Abstract
In this paper, we have developed an interesting imaging method for intracellular ATP molecules with semi-quantitation. While there has been a lot of work in understanding intracellular events, very few can come close to quantitation or semi-quantitation in living cells. In this work, we made an effective use of nanomaterials, graphene oxides, both as a quencher and a carrier for intracellular delivery. In addition, this graphene oxide also serves as the carrier for reference probes for fluorescent imaging. An ATP aptamer molecular beacon (AAMB) is adsorbed on graphene oxide (GO) to form a double quenching platform. The AAMB/GO spontaneously enters cells, and then AAMB is released and opened by intracellular ATP. The resulting fluorescence recovery is used to perform ATP live-cell imaging with greatly improved background and signaling. Moreover, a control ssDNA, which is released non-specifically from GO by non-target cellular proteins, can serve as an internal reference for ATP semi-quantification inside living cells using the intensity ratio of the AAMB and control. This approach can serve as a way for intracellular delivery and quantitative analysis.
Keywords: nonspecific desorption, graphene oxide, aptamer molecular beacon, internal reference, ATP, semi-quantification
Graphene oxide (GO) has been attracting considerable attention in recent years. Due to its rich chemical, optical and mechanical properties, GO has been widely used for sensitive and selective detection of various biomolecules, including small molecules1-3, nucleic acids1-6, and proteins1, 2, 7, both in solution and in living cells8, 9. These methods utilized two important properties of GO. First, GO can strongly adsorb single-stranded DNA (ssDNA) via a π-stacking interaction between the ring structures in the nucleobases and the hexagonal cells of GO10. Second, GO is a super-quencher to a wide range of fluorophores via fluorescence resonance energy transfer or non-radiative dipole–dipole coupling. Therefore, ssDNA labeled with fluorophore can be adsorbed and quenched by GO. Upon addition of cDNA or a specific molecular target, ssDNA will be desorbed from GO and the fluorescence signal will be recovered. This is the basic mechanism for detecting biomolecules by the DNA aptamer/GO system1.
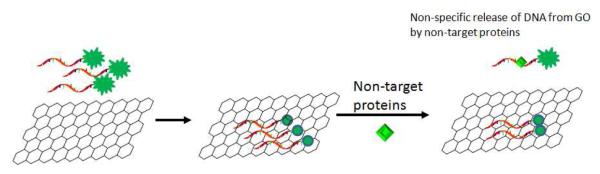
Although this specific desorption of ssDNA from GO by target molecules has been well accepted, the nonspecific desorption of ssDNA from GO by non-target proteins has not been reported. In one of our previous studies7, we discovered a very interesting phenomenon. When the DNA aptamer/GO system was used to detect target insulin in solution, the non-specific signals caused by other non target proteins were relatively high when the concentrations of these proteins exceeded 5 μM. In another paper, Lu et al. reported that when the concentrations of non-target proteins were only 100 nM, the nonspecific signals were low1. We reasoned that nonspecific desorption of ssDNA from GO is concentration dependent; when the concentration of non-target proteins exceeds a certain level, nonspecific desorption increases significantly, resulting in a strong false positive signal (Scheme 1). To confirm this hypothesis, we attempted to use different concentrations of non-target proteins to test the nonspecific desorption of ssDNA from the DNA/GO system.
Scheme 1.
Nonspecific desorption of ssDNA from GO by non-target proteins. Fluorescently labelled ssDNA molecules are adsorbed on GO to form a quenching platform. In the presence of non-target proteins, some of these ssDNAs are released nonspecifically from GO and give false positive signals.
A random 40-mer ssDNA library was generated with a FAM group label on the 5′-end. As shown in Figure 1, upon excitation at 480 nm, the library (100 nM) gave a strong FAM emission at 520 nm. Addition of GO brought the fluorescence to the baseline level. However, when non-target protein, bovine serum albumin (BSA), was added to the reaction solution, the fluorescence was restored, and a significant enhancement occurred when BSA reached 50 μg/mL (750 nM). Furthermore, when the concentration of BAS exceeded 1 mg/mL (15 μM), the enhanced fluorescence reached the maximum value. In addition, we tested the nonspecific desorption assay with a protein mixture, fetal bovine serum (FBS). As shown in Figure S2, the enhancement by FBS was comparable to that of BSA. A significant fluorescence signal was observed with FBS at 100 μg/mL and it approached a maximum value when FBS was 2 mg/mL.
Figure 1.
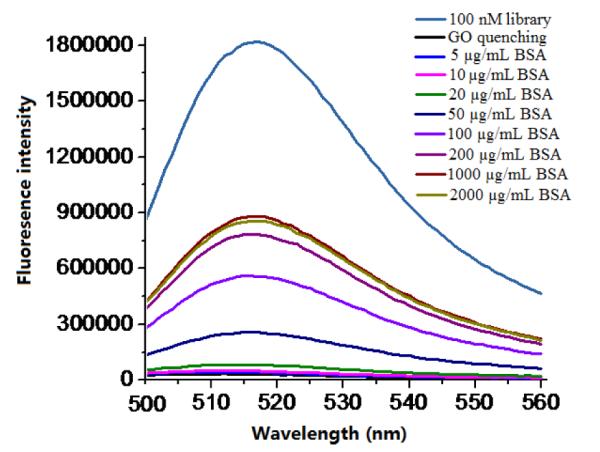
Fluorescence spectra of FAM-labeled ssDNA library only and fluorescence spectra of FAM-labeled ssDNA library/GO complex in the presence of different concentrations of BSA. Excitation: 480 nm.
These data indicated that ssDNAs can be non-specifically desorbed from GO by non-target proteins, especially when they are at relatively high concentrations, resulting in high false positive signals. On the basis of these results, we concluded that this nonspecific effect cannot be ignored when a DNA/GO system is used in a matrix containing high concentrations of non-target proteins, especially inside living cells. Consequently, it is necessary to consider how to reduce this false positive signal when a DNA/GO system is used for intracellular studies. To address this issue, we designed a model study in which an ATP aptamer/GO system was used for in situ ATP detection.
As the primary energy currency, ATP plays important roles in cell signaling and many cellular reactions. Because of its unique importance, many approaches have been developed to detect ATP in solution, including HPLC11, aptamer-based1, enzyme based12, 13 and protein-based methods14. For in situ detection, Wang et al developed an ATP aptamer/GO platform to detect ATP in living cells9. They cultured ATP aptamer/GO with cells in medium containing 10% FBS. When ATP aptamer/GO entered cells, cellular ATP bound to ATP aptamers and released them from the surface of GO. Although their results showed a very clean background, the signal from the sample was rather weak, and therefore the difference between sample and control was small. Because ATP molecules in living cells are usually at very high concentrations, typically 1-10 mM15, assuming most of ATP aptamers were delivered into cells by GO and released from GO by intercellular ATP molecules, the recovered fluorescence signals should be strong. It is questionable why the positive signal was so weak. From our previous nonspecific desorption data we postulated that when aptamer/GO is cultured with cells in the presence of 10% FBS, some of the aptamers would be desorbed from GO by FBS, and only the remaining aptamers could be delivered into cells. As a result, the actual concentration of ATP aptamers on GO inside the cells would be decreased, resulting in a weak positive signal. To confirm this, the ATP aptamer/GO system was cultured with HeLa cells in two different media: one contained 10 % FBS and the other was serum-free. As shown in Figure S3, in the presence of FBS, the fluorescence signal was very weak, while in the absence of FBS, the fluorescence signal was strong. These data confirmed our postulate that some of the ATP aptamers were non-specifically desorbed from the GO by FBS before entering the cells. This result also indicates that when using GO to deliver nucleic acids into cells, a serum-free medium is preferable to a medium containing serum. However, even though a serum-free medium would allow most of the aptamer/GO to be delivered into the cells, the non-specific release of ATP aptamers from GO by intracellular proteins would still occur, leading to false positive signals, as described above. To overcome this problem, we modified the ATP aptamer into an ATP aptamer molecular beacon (AAMB).
An aptamer molecular beacon, AMB, also called an aptamer switch probe16 or an activatable aptamer probe17, is a newly developed molecular beacon which can specifically recognize target molecules, such as ATP16, proteins16 or even cells17. This type of MB is usually composed of three elements (in addition to the fluorophore and quencher on the ends): an aptamer, a short DNA sequence complementary to part of the aptamer, and a linker (e.g., PEG) connecting the former two. Upon binding to their targets, AMBs undergo spontaneous structural reorganization, which opens the hairpin stem, leading to fluorescence recovery. We designed the AAMB based on one of our previous publications16, in which TAMRA was chosen as the fluorophore and Dabcyl was used as the quencher. The activity of AAMB to target ATP was first tested. As shown in Figure S4A, significant fluorescence enhancement was observed after the AAMB was mixed with increasing concentrations of ATP in the reaction buffer. However, there was no fluorescence enhancement when control MB (CMB) was used (Figure S4B), indicating that CMB cannot bind to ATP.
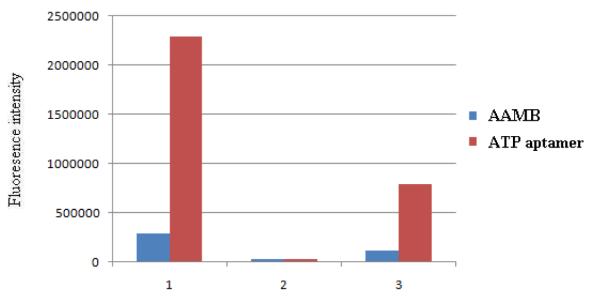
Before we used the AAMB to replace ATP aptamer in GO-mediated cellular delivery, we first tested the non-specific desorption caused by FBS on the ATP aptamer/GO system and the AAMB/GO system in solution. As shown in Figure 2, 100 nM ATP aptamer produced much higher fluorescence than 100 nM AAMB when not adsorbed on GO. When GO was present, the fluorescence from both ATP aptamer and AAMB were severely quenched. But when FBS (1.6 mg/mL) was added, ATP aptamer/GO produced a significant false positive signal, while AAMB/GO did not. This experiment indicated that, although these DNAs can be non-specifically released from GO by non-target proteins, the self-quenching ability of the AAMB results in a much smaller false positive signal.
Figure 2.
Comparison of fluorescence intensities of AAMB and ATP aptamer under different conditions. 1) 100 nM AAMB or ATP aptamer in buffer; 2) AAMB or ATP aptamer treated with1μL of GO (2mg/mL); 3) AAMB/GO or ATP aptamer/GO treated with FBS (1.6mg/mL).
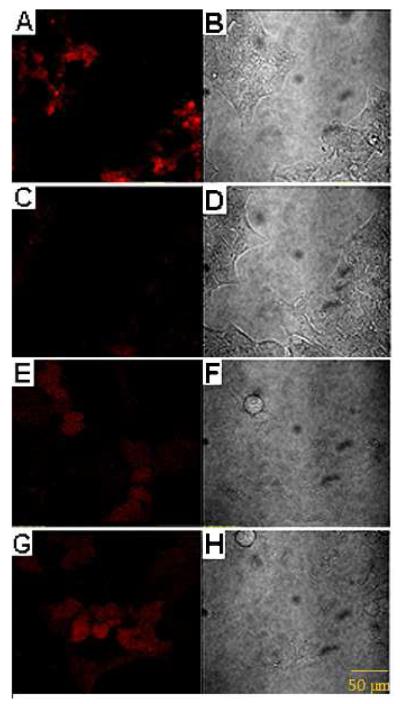
Next, we studied in situ ATP imaging by delivering AAMB or ATP aptamer with their corresponding control DNAs into cells by GO. Figure 3A shows the confocal microscopy images of HeLa cells after incubation with AAMB/GO for 2 h, followed by further culture for another 8 h. Significant TAMRA fluorescence was observed. As a control, HeLa cells treated with CMB/GO under the same conditions showed only very weak fluorescence (Figure 3C). By comparison, the background signal of the control aptamer was visible (Figure 3E), and the difference in fluorescence intensity between the ATP aptamer and the control aptamer was small (Figure 3E, G). These data clearly demonstrated that the AAMB/GO system is much better than the ATP aptamer/GO system for in situ ATP imaging because of: 1) much less false positive signal; and 2) clean background.
Figure 3.
Intracellular imaging of ATP. Confocal microscopy of HeLa cells treated with ATPMB/GO (A, B), CMB/GO (C, D), control ssDNA/GO (E, F), ATP aptamer/GO (G, H). Bright-field images are on the right, and fluorescence images are on the left.
The GO-mediated delivery conditions were further optimized by treating AAMB with different amounts of GO. As shown in Figure S5, 2.5 μg/mL GO is sufficient to deliver 200 nM AAMB into HeLa cells. When the GO concentration was increased to 10 μg/mL, the TAMRA signals decreased significantly (data not shown) due to the following two reasons: 1) the concentration of AAMB complexed per GO particle decreases as the number of GO particles increases; and 2) not all GO particles enter the cell. Thus, the number of GO-delivered AAMBs actually decreases in the cell at 10 μg/mL and above.
The incubation time was also optimized. After incubation with the AAMB/GO complex for 0.5, 1, 2 or 3 h, respectively, HeLa cells were washed and further cultured for 8 h. The 2 h incubation time was found to be sufficient for AAMB/GO complex delivery into cells (data not shown). Moreover, for HeLa cells treated with AAMB/GO, more intense fluorescence was observed when the post-treatment incubation period (after removal of excess AAMB/GO) was increased. As shown in Figure S6, the fluorescence signal from the 8 h incubation group is much stronger than that from the 2 h group.
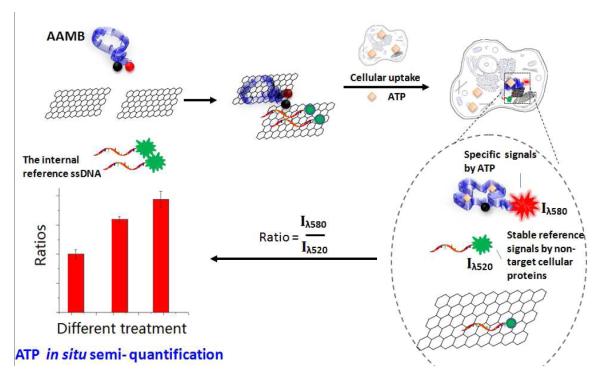
Furthermore, based on the finding of this non-specific desorption, we designed an assay for in situ ATP semi-quantification. As discussed above, the control aptamer/GO system gave a nonspecific signal inside living cells caused by non-target cellular proteins (Fig. 3E). This signal should not be altered by different ATP concentrations inside living cells. Therefore, the control aptamer can be used as an internal reference with AAMB as the probe for in situ ATP semi-quantification. The mechanism is illustrated in Scheme 2. An internal reference was prepared by replacing TAMRA on the control aptamer with a FAM group (See Supporting Information for details.) Then internal reference/GO was mixed with AAMB/GO or CMB/GO for cellular delivery. As shown in Figure S7, internal reference/GO gave almost the same FAM signal in different groups of HeLa cells (Fig. S7B and E, λem = 520nm), as we have assumed before. On the other hand, AAMB gave a much stronger signal compared to CMB (Fig. S7 A and D, λem = 580nm). These data demonstrated that, because of the similar cellular makeup, internal reference/GO would give stable reference signals at 520 nm and AAMB/GO would give signals specific to ATP at 580 nm. Based on this design, we then employed this AAMB-internal reference/GO platform to detect different ATP levels in three groups of HeLa cells treated as follows: medium only, medium + 5 mM Ca2+, medium + 0.1 mM etoposide.
Scheme 2.
Advanced ATP imaging probe design. An ATP aptamer molecular beacon (AAMB) is adsorbed on GO to form a double quenching platform. After the AAMB/GO complex spontaneously enters cells, the AAMB is released and then opened by intracellular ATP. The resulting fluorescence recovery (λem = 580nm) is used to perform ATP live-cell imaging. Moreover, in the presence of an internal reference, i.e., FAM-labeled ssDNA (λem = 520nm), which is released nonspecifically from GO when inside cells, this system can also be used for ATP semi-quantification inside living cells.
We used the advanced imaging probe design to measure intracellular ATP changes upon drug stimulation. Calcium ion and etoposide are both known to induce intracellular ATP concentration enhancement18, 18, 19. As shown in Figure 4, the internal reference signals (Fig. 4 B, E, H) were very similar in the three groups of HeLa cells, but AAMB (Fig. 4 A, D, G) showed different fluorescence signals after Ca2+ or etoposide treatment. The imaging data were collected in triplicate for each group and software Image J was used to measure the signal intensity. The averaged TAMRA signal intensity was divided by the averaged FAM signal intensity to get the quotient value for each group. For the control group, the quotient value was 1.419; for the Ca2+-treated group, it was 2.313; and for the etoposide-treated group, it was 2.753. As shown in Figure 5, semi-quantification of ATP concentration in situ was achieved. If the ATP concentration in the untreated HeLa cells is defined to be 100% (corresponding to an intensity ratio of 1.419), the ATP level in the cells treated with Ca2+ increased to 163% (2.313/1.419), which is in good agreement with the results of others19, and cells treated with etoposide increased to 194% (2.753/1.419). These data clearly demonstrate that the AAMB-internal reference/GO system is a viable tool to perform ATP semi-quantification in living cells.
Figure 4.
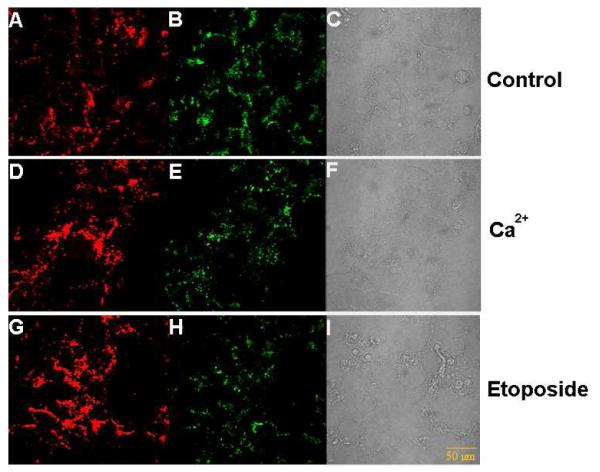
ATP imaging with drug stimulation. Confocal microscopy of HeLa cells incubated with AAMB- internal reference/GO after treatment for 2 h with medium only (top), medium + 5 mM Ca2+ (middle) and medium + 0.1mM etoposide (bottom). A, D, G) fluorescence images of AAMB; B, E, H) fluorescence images of internal reference; C, F, I) bright field images. For FAM, λexc = 480nm, λem = 520nm; for TAMRA, λexc = 565nm, λem = 580nm.
Figure 5.
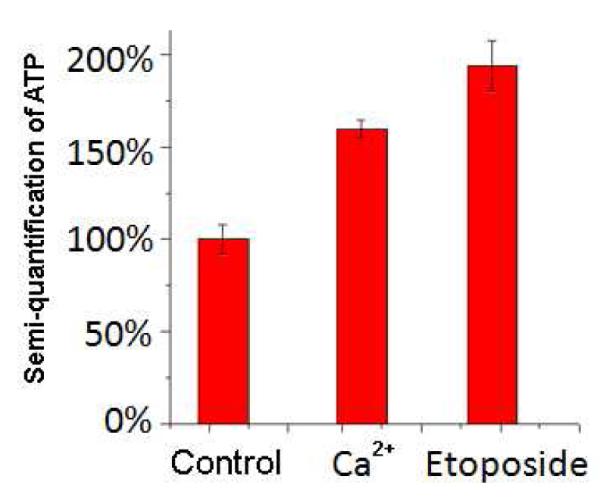
Intracellular imaging quantification. Semi-quantification of ATP in living HeLa cells after treatment with 5 mM Ca2+ or 0.1 mM etoposide, as described in the Figure 3 caption.
Our study offers several advantages. After the discovery of non-specific DNA desorption from the DNA/GO system, we utilized the AAMB/GO, instead of ATP aptamer/GO, for cellular delivery and greatly reduced background for highly sensitive in situ ATP imaging. Moreover, using the non-specific desorption as the internal reference, this platform can be used for a semi-quantitative assay for intracellular ATP imaging, which is currently a challenging task. Finally, and more importantly, our Aptamer MB/GO system has the potential to detect other biomolecules inside living cells, especially proteins having known aptamers. We plan to extend the excellent properties demonstrated in this platform to perform in situ imaging of proteins in living cells. Taken together, we believe our study can serve as a basis for further design and optimization of GO-mediated target detection and DNA delivery.
Experimental section
1. DNAs
AAMB: 5′-TAMRA CACCTGGGGGAGTATTGCGGAGGAAGGTT-PEG6-CCAGGTG-Dabcyl-3′
CMB: 5′-Dabcyl-GCGAGACCGCCGCATTTGATCGATACTCGC-TAMRA-3′
ATP aptamer: 5′-CACCTGGGGGAGTATTGCGGAGGAAGGTT-TAMRA-3′
Control aptamer: 5′-GCGAGACCGCCGCATTTGATCGATA-TAMRA-3′
Internal reference ssDNA: 5′-GCGAGACCGCCGCATTTGATCGATA-FAM-3′
ATP aptamer is indicated by underlining, and the stem part for each MB is indicated in bold
2. Preparation of graphene oxide
Generally, graphite powder (2 g, 325 mesh) was reacted with 12 mL concentrated H2SO4, 3.0 g K2S2O8 and 3.0 g P2O5 at 8° C for 4.5 h with stirring. After cooling to RT, it was diluted with 0.5 L DI water and left overnight. Then the mixture was centrifuged and washed to remove the residual acid and dried overnight under ambient conditions. This pre oxidized graphite was added into 120 mL concentrated H2SO4 (0°C), and 15 g KMnO4 was gradually added with stirring in an ice bath. The reaction was permitted at 35°C for 2 h and then diluted with 250 mL DI water in an ice bath to keep the temperature below 50°C. The mixture was stirred for 2 h and another 0.7 L DI water was added. Afterwards, 20 mL 30% H2O2 was added and a brilliant yellow color was observed along with bubbling. Washed by 1 L 1:10 HCl aqueous solution and by 1 L DI water, the resulting solid was dried in air and diluted to make a GO dispersion (0.5% w/w). Finally, remaining impurities were removed by dialysis in DI water for one week. As shown in Figure S1, the Go is a 2D sheet like material consists of densely packed sp2 hybridized carbon atom network. They have “500 nm - submicrometer” sizes and there are no apparent aggregations.
3. Fluorescence response of AAMB and CMB to ATP in buffer
All fluorescence measurements were performed using a Fluorolog spectrofluorometer (Jobin Yvon Horiba). The MB samples were prepared in 10 mM Tris-HCl buffer containing 6 mM MgCl2. The AAMB and CMB concentrations were both 0.1 μM. ATP concentrations ranged from 0.4 mM to 2.6 mM in the titration experiment. The fluorescence spectra for all samples were measured at 20° C.
4. Live-cell imaging of ATP using GO to deliver DNAs into cells
Generally, 200 nM DNAs were incubated with 2.5 μg/mL GO in 1 mL DMEM medium for 5 min. Then the solution was incubated with HeLa cells for 2 h. Cells were washed with PBS and cultured for another 8 h in fresh DMEM medium, followed by confocal microscopy.
5. In situ semi-quantification of ATP
Generally, 200 nM MBs, or 200 nM internal reference, were incubated with 2.5 μg/mL GO in 1 mL DMEM medium for 5 min, respectively. After combining, the mixture was incubated with HeLa cells for 2 h. Cells were washed with PBS and cultured for another 8 h in fresh DMEM medium. Cells were then detected by confocal microscopy. For Ca2+ or etoposide treatment, the further culture time was 2 h.
Supplementary Material
Acknowledgement
This work was supported by the National Key Scientific Program o f China (2011CB911001, 2011CB911003) and China National Instrumentation Program 2011YQ03012412. This work was also supported by grants awarded by the US National Institutes of Health (GM079359, and CA133086).
Footnotes
Supporting Information: Additional figures are available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- (1).Lu CH, Yang HH, Zhu CL, Chen X, Chen GN. Angew. Chem. Int. Ed. Engl. 2009;48:4785–4787. doi: 10.1002/anie.200901479. [DOI] [PubMed] [Google Scholar]
- (2).Chang H, Tang L, Wang Y, Jiang J, Li J. Anal. Chem. 2010;82:2341–2346. doi: 10.1021/ac9025384. [DOI] [PubMed] [Google Scholar]
- (3).Li F, Huang Y, Yang Q, Zhong Z, Li D, Wang L, Song S, Fan C. Nanoscale. 2010;2:1021–1026. doi: 10.1039/b9nr00401g. [DOI] [PubMed] [Google Scholar]
- (4).Zhou M, Zhai Y, Dong S. Anal. Chem. 2009;81:5603–5613. doi: 10.1021/ac900136z. [DOI] [PubMed] [Google Scholar]
- (5).Guo Y, Deng L, Li J, Guo S, Wang E, Dong S. ACS Nano. 2011;5:1282–1290. doi: 10.1021/nn1029586. [DOI] [PubMed] [Google Scholar]
- (6).Wu W, Hu H, Li F, Wang L, Gao J, Lu J, Fan C. Chem. Commun. (Camb) 2011;47:1201–1203. doi: 10.1039/c0cc04312e. [DOI] [PubMed] [Google Scholar]
- (7).Pu Y, Zhu Z, Han D, Liu H, Liu J, Liao J, Zhang K, Tan W. Analyst. 2011;136:4138–4140. doi: 10.1039/c1an15407a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Lu CH, Zhu CL, Li J, Liu JJ, Chen X, Yang HH. Chem. Commun. (Camb) 2010;46:3116–3118. doi: 10.1039/b926893f. [DOI] [PubMed] [Google Scholar]
- (9).Wang Y, Li Z, Hu D, Lin CT, Li J, Lin Y. J. Am. Chem. Soc. 2010;132:9274–9276. doi: 10.1021/ja103169v. [DOI] [PubMed] [Google Scholar]
- (10).He S, Song B, Li D, Zhu C, Qi W, Wen Y, Wang L, Song S, Fang H, Fan C. Adv. Funct. Mater. 2010;20:453–459. [Google Scholar]
- (11).Viarengo A, Secondini A, Scoppa P, Orunesu M. Experientia. 1986;42:1234–1235. doi: 10.1007/BF01946400. [DOI] [PubMed] [Google Scholar]
- (12).Hara KY, Mori H. J. Biomol. Screen. 2006;11:310–317. doi: 10.1177/1087057105285112. [DOI] [PubMed] [Google Scholar]
- (13).Richard P, Teusink B, Hemker MB, Van Dam K, Westerhoff HV. Yeast. 1996;12:731–740. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C731::AID-YEA961%3E3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- (14).Berg J, Hung YP, Yellen G. Nat. Methods. 2009;6:161–166. doi: 10.1038/nmeth.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Beis I, Newsholme EA. Biochem. J. 1975;152:23–32. doi: 10.1042/bj1520023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Tang Z, Mallikaratchy P, Yang R, Kim Y, Zhu Z, Wang H, Tan W. J. Am. Chem. Soc. 2008;130:11268–11269. doi: 10.1021/ja804119s. [DOI] [PubMed] [Google Scholar]
- (17).Shi H, He X, Wang K, Wu X, Ye X, Guo Q, Tan W, Qing Z, Yang X, Zhou B. Proc. Natl. Acad. Sci. USA. 2011;108:3900–3905. doi: 10.1073/pnas.1016197108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Ainscow EK, Rutter GA. Diabetes. 2002;51(Suppl. 1):S162–170. doi: 10.2337/diabetes.51.2007.s162. [DOI] [PubMed] [Google Scholar]
- (19).Zamaraeva MV, Sabirov RZ, Maeno E, Ando-Akatsuka Y, Bessonova SV, Okada Y. Cell Death Differ. 2005;12:1390–1397. doi: 10.1038/sj.cdd.4401661. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.