 6: 273–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AO, Lam SK, Wong BC, Wong WM, Yuen MF, Yeung YH, Hui WM, Rashid A, Kwong YL 2003. Promoter methylation of E-cadherin gene in gastric mucosa associated with Helicobacter pylori infection and in gastric cancer. Gut 52: 502–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG 2010. Long noncoding RNAs in mammalian cells: What, where, and why? Wiley Interdiscip Rev RNA 1: 2–21 [DOI] [PubMed] [Google Scholar]
- Chen ZX, Riggs AD 2011. DNA methylation and demethylation in mammals. J Biol Chem 286: 18347–18353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Yoza BK, El Gazzar M, Hu JY, Cousart SL, McCall CE 2009. RelB sustains IκBα expression during endotoxin tolerance. Clin Vaccine Immunol 16: 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ghazawi FM, Li Q 2010. Interplay of bromodomain and histone acetylation in the regulation of p300-dependent genes. Epigenetics 5: 509–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Barski A, Zhao K 2010. Epigenomics of T cell activation, differentiation, and memory. Curr Opin Immunol 22: 341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmasso G, Nguyen HT, Yan Y, Laroui H, Charania MA, Ayyadurai S, Sitaraman SV, Merlin D 2011. Microbiota modulate host gene expression via microRNAs. PLoS ONE 6: e19293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SZ, Fischer W, Kaparakis-Liaskos M, Liechti G, Merrell DS, Grant PA, Ferrero RL, Crowe SE, Haas R, Hatakeyama M, et al. 2010a. Helicobacter pylori-induced histone modification, associated gene expression in gastric epithelial cells, and its implication in pathogenesis. PLoS ONE 5: e9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SZ, Goldberg JB, Hatakeyama M 2010b. Helicobacter pylori infection, oncogenic pathways and epigenetic mechanisms in gastric carcinogenesis. Future Oncol 6: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Gazzar M, Yoza BK, Chen X, Hu J, Hawkins GA, McCall CE 2008. G9a and HP1 couple histone and DNA methylation to TNFα transcription silencing during endotoxin tolerance. J Biol Chem 283: 32198–32208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C 2009. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 10: 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassi Fehri L, Koch M, Belogolova E, Khalil H, Bolz C, Kalali B, Mollenkopf HJ, Beigier-Bompadre M, Karlas A, Schneider T, et al. 2010. Helicobacter pylori induces miR-155 in T cells in a cAMP-Foxp3-dependent manner. PLoS ONE 5: e9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehri LF, Rechner C, Janssen S, Mak TN, Holland C, Bartfeld S, Bruggemann H, Meyer TF 2009. Helicobacter pylori-induced modification of the histone H3 phosphorylation status in gastric epithelial cells reflects its impact on cell cycle regulation. Epigenetics 4: 577–586 [DOI] [PubMed] [Google Scholar]
- Fenley AT, Adams DA, Onufriev AV 2010. Charge state of the globular histone core controls stability of the nucleosome. Biophys J 99: 1577–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Watanabe S, Ichimura T, Tsuruzoe S, Shinkai Y, Tachibana M, Chiba T, Nakao M 2003. Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J Biol Chem 278: 24132–24138 [DOI] [PubMed] [Google Scholar]
- Gangaraju VK, Bartholomew B 2007. Mechanisms of ATP dependent chromatin remodeling. Mutat Res 618: 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Dominguez M, Reyes JC 2009. SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim Biophys Acta 1789: 451–459 [DOI] [PubMed] [Google Scholar]
- Garcia-Garcia JC, Barat NC, Trembley SJ, Dumler JS 2009a. Epigenetic silencing of host cell defense genes enhances intracellular survival of the rickettsial pathogen Anaplasma phagocytophilum. PLoS Pathog 5: e1000488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Garcia JC, Rennoll-Bankert KE, Pelly S, Milstone AM, Dumler JS 2009b. Silencing of host cell CYBB gene expression by the nuclear effector AnkA of the intracellular pathogen Anaplasma phagocytophilum. Infect Immun 77: 2385–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI 2010. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev 20: 134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Hahn T, Lee DH, Esworthy RS, Kim BW, Riggs AD, Chu FF, Pfeifer GP 2008. Methylation of polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res 68: 10280–10289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller D, Holt L, Kim SC, Schwabe RF, Sartor RB, Jobin C 2003. Transforming growth factor-β1 inhibits non-pathogenic Gram negative bacteria-induced NF-κB recruitment to the interleukin-6 gene promoter in intestinal epithelial cells through modulation of histone acetylation. J Biol Chem 278: 23851–23860 [DOI] [PubMed] [Google Scholar]
- Hamon MA, Cossart P 2011. K+ efflux is required for histone H3 dephosphorylation by Listeria monocytogenes listeriolysin O and other pore-forming toxins. Infect Immun 79: 2839–2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon MA, Batsche E, Regnault B, Tham TN, Seveau S, Muchardt C, Cossart P 2007. Histone modifications induced by a family of bacterial toxins. Proc Natl Acad Sci 104: 13467–13472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraga A, Miller SI 2003. A Salmonella enterica serovar typhimurium translocated leucine-rich repeat effector protein inhibits NF-κB-dependent gene expression. Infect Immun 71: 4052–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Crabtree GR 2011. ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res 21: 396–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T, Nakayama J 2011. Physiological roles of class I HDAC complex and histone demethylase. J Biomed Biotechnol 2011: 129383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiragami-Hamada K, Shinmyozu K, Hamada D, Tatsu Y, Uegaki K, Fujiwara S, Nakayama J 2011. N-terminal phosphorylation of HP1α promotes its chromatin binding. Mol Cell Biol 31: 1186–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnilicova J, Stanek D 2011. Where splicing joins chromatin. Nucleus 2: 182–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnilicova J, Hozeifi S, Duskova E, Icha J, Tomankova T, Stanek D 2011. Histone deacetylase activity modulates alternative splicing. PLoS ONE 6: e16727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland C, Schmid M, Zimny-Arndt U, Rohloff J, Stein R, Jungblut PR, Meyer TF 2011. Quantitative phosphoproteomics reveals link between Helicobacter pylori infection and RNA splicing modulation in host cells. Proteomics 11: 2798–2811 [DOI] [PubMed] [Google Scholar]
- Hota SK, Dechassa ML, Prasad P, Bartholomew B 2011. Mapping protein-DNA and protein-protein interactions of ATP-dependent chromatin remodelers. Methods Mol Biol 809: 381–409 [DOI] [PubMed] [Google Scholar]
- Hur K, Niwa T, Toyoda T, Tsukamoto T, Tatematsu M, Yang HK, Ushijima T 2011. Insufficient role of cell proliferation in aberrant DNA methylation induction and involvement of specific types of inflammation. Carcinogenesis 32: 35–41 [DOI] [PubMed] [Google Scholar]
- Ichimura T, Watanabe S, Sakamoto Y, Aoto T, Fujita N, Nakao M 2005. Transcriptional repression and heterochromatin formation by MBD1 and MCAF/AM family proteins. J Biol Chem 280: 13928–13935 [DOI] [PubMed] [Google Scholar]
- Imai K, Ochiai K, Okamoto T 2009. Reactivation of latent HIV-1 infection by the periodontopathic bacterium Porphyromonas gingivalis involves histone modification. J Immunol 182: 3688–3695 [DOI] [PubMed] [Google Scholar]
- Imai K, Inoue H, Tamura M, Cueno ME, Takeichi O, Kusama K, Saito I, Ochiai K 2011. The periodontal pathogen Porphyromonas gingivalis induces the Epstein-Barr virus lytic switch transactivator ZEBRA by histone modification. Biochimie 94: 839–846 [DOI] [PubMed] [Google Scholar]
- Izar B, Mannala GK, Mraheil MA, Chakraborty T, Hain T 2012. microRNA response to Listeria monocytogenes infection in epithelial cells. Int J Mol Sci 13: 1173–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner RG, Young RA 2005. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol 3: 281–294 [DOI] [PubMed] [Google Scholar]
- Kaikkonen MU, Lam MT, Glass CK 2011. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res 90: 430–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M 2007. Dysregulation of stem cell signaling network due to germline mutation, SNP, Helicobacter pylori infection, epigenetic change and genetic alteration in gastric cancer. Cancer Biol Ther 6: 832–839 [DOI] [PubMed] [Google Scholar]
- Kay S, Hahn S, Marois E, Hause G, Bonas U 2007. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318: 648–651 [DOI] [PubMed] [Google Scholar]
- Klose RJ, Bird AP 2006. Genomic DNA methylation: The mark and its mediators. Trends Biochem Sci 31: 89–97 [DOI] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120 [DOI] [PubMed] [Google Scholar]
- Lai AY, Wade PA 2011. Cancer biology and NuRD: A multifaceted chromatin remodelling complex. Nat Rev Cancer 11: 588–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham JA, Dent SY 2007. Cross-regulation of histone modifications. Nat Struct Mol Biol 14: 1017–1024 [DOI] [PubMed] [Google Scholar]
- Lebreton A, Lakisic G, Job V, Fritsch L, Tham TN, Camejo A, Mattei P-J, Regnault B, Nahori M-A, Cabanes D, et al. 2011. A bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response. Science 3: 1319–1321 [DOI] [PubMed] [Google Scholar]
- Lebreton A, Cossart P, Bierne H 2012. Bacteria tune interferon responses by playing with chromatin. Virulence 3: 87–91 [DOI] [PubMed] [Google Scholar]
- Lee JS, Smith E, Shilatifard A 2010. The language of histone crosstalk. Cell 142: 682–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Rauch T, Chen ZX, Szabo PE, Riggs AD, Pfeifer GP 2006. The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J Biol Chem 281: 19489–19500 [DOI] [PubMed] [Google Scholar]
- Li H, Xu H, Zhou Y, Zhang J, Long C, Li S, Chen S, Zhou J-M, Shao F 2007. The phosphothreonine lyase activity of a bacterial type III effector family. Science 315: 1000–1003 [DOI] [PubMed] [Google Scholar]
- Licciardi PV, Wong SS, Tang ML, Karagiannis TC 2010. Epigenome targeting by probiotic metabolites. Gut Pathog 2: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Li Y, Tollefsbol TO 2008. Gene-environment interactions and epigenetic basis of human diseases. Curr Issues Mol Biol 10: 25–36 [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T 2010. Regulation of alternative splicing by histone modifications. Science 327: 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KW 2004. Consequences of regulated pre-mRNA splicing in the immune system. Nat Rev Immunol 4: 931–940 [DOI] [PubMed] [Google Scholar]
- Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X 2011a. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol 12: 861–869 [DOI] [PubMed] [Google Scholar]
- Ma KW, Flores C, Ma W 2011b. Chromatin configuration as a battlefield in plant-bacteria interactions. Plant Physiol 57: 535–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, Arii K, Kaneda A, Tsukamoto T, Tatematsu M, et al. 2006. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res 12: 989–995 [DOI] [PubMed] [Google Scholar]
- Maison C, Bailly D, Roche D, Montes de Oca R, Probst AV, Vassias I, Dingli F, Lombard B, Loew D, Quivy JP, et al. 2011. SUMOylation promotes de novo targeting of HP1α to pericentric heterochromatin. Nat Genet 43: 220–227 [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima K, Isomoto H, Inoue N, Nakayama T, Hayashi T, Nakayama M, Nakao K, Hirayama T, Kohno S 2011. MicroRNA signatures in Helicobacter pylori-infected gastric mucosa. Int J Cancer J 128: 361–370 [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV 2005. Small regulatory RNAs in mammals. Hum Mol Genet 14 (Spec No 1): R121–R132 [DOI] [PubMed] [Google Scholar]
- McCall CE, Yoza B, Liu T, El Gazzar M 2010. Gene-specific epigenetic regulation in serious infections with systemic inflammation. J Innate Immun 2: 395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Horng T 2009. Transcriptional control of the inflammatory response. Nat Rev Immunol 9: 692–703 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad HP, Baylin SB 2010. Linking cell signaling and the epigenetic machinery. Nat Biotechnol 28: 1033–1038 [DOI] [PubMed] [Google Scholar]
- Morey C, Avner P 2011. The demoiselle of X-inactivation: 50 years old and as trendy and mesmerising as ever. PLoS Genet 7: e1002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY 2004. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci 13: 1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata M, Azuma Y, Miura K, Rahman MA, Matsutani M, Aoyama M, Suzuki H, Sugi K, Shirai M 2007. Chlamydial SET domain protein functions as a histone methyltransferase. Microbiology 153: 585–592 [DOI] [PubMed] [Google Scholar]
- Nakajima T, Enomoto S, Yamashita S, Ando T, Nakanishi Y, Nakazawa K, Oda I, Gotoda T, Ushijima T 2010. Persistence of a component of DNA methylation in gastric mucosae after Helicobacter pylori eradication. J Gastroenterol 45: 37–44 [DOI] [PubMed] [Google Scholar]
- Niwa T, Tsukamoto T, Toyoda T, Mori A, Tanaka H, Maekita T, Ichinose M, Tatematsu M, Ushijima T 2010. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res 70: 1430–1440 [DOI] [PubMed] [Google Scholar]
- Nora EP, Heard E 2010. Chromatin structure and nuclear organization dynamics during X-chromosome inactivation. Cold Spring Harb Symp Quant Biol 75: 333–344 [DOI] [PubMed] [Google Scholar]
- O’Connell RM, Rao DS, Baltimore D 2011. MicroRNA regulation of inflammatory responses. Ann Rev Immunol 30: 295–312 [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Sun BK, Lee JT 2008. Intersection of the RNA interference and X-inactivation pathways. Science 320: 1336–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda J, Toyotome T, Kataoka N, Ohno M, Abe H, Shimura Y, Seyedarabi A, Pickersgill R, Sasakawa C 2005. Shigella effector IpaH9.8 binds to a splicing factor U2AF35 to modulate host immune responses. Biochem Biophys Res Commun 333: 531–539 [DOI] [PubMed] [Google Scholar]
- Opitz B, Puschel A, Beermann W, Hocke AC, Forster S, Schmeck B, van Laak V, Chakraborty T, Suttorp N, Hippenstiel S 2006. Listeria monocytogenes activated p38 MAPK and induced IL-8 secretion in a nucleotide-binding oligomerization domain 1-dependent manner in endothelial cells. J Immunol 176: 484–490 [DOI] [PubMed] [Google Scholar]
- Park J, Kim KJ, Choi K-S, Grab DJ, Dumler JS 2004. Anaplasma phagocytophilum AnkA binds to granulocyte DNA and nuclear proteins. Cell Microbiol 6: 743–751 [DOI] [PubMed] [Google Scholar]
- Pathak SK, Basu S, Bhattacharyya A, Pathak S, Banerjee A, Basu J, Kundu M 2006. TLR4-dependent NF-κB activation and mitogen- and stress-activated protein kinase 1-triggered phosphorylation events are central to Helicobacter pylori peptidyl prolyl cis-, trans-isomerase (HP0175)-mediated induction of IL-6 release from macrophages. J Immunol 177: 7950–7958 [DOI] [PubMed] [Google Scholar]
- Pennini ME, Pai RK, Schultz DC, Boom WH, Harding CV 2006. Mycobacterium tuberculosis 19-kDa lipoprotein inhibits IFN-γ-induced chromatin remodeling of MHC2TA by TLR2 and MAPK signaling. J Immunol 176: 4323–4330 [DOI] [PubMed] [Google Scholar]
- Pennini ME, Liu Y, Yang J, Croniger CM, Boom WH, Harding CV 2007. CCAAT/enhancer-binding protein β and δ binding to CIITA promoters is associated with the inhibition of CIITA expression in response to Mycobacterium tuberculosis 19-kDa lipoprotein. J Immunol 179: 6910–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennini MSP, Dautry-Varsat A, Subtil A 2010. Histone methylation by NUE, a novel nuclear effector of the intracellular pathogen Chlamydia trachomatis. PLoS Pathog 6: e1000995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrett CA, Lin DY, Zhou D 2011. Interactions of bacterial proteins with host eukaryotic ubiquitin pathways. Front Microbiol 2: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacentini L, Pimpinelli S 2010. Positive regulation of euchromatic gene expression by HP1. Fly (Austin) 4: 299–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier DB, Gribnau J 2011. Xist regulation and function explored. Hum Genet 130: 223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela A, Esteller M 2010. Epigenetic modifications and human disease. Nat Biotechnol 28: 1057–1068 [DOI] [PubMed] [Google Scholar]
- Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, Komatsu Y, Shinkai Y, Cheng X, Jeltsch A 2008. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol 4: 344–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond B, Batsche E, Boutillon F, Wu YZ, Leduc D, Balloy V, Raoust E, Muchardt C, Goossens PL, Touqui L 2009. Anthrax lethal toxin impairs IL-8 expression in epithelial cells through inhibition of histone H3 modification. PLoS Pathog 5: e1000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet D, Cossart P 2010. SUMOylation and bacterial pathogens. Virulence 6: 532–534 [DOI] [PubMed] [Google Scholar]
- Riggs MG, Whittaker RG, Neumann JR, Ingram VM 1977. n-Butyrate causes histone modification in HeLa and Friend erythroleukaemia cells. Nature 268: 462–464 [DOI] [PubMed] [Google Scholar]
- Rivas S 2011. Nuclear dynamics during plant innate immunity. Plant Physiol 158: 87–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde JR, Breitkreutz A, Chenal A, Sansonetti PJ, Parsot C 2007. Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe 1: 77–83 [DOI] [PubMed] [Google Scholar]
- Rother S, Meister G 2011. Small RNAs derived from longer non-coding RNAs. Biochimie 93: 1905–1915 [DOI] [PubMed] [Google Scholar]
- Russo VEA, Martienssen RA, Riggs AD 1996. Epigenetic mechanisms of gene regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Saccani S, Pantano S, Natoli G 2002. p38-Dependent marking of inflammatory genes for increased NF-κB recruitment. Nat Immunol 3: 69–75 [DOI] [PubMed] [Google Scholar]
- Saint-Andre V, Batsche E, Rachez C, Muchardt C 2011. Histone H3 lysine 9 trimethylation and HP1γ favor inclusion of alternative exons. Nat Struct Mol Biol 18: 337–344 [DOI] [PubMed] [Google Scholar]
- Sasai N, Defossez PA 2009. Many paths to one goal? The proteins that recognize methylated DNA in eukaryotes. Int J Dev Biol 53: 323–334 [DOI] [PubMed] [Google Scholar]
- Schmeck B, Beermann W, van Laak V, Zahlten J, Opitz B, Witzenrath M, Hocke AC, Chakraborty T, Kracht M, Rosseau S, et al. 2005. Intracellular bacteria differentially regulated endothelial cytokine release by MAPK-dependent histone modification. J Immunol 175: 2843–2850 [DOI] [PubMed] [Google Scholar]
- Schmeck B, Lorenz J, N’Guessan PD, Opitz B, van Laak V, Zahlten J, Slevogt H, Witzenrath M, Flieger A, Suttorp N, et al. 2008. Histone acetylation and flagellin are essential for Legionella pneumophila-induced cytokine expression. J Immunol 181: 940–947 [DOI] [PubMed] [Google Scholar]
- Schmitz KM, Mayer C, Postepska A, Grummt I 2010. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24: 2264–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte LN, Eulalio A, Mollenkopf HJ, Reinhardt R, Vogel J 2011. Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J 30: 1977–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedarabi A, Sullivan JA, Sasakawa C, Pickersgill RW 2011. A disulfide driven domain swap switches off the activity of Shigella IpaH9.8 E3 ligase. FEBS Lett 584: 4163–4168 [DOI] [PubMed] [Google Scholar]
- Singer AU, Rohde JR, Lam R, Skarina T, Kagan O, Dileo R, Chirgadze NY, Cuff ME, Joachimiak A, Tyers M, et al. 2008. Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nat Struct Mol Biol 15: 1293–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slevogt H, Schmeck B, Jonatat C, Zahlten J, Beermann W, van Laak V, Opitz B, Dietel S, N’Guessan PD, Hippenstiel S, et al. 2006. Moraxella catarrhalis induces inflammatory response of bronchial epithelial cells via MAPK and NF-κB activation and histone deacetylase activity reduction. Am J Physiol Lung Cell Mol Physiol 290: L818–L826 [DOI] [PubMed] [Google Scholar]
- Suganuma T, Workman JL 2011. Signals and combinatorial functions of histone modifications. Annu Rev Biochem 80: 473–499 [DOI] [PubMed] [Google Scholar]
- Sun J 2010. Enteric bacteria and cancer stem cells. Cancers (Basel) 3: 285–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Sugi Y, Nakano K, Tsuda M, Kurihara K, Hosono A, Kaminogawa S 2011. Epigenetic control of the host gene by commensal bacteria in large intestinal epithelial cells. J Biol Chem 286: 35755–35762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ 2007. How chromatin-binding modules interpret histone modifications: Lessons from professional pocket pickers. Nat Struct Mol Biol 14: 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisseur M, Kwapisz M, Morillon A 2011. Pervasive transcription—Lessons from yeast. Biochimie 93: 1889–1896 [DOI] [PubMed] [Google Scholar]
- Tiwari VK, Stadler MB, Wirbelauer C, Paro R, Schubeler D, Beisel C 2011. A chromatin-modifying function of JNK during stem cell differentiation. Nat Genet 44: 94–100 [DOI] [PubMed] [Google Scholar]
- Tolg C, Sabha N, Cortese R, Panchal T, Ahsan A, Soliman A, Aitken KJ, Petronis AB, Auml Gli DJ 2011. Uropathogenic E. coli infection provokes epigenetic downregulation of CDKN2A (p16INK4A) in uroepithelial cells. Lab Invest 91: 825–836 [DOI] [PubMed] [Google Scholar]
- Toyotome T 2001. Shigella protein IpaH9.8 is secreted from bacteria within mammalian cells and transported to the nucleus. J Biol Chem 276: 32071–32079 [DOI] [PubMed] [Google Scholar]
- Tropberger P, Schneider R 2011. Going global: Novel histone modifications in the globular domain of H3. Epigenetics 5: 112–117 [DOI] [PubMed] [Google Scholar]
- Turner AM, Morris KV 2010. Controlling transcription with noncoding RNAs in mammalian cells. Biotechniques 48: x–xvi [DOI] [PubMed] [Google Scholar]
- Ushijima T, Hattori N 2012. Molecular pathways: Involvement of Helicobacter pylori-triggered inflammation in the formation of an epigenetic field defect, and its usefulness as cancer risk and exposure markers. Clin Cancer Res 18: 923–929 [DOI] [PubMed] [Google Scholar]
- van Vliet J, Oates NA, Whitelaw E 2007. Epigenetic mechanisms in the context of complex diseases. Cell Mol Life Sci 64: 1531–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. 2006. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439: 871–874 [DOI] [PubMed] [Google Scholar]
- Wang Y, Curry HM, Zwilling BS, Lafuse WP 2005. Mycobacteria inhibition of IFN-γ induced HLA-DR gene expression by up-regulating histone deacetylation at the promoter region in human THP-1 monocytic cells. J Immunol 174: 5687–5694 [DOI] [PubMed] [Google Scholar]
- Wilson AG 2008. Epigenetic regulation of gene expression in the inflammatory response and relevance to common diseases. J Periodontol 79: 1514–1519 [DOI] [PubMed] [Google Scholar]
- Woodcock CL, Dimitrov S 2001. Higher-order structure of chromatin and chromosomes. Curr Opin Genet Dev 11: 130–135 [DOI] [PubMed] [Google Scholar]
- Wu H, Zhang Y 2011. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev 25: 2436–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A 2011. Gene silencing in X-chromosome inactivation: Advances in understanding facultative heterochromatin formation. Nat Rev Genet 12: 542–553 [DOI] [PubMed] [Google Scholar]
- Xia G, Schneider-Stock R, Diestel A, Habold C, Krueger S, Roessner A, Naumann M, Lendeckel U 2008. Helicobacter pylori regulates p21(WAF1) by histone H4 acetylation. Biochem Biophys Res Commun 369: 526–531 [DOI] [PubMed] [Google Scholar]
- Xu WS, Parmigiani RB, Marks PA 2007. Histone deacetylase inhibitors: Molecular mechanisms of action. Oncogene 26: 5541–5552 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Verma UN, Prajapati S, Kwak YT, Gaynor RB 2003. Histone H3 phosphorylation by IKK-α is critical for cytokine-induced gene expression. Nature 423: 655–659 [DOI] [PubMed] [Google Scholar]
- Yan J, Zhang M, Zhang J, Chen X, Zhang X 2011. Helicobacter pylori infection promotes methylation of WWOX gene in human gastric cancer. Biochem Biophys Res Commun 408: 99–102 [DOI] [PubMed] [Google Scholar]
- Yang SH, Vickers E, Brehm A, Kouzarides T, Sharrocks AD 2001. Temporal recruitment of the mSin3A-histone deacetylase corepressor complex to the ETS domain transcription factor Elk-1. Mol Cell Biol 21: 2802–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG 2011. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147: 773–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Tao H, Park DI, Sepulveda JL, Sepulveda AR 2006. Demonstration and characterization of mutations induced by Helicobacter pylori organisms in gastric epithelial cells. Helicobacter 11: 272–286 [DOI] [PubMed] [Google Scholar]
- Yin L, Chung WO 2011. Epigenetic regulation of human β-defensin 2 and CC chemokine ligand 20 expression in gingival epithelial cells in response to oral bacteria. Mucosal Immunol 4: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Barros SP, Niculescu MD, Moretti AJ, Preisser JS, Offenbacher S 2010. Alteration of PTGS2 promoter methylation in chronic periodontitis. J Dent Res 89: 133–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Kim J, Yuan X, Braun T 2011. Epigenetic modifications of stem cells: A paradigm for the control of cardiac progenitor cells. Circ Res 109: 1067–1081 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Li H, Hu L, Wang J, Zhou Y, Pang Z, Liu L, Shao F 2008. Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nat Struct Mol Biol 15: 1302–1308 [DOI] [PubMed] [Google Scholar]
- Zhu B, Nethery KA, Kuriakose JA, Wakeel A, Zhang X, McBride JW 2009. Nuclear translocated Ehrlichia chaffeensis ankyrin protein interacts with a specific adenine-rich motif of host promoter and intronic Alu elements. Infect Immun 77: 4243–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski DV, Mumy KL, Faherty CS, Mccormick BA, Maurelli AT 2009. Shigella flexneri type III secretion system effectors OspB and OspF target the nucleus to downregulate the host inflammatory response via interactions with retinoblastoma protein. Mol Microbiol 71: 350–368 [DOI] [PMC free article] [PubMed] [Google Scholar]