Abstract
Mammalian neuronal stem cells produce multiple neuron types in the course of an individual’s development. Similarly, neuronal progenitors in the Drosophila brain generate different types of closely related neurons that are born at specific time points during development. We found that in the post-embryonic Drosophila brain, steroid hormones act as temporal cues that specify the cell fate of mushroom body (MB) neuroblast progeny. Chronological regulation of neurogenesis is subsequently mediated by the microRNA (miRNA) let-7, absence of which causes learning impairment due to morphological MB defects. The miRNA let-7 is required to regulate the timing of α′/β′ to α/β neuronal identity transition by targeting the transcription factor Abrupt. At a cellular level, the ecdysone-let-7-Ab signalling pathway controls the expression levels of the cell adhesion molecule Fasciclin II in developing neurons that ultimately influences their differentiation. Our data propose a novel role for miRNAs as transducers between chronologically regulated developmental signalling and physical cell adhesion.
Keywords: differential cell adhesion, Drosophila mushroom body, microRNA let-7, steroid hormone ecdysone, temporal identity switch
Introduction
Cell fate decisions are determined by an activation and repression of lineage-specific genes. In this context microRNAs (miRNAs), small non-coding RNAs that negatively regulate gene expression at the post-transcriptional level, are important factors that maintain the balance between stem cell self-renewal, proliferation and differentiation during embryonic development and adult life (Gangaraju and Lin, 2009). They act in conjunction with transcription factors and epigenetic elements in a regulatory network that is based on feedback-feedforward signalling in order to reduce transcriptional noise and fine-tune gene expression. Tissue-specific miRNAs direct differentiation towards corresponding lineages by suppressing alternative cell fates and ensuring the robustness of cell identity (Ivey et al, 2008; Shi et al, 2010). Under stress and in chronic pathological states, miRNA levels can be misregulated that disrupts tissue regeneration and homeostasis due to miRNA influence on the stem cell proliferation and differentiation program (Christensen and Schratt, 2009; Williams et al, 2009; Leung and Sharp, 2010; Takacs and Giraldez, 2010; Marrone and Shcherbata, 2011). Interestingly, in the Drosophila ovarian germline, miRNAs do not only control stem cell proliferation (Hatfield et al, 2005), but also define developmental, stage-specific requirements for stem cell maintenance and differentiation (Shcherbata et al, 2007a) demonstrating that miRNAs are important components of the temporally and spatially coordinated gene regulation machinery. However, the role of individual miRNAs in the temporal regulation of cell differentiation remains largely unknown.
One of the first discovered miRNAs, let-7 represents a heterochronic gene due to its requirement for the developmental transition between the late larval and adult stages in C. elegans (Reinhart et al, 2000). let-7 is evolutionarily highly conserved among bilaterians (Pasquinelli et al, 2000) and acts as a developmental switch to regulate multiple aspects of cellular processes (reviewed in Ambros, 2011). The mammalian member of the let-7 family, let-7b controls neuronal stem cell proliferation and differentiation, and in C. elegans let-7 is involved in the temporal regulation of neuronal development (Abrahante et al, 2003; Lin et al, 2003; Zhao et al, 2010). Interestingly, in Drosophila the onset of let-7 expression is temporally regulated by hormonal signalling and is coordinated with the progression to the adult state (Sempere et al, 2002, 2003; Bashirullah et al, 2003). let-7 mutants exhibit multiple defects arising during metamorphosis, including defects in the maturation of neuromuscular junctions (Caygill and Johnston, 2008; Sokol et al, 2008). Downregulation of the abrupt (ab) gene encoding a BTB-zinc finger transcription factor that regulates the specificity of neuron-muscle connections by let-7 is essential for the timing of neuromusculature remodelling (Hu et al, 1995; Caygill and Johnston, 2008). Altogether, concurrent studies have identified the Drosophila let-7 miRNA as an evolutionarily conserved (Pasquinelli et al, 2000) ecdysone inducible (Sempere et al, 2002) developmental timer; however, a comprehensive picture of the let-7 signalling cascade has not been provided as yet.
Neuronal diversity is generated by progenitor cells (neuroblasts), which often can produce multiple neuron types throughout the duration of an individual’s development (Pearson and Doe, 2004; Kao and Lee, 2010; Reichert, 2011). Specialized neurons are derived at exact times during development, suggesting a partnership between temporal codes and neuron-intrinsic cell fate determinants (Hirono et al, 2012; Kao et al, 2012; Ulvklo et al, 2012). Hormones are potential candidates for this type of chronological regulation, since they are systemic factors whose release is temporally specified, and they are involved in the coordination of all major developmental steps. In fact, it has been shown that steroid hormones influence human brain organization at critical stages of development, for example, at the early post-embryonic period, puberty and adolescence (McCarthy et al, 2009; Melcangi et al, 2011; Peper et al, 2011). Drosophila mushroom body neuroblasts (MBNs) are similar to mammalian neuronal progenitors in their capacity to generate several closely related neuron types that appear during development with temporal precision (Figure 1A) (Ito and Hotta, 1992; Lee et al, 1999).
Figure 1.
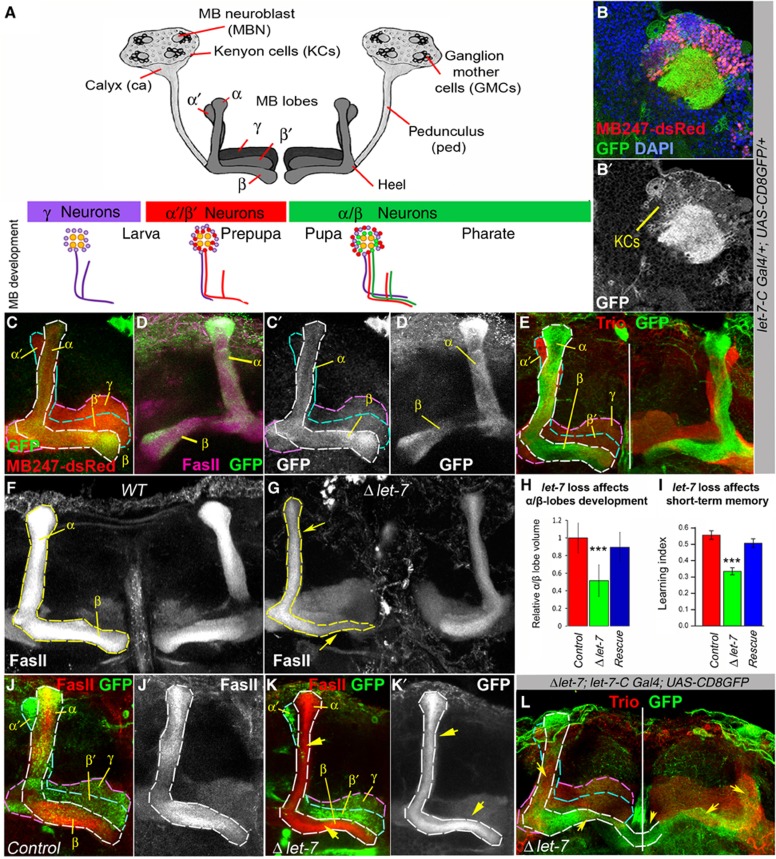
let-7 miRNA is expressed in α/β MB lobes and its loss results in morphological and behavioural defects. (A) Schematic view of the Drosophila mushroom bodies (MBs), which are paired structures originating from four identical MB neuroblasts (MBNs). Within MB cell body cluster, proliferating MBNs give rise to ganglion mother cells (GMCs) that divide one more time to generate intrinsic MB neurons or Kenyon cells (KCs). There are three main types of KCs that each form distinct lobes (γ, α′/β′ and α/β), which connect to the calyx via the pedunculus (ped). The scheme below depicts subsequent birth of γ, α′/β′ and α/β neurons during different stages of Drosophila development. (B) In MBs, let-7-C is detected in a subset of KCs and in the calyx (ca). (C) In the 3- to 6-day-old adult Drosophila brain, high let-7-C expression levels (visualized here by GFP expression, green) are detected in α/β and very weak levels in γ MB lobes; dsRed marks all MB lobes (γ, α′/β′ and α/β). (D) let-7-C expression pattern also coincides with Fas II, which is a marker for α/β lobes. (E) let-7-C expression is not detected in α′/β′ lobes marked with Trio. (F, G) Loss of the miRNA let-7 results in abnormally developed α/β MB lobes in comparison to OregonR control (F). Note the ‘slim-α/β-lobe’ phenotype (G). (H) The volume of α/β lobes is significantly decreased in let-7-deficient brains, which can be rescued via exogenous expression of one copy of let-7 (n=17 MB lobes for Control, n=18 for Δlet-7 and n=8 for Rescue; Supplementary Table 1). (I) Absence of let-7 causes short-term memory impediment, which can be rescued via re-introducing a copy of let-7 (n=16 experiments; Supplementary Table 2); ***P⩽0.001. (J–K) Induction of MARCM clones at the first instar larval stage shows underdeveloped let-7 clonal (GFP positive) α/β lobes as a result of let-7 depletion in MB neurons. Compare outlined α/β lobes in (J) (Control, hsFlp UAS CD8GFP; tubGal80 FRT 40A/FRT 40A; tubGal4/+) and (K) (Δlet-7, hsFlp UAS CD8GFP; tubGal80 FRT 40A/let-7 miR-125 FRT 40A; tubGal4/P{W8, let-7-CΔlet-7}). (L) In let-7 mutants, α/β neurons migrate into α′/β′ lobe as can be seen by co-localization of Trio (red) with let-7-C>GFP (green). Yellow arrows show morphological defects in α/β lobes, white vertical line marks midline in (E) and (L), α/β MB lobes are visualized by the presence of let-7-C Gal4; UAS-CD8GFP (C–E), γ, α/β marker Fas II (D, F, G, J–K), and the absence of γ, α′/β′ marker Trio (E, L).
Here we show that in the post-embryonic Drosophila brain, cell fate of late-born neurons in the mushroom body (MB), a brain region critical for olfactory learning and memory (Heisenberg, 2003; Fiala, 2007) is regulated by the miRNA let-7 that is expressed in response to developmentally regulated steroid pulses. More specifically, ecdysteroid-induced miRNA let-7 controls the neuronal switch from α′/β′ to α/β neurons that occurs at the prepupal to pupal stage, which is one of the many changes occurring at this developmental transition regulated by ecdysone signalling. let-7 is required cell autonomously for proper differentiation of the last-born α/β neurons and its deficiency leads to α/β lobe morphological defects that affect olfactory learning and memory. The cellular effect of steroid hormone-induced let-7 expression is a modulation of levels of the cell adhesion molecule Fasciclin II (Fas II) in differentiating neurons partially via a post-transcriptional regulation of the transcription factor Abrupt (Ab) that we show to be a key factor for establishing α′/β′ neuron identity. The differential adhesion hypothesis (Steinberg, 1975) helps to explain how neurons express different levels of cell adhesion proteins cluster and form complex internal brain structures, for example, Drosophila MBs. Taken together, our data demonstrate that the miRNA let-7 is a steroid hormone-dependent cell fate determinant serving as a temporal code along with spatially controlled lineage cues to specify neuronal cell fate.
Results
Loss of let-7 miRNA affects MB morphology and causes learning defects
To analyse a potential role of let-7 in Drosophila neurogenesis, we first characterized its expression pattern in the adult Drosophila brain. Like in vertebrates, let-7 is transcribed as a part of a polycistronic locus (let-7-Complex, let-7-C), which besides let-7 contains two additional miRNAs, miR-100 and miR-125 (Sempere et al, 2003; Prochnik et al, 2007). Using a transgenic strain that expresses Gal4 under control of the intrinsic let-7-C promoter, broad expression was detected in the optic lobes and central brain structures, for example, the antennal lobes (primary olfactory neuropils), the central complex (a neuropil involved in locomotor and visually guided behaviour) and in parts of the MBs, a brain centre required for associative olfactory learning (Heisenberg, 2003; Fiala, 2007) (Supplementary Figure S1A–D; Figure 1A–E). The same broad expression pattern in the adult brain was detected by in situ hybridization using let-7 locked nucleic acid (LNA) probe (Supplementary Figure S1E and F).
Similarly to mammalian cortical neurons, which are generated in an orderly progression with cells of the deepest layers born first, followed by cells of the middle and last the upper layers (McConnell, 1995; Leone et al, 2008), the Drosophila MBs consist of three main types of neurons with distinct axonal projection patterns (γ, α′/β′ and α/β lobes, Figure 1A and C) that are generated by identical type I neuroblasts (MBNs, Figure 1A) (Ito and Hotta, 1992; Lee et al, 1999). Our detailed expression analysis showed that let-7-C is strongly expressed in α/β and faintly in γ, but is absent from α′/β′ lobes (Figure 1C–E). Since the different MB lobes are constituted of different classes of intrinsic neurons that are sequentially specified during post-embryonic development (Lee et al, 1999) and the miRNAs let-7 has been shown to regulate developmental timing (Caygill and Johnston, 2008; Sokol et al, 2008), we hypothesized a role for this miRNA in the temporal regulation of cell fate determination of particular classes of MB neurons.
Interestingly, let-7 loss of function (Δlet-7 or let-7-CGK1/let-7-CKO1; P{W8, let-7-CΔlet-7}) resulted in the appearance of improperly developed α/β MB lobes; on average, their volume was almost two times smaller than in controls (Figure 1F–H; Supplementary Table 1). This morphological let-7 defect can be restored via exogenous expression of a wild-type let-7 construct (P{W8, let-7-C; let-7-CGK1/let-7-CKO1}, Figure 1H and Supplementary Table 1).
Since the α/β lobes have been shown to be required for the retrieval of long- and short-term memory (McGuire et al, 2001; Akalal et al, 2011), we tested whether the loss of miRNA let-7 has an effect on learning and memory. We performed an associative olfactory conditioning assay (Tully and Quinn, 1985) and observed a significantly reduced olfactory short-term memory in let-7 loss of function mutants, which can be rescued via exogenous let-7 expression (P{W8, let-7-C}; let-7-CGK1/let-7-CKO1, Figure 1I and Supplementary Table 2). The flies’ avoidance of the electric shock is not significantly different between the let-7 mutants and controls (Supplementary Table 2). Since let-7 is also expressed in the antennal lobes we do not exclude potential impairments in olfactory processing. However, the flies’ behavioural response towards the two odorants used for the learning assay is also not significantly different between the let-7 mutants and the controls (Supplementary Table 2), indicating that the pronounced learning phenotype cannot be attributed to a possible impairment of olfactory processing only. These data provide the first evidence for a role of a particular miRNA for the proper development and function of MBs.
Moreover, expression of miRNA let-7 in MB neurons is required cell autonomously for α/β lobe formation, since induction of let-7 loss-of-function clones specifically in MB neurons (hsFlp UAS CD8GFP; tubGal80 FRT 40A/let-7 miR-125 FRT 40A; tubGal4/P{W8, let-7-CΔlet-7}) using MARCM technique (Lee and Luo, 2001) also resulted in the underdeveloped α/β lobe phenotype (Figure 1J–K). On the contrary, let-7-deficient γ lobes developed and remodelled appropriately (Figure 1K; Supplementary Figure S2B and C). The underdeveloped α/β lobe phenotype can result from the requirement for the let-7 miRNA for the neuronal cell survival or differentiation. To discriminate between these possibilities, we counted the number of apoptotic cells in wild-type and let-7 mutant brains and found no significant difference in the amount of Caspase 3-positive cells per MB cell body cluster (6.80±4.16 and 6.40±4.64 for Oregon R and Δlet-7, respectively; P=0.92, n=5 brains per each genotype). However, when we analysed the distribution of let-7-C expressing neurons that normally form α/β lobe marked by the absence of Trio (Figure 1E), we found that in let-7 brains, a part of these neurons projects into α′/β′ lobe and co-localizes with Trio (Figure 1L). These data suggest that let-7 may promote transitions in MB temporal identity.
let-7 expression in MBs is temporally regulated by ecdysteroids
To further test whether let-7 regulates a temporal aspect of chronological MB neuron differentiation, we analysed its expression pattern in MB cell clusters during different developmental stages. Both the let-7-C expression (marked with membrane GFP, Figure 2A) and the miRNA let-7 expression (detected using fluorescent LNA in situ hybridization, Figure 2B) show no signal during early (larval and prepupal) stages of post-embryonic development when early-born γ and α′/β′ neurons are generated (Zhu et al, 2006), but let-7 is present only at later (pupal) stages when late α/β lobe neurons are born. Many steps of Drosophila morphogenesis are dependent on the steroid hormone ecdysone (Riddiford, 1993) and previous in vitro and biochemical studies reported that expression of let-7 miRNA is conditionally dependent on ecdysteroid signalling (Sempere et al, 2002; Garbuzov and Tatar, 2010). Since a cell context-specific role of ecdysone signalling for the control of let-7 has not been shown so far, we analysed the chronological activity of ecdysone signalling in MBs and its influence on let-7 expression.
Figure 2.
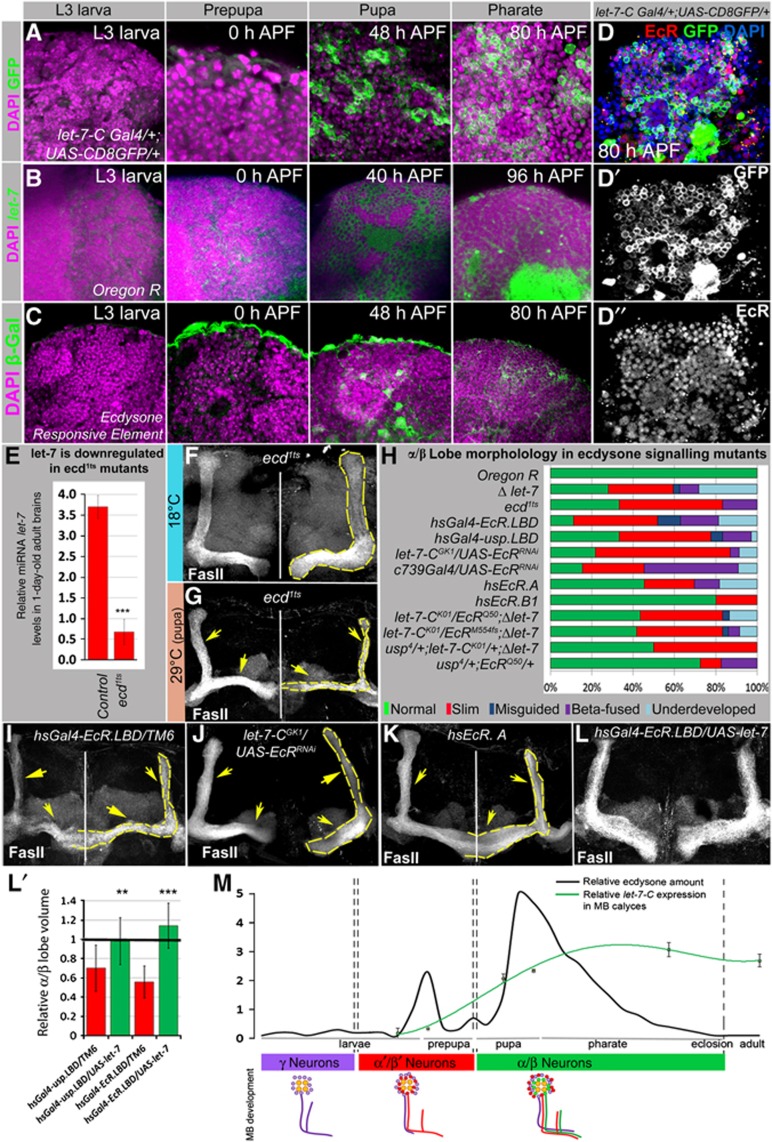
A developmentally regulated ecdysone pulse induces let-7-C expression and affects MB α/β lobe development. (A) let-7-C expression (let-7-CGK1; UAS-CD8GFP:UAS-nLacZ), (B) let-7 LNA in situ hybridization and (C) active ecdysone signalling (EcRE.lacZ) in MB cell body clusters of wild-type animals at different stages of development visualized by anti-GFP, fluorescent in situ hybridization, and anti-β-Gal staining, respectively. Strong let-7-C and let-7 miRNA expression is detected at the pupal stage, which correlates with the ecdysone-signalling peak. Note that let-7-C and let-7 miRNA expression after being induced continues through pharate and adult stages (as seen in Figure 1B–E), while the activity of ecdysone signalling starts at prepupa, peaks at pupa and diminishes at later stages (Supplementary Figure S3). (D–D″) Ecdysone receptor expression pattern shows co-expression of EcR and let-7 in MB cell body clusters. (E) The bar graph represents decreased let-7 expression levels in the newly eclosed adult brains of ecd1ts mutants kept at the restrictive temperature during metamorphosis (n=3 experiments; Supplementary Table 3). (F, G) Abnormally developed α/β lobes were observed in ecd1ts mutants kept at 29°C at the pupal stage (G, yellow arrows), compared to control ecd1ts animals kept at 18°C (F). (H–K) Graph showing the frequency of α/β morphology defects in inducible ecdysone signalling mutants stimulated at the pupal stage (hsGal4-EcRLBD, hsGal4-uspLBD, hs EcR.A, hs EcR.B1; I, K). Note that overexpression of EcR.B1 isoform that has been shown to be specific for γ lobe remodelling (Lee et al, 2000) had less severe effects than overexpression of EcR.A isoform. Similar α/β phenotypes were observed due to cell autonomous downregulation of EcR in α/β lobes (let-7GK1; EcRRNAi and c739Gal4; EcRRNAi), as well as a result of transheterozygous genetic interactions between miRNA let-7 and ecdysone pathway mutants (let-7-CK01/EcRQ50;P{W8, let-7-CΔlet-7}/+, let-7-CK01/EcRM554fs;P{W8, let-7-CΔlet-7}/+, let-7-CK01/usp4;P{W8, let-7-CΔlet-7}/+, usp4/+;EcRQ50/+ (H, J), see also Supplementary Table 5). (L–L′) This defect can be rescued by overexpression of let-7 (hsGal4-EcR-L.B.D./UAS-let-7, hsGal4-usp-L.B.D./UAS-let-7, compare (I) and (L), see Supplementary Table 4). (M) Schematic drawing illustrating developmentally regulated ecdysone pulses (Riddiford, 1993), chronology of different MB lobes formation (Lee et al, 1999), and let-7 complex expression in MB cell body clusters during development. let-7-C expression was analysed via measuring relative GFP intensity in the MB cell body clusters of different stages let-7-CGK1; UAS-CD8GFP animals using ImageJ: L3 larva 0.18±0.19; 0 h APF 1.00±0.13; 48 h APF 2.08±0.16; 58 h APF 2.35±0.05; 80 h APF 3.09±0.24; 1- to 3-day old adults 2.7±0.22; at least three brains for each stage were analysed. Note the correlation between the ecdysone activity, initiation of let-7-C expression in MB neurons, and a transition of their differentiation into α/β type neurons. **P⩽0.01, ***P⩽0.001.
Ecdysone receptor (EcR) expression was detected in MB neuronal cells, including let-7 expressing neurons (Figure 2D) and ecdysone pathway activity, as measured by ecdysone response element expression was found in MB cell body clusters just prior to the onset of let-7-C expression (Figure 2C; Supplementary Figure S3). In ecdysoneless temperature-sensitive mutants (ecd1ts) whose ecdysone production is abolished at restrictive temperatures, let-7 miRNA levels were significantly downregulated (Figure 2E; Supplementary Table 3). These data suggest that the timing of let-7 expression specifically in the brain is dependent on ecdysone levels. Multiple tissues and different cell types respond to systemic hormonal signalling in a particular manner that depends on the differential expression of various components (such as specific receptors, transcriptional co-factors and co-repressors) in individual cell types (Konig et al, 2011). Interestingly, a premature pulse of ecdysteroids achieved via 20-hydroxyecdysone (20E) addition to the food of early larvae failed to induce precocious let-7-C expression in MBs, implying that the temporal expression of let-7 requires the presence of additional components tailored for the specific developmental stage.
Next, we tested if ecdysone signalling in any way influences the development of α/β MB lobes and observed that a reversible disruption of ecdysone production for 24–48 h during different stages of morphogenesis (third instar larva, prepupa, pupa or pharate) had differential effects on α/β lobe formation (Supplementary Table 4). Animals deprived of ecdysone at the prepupal-pupal stages showed distinct α/β lobe defects showing that ecdysone signalling is required for α/β lobe morphogenesis during these stages, which coincides with the timing of the neuronal cell fate transition from α′/β′ to α/β (Figure 2F and G). Similar phenotypes were observed using dominant-negative mutants for EcR and its co-receptor Usp, as well as overexpression of EcR at the pupal stage (Figure 2H–K; Supplementary Table 5). Ecdysone signalling is required for α/β lobe formation cell autonomously, since the ‘slim α/β lobe’ phenotype was also observed when ecdysone signalling was perturbed in α/β neurons using cell-specific downregulation of EcR via RNAi (Figure 2H; Supplementary Table 5). Importantly, we also observed transheterozygous genetic interactions between let-7 and ecdysone signalling pathway mutants, EcR and usp (Figure 2H; Supplementary Table 5). Moreover, forced expression of let-7 rescues α/β morphological defects caused by temporal ecdysone signalling deficit that was generated by keeping temperature-sensitive dominant-negative EcR and usp mutants at the restrictive temperature during the pupal stage (Figure 2I–L; Supplementary Table 4). These data show that the timing of let-7 induction in Drosophila MBs and the temporal establishment of α/β lobe neuron identity correlate with the highest ecdysone peak (Figure 2M), and perturbation in ecdysone signalling or let-7 deficiency causes morphological defects in the α/β lobes. Taken together, these data imply that ecdysone signalling regulates the specification of the late-born MB neurons via the miRNA let-7 that acts as a temporal cue.
Topographic map of let-7 and its two putative target’s expression in the Drosophila MB cell body cluster
Since miRNAs act as negative regulators of gene expression by inhibiting the translation and/or promoting the degradation of target mRNAs (Pauli et al, 2011), we attempted to find let-7 targets in developing MBs. Based on in silico, literature and experimentally derived data (Supplementary Table 6), we selected Apontic (Apt) and Abrupt (Ab) as initial candidates. Both proteins are putative transcription factors that have been implicated in nervous system development (Hu et al, 1995; Takasu-Ishikawa et al, 2001; Sugimura et al, 2004); moreover Apt was detected to be expressed in adult MBs (Kobayashi et al, 2006) and Ab was confirmed as a let-7 target in vitro and in vivo (Burgler and Macdonald, 2005; Caygill and Johnston, 2008). Most importantly, we examined and detected the expression of these two proteins in larval and pupal MB cell body clusters (Figure 3A–D).
Figure 3.
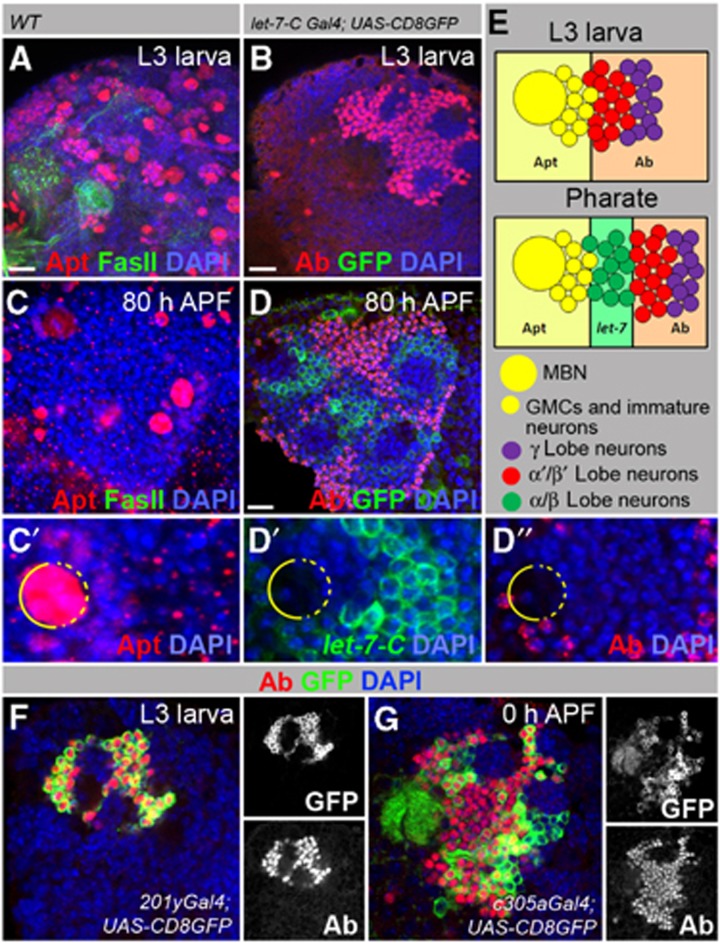
Putative let-7 targets Apt and Ab are expressed in MB cell body clusters. (A, C) In wild-type animals (OregonR), Apt is expressed in the MBNs, GMCs and immature neurons at larval and pupal stages. (B, D) In contrast, the Ab signal is strong in the nuclei of differentiated neurons (α′/β′ and γ lobe neurons). (B) GFP that represents let-7-C expression is clearly absent from MB cell body clusters during the L3 stage (let-7-CGK1; UAS-CD8GFP:UAS-nLacZ). At the pharate stage (80 h APF), neurons located next to GMCs (α/β lobe neurons) express let-7-C but lack Ab (D). (C′, D′, D″) Magnification of the pharate stage MB cell body cluster showing mutually exclusive expression patterns of Apt, Ab and let-7-C. (E) Drawings represent the expression patterns of let-7-C, Apt and Ab in larval and pharate stages. (F, G) Ab is expressed in γ and α′/β′ MB lobes. (F) Larval brain (L3 stage) shows Ab expression in KCs, which form γ lobes marked with GFP driven by the γ lobe-specific driver 201y-Gal4. (G) In the early pupal stage (0 h APF), Ab staining is seen in KCs that form α′/β′ lobes marked with GFP driven by the α′/β′ lobe-specific driver c305a-Gal4. For Gal4 line expression patterns, see Supplementary Figure S5.
We could show that during all post-embryonic developmental stages Apt is expressed in NBs, GMCs and immature neurons (Figure 3A and E; Supplementary Figure S4E). In the pharate brain, only MB NBs continue to divide (Supplementary Figure S4A–C; Siegrist et al, 2010), and Apt staining was restricted to these four NBs and their progeny (Figure 3C; Supplementary Figure S4E). Conversely, Ab marked only terminally differentiated neurons (Supplementary Figure S4D). At earlier stages (third instar larva, L3) Ab-positive neurons were located adjacent to GMCs (Figure 3B and E). These neurons contribute to α′/β′ and γ lobes, as was confirmed using α′/β′ and γ MB-specific Gal4-driver lines and an Ab-specific antibody (Figure 3F and G; Supplementary Figure S5A and B). At later stages (80 h APF), neurons adjacent to GMCs did not stain positively for Ab, but instead expressed let-7-C (Figure 3D; Supplementary Figure S4D). Since let-7-C expression is specific to α/β lobes (Figure 1D and E) we can conclude that last-born neurons forming α/β lobe are Ab negative and let-7 positive. Taken together, these analyses reveal that Apt, let-7 and Ab expression patterns are mutually exclusive (Figure 3E), which prompted us to hypothesize that let-7 might play a role as a negative regulator of apt and/or ab mRNAs.
Abrupt, but not Apontic is a miRNA let-7 target in MB neurons
To test if any of these genes is a let-7 target, we generated populations of clonal cells overexpressing let-7 in larval MB cell body clusters and monitored Apt and Ab protein levels, when compared to non-clonal neighbouring cells and observed that let-7 overexpression decreases Ab (Figure 4A and B), but not Apt levels (Figure 4C). In addition, we analysed Ab and Apt expression patterns in let-7 mutants that, as described above, are characterized by α/β lobe defects and found that Apt expression was not affected in the MB cell body clusters of let-7-deficient flies (Supplementary Figure S4F); however, in the absence of let-7 Ab expression was extended to the nuclei of α/β neurons that are marked by let-7-C Gal4; UAS-CD8GFP (Figure 4D and E; Supplementary Table 7). The number of Ab-positive cells increased 14% due to Ab misexpression in let-7-C Gal4; UAS-CD8GFP marked cells. Since miRNAs normally do not switch ‘on-off’, but just fine-tune gene expression, deletion of let-7 affected Ab levels only to some extent, which indicates that Ab expression is controlled via additional regulators. The presence of Ab-positive cells among the population of late-born neurons marked by let-7-C Gal4; UAS-CD8GFP could affect their identity and be the cause of the undersized α/β lobe phenotype observed in let-7-deficient brains.
Figure 4.
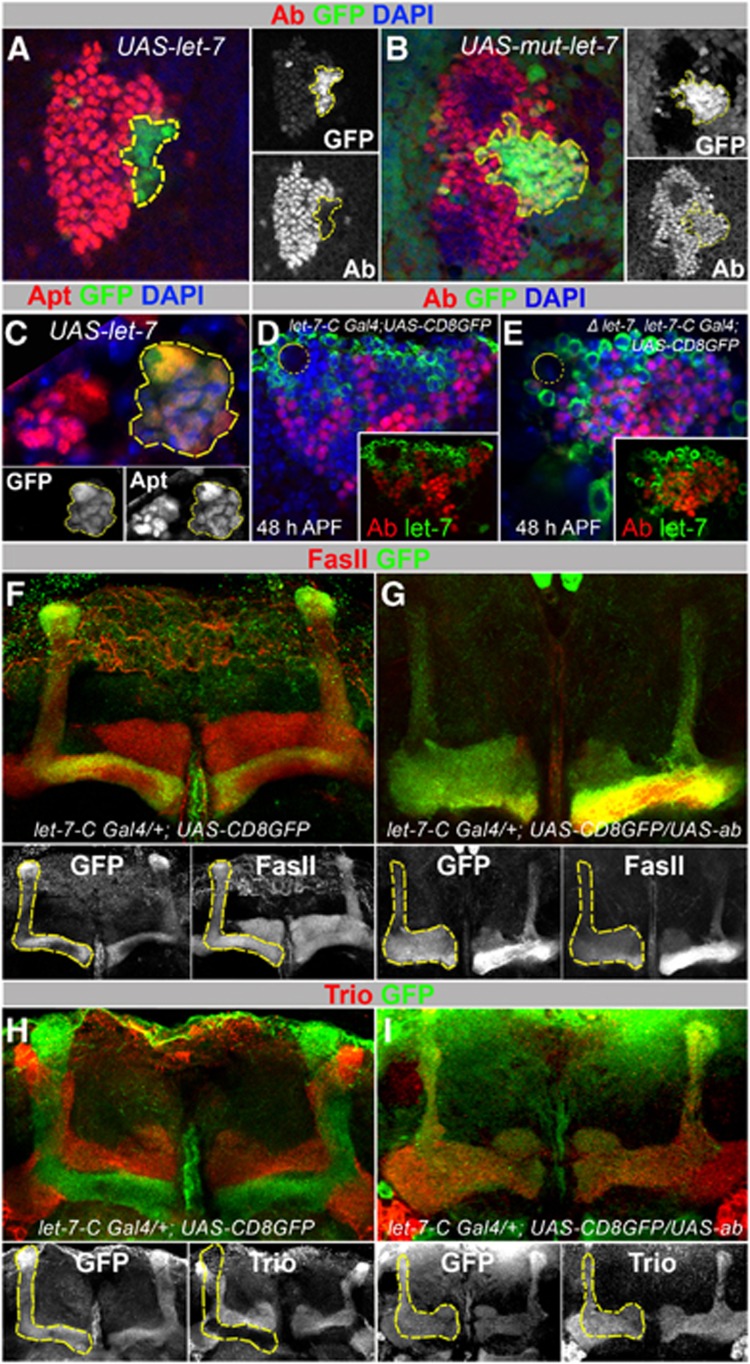
let-7 targets Abrupt in MBs. (A) L3 larval brains with let-7 overexpressing clones (marked with GFP, hsFlp;act>CD2>Gal4;UAS-let-7) show decreased levels of Ab protein levels compared to non-clonal neighbouring cells. (B) Overexpression of the mutant form of let-7 used as a control (hsFlp;act>CD2>Gal4;UAS-mut-let-7) does not affect Ab levels. (C) L3 larval brains with let-7 overexpressing clones do not show significantly changed Apt levels. (D, E) In the let-7 pupal MB cell body cluster (let-7-CGK1/let-7-CK01; P{W8, let-7-CΔlet-7}/UAS-CD8GFP), Ab is detected in let-7-C expressing cells (marked with membrane GFP); compare to control (let-7-CGK1/+; UAS-CD8GFP/+) in (D). On average, 14% more neurons are co-expressing Ab and let-7-C in comparison to control (n=4 MB cell body clusters (11 confocal sections each); Supplementary Table 7). (F, G) Overexpression of Ab in let-7-C expressing neurons causes diminutive α/β lobes. Compare Fas II staining in (F) and (G). (H, I) Overexpression of Ab in let-7-C expressing neurons causes α/β neurons to project and co-localize with Trio-expressing α′/β′ neurons. (A–C) Yellow outlines let-7 overexpressing clones marked with GFP. (D, E) Yellow circles show location of MBNs. (F, G) Yellow outlines let-7-C-expressing cells marked with GFP in control animals and UAS-ab mutants.
Next, to examine if misexpression of Ab would influence the differentiation of α/β neurons, we used the temporally and spatially regulated let-7-C Gal4 driver (Figure 2A) and observed that overexpression of Ab, but not Apt led to α/β lobe defects (Figure 4F–I, Supplementary Figure S4G; Supplementary Table 1). Particularly, α lobes appeared to be more affected indicating possible branching and/or guidance defects that were not observed in let-7 mutants (Figure 4G; Supplementary Figure S6F; Supplementary Table 1), which suggests that forced expression of this putative transcription factor in α/β neurons additionally affects neuron differentiation process. When α/β neurons are forced to express Ab using α/β neuron-specific c739 Gal4 and temporal let-7-C Gal4 drivers, decreased levels of the cell adhesion protein, Fas II are observed (Supplementary Figure S6G and H; Figure 4F and G). Similarly to let-7 mutants (Figure 1L), this also correlates with α/β neurons projecting their axons into α′/β′ lobes, marked by Trio (Figure 4I). These data imply that elimination of Abrupt is required to promote neuronal identity transition and the role of the miRNA let-7 in MBs is to downregulate the BTB-zinc finger protein Ab in order to ensure the transition of MB neuron differentiation from α′/β′ to α/β type during development.
let-7 and Ab are key factors that cell autonomously regulate MB neuronal identity
To prove that let-7 and Ab are temporally required to regulate α′/β′ and α/β neuronal identities in a cell autonomous manner, we performed a chronological lineage analysis using the MARCM technique and considered only single/double cell clones to scrutinize clonal MB neuron identities (Supplementary Figure S2A; Figure 5). When parental control and let-7 mutant clones were induced at third instar larva, we observed 100% of α′/β′ single and double cell clonal neurons in adult brains (Figure 5A and D; Supplementary Table 8). All of the let-7 clonal neurons also projected properly into α′/β′ lobes (Figure 5B–D). However, merely 5% of ab clonal neurons behaved in that manner, instead they migrated into α/β or γ lobes (41 and 54%, respectively) (Figure 5C and D; Supplementary Table 8), this phenotype was even more pronounced in MBN-derived ab clones (Supplementary Figure S6A–E). These data show that the presence of the putative transcription factor Ab is essential for α′/β′ cell fate specification.
Figure 5.
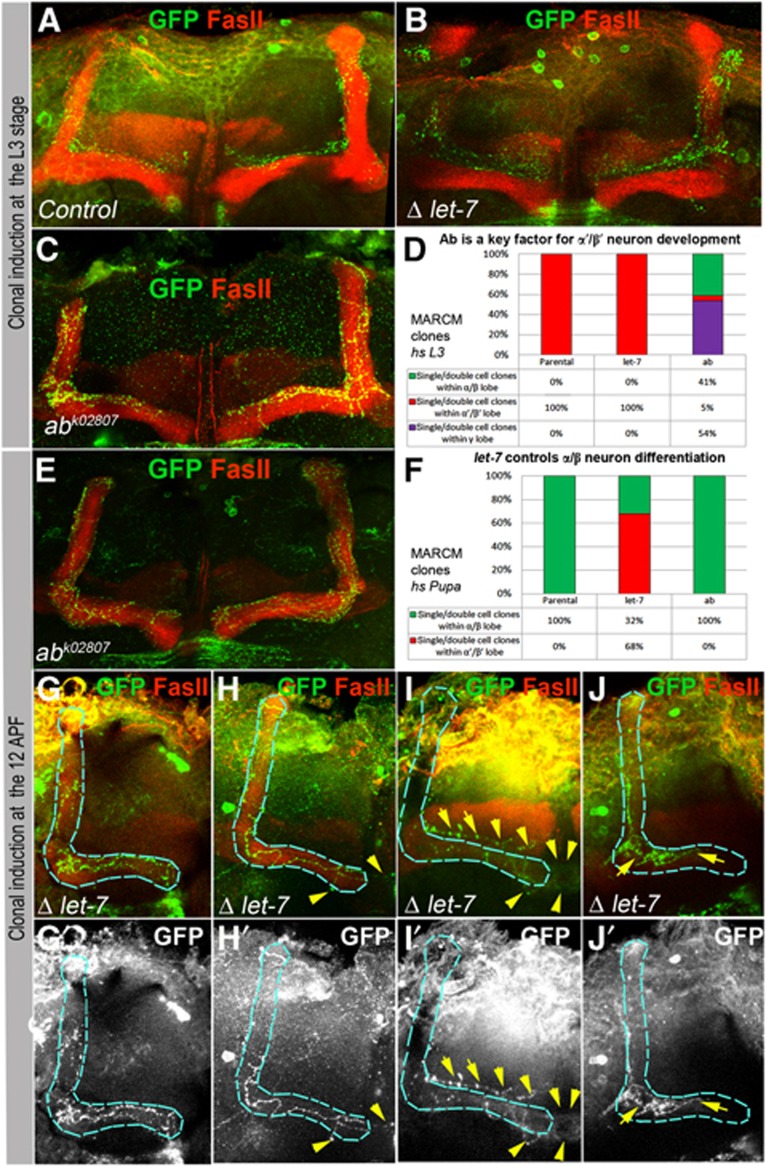
Ab and let-7 are key factors required cell autonomously for MB neuron differentiation. (A, B) At the L3 larval stage induction of clones in MBs resulted in appearance of parental control and let-7 single/double GFP marked α′/β′ neurons in (Control, hsFlp UAS CD8GFP; tubGal80 FRT 40A/FRT 40A; tubGal4/+ and hsFlp UAS CD8GFP; tubGal80 FRT 40A/let-7 miR-125 FRT 40A; tubGal4/P{W8, let-7-CΔlet-7}). (C) While control and let-7 neurons differentiate into α′/β′ neurons, which is in agreement with the time of clonal induction, a part of ab neurons project into α/β neurons which is distinguished here due to co-localization of GFP marked ab neurons with an α/β neuron marker Fas II. (D) The frequency of occurrences of different clonal MB neuronal subtypes induced at L3 (see also Supplementary Table 8). (E) Contrarily, ab loss of function, due to clonal induction at the pupal stage when α/β neurons are specified (12 h APF), is nonessential for differentiation of these neurons; all clonal neurons correctly send their axonal projections to form α/β lobes. (F) The frequency of the properly formed α/β neurons induced at pupal stage (see also Supplementary Table 8). (G-J) let-7 deficiency affects α/β neuron differentiation. While some let-7 neurons form correct axonal projections (G), most of them exhibit random walk and midline crossing (H), projection into α′/β′ lobe (I), and precocious termination (J). Blue outlines α/β lobes, yellow arrows point neuronal defects.
When mutant clones were induced during pupal stages, 100% of ab-deficient neurons chose the correct cell identity that correlated with the time of clonal induction and formed proper α/β lobes (Figure 5E and F; Supplementary Table 8), confirming that Ab is dispensable for α/β cell fate determination. On the contrary, miRNA let-7 absence severely affected α/β neuronal differentiation: only one third of let-7 last-born neurons properly selected α/β identity, while the majority (68%) did not switch their identity and remained as previously originated α′/β′ lobe neurons (Figure 5F and I; Supplementary Table 8). In addition, most let-7 mutant neurons had misguided axonal projections (premature termination, midline crossing or random walk) (Figure 5H–J; Supplementary Table 8). These data show that the temporal expression of the let-7 miRNA is required cell autonomously to regulate α/β specification and differentiation.
let-7 modulated regulation of the cell adhesion molecule Fas II allows for neuron differential specification
Next, we wanted to understand what is the origin of the neuronal identity transition. Cell adhesion molecules are essential factors promoting the direction of neuronal cell differentiation, including neurogenesis, axon pathfinding and brain structure compartmentalization. Particularly, the homophilic cell adhesion molecule Fas II is expressed at different levels in the γ and α/β, but not α′/β′ lobes (Figure 1D and F) and its precise expression is essential for proper MB development (Whitlock, 1993; Kurusu et al, 2002; Fushima and Tsujimura, 2007). In let-7 mutant brains, Fas II mRNA levels were significantly reduced (Figure 6A; Supplementary Table 9) and exogenous expression of let-7 in all MB lobes resulted in Fas II misexpression in γ and α′/β′ lobes (Figure 6B and C). This effect can also be seen at the single-cell level: α′/β′ neurons overexpressing let-7 also have elevated Fas II, demonstrating that let-7 miRNA can modulate Fas II expression (Figure 6D). Moreover, let-7 genetically interacts with Fas II in the process of MB neurogenesis, since let-7/Fas2 transheterozygous mutants displayed the ‘slim α/β lobe’ phenotype (Figure 6E). Additionally, reduction of ab could fully rescue let-7 mutant phenotypes (Figure 6G; Supplementary Table 1) and partially restore normal Fas II mRNA levels (Figure 6A; Supplementary Table 9), suggesting that Fas II is regulated via the transcription factor Abrupt.
Figure 6.
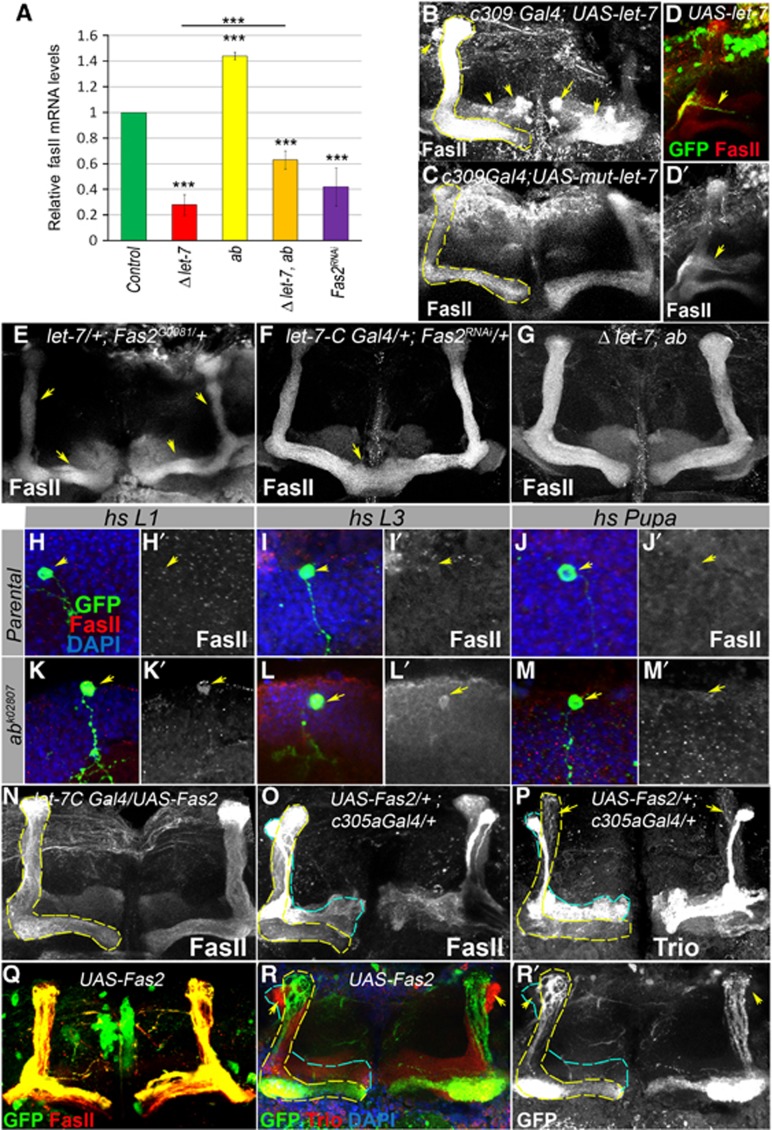
Establishment of Drosophila MBs depends on the level of the cell adhesion molecule Fas II that is temporally regulated by let-7 via the transcription factor Abrupt. (A) Fas II mRNA levels are significantly decreased in the brains of let-7 mutants and increased in Ab hypomorphic mutants. Fas II decrease in let-7 mutants can be partially relieved via introduction of hypomorphic ab mutations. Downregulation of Fas II using Fas II RNAi driven by the let-7 promoter (let-7-CGK1; Fas2RNAi) results in a 60% reduction of Fas II mRNA levels (***P⩽0.001); see also Supplementary Table 9). (B) Overexpression of let-7 miRNA in all MB lobes (c309-Gal4; UAS-let-7, for the c309-Gal4 expression pattern, see Supplementary Figure S5) increases Fas II levels and induces abnormal Fas II expression in γ and α′/β′ lobes (arrows) in comparison to control (C, c309-Gal4; UAS-mut-let-7; Supplementary Table 10). (D) Single GFP marked neuron overexpressing miRNA let-7 (hsFlp; act>CD2>Gal4 UAS-GFP/UAS-let-7; L2 stage clonal induction) has an elevated level of Fas II. (E) Epistatic interaction between let-7 and Fas2 mutants results in the ‘slim α/β lobe’ phenotype (see also Supplementary Table 5). (F) Reduction of Fas II in let-7 expressing neurons results in the β-lobe fusion phenotypes with a frequency of 50%, n=12 MB lobes (see also Supplementary Table 10). (G) let-7 morphological abnormalities are rescued by reduction of Ab levels achieved using the ab amorphic and hypomorphic allelic combination in a let-7 mutant background (let-7 miR-125, ab1/let-7 miR-125, ab1D), n=12 MB lobes. (H–M) Adult MB cell body clusters containing Control (hsFlp UAS CD8GFP; tubGal80 FRT 40A/FRT 40A; tubGal4/+, H–J) and ab loss of function (hsFlp UAS CD8GFP; tubGal80 FRT 40A/abk02807FRT 40A; tubGal4/+, K–M) MARCM clones. Loss of Ab from the early-born KCs (clonal induction at the stage of L1 or L3 larva) that normally have very faint Fas II staining pattern in MB cell body clusters (H, I) caused upregulation of Fas II protein levels (K, L); higher Fas II was not observed when clones were induced in α/β lobe neurons at the 12 h APF stage (M). Arrows point to control or ab mutant clones marked with GFP (H–M) and corresponding Fas II levels (H′–M′). (N) Overexpression of Fas II with let-7-C promoter did not cause noticeable morphological changes in α/β MB lobes, while forced expression of Fas II in α′/β′ lobes (O, P) using c305a Gal4 driver (for the expression pattern, see Supplementary Figure S5) results in abnormal MB morphology resulted from projection of Trio expressing cells into α/β MB lobes. (Q, R) MBN-derived GFP marked neurons overexpressing Fas II (hsFlp; act>CD2>Gal4 UAS-GFP/UAS-Fas2; L2 stage clonal induction) did not project into α′/β′ lobes (marked with arrows, outlined with blue dashed line).
To test this hypothesis, we measured Fas II levels and found that in adult brains of ab hypomorphic mutant the levels of Fas II mRNA are increased (Figure 6A). Interestingly, this Ab-dependant Fas II regulation occurs only in early-born neurons, since ab-deficient neurons born at L1–L3 stages that normally express Abrupt had highly upregulated Fas II protein levels (Figure 6H, I), while deletion of ab at the pupal stage from the normally Ab-negative neurons had no effect on Fas II expression (Figure 6J and M). To additionally ensure the birthdating of clonal cells, we induced MBN-derived ab clones via heat shock in L1 larvae and fed L3 stage animals EdU to mark neurons that are born after EdU treatment. We found that EdU-positive ab clonal cells born at L3 stage express Fas II, while the same EdU-positive non-clonal neurons, that match to α′/β′ neurons based on their birthdate, do not (Supplementary Figure S6I).
To further investigate whether elevated Fas II levels would influence MB neuron differentiation, we overexpressed Fas II using let-7-C Gal4 driver and found that high levels of Fas II do not alter α/β lobe morphology (Figure 6N; Supplementary Table 10). On the contrary, excessive Fas II in α′/β′ neurons has a dramatic effect on α′/β′ lobe shape (Figure 6O and P). Notably, α′/β′ neurons that express Fas II migrate into α/β lobe (arrows, Figure 6P–R, MBN-derived act Gal4; UAS-Fas2 clones induced at L2 and c305a Gal4; UAS-Fas2). Interestingly, we found that not all cell adhesion molecules, for example, DE-Cadherin, affect α/β lobe development (Supplementary Figure S7; Supplementary Table 10), showing that Fas II regulation is rather specific.
These data allow us to conclude that α′/β′ neuron differentiation depends on repression of the cell adhesion protein Fas II by the let-7 target Ab. Nonetheless, a reduction of just one homophilic cell adhesion molecule, Fas II using the chronologically regulated let-7 promoter (let-7-C Gal4; Fas2RNAi, Figure 6F and Supplementary Table 10) did not fully phenocopy the severity of let-7 and ecd1ts MB defects (Figures 1G and 2G; Supplementary Tables 1 and 4), showing that additional factors are involved in the processes of MB neurogenesis. In the process of the α′/β′ to α/β neuronal identity switch, the spatio-temporal steroid-induced miRNA let-7 targets the transcription factor Ab that promotes Fas II expression, which is one of the mechanisms allowing for neuron differential specification.
Taken together, our data demonstrate that during extended neurogenesis let-7 miRNA is required for the chronological establishment of the last-born neurons in the developing Drosophila brain and propose a novel role for miRNAs as transducers between physical cell adhesion and spatiotemporal signalling.
Discussion
The regulation of cell differentiation is a complex process, which requires a coordination of numerous signalling and transcriptional networks that specify cell fate. To maintain normal tissue homeostasis and regeneration, stem cell division and differentiation are likely regulated by local signalling; however, during an organism’s development, maturation and ageing temporal regulation goes into action. The chronological aspect of gene regulation usually involves systemic signalling that stimulates the coordinated response of the whole organism and allows for the diversification of cell types.
Both, in invertebrates as well as in mammals, neuronal progenitor cells are able to generate distinct subtypes of neurons at different times during development. How those temporal identity switches are regulated is an unresolved question of broad biological interest. Here, we show that a developmentally regulated high ecdysteroid hormone pulse initiates the expression of the miRNA let-7 that modulates levels of the cell adhesion molecule Fas II, via the putative transcription factor Ab. Based on our data, we propose a model explaining the mechanism of temporally regulated neuronal identity transition (Figure 7). Temporal cues such as systemic steroid signalling via differentially expressed miRNAs regulate a spatially distributed signalling factor that in turn modulates cell adhesion, putting differential cell adhesion as a key player in fine-tuning the fidelity of neuronal cell differentiation.
Figure 7.
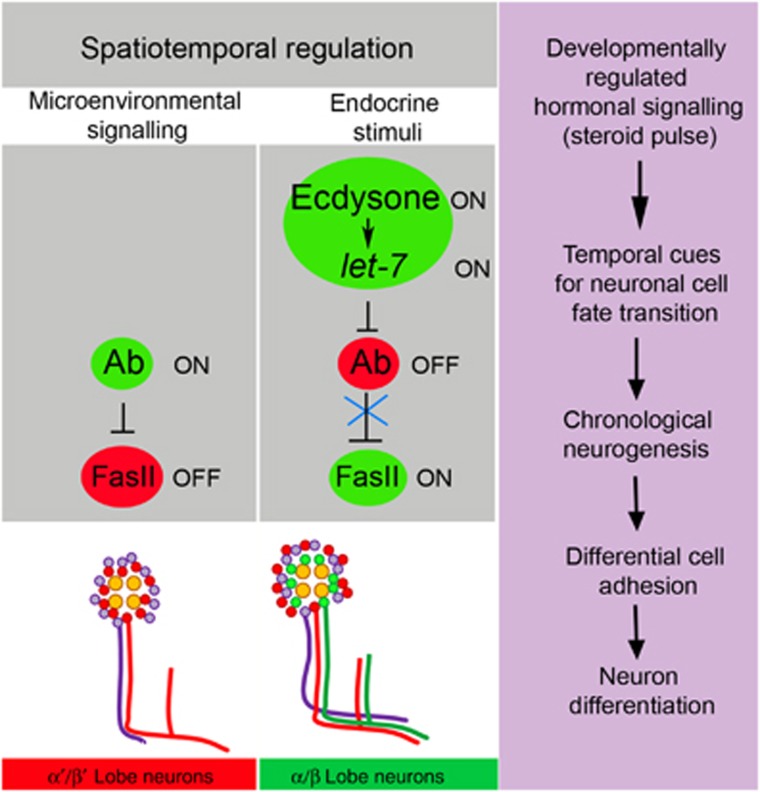
Model of MB neurogenesis regulated by the ecdysone-induced miRNA let-7. Temporal cues such as systemic steroid signalling via the differentially expressed let-7 miRNA regulate a spatially distributed target Abrupt to adjust cell adhesion. During prepupal stages, a cell fate intrinsic determinant Ab is present in the nuclei of early-born α′/β′ neurons. At the pupal stage, the chronologically regulated pulse of ecdysteroids induces let-7 expression. let-7 targets the transcription factor Ab and modulates levels of the cell adhesion molecule Fas II. Since cell adhesion is a critical characteristic of neuronal identity, regulation of cell adhesion levels allows for MB neuron differentiation. Therefore, cell adhesiveness regulated by the ecdysone-let-7 pathway is important for an establishment of neuronal temporal identity.
The differential cell adhesion hypothesis was originally formulated to explain the sorting out and spreading of embryonic tissues based on adhesion energies between cells (Steinberg, 1975). The adhesiveness of the cell is also used to assemble heterotypic cells in order to adopt specific organ anatomies, for example, neurons in the brain are clustered based on the level of cell adhesion molecules, which is essential for establishing brain structure and connectivity during development. Interestingly, the increased cell adhesion of human neurons may have been a crucial factor why the human brain advanced beyond the brains of primates (Prabhakar et al, 2006). Adhesion mechanisms are highly regulated during all stages of an organism’s development and therefore must be tightly coordinated with temporal signalling. Recent studies also connect the accelerated evolution of the human brain with the changes in miRNA expression that shape remodelling of developmental patterns (Somel et al, 2011). Intriguingly, analysis in Drosophila showed that newly evolved genes are often expressed in the brain and all of the MB-positive new genes are expressed in the α/β, but not in more ancestral γ and α′/β′ lobes (Chen et al, 2012). Perhaps, establishment of novel networks between newly originated genes and miRNAs is a general mechanism that might contribute to the phenotypic evolution of behaviour.
Interestingly, both of the predicted let-7 targets that are expressed in MB cell body clusters are associated with the evolutionarily conserved JAK/STAT pathway that plays key roles in multiple developmental and physiological processes in the brain, ranging from the regulation of neurogenesis to memory formation (Bauer et al, 2007; Ngo et al, 2010; Copf et al, 2011). Ab has been shown to be suppressed by the JAK/STAT pathway, whereas JAK/STAT positively upregulates Apt, which via feedback inhibition adjusts JAK/STAT levels (Starz-Gaiano et al, 2008; Jang et al, 2009). JAK/STAT cytokine signalling is well known for its role in the control of immune responses to injury or disease but also in nervous system development, in particular in the regulation of stem cell fate (Bauer et al, 2007). The specificity of JAK/STAT cytokine signalling action depends on the downstream-activated effector genes. For example, the transcription factor Chinmo regulates stem cell self-renewal (Flaherty et al, 2010) and governs neuroblast temporal identity (Zhu et al, 2006). Intriguingly, Chinmo confers all of the transitions in MB temporal identity, except for α/β neurons (Zhu et al, 2006), that, as we show here, is coordinated by the ecdysone peak and let-7 expression. Previous data show that the transcription factor Ab acts as a transdetermination factor that, when misexpressed, is sufficient to potentiate even homeotic arista-leg transformation (Grieder et al, 2007). This implies that regulation of Ab expression should be under strict developmental control to guarantee faithful cell fate determination. Interestingly, a parallel study has shown that let-7-C regulates the temporal identity of MB neurons also via Chinmo (Wu et al, 2012). Both Abrupt and Chinmo belong to the family of evolutionary conserved BTB-zinc finger proteins that are required for a variety of developmental events and the concurrent (Wu et al, 2012) and our studies on the role of let-7 in the Drosophila brain imply that these factors collectively control MB neural differentiation program. Additionally, our data provide evidence that in the developing brain the late-born neuron identity switch is controlled temporally by an ecdysone pulse that activates miRNA let-7 to fine-tune MB neural differentiation program.
Taken together, previous data and our findings demonstrate that neurogenesis is regulated not only by intrinsic factors expressed in progenitors, but also by extrinsic cues and developmentally regulated temporal signals. The interaction between more general developmental and local tissue-specific signalling results in establishment of a robust spatio-temporal pattern of the cell identity specification. Identifying the modulators of temporal codes and the mechanisms of their actions will help to understand how neuronal multiplicity takes place and aid in overcoming age-related obstacles of regenerative therapies that attempt directed neurogenesis.
Materials and methods
Fly strains and genetics
The let-7-CKO1 and let-7-CGK1 mutants contain deletions removing the let-7 complex (Sokol et al, 2008). The let-7-CGK1 transgenic construct also contains Gal4 coding sequences under control of the let-7 complex promoter (let-7-C Gal4). Flies of the P{W8, let-7-C}; let-7-CGK1/let-7-CKO1 genotype (containing the P{W8, let-7-C} rescue transgene for the let-7 complex) (Sokol et al, 2008) will be referred to as Rescue. Animals of the genotype let-7-CGK1/let-7-CKO1; P{W8, let-7-CΔlet-7} (containing the P{W8, let-7-CΔlet-7} transgene that derives from P{W8, let-7-C} but contains a deletion removing the miRNA let-7 sequence) (Sokol et al, 2008), will be referred to as Δlet-7, let-7 miR-125 FRT 40A/CyO (Caygill and Johnston, 2008); abrupt amorphic mutant abk02807 FRT40A/CyO (BDSC). The ecd1ts (BDSC) temperature-sensitive mutation was used to evaluate the influence of ecdysone signalling on MB development. Fly stocks were kept at the permissive temperature (18°C) and to abolish ecdysone production animals of different developmental stages (L3 larva, prepupa, pupa, pharate) were shifted for 1 or 2 days to the restrictive temperature (29°C). As a control, ecd1ts flies were kept at 18°C. In addition to perturb ecdysone signalling pathway hs-GAL4-EcR.LBD, hs-GAL4-usp.LBD, hs-EcR.A and hs-EcR.B1 (BDSC) mutant flies were heat shocked at 37°C water bath three times per day 1 h each for 1 day starting from 12 h APF stage, and brains of the hatched animals were dissected to analyse MB morphology. For the rescue experiments, hs-GAL4-EcR.LBD and hs-GAL4-usp.LBD animals were mated with UAS-let-7/TM6 flies. Their progeny was heat shocked at 37°C water bath three times per day 1 h each for 2 days starting from 12 h APF stage and MB morphology of hs-GAL4-EcR.LBD/UAS-let-7 and hs-GAL4-usp.LBD/UAS-let-7 was compared to MBs of control animals hs-GAL4-EcR.LBD/TM6 and hs-GAL4-usp.LBD/TM6, respectively. For the epistatic interaction analysis EcRM554fs/SM6b, EcRQ50st/SM6b, usp4/FM7a and Fas2G0081FM7c (BDSC) mutants were used.
To test the influence of ecdysone on the onset of let-7-C expression, 1 mM 20-hydroxyecdysone (20E, Sigma-Aldrich) in 5% ethanol was fed to L1–L2 larvae of the let-7-CGK1/UAS-CD8GFP:UAS-nLacZ genotype, then let-7-C expression was analysed in the L3 larval and prepupal brains.
For overexpression experiments, the following lines were used: let-7-CGK1(Sokol et al, 2008), c739-Gal4 (Sinakevitch et al, 2010), c309-Gal4, c305a-Gal4, 201y-Gal4 (BDSC), hsFlp; act>CD2>Gal4 UAS-GFP (Pignoni and Zipursky, 1997), UAS-let-7, UASt-mut-let-7 (Caygill and Johnston, 2008), UAS-ab (BDSC), UAS-tdf (apontic) (Eulenberg and Schuh, 1997), UAS-Fas2 (a gift from Yong Rao), UAS-Fas2RNAi, UAS-shg(DE-Cad)RNAi, UAS-EcRRNAi (VDRC). To generate clones overexpressing miRNA let-7 in L3 larval calyces females of genotypes hsFlp; act>CD2>Gal4 UAS-GFP were mated with males of genotypes UAS-let-7 or UASt-mut-let-7 (control). L1-2 progeny was heat shocked in a 37°C water bath 1 h for 2 consecutive days. To generate clones overexpressing Ab or Fas II, hsFlp; act>CD2>Gal4 UAS-GFP, females were mated with UAS-ab or UAS-Fas2 males. L2 progeny was heat shocked in 37°C water bath for 1 h and adult brains were dissected and analysed.
To visualize the structure of all MB lobes, the MB-247dsRed line was used (Riemensperger et al, 2005). To monitor active ecdysone signalling in the brain EcRE.lacZ (BDSC) flies, kept for 2 days at 29°C, were analysed. The UAS-CD8GFP:UAS-nLacZ line (a gift from Frank Hirth), UAS-CD8GFP on X and third (BDSC) were used to visualize Gal4 expression.
Clonal analysis
We used the MARCM (mosaic analysis with a repressible cell marker; Lee and Luo, 2001) technique to generate GFP marked clonal neuronal cells deficient for miR let-7 or Ab. Females with the genotype hsFlp, UAS-CD8GFP; tubGal80 FRT 40A; tubGal4 were mated to males with the genotype (1) let-7 miR-125 FRT40A/CyO; P{W8, let-7-CΔlet-7} for Δlet-7, (2) abk02807FRT40A/CyO (BDRC) for ab loss of function and (3) FRT40A Parental for the control. In order to generate clones in single γ neurons, progeny from the above crosses was heat shocked for 2 h at 37°C at the first instar larval state; this type of clone induction may also result in all MB lobes being clonal due to recombination in MBNs. In order to generate clones in single late-born neurons, progeny at the third instar larval and at 12–24 h APF stages was heat shocked for the same period of time and the same conditions to generate α′/β′ and α/β lobe clonal neurons, respectively. For the analysis, adult brains from 1- to 3-day-old animals were used. Statistics between MBN-derived versus single/double cell clones were calculated using a two-tailed Student’s t-test and statistics for the frequencies between cell identities of single/double MARCM clonal neurons were calculated using two-way tables and chi-squared test.
Immunohistochemistry
Brains were dissected in PBS and fixed in 4% formaldehyde (Polysciences, Inc.), adult and pupal for 30 min, larval for 15 min. Staining was performed as described (Shcherbata et al, 2007b). The following antibodies were used: mouse anti-Fas II 1:20 (DSHB, marks γ and α/β lobes), anti-Trio 1:20 (DSHB, marks γ and α′/β′ lobes), mouse anti-EcR C1 1:20 (DSHB), rabbit anti-Abrupt 1:500 (Hu et al, 1995), rabbit anti-TDF (Apontic) (Eulenberg and Schuh, 1997), guinea pig anti-Mira 1:1000 (gift from A Wodarz), anti DE-Cadherin 1:50 (DSHB), chicken anti-GFP 1:1000 (Invitrogen), rabbit anti-β-Gal 1:1000 (Invitrogen), anti-Caspase 3 1:200 (Abcam), Alexa 488, 568, or 633 goat anti-mouse, anti-rabbit, anti-guinea pig (1:500, Molecular Probes), goat anti-rat Cy5 (1:250, Jackson Immunoresearch). Images were obtained with a confocal laser-scanning microscope Zeiss LSM700 and processed with Adobe Photoshop.
LNA in situ hybridization
LNA in situ hybridization protocol was modified from the original RNA in situ protocol developed by the Berg laboratory. L3 larvae, pupae and adult brains were dissected in cold 1 × PBS. Brains were fixed in freshly prepared 4% paraformaldehyde (PFA) in 1 × PBS with 0.1% DMSO for 30 min at room temperature and washed afterwards three times in 1 × PBS. After dehydration gradually in 25–50–75–100% ethanol for 10 min each, they were kept at −20°C in ethanol overnight. Brains were then rehydrated in 50–25%-1 × PBS series for 10 min each. Next, brains were treated with Proteinase K (0.05 μg/μl, 50 mM Tris–HCl, 50 mM EDTA) for 1 h. Proteinase K was inactivated by washing two times for 5 min each in 0.2% glycine solution in 1 × PBS followed by 5 min wash in 1 × PBS and two times for 5 min each in PBT. Next, brains were post-fixed in 4% PFA in PBT for 30 min. After fixation brains were washed two times 5 min each in PBT containing 0.1% DEPC followed by additional three washes in PBT for 5 min each. Next, brains were washed with 0.5 × hybridization solution for 5 min (1 × HYB: 50% Formamide, 5 × SSC, 50 μg/ml Heparin, 0.1% Tween-20, 100 μg/ml salmon sperm DNA, 100 μg/ml yeast tRNA). Next, brains were washed with 1 × HYB solution and then prehybridized in 1 × HYB solution for 2–3 h at 54°C on the shaker, then preheated LNA probe was added in 1:1000 dilution ratio in HYB and left overnight on a 59°C shaker. miRCURY LNA probe was ordered from Exiqon company (dme-let-7 #33001-15 5′DIG-ACTATACAACCTACTACCTCA-3′-DIG). After hybridization, brains were washed for 20 min in HYB solution without salmon sperm DNA and yeast tRNA preheated to 59°C followed by washing in 0.5 × HYB (in PBT) for 20 min. Then, brains were washed with PBT five times for 5 min each at 59°C. Next, we blocked samples in Western Block (WB) solution (Sigma Aldrich) for 1 h and incubated with antibodies against DIG conjugated to HRP (Roche, #11207733910) diluted 1:2000 in WB solution for 2 h at the room temperature. Next, FISH-Tyramide Signal Amplification was performed using TSA Cyanine 3 System (Perkin-Elmer©, Inc. #1656398). First, brains were washed in PBT:WB (1:1) solution six times 10 min each and then incubated in streptavidin-HRP solution (diluted 1:100 in PBR:WBR) for 1 h at the room temperature. Brains then were washed in the PBT:WB solution six times 10 min each followed by 10 min wash in PBT and two times in 1 × PBS for 5 min. Then, samples were incubated in solution containing Cyanine 3 Tyramide diluted 1:50 in Amplification Dilutent for 2 h at the room temperature protected from the light. Afterward brains were washed six times 10 min each with 1 × PBS and transferred into mounting medium (70% glycerol, 30% PBS, 3% N-propyl gallate).
For the colour reaction, antibody against DIG (Amersham, 1:2000 in WB solution) was added and samples were placed at 4°C overnight. The next day, brains were washed in PBT four times 20 min each, rinsed once with 0.5 × staining buffer in PBT and two times in 1 × staining buffer (100 mM NaCl, 50 mM MgCl2, 100 mM Tris/HCl pH 9.5, 0.1% Tween-20). The colour reaction was performed by 1:50 NBT/BCIP in staining solution. Colour development was controlled in the dark under a microscope for 10–60 min and stopped with 2–3 times PBT washes before background would develop. Next, tissues were transferred in mounting medium.
Supplementary Material
Acknowledgments
We thank Laura Johnston, Nicholas Sokol, Victor Ambros, Reinhard Schuh, Frank Hirth, Stephen Crews, Andreas Wodarz and Yong Rao for flies and reagents; April Marrone, Andriy Yatsenko, Gerd Vorbrueggen and Roman Shcherbatyy for comments on the manuscript and all members of the Department of Herbert Jäckle and the Shcherbata Laboratory for helpful discussions. This work was supported by the Max Planck Society to MK and HR and the German Research Council via the SPP 1392 (FI 821/2-1) to AF and JB.
Author Contributions: MMK and HRS conceived of and designed the study, conducted experiments, analysed data, and wrote the manuscript. JB and AF designed, conducted and interpreted behavioral experiments; AF discussed the manuscript.
References
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE (2003) The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell 4: 625–637 [DOI] [PubMed] [Google Scholar]
- Akalal DB, Yu D, Davis RL (2011) The long-term memory trace formed in the Drosophila alpha/beta mushroom body neurons is abolished in long-term memory mutants. J Neurosci 31: 5643–5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V (2011) MicroRNAs and developmental timing. Curr Opin Genet Dev 21: 511–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirullah A, Pasquinelli AE, Kiger AA, Perrimon N, Ruvkun G, Thummel CS (2003) Coordinate regulation of small temporal RNAs at the onset of Drosophila metamorphosis. Dev Biol 259: 1–8 [DOI] [PubMed] [Google Scholar]
- Bauer S, Kerr BJ, Patterson PH (2007) The neuropoietic cytokine family in development, plasticity, disease and injury. Nat Rev Neurosci 8: 221–232 [DOI] [PubMed] [Google Scholar]
- Burgler C, Macdonald PM (2005) Prediction and verification of microRNA targets by MovingTargets, a highly adaptable prediction method. BMC Genomics 6: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caygill EE, Johnston LA (2008) Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr Biol 18: 943–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Spletter M, Ni X, White KP, Luo L, Long M (2012) Frequent recent origination of brain genes shaped the evolution of foraging behavior in Drosophila. Cell Rep 1: 118–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen M, Schratt GM (2009) microRNA involvement in developmental and functional aspects of the nervous system and in neurological diseases. Neurosci Lett 466: 55–62 [DOI] [PubMed] [Google Scholar]
- Copf T, Goguel V, Lampin-Saint-Amaux A, Scaplehorn N, Preat T (2011) Cytokine signaling through the JAK/STAT pathway is required for long-term memory in Drosophila. Proc Natl Acad Sci USA 108: 8059–8064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulenberg KG, Schuh R (1997) The tracheae defective gene encodes a bZIP protein that controls tracheal cell movement during Drosophila embryogenesis. EMBO J 16: 7156–7165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala A (2007) Olfaction and olfactory learning in Drosophila: recent progress. Curr Opin Neurobiol 17: 720–726 [DOI] [PubMed] [Google Scholar]
- Flaherty MS, Salis P, Evans CJ, Ekas LA, Marouf A, Zavadil J, Banerjee U, Bach EA (2010) chinmo is a functional effector of the JAK/STAT pathway that regulates eye development, tumor formation, and stem cell self-renewal in Drosophila. Dev Cell 18: 556–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushima K, Tsujimura H (2007) Precise control of fasciclin II expression is required for adult mushroom body development in Drosophila. Dev Growth Differ 49: 215–227 [DOI] [PubMed] [Google Scholar]
- Gangaraju VK, Lin H (2009) MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol 10: 116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbuzov A, Tatar M (2010) Hormonal regulation of Drosophila microRNA let-7 and miR-125 that target innate immunity. Fly 4: 306–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieder NC, Charlafti I, Kloter U, Jackle H, Schafer U, Gehring WJ (2007) Misexpression screen in Drosophila melanogaster aiming to reveal novel factors involved in formation of body parts. Genetics 175: 1707–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H (2005) Stem cell division is regulated by the microRNA pathway. Nature 435: 974–978 [DOI] [PubMed] [Google Scholar]
- Heisenberg M (2003) Mushroom body memoir: from maps to models. Nat Rev Neurosci 4: 266–275 [DOI] [PubMed] [Google Scholar]
- Hirono K, Margolis JS, Posakony JW, Doe CQ (2012) Identification of hunchback cis-regulatory DNA conferring temporal expression in neuroblasts and neurons. Gene Expr Patterns 12: 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Fambrough D, Atashi JR, Goodman CS, Crews ST (1995) The Drosophila abrupt gene encodes a BTB-zinc finger regulatory protein that controls the specificity of neuromuscular connections. Genes Dev 9: 2936–2948 [DOI] [PubMed] [Google Scholar]
- Ito K, Hotta Y (1992) Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev Biol 149: 134–148 [DOI] [PubMed] [Google Scholar]
- Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, Hsiao EC, Schwartz RJ, Conklin BR, Bernstein HS, Srivastava D (2008) MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell 2: 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang AC, Chang YC, Bai J, Montell D (2009) Border-cell migration requires integration of spatial and temporal signals by the BTB protein Abrupt. Nat Cell Biol 11: 569–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CF, Lee T (2010) Birth time/order-dependent neuron type specification. Curr Opin Neurobiol 20: 14–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CF, Yu HH, He Y, Kao JC, Lee T (2012) Hierarchical deployment of factors regulating temporal fate in a diverse neuronal lineage of the Drosophila central brain. Neuron 73: 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Michaut L, Ino A, Honjo K, Nakajima T, Maruyama Y, Mochizuki H, Ando M, Ghangrekar I, Takahashi K, Saigo K, Ueda R, Gehring WJ, Furukubo-Tokunaga K (2006) Differential microarray analysis of Drosophila mushroom body transcripts using chemical ablation. Proc Natl Acad Sci USA 103: 14417–14422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig A, Yatsenko AS, Weiss M, Shcherbata HR (2011) Ecdysteroids affect Drosophila ovarian stem cell niche formation and early germline differentiation. EMBO J 30: 1549–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurusu M, Awasaki T, Masuda-Nakagawa LM, Kawauchi H, Ito K, Furukubo-Tokunaga K (2002) Embryonic and larval development of the Drosophila mushroom bodies: concentric layer subdivisions and the role of fasciclin II. Development 129: 409–419 [DOI] [PubMed] [Google Scholar]
- Lee T, Lee A, Luo L (1999) Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development 126: 4065–4076 [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L (2001) Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci 24: 251–254 [DOI] [PubMed] [Google Scholar]
- Lee T, Marticke S, Sung C, Robinow S, Luo L (2000) Cell-autonomous requirement of the USP/EcR-B ecdysone receptor for mushroom body neuronal remodeling in Drosophila. Neuron 28: 807–818 [DOI] [PubMed] [Google Scholar]
- Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK (2008) The determination of projection neuron identity in the developing cerebral cortex. Curr Opin Neurobiol 18: 28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Sharp PA (2010) MicroRNA functions in stress responses. Mol Cell 40: 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ (2003) The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell 4: 639–650 [DOI] [PubMed] [Google Scholar]
- Marrone AK, Shcherbata HR (2011) Dystrophin orchestrates the epigenetic profile of muscle cells via miRNAs. Front Genet 2: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MM, Wright CL, Schwarz JM (2009) New tricks by an old dogma: mechanisms of the Organizational/Activational Hypothesis of steroid-mediated sexual differentiation of brain and behavior. Horm Behav 55: 655–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell SK (1995) Constructing the cerebral cortex: neurogenesis and fate determination. Neuron 15: 761–768 [DOI] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Davis RL (2001) The role of Drosophila mushroom body signaling in olfactory memory. Science 293: 1330–1333 [DOI] [PubMed] [Google Scholar]
- Melcangi RC, Panzica G, Garcia-Segura LM (2011) Neuroactive steroids: focus on human brain. Neuroscience 191: 1–5 [DOI] [PubMed] [Google Scholar]
- Ngo KT, Wang J, Junker M, Kriz S, Vo G, Asem B, Olson JM, Banerjee U, Hartenstein V (2010) Concomitant requirement for Notch and Jak/Stat signaling during neuro-epithelial differentiation in the Drosophila optic lobe. Dev Biol 346: 284–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G (2000) Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408: 86–89 [DOI] [PubMed] [Google Scholar]
- Pauli A, Rinn JL, Schier AF (2011) Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 12: 136–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ (2004) Specification of temporal identity in the developing nervous system. Annu Rev Cell Dev Biol 20: 619–647 [DOI] [PubMed] [Google Scholar]
- Peper JS, van den Heuvel MP, Mandl RC, Pol HE, van Honk J (2011) Sex steroids and connectivity in the human brain: a review of neuroimaging studies. Psychoneuroendocrinology 36: 1101–1113 [DOI] [PubMed] [Google Scholar]
- Pignoni F, Zipursky SL (1997) Induction of Drosophila eye development by decapentaplegic. Development 124: 271–278 [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Noonan JP, Paabo S, Rubin EM (2006) Accelerated evolution of conserved noncoding sequences in humans. Science 314: 786. [DOI] [PubMed] [Google Scholar]
- Prochnik SE, Rokhsar DS, Aboobaker AA (2007) Evidence for a microRNA expansion in the bilaterian ancestor. Dev Genes Evol 217: 73–77 [DOI] [PubMed] [Google Scholar]
- Reichert H (2011) Drosophila neural stem cells: cell cycle control of self-renewal, differentiation, and termination in brain development. Results Probl Cell Differ 53: 529–546 [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906 [DOI] [PubMed] [Google Scholar]
- Riddiford LM (1993) Hormones and Drosophila development. In The Development of Drosophila melanogaster Bate M, Arias AM (eds), 899–939Plainview: Cold Spring Harbor Laboratory Press [Google Scholar]
- Riemensperger T, Voller T, Stock P, Buchner E, Fiala A (2005) Punishment prediction by dopaminergic neurons in Drosophila. Curr Biol 15: 1953–1960 [DOI] [PubMed] [Google Scholar]
- Sempere LF, Dubrovsky EB, Dubrovskaya VA, Berger EM, Ambros V (2002) The expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster. Dev Biol 244: 170–179 [DOI] [PubMed] [Google Scholar]
- Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V (2003) Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev Biol 259: 9–18 [DOI] [PubMed] [Google Scholar]
- Shcherbata HR, Ward EJ, Fischer KA, Yu JY, Reynolds SH, Chen CH, Xu P, Hay BA, Ruohola-Baker H (2007a) Stage-specific differences in the requirements for germline stem cell maintenance in the Drosophila ovary. Cell Stem Cell 1: 698–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbata HR, Yatsenko AS, Patterson L, Sood VD, Nudel U, Yaffe D, Baker D, Ruohola-Baker H (2007b) Dissecting muscle and neuronal disorders in a Drosophila model of muscular dystrophy. EMBO J 26: 481–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Zhao X, Hsieh J, Wichterle H, Impey S, Banerjee S, Neveu P, Kosik KS (2010) MicroRNA regulation of neural stem cells and neurogenesis. J Neurosci 30: 14931–14936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist SE, Haque NS, Chen CH, Hay BA, Hariharan IK (2010) Inactivation of both Foxo and reaper promotes long-term adult neurogenesis in Drosophila. Curr Biol 20: 643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinakevitch I, Grau Y, Strausfeld NJ, Birman S (2010) Dynamics of glutamatergic signaling in the mushroom body of young adult Drosophila. Neural Dev 5: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol NS, Xu P, Jan YN, Ambros V (2008) Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev 22: 1591–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Liu X, Tang L, Yan Z, Hu H, Guo S, Jiang X, Zhang X, Xu G, Xie G, Li N, Hu Y, Chen W, Paabo S, Khaitovich P (2011) MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol 9: e1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starz-Gaiano M, Melani M, Wang X, Meinhardt H, Montell DJ (2008) Feedback inhibition of Jak/STAT signaling by apontic is required to limit an invasive cell population. Dev Cell 14: 726–738 [DOI] [PubMed] [Google Scholar]
- Steinberg MS (1975) Adhesion-guided multicellular assembly: a commentary upon the postulates, real and imagined, of the differential adhesion hypothesis, with special attention to computer simulations of cell sorting. J Theor Biol 55: 431–443 [DOI] [PubMed] [Google Scholar]
- Sugimura K, Satoh D, Estes P, Crews S, Uemura T (2004) Development of morphological diversity of dendrites in Drosophila by the BTB-zinc finger protein abrupt. Neuron 43: 809–822 [DOI] [PubMed] [Google Scholar]
- Takacs CM, Giraldez AJ (2010) MicroRNAs as genetic sculptors: fishing for clues. Semin Cell Dev Biol 21: 760–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasu-Ishikawa E, Yoshihara M, Ueda A, Rheuben MB, Hotta Y, Kidokoro Y (2001) Screening for synaptic defects revealed a locus involved in presynaptic and postsynaptic functions in Drosophila embryos. J Neurobiol 48: 101–119 [PubMed] [Google Scholar]
- Tully T, Quinn WG (1985) Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol A 157: 263–277 [DOI] [PubMed] [Google Scholar]
- Ulvklo C, MacDonald R, Bivik C, Baumgardt M, Karlsson D, Thor S (2012) Control of neuronal cell fate and number by integration of distinct daughter cell proliferation modes with temporal progression. Development 139: 678–689 [DOI] [PubMed] [Google Scholar]
- Whitlock KE (1993) Development of Drosophila wing sensory neurons in mutants with missing or modified cell surface molecules. Development 117: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR, Olson EN (2009) MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 326: 1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YC, Chen CH, Mercer A, Sokol NS (2012) let-7-Complex MicroRNAs Regulate the Temporal Identity of Drosophila Mushroom Body Neurons via chinmo. Dev Cell 23: 202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Sun G, Li S, Lang MF, Yang S, Li W, Shi Y (2010) MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signalling. Proc Natl Acad Sci USA 107: 1876–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Lin S, Kao CF, Awasaki T, Chiang AS, Lee T (2006) Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell 127: 409–422 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


