Abstract
The structural proteins of the cytoplasmic intermediate filaments (IFs) arise in the nematode Caenorhabditis elegans from eight reported genes and an additional three genes now identified in the complete genome. With the use of double-stranded RNA interference (RNAi) for all 11 C. elegans genes encoding cytoplasmic IF proteins, we observe phenotypes for the five genes A1, A2, A3, B1, and C2. These range from embryonic lethality (B1) and embryonic/larval lethality (A3) to larval lethality (A1 and A2) and a mild dumpy phenotype of adults (C2). Phenotypes A2 and A3 involve displaced body muscles and paralysis. They probably arise by reduction of hypodermal IFs that participate in the transmission of force from the muscle cells to the cuticle. The B1 phenotype has multiple morphogenetic defects, and the A1 phenotype is arrested at the L1 stage. Thus, at least four IF genes are essential for C. elegans development. Their RNAi phenotypes are lethal defects due to silencing of single IF genes. In contrast to C. elegans, no IF genes have been identified in the complete Drosophila genome, posing the question of how Drosophila can compensate for the lack of these proteins, which are essential in mammals and C. elegans. We speculate that the lack of IF proteins in Drosophila can be viewed as cytoskeletal alteration in which, for instance, stable microtubules, often arranged as bundles, substitute for cytoplasmic IFs.
The multigene family of the structural proteins forming the cytoplasmic intermediate filaments (IFs) covers more than 50 members in human and mouse. In stratified epithelia, smooth muscle, and certain neurons, IF proteins are major constituents of the cytoskeleton. A predominantly structural role of IF in cellular resistance to mechanical stress emerged when human epithelial fragility syndromes were attributed to mutations in epidermal keratin genes (1, 2). Today mutations in 16 of the more than 20 keratin genes are associated with human diseases (3). Deletion of murine genes encoding keratin as well as nonkeratin IF proteins results in phenotypes, again emphasizing a structural role for IF (4), although a function as a scaffold allowing specific binding of some cellular constituents cannot be excluded (5).
Cytoplasmic IF proteins are restricted to animals. They probably arose in evolution from a mutated nuclear lamin parallel to metazoan multicellularity (6, 7). IFs are documented by electron microscopy in most invertebrate phyla, and cytoplasmic IF proteins have been cloned from 13 invertebrate phyla (6). Arthropods, however, lack IFs in electron microscopy (8), and the Drosophila genome shows no genes for cytoplasmic IF proteins (9, 10). Hynes and Zhao (11) proposed that this lack of IF proteins may not be surprising, because ablation of murine IF genes frequently results in only subtle cellular defects. This view raises the question of whether IFs present in other invertebrates are essential or nonessential structures.
Nematodes offer several advantages in addressing the functional importance of IFs in an invertebrate. First, IFs are clearly documented in all muscles and epithelia as well as in some axons of the large nematode Ascaris (8, 12), and related but not identical patterns are seen in Caenorhabditis elegans (13). Second, hemidesmosome-linked IFs of the hypodermis participate in the transmission of tension from the muscle cells to the cuticle (13–15). Third, C. elegans has a sizable IF gene family. We cloned eight genes (16), and the complete C. elegans genome documents an additional three genes (see below). Fourth, C. elegans allows inhibition of gene expression by double-stranded RNA (dsRNA)-mediated interference (RNAi) (17). Here we used RNAi analysis on all 11 C. elegans IF genes and obtained phenotypes for five genes (A1, A2, A3, B1, and C2). Embryonal and larval lethal phenotypes document that at least four IF genes (A1, A2, A3, and B1) are essential for C. elegans development.
Materials and Methods
Identification of Three IF Sequences in the C. elegans Database: Sequence Analysis.
blastp and blastn programs with standard parameters were used to search the complete C. elegans databases at the Sanger Centre (www.sanger.ac.uk) in April, 2000. Eight reported C. elegans IF sequences (16) used as queries resulted in the identification of an additional three novel proteins (R04E5.10, F25E2.4, and C43C3.1) with significant similarity to IF proteins. Further searches with the latter three sequences did not find any other IF-related protein. Finally, analyses with several Branchiostoma (18) and human IF proteins (types I–IV) did not identify any additional C. elegans IF protein. Full-length sequences of the 11 IF proteins or their rod domains were aligned with the use of the GCG software program pileup and submitted to phylogenetic [unweighted pair-group method with arithmetic mean (UPGMA)] analysis with the GCG programs distance and growtree.
Cloning of 11 C. elegans IF Sequences and dsRNA Preparation.
C. elegans strain N2 Bristol was routinely cultured and harvested as described (19). Total RNA was prepared from mixed-stage populations of worms by homogenization in Trizol reagent (GIBCO/BRL) according to the manufacturer's instructions. One microgram of poly(A)+ RNA isolated with an Oligotex kit (Qiagen, Chatsworth, CA) was used for cDNA synthesis with a Marathon cDNA amplification kit (CLONTECH). The cDNA library served as a template for PCR amplification of full-length or partial coding sequences of the individual IF cDNAs with the use of Pfu DNA polymerase (Promega) under the conditions described (18). Primer sequences and their locations in the coding regions of their respective cDNAs (16) (C. elegans genome project at http://www.sanger.ac.uk/Projects/C_elegans/wormpep/) were as follows: A1, sense nucleotides 76–101, antisense nucleotides 1779–1753; A2, sense nucleotides 1255–1281, antisense 5′-gatctgtataaagtatttattgttg-3′ (derived from the 3′ untranslated region; ref. 16); A3, sense nucleotides 7–30, antisense nucleotides 848–822; A4, sense nucleotides 1–29, antisense nucleotides 1671–1640; B1a, sense nucleotides 1–28, antisense nucleotides 1677–1651; B2, sense nucleotides 1–29, antisense nucleotides 1632–1602; C1, sense nucleotides 1–33, antisense nucleotides 1521–1491; C2, sense nucleotides 43–71, antisense nucleotides 1494–1466; D1, sense nucleotides 1–28, antisense nucleotides 1707–1677; D2, sense nucleotides 1–30, antisense nucleotides 1332–1303; E1, sense nucleotides 1–28, antisense nucleotides 2331–2298. The PCR fragments were cloned into blunt-TOPO pCR-2.1 (Invitrogen) or pBluescript plasmid vectors, and the inserted nucleotide sequences were confirmed by DNA sequencing. The clones obtained were reamplified with M13 forward-20 and M13 reverse primers. PCR conditions during the use of Expand High Fidelity (Roche Molecular Biochemicals) were as follows: (I) 95°C 4 min, once; (II) 94°C 1 min, 60°C 30 s, 72°C 1 min, 31 times; and (III) 72°C 5 min, once. PCR products were purified on QIAquick silica-gel membrane (Qiagen) and used as templates for in vitro RNA synthesis with Sp6/T7 or T3/T7 RNA polymerases, with the use of RiboMAX large-scale RNA production systems (Promega). RNA was purified with an RNeasy kit (Qiagen) and dissolved in water. Equal amounts of sense and antisense RNA were mixed, denatured (10 min at 68°C), and annealed (30–60 min at 37°C) to prepare dsRNA at concentrations between 0.1 and 0.5 μg/μl. The dsRNA was injected into the gonads of wild-type N2 hermaphrodites, which were transferred after 8 h to individual plates to score for abnormalities of their progeny. Five independent RNAi injections were used for quantification of the resultant phenotypes.
Green Fluorescent Protein (GFP) Reporter Plasmids.
The A1-, A2-, A3-, and B1-promoter/gfp reporter constructs covered the genomic sequences of 2,175, 1,509, 2,656, and 3,404 nt directly upstream of the A1, A2, A3, and B1 coding regions. The A1, A2, and A3 sequences were inserted into pEGFP-1 (CLONTECH) as XhoI/BamHI fragments, and the B1 sequence was inserted as a ClaI/BamHI fragment. The primers used for these PCR reactions were as follows: A1, sense 5′-cattgtgccaaagaatttctcgagcagtttgtaatgc-3′, antisense 5′-ctggtaatctccatcatttttggatccttgattggttaggtac-3′; A2, sense 5′-ctaataaacaacagttagttgctcgagttgtaaaggtggaatagc-3′, antisense 5′-gaatctggatcggtcattatggatccaaattttaaattgttag-3′; A3, sense 5′-cgcagccagcgcaccactcgagttggtttaattacctcgg-3′, antisense 5′-gcggtaggaatctggatcggggatccaaaaggaaactgc-3′; B1, sense 5′-cttaaaatatctgtaaatgagctccctgaagaataacc-3′, antisense 5′-gagacgacatcttctttcggatccttgacgctacctg-3′. Ballistic transformation of the promoter/gfp plasmids into C. elegans (20) was used to make the A1, A3, and B1 unintegrated transgenic lines used for the characterization of GFP expression.
Expression and Purification of Recombinant Proteins: Immunoblotting.
Coding sequences of 10 IF cDNAs were amplified by PCR under the reaction conditions described (16). PCR products of A1, A2, B1a, C1, C2, D1, and E1 were cloned into the pET23 vector (Novagen). A4 and B2 were cloned into pCRT7/CT-TOPO (Invitrogen), and D2 was cloned into pKK388–1 (CLONTECH). Recombinant proteins highly enriched in inclusion body preparations were dissolved in 8 M urea and purified by ion exchange chromatography (18). All recombinant IF proteins were verified by their amino-terminal sequences, obtained by automated Edman degradation. The nucleotide sequence of the cDNA encoding residues 1–278 of the A3 protein was amplified from blunt-TOPO pCR-2.1 vector (see above) and subcloned into the bacterial expression vector pGEX-KT (Amersham Pharmacia) to prepare a glutathione S-transferase (GST)-A3N fusion protein fragment. Expression in Escherichia coli and purification of the recombinant GST fusion protein fragment on glutathione Sepharose 4B followed standard protocols (Amersham Pharmacia). To produce the polypeptide fragment A3C covering residues 279–462 of the A3 protein with a carboxyl-terminal His6 tag, the corresponding coding sequence was amplified by PCR from genomic DNA (16) and subcloned into the bacterial expression vector pET23 (Novagen). Generation and affinity purification of the recombinant polypeptide fragment were performed under denaturing conditions according to the standard protocol (Invitrogen). The 10 purified IF proteins, two affinity-purified A3 fragments, and a total C. elegans extract were used for immunoblotting as described (18).
Antibodies and Immunofluorescence of Worms.
A rabbit B1 antiserum was raised with the synthetic peptide YSIPANVVIKPGKNL (residues 489–503 of B1; see ref. 16) as antigen. The peptide was coupled to hemocyanin via an extra N-terminal cysteine and used for antigen-affinity purification of the B1 antiserum as described (18). The specificity of the resulting antibody was verified in immunoblotting experiments with total C. elegans extracts and the 10 purified recombinant IF proteins A1, A2, A4, B1a, B2, C1, C2, D1, D2, and E1. In addition, two recombinant fragments, GST-A3N and A3C, covering residues 1–278 and residues 279–462 of the A3 IF protein (see above), were used. Indirect immunofluorescence microscopy of embryos and adult C. elegans was performed essentially as described (21). B1 antibody dilution was 1:10. The three murine monoclonal antibodies MH4, MH13, and MH33 were used in a 1:200 dilution of hybridoma supernatant. The mAb IFA was used as a 1:10 diluted supernatant. Antibody-stained animals were mounted in Mowiol 4.88 (Hoechst Pharmaceuticals) and viewed with a Zeiss Axiophot microscope.
Results
The C. elegans Cytoplasmic IF Family.
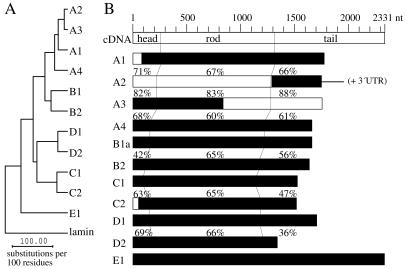
The eight cloned IF genes (16) form, by protein sequence comparison, the three subgroups A (A1–A4), B (B1 and B2), and C (C1 and C2). Fig. 1 shows that this nomenclature is readily extended to the three additional genes, which we identified in the complete C. elegans database Wormpep. Two of them, named R04E5.10 and F25E2.4, form the additional pair D1 and D2, and C43C3.1, now called E1, has the most remote sequence. Genes A2 and A3 are directly adjacent on the X chromosome, which also harbors the A1, A4, C2, D1, D2, and E1 genes. B1 and B2 are adjacent on chromosome II, and C1 is located on chromosome V. Within each subgroup sequence, identity levels of the coding sequences range from 60% (A3 and A4) to 67% (A1 and A2), whereas A2 and A3 share 83% identity.
Figure 1.
(A) Evolutionary tree formed by the rod domains of the 11 cytoplasmic IF proteins and the single nuclear lamin. (B) Schematic presentation of the coding regions of the individual IF cDNAs. Numbers at the top give the size in nucleotides. Full-length coding sequences are boxed; regions used as templates for the preparation of dsRNA are given as filled boxes. Degrees of identity (in %) between the head, rod, or tail encoding sequences of the corresponding A, B, C, and D pairs were calculated with the use of the GCG software program gap. In the case of A2, the dsRNA extended into the 3′ untranslated region. Note the existence of the second splice variant of the B1 protein, named B1b (sequence name F19C1.2B). It contains the unique 42-residue N-terminal end MSSHKESSEYEMQYRSTIQPRTAVRSQSRQSGNYVSGGNGAG attached to serine 12 of the B1a sequence (16).
All 11 proteins show the lamin-like length of the coil 1b domain typical of protostomic IF proteins. The six proteins of the A and B groups also display a lamin homology segment in their tail domains (6, 7, 16).
RNA-Mediated Interference of 11 C. elegans Genes.
Full-length or partial coding sequences of the 11 C. elegans cytoplasmic IF genes were used as templates for in vitro dsRNA synthesis (Fig. 1B and Materials and Methods). The dsRNAs were individually injected into the gonads of hermaphrodites to induce a specific RNA-mediated interference effect. After 8 h of recovery, injected worms were transferred into individual plates, and their progeny were analyzed microscopically and scored for abnormalities. The results are summarized in Table 1 and Figs. 2 and 3.
Table 1.
RNAi phenotypes of the 11 C. elegans IF genes
| Genes | RNAi phenotype |
|---|---|
| A1 | Larval lethality (100%), L1 arrest; wavy and swollen intestine |
| A2 | Early larval lethality (85%), late larval arrest (15%); excretory canals uneven, displaced body muscles, paralysis |
| A3 | Late embryonic lethality (25%), early larval lethality (75%); excretory canals uneven, displaced body muscles, hypodermis visibly detached from the cuticle, paralysis |
| B1 | Late embryonic lethality (100%); from almost normal pretzels with notched hypodermis to severe general morphogenetic defects |
| C2 | Dumpy phenotype, motion slightly reduced (10%). |
| A4, B2, C1, D1, D2, E1 | No phenotype |
Figure 2.
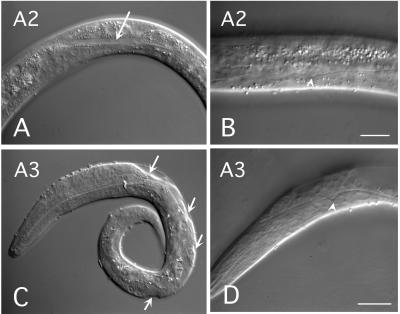
Defects in the development of worms treated with A2 or A3 RNAi. Lateral Nomarski views of A2 RNAi early larva document displaced body wall muscles (arrow in A) and uneven excretory canals (arrowhead in B). A related phenotype (100% lethality, Table 1) with moderately uneven excretory canals (arrowhead in D) and visible detachment of hypodermis from the cuticle (arrows in C) was observed in larva treated with A3 RNAi. [Scale bars: 15 μm (A, C, and D) and 10 μm (B).]
Figure 3.
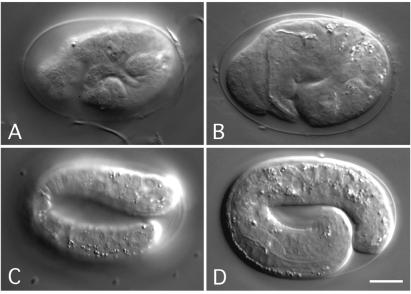
Late embryonic lethality observed with B1 RNAi. Two late-stage embryos affected by RNAi showing severe morphological defects (A and B) are compared with the 3-fold stage wild-type embryo, presented in two focal planes (C and D). (A) The pattern of hypodermal cells is very irregular, resulting in a lumpy surface. (B) The embryo is shorter than normal, and the internal structures show a compressed morphology. (Scale bar: 15 μm.)
Injection of A1 dsRNA caused early larval lethality. The A1 phenotype developed normally but arrested at the L1 stage and died slowly within 3–6 days. Locomotion, morphology, and pharyngeal pumping of the phenotype seemed normal. Arrested larvae did not move away from the E. coli islands, indicating a normal chemotaxis. Compared with the wild type, the intestine was wavy and swollen. RNAi experiments with the A2 gene resulted in early lethality of most larvae (85%). All L1 larvae exhibited uneven excretory canals and displacement of body muscles, and, as a consequence, demonstrated variable deviations in locomotion (Fig. 2 A and B). Approximately 15% of the A2 RNAi larvae slowly matured and arrested before the appearance of the reproductive system. Arrested larvae showed strong paralysis, with only occasional slow movements of the head or tail. Similar effects resulting invariably in early larval lethality were observed for A3 RNAi. This phenotype showed displaced body muscles, and the hypodermis was visibly detached from the cuticle (Fig. 2 C and D). In addition, approximately one-quarter of the eggs laid by A3 dsRNA-treated animals failed to hatch, indicating serious defects in embryonic development.
The B1 RNAi caused a late embryonic lethal phenotype with different morphological abnormalities (Fig. 3), ranging from lumpy pretzels (“end of elongation” stage of the embryogenesis) of normal length with notches in the hypodermis, to shorter embryos with aberrant morphogenesis of the hypodermis. In shorter embryos internal structures were also compressed, which is most likely a secondary effect of the missing elongation of the embryo. During elongation the bean-like embryo is compressed into a worm shape. The last phenotype was identified in animals injected with C2 dsRNA. About 10% of the progeny showed a dumpy morphology and displayed moderate defects in movement (data not shown). However, all C2 animals developed into adults and produced offspring. Finally, worms injected individually with A4, B2, C1, D1, D2, or E1 dsRNA showed no obvious phenotype (Table 1).
Expression Pattern of A1, A3, and B1 IF Genes.
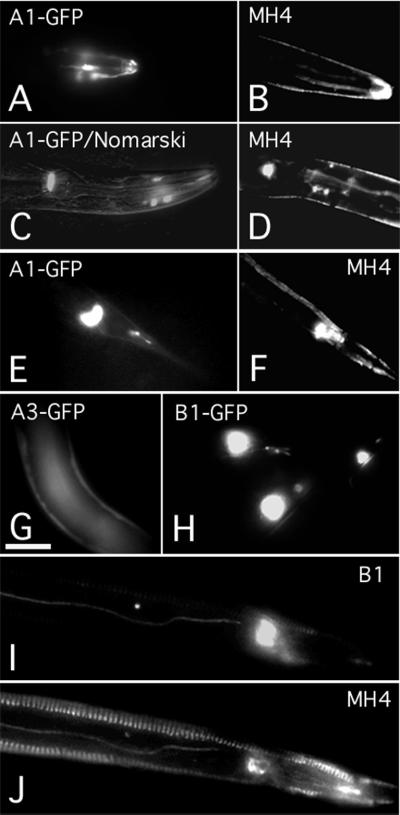
The expression patterns of three IF genes (A1, A3, B1) that showed strong effects in RNAi experiments were analyzed. Genomic sequences of 2,175, 2,656, and 3,404 nt directly upstream of the A1, A3, and B1 coding sequences were fused to gfp and ballistically transformed into worms, and the resulting promoter-driven GFP expression was monitored. The A1 promoter/gfp reporter was expressed in cells associated with amphid sensory neurons, the pharyngeal-intestinal valve, the vulva, a few yet unidentified neurons, as well as in the rectum and some neurons of the tail (Fig. 4 A, C, and E). In addition, there was some mosaicism for small and as yet unidentified muscles of the pharynx. Not all A1 promoter/gfp worms showed GFP expression in these cells. The A3 promoter/gfp reporter was detected in the embryonic and larval hypodermis and sometimes also in the ventral nerve cord of larvae, but no expression was detected in adults (Fig. 4G). The loss of A3 expression in the adult is in line with Northern blots from a mixed population that showed a barely detectable signal for A3 compared with seven other IF messenger RNAs (16). The B1 promoter-driven GFP staining was seen in cells associated with the amphid sensory neurons, the excretory cells, the vulva, and the uterus (data not shown) and, finally, in the rectum and some neurons of the tail (Fig. 4H). In the pharynx, staining localized to the marginal cells and the pharyngeal-intestinal valve. The same structures were seen in immunofluorescence experiments with a B1-specific antibody (Fig. 4I; see Materials and Methods). This antibody recognized in immunoblots the recombinant protein B1a and two closely spaced bands in a total nematode extract (Fig. 5). They probably correspond to the two variants of B1 protein (Fig. 1). Thus the A1 and B1 expression patterns show certain similarities but are not identical.
Figure 4.
Expression of A1-, A3-, and B1-promoter/gfp constructs and the IF protein B1. Expression of the A1 promoter/gfp reporter in cells associated with amphid sensory neurons (A), the pharyngeal-intestinal valve (C), the rectum, and some neurons of the tail (E). Similar structures were immunostained with antibody MH4 which, however, also decorates the hypodermis (B, D, F, and J). The A3 promoter/gfp reporter is primarily expressed in the embryonal and larval hypodermis (G). The B1 promoter/gfp is expressed in the rectum and some unidentified neurons in mixed-stage animals (H). Whole-mount worms were double-labeled with affinity-purified rabbit antibody specific for IF protein B1 (I) and with antibody MH4 (J). [Scale bars: 50 μm (A, C, E, and H) and 25 μm (B, D, F, G, I, and J).]
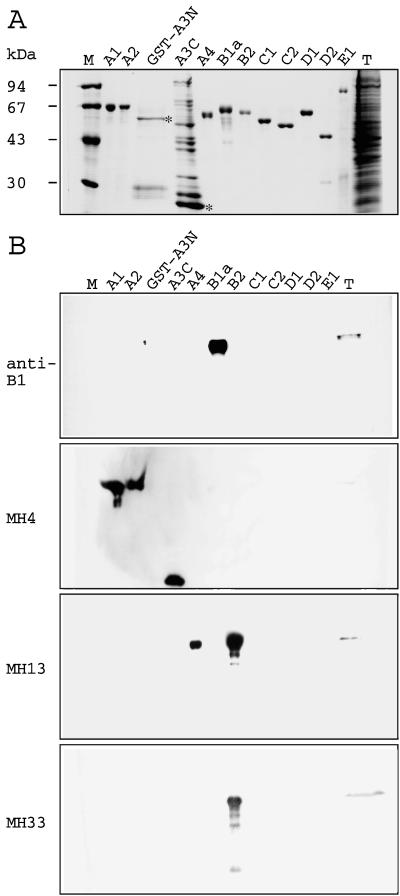
Figure 5.
Immunoblot analyses of C. elegans recombinant IF proteins and total extract. Equal amounts of recombinant IF proteins A1, A2, A4, B1a, B2, C1, C2, D1, D2, and E1; the two recombinant fragments GST-A3N and A3C of A3 (see Materials and Methods); and a total protein extract (T) were separated by SDS/PAGE (10%) and stained with Coomassie (A) or blotted on nitrocellulose membranes (B). Blots were incubated with affinity-purified rabbit anti-B1 antibody and the murine mAbs MH4, MH13, and MH33. The B1 antibody recognized the recombinant protein B1a and two closely spaced bands in the total nematode extract. They probably correspond to the two variants of the B1 protein (see Fig. 1). MH4 reacts strongly with A1, A2, and A3C and weakly detects comigrating band in the total nematode extract. MH13 recognized recombinant proteins A4 and B2, and comigrating band in the total nematode extract. MH33 recognized recombinant protein B2 and its degradation products as well as a single band of the expected molecular mass in the total C. elegans extract. The asterisks denote the positions of the recombinant fragments GST-A3N and A3C of protein A3. Marker proteins and an approximate molecular mass standard in kDa are given at the left (M).
The expression patterns of the A1, A3, and B1 genes relate to the staining patterns reported for the murine monoclonal antibody MH4, which detects one or more C. elegans IF proteins (refs. 13 and 14; see also Fig. 4 B, D, F, and J). To further characterize the MH4 epitope, the 10 IF proteins A1, A2, A4, B1a, B2, C1, C2, D1, D2, and E1 and two peptides covering residues 1–278 and 279–462 of A3 were expressed in E. coli and purified. Immunoblotting of the recombinant proteins showed that MH4 reacts strongly with A1 and A2 (Fig. 5) and detects comigrating bands in the total nematode extract. Strong reaction of MH4 also was observed with the purified recombinant fragment A3C, which covers residues 279–462 of the IF protein A3 (Fig. 5). We also noted a rather weak reaction of MH4 with recombinant A4 and B1a proteins visualized after overnight incubation at 4°C. Whether the weak MH4 reactivity of A4 and B1 detected on immunoblots by the highly sensitive enhanced chemiluminescence system would be sufficient for detection by immunofluorescence microscopy is questionable. Thus the prime targets of MH4 are the IF proteins A1–A3. We also tested the mAbs MH13, MH33 (13), and IFA (22). Antibody IFA showed the same reactivity pattern as MH4. Antibody MH13 recognized proteins A4 and B2, and MH33 reacted exclusively with the recombinant IF protein B2 (Fig. 5).
Discussion
With the use of RNAi analysis, we found that four of the 11 genes encoding cytoplasmic IF proteins (A1, A2, A3, and B1) have essential roles in C. elegans development. Inhibition of expression of a fifth gene (C2) resulted only in a mild phenotype, with about 10% of the progeny showing a dumpy appearance and reduced locomotion (Table 1). Not all C. elegans genes are susceptible to RNAi, which seems less potent in silencing gene expression during later development (17). Whereas embryonic lethal phenotypes were detected with similar efficiency by injection and feeding of RNAi (23), the latter application of RNAi seemed to detect over 50% more postembryonic phenotypes than injection (24). Thus it remains to be seen whether some of the six IF genes (A4, B2, C1, D1, D2, and E1; Table 1) that so far are unaffected by RNAi introduced by injection become targeted when another application form is used.
The displaced body muscles and the paralysis observed with the lethal larval phenotypes of A2 and A3 could be due to a muscle or a hypodermal defect, inasmuch as C. elegans locomotion is thought to involve a transmission of contractile force from the muscle dense bodies to the cuticle via the basal lamina and the fibrous organelles of the syncitial hypodermis, i.e., the hemidesmosomes on the basal and apical membrane of the hypodermis plus the connecting IF. In addition, during development the assembly of hypodermal hemidesmosomes is spatially and temporally coordinated with the assembly of the myofibrillar organization in the muscle cell (13–15). Thus, for instance, C. elegans mutants lacking myotactin, a hypodermal transmembrane protein of the basal hemidesmosomes, show detachment of muscle cells (15). Because hypodermal IFs as well as IF at some other sites of the body, but not muscle IFs, react with the mAb MH4 (ref. 13; see also Fig. 4), we characterized this antibody in more detail. To decide which IF proteins carry the MH4 epitope, we generated 10 recombinant proteins and two recombinant fragments (A3) in E. coli. Immunoblotting of the purified proteins showed that MH4 reacts very strongly with IF proteins A1, A2, and A3 (Fig. 5). In contrast, the mAb MH33, which is found in the intestine (13), was specific for the B2 IF protein (Fig. 5). Additional information on the expression patterns was obtained with gfp-promoter constructs, which showed that A1 is expressed at many body sites but not in hypodermis (Fig. 4), whereas A3 is expressed in the embryonic and larval hypodermis but not in the adult. Thus to account for the strong MH4 reactivity of the adult hypodermis, we must assume that A2 is a major IF protein of the hypodermis. Direct proof by a gfp-promoter construct was not obtained. We assume that the A2 gene contains regulatory elements also outside the promoter region covered in our construct. Our conclusion that A2 is a major hypodermal IF protein is not influenced by the weak reactivity of MH4 on B1 and A4. The B1-gfp promoter construct and a B1-specific antibody both show that B1 is not expressed in the hypodermis (see Fig. 4 and Results). Similarly, an A4-gfp promoter construct and an antibody specific for A4 showed A4 expression in the H-shaped excretory cells and some other not yet identified interior regions of C. elegans, but not in the hypodermis (data not shown). Thus the combined evidence points to a reduction of hypodermal IF in the lethal larval RNAi phenotypes obtained for A2 and A3. The detachment of the hypodermis from the cuticle observed in the A3 phenotype fits this interpretation. The moderate enlargement and uneven appearance of the excretory canals also may be the result of the disturbed hypodermal IF, because the hypodermis and excretory canals are in close contact (25) and the position of the canals is marked by MH4 staining (ref. 13; see also Fig. 4J).
The late embryonic lethal B1 phenotype as well as the L1 arrested A1 phenotype are currently not understood and may require a detailed electron microscopic analysis to get additional information. It is interesting to compare the results on IF gene silencing in mice and C. elegans. The RNAi phenotypes obtained for four IF genes in C. elegans are severe, given their embryonic and early larval lethality. In contrast, individual ablation of a large number of murine IF genes has not resulted in an embryonic lethal phenotype (4). This case was only recently observed in the double knockouts of either keratins K18 plus K19 or keratins K8 plus K19 (26, 27).
The last member of the C. elegans IF protein family is a nuclear lamin (28). RNAi showed that the single C. elegans lamin is an essential embryonic gene. The lamin protein is involved in nuclear organization, cell cycle progression, and the spatial organization of nuclear pore complexes (21). The embryonic lethal phenotype also emerged in the systematic RNA interference analysis of chromosome I (24).
Because the Drosophila genome lacks IF genes (9), the original aim of our study was to determine whether IF genes of C. elegans have essential functions. As this question is now answered positively for at least four genes, the problem can be rephrased: How does Drosophila compensate for the lack of IF proteins? An ultrastructural survey of many different invertebrates revealed that arthropods, including Drosophila, differ from other invertebrate phyla in their lack of cytoplasmic IF. Often massive bundles of microtubules seem to substitute for the IF bundles used by other phyla in related cell types. For instance, in the esophageal epithelium of crayfish and Limulus, such bundles contain in cross sections more than 100 microtubules. Axons of various arthropods show cross-linked microtubular profiles in the absence of neurofilaments (ref. 8 and references therein). The cell surface-associated transcellular bundles of Drosophila wing epidermal cells contain 900 microtubules and intermicrotubular actin filaments of uniform polarity. Curiously, these microtubules are composed of 15 rather than the usual 13 protofilaments (29, 30). Adhesion between and within cell layers of Drosophila requires kakapo, a large cytoskeletal linker protein related to mammalian plektin but without plektins's IF binding domain. Kakapo is thought to bind to F-actin and microtubules and to provide membrane anchorage in the absence of IFs (31). Thus, the lack of cytoplasmic IF genes in the Drosophila genome (9) can be understood from the ultrastructural results, which indicate that specialized stable microtubular bundles provide for the function served by IFs in other metazoan phyla. These results also seem to be compatible with the recent finding that in mammalian cells IFs assemble along microtubule tracks in a kinesin-dependent manner (32).
Acknowledgments
We thank Wolfgang Berning-Koch, Jürgen Schünemann, and Cathrin Struck for expert technical assistence. We also thank Alan Coulson for providing cosmids and Robert H. Waterston for mAbs MH4, MH13, and MH33.
Abbreviations
- dsRNA
double-stranded RNA
- IF
intermediate filament
- RNAi
RNA interference
- GST
glutathione S-transferase
- GFP
green fluorescent protein
Footnotes
See commentary on page 7659.
References
- 1.Fuchs E, Weber K. Annu Rev Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- 2.Parry D A D, Steinert P M. Intermediate Filament Structure. New York: Springer; 1995. [DOI] [PubMed] [Google Scholar]
- 3.Irvine A D, McLean W H I. Br J Dermatol. 1999;140:815–828. doi: 10.1046/j.1365-2133.1999.02810.x. [DOI] [PubMed] [Google Scholar]
- 4.Magin T M, Hesse M, Schröder R. Protoplasma. 2000;211:140–150. [Google Scholar]
- 5.Ku N O, Liao J, Omary M B. EMBO J. 1998;17:1892–1906. doi: 10.1093/emboj/17.7.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erber A, Riemer D, Bovenschulte M, Weber K. J Mol Evol. 1998;47:751–762. doi: 10.1007/pl00006434. [DOI] [PubMed] [Google Scholar]
- 7.Weber K, Plessmann U, Ulrich W. EMBO J. 1989;8:3221–3227. doi: 10.1002/j.1460-2075.1989.tb08481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartnik E, Weber K. Eur J Cell Biol. 1989;50:17–33. [Google Scholar]
- 9.Rubin G M, Yandell M D, Wortman J R, Gabor Miklos G L, Nelson C R, Hariharan I K, Fortini M E, Li P W, Apweiler R, Fleischmann W, et al. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein L S B, Gunawardena S. J Cell Biol. 2000;150:F63–F68. doi: 10.1083/jcb.150.2.f63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hynes R O, Zhao Q. J Cell Biol. 2000;150:F89–F96. doi: 10.1083/jcb.150.2.f89. [DOI] [PubMed] [Google Scholar]
- 12.Bartnik E, Osborn M, Weber K. J Cell Biol. 1986;102:2033–2041. doi: 10.1083/jcb.102.6.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francis R, Waterston R H. J Cell Biol. 1991;114:465–479. doi: 10.1083/jcb.114.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hresko M C, Williams B D, Waterston R H. J Cell Biol. 1994;124:491–506. doi: 10.1083/jcb.124.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hresko M C, Schriefer L A, Shrimankar P, Waterston R H. J Cell Biol. 1999;146:659–672. doi: 10.1083/jcb.146.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dodemont H, Riemer D, Ledger N, Weber K. EMBO J. 1994;13:2625–2638. doi: 10.1002/j.1460-2075.1994.tb06553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S E, Mello C C. Nature (London) 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 18.Karabinos A, Riemer D, Panopoulou G, Lehrach H, Weber K. Eur J Cell Biol. 2000;79:17–26. doi: 10.1078/S0171-9335(04)70003-5. [DOI] [PubMed] [Google Scholar]
- 19.Sulston J, Hodgkin J. In: The Nematode Caenorhabditis elegans. Wood W B, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 587–606. [Google Scholar]
- 20.Wilm T, Demel P, Koop U, Schnabel R. Gene. 1999;229:31–35. doi: 10.1016/s0378-1119(99)00043-8. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Ben-Shahar T R, Riemer D, Treinin M, Spann P, Weber K, Fire A, Gruenbaum Y. Mol Biol Cell. 2000;11:3937–3947. doi: 10.1091/mbc.11.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruss R M, Mirsky R, Raff M C, Thorpe R, Dowding A J, Anderton B H. Cell. 1981;27:419–428. doi: 10.1016/0092-8674(81)90383-4. [DOI] [PubMed] [Google Scholar]
- 23.Timmons L, Fire A. Nature (London) 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 24.Fraser A G, Kamath R S, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Nature (London) 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 25.Buechner M, Hall D H, Bhatt H, Hedgecock E M. Dev Biol. 1999;214:227–241. doi: 10.1006/dbio.1999.9398. [DOI] [PubMed] [Google Scholar]
- 26.Hesse M, Franz T, Tamai Y, Taketo M M, Magin T M. EMBO J. 2000;19:5060–5070. doi: 10.1093/emboj/19.19.5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamai Y, Ishikawa T, Rösl M R, Mori M, Nozaki M, Baribault H, Oshima R G, Taketo M M. J Cell Biol. 2000;151:563–572. doi: 10.1083/jcb.151.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riemer D, Dodemont H, Weber K. Eur J Cell Biol. 1993;62:214–223. [PubMed] [Google Scholar]
- 29.Mogensen M M, Tucker J B. J Cell Sci. 1987;88:95–107. doi: 10.1242/jcs.88.1.95. [DOI] [PubMed] [Google Scholar]
- 30.Mogensen M M, Tucker J B. J Cell Sci. 1988;91:431–438. doi: 10.1242/jcs.91.3.431. [DOI] [PubMed] [Google Scholar]
- 31.Gregory S L, Brown N H. J Cell Biol. 1998;143:1271–1282. doi: 10.1083/jcb.143.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prahlad V, Yoon M, Moir R D, Vale R D, Goldman R D. J Cell Biol. 1998;143:159–170. doi: 10.1083/jcb.143.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]