Abstract
In enterics, the transcription factor SoxR triggers a global stress response by sensing a broad spectrum of redox-cycling compounds. In the non-enteric bacteria Pseudomonas aeruginosa and Streptomyces coelicolor, SoxR is activated by endogenous redox-active small molecules and only regulates a small set of genes. We investigated if the more general response in enterics is reflected in the ability of SoxR to sense a wider range of redox-cycling compounds. Indeed, while Escherichia coli SoxR is tuned to structurally diverse compounds that span a redox range of −450 to +80 mV, P. aeruginosa and S. coelicolor SoxR are less sensitive to viologens, which have redox potentials below −350 mV. Using a mutagenic approach, we pinpointed three amino acids that contribute to the reduced sensitivity of P. aeruginosa and S. coelicolor SoxR. Notably these residues are not conserved in homologs of the Enterobacteriaceae. We further identified a motif within the sensor domain that tunes the activity of SoxR from enterics – inhibiting constitutive activity while allowing sensitivity to drugs with low redox potentials. Our findings highlight how small alterations in structure can lead to the evolution of proteins with distinct specificities for redox-active small molecules.
Keywords: redox-active molecules, Fe-S clusters, SoxR, viologens, paraquat
INTRODUCTION
Iron-sulfur clusters (Fe-S) are remarkably diverse in structure and chemistry. Different cluster types span a wide range of redox potentials and the redox potential of a single cluster type can be further tuned by changing its molecular environment (Beinert, 2000). These features enabled the evolution of Fe-S proteins that perform crucial and versatile functions as metabolic enzymes, components of electron transport chains, and redox-sensing regulators of gene expression. The latter act as molecular switches that are either activated or inactivated by specific redox signals (such as oxygen, hydrogen peroxide, superoxide, nitric oxide, or redox-active small molecules) to regulate important aspects of bacterial development and physiology. For example, FNR, a [4Fe-4S] protein, controls the switch between aerobic and anaerobic metabolism in Escherichia coli in response to molecular oxygen (Khoroshilova et al., 1997); IscR, a [2Fe-2S] protein, increases the production of Fe-S cluster biogenesis machinery under conditions of oxidative stress (Zheng et al., 2001; Yeo et al., 2006); and SoxR, also a [2Fe-2S] protein, mediates an oxidative stress response to redox-cycling drugs in the enteric bacteria E. coli and Salmonella enterica (Hidalgo and Demple, 1996). Within this group of redox-sensing transcription factors SoxR is unique in that, unlike the other proteins that are regulated by assembly/disassembly of their Fe-S clusters, the activity of SoxR is modulated by reversible one-electron oxidation-reduction of its [2Fe-2S] clusters (Ding et al., 1996; Gaudu and Weiss, 1996; Ding and Demple, 1997; Gaudu et al., 1997).
In E. coli (and related enteric bacteria), SoxR senses redox stress imposed by a broad collection of redox-active compounds including viologens, phenazines and quinones (Table 1; Gu and Imlay, 2011). SoxR transduces these redox signals into a global defense program via a second transcription factor, SoxS. SoxR is a constitutively expressed regulator bound to the soxS promoter poised to detect stress. In the absence of oxidants, SoxR exists in a quiescent state with reduced [2Fe-2S] clusters and soxS is not expressed. Exposure to redox-cycling drugs causes oxidation of SoxR’s [2Fe-2S] centers, and the oxidized protein activates soxS expression by mediating structural changes in the promoter DNA that allow RNA polymerase to initiate transcription (Hidalgo et al., 1995). SoxS in turn recruits RNA polymerase to transcribe >100 genes, some of which encode proteins that reestablish redox balance and repair oxidant-induced damage (Pomposiello et al., 2001). The SoxRS system in enterics allows for rapid amplification of the stress signal into a stress response geared towards oxidants.
Table 1.
Redox drugs used in this work*
| Class | Drug | Structure | Redox potential (mV) |
|---|---|---|---|
| Viologen | Paraquat (PQ) | 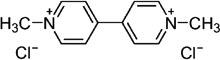 |
−440 (Steckhan and Kuwana, 1974) |
| Viologen | Diquat (DQ) | 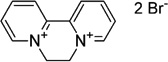 |
−361 (Steckhan and Kuwana, 1974) |
| Phenazine | Phenazine-1-carboxylic acid (PCA) | 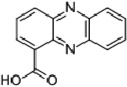 |
−177 (Price-Whelan et al., 2006) |
| Napthoquinone | Plumbagin (PB) | 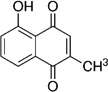 |
−135 (Hakura et al., 1994) |
| Phenazine | Pyocyanin (Pyo) | 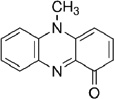 |
−34 (Friedheim and Michaelis, 1931) |
| Phenothiazine | Methylene blue (MB) | 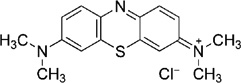 |
+11 (Kamat et al. 1987) |
| Quinoline | 4-Nitroquinoline-N-oxide (4NQO) | 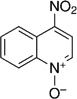 |
+74 (Biaglow et al. 1978) |
| Phenazine | Phenazine methosulfate (PMS) | 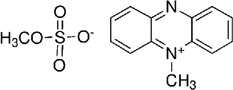 |
+80 (Moffet et al., 2003) |
| Anthraquinone | γ-Actinorhodin (Act) | 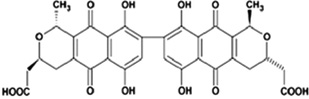 |
Unknown |
Drugs are arranged in order of increasing midpoint redox potential. The redox potentials are reported versus the normal hydrogen electrode (NHE).
The SoxRS regulon is unique to enterics. Although soxR is widely distributed (and highly similar at the amino acid level) across the Gram-negative Proteobacteria and the Gram-positive Actinobacteria, soxS is present exclusively in enterobacteria. An extensive bioinformatic analysis of soxS-deficient genomes predicted that in non-enterics SoxR directly regulates a relatively small set of genes that encode putative oxygenases, oxidoreductases, or transporters (Dietrich et al., 2008). This has been verified for the γ-Proteobacterium Pseudomonas aeruginosa and the Actinomycete Streptomyces coelicolor, both soil-dwelling organisms notable for producing redox-active secondary metabolites. The SoxR regulon in P. aeruginosa consists of a Resistance-Nodulation-Division (RND) efflux pump MexGHI-OmpD (PA4205-4208), a major facilitator superfamily (MFS) transporter (PA3718), and a monooxygenase (PA2274) (Palma et al., 2005). In S. coelicolor, SoxR regulates a monooxygenase (SCO1909) with homology to PA2274, two oxidoreductases (SCO2478, SCO4266), an epimerase/dehydratase (SCO1178), and an ABC transporter (SCO7008) (Dela Cruz et al., 2010; Shin et al., 2011). In these bacteria SoxR-regulated genes are induced in stationary phase during the production and secretion of redox-active metabolites – phenazines in the case of P. aeruginosa and the benzochromanequinone polyketide actinorhodin (Act) in the case of S. coelicolor (Dietrich et al., 2006; Dela Cruz et al., 2010; Shin et al., 2011). This is not a mere correlation as expression of each SoxR regulon is dependent on production of the redox-active compounds by the microbe (Dietrich et al., 2006; Dela Cruz et al., 2010; Shin et al., 2011). These observations support the view that SoxR evolved to regulate the machinery that processes/transports endogenous redox-active metabolites in producer organisms. The enterobacteria (which do not produce redox-active secondary metabolites) are unique in that SoxR regulates only one gene, soxS. They may have acquired soxR via lateral gene transfer, taking advantage of its redox-sensing abilities to regulate a generalized stress response (SoxS regulon) against toxic redox-cycling compounds.
Given that SoxR performs distinct functions in different bacteria, we asked if the differences in SoxR functionality are manifested only by its regulons, or if SoxR from different species also sense different inputs. We hypothesized that SoxR from P. aeruginosa and S. coelicolor sense redox molecules that resemble their endogenous activators (phenazines and anthraquinones, respectively), while E. coli SoxR, given its involvement in a general stress response, senses a broader spectrum of redox-active compounds. Here we report that the non-enteric SoxR proteins are indeed more restricted in the range of molecules they sense compared to their E. coli counterpart, and we have identified key features that contribute to the differential sensitivities. This study provides insight into the evolutionary fine-tuning of this redox-sensing transcription factor that adapted it to serve the needs of organisms with different physiologies.
RESULTS
SoxR protects E. coli, but not P. aeruginosa or S. coelicolor, against redox-cycling drugs
In E. coli and related enterobacteria, SoxR mediates a general stress response against redox-cycling compounds by activating the SoxS regulon. By contrast, non-enterics lack a SoxS regulon. Instead, SoxR directly regulates a small set of genes, making a general stress response unlikely. To test this, we exposed soxR deletion mutants of E. coli (which contains soxS), the Gram-negative P. aeruginosa PA14, and the Gram-positive S. coelicolor M145 (both of which lack soxS) to a diverse array of redox-cycling compounds using a filter disk assay. In agreement with previous reports, an E. coli ΔsoxR mutant was more sensitive to most tested redox-cycling agents such as pyocyanin (Pyo), plumbagin (PB), and 4-nitroquinoline-N-oxide (4NQO), compared to wild type (Greenberg et al. 1990; Tsaneva and Weiss, 1990; Fig. 1A). The E. coli ΔsoxR mutant was no more sensitive to the viologen diquat (DQ) than wild type, and a previous study had shown this to also be true for paraquat (PQ), another viologen (Greenberg et al., 1990). This appears to be a strain-specific phenomenon, since a different E. coli ΔsoxR mutant strain was more sensitive to PQ than wild type (Tsaneva and Weiss, 1990). In contrast to E. coli, the S. coelicolor ΔsoxR mutant and wild type were equally sensitive to all tested compounds (Fig. 1B). P. aeruginosa was generally more resistant to all drugs tested in this study. The wild type and ΔsoxR mutant were resistant to Pyo, PB, and 4NQO, and were equally sensitive to DQ (Fig. 1C). While SoxR does not contribute to S. coelicolor or P. aeruginosa’s resistance to redox-cycling drugs under the conditions tested here, it is worth noting that it might do so under other experimental conditions. It is also worth noting that the P. aeruginosa ΔsoxR mutant has a colony morphology phenotype (Dietrich et al., 2008), which can be reverted by complementation with E. coli or P. aeruginosa SoxR (Fig. S1).
Figure 1. S. coelicolor and P. aeruginosa ΔsoxR mutants are not hypersensitive to superoxide-generating agents.
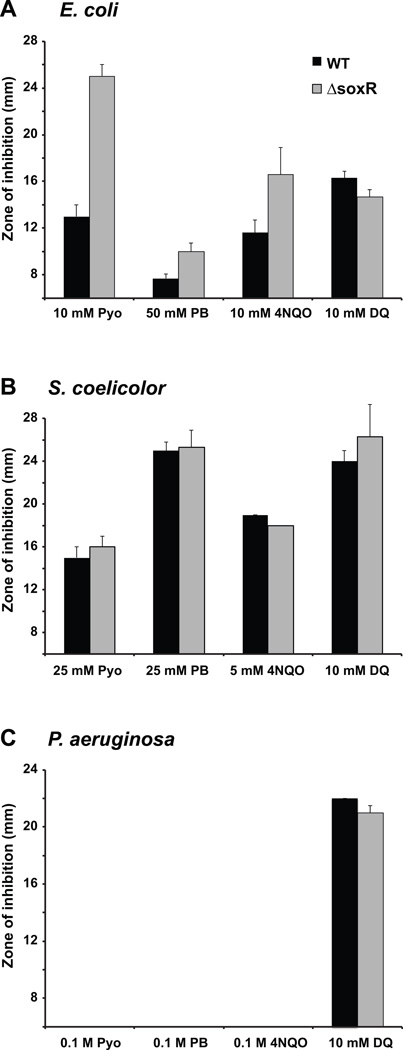
Paper disks soaked with solutions of the indicated compounds were placed on bacterial lawns of wild type (black columns) or ΔsoxR mutant (grey columns) growing on nutrient agar plates. Zones of growth inhibition around the disks were recorded after 24 h at 37°C for E. coli (A) or 48 h at 30°C for S. coelicolor (B) and P. aeruginosa (C). A zone of inhibition of 6 mm corresponds to the diameter of the disk and indicates that there was no detectable inhibition zone. The data represent the means of 3 to 5 replicates ± standard deviations (bars; some not visible).
P. aeruginosa and S. coelicolor SoxRs sense a narrower spectrum of redox drugs than E. coli SoxR
Our finding that SoxR did not contribute to resistance against redox-cycling agents in P. aeruginosa and S. coelicolor is consistent with the notion that the enteric-specific SoxS regulon governs a general stress response. To explore the hypothesis that the SoxR regulons in P. aeruginosa and S. coelicolor may be specific to phenazines and Act, respectively, we posited that SoxR itself might be optimized to sense specific redox inputs. Given the role of SoxR in E. coli as a general stress-response regulator, we predicted this protein would respond to a broad spectrum of redox-cycling molecules, while P. aeruginosa and S. coelicolor SoxRs would only respond to molecules that resemble their endogenous activators, i.e. phenazines and anthraquinones, respectively.
To quantify the SoxR response to a wide spectrum of redox-cycling compounds, we employed a β-galactosidase assay using a soxS promoter-lacZ reporter in E. coli. SoxR orthologs from E. coli, P. aeruginosa, and S. coelicolor have very similar DNA-binding domains, and all three proteins bind to the soxS promoter with high affinity in vitro (Fig. S2C). Therefore, using the soxS promoter-lacZ reporter seemed a reasonable approach. The three soxR genes (each with an N-terminal histidine-tag) were transformed into an E. coli ΔsoxRS mutant lysogenized with a λ[soxS promoter-lacZ reporter] (Table 2). Transformed cells were grown to exponential phase and then treated with representative drugs that span a wide range of redox potentials and belong to different structural classes (Table 1). The phenazines Pyo and phenazine-1-carboxylic acid (PCA) are produced by P. aeruginosa, while phenazine methosulfate (PMS) is synthetic.
Table 2.
Bacterial strains and plasmids used in this study
| Strain/plasmid | Genotype/description | Source/reference |
|---|---|---|
| E. coli | ||
| GC4468 | K12 rpsL thi soxR+ soxS+ | Greenberg et al. (1990) |
| DJ901 | Δ(soxRS) derivative of GC4468 | Greenberg et al. (1990) |
| EH46 | DJ901 lysogenized with λ(soxS promoter-lacZ) | Hidalgo and Demple (1997) |
| EH86 | DJ901 lysogenized with λ(16-bp spacer mutant soxS promoter-lacZ) | Hidalgo and Demple (1997) |
| P. aeruginosa | ||
| PA14 | Clinical isolate UCBPP-PA14 | Rahme et al. (1995) |
| PA14ΔsoxR | PA14 with a deletion in soxR | Dietrich et al. (2006) |
| WTpmexgfp | PA14 with insert of mexG promoter fused to gfp reporter gene | This study |
| ΔsoxRpmexgfp | PA14ΔsoxR with insert of mexG promoter fused to gfp reporter gene | This study |
| S. coelicolor | ||
| M145 | SCP1−, SCP2− | Kieser et al. (2000) |
| M145-1A | M145 with a deletion in soxR | Dela Cruz et al. (2010) |
| M511 | ΔactII-ORF4 derivative of M145 | Floriano and Bibb (1996) |
| M511ΔsoxR | ΔsoxR derivative of M511 | Dela Cruz et al. (2010) |
| Plasmids | ||
| pSE380 | trc promoter-containing plasmid with lacIq gene (Ampicillinr) | Invitrogen |
| pSE380:H-ECO | N-terminally histidine-tagged E. coli soxR gene in pSE380 | This study |
| pSE380:H-PA | N-terminally histidine-tagged P. aeruginosa soxR gene in pSE380 | This study |
| pSE380:H-SCO | N-terminally histidine-tagged S. coelicolor soxR gene in pSE380 | This study |
| pSET152 | Apramycinr lacZα MCS reppUC | Bierman et al. (1992) |
| pSET152:H-SCO | N-terminally histidine-tagged S. coelicolor soxR gene in pSET152 | This study |
| pSET152:H-ΔC | N-terminally histidine-tagged truncated S. coelicolor soxR gene in pET152 | This study |
| pUCp18 | Carbenicillinr, pUCP18 vector containing a P. aeruginosa origin of replication | Schweizer (1991) |
| pUC:ECsoxR | N-terminally histidine-tagged E. coli soxR gene in pUCp18 | This study |
| pUC:PAsoxR | N-terminally histidine-tagged P. aeruginosa soxR gene in pUCp18 | This study |
| pYL122 | Ampicillinr, rhlA-gfp transcription fusion in mini-CTX-lacZ | Lequette and Greenberg (2005) |
E. coli SoxR was strongly activated (albeit to different extents) by all nine drugs tested (Fig. 2A). P. aeruginosa SoxR was activated at levels comparable to E. coli SoxR by PCA, PB, Pyo, methylene blue (MB), 4NQO, PMS, and Act, but at significantly lower levels by the viologens PQ and DQ (Fig. 2A). PQ elicited 15-fold lower β-galactosidase activity in cells expressing P. aeruginosa SoxR compared with E. coli SoxR. Ethyl viologen, which has a similar midpoint redox potential as PQ (−480 mV), was also a weak inducer of P. aeruginosa SoxR activity (data not shown). The response to DQ, which has a higher redox potential than PQ (−360 mV) was more robust (2200 Miller units), but still only about half that of E. coli SoxR (4600 Miller units). Thus P. aeruginosa SoxR has low sensitivity to the viologens that have redox potentials more negative than −350 mV (Fig. 2B).
Figure 2. P. aeruginosa and S. coelicolor SoxRs sense a narrower spectrum of redox-active compounds than E. coli SoxR.
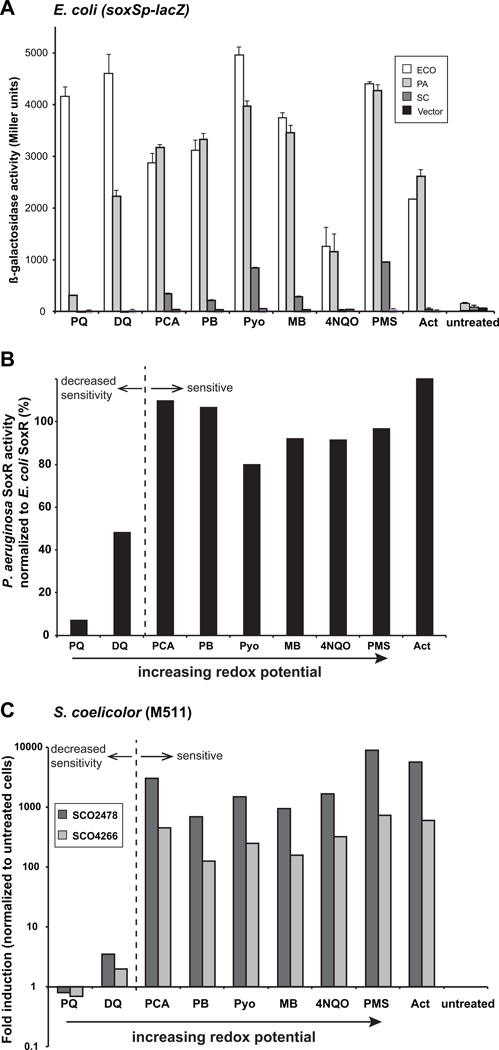
A. E. coli strain EH46 (ΔsoxRS lysogenized with λ[soxS promoter-lacZ reporter) expressing histidine-tagged E. coli SoxR (white columns), P. aeruginosa SoxR (light grey columns), S. coelicolor SoxR (dark grey columns) or empty vector (black columns) were untreated or treated with 200 µM PQ, 200 µM DQ, 500 µM PCA, 25 µM PB, 20 µM Pyo, 25 µM MB, 50 µM 4NQO, 20 µM PMS or 25 µM Act for 1 h before the assay for β-galactosidase activity. The results shown represent the means and standard errors (bars; some not visible at this scale) of three independent experiments. Dose-response curves for most of these drugs are shown in Figure S3.
B. P. aeruginosa SoxR activity normalized to that of E. coli SoxR indicates that the former displays reduced sensitivity to drugs with redox potentials below −300 mV. The data in this figure is the same as the P. aeruginosa SoxR data in Fig. 2A.
C. The Act-deficient S. coelicolor strain M511 was grown for 20 h in R5− medium, then exposed for 30 min to 1 mM PQ, 1 mM DQ, 500 µM PCA, 100 µM PB, 10 µM Pyo, 25 µM MB, 1 mM 4NQO, 10 µM PMS or 10 µM Act, or left untreated. qRT-PCR was performed on RNA extracted from these cells to detect induction of SoxR-target genes SCO2478 (dark grey columns) and SCO4266 (light grey columns). Signals were standardized to the level of the housekeeping sigma factor, hrdB, and fold-induction was normalized to untreated M511 cells. Note that the y-axis is shown as a logarithmic scale.
In stark contrast to the high levels of β-galactosidase activity produced by E. coli and P. aeruginosa SoxR, S. coelicolor SoxR produced relatively low signals in response to all tested drugs (Fig. 2A). As such we were unable to draw any meaningful conclusions about S. coelicolor SoxR activation using the heterologous E. coli system. We therefore investigated this transcription factor’s activity in its native background. Because this protein is activated by the endogenous metabolite Act, it was necessary to monitor its response to exogenous drugs in S. coelicolor M511, a strain that does not synthesize Act (Table 2). Cells were grown for 20 h in liquid culture before a 30 min exposure to the redox-cycling drugs listed in Table 1. SoxR activity was assessed by monitoring the expression levels of two of its target genes, SCO2478 and SCO4266, by quantitative real-time PCR (qRT-PCR). In addition to Act, PCA, PB, Pyo, MB, 4NQO and PMS induced SoxR-target gene expression to high levels over background (Fig. 2C). Drug-induced expression of SCO2478 and SCO4266 was SoxR-dependent since these mRNAs were not detectable in M511ΔsoxR cells that were similarly treated (data not shown). Similar to P. aeruginosa SoxR, only the viologens PQ and DQ failed to activate SoxR to any appreciable level (Fig. 2C). Thus, S. coelicolor and P. aeruginosa SoxR sense redox-active molecules in the same range of redox potential.
The inability of S. coelicolor SoxR to complement an E. coli ΔsoxR mutant could result from inefficient protein expression or failure to effect soxS transcription. We confirmed that this protein is stably expressed in E. coli by immunoblot analysis (Fig. S2A). Furthermore, S. coelicolor SoxR binds efficiently to the E. coli soxS promoter in vitro (Fig. S2C) and in vivo (Fig. S2B), and stimulates transcription of the soxS gene in vitro (Fig. S2D). Interestingly, the C-terminus of S. coelicolor SoxR has an additional 22-residues not present in homologs from members of the Enterobacteriaceae or pseudomonads (Fig. 3A). In fact, an extended C-terminal region is found in SoxR proteins from several other Streptomyces species (data not shown) and is peculiar to this genus. Given that this is the most obvious structural difference between S. coelicolor SoxR and its E. coli and P. aeruginosa counterparts, we asked if this region could be involved in the regulation of S. coelicolor SoxR. To test this, we constructed a mutant that lacks the extreme C-terminal 22-residues by engineering a stop codon at position 154 (see Fig. 3A). We confirmed that the mutant protein is expressed in E. coli and interacts with the soxS promoter (Fig. S2B). The transcriptional activity of this protein in response to PMS was measured in the E. coli ΔsoxRS mutant lysogenized with λ[soxS promoter-lacZ reporter] (Table 2). Figure 4A shows that PMS induced similar (low) β-galactosidase levels in cells expressing wild type or truncated S. coelicolor SoxR proteins. Deletion of the C-terminus also did not affect the activity of this protein when expressed in a S. coelicolor ΔsoxR strain. We introduced the truncated gene into the ΔsoxR strain via the pSET152 vector and monitored the transcription of two SoxR target genes, SCO4266 and SCO1178, by qRT-PCR. The results indicated in Fig. 4B show that, in response to the endogenous activator Act, both wild type SoxR and the truncated mutant protein activated SCO4266 and SCO1178 expression to levels similar to those observed in wild type cells (WT/pSET152). Thus, the extreme C-terminal region is dispensible for S. coelicolor SoxR function, and at this point we have no ready explanation for why S. coelicolor SoxR failed to complement the E. coli ΔsoxR mutant.
Figure 3. Sequence comparison of SoxR homologs.
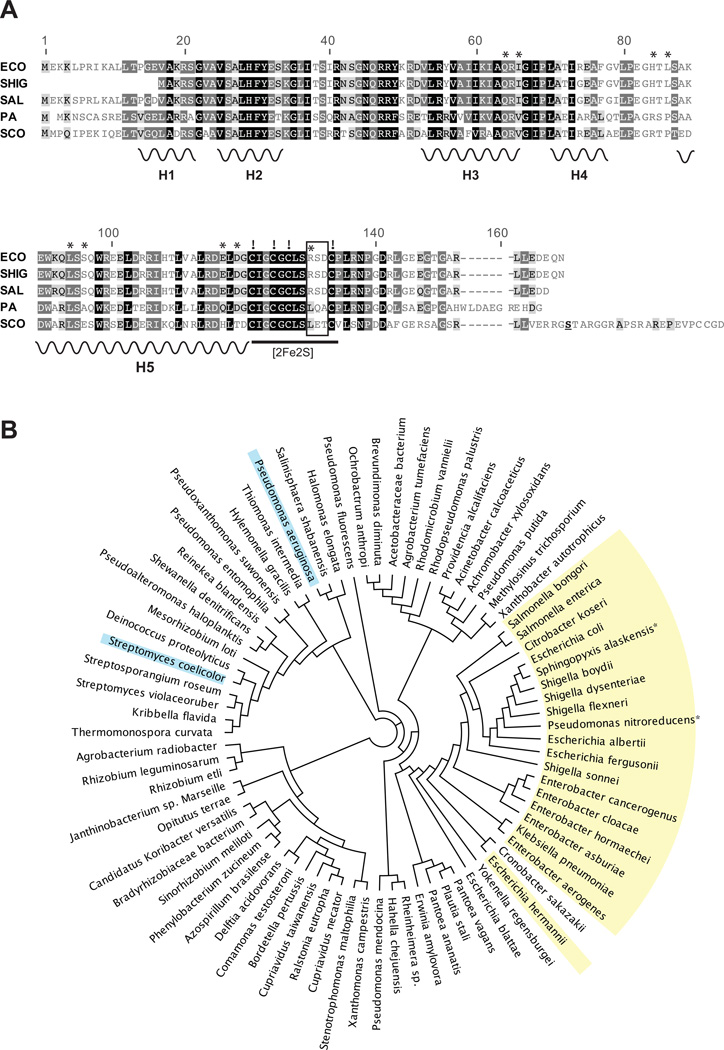
A. A BLAST analysis was performed for E. coli SoxR against all available bacterial genomes. The 250 closest homologs were aligned using ClustalW, and shown is an alignment of five of these SoxR proteins from the enterics E. coli (ECO), Shigella flexneri (SHIG), Salmonella enterica (SAL), and the non-enterics P. aeruginosa (PA), and S. coelicolor (SCO). Black, dark grey or light grey boxes surrounding residues indicate 100%, 80–100% or 60–80% similarity between all 250 SoxRs (Blosum62 score matrix with threshold of 1). Secondary structural motifs are based on the crystal structure of E. coli SoxR (Watanabe et al., 2008): H1 and H2 indicate the helix-turn-helix motif that makes specific contacts with the soxS promoter; H3 and H4 form a second helix-turn-helix motif within the DNA binding domain; H5 indicates the dimerization helix. The four conserved cysteine residues that anchor the [2Fe-2S] cluster are indicated by exclamation marks (!). A three-residue, hypervariable motif in the [2Fe-2S] region is indicated by a box. In most species of the Enterobacteriaceae this motif is RSD. Other residues that influence the redox-sensing properties of E. coli SoxR are marked by asterisks (Chander and Demple, 2004). The serine residue that was changed to a stop codon to construct the C-terminal truncated S. coelicolor SoxR mutant is underlined.
B. Based on the alignment of 250 SoxR proteins a tree was generated with Geneious Pro 5.6 using the neighbor-joining method (Saitou and Nei, 1987; Drummond, et al., 2011). For clarity only one member of each represented species is shown. Species that contain the RSD motif in SoxR are highlighted in yellow; these all belong to the Enterobacteriaceae excepting Pseudomonas nitroreducens and Sphingopyxis alaskensis. P. aeruginosa and S. coelicolor SoxR are highlighted in blue.
Figure 4. The extended C-terminal region of S. coelicolor SoxR is not important for function.
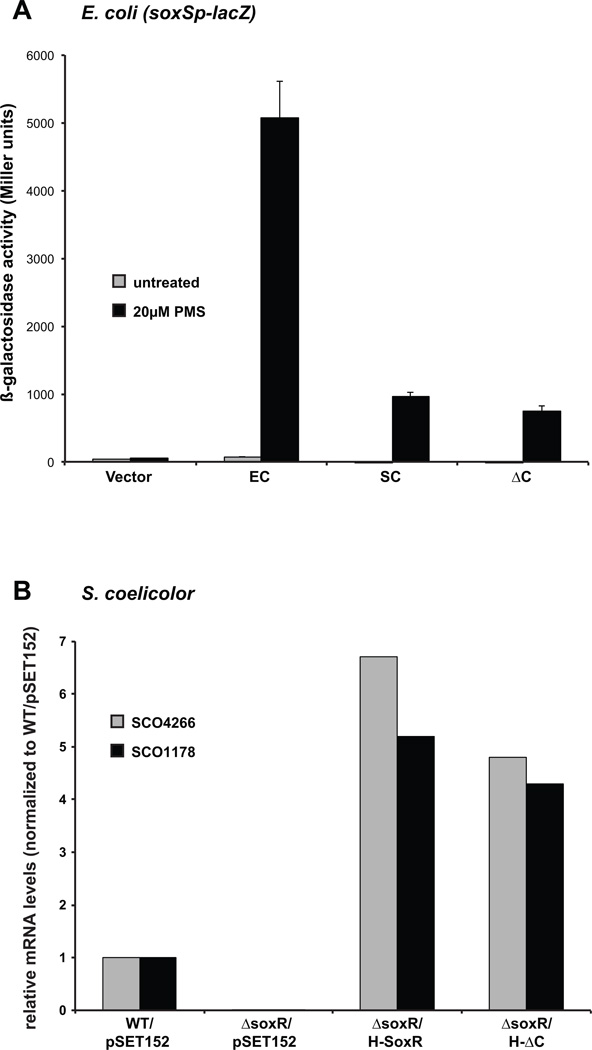
A. E. coli cells (strain EH46) expressing histidine-tagged wild type E. coli or S. coelicolor soxR alleles, or the S. coelicolor C-terminal truncated mutant from pSE380-based plasmids were either untreated (grey columns) or treated with 20 µM PMS (black columns) for 1 h before the assay for β-galactosidase activity. The results represent the means and standard errors (bars; some not visible on this scale) of three independent experiments.
B. qRT-PCR was performed on RNA isolated from the following S. coelicolor strains: WT/pSET152, ΔsoxR/pSET152, and a ΔsoxR strain complemented with wild type soxR (H-SoxR) or the C-terminal truncated mutant (H-ΔC), grown in R5− liquid medium for 3 days at which point Act was robustly produced. The expression levels of SoxR target genes, SCO4266 (grey columns) and SCO1178 (black columns) were standardized to the level of hrdB and normalized to expression in WT/pSET152.
Mutations in specific residues of P. aeruginosa SoxR alters its specificity for redox-active molecules
The transcriptional assays described in Figure 2 demonstrated that P. aeruginosa and S. coelicolor SoxR were more selective than E. coli SoxR, with reduced sensitivity to compounds with low redox potentials (viologens). The activation profiles for P. aeruginosa and S. coelicolor SoxR (responsive to PMS but not PQ) were reminiscent of E. coli SoxR mutant proteins that were reported several years ago (Chander et al. 2003; Chander and Demple, 2004). In those studies, nine residues were identified that, when individually changed, rendered E. coli SoxR insensitive to PQ, but fully responsive to PMS (Fig. 5A). It was suggested that changes in these residues alter the redox-reactivity of SoxR, rendering the protein hyposensitive to certain redox signals. Thus, while these mutant proteins are still activated by the strongly oxidizing drug PMS (redox potential of +80 mV), they are unresponsive to the less oxidizing drug PQ (redox potential of −440 mV). Of these nine residues, only two are conserved in S. coelicolor SoxR (Q63, L93) and three are conserved in P. aeruginosa SoxR (Q62, L92, D115) (Fig. 3A). We hypothesized that changing the non-conserved residues in P. aeruginosa or S. coelicolor SoxR to those found in E. coli SoxR might decrease their drug-selectivity, i.e. that the mutant proteins would respond to PQ. To exclude any SoxR-independent differences between the species, such as drug uptake, we expressed the mutant proteins in E. coli. Because S. coelicolor SoxR is only weakly active in E. coli, we focused our comparison on P. aeruginosa and E. coli SoxR.
Figure 5. Mutations that alter drug-selectivity of P. aeruginosa and E. coli SoxR.
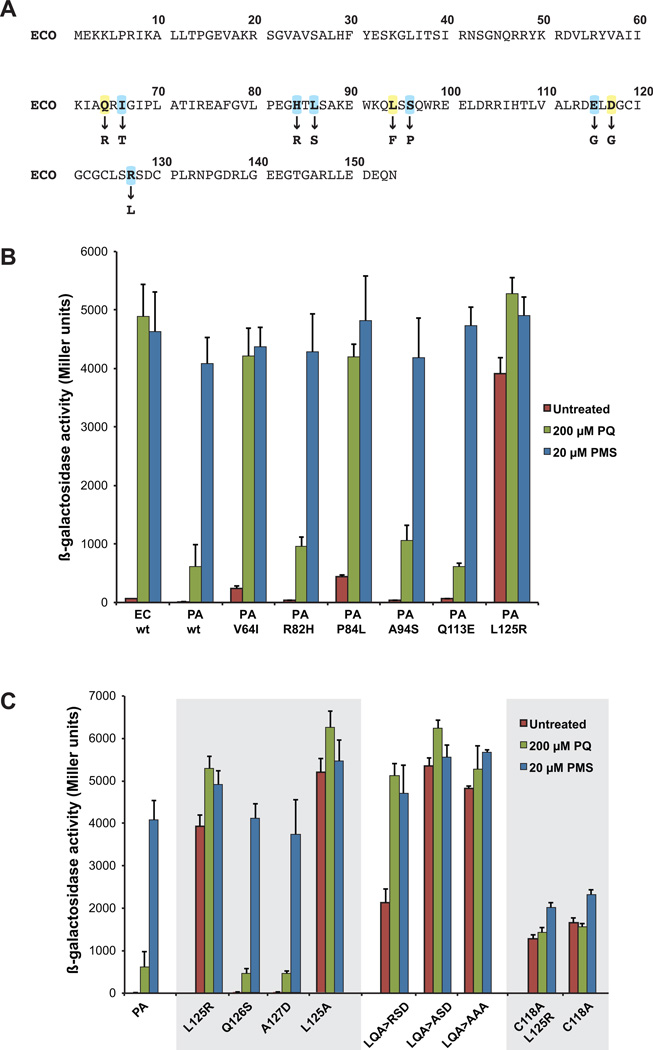
A. The amino acid sequence of E. coli SoxR is shown. The nine residues that had previously been shown to influence the redox-sensing properties of SoxR are in bold type; the substitutions that caused reduced sensitivity to PQ are indicated below the wild type sequence (Chander and Demple, 2004). Two of these residues (Q64 and L94) are conserved in P. aeruginosa SoxR; the six residues highlighted in blue are not conserved between P. aeruginosa and E. coli SoxR, and were targeted for mutagenesis in the former.
B. EH46 cells expressing wild type E. coli or P. aeruginosa soxR alleles, or P. aeruginosa soxR mutant alleles from pSE380-based plasmids were untreated (red columns), or treated with 200 µM PQ (green columns) or 20 µM PMS (blue columns) for 1 h before the assay for β-galactosidase activity. The results represent the means and standard errors of three independent experiments.
C. EH46 cells expressing wild type or mutant P. aeruginosa soxR alleles from pSE380 plasmids were treated as in (B). The mutations in these alleles are all within the [2Fe-2S] cluster domain. The SoxR variants L125R, C118A, and C118AL125R are expressed at similar levels in EH46 cells as assessed by immunoblot analysis (data not shown).
We individually mutated six of the aforementioned residues in P. aeruginosa SoxR (V64I, R82H, P84L, A94S, Q113E, L125R) and analyzed the resulting variants using the β-galactosidase assay described previously. As shown before, while E. coli SoxR was activated with similar efficiency by both PQ and PMS, wild type P. aeruginosa SoxR was strongly activated by PMS but only weakly by PQ (Fig. 5B). Mutant proteins R82H, A94S and Q113E resembled wild type P. aeruginosa SoxR (Fig. 5B). Two amino acid substitutions, V64I and P84L, conferred PQ-sensitivity to P. aeruginosa SoxR, essentially converting this protein into its E. coli counterpart (Fig. 5B). The L125R substitution, which alters a residue in the [2Fe-2S] cluster region, rendered P. aeruginosa SoxR constitutively active; in untreated cells, this variant displayed ~80% of the activity obtained in the presence of PQ or PMS (Fig. 5B). To determine if the constitutive activity of the L125R variant depends on the [2Fe-2S] clusters, we introduced a C118→A mutation into this background (C118AL125R). The equivalent cysteine→alanine mutation in E. coli and S. coelicolor SoxRs results in cluster-deficient proteins (Bradley et al., 1997; Dela Cruz et al., 2010). The C118AL125R variant showed a ~3-fold reduction in constitutive activity compared to the L125R single variant, indicating the importance of the [2Fe-2S] clusters (Fig. 5C). The relatively high basal level activity of the C118AL125R protein was also produced by the C118A single mutant protein (Fig. 5C) and had been previously observed with cluster-deficient E. coli SoxR (Bradley et al., 1997).
The constitutive activity displayed by the L125R P. aeruginosa SoxR variant was unexpected, given that E. coli SoxR (which has an arginine in this position) is not constitutive. It is interesting that SoxR homologs from enteric species contain an arginine in this position, which is replaced by a hydrophobic residue (most often leucine) in SoxR from almost all non-enteric species analyzed (Fig. 3A and Fig. S4A). Our finding that the presence of arginine within the [2Fe-2S] domain makes SoxR constitutively active, as was observed with the L125R P. aeruginosa SoxR variant, conflicted with the fact that E. coli SoxR is not constitutive. We hypothesized that this difference might be attributed to other amino acids in the vicinity of the [2Fe-2S] cluster that modulate E. coli SoxR activity, tuning it so that it is only active in the presence of redox-active drugs. A closer examination of the SoxR sequences from enteric and non-enteric bacteria revealed that while the [2Fe-2S] cluster domain is highly conserved, it contains a three-residue hypervariable sequence (Fig. S4A). In most species of the Enterobacteriaceae it is comprised of a hydrophilic “RSD” motif. However, it is not conserved in SoxRs from other bacteria, including P. aeruginosa, which instead contains the sequence LQA (Figs. 3A, S4A). In fact, we found only two species not belonging to the Enterobacteriaceae, Pseudomonas nitroreducens and Sphingopyxis alaskensis, that contain the RSD motif (Fig. 3B). We speculated that the RSD motif is crucial for the E. coli-specific activation of SoxR. First, we asked if it is the presence of the arginine residue that renders the L125R P. aeruginosa SoxR mutant constitutively active, or the lack of its native leucine. For this we generated an L125A mutant, which proved to be constitutively active (Fig. 5C). This suggested that the absence of L125 was sufficient to make P. aeruginosa SoxR constitutively active. Next we tested if the serine and aspartate residues within this motif are responsible for preventing constitutive activity. Introduction of mutations Q126S or A127D alone had no effect on P. aeruginosa SoxR activity (Fig. 5C). In contrast, introduction of both mutations into the L125R background (LQA→RSD), such that the [2Fe-2S] cluster domain was now an exact replica of E. coli SoxR, dampened the constitutive phenotype significantly, while still allowing sensitivity to PQ (Fig. 5C). Interestingly, changing QA→SD in the L125A background (LQA→ASD) had no attenuating effect on the constitutive activity of the single mutant protein (Fig. 5C). Similarly, the LQA→AAA triple mutant protein also demonstrated high constitutive activity (Fig. 5C). This showed that, while rendering P. aeruginosa SoxR constitutively active could be achieved by a variety of substitutions of L125, the “dampening effect” required a specific combination of residues in the hypervariable region (Fig. 5C).
Having demonstrated that a three-residue substitution (LQA→RSD) in the [2Fe-2S] region is sufficient to transform the activity of P. aeruginosa SoxR to mimic that of E. coli SoxR, we tested if the same would hold true in reverse. We changed RSD→LQA in E. coli SoxR, such that its [2Fe-2S] region replicated that of P. aeruginosa SoxR, and found that this change did not affect its activity; this mutant protein retained sensitivity to PQ (Fig. S4B). Interestingly, a mere substitution of R127→ L rendered E. coli SoxR insensitive to PQ, highlighting the importance of this position in the regulation of SoxR activity (Fig. S4B, Chander and Demple, 2004).
DISCUSSION
SoxR regulates a global stress response against redox-cycling drugs in E. coli. As such this protein is engineered to sense and respond to a wide spectrum of redox-active molecules that vary in structure and redox potentials. This is not the case for P. aeruginosa and S. coelicolor in which SoxR performs a more specific role as suggested by the small number of genes it regulates in response to endogenous redox-active signals. Given the functional differences of SoxR homologs across species, we asked if the sensitivities of P. aeruginosa and S. coelicolor SoxR were tuned towards compounds that resemble phenazines and Act, respectively. We found this to be partially true. While E. coli SoxR was activated by structurally distinct drugs that span a range in redox potentials from ~−450 to ~+80 mV, P. aeruginosa and S. coelicolor SoxR appeared to be less sensitive to the viologens that have redox potentials below ~−300 mV. Although SoxR appears to be tuned to sense drugs based on their redox potentials, we cannot rule out other contributing factors, such as structural features, charge and hydrophobicity.
What is the mechanism underlying SoxR’s differential selectivity for drugs? An earlier study on E. coli SoxR had identified residues that, when mutated, reduce the protein’s reactivity to drugs with low redox potentials (such as PQ). Six of these residues (Ile66, His84, Leu86, Ser96, Glu115, Arg127 in E. coli) are conserved in SoxR homologs from enterics (which display broad drug selectivity), but not in those from non-enterics (which show narrower drug selectivity). Changing the corresponding residues in P. aeruginosa SoxR to those found in E. coli SoxR revealed that three of the six residues individually affected drug-sensitivity. Substitutions V64→Ile and Pro84→Leu both increased the sensitivity of P. aeruginosa SoxR to PQ, and the variants were indistinguishable from E. coli SoxR (Fig. 5B). The Leu125→Arg change resulted in constitutive activity (Fig. 5B). These amino acids are conserved in S. coelicolor SoxR (V65, P85, L126; Fig. 3A), which like P. aeruginosa SoxR showed reduced sensitivity to viologens (Fig. 2C). These findings emphasize the importance of these amino acids in SoxR sensitivity to redox-active small molecules, and suggest structural changes that E. coli SoxR may have evolved if its soxR gene was acquired by horizontal gene transfer.
SoxR forms a homodimer. Each subunit contains three distinct domains: a DNA binding domain composed of four helices (h1–h4), a coiled-coil dimerization helix (h5) and the C-terminal sensor domain that contains the [2Fe-2S] clusters (Fig. 6). The crystal structure for the oxidized SoxR dimer bound to DNA shows that helices 3 and 4 within the DNA binding domain make hydrophobic contacts with helix 5 within the same subunit (Watanabe et al., 2008). Furthermore, the metal binding domain of one subunit is stabilized by interactions with helices 3, 4 and 5 of the other monomer. The structure of reduced SoxR is unknown, but using Raman spectroscopy, Kobayashi and colleagues (2011) showed that the relative orientations of helices 3 and 4 (in the DNA binding domain) and helix 5 (dimerization domain) depend on the redox state of SoxR. It is tempting to speculate that transmission of oxidative signals from the [2Fe-2S] clusters to the DNA involves an orchestrated rearrangement of the metal binding, dimerization and DNA binding domains, thereby explaining how the redox signal may be propagated from the [2Fe-2S] clusters to the DNA. Conversely, potential structural changes that result from DNA binding of SoxR have dramatic effects on the redox potential of its [2Fe-2S] cluster, highlighting the fine-tuned feedback between the DNA binding and sensory domains (Gorodetsky et al., 2008). It would therefore not be surprising that even small changes in the protein structure impact sensing and activation. It is peculiar that previous measurements of SoxR redox potentials gave almost identical results for E. coli and P. aeruginosa SoxR in solution (−280 mV; Ding, et al., 1996; Gaudu and Weiss, 1996; Kobayashi and Tagawa, 2004), and in the DNA-bound forms (+200 mV; Gorodetsky et al., 2008). A potential caveat of these analyses is that the redox potential of SoxR was not measured in vivo (i.e. bound to DNA within a cell). A previous study by Koo and colleagues (2003) suggests that SoxR may be part of a transient protein complex. We speculate that these interactions may differ for SoxRs from different species and could affect their redox potentials differentially in vivo. A recent paper (Fujikawa et al., 2012) also demonstrated that E. coli SoxR was more readily oxidized by superoxide than was P. aeruginosa SoxR, hinting at differences in their redox potentials. At present, we can only speculate that the tuning of SoxR’s specificity towards redox-active compounds is based on its redox potential. However, other factors (such as differences in structure and charge) may also contribute to the differential recognition of small molecules. Independent of the precise mechanism, we identified key residues that regulate SoxR specificity: Ile66, His84, Leu86, Ser96, Glu115, Arg127 in E. coli SoxR. Ile66, His84 and Leu86 are located at the interface between helix 3 in the DNA binding domain and the dimerization helix 5 (Fig. 6), which makes them good candidates for mediating the functional interaction between DNA binding and sensing. Leu86, for example, which is located just upstream of helix 5, forms hydrophobic interactions with Tyr56 and Ile59 in helix 3 (Watanabe et al., 2008). Interestingly, helix 3 also interacts with the sensor domain of the second dimer subunit (Fig. 6). Our findings demonstrate that even point mutations can change the sensitivity towards specific compounds.
Figure 6. Location of key residues in E. coli SoxR.
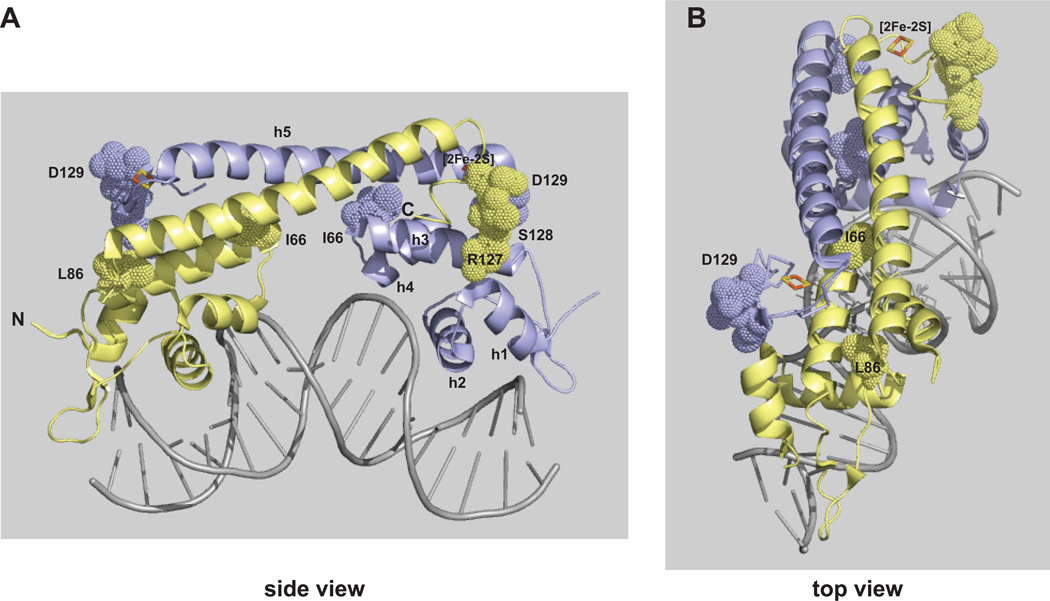
The structure of E. coli SoxR protein complexed with the soxS promoter is depicted side-on (A) or from the top (B) (Watanabe et al., 2008; pdb 2ZHG). The amino- and carboxy- termini are indicated by N and C, respectively, on one of the monomers. Helices labeled h1–h4 comprise the DNA binding domain; h5 is the dimerization domain; the [2Fe-2S] cluster in one monomer is labeled. Residues (I66, L86, R127, S128, D129) identified as playing an important role in tuning the redox-reactivity of SoxR are shown. The images were created using MacPyMOL Molecular Graphics System, Version 1.5, Schrodinger, LLC.
A particularly intriguing example is the [2Fe-2S] binding site itself. Although it is remarkably conserved among SoxR homologs, it contains a hypervariable stretch of three residues (Fig. 3A, S3A). Strikingly, within almost all enterics we found it to be conserved as the charged RSD motif (Fig. 3B, S4A). In contrast, in P. aeruginosa it is replaced by LQA (Fig. 3A). When P. aeruginosa SoxR was mutated to replace the LQA motif with RQA, the resulting variant displayed strong constitutive activity (Fig. 5B). Thus having an arginine residue within the [2Fe-2S] domain makes SoxR constitutively active. However, when the original LQA motif was mutated to RSD (so that the [2Fe-2S] cluster was now identical to that in E. coli SoxR), the level of constitutive activity significantly decreased, but the protein still retained the ability to respond to PQ (Fig. 5C). Thus, the RSD motif in SoxRs from Enterobacteriaceae is essential for fine-tuning the protein’s specificity – preventing constitutive activity while retaining low selectivity for drugs.
Considering that the RSD motif is almost exclusively found in the Enterobacteriaceae, it is tempting to speculate that it is an evolutionary adaptation specific to this bacterial family (Figure S4A). In this context, it is interesting that an LQA→RSD substitution in P. aeruginosa SoxR mimicked E. coli SoxR but the analogous RSD→LQA E. coli mutant remained sensitive to PQ (Fig. S4B). The idea that protein evolution from one functional state to another may not be reversible due to the occurrence of secondary mutations has recently been demonstrated (Bridgham et al, 2009). The fact that the LQA motif has different effects on PQ sensitivity in E. coli and P. aeruginosa SoxR indicates that specific residues outside of the [2Fe-2S] region contribute to the regulatory function of the hypervariable region.
Our findings give insight into the diversity of SoxR proteins with respect to their ability to sense redox-active compounds. They demonstrate how minor changes in the primary sequence can lead to the evolution of SoxR proteins with narrow- or broad-range sensing capacities.
EXPERIMENTAL METHODS
Bacterial strains and plasmids
Bacterial strains and plasmids that were utilized or constructed in this study are listed in Table 2.
Redox-cycling drugs
The redox-cycling drugs used in this study are listed in Table 1 along with their chemical structures and midpoint redox potentials. All chemicals were purchased from Sigma, with the exception of PCA which was purchased from Princeton Biomolecular Research, and γ-Act which was extracted from S. coelicolor cells as described by Bystrykh et al (1996). PQ, DQ, MB, and PMS were dissolved in water; PB, Pyo, PCA and Act in dimethylsulfoxide; 4NQO in acetone.
Drug susceptibility tests
The effects of various redox active drugs on the growth of wild type and ΔsoxR E. coli, P. aeruginosa, and S. coelicolor cells were determined using a disk diffusion assay (strains are listed in Table 2). E. coli and P. aeruginosa cells were grown for 16 h at 37°C in LB medium, 100 µL added to 4 mL of melted soft nutrient agar (Difco), then plated on nutrient agar plates (Difco). S. coelicolor spores (~108) were similarly plated. Six-millimeter Whatman paper disks impregnated with 15 µL of drug were placed onto the agar. E. coli plates were incubated at 37°C for 24 h, and P. aeruginosa and S. coelicolor plates at 30°C for 48 h, after which the zone of growth inhibition around each disk was recorded.
Cloning of his-tagged soxR genes for complementation analysis in E. coli and P. aeruginosa
For complementation analysis in E. coli, the soxR alleles from E. coli, P. aeruginosa, and S. coelicolor were expressed as N-terminally histidine-tagged proteins from the plasmid pSE380 under the control of the trc promoter (Table 2). The coding region of the soxR alleles (including the histidine-tag) was PCR-amplified from pET16b-based vectors (Chander and Demple, 2004, Gorodetsky et al., 2008, Dela Cruz et al., 2010) using primers pET-F and pET-R (Table S1) and Pfu Polymerase (Stratagene). The PCR fragments were digested with BamHI and SalI and ligated into pSE380. The resulting plasmids containing soxR alleles with a 10-histidine tag attached to the N-terminus were sequenced on both strands and transformed into E. coli strain EH46 or EH86 for β-galactosidase assays (Table 2).
For complementation analysis in P. aeruginosa, the histidine-tagged soxR proteins were subcloned from pSE380 into the BamHI/SalI site of the vector, pUCp18 (Table 2), and expressed under the control of the lac promoter. The resulting clones were sequenced on both strands and transformed into P. aeruginosa strain PA14ΔsoxR (Table 2).
Construction of soxR mutant alleles
Mutations in the P. aeruginosa or E. coli soxR genes were generated using the GENEART site-directed mutagenesis kit from Invitrogen according to the manufacturer’s instructions. Mutations in the S. coelicolor soxR gene were generated using the QuikChange site-directed mutagenesis kit from Stratagene following manufacturer’s recommendations. Plasmid pSE380, containing the histidine-tagged soxR genes from P. aeruginosa, E. coli, or S. coelicolor were used as templates for mutagenesis along with the mutagenic primers listed in Table S1. All mutations were verified by DNA sequence analysis.
For expression of the histidine-tagged C-terminal truncated soxR gene in S. coelicolor, the coding region (along with the 10-histidine tag) was PCR-amplified from pSE380 using Pfu polymerase and primers 380F-Bam and 380R-Bam (Table S1), and subcloned into the BamHI site of the integrating vector pSET152, to yield H-ΔC. The histidine-tagged WT S. coelicolor soxR gene was similarly constructed to yield H-SoxR. The pSET152-based plasmids were introduced into the S. coelicolor ΔsoxR strain M145-1A by intergenic conjugation from E. coli ET12567/pUZ8002.
β-galactosidase assay to measure complementation in an E. coli ΔsoxR mutant
The ability of the various SoxR homologs (and mutant derivatives) to complement an E. coli ΔsoxR strain was assessed by measuring β-galactosidase activity in EH46 cells (Table 2) expressing the various histidine-tagged SoxR proteins from pSE380-based plasmids as previously described (Chander et al., 2003). Cells were treated with various redox-active drugs for 1 h with shaking at 220 rpm.
β-galactosidase assays were also used to analyze the stable production and soxS promoter binding ability of the various SoxR proteins in vivo. Strain EH86 (Table 2) was transformed with the aforementioned plasmids and grown for 2.5 h in the absence of oxidative stress before the lysates were assayed for β-galactosidase activity.
qRT-PCR assay in S. coelicolor
Liquid R5− medium (Huang et al. 2001) was inoculated with 107 S. coelicolor spores mL−1 and grown at 30°C with shaking at 220 rpm for the indicated times. Cells were harvested by incubating with RNAprotect bacterial reagent (Qiagen) for 5 min at room temperature, centrifuging for 10 min at 5,000 × g, and frozen at −80°C. Total RNA was extracted and qRT-PCR assays conducted as previously described (Dela Cruz et al., 2010). The primers used for qRT-PCR are listed in Table S1.
Construction of P. aeruginosa mexG-gfp reporter strains and Gfp fluorescence quantification
The mexG promoter region was PCR-amplified from PA14 genomic DNA using primers pmexG-F and pmexG-R (Table S1), and cloned into the HindIII/EcoRI site of the vector pYL122 (Table 2). The pmexG-gfp reporter fusion was integrated into the attB site of P. aeruginosa PA14 or PA14ΔsoxR using a previously described protocol (Lequette and Greenberg, 2005).
To quantify Gfp fluorescence, the pmexG-gfp reporter strains expressing histidine-tagged E. coli or P. aeruginosa soxR from pUCp18, were grown in LB medium supplemented with carbenicillin (300 µg mL−1) for 16 h at 37°C. Cultures were then diluted 100-fold and grown for an additional 3 h (to logarithmic phase), before finally diluting to an optical density of 0.05 at 500 nm into a 96-well plate (Costar). The optical density and fluorescence was monitored for 19 h using a Synergy 4-plate reader (BioTek). The excitation wavelength was 488 nm; emission wavelength was 520 nm. Data was acquired using the Gen5 program.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by an AREA grant from the National Institute of General Medical Sciences (R15GM093366) to MC, and a start-up grant from Columbia University to LEPD.
We would like to thank Rica Dela Cruz for conducting EMSAs, Lilian Hsu for providing the facilities to conduct in vitro transcription assays, Jessica Watkins for assisting in the design of mutagenic primers, and Joanne Willey, Alexa Price-Whelan and Michelle Wien for valuable feedback on the manuscript.
Footnotes
Supplementary Information
The following supplementary material is available for this article online:
Figure S1. Complementation of a P. aeruginosa ΔsoxR mutant by E. coli or P. aeruginosa SoxR.
Figure S2. SoxR protein expression, soxS promoter binding, and in vitro transcription of the soxS gene.
Figure S3. Comparison of the transcriptional response of E. coli and P. aeruginosa SoxR to varying doses of redox-active molecules.
Figure S4. The [2Fe-2S] cluster domain of SoxR has a hypervariable stretch of three amino acids.
Table S1. Primers used in this study.
References
- Beinert H. Iron-sulfur proteins: ancient structures, still full of surprises. J. Biol. Inorg. Chem. 2000;5:2–15. doi: 10.1007/s007750050002. [DOI] [PubMed] [Google Scholar]
- Biaglow JE, Jacobson B, Varnes M, Koch C. The oxidation of ascorbate by electron affinic drugs and carcinogens. Photochem. Photobiol. 1978;28:869–876. doi: 10.1111/j.1751-1097.1978.tb07034.x. [DOI] [PubMed] [Google Scholar]
- Bierman M, Logan R, O’Brien K, Seno ET, Nagaraja Rao R, Schoner BE. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- Bradley TM, Hidalgo E, Leautaud V, Ding H, Demple B. Cysteine-to-alanine replacements in the Escherichia coli SoxR protein and the role of the [2Fe-2S] centers in transcriptional activation. Nucl. Acids Res. 1997;8:1469–1475. doi: 10.1093/nar/25.8.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgham JT, Ortlund EA, Thornton JW. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature. 2009;461:515–519. doi: 10.1038/nature08249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystrykh LV, Fernandez-Moreno MA, Herrema JK, Malpartida F, Hopwood DA, Dijkhuizen L. Production of actinorhodin “blue pigments” by Streptomyces coelicolor A3(2) J. Bacteriol. 1996;178:2238–2244. doi: 10.1128/jb.178.8.2238-2244.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chander M, Raducha-Grace L, Demple B. Transcription-defective soxR mutants of Escherichia coli: isolation and in vivo characterization. J. Bacteriol. 2003;185:2441–2450. doi: 10.1128/JB.185.8.2441-2450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chander M, Demple B. Functional analysis of SoxR residues affecting transduction of oxidative stress signals into gene expression. J. Biol. Chem. 2004;279:41603–41610. doi: 10.1074/jbc.M405512200. [DOI] [PubMed] [Google Scholar]
- Dela Cruz R, Gao Y, Penumetcha S, Sheplock R, Weng K, Chander M. Expression of the Streptomyces coelicolor SoxR regulon is intimately linked with actinorhodin production. J. Bacteriol. 2010;192:6428–6438. doi: 10.1128/JB.00916-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich LEP, Price-Whelan A, Petersen A, Whitely M, Newman DK. The phenazine pyocyanin is a terminal signaling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 2006;61:1308–1321. doi: 10.1111/j.1365-2958.2006.05306.x. [DOI] [PubMed] [Google Scholar]
- Dietrich LEP, Teal TK, Price-Whelan A, Newman DK. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science. 2008;321:1203–1206. doi: 10.1126/science.1160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Hidalgo E, Demple B. The redox state of the [2Fe-2S] clusters in SoxR protein regulates its activity as a transcription factor. J. Biol. Chem. 1996;271:33173–33175. doi: 10.1074/jbc.271.52.33173. [DOI] [PubMed] [Google Scholar]
- Ding H, Demple B. In vivo kinetics of a redox-regulated transcriptional switch. Proc. Natl. Acad. Sci. USA. 1997;94:8445–8449. doi: 10.1073/pnas.94.16.8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A. Geneious v5.4. 2011 http://www.geneious.com. [Google Scholar]
- Floriano B, Bibb M. afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A(3)2. Mol. Microbiol. 1996;21:385–396. doi: 10.1046/j.1365-2958.1996.6491364.x. [DOI] [PubMed] [Google Scholar]
- Friedheim E, Michaelis L. Potentiometric study of pyocyanine. J. Biol. Chem. 1931;91:355–368. [Google Scholar]
- Fujikawa M, Kobayashi K, Kozawa T. Direct oxidation of the [2Fe-2S] cluster in SoxR by superoxide: distinct differential sensitivity to superoxide-mediated signal transduction. J. Biol. Chem. 2012;287:35702–35708. doi: 10.1074/jbc.M112.395079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudu P, Weiss B. SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc. Natl. Acad. Sci. USA. 1996;93:10094–10098. doi: 10.1073/pnas.93.19.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudu P, Moon N, Weiss B. Regulation of the soxRS oxidative stress regulon. Reversible oxidation of the Fe-S centers of SoxR in vivo. J. Biol. Chem. 1997;272:5082–5086. doi: 10.1074/jbc.272.8.5082. [DOI] [PubMed] [Google Scholar]
- Gorodetsky AA, Dietrich LEP, Lee PE, Demple B, Newman DK, Barton JK. DNA binding shifts the redox potential of the transcription factor SoxR. Proc. Natl. Acad. Sci. USA. 2008;105:3684–3689. doi: 10.1073/pnas.0800093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg JT, Monach P, Chou JH, Josephy PD, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Imlay JA. The SoxRS response of Escherichia coli is directly activated by by redox-cycling drugs rather than by superoxide. Mol. Microbiol. 2011;79:1136–1150. doi: 10.1111/j.1365-2958.2010.07520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakura A, Mochida H, Tsutsui Y, Yamatsu K. Mutagenicity and cytotoxicity of napthoquinones for Ames Salmonella tester strains. Chem. Res. Toxicol. 1994;7:559–567. doi: 10.1021/tx00040a012. [DOI] [PubMed] [Google Scholar]
- Hidalgo E, Bollinger JM, Bradley TM, Walsh CT, Demple D. Binuclear [2Fe-2S] clusters in the Escherichia coli SoxR protein and role of the metal centers in transcription. J. Biol. Chem. 1995;270:20908–20914. doi: 10.1074/jbc.270.36.20908. [DOI] [PubMed] [Google Scholar]
- Hidalgo H, Demple B. In: Regulation of gene expression in Escherichia coli. Lin ECC, Lynch AS, editors. Austin, TX: R.G. Landes Co; 1996. [Google Scholar]
- Hidalgo E, Demple B. Spacing of promoter elements regulates the basal expression of the soxS gene and converts SoxR from a transcriptional activator into a repressor. EMBO J. 1997;13:138–146. doi: 10.1093/emboj/16.5.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Lih CJ, Pan H, Cohen SN. Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes Dev. B. 2001;15:3183–3192. doi: 10.1101/gad.943401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamat P, Mimitijevic N, Fessenden R. Photoelectrochemistry in particulate systems: electron-transfer reactions of small CdS colloids in acetonitrile. J. Phys. Chem. 1987;91:396–401. [Google Scholar]
- Khoroshilova N, Popescu C, Münck E, Beinert H, Kiley PJ. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc. Natl. Acad. Sci. USA. 1997;94:6087–6092. doi: 10.1073/pnas.94.12.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces coelicolor genetics. Norwich, England: John Innes Foundation; 2000. [Google Scholar]
- Kobayashi K, Tagawa S. Activation of SoxR-dependent transcription in Pseudomonas aeruginosa. J. Biochem. 2004;136:607–615. doi: 10.1093/jb/mvh168. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Mizuno M, Fujikawa M, Mizutani Y. Protein conformational changes of the oxidative stress sensor, SoxR, upon redox changes of the [2Fe-2S] cluster probed with ultraviolet resonance Raman spectroscopy. Biochemistry. 2011;50:9468–9474. doi: 10.1021/bi201526y. [DOI] [PubMed] [Google Scholar]
- Koo M-S, Lee J-H, Rah S-Y, Yeo W-S, Lee J-W, Lee K-L, Koh Y-S, Kang S-A, Roe J-H. A reducing system of the superoxide sensor SoxR in Escherichia coli. EMBO J. 2003;22:2614–2622. doi: 10.1093/emboj/cdg252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lequette Y, Greenberg EP. Timing and localization of rhamnolipid synthesis gene expression in Pseudomonas aeruginosa biofilms. J. Bacteriol. 2005;187:37–44. doi: 10.1128/JB.187.1.37-44.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffet DA, Foley J, Hecht M. Midpoint reduction potentials and heme binding stoichiometries of de novo proteins from designed combinatorial libraries. Biophys. Chem. 2003;105:231–239. doi: 10.1016/s0301-4622(03)00072-3. [DOI] [PubMed] [Google Scholar]
- Palma M, Zurita J, Ferreras JA, Worgall S, Larone DH, Shi L, Campagne F, Quadri LEN. Pseudomonas aeruginosa SoxR does not conform to the archetypal paradigm for SoxR-dependent regulation of the bacterial oxidative stress adaptive response. Infect. Immun. 2005;73:2958–2966. doi: 10.1128/IAI.73.5.2958-2966.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomposiello PJ, Bennik MH, Demple B. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 2001;183:3890–3902. doi: 10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price-Whelan A, Dietrich LEP, Newman DK. Rethinking ‘secondary’ metabolism: physiological roles for phenazine antibiotics. Nature Chem. Biol. 2006;2:71–78. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schweizer HP. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991;97:109–121. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- Shin JH, Singh AK, Cheon DJ, Roe JH. Activation of the SoxR regulon in Streptomyces coelicolor by the extracellular form of the pigmented antibiotic actinorhodin. J. Bacteriol. 2011;193:75–81. doi: 10.1128/JB.00965-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckhan E, Kuwana T. Spectrochemical study of mediators. I Bipyridylium salts and their electron transfer rates to cytochrome C. Ber. Bunsenges. Phys. Chem. 1974;78:253–258. [Google Scholar]
- Tsaneva IR, Weiss B. soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J. Bacteriol. 1990;172:4197–4205. doi: 10.1128/jb.172.8.4197-4205.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Kita A, Kobayashi K, Miki K. Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA. Proc. Natl. Acad. Sci. USA. 2008;105:4121–4126. doi: 10.1073/pnas.0709188105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo W-S, Lee J-H, Lee K-C, Roe J-H. IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Mol. Microbiol. 2006;61:206–218. doi: 10.1111/j.1365-2958.2006.05220.x. [DOI] [PubMed] [Google Scholar]
- Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 2001;183:4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


