Abstract
Amyloid formation, a complex process involving many intermediate states, is proposed to be the driving force for amyloid-related toxicity in common degenerative diseases. Unfortunately, the details of this process have been obscured by the limitations in the methods that can follow this reaction in real-time. We show that alternative pathways of aggregation can be distinguished by using 19F NMR to monitor monomer consumption along with complementary measurements of fibrillogenesis. The utility of this technique is demonstrated by tracking amyloid formation in the diabetes-related islet amyloid polypeptide (IAPP). Using this technique, we show IAPP fibrillizes without an appreciable build up of non-fibrillar intermediates, in contrast to the well-studied Aβ and α-synuclein proteins. To further develop the usage of 19F NMR, we have tracked the influence of the polyphenolic amyloid inhibitor epigallocatechin gallate (EGCG) on the aggregation pathway. Polyphenols have been shown to strongly inhibit amyloid formation in many systems. However, spectroscopic measurements of amyloid inhibition by these compounds can be severely compromised by background signals and competitive binding with extrinsic probes. Using 19F NMR, we show that thioflavin T strongly competes with EGCG for binding sites on IAPP fibers. By comparing the rates of monomer consumption and fiber formation, we are able to show that EGCG stabilizes non-fibrillar large aggregates during fibrillogenesis.
Keywords: IAPP, fluorine NMR, kinetics, mechanism, EGCG, inhibitor
Introduction
The formation of amyloid fibers is associated with a wide range of pathologies including Alzheimer’s disease, Parkinson’s disease, and type II diabetes.1 Under pathological conditions, amyloidogenic proteins self-assemble into long fibrillar structures with a characteristic β-sheet core.2-4 Although a detailed mechanism is lacking for most amyloidogenic proteins, amyloidogenesis is believed to be a complex, multi-step process, typically involving the formation of energetically unfavorable intermediates before the formation of the final amyloid product.5-8
Because amyloid formation is such a complex process, a full understanding of the pathology caused by amyloid formation requires establishing both the identity of intermediates along the amyloid pathway and the kinetics of their formation. The actual identity of the species responsible for toxicity is elusive and has been the subject of much debate since the discovery of amyloid plaques more than a century ago.7-9 A current hypothesis holds that small to intermediate size (~5-6 nm in diameter) oligomers may be responsible for much of the toxicity of amyloid proteins.6, 8 For the Aβ protein,1 probably the best studied amyloidogenic protein, oligomers of this type have been isolated both in vitro and in situ from tissue samples of Alzheimer’s patients. While a large body of literature exists linking small oligomers of this type to the pathological action of other amyloidogenic proteins besides Aβ, their actual existence in many other amyloidogenic proteins has largely been inferred rather than directly tested.7 For islet amyloid polypeptide (IAPP) in particular, the existence of small oligomeric species of this type is controversial.7, 8 Most attempts at detection has used the conformation-specific antibody A11, whose specificity towards non-Aβ oligomers has been called into question,7 as it appears to give both false positive and false negative under some conditions.7, 10-13 Attempts at direct detection of these oligomers have largely been negative, although these experiments have used an equilibrium approach that may not detect transient oligomers.14, 15 In the absence of well-defined targets, research attempting to attenuate amyloid-linked toxicity has been substantially impeded.
Towards this end, various methods have recently been exploited to monitor the kinetics of amyloid fibril formation including fluorescence,16-19 Nuclear Magnetic Resonance (NMR)20, 21, 2D-infrared,22, 23 and mass spectroscopy.24, 25 The most common method for measuring amyloid formation exploits the increase in fluorescence when the dye thioflavin T (ThT) binds to the fibers. Although the ThT assay is simple and relatively sensitive, it does not accurately distinguish between oligomeric species.26 Therefore, it is not particularly useful by itself to investigate amyloidogenic intermediates. Similarly, reporters of secondary structures such as circular dichroism (CD), Fourier transform infrared and multidimensional NMR spectroscopy have difficulty in detecting the underlying spectral signature of the intermediate species, which may be transient and not form a majority of the total population.
On the other hand, real-time 1H NMR measurements, in theory, allow the signal from each species to be resolved and analyzed independently. Unfortunately, 1H NMR is relatively insensitive to changes in structure due to small chemical shift dispersion associated with the 1H nucleus. Whereas 13C and 15N experiments are more sensitive to environmental changes, the low sensitivity of these nuclei makes it difficult to acquire data sufficiently rapidly to track intermediates during aggregation. In contrast, 19F combines both excellent sensitivity and large chemical shift dispersion, making it an excellent reporter for changes in chemical environment.27, 28 Thus 19F NMR has been broadly applied to investigate protein conformational changes and dynamics as well as protein-ligand and protein-membrane interactions in biological systems.29-43 Whereas the properties of 19F NMR spectroscopy are attractive for amyloid research, applications to amyloid proteins have been relatively limited.44, 45 In particular, detailed time-resolved studies of protein aggregation or inhibition assays using 19F NMR have not been performed.
In this report we demonstrate the utility of 19F NMR to provide a direct, sensitive and real-time measurement of amyloid formation by the highly amyloidogenic peptide, IAPP, which is achieved by monitoring the consumption of monomeric protein rather than the formation of fibrils by dye binding. IAPP is a 37-residue peptide hormone secreted from pancreatic β-cells along with insulin to contribute glycemic control.46 However, an insoluble fibril form of IAPP is found in the pancreas of up to 90% of patients with type II diabetes that may contribute to the pathology of the disease.7, 8, 47, 48 Using real-time 19F NMR in combination with other techniques to measure conformational changes during the aggregation pathway, we show IAPP forms fibers from monomeric IAPP at pH 7.3 without an appreciable buildup of non-fibrillar intermediates during aggregation, in contrast to most amyloidogenic proteins.6 However, this pathway can be altered towards the formation of non-fibrillar intermediates using inhibitors such as epigallocatechin gallate (EGCG).
Materials and Methods
Peptide Synthesis
Peptide (IAPP-tfmF23) was synthesized manually in the free acid form by solid-phase 9-fluorenylmethoxycarbonyl (Fmoc)-based chemistry using pseudoproline dipeptides to disrupt aggregation during synthesis, as described previously.49 4-trifluoromethylphenylalanine (tfmF) was used to replace Phe at position 23. The disulfide bridge between Cys2 and Cys7 in IAPP was formed on the resin using thallium (III) trifluoroacetate as a mild oxidant, and cleaved from resin using 95% trifluoroacetic acid.49 Crude peptides were dissolved in 35% acetic acid (v/v) and purified on a reverse-phase HPLC using a Waters semi-preparative C18 column equilibrated in 0.045% HCl. The peptides were eluted with a linear gradient of 0% to 80% acetonitrile at a flow rate of 10 ml/min. Peptide identity was confirmed using matrix-assisted laser desorption ionization mass spectroscopy.
Sample preparation
To remove preformed aggregates, the purified peptide was dissolved in hexafluoroisopropanol followed by the removal of the solvent by lyophilization. A 850 μM stock solution was made before each set of experiments by first dissolving the lyophilized peptide in 0.1 mM HCl (pH 4.0). The stock solution was then passed though a 0.22 μm filter before dilution into the final buffer solution immediately before the start of each experiment. The concentrations of peptide stock solutions were determined by the absorbance of Y37 (ε = 1215 L mol−1 cm−1 at 280 nm).50 All experiments were performed at a constant peptide concentration of 85 μM. The final solutions contained 20 mM sodium phosphate, 50 mM NaCl (pH 7.3), 10% D2O, and various concentrations of EGCG, ThT, and EDC/Sulfo-NHS as indicated in the figure captions. For NMR measurements, 19F chemical shifts were referenced to an internal standard of trifluoroethanol (100 μM TFE, at 0 ppm).
Kinetic studies using 19F NMR spectroscopy
All 19F NMR experiments were performed using a Varian VNMRS 500 MHz NMR spectrometer equipped with a double-tuned 1H-19F probe. After the addition of peptide into the NMR solution, a series of 19F spectra were recorded without spinning. Each 19F spectrum takes 116 s using 64 transients (25°C and 37 °C) or 232 s using 128 transients (10 °C) with a 1.0 s pulse delay between each transient. Line broadening of 3.0 Hz was used to process the final spectra. All of the samples were pre-equilibrated at the indicated temperatures before mixing with peptide stock solution. Origin 8.5 was used for plotting and fitting the data.51
Kinetic Studies Using Thioflavin Fluorescence
The kinetics of IAPP amyloid formation was measured using the increase in fluorescence intensity observed when the amyloid specific dye ThT binds to the amyloid fibrils. The samples were prepared as described above for the NMR solution except that 100 μM ThT was premixed with buffer solution before the addition of peptide. Time traces were recorded with a FluoroMax-2 spectrofluorometer using a 440 nm excitation filter and a 485 nm emission filter at a constant temperature without shaking.
Circular Dichroism
CD measurements were performed with an Aviv 62DS spectropolarimeter using a 0.1 cm pathlength cell. The samples were prepared as described above for the NMR solution. Mean residue ellipticities, [θ], were calculated using [θ] = θobsd/(10*l*c*n) where θobsd is the ellipticity measured in millidegrees, c is the molar concentration, l is the cell path length in centimeters, and n is the number of residues in the peptide. Full spectra were smoothed by Savitzky-Golay smoothing (5 point window).
Electron Microscopy
Aliquots were directly taken from the NMR samples at the time-points indicated. 6 μL aliquots were incubated onto Formvar-coated copper grids (Ernest F. Fullam, Inc., Latham, NY) for 2 min, washed five times with 6 μL of deionized water, and then negatively stained for 1 min with 2% uranyl acetate. Samples were imaged using a Philips CM10 Transmission Electron Microscope.
Results and Discussion
19F NMR sensitively measures monomer consumption during aggregation
To study fibril formation using 19F NMR, we synthesized a 19F-labeled IAPP (IAPP-tfmF23) in which Phe-23 is substituted by 4-trifluoromethyl phenylalanine (tfmF). Phe-23 is solvent exposed in the unstructured monomeric peptide,19, 52 but is believed to be become buried when the peptide forms aggregates.2, 19, 53, 54 The 19F chemical shifts and line-widths of the trifluoromethyl groups should change significantly as the peptide changes its secondary structure and oligomerization state, as has been observed for other fluorinated peptides and proteins.38-40 In particular, if oligomeric intermediates on the amyloid-forming pathway accumulate to an appreciable extent they should be detectable by NMR, provided they are not so large that their line-widths are broadened excessively, as is the case for amyloid fibers.20 Furthermore, the 19F signal is sufficiently sensitive to monitor peptide aggregation at low concentrations that allows a direct comparison with other methods for following aggregation such as CD or fluorescence spectroscopy.
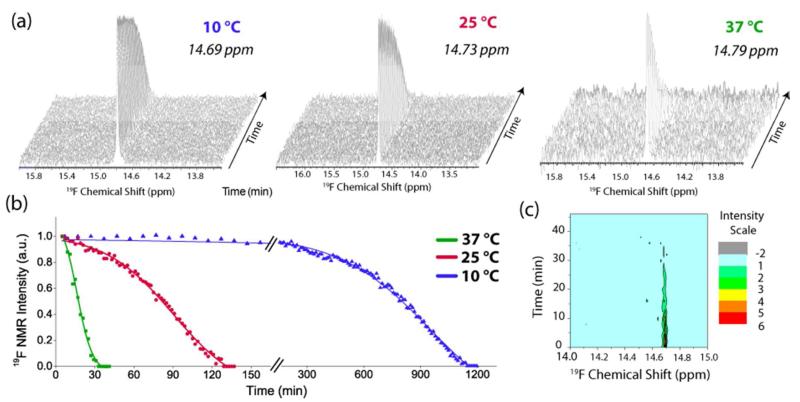
We first checked the sensitivity of the 19F chemical shift of the tfmF23 group to changes in environment (Figure S1) by examining the 19F spectra in different environments. A detectable change was noticed for almost all changes in environment, including a substantial change in the chemical shift in urea, even though IAPP is considered to be natively unfolded.52The consumption of monomeric IAPP-tfmF23 (initial concentration 85 μM in 20 mM sodium phosphate, 50 mM NaCl pH 7.3) during amyloid fibrillogenesis was monitored by 19F NMR (Figure 1). The monomeric peptide exhibits a single sharp peak near 14.7 ppm in the NMR spectrum. As expected, the signal intensity due to the monomeric peptide decreased in intensity over time in the characteristic sigmoidal manner observed in many studies of amyloid fiber formation.55-57 No signal was observed due to the large amyloid fibers as the peaks are broadened beyond detection. To determine the sensitivity of this method, the rate of monomer consumption was detected by measuring changes in 19F NMR signal intensity changes at three different temperatures: 10, 25 and 37 °C (Figure 1). As expected, both the lag time and elongation rate of fibril formation were temperature dependent.14, 15, 58 Even at higher temperatures (37 °C) and accelerated aggregation rates, we were able to monitor monomer consumption during aggregation efficiently using this technique.
Figure 1. Kinetics of the disappearance the IAPP-tfmF23 monomer followed by 19F NMR.
(A) Stacked plots of 19F NMR spectra (TFE = 0.0 ppm). The experiment was conducted at 10 °C (left), 25 °C (center), and 37 °C (right) at pH 7.3 (B) Plots of peak intensity of the main resonance from (A) as a function of time. Solid lines represent sigmoidal fits to the 19F data. (C) Contour plot of the 19F NMR spectra at 37 °C showing the absence of new peaks.
IAPP-tfmF23 forms fibers from monomers without accumulation of non-fibrillar intermediates
No additional peaks were observed in the 19F spectrum of the peptide during the time-course of the experiment, even at 10 °C where fibril formation is slowest (Figure 1). The position of the main resonance also did not change over the course of the experiment (Figure 1c), consistent with fluorescence studies that show Phe-23 remains exposed during the lag-phase of aggregation.19 We also confirmed that tfmF23 is actually sensitive to the formation of small oligomers, were they to be formed in significant concentrations by cross-linking IAPP-tfmF23 with EDC/sulfo-NHS.59 In contrast to IAPP-tfmF23 alone, additional peaks can clearly be seen downfield of the main peak when IAPP-tfmF23 is incubated with 40 mM EDC/sulfo-NHS (Figure S2). The intensity of the additional peaks increase with time as the main peak decreases (Figure S3), confirming its presence is the result of the cross-linking reaction.
It is also important to note that even if an oligomer population is not directly detectable by 19F NMR because it is either structurally inhomogeneous (which would broaden the signal), too large to be detected by NMR, or the IAPP-tfmF23 19F NMR shift is unaffected by oligomerization, it still may be indirectly detected by a comparison of the monomer loss curves observed by 19F NMR and the ThT and CD measurements of amyloid formation. If large oligomers or undetectable oligomers accumulate to a significant extent, a delay between the disappearance of monomer (as measured by NMR) and the appearance of fibrils (as measured by ThT fluorescence and CD) would be observed.21, 60
To test this possibility, we followed the aggregation of similarly prepared samples of IAPP-tfmF23 by ThT fluorescence, CD spectroscopy, and 19F NMR (Figure 2, parameters from the sigmoidal fit can be found in Table S1). The rate of monomer consumption of IAPP-tfmF23 closely matches the rate of amyloid formation when performed under identical buffer and peptide concentrations at 25 °C and 37 °C (Figure 2 and S4). The ThT and NMR curves match well at 37°C (t1/2=18.8 ± 0.8 and 16.8 ± 1.6, respectively, p=0.30) and the CD and NMR curves match very closely at 25°C (t1/2=91.8 ± 6.3 and 91.9 ± 3.1, respectively, p=0.99). A relatively small in this context but statistically significant (p=0.02) difference was observed between the ThT and NMR curves at 25 °C (t1/2=64.3 ± 6.9 and 91.9 ± 3.1, respectively). This small difference in the kinetics between the two measurements may be caused by differences between the shape and size of the NMR tube and fluorescence cuvette, as interactions between sample and container surfaces are known to nucleate amyloid formation.61-63 This difference is magnified at the slower nucleation rates occurring at lower temperatures.61
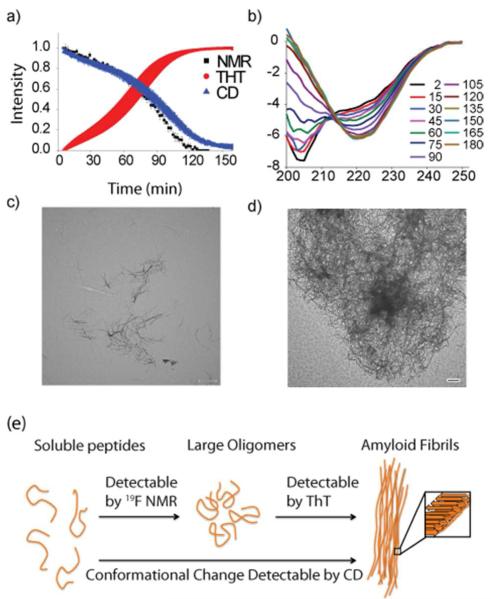
Figure 2. Comparison of the rates of monomer consumption and fiber formation.
(a) Overlay of kinetic traces from obtained 19F NMR (black), ThT fluorescence (red), and CD (blue). Solid lines represent sigmoidal fits to the 19F and CD data. Error bars indicate standard error of measurement (n=4 for NMR and CD and n=5 for ThT). The close correspondence between the curves suggests fiber formation closely follows monomer consumption. (b) CD spectra showing the time evolution of secondary structure. (c, d) TEM images of IAPP-tfmF23 after 1/2 (c) and complete (d) depletion of the 19F signal intensity at 25 °C. Scale bars represent 500 nm. (e) Cartoon showing the sensitivities of each method.
The fact that the NMR, CD, and ThT curves closely track each other suggests the monomeric peptide is converted into a large, mostly β-sheet, ThT-reactive species without the accumulation of non-fibrillar intermediates. The absence of a substantial population of non-fibrillar oligomeric species along the aggregation pathway is notably different from what has been observed for many other amyloidogenic proteins.16, 64, 65 For example, many intermediate species are formed during the aggregation of Aβ, including a distinct α-helical intermediate.11, 16, 64-66 Systems that are known to have non-fibrillar oligomeric intermediates typically show a much larger difference in rates of monomer depletion and fiber formation. For example, the midpoints of the monomer consumption and fiber formation curves of Aβ can differ by a order of magnitude.16 Similarly, monomer consumption and fiber formation curves of CsgA differ by a factor of 2.21 The correspondence between the curves suggests any transient oligomers that are formed by IAPP-tfmF23 comprise only a minor population of the total at any given point in time.
The close coincidence of the midpoints of aggregation determined by the 19F NMR and the ThT and CD measurements impose fairly stringent requirements on the type of non-fibrillar oligomers that can be formed. However, the data does not distinguish between certain kinetic mechanisms. For example, our data cannot exclude the presence of large, off-pathway oligomers whose concentration does not change appreciably in the lag-phase of aggregation.14, 15 In addition, very transient oligomers that do not accumulate to a significant extent will not be observed in the experiment.15
TEM images acquired at the mid- and end-points of aggregation also support this conclusion (Figure 2c, 2d and Figure S5). Images taken at the mid-point of aggregation show fibers that are similar in morphology to those found in at the end-point but with a smaller density of fibers on the grid (Figure 2c and S5a). Non-fibrillar aggregates large enough to be resolved in the TEM image, such as the spherical and annular intermediates frequently detected for α synuclein and Aβ and implicated in toxicity, are noticeably absent.6, 66
19F NMR measurement of the interaction of polyphenolic inhibitors with IAPP without interference from external probes
Having demonstrated the utility of 19F NMR to follow the kinetics of amyloid formation, we used it to investigate the mechanism by which EGCG (Figure 3a) inhibits IAPP-tfmF23 amyloid formation. EGCG is the most abundant catechin found in tea and is a potent antioxidant, making it a potential therapeutic for many disorders. It has been reported that EGCG inhibits fiber formation in vitro for many amyloidogenic proteins including Aβ, α-synuclein, and polyglutamine peptides.67-72 Meng et al. recently showed that EGCG not only inhibits IAPP amyloid formation effectively but is one of the few small molecule compounds that also disaggregates IAPP amyloid fibrils in vitro.69 However, the mechanism of inhibition by EGCG is not known for IAPP.69 Optical measurements of fiber inhibition by EGCG and other polyphenols are complicated by the background signal from these compounds and, potentially, by competitive binding with extrinsic probes like ThT.68, 73-76 In contrast, measurements of the rate of monomer consumption by 19F NMR are easily quantified and are not affected by the nature of the inhibitor.
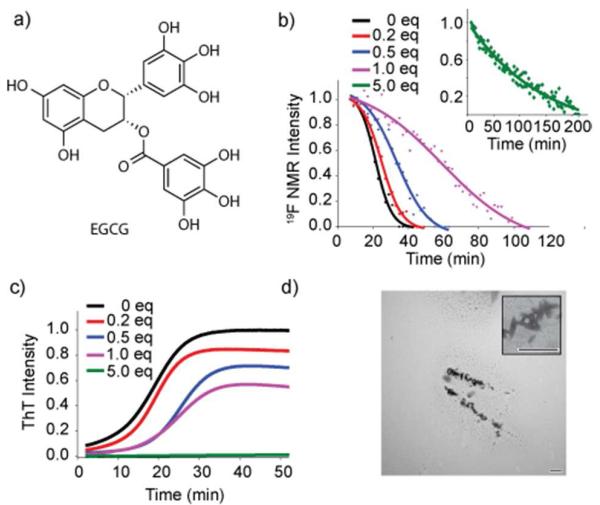
Figure 3. EGCG inhibits IAPP-tfmF23 amyloid fibrillogenesis.
(a) Structure of EGCG. (b, c) Time course for peptide aggregation followed by 19F NMR (b) and ThT fluorescence (c) of IAPP-tfmF23 alone (85 μM, black) and at 1:5 (red) 1:2 (blue), 1:1 (magenta), 5:1 (green) molar ratios of EGCG to IAPP at pH 7.3, 37°C. Solid lines represent sigmoidal fits to the 19F data. 100 μM ThT is present in the ThT samples (c) but is absent in the NMR samples (b). (d) TEM images of a 1:5 mixture of IAPP and EGCG after the complete depletion of the 19F signal intensity. Scale bar represents 500 nm.
The kinetics of IAPP-tfmF23 aggregation were studied in the presence of 0.2, 0.5, 1.0, and 5.0 molar equivalents of EGCG by 19F NMR and ThT fluorescence (Figure 3b and c). It is apparent from both the 19F and ThT measurements that EGCG inhibits fiber formation in a dose-dependent manner. However, a comparison of the data in Figures 3b and 3c reveals some important differences between the ThT and 19F NMR measurements of the apparent effects of EGCG. When IAPP is incubated with an excess of EGCG (green lines in Figures 3b and c), the ThT assay shows a complete inhibition of fiber formation while the 19F NMR measurement shows a slow but steady decrease at longer time-periods. This delay between the disappearance of monomer and the appearance of fibrils is consistent with the production of large non-fibrillar aggregates that are not fluorescent when incubated with ThT.67
This interpretation is supported by the analysis of TEM images taken at the end-points of aggregation, which show the appearance of large non-fibrillar aggregates at higher concentrations of EGCG but with a greatly reduced density of fibers (Figure 3d and S6). However, the TEM images are not definitive by themselves as EGCG by itself polymerizes over time to form aggregates with a similar morphology as the aggregates seen in Figure S6 (Figure S7). 16 While the delay between the disappearance of monomer and the appearance of fibrils is indicative of the formation of large non-fibrillar aggregates, sub-stoichiometric amounts of EGCG shift the 19F NMR curve to later times relative to ThT measurements (blue and pink curves in Figure 3b and 3c, 0.5 and 1 equivalent of EGCG respectively). This result seems to imply that the monomer disappears after the appearance of fibrils at low concentrations of EGCG. We believe that this apparent contradiction is due to interfence by EGCG with ThT binding assay as discussed below.
Thioflavin T and EGCG competitively bind to IAPP fibers
In Figure 3, 100 μM ThT is present during the ThT assay but not in the NMR measurements. To determine more accurately the effect of ThT on EGCG inhibition, we first determined if ThT by itself has any effect on amyloid aggregation. The rate of disappearance of monomer was not significantly changed by the presence of ThT (Figure 4a), indicating that ThT by itself does not affect the kinetics of fiber formation.
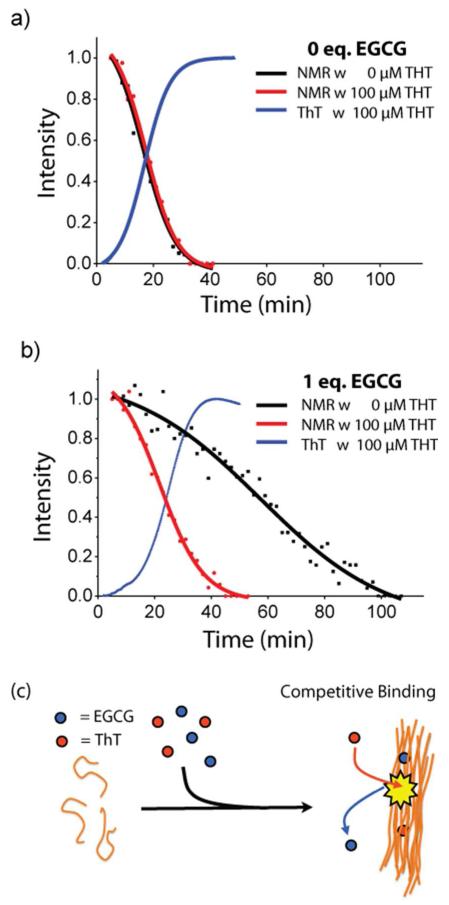
Figure 4. Competition for amyloid binding between ThT and EGCG.
(a) Overlay of kinetic traces from 19F NMR (black: 0 μM ThT and red: 100 μM ThT) and ThT fluorescence (blue: 100 μM ThT) without EGCG. Solid lines represent sigmoidal fits to the 19F data. (b) Kinetic traces obtained under the same conditions but performed in the presence of 1 equivalents of EGCG to peptide. Kinetic traces in the presence of 0.5 equivalents of EGCG can be found in Fig. S8. (c) Cartoon schematic of competitive binding effect.
However, although ThT has little effect on the rate of monomer depletion on its own, it may indirectly influence the rate of aggregation by significantly attenuating the inhibitory effect of EGCG. If ThT competes for the same binding site as EGCG on the amyloid fiber, the decrease in the amount of bound EGCG in the ThT assay would be reflected in lower levels of inhibition. This effect would not be observable when EGCG is in great excess of ThT (green lines Figure 3b and 3c).
To check for this possibility, we performed the 19F measurements using the same concentration of ThT as in the ThT assay (100 μM, Figure 4b and S8). In samples with EGCG, the addition of ThT causes a substantial shift of the NMR curves to earlier times, indicating monomer depletion happens at a faster rate in samples with EGCG when ThT is present (Figure 4b and S8). This shift is not observed in samples without EGCG (Figure 4a). This finding indicates that while ThT has little effect on the rate of aggregation by itself, it significantly attenuates the inhibitory effect of EGCG, consistent with ThT effectively competing with EGCG for binding to IAPP fibrils. Similarly, the observation that high concentrations of EGCG prevent amyloid fiber formation (Figure 3c) may be attributed to effective competition of EGCG for ThT binding sites when it is in sufficient excess over ThT.
EGCG diverts amyloid aggregation to non-fibrillar aggregates
Given the many proposed modes of action of polyphenolic compounds against amyloidogenic proteins, it is difficult to determine the precise mechanism by which EGCG inhibits IAPP amyloid formation. The chemical shift of IAPP-tfmF23 was unchanged by the presence of either EGCG or ThT, indicating neither compound binds to the monomeric peptide, as has been proposed to happen for some amyloidogenic proteins (Figure S9).68, 72, 77, 78
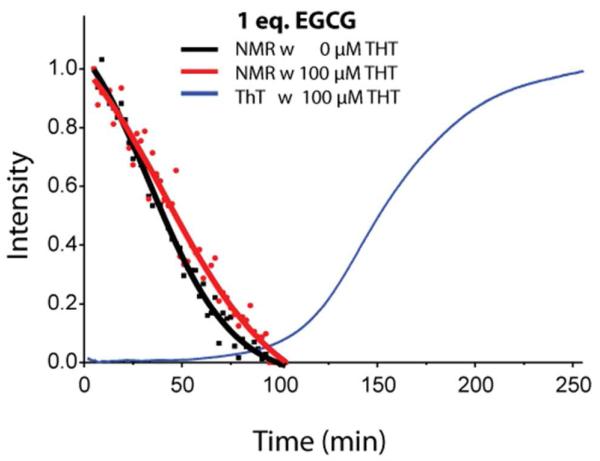
As shown in Figure 4, competition between EGCG and ThT introduces significant artifacts when the ThT concentration is close to the peptide concentration. This is an important observation, because ThT is commonly used at relatively high concentrations in aggregation studies. However, when the concentration of ThT was lowered to 10 μM, competition between ThT and EGCG was lessened and it was possible to obtain accurate results for both the rate of fiber formation and monomer consumption (Figure 5). From the 19F results, it is apparent that the monomer is almost completely consumed in the presence of EGCG during the lag-time before the formation of fibers (Figure 5). This result is striking contrast to the kinetics observed in the absence of EGCG, where fiber formation and monomer consumption occur simultaneously.
Figure 5. Competition between ThT and EGCG is decreased at lower ThT concentrations.
Overlay of kinetic traces from 19F NMR (black: 0 μM ThT and red: 10 μM ThT) and ThT fluorescence (blue: 10μM) with 1eq. of EGCG. Solid lines represent sigmoidal fits to the 19F data.
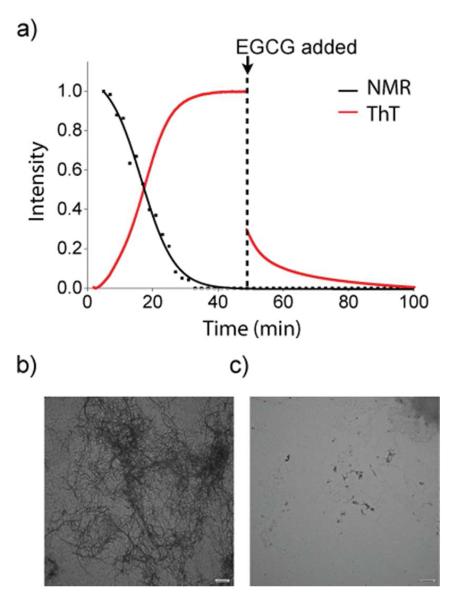
The delay between monomer consumption and fiber formation suggests that EGCG stabilizes the formation of non-fibrillar aggregates that are too large to be detected by NMR.67 To test this possibility, we examined the reverse reaction by testing the ability of EGCG to disaggregate IAPP-tfmF23 amyloid fibrils using 19F NMR and ThT fluorescence. As Figure 6a shows, the addition of a 5-fold molar excess of EGCG after the formation of IAPP amyloid fibrils caused a significant decrease in ThT fluorescence suggestive of the breakup of fibrils. However, no signals due to IAPP-tfmF23 were observed by 19F NMR after the addition of EGCG (Figure 6c), as would be expected if the fibers were completely broken up by EGCG to the monomeric peptide or small oligomers. An examination of the TEM images of supports this conclusion (Figures 6b and S10). TEM images taken at the end-point of aggregation show small aggregates that are much shorter in length (Figures 6b and S6) than the regular fibers formed by IAPP (Figure 2 and S5) that are similar to those found with Aβ and α-synuclein incubated with EGCG. Instead of stabilizing the monomeric state, EGCG apparently diverts aggregation of IAPP towards large oligomers with a different morphology than the amyloid fiber. In this respect, the interaction of EGCG with IAPP is similar to many but not all amyloidogenic proteins.67, 79, 80
Figure 6. Disaggregation of amyloid fibrils due to the addition of EGCG.
(a) Kinetic traces from 19F NMR (black) and ThT fluorescence (red) measurements at 37 °C. Disaggregation of amyloid fibril formation was complete by the addition of 5 equivalents of EGCG at the time point indicated. Solid lines represent sigmoidal fits to the 19F data. (b) TEM image just before the addition of EGCG (c) TEM image after complete depletion of ThT fluorescence intensity (100 minutes). Scale bars represent 500 nm.
Conclusions
These studies demonstrate the utility of using 19F NMR to follow fibrillogenesis in real-time by observing the rate of monomer consumption. Most of the methods typically used for monitoring the kinetics of aggregation measure the rate of appearance of the final product. By providing an additional independent measurement of the starting material, 19F NMR can help distinguish between alternative pathways of aggregation, a task that is difficult to accomplish by other methods. Since fluorine is not found in most biological systems, there is no competition from background signals, a problem that often afflicts measurements using 1H, 13C, and 15N NMR. The absence of a fluorophore simplifies detection and analysis of amyloid formation by other techniques. Therefore, the use of 19F NMR should be applicable for automated screening of amyloid inhibitors without the false negative and positives that might be present with other techniques.
Supplementary Material
Acknowledgments
Funding Information
This work was supported by grants from the National Institutes of Health (GM GM084018 and GM095640to A.R.) and the Department of Defense Multidisciplinary University Research Initiative (DoD 59743-CH-MUR to E.N.G.M.).
Footnotes
The authors declare no competing financial interest.
Abbreviations: Aβ: amyloid-beta, IAPP: islet amyloid polypeptide, ThT : thioflavin T; CD: circular dichroism, EGCG: (-)-epigallocatechin gallate, tfmF: trifluoromethylphenylalanine, Fmoc: fluorenylmethyloxycarbonyl, EDC: 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide, Sulfo-NHS: N-Hydroxysulfosuccinimide, TFE: trifluoroethanol, TEM: transmission electron microscopy
Supporting information
Complete reference 78. Additional figures illustrating the kinetics of amyloid formation, 19F spectra showing the environmental sensitivity of the tfmF23 group, 19F spectra showing the formation of oligomers when incubated with a cross-linking reagent, electron micrographs showing the morphologies of the IAPP aggregates in the presence and absence of EGCG, and a table of parameters derived from the sigmoidal fits are available as supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Luca S, Yau WM, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: Constraints from solid-state NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson R, Eisenberg D. Recent atomic models of amyloid fibril structure. Curr. Opin. Struct. Biol. 2006;16:260–265. doi: 10.1016/j.sbi.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Tycko R. Solid-state NMR studies of amyloid fibril structure. Annu. Rev. Phys. Chem. 2011;62:279–299. doi: 10.1146/annurev-physchem-032210-103539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrison RS, Sharpe PC, Singh Y, Fairlie DP. Amyloid peptides and proteins in review. Rev. Physiol. Biochem. Pharmacol. 2007;159:1–77. doi: 10.1007/112_2007_0701. [DOI] [PubMed] [Google Scholar]
- 6.Ferreira ST, Vieira MNN, De Felice FG. Soluble protein oligomers as emerging toxins in Alzheimer’s and other amyloid diseases. IUBMB Life. 2007;59:332–345. doi: 10.1080/15216540701283882. [DOI] [PubMed] [Google Scholar]
- 7.Zraika S, Hull RL, Verchere CB, Clark A, Potter KJ, Fraser PE, Raleigh DP, Kahn SE. Toxic oligomers and islet beta cell death: Guilty by association or convicted by circumstantial evidence? Diabetologia. 2010;53:1046–1056. doi: 10.1007/s00125-010-1671-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haataja L, Gurlo T, Huang CJ, Butler PC. Islet amyloid in type 2 diabetes, and the toxic oligomer hypothesis. Endocr. Rev. 2008;29:302–316. doi: 10.1210/er.2007-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lansbury PT, Lashuel HA. A century-old debate on protein aggregation and neurodegeneration enters the clinic. Nature. 2006;443:774–779. doi: 10.1038/nature05290. [DOI] [PubMed] [Google Scholar]
- 10.Capone R, Quiroz FG, Prangkio P, Saluja I, Sauer AM, Bautista MR, Turner RS, Yang J, Mayer M. Amyloid-beta-induced ion flux in artificial lipid bilayers and neuronal cells: Resolving a controversy. Neurotox. Res. 2009;16:1–13. doi: 10.1007/s12640-009-9033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chimon S, Shaibat MA, Jones CR, Calero DC, Aizezi B, Ishii Y. Evidence of fibril-like beta-sheet structures in a neurotoxic amyloid intermediate of alzheimer’s beta-amyloid. Nat. Struct. Mol. Biol. 2007;14:1157–1164. doi: 10.1038/nsmb1345. [DOI] [PubMed] [Google Scholar]
- 12.Yoshiike Y, Minai R, Matsuo Y, Chen YR, Kimura T, Takashima A. Amyloid oligomer conformation in a group of natively folded proteins. Plos One. 2008;3:e3235. doi: 10.1371/journal.pone.0003235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masuda M, Hasegawa M, Nonaka T, Oikawa T, Yonetani M, Yamaguchi Y, Kato K, Hisanaga S, Goedert M. Inhibition of alpha-synuclein fibril assembly by small molecules: Analysis using epitope-specific antibodies. FEBS Lett. 2009;583:787–791. doi: 10.1016/j.febslet.2009.01.037. [DOI] [PubMed] [Google Scholar]
- 14.Soong R, Brender JR, Macdonald PM, Ramamoorthy A. Association of highly compact type II diabetes related islet amyloid polypeptide intermediate species at physiological temperature revealed by diffusion NMR spectroscopy. J. Am. Chem. Soc. 2009;131:7079–7085. doi: 10.1021/ja900285z. [DOI] [PubMed] [Google Scholar]
- 15.Vaiana SM, Ghirlando R, Yau WM, Eaton WA, Hofrichter J. Sedimentation studies on human amylin fail to detect low-molecular-weight oligomers. Biophys. J. 2008;94:L45–L47. doi: 10.1529/biophysj.107.125146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, Culyba EK, Powers ET, Kelly JW. Amyloid-beta forms fibrils by nucleated conformational conversion of oligomers. Nat. Chem. Biol. 2011;7:602–609. doi: 10.1038/nchembio.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groenning M. Binding mode of thioflavin T and other molecular probes in the context of amyloid fibrils-current status. J. Chem. Biol. 2009;3:1–18. doi: 10.1007/s12154-009-0027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marek P, Gupta R, Raleigh DP. The fluorescent amino acid p-cyanophenylalanine provides an intrinsic probe of amyloid formation. Chembiochem. 2008;9:1372–1374. doi: 10.1002/cbic.200800052. [DOI] [PubMed] [Google Scholar]
- 19.Marek P, Mukherjee S, Zanni MT, Raleigh DP. Residue-specific, real-time characterization of lag-phase species and fibril growth during amyloid formation: A combined fluorescence and IR study of p-cyanophenylalanine analogs of islet amyloid polypeptide. J. Mol. Biol. 2010;400:878–888. doi: 10.1016/j.jmb.2010.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mishra R, Geyer M, Winter R. NMR spectroscopic investigation of early events in iapp amyloid fibril formation. Chembiochem. 2009;10:1769–1772. doi: 10.1002/cbic.200900237. [DOI] [PubMed] [Google Scholar]
- 21.Horvath I, Weise CF, Andersson EK, Chorell E, Sellstedt M, Bengtsson C, Olofsson A, Hultgren SJ, Chapman M, Wolf-Watz M, Almqvist F, Wittung-Stafshede P. Mechanisms of protein oligomerization: Inhibitor of functional amyloids templates alpha-synuclein fibrillation. J. Am. Chem. Soc. 2012;134:3439–3444. doi: 10.1021/ja209829m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shim SH, Gupta R, Ling YL, Strasfeld DB, Raleigh DP, Zanni MT. Two-dimensional IR spectroscopy and isotope labeling defines the pathway of amyloid formation with residue-specific resolution. Proc. Natl. Acad. Sci. U. S. A. 2009;106:6614–6619. doi: 10.1073/pnas.0805957106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Middleton CT, Singh S, Reddy AS, Woys AM, Strasfeld DB, Marek P, Raleigh DP, de Pablo JJ, Zanni MT, Skinner JL. 2DIR spectroscopy of human amylin fibrils reflects stable beta-sheet structure. J. Am. Chem. Soc. 2011;133:16062–16071. doi: 10.1021/ja204035k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larson JL, Ko E, Miranker AD. Direct measurement of islet amyloid polypeptide fibrillogenesis by mass spectrometry. Protein Sci. 2000;9:427–431. doi: 10.1110/ps.9.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cole HL, Kalapothakis JMD, Bennett G, Barran PE, MacPhee CE. Characterizing early aggregates formed by an amyloidogenic peptide by mass spectrometry. Angew. Chem. Int. Ed. 2010;49:9448–9451. doi: 10.1002/anie.201003373. [DOI] [PubMed] [Google Scholar]
- 26.Maezawa I, Hong HS, Liu R, Wu CY, Cheng RH, Kung MP, Kung HF, Lam KS, Oddo S, LaFerla FM, Jin LW. Congo red and thioflavin-T analogs detect a beta oligomers. J. Neurochem. 2008;104:457–468. doi: 10.1111/j.1471-4159.2007.04972.x. [DOI] [PubMed] [Google Scholar]
- 27.Cobb SL, Murphy CD. 19F NMR applications in chemical biology. J. Fluorine Chem. 2009;130:132–143. [Google Scholar]
- 28.Salwiczek M, Nyakatura EK, Gerling UIM, Ye S, Koksch B. Fluorinated amino acids: Compatibility with native protein structures and effects on protein-protein interactions. Chem. Soc. Rev. 2012;41:2135–2171. doi: 10.1039/c1cs15241f. [DOI] [PubMed] [Google Scholar]
- 29.Bann JG, Pinkner J, Hultgren SJ, Frieden C. Real-time and equilibrium F-19-NMR studies reveal the role of domain-domain interactions in the folding of the chaperone papd. Proc. Natl. Acad. Sci. U.S.A. 2002;99:709–714. doi: 10.1073/pnas.022649599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li HL, Frieden C. Observation of sequential steps in the folding of intestinal fatty acid binding protein using a slow folding mutant and F-19 NMR. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11993–11998. doi: 10.1073/pnas.0705253104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Frieden C. Comparison of C40/82A and P27A C40/82A barstar mutants using F-19 NMR. Biochemistry. 2007;46:4337–4347. doi: 10.1021/bi6026083. [DOI] [PubMed] [Google Scholar]
- 32.Schlesinger AP, Wang YQ, Tadeo X, Millet O, Pielak G. Macromolecular crowding fails to fold a globular protein in cells. J. Am. Chem. Soc. 2011;133:8082–8085. doi: 10.1021/ja201206t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderluh G, Razpotnik A, Podlesek Z, Macek P, Separovic F, Norton RS. Interaction of the eukaryotic pore-forming cytolysin equinatoxin II with model membranes: F-19 NMR studies. J. Mol. Biol. 2005;347:27–39. doi: 10.1016/j.jmb.2004.12.058. [DOI] [PubMed] [Google Scholar]
- 34.Glaser RW, Sachse C, Durr UHN, Wadhwani P, Ulrich AS. Orientation of the antimicrobial peptide PGLa in lipid membranes determined from F-19-NMR dipolar couplings of 4-CF3-phenylglycine labels. J. Magn. Reson. 2004;168:153–163. doi: 10.1016/j.jmr.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Afonin S, Grage SL, Ieronimo M, Wadhwani P, Ulrich AS. Temperature-dependent transmembrane insertion of the amphiphilic peptide PGLa in lipid bilayers; observed by solid state 19F NMR spectroscopy. J. Am. Chem. Soc. 2008;130:16512–16514. doi: 10.1021/ja803156d. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki Y, Buer BC, Al-Hashimi HM, Marsh ENG. Using fluorine nuclear magnetic resonance to probe changes in the structure and dynamics of membrane-active peptides interacting with lipid bilayers. Biochemistry. 2011;50:5979–5987. doi: 10.1021/bi200639c. [DOI] [PubMed] [Google Scholar]
- 37.Buer BC, Chugh J, Al-Hashimi HM, Marsh ENG. Using fluorine nuclear magnetic resonance to probe the interaction of membrane-active peptides with the lipid bilayer. Biochemistry. 2010;49:5760–5765. doi: 10.1021/bi100605e. [DOI] [PubMed] [Google Scholar]
- 38.Li CG, Wang GF, Wang YQ, Creager-Allen R, Lutz EA, Scronce H, Slade KM, Ruf RAS, Mehl RA, Pielak GJ. Protein 19F NMR in escherichia coli. J. Am. Chem. Soc. 2010;132:321–327. doi: 10.1021/ja907966n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson JC, Hammill JT, Mehl RA. Site-specific incorporation of a 19F-amino acid into proteins as an NMR probe for characterizing protein structure and reactivity. J. Am. Chem. Soc. 2007;129:1160–1166. doi: 10.1021/ja064661t. [DOI] [PubMed] [Google Scholar]
- 40.Shi P, Wang H, Xi ZY, Shi CW, Xiong Y, Tian CL. Site-specific 19F NMR chemical shift and side chain relaxation analysis of a membrane protein labeled with an unnatural amino acid. Protein Sci. 2011;20:224–228. doi: 10.1002/pro.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu JJ, Horst R, Katritch V, Stevens RC, Wuthrich K. Biased signaling pathways in β2-adrenergic receptor characterized by 19F-NMR. Science. 2012;335:1106–1110. doi: 10.1126/science.1215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackel C, Salwiczek M, Koksch B. Fluorine in a native protein environment - how the spatial demand and polarity of fluoroalkyl groups affect protein folding. Angew. Chem. Int. Ed. 2006;45:4198–4203. doi: 10.1002/anie.200504387. [DOI] [PubMed] [Google Scholar]
- 43.Salwiczek M, Mikhailiuk PK, Afonin S, Komarov IV, Ulrich AS, Koksch B. Compatibility of the conformationally rigid CF3-Bpg side chain with the hydrophobic coiled-coil interface. Amino Acids. 2010;39:1589–1593. doi: 10.1007/s00726-010-0581-8. [DOI] [PubMed] [Google Scholar]
- 44.Li CG, Lutz EA, Slade KM, Ruf RAS, Wang GF, Pielak GJ. 19F NMR studies of alpha-synuclein conformation and fibrillation. Biochemistry. 2009;48:8578–8584. doi: 10.1021/bi900872p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maisch D, Wadhwani P, Afonin S, Boettcher C, Koksch B, Ulrich AS. Chemical labeling strategy with (r)- and (s)-trifluoromethylaianine for solid state 19F NMR analysis of peptaibols in membranes. J. Am. Chem. Soc. 2009;131:15596. doi: 10.1021/ja9067595. [DOI] [PubMed] [Google Scholar]
- 46.Westermark P, Andersson A, Westermark GT. Islet amyloid polypeptide, islet amyloid, and diabetes mellitus. Physiol. Rev. 2011;91:795–826. doi: 10.1152/physrev.00042.2009. [DOI] [PubMed] [Google Scholar]
- 47.Hoppener JWM, Ahren B, Lips CJM. Islet amyloid and type 2 diabetes mellitus. N. Engl. J. Med. 2000;343:411–419. doi: 10.1056/NEJM200008103430607. [DOI] [PubMed] [Google Scholar]
- 48.Brender JR, Salamekh S, Ramamoorthy A. Membrane disruption and early events in the aggregation of the diabetes related peptide IAPP from a molecular perspective. Acc. Chem. Res. 2012;45:454–462. doi: 10.1021/ar200189b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Page K, Hood CA, Patel H, Fuentes G, Menakuru M, Park JH. Fast Fmoc synthesis of hAmylin(1-37) with pseudoproline assisted on-resin disulfide formation. J. Pept. Sci. 2007;13:833–838. doi: 10.1002/psc.909. [DOI] [PubMed] [Google Scholar]
- 50.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption-coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brender JR, Hartman K, Nanga RP, Popovych N, de la Salud Bea R, Vivekanandan S, Marsh EN, Ramamoorthy A. Role of zinc in human islet amyloid polypeptide aggregation. J. Am. Chem. Soc. 2010;132:8973–8983. doi: 10.1021/ja1007867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yonemoto IT, Kroon GJ, Dyson HJ, Balch WE, Kelly JW. Amylin proprotein processing generates progressively more amyloidogenic peptides that initially sample the helical state. Biochemistry. 2008;47:9900–9910. doi: 10.1021/bi800828u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiltzius JJW, Sievers SA, Sawaya MR, Cascio D, Popov D, Riekel C, Eisenberg D. Atomic structure of the cross-beta spine of islet amyloid polypeptide (amylin) Protein Sci. 2008;17:1467–1474. doi: 10.1110/ps.036509.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiltzius JJ, Sievers SA, Sawaya MR, Eisenberg D. Atomic structures of iapp (amylin) fusions suggest a mechanism for fibrillation and the role of insulin in the process. Protein Sci. 2009;18:1521–1530. doi: 10.1002/pro.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Linse B, Linse S. Monte carlo simulations of protein amyloid formation reveal origin of sigmoidal aggregation kinetics. Mol. Biosyst. 2011;7:2296–2303. doi: 10.1039/c0mb00321b. [DOI] [PubMed] [Google Scholar]
- 56.Kodaka M. Requirements for generating sigmoidal time-course aggregation in nucleation-dependent polymerization model. Biophys. Chem. 2004;107:243–253. doi: 10.1016/j.bpc.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 57.Jarrett JT, Lansbury PT. Seeding one-dimensional crystallization of amyloid - a pathogenic mechanism in Alzheimers-disease and scrapie. Cell. 1993;73:1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 58.Kayed R, Bernhagen J, Greenfield N, Sweimeh K, Brunner H, Voelter W, Kapurniotu A. Conformational transitions of islet amyloid polypeptide (iapp) in amyloid formation in vitro. J. Mol. Biol. 1999;287:781–796. doi: 10.1006/jmbi.1999.2646. [DOI] [PubMed] [Google Scholar]
- 59.Schulz DM, Ihling C, Clore GM, Sinz A. Mapping the topology and determination of a low-resolution three-dimensional structure of the calmodulin-melittin complex by chemical cross-linking and high-resolution fticrms: Direct demonstration of multiple binding modes. Biochemistry. 2004;43:4703–4715. doi: 10.1021/bi036149f. [DOI] [PubMed] [Google Scholar]
- 60.Powers ET, Powers DL. The kinetics of nucleated polymerizations at high concentrations: Amyloid fibril formation near and above the “Supercritical concentration”. Biophys. J. 2006;91:122–132. doi: 10.1529/biophysj.105.073767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giehm L, Otzen DE. Strategies to increase the reproducibility of protein fibrillization in plate reader assays. Anal. Biochem. 2010;400:270–281. doi: 10.1016/j.ab.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 62.Jean L, Lee CF, Lee C, Shaw M, Vaux DJ. Competing discrete interfacial effects are critical for amyloidogenesis. FASEB J. 2010;24:309–317. doi: 10.1096/fj.09-137653. [DOI] [PubMed] [Google Scholar]
- 63.Pronchik J, He XL, Giurleo JT, Talaga DS. In vitro formation of amyloid from alpha-synuclein is dominated by reactions at hydrophobic interfaces. J. Am. Chem. Soc. 2010;132:9797–9803. doi: 10.1021/ja102896h. [DOI] [PubMed] [Google Scholar]
- 64.Kirkitadze MD, Condron MM, Teplow DB. Identification and characterization of key kinetic intermediates in amyloid beta-protein fibrillogenesis. J. Mol. Biol. 2001;312:1103–1119. doi: 10.1006/jmbi.2001.4970. [DOI] [PubMed] [Google Scholar]
- 65.Ding H, Wong PT, Lee EL, Gafni A, Steel DG. Determination of the oligomer size of amyloidogenic protein beta-amyloid(1-40) by single-molecule spectroscopy. Biophys. J. 2009;97:912–921. doi: 10.1016/j.bpj.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, Morgan TE, Rozovsky I, Trommer B, Viola KL, Wals P, Zhang C, Finch CE, Krafft GA, Klein WL. Diffusible, nonfibrillar ligands derived from Aβ(1-42) are potent central nervous system neurotoxins. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ehrnhoefer DE, Bieschke J, Boeddrich A, Herbst M, Masino L, Lurz R, Engemann S, Pastore A, Wanker EE. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat. Struct. Mol. Biol. 2008;15:558–566. doi: 10.1038/nsmb.1437. [DOI] [PubMed] [Google Scholar]
- 68.Popovych N, Brender JR, Soong R, Vivekanandan S, Hartman K, Basrur V, Macdonald PM, Ramamoorthy A. Site specific interaction of the polyphenol egcg with the SEVI amyloid precursor peptide PAP(248-286) J. Phys. Chem. B. 116:3650–3658. doi: 10.1021/jp2121577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meng FL, Abedini A, Plesner A, Verchere CB, Raleigh DP. The flavanol (−)-epigallocatechin 3-gallate inhibits amyloid formation by islet amyloid polypeptide, disaggregates amyloid fibrils, and protects cultured cells against iapp-induced toxicity. Biochemistry. 2010;49:8127–8133. doi: 10.1021/bi100939a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ferreira N, Cardoso I, Domingues MR, Vitorino R, Bastos M, Bai GY, Saraiva MJ, Almeida MR. Binding of epigallocatechin-3-gallate to transthyretin modulates its amyloidogenicity. FEBS Lett. 2009;583:3569–3576. doi: 10.1016/j.febslet.2009.10.062. [DOI] [PubMed] [Google Scholar]
- 71.Hauber I, Hohenberg H, Holstermann B, Hunstein W, Hauber J. The main green tea polyphenol epigallocatechin-3-gallate counteracts semen-mediated enhancement of HIV infection. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9033–9038. doi: 10.1073/pnas.0811827106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang R, Vivekanandan S, Brender JR, Abe Y, Naito A, Ramamoorthy A. NMR characterization of monomeric and oligomeric conformations of human calcitonin and its interaction with EGCG. J. Mol. Biol. 2012;416:108–120. doi: 10.1016/j.jmb.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hudson SA, Ecroyd H, Kee TW, Carver JA. The thioflavin T fluorescence assay for amyloid fibril detection can be biased by the presence of exogenous compounds. FEBS J. 2009;276:5960–5972. doi: 10.1111/j.1742-4658.2009.07307.x. [DOI] [PubMed] [Google Scholar]
- 74.Chandrashekaran IR, Adda CG, MacRaild CA, Anders RF, Norton RS. EGCG disaggregates amyloid-like fibrils formed by plasmodium falciparum merozoite surface protein 2. Arch. Biochem. Biophys. 2011;513:153–157. doi: 10.1016/j.abb.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu KN, Wang HY, Chen CY, Wang SSS. L-arginine reduces thioflavin T fluorescence but not fibrillation of bovine serum albumin. Amino Acids. 2010;39:821–829. doi: 10.1007/s00726-010-0536-0. [DOI] [PubMed] [Google Scholar]
- 76.Meng F, Marek P, Potter KJ, Verchere CB, Raleigh DP. Rifampicin does not prevent amyloid fibril formation by human islet amyloid polypeptide but does inhibit fibril thioflavin-T interactions: Implications for mechanistic studies of beta-cell death. Biochemistry. 2008;47:6016–6024. doi: 10.1021/bi702518m. [DOI] [PubMed] [Google Scholar]
- 77.Hudson SA, Ecroyd H, Dehle FC, Musgrave IF, Carver JA. (−)-epigallocatechin-3-gallate (egcg) maintains kappa-casein in its pre-fibrillar state without redirecting its aggregation pathway. J. Mol. Biol. 2009;392:689–700. doi: 10.1016/j.jmb.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 78.Miyata, et al. The crystal structure of the green tea polyphenol (−)-epigallocatechin gallate-transthyretin complex reveals a novel binding site distinct from the thyroxine binding site. Biochemistry. 2010;49:6104–6114. doi: 10.1021/bi1004409. [DOI] [PubMed] [Google Scholar]
- 79.Bieschke J, Russ J, Friedrich RP, Ehrnhoefer DE, Wobst H, Neugebauer K, Wanker EE. EGCG remodels mature alpha-synuclein and amyloid-beta fibrils and reduces cellular toxicity. Proc. Natl. Acad. Sci. U. S. A. 2010;107:7710–7715. doi: 10.1073/pnas.0910723107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.He J, Xing YF, Huang B, Zhang YZ, Zeng CM. Tea catechins induce the conversion of preformed lysozyme amyloid fibrils to amorphous aggregates. J. Agric. Food Chem. 2009;57:11391–11396. doi: 10.1021/jf902664f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.