Abstract
Accelerating crop improvement in sorghum, a staple food for people in semiarid regions across the developing world, is key to ensuring global food security in the context of climate change. To facilitate gene discovery and molecular breeding in sorghum, we have characterized ∼265,000 single nucleotide polymorphisms (SNPs) in 971 worldwide accessions that have adapted to diverse agroclimatic conditions. Using this genome-wide SNP map, we have characterized population structure with respect to geographic origin and morphological type and identified patterns of ancient crop diffusion to diverse agroclimatic regions across Africa and Asia. To better understand the genomic patterns of diversification in sorghum, we quantified variation in nucleotide diversity, linkage disequilibrium, and recombination rates across the genome. Analyzing nucleotide diversity in landraces, we find evidence of selective sweeps around starch metabolism genes, whereas in landrace-derived introgression lines, we find introgressions around known height and maturity loci. To identify additional loci underlying variation in major agroclimatic traits, we performed genome-wide association studies (GWAS) on plant height components and inflorescence architecture. GWAS maps several classical loci for plant height, candidate genes for inflorescence architecture. Finally, we trace the independent spread of multiple haplotypes carrying alleles for short stature or long inflorescence branches. This genome-wide map of SNP variation in sorghum provides a basis for crop improvement through marker-assisted breeding and genomic selection.
Keywords: Sorghum bicolor, quantitative trait locus, adaptation
Agricultural production and food security in the developing world face numerous threats, particularly in semiarid regions, which are acutely vulnerable to climate change (1). Sorghum [Sorghum bicolor (L.) Moench.] is an important crop species for farmers in semiarid and arid regions because relative to other cereal crops it can sustain high yields where precipitation is low or erratic. Thus, sorghum has become the major cereal crop in semiarid regions and a dietary staple for more than 500 million people, predominantly in sub-Saharan Africa and south Asia (2). Worldwide, sorghum is grown for food (grain and syrup), animal feed, fiber, and fuel in both subsistence and commercial agriculture systems. Because rising temperatures and reduced precipitation due to climate change make some areas unsuitable for maize and rice production, the importance of drought-tolerant crops like sorghum is likely to increase (1). Current breeding priorities in sorghum seek to mitigate climate-dependent stressors, both abiotic (e.g., drought and acid soils) and biotic (e.g., insect pests and fungal diseases) (2). To meet the projected doubling of global food demand over the next few decades in the context of global change, the pace of crop improvement must be accelerated (3).
Sorghum has a wide range of adaptation, and traditional varieties from across Africa and Asia provide a rich source of morphological and physiological traits for crop improvement (4–6). The primary domestication of sorghum occurred near present-day Sudan approximately 10,000 y ago, and diffusion occurred to diverse climates across Africa, India, the Middle East, and east Asia between 8,000 and 1,500 y ago (7). Because of this ancient origin and diffusion, adaptation to local climates and cultural practices is reflected in morphological and physiological variation among and within the five major types (races) of domesticated sorghum (8). For instance, in parts of West Africa where rainy periods are long and erratic, open panicle guinea types are preferred to reduce grain mold and insect damage. Conversely, in parts of South and East Africa where rainy seasons are relatively short and predictable, dense panicle kafir and durra types are preferred to increase grain yield per plant (2). Further natural and human selection has occurred in the United States over the past ∼150 y as temperate and tropical germplasm from Africa and Asia has been adapted for use in combine-harvested commercial agriculture (9).
Genomic analysis of diverse populations is increasingly being used to uncover the genetic basis of complex traits, including agroclimatic traits of crop species. Genome-wide single nucleotide polymorphism (SNP) scans of population genetic parameters in crops have been used to identify loci under selection (10, 11) and dissect quantitative traits (11). In addition, genome-wide association studies (GWAS) have been used to elucidate the genetic basis of agronomic traits in rice (12) and maize (10). Nucleotide diversity scans (13, 14) and association studies (5, 15) have been carried out in sorghum, but the resolution and sensitivity of these studies has been limited by the small number of markers (14). Thus, compared with maize and rice, less is known about the genetic basis of agronomic traits in sorghum. Among the four classical dwarfing loci that have been studied in sorghum for more than 70 y (9), only one has been cloned (Dw3/SbPGP1) (16). Recently, it has become feasible to genotype thousands of markers rapidly and at low cost through the application of barcode multiplexing and high-throughput sequencing (17). To better understand the diversity of sorghum, facilitate the genetic dissection of agroclimatic traits, and accelerate marker-assisted breeding, we characterized 971 sorghum accessions at 265,487 SNPs by using genotyping-by-sequencing (GBS). Here, we describe a genome-wide map of SNP variation, trace patterns of crop diffusion to diverse agroclimatic regions, and use GWAS to identify genes underlying natural variation in agroclimatic traits.
Results and Discussion
Genome-Wide Map of SNP Variation.
To represent the genetic, geographic, and morphological diversity of sorghum, we used 971 accessions from the world germplasm collections, combining three previously defined sorghum diversity panels (Dataset S1) (4–6). The majority of these accessions consist of source-identified landraces or traditional cultivars from across Africa and Asia (Fig. 1A). Of these accessions, 238 are landrace-derived sorghum conversion lines, in which alleles for short stature and early maturity were introgressed into tropical landraces to facilitate the use of tropical germplasm in temperate breeding programs (18). The remainder consists of wild/weedy relatives or elite lines and breeding materials, many of which have unknown geographic origin and/or mixed ancestry. For each accession, we constructed ApeKI-reduced representation libraries and generated a total ∼21 Gbp of sequence on the Illumina Genome AnalyzerIIx/HiSeq by using GBS (Dataset S2) (17). In total, 6.13 million unique 64-bp tags were identified across all sorghum accessions. Eighty-five percent of these tags aligned to the reference sorghum genome (19), and 384,561 putative SNPs were identified. After filtering for local linkage disequilibrium and tag coverage (>10% of taxa), 265,487 SNPs were retained, with an average density of one SNP per 2.7 kbp. Of 27,412 annotated genes in the reference sorghum genome, 72% were tagged by a SNP within the gene and 99% were tagged by a SNP within 10 kb. Importantly, this genome-wide map of SNP variation is of sufficient resolution for GWAS in sorghum, given >100,000 SNPs are estimated to be required (14). Additionally, because of simultaneous SNP discovery and genotyping, this sequencing-based SNP map will have little ascertainment bias and greater power for mapping studies (20).
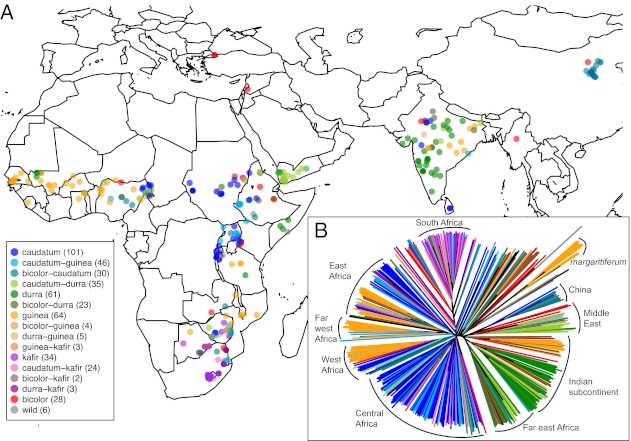
Fig. 1.
Germplasm origin and genetic relationships among worldwide sorghum accessions. (A) Geographic origin for 469 of 971 worldwide accessions, for which source location is known, color-coded by morphological type. (B) Genetic relatedness among the same 469 accessions assessed by neighboring joining method, with the predominant region of origin for each cluster noted, or in one case, the cluster containing guinea margaritiferum types. Worldwide sorghum populations show structuring by morphological type within regions.
Linkage Disequilibrium and Recombination Rates.
Characterizing patterns of linkage disequilibrium (LD) is critical for the design of association studies (21, 22), interpretation of association peaks (12), and the transfer of alleles in marker-assisted selection (23). To characterize the mapping resolution for genome scans and GWAS, we quantified the average extent of LD decay and localized patterns of LD for each chromosome (Table S1 and Fig. S1). On average, LD decays to 50% of its initial value by 1 kb and to background levels (r2 < 0.1) within 150 kb. These LD decay estimates are higher than previously published values in sorghum of 15–20 kb (24) and 50–100 kb (14). This difference may be attributed to low genome coverage of markers and fewer genotypes in previous studies. Because sorghum is a predominantly selfing species, but readily outcrosses, we expect a greater extent of LD than in out-crossing species (25). Accordingly, the extent of LD in sorghum is similar to that in rice (∼75–150 kb) (22), another self-pollinated crop, but much greater than in maize (∼2 kb) (26), which is an outcrosser. Sliding window (1 Mb) estimates of pairwise LD show that telomeric regions have lower LD than centromeric regions (Fig. S1B). This pattern is likely due to higher historical recombination rates in telomeric regions compared with centromeric regions (Fig. 2C). The average recombination rate in sorghum (1.4 ρ/kb) is intermediate relative to recent estimates in plants such as Arabidopsis (0.8 ρ/kb) (21) and maize (2.2 ρ/kb) (26). Based on these results, we expect mapping resolution to range widely across the genome, from single-gene resolution in some telomeric regions to megabase-level resolution near the centromeres.
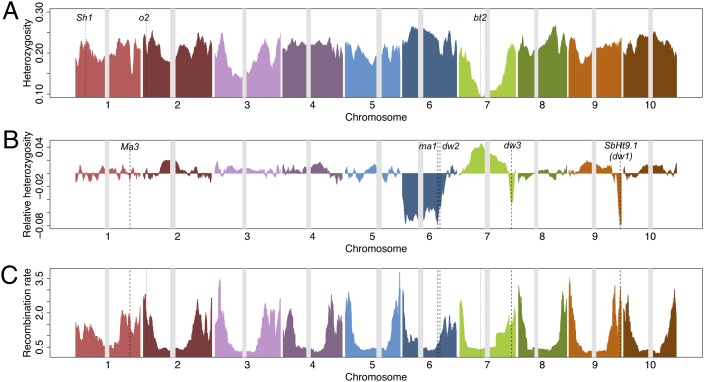
Fig. 2.
Genome-wide patterns of SNP variation. (A) Genome-wide variation of expected heterozygosity for sorghum landraces, smoothed with a 2000 SNP moving average. The location of the centromeres are noted by the gray bars. Dotted vertical lines indicate the Shattering1 (Sh1) domestication locus and two orthologs of starch-related domestication loci from maize (opaque2 and brittle endosperm2) that colocalize with regions of reduced diversity. (B) The relative heterozygosity in sorghum conversion lines compared with landraces. The reduction in heterozygosity in conversion lines is due to introgressions of short stature and early maturity alleles, with known dwarfing (dw) and maturity (ma) loci noted. (C) Genome-wide variation in historical recombination rates averaged across 10 subpopulations. Wider regions of reduced heterozygosity occur in regions near centromeres with low recombination rates.
Population Structure and Geographic Differentiation.
To understand the geographic structuring of genetic diversity, we contrasted genome relatedness among 971 sorghum accessions to the stated location of origin and morphological descriptors in the worldwide germplasm database. The resulting neighbor-joining trees (Fig. 1B) and Bayesian clustering analysis (Fig. S2 and Dataset S3) show population structuring along both morphological type and geographic origin, confirming previous analyses (4, 14) and providing additional insights into the fine-scale patterns of ancestry resulting from crop diffusion. Of the five morphological types, the kafir sorghums that predominate in southern Africa show the strongest pattern of population subdivision relative to other races (Fig. 1B and Fig. S2). Durra type sorghums, found in warm semiarid or warm desert climates of the Horn of Africa, Sahel, Arabian peninsula, and west central India, form a distinct cluster that is further differentiated according to geographic origin. Bicolor types are not notably clustered, except those from China (known as kaoliang), which forms a distinct subgroup and shows genetic similarity to durra types, particularly those from Yemen. Caudatum types, which are primarily found in tropical savanna climates of central Africa, are diverse and show only modest clustering according to geographic distribution. Finally, guinea types, which are widely distributed in tropical savanna climates, show five distinct subgroups, four of which cluster according their geographic origin (far west Africa, west Africa, eastern Africa, and India). A fifth guinea subgroup, which includes guinea margaritiferum types, forms a separate cluster along with wild genotypes from western Africa (Fig. 1B) and may represent an independent domestication (4). In the neighbor-joining analysis but not the Bayesian clustering, Indian guinea types cluster with durra types, likely due to admixture with sympatric Indian durra populations (Fig. S2).
The structure of sorghum populations provides insight into historical processes of crop diffusion within and across agroclimatic zones of Africa and Asia. Diffusion across agroclimatic zones is expected to be rare relative to diffusion within agroclimatic zones (27). Indeed, the patterns of relatedness among sorghum populations suggest that agroclimatic constraints have been at least as important as geographic isolation in shaping the diffusion process (Fig. S3). Among the four phylogenetically supported sorghum types (kafir, durra, guinea, and caudatum), there is the least population structure among caudatum types, which range primarily in the ancestral region of domestication or adjacent areas with similar climate (Fig. 1A). The one geographically structured subpopulation of caudatum is the latitudinal-diffused subpopulation from highland areas in east Africa. Although durra types diffused widely across Africa and Asia, they are restricted to regions with semiarid and desert climates (Fig. S3). This diffusion included kaoliang sorghums, which are likely derived from durra populations of the Middle East, but are found in cold semiarid regions of northern China (Fig. 1 and Figs. S2 and S3). Similarly, although guinea types have diffused over long distances, from western Africa to southeastern Africa and eastern India, they remain restricted to tropical savanna climates. Interestingly, Bayesian clustering analysis suggest that the temperate/subtropical-adapted kafir type is derived from (or at least shares ancestry with) guinea types of east African populations (k = 3 through k = 6; Fig. S2). In this case, the kafir type may represent major phenotypic divergence and genetic bottlenecking resulting from a shift to a contrasting agroclimatic zone.
Genomic Patterns of Nucleotide Variation.
To investigate the genomic signatures of domestication and diversification in sorghum, we quantified genome-wide nucleotide variation across sorghum landraces. Overall, average nucleotide diversity (π) was 0.00037/kb, θ = 0.00017/kb, and Tajima’s D value of 3.6. A scan of expected heterozygosity (He) values across the genome revealed many megabase-scale regions of low heterozygosity including a ∼40-Mb region of reduced nucleotide variation around the centromere of chromosome 7 (Fig. 2A). Of six starch-related genes previously studied as a priori candidates for domestication loci (15, 28), two are found at regions with notably low heterozygosity (Fig. 2A). The starch biosynthesis enzyme brittle endosperm 2 (bt2) gene, which has been shown to be a likely domestication locus in maize (28) and sorghum (15), is at the base of the low heterozygosity region on chromosome 7. The size of this low diversity region and extensive LD (Fig. S1) may be due to low recombination rates in this pericentromeric region (Fig. 2C) or the presence of additional loci under selection. Another a priori domestication candidate from starch metabolic pathways, transcription factor opaque2 (15), is found at the base of a low heterozygosity region on chromosome 2. Lastly, another recently identified domestication locus, the shattering gene Sh1, is found at the edge of a region with moderately, but not strikingly, lower heterozygosity, consistent with the observation that nonshattering alleles are found on at least three haplotypes in domesticated sorghums (29). The large footprints of selection we observe here (up to several megabases) are consistent with the predominance of inbreeding in sorghum. Selective sweeps in out-crossing maize left smaller footprints (<100 kb) (30) than in self-pollinating rice (250 kb to 1 Mb) (31).
In sorghum conversion lines that carry introgressions of early maturity and short stature alleles, we also observed major reduction in heterozygosity in several genomic regions (Fig. 2B). These regions colocalize with previously mapped height [Dw2 (32), Dw3 (16), and Dw1/SbHT9.1 (33)] and maturity loci [Ma1/SbPRR37 (34)] that are recessive in the introgression donor BTx406. In contrast, another classical maturity locus, Ma3/phyB (35), which is wild type in BTx406 and therefore was not under selection during the conversion process, shows no such reduction in heterozygosity. On chromosome 6, the low heterozygosity region extends from approximately 6.6 Mb to 42 Mb (the Ma1/Dw2 locus), suggesting that another height or maturity locus may be localized at 6.6 Mb (SI Results and Discussion). As was seen in the landraces, we find that large LD blocks result when selection occurs in low recombination regions around the centromere (Fig. 2 B and C).
GWAS.
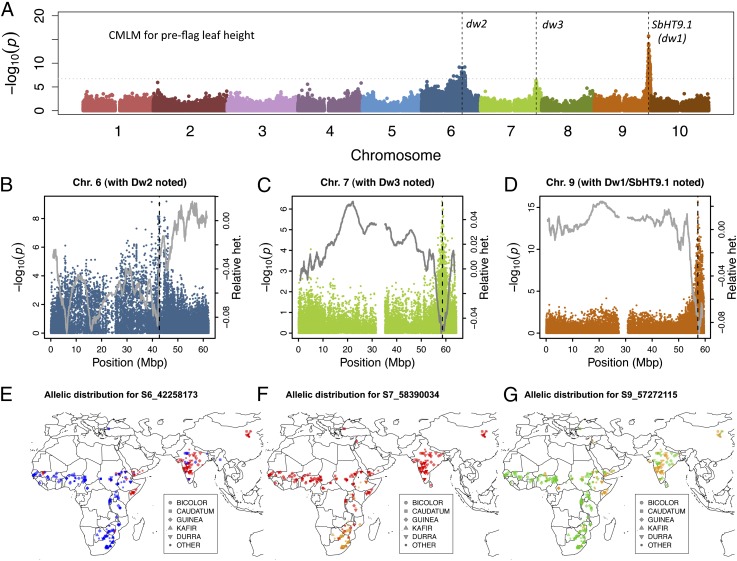
The genome-wide map of SNP variation we generated permits the dissection of complex traits in sorghum by using GWAS. To elucidate the genetic basis of plant height in sorghum, we determined associations between SNPs and plant height components by using data from 336 lines in the sorghum association panel (SAP; Fig. 3 and Fig. S4) (5, 33). Plant height is an important component for many agroclimatic traits such as competitive growth with weeds, resistance to lodging, and, in the case of temperate-adapted grain sorghums, the efficiency of combine harvest (2). Because this panel incorporates a large fraction of sorghum conversion lines with introgressions of dwarfing alleles, we know that much of the variation for height in this panel has a common genetic basis. We identified SNPs associated with total plant height, and two height components: preflag height, which quantifies elongation in the lower portion of the stem, and flag-to-apex length, which quantifies elongation in the upper portion of the stem (Fig. S4). The Dw3/SbPGP1 gene provides a positive control for GWAS (16). It is known that the reduced height of dw3 mutants is due to reduced elongation of lower internodes, therefore we considered preflag leaf height as a measure of lower internode elongation (33). The third most significant association peak for preflag height is found at the dw3 locus [within 12 kb for general linear model (GLM) and 22 kb for compressed mix linear model (CMLM)]. We also refined the mapping location of dw1 and dw2 and identified a possible location of dw4 (SI Results and Discussion).
Fig. 3.
GWAS of preflag leaf height using landraces and introgression lines. (A) Manhattan plot for compressed mixed linear model with known dwarfing loci indicated. (B–D) GWAS peaks for height colocalize with reductions in heterozygosity in the sorghum association panel due to introgression of short stature and early maturity alleles. (E–G) Geographic distribution of alleles at dw2, dw3, and dw1/SbHT9.1, respectively, color-coded by allele (A, green; C, blue; T, red; G, orange).
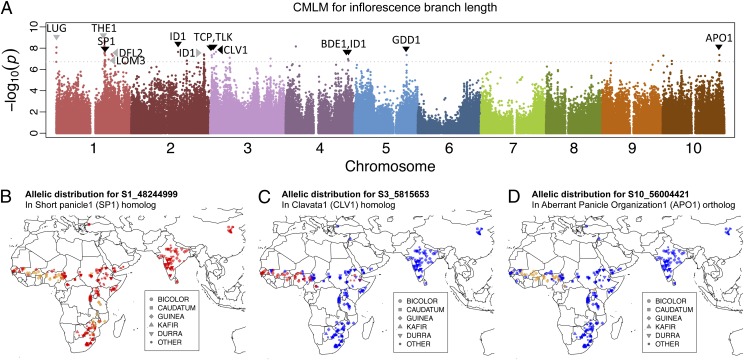
Because the SAP includes a large fraction of converted lines, with large introgressions around height and maturity loci, the previous analysis does not reflect a typical GWAS case. To validate the broader applicability of GWAS in sorghum, we also sought to dissect a trait that was not a target of selection in the sorghum conversion program. Inflorescence architecture is a major agroclimatic trait that, in part, defines the major morphological types in sorghum (8). Moreover, because the genetic basis of inflorescence architecture is well-studied in maize, rice, and Arabidopsis, there are many a priori candidate genes that can be considered to evaluate the mapping approach. Indeed, several of the significant association peaks for inflorescence branch length were located in or near a priori candidate genes for inflorescence architecture, which are homologous to known maize, rice, or Arabidopsis floral regulators (Fig. 4A and Table S2). For example, two peaks are in, and one is near (47 kb), C2H2 zinc finger transcription factors homologous to the classical maize floral development gene INDETERMINATE 1 (ID1) (Fig. 4A and Table S2) (36). Another association peak was found in a sorghum ortholog of Arabidopsis UNUSUAL FLORAL ORGAN (UFO) and rice ABERRANT PANICLE ORGANIZATION 1 (APO1). In rice, apo1 mutants exhibit small panicles and fewer branches (37).
Fig. 4.
GWAS on inflorescence branch-length and geographic distribution QTL alleles. (A) Compressed mixed linear model using first three principal components of population structure as covariates. Candidate genes at peaks are indicated (Table S2), with association peaks in the given gene denoted by black triangles and outside the given gene by gray triangles. (B–D) Worldwide distribution of alleles at three branch-length QTL illustrate the spread of haplotypes associated with variation in branch length, color-coded by allele (A, green; C, blue; T, red; G, orange).
In sorghum, strong population structure among the morphological types presents a challenge for mapping the genetic basis of the inflorescence architecture and other population-associated traits. Although statistical controls for population structure have proven effective here, the effects of population structure can be better addressed by the experimental design of mapping populations, using regional mapping (20) or nested-association mapping (NAM) (38) approaches. Because of the use of sorghum conversion lines, the SAP captures some aspects of the NAM approach. The introgressions reduce the confounding effects of maturity differences in diverse germplasm (5) and, for height and maturity loci, improve mapping power by increasing the frequency of rare alleles (33). However, because the introgressions originate from the same donor line (BTx406), the large blocks of linked noncausative variation reduces the resolution of the association analysis (Fig. 2B). Also, the low diversity around height and maturity loci on chromosomes 6, 7, and 9 may prevent the mapping of QTL for other traits in these regions, especially on chromosome 6 where most of the chromosome has been introgressed in sorghum conversion lines (Fig. 2B). In some cases, therefore, mapping populations without converted lines will be more effective for association studies.
Geographic Distribution of Haplotypes.
To gain further insight into the origin and spread of haplotypes linked to agroclimatic traits, we characterized the geographic distribution for SNPs of interest in 330 source-identified landraces that are independent of the lines used for GWAS. As expected given the role of Ma1/SbPRR37 in temperate zone adaptation (34), a previously identified functional variant (K162N) is near fixation in high-latitude kafir accessions from southern Africa and rare (<5%) elsewhere (Fig. S5). The three major height QTL identified by genome scan and GWAS, dw2, dw3, and SbHT9.1, have distinct allelic distributions across Africa and Asia (Fig. 3 E–G). One of the alleles at the SNP closest to the putative SbHT9.1 causative gene (GA2-oxidase) (39) is found at high frequency (>90%) in East African durra, Indian durra, and Chinese accessions and low frequency (<10%) in all other accessions (Fig. 3G). Likewise, one allele at the Dw2 QTL peak is common in northeast Africa and Asia and rare elsewhere (Fig. 3E). This pattern is consistent with a common genetic basis for semidwarfism in east African and Asian sorghums, conferred by at least two causative polymorphisms, and originating from ancestral East African durra populations. Taken together, the evidence suggests that the classical dwarfing alleles were likely selected from standing variation in durra (dw1, dw2, dw4) and kafir (dw3) landraces (SI Results and Discussion).
The geographic distribution of alleles at inflorescence branch length QTL also reveals evidence of the independent spread of multiple alleles controlling branch length (Fig. 4 B–D). In general, the minor allele associated with longer branches is found at high frequency in West African guinea populations, and in a number of cases it also is found in other geographically distant populations. Interestingly, none of the alleles at top branch length association peaks were restricted to durra accessions (Table S2 and Fig. 4 B–D), suggesting that we were able to identify QTL for the long-branch phenotype in guinea types, but not QTL for the short-branch phenotype in durra types. This result may be attributed to stronger population structuring of durra populations, compared with guinea, which can confound mapping of traits (20). Given the statistical correction for population structure, we did not map QTL underlying the branch length differences among major morphological types, rather we mapped QTL for branch length segregating within the morphological types, which are globally rare but locally common (10) (Table S2 and Fig. 4 B–D).
Conclusion
A better understanding of genetic diversity in sorghum will support in situ conservation efforts, enhance the use of germplasm collections, and guide ongoing collection efforts (40). This genome-wide map of SNP variation will accelerate molecular breeding by expanding the diversity of germplasm accessible to crop improvement programs and increasing the resolution of GWAS, marker-assisted selection, and genomic selection (23, 41). By facilitating crop improvement in locally adapted and locally improved cultivars, genomic analysis of diverse crop germplasm can play an important role in supporting sustainable agriculture in Africa, Asia, and semiarid regions worldwide.
Materials and Methods
Plant Materials.
Diverse sorghum germplasm from worldwide collections were used, combining three diversity panels: the US sorghum association panel (SAP) (5), the sorghum mini core collection (MCC) (6) and the Generation Challenge Program sorghum reference set (RS) (www.icrisat.org/what-we-do/crops/sorghum/Sorghum_Reference.htm). We were able to obtain appropriate plant material for 971 accessions (Dataset S1). The SAP was obtained from GRIN (www.ars-grin.gov). Country of origin, and approximate latitude and longitude for source-identified accessions, were obtained from the SINGER crop germplasm database (www.genesys-pgr.org).
Genotyping by Sequencing.
DNA from MCC and RS lines (5–6 plants per accession) was isolated from 12-d-old seedlings by using the CTAB protocol (42). SAP DNAs were isolated by using DNeasy Plant Mini Kit (Qiagen). Genomic DNAs were digested individually with ApeKI (recognition site: G|CWCG), and 96- or 384-plex GBS libraries were constructed (Dataset S2) (17). DNA sequencing was performed either on the Illumina Genome Analyzer IIx or HiSeq2000. Sequences were mapped to the BTx623 sorghum reference genome (19) by using BWA (43), and SNPs were called with the TASSEL 3.0 GBS pipeline www.maizegenetics.net/tassel/). Sequence tags, 64-bp sequences that included a leading 4-bp C[T/A]GC signature from the cut site, were identified, and tags with at least 10× total coverage were retained. Missing data were imputed with NPUTE (44).
Population Genetic Analysis.
Hierarchical population structure was estimated by using the ADMIXTURE program (45), a model-based estimation of ancestry in unrelated individuals using maximum-likelihood method. The neighbor-joining trees were built and heterozygosity calculated by using the ape package in R (46). Pairwise LD was calculated (r2) separately for each chromosome by using TASSEL 3.0 (47). Recombination rates were inferred by using Bayesian reversible-jump MCMC under the cross-over model of rhomap in LDhat (48) with 1 million iterations and 1 million burn-ins. Rates were estimated separately for each subgroup identified by ADMIXTURE at k = 10. To avoid confounding effects of shared introgressions, SAP lines were not included in recombination and LD analyses.
GWAS.
Published phenotypes for plant height components and inflorescence branch length for the SAP were used for GWAS (33). GWAS was carried out in Genomic Association and Prediction Integrated Tool (49) by using a (i) GLM; or, to control for population structure, (ii) CMLM with population parameters previously determined (50) with the first three principal components as covariates. Bonferroni correction was used to identify significant associations.
Supplementary Material
Acknowledgments
We thank two anonymous reviewers for helpful comments. We also thank National Science Foundation for providing funds to carry out this research under Basic Research to Enable Agricultural Development Project IOS-0965342. This work was also supported by Department of Agriculture-National Institute of Food and Agriculture Plant Feedstock Genomics for Bioenergy Program Grant 2011-03502 (to S.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Sequence Read Archive database, www.ncbi.nlm.nih.gov/sra (accession no. SRA062716).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215985110/-/DCSupplemental.
References
- 1.Lobell DB, et al. Prioritizing climate change adaptation needs for food security in 2030. Science. 2008;319(5863):607–610. doi: 10.1126/science.1152339. [DOI] [PubMed] [Google Scholar]
- 2.National Research Council 1996. Lost Crops of Africa: Volume I: Grains (Natl Acad Press, Washington, DC)
- 3.Foley JA, et al. Solutions for a cultivated planet. Nature. 2011;478(7369):337–342. doi: 10.1038/nature10452. [DOI] [PubMed] [Google Scholar]
- 4.Deu M, Rattunde F, Chantereau J. A global view of genetic diversity in cultivated sorghums using a core collection. Genome. 2006;49(2):168–180. doi: 10.1139/g05-092. [DOI] [PubMed] [Google Scholar]
- 5.Casa AM, et al. Community resources and strategies for association mapping in sorghum. Crop Sci. 2008;48(1):30–40. [Google Scholar]
- 6.Upadhyaya HD, et al. Developing a mini core collection of sorghum for diversified utilization of germplasm. Crop Sci. 2009;49(5):1769–1780. [Google Scholar]
- 7.Kimber CT. In: Sorghum: Origin, History, Technology, and Production. Smith CW, Frederiksen RA, editors. 2000. (John Wiley and Sons, New York), pp 3–98. [Google Scholar]
- 8. Harlan JR, Wet de, JMJ (1972) A simplified classification of cultivated sorghum. Crop Sci 12(2):172–176.
- 9.Quinby JR. The genetics of sorghum improvement. J Hered. 1975;66(2):56–62. [Google Scholar]
- 10.Jiao Y, et al. Genome-wide genetic changes during modern breeding of maize. Nat Genet. 2012;44(7):812–815. doi: 10.1038/ng.2312. [DOI] [PubMed] [Google Scholar]
- 11.Hufford MB, et al. Comparative population genomics of maize domestication and improvement. Nat Genet. 2012;44(7):808–811. doi: 10.1038/ng.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang X, et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 2010;42(11):961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- 13.Casa AM, et al. Evidence for a selective sweep on chromosome 1 of cultivated sorghum. Crop Sci. 2006;46(S1):S27–S40. [Google Scholar]
- 14.Bouchet S, et al. Genetic structure, linkage disequilibrium and signature of selection in Sorghum: Lessons from physically anchored DArT markers. PLoS ONE. 2012;7(3):e33470. doi: 10.1371/journal.pone.0033470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Alencar Figueiredo L, et al. Variability of grain quality in sorghum: Association with polymorphism in Sh2, Bt2, SssI, Ae1, Wx and O2. Theor Appl Genet. 2010;121(6):1171–1185. doi: 10.1007/s00122-010-1380-z. [DOI] [PubMed] [Google Scholar]
- 16.Multani DS, et al. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science. 2003;302(5642):81–84. doi: 10.1126/science.1086072. [DOI] [PubMed] [Google Scholar]
- 17.Elshire RJ, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE. 2011;6(5):e19379. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stephens JC, Miller FR, Rosenow DT. Conversion of alien sorghums to early combine genotypes. Crop Sci. 1967;7(4):396. [Google Scholar]
- 19.Paterson AH, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457(7229):551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 20.Brachi B, Morris GP, Borevitz JO. Genome-wide association studies in plants: The missing heritability is in the field. Genome Biol. 2011;12(10):232. doi: 10.1186/gb-2011-12-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S, et al. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet. 2007;39(9):1151–1155. doi: 10.1038/ng2115. [DOI] [PubMed] [Google Scholar]
- 22.Mather KA, et al. The extent of linkage disequilibrium in rice (Oryza sativa L.) Genetics. 2007;177(4):2223–2232. doi: 10.1534/genetics.107.079616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomson MJ, Ismail AM, McCouch SR, Mackill DJ. In: Abiotic Stress Adaptation in Plants. Pareek A, Sopory SK, Bohnert HJ, editors. Dordrecht, The Netherlands: Springer Netherlands; 2009. pp. 451–469. [Google Scholar]
- 24.Hamblin MT, et al. Equilibrium processes cannot explain high levels of short- and medium-range linkage disequilibrium in the domesticated grass Sorghum bicolor. Genetics. 2005;171(3):1247–1256. doi: 10.1534/genetics.105.041566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flint-Garcia SA, Thornsberry JM, Buckler ES. Structure of linkage disequilibrium in plants. Annu Rev Plant Biol. 2003;54(1):357–374. doi: 10.1146/annurev.arplant.54.031902.134907. [DOI] [PubMed] [Google Scholar]
- 26.Yan J, et al. Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers. PLoS ONE. 2009;4(12):e8451. doi: 10.1371/journal.pone.0008451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418(6898):700–707. doi: 10.1038/nature01019. [DOI] [PubMed] [Google Scholar]
- 28.Whitt SR, Wilson LM, Tenaillon MI, Gaut BS, Buckler ES. Genetic diversity and selection in the maize starch pathway. Proc Natl Acad Sci USA. 2002;99(20):12959–12962. doi: 10.1073/pnas.202476999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Z, et al. Parallel domestication of the Shattering1 genes in cereals. Nat Genet. 2012;44(6):720–724. doi: 10.1038/ng.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian F, Stevens NM, Buckler ES. Tracking footprints of maize domestication and evidence for a massive selective sweep on chromosome 10. Proc Natl Acad Sci USA. 2009;106(Suppl 1):9979–9986. doi: 10.1073/pnas.0901122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sweeney MT, et al. Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genet. 2007;3(8):e133. doi: 10.1371/journal.pgen.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin YR, Schertz KF, Paterson AH. Comparative analysis of QTLs affecting plant height and maturity across the Poaceae, in reference to an interspecific sorghum population. Genetics. 1995;141(1):391–411. doi: 10.1093/genetics/141.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown PJ, Rooney WL, Franks C, Kresovich S. Efficient mapping of plant height quantitative trait loci in a sorghum association population with introgressed dwarfing genes. Genetics. 2008;180(1):629–637. doi: 10.1534/genetics.108.092239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy RL, et al. Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc Natl Acad Sci USA. 2011;108(39):16469–16474. doi: 10.1073/pnas.1106212108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Childs KL, et al. The sorghum photoperiod sensitivity gene, Ma3, encodes a phytochrome B. Plant Physiol. 1997;113(2):611–619. doi: 10.1104/pp.113.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colasanti J, Yuan Z, Sundaresan V. The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell. 1998;93(4):593–603. doi: 10.1016/s0092-8674(00)81188-5. [DOI] [PubMed] [Google Scholar]
- 37.Ikeda K, Nagasawa N, Nagato Y. ABERRANT PANICLE ORGANIZATION 1 temporally regulates meristem identity in rice. Dev Biol. 2005;282(2):349–360. doi: 10.1016/j.ydbio.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 38.Jordan DR, Mace ES, Cruickshank AW, Hunt CH, Henzell RG. Exploring and exploiting genetic variation from unadapted sorghum germplasm in a breeding program. Crop Sci. 2011;51(4):1444. [Google Scholar]
- 39.Wang Y-H, Bible P, Loganantharaj R, Upadhyaya H. Identification of SSR markers associated with height using pool-based genome-wide association mapping in sorghum. Mol Breed. 2011;30(11):281–292. doi: 10.1139/g11-055. [DOI] [PubMed] [Google Scholar]
- 40.Ramanatha Rao V, Hodgkin T. Genetic diversity and conservation and utilization of plant genetic resources. Plant Cell Tissue Organ Cult. 2002;68(1):1–19. [Google Scholar]
- 41.Morrell PL, Buckler ES, Ross-Ibarra J. Crop genomics: Advances and applications. Nat Rev Genet. 2011;13(2):85–96. doi: 10.1038/nrg3097. [DOI] [PubMed] [Google Scholar]
- 42.Mace E, Buhariwalla K, Buhariwalla H, Crouch J. A high-throughput DNA extraction protocol for tropical molecular breeding programs. Plant Mol Biol Rep. 2003;21(4):459–460. [Google Scholar]
- 43.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts A, et al. Inferring missing genotypes in large SNP panels using fast nearest-neighbor searches over sliding windows. Bioinformatics. 2007;23(13):i401–i407. doi: 10.1093/bioinformatics/btm220. [DOI] [PubMed] [Google Scholar]
- 45.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20(2):289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 47.Bradbury PJ, et al. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23(19):2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 48.Auton A, McVean G. Recombination rate estimation in the presence of hotspots. Genome Res. 2007;17(8):1219–1227. doi: 10.1101/gr.6386707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lipka AE, et al. GAPIT: Genome association and prediction integrated tool. Bioinformatics. 2012;28(18):2397–2399. doi: 10.1093/bioinformatics/bts444. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Z, et al. Mixed linear model approach adapted for genome-wide association studies. Nat Genet. 2010;42(4):355–360. doi: 10.1038/ng.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.