Summary
The expression of the spalt genes is regulated by the Decapentaplegic signalling pathway in the Drosophila wing. These genes participate in the patterning of the longitudinal wing veins by regulating the expression of vein-specific genes, and in the establishment of cellular affinities in the central region of the wing blade epithelium. The Spalt proteins act as transcription factors, most likely regulating gene expression by repression, but the identity of their target genes in the wing is still unknown. As a preliminary step to unravel the genetic hierarchy controlled by the Spalt proteins, we have analysed their requirements during wing development, and addressed to what extent they mediate all the functions of the Decapentaplegic pathway in this developmental system. We identify additional functions for Spalt in cell division, survival, and maintenance of epithelial integrity. Thus, Spalt activity is required to promote cell proliferation, acting in the G2/M transition of the cell cycle. The contribution of Spalt to cell division is limited to the central region of the wing blade, as they do not mediate the extra growth triggered by Decapentaplegic signalling in the peripheral regions of the wing disc. In addition, Spalt function is required to maintain cell viability in cells exposed to high levels of Decapentaplegic signalling. This aspect of Spalt function is related to the repression of JNK signalling in the spalt domain of expression. Finally, we further characterise the requirements of Spalt to maintain epithelial integrity by regulating cellular affinities between cells located in the central wing region. Our results indicate that Spalt function mediates most of the requirements identified for Decapentaplegic signalling, contributing to establish the cellular qualities that differentiate central versus peripheral territories in the wing blade.
Key words: Drosophila, Imaginal discs, Wing, Cell death, Dpp signalling, Spalt
Introduction
The Drosophila spalt complex contains two genes, spalt major (salm) and spalt related (salr), both encoding nuclear proteins which main feature is the presence of several Zn-finger domains of the C2H2 class (de Celis and Barrio, 2009). These proteins are conserved through evolution, and play important roles during animal development in a variety of processes such as cell fate specification, pattern formation and organogenesis (Sweetman and Münsterberg, 2006; de Celis and Barrio, 2009). Two of the four human orthologues of Sal are associated to the genetic diseases Townes–Brocks syndrome (Sall1) and Okihiro syndrome (Sall4) (Domingos et al., 2004; de Celis and Barrio, 2009). Sall4 is also involved in embryonic stem cells maintenance interacting with Nanog (Wu et al., 2006). The molecular characteristics of Sal proteins, and the multitude of functional requirements identified for sal genes in different organisms, suggest that these proteins act as transcriptional regulators in a context-dependent manner.
The salm and salr genes have been extensively characterised during embryonic and imaginal disc development (Kühnlein et al., 1994; Kühnlein and Schuh, 1996; de Celis and Barrio, 2000; Rusten et al., 2001; Cantera et al., 2002). Thus, sal function is required for the specification of several cell types including the oenocytes (Gould et al., 2001), the R7 and R8 photoreceptors (Mollereau et al., 2001) and sensory organ precursors of the thorax (de Celis et al., 1999), and they also participate in the formation of the tracheal system, the migration of the tracheal dorsal trunk cells (Kühnlein and Schuh, 1996), neural development (Cantera et al., 2002), Johnston organ formation (Dong et al., 2003) and wing imaginal disc development (de Celis et al., 1996). In this last process, the Decapentaplegic (Dpp) signalling pathway regulates the expression of sal genes in the wing blade region of the disc, but it is not yet clear to what extent these genes mediate the function of this pathway.
The wing imaginal disc is an epithelial tissue that grows by cell proliferation during larval development and differentiates the wing and half of the thorax during pupal development (de Celis, 2003). The growth of the epithelium is linked to the progressive specification of spatial territories with different genetic identities and cell fates. Several signalling pathways play a fundamental role in these processes, in part by regulating the expression of transcription factors. Of paramount importance for the growth and patterning of the wing disc is the Dpp signalling pathway (Affolter and Basler, 2007). The ligand of the pathway, Dpp, is expressed in a narrow stripe of anterior cells abutting the anterior–posterior compartment boundary of the disc. Dpp secreted from these cells binds and activates its receptor complex, formed by the transmembrane kinases Thickvein (Tkv) and Punt (Put), leading to the activation of the pathway in a broad stripe of anterior and posterior cells occupying most of the presumptive central region of the wing (Affolter and Basler, 2007). The domain of Dpp signalling accumulates phosphorylated Mad (PMad), a direct target of the Tkv/Put kinase (Affolter and Basler, 2007). Once phosphorylated, PMad forms a complex with Medea (Med) that enters the nucleus and regulates the expression of target genes. Mad and Med are conserved through evolution, being the orthologues of vertebrate Smad3 and Smad4 proteins, respectively (Massagué and Wotton, 2000). Several targets and additional components of the transcriptional regulation events triggered by Dpp signalling have being identified in Drosophila (Affolter and Basler, 2007), but it is not clear which of the different functions of the pathway are mediated by these known targets. These functions include the promotion of cell proliferation throughout the wing blade primordium, the patterning and differentiation of the longitudinal and transverse veins during larval and pupal development and the maintenance of epithelial integrity in the wing disc epithelium (Affolter and Basler, 2007). Because the sal genes are downstream components of the Dpp pathway in the wing blade (Lecuit et al., 1996; Nellen et al., 1996), and are also required for wing patterning (de Celis et al., 1996), it is likely that they mediate some of the requirements of Dpp during the development of the wing blade.
In this work we have dissected the functions of Sal proteins during the development of the wing blade, aiming to relate its requirements to the known functions of Dpp signalling. We find that Salm/Salr are required for the proliferation of cells located in its domain of expression, promoting the transition between G2 and Mitosis. Consequently, wing blade cells deprived of sal function accumulate in the G2 phase of the cell cycle in the developing disc. We also find that loss of salm/salr in the wing disc results in apoptosis, indicating an unsuspected pro-survival function of these genes in the central region of the wing blade. In addition to a requirement for cell proliferation and survival, salm/salr functions are also needed to maintain epithelial integrity in the wing disc and pupal wing. Thus, the boundaries between sal-expressing and non-expressing cells lead to the formation of grooves in the disc epithelium, and subsequently during pupal development to the extrusion of mutant cells. Finally, we find that loss of salm/salr and gain of salm expression can rescue some of the effects caused by excess and loss of dpp function, respectively, indicating that Sal proteins mediate in part the function of Dpp signalling during wing development.
Materials and Methods
Genetic strains
We used the Gal4 lines dpp-Gal4, en-Gal4 and salEPv-Gal4 (Cruz et al., 2009), the tub-Gal80ts line, and the UAS lines UAS-GFP (Ito et al., 1997), UAS-salm (de Celis et al., 1996), UAS-brk (Jaźwińska et al., 1999), UAS-tkvQD (Nellen et al., 1996), UAS-dppGFP (Teleman and Cohen, 2000), UAS-puc (Martín-Blanco et al., 1998), UAS-stg and UAS-dicer2 (Dietzl et al., 2007). The expression of dpp-Gal4 is restricted to the anterior–posterior compartment boundary of the wing imaginal disc. The expression of en-Gal4 is restricted to the posterior compartment of all imaginal discs, and the expression of salEPv-Gal4 occurs in the central region of wing imaginal disc between the vein L2 and intervein L4–L5. We also used the following UAS lines to express interference RNA: UAS-salm-i (ID 3029 VDRC), UAS-salr-i (ID 28386 VDRC), UAS-Mad-i (ID 12399-R2 NIG-FLY) and UAS-CycA-i (ID 32421 VDRC), and the loss of function alleles Df(2L)32FP5 (Barrio et al., 1999) and tkva12 (Burke and Basler, 1996). Unless otherwise stated, crosses were done at 25°C. Lines not described in the text can be found in FlyBase (Gelbart et al., 1997).
Induction of mitotic recombination
Marked clones were generated by FLP-mediated recombination (Xu and Rubin, 1993) by 1 hour heat shock at 37°C 48–72 h and 72–96 h after egg-laying. Clones were visualised in third instar larvae by the absence of β-gal expression or by the presence or absence of GFP expression, and in the adult wing by the cell marker forked (f). Clones were induced in larvae of the following genotypes:
hsFLP1.22/+; FRT40 tkva12/FRT40 P[armlacZ]
hsFLP1.22/+; FRT40 Df(2L)32FP5/FRT40 P[armlacZ]
y w hsFLP1.22 P[tub-Gal4] UAS-GFP/+; FRT40A P[tub-Gal80]/FRT40 UbiGFP; UAS-salm/+
y w hsFLP1.22 P[tub-Gal4] UAS-GFP/+; FRT40A P[tub-Gal80]/FRT40 tkva12; UAS-salm/+
y w hsFLP1.22 P[tub-Gal4] UAS-GFP/+; FRT40A P[tub-Gal80]/FRT40A Df(2L)32FP5; UAS-tkvQ253D/+
f36a hsFLP1.22; FRT40A Df(2L)32FP5/FRT40A ck P[f+]32F.
sal Minute+ clones were induced by X-rays as described by de Celis et al. (de Celis et al., 1996) in larvae of f36a; Df(2L)32FP5/P[f+]32F M(2)z genotype. Homozygous sal mutant cells were labelled with the cell marker forked (f). We quantified the anterior or posterior area of pairs of wings from the same fly bearing either anterior or posterior sal− Minute+ clones, and calculated the average value of the ratios between the sizes of mutant anterior compartment and control anterior compartment of the contralateral wing, and between the sizes of mutant posterior compartment and control posterior compartment of the contralateral wing.
Mitotic index
We quantified the ratio between the number of cells in mitosis in the central region of the wing blade, detected by the expression of Phospho-Histone3 (PH3), and the size of this region, and calculated a mitotic index as the average value of these ratios. The images used to make the quantifications were projections of at least 10 confocal planes including the entire height of the wing blade epithelium. We did these measures in third instar wing discs of the following genotypes: UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ and UAS-dicer2/+; salEPv-Gal4 UAS-GFP/+. The expression of GFP was used to label the central region.
Fluorescence Activated Cell Sorting (FACS)
We incubated wing imaginal discs of UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ and UASdicer2/+; salEPv-Gal4 UAS-GFP/+ (control discs) in 300 µl of trypsin solution (Trypsin–EDTA Sigma T4299) and 0.5 µl Hoescht (Hoescht33342, Trihydrochloride Trihydrate H3570, Molecular ProbesTM) at 28°C during 40 minutes. We stopped the trypsin reaction with 200 µl of 1% FCS (Fetal Bovine Serum SigmaTM 9665). The cells were suspended in 300 µl of 1% FCS and the cell cycle profiles of GFP positive and GFP negative cells was quantified with a FACS Vantage 2 (Becton DickinsonTM). The cell cycle profile of 5 independent experiments for each genotype was analysed using FloJow 7.5. We quantified for each genotype the G1/G2 ratio in the central region (GFP positive) and in the rest of the wing imaginal disc (GFP negative).
Immunocytochemistry and in situ hybridization
We used rabbit anti-Salm and rat anti-Salm (de Celis et al., 1999), rabbit anti-activated Cas3 (Cell Signalling Technology) and anti-Phosphorylated Histone 3 (PH3; Cell Signalling Technology), mouse anti-βGalactosidase (Promega), TO-PRO (Invitrogen) and Phalloidine (Sigma). From the Hybridoma bank at Iowa University we used monoclonals anti-Wg, anti-FasIII, anti-Dlg and anti-Dl. Primary antibodies were used at a 1:50 dilution, except anti-Dl (1:5). Secondary antibodies were from Jackson Immunological Laboratories (used at 1/200 dilution). Imaginal wing discs and wing pupal were dissected, fixed and stained as previously described by de Celis and Bray (de Celis and Bray, 1997). Confocal images were captured using a LSM510 confocal microscope. In situ hybridization with stg RNA probes was carried out as described by de Celis and Bray (de Celis and Bray, 1997).
Statistical analysis
We analysed at least 10 wings from females of each genotype. Wing size, imaginal disc size, cell size, cell number data and ratios between number of cells were expressed as means ± standard error of the mean (SEM) and were compared using a T-test. All data were collected and processed in Microsoft Excel (Microsoft Inc.). We consider a significant p-value lower than 0.005.
Results
Wing phenotypes resulting from modifications in salm and/or salr expression
The expression of salm and salr is regulated by Dpp signalling in the wing pouch and occupies a broad stripe of cells centred along the anterior–posterior compartment boundary (Fig. 1H,H′). This territory corresponds in the wing to a region including the vein L2 and extending to the L4/L5 intervein (Fig. 1A,J). The salEPv-Gal4 driver reproduces the domain of sal expression in the wing blade region of the imaginal disc (Cruz et al., 2009) (Fig. 1I, compare with Fig. 1H,H′), and this domain can also be visualised in pupal wings (Fig. 1J). The reduction of only salm or salr expression in the domain of salEPv-Gal4 has a modest phenotype in adult wings, which are slightly reduced in size and display ectopic vein stretches in the region between the veins L2 and L3 (Fig. 1B,C,E; UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i and UAS-dicer2/+; salEPv-Gal4 UAS-GFP/+; UAS-salr-i/+). As expected, wing discs expressing salm and salr RNAi show very much reduced levels of Salm (Fig. 1K′) and Salr proteins (not shown), respectively. The over-expression of salm in its normal domain of expression (salEPv-Gal4/+; UAS-salm/+) results in a phenotype similar to the loss of only salm or salr (Fig. 1G). This is in contrast to the consequences of ectopic and generalised expression of either salm or salr, which results in very small wings with severe defects in the pattern of veins (de Celis et al., 1996). When both salm and salr expression are reduced, the wing shows a phenotype which strength depends on the particular genetic combination used and temperature. The stronger version of this phenotype consists in wings losing most of its central region and the veins L2 and L4 (Fig. 1D,E; UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+). The corresponding discs show a domain of salEPv-Gal4 expression reduced in size (Fig. 1K), and this reduction is even stronger in pupal wings (Fig. 1L, compare with Fig. 1J). The extreme phenotype caused by a reduction in salm and salr expression is also observed in wings homozygous for a genetic deficiency that removes the salm and salr genes (Fig. 1F; 638-Gal4/+; FRT40 Df(2L)32FP5/FRT40 M(2)z; UAS-FLP/+). In this manner, the central region of the wing fails to grow in the absence of both salm and salr, but is very much insensitive to the increase in the expression of these proteins, as far as this is restricted to the normal domain of expression.
Fig. 1. Phenotype of salm and salr mutant wings and development of the Sal domain of expression in wild type and sal mutant discs and pupal wings.
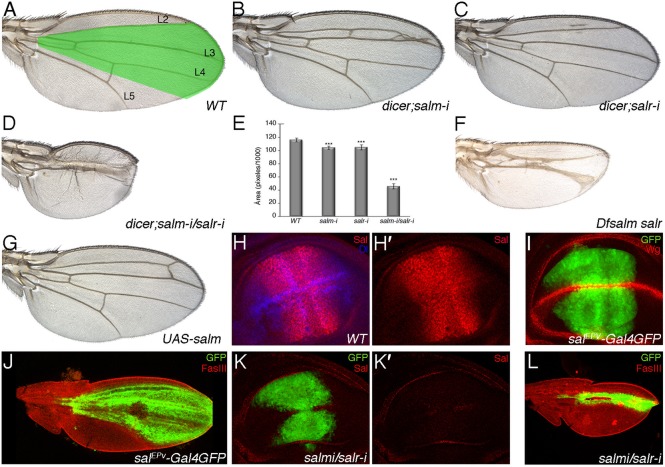
(A) Wild type wing showing the territory of salm/salr expression (green shadowing) and the longitudinal veins L2 to L5. (B,C) Phenotype of loss of salm and salr in flies of genotype UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i (salm-i in B) and UAS-dicer2/+; salEPv-Gal4 UAS-GFP/+; UAS-salr-i/+ (salr-i in C). (D) Loss of both genes (UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+) results in a much stronger phenotype of wing size reduction and in the loss of the L2 and L4 veins. (E) Quantification of wing size (measured in pixels/1000) in 10 wings of WT, UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i, UAS-dicer2/+; salEPv-Gal4 UAS-GFP/+; UAS-salr-i/+ and UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+. Bars represent mean ± SEM. ***p-value<0.005. (F) Mosaic wing of 638-Gal4/+; FRT40A Df(2L)32FP5/FRT40A M(2)z; UAS-FLP/+ genotype. In these wings most of the wing blade is homozygous for the salm and salr deficiency. (G) Wing of salEPv-Gal4; UAS-salm/+ genotype, showing that over-expression of salm in its normal domain of expression only causes small perturbations in the pattern of the L2 vein. (H) Expression of Salm (red) and Dl (blue) in a wild type third instar wing imaginal disc. The expression of only Salm is shown in H′. (I) Expression of GFP (green) and Wg (red) in a UAS-dicer2/+; salEPv-Gal4 UAS-GFP/+ third instar wing imaginal disc. (J) Expression of GFP (green) and FasIII (red) in a UAS-dicer2/+; salEPv-Gal4 UAS-GFP/+ pupal wing 36–40 hours APF. (K,K′) Expression of GFP (green) and Salm (red) in a UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ third instar wing imaginal disc. The expression of Salm is totally lost (red channel shown in K′). (L) Expression of GFP (green) and FasIII (red) in a UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ pupal wing 36–40 hours APF.
The development of the central wing blade region in absence of salm and salr function
We followed the development of the central region of the wing blade in wing discs and pupal wings of UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ genotype. The sal domain of expression in third instar discs, labelled by GFP, is smaller than normal in mutant discs, and in general the appearance of the epithelium is normal (Fig. 2A–A″, compare with Fig. 1I and Fig. 2C–C″). We also observed the presence of activated-Cas3 positive cells located in the basal part of the epithelium of the Sal domain (Fig. 2B–B″), and a variable formation of epithelial folds (Fig. 2B′,B″) that is never observed in control discs (Fig. 2C–C″). The expression of FasIII, which is mostly localised along the baso-lateral side of the epithelium in wild type discs, is very much reduced in sal mutant cells compared to wild type cells (Fig. 2D,E). The mutant salEPv-Gal4 expressing cells are also present during pupal development (24–40 h APF), although at these stages they have an abnormal cell size, being much larger than normal cells (Fig. 2F,G). We also found that the integrity of the epithelium is compromised in 24–30 h APF pupae (Fig. 2H–K), and that the contacts between the dorsal and ventral wing surfaces appear abnormal (Fig. 2H,I, Fig. 2H′,I′). We observed groups of cells that seem extruded from the epithelium (Fig. 2I′), but detected very few apoptotic cells (see below). In late pupa 36–40 h APF, the central domain of the wing blade can form abnormal blisters (Fig. 2K). In all cases we find a large number of GFP-negative cells located between the dorsal and ventral wing surfaces, which likely correspond to circulating haemocytes (Fig. 2I–I‴, Fig. 2K–K‴). These cells are normally localised in the veins (Fig. 2H,H′), but in sal mutant pupa they also appear intercalated within the epithelium (Fig. 2I–I‴, Fig. 2K–K‴). In this manner, the domain of sal expression grows to a smaller than normal size during imaginal development, and some cells are extruded from the epithelium. The extrusion of cells is more dramatic during pupal development, and after this stage only a small fraction of the central domain remains in the adult wing.
Fig. 2. Cellular effects of loss of salm and salr expression.
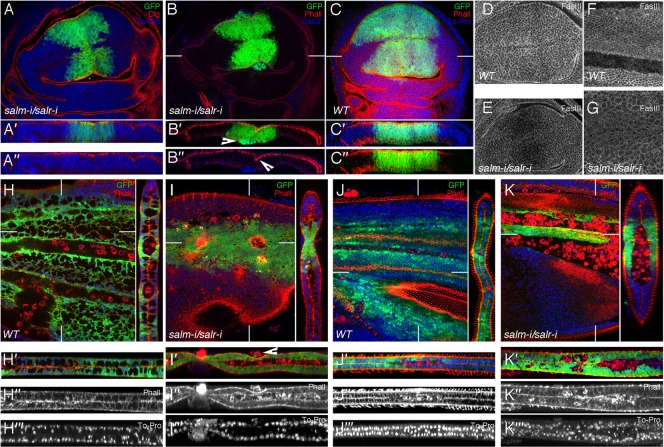
(A–A″) Third instar wing disc of UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ genotype showing the expression of GFP (green), Dlg (red) and TO-PRO (blue). A′ and A″ are transversal sections showing the three channels (A′) and the red and blue channels (A″). (B–B″) Expression of activated Cas3 (blue), GFP (green) and Phalloidin (red) in UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+. The arrowhead in B′ indicates the localization of activated Cas3 cells that also express GFP. The arrowhead in B″ indicates the epithelial fold. (C–C″) salEPv-Gal4 UAS-GFP/+ third instar control wing disc showing the expression of GFP (green), Phalloidin (red) and To-Pro (blue). C′ and C″ are transversal sections showing the three channels (C′) and the red and green channels (C″). (D,E) Expression of FasIII in wild type (D) and UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ (E) third instar discs. Note the reduction in FasIII expression in discs where Sal expression is reduced. (F,G) Wild type pupal wing of 36–40 hours APF (F) and pupal wing of the same age of UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ genotype (G). Sal mutant cells show larger size than wild type cells, and a reduced expression of FasIII. (H–I‴) Expression of GFP (green), Phalloidin (red) and TO-PRO (blue) in UAS-dicer2/+; salEPv-Gal4 UAS-GFP/+ control pupal wings (H–H‴) and UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ (I–I‴) pupal wings 24–30 hours APF. H′–H‴ and I′–I‴ are tangential sections showing the expression of these three markers (H′ and I′), and the single channels with Phalloidin (H″ and I″) and TO-PRO (H‴ and I‴) expression. The arrowhead in I′ indicates extruded cells. (J–K‴) Expression of Phalloidin (red), GFP (green) and TO-PRO (blue) in UAS-dicer2/+; salEPv-Gal4 UAS-GFP/+ control pupal wings (J–J‴) and UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ (K–K‴) pupal wings 36–40 hours APF. Cells belonging to the sal domain are still present (labelled in green in H,I,J,K) and the epithelium show a strong phenotype of loss of integrity which is better appreciated in the sagittal sections shown in I′–I‴ and K′–K‴. Cell morphology is also strongly altered, and the wings can display indentations (I–I‴). The GFP-negative cells located between the dorsal and ventral wing surfaces, which correspond to circulating haemocytes, are marked with Phalloidin (red) in I″ and K″. White lines in B,C,H–K label the sections shown to the right and to the bottom of each panel.
A role of sal in cell proliferation controlling the G2 to M transition
The small size of the mutant sal domain of expression suggests a role for salm and salr in the regulation of cell proliferation and/or cell survival. Clonal analysis of sal mutant cells confirms a cell-autonomous requirement for these genes in the central region, where sal mutant clones are rounder and smaller than their corresponding twins in both wing discs and in adult wings, and a considerable fraction of wild type twins (70%) appear without associated salm/salr mutant clone (de Celis et al., 1996; Milán et al., 2002) (Fig. 3A,B; supplementary material Fig. S1B–D). In addition, the size of anterior or posterior compartments formed by salm/salr mutant cells is strongly reduced, whereas the opposite compartment, formed by wild type cells, is not reduced in size (Fig. 3C–F; supplementary material Fig. S1). To identify which aspects of cell proliferation are affected in salm/salr mutant cells, we first measured the fraction of mitotic cells present in the salEPv-Gal4 domain of expression of wing discs growing with reduced levels of salm and salr. We found that the fraction of mitotic cells is strongly reduced in mutant (UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+; Fig. 3H,H′,I) compared to wild type discs (UAS-dicer2/+; salEPv-Gal4 UAS-GFP/+; Fig. 3G,G′,I), indicating that entering into mitosis is affected in cells with reduced expression of sal located in the central domain of the wing blade. We also measured the fraction of cells with reduced levels of salm/salr in the different phases of the cell cycle, and found a significant accumulation of salm/salr knockdown cells in G2 compared to wild type cells of the same disc and with wild type cells of control discs (Fig. 3J–K″). In this manner, we suggest that Sal promotes the progression from G2 to Mitosis, and consequently, cells in the sal domain of expression growing with reduced levels of sal expression accumulate in G2, causing a depletion of mitotic cells. The activity of the phosphatase String (stg, orthologous of cdc25) is a key determinant of the G2 to M transition in the wing disc (Edgar and O'Farrell, 1989; O'Farrell et al., 1989). We studied the expression of stg in sal mutant discs by in situ hybridization, and found a reduction of stg expression in the central domain of sal mutant discs (UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+; Fig. 3L′,M) compared to control discs (salEPv-Gal4/+; Fig. 3L,M). However, the pattern of stg expression is very variable from disc to disc, both in wild type and in sal mutant discs (data not shown). When we quantified the ratio of stg-expressing cells relative to the area of the disc we found a 15% reduction in sal knockdown discs, but the dispersion of the mean was very high (Fig. 3M). Ectopic expression of stg in the central domain of the wing disc (salEPv-Gal4 UAS-salm-i/+; UAS-salr-i/UAS-stg; Fig. 3N′,O) did not rescue the size of wings growing with reduced levels of sal expression (salEPv-Gal4 UAS-salm-i/+; UAS-salr-i/UAS-GFP; Fig. 3N,O). In this manner, we interpret that the accumulation of sal mutant cells in G2 is not caused by defects in stg expression, and that the variable reduction in the number of cells expressing stg might be related to the stoppage in G2 of sal mutant cells before they initiate stg expression.
Fig. 3. Requirements of sal genes for cell proliferation and cell cycle progression.
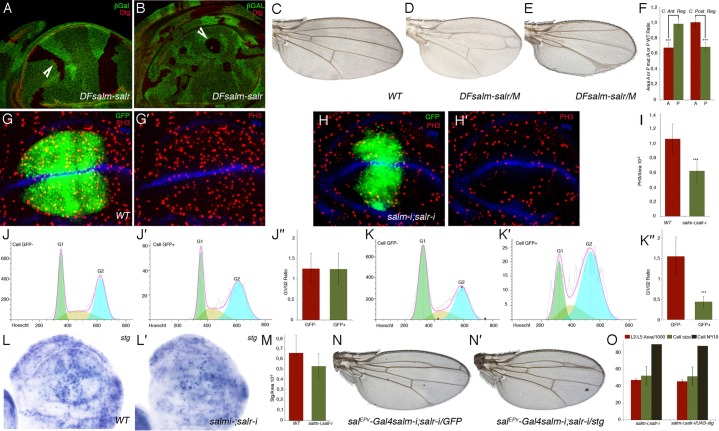
(A,B) Overall appearance of sal mutant clones and their twin spots in lateral regions of the disc (arrowhead in A) and in the central region of the disc (arrowhead in B). Clones are labelled by the absence of βGal (in green) and the expression of Dlg, localized in the apical part of the cells, is in red. (C–E) Wild type wing (C) and mosaic wings bearing large salm/salr M+ clones in the anterior (D) and posterior compartments (E). (F) Quantification of size reductions in mosaic wings. Bars represent mean size ± SEM of the anterior (red bars) and posterior (green bars) compartments. Wings with clones in the anterior compartment are represented in the two left columns, and wings with posterior clones in the two right columns. (G,G′) Expression of PH3 (red) in UAS-dicer2/+; salEPv-Gal4 UAS-GFP/+ control disc with the domain of Sal expression labelled by GFP (green). The expression of Wingless is in blue. (H,H′) Expression of PH3 (red) in UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+. The domain of salEPv-Gal4 expression is labelled by GFP (green) and the expression of Wingless is in blue. (I) Mitotic index of control (left column) and UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ (right column) discs. (J–J″) Cell cycle profiles of imaginal cells located outside the salEPv-Gal4 domain of expression (GFP negative cells, J) and cells located in this domain (labelled by GFP, J′). The average G1/G2 fractions of five independent experiments is shown in J″. Left column GFP negative cells and right column GFP cells. Imaginal discs were of UAS-dicer2/+; salEPv-Gal4 UAS-GFP/+ genotype. (K–K″) Cell cycle profiles of sal mutant imaginal cells located outside the salEPv-Gal4 domain of expression (GFP negative cells, K) and cells located in this domain (labelled by GFP, K′). The average G1/G2 fractions of five independent experiments is shown in K″. Left column GFP negative cells and right column GFP cells. Imaginal discs were of UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ genotype. (***) in F,H,K″ represents a p-value<0.005. (L,L′) In situ hybridization of stg RNA probes in wild type (L) and UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ (L′) third instar discs. Note the reduction in stg RNA in discs where Sal expression is reduced. (M) Quantification of ratio of stg-expressing cells relative to the area of the wing blade in control (left column; 17 wing discs) and UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ (right column; 22 wing discs) discs. The average ratio is a 15% reduced in UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ disc, but the dispersion of the mean is very high. (N,N′) Wings of genotype salEPv-Gal4 UAS-salm-i/+; UAS-salr-i/UAS-GFP. (N) and salEPv-Gal4 UAS-salm-i/+; UAS-salr-i/UAS-stg (N′). (O) Quantification of L2–L5 area/1000 (red columns), cell size (green columns) and cell number/10 (grey columns) in salEPv-Gal4 UAS-salm-i/+; UAS-salr-i/UAS-GFP (left columns) and salEPv-Gal4 UAS-salm-i/+; UAS-salr-i/UAS-stg (right columns) wings. Bars represent mean ± SEM in red and green columns, and the cell number.
Sal function in the central domain of the wing prevents cell death
The smaller than normal size of sal mutant clones, and the reduction of the size of the sal mutant central domain of the wing blade can also result from cell death, which is a rare occurrence in wild type discs (Milán et al., 1997). We found small numbers of wing cells expressing activated Cas3 in central domain of the wing blade of salm/salr knockdown wing discs (UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+; Fig. 4A). These cells were located mostly in the periphery of the central domain, and in general they are positioned in the basal side of the epithelium (Fig. 4A–A‴). In contrast, cell death is very rare during pupal development (Fig. 4B–B‴), indicating that the observed epithelial disruption occurring at this stage is not related to apoptosis. We also looked at cell death in a situation where sal mutant and non-mutant cells are confronted along the central domain of sal expression. In wing discs where the expression of salm/salr is reduced in the posterior compartment (en-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+) we observed a robust induction of cell death located mainly along the boundary confronting sal mutant and not mutant cells (Fig. 4C,C′). However, we did not observe any significant rescue of the sal mutant phenotype in genetic backgrounds in which cell death is prevented, suggesting that the contribution of cell death to the sal phenotype is minor (supplementary material Fig. S2). The activity of the JNK signalling pathway is related to cell death in the wing disc. We looked at the reported of JNK activity puckered (puc) (Martín-Blanco et al., 1998), and found a robust and cell autonomous induction of puc expression in sal mutant discs (en-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/puc-lacZ (Fig. 4D,D′) and salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/puc-lacZ (Fig. 4F,F′)). The suppression of JNK activation in the salm/salr knockdown genetic background does not correct the sal mutant phenotype (salEPv-Gal4/UAS-salm-i; UAS-salr-i/UAS-puc (Fig. 4H–I′), compare with salEPv-Gal4/UAS-salm-i; UAS-salr-i/+ (Fig. 4E–G′)). In fact, suppressing JNK signalling by puc over-expression aggravates the phenotype of sal mutant wings (compare Fig. 4E, Fig. 4H), perhaps because mis-specified cells are not removed from the epithelium during wing disc development.
Fig. 4. Relationships of the sal genes with JNK activity and cell death.
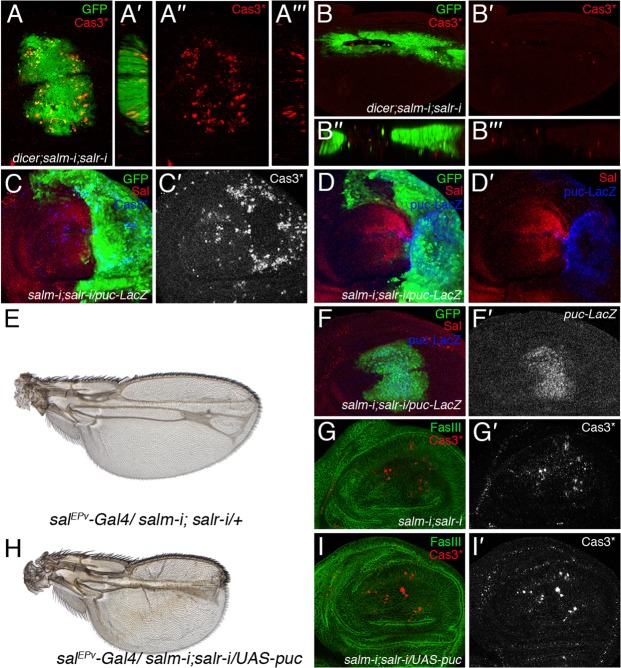
(A–A‴) Expression of activated Cas3 (Cas3*; red) in UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ third instar discs. Cell death is observed in the Sal domain, labelled in green, and is particularly prominent in its periphery. The red channels (activated Cas3) are shown in A″ and A‴. (B–B‴) Expression of activated Cas3 in a pupal wing of UAS-dicer2/+; salEPv-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ genotype 36–40 hours APF. (C,C′) Expression of activated Cas3 in en-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+. The expression of activated Cas3 is detected along the anterior–posterior compartment boundary, in the most posterior cells, and in scattered cells located basally in the anterior compartment. (D,D′) Activation of JNK signalling in the posterior compartment of en-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/puc-lacZ. The expression of puc-lacZ (blue in D and D′) is detected through the posterior compartment (labelled in green). The expression of Salm is shown in red. (E) salEPv-Gal4/UAS-salm-i; UAS-salr-i/+ wing. (F,F′) Third instar wing imaginal disc of salEPv-Gal4/UAS-salm-i; UAS-salr-i/puc-lacZ genotype showing the expression of GFP (green in F) and puc-lacZ (single channel in F′). (G,G′) Expression of activated Cas3 (red in G and single channel in G′) in salEPv-Gal4/UAS-salm-i; UAS-salr-i/+ third instar discs. The expression of FasIII is shown in green. (H–I′) salEPv-Gal4/UAS-salm-i; UAS-salr-i/UAS-puc wing (H) and corresponding imaginal disc showing the expression of activated Cas3 (red in I and single channel in I′) and FasIII (green in I). The expression of activated Cas3 is not prevented by the over-expression of the JNK negative regulator Puc (compare with G′ and I′), but the resulting wing shows an even stronger sal loss-of-function phenotype (compare E with H).
The confrontation of sal-expressing and non-expressing cells generate epithelial grooves
The shape of sal mutant clones in the central region of the wing blade is rounded, suggesting that sal mutant cells tend to minimize their contacts with wild type cells (Fig. 3B, Fig. 5A) (see also Milán et al., 2002). In addition, we also observed that the boundary of these clones form grooves separating mutant and normal cells (Fig. 5A′,A″). The formation of grooves separating sal mutant and non-mutant cells is always induced when salm and salr expression is reduced within its normal domain of expression. For example, reduction of salm/salr in the domain of dpp expression (UAS-salm-i/+; dpp-Gal4 UAS-GFP/UAS-salr-i), generates a strong groove that also affects the neighbouring wild type cells (Fig. 5B–C″). Similarly, knockdown of salm/salr in the posterior compartment (en-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+) also generates a deep groove separating anterior and posterior cells (Fig. 5D–D″). We followed the temporal sequence of groove formation in discs of en-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/tub-Gal80ts genotype. We grow larvae of this genotype at 17°C, preventing the action of the Gal4 protein, and transferred them to 29°C at the beginning of the third larval instar. We observed that Gal4 activity, revealed by the expression of GFP, becomes apparent at 6 hours after the temperature shift (Fig. 5E,E′). At this time the expression of Salm is only moderately reduced, and the epithelium in normal (Fig. 5E′,E″). The expression of GFP together with the loss of Salm protein is very robust 24 h after the temperature shift (Fig. 5F). In these discs, the epithelium still maintains its continuity without any indication of groove formation (Fig. 5F″). Groove formation becomes apparent in discs 48 hours after the temperature shift (Fig. 5G–G‴). At this time, the posterior compartment is reduced in size and is separated from the anterior compartment by a continuous epithelial groove (Fig. 5G–G‴). The formation of grooves is not the result of a discrepancy in size between the posterior and anterior compartments, because when cell proliferation is strongly reduced in posterior cells (en-Gal4 UAS-GFP/UAS-CycA-i; tub-Gal80ts/+), the boundary between the anterior and posterior compartments is smooth (Fig. 5H–H‴).
Fig. 5. Cellular consequences of confrontations between cells expressing and non-expressing sal genes.
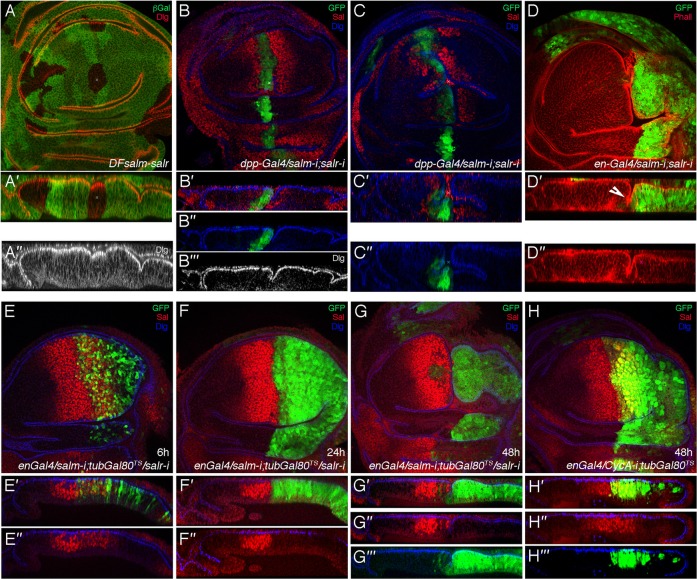
(A–A″) Clones of sal mutant cells, induced in hsFLP; FRT40A Df(2L)32FP5/FRT40A armlacZ larvae, are rounded in the central region of the wing blade, and form small epithelial indentations with neighbouring wild type cells. Clones are labelled by the absence of lacZ (in green) and the expression of Dlg, localized in the apical part of the cells, is in red. A′ and A″ are tangential sections showing both channels (A′), or only the red channel (A″). (B–C″) Third instar wing imaginal discs of UAS-salm-i/+; dpp-Gal4 UAS-GFP/UAS-salr-i showing the expression of GFP (green), Salm (red) and Dlg (blue). The expression and apical localization of Dlg is normal (see B″,C″). Cells with reduced expression of sal genes generate deep indentations (see transversal sections in B′–B‴,C′,C″), which in some extreme cases also affect the surrounding wild type cells (C′,C″). (D–D″) Third instar wing imaginal discs of en-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/+ showing the expression of GFP (green) and Phalloidin (red). The posterior compartment is always reduced in size and the boundary between anterior and posterior cells form a strong epithelial indentation (arrow in D″). The overall distribution of F-actin is normal (D″). (E–G‴) Third instar wing imaginal discs of en-Gal4 UAS-GFP/UAS-salm-i; UAS-salr-i/tub-Gal80ts in a temporal sequence after of the activation of Gal4: 6 hours (E), 24 hours (F) and 48 hours (G), showing the expression of GFP (green), Salm (red) and Dlg (blue). The Gal4 activity, revealed by the expression of GFP, becomes apparent at 6 hours (E,E′) and is robust at 24 (F,F′). The expression of Salm is moderately reduced at 6 hours (E″), and disappears at 24 hours (F″). At both time intervals the appearance of the epithelium is normal (E″–F″). (G–G‴) The formation of an epithelial groove becomes apparent after 48 hours at the restrictive temperature, and the size of posterior compartment is reduced (G). (H–H‴) Third instar wing imaginal discs of en-Gal4 UAS-GFP/UAS-CycA-i; tub-Gal80ts/+ showing expression of GFP (green), Salm (red) and Dlg (blue). Although cell division is impaired and the cells are larger than normal, the structure of the epithelium is normal (H′).
Genetic relationships between Sal function and dpp signalling in the wing
From the previous analysis, we conclude that Salm/Salr function is required for cell proliferation and survival, and for the maintenance of epithelial integrity, perhaps determining cell affinities in the central region of the wing disc. These requirements are similar to those reported for different components of the Dpp pathway, and consequently we analysed whether Salm/Salr not only are transcriptional targets of Dpp, but whether they also mediate the functions of this pathway during wing development. We first aimed to rescue the consequences of loss of dpp function by ectopic expression of Salm. The expression of interference RNA against Mad, the transducer of the Dpp pathway, in the central region of the wing disc (salEPv-Gal4 UAS-GFP/+; UAS-Mad-i) results in smaller than normal wings that fail to differentiate the veins L2, L3 and L4 (Fig. 6A,B). The corresponding wing imaginal discs show, as expected, very much reduced levels of Salm protein (Fig. 6F, compare with Fig. 6D), and also the loss of Delta expression in the presumptive veins L3 and L4 (Fig. 6G, compare with Fig. 6E). The expression of Salm in this background of reduced Mad (salEPv-Gal4/+; UAS-Mad-i/UAS-salm) rescues to a large extent the wing phenotype and the expression of Delta (Fig. 6C,I), despite a patchy accumulation of Sal protein (Fig. 6H). Over-expression of Brinker (Brk) in the central domain of the wing disc (salEPv-Gal4/+; UAS-brk/+) generates a strong reduction of wing size and the loss of L2 and L4 veins (Fig. 6J). In the corresponding discs the expression of Salm is entirely lost (Fig. 6K). The presence of Salm in salEPv-Gal4/+; UAS-brk/UAS-salm discs does not rescue the growth and pattern defects caused by brk over-expression (Fig. 6L). However, we find that Salm expression is barely detected in these mutant discs (Fig. 6M), most likely because the salEPv-Gal4 driver is repressed upon Brk over-expression. We aimed to prevent the effects of Brk and Mad on the regulation of the salEPv-Gal4 driver in discs of salEPv-Gal4/UAS-salm; tub<FRT-GFP-FRT>Gal4 UAS-FLP/UAS-Mad-i and salEPv-Gal4/UAS-salm; tub<FRT-GFP-FRT>Gal4 UAS-FLP/UAS-brk genotypes. In these discs, the expression of Gal4 in the central region of the wing disc is first initiated by the salEPv-Gal4 driver and then clonally sustained by the tub-Gal4 driver, once the FRT cassette is removed by FLP-mediated recombination. Unfortunately, we found that the over-expression of salm using the tub-FRT-Gal4 driver results in the generation of clones of cells expressing high levels of salm, leading to the formation of small wings with a severe phenotype of epithelial extrusion (data not shown). The behaviour of clones of cells over-expressing Salm prevented us from checking its ability to rescue the brk and Mad-i phenotypes.
Fig. 6. Genetic relationships between Sal and the Dpp pathway.
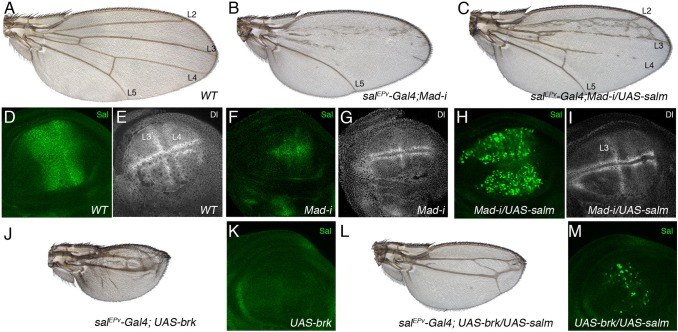
(A) Wild type wing. (B) salEPv-Gal4/+; UAS-Mad-i/UAS-GFP. The reduction in Mad expression in the salEPv-Gal4 domain of expression results in smaller wings that loss most of the L2, L3 and L4 veins. (C) salEPv-Gal4/+; UAS-Mad-i/UAS-salm. The expression of Salm under UAS control in wings with a reduction in Mad expression rescues the differentiation of the L2, L3 and part of the L4 veins. (D–I) Expression of Salm (Salm, green in D,F,H) and Dl (Dl, white in E,G,I) in wild type (D,E), salEPv-Gal4/+; UAS-Mad-i/UAS-GFP (F,G) and salEPv-Gal4/+; UAS-Mad-i/UAS-salm third instar imaginal wing discs (H,I). Note that the expression of both Salm and Dl is strongly reduced in the central region of the wing blade in F and G, the expression of Salm is strong in H, and the expression of Dl is rescued in the L3 vein in I. (J–M) No rescue by Salm of the consequences of Brk expression in the central region of the wing blade. The phenotype of salEPv-Gal4/UAS-GFP; UAS-brk/+ wings consists in a strong reduction of wing size and loss of the L2 and L4 veins (J). In the corresponding wing imaginal disc the expression of Salm is strongly reduced (K). The phenotype of ectopic brk expression is weakly rescued by the expression of Salm in flies of salEPv-Gal4/+; UAS-brk/UAS-salm genotype (L). In the corresponding wing disc, however, Salm is only expressed in a patchy pattern (M), probably due to the effects of Brk on the Gal4 driver.
We reason that some consequences of Dpp hyper-activation might be due to the ectopic or increased expression of the sal genes. For example, wing discs expressing the constitutively activated version of the receptor Tkv in the central region of the wing (salEPv-Gal4/+; UAS-tkvQD/UAS-GFP) show higher than normal levels of Sal and a strong disruption of the epithelium compared to wild type discs (Fig. 7A′–B‴). The adult wings of this genotype display a non-autonomous loss of anterior and posterior wing tissue (Fig. 7B). Loss of salm/salr in the TkvQD background (salEPv-Gal4/UAS-salm-i; UAS-tkvQD/UAS-salr-i) entirely rescues all components of the TkvQD phenotype (Fig. 7C), and the resulting wing discs and adult wing show the characteristic defects of salm/salr loss of function (Fig. 7C′–C‴). Ectopic expression of Dpp in the central region of the wing blade (salEPv-Gal4 UAS-dpp-GFP) results in wings that differentiate extra-vein tissue in the L2/L3 and L4/L5 interveins (Fig. 7D). Loss of salm/salr in this background (salEPv-Gal4 UAS-dpp-GFP/UAS-salm-i; UAS-salr-i/+) results in smaller wings that also differentiate extra-veins in the same wing regions (Fig. 7F, compare with Fig. 7D,E). In this manner, it seems that the promotion of vein differentiation caused by Dpp does not require Sal function, whereas the growth-promoting function of Sal is not rescued by ectopic Dpp signalling.
Fig. 7. Correction of increased Dpp signalling by loss of salm/salr expression.
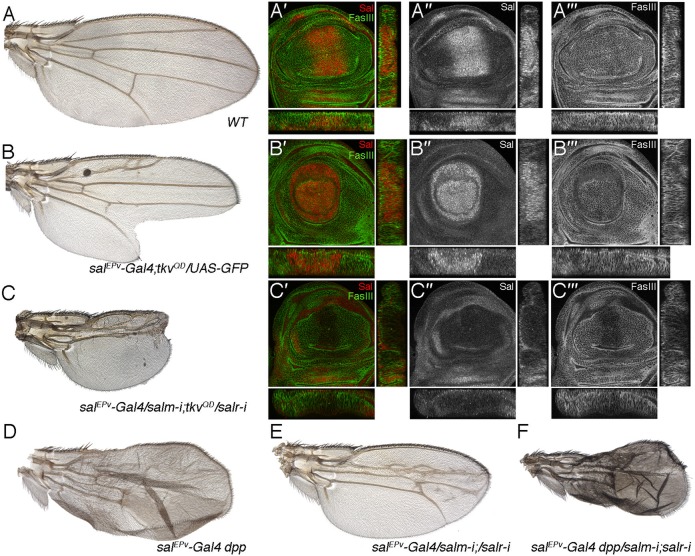
(A–A‴) Wild type wing (A) and third instar wing imaginal disc showing the expression of Salm (red in A′ and single channel in A″) and FasIII (green in A′ and single channel in A‴). Below and to the right of each image are the transversal and longitudinal sections of each disc. (B–B‴) Wing of salEPv-Gal4/+; UAS-tkvQD/UAS-GFP genotype (B) and third instar wing imaginal disc of the same genotype showing the expression of Salm (red in B′ and single channel in B″) and FasIII (green in B′ and single channel in B‴). Below and to the right of each image are the transversal and longitudinal sections of each disc. (C–C‴) Wing of salEPv-Gal4/UAS-salm-i; UAS-salr-i/UAS-tkvQD (C) and corresponding third instar wing disc showing the expression of Salm (red in C′ and single channel in C″) and FasIII (green in C′ and single channel in C‴). Below and to the right of each image are the transversal and longitudinal sections of each disc. The phenotype of these wings, and the expression of FasIII are similar to those observed in wings and discs where only the expression of sal genes is reduced. (D–F) Wings of genetic combinations between overexpression of Dpp (salEPv-Gal4 UAS-dppGFP/+; D) and reduction of salm/salr (salEPv-Gal4/UAS-salm-i; UAS-salr-i/+; E). The combination of these two conditions (salEPv-Gal4 UAS-dppGFP/UAS-salm-i; UAS-salr-i/+; F) results in wings of reduced size that differentiates the same pattern of extra-veins typical of wings over-expressing Dpp (shown in D).
We also studied the contribution of Sal to the cell proliferation effects caused by loss or gain of Tkv function. The behaviour of TkvQD-expressing clones depends on the position of these clones along the anterior–posterior axes (Burke and Basler, 1996; Rogulja and Irvine, 2005). Thus, clones in the periphery of the wing blade cause extreme overgrowths, due to the repression of brk transcription (Schwank et al., 2008) (Fig. 8A–B′). Clones located in the central region of the wing blade also behave abnormally, as they are also larger than normal and are surrounded by epithelial grooves (Fig. 8A–B′). The loss of sal function in cells expressing TkvQD has very different effects depending on their location in the wing blade. Thus TkvQD/Dfsal clones in the central region of the wing blade are smaller than normal and behave as Dfsal clones (Fig. 8C–D′). In contrast, clones in the periphery of the disc still cause extreme overgrowth, indicating that Sal only mediates the functions of Tkv in the central region of the wing, but does not contribute to the overgrowth caused by ectopic pathway activation in the peripheral regions. Complementary, we also observed a rescue of the cell lethality of tkv mutant clones in the central region of the wing blade. Thus, tkva12 clones fail to grow in the wing blade (Burke and Basler, 1996; data not shown), but when these cells express Salm they can grow in the centre of the wing, forming clones that tend to extrude from the epithelium (Fig. 8G–H′). As mentioned before, clones of cells over-expressing sal also cause epithelial defects and clone extrusion (Fig. 8E–F′).
Fig. 8. Genetic interactions between the Dpp receptor Tkv and Sal.
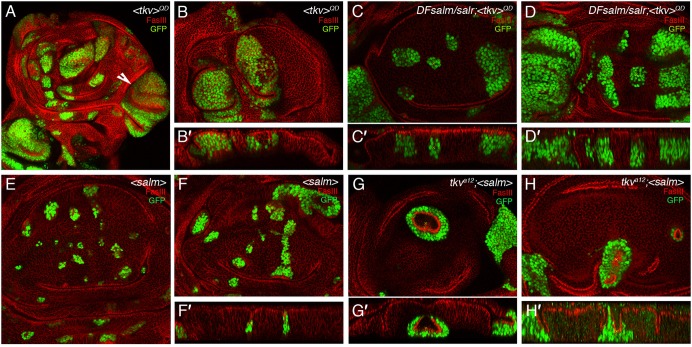
(A–B′) Examples of third instar wing discs bearing clones of cells expressing an activated version of the receptor Tkv (TkvQD). Clones are labelled in green and FasIII is labelled in red. TkvQD clones cause large overgrowth in the lateral regions of the wing blade (white arrowhead in A) and epithelial folding in the central region of the wing blade (see sagittal section in B′ of the clones shown in B). (C–D′) Examples of clones homozygous for the sal deficiency Df(2L)32FP5 and expressing TkvQD. The clones in the lateral regions still show overgrowths, but the epithelial phenotype and growth of clones located in the central region of the wing blade is more similar to that of sal mutant clones. (E–F′) Clones of cells over-expressing Salm (labelled in green) are smaller than expected and cause small epithelial invaginations (see transversal section in F′). (G–H′) Clones of cells homozygous for the tkv allele tkva12 and over-expressing Salm under the control of UAS sequences. The expression of Salm rescues the proliferation defect of tkva12 cells in the wing blade, and these clones are surrounded by strong epithelial indentations.
Discussion
In this work we describe the requirements of the Sal transcription factors during the development of the wing blade. This description extends the analysis of Sal activity in regulating pattern formation and cell affinities (de Celis and Barrio, 2000; Milán et al., 2002), and identifies additional functions in cell survival, division and maintenance of epithelial integrity. We also show that some of the effects of Dpp signalling during the growth and patterning of the wing disc are mediated by Sal activity. The salm and salr genes were among the first identified transcriptional targets of Dpp signalling (de Celis et al., 1996), and given the key roles this pathway plays during imaginal development, it is important to understand whether its functions are mediated by the Salm and Salr proteins.
Sal function and cellular affinities
The analysis of sal mutant cells, and of wing discs growing with reduced levels of Sal proteins, identifies several requirements for these genes. First, Sal expression in the central domain of the wing blade seems to confer these cells with some affinity label that makes them different from other cells non-expressing the genes. This is reflected in the shape of sal mutant clones, which are rounder than normal (Milán et al., 2002), and in the formation of epithelial folds at the place of confrontation between sal-expressing and sal-non-expressing cells located in the central region of the wing. The nature of this affinity label seems to be quantitative, because clones of cells over-expressing sal also manifest an aberrant behaviour and tend to sort-out from the epithelium, whereas uniform increase in sal expression in its normal domain does not affect epithelial continuity. This function of Sal proteins was related to the regulation of the cell-membrane proteins Capricious and Tartan (Milán et al., 2002). In addition, we also identified FasIII as a membrane protein which expression depends on Sal in the central region of the wing blade. In this manner, it seems that Sal proteins regulate, by repression and activation, the transcription of several cell-adhesion proteins imposing an affinity marker that distinguish the central versus the peripheral regions of the wing blade (Cap and Tartan), and that also might contribute to cell adhesion (FasIII). Intriguingly, there are normal confrontations between sal expressing and not expressing cells in wild type discs, which are located in the anterior and posterior boundaries of the sal domain of expression. These confrontations are not associated to epithelial folds, indicating that wild type central (sal-expressing cells) and peripheral cells associate normally to each other. Nonetheless, some differences exist between these populations of cells, because the limits of sal expression correspond to weak clonal restrictions boundaries during normal development (Resino et al., 2002), indicating that the descendants of cells born in the sal domain of expression tend to remain in this domain. Therefore, it seems that the central and peripheral domains of the wing blade display some difference in affinity that is sufficient to promote a clonal restriction, but not able to trigger epithelial sorting during normal development. It is only when sal mutant cells, or cell expressing abnormally high levels of Sal, grow within the central domain that these differences became sufficient to trigger epithelial sorting. It has been argued that the central and peripheral domains of the wing blade have an intrinsic difference with respect to the response to Dpp signalling, and that this difference, of unknown nature, is set independently of Dpp signalling (Schwank et al., 2008). The distinct behaviour of the central domain with respect to abnormal confrontations between cells expressing different levels of Sal proteins agrees with this proposal, and suggests that Sal function contributes within this central territory to balance the expression of other undefined cell affinity markers involved in the differentiation of the central and peripheral territories.
Sal function in cell division
A second role of Sal proteins is related to the promotion of cell proliferation within the central region of the wing blade. Thus, sal mutant clones are smaller than their twins, and when entire anterior or posterior compartments are formed by sal mutant cells their sizes are smaller than normal. We also found that mutant cells located in the central domain of the wing blade growing with lower than normal levels of Sal proteins tend to accumulate in the G2 phase of the cell cycle, and enter into mitosis at a lower rate than wild type cells. These mutant cells are also larger in size than wild type cells during pupal development. We speculate that Sal affects some aspect of the G2/M transition. The G2/M transition is regulated by String (stg), a phosphatase orthologous to cdc25 that de-phosphorylates the complex CycB/Cdk1. We observed that the expression of stg seems reduced in the central region of salm/salr mutant discs, but that forced stg expression does not rescue the proliferation defects of sal mutant cells, indicating that Sal proteins regulate other aspect than stg expression during G2/M progression. The Dpp pathway regulates growth by repressing brk expression, and it has been proposed that the Dpp/Brk system mostly limits proliferation in lateral areas of the wing disc (Schwank et al., 2008). We find that this aspect of Dpp/Brk function is independent of Sal activity, because clones of cells expressing TkvQD, which lack brk and have ectopic expression of Sal genes, still overgrow in peripheral regions of the disc in the absence of Sal function. In contrast Sal can rescue the proliferative defect of tkv mutant cells in the centre of the wing blade, suggesting that in this territory Sal mediates Dpp/Tkv activity. The analysis of tkv mutant cells suggested that they show a severe reduction in the number of cells in S and G2, and an increase in the number of cells in G1 (Martín-Castellanos and Edgar, 2002). This is in contrast to the behaviour of sal mutant cells, which tend to accumulate in the G2 phase of the cell cycle. To reconcile these apparently conflicting results, we suggests that the Dpp pathway regulates the cell cycle both at the G1/S and G2/M transitions, and does so using different mechanisms. In this scenario, Dpp/Tkv would promote G1/S independently of Sal, and G2/M in a Sal-dependent manner. In the absence of tkv, cells retaining some Sal expression would go through G2/M and accumulate at G1/S, and in the absence of Sal, cells would tend to accumulate at G2/M.
Sal functions in cell survival
In addition to cell proliferation and cellular affinities, Sal function is also required for cell viability in the central region of the wing blade. Thus, the reduction in sal expression is associated to the activation of JNK signalling and to the induction of cell death, although these two events have a minor contribution in the generation of the sal mutant phenotype. In this manner, when cell death or JNK signalling is prevented in the absence of sal, the wings are still smaller than normal and display similar pattern defects as sal mutant wings. We don't know the mechanisms linking Sal with JNK signalling and cell death. It is well know that epithelial discontinuities in Tkv activity lead to JNK activation and cell death (Gibson and Perrimon, 2005), but in our hands JNK activity, as visualized by puc expression, occurs in situations where the appearance of the epithelium is entirely normal. Thus, wing discs in which sal expression is reduced in its entire domain show a normal epithelium with a robust induction of puc expression. In this situation, cell death is mostly observed in the periphery of the sal domain of expression.
The activities of Sal in regulating cell division, viability, epithelial integrity and vein positioning suggest that these proteins regulate most aspects of the patterning of the central domain of the wing blade, and they do so acting as downstream mediators of Dpp signalling. Salm and Salr belong to a family of transcription factors containing several Zn-fingers domains, and recent biochemical evidence suggests that they might be acting as transcriptional repressors (Sánchez et al., 2011). This is compatible with the function as a repressor of vertebrate Sall1, which interacts with histone deacetylase and other components of the chromatin remodelling complex NuRD (Lauberth et al., 2007). The understanding of the genetic hierarchy acting downstream of the Dpp/Brk/Sal system requires the identification of the target genes regulated by the Brk and Sal transcription factors, which likely would include cell cycle regulators, cell adhesion proteins and cytoskeleton components. We think that the identification and cellular characterization of Sal functional requirements constitutes a necessary step towards the identification of their target genes, and consequently to the reconstruction of the cascade of events triggered by Dpp signalling in the Drosophila wing.
Supplementary Material
Acknowledgments
We are very grateful to Ana López-Varea and Verónica Flores for their skilful technical help, and Antonio Baonza, Carlos Estella and Cristina Molnar for criticism that greatly improved the manuscript. We thank the Hybridome Bank at Iowa University, NIG in Japan, Bloomington Stock Center and other colleagues for providing the tools necessary for this work. Our work is supported by grants BFU2009-09403 and CSD-2007-00008 from the Spanish Ministerio de Economia y competitividad. The CBMSO enjoins the institutional support of the Ramón Areces foundation.
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Affolter M., Basler K. (2007). The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nat. Rev. Genet. 8, 663–674 10.1038/nrg2166 [DOI] [PubMed] [Google Scholar]
- Barrio R., de Celis J. F., Bolshakov S., Kafatos F. C. (1999). Identification of regulatory regions driving the expression of the Drosophila spalt complex at different developmental stages. Dev. Biol. 215, 33–47 10.1006/dbio.1999.9434 [DOI] [PubMed] [Google Scholar]
- Burke R., Basler K. (1996). Dpp receptors are autonomously required for cell proliferation in the entire developing Drosophila wing. Development 122, 2261–2269. [DOI] [PubMed] [Google Scholar]
- Cantera R., Lüer K., Rusten T. E., Barrio R., Kafatos F. C., Technau G. M. (2002). Mutations in spalt cause a severe but reversible neurodegenerative phenotype in the embryonic central nervous system of Drosophila melanogaster. Development 129, 5577–5586 10.1242/dev.00158 [DOI] [PubMed] [Google Scholar]
- Cruz C., Glavic A., Casado M., de Celis J. F. (2009). A gain-of-function screen identifying genes required for growth and pattern formation of the Drosophila melanogaster wing. Genetics 183, 1005–1026 10.1534/genetics.109.107748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis J. F. (2003). Pattern formation in the Drosophila wing: The development of the veins. Bioessays 25, 443–451 10.1002/bies.10258 [DOI] [PubMed] [Google Scholar]
- de Celis J. F., Barrio R. (2000). Function of the spalt/spalt-related gene complex in positioning the veins in the Drosophila wing. Mech. Dev. 91, 31–41 10.1016/S0925-4773(99)00261-0 [DOI] [PubMed] [Google Scholar]
- de Celis J. F., Barrio R. (2009). Regulation and function of Spalt proteins during animal development. Int. J. Dev. Biol. 53, 1385–1398 10.1387/ijdb.072408jd [DOI] [PubMed] [Google Scholar]
- de Celis J. F., Bray S. (1997). Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development 124, 3241–3251. [DOI] [PubMed] [Google Scholar]
- de Celis J. F., Barrio R., Kafatos F. C. (1996). A gene complex acting downstream of dpp in Drosophila wing morphogenesis. Nature 381, 421–424 10.1038/381421a0 [DOI] [PubMed] [Google Scholar]
- de Celis J. F., Barrio R., Kafatos F. C. (1999). Regulation of the spalt/spalt-related gene complex and its function during sensory organ development in the Drosophila thorax. Development 126, 2653–2662. [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S.et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Domingos P. M., Brown S., Barrio R., Ratnakumar K., Frankfort B. J., Mardon G., Steller H., Mollereau B. (2004). Regulation of R7 and R8 differentiation by the spalt genes. Dev. Biol. 273, 121–133 10.1016/j.ydbio.2004.05.026 [DOI] [PubMed] [Google Scholar]
- Dong P. D., Todi S. V., Eberl D. F., Boekhoff–Falk G. (2003). Drosophila spalt/spalt-related mutants exhibit Townes-Brocks' syndrome phenotypes. Proc. Natl. Acad. Sci. USA 100, 10293–10298 10.1073/pnas.1836391100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B. A., O'Farrell P. H. (1989). Genetic control of cell division patterns in the Drosophila embryo. Cell 57, 177–187 10.1016/0092-8674(89)90183-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart W. M., Crosby M., Matthews B., Rindone W. P., Chillemi J., Russo Twombly S., Emmert D., Ashburner M., Drysdale R. A., Whitfield E.et al. (1997). FlyBase: a Drosophila database. Nucleic Acids Res. 25, 63–66 10.1093/nar/25.1.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson M. C., Perrimon N. (2005). Extrusion and death of DPP/BMP-compromised epithelial cells in the developing Drosophila wing. Science 307, 1785–1789 10.1126/science.1104751 [DOI] [PubMed] [Google Scholar]
- Gould A. P., Elstob P. R., Brodu V. (2001). Insect oenocytes: a model system for studying cell-fate specification by Hox genes. J. Anat. 199, 25–33 10.1046/j.1469-7580.2001.19910025.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Awano W., Suzuki K., Hiromi Y., Yamamoto D. (1997). The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development 124, 761–771. [DOI] [PubMed] [Google Scholar]
- Jaźwińska A., Kirov N., Wieschaus E., Roth S., Rushlow C. (1999). The Drosophila gene brinker reveals a novel mechanism of Dpp target gene regulation. Cell 96, 563–573 10.1016/S0092-8674(00)80660-1 [DOI] [PubMed] [Google Scholar]
- Kühnlein R. P., Schuh R. (1996). Dual function of the region-specific homeotic gene spalt during Drosophila tracheal system development. Development 122, 2215–2223. [DOI] [PubMed] [Google Scholar]
- Kühnlein R. P., Frommer G., Friedrich M., Gonzalez–Gaitan M., Weber A., Wagner–Bernholz J. F., Gehring W. J., Jäckle H., Schuh R. (1994). spalt encodes an evolutionarily conserved zinc finger protein of novel structure which provides homeotic gene function in the head and tail region of the Drosophila embryo. EMBO J. 13, 168–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauberth S. M., Bilyeu A. C., Firulli B. A., Kroll K. L., Rauchman M. (2007). A phosphomimetic mutation in the Sall1 repression motif disrupts recruitment of the nucleosome remodeling and deacetylase complex and repression of Gbx2. J. Biol. Chem. 282, 34858–34868 10.1074/jbc.M703702200 [DOI] [PubMed] [Google Scholar]
- Lecuit T., Brook W. J., Ng M., Calleja M., Sun H., Cohen S. M. (1996). Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing. Nature 381, 387–393 10.1038/381387a0 [DOI] [PubMed] [Google Scholar]
- Martín–Blanco E., Gampel A., Ring J., Virdee K., Kirov N., Tolkovsky A. M., Martinez–Arias A. (1998). puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 12, 557–570 10.1101/gad.12.4.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín–Castellanos C., Edgar B. A. (2002). A characterization of the effects of Dpp signaling on cell growth and proliferation in the Drosophila wing. Development 129, 1003–1013. [DOI] [PubMed] [Google Scholar]
- Massagué J., Wotton D. (2000). Transcriptional control by the TGF-β/Smad signaling system. EMBO J. 19, 1745–1754 10.1093/emboj/19.8.1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milán M., Campuzano S., García–Bellido A. (1997). Developmental parameters of cell death in the wing disc of Drosophila. Proc. Natl. Acad. Sci. USA 94, 5691–5696 10.1073/pnas.94.11.5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milán M., Pérez L., Cohen S. M. (2002). Short-range cell interactions and cell survival in the Drosophila wing. Dev. Cell 2, 797–805 10.1016/S1534-5807(02)00169-7 [DOI] [PubMed] [Google Scholar]
- Mollereau B., Dominguez M., Webel R., Colley N. J., Keung B., de Celis J. F., Desplan C. (2001). Two-step process for photoreceptor formation in Drosophila. Nature 412, 911–913 10.1038/35091076 [DOI] [PubMed] [Google Scholar]
- Nellen D., Burke R., Struhl G., Basler K. (1996). Direct and long-range action of a DPP morphogen gradient. Cell 85, 357–368 10.1016/S0092-8674(00)81114-9 [DOI] [PubMed] [Google Scholar]
- O'Farrell P. H., Edgar B. A., Lakich D., Lehner C. F. (1989). Directing cell division during development. Science 246, 635–640 10.1126/science.2683080 [DOI] [PubMed] [Google Scholar]
- Resino J., Salama–Cohen P., García–Bellido A. (2002). Determining the role of patterned cell proliferation in the shape and size of the Drosophila wing. Proc. Natl. Acad. Sci. USA 99, 7502–7507 10.1073/pnas.072208199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogulja D., Irvine K. D. (2005). Regulation of cell proliferation by a morphogen gradient. Cell 123, 449–461 10.1016/j.cell.2005.08.030 [DOI] [PubMed] [Google Scholar]
- Rusten T. E., Cantera R., Urban J., Technau G., Kafatos F. C., Barrio R. (2001). Spalt modifies EGFR-mediated induction of chordotonal precursors in the embryonic PNS of Drosophila promoting the development of oenocytes. Development 128, 711–722. [DOI] [PubMed] [Google Scholar]
- Sánchez J., Talamillo A., González M., Sánchez–Pulido L., Jiménez S., Pirone L., Sutherland J. D., Barrio R. (2011). Drosophila Sal and Salr are transcriptional repressors. Biochem. J. 438, 437–445. [DOI] [PubMed] [Google Scholar]
- Schwank G., Restrepo S., Basler K. (2008). Growth regulation by Dpp: an essential role for Brinker and a non-essential role for graded signaling levels. Development 135, 4003–4013 10.1242/dev.025635 [DOI] [PubMed] [Google Scholar]
- Sweetman D., Münsterberg A. (2006). The vertebrate spalt genes in development and disease. Dev. Biol. 293, 285–293 10.1016/j.ydbio.2006.02.009 [DOI] [PubMed] [Google Scholar]
- Teleman A. A., Cohen S. M. (2000). Dpp gradient formation in the Drosophila wing imaginal disc. Cell 103, 971–980 10.1016/S0092-8674(00)00199-9 [DOI] [PubMed] [Google Scholar]
- Wu Q., Chen X., Zhang J., Loh Y. H., Low T. Y., Zhang W., Zhang W., Sze S. K., Lim B., Ng H. H. (2006). Sall4 interacts with Nanog and co-occupies Nanog genomic sites in embryonic stem cells. J. Biol. Chem. 281, 24090–24094 10.1074/jbc.C600122200 [DOI] [PubMed] [Google Scholar]
- Xu T., Rubin G. M. (1993). Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117, 1223–1237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


