Abstract
Background
The spread of MRSA strains at hospitals as well as in the community are of great concern worldwide. We characterized the MRSA clones isolated at Tunisian hospitals and in the community by comparing them to those isolated in other countries.
Results
We characterized 69 MRSA strains isolated from two Tunisian university hospitals between the years 2004-2008. Twenty-two of 28 (79%) community-associated MRSA (CA-MRSA) strains and 21 of 41 (51%) healthcare-associated MRSA (HA-MRSA) strains were PVL-positive. The PVL-positive strains belonged to predicted founder group (FG) 80 in MLST and carried either type IVc SCCmec or nontypeable SCCmec that harbours the class B mec gene complex. In contrast, very diverse clones were identified in PVL-negative strains: three FGs (5, 15, and 22) for HA-MRSA strains and four FGs (5, 15, 45, and 80) for CA-MRSA strains; and these strains carried the SCCmec element of either type I, III, IVc or was nontypeable. The nucleotide sequencing of phi7401PVL lysogenized in a CA-MRSA strain JCSC7401, revealed that the phage was highly homologous to phiSA2mw, with nucleotide identities of more than 95%. Furthermore, all PVL positive strains were found to carry the same PVL phage, since these strains were positive in two PCR studies, identifying gene linkage between lukS and mtp (major tail protein) and the lysogeny region, both of which are in common with phi7401PVL and phiSa2mw.
Conclusions
Our experiments suggest that FG80 S. aureus strains have changed to be more virulent by acquiring phi7401PVL, and to be resistant to β-lactams by acquiring SCCmec elements. These novel clones might have disseminated in the Tunisian community as well as at the Tunisian hospitals by taking over existing MRSA clones.
Background
The spread of antibiotic resistance among Staphylococcus aureus strains is of great concern in the treatment of Staphylococcal infections. Since the first Methicillin Resistant Staphylococcus aureus (MRSA) strain was reported in England in 1961 [1], MRSA has become one of the most prevalent pathogens that cause nosocomial infections throughout the world. Recent reports suggest that it has become increasingly prevalent in the community as well since the 1990s [2-5]. In the 2000s, outbreaks of community-associated MRSA (CA-MRSA) strains were observed worldwide as causative agents of community-associated infections, e.g., superficial skin and soft tissue infections, urinary tract infections and pneumonia [6-9].
Methicillin resistance in MRSA is encoded by the mecA gene, which is carried by the SCCmec element, a mobile genetic element that carries methicillin resistance [10,11]. The structures of SCCmec elements are divergent. At least 11 types of SCCmec elements have been identified [12-14]. Accordingly, MRSA clones are defined by the combination of the genotype of the S. aureus strain and the type of SCCmec[15]. By using molecular epidemiological techniques, it became evident that CA-MRSA strains were distinct from those of healthcare-associated MRSA (HA-MRSA) strains. The majority of CA-MRSA strains harbour small-sized type IV or type V SCCmec elements and are susceptible to many antibiotics [16-18]. In contrast, HA-MRSA isolates carry one of the three types of SCCmec (types I, II or III) or occasionally types IV and V, and are generally multidrug resistant [6,19].
Interestingly, the majority of CA-MRSA strains that have emerged worldwide carried the lukS-PV and lukF-PV genes encoding Panton Valentine Leukocidine. Characteristic PVL-positive MRSA clones have been disseminated in each district or continent. In the United States, the ST8-SCCmecIVa (USA300) clone and ST1-SCCmecIVa (USA400) clone have been predominant. In Europe and some Asian countries, the ST80-type IVa SCCmec and ST59-SCCmecV(5C2&5) clones have been predominant, respectively. The lukS-PV and lukF-PV genes are located on bacteriophages. Since the first report of the PVL phage, the nucleotide sequences of several PVL phages have been reported [16,20-24]. Three structurally distinct PVL phages belonging to groups 1-3, have been identified to date.
We characterized the MRSA clones disseminated in Tunisian hospitals and the community. In this study, we conducted a retrospective analysis of the HA-MRSA and CA-MRSA strains isolated from two Tunisian hospitals between the years of 2004 and 2008. In order to characterize the MRSA strains, several different molecular typing methods were used: mecA gene PCR, SCCmec typing, the carriage of PVL gene and the genotyping using the agr locus typing, spa-typing and Multilocus Sequences Typing (MLST). Furthermore, the nucleotide sequence of the PVL phage carried by one strain was determined.
Results
Antimicrobial susceptibility
The CA-MRSA strains were resistant to gentamicin (7%), kanamycin (89%), amikacin (86%), tobramycin (18%), tetracyclines (75%), ofloxacine (11%), ciprofloxacin (36%), erythromycin (46%), clindamycin (14%) and rifampicin (4%). All strains were susceptible to pristinamycin, vancomycin, teicoplanin, trimethoprime-sulfamethoxazole and chloramphenicol. The HA-MRSA strains were resistant to gentamicin (38%), kanamycin (90%), amikacin (90%), tobramycin (26%), tetracyclines (88%), ofloxacine (30%), ciprofloxacin (45%), erythromycin (55%), trimethoprim-sulfamethoxazole (15%), chloramphenicol (7.5%), clindamycin (18%), rifampicin (32%) and fosfomycine (10%). All strains were sensitive to pristinamycin, vancomycin and teicoplanin.
Characteristics of HA-MRSA clones
The characteristics of 41 HA-MRSA strains are summarized in Table 1. Twenty-one strains were PVL positive, while 20 strains were PVL negative. All PVL-positive strains belonged to the predicted founder group (FG, formerly called the “clonal complex”) 80 in the MLST genotype (ST80, 20 strains and ST1440, 1 strain). All strains belonged to agr group III, and four spa-types (70, 346, 435, and new) were identified among them. All PVL-positive strains carried the type IVc SCCmec element. In contrast, the PVL-negative clones were very diverse. Eight STs, three agr groups, and more than nine spa types were identified (Table 1). These strains carried SCCmec elements of type I, III, IVc, or were nontypeable (NT). Accordingly, at least eight MRSA clones (ST247-SCCmecI, ST1819-SCCmecI, ST239-SCCmecIII, ST241-SCCmecIII, ST5-SCCmecIVc, ST1-SCCmecNT, ST22-SCCmecNT and ST97-SCCmecNT) were identified in the 20 PVL-negative HA-MRSA strains, whereas there were only two MRSA clones, ST80-SCCmecIVc and ST1440-SCCmecIVc in PVL-positive HA-MRSA strains.
Table 1.
Characteristics of the MRSA clones isolated from Tunisian hospitals and the community
| ST | Predicted founder group (old clonal complex) | agr type | spa type | SCC mec type | |
|---|---|---|---|---|---|
| HA-MRSA (n=41) |
|
|
|
|
|
| PVL-positive (n=21) |
ST80(n=20) |
80 |
III |
70(n=16) |
IVc(n=16) |
| 346(n=1) |
IVc(n=1) |
||||
| 435(n=2) |
IVc(n=2) |
||||
| new(n=1) |
IVc(n=1) |
||||
| ST1440(n=1) |
80 |
III |
70(n=1) |
IVc(n=1) |
|
| PVL-negative (n=20) |
ST1(n=1) |
15(CC1) |
III |
35(n=1) |
bNT-1(n=1) |
| ST5(n=3) |
5(CC5) |
II |
45(n=2) |
IVc(n=1) |
|
| |
|
|
|
NT-A(n=1) |
|
| |
|
|
335(n=1) |
IVc(n=1) |
|
| ST22(n=1) |
22 |
II |
998(n=1) |
NT-N(n=1) |
|
| ST97(n=2) |
15 |
I |
3(n=1) |
NT-B(n=1) |
|
| |
|
I |
new(n=1) |
NT-B(n=1) |
|
| ST239(n=4) |
5(CC8) |
I |
3(n=4) |
III(n=3) |
|
| ST241(n=3) |
5(CC8) |
I |
125(n=2) |
III(n=2) |
|
| |
|
|
4(n=1) |
III(n=1) |
|
| ST247(n=3) |
5(CC8) |
I |
40(n=3) |
I(n=3) |
|
| ST1819(n=3) |
5(CC8) |
I |
new(n=3) |
I(n=3) |
|
| CA-MRSA(n=28) |
|
|
|
|
|
| PVL-positive(n=22) |
ST80(n=19) |
80 |
III(n=19) |
70(n=17) |
IVc(n=15) |
| |
|
|
|
|
NT-B(n=2) |
| |
|
|
346(n=1) |
IVc(n=1) |
|
| |
|
|
new(n=1) |
IVc(n=1) |
|
| ST153(n=2) |
80 |
III |
new(n=1) |
NT-B(n=1) |
|
| |
|
|
70(n=1) |
IVc(n=1) |
|
| ST2563(n=1) |
80 |
III |
70(n=1) |
IVc(n=1) |
|
| PVL-negative(n=6) | ST1(n=1) |
15(CCI) |
III |
35(n=1) |
NT-Bc(n=1) |
| ST5(n=2) |
5 |
II |
381(n=1) |
I(n=1) |
|
| |
|
|
1021(n=1) |
IVc(n=1) |
|
| ST45(n=1) |
45 |
I |
aND(n=1) |
NT-B(n=1) |
|
| ST80(n=2) |
80 |
II |
1021(n=1) |
IVc(n=1) |
|
| III | ND(n=1) | IVc(n=1) |
aND: could not be detected.
bNT: non typeable.
Characteristics of CA-MRSA strains
The characteristics of the 28 isolated CA-MRSA strains are summarized in Table 1. Twenty-two strains (79%) were PVL-positive and six strains (21%) were PVL-negative. All PVL-positive strains belonged to FG80 and agr group III, and carried the type IVc or NT SCCmec element similar to the cases of PVL-positive HA-MRSA strains. Three spa-types (70, 346, and new) were identified among them. The PVL-negative strains belonged to four FGs (5, 15, 45, and 80), three agr groups, I- III, and there were more than four spa types (35, 381, 1021, and new). These strains carried SCCmec elements of type IVc or NT. As a result, five MRSA clones (ST1-SCCmecNT, ST5-SCCmecI, ST5-SCCmecIVc, ST45-SCCmecNT and ST80-SCCmecIVc) were identified in six PVL-negative CA-MRSA strains.
SCCmec elements identified in Tunisian MRSA
As listed in Table 1, the SCCmec type of 59 out of 69 MRSA strains were classified by one of the extant types. All PVL-positive HA-MRSA strains and the majority of CA-MRSA strains carried type IV SCCmec of subtype c. Three PVL-positive CA-MRSA strains carried class B mec, but no ccr genes were identified so far. We expressed this as “NT-B”. SCCmecNT-B was identified in three PVL-negative strains belonging to ST45 and ST97. The SCCmec elements of the other four strains were expressed as follows: NT-1 (type 1 ccr positive, the class of the mec gene complex could not be determined), NT-A (ccr genes were not identified, but it carried the class A mec gene complex), NT-N (neither the ccr genes nor the mec gene complex could be identified), and NT-Bc (two ccr genes, types 1 and 2, were identified, and the class B mec gene complex was identified).
The characteristics of lysogenized PVL phage
We determined the nucleotide sequence of a PVL phage lysogenized in a PVL-positive CA-MRSA strain, JCSC7401, isolated in 2006. The strain belonged to ST80 and carried nontypeable SCCmec (NT-B). This phi7401PVL was 45,334 bp in length from the rightmost phage attachment site (attP-R) to the leftmost site (attP-L), in which 44 predicted ORFs larger than 99 bp were identified. The core sequences of 29 nucleotides were located at both ends of phi7401PVL. The G+C content of phi7401PVL was 33.2%, and was comparable to other staphylococcal phages. The overall organization of phi7401PVL was the same as that of previously-reported PVL phages, which consisted of five regions relating to 1) lysogeny, 2) DNA replication/transcriptional regulation, 3) structural modules (the packaging/head and tail), 4) the lysis module, and 5) lukS-PV and lukF-PV (Figure 1a). The phage was highly homologous to phiSa2mw, which belongs to group 2 of sfi21-like Siphoviridae (Figure 1a and 1b). The entire genome of the phage showed nucleotide identity of more than 95% to that of phiSa2mw. Forty-two of the 44 ORFs were highly homologous to those of phiSa2mw, with the nucleotide identities ranging from 91-100% (Additional file 1: Table S1). The int gene was truncated, although it was highly homologous to extant PVL phages. Two ORFs, TUP03 encoding Na/K ATPase and TUP16 encoding dUTPase, were less homologous to phiSa2mw.
Figure 1.
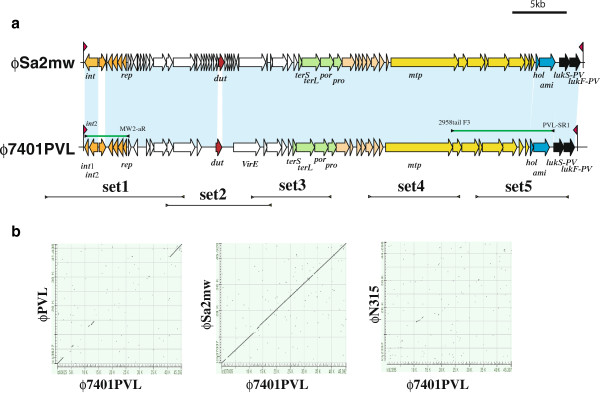
a. Structural comparisons of the PVL phages. Structures of phi7401PVL and phiSa2mw are illustrated based on the nucleotide sequences deposited in databases DDBJ/EMBL/GenBank under accession nos. BA000033 for phiSa2mw and AP012341 for phi7401PVL. Red arrowhead indicates the location of attP. Black bars indicate the locus of amplified DNA fragments using 5 sets of primers. Green bars indicate the locus of amplified DNA fragment identifying the carriage of gene linkages in phi7401PVL. ORFs are colored as follows: orange, ORFs related to lysogeny; red, a ORF in DNA replication/recombination region with assigned functions; bright green, ORFs related to capsid formation; yellowish orange, ORFs related to head formation; yellowish green, ORFs related to tail formation; blue, ORFs related to cell lysis; black, lukS-PV and lukF-PV. The locations of the primers are indicated in lines flanked by arrow heads. Nucleotide sequences of the primers are listed in Additional file 2: Table S2. b. Comparisons of the two phage genomes with a dot plot analysis. The genome sequence of phi7401PVL was compared to those of phiPVL (group 1 cos-site Siphoviridae), phiSa2mw (group 2 cos-site Siphoviridae), and phiN315 (group 3 cos-site Siphoviridae) using a specialized BLAST at NIBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Ordinate indicates the genome phi7401PVL. Abscissa indicates the genomes of three phages, phiPVL, phiSa2mw, and phiN315, in dot plots A, B, and C, respectively.
In order to determine whether the Tunisian PVL positive strains also carried the same PVL phage as phi7401PVL, we conducted two PCR studies to identify the regions in common with two PVL phages (phi7401PVL and phiSA2mw): a long range PCR study identifying gene linkage lukS and the tail gene that can identify PVL phages of the elongated head type and another PCR study identifying the region related to lysogeny (Additional file 1: Table S1 and Figure 1a). In our experiments, all the PVL positive strains were positive in both PCR studies.
Discussion
Antibiotic resistance to agents other than β-lactams
The majority of the HA-MRSA isolates were resistant to kanamycin, amikacin and tetracycline. Although the ratio was slightly low (25~55%), these strains were also frequently resistant to tobramycin, gentamicin, erythromycin, quinolones and rifampicin. Recently, it has been reported that rifampicin resistance is related to glycopeptides resistance [25,26]. Since the ratio of rifampicin resistant strains was relatively high, there is a possibility that there might be glycopeptides related to low resistance strains, e.g., hetero-VISA strains. However, glycopeptide resistance is beyond the focus of this study, so we did not examine the details for these findings.
Similar to HA-MRSA isolates, the majority of CA-MRSA isolates were resistant to kanamycin, amikacin and tetracycline, but were susceptible to other antibiotics, except for erythromycin and ciprofloxacin. These data suggest that Tunisian CA-MRSA strains were more resistant to kanamycin, tetracycline and erythromycin than U.S. and Oceanian isolates [27]. In our study, only four CA-MRSA strains were resistant to clindamycin, thus suggesting that clindamycin can be used for the treatment of CA-MRSA infections in Tunisia.
The PVL phage carried by Tunisian MRSA
The phi7401 carried by a ST80 Tunisian MRSA was highly homologous to phiSa2mw carried by ST1-SCCmecIVa MRSA. Only two ORFs, TUP03 and TUP16, showed a lower identity to those of phiSa2mw. Interestingly, TUP03 was identical to ORFs in phi12, phi13, and the bacteriophage in MRSA strains JH1 and JH9, and TUP16 was highly homologous to dUTPase in phiSLT and phi108PVL, with nucleotide identities of 97%. These data suggest that the components of phages were chimeric. Numerous lysogenized phages were induced from the cells of four strains, including JCSC7401 by mitomycin C induction. However, a hybridization experiment with a PVL probe showed that no plaque of the PVL phase was observed. This might have been due to the carriage of a truncated int. It seems that lysogenization of the phage occurred early to thus cause a mutation in the phage genome or that the ST80 strains might have an ability to cause a mutation in the int to keep the inserted phage genome in the chromosome in a stable form.
Characteristics of Tunisian MRSA
We determined MLST genotype of all 69 MRSA strains. Our data clearly indicated that all PVL-positive MRSA strains belonged to predicted founder group (FG) 80, which was previously indicated as clonal complex (CC) 80 at the MLST website. In contrast, the PVL-negative MRSA strains belonged to diverse FGs. In this study, we used the FG, which is used at present in the eBurst system on the MLST website. However, by using the old CC system, we can distinguish some lineages more clearly, e.g., ST239 that carries type III SCCmec as CC8 and ST5 that carries type II SCCmec as CC5, both of which belonged to FG5. Therefore, we listed both the present and former grouping systems in Table 1.
The agr types were well correlated with the MLST genotypes; group I, STs 45, 97, 239, 241, 247, and 1819; group II, STs 5 and 22; group III, STs 1, 80, 153, 1440, and new. There was only one exceptional case of a ST80 strain belonging to the agr group II. Further experiments including nucleotide sequence determination will be needed to clarify this discrepancy.
The SCCmec types of the strains were further determined by multiplex PCR studies, leaving 10 strains still nontypeable. The type IVc SCCmec was the most representative one in Tunisia. It was identified both in CA-MRSA (79%) and HA-MRSA (56%). PVL-positive MRSA strains carried SCCmec IVc and NT-B, which was supposed to be a novel SCCmec type.
The characteristics of Tunisian MRSA strains were also reported by Ben Nejma et al [28]. It has also been reported that the CC80 CA-MRSA strains were predominant clones in Tunisia, similar to many Europeans countries like France, Belgium, and Switzerland [27,29]. The predominance of the type IVc SCCmec stain was also reported.
The majority of our CA-MRSA (79%) and HA-MRSA (51%) isolates were pvl-positive and belonged to FG80. Our study suggested that the PVL-positive MRSA strains disseminated in Tunisia might be unique to Tunisia or the surrounding countries. Although CC80 PVL positive MRSA strains have been identified in European countries [30], the majority of them carried a type IVa SCCmec element or their SCCmec subtype was not determined. While two CA-MRSA isolates from Belgium [29] were reported to belonged to ST153-MRSA-IV, the report did not show its subtype.
According to previous studies, PVL-positive MRSA isolates were reported to be associated with an agr group III background [27,28,31]. Among our CA-MRSA isolates, the most predominant agr group was group III, followed by group II, then group I.
The PVL-positive MRSA clones disseminated in other countries belonged to ST1, ST8, ST22, ST30 and ST59, and carried distinct SCCmec elements. Recently, ST30 has been associated with CA-MRSA strains in the United States and in Ireland [27,31] and the ST93 and ST772 strains have been reported in Australia and India, respectively [32,33]. These data suggest that the possibility of simultaneous co-evolution of CA-MRSA organisms in different locations [27] is higher than the possibility of dissemination of a single CA-MRSA clone all over the world. PVL positive strains might therefore have emerged elsewhere and spread in the community and at hospitals.
It is interesting that the PVL-negative MRSA clones were the same MRSA strains isolated in other countries. Two other CA-MRSA isolates belonged to ST5-MRSA-IV which is one of predominant clones in the Netherlands [34]. Concerning the HA-MRSA, the agr group I was predominant, as reported previously in Tunisian MRSA [27]. The predominance of a group I background was also reported in United States and in Korea [35,36]. Similar results were obtained in European countries such as Germany and Belgium [36]. Three isolates belonged to the clone ST241-SCCmecIII. Two belonged to the ST247-SCCmecI (Iberian) clone, which is one of predominant clones in Poland [37]. Two other isolates belonged to ST239-SCCmecIII (Hungarian) clone, which is predominant in Turkey [38].
Conclusion
Tunisian PVL positive MRSA strains carried the PVL phage, which was highly homologous to phiSa2mw, but distinct in two ORFs. They belonged to FG80 and agr group III, and carried type IVc or nontypeable SCCmec. Such strains disseminated in the community and might have spread at the Tunisian hospitals by taking over existing MRSA clones, e.g., CC8-SCCmecI and CC8-SCCmecIII.
Methods
Bacterial strains
One hundred and fifty-four non-replicated HA-MRSA strains were isolated from 1999 through 2008 at Charles Nicolle Hospital of Tunis. Among them, 41 strains isolated from 2004 through 2008 were chosen based on their resistance profiles. HA-MRSA strains were isolated from mucous pus and blood cultures, puncture fluids, urine, and biomaterials of inpatients.
A total of 28 non-replicated CA-MRSA strains were isolated from January 2004 through June 2008 in two Tunisian hospitals (Charles Nicolle Hospital and Habib Bourguiba Hospital). CA-MRSA strains were isolated from the specimens of the patients with MRSA infections who had not been recently (¬within the past year) hospitalized or undergone a medical procedure (such as dialysis, surgery, catheterization). The CA-MRSA strains were generally recovered from mucous pus, puncture fluids, urine and biomaterials from outpatients. Some MRSA strains isolated from patients within 48 h of hospitalization, e.g., after surgery, in the intensive care unit, in the departments of nephrology, otorhinolaryngology and gynecology, were also included.
Strain identification
The isolates were identified by the conventional methods (Gram-positive cocci, catalase positive, mannitol fermenting and DNase-positive) and were confirmed to be S. aureus by their ability to coagulate rabbit plasma (bioMérieux, Marcy l’Etoile, France) and to produce clumping factor (Staphyslide test, bioMérieux). The biotypes were determined using Api20 Staph (bioMérieux, Marcy l’Etoile, France).
Isolation of MRSA strains
Strains were purified twice, first on MRSA BD select agar (from Baird Dickinson) and then on the Difco Tryptic Soy Agar (TSA) supplemented with an antimicrobial agent (ceftizoxime sodium) from Astellas Pharma Inc, Tokyo, Japan. The detection of the mecA gene by PCR was conducted as described previously [39].
The MIC of oxacilline
The MIC of oxacilline was determined by the agar dilution method according to the Clinical and Laboratory Standards Institute guidelines (CLSI) [40]. S. aureus ATCC 29213 was used as a reference strain.
Antimicrobial susceptibility testing
The antibiotic susceptibility of the isolates was assessed using the disk diffusion method according to the CLSI guidelines, except for pristinamycine, which was used according to the CA-SFM guidelines. The following antimicrobial disks were tested: penicillin G (10UI), oxacillin (1 μg), ampicillin (10 μg), amoxicillin + clavulanic acid (20/10 μg), cephalotin (30 μg), cefoxitin (30 μg), kanamycin (30 μg), gentamicin (10 μg), tobramycin (10 μg), tetracyclines (30 μg), chloramphenicol (30 μg), ofloxacin (5 μg), ciprofloxacin (5 μg), trimethoprim + sulfamethoxazole (1.25/23.75 μg), erythromycin (15 μg), clindamycin (2 μg), vancomycin (10 μg), teicoplanin (30 μg), rifampicin (5 μg), fosfomycin (5 μg) and pristinamycine (15 μg).
Culture and DNA extraction
The strains were grown on TSB culture at 37°C overnight with shaking. Genomic DNA used as a target for PCR assays was extracted by using a Qiagene kit (QIAamp DNA Mini Kit (250) QUIAGEN. Sciences - US) according to the manufacturer’s instructions.
SCCmec typing
The SCCmec elements were typed using two multiplex PCR strategies (M-PCR1 and M-PCR2) which are used for SCCmec typing assignment, M-PCR3 was used for the J1 region difference-based subtyping, as described previously [41]. The reference strains used were as follows: NCTC10442(type I), N315(type II), 85/2082(type III), CA05(type IVa), 8/6-3P(type IVb), and 81/108(type IVc).
Detection of the Panton-valentine leukocidin gene
The carriage of lukF-PV and lukS-PV genes encoding PVL was examined by PCR as described previously [42].
agr typing
The presence of the accessory gene regulator, agr, was determined by multiplex PCR amplifying the hypervariable domain of the agr locus, as described previously [43]. PCR amplification was performed in a 50 μl reaction mixture composed of 2U of Ex Taq (Takara Shuzo Co., Ltd., Kyoto, Japan), 10 pmol of each primer, 0.2 mM deoxynucleoside triphosphate mixture, 10 ng of chromosomal DNA, 1X reaction buffer (Takara Shuzo Co., Ltd.) and H2O. Thermal cycling was set at 30 cycles (30s for denaturation at 95°C, 1 min for annealing at 55°C, and 2 min elongation at 72°C) and was performed with a Gene Amp PCR system 9600 (Perkin-Elmer, Wellesley, Massachusetts).
MLST
The genotypes were determined by Multilocus Sequence Typing (MLST) according to the procedure used by Enright et al [44]. The alleles of each locus were compared, and sequence types (STs) were assigned based on the S. aureus MLST database (http://saureus.mlst.net/).
Spa typing
The typing of the polymorphic region of the protein A gene (spa) was performed as described previously [45]. Purified spa PCR products were sequenced, and short sequence repeats (SSRs) were assigned using the spa database website (http://www.tools.egenomics.com/).
Determination of nucleotide sequences
Genomic DNA of strain JCSC7401 was extracted with phenol/chloroform and the nucleotide sequences were determined using a 454 genetic analyzer. PCR studies were conducted to amplify the DNA fragment covering the gap of the contigs obtained by the 454 genetic analyzer. The nucleotide sequence of PCR products amplified by long-range PCRs with primer’s pairs listed in Additional file 2 were determined using an ABI sequencer. The nucleotide sequence of phi7401PVL was deposited to the DDBJ/EMBL/GenBank databases under accession no. AP012341.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JM, BI, and HA collected strains at Tunisian hospitals. TI, JM, and BI designed the research and prepared the manuscript. KH and HA add the suggestions for the research and preparing the manuscript. JM, MZ, JJ, SL, and HX performed experiments. MZ, JJ and TI contributed for the nucleotide sequencing and data analysis of the PVL phage. All authors read and approved the final manuscript.
Supplementary Material
Table S1. ORFs in and around phi7401PVLand their similarities to phiSa2mw.
Table S2. List of primers used in this experiment.
Contributor Information
Ben Jomàa-Jemili Mariem, Email: jemilimariem@yahoo.fr.
Teruyo Ito, Email: teruybac@juntendo.ac.jp.
Meng Zhang, Email: mchou@juntendo.ac.jp.
Jingxun Jin, Email: isao@kanehirocorp.com.
Shanshuang Li, Email: lishsh@sjtu.edu.cn.
Boutiba-Ben Boubaker Ilhem, Email: ilhem.boutiba@rns.tn.
Hammami Adnan, Email: adnene.hammami@rns.tn.
Xiao Han, Email: hx2005@juntendo.ac.jp.
Keiichi Hiramatsu, Email: khiram06@juntendo.ac.jp.
Acknowledgement
This work was supported by the Oyama Health Foundation, a Grant-in-Aid from MEXT (Ministry of Education, Culture, Sports,Science and Technology) - Supported Program for the Strategic Research Foundation at Private Universities and the ministry of Scientific Research, Technology and Competence Development of Tunisia.
References
- Jevons MP. "Celbenin"-resistant staphylococci. Br Med J. 1961;124:124–125. [Google Scholar]
- Udo EE, Pearman JW, Grubb WB. Genetic analysis of community isolates of methicillin-resistant Staphylococcus aureus in Western Australia. J Hosp Infect. 1993;25:97–108. doi: 10.1016/0195-6701(93)90100-E. [DOI] [PubMed] [Google Scholar]
- Salgado CD, Farr BM, Calfee DP. Community-acquired methicillin-resistant Staphylococcus aureus: a meta-analysis of prevalence and risk factors. Clin Infect Dis. 2003;36:131–139. doi: 10.1086/345436. [DOI] [PubMed] [Google Scholar]
- Hiramatsu K, Okuma K, Ma XX, Yamamoto M, Hori S. et al. New trends in Staphylococcus aureus infections: glycopeptide resistance in hospital and methicillin resistance in the community. Curr Opin Infect Dis. 2002;15:407–413. doi: 10.1097/00001432-200208000-00009. [DOI] [PubMed] [Google Scholar]
- Chambers HF. The changing epidemiology of Staphylococcus aureus? Emerg Infect Dis. 2001;7:178–182. doi: 10.3201/eid0702.010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SK, Stemper ME, Ramaswamy SV, Conradt JM, Reich R. et al. Molecular characteristics of nosocomial and Native American community-associated methicillin-resistant Staphylococcus aureus clones from rural Wisconsin. J Clin Microbiol. 2004;42:3752–3757. doi: 10.1128/JCM.42.8.3752-3757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XX, Ito T, Tiensasitorn C, Jamklang M, Chongtrakool P. et al. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother. 2002;46:1147–1152. doi: 10.1128/AAC.46.4.1147-1152.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Roth E, Lorenzo-Diaz F, Batista N, Moreno A, Mendez-Alvarez S. Tracking methicillin-resistant Staphylococcus aureus clones during a 5-year period (1998 to 2002) in a Spanish hospital. J Clin Microbiol. 2004;42:4649–4656. doi: 10.1128/JCM.42.10.4649-4656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liassine N, Auckenthaler R, Descombes MC, Bes M, Vandenesch F. et al. Community-acquired methicillin-resistant Staphylococcus aureus isolated in Switzerland contains the Panton-Valentine leukocidin or exfoliative toxin genes. J Clin Microbiol. 2004;42:825–828. doi: 10.1128/JCM.42.2.825-828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Katayama Y, Hiramatsu K. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob Agents Chemother. 1999;43:1449–1458. doi: 10.1128/aac.43.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcal cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44:1549–1555. doi: 10.1128/AAC.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother. 2009;53:4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Skov RL, Han X, Larsen AR, Larsen J. et al. Novel types of staphylococcal cassette chromosome mec elements identified in clonal complex 398 methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother. 2011;55:3046–3050. doi: 10.1128/AAC.01475-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alvarez L, Holden MT, Lindsay H, Webb CR, Brown DF. et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA) Proc Natl Acad Sci. 2002;99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K. et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/S0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- Ito T, Ma XX, Takeuchi F, Okuma K, Yuzawa H. et al. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob Agents Chemother. 2004;48:2637–2651. doi: 10.1128/AAC.48.7.2637-2651.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eady EA, Cove JH. Staphylococcal resistance revisited: community-acquired methicillin resistant Staphylococcus aureus–an emerging problem for the management of skin and soft tissue infections. Curr Opin Infect Dis. 2003;16:103–124. doi: 10.1097/00001432-200304000-00007. [DOI] [PubMed] [Google Scholar]
- Shore A, Rossney AS, Keane CT, Enright MC, Coleman DC. Seven novel variants of the staphylococcal chromosomal cassette mec in methicillin-resistant Staphylococcus aureus isolates from Ireland. Antimicrob Agents Chemother. 2005;49:2070–2083. doi: 10.1128/AAC.49.5.2070-2083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XX, Ito T, Chongtrakool P, Hiramatsu K. Predominance of clones carrying Panton-Valentine leukocidin genes among methicillin-resistant Staphylococcus aureus strains isolated in Japanese hospitals from 1979 to 1985. J Clin Microbiol. 2006;44:4515–4527. doi: 10.1128/JCM.00985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko J, Kimura T, Kawakami Y, Tomita T, Kamio Y. Panton-valentine leukocidin genes in a phage-like particle isolated from mitomycin C-treatedStaphylococcus aureus V8 (ATCC 49775) Biosci Biotechnol Biochem. 1997;61:1960–1962. doi: 10.1271/bbb.61.1960. [DOI] [PubMed] [Google Scholar]
- Kaneko J, Kimura T, Narita S, Tomita T, Kamio Y. Complete nucleotide sequence and molecular characterization of the temperate staphylococcal bacteriophage phiPVL carrying Panton-Valentine leukocidin genes. Gene. 1998;215:57–67. doi: 10.1016/S0378-1119(98)00278-9. [DOI] [PubMed] [Google Scholar]
- Diep BA, Gill SR, Chang RF, Phan TH, Chen JH. et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- Zhang K, McClure JA, Elsayed S, Conly JM. Novel staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53:531–540. doi: 10.1128/AAC.01118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo M, Hishinuma T, Katayama Y, Cui L, Kapi M. et al. Mutation of RNA polymerase beta subunit (rpoB) promotes hVISA-to-VISA phenotypic conversion of strain Mu3. Antimicrob Agents Chemother. 2011;55:4188–4195. doi: 10.1128/AAC.00398-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Isii T, Fukuda M, Ochiai T, Neoh HM. et al. A RpoB Mutation Confers Dual Hetero-Resistance to Daptomycin and Vancomycin in Staphylococcus aureus. Antimicrob Agents Chemother. 2010;54:5222–5233. doi: 10.1128/AAC.00437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenesch F, Naimi T, Enright MC, Lina G, Nimmo GR. et al. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis. 2003;9:978–984. doi: 10.3201/eid0908.030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben M, Nejma MM, Frih S, Sakly N, Ben Y, Salem M. Nour Molecular charcterization of Methicillin-resistant Staphylococcus aureus isoleted in Tunisia. Diag Microbiol Infect Dis. 2006;55:324–328. doi: 10.1016/j.diagmicrobio.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Denis O, Deplano A, De Beenhouwer H, Hallin M, Huysmans G. et al. Polyclonal emergence and importation of community-acquired methicillin-resistant Staphylococcus aureus strains harbouring Panton-Valentine leucocidin genes in Belgium. J Antimicrob Chemother. 2005;56:1103–1106. doi: 10.1093/jac/dki379. [DOI] [PubMed] [Google Scholar]
- Holmes A, Ganner M, McGuane S, Pitt TL, Cookson BD. et al. Staphylococcus aureus isolates carrying Panton-Valentine leucocidin genes in England and Wales: frequency, characterization, and association with clinical disease. J Clin Microbiol. 2005;43:2384–2390. doi: 10.1128/JCM.43.5.2384-2390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossney AS, Shore AC, Morgan PM, Fitzgibbon MM, O'Connell B. et al. The emergence and importation of diverse genotypes of methicillin-resistant Staphylococcus aureus (MRSA) harboring the Panton-Valentine leukocidin gene (pvl) reveal that pvl is a poor marker for community-acquired MRSA strains in Ireland. J Clin Microbiol. 2007;45:2554–2563. doi: 10.1128/JCM.00245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs GW, Goering RV, Chua KY, Monecke S, Howden BP. et al. The molecular epidemiology of the highly virulent ST93 Australian community Staphylococcus aureus strain. PLoS One. 2012;7:e43037. doi: 10.1371/journal.pone.0043037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shambat S, Nadig S, Prabhakara S, Bes M, Etienne J. et al. Clonal complexes and virulence factors of Staphylococcus aureus from several cities in India. BMC Microbiol. 2012;12:64. doi: 10.1186/1471-2180-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deurenberg RH, Nulens E, Valvatne H, Sebastian S, Driessen C. et al. Cross-border dissemination of methicillin-resistant Staphylococcus aureus, Euregio Meuse-Rhin region. Emerg Infect Dis. 2009;15:727–734. doi: 10.3201/eid1505.071618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen W, van Nieuwenhuizen W, Gijzen C, Verbrugh H, van Belkum A. Population studies of methicillin-resistant and -sensitive Staphylococcus aureus strains reveal a lack of variability in the agrD gene, encoding a staphylococcal autoinducer peptide. J Bacteriol. 2000;182:5721–5729. doi: 10.1128/JB.182.20.5721-5729.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HJ, Choi JY, Lee K, Yong D, Kim JM. et al. Accessory gene regulator group polymorphisms in methicillin-resistant Staphylococcus aureus: an association with clinical significance. Yonsei Med J. 2007;48:176–183. doi: 10.3349/ymj.2007.48.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luczak-Kadlubowska A, Sulikowska A, Empel J, Piasecka A, Orczykowska M. et al. Countrywide molecular survey of methicillin-resistant Staphylococcus aureus strains in Poland. J Clin Microbiol. 2008;46:2930–2937. doi: 10.1128/JCM.00869-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alp E, Klaassen CH, Doganay M, Altoparlak U, Aydin K. et al. MRSA genotypes in Turkey: persistence over 10 years of a single clone of ST239. J Infect. 2009;58:433–438. doi: 10.1016/j.jinf.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H. et al. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29:2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and laboratory standard institute Performance standards for antimicrobial susceptibility testing. Wayne, PA, USA; 2006. (16th informational supplement M100-S16 CLSI). [Google Scholar]
- Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN. et al. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51:264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XX, Galiana A, Pedreira W, Mowszowicz M, Christophersen I. et al. Community-acquired methicillin-resistant Staphylococcus aureusn Uruguay. Emerg Infect Dis. 2005;11:973–976. doi: 10.3201/eid1106.041059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopsin B, Mathema B, Alcabes P, Said-Salim B, Lina G. et al. Prevalence of agr specificity groups among Staphylococcus aureus strains colonizing children and their guardians. J Clin Microbiol. 2003;41:456–459. doi: 10.1128/JCM.41.1.456-459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M. et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37:3556–3563. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. ORFs in and around phi7401PVLand their similarities to phiSa2mw.
Table S2. List of primers used in this experiment.


