SUMMARY
Modifications and loss of bases are frequent types of DNA lesions, often handled by the base excision repair (BER) pathway. BER is initiated by DNA glycosylases, generating abasic (AP) sites that are subsequently cleaved by AP endonucleases, which further pass on nicked DNA to downstream DNA polymerases and ligases. The coordinated handover of cytotoxic intermediates between different BER enzymes is most likely facilitated by the DNA conformation. Here we present the atomic structure of Schizosaccharomyces pombe Mag2 in complex with DNA to reveal an unexpected structural basis for non-enzymatic AP site recognition with an unflipped AP site. Two surface exposed loops intercalate and widen the DNA minor groove to generate a DNA conformation previously only found in the mismatch repair MutS-DNA complex. Consequently, the molecular role of Mag2 appears to be AP site recognition and protection while possibly facilitating damage signaling by structurally sculpting the DNA substrate.
INTRODUCTION
Cellular DNA is constantly subjected to modifications by intracellular and extracellular chemicals, which can result in covalent changes of the genome (Lindahl, 1993). One group of such chemicals is alkylating agents, which exist endogenously, but are also prevalent in our environment and used as anticancer compounds in the clinical setting (Hecht, 1999; Hurley, 2002; Rajski and Williams, 1998; Rydberg and Lindahl, 1982; Taverna and Sedgwick, 1996; Vaughan et al., 1993). Further, reactive oxygen species (ROS) form a series of oxidized purine and pyrimidine derivatives (David et al., 2007; Neeley and Essigmann, 2006). The multistep base excision repair (BER) pathway is the major cellular process for correction of non-bulky single-base lesions resulting from alkylation, oxidation or deamination of the nucleobases in DNA (Dalhus et al., 2009; Huffman et al., 2005; Robertson et al., 2009). The pathway is initiated by DNA glycosylases that recognize and remove the chemically altered bases by cleavage of the N-glycosylic bond between the base and deoxyribose to form a cytotoxic apurinic/apyrimidinic (AP) site (Stivers and Jiang, 2003; Berti and McCann, 2006). AP sites may also be formed by spontaneous hydrolysis of the N-glycosylic bond (Lindahl, 1993). Since AP sites are non-instructional and may be bypassed by the translesion DNA polymerases, they can lead to mutagenesis if left unrepaired (Boiteux and Guillet, 2004; Loeb, 1985). These spontaneously or enzymatically generated AP sites are substrates for AP endonucleases (Ribar et al., 2004; Dianov et al., 2003). The BER pathway is completed by a template-guided restoration of the damaged DNA strand by means of the sequential action of phosphodiesterases, DNA polymerases and DNA ligases by either short patch or FEN1-dependent long patch repair (Frosina et al., 1996; Kim et al., 1998; Robertson et al., 2009). Additional accessory factors are important for coordination and regulation of the different steps of BER in vivo (Caldecott et al., 1994; Frosina et al., 1996; Kim et al., 1998; Kubota et al., 1996; Matsumoto et al., 1994). In addition to BER, epistasis analyses in Escherichia coli and yeast have revealed that nucleotide excision repair (NER) and recombination repair (RR) also contribute to cellular resistance to alkylating agents (Alseth et al., 2005; Memisoglu and Samson, 2000; Xiao and Chow, 1998).
Multiple forms of base lesions are removed by diverse DNA glycosylases to form AP sites. Endogenous agents like nitric oxide, which are regulated by synthesis (Crane et al., 1997) also form nitrosatively induced AP sites that must be detected and repaired. Moreover, AP sites are themselves lesions at least as toxic and mutagenic as many types of base damage as their instability leads to strand breaks (Guillet and Boiteux, 2002). The central role of AP sites in genome integrity and their instability raises the question as to whether cells may have evolved other mechanisms besides canonical BER and NER to recognize, protect, and process this lesion.
All DNA glycosylases have high affinity for the cytotoxic abasic product in addition to the damaged substrate DNA. Remarkably, both DNA glycosylases and the downstream AP endonucleases have rigid, preformed DNA binding surfaces that invade the DNA helix to hold it in a severely distorted conformation with a flipped AP site (Bruner et al., 2000; Doublie et al., 2004; Hollis et al., 2000a; Lau et al., 2000; Mol et al., 2000a; Mol et al., 2000b; Slupphaug et al., 1996). It has been suggested that this similarity in DNA shape is the molecular basis for an efficient handover of the abasic product (Mol et al., 2000b; Wilson and Kunkel, 2000). This hypothesis matches with observations of physical interactions between human DNA glycosylases and human AP endonuclease APE1. In particular, APE1 can actively replace the 8-oxoguanine DNA glycosylase OGG1 and thereby increase the turnover of the glycosylase, and possibly displace OGG1 in a processive mode, with OGG1 remaining on DNA but sliding away in search for a new lesion (Sidorenko et al., 2007). A similar mechanism has been observed for the human thymine DNA glycosylase TDG, where APE1 significantly increases the dissociation rate of TDG from AP sites (Waters et al., 1999).
Recently, a completely different abasic DNA capture mechanism was described for the AlkD HEAT-like repeat DNA glycosylase family (Rubinson et al., 2010). In the structure of the AlkD-DNA complex, the DNA is distorted in a surprisingly different way compared to other DNA glycosylase complexes. The crystal structures of the S. pombe Mag2 in complex with abasic DNA, presented in this work, reveal yet another AP site recognition mechanism, thus representing a third configuration for shaping of abasic DNA, with the first example of an unflipped AP site combined with an unusual widening of the minor groove. This novel non-enzymatic DNA sculpting by Mag2 is induced by two loops intercalating into and widening the DNA minor groove flanking the AP site to mimic the DNA conformation previously only found in the mismatch repair protein MutS in complex with DNA. Furthermore, kinetics binding analyses reveal a dynamic interaction suitable to rapidly recognize and release AP sites providing a comparative basis to understand mechanisms of stable versus transient interactions with DNA lesions. Collectively, structural, biochemical, and genetic results establish Mag2 as a novel AP site recognition protein that operates separately from known BER processing pathway mechanisms.
RESULTS
S. pombe Mag2 is a DNA Damage Inducible Nuclear Protein
The S. pombe genome sequence contains two genes, mag1 and mag2, encoding putative 3mA DNA glycosylases with 43% sequence identity and 65% sequence similarity, indicating a close genetic and structural relationship between these two proteins (Figure 1A). Whereas Mag1 is involved in repair of alkylated bases (Adhikary and Eichman, 2011; Alseth et al., 2005), the function of Mag2 is not clearly ascribed. Northern blot analysis showed no expression of mag2 in untreated exponentially growing yeast cells, however, transcription was strongly induced following exposure of S. pombe cells to the alkylating agent methyl methanesulfonate (MMS) and oxidizing H2O2 (Figure 1B). Fluorescence microscopy of cells expressing Mag2 fused in front of green fluorescent protein (GFP) revealed that Mag2 was sorted to the nucleus (Figure 1C).
Figure 1. Characterization of the mag2 Gene Function.
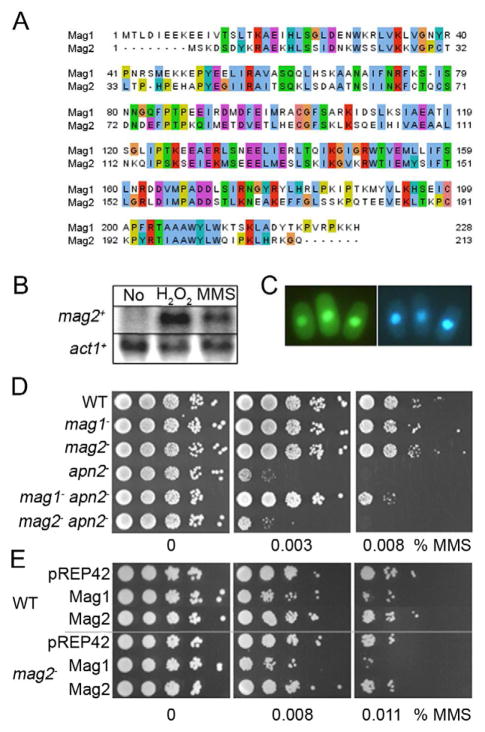
(A) Multiple-sequence alignment of S. pombe Mag1 and Mag2. Panel made using Jalview (Waterhouse et al., 2009) (B) Northern blot analysis of mag2 gene expression. Total RNA was isolated from untreated control (No) and from wildtype cells exposed to H2O2 or MMS and the filter was hybridized with a 642 bp mag2 probe. A β-a ctin (act1) probe was used as control. (C) Fluorescence microscopy of S. pombe cells expressing fusions of Mag2 to GFP. Yeast cells were transformed by DNA constructs expressing Mag2-GFPc (left panel) and stained with Hoechst 33342 as a nuclear marker (right panel). (D) MMS sensitivity of S. pombe BER mutant strains. S. pombe wild type (WT), mag1−, mag2−, apn2−, mag1− apn2− and mag2− apn2− mutant cells were spread on solid media containing increasing doses of MMS and cell survival was evaluated relative to plates without MMS. (E) Overexpression of Mag1, but not Mag2 is toxic for the fission yeast cells. S. pombe wild type (WT) and mag2− mutant strains overexpressing Mag1 or Mag2 from pREP42 were spread on minimal media containing increasing doses of MMS and cell survival was evaluated relative to plates without MMS. Cells with empty vector (pREP42) were used as control. See also Figure S1.
Biochemical and Epistasis Analysis Suggest Mag2 is Not a Classical DNA Glycosylase
Given the strong homology to the BER glycosylase Mag1, we investigated whether recombinant Mag2 could excise alkylated bases from DNA. Unexpectedly, Mag2 showed no DNA glycosylase activity for alkylated bases, even at very high enzyme concentration and under different assay conditions (titration of NaCl, Mg2+, ATP; data not shown). Further, heterologous expression of Mag2 did neither rescue the extreme alkylation sensitive phenotype of an E. coli mutant lacking the two 3mA DNA glycosylases AlkA and Tag, nor the corresponding Saccharomyces cerevisiae mag1 deletion mutant (data not shown). Mag2 was assayed for activity towards a variety of different base lesions which are known substrates for other DNA glycosylases including alkylated, oxidized and deaminated bases, base mismatches and AP sites; however no enzymatic activity was observed for any of the lesions tested (Table S1).
Neither the mag2− single mutant nor the mag1− mag2− double mutant are sensitive to MMS exposure (Kanamitsu et al., 2007) which together with our biochemical analysis suggests that Mag2 is not overlapping with Mag1 in the repair of alkylated bases. Further, deletion of mag2 does not suppress the MMS sensitive phenotype of the apn2− strain (Figure 1D), which contrasts the alleviating effect of mag1 deletion on the apn2 deficient cells (Alseth et al., 2005). Also the MMS sensitivity of the nth1− mutant was suppressed by the deletion of mag1 but not mag2 (Figure S1), which contrasts previous results (Kanamitsu et al., 2007). We do not know the reason for this discrepancy, but it could be because of different strain backgrounds, as the strains used by Kanamisu and co-workers tolerate much higher MMS doses (0.03% versus 0.007% in our experiments). Finally, whereas overexpression of Mag1 in S. pombe or S. cerevisiae (Glassner et al., 1998) is toxic, overexpression of Mag2 had no effect (Figure 1E). Indeed, it has been shown that alkylation repair must be carefully balanced to avoid toxicity from removal of normal bases (Fu et al., 2012). Taken together, our in vivo and in vitro results suggest that Mag2 is not involved in removal of alkylated DNA bases.
Mag2 Recognizes and Binds to Abasic Sites, But is Not Involved in Processing of Such Lesions Via BER, NER or RR in vivo
Many DNA glycosylases remain bound to the AP site product until acted upon by an AP endonuclease or an AP lyase (Hitomi et al., 2007). In contrast, electrophoretic mobility shift assay (EMSA) experiments revealed no binding of Mag2 to abasic DNA, nor did Mag2 bind undamaged DNA (data not shown). Unlike gel based EMSAs that monitor stable molecular interactions, surface plasmon resonance (SPR) can monitor transient interactions in real-time under native conditions. The binding characteristic of Mag2 by SPR showed a rapid association followed by rapid dissociation indicating a transient binding to the DNA. Mag2 binds stronger to the abasic oligonucleotide than to non-damaged DNA (Figure 2), with a dissociation rate constant more than 15 times higher for non-damaged DNA (kd 26 × 10−3 s−1) as compared to DNA containing the AP site analogue tetrahydrofuran (THF) (kd 1.6 × 10−3 s−1). Mag2 injected on sensor chips coated with oligonucleotides containing a single ethenoadenine or 8-oxoguanine lesion showed the same resonance levels as non-damaged DNA (data not shown), demonstrating that Mag2 preferentially binds to AP sites in DNA.
Figure 2. SPR Analyses of Mag2 wt and K53G Mutant Binding to Abasic DNA.
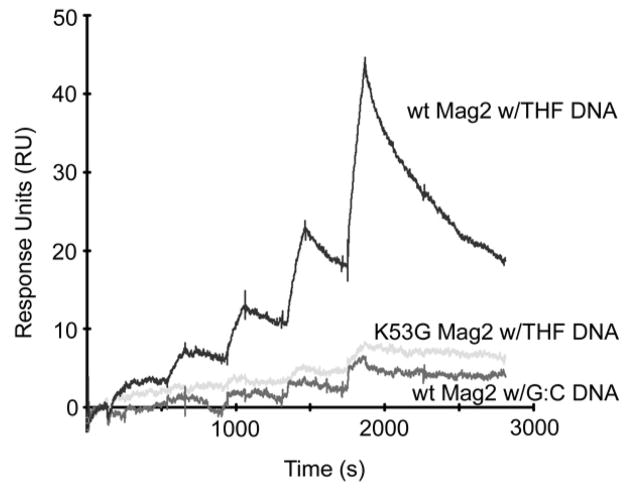
Increasing amounts (25, 50, 100, 200 and 400 nM) of Mag2 wt (dark grey) and K53G mutant (light gray) were injected on chip containing the abasic site analogue THF, or WT Mag2 on non-damaged DNA (intermediate grey).
Establishing that Mag2 binds AP sites, we tested by epistasis analysis whether Mag2 is involved in their processing in vivo. MMS treatment induces high amounts of AP sites as a consequence of Mag1 alkylbase DNA glycosylase activity. In S. pombe, AP sites are repaired by nucleotide excision repair and recombination repair in addition to base excision repair (Alseth et al., 2005). Double mutants of mag2− with RR (rhp55−) (Khasanov et al., 1999) and NER (rad16− = S. cerevsiae Rad1/human XPF) (Carr et al., 1994) were generated and the strains challenged to MMS. Disruption of mag2 in the rad16− or rhp55− mutants had no effect on the MMS hypersensitivity of the respective single mutant strains (Figure S1). From these data we conclude that Mag2 is not directly involved in alleviating the cytotoxic effects of AP sites in vivo suggesting other types of transient transactions involving abasic DNA.
Crystal Structure of Mag2 in Complex With DNA Reveals Novel Non-Enzymatic AP Site Recognition and DNA Sculpting
Having established that the damage inducible Mag2 binds rapidly and specifically to abasic DNA, we attempted to crystallize Mag2 in complex with DNA. The rapid association of Mag2 with abasic DNA may allow crystal nucleation, and that once formed, the crystal lattice interactions would slow the rapid dissociation observed in solution to allow the formation of Mag2-DNA crystals for structure determination. We were able to successfully grow DNA complex crystals and solve the atomic resolution structure of the Mag2-DNA complex by X-ray diffraction. The structure of Mag2 in complex with abasic DNA at 1.45 Å resolution reveals an all α-class protein with a fold reminiscent of that of a classical helix-hairpin-helix (HhH) DNA glycosylase (Figures 3A and 4A–F), as exemplified by E. coli endonuclease III (Nth) (Thayer et al., 1995), E. coli 3mA DNA glycosylase II (AlkA) (Hollis et al., 2000a), Helicobacter pylori 3mA DNA glycosylase MagIII (Eichman et al., 2003), Bacillus stearothermophilus endonuclease III (Nth) (Fromme and Verdine, 2003) and E. coli and B. stearothermophilus adenine DNA glycosylase (MutY) (Fromme et al., 2004; Guan et al., 1998). Mag2 binds to the minor groove of DNA and with a large distortion of the helical axis (Figure 3A). The hydrogen bond interactions between Mag2 and the DNA, predominantly on the 3′ side of the AP site, are mainly involving protein residues in the HhH segment (Figure 5A). Further, both polar and hydrophobic side-chains intercalate into the DNA (Figures 5A and 5B). Mag2 thus reveal a non-enzymatic application for the HhH motif and other features frequently employed by DNA glycosylase enzymes. Such non-enzymatic binding provides the framework to dissect specific binding versus lesion processing features.
Figure 3. Overall Structure and DNA Conformation in the S. pombe Mag2-DNA Complex.
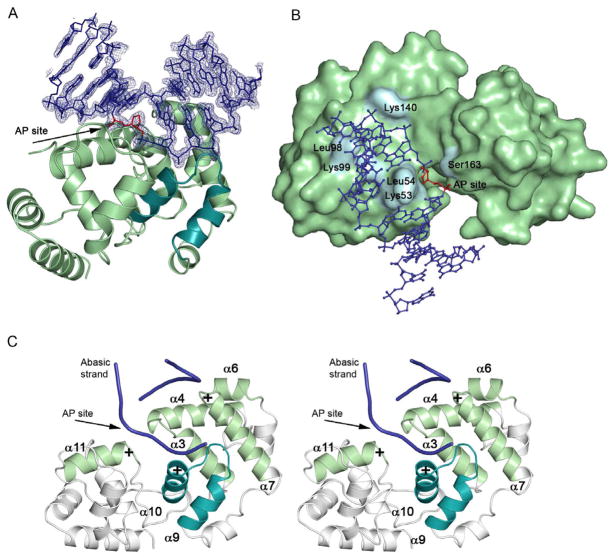
(A) Ribbon display of Mag2 in complex with abasic DNA (blue DNA with simulated annealing composite omit map) showing the distortion of the bound DNA. The canonical helix-hairpin-helix (HhH) motif is highlighted (cyan) and the unflipped AP site analogue is coloured red. (B) Surface representation of Mag2 with DNA in ball-and-stick. (C) Stereo-view of the various helices and loops involved in DNA bending, unwinding and strand separation. Helices α10 and α11 both have their positive dipole toward the negatively charged phosphate backbone, and pull and stretch the DNA strand containing the abasic site. Helices α6 and α7 form a “push-and-pull” pair as they have opposite charged dipoles close to the undamaged strand. The loop between helix α3 and α4 include residues Lys53 and Leu54, both of which intercalate with the DNA. Helix α7 contains Leu98 and Lys99, whereas the last interacting loop between helix α9 and α10 is part of the helix-hairpin-helix motif.
Figure 4. Protein Folding of Selected DNA Glycosylases and Trp-Asp-Trp Triad in S. pombe Mag2 and E. coli AlkA.
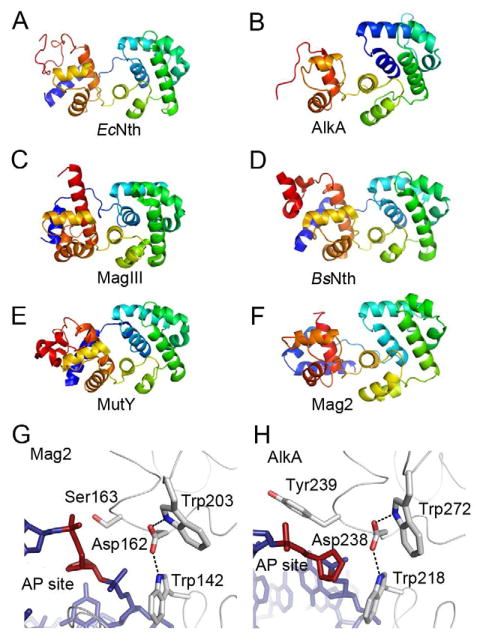
(A)–(F) Protein folding of (A) E. coli endonuclease III (Nth) (Thayer et al., 1995), (B) E. coli 3mA DNA glycosylase II (AlkA) (Hollis et al., 2000a), (C) H. pylori 3mA DNA glycosylase MagIII (Eichman et al., 2003), (D) B. stearothermophilus endonuclease III (Nth) (Fromme and Verdine, 2003), (E) B. stearothermophilus adenine DNA glycosylase (MutY) (Fromme et al., 2004), and (F) S. pombe Mag2. Color spectrum goes from N-terminal blue to C-terminal red. Only the C-terminal α-helical part of AlkA and the N-terminal part of B. stearothermophilus Nth are shown. (G) and (H) Close-up view of the AP site binding region in (G) S. pombe Mag2 and (H) E. coli AlkA (Hollis et al., 2000a) with stick models of the DNA (blue with abasic site in red). Both structures contain a Trp-Asp-Trp motif. The bulky side chain of Tyr239 in AlkA (H) pushes on the 5′ phosphate of the AP site as part of the flipping mechanism, while the less bulky Ser163 in Mag2 (G) allow the accommodation of a non-flipped AP site.
Figure 5. Protein-DNA Contacts in the S. pombe Mag2-DNA Complex.
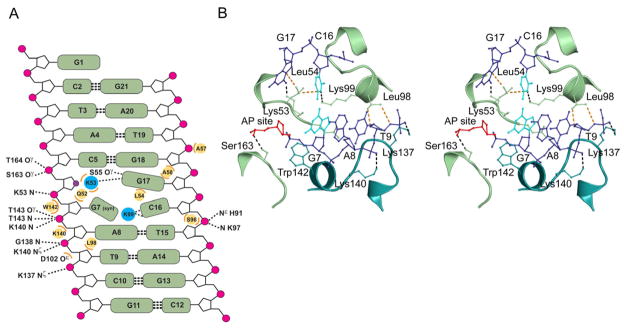
(A) Diagram showing all contacts between Mag2 and the abasic DNA. Hydrogen bond/ionic interactions < 3.35Å are shown with dashed lines, wherein symbol N denotes main chain amide. Steric interactions < 3.75Å are shown as orange arcs with the respective amino acids as yellow circles. Leu54 is wedged in between Cyt16 and the orphan Gua17, and together with Lys99 dictate the spatial orientation of Cyt16. Gua7 has the unusual syn conformation. The abasic site analogue is shown as a violet circle. (B) Stereo view of the intercalating loops in Mag2, showing key interactions between the protein and the abasic DNA. Only the central part of the bound DNA (blue) with the abasic site analogue (AP site, red) and the distorted base pair Gua7:Cyt16 (cyan) is shown. Amino acid side chains involved in DNA-bending, strand separation and widening of the DNA minor groove are shown in ball-and-stick. Lys53 occupies the empty space close to the abasic site in the DNA-ladder, splitting the two DNA strands. The side chain of Leu54 is wedged in between the C16 and G17 bases. Gln52 (partly hidden behind nucleotides G7 and A8) and Lys99 protrude into the DNA helix, forcing G7 to adopt a syn-conformation, abolishing normal Watson-Crick base pairing with C16. Leu98 contacts the DNA backbone near the ribose of Thy9, pushing the DNA toward Lys140 and Lys137 in the helix-hairpin-helix motif. Trp142 form a platform onto which the 5′ phosphate of G7 can rest. Finally, Ser163 form a hydrogen bond to the 5′ phosphate of the AP site. See also Figure S3.
Release of the damaged base by the DNA glycosylases is normally dependent on the presence of a general base (aspartate or glutamate) promoting the hydrolytic cleavage of the N-glycosylic bond by activating a water molecule for nucleophilic attack or charge-stabilization of a reaction intermediate (Hollis et al., 2000b; Norman et al., 2001; Norman et al., 2003; Sartori et al., 2002). The spatial location of Asp162 in Mag2 matches with the corresponding catalytic residues in enzymatically active DNA glycosylases. Also present in Mag2, interacting with the aspartate, are two tryptophan residues (Trp142 and Trp203), whose AlkA counterparts participate in stabilization of the protein-DNA complex and π-cation stacking with the damaged, extrahelical bases carrying a positive charge due to alkylation (Hollis et al., 2000a). Together, the Asp and Trp residues form a conserved triad in both AlkA and Mag2 (Figures 4G and 4H). A similar triad was recently discovered in the active S. pombe Mag1 homologue (Adhikary and Eichman, 2011). Strikingly, despite the similarity in the active site configuration with enzymatically functional DNA glycosylases, Mag2 has no activity for removal of alkylated bases in DNA, suggesting that the Mag2 protein is not able to flip alkylated bases out of the DNA duplex and stabilize the extrahelical base by aromatic stacking.
Indeed, the co-crystallized double-stranded DNA 11-mer is bound to Mag2 in an unflipped configuration (Figures 3A, 4G and 5), contrasting the universal feature of the DNA in the complexes of both HhH DNA glycosylases (Adhikary and Eichman, 2011; Bruner et al., 2000; Eichman et al., 2003; Fromme and Verdine, 2003; Fromme et al., 2004; Hollis et al., 2000a) and the AAG (Lau et al., 2000), Nei/NEIL/Fpg (Fromme and Verdine, 2002; Gilboa et al., 2002; Imamura et al., 2009; Zharkov et al., 2002) and UNG (Slupphaug et al., 1996) structural families of DNA glycosylases, as well as AP endonuclease (Mol et al., 2000b). In all these complexes, flipping of the damaged nucleotide or abasic deoxyribose into a base lesion/AP site binding pocket is a common signature and integral part of catalysis (Dalhus et al., 2009; Huffman et al., 2005). This flipping is achieved by way of a bending and widening of the minor groove of the substrate DNA, combined with an intercalating amino acid side chain, replacing the flipped base in the DNA double helix to prevent rapid back-flipping. In the complex of Mag2 bound to abasic DNA, the AP site analogue is strikingly not flipped, making Mag2 the first known HhH protein lacking the ability to flip bases/AP sites (Figures 3A, 4G and 5).
In addition to an intercalating residue preventing back-flipping in AlkA, the bulky phenolic ring of Tyr239 pushes on the 5′ phosphate of the extrahelical abasic site, aiding the flipping of the modified base into the binding pocket by a switch-like motion. This notion is suggested by comparing the unflipped DNA from the present structure of Mag2 with the structure of AlkA (Figures 4G and 4H). A similar tyrosine-mediated phosphate rotation was first realized from the structure of human AGT (Daniels et al., 2004) and is believed to be a widespread, yet not indispensable principle. In Mag2, the much smaller side-chain of Ser163 instead forms a hydrogen bond to the 5′ phosphate without steric obstruction of the abasic ribose in an unflipped conformation (Figures 4G and 5). In S. pombe Mag1 alkylpurine DNA glycosylase, the base flipping is assisted by Leu 171 (Adhikary and Eichman, 2011).
The bend in the DNA in various DNA glycosylases typically varies between ~20° for human AAG in complex with ethenoadenine (Lau et al., 2000) all the way to ~65° for AlkA in complex with abasic DNA (Hollis et al., 2000b) and ~70° for OGG1 in complex with the 8-oxoG lesion (Bruner et al., 2000). In the abasic DNA complex of the downstream AP endonuclease (Mol et al., 2000b), the bending angle takes on an intermediate value of ~35°. Mag2 severely distorts the DNA with a bending angle beyond the typical range for DNA glycosylases (Figures 3A and S2). Further, Mag2 introduce a large widening of the minor groove, far exceeding that of active DNA glycosylases and AP endonucleases (Figure 6). The large distortion of the abasic DNA by Mag2 is caused by the insertion of two loops in the minor groove, both of which contain Leu and Lys residues that seem to play major roles in shaping of the DNA. The first loop includes residues Gln52, Lys53 and Leu54 between helix α3 and α4 (Figures 3C and 5B). Lys53 intercalates DNA in the vacant base position at the AP site and separates the two DNA strand (Figures 3B and 5). The amino group of Lys53 forms a hydrogen bond to the estranged Gua17, while Leu54 intercalates between Cyt16 and Gua17, pushing on the adjacent faces of these two bases (Figure 5B). The role of Lys53 in AP site recognition is underscored by SPR analysis of the K53G mutant, which has just about lost its ability to bind abasic DNA (Figure 2). Steric limitations around Gln52 probably further cause Gua7 to flip to the less preferred syn-conformation, eliminating normal Watson-Crick base pairing with the partner Cyt16 (Figure 5).
Figure 6. Comparison of Minor Groove Widening in Selected Protein-DNA Complexes.
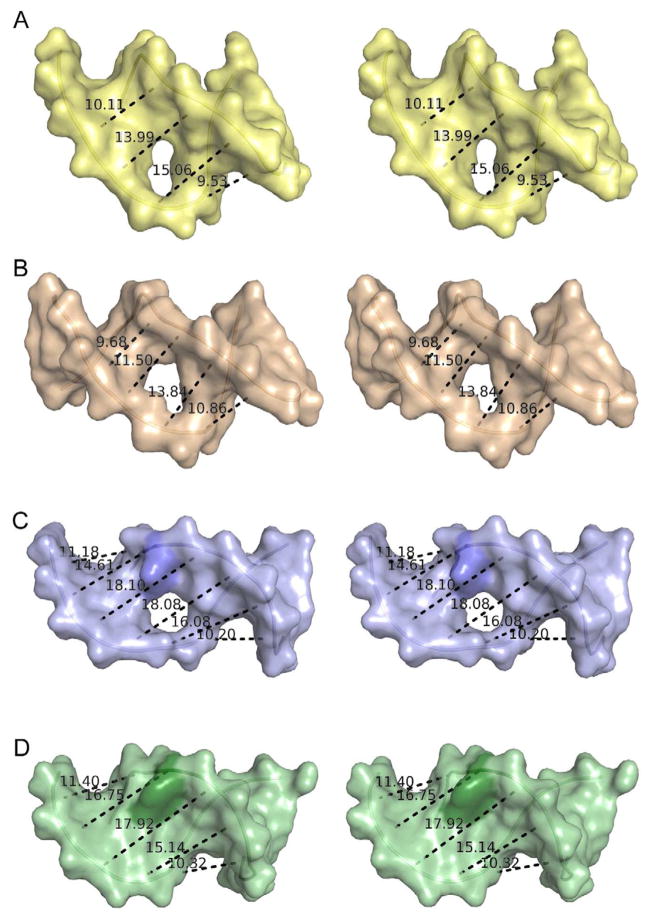
(A) Stereo view of DNA in the structure of E. coli 3mA DNA glycosylase AlkA (Hollis et al., 2000a). (B) Stereo view of DNA in the structure of human AP endonuclease APE1 (Mol et al., 2000b). (C) Stereo view of DNA in the structure of S. pombe Mag2. (D) Stereo view of DNA in the structure of E. Coli MutS bound to DNA harbouring an A:A mismatch (Natrajan et al., 2003). The displayed distances correspond to the C4′-C4′ distance between nucleotide n and (n + 2). The protruding Cyt16 in the DNA bound to Mag2 and the corresponding Ade in the MutS-DNA complex are coloured in a darker shade. The holes in the middle of the DNA show the position of the abasic sites. The DNA from the AlkA and MutS complexes has been reduced to 11 base pairs for the sake of clarity. See also Figures S2 and S3.
The loop between helix α6 and α7, comprising residues Ser96, Leu98 and Lys99, form the second minor groove wedging loop (Figures 3C and 5B). Comparison with structures of various DNA glycosylases reveals that the corresponding loops in the active enzymes are more extensive, and will make severe steric clashes with an unflipped DNA ligand with a conformation as found in the structure of Mag2 (Figure 7). In the Mag2 structure, the hydroxyl group of Ser96 caps the α7 helix and coordinates to the sugar-phosphate backbone around Cyt16. Leu98 is engaged in steric interactions with the DNA backbone of the abasic strand close to Thy9. Lys99 projects in-between Gua7 and Cyt16, forming a hydrogen bond to N3 in Cyt16, further preventing normal base paring between Gua7 and Cyt16 next to the AP site (Figure 5B). In addition, there is no helical base-stacking between Cyt16 and Thy15 (Figure S3A). In sharp contrast to the large distortion of the Gua7-Cyt16 base pair in the present structure of Mag2 (Figures 5B and S3A), regular base-pairing and π–π stacking is observed adjacent to the DNA lesion in complexes of DNA glycosylases and AP endonucleases. Thus, the two minor-groove wedging loops in Mag2 (Figures 3C and 5B) seem to play key roles in the formation and maintenance of an irregular and distorted base pair next to the lesion in a surprisingly wide DNA minor groove. This steric remodeling of the DNA adds to our knowledge of how proteins may rapidly and specifically recognize AP sites with the surprising observation that increased DNA bending and groove widening is consistent with rapid recognition.
Figure 7. Close-up View of the Wedging Loop Between Helix α6 and α7 in Mag2 and Corresponding Loops in Selected 3mA Glycosylases, AP Endonuclease and MutY, Showing Steric Conflicts With the Mag2 DNA Conformation.
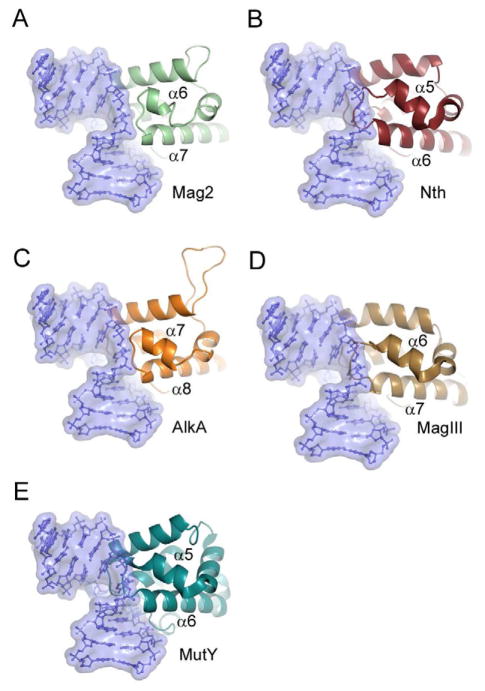
(A) S. pombe Mag2, (B) E. coli endonuclease III (Nth) (Thayer et al., 1995), (C) E. coli 3mA DNA glycosylase II (AlkA) (Hollis et al., 2000a), (D) H. pylori 3mA DNA glycosylase MagIII (Eichman et al., 2003) and (E) B. stearothermophilus adenine DNA glycosylase MutY (Fromme et al., 2004). DNA from Mag2 is shown in blue ball-and-stick representation within a semi-transparent surface boundary in all five panels. The different proteins have been superimposed onto the Cα-trace of Mag2, with an r.m.s.d of less than 3.3 Å for a selection of 53 – 103 atoms in a core domain comprising residues 40 – 170 in Mag2.
To test the sequence-dependence of the formation of an irregular base pair next to the AP site, structures of Mag2 in complex with DNA harboring the other Watson-Crick base combinations in position 7 and 16 were examined. All the Mag2-DNA complexes share the large distortion and widening of the DNA. In particular, an unpaired, minor-groove protruding base is found in both structures with pyrimidine in position 16, and there is no base stacking between residue 15 and 16 in the non-damaged strand (Figure S3). The Thy7-Ade16 and the Cyt7-Gua16 base pairs, on the other hand, form normal Watson-Crick base pairs, but the bases are inclined at ~40° and without proper π–π stacking between Ade/Gua16 and Thy15. In conclusion, a distorted base pair is formed for all possible base pair combinations next to the AP site.
Mag2 Stimulates AP Site Binding to Mismatch Repair Protein MutS
Interestingly, the DNA conformation in the Mag2 complex resembles that found in the crystal structures of the prokaryotic MMR protein MutS and the corresponding eukaryotic MSH2/MSH3 heterodimer (MutSβ) bound to various mismatches, both in the context of repair (Lamers et al., 2000; Natrajan et al., 2003; Obmolova et al., 2000) (Figure 6) and cell death response (Salsbury, Jr. et al., 2006). An efficient, secure and specific handover of partially processed DNA intermediates from one enzyme to the next could in part rely on the ability of one protein to shape and maintain a DNA conformation that will match the next enzyme in the pathway with only minor adjustments in the product structure (Mol et al., 2000b; Wilson and Kunkel, 2000). In line with this concept of DNA shaping as a structural determinant in efficient product handover, we performed a DNA binding assay to investigate if Mag2 could stimulate binding of MutSβ to AP sites in DNA by a putative product handover mechanism based on the similarity in DNA shape (Figures 6C and 6D). EMSA of Mag2 alone showed no binding neither to abasic DNA nor undamaged DNA (Figure 8B, lanes 5 and 11), however our SPR data (Figure 2) showed that Mag2 forms a transient complex with abasic DNA. Next, human MutSβ was incubated with abasic DNA or undamaged DNA without or in presence of Mag2 (Figure 8). Formation of a MutSβ-AP DNA complex was monitored in presence of increasing concentrations Mag2. These data showed 5 and 15 fold stimulation in MutSβ binding to abasic DNA in presence of 400 nM and 1600 nM of Mag2, respectively (Figure 8B), supporting our hypothesis that Mag2 can shape abasic DNA for efficient handover to MutSβ. MutSα was also tested in EMSAs, but had no affinity to AP sites, nor did Mag2 have any stimulating effect (data not shown). Further, EMSA of MutSβ and abasic DNA in presence of increasing concentrations of human NTH1, which is homologous to Mag2 but flips AP sites like any other HhH DNA glycosylase, showed no increase in MutSβ binding to abasic DNA (data not shown), underscoring the role of the DNA conformation as the governing factor for product handover. Notably, the K53G mutant of Mag2, found to be deficient in AP site recognition (Figure 2), was severely compromised in stimulating MutSβ to bind to abasic DNA (Figure S4), consistent with the hypothesis of DNA shaping as a determinant factor in product handover.
Figure 8. Electrophoretic Mobility Shift Assay With Mag2 and Human MutSβ.
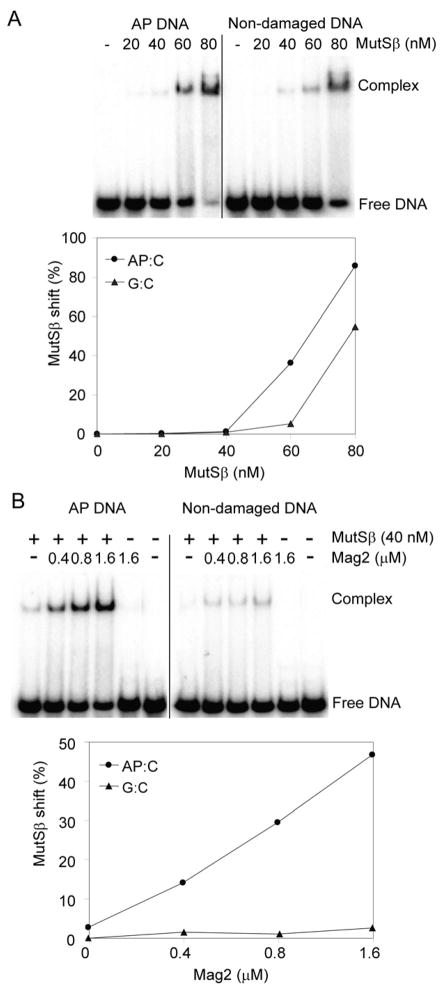
(A) Duplex DNA containing a single AP site or non-damaged DNA (5 nM) was incubated with increasing concentrations of MutSβ (20–80 nM) in a buffer containing 20 mM HEPES, pH 7.5, 2 mM MgCl2, 100 mM NaCl, 5% glycerol, 0.1 mg/ml BSA and 1 mM DTT. Formation of a MutSβ-DNA complex was observed, both for AP DNA and undamaged DNA. (B) Specific binding of MutSβ to AP DNA was stimulated >10 fold by increasing the concentration of Mag2 (0.4–1.6 μM). The amounts of free DNA and DNA in complex with MutSβ were analysed on 10% polyacrylamide gels, visualised using PhosphoImager and quantified using ImageQuant. These data represent a typical result of three independent experiments. See also Figure S4.
DISCUSSION
Enzymes counteracting alkylation damage to the DNA are widespread and ubiquitous in nature and most organisms have several repair mechanisms to cope with such threats. For the removal of alkylated bases, the E. coli genome encodes two 3-methyl adenine DNA glycosylases, Tag and AlkA. Mag1 is the AlkA homologue in S. cerevisiae, and in humans this function is replaced with the AAG DNA glycosylase (Hollis et al., 2000b; Samson et al., 1991). Both Mag1 and AAG are the sole alkylbase DNA glycosylases in these organisms. Common for E. coli, yeast and mice embryonic fibroblasts, is that cells lacking the 3mA DNA glycosylases are extremely sensitive to MMS (Chen et al., 1990; Engelward et al., 1997; Karran et al., 1980). In contrast, the S. pombe mag1− mutant is only moderate sensitive to the lethal effect of MMS (Memisoglu and Samson, 2000; Alseth et al., 2005) suggesting that backup enzymes exist. Indeed, the S. pombe genome encodes a second open reading frame (mag2) with homology to Mag1. Here we show that Mag2 is a damage inducible nuclear protein, but unexpectedly, a mag2 disruption mutant displays no MMS sensitivity. Moreover, deletion of mag2 in the mag1− background does not increase the MMS sensitivity of the mag1− single mutant cells. Further, recombinant Mag2 appears to be devoid of alkylbase DNA glycosylase activity despite strong similarities in the active site structure with functional DNA glycosylases like AlkA (Hollis et al., 2000a) and Mag1 (Adhikary and Eichman, 2011). Mag2 expression is also induced by exposure to H2O2. Notably, the AP site is a common product for partially processed MMS- and H2O2-induced DNA damages. Mag2 forms specific, yet transient, complexes with abasic DNA. Nonetheless, epistasis analysis of mag2− double mutants shows that Mag2 is not participating in processing of abasic sites via any of the BER, NER or RR pathways. Mag2 thus provides a novel example of non-enzymatic AP site recognition suitable for protecting labile AP sites while allowing efficient handoffs rather than stable binding associated with repair. In humans, PARP-1 evidently binds and protects AP sites, which are in part toxic as they block transcription (Cistulli et al., 2004).
In order to shed light on the role of the DNA damage inducible Mag2 in abasic DNA recognition, we solved the crystal structure of Mag2 in complex with DNA containing the tetrahydrofurane AP site analogue. The structure reveals a novel mechanism for AP site recognition and DNA sculpting (Figures 3, 5 and 6). Mag2 shapes the abasic DNA distinctly different from the DNA in all known structures of HhH DNA glycosylases and AP endonucleases in complex with DNA (Figure 6). Mag2 uses various structural elements, including residues in several protein loops wedged into the minor groove (Figure 5B), to shape the bound DNA to comprise an unflipped abasic site, a highly distorted base pair next to the abasic site and a wide minor groove (Figure 6). The Mag2 structure also reveals novel functional features of the HhH motif in which the highly conserved glycine in the DNA glycosylases is replaced by the Lys140 residue, which forms a ridge on the protein surface (Figure 3B). Together with residues Leu98 and Lys137, residue Lys140 forms a distinct cleft directing the DNA backbone 3′ of the AP site and contributes to a particularly wide DNA minor groove (Figures 5 and 6). The minor groove width increases from ~10 Å in undistorted B-DNA to a maximum of ~15 Å when bound to a DNA glycosylase or AP endonuclease (Figures 6A and 6B). However, the widening extends only over a few base pairs. In Mag2, on the other hand, the minor groove is substantially wider, with a maximum of about ~18 Å, spanning approximately four base pairs (Figure 6C). Comparison of mismatch-containing DNA in the DNA complexes of the MMR protein MutS (Lamers et al., 2000; Natrajan et al., 2003; Obmolova et al., 2000) with abasic DNA bound to Mag2, reveal a remarkable similarity in sugar-phosphate conformation and overall shape (Figures 6C and 6D). This discovery of Mag2’s transient-specific interaction with AP sites provides a framework to compare structural mechanisms promoting stable versus transient-specific binding to DNA lesions to protect and control possible outcomes rather than allowing naked lesions to be processed or decomposed randomly.
The structures of the MutS/MutSβ dimer bound to DNA harbouring various base mismatches or single nucleotide inserts have been thoroughly investigated in the context of DNA repair (Lamers et al., 2000; Natrajan et al., 2003; Obmolova et al., 2000) and in relation to cell death response (Salsbury, Jr. et al., 2006). The present comparative analysis of the conformation of DNA in complex with Mag2, MutS, AlkA and APE1 reveal that the DNA structure in the Mag2 complex closely resembles that found in all known complexes between the MMR protein MutS/MutSβ and DNA with various base mismatches, be it either simple base-base mismatches (Lamers et al., 2000; Obmolova et al., 2000) or single base insertions (Natrajan et al., 2003; Obmolova et al., 2000; Salsbury, Jr. et al., 2006). It has been suggested that the similarity in DNA shape between DNA glycosylases, AP endonucleases and downstream DNA polymerase β is the molecular basis for an efficient handover of repair intermediates in the BER pathway (Mol et al., 2000b; Wilson and Kunkel, 2000). The similarity in DNA shape between abasic DNA bound to Mag2 and MMR protein MutS/MutSβ is underscored by the fact that Mag2 is able to stimulate abasic DNA binding by MutS/MutSβ (Figure 8). Another striking example of the role of DNA conformation/shape in pathway selection, is the binding of alkylated DNA by alkyltransferase-like proteins (ATLs) to create an intersection between base damage processing and nucleotide excision repair (Tubbs et al., 2009; Tubbs and Tainer, 2010). Here, ATL binds to the weakly distorted O6-alkylguanine lesion and presents the damage to the NER pathway by non-enzymatic nucleotide flipping.
The present Mag2 structure in complex with DNA reveals a novel non-enzymatic and transient DNA sculpting of abasic DNA that is strikingly different from the canonical DNA glycosylases and AP endonucleases with a non-flipped AP site (Figures 3A and 4G) and a particularly wide minor groove (Figure 6). A third configuration of abasic DNA binding was recently reported for the HEAT-like repeat family of DNA glycosylases (Rubinson et al., 2010). In the structure of AlkD in complex with abasic DNA, the DNA is distorted in an extraordinarily different way than in any other DNA glycosylase complex. The DNA helix is linear, and both the AP site and the opposing nucleotide are flipped out of the DNA helix, which is partially collapsed to preserve base-stacking across the extrahelical base pair.
A distinct but somewhat analogous damage detection system whereby XPB may bind damaged DNA bases based upon a structural similarity to the MutS damage-recognition domain was recently proposed (Fan et al., 2006), but no XPB-DNA structure is yet available for comparison to the MMR-like complex shown here experimentally for Mag2. The current results implicating an AP site protection protein thus build upon and expand our understanding of base repair pathways as well as the structural basis for pathway selection and the coordination of distinct processes that respond to DNA lesions. Such protection processes have recently been identified as important for stalled replication forks (Schlacher et al., 2011) but no structures for fork protection complexes are yet available. Together these results on replication fork protection and those presented here with structures revealing AP site binding without processing thus have potentially broad implications for the fundamental understanding of genetic integrity in biology and its breakdown in many cancer prone disorders. For lesions such as AP sites, that can be generated in relatively large numbers in cells, proteins that rapidly bind and protect the lesion while allowing efficient exchange with appropriate response partners may provide advantages over a specific repair pathway. Thus, we propose Mag2 may represent a new class of versatile lesion protection proteins that bind lesions selectively with rapid kinetics but protect rather than processes them until suitable multi-protein damage responses can be coordinated.
EXPERIMENTAL PROCEDURES
Crystal data, data collection and refinement statistics are summarized in Table 1. Information on cell culture conditions, strains, plasmids, cloning and disruption of mag2, epistasis analysis, functional complementation, survival assays, northern blot analysis, fluorescence microscopy, site-specific mutagenesis, enzymatic assays, SPR, protein expression and purification, DNA binding assays, crystallization, data collection and processing, phasing and model refinement is available in the Supplemental Experimental Procedures.
Table 1.
Summary of X-ray Data Collection and refinement Statistics for Mag2-DNA Complexes
| Crystal | Gua7:Cyt16 | 5-BrU derivative | Cyt7:Gua16 | Ade7:Thy16 | Thy6:Ade16 | ||
|---|---|---|---|---|---|---|---|
| PDB code | 4b21 | 4b22 | 4b23 | 4b24 | |||
| Data collection | |||||||
| Space group | I222 | I222 | I222 | I222 | I222 | ||
| Cell dimensions (Å) | a = 68.93 | a = 67.66 | a = 68.48 | a = 68.88 | a = 68.65 | ||
| b = 84.75 | b = 84.12 | b = 84.52 | b = 84.60 | b = 84.65 | |||
| c = 126.71 | c = 125.41 | c = 125.75 | c = 125.58 | c = 125.96 | |||
| Peak | Inflection | Remote | |||||
| Wavelength (Å) | 1.11 | 0.9203 | 0.9205 | 0.9558 | 0.9200 | 1.5418 | 1.5418 |
| Resolution (Å) | 50 - 1.45 | 50 – 2.8 | 50 – 2.8 | 50 – 2.8 | 50 – 1.9 | 50 – 2.0 | 50.0 – 2.3 |
| Wilson B-factor | 28.2 | 22.7 | 29.8 | 27.3 | |||
| Rsyma | 0.075 (0.44) | 0.047 (0.10) | 0.047 (0.10) | 0.042 (0.076) | 0.061 (0.315) | 0.089 (0.21) | 0.13 (0.36) |
| I/σI | 15.2 (4.3) | 23.6 (11.2) | 23.9 (11.4) | 27.6 (14.9) | 16.2 (4.3) | 8.4 (2.7) | 4.9 (1.8) |
| Completeness (%) | 100 (100) | 99.5 (100) | 99.5 (100) | 99.5 (100) | 99.3 (96.0) | 94.4 (93.9) | 99.5 (99.3) |
| Redundancy | 6.9 (4.6) | 4.3 (4.3) | 4.3 (4.3) | 4.3 (4.3) | 4.3 (3.8) | 3.7 (3.7) | 3.4 (3.4) |
| Figure of merit, Resolve | 0.68 | ||||||
| Refinement | |||||||
| Resolution (Å) | 50 – 1.45 | 50 – 1.9 | 50 – 2.0 | 50 – 2.3 | |||
| No. reflections | 59374 | 26375 | 21185 | 15169 | |||
| Rwork/Rfreeb | 0.21/0.22 | 0.20/0.23 | 0.25/0.29 | 0.23/0.27 | |||
| No. atoms | |||||||
| Protein | 1688 | 1637 | 1646 | 1646 | |||
| DNA | 418 | 437 | 437 | 418 | |||
| Water | 255 | 245 | 208 | 146 | |||
| Phosphate | 15 | - | - | - | |||
| Glucose | 12 | - | - | - | |||
| B-factors | |||||||
| Protein | 23.2 | 21.5 | 27.2 | 30.6 | |||
| DNA | 30.1 | 31.7 | 35.1 | 35.1 | |||
| Water | 40.6 | 33.0 | 36.4 | 37.0 | |||
| R.m.s. deviation from ideal geometry | |||||||
| bond lengths (Å) | 0.004 | 0.005 | 0.005 | 0.008 | |||
| bond angles (°) | 1.3 | 1.1 | 1.1 | 1.3 | |||
| Ramachandran plot (%) | |||||||
| Best regions | 96.3 | 96.6 | 96.1 | 93.6 | |||
| Disallowed | 0.0 | 0.5 | 0.5 | 0.5 | |||
Values in parentheses refer to the highest resolution shells. These are 1.50 - 1.45, 2.89 – 2.8, 2.0 – 1.9, 2.07 – 2.0, and 2.38 – 2.3 Å, respectively.
4.8, 4.6, 4.3 and 4.1%, respectively, of randomly distributed data over the full resolution range, were flagged as belonging to the Rfree cross validation set not to be used in the refinement.
Supplementary Material
Highlights.
Unflipped AP Site Stabilized by Helix-hairpin-helix DNA Glycosylase Homolog Mag2
Non-Enzymatic AP Site Recognition and DNA Sculpting
Sculpting of DNA Conformation Enhances Substrate Transfer Between Proteins
Acknowledgments
We would like to thank Fekret Osman (Department of Biochemistry, University of Oxford, UK) for providing all the FO strains and Oliver Fleck (School of Biological Sciences, University of Wales, UK) for the MMR strains (RU 39, RU106, OL712 and MAB 032). We thank the technical support at beam lines 8.3.1/ALS (Berkeley) and ID14-4/ESRF (Grenoble). Figure panels prepared with PyMOL (www.pymol.org).
FUNDING
The work in the Bjørås laboratory was supported by the Research Council of Norway and the Norwegian Cancer Society. DNA base repair research in the Tainer laboratory is supported by the National Institutes of Health (grant GM46312). CTM and JAT were also supported by The National Cancer Institute, Program for Structural Biology of DNA Repair (SBDR). BD, PHB and MB were supported by the South-Eastern Norway Regional Health Authority (grants no. 2009100 and 2011040) for establishing the Regional Core Facility for Structural Biology and Bioinformatics.
Footnotes
ACCESSION NUMBERS
Atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 4b21.pdb (Mag2 G:C), 4b22.pdb (Mag2 C:G), 4b23.pdb (Mag2 A:T) and 4b24.pdb (Mag2 T:A).
Supplemental Information includes six figures, two tables, Supplemental Experimental Procedures and Supplemental References and can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Adhikary S, Eichman BF. Analysis of substrate specificity of Schizosaccharomyces pombe Mag1 alkylpurine DNA glycosylase. EMBO Rep. 2011;12:1286–1292. doi: 10.1038/embor.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alseth I, Osman F, Korvald H, Tsaneva I, Whitby MC, Seeberg E, Bjoras M. Biochemical characterization and DNA repair pathway interactions of Mag1-mediated base excision repair in Schizosaccharomyces pombe. Nucleic Acids Res. 2005;33:1123–1131. doi: 10.1093/nar/gki259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berti PJ, McCann JA. Toward a detailed understanding of base excision repair enzymes: transition state and mechanistic analyses of N-glycoside hydrolysis and N-glycoside transfer. Chem Rev. 2006;106:506–555. doi: 10.1021/cr040461t. [DOI] [PubMed] [Google Scholar]
- Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amst) 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- Caldecott KW, McKeown CK, Tucker JD, Ljungquist S, Thompson LH. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol Cell Biol. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr AM, Schmidt H, Kirchhoff S, Muriel WJ, Sheldrick KS, Griffiths DJ, Basmacioglu CN, Subramani S, Clegg M, Nasim A. The rad16 gene of Schizosaccharomyces pombe: a homolog of the RAD1 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:2029–2040. doi: 10.1128/mcb.14.3.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Derfler B, Samson L. Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. EMBO J. 1990;9:4569–4575. doi: 10.1002/j.1460-2075.1990.tb07910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cistulli C, Lavrik OI, Prasad R, Hou E, Wilson SH. AP endonuclease and poly(ADP-ribose) polymerase-1 interact with the same base excision repair intermediate. DNA Repair (Amst) 2004;3:581–591. doi: 10.1016/j.dnarep.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Crane BR, Arvai AS, Gachhui R, Wu C, Ghosh DK, Getzoff ED, Stuehr DJ, Tainer JA. The structure of nitric oxide synthase oxygenase domain and inhibitor complexes. Science. 1997;278:425–431. doi: 10.1126/science.278.5337.425. [DOI] [PubMed] [Google Scholar]
- Dalhus B, Laerdahl JK, Backe PH, Bjoras M. DNA base repair--recognition and initiation of catalysis. FEMS Microbiol Rev. 2009;33:1044–1078. doi: 10.1111/j.1574-6976.2009.00188.x. [DOI] [PubMed] [Google Scholar]
- Daniels DS, Woo TT, Luu KX, Noll DM, Clarke ND, Pegg AE, Tainer JA. DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat Struct Mol Biol. 2004;11:714–720. doi: 10.1038/nsmb791. [DOI] [PubMed] [Google Scholar]
- David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianov GL, Sleeth KM, Dianova II, Allinson SL. Repair of abasic sites in DNA. Mutat Res. 2003;531:157–163. doi: 10.1016/j.mrfmmm.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Doublie S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc Natl Acad Sci U S A. 2004;101:10284–10289. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichman BF, O’Rourke EJ, Radicella JP, Ellenberger T. Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases. EMBO J. 2003;22:4898–4909. doi: 10.1093/emboj/cdg505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelward BP, Weeda G, Wyatt MD, Broekhof JL, de WJ, Donker I, Allan JM, Gold B, Hoeijmakers JH, Samson LD. Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc Natl Acad Sci U S A. 1997;94:13087–13092. doi: 10.1073/pnas.94.24.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Arvai AS, Cooper PK, Iwai S, Hanaoka F, Tainer JA. Conserved XPB core structure and motifs for DNA unwinding: implications for pathway selection of transcription or excision repair. Mol Cell. 2006;22:27–37. doi: 10.1016/j.molcel.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Fromme JC, Banerjee A, Huang SJ, Verdine GL. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature. 2004;427:652–656. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- Fromme JC, Verdine GL. Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nat Struct Biol. 2002;9:544–552. doi: 10.1038/nsb809. [DOI] [PubMed] [Google Scholar]
- Fromme JC, Verdine GL. Structure of a trapped endonuclease III-DNA covalent intermediate. EMBO J. 2003;22:3461–3471. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. J Biol Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer. 2012;12:104–120. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa R, Zharkov DO, Golan G, Fernandes AS, Gerchman SE, Matz E, Kycia JH, Grollman AP, Shoham G. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J Biol Chem. 2002;277:19811–19816. doi: 10.1074/jbc.M202058200. [DOI] [PubMed] [Google Scholar]
- Glassner BJ, Rasmussen LJ, Najarian MT, Posnick LM, Samson LD. Generation of a strong mutator phenotype in yeast by imbalanced base excision repair. Proc Natl Acad Sci U S A. 1998;95:9997–10002. doi: 10.1073/pnas.95.17.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Manuel RC, Arvai AS, Parikh SS, Mol CD, Miller JH, Lloyd S, Tainer JA. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat Struct Biol. 1998;5:1058–1064. doi: 10.1038/4168. [DOI] [PubMed] [Google Scholar]
- Guillet M, Boiteux S. Endogenous DNA abasic sites cause cell death in the absence of Apn1, Apn2 and Rad1/Rad10 in Saccharomyces cerevisiae. EMBO J. 2002;21:2833–2841. doi: 10.1093/emboj/21.11.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht SS. DNA adduct formation from tobacco-specific N-nitrosamines. Mutat Res. 1999;424:127–142. doi: 10.1016/s0027-5107(99)00014-7. [DOI] [PubMed] [Google Scholar]
- Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair. DNA Repair (Amst) 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Hollis T, Ichikawa Y, Ellenberger T. DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J. 2000a;19:758–766. doi: 10.1093/emboj/19.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis T, Lau A, Ellenberger T. Structural studies of human alkyladenine glycosylase and E. coli 3-methyladenine glycosylase. Mutat Res. 2000b;460:201–210. doi: 10.1016/s0921-8777(00)00027-6. [DOI] [PubMed] [Google Scholar]
- Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Hurley LH. DNA and its associated processes as targets for cancer therapy. Nat Rev Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- Imamura K, Wallace SS, Doublie S. Structural characterization of a viral NEIL1 ortholog unliganded and bound to abasic site-containing DNA. J Biol Chem. 2009;284:26174–26183. doi: 10.1074/jbc.M109.021907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamitsu K, Tanihigashi H, Tanita Y, Inatani S, Ikeda S. Involvement of 3-methyladenine DNA glycosylases Mag1p and Mag2p in base excision repair of methyl methanesulfonate-damaged DNA in the fission yeast Schizosaccharomyces pombe. Genes Genet Syst. 2007;82:489–494. doi: 10.1266/ggs.82.489. [DOI] [PubMed] [Google Scholar]
- Karran P, Lindahl T, Ofsteng I, Evensen GB, Seeberg E. Escherichia coli mutants deficient in 3-methyladenine-DNA glycosylase. J Mol Biol. 1980;140:101–127. doi: 10.1016/0022-2836(80)90358-7. [DOI] [PubMed] [Google Scholar]
- Khasanov FK, Savchenko GV, Bashkirova EV, Korolev VG, Heyer WD, Bashkirov VI. A new recombinational DNA repair gene from Schizosaccharomyces pombe with homology to Escherichia coli RecA. Genetics. 1999;152:1557–1572. doi: 10.1093/genetics/152.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Biade S, Matsumoto Y. Involvement of flap endonuclease 1 in base excision DNA repair. J Biol Chem. 1998;273:8842–8848. doi: 10.1074/jbc.273.15.8842. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de WN, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T. Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase. AAG Proc Natl Acad Sci U S A. 2000;97:13573–13578. doi: 10.1073/pnas.97.25.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Loeb LA. Apurinic sites as mutagenic intermediates. Cell. 1985;40:483–484. doi: 10.1016/0092-8674(85)90191-6. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Kim K, Bogenhagen DF. Proliferating cell nuclear antigen-dependent abasic site repair in Xenopus laevis oocytes: an alternative pathway of base excision DNA repair. Mol Cell Biol. 1994;14:6187–6197. doi: 10.1128/mcb.14.9.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memisoglu A, Samson L. Contribution of base excision repair, nucleotide excision repair, and DNA recombination to alkylation resistance of the fission yeast Schizosaccharomyces pombe. J Bacteriol. 2000;182:2104–2112. doi: 10.1128/jb.182.8.2104-2112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol CD, Hosfield DJ, Tainer JA. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3′ ends justify the means. Mutat Res. 2000a;460:211–229. doi: 10.1016/s0921-8777(00)00028-8. [DOI] [PubMed] [Google Scholar]
- Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature. 2000b;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- Natrajan G, Lamers MH, Enzlin JH, Winterwerp HH, Perrakis A, Sixma TK. Structures of Escherichia coli DNA mismatch repair enzyme MutS in complex with different mismatches: a common recognition mode for diverse substrates. Nucleic Acids Res. 2003;31:4814–4821. doi: 10.1093/nar/gkg677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- Norman DP, Bruner SD, Verdine GL. Coupling of substrate recognition and catalysis by a human base-excision DNA repair protein. J Am Chem Soc. 2001;123:359–360. doi: 10.1021/ja003144m. [DOI] [PubMed] [Google Scholar]
- Norman DP, Chung SJ, Verdine GL. Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase. Biochemistry. 2003;42:1564–1572. doi: 10.1021/bi026823d. [DOI] [PubMed] [Google Scholar]
- Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- Rajski SR, Williams RM. DNA Cross-Linking Agents as Antitumor Drugs. Chem Rev. 1998;98:2723–2796. doi: 10.1021/cr9800199. [DOI] [PubMed] [Google Scholar]
- Ribar B, Izumi T, Mitra S. The major role of human AP-endonuclease homolog Apn2 in repair of abasic sites in Schizosaccharomyces pombe. Nucleic Acids Res. 2004;32:115–126. doi: 10.1093/nar/gkh151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinson EH, Gowda AS, Spratt TE, Gold B, Eichman BF. An unprecedented nucleic acid capture mechanism for excision of DNA damage. Nature. 2010;468:406–411. doi: 10.1038/nature09428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydberg B, Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1:211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salsbury FR, Jr, Clodfelter JE, Gentry MB, Hollis T, Scarpinato KD. The molecular mechanism of DNA damage recognition by MutS homologs and its consequences for cell death response. Nucleic Acids Res. 2006;34:2173–2185. doi: 10.1093/nar/gkl238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson L, Derfler B, Boosalis M, Call K. Cloning and characterization of a 3-methyladenine DNA glycosylase cDNA from human cells whose gene maps to chromosome 16. Proc Natl Acad Sci U S A. 1991;88:9127–9131. doi: 10.1073/pnas.88.20.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Fitz-Gibbon S, Yang H, Miller JH, Jiricny J. A novel uracil-DNA glycosylase with broad substrate specificity and an unusual active site. EMBO J. 2002;21:3182–3191. doi: 10.1093/emboj/cdf309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko VS, Nevinsky GA, Zharkov DO. Mechanism of interaction between human 8-oxoguanine-DNA glycosylase and AP endonuclease. DNA Repair (Amst) 2007;6:317–328. doi: 10.1016/j.dnarep.2006.10.022. [DOI] [PubMed] [Google Scholar]
- Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- Stivers JT, Jiang YL. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- Taverna P, Sedgwick B. Generation of an endogenous DNA-methylating agent by nitrosation in Escherichia coli. J Bacteriol. 1996;178:5105–5111. doi: 10.1128/jb.178.17.5105-5111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs JL, Latypov V, Kanugula S, Butt A, Melikishvili M, Kraehenbuehl R, Fleck O, Marriott A, Watson AJ, Verbeek B, McGown G, Thorncroft M, Santibanez-Koref MF, Millington C, Arvai AS, Kroeger MD, Peterson LA, Williams DM, Fried MG, Margison GP, Pegg AE, Tainer JA. Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature. 2009;459:808–813. doi: 10.1038/nature08076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs JL, Tainer JA. Alkyltransferase-like proteins: molecular switches between DNA repair pathways. Cell Mol Life Sci. 2010;67:3749–3762. doi: 10.1007/s00018-010-0405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan P, Lindahl T, Sedgwick B. Induction of the adaptive response of Escherichia coli to alkylation damage by the environmental mutagen, methyl chloride. Mutat Res. 1993;293:249–257. doi: 10.1016/0921-8777(93)90076-s. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters TR, Gallinari P, Jiricny J, Swann PF. Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J Biol Chem. 1999;274:67–74. doi: 10.1074/jbc.274.1.67. [DOI] [PubMed] [Google Scholar]
- Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat Struct Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- Xiao W, Chow BL. Synergism between yeast nucleotide and base excision repair pathways in the protection against DNA methylation damage. Curr Genet. 1998;33:92–99. doi: 10.1007/s002940050313. [DOI] [PubMed] [Google Scholar]
- Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002;21:789–800. doi: 10.1093/emboj/21.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


