Summary
E. coli NusA and NusB proteins bind specific sites, such as those in the leader and spacer sequences that flank the 16S region of the ribosomal RNA transcript, forming a complex with RNA polymerase that suppresses Rho-dependent transcription termination. Although antitermination has long been the accepted role for Nus factors in rRNA synthesis, we propose that another major role for the Nus modified transcription complex in rrn operons is as an RNA chaperone insuring coordination of 16S rRNA folding and RNase III processing that results in production of proper 30S ribosome subunits. This contrarian proposal is based on our studies of nusA and nusB cold-sensitive mutations that have altered translation and at low temperature accumulate 30S subunit precursors. Both phenotypes are suppressed by deletion of RNase III. We argue that these results are consistent with the idea that the nus mutations cause altered rRNA folding that leads to abnormal 30S subunits and slow translation. According to this idea, functional Nus proteins stabilize an RNA loop between their binding sites in the 5′ RNA leader and on the transcribing RNA polymerase, providing a topological constraint on the RNA that aids normal rRNA folding and processing.
Keywords: transcription antitermination, 16S rRNA, NusA, NusB, SecY, rnc
INTRODUCTION
Bacterial ribosome synthesis consumes a major fraction of the energy of the cell, requiring tight control of the synthesis, cleavage, and maturation of ribosomal RNA (rRNA) and assembly of ribosomal proteins. Ribosomal RNA (rRNA) transcription and ribosome assembly are uniquely coordinated in these processes (Kaczanowska & Ryden-Aulin, 2007, Woodson, 2008, Nierhaus, 2004, Noller, 1993). There are seven E. coli rRNA operons (rrn) each containing a 16S, 23S, and 5S rRNA gene. The rRNA operons contain a dual promoter region (P1 and P2), a leader RNA upstream of 16S, and a spacer region between 16S and 23S genes encoding one or more tRNAs (Fig. 1A). The leader and spacer RNAs (Fig. 1) each contain a BOXA site involved in transcription antitermination, and complementary sequences that form a dsRNA, which is recognized and processed by RNase III during nascent rRNA transcription (Li et al., 1984, Young & Steitz, 1978).
Fig. 1.
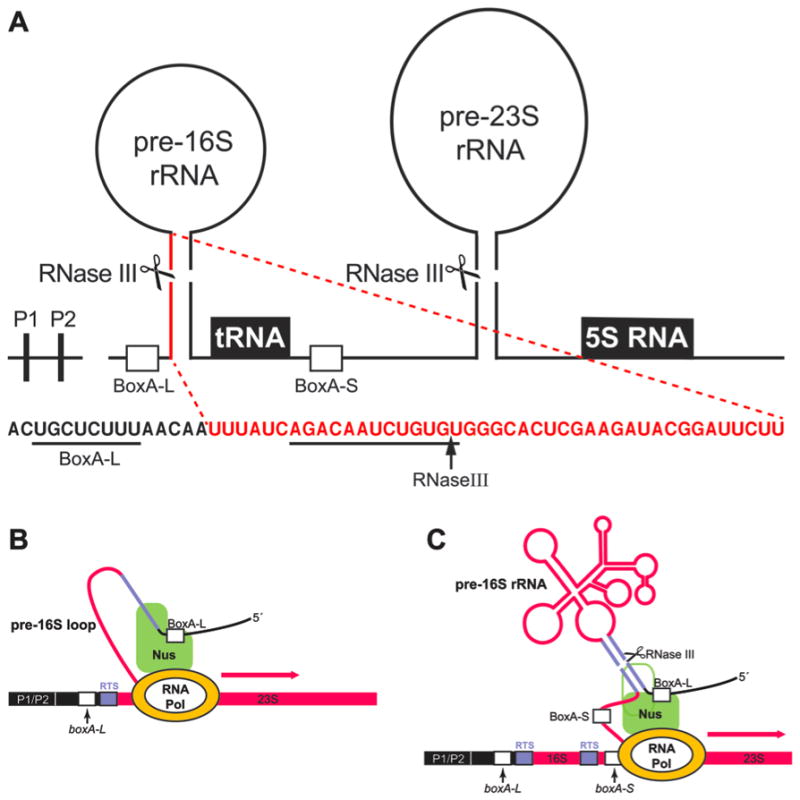
Dynamics of transcription, co-transcriptional folding, and processing of rrn operons. A. The primary rRNA transcript is expressed from two promoters P1 and P2, and consists of a leader sequence, defined as the 178 nucleotide region between the P2 promoter and the first nucleotide of the mature 16S rRNA, the 16S, 23S and 5S rRNA genes, and a spacer region separating the 16S and 23S rRNA genes and containing one or more t-RNA genes. Leader and spacer regions each contain conserved boxA DNA motifs (Olson et al., 1984), boxA-L and boxA-S, respectively. The stems of the pre-16S and pre-23S rRNA stem-loop structures contain sites for RNase III cleavage (gaps associated with scissors). Shown directly below is part of the leader RNA sequence with the conserved BOXA-L (UGCUCUUU) underlined. This conserved RNA sequence binds the NusB/NusE protein complex (Stagno et al., 2011). A third protein, NusA, binds to the leader RNA sequence shown underlined and downstream of BOXA-L (Beuth et al., 2005). The 43bp RNase III stem flanking the 16S rRNA loop includes part of the leader RNA just distal to BOXA-L as shown here in red. Dashed lines in red at each end of the stem define the corresponding red sequence just beyond BOXA-L. A vertical arrow designates the position of RNase III cleavage on the leader sequence (Young & Steitz, 1978). B. RNAP is shown after transcribing the leader and entering the 16S gene sequence. The Nus factors, including NusB/NusE bound at BOXA-L and NusA bound downstream, are shown as a green complex on the RNA. The RTS encodes one strand of the RNase III sensitive stem, shown in blue. The Nus-RNA complex bound from the BOXA-L site through the blue segment is also bound to RNAP, generating a growing loop of pre-16S rRNA as transcription ensues. C. RNAP has transcribed beyond boxA-S of the spacer. The DNA representations are written in italics (boxA-L and boxA-S) whereas their transcript products are written as BoxA-L and BoxA-S. The NusA protein (green outline) has been displaced from its binding site by the annealing of the dsRNA at the RNase III site. The rest of the Nus complex will be released to bind the spacer RNA including BOXA-S thereby generating another looped complex for 23S rRNA transcription. The pre-16S rRNA is shown as a folded, assembly intermediate released by RNase III processing of the primary transcript.
RNA polymerase (RNAP) is modified to suppress Rho-dependent transcription termination as it transcribes the leader and spacer regions of rrn operons (Li et al., 1992, Condon et al., 1995, Heinrich et al., 1995, Pfeiffer & Hartmann, 1997). This modification occurs at the BOXA RNA sites in these regions by the assembly of a transcription antitermination complex composed of at least the following proteins: NusA, NusB, NusG, and NusE, as well as S4 and additional ribosomal proteins (Li et al., 1984, Quan et al., 2005, Sharrock et al., 1985, Squires et al., 1993, Torres et al., 2004, Torres et al., 2001). NusE (Friedman et al., 1981) is the S10 protein of the small ribosome subunit and binds the antitermination complex only when not bound to the ribosome; hence the level of free S10 may regulate activity of the antitermination complex (Luo et al., 2008).
The double-strand specific endoribonuclease, RNase III, generates the initial rRNA cleavage events in the leader and spacer RNAs as RNAP transcribes the 16S and 23S genes (Young & Steitz, 1978, French & Miller, 1989, Srivastava & Schlessinger, 1990, Dunn & Studier, 1973). Because of this close coupling of transcription and RNase III cleavage (Fig. 1C), the full-length 30S rRNA precursor is never detected except in rnc mutants defective for RNase III (Gegenheimer & Apirion, 1975). Cleavage by RNase III generates precursor forms of 16S and 23S rRNA (Fig. 1), which are further processed by other ribonucleases to generate mature 16S and 23S rRNA (Deutscher, 2009). Assembly of the small (30S) and large (50S) ribosome subunits also occurs during transcription, folding, and processing of the rRNA precursors (de Narvaez & Schaup, 1979, Tu et al., 2009). In the absence of RNase III, a full-length 30S transcript, including both 16S and 23S, forms and is eventually processed by the same nucleases that normally generate the mature rRNA (Gegenheimer & Apirion, 1978, Court, 1993).
Much of our understanding of the role of Nus factors in transcription antitermination derives from in vivo and in vitro studies of the phage λ N transcription antitermination system, in which the co-assembly of four E. coli Nus proteins with λN protein forms a complex that suppresses termination and accelerates the rate of transcription elongation (Court et al., 2007, Das, 1993, Greenblatt et al., 1993). Those studies have provided valuable insights into the less well-characterized rrn system. We know from λ studies that NusA and NusG bind to core RNAP; NusA also binds to rRNA just downstream to BOXA (Beuth et al., 2005, Prasch et al., 2009). NusB and NusE bind to each other and to BOXA RNA (Stagno et al., 2011). Excess levels of NusE bypass the requirement for NusB, suggesting that a major role of NusB is to enhance the NusE binding to BOXA RNA and the RNA polymerase complex (Luo et al., 2008, Mason et al., 1992, Nodwell & Greenblatt, 1993, Squires et al., 1993, DeVito & Das, 1994).
Studies of transcription in the rrn operons reveal similarities and differences with the λ N system. Consistent with the λ N system paradigm, NusB and NusE bind to each other and to BOXA sites in the nascent rRNA leader and spacer (Mason et al., 1992, Nodwell & Greenblatt, 1993, Greive et al., 2005). As they do in the λ N system, Nus antitermination factors reduce pausing by RNAP and thereby increase the speed of transcription elongation two- to three-fold (Vogel & Jensen, 1997, Zellars & Squires, 1999, Klumpp & Hwa, 2008, Roberts et al., 2008). In NusA or NusB mutants or when BOXA-L or BOXA-S is mutant, transcription elongation rates are reduced, as is expression of 16S and/or 23S rRNA (Sharrock et al., 1985, Heinrich et al., 1995, Pfeiffer & Hartmann, 1997). In contrast to the λ N system, which antiterminates intrinsic and Rho-dependent terminators, the Nus complex assembled in rrn operons suppresses only the latter (Albrechtsen et al., 1990). Studies with nusA and nusB cold-sensitive (cs) mutants provide evidence for antitermination within rrn operons. These mutants have more mature 30S than 50S ribosome subunits, consistent with transcriptional polarity. Correspondingly, the 16S/23S RNA ratio is increased in the mutants, with reduced RNAP occupancy of distal parts of the rrn operons (Quan et al., 2005, Sharrock et al., 1985).
There is evidence suggesting that the Nus antitermination complex may be important in rRNA folding and ribosome activity. It has been shown that nusB cs mutations that affect rRNA transcription also reduce ribosome translation elongation speed at 37° and 42° (Taura et al., 1992), and that moreover, these nusB cs mutations suppress thermal sensitive (ts) secretion defects caused by sec mutations (Rajapandi & Oliver, 1994, Shiba et al., 1986b, Taura et al., 1992). A number of other cs mutations also suppress these same secretion defects. These ssy mutations (suppressors of secY) are primarily in genes encoding proteins involved in translation (Shiba et al., 1986b). SecY is a component of the E. coli protein membrane translocation machinery (Frauenfeld et al., 2011). By reducing the translation rate, the ssy mutations are thought to coordinate slowed membrane translocation caused by secY mutations with slow translation of membrane proteins (Rajapandi & Oliver, 1994, Lee & Beckwith, 1986). These phenotypes of nusB mutants indicate that NusB plays a role in ribosome assembly and/or function, possibly due to its effects on rRNA transcription. Whether other components of the antitermination complex have similar effects has not been established.
The nusAcs10 mutation, like nusB cs mutations, alters rrn transcription (Vogel & Jensen, 1997, Sharrock et al., 1985). We show here that nusAcs10 suppresses the secYts24 mutation, and that both nusB and nusA cs mutations affect ribosome maturation. Importantly, we demonstrate that abrogation of RNase III activity, which initiates rRNA processing, reverses the translational defects of the nusA and nusB cs mutants. To explain these results, we propose that the principal role of the Nus complex in the rrn operons is to coordinate transcription with 16S rRNA folding and processing during 30S subunit maturation.
RESULTS
nusA and nusB cs mutations suppress secYts24
Mutations in ssy genes whose products are involved in translation suppress the secYts24 defect at 42°. Suppressor ssyF and ssyE genes encode small 30S subunit ribosomal proteins S1 (rpsA) and S5 (rpsE), respectively. A third suppressor, ssyG, encodes translation initiation factor IF2 (Shiba et al., 1986a). All ssy mutations are cs, and fail to grow at 23°, independent of the secYts24 allele (Tables 1 and 2). The mutations are thought to suppress secYts24 by slowing the rate of protein synthesis (Shiba et al., 1986b).
Table 1.
To study suppression of secY24ts
| strain 1 | nus or ssy allele | gene | secY24ts 2 | ||
|---|---|---|---|---|---|
|
| |||||
| 23° | 37° | 42° | |||
|
|
|||||
| NB50 | none | − | + | + | M |
| NB60 | ssyB63 | nusB | − | + | S |
| NB61 | nusB< >cat | nusB | − | + | S |
| NB23 | nusAcs10 | nusA | − | + | S |
| NB24 | nusA1 | nusA | + | + | M |
| NBC971 | nusE71 | rpsJ | + | + | M |
| NB56 | ssyE36 | rpsE | − | + | S |
| NB54 | ssyF29 | rpsA | − | + | S |
| NB58 | ssyG40 | infB | − | + | S |
The strains are all derived from NB50 that is MC4100 secY24ts
Colony formation is shown as + for colonies at 2 days or − for no colony formation after 2 days. M indicates that a colony forms with an abnormal mutant phenotype at 42° that is caused by secY24ts. S indicates suppression of secY24ts to yield a normal colony morphology at 42° that is similar to those (+) at 37°.
Table 2.
Absence of RNase III suppresses the cold-sensitive nus mutant phenotype
| strain 1 | Growth at 23° | |||
|---|---|---|---|---|
|
| ||||
| rnc+ | Δrnc 2 | nus or ssy allele | rnc+ | Δrnc |
| MC4100 | NB97 | wild-type | + | + |
| IQ527 | NB778 | ssyB63 (nusB63) | − | + |
| NB747 | NB853 | nusB<>cat | − | + |
| K1914 | NB6 | nusAcs10 | − | + |
| IQ717 | NB83 | ssyE36 | − | − |
| IQ626 | NB3 | ssyF29 | − | − |
| IQ607 | NB4 | ssyG40 | − | − |
Strain MC4100 and derivatives with nus and ssy cold sensitive mutations and either rnc+ or Δrnc.
Δrnc indicates deletion-substitutions constructed with spc or cat antibiotic resistance cassettes. The cassettes replace the open reading frame of rnc from codon 1 through 224 and make gene fusions with the last 3 codons of rnc (Yu et al., 2000).
Colony formation is shown as + for colonies at 2 days or − for no colony formation after 2 days.
A mutation in the gene encoding the transcription antitermination factor, NusB (ssyB63) was isolated in the ssy suppressor collection (Shiba et al., 1986b). The cs nusB mutation, nusB5, was also reported to suppress secYts24 (Taura et al., 1992). We show in Table 1 that the cs point mutation in nusA, nusAcs10 (Craven & Friedman, 1991), and the cs deletion mutation of nusB, nusB< >cat (Bubunenko et al., 2007), both suppress the temperature-sensitive phenotype of secYts24 (Table 1). Other nus alleles tested, nusA1 and nusE71, isolated as defective for λ N antitermination (Friedman & Court, 1995), are not cs, had no effect on rRNA antitermination (Sharrock et al., 1985), and did not suppress secYts24 (Table 1). Conversely, the nusAcs10 mutation, which inhibits rRNA antitermination (Vogel & Jensen, 1997, Sharrock et al., 1985), has very little effect on λ N antitermination (Craven & Friedman, 1991). The Ssy phenotype of the cs nus mutations raises the possibility that they might affect translation efficiency. We present evidence below supporting this notion.
RNase III mutants suppress the cold-sensitive phenotype of nusAcs10 and nusB< >cat
RNase III processes the primary ribosomal precursor RNA. The rRNA leader site that is cleaved by RNase III is just downstream of BOXA-L (Fig. 1A) (Beuth et al., 2005). In the phage λ genome, BOXA and a downstream RNase III site are closely linked and near the N gene. These observations prompted us to examine whether RNase III affected the cold-sensitivity of nusAcs10 and nusB cs mutants. Table 2 shows that an rnc knockout mutation suppressed the cold-sensitivity of the nusA and nusB cs mutants (lines 2–4), but not that of ssyE, ssyF or ssyG cs mutants (lines 5–7).
RNase III deletion suppresses the Ssy phenotype of the nus mutants
To determine whether RNase III also plays a role in ssy suppression of the secYts24 mutation, an rnc knockout was introduced into the secYts24 mutant and ssy derivatives of that strain. The resultant constructs were analyzed for colony phenotypes at both 23° and 42° (Table 3). At 23° the rnc knockout suppressed the cold-sensitivity of the nusBcs63, nusB<>cat, and nusAcs10 alleles (Table 3). At 42°, the rnc knockout reversed nusA and nusB suppression of secYts24. In contrast, the rpsA, rpsE or infB alleles (ssyF, ssyE, or ssyG, respectively) retain the Ssy phenotype in the absence of RNase III (Table 3). Thus, the rnc knockout specifically suppresses the effects of the nus cold-sensitive mutants but by itself had no effect on secYts24 at any temperature (Table 3).
Table 3.
Absence of RNase III reverses nus-mediated suppression of secY24ts
| Strain1 | suppressor allele | secY24ts 2 | ||||
|---|---|---|---|---|---|---|
| Growth at 23°C | Growth at 42°C | |||||
| rnc+ | Δrnc | rnc+ | Δrnc | rnc+ | Δrnc | |
| NB50 | NB2 | none | + | + | M | M |
| NB60 | NB70 | ssyB63 (nusB63) | − | + | S | M |
| NB61 | NB75 | nusB< >cat | − | + | S | M |
| NB23 | NB74 | nusAcs10 | − | + | S | M |
| NB56 | NB73 | ssyE36 | − | − | S | S |
| NB54 | NB72 | ssyF29 | − | − | S | S |
| NB58 | NB71 | ssyG40 | − | − | S | S |
Strains are all derived from NB50 (MC4100 secY24ts).
Colony phenotypes are as described in Table 1.
Ribosome biogenesis is defective in nusB and nusA mutants
We examined ribosome biogenesis using sucrose gradient sedimentation to profile ribosome subunits in which rRNA had been labeled with [3H]-uridine. Because the nus mutants are cs, we assumed that any alteration in ribosome subunit assembly would be most evident at lower temperatures, e.g. 20°. Bacteria were initially grown at 37° followed by incubation at 20° prior to labeling for an hour with [3H]-uridine (see Methods). Crude extracts were prepared and centrifuged on sucrose gradients under ionic conditions that completely dissociate 70S particles into 30S and 50S subunits. To compare the labeled subunits in each run, a wild-type preparation of unlabeled 30S and 50S subunits, monitored by A260 was included in each gradient as standard markers.
Panels A & B of Fig. 2 shows the effect of the nusB<>cat mutation on the distribution of ribosomal subunits in strains labeled with [3H]-uridine at 20°. Instead of the sharp 30S peak observed with the wild-type control (A), a broader peak with a large component shifted to the right was observed in the nusB<>cat mutant NB421 (B), whereas an observable difference between the 50S peaks could not be readily discerned. This was not limited to the W3110 strain background since strain NB747, a derivative of MC4100 carrying the nusB<>cat mutation, exhibited the same sedimentation profiles as NB421 (data not shown).
Fig. 2.
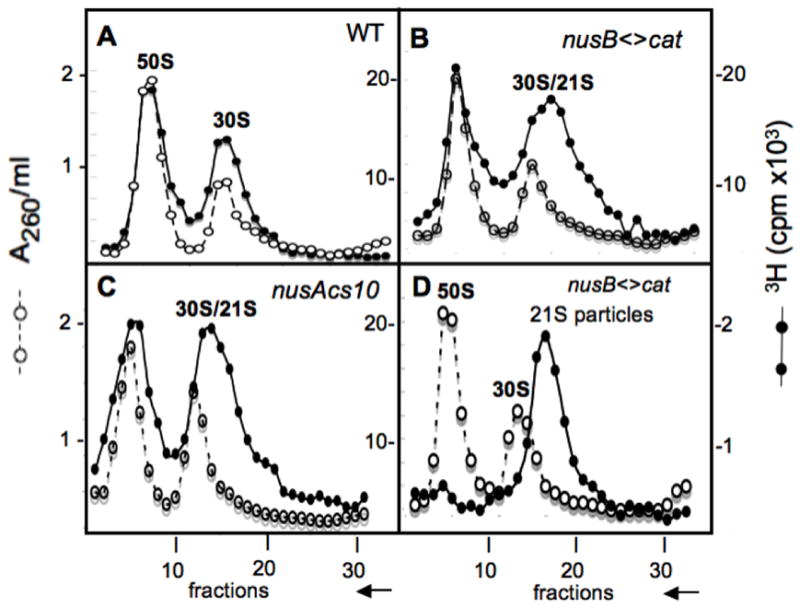
Identification of 21S ribosomal particles in NusA and NusB cold-sensitive mutants at 20°. Sucrose gradients were used to analyze ribosome subunits. The 50S and 30S subunits from wild-type bacteria serve as markers and were monitored in each fraction by A260 (left Y-axis). Ribosome subunits from cells whose RNA was labeled with [3H]-uridine for 1 hr at 20° before harvesting, monitored by [3H] (right Y-axis). Profiles of ribosome subunits prepared from the strain W3110 (panel A), the nusB< >cat derivative NB421 (panel B), and the nusAcs10 derivative NB755 (panel C) are shown as sedimenting from right to left as indicated by the arrow on the X-axis. Fraction 15 taken from the right shoulder of the 30S peak shown in panel B was mixed with unlabeled 50S and 30S subunits from a wild type strain and analyzed by sucrose gradient (see panel D), where a [3H]-21S peak is observed. Bacterial crude extracts were prepared and sedimented on sucrose gradients as described in Methods.
To examine further the broad fraction associated with the small subunit observed in the gradient from the nusB<>cat strain (Fig. 2B), we reanalyzed a fraction from the lighter-sedimenting edge of the “30S peak” by sucrose gradient. This rerun fraction yielded a sharp peak at 21S (Fig. 2D), suggesting that the broad peak in the 30S region was a mixture of mature 30S and precursor 21S particles (Lindahl, 1975). To confirm that the production of abnormal small ribosome subunits results from lack of NusB, we complemented the nusB<>cat mutant with wild-type nusB expressed from pAB36. The resulting strain, NB837, was not cs and exhibited a normal ribosome profile at 20°, in contrast to a nusB<>cat strain carrying the empty vector pACYC177 (compare panels B and A of Supplemental Fig. S1).
E. coli generates 21S particles as precursor intermediates during the synthesis and assembly of the 30S subunit (Lindahl, 1975). These precursor intermediates are only detected by very short (one min) pulse-labeling experiments in wild type cells because they very rapidly convert to 30S particles. To determine whether the 21S particles that accumulated in the absence of NusB at 20° were dead-end products or just slow to convert to 30S subunits, the nusB<>cat strain NB421 was labeled as described above at 20°, chased with an excess of unlabeled uridine (0.5 mM), treated with rifampicin (500 μg/ml), and then incubated at 20°. Samples were withdrawn after 0, 10, 20, 30, and 60 min for sucrose gradient sedimentation. Aligning the positions of the 3H and A260 peaks in panels A and D (Supplemental Fig. S2) revealed a shift of 21S particles to 30S subunits by 20 min after the chase. Since rifampicin blocks new rRNA transcription, we conclude that the shift results from conversion of the accumulated 21S particles to 30S subunits. Hence, the 21S particles are bona fide precursors of 30S ribosome subunits. These experiments indicate that NusB stimulates the rate of this conversion. Consistent with our findings, Sharrock (Sharrock et al., 1985) showed that when wild type and a NusB mutant were compared by labeling at 37° for 5 min, only 30S precursor intermediates from the NusB mutant could be detected under those experimental conditions. We propose that the 30S subunits and the resulting 70S ribosomes in the nusB mutant are altered at all temperatures, possibly because the 16S rRNA is improperly folded. Altered ribosome function offers the explanation for slowed translation elongation at 37° and suppression of secYts24 at 42° by nusBcs mutants (Shiba et al., 1986b, Taura et al., 1992).
The ribosome subunits from the nusAcs10 mutant NB755 incubated at 20° also showed a significant broadening of the sedimentation profile of the small subunit component toward 21S (compare Fig. 2, A and C). We confirmed that the defect in 30S assembly was due to the nusAcs10 allele by complementing with plasmid pA2-1, which expresses wild-type NusA. The plasmid fully restored 30S subunit assembly in the nusAcs10 strain (Supplemental Fig. S1D), in contrast to the empty vector, pACYC184 (Supplemental Fig. S1C).
The rRNA in 21S intermediate particles has been shown to be a precursor to mature 16S rRNA. During conversion to mature 30S subunit particles, the precursor RNA is processed by removal of 115 and 33 nucleotides respectively from its 5′ and 3′ ends (Srivastava and Schlessinger, 1990; Al Refaii and Alix, 2009). We show here that during growth at 37°, the nus mutants produce 30S ribosomal subunits indistinguishable from those produced in cells wild type at the nus loci (Fig. S3). We used primer extension (Fig. S4) to examine the 5′ end of 16S rRNA isolated from both mutant and wild type cells grown at 37°. To enhance the possibility of detecting a processing difference, we tested E. coli carrying both the nusAcs10 and nusB<>amp mutations. We found that the 16S rRNA from the nus mutant was processed normally at the 5′ end (Fig. S4). The result is compatible with the proposition that the ribosome is partially defective not by a 16S rRNA processing abnormality but by an RNA folding problem.
Elimination of RNase III suppresses the 30S subunit biogenesis defect of nusB< >cat and nusAcs10 mutants at 20°
The observation that an rnc knockout suppresses the growth phenotypes of the nusB and nusAcs10 mutants (Tables 2 & 3) prompted us to consider whether an rnc mutation also suppresses the ribosome biogenesis defects in nus mutants at low temperatures. We reasoned that the primary precursor 16S rRNA transcript in nus mutants was configured abnormally, and that RNase III cleavage of that abnormal precursor might permanently stabilize the misfolded configuration and result in abnormal 21S intermediates that generate partially defective 30S particles. Eliminating RNase III might allow proper 16S folding and formation of fully active 30S particles. Confirming our assumption, sedimentation profiles of ribosome subunits from rnc mutant derivatives of the nusB<>cat and nusAcs10 strains showed some, but significantly less, 21S material than the corresponding nus rnc+ strains (compare Fig. 3B with Fig. 2B and Fig. 3C with Fig. 2C and Supplemental Fig. S1C). The rnc mutation by itself had little effect on the ribosome sedimentation profile save for a small, light shoulder on the 30S peak (Fig. 3A). Therefore, an rnc mutation substantially suppressed the biogenesis defects of the nus mutants (compare gradients in Figs. 2 and 3).
Fig. 3.
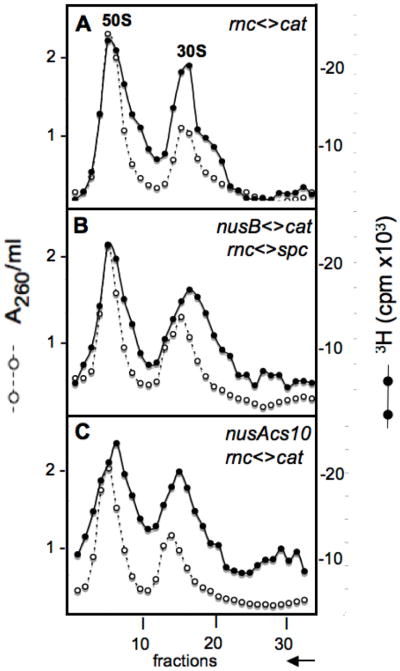
Elimination of RNase III suppresses the Nus cold-sensitive ribosome subunit phenotype. Shown are sedimentation profiles of ribosomal subunits prepared from W3110 derivatives NB478 (rnc < >cat) (panel A), NB840 (nusB < >cat, rnc< >spc) (panel B), and NB876 (nusAcs10, rnc< >cat) (panel C) labeled with [3H]-uridine at 20°C. Cells were labeled and extracts prepared and run on gradients as shown for Fig. 2 and described in Methods.
Ribosomal RNA levels in nus mutants and the effect of RNase III
Previous studies (Sharrock et al., 1985, Quan et al., 2005) demonstrated a ~60% reduction in the level of 23S rRNA with respect to 16S rRNA in cs nusB and nusA mutants. This was attributed to transcription polarity on expression of the downstream 23S rRNA. We have confirmed and extended this finding. We quantified the relative amount of [3H]-uridine incorporated into the 30S and 50S subunits at 20° in the various mutant strains. The gradient data from Figs. 2 and 3, compiled in Table 4, show a 1.6- to 1.8-fold reduction in 50S relative to 30S particles in the nusA and nusB cs mutants as compared to wild-type bacteria. The decrease in 50S concentration was largely reversed by elimination of RNase III (Table 4).
Table 4.
Quantitative analysis of sucrose gradients 1
| Strain | Alleles | 30S/50S 2 |
|---|---|---|
| W3110 | WT | 1.00 |
| NB421 | nusB<>cat | 1.80 |
| NB755 | nusAcs10 | 1.64 |
| NB478 | rnc<>cat | 1.13 |
| NB840 | nusB<>cat rnc<>spc | 1.21 |
| NB876 | nusAcs10 rnc<>cat | 1.07 |
Areas under the 50S and 30S (including 21S) peaks as delineated by the [3H]-cpm shown in Figs. 2A (W3110 strain = WT), 2B (NB421 = nusB<>cat), 2C (NB755 = nusAcs10), 3A (NB478 = rnc<>cat), 3B (NB840 = nusB<>cat, rnc<>spc), and 3C (NB876 = nusAcs10, rnc<>cat) were quantified using the graphics program ORIGIN developed by OriginLab.
The ratio of the 30S (including 21S) peak area to the 50S peak area was determined for each gradient indicated in footnote 1. Each ratio (30S/50S) was divided by the ratio of 0.86 found for wild-type in order to normalize to a 1:1 molar ratio of 30S to 50S subunits expected in wild-type.
We next examined the roles of NusA and NusB on polarity in one of the rrn operons, rrnH. We constructed luciferase reporter fusions to rrnH-16S and rrnH-aspU; the latter fusion junction is just distal to the 23S rRNA gene. As shown in Supplemental Table S1, the 16S/23S luciferase ratio was increased in the nusAcs10 and the nusB<>cat mutants by 20% and 60%, respectively. Importantly, this ratio was restored to wild-type levels when an rnc mutation was introduced into the nus mutants. Thus, by either measuring labeled rRNA levels directly (Table 4) or by a reporter assay (Table S1), the reduction in 23S rRNA levels is reversed in an RNase III mutant.
DISCUSSION
Our studies provide evidence for an additional role for the Nus factors in the synthesis of rRNA. In addition to the long-held view that Nus factors suppress transcription termination, we propose that these proteins play a critical role in biogenesis of the 30S subunit and, ultimately, translation. Further, we propose that the Nus factors promote proper 16S rRNA folding, and as a corollary, suggest that the role of Nus factors in rRNA transcription antitermination may be less significant than previously thought.
We summarize the results from our experiments with nus cs mutants that lead to these conclusions: 1) Sucrose gradient analyses demonstrate that 30S ribosome subunit assembly is altered with an accumulation of 21S precursors. 2) Suppression of secY24ts mutations by the cs nusA and nusB mutations indicates a defect in ribosome function in these mutants during translation. 3) The ratio of [3H] rRNA in 30S/21S subunits compared to 50S subunits is increased. 4) A knockout of the gene encoding RNase III,Δrnc, reverses the cold-sensitivity of the nus mutations as well as the other phenotypes caused by these mutations, including, to a large extent, the accumulation of 21S-like particles.
Suppression by Δrnc dictated a reevaluation of the role of the Nus complex in rRNA synthesis; namely, how to explain RNase III modulation of the multiple phenotypes caused by the cs nus mutations. The current antitermination model, in which Nus complex modification of RNAP promotes transcription elongation, cannot explain our results. Importantly, previous work shows that RNase III does not stimulate Rho-dependent transcription termination (Wilson et al., 2002); thus suppression of polarity by Δrnc in the nus mutants is unrelated to transcription termination.
RNase III processing, the first event in rRNA processing, occurs very rapidly, cleaving the dsRNA site within seconds after 16S rRNA sequences are transcribed and releasing the 16S precursor rRNA before transcription of 23S rRNA (French & Miller, 1989). Just prior to RNase III processing, the complementary RNA sequences flanking the 16S rRNA anneal to form a 43 bp stem (Young & Steitz, 1978) and a loop that contains the entire 16S rRNA (Fig. 1C). The 16S rRNA precursor, produced by RNase III cleavage of this dsRNA stem, is rapidly trimmed by other RNases, generating the mature 16S rRNA (Deutscher, 2009, Srivastava & Schlessinger, 1990). The 5′ side of the 43bp dsRNA stem is in the leader RNA, and its 3′ side in the spacer downstream of 16S rRNA. Rapid rRNA processing also occurs in nus cs mutants, indicating that Nus-RNAP-rRNA tethering is not essential for this process (Quan et al., 2005). When RNase III is absent, rRNA processing occurs much more slowly. The entire rRNA operon is transcribed before any processing occurs; this generates a 30S-sized transcript including the leader and the entire 16S, tRNA, 23S, and 5S rRNA regions (Gegenheimer & Apirion, 1975). Since RNAP would take more than 1 min to transcribe the entire operon beyond the 16S gene (Condon et al., 1993), processing must be delayed by at least this length of time in the rnc mutant. RNase III processing of rRNA is not essential for ribosome biogenesis because other RNA processing enzymes substitute, albeit at a much slower rate, to yield mature 16S rRNA and a functional ribosome (Deutscher, 2009). In the absence of RNase III, RNase P processes the full length 30S transcript at the tRNA(s) in the spacer regions, thereby generating novel precursor 16S rRNA species at a slower rate with respect to the precursors that are generated by RNase III (Gegenheimer & Apirion, 1978). RNase E and RNase G and a still unknown RNase then generate the mature 16S rRNA, which is assembled into functional ribosomes (Deutscher, 2009). This type of alternative processing in rnc mutants presumably occurs independently of the Nus system.
Essential to our understanding of how Nus factors direct ribosomal maturation is the observation that the nascent 16S rRNA transcript folds as it is transcribed (Nierhaus, 2004). Results of our experiments lead us to propose that rapid and proper folding requires that the 5′ and 3′ ends of the transcript be held together during transcription. This is achieved by tethering the leader 5′ RNA end to RNAP by the Nus proteins. This chaperone activity of the Nus proteins would constrain the 16S rRNA in a growing loop (Fig. 1B, C). In the absence of the Nus complex, this 16S loop would not form properly during transcription. Misfolded or partially-folded rRNA incorporated into 21S intermediates would explain the very slow maturation into 30S particles observed at the non-permissive temperature of 20° but not seen at higher temperatures under the same experimental conditions. However, at these higher temperatures, 37° or 42°, nus mutants are defective in translation, indicating that Nus factors are required to generate fully active ribosomes even at temperatures permissive for growth. Ablating RNase III activity compensates for the absence of a functional Nus complex. By leaving the 43 bp dsRNA uncut, the precursor 16S rRNA remains with its catalytic leader RNA and a stem-loop is formed by the dsRNA (Fig. 1). We speculate that this stem-loop simulates the loop generated during transcription by a tethered RNAP-Nus complex, and induces similar constraints on folding that guide normal folding of the 16S rRNA and maturation of the 30S subunit.
Our idea is supported by previous reports: 1) point mutations in the rrn leader cause defects in 16S rRNA folding (Theissen et al., 1990, Balzer & Wagner, 1998, Besancon & Wagner, 1999, Schäferkordt & Wagner, 2001) and 2) Nus factors can assist in RNA folding (Wong et al., 2005, Pan et al., 1999) 3) An intact leader sequence is known to be critical for proper 16S rRNA folding and ribosome biogenesis (Kaczanowska & Ryden-Aulin, 2007). Morgan (Morgan, 1986) first suggested that an RNAP-Nus complex bound to BOXA in the nascent leader might act as an RNA chaperone. Accordingly, defective growth of the nus cs mutants would reflect the absence of this RNA chaperone activity, resulting in a failure of the proper and timely folding of rRNA. Aberrant folding leads to defective ribosome assembly and 30S maturation with resulting translation defects. Further, the tethering of the BOXA-L region of the leader to the Nus-RNAP complex is similar to what occurs during N-dependent transcription-antitermination in phage λ (Wilson et al., 2004, Conant et al., 2008). Although there is no direct evidence for rRNA tethering by Nus factors during rRNA synthesis, as has been observed during λ transcription, we think it likely to occur based on the similarities between the structure of the two systems. NusB and NusE bind tightly to the rRNA leader BOXA-L (Stagno et al., 2011), which is synthesized soon after transcription initiation. NusE interacts with RNAP and tethers RNAP to BOXA-L (Nodwell & Greenblatt, 1993, Luo et al., 2008). NusA also binds and links the rRNA leader (Beuth et al., 2005, Prasch et al., 2009) and RNAP-Nus complex (Zhou et al., 2001). Thus, constrained in a trailing loop between the leader RNA and the RNAP-Nus complex, the nascent rRNA would be rapidly guided in its RNA:RNA interactions and limited in the types of RNA folding events it could undergo. In this state, the RNAP-Nus complex would act to chaperone rapid and proper rRNA folding (Fig. 1C) (Wong et al., 2005, Pan et al., 1999) and ribosome maturation coincident with transcription of the 16S rRNA precursor. We propose that the nus cs mutations weaken or abolish tethering and looping, leading to rRNA misfolding and slower ribosome maturation.
We also demonstrated that an rnc mutation suppresses the abnormal 16S/23S rRNA ratios in both the nusA and nusB mutants. Since RNase III is not involved in known Rho-dependent transcription termination (Wilson et al., 2002), this observation raises the question of whether the high 16S to 23S rRNA ratio in nus mutants really indicates transcription polarity. One possibility, that abnormal 16S rRNA synthesis destabilizes downstream RNA, has been ruled out by short pulse-chase experiments (Sharrock et al., 1985). As of now, therefore, we do not have a satisfactory explanation for this result.
We have shown here by genetic and biochemical methods that a functional interaction of NusA and NusB factors during rRNA transcription is important for proper 30S ribosomal subunit biogenesis. Our results suggest that Nus modified transcription coordinates with RNase III processing to assure proper rRNA folding, ribosome maturation, and, ultimately, translation.
Experimental Procedures
Bacteriological techniques
Standard bacteriological techniques were used (Miller, 1972). Cells were grown in LB medium containing either 25 μg/ml kanamycin, 10 μg/ml chloramphenicol, 75 μg/ml spectinomycin or 100 μg/ml ampicillin, when needed.
Strains and plasmids
Strains and plasmids are listed in Supplemental Table S2.
Construction of ssy-nus-rnc double and triple mutants
All strains were constructed using P1 transduction (Miller, 1972). The original secYts24 and ssy strains provided by Dr. K. Ito contained a tetR-determinant Tn10, which was removed from the secYts24 cells by the chlortetracycline method (Bochner et al., 1980) to construct the NB50 secYts24 tetracycline sensitive cells. The ssy, nus and rnc mutations were then transduced into NB50 to obtain the respective double mutants. Triple mutants were contructed by transducing rnc<>cat or rnc<>spc knockouts into the double secYts24-ssy/nus mutants.
Generation of gene knockouts by recombineering
The rnc gene was disrupted with a spectinomycin resistance-drug cassette using recombineering as described for rnc<>cat (Yu et al., 2000). The nusB<>cat knockout was described previously (Bubunenko et al., 2007).
Gene cloning for complementation analysis
The nusB open reading frame was amplified from the E. coli W3110 chromosome using the Expand PCR kit (Roche Applied Science, Indianapolis, IN) using a pair of primers: the forward primer CAGAAGGAGATATTCATATGAAACCTGCTGCTCGTCGCCGC and the reverse primer TTGCATGCGCATCTAGATTATCACTTTTTGTTAGGGCGAATC. A number of nucleotide changes was introduced in the underlined regions of the forward primer to improve the nusB Shine-Dalgarno region and to change its native UUG initiation codon to AUG to maximize the expression of the cloned NusB. The nusB PCR fragment was blunt-ended and cloned into the pCR-Script Cam vector (Stratagene, La Jolla, CA). Plasmids with the correct orientation of the nusB insert relative to the lac promoter were screened by restriction analysis and verified by DNA sequencing. The SacII-PstI fragment of nusB from the resulting pAB31 plasmid was isolated from an agarose gel and subcloned into the HincII-PstI sites of pACYC177 to generate pAB36. The plasmids were screened by restriction analysis and the correct pAB36 clones verified by DNA sequencing. The subcloning of nusA into pA2-1 was described previously (Plumbridge & Springer, 1983). For growth complementation studies, mutant cells were transformed with the respective plasmids by electroporation and grown on LB agar plates or in LB medium containing the appropriate antibiotic.
Construction of rRNA 16S-luc and aspU-luc gene fusions
The 16S-luc and aspU-luc gene fusions were constructed in the rrnH operon of the E. coli chromosome to assay the transcription activity within rRNA gene operons in vivo. The construction of the fusions was done in two steps using recombineering as described in detail for the rRNA 16S-luc fusion (Schweimer et al., 2011). Briefly, the luc/amp gene cassette was first assembled in the E. coli W3110 chromosome by placing the amp gene downstream to luc in the galK<> luc chromosomal locus in the recombinogenic DY330 cells to make NB371. Then, the luc/amp cassette was amplified from NB371 cells and the PCR product was used to construct both of the rrnH-luc fusions described below in NB363. The NB363 strain is a W3110 derivative containing a mini-λ-tet expressing the Red recombination genes (Court et al., 2003).
To make the rrnH 16S-luc and aspU-luc fusions, the luc/amp cassette was amplified by PCR using the forward primer AATTCATTACAAAGTTTAATTCTTTGAGCATCAAACTTTTGAAGGAGATATTCATATGGAAGACGCCAAAAACATAAAG and the forward primer GTAAGCCGGTCATAAAACCGGTGGTTGTAAAAGAATTCGGTGAAGGAGATATTCATATGGAAGACGCCAAAAACATAAAG, respectively. The universal reverse primer CTTATTAAGAAGCCTCGAGTTAACGCTCGAGGTTTTTTTTCGTCTTTACCAATGCTTAATCAGTGAGGC was used for amplification of both luc/amp cassettes. In these bipartite primers, the last 24 bases (shown in boldface) provided the regions for amplification of luc/amp. The first 55 bases of the forward primers and 45 bases of the reverse primer provided the regions of homology to the respective target regions of the chromosomal rrnH rRNA gene operon. The PCR-amplified fragments were purified and used for recombineering in NB363 as described. The cells were plated on L plates with ampicillin and recombinants screened for luc-amp insertions by PCR using a pair of primers flanking the rrnH-aspU region, TACCAAGTCTCAAGAGTGAACACG and GCAGGGATAGCCATAATATGCCTC or the aspU gene, CGCCGAAGCTGTTTTGGCGGATTG and GCAGGGATAGCCATAATATGCCTCrespectively. The mini-λ-tet was removed from the final NB375 (W3110 rrnH 16S-luc/amp) and NB377 (W3110 rrnH aspU-luc/amp) strains by growing cells at 37° and screening for tetracycline sensitive colonies. The position of the luc-fusions in the rrnH operon of the positive clones was verified by sequencing.
The rrnH 16S-luc and the aspU-luc-fusions were transduced into W3110, and derivatives containing various combinations of the fusions with the nusB<>cat, nusAcs10, and rnc<>spc mutations were constructed (see Supplemental Table S2). The luciferase activity of these fusions was assayed at 37° as previously described for the rrnH aspU-luc/amp fusion (Schweimer et al., 2011) and in the Promega Technical Bulletin # TB281,3/05.
Cell labeling with [3H]-uridine for ribosome analysis in sucrose gradients
Cells were labeled as described previously (Al Refaii & Alix, 2009) with changes described below. The W3110 or MC4100 cells containing nusAcs10 and nusB<>cat mutations or their combinations with rnc knockouts (see legends to Figs. 2 & 3 and Supplementary Figs. S1, S2 & S3 for the specific E. coli strains) were plated on minimal medium A supplemented with 0.2% glucose, 0.2% casamino acids and the respective antibiotic at 37°. A single colony was put into 10-ml medium A supplemented with 0.4% glucose, 0.2% casamino acids, 1 μg/ml thiamine, and 0.1 mM L-tryptophan and incubated overnight at 37°. Next day, 200 ml of the same supplemented medium A was inoculated with 1/200 volume of the overnight culture and was shaken at 37° until the optical density of the culture was A600 = 0.2, the culture was shifted to 20° for 2 or 4 hrs for nusB<>cat or nusAcs10, respectively, and 1 μC/ml [5-3H]-uridine (Amersham, TRK178, 28 Ci/mmol) diluted with non-radioactive uridine at 3 μM, (Sigma, U3750) was added. After an additional incubation for 1 hr, the cells were harvested by centrifugation and bacterial pellets kept at −20°. To label cells at 37° in the same culture conditions (see Fig. S3), cells were kept at 37° and RNA was labeled as above during a 1 hr growth period.
For chase labeling experiments (Fig. S2), cell cultures were grown as above, but following incubation with 3 μM, 1 μC/ml [5-3H]-uridine, they were supplemented with 0.5mM non-radioactive uridine and 0.5 mg/ml rifampicin, with continued incubation at 20°. Cell culture aliquots were withdrawn at 0, 10, 20, 30 and 60 min of incubation and processed as above.
Sucrose gradient analysis of 3H-labeled ribosomal particles
Frozen cell pellets were ground in ice with Al2O3 (Alcoa, A305) in TMNSH buffer (10 mM Tris-HCl pH7.4, 10 mM MgCl2, 60 mM NH4Cl, 1 mM DTT). Crude extracts were cleared by centrifugation at 12,000×g, 20 min, 3°. Supernatants were added with trace amount of non-radioactive wild type E. coli ribosomes (5 A260 units), adjusted to 0.4 M NaCl to dissociate 70S ribosomes and immediately layered onto (35 ml) 10–30% sucrose gradient prepared in TMNSH buffer with 0.4 M NaCl. The gradients were centrifuged at 27,000 rpm, 3° for 17 hrs in SW28 rotor (Beckman) and fractionated with a peristaltic pump from the bottom. The A260 optical density of each gradient fraction was measured. Each entire gradient fraction was precipitated with 5% trichloracetic acid to measure [3H]-radioactivity of the acid-insoluble material by liquid scintillation counting.
Primer extension analysis of rRNA
Cultures of wild type strain W3110 and nus cold sensitive mutant strain W3110 nusAcs10 nusB< >cat (NB452) were grown in LB at 37° to OD600=0.6. Total RNA was isolated using Qiagen RNA isolation kit, and elution was done in RNase free DEPC water and quantified using nanodrop. The RNA preparation was used for the primer extension experiment described in Fig. S4 as has been described previously (Rene & Alix, 2011).
Supplementary Material
Acknowledgments
We thank Grigory Mogilnitskiy for excellent technical assistance, Nina Costantino and Lynn Thomason for frequent consultation, and Maria Kireeva for providing the graphics program ORIGIN developed by ORIGINLAB. Dr. K. Ito provided ssy and secY mutant strains. DLC was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, and in part by a Trans National Institutes of Health/Food and Drug Administration Intramural Biodefense Program Grant of National Institutes of Allergy and Infectious Disease. DIF was supported by Public Health Service Grant AI11459-10. A. Al Refaii was a recipient of a fellowship from the Syrian Government. J-H. A. was supported by the UPR 9073 of the Centre National de la Recherche Scientifique (C.N.R.S.) and the Université Paris 7-Diderot. MEG was supported by NIH grant GM37219.
Abbreviations
- rRNA
ribosomal RNA
- rrn
ribosomal RNA operon
- ts
thermosensitive
- cs
cold-sensitive
- RNAP
RNA Polymerase
- dsRNA
double-strand RNA
- bp
base pair(s)
- WT
wild type
References
- Al Refaii A, Alix JH. Ribosome biogenesis is temperature-dependent and delayed in Escherichia coli lacking the chaperones DnaK or DnaJ. Mol Microbiol. 2009;71:748–762. doi: 10.1111/j.1365-2958.2008.06561.x. [DOI] [PubMed] [Google Scholar]
- Albrechtsen B, Squires CL, Li S, Squires C. Antitermination of characterized transcriptional terminators by the Escherichia coli rrnG leader region. J Mol Biol. 1990;213:123–134. doi: 10.1016/S0022-2836(05)80125-1. [DOI] [PubMed] [Google Scholar]
- Balzer M, Wagner R. Mutations in the leader region of ribosomal RNA operons cause structurally defective 30 S ribosomes as revealed by in vivo structural probing. J Mol Biol. 1998;276:547–557. doi: 10.1006/jmbi.1997.1556. [DOI] [PubMed] [Google Scholar]
- Besancon W, Wagner R. Characterization of transient RNA-RNA interactions important for the facilitated structure formation of bacterial ribosomal 16S RNA. Nucleic Acids Res. 1999;27:4353–4362. doi: 10.1093/nar/27.22.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuth B, Pennell S, Arnvig KB, Martin SR, Taylor IA. Structure of a Mycobacterium tuberculosis NusA-RNA complex. Embo J. 2005;24:3576–3587. doi: 10.1038/sj.emboj.7600829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner BR, Huang HC, Schieven GL, Ames BN. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980;143:926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubunenko M, Baker T, Court DL. Essentiality of ribosomal and transcription antitermination proteins analyzed by systematic gene replacement in Escherichia coli. J Bacteriol. 2007;189:2844–2853. doi: 10.1128/JB.01713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant CR, Goodarzi JP, Weitzel SE, von Hippel PH. The antitermination activity of bacteriophage lambda N protein is controlled by the kinetics of an RNA-looping-facilitated interaction with the transcription complex. J Mol Biol. 2008;384:87–108. doi: 10.1016/j.jmb.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C, French S, Squires C, Squires CL. Depletion of functional ribosomal RNA operons in Escherichia coli causes increased expression of the remaining intact copies. Embo J. 1993;12:4305–4315. doi: 10.1002/j.1460-2075.1993.tb06115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C, Squires C, Squires CL. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995;59:623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court DL. RNA processing and degradation by RNase III. In: Belasco J, Brawerman G, editors. Control of Messenger RNA Stability. New York: Academic Press; 1993. pp. 71–116. [Google Scholar]
- Court DL, Oppenheim AB, Adhya SL. A new look at bacteriophage lambda genetic networks. J Bacteriol. 2007;189:298–304. doi: 10.1128/JB.01215-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court DL, Swaminathan S, Yu D, Wilson H, Baker T, Bubunenko M, Sawitzke J, Sharan SK. Mini-lambda: a tractable system for chromosome and BAC engineering. Gene. 2003;315:63–69. doi: 10.1016/s0378-1119(03)00728-5. [DOI] [PubMed] [Google Scholar]
- Craven MG, Friedman DI. Analysis of the Escherichia coli nusA10(Cs) allele: relating nucleotide changes to phenotypes. J Bacteriol. 1991;173:1485–1491. doi: 10.1128/jb.173.4.1485-1491.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A. Control of transcription termination by RNA-binding proteins. Annual Review of Biochemistry. 1993;62:893–930. doi: 10.1146/annurev.bi.62.070193.004333. [DOI] [PubMed] [Google Scholar]
- de Narvaez CC, Schaup HW. In vivo transcriptionally coupled assembly of Escherichia coli ribosomal subunits. J Mol Biol. 1979;134:1–22. doi: 10.1016/0022-2836(79)90411-x. [DOI] [PubMed] [Google Scholar]
- Deutscher MP. Maturation and degradation of ribosomal RNA in bacteria. Progress Mol Biol Transl Sci. 2009;85:369–391. doi: 10.1016/S0079-6603(08)00809-X. [DOI] [PubMed] [Google Scholar]
- DeVito J, Das A. Control of transcription processivity in phage λ: Nus factors strengthen the termination-resistant state of RNA polymerase induced by N antiterminator. Proc Natl Acad Sci U S A. 1994;91:8660–8664. doi: 10.1073/pnas.91.18.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JJ, Studier FW. T7 early RNAs and Escherichia coli ribosomal RNAs are cut from large precursor RNAs in vivo by ribonuclease III. Proc Natl Acad Sci U S A. 1973;70:3296–3300. doi: 10.1073/pnas.70.12.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauenfeld J, Gumbart J, Sluis EO, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, Beckmann R. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat Struct Mol Biol. 2011;18:614–621. doi: 10.1038/nsmb.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SL, Miller OL., Jr Transcription mapping of the Escherichia coli chromosome by electron microscopy. J Bacteriol. 1989;171:4207–4216. doi: 10.1128/jb.171.8.4207-4216.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DI, Court DL. Transcription antitermination: the lambda paradigm updated. Mol Microbiol. 1995;18:191–200. doi: 10.1111/j.1365-2958.1995.mmi_18020191.x. [DOI] [PubMed] [Google Scholar]
- Friedman DI, Schauer AT, Baumann MR, Baron LS, Adhya SL. Evidence that ribosomal protein S10 participates in control of transcription termination. Proc Natl Acad Sci U S A. 1981;78:1115–1118. doi: 10.1073/pnas.78.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegenheimer P, Apirion D. Escherichia coli ribosomal ribonucleic acids are not cut from an intact precursor molecule. J Biol Chem. 1975;250:2407–2409. [PubMed] [Google Scholar]
- Gegenheimer P, Apirion D. Processing of rRNA by RNAase P: spacer tRNAs are linked to 16S rRNA in an RNAase P RNAase III mutant strain of E. coli. Cell. 1978;15:527–539. doi: 10.1016/0092-8674(78)90021-1. [DOI] [PubMed] [Google Scholar]
- Greenblatt J, Nodwell JR, Mason SW. Transcriptional antitermination. Nature. 1993;364:401–406. doi: 10.1038/364401a0. [DOI] [PubMed] [Google Scholar]
- Greive SJ, Lins AF, von Hippel PH. Assembly of an RNA-protein complex. Binding of NusB and NusE (S10) proteins to boxA RNA nucleates the formation of the antitermination complex involved in controlling rRNA transcription in Escherichia coli. J Biol Chem. 2005;280:36397–36408. doi: 10.1074/jbc.M507146200. [DOI] [PubMed] [Google Scholar]
- Heinrich T, Condon C, Pfeiffer T, Hartmann RK. Point mutations in the leader boxA of a plasmid-encoded Escherichia coli rrnB operon cause defective antitermination in vivo. J Bacteriol. 1995;177:3793–3800. doi: 10.1128/jb.177.13.3793-3800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczanowska M, Ryden-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp S, Hwa T. Stochasticity and traffic jams in the transcription of ribosomal RNA: Intriguing role of termination and antitermination. Proc Natl Acad Sci U S A. 2008;105:18159–18164. doi: 10.1073/pnas.0806084105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Beckwith J. Cotranslational and posttranslational protein translocation in prokaryotic systems. Annu Rev Cell Biol. 1986;2:315–336. doi: 10.1146/annurev.cb.02.110186.001531. [DOI] [PubMed] [Google Scholar]
- Li J, Horwitz R, McCracken S, Greenblatt J. NusG, a new Escherichia coli elongation factor involved in transcriptional antitermination by the N protein of phage lambda. J Biol Chem. 1992;267:6012–6019. [PubMed] [Google Scholar]
- Li SC, Squires CL, Squires C. Antitermination of E. coli rRNA transcription is caused by a control region segment containing lambda nut-like sequences. Cell. 1984;38:851–860. doi: 10.1016/0092-8674(84)90280-0. [DOI] [PubMed] [Google Scholar]
- Lindahl L. Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes. J Mol Biol. 1975;92:15–37. doi: 10.1016/0022-2836(75)90089-3. [DOI] [PubMed] [Google Scholar]
- Luo X, Hsiao HH, Bubunenko M, Weber G, Court DL, Gottesman ME, Urlaub H, Wahl MC. Structural and functional analysis of the E. coli NusB-S10 transcription antitermination complex. Mol Cell. 2008;32:791–802. doi: 10.1016/j.molcel.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason SW, Li J, Greenblatt J. Direct interaction between two Escherichia coli transcription antitermination factors, NusB and ribosomal protein S10. J Mol Biol. 1992;223:55–66. doi: 10.1016/0022-2836(92)90715-v. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, N. Y: 1972. [Google Scholar]
- Morgan EA. Antitermination mechanisms in rRNA operons of Escherichia coli. J Bacteriol. 1986;168:1–5. doi: 10.1128/jb.168.1.1-5.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierhaus KH. Assembly of the prokaryotic ribosome. In: Nierhaus KH, Wilson DN, editors. Protein Synthesis and Ribosome Structure. 2004. pp. 85–105. [Google Scholar]
- Nodwell JR, Greenblatt J. Recognition of boxA antiterminator RNA by the E. coli antitermination factors NusB and ribosomal protein S10. Cell. 1993;72:261–268. doi: 10.1016/0092-8674(93)90665-d. [DOI] [PubMed] [Google Scholar]
- Noller HF. Peptidyl transferase: protein, ribonucleoprotein, or RNA? J Bacteriol. 1993;175:5297–5300. doi: 10.1128/jb.175.17.5297-5300.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson ER, Tomich CS, Friedman DI. The NusA recognition site. Alteration in its sequence or position relative to upstream translation interferes with the action of the N antitermination function of phage lambda. J Mol Biol. 1984;180:1053–1063. doi: 10.1016/0022-2836(84)90270-5. [DOI] [PubMed] [Google Scholar]
- Pan T, Artsimovitch I, Fang XW, Landick R, Sosnick TR. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc Natl Acad Sci U S A. 1999;96:9545–9550. doi: 10.1073/pnas.96.17.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer T, Hartmann RK. Role of the spacer boxA of Escherichia coli ribosomal RNA operons in efficient 23 S rRNA synthesis in vivo. J Mol Biol. 1997;265:385–393. doi: 10.1006/jmbi.1996.0744. [DOI] [PubMed] [Google Scholar]
- Plumbridge JA, Springer M. Organization of the Escherichia coli chromosome around the genes for translation initiation factor IF2 (infB) and a transcription termination factor (nusA) J Mol Biol. 1983;167:227–243. doi: 10.1016/s0022-2836(83)80333-7. [DOI] [PubMed] [Google Scholar]
- Prasch S, Jurk M, Washburn RS, Gottesman ME, Wohrl BM, Rosch P. RNA-binding specificity of E. coli NusA. Nucleic Acids Res. 2009;37:4736–4742. doi: 10.1093/nar/gkp452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan S, Zhang N, French S, Squires CL. Transcriptional polarity in rRNA operons of Escherichia coli nusA and nusB mutant strains. J Bacteriol. 2005;187:1632–1638. doi: 10.1128/JB.187.5.1632-1638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapandi T, Oliver D. ssaD1, a suppressor of secA51(Ts) that renders growth of Escherichia coli cold sensitive, is an early amber mutation in the transcription factor gene nusB. J Bacteriol. 1994;176:4444–4447. doi: 10.1128/jb.176.14.4444-4447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rene O, Alix JH. Late steps of ribosome assembly in E. coli are sensitive to a severe heat stress but are assisted by the HSP70 chaperone machine. Nucleic Acids Res. 2011;39:1855–1867. doi: 10.1093/nar/gkq1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JW, Shankar S, Filter JJ. RNA polymerase elongation factors. Annu Rev Microbiol. 2008;62:211–233. doi: 10.1146/annurev.micro.61.080706.093422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäferkordt J, Wagner R. Effects of base change mutations within an Escherichia coli ribosomal RNA leader region on rRNA maturation and ribosome formation. Nucleic Acids Res. 2001;29:3394–3403. doi: 10.1093/nar/29.16.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweimer K, Prasch S, Sujatha PS, Bubunenko M, Gottesman ME, Rosch P. NusA interaction with the α subunit of E. coli RNA polymerase is via the UP element site and releases autoinhibition. Structure. 2011;19:945–954. doi: 10.1016/j.str.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Gourse RL, Nomura M. Defective antitermination of rRNA transcription and derepression of rRNA and tRNA synthesis in the nusB5 mutant of Escherichia coli. Proc Natl Acad Sci U S A. 1985;82:5275–5279. doi: 10.1073/pnas.82.16.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba K, Ito K, Nakamura Y, Dondon J, Grunberg-Manago M. Altered translation initiation factor 2 in the cold-sensitive ssyG mutant affects protein export in Escherichia coli. Embo J. 1986a;5:3001–3006. doi: 10.1002/j.1460-2075.1986.tb04598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba K, Ito K, Yura T. Suppressors of the secY24 mutation: identification and characterization of additional ssy genes in Escherichia coli. J Bacteriol. 1986b;166:849–856. doi: 10.1128/jb.166.3.849-856.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires CL, Greenblatt J, Li J, Condon C, Squires CL. Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc Natl Acad Sci U S A. 1993;90:970–974. doi: 10.1073/pnas.90.3.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AK, Schlessinger D. Mechanism and regulation of bacterial ribosomal RNA processing. Annu Rev Microbiol. 1990;44:105–129. doi: 10.1146/annurev.mi.44.100190.000541. [DOI] [PubMed] [Google Scholar]
- Stagno JR, Altieri AS, Bubunenko M, Tarasov SG, Li J, Court DL, Byrd RA, Ji X. Structural basis for RNA recognition by NusB and NusE in the initiation of transcription antitermination. Nucleic Acids Res. 2011;39:7803–7815. doi: 10.1093/nar/gkr418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taura T, Ueguchi C, Shiba K, Ito K. Insertional disruption of the nusB (ssyB) gene leads to cold- sensitive growth of Escherichia coli and suppression of the secY24 mutation. Mol Gen Genet. 1992;234:429–432. doi: 10.1007/BF00538702. [DOI] [PubMed] [Google Scholar]
- Theissen G, Behrens SE, Wagner R. Functional importance of the Escherichia coli ribosomal RNA leader boxA sequence for post-transcriptional events. Mol Microbiol. 1990;4:1667–1678. doi: 10.1111/j.1365-2958.1990.tb00544.x. [DOI] [PubMed] [Google Scholar]
- Torres M, Balada JM, Zellars M, Squires C, Squires CL. In vivo effect of NusB and NusG on rRNA transcription antitermination. J Bacteriol. 2004;186:1304–1310. doi: 10.1128/JB.186.5.1304-1310.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M, Condon C, Balada JM, Squires C, Squires CL. Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in both non-ribosomal and ribosomal RNA antitermination. Embo J. 2001;20:3811–3820. doi: 10.1093/emboj/20.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C, Zhou X, Tropea J, Austin B, Waugh D, Court D, Ji X. Structure of ERA in complex with the 3′ end of 16S rRNA: implications for ribosome biogenesis. Proc Natl Acad Sci U S A. 2009;106:14843–14848. doi: 10.1073/pnas.0904032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel U, Jensen KF. NusA is required for ribosomal antitermination and for modulation of the transcription elongation rate of both antiterminated RNA and mRNA. J Biol Chem. 1997;272:12265–12271. doi: 10.1074/jbc.272.19.12265. [DOI] [PubMed] [Google Scholar]
- Wilson HR, Yu D, Peters HK, 3rd, Zhou JG, Court DL. The global regulator RNase III modulates translation repression by the transcription elongation factor N. Embo J. 2002;21:4154–4161. doi: 10.1093/emboj/cdf395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson HR, Zhou JG, Yu D, Court DL. Translation repression by an RNA polymerase elongation complex. Mol Microbiol. 2004;53:821–828. doi: 10.1111/j.1365-2958.2004.04170.x. [DOI] [PubMed] [Google Scholar]
- Wong T, Sosnick TR, Pan T. Mechanistic insights on the folding of a large ribozyme during transcription. Biochemistry. 2005;44:7535–7542. doi: 10.1021/bi047560l. [DOI] [PubMed] [Google Scholar]
- Woodson SA. RNA folding and ribosome assembly. Curr Opin Chem Biol. 2008;12:667–673. doi: 10.1016/j.cbpa.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA, Steitz JA. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc Natl Acad Sci U S A. 1978;75:3593–3597. doi: 10.1073/pnas.75.8.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellars M, Squires CL. Antiterminator-dependent modulation of transcription elongation rates by NusB and NusG. Mol Microbiol. 1999;32:1296–1304. doi: 10.1046/j.1365-2958.1999.01442.x. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Mah TF, Yu YT, Mogridge J, Olson ER, Greenblatt J, Friedman DI. Interactions of an Arg-rich region of transcription elongation protein NusA with NUT RNA: implications for the order of assembly of the lambda N antitermination complex in vivo. J Mol Biol. 2001;310:33–49. doi: 10.1006/jmbi.2001.4722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


