Abstract
We present a general method for rigorously identifying correlations between variations in large-scale molecular profiles and outcomes and apply it to chromosomal comparative genomic hybridization data from a set of 52 breast tumors. We identify two loci where copy number abnormalities are correlated with poor survival outcome (gain at 8q24 and loss at 9q13). We also identify a relationship between abnormalities at two loci and the mutational status of p53. Gain at 8q24 and loss at 5q15-5q21 are linked with mutant p53. The 9q and 5q losses suggest the possibility of gene products involved in breast cancer progression. The analytical techniques are general and also are applicable to the analysis of array-based expression data.
Techniques for in vitro genomic and proteomic analysis are generating vast amounts of quantitative biological data. As an example, high-density DNA microarrays are capable of producing 30,000 measurements from a single sample of RNA (1). In the area of cancer, such data offer fertile ground for systematic computational analyses to help identify new cancer targets or potential therapeutics. Chromosomal comparative genomic hybridization (CGH) has been applied extensively to human tumor specimens, and array-based CGH methods are beginning to generate higher-density data (2, 3). For such techniques to be most useful, computational methods must generate conclusions that are supportable quantitatively in a rigorous statistical sense, and not provide just a means of visualization.
The challenge arises when the ratio between the number of measurements to the number of experimental samples is high. In this case, false patterns often emerge. For example, suppose we measure expression levels for several thousand mRNAs in 10 cell lines, 5 of which exhibit phenotype A and 5 that exhibit phenotype B. The expression ratios for each gene will show some variation regardless of correlation with phenotype. The apparent correlation to cell-line phenotype resulting from a naïve computation of correlation over all genes will be distributed approximately normally, and some genes may show an apparently significant correlation. In fact, because there are only 252 [10!/(5!(10-5)!)] ways of labeling 10 cell lines with 5 each of phenotypes A and B, it is extremely likely that many genes of the several thousand will show an apparently perfect correlation with phenotype, even if there is no true relationship between any observed genes' expression and phenotype.
We present a method for rigorously identifying correlations between large-scale multivariate measurements and outcomes and apply it to chromosomal CGH data from a set of 52 human breast tumors. We identify two loci (8q24 and 9q13) where copy number abnormalities are correlated with poor survival outcome and also identify a relationship between two loci (8q24 and 5q15-5q21) and the mutational status of p53. The techniques are applicable generally and also are used easily in the analysis of array-based expression data.
Materials and Methods
Tumor Specimens.
Fifty-two samples from breast tumors were obtained from three series of surgical specimens (35, 6, and 11 from refs. 4–6, respectively). Material was frozen promptly at −70°C until DNA isolation. Samples were trimmed to avoid normal cell contamination, and DNA was isolated by standard phenol/chloroform extraction. The tumors had been analyzed previously for TP53 gene mutation by using constant denaturant gel electrophoresis (CDGE) followed by sequencing as described (7). The 52 samples were selected from the 3 series based on the TP53 status—25 tumors with missense mutation, 3 tumors with deletions, and 24 tumors without mutation.
Comparative Genomic Hybridization.
Genome copy number was assessed by using CGH as described (8). Briefly, DNA samples isolated from normal human lymphocytes and tumor samples were labeled by nick translation with fluorescein-12-dUTP and Texas red-dUTP, respectively. DNA probes (200 ng) were mixed with 20 μg of unlabeled Cot-1 DNA and were hybridized with normal lymphocyte metaphase spreads for 3 days. The preparations were washed to remove nonspecific bound DNA and counterstained with 4′,6-diamidoino-2-phenylindol (DAPI) for chromosome identification.
Digital Image Analysis.
Fluorescein, 4′,6-diamidoino-2-phenylindol (DAPI), and Texas red images were acquired from several metaphases for each hybridization by using a Quantitative Image Processing System (QUIPS) as described (9). Chromosomes were segmented based on the DAPI image, and green–red ratio profiles along the segmented images were calculated for each chromosome. The results from 8–10 chromosomes of each type for each hybridization were combined to determine a mean (±1σ) for each chromosome type. Mean profiles for the 23 chromosome types (results were not calculated for the Y chromosomes because all samples were female) were arranged from short arm to long arm and from chromosome 1 to 22 and then X to produce a genome profile comprised of 1,225 bins.
Our expectation when comparing two normal samples is that all ratios should be 1 and that deviations from 1 are the result of experimental noise or experimental artifact. The distribution of values when logarithm (log)-transformed is very close to normal, with untransformed data exhibiting skew to the left (data not shown). This skew is expected in ratio measurements where the numerator and denominator both have normally distributed noise, as is the case here. We have used log-transformed data uniformly in our analyses.
Statistical Analyses.
We used Kendall's Tau in our analyses, a rank-based nonparametric statistic that compares all pairs of observations within two data series, assigning a score of 1 to pairs with the same rank relationship (i.e., item 1 is greater than item 2 in each pair), a score of 0 to ties, and a score of −1 to pairs that are mismatched. Kendall's Tau is normalized for the number of comparisons, yielding values from −1 to 1 with 1 indicating perfect correlation of ranks, −1 indicating perfect anti-correlation, and 0 indicating no correlation. The choice of Kendall's Tau is not critical to the analyses; many statistics work well.
To assess statistical significance given our unfavorable ratio of variables (1,225) to samples (52), we use permutation analysis. The procedure is simple.
I. Do 10,000 times:
A. Randomly permute the index of tumor outcome to CGH profile.
B. Compute the correlation between copy number and survival.
C. Record the maximum magnitude correlation across the entire profile.
II. Accumulate the maximum magnitude correlations into a cumulative histogram.
III. Plot the histogram and determine the correlation threshold corresponding to a selected P value.
The correlation threshold selected in this manner is conservative. Suppose we select a P value of 0.05 and find the corresponding threshold. If any locus in our data shows a better correlation, we can be confident that we would not have observed such a correlation more often than 1 in 20 times. Note that it is safe to ascribe significance to any locus (say the Mth best) that exceeds the threshold, because recomputation of the threshold using the Mth highest magnitude correlation for each permutation would result in a more permissive threshold than using the maximum magnitude correlation. This method has the advantage of being nonparametric and applicable to any correlation metric. Further, the method accounts for distributional irregularities in the data, because the permutation happens in the indexing of outcomes to CGH data. If, for example, there are strong crosscorrelations between different parts of our data, the correlation threshold we select will not be “penalized” by redundant information in the profiles.
Results and Discussion
We performed chromosomal CGH on 52 primary breast tumors and on 8 samples of normal tissue. Fig. 1 shows an example of CGH profiles for a normal control and for a tumor. Note that the magnitudes of some of the CGH ratios in the tumor vastly exceed the magnitudes of those in the normal control.
Figure 1.
Full CGH profile for a normal (dashed line) and a tumor (solid line). The tumor shows marked abnormalities on chromosomes 1, 8, and 9 in addition to several other significant deviations.
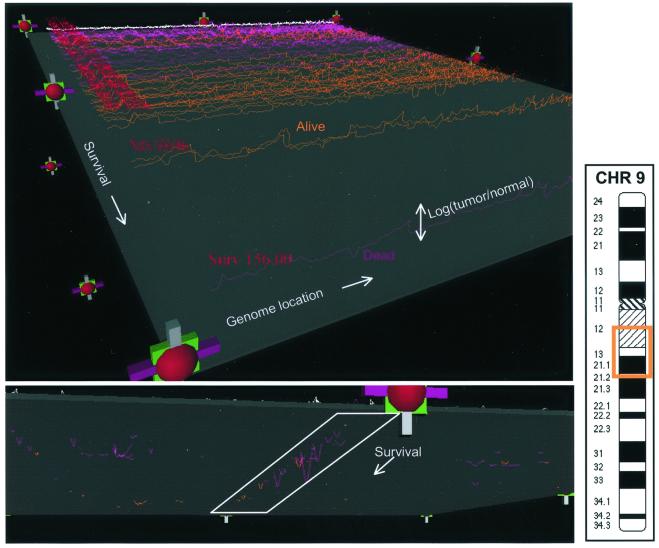
We explored the statistical correlation between CGH ratio measurements and patient outcome (e.g., survival) and tumor phenotype (e.g., p53 mutational status). Before discussing the statistical conclusions, it is instructive to inspect the entire collection of data to look for obvious patterns or problems. Although it is relatively easy to visualize a single CGH profile, it becomes difficult to simultaneously visualize many such profiles. We have implemented a simple three-dimensional visualization method for real-time inspection of the data and associated outcomes. The display is made by using vrml (http://www.vrml.org/Specifications/VRML97; ref. 10), using primitives such as colored lines, spheres, and blocks that can be rendered within a standard Internet browser. Fig. 2 shows two snapshots from this display.
Figure 2.
VMRL display of 52 CGH profiles and 8 normal profiles. Each CGH profile is displayed along the x axis, with the y axis being log (relative copy number), z being overall patient survival, and the color of the lines encoding patient status (purple is deceased, orange is alive, and white indicates a normal control). The top view shows all profiles, with the lowest survival furthest from the viewer. The bottom view shows the data thresholded by using the gray plane, to eliminate the variation caused by noise, and highlights a region on chromosome 9 that appears to be linked with poor survival.
By manipulating the display through the handles (rotation, translation, scaling, and y axis motion of the gray plane), we can explore relationships between CGH ratio variation and the patient-related outcomes displayed. The view (Lower) in Fig. 2 is oriented such that low-surviving patients are near the viewer. We have highlighted a region on chromosome 9 that appears to have a preponderance of deletions in tumors from low-surviving patients who died during follow-up. The region, 9q13, is highlighted in the ideogram. As with all visualization methods, the appearance of a correlation must be verified by statistical examination. The following section explores the genome-wide correlations between CGH ratio variation and patient outcome as well as tumor phenotype.
Correlation with Patient Survival.
There are many statistical metrics that can be used to measure a correlation between copy number and outcome or phenotype. In this work, we have used Kendall's Tau, although the choice is not critical (see Materials and Methods for details). With this metric, loss at a particular locus that is correlated with poor survival will show a positive correlation, gain correlated with poor survival will show a negative correlation, etc. Any deviation from a random relationship (i.e., 0.0) at any locus suggests a possible linkage between local copy number and the outcome under study.
Fig. 3 (Upper Left) shows the magnitude of correlation of CGH ratio to patient survival for each of the 1,225 bins in the CGH profiles, ignoring the effects of data censorship. (Kendall's Tau can be modified to take censorship into account, but in this case it does not change the outcome of the analysis. Kaplan–Meier survival analysis is used to verify the results of the initial correlation computations.) There is a wide range of apparent correlation, but recall from the previous discussion that we expect a wide apparent distribution of correlations even if there is no true correlation.
Figure 3.
(Upper) Correlation magnitude of copy-number variation with survival (Left). Cumulative distribution of correlation magnitudes under the null hypothesis (Right). None of the loci are significant at P = 0.05. (Lower) CGH variation energy across the genome (Left). Cumulative distribution of correlation magnitudes by using energy cutoffs of 0.0, 3.0, and 6.0 (Right). The P = 0.05 correlation thresholds are 0.35, 0.31, and 0.18, respectively.
To assess statistical significance, we used permutation analysis, as described previously. Fig. 3 (Upper Right) shows the cumulative histogram of the maximal correlation magnitudes for the 52 tumor profiles. For P = 0.05, we need to observe a correlation of greater than 0.35. Unfortunately, none of the loci meet that strict criterion despite the observations highlighted in Fig. 2. However, what this analysis has ignored is the magnitude of deviation from normalcy at the loci as well as the frequency with which a significant deviation is observed. Small, random variations in CGH ratio are almost certainly noise, but they contribute to the computation of maximal correlation in the permutation test by virtue of their sheer numbers. Intuitively, we would like to focus on CGH ratio variations that are either large in magnitude, and thus not likely to be noise, or that are moderate in magnitude but seen so often that they are not likely to be noise.
For each locus, we compute the sum of the absolute value of the log-transformed CGH ratio over all tumor profiles. In our data, the deviations of log ratio observed in normal controls is distributed normally with mean zero, thus this measure of variability (termed energy) identifies loci that fit the characteristics discussed previously—loci with frequent and/or large magnitude copy-number deviations. More generally, we can define any measure of variability over experimental cases and use it as a basis for filtering, as long as we do not make use of data to which we want to test a correlation. With CGH data, deviation from normality, instead of sample variance at each locus, is advantageous. With the latter, a very common aberration may exhibit relatively low variance but it will have high energy. In spirit, this approach is taking the prior probability of an observation into account, independently of the observation's correlation with outcome.
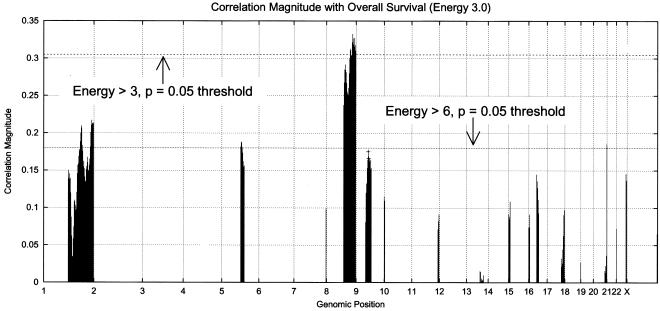
Fig. 3 (Lower Left) shows the energy for the 52 tumor profiles across the genome. Chromosomes 8 and 9 show the maximum, with chromosomes 1, 17, and 20 showing the next highest energies. Fig. 3 (Lower Right) shows the results of permutation analysis, as above, with three energy thresholds: 0.0, 3.0, and 6.0. The permutation procedure is modified: step IB now restricts the computation of correlation to those loci whose energy exceeds the threshold selected. By eliminating a great deal of (presumably) random variation from the permutation analysis through reduction of the number of bins under consideration, we see lower correlation magnitude thresholds corresponding to P = 0.05. Fig. 4 shows the correlation magnitude for loci exceeding energies 3.0 and 6.0. The locus at 8q24 exceeds the threshold for energy 3.0 at P = 0.05. The locus previously identified as 9q13 (from Fig. 2) nearly meets the P = 0.05 threshold for energy 6.0. Note that the permutation test, as constructed, is performing a two-tailed significance test. The 9q13 locus passes a single-sided test at P = 0.05.
Figure 4.
Correlation magnitude of copy-number variation with survival (energy greater than 3.0). (Energy greater than 6.0 is marked with a +.) The 8q24 locus meets the P = 0.05 significance test, and the 9q13 locus meets a single-sided test.
Fig. 5 shows Kaplan–Meier plots of survival for patients with amplifications at 8q24 (CGH ratio > 1.3) vs. normal at 8q24, and for patients with deletions at 9q13 (CGH ratio < 0.7) vs. normal at 9q13. The P values associated with this survival analysis for each locus are less than 0.01 (with the 8q24 locus slightly more significant). The locus at 8q24 contains c-myc, which has been well established in many cancers as an indicator of poor prognosis, when amplified. The locus at 9q13 has not been associated previously with poor outcome in breast cancer. In chromosomal CGH, the centromeric region of chromosome 9 is known to yield variable results. However, there is no reason to expect that spuriously observed deletions should be correlated with patient outcome, nor were correlations observed for other “problematic” regions on chromosomes 1, 16, and 22. Of note, the 8q24 gain is correlated with tumor size and disease stage, but the 9q13 loss is correlated with neither in a statistically significant manner.
Figure 5.
Kaplan–Meier plot of patient survival for those with normal (dashed lines) vs. increased (solid lines) copy number at 8q24 (Upper), and normal vs. deleted copy number at 9q13 (Lower). Both aberrations yield statistically significant survival differences (P < 0.01).
The tumor samples were taken from patients from three series (as detailed in Materials and Methods), with the patients having undergone heterogeneous treatment regimes. In the previous analysis, the three groups were pooled, and care must be exercised in interpreting correlations with survival in this circumstance. In particular, if there is some correlation between CGH genotype and series, and if there is a correlation between survival and series, conclusions involving the linkage of CGH genotype to survival may be suspect. However, in this set of tumors, although there are survival differences between the three series, there is no significant correlation between CGH genotype and series (assessed by using the permutation-based methods described previously). Further, in the case of the 8q24 locus, there are sufficient samples of both normal and amplified DNA from the series with the largest number of tumors to demonstrate a significant correlation with survival (P = 0.01 based on Kaplan–Meier analysis). The 9q13 locus is observed only as significant in the pooled analysis.
Correlation with P53 Status.
Fig. 6 shows a hierarchical clustering of the copy-number data for chromosomes 8 and 9 (both implicated as predictors of patient survival). Patient survival and the p53 status of each tumor are shown also. The top block, subordinate to node 31, is characterized by largely normal CGH ratios with a group of tumors having deletions in the 9q13 region. The bottom block, subordinate to node 49, is characterized by amplifications in 8q (particularly distal) and deletions centered on the 9q13 locus. The block subordinate to node 45 shows a mixture of 8q gains with some 9p losses.
Figure 6.
Hierarchical clustering of tumors based on copy number across chromosomes 8 and 9. The CGH data are presented on a red to green (deletion to amplification) color scale. Patient survival also is displayed (black is low survival and green is high on a linear scale), and the p53 status of each tumor also is shown [black is wild type (WT) and green is mutant]. Centromeres are indicated with arrows. Both patient survival and p53 status appear to be related to the clustering. The clustering was single-linkage agglomerative clustering, using all 1,225 bins of log-transformed CGH data in computing distances.
The clustering shows an interesting pattern. The top block of patients have relatively long survival and also show a preponderance of wild-type p53. The bottom two blocks show the reverse. Although we expect that clustering based on copy number for chromosomes 8 and 9 will show an enrichment of similar survival in clusters, it is not clear that there should be a strong enrichment for similar p53 status. This observation may be explained by a correlation between p53 status and survival, which has been shown elsewhere (e.g., refs. 11 and 12), but in these data, the correlation between p53 status and survival is not statistically significant.
Fig. 7 shows the direct correlation between copy number and p53 mutational status for loci exceeding energy level 3.0. Copy number at two loci (5q15-5q21 and 8q24) show sufficiently strong correlations to exceed the appropriate thresholds for P = 0.05 (the 5q locus exceeds P = 0.01). We also assessed correlations between copy number and estrogen and progesterone receptor status for these tumors, but no correlation was found with copy numbers that exceeded significance thresholds estimated by permutation analysis.
Figure 7.
Correlation of p53 status with copy number. There is a significant association between mutations in p53 and both 5q15-21 loss and 8q24 gain.
The correlation of copy-number gain at 8q24 with poor survival is not surprising given what is known about the c-myc gene product and its effects on breast cancer (13, 14). The correlation between loss at 9q13 and poor survival is more interesting. This region of chromosome 9 has few identified genes, and none that suggest a significant tumor-suppressor function. We propose that this may be an interesting area for gene discovery, pending prospective validation of the correlative observation on a larger set of matched tumor specimens.
The correlation of copy-number variations on 5q and 8q with genetic alterations in p53 status are also potentially interesting. The probability that the correlations are not real is low, given the permutation analyses, particularly for the 5q13-21 locus. There are, of course, many potential explanations of such correlations that do not involve direct causal effects between the loci and the p53 pathway. However, in the case of the 8q24 locus, it is known, for example, that the p53 promoter is transactivated by c-myc (15). Thus, in a tumor that has amplified c-myc genomically, there is potentially disproportionate advantage to losing p53 vs. tumors that have no such amplification.
In the case of the 5q deletion, for the p53 mutant tumors, 18/28 have a significant copy-number loss (based on a cutpoint estimated from the normal/normal distribution), and for the p53 wild-type tumors, only 1/24 have a significant copy-number loss. We hypothesize that further characterization of this region may elucidate a link to the p53 pathway.
The computational methods used in this work are applicable to many types of data, including gene expression quantitated by DNA microarrays. Permutation analysis over any data analysis procedure may be used to estimate statistical significance of an observation. Permutation-based procedures require no assumptions about the underlying distributional characteristics of the data. By coupling permutation analysis with systematic data reduction based on prior probabilities of observations, it is possible to identify significant correlations where the number of measurements vastly exceeds the number of experimental samples. In this case, the ratio of measurements to samples was about 20, but we have successfully used the methods on preliminary data where the ratio exceeds 1,500.
Acknowledgments
This work was supported in part by a grant from the National Cancer Institute (CA 58207) and grants from the Norwegian Cancer Society.
Abbreviations
- CGH
comparative genomic hybridization
- log
logarithm
References
- 1.Alizadeh A, Eisen M, Botstein D, Brown P O, Staudt L M. J Clin Immunol. 1998;18:373–379. doi: 10.1023/a:1023293621057. [DOI] [PubMed] [Google Scholar]
- 2.Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi O P. Genes Chromosomes Cancer. 1998;21:177–184. [PubMed] [Google Scholar]
- 3.Pinkel D, Segraves R, Sudar S, Clark S, Poole I, Kowbel D, Collins C, Kuo W-L, Chen C, Zhai Z, et al. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 4.Andersen T I, Holm R, Nesland J M, Heimdal K R, Ottestad L, Børresen-Dale A-L. Br J Cancer. 1993;68:540–548. doi: 10.1038/bjc.1993.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukholm I K, Nesland J M, Karesen R, Jacobsen U, Børresen-Dale A-L. J Pathol. 1997;181:140–145. doi: 10.1002/(SICI)1096-9896(199702)181:2<140::AID-PATH745>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 6.Aas T, Børresen-Dale A-L, Geisler S, Smith-Sørensen B, Johnsen H, Varhaug J E, Akslen L A, Lønning P E. Nat Med. 1996;2:811–814. doi: 10.1038/nm0796-811. [DOI] [PubMed] [Google Scholar]
- 7.Andersen T I, Børresen-Dale A-L. Diagn Mol Pathol. 1995;4:203–211. doi: 10.1097/00019606-199509000-00008. [DOI] [PubMed] [Google Scholar]
- 8.DeVries S, Gray J W, Pinkel D, Waldman F M, Sudar D. In: Current Protocols in Human Genetics, Supplement 6. Dracopoli N C, Haines J L, Korf B R, Morton C C, Seidman P E, Seidman J G, Smith D R, editors. New York: Wiley; 1995. , Unit 4.6, pp. 1–18. [Google Scholar]
- 9.Piper J, Rutovitz D, Sudar D, Kallioniemi A, Kallioniemi O P, Waldman F M, Gray J W, Pinkel D. Cytometry. 1995;19:10–26. doi: 10.1002/cyto.990190104. [DOI] [PubMed] [Google Scholar]
- 10.Carey R, Bell G, Marrin C. vrml, Virtual Reality Modeling Language. San Francisco: Aereal; 1997. [Google Scholar]
- 11.Kovach J S, Hartmann A, Blaszyk H, Cunningham J, Schaid D, Sommer S S. Proc Natl Acad Sci USA. 1996;93:1093–1096. doi: 10.1073/pnas.93.3.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geisler S, Lønning P E, Aas T, Johnsen H, Fluge Ø, Haugen D F, Liilehaug J R, Akslen L A, Børresen-Dale A-L. Cancer Res. 2001;61:2505–2512. [PubMed] [Google Scholar]
- 13.Bonilla M, Ramirez M, Lopez-Cueto J, Gariglio P. J Natl Cancer Inst. 1988;80:665–671. doi: 10.1093/jnci/80.9.665. [DOI] [PubMed] [Google Scholar]
- 14.Berns E M, Klijn JG, van Putten W L, van Staveren I L, Portengen H, Foekens J A. Cancer Res. 1992;52:1107–1113. [PubMed] [Google Scholar]
- 15.Roy B, Beamon J, Balint E, Reisman D. Mol Cell Biol. 1994;14:7805–7815. doi: 10.1128/mcb.14.12.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]