Abstract
The Ccr4-Not complex is a highly conserved nine-subunit protein complex that has been implicated in virtually all aspects of gene control, including transcription, mRNA decay and quality control, RNA export, translational repression and protein ubiquitylation. Understanding its mechanisms of action has been difficult due to the size of the complex and the fact that it regulates mRNAs and proteins at many levels in both the cytoplasm and the nucleus. Recently, biochemical and genetic studies on the yeast Ccr4-Not complex have revealed insights into its role in promoting elongation by RNA polymerase II. This review will describe what is known about the Ccr4-Not complex in regulating transcription elongation and its possible collaboration with other factors traveling with RNAPII across genes.
Keywords: Ccr4-Not, transcription elongation, histone methylation, Not4, TFIIS
Introduction
The Ccr4-Not complex was discovered in budding yeast and it is composed of nine “core” subunits: Not1, Not2, Not3, Not4/Mot2, Not5, Caf130, Caf40, Pop2/Caf1, and Ccr4 [1–3]. A larger version of the complex (>1.9 MDa) was identified by crude fractionation schemes of whole cell extracts on size exclusion columns that may contain Dhh1, Dbf2, Caf4, Caf16, and Btt1 in addition to the core subunits [4–7]. It is still unclear if the >1.9 MDa complex is a distinct entity since highly purified Ccr4-Not only contains the nine core subunits. The complex is conserved among eukaryotes, and metazoans contain multiple isoforms of the corresponding yeast subunits [1–3, 8, 9]. Human Ccr4-Not elutes at a molecular weight of ~1.0 MDa, but a >1.9 MDa version was detected also [9, 10]. The larger version of the human complex has not been characterized, and the predominant form is the smaller core complex.
Ccr4-Not was first considered a regulator of TFIID and transcription initiation, which was supported by a plethora of genetic, function and physical interactions with initiation factors (reviewed in [2, 3, 8]). For years, the only known function of Ccr4-Not was controlling transcription initiation. However, two groups made the surprising discovery that Ccr4 and Pop2/Caf1 are the major cytoplasmic mRNA deadenylases [11–13]. This was followed shortly after by studies confirming that this function is conserved in metazoans [14–16]. Now, new lines of evidence suggest it may play roles in controlling chromatin modifications, elongation, RNA export, nuclear mRNA surveillance, transcription coupled repair, translation and protein degradation (for review see [1, 2]). Remarkably, Ccr4-Not has been tied to just about every step in the regulation of a gene product, from mRNA synthesis to protein destruction. It is still not clear if Ccr4-Not is directly involved in all of these processes, however. Here, I will discuss its role in regulating different aspects of elongation and summarize known genetic and physical interactions that suggest it exists as a component of RNAPII elongation complexes in cells.
Genetic evidence for Ccr4-Not’s function in transcription elongation
The involvement of Ccr4-Not in the process of elongation emerged from genetic and phenotypic analysis of Ccr4-Not mutants by Clyde Denis and colleagues. Strains harboring mutations in CCR4, NOT1, NOT2, NOT4 and NOT5 were sensitive to the elongation inhibitors 6-azauracil (6-AU) and mycophenolic acid [17]. The sensitivity of mutants of complex subunits has been confirmed by others [18, 19]. Importantly, mutations in Xrn1 or exosome subunits, the major exonucleases used in mRNA decay, do not result in sensitivity to elongation inhibitors, suggesting that impaired mRNA decay does not account for the phenotype [17].
Sensitivity to elongation inhibitors is not considered sufficient evidence for impaired elongation because mutants in many genes not involved in transcription are sensitive to these drugs [20, 21]. Genetic interactions with known elongation factors, combined with drug sensitivity can make a compelling case for a gene’s function in elongation. Many elongation factor mutants have weak phenotypes on their own, but display lethality or enhanced phenotypes in combination with mutations in other elongation factors [22]. For simplicity, this will be referred to below as synthetic lethal interactions. Ccr4-Not mutants show synthetic lethal interactions with mutants of the elongation factors Paf1c, FACT (SPT16), TFIIS (DST1), Bur1/2 kinase and DSIF (SPT4 and SPT5) [17, 19, 23–25]. Furthermore, mutants of RNAPII, such as rpb9Δ and elongation deficient alleles of the two large subunits, RPB1 and RPB2, also display synthetic lethal interactions with Ccr4-Not mutations [17](Libert and Reese, unpublished). Furthermore, deleting CCR4 suppressed a cold sensitive allele of SPT5, spt5-242 [17]. The spt5-242 allele is unique in that it is suppressed by mutations in RNAPII and DST1 (TFIIS) that impede the rate of elongation [26]; thus, the suppression of the spt5-242 allele by deleting CCR4 implies Ccr4-Not promotes elongation in vivo.
Evidence for a role in promoting elongation in vivo
The role of Ccr4-Not in elongation has been examined only in budding yeast up to this point. Saccharomyces cerevisiae has small intron-less genes, compared to their metazoan counterparts, making it difficult to detect elongation defects at natural loci in living cells. To overcome this, reporter gene assays for measuring elongation were developed. Based on the premise that transcription through long-GC-rich regions is particularly sensitive to perturbations in the elongation process, the gene length-dependent accumulation of mRNA, or GLAM assay, was developed [27]. Another assay to detect elongation defects that has gained popularity in the field uses a long yeast gene driven by the GAL1 promoter, GAL1p-YLR454w [28]. RNAPII density is monitored by chromatin immunoprecipitation (ChIP) across the reporter under steady state (galactose) conditions and temporally after repression of the promoter by the addition of dextrose. Monitoring the “last wave” of RNAPII across the gene gives an estimation of elongation rate. However, a word of caution about relying only on these reporter assays is that both of these systems are not fool proof as mutation of some known elongation factors, such as TFIIS, do not display altered GLAM ratios or altered RNAPII density at GAL1p-YLR454w.
Using the GLAM assay, Aguilera and colleagues identified multiple Ccr4-Not complex mutants with altered GLAM ratios [18], suggesting an elongation defect. The GC-rich regions contained in the GLAM reporter genes impede elongation by forming R-loops, regions of RNA-DNA hybrids that occur during transcription [29–32]. Mutations in factors traveling with RNAPII during elongation, such as Hpr1 and other subunits of the TRO/TREX and Paf1c complexes show impaired elongation through these reporters similar to Ccr4-Not mutations [18]. As will be discussed below, Ccr4 co-purifies with Hpr1 and Paf1c subunits [24]. The THO/TREX complex is believed to suppress R-loop formation by binding to the transcript or assembling hnRNPs onto the RNA, and thus, preventing the RNA-DNA hybrids from forming [31]. Since Ccr4-Not binds to the transcript in elongation complexes [33], it is possible that Ccr4-Not promotes elongation through GC-rich regions by preventing R-loop formation during transcription. R-loop formation during transcription causes a phenomenon called transcription-associated recombination, TAR [30]. Mutation of genes suppressing R-loop formation during transcription show a hyperrecombination phenotype, including Paf1c and Hpr1 mutants; however, deleting CCR4 did not cause a hyperrecombination phenotype [24]. This might suggest that Ccr4-Not is not required to prevent the formation of R-loops during transcription. However, R-loop formation in Ccr4-Not mutants should be examined directly to determine if the impaired elongation in Ccr4-Not mutants is caused, at least in part, by R-loop formation.
Evidence for a role of Ccr4-Not in elongation comes from the analysis of RNAPII density across genes. Ccr4-Not mutants ccr4Δ, dhh1Δ (associates with Ccr4-Not complex) and not4Δ displayed a shift in RNAPII density across the GAL1p-YLR454w reporter gene consistent with an elongation defect [33]. RNAPII density was significantly higher at the 3′ end of the gene in the steady state, suggesting a piling up of RNAPII at the 3′ end. In addition, the last wave of RNAPII cleared the gene after transcriptional repression more slowly in Ccr4-Not mutants. This suggests that Ccr4-Not is required to promote RNAPII transcription through natural blocks in elongation. An important question is if the increase in RNAPII over the 3′ end of GAL1p-YLR454w is unique to the reporter gene or is prevalent throughout the genome. Evidence for the latter was provided by Chavez and colleagues who monitored RNAPII density across the open reading frame of 377 yeast genes in elongation factor mutants. Using a combined transcription run-on-DNA macroarray technique they found a modest, but statistically significant, accumulation of run-on signal at the 3′ ends of genes in a ccr4Δ mutant [34]. This indicates that RNAPII accumulated at the 3′ ends of genes in this mutant. In contrast, a significant difference in RNAPII density in the not5Δ mutant was not observed. The reason for the difference between the ccr4Δ and not5Δ mutant is not clear. Nonetheless, this study, like the reporter gene assays, suggests that Ccr4-Not affects the elongation of RNAPII in vivo.
Physical interaction with RNAPII during elongation
Ccr4p was found to co-purify with RNAPII, Hpr1 and subunits of the Paf1c complex in crude extracts on GST-Cdc73 and GST-Paf1 beads [24]. This was initially thought to be a distinct form of RNAPII that lacked mediator. However, an alternative interpretation is that these subcomplexes or individual interactions may have arisen from fragmenting native elongation complexes. Most notably, other subunits of the Ccr4-Not, such as the NOT proteins, did not copurify with Paf1c and Hpr1. Nonetheless, this was an important observation because it suggested that Ccr4-Not components bind to Paf1c and other elongation factors. The interaction between Ccr4-Not and RNAPII was revisited recently. Multiple Ccr4-Not subunits and Dhh1 was shown to immunoprecipitate with RNAPII in an RNA-independent manner [33]. The binding of Dhh1 to RNAPII required an intact Ccr4-Not complex, because the interaction was lost in extracts from Ccr4-Not mutants. Furthermore, the form of RNAPII co-immunoprecipitated with Ccr4-Not was distinct from the version described by Chang and colleagues that contained Hpr1 and Paf1c subunits, because deleting two integral subunits of the Paf1c complex [25], Cdc73 and Paf1, did not affect the interaction between Ccr4-Not and RNAPII [33]. These results reinforced the notion that the Ccr4-Not complex, and not just Ccr4, interacts with RNAPII in solution.
Co-immunoprecipitation studies indicate that Ccr4-Not co-purifies with RNAPII in crude extracts, but this does not mean the interaction is direct. In addition, interaction in solution does not mean that it can bind RNAPII assembled into elongation complexes (ECs). Kruk and colleagues used a strategy to prepare radiolabeled ECs from highly purified yeast RNAPII to show that Ccr4-Not directly interacted with functional elongation complexes [33]. The interaction was not dependent on nucleic acids, because Ccr4-Not could not bind to ECs built from Drosophila or archeal RNAP. This suggests that that Ccr4-Not is making direct contact with subunits of RNAPII.
The next question is what features of RNAPII recruit Ccr4-Not into the EC. Many factors that associate with RNAPII during elongation do so by binding the carboxy-terminal domain (CTD) of the large subunit. However, using a strain co-expressing a CTD-less version of Rpb1, it was shown that the CTD is not required for the interaction [33]. An attractive candidate to consider is the Rpb4/7 module of RNAPII. Ccr4-Not and rpb4Δ mutants share many phenotypes, such as stress sensitivity, 6-AU sensitivity, impaired transcription through long GC-rich genes (GLAM phenotypes), impaired transcription coupled (TCR) repair and reduced deadenylation and decay of mRNAs (for review see [2, 35]). Furthermore, evidence is mounting that Rpb4/7 is important for linking the processes of mRNA synthesis and decay [36–38]. Initially, the involvement of the Rpb4/7 module in the association between Ccr4-Not and RNAPII was addressed in a crude manner by analyzing the ability of Ccr4-Not to co-immunoprecipitiate with RNAPII in an rpb4Δ strain, which suggested that contact with the Rpb4/7 module is not important for its association with polymerase [33]. However, it is possible that Rpb7 associates with RNAPII in crude extracts in rpb4Δ mutant [39] or that contacts with the Rpb4/7 module is more important for recruiting Ccr4-Not into ECs but not for their interaction in solution. The requirement for the Rpb4/7 module was revisited recently in a more sophisticated way. The ability of Ccr4-Not to bind to elongation complexes (ECs) formed from highly purified RNAPII lacking the Rpb4/7 module was examined using a gel mobility shift assay, and the results indicate that the Rpb4/7 module is required for the stable association of Ccr4-Not with the EC (Babbarwal and Reese, unpublished). The involvement of Rpb4/7 in recruiting Ccr4-Not into the EC is further suggested by the observation that a subunit of Ccr4-Not crosslinks to the transcript within the EC [33]. The Rpb4/7 module is located near the RNA exit channel and Rpb7 crosslinks to RNA [40–42]. This places Ccr4-Not and Rpb4/7 in close proximity to each other in the EC. This interesting connection between Ccr4-Not and the Rpb4/7 dimer may provide a mechanism for how the synthesis and decay of mRNAs are linked [36, 38, 43].
Ccr4-Not subunits are recruited to transcriptionally active genes, suggesting their direct involvement in the transcription process [19, 33, 44, 45]. At PYK1 and ARG1, recruitment was observed over the promoter or UAS regions [19, 44]. In the latter case, it was proposed that the activator Gcn4 recruits Ccr4-Not, because Ccr4-Not subunits were retained from cell extracts on GST-Gcn4 affinity columns [44]. On the other hand, Ccr4-Not crosslinks strongly to the open reading frame of the GAL1 and RNR3 genes, but not to the upstream regulatory regions, suggesting its recruitment is dependent on RNAPII and not the activators that bind upstream of the start site of transcription [33]. The model that RNAPII recruits Ccr4-Not to genes is consistent with the ability of Ccr4-Not to bind functional ECs in vitro. In the case of RNR3, it was uniformly recruited across the ORF and did not display obvious polarity that would indicate it is preferentially recruited by a specific form of modified RNAPII that is generated during the “elongation cycle”. It is possible that Ccr4-Not can be recruited to genes by both activators and RNAPII elongation complexes in a gene specific manner. However, a ChIP-seq study concluded that Ccr4-Not is crosslinked to the open reading frames of genes similar to other elongation factors on a genome-wide scale [45]. Thus, the localization of Ccr4-Not across genes is consistent with it acting in elongation and its recruitment to genes is likely due to a direct interaction with RNAPII or other factors in the elongation complex.
RNAPII is sufficient to recruit Ccr4-Not into ECs in vitro [33]. However, this does not preclude the possibility that other elongation factors affect its recruitment to actively transcribed genes in cells. As discussed above, Ccr4 copurified with subunits of the Paf1c and THO/TREX complexes [24]. Furthermore, a two hybrid screen using a fragment of Ccr4 as bait identified Spt5 as an interacting protein, and a direct interaction between purified Spt5 and Ccr4 and Caf1/Pop2 was observed [46]. Spt4/5 (DSIF) is known to serve as platform to recruit many RNA processing factors to the EC during transcription (for review [47]). In addition, it has been implicated recently in recruiting RNA binding proteins during transcription, potentially “marking” mRNAs for control in the cytoplasm [48]. Ccr4-Not has functions in the nucleus and cytoplasm, and it has been proposed that it marks mRNAs for future regulation in the cytoplasm [2, 35]. Thus, the idea that Spt4/5 participates in the loading of Ccr4-Not onto genes is worth considering.
An interesting genetic connection between Not4 and Spt5 can be made through the Bur1/2 kinase. Bur1/2 phosphorylates Spt5 [49–51] and Not4 and Bur1/2 function in parallel pathways to regulate stress resistance and histone H3 K4 methylation [19, 49]. Even though Not4 is a phosphoprotein, this connection does not involve Bur1/2-dependent phosphorylation of Not4; thus, additional biochemical and molecular analysis will be necessary to reveal the nature of the interactions between Not4, Bur1/2 and Spt5.
Direct biochemical evidence suggests Ccr4-Not prevents arrest or reactivates backtracked RNAPII complexes
Studies utilizing the ChIP assay indicate that many protein complexes track with RNAPII across genes in vivo. Only a subset of these directly affect RNAPII elongation, while others may be involved in modifying chromatin, RNA processing, packaging the RNA into mRNPs and export. Dissecting the contributions of these proteins in elongation control is not trivial, but the development of biochemical assays to study elongation is starting to provide the means to do so. Affinity purification strategies, such as tandem-affinity purification (TAP-tag), have facilitated biochemical studies on elongation by providing a method to easily isolate highly purified factors from source. Using a simplified biochemical elongation system, a case has been made that Ccr4-Not is a direct regulator of RNAPII elongation. The addition of Ccr4-Not rescued backtracked RNAPII arrested at the end of a G-less cassette [33]. Interestingly, by changing the order of addition of various reaction components, the authors provided evidence that Ccr4-Not had no detectable effect on the transcription rate of unarrested RNAPII, suggesting that it specifically acts on arrested complexes. However, it is possible that Ccr4-Not has a subtle affect on the elongation rate of RNAPII that was not detected under the experimental conditions utilized in that study.
The ability of Ccr4-Not to rescue backtracked RNAPII complexes is similar to that of the well-studied elongation factor TFIIS. Backtracking causes the displacement of the 3′ OH group on the growing RNA chain out of the active site of RNAPII, causing it to thread into the secondary channel or “funnel”[52]. TFIIS reactivates backtracked complexes by inserting its acidic C-terminal domain into the secondary channel, stimulating the intrinsic nucleolytic activity of RNAPII and causing the removal of the displaced RNA strand [22, 52, 53]. The most straightforward mechanism of action of Ccr4-Not would be to stimulate transcript cleavage like TFIIS. However, this appears not to be the case. Instead, a model was proposed where Ccr4-Not realigns the 3′ end of the RNA into the active site by trapping the transient forward excursions of RNAPII over the arrest site by reiterative rounds of transcript release and binding. The binding of Ccr4-Not to the transcript restricts the movement of the RNA through the exit channel, moving RNAPII forward in a ratchet-like motion (Figure 1). This model was supported by two observations. The first is that crosslinking studies showed that a subunit of Ccr4-Not binds to the transcript in the EC. The second is that the ability of Ccr4-Not to rescue backtracked RNAPII was dependent upon the length of the transcript, suggesting that reducing the contact between Ccr4-Not and the transcript impaired the reactivation of the arrested RNAPII.
Figure 1. Ccr4-Not rescues RNAPII from arrest.
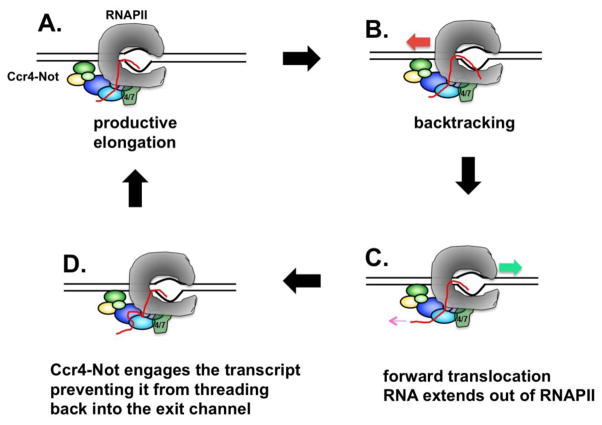
RNAPII in the productive elongation phase (A) encounters a block in transcription resulting in backtracking (B). RNAPII undergoes transient forward excursions over the arrest site, causing movement of the transcription bubble and the threading of RNA out of the exit channel (C). Reiterative transcript binding and release by Ccr4–Not causes realignment of the 3′ end of the RNA into the active site and promotes the resumption of elongation (D).
While engaging the transcript is an attractive model for how Ccr4-Not reactivates backtracked complexes, other actions on the ECs cannot be ruled out. Ccr4-Not could change the structure of RNAPII into a form that favors forward translocation. It was noted that the addition of Ccr4-Not to ECs increased the crosslinking of Rpb1 to the transcript. Whether or not the increased crosslinking of Rpb1 in this assay reflects a conformational change in RNAPII that enhances its association with the transcript remains to be seen. There is still much to be learned about how Ccr4-Not affects the RNAPII ECs. Nonetheless, Ccr4-Not joins a surprisingly small number of putative elongation factors that has been shown to directly stimulate elongation, especially those capable of rescuing arrested RNAPII complexes.
Ccr4-Not regulates histone H3 K4 methylation through Jhd2
The Ccr4-Not complex has been connected to the process of elongation through histone H3 K4 trimethylation (H3 K4me3). H3 K4me3 is a co-transcriptional modification that correlates with transcription levels and it is considered a signature of transcription elongation [54, 55]. The Timmer’s group first reported that deletion of NOT group genes, especially NOT4, led to global reductions in H3K4me3 [19]. This was shortly followed by another story published by the Strahl group, which also linked the proteasome to the Ccr4-Not complex [56]. Earlier biochemical studies determined that the Ccr4-Not complex is modular, separated into the “Ccr4-group” of genes, defined as CCR4 and CAF1, and the “Not-group” (for review see [3, 57]). Interestingly, the reduction in H3K4me3 is a phenotype specific to the NOT group mutants, because deleting CCR4 or CAF1 did not reduce H3K4me3 [19]. This also suggests that the reduced histone methylation in the not4Δ mutant is not the result of impaired deadenylation and mRNA decay because Ccr4 is the catalytic subunit required for the mRNA decay functions of Ccr4-Not. Importantly, evidence was provided that deleting NOT4 did not affect the levels of the subunits or activity of COMPASS, the H3 K4 histone methyltransferase complex in yeast [19, 56].
Both groups proposed different mechanisms for how Ccr4-Not regulates H3 K4me3. Timmers and colleagues attributed the H3K4me3 defect to reduced Paf1c recruitment and histone H2b ubiquitylation (H2b-Ub), a prerequisite for transcription-linked histone modifications [19]. On the other hand, the Strahl group did not observe reduced Paf1c recruitment and, if anything, slightly increased H2B-Ub levels in a not4Δ mutant [56]. Instead, they attributed the regulation of H3K4me3 by Ccr4-Not to its association with the proteasome. The proteasome had previously been suggested to regulate H3K4me3 [58]. The regulation of H3 K4me3 by Ccr4-Not thorough the Paf1c complex is consistent with physical and genetic interactions between the Ccr4-Not and Paf1c complex subunits, but contrary to the result that NOT mutants do not display defects in H3 K79 methylation, which is also dependent upon Paf1c and H2B-Ub. The differences between the two studies have not been resolved.
While the aforementioned mechanisms cannot be ruled out, a very attractive model has emerged recently for how Not4 regulates H3 K4me3 through protein destruction. Not4 is an E3 RING-domain ubiquitin ligase [59–62]. The E2 enzymes that function with Not4 are Ubc4 and Ubc5, which appear to function redundantly [61]. Only a few substrates of Not4 are known, but include the transcriptional activator Yap1, cyclin C (Srb 11 mediator subunit), DNA polymerase alpha, NAC and Jhd2 [2, 59, 63–66].
Jhd2 is the only known H3 K4me3-specific histone demethylase in yeast [67–69]. Strong genetic and biochemical evidence indicates that Ccr4-Not, especially Not4, controls H3 K4me3 by regulating the stability of Jhd2 [59, 70]. Jhd2 levels increased in a not4Δ mutant, resulting in enhanced demethylation of H3 K4me3. This mechanism was supported by the genetic observation that deleting JHD2 suppressed the reduced methylation in a not4Δ background [59]. Mersman and colleagues showed that human Not4, CNOT4, can polyubiquitylate JARID1C, the human version of Jhd2 [59], indicating that this function is conserved in eukaryotes. Using a mutant of Not4 lacking the entire RING domain (not4-RINGΔ), it was determined that the regulation of H3K4me3 and Jhd2 turnover requires the RING domain, and thus, its E3 ubiquitin ligase activity [56, 59]. However, another study reported that a strain containing a single point mutant in the RING domain of Not4 (L35A) did not display reduced H3K4me3 levels like a not4Δ or a not4-RINGΔ mutant [61]. The first thought one might have is that the single mutation did not destroy ubiquitin ligase activity. However, the single point mutation abolished the interaction between Not4 and the E2 ligases Ubc4 and 5 in a two-hybrid assay and inactivated the auto-ubiquitylation activity of Not4 [61]. The L35A allele displays weaker phenotypes compared to those of the deletion mutation, and a plausible explanation for this is that the L35A mutation is a hypomorphic allele that retains some functions in cells. On the other hand, deleting the entire RING domain could impact more than the ubiquitin ligase activity of Not4. The differences between the RING deletion and point mutants should be examined further. Nonetheless, there is solid evidence that Ccr4-Not regulates elongation-linked histone modifications by regulating Jhd2 stability.
Since Ccr4-Not associates with elongating RNAPII [33], it may ubiquitylate and destroy Jhd2 locally during transcription (Figure 2). This would increase H3 K4me3 levels and promote the recruitment of factors that recognize the trimethyl lysine modifcation. The role of H3 K4me3 in yeast is still not clear, although it correlates with transcription levels (for a recent review see [71]). Therefore, it is difficult to predict the consequences of the increased H3 K4me3 resulting from Jhd2 destruction.
Figure 2. Ccr4-Not promotes transcription through nucleosomes by regulating histone modifications and rescuing arrested RNAPII.
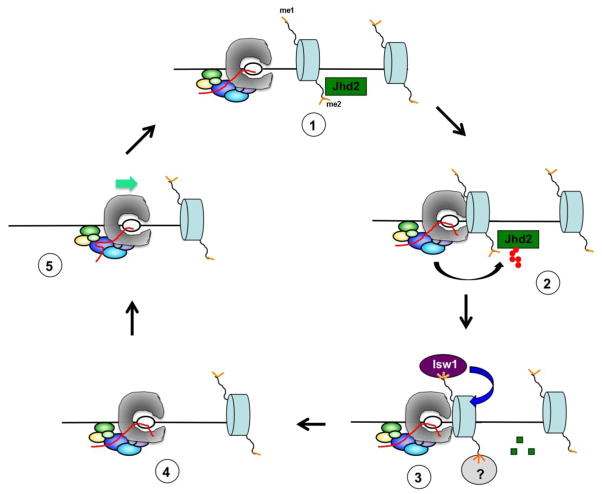
The elongation complex encounters a nucleosome with low H3 K4me3 levels caused by the presence of Jhd2. Jhd2 is polyubiquitylated by Not4 and ultimately destroyed by the proteasome. This leads to an increase in H3K4me3 and recruitment of nucleosome remodeling activities, removing the nucleosomal barrier. The collision of RNAPII with the nucleosome caused backtracking, which is then relieved by Ccr4-Not.
It should not be overlooked that Not4 has substrates other than Jhd2 and may target other proteins for destruction during transcription. For example, the stress-induced destruction of cyclin C (Srb11), a subunit of the mediator, requires Not4 [65]. An interaction between Cdk8/CycC (Srb10/11) and Ccr4-Not has been described [6]. Recently, mediator has been shown to participate in elongation in yeast and metazoans [72, 73]. Thus, the ability of Not4 to target CycC may have consequences on elongation. Furthermore, since Ccr4-Not is required to activate arrested RNAPII and has an undefined role in transcription-coupled repair [18, 33], Not4-dependent ubiquitylation may be involved in these processes too. It would be interesting to test if Not4 can ubiquitylate RNAPII or other elongation factors in response to cellular stress.
Conclusions
Ccr4-Not displays many genetic and physical interactions with a number of elongation factors (see above and [1, 2]). The next challenge is to untangle the genetic interactions and confirm that the physical associations are functionally relevant. Many of the interactions that have been described between Ccr4-Not and subunits of known elongation and mRNA processing factors may originate from their residence within a large super-elongation complex transcribing across genes with RNAPII. Other interactions, such as those with hnRNPs and nuclear pore complex protein, may represent transient interactions between Ccr4-Not, RNAPII and the machineries that package mRNA for export into the cytoplasm [74, 75]. It will be especially difficult to trace back the network of the interactions between Ccr4-Not and other elongation factors in cells. The predominant approach used in the field is to examine the recruitment of a particular protein to genes using ChIP in a mutant of another elongation factor. However, most of these mutations reduce RNAPII levels at genes and it is difficult to determine if the loss of Ccr4-Not recruitment is due to absence of a particular elongation factor or reduced RNAPII. Reconstitution assays using purified factors will be required to address these questions. But even here, the challenge will be to resolve and isolate huge macromolecular machineries formed from large multi-subunit complexes.
Based on the observations that Ccr4-Not rescues arrested RNAPII in vitro and mutants of the complex show impaired elongation in vivo, it seems feasible that Ccr4-Not is required to prevent elongation arrest across genes, especially at nucleosomes. Application of a novel nascent transcript sequencing (NET-Seq) method confirmed biochemical studies indicating that the nucleosome is a major barrier causing transcription arrest in cells [76, 77]. Furthermore, it was shown that preventing RNAPII backtracking by hybridizing an antisense oligonucleotide to the emerging transcript promoted transcription through a nucleosome [77]. The RNA-DNA duplex prevented the movement of the RNA transcript from threading back into RNAPII. The binding of Ccr4-Not to the transcript (see above) would likewise prevent the RNA from being threaded into the exit channel and prevent backtracking or favor forward translocation of RNAPII in a similar fashion.
If Ccr4-Not is required to resume RNAPII elongation after nucleosome-induced arrest, a model can be constructed to connect its regulation of H3 K4me to its ability to transcribe through nucleosomes. H3 K4me stimulates the recruitment of the ATP-dependent chromatin remodeler Isw1 to genes, and Isw1 remodels nucleosomes within the coding region during elongation [78–81]. As the EC containing Ccr4-Not and RNAPII encounters a nucleosome, Not4 ubiquitylates Jhd2. Ubiquitylation of Jhd2 leads to its destruction and results in increased H3 K4me3 in the region. This in turn, recruits the Isw1 complex, or another transcription factor complex, to the nucleosome to promote remodeling. Ccr4-Not remains engaged with the transcript in the EC to promote resumption of elongation after the nucleosome has been remodeled or removed (Figure 2).
Highlights.
Ccr4-Not has many genetic and physical interactions with RNAPII elongation factors
Ccr4-Not associates with RNAPII in solution and within functional elongation complexes
Ccr4-Not promotes elongation through genes in vivo
Ccr4-Not directly activates arrested RNAPII
Not4 regulates co-transcriptional histone modifications through the destruction of Jhd2
Acknowledgments
Jason Miller is acknowledged for critically reading this work. This research is the Reese lab is supported by funds from National Institutes of Health (GM58672) to J.C.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Collart MA, Panasenko OO. The Ccr4-Not complex. Gene. 2011 doi: 10.1016/j.gene.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 2.Miller JE, Reese JC. Ccr4-Not complex: the control freak of eukaryotic cells. Critical reviews in biochemistry and molecular biology. 2012;47:315–333. doi: 10.3109/10409238.2012.667214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denis CL, Chen J. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog Nucleic Acid Res Mol Biol. 2003;73:221–250. doi: 10.1016/s0079-6603(03)01007-9. [DOI] [PubMed] [Google Scholar]
- 4.Maillet L, Collart MA. Interaction between Not1p, a component of the Ccr4-not complex, a global regulator of transcription, and Dhh1p, a putative RNA helicase. J Biol Chem. 2002;277:2835–2842. doi: 10.1074/jbc.M107979200. [DOI] [PubMed] [Google Scholar]
- 5.Liu HY, Toyn JH, Chiang YC, Draper MP, Johnston LH, Denis CL. DBF2, a cell cycle-regulated protein kinase, is physically and functionally associated with the CCR4 transcriptional regulatory complex. Embo J. 1997;16:5289–5298. doi: 10.1093/emboj/16.17.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu HY, Chiang YC, Pan J, Chen J, Salvadore C, Audino DC, Badarinarayana V, Palaniswamy V, Anderson B, Denis CL. Characterization of CAF4 and CAF16 reveals a functional connection between the CCR4-NOT complex and a subset of SRB proteins of the RNA polymerase II holoenzyme. J Biol Chem. 2001;276:7541–7548. doi: 10.1074/jbc.M009112200. [DOI] [PubMed] [Google Scholar]
- 7.Cui Y, Ramnarain DB, Chiang YC, Ding LH, McMahon JS, Denis CL. Genome wide expression analysis of the CCR4-NOT complex indicates that it consists of three modules with the NOT module controlling SAGA-responsive genes. Mol Genet Genomics. 2008;279:323–337. doi: 10.1007/s00438-007-0314-1. [DOI] [PubMed] [Google Scholar]
- 8.Collart MA, Timmers HT. The eukaryotic Ccr4-not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog Nucleic Acid Res Mol Biol. 2004;77:289–322. doi: 10.1016/S0079-6603(04)77008-7. [DOI] [PubMed] [Google Scholar]
- 9.Lau NC, Kolkman A, van Schaik FM, Mulder KW, Pijnappel WW, Heck AJ, Timmers HT. Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem J. 2009;422:443–453. doi: 10.1042/BJ20090500. [DOI] [PubMed] [Google Scholar]
- 10.Morel AP, Sentis S, Bianchin C, Le Romancer M, Jonard L, Rostan MC, Rimokh R, Corbo L. BTG2 antiproliferative protein interacts with the human CCR4 complex existing in vivo in three cell-cycle-regulated forms. J Cell Sci. 2003;116:2929–2936. doi: 10.1242/jcs.00480. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Rappsilber J, Chiang YC, Russell P, Mann M, Denis CL. Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J Mol Biol. 2001;314:683–694. doi: 10.1006/jmbi.2001.5162. [DOI] [PubMed] [Google Scholar]
- 12.Liu HY, Badarinarayana V, Audino DC, Rappsilber J, Mann M, Denis CL. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. Embo J. 1998;17:1096–1106. doi: 10.1093/emboj/17.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 14.Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. Embo J. 2004;23:2862–2871. doi: 10.1038/sj.emboj.7600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang TC, Yamashita A, Chen CY, Yamashita Y, Zhu W, Durdan S, Kahvejian A, Sonenberg N, Shyu AB. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes Dev. 2004;18:2010–2023. doi: 10.1101/gad.1219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 17.Denis CL, Chiang YC, Cui Y, Chen J. Genetic evidence supports a role for the yeast CCR4-NOT complex in transcriptional elongation. Genetics. 2001;158:627–634. doi: 10.1093/genetics/158.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaillard H, Tous C, Botet J, Gonzalez-Aguilera C, Quintero MJ, Viladevall L, Garcia-Rubio ML, Rodriguez-Gil A, Marin A, Arino J, Revuelta JL, Chavez S, Aguilera A. Genome-wide analysis of factors affecting transcription elongation and DNA repair: a new role for PAF and Ccr4-not in transcription-coupled repair. PLoS Genet. 2009;5:e1000364. doi: 10.1371/journal.pgen.1000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulder KW, Brenkman AB, Inagaki A, van den Broek NJ, Timmers HT. Regulation of histone H3K4 tri-methylation and PAF complex recruitment by the Ccr4-Not complex. Nucleic Acids Res. 2007;35:2428–2439. doi: 10.1093/nar/gkm175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riles L, Shaw RJ, Johnston M, Reines D. Large-scale screening of yeast mutants for sensitivity to the IMP dehydrogenase inhibitor 6-azauracil. Yeast. 2004;21:241–248. doi: 10.1002/yea.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desmoucelles C, Pinson B, Saint-Marc C, Daignan-Fornier B. Screening the yeast “disruptome” for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid. J Biol Chem. 2002;277:27036–27044. doi: 10.1074/jbc.M111433200. [DOI] [PubMed] [Google Scholar]
- 22.Arndt KM, Kane CM. Running with RNA polymerase: eukaryotic transcript elongation. Trends Genet. 2003;19:543–550. doi: 10.1016/j.tig.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Biswas D, Yu Y, Mitra D, Stillman DJ. Genetic interactions between Nhp6 and Gcn5 with Mot1 and the Ccr4-Not complex that regulate binding of TATA-binding protein in Saccharomyces cerevisiae. Genetics. 2006;172:837–849. doi: 10.1534/genetics.105.050245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang M, French-Cornay D, Fan HY, Klein H, Denis CL, Jaehning JA. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol Cell Biol. 1999;19:1056–1067. doi: 10.1128/mcb.19.2.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaehning JA. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim Biophys Acta. 2010;1799:379–388. doi: 10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morillo-Huesca M, Vanti M, Chavez S. A simple in vivo assay for measuring the efficiency of gene length-dependent processes in yeast mRNA biogenesis. FEBS J. 2006;273:756–769. doi: 10.1111/j.1742-4658.2005.05108.x. [DOI] [PubMed] [Google Scholar]
- 28.Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 29.Dominguez-Sanchez MS, Barroso S, Gomez-Gonzalez B, Luna R, Aguilera A. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet. 2011;7:e1002386. doi: 10.1371/journal.pgen.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Molecular cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 31.Luna R, Rondon AG, Aguilera A. New clues to understand the role of THO and other functionally related factors in mRNP biogenesis. Biochim Biophys Acta. 2012;1819:514–520. doi: 10.1016/j.bbagrm.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 32.Rondon AG, Jimeno S, Aguilera A. The interface between transcription and mRNP export: from THO to THSC/TREX-2. Biochim Biophys Acta. 2010;1799:533–538. doi: 10.1016/j.bbagrm.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 33.Kruk JA, Dutta A, Fu J, Gilmour DS, Reese JC. The multifunctional Ccr4-Not complex directly promotes transcription elongation. Genes Dev. 2011;25:581–593. doi: 10.1101/gad.2020911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez-Gil A, Garcia-Martinez J, Pelechano V, de Munoz-Centeno LM, Geli V, Perez-Ortin JE, Chavez S. The distribution of active RNA polymerase II along the transcribed region is gene-specific and controlled by elongation factors. Nucleic Acids Res. 2010;38:4651–4664. doi: 10.1093/nar/gkq215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choder M. mRNA imprinting: Additional level in the regulation of gene expression. Cell Logist. 2011;1:37–40. doi: 10.4161/cl.1.1.14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dori-Bachash M, Shema E, Tirosh I. Coupled evolution of transcription and mRNA degradation. PLoS Biol. 2011;9:e1001106. doi: 10.1371/journal.pbio.1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shalem O, Groisman B, Choder M, Dahan O, Pilpel Y. Transcriptome kinetics is governed by a genome-wide coupling of mRNA production and degradation: a role for RNA Pol II. PLoS Genet. 2011;7:e1002273. doi: 10.1371/journal.pgen.1002273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez-Ortin JE, de Miguel-Jimenez L, Chavez S. Genome-wide studies of mRNA synthesis and degradation in eukaryotes. Biochim Biophys Acta. 2012;1819:604–615. doi: 10.1016/j.bbagrm.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 39.Sheffer A, Varon M, Choder M. Rpb7 can interact with RNA polymerase II and support transcription during some stresses independently of Rpb4. Mol Cell Biol. 1999;19:2672–2680. doi: 10.1128/mcb.19.4.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cramer P, Armache KJ, Baumli S, Benkert S, Brueckner F, Buchen C, Damsma GE, Dengl S, Geiger SR, Jasiak AJ, Jawhari A, Jennebach S, Kamenski T, Kettenberger H, Kuhn CD, Lehmann E, Leike K, Sydow JF, Vannini A. Structure of eukaryotic RNA polymerases. Annu Rev Biophys. 2008;37:337–352. doi: 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- 41.Ujvari A, Luse DS. RNA emerging from the active site of RNA polymerase II interacts with the Rpb7 subunit. Nat Struct Mol Biol. 2006;13:49–54. doi: 10.1038/nsmb1026. [DOI] [PubMed] [Google Scholar]
- 42.Chen CY, Chang CC, Yen CF, Chiu MT, Chang WH. Mapping RNA exit channel on transcribing RNA polymerase II by FRET analysis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:127–132. doi: 10.1073/pnas.0811689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwalb B, Schulz D, Sun M, Zacher B, Dumcke S, Martin DE, Cramer P, Tresch A. Measurement of genome-wide RNA synthesis and decay rates with Dynamic Transcriptome Analysis (DTA) Bioinformatics. 2012;28:884–885. doi: 10.1093/bioinformatics/bts052. [DOI] [PubMed] [Google Scholar]
- 44.Swanson MJ, Qiu H, Sumibcay L, Krueger A, Kim SJ, Natarajan K, Yoon S, Hinnebusch AG. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol Cell Biol. 2003;23:2800–2820. doi: 10.1128/MCB.23.8.2800-2820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck-Walsh C, Pugh BF. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Molecular cell. 2011;41:480–492. doi: 10.1016/j.molcel.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui Y-CCY, Viswanathan Palaniswamy, Lee Darren J, Denis Clyde L. SPT5 affects the rate of mRNA degradation and physically interacts with CCR4 but does not control mRNA deadenylation. American Journal of Molecular Biology. 2012;2:11–20. doi: 10.4236/ajmb.2012.21002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Werner F. A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J Mol Biol. 2012;417:13–27. doi: 10.1016/j.jmb.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen Z, St-Denis A, Chartrand P. Cotranscriptional recruitment of She2p by RNA pol II elongation factor Spt4-Spt5/DSIF promotes mRNA localization to the yeast bud. Genes Dev. 2010;24:1914–1926. doi: 10.1101/gad.1937510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lau NC, Mulder KW, Brenkman AB, Mohammed S, van den Broek NJ, Heck AJ, Timmers HT. Phosphorylation of Not4p functions parallel to BUR2 to regulate resistance to cellular stresses in Saccharomyces cerevisiae. PLoS One. 2010;5:e9864. doi: 10.1371/journal.pone.0009864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou K, Kuo WH, Fillingham J, Greenblatt JF. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6956–6961. doi: 10.1073/pnas.0806302106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y, Warfield L, Zhang C, Luo J, Allen J, Lang WH, Ranish J, Shokat KM, Hahn S. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol Cell Biol. 2009;29:4852–4863. doi: 10.1128/MCB.00609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 53.Fish RN, Kane CM. Promoting elongation with transcript cleavage stimulatory factors. Biochim Biophys Acta. 2002;1577:287–307. doi: 10.1016/s0167-4781(02)00459-1. [DOI] [PubMed] [Google Scholar]
- 54.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 55.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 56.Laribee RN, Shibata Y, Mersman DP, Collins SR, Kemmeren P, Roguev A, Weissman JS, Briggs SD, Krogan NJ, Strahl BD. CCR4/NOT complex associates with the proteasome and regulates histone methylation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:5836–5841. doi: 10.1073/pnas.0607996104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collart MA. Global control of gene expression in yeast by the Ccr4-Not complex. Gene. 2003;313:1–16. doi: 10.1016/s0378-1119(03)00672-3. [DOI] [PubMed] [Google Scholar]
- 58.Ezhkova E, Tansey WP. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Molecular cell. 2004;13:435–442. doi: 10.1016/s1097-2765(04)00026-7. [DOI] [PubMed] [Google Scholar]
- 59.Mersman DP, Du HN, Fingerman IM, South PF, Briggs SD. Polyubiquitination of the demethylase Jhd2 controls histone methylation and gene expression. Genes Dev. 2009;23:951–962. doi: 10.1101/gad.1769209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winkler GS, Albert TK, Dominguez C, Legtenberg YI, Boelens R, Timmers HT. An altered-specificity ubiquitin-conjugating enzyme/ubiquitin-protein ligase pair. J Mol Biol. 2004;337:157–165. doi: 10.1016/j.jmb.2004.01.031. [DOI] [PubMed] [Google Scholar]
- 61.Mulder KW, Inagaki A, Cameroni E, Mousson F, Winkler GS, De Virgilio C, Collart MA, Timmers HT. Modulation of Ubc4p/Ubc5p-mediated stress responses by the RING-finger-dependent ubiquitin-protein ligase Not4p in Saccharomyces cerevisiae. Genetics. 2007;176:181–192. doi: 10.1534/genetics.106.060640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Albert TK, Hanzawa H, Legtenberg YI, de Ruwe MJ, van den Heuvel FA, Collart MA, Boelens R, Timmers HT. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. Embo J. 2002;21:355–364. doi: 10.1093/emboj/21.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Panasenko OO, David FP, Collart MA. Ribosome association and stability of the nascent polypeptide-associated complex is dependent upon its own ubiquitination. Genetics. 2009;181:447–460. doi: 10.1534/genetics.108.095422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gulshan K, Thommandru B, Moye-Rowley WS. Proteolytic degradation of the Yap1 transcription factor is regulated by subcellular localization and the E3 ubiquitin ligase Not4. J Biol Chem. 2012 doi: 10.1074/jbc.M112.384719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cooper KF, Scarnati MS, Krasley E, Mallory MJ, Jin C, Law MJ, Strich R. Oxidative-stress-induced nuclear to cytoplasmic relocalization is required for Not4-dependent cyclin C destruction. J Cell Sci. 2012;125:1015–1026. doi: 10.1242/jcs.096479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haworth J, Alver RC, Anderson M, Bielinsky AK. Ubc4 and Not4 regulate steady-state levels of DNA polymerase-alpha to promote efficient and accurate DNA replication. Mol Biol Cell. 2010;21:3205–3219. doi: 10.1091/mbc.E09-06-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liang G, Klose RJ, Gardner KE, Zhang Y. Yeast Jhd2p is a histone H3 Lys4 trimethyl demethylase. Nature structural & molecular biology. 2007;14:243–245. doi: 10.1038/nsmb1204. [DOI] [PubMed] [Google Scholar]
- 68.Seward DJ, Cubberley G, Kim S, Schonewald M, Zhang L, Tripet B, Bentley DL. Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nature structural & molecular biology. 2007;14:240–242. doi: 10.1038/nsmb1200. [DOI] [PubMed] [Google Scholar]
- 69.Tu S, Bulloch EM, Yang L, Ren C, Huang WC, Hsu PH, Chen CH, Liao CL, Yu HM, Lo WS, Freitas MA, Tsai MD. Identification of histone demethylases in Saccharomyces cerevisiae. J Biol Chem. 2007;282:14262–14271. doi: 10.1074/jbc.M609900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang F, Chandrasekharan MB, Chen YC, Bhaskara S, Hiebert SW, Sun ZW. The JmjN domain of Jhd2 is important for its protein stability, and the plant homeodomain (PHD) finger mediates its chromatin association independent of H3K4 methylation. J Biol Chem. 2010;285:24548–24561. doi: 10.1074/jbc.M110.117333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kremer SB, Kim S, Jeon JO, Moustafa YW, Chen A, Zhao J, Gross DS. Role of Mediator in Regulating Pol II Elongation and Nucleosome Displacement in Saccharomyces cerevisiae. Genetics. 2012;191:95–106. doi: 10.1534/genetics.111.135806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, Florens L, Seidel CW, Lin C, Smith ER, Shilatifard A, Conaway RC, Conaway JW. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 2011;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Azzouz N, Panasenko OO, Colau G, Collart MA. The CCR4-NOT complex physically and functionally interacts with TRAMP and the nuclear exosome. PLoS One. 2009;4:e6760. doi: 10.1371/journal.pone.0006760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kerr SC, Azzouz N, Fuchs SM, Collart MA, Strahl BD, Corbett AH, Laribee RN. The Ccr4-Not complex interacts with the mRNA export machinery. PLoS One. 2011;6:e18302. doi: 10.1371/journal.pone.0018302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell. 2005;18:97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 78.Santos-Rosa H, Schneider R, Bernstein BE, Karabetsou N, Morillon A, Weise C, Schreiber SL, Mellor J, Kouzarides T. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Molecular cell. 2003;12:1325–1332. doi: 10.1016/s1097-2765(03)00438-6. [DOI] [PubMed] [Google Scholar]
- 79.Tirosh I, Sigal N, Barkai N. Widespread remodeling of mid-coding sequence nucleosomes by Isw1. Genome Biol. 2010;11:R49. doi: 10.1186/gb-2010-11-5-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gkikopoulos T, Schofield P, Singh V, Pinskaya M, Mellor J, Smolle M, Workman JL, Barton GJ, Owen-Hughes T. A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science. 2011;333:1758–1760. doi: 10.1126/science.1206097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. Genome-wide Nucleosome Specificity and Directionality of Chromatin Remodelers. Cell. 2012;149:1461–1473. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]


