Abstract
In all domains of life, elongating RNA polymerases require the assistance of accessory factors to maintain their processivity and regulate their rate. Among these elongation factors, the Spt5/NusG factors stand out. Members of this protein family appear to be the only transcription accessory proteins that are universally conserved across all domains of life. In archaea and eukaryotes, Spt5 associates with a second protein, Spt4. In addition to regulating elongation, the eukaryotic Spt4-Spt5 complex appears to couple chromatin modification states and RNA processing to transcription elongation. This review discusses the experimental bases for our current understanding of Spt4-Spt5 function and recent studies that are beginning to elucidate the structure of Spt4-Spt5/RNA polymerase complexes and mechanism of Spt4-Spt5 action.
Keywords: Spt4, Spt5, DSIF, NELF, P-TEFb, transcription elongation
1. The Spt4-Spt5 complex, a universally conserved transcription elongation factor
Transcription elongation by RNA polymerase II (RNAPII) is a dynamic and highly regulated step in the gene expression cycle. In vitro, RNAPII is capable of transcribing naked DNA at rates that rival those observed in vivo [1]. However, RNAPII cannot efficiently transcribe chromatin templates in vitro, suggesting that transcription elongation accessory factors and/or chromatin remodeling plays an essential role in facilitating elongation in vivo [1, 2]. Over the past decade, a wealth of work has established that both of these mechanisms are employed to facilitate transcription elongation (reviewed in [3]). Furthermore, it is now clear that in addition to being a target for regulation, elongating RNAPII also serves as an interaction platform for factors that couple elongation to chromatin remodeling and RNA processing activities.
The multisubunit RNA polymerases that are responsible for the bulk of transcription across the three domains of life are conserved at the levels of sequence, structure and mechanism [4–7]. Genomic analysis suggests that other than RNA polymerase subunits, the only transcription regulator universally conserved across all domains of life is the Spt5/NusG family of proteins [8]. Thus, it is likely that the function encoded by these proteins is ancient, i.e., that it was already present prior to divergence of the eukaryotic, archaeal and bacterial lineages. Consistent with the idea that they play a central role in transcription, Spt5/NusG proteins are essential for life and they regulate transcription elongation in all 3 domains of life. Furthermore, in eukaryotes they also coordinate elongation with chromatin states and pre-mRNA processing.
In the past several years, a confluence of structural, biochemical, genetic and genome-wide approaches have considerably expanded our understanding of Spt4-Spt5 function. This review will describe the experimental bases for our current conception of Spt4-Spt5 function and will also point out outstanding issues regarding the structure and mechanisms of Spt4-Spt5 in transcription elongation.
2. Discovery of Spt4-Spt5 in yeast and humans
SPT4 and SPT5 were originally discovered in a genetic screen, performed in S. cerevisiae, for mutations that suppressed the transcriptional defects caused by particular set of insertions in the promoter of the HIS4 gene [9]. Subsequent analysis showed that spt4 and spt5 mutations shared this phenotype with several histone mutations, that the spt4 and spt5 mutations affected transcription, and that spt5 mutations could genetically suppress mutations affecting the Snf/Swi complex [10–12]. Collectively, these observations suggested that Spt4 and Spt5 might influence chromatin and promoter function.
A pair of studies published in 1998 indicated that Spt4 and Spt5 form a protein complex that regulates transcription elongation. In the first, selection for genetic suppressors of a cold-sensitive spt5 allele yielded S. cerevisiae strains with mutations in catalytic subunits of RNAPII [13]. One of these polymerase mutations was known to decrease the rate of elongation by RNA polymerase II and several others were predicted to do so, suggesting that Spt5’s function is related to the rate of elongation. Consistent with this, spt4 and spt5 mutations exhibited growth defects when combined with deletions in elongation factor TFIIS, which functions to rescue RNAPII from transcription arrest [14], suggesting that spt4 and spt5 mutations increase the probability that RNAPII will undergo transcription arrest. Treatment of cells with 6-azauracil or mycophenolic acid, inhibitors of nucleotide biosynthesis that may interfere with elongation rate and processivity of RNA polymerase II [15, 16], suppressed the growth defects of the cold-sensitive spt5 mutant. The suppression of the spt5 cold-sensitive defects in RNAPII or inhibitors of elongation was allele-specific; although other spt4 and spt5 mutants display enhanced growth defects when combined with deletions of the gene for TFIIS, 6-azauracil and mycophenolic acid enhances rather than suppresses their growth defects and they are not phenotypically suppressed by elongation defective mutations in RNAPII ([13]; GAH, unpublished). Mutations that cause cold-sensitivity have been implicated in defective assembly of protein complexes, defects in protein activity and in defects in proteins that interact with RNA [17]. Thus, in the spt5 cold sensitive mutant, a decreased rate of transcription elongation may allow the defective Spt5 protein more time to execute its function or to associate with an interaction partner. Overall, these observations suggest that Spt4-Spt5 acts during transcription elongation, that it may prevent pausing or arrest of the elongating RNAPII, and that Spt4-Spt5 function and RNA polymerase II elongation rate must be coordinated for normal growth.
The second study examined the mechanism of action of 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB), which inhibits transcription elongation, but not initiation [18]. Wada and colleagues identified a fraction of a HeLa cell nuclear extract which supported RNAPII transcription in vitro, but that was insensitive to DRB[19]. They then purified a protein complex which, when added back to their in vitro transcription reactions, restored DRB sensitivity. This complex, DRB Sensitivity Inducing Factor (DSIF), was composed of the human homologs of Spt4 and Spt5. Interestingly, although DSIF was purified as an inhibitor of elongation, it was also found to stimulate elongation over the body of a gene in vitro, although only when nucleotides were present at limiting concentrations [19, 20]. As will be discussed in more detail below, Spt4-Spt5’s inhibitory activity has only been demonstrated in a few organisms. Thus, the term DSIF will be used here only in reference to systems in which this negative activity is known to occur.
3. Organization of Spt4 and Spt5 proteins
Spt5 is a large, highly conserved, multi-domain protein consisting of an N-terminal acidic domain, a NusG N-terminal (NGN) domain, multiple Kyprides, Ouzounis, Woese (KOW) domains and a set of short repeats at its C-terminus (CTR) (Figure 1A;[21]). Some metazoan and plant Spt5 proteins include an additional stretch of ~100 amino-acids at their extreme C-terminus which may include a poorly conserved KOW domain. Although the NGN domain appears to be unique, KOW domains are a subset of a larger family of domains called Tudor (Figure 1B), which are thought to mediate protein-protein and protein-nucleic acid recognitions [22–24]. Sequences of the repeats within the CTR vary across species, but typically contain residues that can be phosphorylated (tyrosine, serine, threonine). Archaeal Spt5 proteins and their bacterial homologs, NusG proteins, are much smaller than eukaryotic Spt5, consisting of an NGN and single KOW domain. SPT5/NusG genes appear to be essential for life in bacteria, yeasts, Drosophila, Zebra fish and mammalian cells [11, 25–29].
Figure 1.
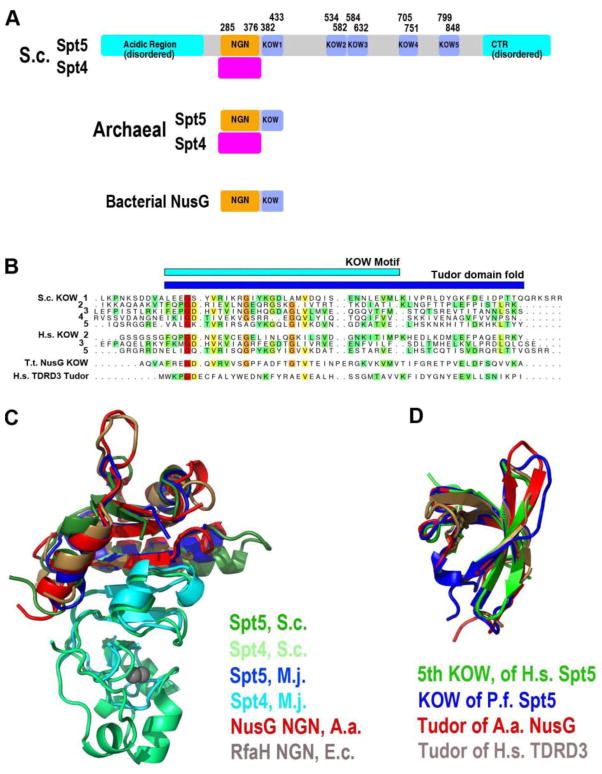
(A) Domain organization of Spt4 and Spt5 proteins. Domain boundaries are drawn to proportions according to their polypeptide lengths. (B) Alignment of sequence segments from Spt5 and NusG proteins that encompass the KOW motif. Note a Tudor domain sequence of TDRD (Tudor domain-containing protein 3) is also included. Color variation (from red to green) reflects degree of conservation (invariant to similar). (C) Structural superposition of homologous Spt4 and Spt5 proteins. Protein IDs are indicated with the different colors. (D) Structural superposition of KOW domains of Spt5 and NusG together the Tudor domain of TDRD3. Abbreviations: S.c. - Saccharomyces_cerevisiae; M.j. - Methanococcus jannaschii; A.a. - Aquifex aeolicus; P.f. - Pyrococcus furiosus; T.t. - Thermus thermophilus; E.c. - Escherichia coli; H.s. - Homo sapiens; and TDRD3 - Tudor domain-containing protein 3.
In contrast to Spt5, Spt4 is a small zinc finger protein [10]. Spt4 proteins are conserved across the eukaryotic and archaeal lineages, but are not found in bacteria [21]. Mutation of any one of the 4 cysteines of the zinc finger of Spt4 causes severe phenotypes similar to those of null alleles [10, 30, 31]. Although spt4 null mutants of S. cerevisiae are viable [10], SPT4 may be essential in Drosophila [32]. As discussed in a section below, crystallographic studies have shown that Spt4 interacts intimately with the NGN domain of Spt5 in forming the Spt4-Spt5 heterodimeric complex. Consistent with its sequence conservation [33], human SPT4 can complement spt4 null mutation in yeast, attesting to functional conservation and probably structural conservation as well [34].
4. Spt4-Spt5 plays pervasive role in transcription elongation
The initial indications that Spt4 and Spt5 form a complex that regulates elongation are now supported by an abundance of other data. The overwhelming majority of Spt4 and Spt5 proteins are found in the Spt4-Spt5 complex [35], and Spt4-Spt5 tightly associates with RNAPII in a transcription-dependent manner [36]. In vitro transcription experiments [37, 38] show that this association begins just downstream of the transcription start site. Multi-gene and genome-wide ChIP studies also show that association of Spt4-Spt5 with genes begins downstream of transcription start sites, that it persists until around the site of termination and that it largely mirrors the distribution of RNAPII [39–44]. Consistent with this, staining of Drosophila polytene chromosomes shows that Spt5 extensively co-localizes with the elongating, Ser2-phosphorylated form of RNAPII [45, 46]. Furthermore, using a ChIP assay to follow the migration of RNAPII elongation complexes across a long (~8kb) gene in yeast [16], several groups have shown that spt4 and spt5 mutations affect the translocation rate and processivity of the elongating RNAPII in vivo [16, 47, 48]. An in vitro transcription assay with spt4-null yeast whole cell extracts also found a positive role for the yeast factor in RNAPII transcription [49]. More recently, Spt4-Spt5 has been shown to required for normal transcription of a variety of long homopolymeric sequences including the expanded poly-CAG repeats which are found in disease associated variants of the Huntington’s gene and several other neurological disease genes [50].
Although genome-wide studies suggest that Spt4-Spt5 likely acts at all genes, in some cases it may also act in a gene-specific manner. DSIF appears to play a role in TAT-mediated regulation of transcription elongation across the HIV genome [37, 38, 51]. In addition, DSIF represses transcription of the NF-κB-responsive A20 gene in an activator and core promoter-dependent fashion [52, 53].
Consistent with the idea that Spt5’s function arose early in evolution, Spt4-Spt5 also associates with and appears to regulate transcription elongation by RNA polymerase I [54–56], suggesting that Spt4-Spt5 existed prior to divergence of the nuclear RNA polymerases of eukaryotes. In contrast to RNAPII and RNAPI, there is no evidence that Spt4-Spt5 associates with or regulates RNA polymerase III. In addition, the bacterial homolog of Spt5, NusG, also regulates transcription elongation [57–59]. Like Spt5, NusG associates with elongating polymerases in vitro [60] and ChIP analysis suggest that NusG associates with the majority of transcription units in vivo, joining polymerase downstream of the initiation site [61]. Finally, in vitro transcription assays show that the archaeal Spt4-Spt5 complex also associates with RNA polymerase and can stimulate elongation of a 10 subunit (rather than the normal 12) form of RNA polymerase that is less processive than the complete complex under low-nucleotide conditions [62], demonstrating that Spt5 functions are universally conserved.
5. What mechanisms underlie Spt4-Spt5’s function?
As is the case for several other elongation factors (reviewed in [63]), including NusG and RfaH [64, 65], Spt4-Spt5 appears to promote elongation by reducing the frequency of transcription pausing or arrest [37, 66]. This may explain the multiple observations that Spt4-Spt5 only promotes transcription elongation in vitro when nucleotides are limiting, a condition that increases the frequency of pausing and arrest. These in vitro results are consistent with the genetic interactions of mutations affecting both Spt4-Spt5 and TFIIS described above. Although nucleotide concentrations are not generally limiting in vivo, Spt4-Spt5 may protect elongation complexes from other triggers of pausing and arrest, such as nucleosomes (see below). Spt4-Spt5 may also facilitate elongation indirectly by interacting with the nascent transcript and by assisting cotranscriptional RNA processing events including 5′ capping (discussed below), which has been suggested to have a positive role in early elongation [67].
Further work on DSIF has yielded new insights into the mechanism of its inhibitory activity in human cells. First, a second multisubunit complex, Negative Elongation Factor (NELF), is also required for DSIF activity [68]. Curiously, although Spt4-Spt5 is found in all eukaryotes, NELF is not found in plant, yeast or nematode genomes [69]. Whether or not Spt4-Spt5 inhibits early elongation in organisms that lack NELF is an open question.
A second set of insights into Spt4-Spt5 function derives from the observation that DRB inhibits P-TEFb, the protein kinase that phosphorylates the serine-2 position of the RNAPII CTD repeats [70]. Consistent with the idea that DRB’s effects on transcription are mediated through its inhibition of P-TEFb, flavopiridol, an inhibitor of P-TEFb unrelated to DRB, also inhibits CTD phosphorylation and prevents RNAPII from leaving promoter proximal locations and entering into productive elongation [71, 72]. Furthermore, In addition to modifying the CTD, P-TEFb and its yeast homolog, Bur1, also phosphorylate a conserved serine (S. cerevisiae; [73]) or threonine (S. Pombe and humans; [29, 74, 75]) in the C-terminal repeats of Spt5. Deletion analysis of the Spt5 CTR suggests that it is necessary for Spt5’s ability to both repress and activate transcription [75, 76]. Phosphorylation of Spt5’s CTR prevents its inhibitory function and stimulates its positive function in elongation [29, 77].
The biochemical mechanisms that underlie the function of Spt5’s CTR remain to be determined. Genetic interactions suggest a functional overlap between Spt4-Spt5 and the RNAPII CTD [78, 79], DSIF can bind RNAPII independently of the RNAPII CTD [80]. The phosphorylation states of the RNAPII CTD and that of the Spt5 CTR by P-TEFb do not affect RNAPII-DSIF binding [80, 81]. While CTR phosphorylation is critical for DSIF to exert a positive role on elongation, it is not involved in the repressive activity of DSIF/NELF as assayed in vitro [29]. Furthermore, in contrast to bur1 and spt5 null mutations, deletion of the Spt5 CTR is not a lethal mutation in budding and fission yeast [73, 78, 82, 83], and does not prevent association of human Spt5 with RNAPII [75, 76]. Thus, neither the phosphorylation state of Spt5’s CTR nor that of the RNAPII CTD controls Spt4-Spt5/RNAPII association.
Our understanding of transcription elongation and its role in transcriptional regulation has recently been transformed by the discovery that, in at least some eukaryotes, including Drosophila, mice and humans, RNAPII is frequently found just downstream from the transcription start site of genes in a transcriptionally engaged, but paused state (reviewed in [84]). Significantly, this 5′ transcriptional pausing depends upon DSIF and NELF, and release from the pause appears to be mediated by P-TEFb in a process that, in at least some cases, involves transcription activators such as c-Myc [43, 85, 86]. In line with such a pause release mechanism, recent studies have identified associations of P-TEFb and other elongation factors with components of the RNAPII coactivator complex, Mediator, and revealed functional involvement of Mediator in the transition of RNAPII from pre-initiation complex (PIC) into the elongation phase of transcription [87, 88]. This model offers a basis for understanding a functional interplay between DSIF and Mediator observed in an earlier biochemical analysis [89]. Yeast genetic data also indicate a role of Mediator in regulating the post-recruitment RNAPII activity on inducible yeast genes such as CYC1 and HSP82 [90, 91], although it remains to be characterized whether the mechanisms that withhold RNAPII at these yeast promoters also involves Spt5, the yeast DSIF protein.
Given its inherent structural adaptability [92, 93], the Mediator complex seems well suited for relaying induction-specific inputs to regulate the early elongation activities of RNAPII, subsequent to the formation of PIC. As has been shown for NF-κB target genes, induction-specific regulation of RNAPII elongation can be determined by enhancer sequences in conjunction with the type of core promoter, TATA-less (e.g. A20 gene) vs. TATA-containing (e.g. IL-8) [94]. In this case, DSIF inhibits A20 gene elongation in the uninduced state with the aid of USF1 binding to an upstream DNA sequence [53].
The relationship between Spt5’s positive and negative functions is not clear. Only a few mutations have been reported to selectively remove either the positive or negative functions of Spt5, and these have yet to yield insights into the mechanism(s) of Spt4-Spt5 function [20,29]. Paused polymerases have not been clearly demonstrated in either Saccharomyces cerevisiae or C. elegans, which lack NELF. However, polymerases do appear to accumulate over promoters in these organisms under conditions of nutrient limitation [95, 96], suggesting the potential for an alternative mechanism for 5′ arrest in organisms that lack NELF. Furthermore, S. cerevisiae clearly possesses promoters that constitutively recruit RNAPII, TBP and other basal factors, but are regulated at a step after the assembly of these basal transcription components [97–99]. In a high-resolution genome-wide mapping, yeast genes with promoter-proximally enriched RNAPII were identified as one of the three major classes of genes that each display a different RNAPII occupancy profile: enriched at promoters, immediately downstream of promoters, or biased toward 3′ ends [100]. These data highlight the importance to understand the detailed mechanism in yeast that underlies the RNAPII withholding at and release from promoter proximal regions.
6. Spt4-Spt5 coordinates transcription elongation with chromatin states
Several sets of observations suggest that Spt4-Spt5 may coordinate chromatin remodeling and histone modification with transcription elongation. First, in addition to the RNAPII mutations mentioned above, genetic suppressors of the cold-sensitive spt5 allele in S. cerevisiae reveal an extensive set of interactions between Spt4-Spt5 and chromatin. These chromatin suppressors of spt5 include: mutations in the ATP-dependent chromatin remodeler Chd1 [101]; the Paf1 complex [102] which associates with elongating RNAPII and coordinates a variety of histone modifications [103]; substitutions of histone H3 lysines 4 and 36 as well as mutations that inactivate their cognate methyltransferases Set1 and Set2 [48]; and subunits of the Rpd3S histone deacetylase complex [48], whose function depends upon H3K36 methylation [44]. Furthermore, mutations that inactivate the SAGA histone acetyltransferase complex also strongly enhance phenotypes of spt4 and spt5 mutations [48].
Interestingly, the chromatin suppressors of the spt5 cold-sensitive mutation also alleviate the synthetic growth defects observed in spt4 dst1 and spt5 dst1 (DST1 encodes TFIIS) double mutants [48]. Nucleosomes are known to provoke transcription arrest by elongating RNAPII [104]. Thus, one intriguing interpretation of this result is that Spt4-Spt5 protects elongating RNAPII from transcription arrest events provoked by nucleosomes, and that the chromatin suppressors of spt5 identify perturbations of chromatin that decrease the frequency of nucleosome-provoked transcription arrest. These data suggest that in addition to decreasing the frequency of pausing by RNAPII, Spt4-Spt5 likely also functions to help elongating RNAPII to overcome nucleosomal barriers to elongation. This model suggests that there are two mechanisms for suppression of the spt5 cold sensitive mutation. In the first, a slow polymerase gives the partially defective Spt5 protein additional time to act before RNAPII collides with a nucleosome, provoking a pause or arrest. In the second model, defective chromatin modification or remodeling lowers the nucleosomal barrier to elongation so that the probability of arrest and the requirement for Spt4-Spt5 function is decreased. Deeper examination of these ideas will likely require in vitro analysis of Spt4-Spt5 on chromatin templates.
A second set of observations suggests a mechanism by which Spt4-Spt5 may coordinate transcription with chromatin states. In S. cerevisiae, the P-TEFb homolog, Bur1 [105, 106], Spt4 [107] and the Spt5 CTR [73, 83] are required for recruitment of the Paf1 complex to elongating RNAPII, and for methylation of histone H3 lysine 4, one of the histone modifications regulated by the Paf1 complex [103]. Mutations of the Spt5 CTR that substitute alanines for the serine targets of Bur1 also abolish Paf1 recruitment [73]. In addition, Bur1, Spt4, and the Spt5 CTR also regulate association of the Rpd3S complex with chromatin [44]. Thus, like the RNAPII CTD, Spt5’s CTR appears to act as a phosphorylation-state dependent regulator of recruitment of transcription-associated chromatin modifying activities. It is appealing to speculate that the dependencies of Rpd3S and Paf1 complex chromatin-association on Spt4-Spt5 reflect direct and phospho-regulated binding of these complexes to the Spt5 CTR. However, forms of Spt5 lacking its CTR or that have been rendered phosphorylation-defective by threonine to alanine substitutions in the CTR can be co-precipitated with the Paf1 complex [77]. Thus the CTR may act indirectly to control factor recruitment.
7. Spt4-Spt5 and pre-mRNA processing
Spt4-Spt5 has also been implicated in a second set of co-transcriptional events, RNA processing. For example, the RNA capping enzymes of budding and fission yeast bind to Spt5 [25, 108]. This interaction, which depends upon Spt5’s CTR, may supply an overlapping function in addition to that mediated by RNAPII CTD-phosphorylation and the foot domain of RNAPII that assists the recruitment of capping enzyme [78, 109]. Furthermore, in S. cerevisiae, spt4 and spt5 mutations display synthetic growth defects with capping enzyme mutations, cause splicing defects, affect poly-adenylation site choice, and suppress certain mRNA export-defective mutations [108, 110–114]. Spt4-Spt5 interacts with the RNA export factor She2 and attracts She2 to the elongating RNAPII, thereby facilitating the recognition by She2 of the specific bud-localization signal in nascent RNA [115]. Interestingly, spt4 and spt5 mutations also alter rRNA processing, consistent with Spt4-Spt5’s association with RNA polymerase I [54, 56, 116].
These diverse roles for Spt4-Spt5 in RNA processing in yeast likely reflect similar functions in metazoans. In Drosophila, Spt5 interacts with the exosome, suggesting a potential role for Spt4-Spt5 in RNA degradation [117]. A recent RNA-seq analysis showed that knockdown of Spt4 also changes splicing patterns in mammalian cells [50]. Finally, human Spt5 interacts with the capping enzyme and stimulates capping activity in vitro [118].
An intriguing set of observations suggests that Spt4-Spt5 may influence RNA processing via direct interactions with pre-mRNAs. First, purified Spt4-Spt5 complexes from yeast bind RNA in a gel mobility assay [55, 119]. Second, using a native gel assay to examine transcription elongation complexes in vitro, Price and colleagues found that DSIF is recruited more efficiently to elongation complexes with long RNA transcripts [119]. Subsequently, Gilmour and colleagues used a similar analytical strategy to show that DSIF does not associate with elongation complexes whose nascent RNA transcripts are 18-nucleotide or shorter [120]. They also showed that Spt5 in these complexes could be crosslinked to nascent transcripts that were 22-nucleotide or longer, but not those which were 18 nucleotides long. Although not in agreement with earlier data suggesting that Spt4-Spt5/RNA polymerase interactions are RNAase resistant [108, 121], these data suggest that interactions between Spt5 and RNA are an important determinant of Spt4-Spt5’s interactions with RNAPII elongation complex. One intriguing possibility is that, by binding to nascent transcripts in elongation complexes, Spt4-Spt5 restricts the ability of RNAPII to backtrack and arrest, which necessarily requires that the nascent RNA be fed back into the elongation complex as it moves backwards on the template. This mechanism for an anti-pausing and -arrest activity has previously been suggested for U2AF65, which binds RNA, associates with elongating RNAPII and stimulates elongation in vitro under conditions of limiting nucleotides [122]. A more definitive test of this model awaits identification of RNA binding-defective forms of Spt5.
How might Spt4-Spt5 influence pre-mRNA processing? One possibility is that Spt4-Spt5 acts indirectly, influencing processing factor recruitment via its effects on the processivity and rate of RNAPII elongation. Consistent with this possibility, studies of splicing in yeast and metazoan systems show that elongation rate influences alternative splicing patterns (reviewed in [123]), and elongation-defective yeast strains, including spt4 and spt5 mutants, show upstream shifts in poly(A) site selection [112–114]. A second possibility is that Spt5’s CTR participates directly in recruiting processing factors to elongation complexes. This seems to be the case in capping. Moreover, ChIP analysis indicates a contributing role for the Spt5 CTR in RNA cleavage factor I in yeast [124]. Whether or not recruitment of other pre-mRNA processing factors depends upon Spt5’s CTR remains to be determined. However, the observation that bur1 mutations interfere with the splicing of a significant subset of yeast genes suggests that this question merits experimental analysis [111]. Finally, observations of the Spt5-RNA interaction suggest that Spt4-Spt5 may influence pre-mRNA processing reactions by binding to nascent transcripts soon after they emerge from the elongating RNAPII and possibly serving as a ‘gate keeper’ to regulate access of other processing factors to the nascent transcript.
8. Other functions for Spt4-Spt5?
A recent series of papers provide additional support for and insights into roles of Spt5 in regulating RNA processing. In plants, siRNA-mediated DNA methylation and gene silencing depends upon atypical RNA polymerases that are related to, but distinct from RNAPII [125]. One of these, RNA Polymerase V (RNAPV), is required for some, but perhaps not all instances of Ago4-mediated gene silencing [125, 126]. Interestingly, RNAPV associates with an Spt5 homolog, KTF1, also known as Spt5 Like (SPT5L) [127]. Like canonical Spt5, Spt5L is a large protein with a highly charged N-terminus followed by an NGN domain, several KOW domains and C-terminal repeats, in this case containing WG/GW dipeptides [128, 129]. These repeats mediate Spt5L’s association with Ago4, and they resemble WG/GW repeats found in other Ago4 binding proteins [128, 129]. ChIP assays demonstrate that Spt5L associates with chromatin, and Spt5L’s stability depends upon RNAPV, suggesting that it is recruited to chromatin by and functions in the context of its association with RNAPV [126]. Finally, like canonical Spt5, Spt5L binds RNA in vitro and in vivo [129].
Spt4-Spt5 has also been implicated in transcription-coupled DNA repair [82, 130, 131], a function it may execute as part of RNAPII elongation complexes. Immunoglobulin class-switch recombination and somatic hypermutation also requires mammalian Spt4-Spt5 [42, 132]. Mechanistically, Spt4-Spt5 facilitates the targeting of this pathway by specifically interacting with activation-induced cytidine deaminase (AID), a key enzyme that initiates these processes, and recruits AID to transcription sites where RNAPII dwells (or stalls). Since AID does not directly interact with RNAPII, Spt4-Spt5 most likely functions as an adaptor between AID and the RNAPII apparatus [42]. In addition, spt4 mutations affect chromosome segregation [30], and Spt4-Spt5 is required to restrict spread of the histone H3 variant Cse4 away from centromeres [31]. Interestingly, ChIP studies show that Spt4-Spt5 can be found in the vicinity of centromeres, and that this localization may not depend upon RNAPII [31].
As with other functions of Spt4-Spt5, it is likely that its ability to facilitate co-transcriptional events is an ancient function; in bacteria, NusG is required for Rho-dependent termination [133], and may couple transcription and translation via an interaction of its KOW domain and ribosomal protein S10 [134].
9. Structure of the Spt4-Spt5 complex
Several groups have recently made progress on determining the structure of the Spt4-Spt5 complex. Guo et al. solved the x-ray crystal structure of an Spt4-Spt5 NGN domain fusion protein [135]. This fusion protein formed domain-swapped dimers in which the Spt4 domain from one monomeric unit of the dimer bound to the NGN domain found in the other monomer. The central feature of the Spt4-Spt5 interface is a large beta-sheet formed by alignment of 4 anti-parallel strands from each of Spt4 and Spt5, creating a large hydrophobic surface [135]. Surrounding this surface are alpha helices, which contribute charged and polar interactions that appear to hold Spt4 and Spt5 in register. Mutations targeting predicted contacts between Spt4 and Spt5 disrupted Spt4-Spt5 interactions both in vitro and in vivo, suggesting that the structure provided an accurate model of in vivo interactions of Spt4 and Spt5.
Subsequent structural analysis of the human [136, 137] and several archaeal [62, 138–140] Spt4-Spt5 complexes support the model of Guo et al. The structures of these different Spt4-Spt5 complexes superimpose on top of one another with very small deviations (Figure 1C–D). Importantly, one of the archaeal Spt4-Spt5 structures includes the single KOW domain found in archaeal members of the Spt5 protein family [138], and an NMR structure of the fifth KOW domain of human Spt5 displays a similar structure (PDB ID: 2E70).
10. Structural analysis of Spt4-Spt5/RNA polymerase interactions
It is likely that all of the diverse functions of Spt4-Spt5 are mediated in the context of its physical interactions with RNAPII. Protein mapping analysis in the human system showed that RNAPII binding involves a middle region of Spt5 encompassing the multiple KOW domains, while Spt4 binding involves the NGN domain of Spt5 [75, 76]. The RNAPII–interacting domains of Spt4-Spt5 were further mapped in archaea by Werner and collaborators. They found that in addition to binding Spt4, the Spt5 NGN domain also interacts with RNA polymerase, and is required for the transcription stimulation function of Spt4-Spt5 in vitro [62]. Furthermore, they observed that the affinity of the Spt5 NGN alone for polymerase was not as strong as that of the intact Spt4-Spt5 complex and that the NGN’s affinity for RNA polymerase can be enhanced by including either Spt4 or recombinant Spt5 proteins with both the NGN and KOW domains in the binding reactions [62]. Thus, although direct Spt4/polymerase interactions have not been observed, Spt4 may influence Spt5/polymerase interactions.
Spt4 may also regulate the stability of Spt5. Spt5 proteins levels drop to about 1/3 of normal in yeast cells that lack Spt4 [82] and Spt4 enhances the thermal stability of archaeal Spt5 [62]. These data may explain the temperature sensitivity of some yeast spt4 mutants [30]. Collectively, these data indicate a distributed, multi-domain interaction between Spt5 and the polymerase that is modulated by Spt4. The complexity of the interface drives home the point that we do not yet fully understand the regulation and functional importance of Spt4-Spt5/polymerase interactions.
Further structural and mutagenesis data from the bacterial and archaeal systems have identified the Clamp domain of RNA polymerase as the binding target of the NusG/Spt5 proteins [61, 62, 141]. More precisely, this interface is formed between a concave patch of hydrophobic residues on the NGN domain of NusG/Spt5 and the coiled-coil (CC) motif of the Clamp domain of the largest subunit of RNA polymerases. These structural features are also found in eukaryotic Spt5 and RNAPII, suggesting a universally conserved interaction surface.
A detailed description of the mechanisms by which Spt4-Spt5 executes its diverse functions will require a clear understanding of the structure of the entire Spt4-Spt5/RNAPII complex. However, Spt5’s multi-domain structure and the likely unstructured nature of its N- and C-termini present obstacles to obtaining crystals of this large complex. Still, clever alternatives to obtaining tertiary information have been taken successfully with the archaeal homologs. Recent reviews describe much of the structural and biochemical findings in both the archaeal and bacterial systems with a reasonable extrapolation to the eukaryotic complex [142, 143]. As such, we will provide a brief summary of the main features in the current model for tertiary organization of the Spt4-Spt55 complex with RNAPII.
Initial clues to the organization of Spt5/NusG interactions with RNA polymerase came from extensive biochemical and molecular genetic analyses of RfaH, an unusual bacterial member of the NusG family. Unlike NusG, which operates at all genes, RfaH is recruited to particular genes by binding to a specific DNA sequence in the non-transcribed strand of target genes [144]. Artsimovitch and colleagues identified a potential RfaH/RNA polymerase interaction surface in a hydrophobic pocket of the NGN domain of RfaH and in the conserved CC motif of the RNA polymerase clamp domain [141]. Mutations in these surfaces appear to disrupt RfaH/RNA polymerase interactions [141, 145, 146]. These data led to a model in which NusG spans the central cleft of elongating RNA polymerase, sealing nucleic acids in the elongation complex and thereby promoting processivity. Consistent with this model, RfaH makes a second, possibly dynamic, contact with RNA polymerase at the gate loop (GL) element of the β subunit on the other side of the central cleft, so that it completely seals the central cleft [147]. Interestingly, although the RfaH/GL interaction does not appear to make a significant contribution to the strength of the RfaH/RNA polymerase interaction, it is essential for RfaH’s (and NusG’s) ability to prevent pausing [145]. Importantly, the GL, CC domain of the Clamp and the hydrophobic pocket of Spt5 are all conserved features in archaea and eukaryotes.
This view of the association of NusG with RNA polymerase has subsequently been supported by structural studies of Spt5/RNA polymerase interactions in archaea. To circumvent the difficulty of crystallizing a complete Spt5/polymerase complex, Cramer’s group crystallized a Pyrococcus furiosus archaeal complex formed between Spt4-Spt5 and elements of the Clamp domain of the cognate RNA polymerase [139]. Their structure, determined to 3.3 Å resolution, verified the predicted Spt5-Clamp interface described above and, when superimposed over multiple aligned sequences of NGN of various organisms, verified the conservation of this interface. Based on this conservation and the available 3-D structure of another archaeal (Sulfolobus sulfataricus) RNA polymerase, the researchers formulated a 3-D model of the archaeal Spt5/polymerase complex. Applying the same modeling procedure, they also modeled the yeast Spt4-Spt5/RNAPII (Figure. 2A) and bacterial NusG/polymerase (Figure. 2B) complexes.
Figure 2.
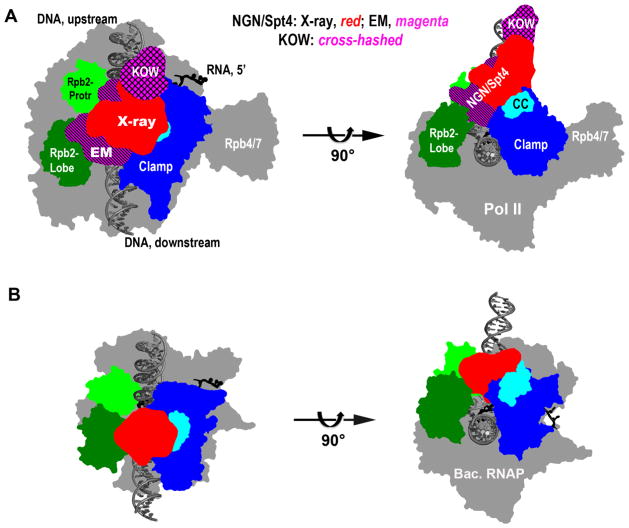
Quaternary organization of (A) Spt4-Spt5-RNA polymerase elongation complex of archaea and eukaryotes and (B) bacterial NusG-polymerase elongation complex. Left hand images present a top view of the complexes and right hand images a front view. Significant structural domains are marked and colored differently. Two locations of the Spt5 NGN domain are included in A, with the X-ray and EM results rendered in red and magenta, respectively.
In these semi-crystallographic models, the NGN/Spt4 complex (Figure 2A, red) binds to the Clamp (blue) near the tip of the CC motif (cyan) of Clamp, and sits over the main nucleic acid cleft in the polymerase, enclosing the DNA/RNA hybrid in RNAPII (Figure 2A). As with the bacterial complex [141, 147] these models place the NGN domain near the non-coding DNA strand in transcription bubble. This raises the possibility that Spt5/NusG may promote polymerase processivity by helping maintain the downstream edge of the transcription bubble by withholding the non-coding DNA strand through non sequence-specific interactions. Consistent with this idea, RfaH can be crosslinked to the non-transcribed strand in elongation complexes [144]. Although the X-ray data did not provide information on the location of the sole KOW domain in archaeal Spt5, a consideration of the linker length from NGN to KOW would argue against a placement of the KOW anywhere near the RNA exit pore on RNAPII [139].
The crystallographic data described above are in general agreement with a low-resolution (13 Å) structure determined by cryo-EM single particle analysis of a complete Spt4/5-polymerase complex also from Pyrococcus furiosus [138]. However, the EM data indicated a shift of the NGN/Spt4 structure (Figure 2A, magenta) toward the opposite side of the nucleic acid cleft, making contacts with domains that are correspondingly named Protrusion (light green) and Lobe (dark green) in the RNAPII structure. While the KOW was not observed in the crystal structure, the EM map revealed a region of density (Figure 2A, cross-hashed) that could be docked with a KOW structure. This places the archaeal KOW near the upstream end of the transcription bubble, but its weak connection to the NGN/Spt4 density dampens the confidence of its identification.
Could the difference between the crystallographic and EM results reflect different aspects of the same complex? A closer examination of the EM structure indicates that the site of NGN contact on the Lobe domain is located at the tip of Lobe, which shares similarity to the flexible GL element in the bacterial RNA polymerase. Two possible mechanisms consistent with the current data of GL activity can be envisaged: (i) the flexible GL structure may serve as a dynamic gate to ‘seal’ off the gap left at the top of the nucleic acid cleft after the binding of an NGN domain-containing elongation factor; and/or (ii) the edge of the bound NGN domain may contact the GL in a dynamic fashion to close the remaining space and/or alter the intrinsic activity of RNA polymerase [147]. Either of these scenarios will yield a population-averaged structure similar to the EM image of Klein et al. in which the protein mass will appear to be continuous. In this sense, the GL, Clamp and NGN domains may associate in multiple modes, resulting in visualization of complexes with ambiguous structures due to superposition of multiple related but subtly distinct structures, a result obtained often in structural analysis of transcription and signaling networks [109, 148, 149].
11. Mechanistic Insights
The current structural model for Spt4-Spt5 bound to RNA polymerase offers a simple mechanism to explain their functions in transcription elongation. The binding of Spt4-Spt5 to the RNAPII elongation complexes will completely encircle the DNA template and RNA/DNA hybrid. The NGN domain may also affect the dynamics of the Clamp. The Clamp is a major structural element that RNAPII uses to engage DNA and to hold the 8–9 base pair DNA/RNA hybrid before the strands separate [150]. Five loop structures (named Switch 1–5) are located near the base of this mobile domain and are seen in the ternary complex making intimate interactions with nucleic acids adjacent to the NTP addition site [150, 151]. Mutations of Clamp residues conceivably impact transcription functions such as the processivity during early stages of elongation, as has been demonstrated for a Switch-2 region mutant [152]. Either by directly restraining the DNA and RNA in the cleft, or allosterically affecting the Switch conformations, or combined, these mechanisms may altogether prevent disassembly of elongation complexes and thus ensure processivity.
Spt4-Spt5 appears to interact exclusively with structural elements on the surface of RNAPII and not directly probe the active site of the polymerase. The best current data indicate the Spt4-Spt5’s closest point of contact with the substrate is its interactions with the non-coding DNA strand, and this is located over the top of the cleft, away from the catalytic center. If Spt4-Spt5 exerts a conformational influence on RNAPII active site, it is most likely achieved via allosteric mechanisms. This stands in contrast to the direct mechanism used by elongation factor TFIIS (or SII). Extensive biochemical and structural analyses show that TFIIS binds RNAPII and inserts its Zn-ribbon domain (domain III) into the secondary channel (also called Funnel) of the polymerase, and, in an almost acrobatic fashion, projects its β hairpin of Zn-ribbon domain close to the polymerase catalytic site [153]. The net result of this binding is a precise placement of three charged groups (R287, D290 and E291) at the tip of the hairpin right next to the polymerase catalytic center where phosphodiester bond formation (and its reversal hydrolysis) occurs. This structural model satisfactorily explains the TFIIS function in stimulating the intrinsic RNA cleavage activity and proofreading function of RNAPII [154, 155]. Chiefly, the charged loop-tip residues of TFIIS augment the RNAPII catalytic center in its capacity to stabilize the second Mg ion and also a water molecule that attacks the scissile phosphodiester bond [156, 157]. Additionally, the inserted TFIIS hairpin appears to help maintain the trigger-loop of the largest subunit in a catalytically functional conformation.
Interestingly, like Spt4-Spt5, the general transcription factor TFIIF also appears to use indirect mechanisms to regulate polymerase elongation. (Whether its other functions during PIC assembly involve direct interactions with partner factors such as TFIIB is currently unclear.) Stimulation of elongation has been well demonstrated for metazoan TFIIF [158–160], although not for its fungal counterpart. Biochemical mapping using protein cleavage and crosslinking-coupled mass spectrometry has demarcated TFIIF’s binding sites on RNAPII: the two large subunits of yeast TFIIF interact with the Rpb2 Lobe and Protrusion domains that form one side of the DNA/RNA cleft [161, 162]. The binding area seems to extend toward the Rpb1 Jaw as revealed in the latest structural visualization using EM [163]. All the binding sites that have been elucidated to date are distributed on the surfaces that are immediately outside of the DNA/RNA cleft; current data do not indicate a close approach of any TFIIF element into the cleft or catalytic center. This binding strategy likely explains the functional competition of TFIIF with the newly identified negative regulator Gdown1 of mammals [163–165]. Since mammalian TFIIF also functionally competes with DSIF [166], the question arises: does TFIIF also compete with Spt4-Spt5 for a common binding site(s)? Observing functional interactions between a TFIIF mutation and a Switch-2 mutation, Ponticelli and coworkers proposed an allosteric mechanism in which TFIIF can indirectly affect the conformation around the polymerase cleft and hence the stability of early elongation complexes having short transcripts (e.g. 5-mer) [167]. It remains to be tested whether Spt4-Spt5 can ‘remotely’ modulate RNAPII conformation at interiors of the cleft (e.g. the Switches) or even the active site.
These structural models suggest that Spt4-Spt5/NusG proteins bind polymerase at sites that are also bound by transcription initiation factors. In bacteria, sigma factor binds to the CC domain in the Clamp [168, 169], and RfaH functionally competes with sigma factor [147, 170]. In eukaryotes, initiation factor TFIIE binds to the Clamp of RNAPII [162] at a site that appears to overlap the NGN binding site. This conflict suggests that TFIIE and Spt4-Spt5 may compete for access to the Clamp. Werner and coworkers used single-molecule FRET to directly address this problem in archaea [171]. Their results confirm (i) an overlap (e.g. the shared CC motif) between the RNA polymerase binding sites of the archaeal TFIIE and Spt4-Spt5, and (ii) the predicted binding competition between these factors [171]. More interestingly, their transcription activity assay reveals that the relative abilities of Spt4-Spt5 and TFE to compete for polymerase depends upon the functional state of the polymerase complex: TFE out-competes the inhibitory effect of Spt4-Spt5 on pre-initiation complex assembly; whereas Spt4-Spt5 displaces TFE from the elongation complex, and hence may exert a positive effect on transcription by facilitating the factor handover process during the transition from initiation to elongation [41, 172].
Factor competition during the initiation-to-elongation transition may also be coordinated with progression of the early elongation complex, particularly in terms of accumulated length of the transcript. Cheng and Price have shown that DSIF enjoys an increased affinity toward complexes with longer RNAs (>35 nucleotides) [119], consistent with the RNA gel-shift data mentioned earlier, RNA-Spt5 crosslinking within an elongation complex [120], and the proposal that Spt4-Spt5 directly interacts with the nascent transcript emerging from the RNA exit pore. If so, one may speculate that Spt4-Spt5 bridges the Rpb1 Clamp and the 5′ RNA to form a structural relay that could coordinate the status of 5′ end of the transcript with events occurring inside the central cleft or even at the 3′ end of the transcript. It might also be possible that such bridging could serve as a ratchet to help prevent the 5′ RNA from retrograde movement and hence reduce pausing. A similar binding organization on RNAPII has been demonstrated for the RNA splicing factor U2AF65 as mentioned earlier. Progressive change of RNAPII conformation along the transcription coordinate (from initiation to promoter clearance to elongation) might be another parameter that influences Spt4-Spt5 interactions [66]. This mechanism might be especially significant when RNAPII traverses from ~20 to ~30 nucleotides during which range the polymerase displays a change of behaviors in terms of its tendency to backtrack [173] and slip over repetitive sequences [174].
Several additional difficulties may be anticipated for efforts to solve the complete structure of eukaryotic Spt4-Spt5 complexes. First, these Spt5 proteins contain five to six KOW domains separated by linker sequences that are predicted to be disordered (Fu, unpublished). It is reasonable to expect that some of these domains directly participate in coupling RNAPII elongation with various other nuclear pathways including nucleosome modification, RNA processing and DNA modifications. However, it is not known if the different KOW domains serve distinct or overlapping functions. Furthermore, if two or more of the KOW domains can interact with the same surface on RNAPII, it may be difficult to obtain a uniform population of complexes for structural studies of Spt4-Spt5/RNAPII interactions. In addition to problems presented by the KOW domains of eukaryotic Spt5, protein disorder prediction algorithms indicate that both the N-terminal acidic and the CTR regions do not adopt a stable fold (Fu, unpublished). Thus, as is the case for the RNAPII CTD [175], Spt5’s N- and C-termini are likely not amenable to crystallographic analysis.
How could Spt4/5’s association with RNAPII be regulated by other cellular factors? Gaynor and coworkers have identified consensus arginine motifs between human KOW domains 3 and 4 that are recognized by protein arginine methyltransferase (PRMT) 1 and 5 [176]. Inhibition of methylation, either by mutating the targeted residues or by using a methylation inhibitor, enhanced Spt4-Spt5 activity in terms of both DRB-induced inhibition and Tat-induced activation. Further, they have found that methylation inhibition is associated with increased association of Spt4-Spt5 with the IL-8 promoter as assayed by ChIP and elevated association with RNAPII as measured by immunoprecipitation and Western blotting. The broader significance of these findings has yet to be tested in a genome-wide study. However, it is conceivable that PRMT-mediated methylation of Spt5 may modulate the stability of elongation complexes and promote their turnover during termination; facilitated removal of Spt4-Spt5 may allow RNAPII to disengage from the DNA template and be prepared for reinitiation by leaving an un-obstructed nucleic acid cleft. An understanding of how methylation of Spt5 modulates its association with RNAPII will require structural information as to where KOW 3 and 4 are placed on the surface of RNAPII.
12. Perspectives
Tremendous progress has been made in elucidation of the structure and function of Spt4-Spt5 complexes. We now have a clear view of the deep structural and functional conservation shared among members of the Spt5/NusG family of proteins. However, a comprehensive structural framework is not yet available for understanding how Spt4-Spt5 engages and modulates the properties of elongating RNAPII nor for understanding how other nuclear functions are temporally and mechanistically linked to transcription elongation. Future challenges include: obtaining more complete structures for Spt4-Spt5/RNA polymerase complexes; determining how Spt4-Spt5 impacts the enzymatic mechanisms of RNAPII and its translocation down a DNA template; understanding how Spt4-Spt5 may participate in regulatory transitions from initiation to elongation and elongation to termination; and more deeply examining Spt4-Spt5’s roles in RNA processing and chromatin remodeling.
Highlights.
Spt4-Spt5 is a positive and negative regulator of transcription elongation.
Spt5/NusG proteins are found in all organisms and are essential for life.
Spt4-Spt5 links transcription elongation to chromatin remodeling and RNA processing.
Structural studies are beginning to provide insights into Spt4-Spt5 function.
Acknowledgments
This work was supported in part by grants to GAH and JF from the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Grant A Hartzog, Email: hartzog@ucsc.edu.
Jianhua Fu, Email: jfu@mcw.edu.
References
- 1.Izban MG, Luse DS. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. The Journal of biological chemistry. 1992;267:13647–13655. [PubMed] [Google Scholar]
- 2.Izban MG, Luse DS. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes & development. 1991;5:683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- 3.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 4.Werner F, Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol. 2011;9:85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- 5.Allison LA, Moyle M, Shales M, Ingles CJ. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985;42:599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- 6.Korzheva N, Mustaev A. Transcription elongation complex: structure and function. Curr Opin Microbiol. 2001;4:119–125. doi: 10.1016/s1369-5274(00)00176-4. [DOI] [PubMed] [Google Scholar]
- 7.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 8.Harris JK, Kelley ST, Spiegelman GB, Pace NR. The genetic core of the universal ancestor. Genome Res. 2003;13:407–412. doi: 10.1101/gr.652803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winston F, Chaleff DT, Valent B, Fink GR. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics. 1984;107:179–197. doi: 10.1093/genetics/107.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malone EA, Fassler JS, Winston F. Molecular and genetic characterization of SPT4, a gene important for transcription initiation in Saccharomyces cerevisiae. Mol Gen Genet. 1993;237:449–459. doi: 10.1007/BF00279450. [DOI] [PubMed] [Google Scholar]
- 11.Swanson MS, Malone EA, Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Molecular and cellular biology. 1991;11:4286. doi: 10.1128/mcb.11.8.4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swanson MS, Winston F. SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics. 1992;132:325–336. doi: 10.1093/genetics/132.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes & development. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fish RN, Kane CM. Promoting elongation with transcript cleavage stimulatory factors. Biochimica et biophysica acta. 2002;1577:287–307. doi: 10.1016/s0167-4781(02)00459-1. [DOI] [PubMed] [Google Scholar]
- 15.Exinger F, Lacroute F. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1992;22:9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- 16.Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Molecular cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 17.Noble SM, Guthrie C. Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics. 1996;143:67–80. doi: 10.1093/genetics/143.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chodosh LA, Fire A, Samuels M, Sharp PA. 5,6-Dichloro-1-beta-D-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. The Journal of biological chemistry. 1989;264:2250–2257. [PubMed] [Google Scholar]
- 19.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes & development. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo S, Yamaguchi Y, Schilbach S, Wada T, Lee J, Goddard A, French D, Handa H, Rosenthal A. A regulator of transcriptional elongation controls vertebrate neuronal development. Nature. 2000;408:366–369. doi: 10.1038/35042590. [DOI] [PubMed] [Google Scholar]
- 21.Ponting CP. Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic acids research. 2002;30:3643–3652. doi: 10.1093/nar/gkf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charier G, Couprie J, Alpha-Bazin B, Meyer V, Quemeneur E, Guerois R, Callebaut I, Gilquin B, Zinn-Justin S. The Tudor tandem of 53BP1: a new structural motif involved in DNA and RG-rich peptide binding. Structure. 2004;12:1551–1562. doi: 10.1016/j.str.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Ponting CP. Tudor domains in proteins that interact with RNA. Trends Biochem Sci. 1997;22:51–52. doi: 10.1016/s0968-0004(96)30049-2. [DOI] [PubMed] [Google Scholar]
- 24.Lasko P. Tudor domain. Curr Biol. 2010;20:R666–667. doi: 10.1016/j.cub.2010.05.056. [DOI] [PubMed] [Google Scholar]
- 25.Pei Y, Shuman S. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. The Journal of biological chemistry. 2002;277:19639–19648. doi: 10.1074/jbc.M200015200. [DOI] [PubMed] [Google Scholar]
- 26.Keegan BR, Feldman JL, Lee DH, Koos DS, Ho RK, Stainier DY, Yelon D. The elongation factors Pandora/Spt6 and Foggy/Spt5 promote transcription in the zebrafish embryo. Development (Cambridge, England) 2002;129:1623–1632. doi: 10.1242/dev.129.7.1623. [DOI] [PubMed] [Google Scholar]
- 27.Jennings BH, Shah S, Yamaguchi Y, Seki M, Phillips RG, Handa H, Ish-Horowicz D. Locus-specific requirements for Spt5 in transcriptional activation and repression in Drosophila. Curr Biol. 2004;14:1680–1684. doi: 10.1016/j.cub.2004.08.066. [DOI] [PubMed] [Google Scholar]
- 28.Shim EY, Walker AK, Shi Y, Blackwell TK. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes & development. 2002;16:2135–2146. doi: 10.1101/gad.999002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Molecular cell. 2006;21:227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 30.Basrai MA, Kingsbury J, Koshland D, Spencer F, Hieter P. Faithful chromosome transmission requires Spt4p, a putative regulator of chromatin structure in Saccharomyces cerevisiae. Molecular and cellular biology. 1996;16:2838–2847. doi: 10.1128/mcb.16.6.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crotti LB, Basrai MA. Functional roles for evolutionarily conserved Spt4p at centromeres and heterochromatin in Saccharomyces cerevisiae. The EMBO journal. 2004;23:1804–1814. doi: 10.1038/sj.emboj.7600161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deuring R, Fanti L, Armstrong JA, Sarte M, Papoulas O, Prestel M, Daubresse G, Verardo M, Moseley SL, Berloco M, Tsukiyama T, Wu C, Pimpinelli S, Tamkun JW. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Molecular cell. 2000;5:355–365. doi: 10.1016/s1097-2765(00)80430-x. [DOI] [PubMed] [Google Scholar]
- 33.Chiang PW, Wang SQ, Smithivas P, Song WJ, Crombez E, Akhtar A, Im R, Greenfield J, Ramamoorthy S, Van Keuren M, Blackburn CC, Tsai CH, Kurnit DM. Isolation and characterization of the human and mouse homologues (SUPT4H and Supt4h) of the yeast SPT4 gene. Genomics. 1996;34:368–375. doi: 10.1006/geno.1996.0299. [DOI] [PubMed] [Google Scholar]
- 34.Hartzog GA, Basrai MA, Ricupero-Hovasse SL, Hieter P, Winston F. Identification and analysis of a functional human homolog of the SPT4 gene of Saccharomyces cerevisiae. Molecular and cellular biology. 1996;16:2848–2856. doi: 10.1128/mcb.16.6.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim DK, Inukai N, Yamada T, Furuya A, Sato H, Yamaguchi Y, Wada T, Handa H. Structure-function analysis of human Spt4: evidence that hSpt4 and hSpt5 exert their roles in transcriptional elongation as parts of the DSIF complex. Genes Cells. 2003;8:371–378. doi: 10.1046/j.1365-2443.2003.00638.x. [DOI] [PubMed] [Google Scholar]
- 36.Tardiff DF, Abruzzi KC, Rosbash M. Protein characterization of Saccharomyces cerevisiae RNA polymerase II after in vivo cross-linking. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19948–19953. doi: 10.1073/pnas.0710179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bourgeois CF, Kim YK, Churcher MJ, West MJ, Karn J. Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Molecular and cellular biology. 2002;22:1079–1093. doi: 10.1128/MCB.22.4.1079-1093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ping YH, Chu CY, Cao H, Jacque JM, Stevenson M, Rana TM. Modulating HIV-1 replication by RNA interference directed against human transcription elongation factor SPT5. Retrovirology. 2004;1:46. doi: 10.1186/1742-4690-1-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nature structural & molecular biology. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nature structural & molecular biology. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 42.Pavri R, Gazumyan A, Jankovic M, Di Virgilio M, Klein I, Ansarah-Sobrinho C, Resch W, Yamane A, Reina San-Martin B, Barreto V, Nieland TJ, Root DE, Casellas R, Nussenzweig MC. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drouin S, Laramee L, Jacques PE, Forest A, Bergeron M, Robert F. DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS genetics. 2010;6:e1001173. doi: 10.1371/journal.pgen.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andrulis ED, Guzman E, Doring P, Werner J, Lis JT. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes & development. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaplan CD, Morris JR, Wu C, Winston F. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes & development. 2000;14:2623–2634. doi: 10.1101/gad.831900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morillon A, Karabetsou N, O’Sullivan J, Kent N, Proudfoot N, Mellor J. Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell. 2003;115:425–435. doi: 10.1016/s0092-8674(03)00880-8. [DOI] [PubMed] [Google Scholar]
- 48.Quan TK, Hartzog GA. Histone H3K4 and K36 methylation, Chd1 and Rpd3S oppose the functions of Saccharomyces cerevisiae Spt4-Spt5 in transcription. Genetics. 2010;184:321–334. doi: 10.1534/genetics.109.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rondon AG, Garcia-Rubio M, Gonzalez-Barrera S, Aguilera A. Molecular evidence for a positive role of Spt4 in transcription elongation. The EMBO journal. 2003;22:612–620. doi: 10.1093/emboj/cdg047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu CR, Chang CR, Chern Y, Wang TH, Hsieh WC, Shen WC, Chang CY, Chu IC, Deng N, Cohen SN, Cheng TH. Spt4 is selectively required for transcription of extended trinucleotide repeats. Cell. 2012;148:690–701. doi: 10.1016/j.cell.2011.12.032. [DOI] [PubMed] [Google Scholar]
- 51.Wu-Baer F, Lane WS, Gaynor RB. Role of the human homolog of the yeast transcription factor SPT5 in HIV-1 Tat-activation. Journal of molecular biology. 1998;277:179–197. doi: 10.1006/jmbi.1997.1601. [DOI] [PubMed] [Google Scholar]
- 52.Ainbinder E, Amir-Zilberstein L, Yamaguchi Y, Handa H, Dikstein R. Elongation inhibition by DRB sensitivity-inducing factor is regulated by the A20 promoter via a novel negative element and NF-kappaB. Molecular and cellular biology. 2004;24:2444–2454. doi: 10.1128/MCB.24.6.2444-2454.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amir-Zilberstein L, Dikstein R. Interplay between E-box and NF-kappaB in regulation of A20 gene by DRB sensitivity-inducing factor (DSIF) The Journal of biological chemistry. 2008;283:1317–1323. doi: 10.1074/jbc.M706767200. [DOI] [PubMed] [Google Scholar]
- 54.Anderson SJ, Sikes ML, Zhang Y, French SL, Salgia S, Beyer AL, Nomura M, Schneider DA. The transcription elongation factor Spt5 influences transcription by RNA polymerase I positively and negatively. The Journal of biological chemistry. 2011;286:18816–18824. doi: 10.1074/jbc.M110.202101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Viktorovskaya OV, Appling FD, Schneider DA. Yeast transcription elongation factor Spt5 associates with RNA polymerase I and RNA polymerase II directly. The Journal of biological chemistry. 2011;286:18825–18833. doi: 10.1074/jbc.M110.202119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schneider DA, French SL, Osheim YN, Bailey AO, Vu L, Dodd J, Yates JR, Beyer AL, Nomura M. RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12707–12712. doi: 10.1073/pnas.0605686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burova E, Hung SC, Sagitov V, Stitt BL, Gottesman ME. Escherichia coli NusG protein stimulates transcription elongation rates in vivo and in vitro. J Bacteriol. 1995;177:1388–1392. doi: 10.1128/jb.177.5.1388-1392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burns CM, Richardson LV, Richardson JP. Combinatorial effects of NusA and NusG on transcription elongation and Rho-dependent termination in Escherichia coli. Journal of molecular biology. 1998;278:307–316. doi: 10.1006/jmbi.1998.1691. [DOI] [PubMed] [Google Scholar]
- 59.Zellars M, Squires CL. Antiterminator-dependent modulation of transcription elongation rates by NusB and NusG. Mol Microbiol. 1999;32:1296–1304. doi: 10.1046/j.1365-2958.1999.01442.x. [DOI] [PubMed] [Google Scholar]
- 60.Li J, Horwitz R, McCracken S, Greenblatt J. NusG, a new Escherichia coli elongation factor involved in transcriptional antitermination by the N protein of phage lambda. The Journal of biological chemistry. 1992;267:6012–6019. [PubMed] [Google Scholar]
- 61.Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R. Regulator trafficking on bacterial transcription units in vivo. Molecular cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hirtreiter A, Damsma GE, Cheung AC, Klose D, Grohmann D, Vojnic E, Martin AC, Cramer P, Werner F. Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic acids research. 2010;38:4040–4051. doi: 10.1093/nar/gkq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shilatifard A. Transcriptional elongation control by RNA polymerase II: a new frontier. Biochimica et biophysica acta. 2004;1677:79–86. doi: 10.1016/j.bbaexp.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 64.Svetlov V, Belogurov GA, Shabrova E, Vassylyev DG, Artsimovitch I. Allosteric control of the RNA polymerase by the elongation factor RfaH. Nucleic acids research. 2007;35:5694–5705. doi: 10.1093/nar/gkm600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu W, Wada T, Okabe S, Taneda T, Yamaguchi Y, Handa H. DSIF contributes to transcriptional activation by DNA-binding activators by preventing pausing during transcription elongation. Nucleic acids research. 2007;35:4064–4075. doi: 10.1093/nar/gkm430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim HJ, Jeong SH, Heo JH, Jeong SJ, Kim ST, Youn HD, Han JW, Lee HW, Cho EJ. mRNA capping enzyme activity is coupled to an early transcription elongation. Molecular and cellular biology. 2004;24:6184–6193. doi: 10.1128/MCB.24.14.6184-6193.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 69.Narita T, Yamaguchi Y, Yano K, Sugimoto S, Chanarat S, Wada T, Kim DK, Hasegawa J, Omori M, Inukai N, Endoh M, Yamada T, Handa H. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Molecular and cellular biology. 2003;23:1863–1873. doi: 10.1128/MCB.23.6.1863-1873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Molecular cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 71.Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. The Journal of biological chemistry. 2001;276:31793–31799. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- 72.Ni Z, Saunders A, Fuda NJ, Yao J, Suarez JR, Webb WW, Lis JT. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Molecular and cellular biology. 2008;28:1161–1170. doi: 10.1128/MCB.01859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu Y, Warfield L, Zhang C, Luo J, Allen J, Lang WH, Ranish J, Shokat KM, Hahn S. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Molecular and cellular biology. 2009;29:4852–4863. doi: 10.1128/MCB.00609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pei Y, Shuman S. Characterization of the Schizosaccharomyces pombe Cdk9/Pch1 protein kinase: Spt5 phosphorylation, autophosphorylation, and mutational analysis. The Journal of biological chemistry. 2003;278:43346–43356. doi: 10.1074/jbc.M307319200. [DOI] [PubMed] [Google Scholar]
- 75.Ivanov D, Kwak YT, Guo J, Gaynor RB. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Molecular and cellular biology. 2000;20:2970–2983. doi: 10.1128/mcb.20.9.2970-2983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H. Structure and function of the human transcription elongation factor DSIF. The Journal of biological chemistry. 1999;274:8085–8092. doi: 10.1074/jbc.274.12.8085. [DOI] [PubMed] [Google Scholar]
- 77.Chen Y, Yamaguchi Y, Tsugeno Y, Yamamoto J, Yamada T, Nakamura M, Hisatake K, Handa H. DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes & development. 2009;23:2765–2777. doi: 10.1101/gad.1834709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schneider S, Pei Y, Shuman S, Schwer B. Separable functions of the fission yeast Spt5 carboxyl-terminal domain (CTD) in capping enzyme binding and transcription elongation overlap with those of the RNA polymerase II CTD. Molecular and cellular biology. 2010;30:2353–2364. doi: 10.1128/MCB.00116-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lindstrom DL, Hartzog GA. Genetic interactions of Spt4-Spt5 and TFIIS with the RNA polymerase II CTD and CTD modifying enzymes in Saccharomyces cerevisiae. Genetics. 2001;159:487–497. doi: 10.1093/genetics/159.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Z, Wu CH, Gilmour DS. Analysis of polymerase II elongation complexes by native gel electrophoresis. Evidence for a novel carboxyl-terminal domain-mediated termination mechanism. The Journal of biological chemistry. 2004;279:23223–23228. doi: 10.1074/jbc.M402956200. [DOI] [PubMed] [Google Scholar]
- 81.Cheng B, Price DH. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. The Journal of biological chemistry. 2007;282:21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- 82.Ding B, LeJeune D, Li S. The C-terminal repeat domain of Spt5 plays an important role in suppression of Rad26-independent transcription coupled repair. The Journal of biological chemistry. 2010;285:5317–5326. doi: 10.1074/jbc.M109.082818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou K, Kuo WH, Fillingham J, Greenblatt JF. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6956–6961. doi: 10.1073/pnas.0806302106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gilchrist DA, Adelman K. Coupling polymerase pausing and chromatin landscapes for precise regulation of transcription. Biochimica et biophysica acta. 2012 doi: 10.1016/j.bbagrm.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eberhardy SR, Farnham PJ. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. The Journal of biological chemistry. 2002;277:40156–40162. doi: 10.1074/jbc.M207441200. [DOI] [PubMed] [Google Scholar]
- 86.Kanazawa S, Soucek L, Evan G, Okamoto T, Peterlin BM. c-Myc recruits P-TEFb for transcription, cellular proliferation and apoptosis. Oncogene. 2003;22:5707–5711. doi: 10.1038/sj.onc.1206800. [DOI] [PubMed] [Google Scholar]
- 87.Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nature structural & molecular biology. 2010;17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, Florens L, Seidel CW, Lin C, Smith ER, Shilatifard A, Conaway RC, Conaway JW. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 2011;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Malik S, Barrero MJ, Jones T. Identification of a regulator of transcription elongation as an accessory factor for the human Mediator coactivator. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6182–6187. doi: 10.1073/pnas.0608717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee SK, Fletcher AG, Zhang L, Chen X, Fischbeck JA, Stargell LA. Activation of a poised RNAPII-dependent promoter requires both SAGA and mediator. Genetics. 2010;184:659–672. doi: 10.1534/genetics.109.113464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kremer SB, Kim S, Jeon JO, Moustafa YW, Chen A, Zhao J, Gross DS. Role of Mediator in Regulating Pol II Elongation and Nucleosome Displacement in Saccharomyces cerevisiae. Genetics. 2012;191:95–106. doi: 10.1534/genetics.111.135806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meyer PA, Fu J. Mutual remodeling and conformation grid: a mediator code? Structure. 2012;20:755–757. doi: 10.1016/j.str.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Amir-Zilberstein L, Ainbinder E, Toube L, Yamaguchi Y, Handa H, Dikstein R. Differential regulation of NF-kappaB by elongation factors is determined by core promoter type. Molecular and cellular biology. 2007;27:5246–5259. doi: 10.1128/MCB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Radonjic M, Andrau JC, Lijnzaad P, Kemmeren P, Kockelkorn TT, van Leenen D, van Berkum NL, Holstege FC. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Molecular cell. 2005;18:171–183. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 96.Baugh LR, Demodena J, Sternberg PW. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science (New York, NY. 2009;324:92–94. doi: 10.1126/science.1169628. [DOI] [PubMed] [Google Scholar]
- 97.Chen J, Ding M, Pederson DS. Binding of TFIID to the CYC1 TATA boxes in yeast occurs independently of upstream activating sequences. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:11909–11913. doi: 10.1073/pnas.91.25.11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Martens C, Krett B, Laybourn PJ. RNA polymerase II and TBP occupy the repressed CYC1 promoter. Mol Microbiol. 2001;40:1009–1019. doi: 10.1046/j.1365-2958.2001.02445.x. [DOI] [PubMed] [Google Scholar]
- 99.Stargell LA, Struhl K. A new class of activation-defective TATA-binding protein mutants: evidence for two steps of transcriptional activation in vivo. Molecular and cellular biology. 1996;16:4456–4464. doi: 10.1128/mcb.16.8.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Venters BJ, Pugh BF. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009;19:360–371. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. The EMBO journal. 2003;22:1846–1856. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, Hartzog GA. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. The EMBO journal. 2002;21:1764–1774. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jaehning JA. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochimica et biophysica acta. 2010;1799:379–388. doi: 10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M. Nature of the nucleosomal barrier to RNA polymerase II. Molecular cell. 2005;18:97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 105.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Molecular cell. 2005;20:589–599. doi: 10.1016/j.molcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 106.Laribee RN, Krogan NJ, Xiao T, Shibata Y, Hughes TR, Greenblatt JF, Strahl BD. BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr Biol. 2005;15:1487–1493. doi: 10.1016/j.cub.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 107.Qiu H, Hu C, Wong CM, Hinnebusch AG. The Spt4p subunit of yeast DSIF stimulates association of the Paf1 complex with elongating RNA polymerase II. Molecular and cellular biology. 2006;26:3135–3148. doi: 10.1128/MCB.26.8.3135-3148.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, McCleery JA, Yates JR, 3rd, Hartzog GA. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Molecular and cellular biology. 2003;23:1368–1378. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Suh MH, Meyer PA, Gu M, Ye P, Zhang M, Kaplan CD, Lima CD, Fu J. A dual interface determines the recognition of RNA polymerase II by RNA capping enzyme. The Journal of biological chemistry. 2010;285:34027–34038. doi: 10.1074/jbc.M110.145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xiao Y, Yang YH, Burckin TA, Shiue L, Hartzog GA, Segal MR. Analysis of a splice array experiment elucidates roles of chromatin elongation factor Spt4-5 in splicing. PLoS Comput Biol. 2005;1:e39. doi: 10.1371/journal.pcbi.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Burckin T, Nagel R, Mandel-Gutfreund Y, Shiue L, Clark TA, Chong JL, Chang TH, Squazzo S, Hartzog G, Ares M., Jr Exploring functional relationships between components of the gene expression machinery. Nature structural & molecular biology. 2005;12:175–182. doi: 10.1038/nsmb891. [DOI] [PubMed] [Google Scholar]
- 112.Bucheli ME, Buratowski S. Npl3 is an antagonist of mRNA 3′ end formation by RNA polymerase II. The EMBO journal. 2005;24:2150–2160. doi: 10.1038/sj.emboj.7600687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cui Y, Denis CL. In vivo evidence that defects in the transcriptional elongation factors RPB2, TFIIS, and SPT5 enhance upstream poly(A) site utilization. Molecular and cellular biology. 2003;23:7887–7901. doi: 10.1128/MCB.23.21.7887-7901.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kaplan CD, Holland MJ, Winston F. Interaction between transcription elongation factors and mRNA 3′-end formation at the Saccharomyces cerevisiae GAL10-GAL7 locus. The Journal of biological chemistry. 2005;280:913–922. doi: 10.1074/jbc.M411108200. [DOI] [PubMed] [Google Scholar]
- 115.Shen Z, St-Denis A, Chartrand P. Cotranscriptional recruitment of She2p by RNA pol II elongation factor Spt4-Spt5/DSIF promotes mRNA localization to the yeast bud. Genes & development. 2010;24:1914–1926. doi: 10.1101/gad.1937510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lepore N, Lafontaine DL. A functional interface at the rDNA connects rRNA synthesis, pre-rRNA processing and nucleolar surveillance in budding yeast. PLoS One. 2011;6:e24962. doi: 10.1371/journal.pone.0024962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- 118.Wen Y, Shatkin AJ. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes & development. 1999;13:1774–1779. doi: 10.1101/gad.13.14.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cheng B, Price DH. Analysis of factor interactions with RNA polymerase II elongation complexes using a new electrophoretic mobility shift assay. Nucleic acids research. 2008;36:e135. doi: 10.1093/nar/gkn630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Missra A, Gilmour DS. Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11301–11306. doi: 10.1073/pnas.1000681107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ping YH, Rana TM. DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. The Journal of biological chemistry. 2001;276:12951–12958. doi: 10.1074/jbc.M006130200. [DOI] [PubMed] [Google Scholar]
- 122.Ujvari A, Luse DS. Newly Initiated RNA encounters a factor involved in splicing immediately upon emerging from within RNA polymerase II. The Journal of biological chemistry. 2004;279:49773–49779. doi: 10.1074/jbc.M409087200. [DOI] [PubMed] [Google Scholar]
- 123.Perales R, Bentley D. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Molecular cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mayer A, Schreieck A, Lidschreiber M, Leike K, Martin DE, Cramer P. The spt5 C-terminal region recruits yeast 3′ RNA cleavage factor I. Molecular and cellular biology. 2012;32:1321–1331. doi: 10.1128/MCB.06310-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lahmy S, Bies-Etheve N, Lagrange T. Plant-specific multisubunit RNA polymerase in gene silencing. Epigenetics. 2010;5:4–8. doi: 10.4161/epi.5.1.10435. [DOI] [PubMed] [Google Scholar]
- 126.Rowley MJ, Avrutsky MI, Sifuentes CJ, Pereira L, Wierzbicki AT. Independent chromatin binding of ARGONAUTE4 and SPT5L/KTF1 mediates transcriptional gene silencing. PLoS genetics. 7:e1002120. doi: 10.1371/journal.pgen.1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huang L, Jones AM, Searle I, Patel K, Vogler H, Hubner NC, Baulcombe DC. An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nature structural & molecular biology. 2009;16:91–93. doi: 10.1038/nsmb.1539. [DOI] [PubMed] [Google Scholar]
- 128.Bies-Etheve N, Pontier D, Lahmy S, Picart C, Vega D, Cooke R, Lagrange T. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 2009;10:649–654. doi: 10.1038/embor.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.He XJ, Hsu YF, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, Liu HL, Wang CS, Jin H, Zhu JK. An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jansen LE, den Dulk H, Brouns RM, de Ruijter M, Brandsma JA, Brouwer J. Spt4 modulates Rad26 requirement in transcription-coupled nucleotide excision repair. The EMBO journal. 2000;19:6498–6507. doi: 10.1093/emboj/19.23.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jansen LE, Belo AI, Hulsker R, Brouwer J. Transcription elongation factor Spt4 mediates loss of phosphorylated RNA polymerase II transcription in response to DNA damage. Nucleic acids research. 2002;30:3532–3539. doi: 10.1093/nar/gkf475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Stanlie A, Begum NA, Akiyama H, Honjo T. The DSIF subunits Spt4 and Spt5 have distinct roles at various phases of immunoglobulin class switch recombination. PLoS genetics. 2012;8:e1002675. doi: 10.1371/journal.pgen.1002675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li J, Mason SW, Greenblatt J. Elongation factor NusG interacts with termination factor rho to regulate termination and antitermination of transcription. Genes & development. 1993;7:161–172. doi: 10.1101/gad.7.1.161. [DOI] [PubMed] [Google Scholar]
- 134.Burmann BM, Schweimer K, Luo X, Wahl MC, Stitt BL, Gottesman ME, Rosch P. A NusE:NusG complex links transcription and translation. Science (New York, NY. 2010;328:501–504. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- 135.Guo M, Xu F, Yamada J, Egelhofer T, Gao Y, Hartzog GA, Teng M, Niu L. Core structure of the yeast spt4-spt5 complex: a conserved module for regulation of transcription elongation. Structure. 2008;16:1649–1658. doi: 10.1016/j.str.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wenzel S, Martins BM, Rosch P, Wohrl BM. Crystal structure of the human transcription elongation factor DSIF hSpt4 subunit in complex with the hSpt5 dimerization interface. Biochem J. 2010;425:373–380. doi: 10.1042/BJ20091422. [DOI] [PubMed] [Google Scholar]
- 137.Wenzel S, Schweimer K, Rosch P, Wohrl BM. The small hSpt4 subunit of the human transcription elongation factor DSIF is a Zn-finger protein with alpha/beta type topology. Biochem Biophys Res Commun. 2008;370:414–418. doi: 10.1016/j.bbrc.2008.03.080. [DOI] [PubMed] [Google Scholar]
- 138.Klein BJ, Bose D, Baker KJ, Yusoff ZM, Zhang X, Murakami KS. RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:546–550. doi: 10.1073/pnas.1013828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. The EMBO journal. 2011;30:1302–1310. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhou H, Liu Q, Gao Y, Teng M, Niu L. Crystal structure of NusG N-terminal (NGN) domain from Methanocaldococcus jannaschii and its interaction with rpoE″. Proteins. 2009;76:787–793. doi: 10.1002/prot.22465. [DOI] [PubMed] [Google Scholar]
- 141.Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG, Artsimovitch I. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Molecular cell. 2007;26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hartzog GA, Kaplan CD. Competing for the clamp: promoting RNA polymerase processivity and managing the transition from initiation to elongation. Molecular cell. 2012;43:161–163. doi: 10.1016/j.molcel.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 143.Werner F. A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. Journal of molecular biology. 2012;417:13–27. doi: 10.1016/j.jmb.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- 145.Belogurov GA, Sevostyanova A, Svetlov V, Artsimovitch I. Functional regions of the N-terminal domain of the antiterminator RfaH. Mol Microbiol. 2010;76:286–301. doi: 10.1111/j.1365-2958.2010.07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mooney RA, Schweimer K, Rosch P, Gottesman M, Landick R. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. Journal of molecular biology. 2009;391:341–358. doi: 10.1016/j.jmb.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sevostyanova A, Belogurov GA, Mooney RA, Landick R, Artsimovitch I. The beta subunit gate loop is required for RNA polymerase modification by RfaH and NusG. Molecular cell. 2011;43:253–262. doi: 10.1016/j.molcel.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Brzovic PS, Heikaus CC, Kisselev L, Vernon R, Herbig E, Pacheco D, Warfield L, Littlefield P, Baker D, Klevit RE, Hahn S. The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex. Molecular cell. 2011;44:942–953. doi: 10.1016/j.molcel.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 150.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science (New York, NY. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 151.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Molecular cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 152.Majovski RC, Khaperskyy DA, Ghazy MA, Ponticelli AS. A functional role for the switch 2 region of yeast RNA polymerase II in transcription start site utilization and abortive initiation. The Journal of biological chemistry. 2005;280:34917–34923. doi: 10.1074/jbc.M502932200. [DOI] [PubMed] [Google Scholar]
- 153.Kettenberger H, Armache KJ, Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- 154.Rudd MD, Izban MG, Luse DS. The active site of RNA polymerase II participates in transcript cleavage within arrested ternary complexes. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:8057–8061. doi: 10.1073/pnas.91.17.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Weilbaecher RG, Awrey DE, Edwards AM, Kane CM. Intrinsic transcript cleavage in yeast RNA polymerase II elongation complexes. The Journal of biological chemistry. 2003;278:24189–24199. doi: 10.1074/jbc.M211197200. [DOI] [PubMed] [Google Scholar]
- 156.Wang D, Bushnell DA, Huang X, Westover KD, Levitt M, Kornberg RD. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science (New York, NY. 2009;324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 158.Funk JD, Nedialkov YA, Xu D, Burton ZF. A key role for the alpha 1 helix of human RAP74 in the initiation and elongation of RNA chains. The Journal of biological chemistry. 2002;277:46998–47003. doi: 10.1074/jbc.M206249200. [DOI] [PubMed] [Google Scholar]
- 159.Tan S, Aso T, Conaway RC, Conaway JW. Roles for both the RAP30 and RAP74 subunits of transcription factor IIF in transcription initiation and elongation by RNA polymerase II. The Journal of biological chemistry. 1994;269:25684–25691. [PubMed] [Google Scholar]
- 160.Kephart DD, Wang BQ, Burton ZF, Price DH. Functional analysis of Drosophila factor 5 (TFIIF), a general transcription factor. The Journal of biological chemistry. 1994;269:13536–13543. [PubMed] [Google Scholar]
- 161.Eichner J, Chen HT, Warfield L, Hahn S. Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. The EMBO journal. 2010;29:706–716. doi: 10.1038/emboj.2009.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chen HT, Warfield L, Hahn S. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nature structural & molecular biology. 2007;14:696–703. doi: 10.1038/nsmb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wu YM, Chang JW, Wang CH, Lin YC, Wu PL, Huang SH, Chang CC, Hu X, Gnatt A, Chang WH. Regulation of mammalian transcription by Gdown1 through a novel steric crosstalk revealed by cryo-EM. The EMBO journal. 2012 doi: 10.1038/emboj.2012.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cheng B, Li T, Rahl PB, Adamson TE, Loudas NB, Guo J, Varzavand K, Cooper JJ, Hu X, Gnatt A, Young RA, Price DH. Functional association of Gdown1 with RNA polymerase II poised on human genes. Molecular cell. 2012;45:38–50. doi: 10.1016/j.molcel.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Jishage M, Malik S, Wagner U, Uberheide B, Ishihama Y, Hu X, Chait BT, Gnatt A, Ren B, Roeder RG. Transcriptional regulation by Pol II(G) involving mediator and competitive interactions of Gdown1 and TFIIF with Pol II. Molecular cell. 2012;45:51–63. doi: 10.1016/j.molcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Renner DB, Yamaguchi Y, Wada T, Handa H, Price DH. A highly purified RNA polymerase II elongation control system. The Journal of biological chemistry. 2001;276:42601–42609. doi: 10.1074/jbc.M104967200. [DOI] [PubMed] [Google Scholar]
- 167.Khaperskyy DA, Ammerman ML, Majovski RC, Ponticelli AS. Functions of Saccharomyces cerevisiae TFIIF during transcription start site utilization. Molecular and cellular biology. 2008;28:3757–3766. doi: 10.1128/MCB.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Arthur TM, Anthony LC, Burgess RR. Mutational analysis of beta ‘260-309, a sigma 70 binding site located on Escherichia coli core RNA polymerase. The Journal of biological chemistry. 2000;275:23113–23119. doi: 10.1074/jbc.M002040200. [DOI] [PubMed] [Google Scholar]
- 169.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 170.Sevostyanova A, Svetlov V, Vassylyev DG, Artsimovitch I. The elongation factor RfaH and the initiation factor sigma bind to the same site on the transcription elongation complex. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:865–870. doi: 10.1073/pnas.0708432105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Grohmann D, Hirtreiter A, Werner F. RNAP subunits F/E (RPB4/7) are stably associated with archaeal RNA polymerase: using fluorescence anisotropy to monitor RNAP assembly in vitro. Biochem J. 2009;421:339–343. doi: 10.1042/BJ20090782. [DOI] [PubMed] [Google Scholar]
- 172.Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Molecular cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 173.Pal M, McKean D, Luse DS. Promoter clearance by RNA polymerase II is an extended, multistep process strongly affected by sequence. Molecular and cellular biology. 2001;21:5815–5825. doi: 10.1128/MCB.21.17.5815-5825.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pal M, Luse DS. The initiation-elongation transition: lateral mobility of RNA in RNA polymerase II complexes is greatly reduced at +8/+9 and absent by +23. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5700–5705. doi: 10.1073/pnas.1037057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science (New York, NY. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 176.Kwak YT, Guo J, Prajapati S, Park KJ, Surabhi RM, Miller B, Gehrig P, Gaynor RB. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Molecular cell. 2003;11:1055–1066. doi: 10.1016/s1097-2765(03)00101-1. [DOI] [PubMed] [Google Scholar]


