Small interfering RNAs (siRNAs) direct Argonaute proteins, the core components of the RNA-induced silencing complex (RISC), to cleave complementary target RNAs. Here, the authors describe a method to purify active RISC containing a single, unique small RNA guide sequence. They begin by capturing RISC using a complementary 2′-O-methyl oligonucleotide tethered to beads. Unlike other methods that capture RISC but do not allow its recovery, their strategy purifies active, soluble RISC in good yield. They use this strategy to purify fly Ago1- and Ago2-RISC, as well as mouse AGO2-RISC. The method can discriminate among RISCs programmed with different guide strands, making it possible to deplete and recover specific RISC populations. Endogenous microRNA:Argonaute complexes can also be purified from cell lysates. The method scales readily and takes less than a day to complete.
Keywords: Ago2, Argonaute, miRNA, RISC, RNAi, siRNA
Abstract
Small interfering RNAs (siRNAs) direct Argonaute proteins, the core components of the RNA-induced silencing complex (RISC), to cleave complementary target RNAs. Here, we describe a method to purify active RISC containing a single, unique small RNA guide sequence. We begin by capturing RISC using a complementary 2′-O-methyl oligonucleotide tethered to beads. Unlike other methods that capture RISC but do not allow its recovery, our strategy purifies active, soluble RISC in good yield. The method takes advantage of the finding that RISC partially paired to a target through its siRNA guide dissociates more than 300 times faster than a fully paired siRNA in RISC. We use this strategy to purify fly Ago1- and Ago2-RISC, as well as mouse AGO2-RISC. The method can discriminate among RISCs programmed with different guide strands, making it possible to deplete and recover specific RISC populations. Endogenous microRNA:Argonaute complexes can also be purified from cell lysates. Our method scales readily and takes less than a day to complete.
INTRODUCTION
Small interfering RNAs (siRNAs) and microRNAs (miRNAs) mediate the post-transcriptional repression of complementary target RNAs (Bartel 2009; Ghildiyal and Zamore 2009). siRNAs typically guide the cleavage of extensively complementary RNAs (Hammond et al. 2000; Zamore et al. 2000; Elbashir et al. 2001a,b), a phenomenon called RNA interference (RNAi). In contrast, most animal miRNAs target partially complementary RNAs, triggering their destruction (Baek et al. 2008; Selbach et al. 2008; Ingolia et al. 2009; Guo et al. 2010) and, in some cases, repressing their translation into protein (Hendrickson et al. 2009; Bazzini et al. 2012). Both siRNAs and miRNAs assemble with Argonaute proteins to form the RNA-induced silencing complex (RISC). Assembly of siRNA or miRNA/miRNA* duplexes into RISCs is facilitated by proteins that orient the siRNA, such as the Dicer-2/R2D2 heterodimer in insects, and by proteins thought to allow conformational rearrangement of Argonaute, such as HSP90 and Hsc70 chaperones (Tahbaz et al. 2001; Iki et al. 2010; Iwasaki et al. 2010; Johnston et al. 2010; Miyoshi et al. 2010; Olivieri et al. 2012; Preall et al. 2012; Xiol et al. 2012).
In Drosophila melanogaster, miRNAs and siRNAs are sorted between Argonaute1 (Ago1) and Argonaute2 (Ago2) according to their duplex structure and the identity of their first nucleotide (Forstemann et al. 2007; Tomari et al. 2007; Czech et al. 2008; Ghildiyal et al. 2008; Okamura et al. 2008; Seitz et al. 2011). The extensively double-stranded structure of siRNA sends them into Ago2, while the presence of specific mismatches within the miRNA/miRNA* duplex directs miRNAs into Ago1. In mammals, whose four Argonaute proteins are more closely related to fly Ago1 than to fly Ago2 (Tolia and Joshua-Tor 2007), no sorting mechanism has thus far been detected, and miRNAs and siRNAs assemble into AGO1, AGO2, AGO3, and AGO4. In mice and humans, only AGO2 can catalyze target cleavage (Liu et al. 2004; Meister et al. 2004b).
Chemically modified, complementary oligonucleotides that bind siRNAs and miRNAs block their activity in vitro and in vivo (Hutvágner et al. 2004; Meister et al. 2004a; Krutzfeldt et al. 2005; Leaman et al. 2005; Elmen et al. 2008; Lanford et al. 2010; Obad et al. 2011). The most widely used anti-miRNA oligonucleotides use 2′-O-methyl ribose modifications to block degradation of the oligonucleotide. Such 2′-O-methyl anti-miRNA oligonucleotides not only inhibit miRNA function in cultured cell lines, cell lysates, and Caenorhabditis elegans (Hutvágner et al. 2004; Zheng et al. 2010), but also can be used to capture Argonaute complexes programmed with a specific small RNA sequence (Hutvágner et al. 2004). Unfortunately, RISC captured with a fully complementary 2′-O-methyl oligonucleotides tethered to beads cannot be recovered under native conditions, precluding its further study.
Despite great advances in understanding the miRNA and siRNA pathways, the biochemical details of how Argonaute proteins function remain incomplete. A key limitation in the study of Argonaute protein function is the lack of methods to purify RISC complexes assembled through natural pathways and that contain a single, unique guide sequence. Purification of RISC using antibodies against endogenous or epitope-tagged Argonautes allows the selection of specific Argonaute proteins, but these contain a complex mixture of siRNA and miRNA guides (Hutvágner and Zamore 2002; Mourelatos et al. 2002; Liu et al. 2004; Meister et al. 2004a; Ikeda et al. 2006; Beitzinger et al. 2007; Azuma-Mukai et al. 2008). RISC has also been purified using a guide strand with a 3′ biotin joined to the siRNA through a UV-sensitive linker, which was cleaved by photolysis (Martinez et al. 2002; Martinez and Tuschl 2004; Ameres et al. 2007) and using tethered siRNAs from which proteins were recovered with denaturing buffers (Gerbasi et al. 2010). However, photolysis is inefficient, and recovery by denaturation, of course, fails to preserve RISC activity. Both approaches can only be used to isolate experimentally programmed RISC, whereas our method permits purification of endogenous complexes from cell and tissue extracts.
Recombinant eukaryotic Argonaute proteins have been produced in bacteria, and in insect, yeast, human, and lepidopteran cells (Rivas et al. 2005; MacRae et al. 2008; Ye et al. 2011; Elkayam et al. 2012; Nakanishi et al. 2012; Schirle and MacRae 2012). Recombinant Argonaute proteins can be partially inactive (Rivas et al. 2005) and often contain endogenous RNAs (Nakanishi et al. 2012; Schirle and MacRae 2012). Furthermore, recombinant Argonaute proteins must be loaded with single-stranded RNA, a pathway not thought to exist in vivo, and lack associated proteins that may modify Argonaute function.
Here, we describe a method to purify active Argonaute complexes (RISCs) containing a single, unique small RNA guide. Like previous techniques, our method begins by capturing RISC using a complementary 2′-O-methyl oligonucleotide tethered to paramagnetic beads (the capture oligo). The strategy takes advantage of the finding that fly Ago2-RISC dissociates from a target RNA more than 300 times more rapidly when its guide strand is partially paired than when it is fully paired to a target (Wee et al. 2012). We can therefore elute purified, active, soluble RISC from the capture oligo in good yield in native conditions. We use this strategy to purify Drosophila Ago2-RISC bearing an siRNA and Ago1-RISC loaded with an miRNA, as well as an siRNA assembled into mouse AGO2 in lysates from immortalized embryonic fibroblasts. Finally, we show that active RISC can be selectively purified from a population of RISCs containing different guide strands. This method scales readily and takes no more than a single day to complete.
RESULTS
miRNA-like pairing allows efficient capture and release of RISC
We incubated an siRNA duplex in Drosophila embryo lysate to assemble the guide strand into Ago2-RISC. We captured the RISC containing the siRNA guide using a 31-nt 2′-O-methyl oligonucleotide tethered via a 5′ biotin to streptavidin-paramagnetic beads. RISC, captured on the oligo, was then washed in buffer containing 2 M potassium acetate and eluted with a competitor DNA oligonucleotide fully complementary to the capture oligo in buffer containing 1 M potassium acetate. Figure 1 illustrates the method. Three types of capture oligos were tested: an oligo fully complementary to the siRNA guide; an oligo complementary only to the seed of the siRNA (positions 2–8); and an oligo complementary to the seed and four nucleotides in the 3′ supplementary region of the siRNA. The amount of capture oligo used (5 µM) was greater than the concentration of 2′-O-methyl oligonucleotide (1–2 µM) that fully blocks target cleavage directed by this siRNA in Drosophila embryo lysate (Supplemental Fig. S1).
FIGURE 1.
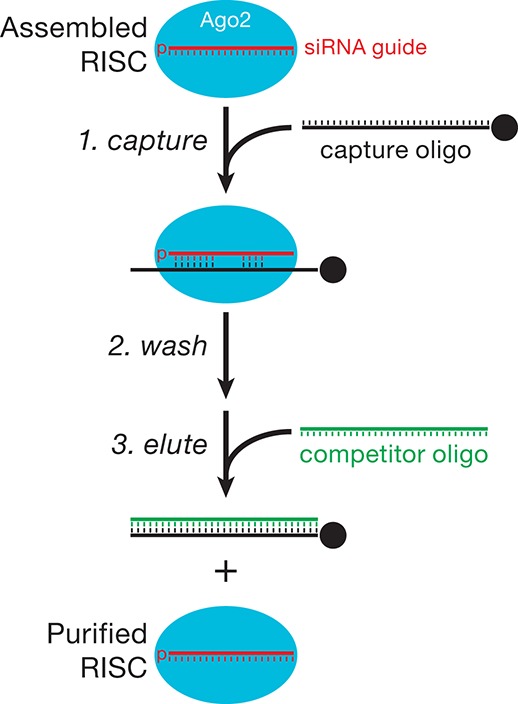
Strategy to purify active RISC.
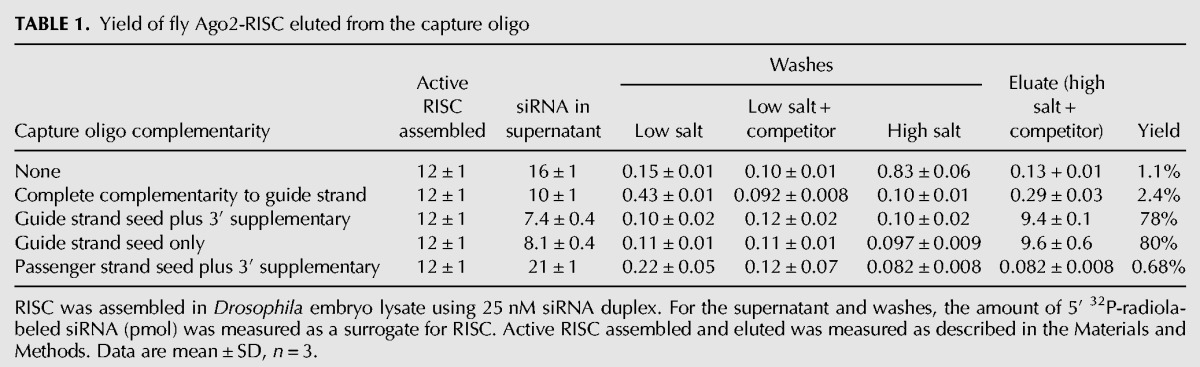
To monitor the efficiency of binding and recovery of siRNA-programmed RISC for each of the three types of capture oligos, we assayed cleavage of a 5′ 32P-radiolabeled target RNA (Fig. 2). In parallel, we followed a 5′ 32P-radiolabeled guide siRNA through the purification procedure (Table 1). The fully complementary capture oligo, the capture oligo with seed plus 3′ supplementary pairing, and the capture oligo pairing only with the seed all effectively depleted the programmed RISC from the lysate: We detected <2% of the original RISC cleaving activity in the supernatant of an assembly reaction incubated with any of the capture oligos tethered to paramagnetic beads or in the subsequent, pooled washes (100 mM and 2 M potassium acetate). For the fully complementary capture oligo, addition of a competitor oligonucleotide (the competitor) complementary to the capture oligo failed to release any detectable target-cleaving RISC activity (Table 1). In contrast, a competitor efficiently released active RISC from the capture oligo complementary to only the seed plus the 3′ supplementary region or the seed alone. We note that high salt (1 M potassium acetate) was essential: At lower salt concentrations, addition of a competitor complementary to the capture oligo failed to release any detectable target-cleaving RISC activity (Fig. 2) or 32P-radiolabeled guide siRNA (Table 1, cf. “low salt + competitor” to “eluate”).
FIGURE 2.
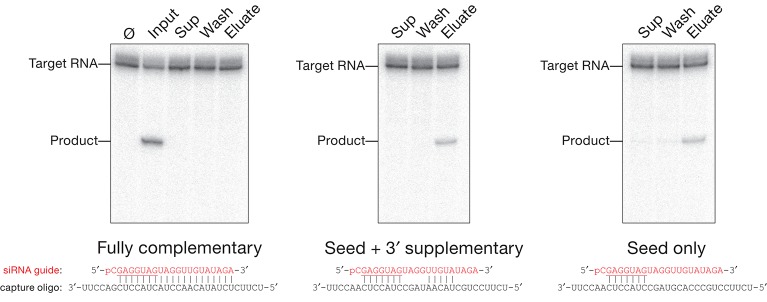
Active RISC can be eluted from a capture oligo partially complementary to the small RNA guide. let-7-programmed Drosophila Ago2-RISC, assembled in Drosophila embryo lysate, was incubated with a capture oligo fully complementary to let-7, to the let-7 seed plus 3′ supplementary region, or only to the let-7 seed sequence. The RISC assembly reaction (Input), the supernatant after capture (Sup), the washes (five with 100 mM and five with 2 M potassium acetate, pooled and concentrated to the volume of the original RISC assembly reaction, 100 µL; Wash), and the eluate from the capture oligos were incubated with 100 nM target RNA for 5 min at 25°C to detect Ago2-RISC activity. (Ø) No incubation.
TABLE 1.
Yield of fly Ago2-RISC eluted from the capture oligo
The RISC eluted by this method contains competitor DNA that is partially complementary to an RNA target fully matching the siRNA guide. In practice, the contaminating competitor DNA does not detectably interfere with target cleavage. However, the competitor can be readily removed by subsequent ion exchange chromatography (see Materials and Methods).
Purification of Ago1 programmed with an miRNA
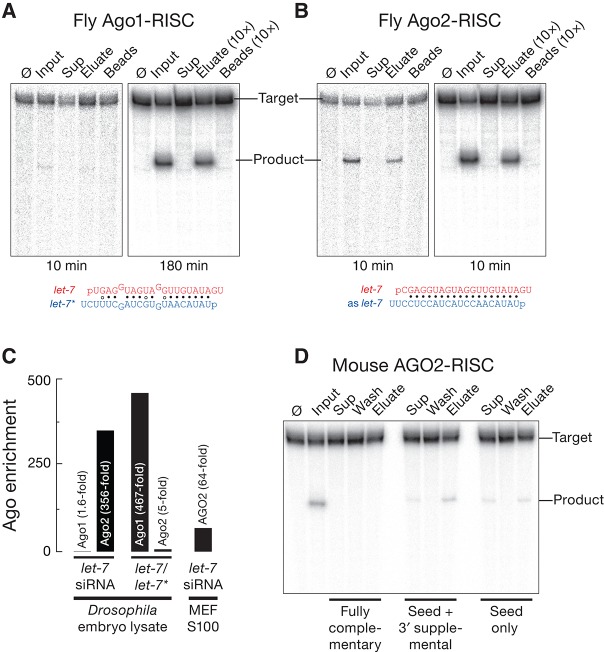
In flies, canonical miRNA/miRNA* duplexes load Ago1 rather than Ago2 (Forstemann et al. 2007; Tomari et al. 2007). To test whether the method could be used to selectively purify Ago1-RISC, we incubated a let-7/let-7* duplex with Drosophila embryo lysate, and then purified the let-7-programmed Ago1-RISC using the capture oligo complementary to the seed plus the 3′ supplementary region. Although Ago 1-RISC is inherently less catalytically active than Ago2 (kcat/KM for Ago2 is >60-fold greater than for Ago1) and target cleavage is unlikely to play an important role in Ago1-mediated mRNA repression (Forstemann et al. 2007), Ago1 can be followed by target cleaving activity (Fig. 3A). Target cleavage assays of let-7-programmed Ago1-RISC detected activity in the input and eluate samples (Fig. 3A). Quantitative mass spectrometry (Fig. 3C) revealed that the eluate was more enriched for Ago1 (467-fold over background) than for Ago2 (fivefold enrichment). In contrast, RISC assembled with a let-7 siRNA (Fig. 3B,C; Supplemental Tables S1, S2) contained mainly Ago2 (356-fold enrichment over background) rather than Ago1 (1.6-fold enrichment). Importantly, we used the same capture oligo to purify each Argonaute complex, changing only the let-7 duplex used to assemble RISC. Flies do not produce let-7 during embryogenesis (Pasquinelli et al. 2000), thus all let-7 RISC activity reflects Argonaute programmed in the lysate by the let-7 siRNA or miRNA/miRNA* duplex.
FIGURE 3.
Drosophila Ago1- and Ago2-RISC, as well as mouse AGO2-RISC, can be purified using a partially complementary capture oligo. (A) A let-7/let-7* duplex was loaded into Drosophila Ago1 or (B) a let-7 siRNA was loaded into Drosophila Ago2 by incubation in Drosophila embryo lysate and purified with a partially complementary capture oligo. The RISC assembly reaction (Input), the supernatant after capture (Sup), the first wash, and the eluates were incubated with 100 nM let-7 complementary target RNA for 10 min (Ago1 and Ago2) and 180 min (Ago1) at 25°C. (C) Tenfold concentrated, purified Drosophila Ago1- and Ago2-RISC, mouse AGO2, and control samples were analyzed by quantitative mass spectrometry to determine their protein composition. The enrichment of each Argonaute protein in the purified RISC was calculated as the ratio of let-7-programmed, purified samples to control samples in which the small RNA duplex was omitted (background). (D) let-7-programmed mouse AGO2-RISC was assembled in S100 cytosolic extract from Ago2−/− MEFs overexpressing mouse AGO2, then purified using either a fully or partially complementary capture oligo. Target cleaving activity was tested using 100 nM target RNA for 5 min at 37°C.
Purification of mouse AGO2-RISC
Our method also effectively purifies mammalian AGO2-RISC. We prepared cytosolic S100 extract from immortalized, Ago2−/− mouse embryonic fibroblasts (MEFs) (O’Carroll et al. 2007; Broderick et al. 2011) that stably express ∼30-fold more Ago2 mRNA than wild-type MEFs, and incubated the S100 with a let-7 siRNA duplex to program RISC. We used the same capture oligo and protocol as for fly RISC and then tested the eluate for target cleaving activity (Fig. 3D). Again, target cleaving activity was detected in the eluate when we used a capture oligo complementary to seed plus the 3′ supplementary region or to the seed alone, but not the fully complementary capture oligo.
Although MEFs express at least three of the four mammalian Argonaute proteins (Broderick et al. 2011), quantitative mass spectrometry detected in the eluate only mouse AGO2 (Supplemental Table S3), which was 64-fold enriched over the background control (Fig. 3C). Perhaps the endogenous mouse AGO1, AGO3, and AGO4 proteins in the S100 are preoccupied with endogenous miRNAs and therefore unavailable for loading with exogenous siRNA.
Separation of a mixture of two RISCs
As a test of specificity, we asked whether our method can resolve RISCs programmed with different siRNAs. We separately assembled let-7 and luciferase siRNAs into RISC using fly embryo lysate. Next, we mixed the assembly reactions and then used a let-7-specific capture oligo to purify RISC from one-half of the mixture. We used a luciferase-specific capture oligo to purify luciferase-siRNA-programmed RISC from the other half. Finally, we tested the eluates for their ability to cleave either let-7 or luciferase target RNAs. RISC purified using the let-7-specific capture oligo only cleaved the let-7 target (Fig. 4). Conversely, RISC purified using the luciferase-specific capture oligo only cleaved the luciferase target. Thus, the method can separate a mixture of RISCs programmed with different small RNA sequences.
FIGURE 4.
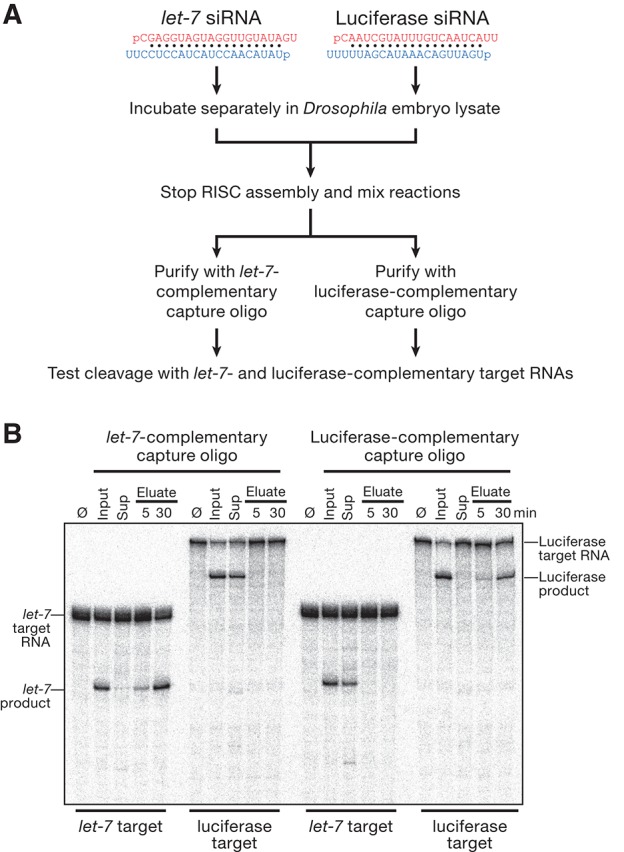
The purification method separates RISCs programmed with different siRNA guides. (A) Experimental strategy. (Red) Guide; (blue) passenger strand. (B) The activity of the samples was measured by incubating them with target RNAs (100 nM) complementary to let-7 (186 nt) and luciferase (506 nt) guide siRNA strands for 5 min (Input, Sup, Eluate) or 30 min (Eluate) at 25°C. (Ø) No incubation.
Efficient purification with good yield
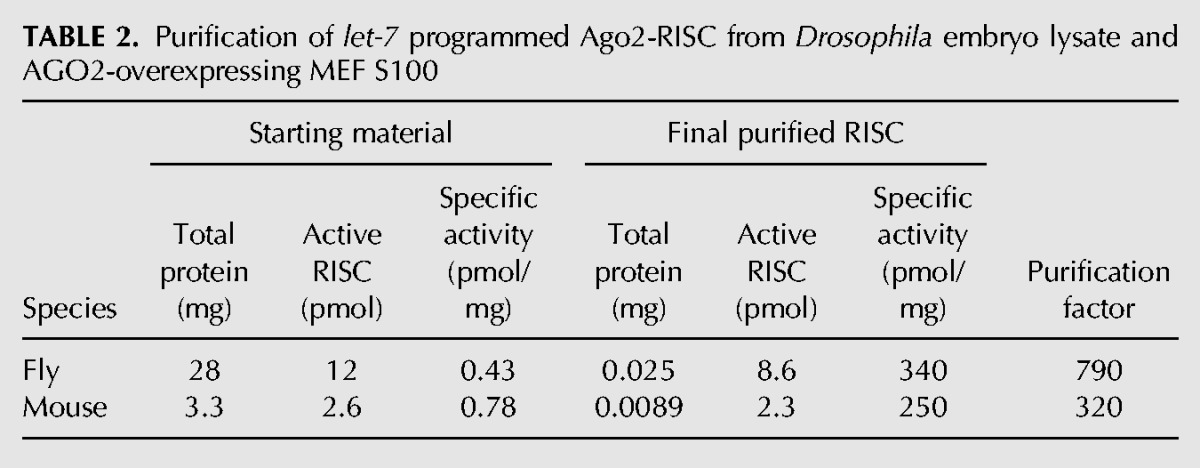
Both fully complementary and partially complementary capture oligos depleted all (Figs. 2, 3A,B) or most (Fig. 3D) detectable RISC activity from the lysate, but RISC was recovered efficiently only from the partially complementary capture oligo. To estimate the yield for our purification method, we measured both the amount of active purified RISC recovered in the eluate by pre-steady-state kinetic analysis and the differential loss of the passenger and guide strands (Supplemental Fig. S2A,C,D). Nearly all the active RISC assembled was recovered from the partially complementary capture oligo: 78% for the capture oligo complementary to the seed plus the 3′ supplementary region and 80% for the capture oligo complementary to the seed alone (Table 1). Overall, the specific activity (pmol of active RISC/milligram of protein) of fly Ago2-RISC increased 790-fold between the assembly reaction and the eluate of the capture oligo (Table 2). For mouse AGO2-RISC, the specific activity increased 320-fold.
TABLE 2.
Purification of let-7 programmed Ago2-RISC from Drosophila embryo lysate and AGO2-overexpressing MEF S100
Purification of endogenously loaded miRNAs
We tested whether our method can also be used to purify RISC programmed with endogenous miRNAs. miR-21 is the most abundant miRNA in MEFs (JA Broderick and PD Zamore, unpubl.). We used a capture oligo complementary to the miR-21 seed plus 3′ supplementary region to purify miR-21-RISC from MEF S100 lysate, and then tested its ability to cleave a fully complementary miR-21 target RNA. Active miR-21-programmed RISC was recovered in the eluate (Fig. 5A).
FIGURE 5.
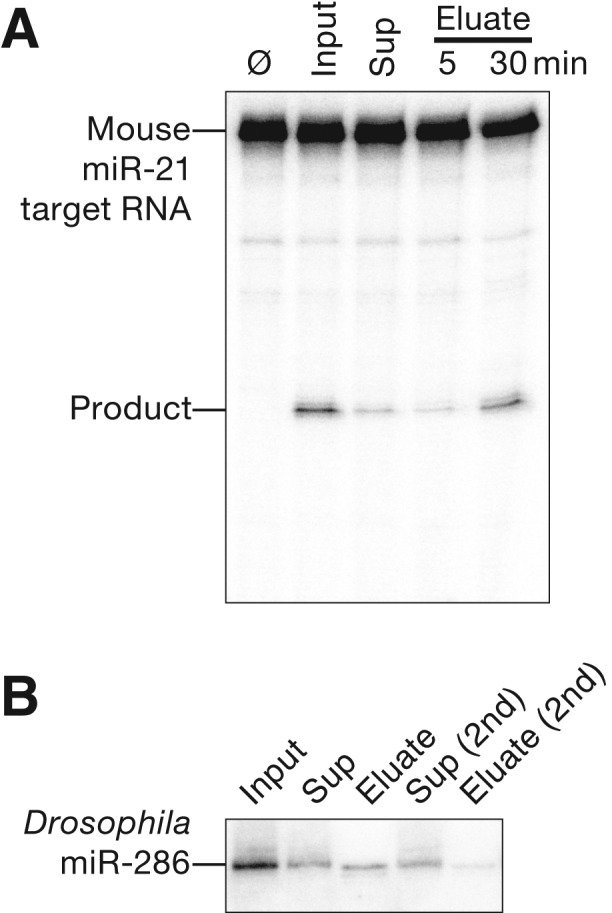
Purification of endogenous miRNA-RISC complexes. (A) S100 cytosolic extract from Ago2−/− MEFs overexpressing mouse AGO2 was concentrated 10-fold and then purified using a capture oligo partially complementary to mouse miR-21. Target cleavage assays were performed by incubating samples with 100 nM target RNA for 5 min (Input, Sup, Eluate) or 30 min (Eluate) at 25°C. (Ø) No incubation. (B) Drosophila miR-286 was purified from embryo lysate using a partially complementary capture oligo. Total RNA from each sample was resolved by denaturing electrophoresis and miR-286 was detected by Northern hybridization.
miR-286 is one of the most abundant miRNAs in 2- to 6-h Drosophila embryos (Ruby et al. 2007; Chung et al. 2008). We used a capture oligo complementary to the miR-286 seed plus 3′ supplementary region to purify miR-286-RISC from embryo lysate. Northern hybridization demonstrated that we could successfully recover miR-286 in the eluate of the capture oligo (Fig. 5B).
Unlike purification of exogenously programmed RISC from fly embryo or mouse cell lysate, the capture oligo did not fully deplete the targeted miRNA from the lysate for either of the endogenous miRNAs. The failure to deplete miR-21- or miR-286-RISC from the lysate cannot be explained by an insufficient amount of capture oligo, because a second round of incubation with fresh capture oligo failed to deplete the remaining Drosophila miR-286 (Fig. 5B). The simplest explanation is that as much as half of the miR-286 in the embryo lysate is bound to its mRNA targets and is therefore unavailable to bind the capture oligo.
DISCUSSION
We have described a simple method to purify from crude extracts active RISC complexes containing a single guide RNA sequence. Our procedure preserves the activity of target-cleaving Argonaute proteins, but should also find use purifying Argonaute:small RNA complexes that do not catalyze endonucleolytic cleavage of their target RNAs.
Our method offers the advantage of purifying freely diffusing RISC complexes that can be used to study small RNA-directed Argonaute enzyme kinetics (Wee et al. 2012). Free diffusion is essential for quantitative population or single-molecule analysis of target binding by RISC.
Our method isolates only fully assembled, mature RISC containing a single-stranded RNA guide (Table 1). Thus, the proteins that copurify with RISC should be only those present at the encounter with target. Our mass spectrometry analyses support this view, because we did not find proteins that participate in small RNA biogenesis or RISC assembly. We detected no peptides from Dicer-1, Dicer-2, R2D2, Loquacious, or C3PO (Jiang et al. 2005; Liu et al. 2006, 2007, 2009) copurifying with Drosophila Ago1- or Ago2-RISC, nor did we detect Dicer, TRBP, or PACT (Hutvágner et al. 2001; Chendrimada et al. 2005; Haase et al. 2005; Lee et al. 2006) copurifying with mouse AGO2-RISC (Supplemental Fig. S3; Supplemental Tables S1–S3).
Heat shock proteins such as Hsp90 and Hsc70 also function in RISC assembly (Iki et al. 2010; Iwasaki et al. 2010; Johnston et al. 2010; Miyoshi et al. 2010). Hsc70-4 was enriched over background for Drosophila Ago1-RISC purified by our method (Supplemental Tables S1–S3). Hsc70-4 associates with Argonaute during its assembly into RISC, but Hsc70 and Hsp90 inhibitors had no detectable affect on target cleavage by Ago2 (Iwasaki et al. 2010); our data suggest that Hsc70-4 may nonetheless play a role for Ago1 in target finding, target cleavage, product release, or enzyme regeneration after target cleavage. Whereas Ago2 was the most enriched protein in the Ago2 sample, other proteins were highly enriched in the Ago1 sample (Supplemental Table S1); ribosomal protein L7A was the only protein enriched in both samples. We do not yet know if these Argonaute-associated proteins play a role in RISC function.
Although the siRNAs and miRNAs used here contained no chemical modifications, we have used the method to purify RISC programmed with fluorescently labeled siRNA guide strands (CF Flores-Jasso and PD Zamore, unpubl.). We envision that our method will prove useful in the biochemical dissection of the RNAi and miRNA pathways and in the identification of proteins that modify and extend RISC function.
MATERIALS AND METHODS
General methods
Synthetic DNA (IDT) and RNA (Dharmacon) oligonucleotides were gel-purified before use. RNA oligonucleotides were synthesized containing a 5′ phosphate. The strands of the let-7 and luciferase siRNA duplexes were annealed in lysis buffer (30 mM HEPES-KOH at pH 7.4, 100 mM potassium acetate, 2 mM magnesium acetate) using a 1:1.3 molar ratio of guide to passenger. The let-7/let-7* duplex was annealed using a 1:5 molar ratio of miRNA to miRNA*.
Drosophila embryo lysate and MEF S100 cytosolic extract
Drosophila embryo lysate from 0- to 8-h embryos and RISC assembly were as described (Haley et al. 2003). The protein concentration of the embryo lysate was ∼30 mg/mL.
S100 extract was generated from SV40 large T-antigen immortalized Ago2−/− MEFs that stably overexpress mouse Ago2 mRNA (O’Carroll et al. 2007). Cells were grown to confluence in 5% CO2 at 37°C in DMEM (Invitrogen) supplemented with 15% fetal bovine serum (PAA Laboratories) and 50 units/mL penicillin and streptomycin (Invitrogen). S100 extract was prepared as described (Dignam et al. 1983) except that the cell pellet was washed three times in ice-cold PBS and once in Buffer A (10 mM HEPES-KOH at pH 7.9, 10 mM potassium acetate, 1.5 mM magnesium acetate, 0.5 mM DTT, EDTA-free protease inhibitor cocktail). The supernatant was removed, and 0.11 cell pellet volumes of Buffer B (300 mM HEPES-KOH at pH 7.9, 1.4 M potassium acetate, 30 mM magnesium acetate, 0.5 mM DTT, EDTA-free protease inhibitor cocktail) was added, followed by centrifugation at 100,000g for 20 min at 4°C. Ice-cold 80% (w/v) glycerol was then added to achieve a 13% (w/v) final glycerol concentration, followed by gentle inversion to mix. S100 was aliquoted, frozen in liquid nitrogen, and stored at −80°C. S100 protein concentration was ∼3–4 mg/mL.
Capture oligonucleotides and siRNAs
Streptavidin paramagnetic beads (100 µL; Dynabeads MyOne Streptavidin T1 10 mg/mL; Invitrogen) were washed and incubated with 5′ biotinylated, 2′-O-methyl capture oligonucleotides (500 pmol) according to the manufacturer’s instructions, then resuspended in 100 µL of lysis buffer containing 2 mM dithiothreitol (DTT) and kept on ice for RISC purification. The capture oligonucleotides (entirely 2′-O-methyl ribose) were 5′-biotin-UCUUCACUAUACAACCUACUACCUCAACCUU-3′ (fully complementary to let-7), 5′-biotin-UCUUCCUGCGACAAUAGCC UACCUCAACCUU-3′ (seed plus 3′ supplementary pairing for let-7); 5′-biotin-UCUUCCUGCGCACCAAGCCUACCUCAACCUU-3′ (seed pairing to let-7); 5′-biotin-UCUGACGCACUUGAUUCU UACGAUUUAUCUA-3′ (seed plus 3′ supplementary pairing for luciferase siRNA guide); 5′-biotin-GAUGAACCACUCAGAGACA UAAGCUAAUCUA-3′ (seed plus 3′ supplementary pairing for mmus-miR-21); 5′-biotin-UCUGACAACGUUGUGUAACCUC UAGUCCAUCU-3′ (seed plus 3′ supplementary pairing for Drosophila miR-286). siRNA sequences were 5′-pCGAGGUAGU AGGUUGUAUAGU-3′ (used as let-7 siRNA guide in Drosophila embryo lysate) or 5′-pUGAGGUAGUAGGUUGUAUAGU-3′ (used as let-7 siRNA guide in MEF lysate); 5′-pUAUACAACC UACUACCUCCUU-3′ (let-7 passenger); 5′-pCAAUCGUAUUU GUCAAUCAGA-3′ (luciferase guide); 5′-pUGAUUGACAAAUA CGAUUUUU-3′ (luciferase passenger). miRNA sequences were 5′-pUGAGGUAGUAGGUUGUAUAGU-3′ (let-7); 5′-pUAUACA AUGUGCUAGCUUUCU-3′ (let-7*).
RISC purification
The buffer from the washed capture oligo paramagnetic beads (500 pmol of capture oligo on 1 mg of beads) was removed and replaced with 100 µL of RISC assembly reaction, then incubated with gentle rotation for 30 min at room temperature. The supernatant was removed, and the beads were washed five times with lysis buffer containing 2 mM DTT and 0.01% (v/v) Triton X-100, followed by five washes with lysis buffer containing 2 M potassium acetate, DTT, and Triton X-100. RISC was eluted by resuspending the beads in 100 µL of lysis buffer containing 1 M potassium acetate, DTT, Triton X-100, and 1 nmol (10 µM f.c.) competitor oligo and incubating with gentle rotation for 120 min at room temperature. Competitor sequences were 5′-AAGGTTGAGGTAGTAGGTTGTATAGTGAAGA-3′ (for the capture oligo fully complementary to let-7); 5′-AAGGTTGAGGTAGGCTATTGTCGCAGGAAGA-3′ (for the capture oligo with seed plus 3′ supplementary pairing to let-7); 5′-AAGGTTGAGGTAGGCTTGGTGCGCAGGAAGA-3′ (for seed pairing to let-7); 5′-TAGATAAATCGTAAGAATCAAGTGCG TCAGA-3′ (for seed plus 3′ supplementary to luciferase). 5′-AGATGGACTAGAGGTTACACAACGTTGTCAGA-3′, (for seed plus 3′ supplementary pairing to Drosophila miR-286); 5′-UAGATTAGCUTAUGTCTCTGAGUGGTTCATC-3′, (for seed plus 3′ supplementary pairing to mouse miR-21). Finally, all samples were dialyzed at 4°C against three changes (2 h each) of a 2000-fold excess of lysis buffer containing 2 mM DTT. For quantitative analysis of yield (Table 1), guide strands were 5′ 32P-radiolabeled.
Ion exchange chromatography on Mono S 5/50 GL (GE Healthcare) was used to remove competitor DNA. The column was equilibrated in lysis buffer containing 2 mM DTT. After loading, the column was washed with five column volumes of lysis buffer containing 2 mM DTT. RISC was eluted with lysis buffer containing 2 mM DTT and 500 mM potassium acetate. The competitor DNA eluted in the wash. When necessary, samples were concentrated by centrifugal ultrafiltration (10 kDa cutoff; Amicon).
RISC activity and concentration
Target cleavage assays were as described (Haley et al. 2003). Target RNAs were transcribed with T7 RNA polymerase from templates generated by PCR of pGL2 plasmid (Promega) using these primers: 5′-GCGTAATACGACTCACTATAGGGTCACATCTCATCTACCT CC-3′ (let-7 forward); 5′-CCCATTTAGGTGACACTATAGATTTACATCGCGTTGAGTGTAGAACGGTTGTATAAAAGGTTGAGGTAGTAGGTTGTATAGTATCCAGAGGAATTCATTATCAGTG-3′ (let-7 reverse); 5′-GATGCGTAATACGACTCACTATAGGGT TCCGCATAGAACTGCCTGCGTCA-3′ (luciferase forward); 5′-TCCAGATCCACAACCTTCGCTTCA-3′ (luciferase reverse); 5′-GCGTAATACGACTCACTATAGGGGTCCTTTGATCGTGACAAAACAAT-3′ (mouse miR-21 forward); 5′-CCCATTTAGGTGACACTATAGATTTACATCTAGTTGAGGTGCGGAACTGTGTATAAAAGGTTAGCTTATCAGACTGATGTTGAATCCAGAGGAATTCATTATCAGTG-3′ (mouse miR-21 reverse). In Supplemental Figure S1, reaction products were resolved by denaturing polyacrylamide gel electrophoresis, and the amount of cleaved target was quantified using an FLA-9000 phosphorimager (GE Healthcare) and ImageGauge software (Fuji Life Sciences). IC50 values were obtained by fitting (Igor Pro 6.22, WaveMetrics) the data to the Hill equation assuming noncooperative behavior. Pre-steady-state kinetic data were fit in Igor Pro to the burst-and-steady-state equation,
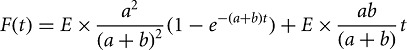 |
where F(t) is target cleaved with time, E is the enzyme concentration, and a and b are rate constants according to the following scheme:
The concentration of RISC assembled in Drosophila embryo lysate (Supplemental Fig. S2) was estimated by measuring the loss of 5′ 32P-radiolabeled passenger and guide strands. Control reactions, in which the lysate was pre-incubated with 1 mM (f.c.) N-ethylmaleimide (NEM) for 10 min at 4°C to prevent RISC assembly (Nykanen et al. 2001), demonstrated that differential loss of passenger and guide strands required RISC assembly. Unreacted NEM was quenched with 1.2 mM (f.c.) DTT. Reaction products were resolved by denaturing 20% polyacrylamide gel electrophoresis and detected as described. The rate of siRNA decay was determined separately for each replicate, and the significance determined using a Student’s two-sample t-test (GraphPad Prism 5; GraphPad Software).
Northern hybridization
Total RNA (5 µg), isolated from 2- to 6-h fly embryo lysates using TRIzol (Invitrogen) according to the manufacturer’s instructions, was resolved on a 0.4-mm thick, 15% denaturing polyacrylamide sequencing gel. After electrophoresis, the gels were blotted at 20 V for 1 h to a Hybond-NX membrane (Amersham Biosciences) in 0.5× TBE (Tris-Borate-EDTA) buffer by semidry transfer (Trans-Blot SD; Bio-Rad). The RNA was cross-linked to the membrane with freshly prepared 140 mM l-ethyl-3-(3-dimethylaminopropyl) carbodiimide (Sigma-Aldrich) in 130 mM 1-methylimidazole (Sigma-Aldrich) (pH 8) for 1 h at 60°C. The DNA oligonucleotide probe, Drosophila miR-286 5′-AGCACGAGTGTTCGGTCTAG TCA-3′ (25 pmol), was 5′-radiolabeled with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (450 μCi per reaction, specific activity ∼6000 Ci/mmol; PerkinElmer). After labeling, unincorporated nucleotides were removed (Sephadex G-25 spin column; GE Healthcare), and the probes were added to the Church buffer (Church and Gilbert 1984) and hybridized overnight at 37°C. Membranes were washed three times for 20 min in 0.01× SSC containing 0.1% SDS (w/v) at 37°C and exposed to storage phosphor screens (Fuji).
Mass spectrometry
For mass spectrometry, RISC was purified from a 2-mL assembly reaction and then concentrated 10-fold. Proteins were separated from low-molecular-weight contaminants by a brief period of SDS polyacrylamide gel electrophoresis, then a single gel slice containing all protein was excised, eluted, and digested with trypsin. For comparison, control reactions in which the small RNA duplex was omitted were included (Fig. 3C, background).
Tryptic peptides were dissolved in 0.1% (v/v) trifluoroacetic acid, loaded onto a fused silica trap column (2 cm × 100 µm C18), and then fractionated on a fused silica column (25 cm × 75 µm C18) developed with a linear gradient from 100% solvent A (0.1% formic acid, 5% acetonitrile in water) to 35% solvent B (0.1% formic acid in water) at a flow rate of 300 nL/min over 90 min using a nano LC: EASY system (Proxeon) directly coupled to an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific). Data-dependent acquisitions from MS scans (350–2000 m/z) in the Orbitrap at resolution, r = 60,000 were followed by 10 MS/MS scans acquired in the LTQ ion trap instrument.
Raw data files were processed with Mascot Distiller (Matrix Science) or Extract-MSn (Thermo Scientific) to generate peak lists and then searched against the Drosophila melanogaster (NCBInr) or Mus musculus (UniProtKB/SWISS-PROT) index using Mascot Search engine 2.3.02 (Matrix Science) with 10 ppm parent mass and 0.5 Da fragment mass tolerances. Amino acid modifications considered were acetyl (for the amino terminus of the protein), pyroglutamic acid (for amino-terminal glutamine), propionamide and carbamidomethylation modification of cysteine, and oxidation of methionine. Label-free quantification using extracted ion chromatograms (XIC) was performed using both the replicate and average methods in the Mascot Distiller quantitation software (Matrix Science). Mascot searches were also loaded into Scaffold3 software (Proteome Software) for further comparative analyses and filtering.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Dónal O’Carroll (EMBL Monterotondo, Italy) for ago2−/− MEFs overexpressing mouse AGO2; John Leszyk and the UMass Medical School Proteomics and Mass Spectrometry Facility for mass spectrometry; Alicia Boucher and Cindy Tipping for fly husbandry; and members of the Zamore laboratory for help, discussions, advice, and comments on the manuscript. This work was supported in part by grants from the National Institutes of Health to P.D.Z. (GM62862 and GM65236) and a CONACyT Postdoctoral Fellowship to C.F.F-J.
REFERENCES
- Ameres S, Martinez J, Schroeder R 2007. Molecular basis for target RNA recognition and cleavage by human RISC. Cell 130: 101–112 [DOI] [PubMed] [Google Scholar]
- Azuma-Mukai A, Oguri H, Mituyama T, Qian Z, Asai K, Siomi H, Siomi M 2008. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci 105: 7964–7969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo F, Gygi S, Bartel D 2008. The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini A, Lee M, Giraldez A 2012. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336: 233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitzinger M, Peters L, Zhu J, Kremmer E, Meister G 2007. Identification of human microRNA targets from isolated Argonaute protein complexes. RNA Biol 4: 76–84 [DOI] [PubMed] [Google Scholar]
- Broderick J, Salomon W, Ryder S, Aronin N, Zamore P 2011. Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. RNA 17: 1858–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada T, Gregory R, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W, Okamura K, Martin R, Lai E 2008. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol 18: 795–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G, Gilbert W 1984. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Proc Natl Acad Sci 81: 1991–19956326095 [Google Scholar]
- Czech B, Malone C, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel J, Sachidanandam R, et al. 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453: 798–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J, Lebovitz R, Roeder R 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S, Lendeckel W, Tuschl T 2001a. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15: 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T 2001b. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J 20: 6877–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkayam E, Kuhn C, Tocilj A, Haase A, Greene E, Hannon G, Joshua-Tor L 2012. The structure of human Argonaute-2 in complex with miR-20a. Cell 150: 100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen H, Berger U, et al. 2008. LNA-mediated microRNA silencing in non-human primates. Nature 452: 896–899 [DOI] [PubMed] [Google Scholar]
- Forstemann K, Horwich M, Wee L, Tomari Y, Zamore P 2007. Drosophila microRNAs are sorted into functionally distinct Argonaute complexes after production by Dicer-1. Cell 130: 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbasi V, Golden D, Hurtado S, Sontheimer E 2010. Proteomics identification of Drosophila small interfering RNA–associated factors. Mol Cell Proteomics 9: 1866–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore P 2009. Small silencing RNAs: An expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich M, Li C, Du T, Lee S, Xu J, Kittler E, Zapp M, Weng Z, et al. 2008. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320: 1077–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia N, Weissman J, Bartel D 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase A, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W 2005. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep 6: 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B, Tang G, Zamore P 2003. In vitro analysis of RNA interference in Drosophila melanogaster. Methods 30: 330–336 [DOI] [PubMed] [Google Scholar]
- Hammond S, Bernstein E, Beach D, Hannon G 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296 [DOI] [PubMed] [Google Scholar]
- Hendrickson D, Hogan D, Mccullough H, Myers J, Herschlag D, Ferrell J, Brown P 2009. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol 7: e1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, Zamore P 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060 [DOI] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli A, Bálint É, Tuschl T, Zamore P 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838 [DOI] [PubMed] [Google Scholar]
- Hutvágner G, Simard M, Mello C, Zamore P 2004. Sequence-specific inhibition of small RNA function. PLoS Biol 2: e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Satoh M, Pauley K, Fritzler M, Reeves W, Chan E 2006. Detection of the Argonaute protein Ago2 and microRNAs in the RNA induced silencing complex (RISC) using a monoclonal antibody. J Immunol Methods 317: 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iki T, Yoshikawa M, Nishikiori M, Jaudal M, Matsumoto-Yokoyama E, Mitsuhara I, Meshi T, Ishikawa M 2010. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol Cell 39: 282–291 [DOI] [PubMed] [Google Scholar]
- Ingolia N, Ghaemmaghami S, Newman J, Weissman J 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, Tomari Y 2010. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell 39: 292–299 [DOI] [PubMed] [Google Scholar]
- Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q 2005. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev 19: 1674–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Geoffroy M, Sobala A, Hay R, Hutvagner G 2010. HSP90 protein stabilizes unloaded Argonaute complexes and microscopic P-bodies in human cells. Mol Biol Cell 21: 1462–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev K, Tuschl T, Manoharan M, Stoffel M 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438: 685–689 [DOI] [PubMed] [Google Scholar]
- Lanford R, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk M, Kauppinen S, Orum H 2010. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327: 198–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaman D, Chen P, Fak J, Yalcin A, Pearce M, Unnerstall U, Marks D, Sander C, Tuschl T, Gaul U 2005. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell 121: 1097–1108 [DOI] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park S, Kim Y, Suh M, Kim V 2006. The role of PACT in the RNA silencing pathway. EMBO J 25: 522–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell M, Rivas F, Marsden C, Thomson J, Song J, Hammond S, Joshua-Tor L, Hannon G 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang F, Kalidas S, Smith D, Liu Q 2006. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA 12: 1514–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Park J, Jiang F, Liu Y, McKearin D, Liu Q 2007. Dicer-1, but not Loquacious, is critical for assembly of miRNA-induced silencing complexes. RNA 13: 2324–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ye X, Jiang F, Liang C, Chen D, Peng J, Kinch L, Grishin N, Liu Q 2009. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science 325: 750–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae I, Ma E, Zhou M, Robinson C, Doudna J 2008. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci 105: 512–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Tuschl T 2004. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev 18: 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Lührmann R, Tuschl T 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Dorsett Y, Tuschl T 2004a. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA 10: 544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T 2004b. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Takeuchi A, Siomi H, Siomi M 2010. A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat Struct Mol Biol 17: 1024–1026 [DOI] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G 2002. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev 16: 720–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Weinberg D, Bartel D, Patel D 2012. Structure of yeast Argonaute with guide RNA. Nature 486: 368–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykanen A, Haley B, Zamore P 2001. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107: 309–321 [DOI] [PubMed] [Google Scholar]
- Obad S, Dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup E, et al. 2011. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet 43: 371–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Carroll D, Mecklenbrauker I, Das P, Santana A, Koenig U, Enright A, Miska E, Tarakhovsky A 2007. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev 21: 1999–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung W, Ruby J, Guo H, Bartel D, Lai E 2008. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453: 803–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D, Senti K, Subramanian S, Sachidanandam R, Brennecke J 2012. The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Mol Cell 47: 954–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli A, Reinhart B, Slack F, Martindale M, Kuroda M, Maller B, Hayward D, Ball E, Degnan B, Muller P, et al. 2000. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408: 86–89 [DOI] [PubMed] [Google Scholar]
- Preall J, Czech B, Guzzardo P, Muerdter F, Hannon G 2012. shutdown is a component of the Drosophila piRNA biogenesis machinery. RNA 18: 1446–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas F, Tolia N, Song J, Aragon J, Liu J, Hannon G, Joshua-Tor L 2005. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 12: 340–349 [DOI] [PubMed] [Google Scholar]
- Ruby J, Stark A, Johnston W, Kellis M, Bartel D, Lai E 2007. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 17: 1850–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle N, MacRae I 2012. The crystal structure of human Argonaute2. Science 336: 1037–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H, Tushir J, Zamore P 2011. A 5′-uridine amplifies miRNA/miRNA* asymmetry in Drosophil a by promoting RNA-induced silencing complex formation. Silence 2: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Tahbaz N, Carmichael J, Hobman T 2001. GERp95 belongs to a family of signal-transducing proteins and requires Hsp90 activity for stability and Golgi localization. J Biol Chem 276: 43294–43299 [DOI] [PubMed] [Google Scholar]
- Tolia N, Joshua-Tor L 2007. Slicer and the Argonautes. Nat Chem Biol 3: 36–43 [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore P 2007. Sorting of Drosophila small silencing RNAs. Cell 130: 299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee L-M, Flores-Jasso CF, Salomon WE, Zamore PD 2012. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell 151: 1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiol J, Cora E, Koglgruber R, Chuma S, Subramanian S, Hosakawa M, Reuter M, Yang Z, Berninger P, Palencia A, et al. 2012. A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol Cell 47: 970–979 [DOI] [PubMed] [Google Scholar]
- Ye X, Huang N, Liu Y, Paroo Z, Huerta C, Li P, Chen S, Liu Q, Zhang H 2011. Structure of C3PO and mechanism of human RISC activation. Nat Struct Mol Biol 18: 650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P, Tuschl T, Sharp P, Bartel D 2000. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33 [DOI] [PubMed] [Google Scholar]
- Zheng G, Ambros V, Li W 2010. Inhibiting miRNA in Caenorhabditis elegans using a potent and selective antisense reagent. Silence 1: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]