Abstract
Introduction
MicroRNAs (miRNAs) are being increasingly studied in relation to energy metabolism and body composition homeostasis. Indeed, the quantitative analysis of miRNAs expression in different adiposity conditions may contribute to understand the intimate mechanisms participating in body weight control and to find new biomarkers with diagnostic or prognostic value in obesity management.
Objective
The aim of this study was the search for miRNAs in blood cells whose expression could be used as prognostic biomarkers of weight loss.
Methods
Ten Caucasian obese women were selected among the participants in a weight-loss trial that consisted in following an energy-restricted treatment. Weight loss was considered unsuccessful when <5% of initial body weight (non-responders) and successful when >5% (responders). At baseline, total miRNA isolated from peripheral blood mononuclear cells (PBMC) was sequenced with SOLiD v4. The miRNA sequencing data were validated by RT-PCR.
Results
Differential baseline expression of several miRNAs was found between responders and non-responders. Two miRNAs were up-regulated in the non-responder group (mir-935 and mir-4772) and three others were down-regulated (mir-223, mir-224 and mir-376b). Both mir-935 and mir-4772 showed relevant associations with the magnitude of weight loss, although the expression of other transcripts (mir-874, mir-199b, mir-766, mir-589 and mir-148b) also correlated with weight loss.
Conclusions
This research addresses the use of high-throughput sequencing technologies in the search for miRNA expression biomarkers in obesity, by determining the miRNA transcriptome of PBMC. Basal expression of different miRNAs, particularly mir-935 and mir-4772, could be prognostic biomarkers and may forecast the response to a hypocaloric diet.
Introduction
The discovery and characterization of small non-coding molecules of RNA with approximately 20–30 nucleotides has radically contributed to reinterpret the function of RNA and the regulation of gene expression [1]. MicroRNAs (miRNAs) can modulate (generally inhibiting) the expression of over one-third of protein-coding genes by complementary binding to specific nucleotide regions of mRNAs [2]. In this sense, in the version 19 of miRBase database (http://www.mirbase.org), 25,141 mature miRNAs have been identified in 193 species, and each one can specifically target hundreds of genes (about 60% of mammalian genes), being their dysregulation putatively involved in the pathogenesis of many chronic diseases including cancer, cardiovascular disease, type 2 diabetes and obesity [3].
Regarding obesity, miRNAs seem to participate in the regulation of several important biological processes, such as adipocyte differentiation, metabolic integration, fat metabolism and insulin sensitivity [4]. Thus, different investigations have compared the miRNA expression patterns between the undifferentiated preadipocytes and the differentiated adipocytes, both in 3T3-L1 murine cells [5] and in human subcutaneous preadipocytes [6]. These studies suggest that the miRNA expression patterns play an important role in the regulation of cell differentiation during the adipogenic process, accelerating or inhibiting the process, and hence regulating fat cell development. However, as occurs with many adipogenic genes whose expression increases during adipogenesis but decreases in adipocytes from ob/ob and diet-induced obese mice [7], many miRNAs show inverse regulatory patterns during adipogenesis and in obese patients [5]. These outcomes have been explained by the interference of hypoxia, macrophage infiltration and proinflammatory cytokines like TNF-α in the process [8]. Also, the miRNA expression patterns of omental and subcutaneous adipose tissue show some disparities that may contribute to intrinsic differences between both depots and that eventually could influence the development of some of the comorbidities associated to obesity [9].
The recent advances in sequencing and omics technologies have enabled the characterization of all the miRNAs that are expressed in a tissue or cell culture [10], allowing to identify new biomarkers of diagnosis and prognosis, but also to search for novel gene regulators for a number of chronic conditions, including obesity. In this context, the search for obesity-related biomarkers or prognostic markers of weight loss has been until now focused on genetics (SNP identification) or transcriptomics (gene expression), being whole blood or white cells the preferred biological sources because of the less invasive method of extraction. Thus, several SNPs have been identified in relation to the success of a weight loss program [11], whereas mRNA microarrays have been also used to early differentiate the populations of high and low responders to a hypocaloric diet [12]. In relation to epigenetics, which usually encompasses mitotically heritable but potentially reversible changes in DNA methylation, histone modifications and miRNA expression [13], most of the existing research has been focused on DNA methylation, studying the differences between obese and non-obese rodents [14]–[17] and humans [18], [19], but also between high and low responders to a low calorie diet [20]–[23] trying to search for epigenetic biomarkers of response to the diet. Concerning miRNAs, another epigenetic-related mechanism of gene expression regulation that is influenced by dietary manipulation [24], some studies have compared the expression differences between obese and non-obese individuals in both mice [25]–[27] and humans [6], [28], demonstrating that some miRNAs are altered in adipose tissue and circulation in obesity. Variations of miRNA expression seem also to be involved in the severity and susceptibility to nonalcoholic steatohepatitis (NASH) [29]. However, no studies have been focused on the early prediction of the weight loss outcome after a dietary treatment, which can be of great interest in the design of personalized nutrition.
The aim of the present research is to characterize the miRNA expression pattern of subjects with a different outcome to a hypocaloric dietary treatment by high-throughput sequencing in peripheral blood mononuclear cells (PBMC), in order to find prognostic biomarkers of response to the diet.
Subjects and Methods
Study Population and Ethics Statement
Ten Caucasian women with excess body weight (BMI = 35.6 kg/m2) were selected among the participants in a weight-loss trial that consisted in following an energy-restricted treatment according to a food exchange system (8-week low-calorie diet providing 800–880 kcal/day), as reported elsewhere [20]. The diet supplied 15% of energy as proteins, 55% as carbohydrates and 30% as fat [20]. After a detailed explanation of the study, a written informed acceptance was obtained from all participants in agreement with the Helsinki Declaration. This consent document and the study protocol were previously approved by the Ethical Committee of the Clínica Universidad de Navarra (Ref.090/2008). At the baseline, a screening including physical examination, medical history, activity patterns and fasting blood profile was performed. Body weight was recorded with a digital balance accurate to 0.1 kg (Seca 767, Vogel & Halke, Hamburg, Germany), and height was measured with a calibrated stadiometer (Seca 220, Vogel & Halke). Measurements were carried out in underwear after an overnight fast. Waist circumference was recorded at the midpoint between the superior iliac crest and lower costal margin whereas hip circumference was measured at the maximum protuberance of the buttocks. Body mass index (BMI) was calculated as body mass (kg) divided by height (m) squared. Total fat mass was measured by bioelectrical impedance with a Quadscan 4000 equipment (Bodystat, UK). Baseline circulating glucose was quantified by a specific colorimetric assay (Horiba ABX Diagnostics, Montpellier, France) using an automatized system (COBAS MIRA, Roche, Basel, Switzerland). Blood pressure was recorded with a standard mercury sphygmomanometer (Minimus II, Riester, Junginger, Germany) after at least 5 minutes of rest in a sitting position, following standardized procedures according to WHO criteria. Leisure time physical activity was estimated in MET hours by a validated questionnaire [30], and the number of hours devoted to different activities was multiplied by the specific MET score of each activity.
Weight loss was considered unsuccessful when it was ≤5% of initial body weight and successful when it was ≥5%. Based on this, the sample was categorized as unsuccessful (n = 5) or successful (n = 5) weight-loss subjects.
miRNA Sequencing by SOLiD v4
At baseline, PBMCs were isolated by differential centrifugation using Polymorphprep (Axis Shield PoC AS, Oslo, Norway). The PBMC band was harvested with a Pasteur pipette and, after washing with phosphate-buffered saline (PBS), total RNA was extracted by using a commercial kit (mirVana, Ambion, Austin, TX, USA). RNA quality was assessed by measuring 1 µl of 100 ng/µl total RNA in a 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) by using a RNA 6000 Nano kit (Agilent). The percentage of miRNAs (10–40 nucleotides) was estimated with the same equipment by also analysing 1 µl total RNA in a Small RNA Chip (Agilent). The small RNAs were purified with the PureLink miRNA Isolation Kit (Invitrogen, Carlsbad, CA, USA) and samples were concentrated with a speed vacDNA. 3 µl (10 ng approximately) of miRNA were used for next-generation sequencing. Quality of RNA was assessed by 260 nm absorbance with a Nanodrop ND-1000 spectrophotometer (NanoDrop Technologies Inc, Wilmington, DE, USA), by using Qubit fluorescence (Invitrogen), and by capillary electrophoresis with an Agilent 2100 Bioanalyzer (Agilent Technologies). Fragment libraries compatible with the SOLiD platform were obtained from small RNA by following the recommendations provided by Life Technologies for SOLiDv4 sequencing (http://www.umassmed.edu/uploadedFiles/nemo/STaR-smallRNAs-QRC.PDF). The quality and quantity of the libraries were assessed by Agilent 2100 Bioanalyzer and Qubit.
The equimolar mixture of the libraries was subjected to emulsion PCR process for clonal amplification of the fragments, followed by an enrichment process and chemical modification to generate microspheres and allow loading in the reaction chamber, as indicated by Life Technologies for SOLiD v4 sequencing. The quality and quantity of the microspheres obtained from the pool of libraries have been estimated taking into account the parameters obtained from the workflow analysis. The reactions to obtain sequences of 35 nucleotides +10 nucleotides have been subsequently carried out (Barcode in SOLiD v4). The quality of the data obtained has been estimated using parameters provided by the SETS software (SOLiD Experimental Tracking System). Mass sequencing reactions of miRNAs and the bioinformatic analysis were carried out by Sistemas Genómicos (Spain) [http://www. sistemasgenomicos.com] and the results are shown as a supplementary table (table S1).
Reads were mapped against the latest version of the human genome (version GRch37/hg19) using the BWA software [31]. Known miRNAs were identified and quantified using BedTools [32] and miRBase (v17). Novel miRNAs were discovered by secondary structure predictions with the mirDeep2 package [33].
Reverse-Transcription and Quantitative Real-time PCR (qRT-PCR) of miRNAs
Total RNA (3 ng) was reverse-transcribed using the TaqMan® MicroRNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA) and the miRNA-specific reverse-transcription primers provided with the TaqMan® MicroRNA Assay (Applied Biosystems, Foster City, CA, USA). For the reverse transcription iCycler™ Thermal cycler (Applied Biosystems, Foster City, CA, USA) was used with the following conditions: 16°C for 30 min; 42°C for 30 min and 85°C for 5 min. 1.33 µl of miRNA-specific cDNA from this reaction was amplified with the TaqMan Universal PCR master mix and the respective specific probe provided in the TaqMan® MicroRNA Assay (Applied Biosystems, Foster City, CA, USA). The targeted miRNA assay sequences were as follows: miR-27b 5′-UUCACAGUGGCUAAGUUCUGC-3′, miR-223’- 5′UGUCAGUUUGUCAAAUACCCCA-3′, miR-224 5′-CAAGUCACUAGUGGUUCCGUU-3′, miR-4772-3p 5′-CCUGCAACUUUGCCUGAUCAGA-3′, and miR- 935 5′-CCAGUUACCGCUUCCGCUACCGC-3′. PCR was performed in an iCycler™–MyiQ™ Real-time PCR Detection System (Applied Biosystems, Foster City, CA, USA). Amplification was performed at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. miRNA-148a (sequence 5′-UCAGUGCACUACAGAACUUUGU-3′), a miRNA with a fold change close to 1 in mass sequencing, was used as an endogenous control. All mRNA levels were normalized to the values of miRNA-148a and the results expressed as fold changes of threshold cycle (Ct) value relative to controls using the 2−ΔΔCt method [34].
Statistical Analyses
The Mann-Whitney U test was used to analyse differences between both responder and non-responder groups regarding anthropometric and biochemical variables. For this purpose, SPSS 15.0 software (SPSS, Chicago, IL) was used. SPSS was also used to perform bivariate correlations and calculate Pearson’s r.
Statistical analyses of high-throughput sequencing results were performed with the DESeq software [35] comparing the expression of each miRNA in the responder and non-responder subjects. An adjusted value of probability was achieved by using the Benjamini-Hochberg method for False Discovery Rate (FDR) correction. In the results section, differences in miRNA expression with an adjusted P-value<0.05 were interpreted as significant, whereas those with adjusted P-value<0.10 were considered as trends towards significance.
Those miRNAs with a significant fold change of 2 or greater (in terms of absolute value) were mapped by chromosome. Moreover, chromosome X and 14 were clustered based on inter-miRNA distance <100000 (http://mirbase.org) [36]. These results are shown in tables S2 and S3.
Prediction of targets for those miRNAs that showed significant differential expression between responders and non-responders was undertaken by means of two widely used algorithms: miRanda and TargetScan. The software MicroCosm Targets Version 5 [36] and the prediction source http://microRNA.org [37] were used for calculating miRanda algorithm, and the software TargetScanHuman 6.0 in the case of TargetScan algorithm [38]. At the time of writing, miR-4772 was not in the MicroCosm Targets Version 5 and TargetScanHuman 6.0 databases.
Results
Body Weight after Hypocaloric Diet Treatment
A statistically higher body weight reduction in the responder group (subjects who lost ≥5% body weight) was observed when compared to the non-responders (subjects who lost ≤5% body weight), being the physical activity pattern similar between both groups (table 1).
Table 1. Characteristics of the population at baseline and after the 8-week intervention categorized according to the outcome (responders and non-responders to the energy restriction).
| NON RESPONDERS (n = 5) | RESPONDERS (n = 5) | P-value | |
| Age (years) | 42±2 | 38±2 | 0.31 |
| Initial Body Weight (kg) | 90.3±6.2 | 95.8±3.9 | 0.55 |
| Initial Body Mass Index (BMI) | 34.8±2.1 | 36.4±1.7 | 0.69 |
| Initial % Fat (Impedance) | 46.2±2.1 | 46.9±1.5 | 1.00 |
| Waist Circumference (cm) | 105±5 | 107±4 | 0.84 |
| Hip Circumference (cm) | 120±5 | 125±4 | 0.58 |
| Waist/Hip ratio | 0.88±0.04 | 0.86±0.01 | 0.69 |
| Physical activity (MET h/week) | 12.6±7.5 | 12.1±8.4 | 0.85 |
| Fasting Glucose (mmol/L) | 4.94±0.21 | 4.56±0.27 | 0.31 |
| Systolic Blood Pressure (mm Hg) | 122±5 | 123±4 | 1.00 |
| Diastolic Blood Pressure (mm Hg) | 70±4 | 62±2 | 0.15 |
| Weight Loss (kg) | 3.1±0.7 | 10.8±0.6 | 0.008 |
Comparisons between groups were made using Mann-Whitney U test. Values represent mean ± SEM.
miRNA Expression Profile in PBMC Cells
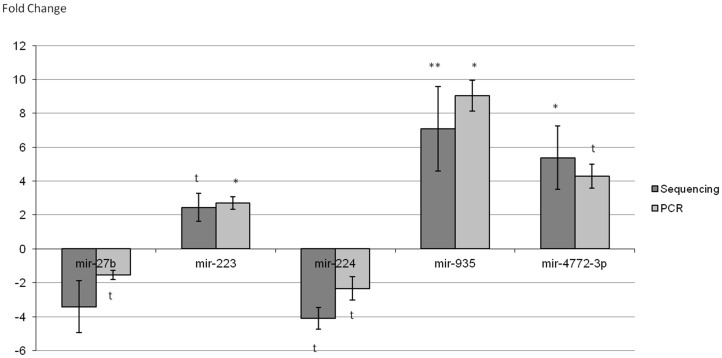
Two miRNAs were up-regulated in the non-responder group according to high-throughput sequencing at baseline (figures 1 and 2): mir-935, with a fold change of 7.1 (adjusted P -value of 0.008 and a fold change value obtained by qRT-PCR of 9.05, P = 0.02), and mir-4772, with a fold change of 5.3 (adjusted P-value of 0.022 and a fold change value obtained by qRT-PCR of 4.29, P = 0.09). Three other miRNAs were down-regulated but showed an adjusted P-value lower than 0.1 and a fold change higher than two or lower than minus two (figures 1 and 2): mir-223, with a fold change of 2.5 (the fold change value obtained by qRT-PCR was 2.72, P = 0.03); mir-224, with a fold change of −4.1 (the fold change value obtained by qRT-PCR was −2.33, P = 0.08); and mir-376b, with a fold change of −4.2.
Figure 1. miRNAs differentially expressed between responders and non-responders at baseline (adjusted P-value<0.1).
The data correspond to the mean of the transcripts sequenced in each group ± SEM.
Figure 2. Comparison between the two technologies applied in this work, high-throughput sequencing (dark columns) and RT-PCR (light columns).
The expression of mir-27b, mir-223, mir-224, mir-935 and mir-4772-3p is represented as fold change of non-responders with respect to responders at baseline. The data correspond to the mean ± SEM. Comparisons between groups were made using adjusted P-value (mass sequencing) and Mann-Whitney U test (RT-PCR). *, p<0.05; t, p<0.1.
Six other transcripts are shown in table 2 because, according to high-throughput sequencing, they presented differential expression between responders and non-responders before using the Benjamini-Hochberg correction for multiple tests (P-value<0.005 and fold change of 2.9 or greater in terms of absolute value, but an adjusted P-value higher than 0.1). Two of them were overexpressed, being mir-183 the most up-regulated with a fold change of 9.0. Other four miRNAs were down-regulated, showing mir-433 a fold change of −9.6 and mir-27b a fold change of −3.41 (the fold change value obtained by qRT-PCR for mir-27b was −1.54, P = 0.09; figure 2).
Table 2. miRNA transcripts showing differential miRNA expression between responders and non-responders by using the mass sequencing approach (non-adjusted P-value<0.005 and fold change>2.9), but whose adjusted P-values were not significant.
| miRNAs | Responders | Non-responders | Fold Change | Non-adjusted P-value |
| mir-183 | 4±4 | 36±13 | 8.96 | 0.004 |
| mir-542 | 148±24 | 555±158 | 3.75 | 0.003 |
| mir-433 | 61±16 | 6±6 | −9.56 | 0.003 |
| mir-154 | 269±105 | 79±37 | −3.42 | 0.005 |
| mir-27b | 1655±550 | 485±217 | −3.41 | 0.003 |
| mir-409 | 4744±1184 | 1609±722 | −2.95 | 0.005 |
The data report the mean of the number of transcripts sequenced in each group ± SEM.
Table S4 shows the most expressed miRNAs in PBMC cells based on read counts. The transcript mir-223, which was differentially expressed when comparing responder and non-responder subjects, is the most abundant in this type of cells, followed by mir-150 and mir-126.
Novel miRNAs
From a total of 102 detected miRNAs with a significant fold change of −2 or greater, fourteen not identified miRNAs (according to miRBase database) were found. Although none of these novel miRNAs showed significant differences between responders and non-responders (table S5), and despite their relatively low expression levels, they deserve a study in the future.
Prediction of Targets for miRNAs
The predicted gene targets for miRNAs were carried out for miR-935, miR-223, miR-224, miR-376b, miR-542, miR-433, miR-154 miR-27b, miR-409, miR-183, miR-542-3p and miR-4772-3p with miRanda and TargetScan algorithms. With the exception of miR-542, predicted targets related to obesity and its co-morbidities were found in these miRNAs (table 3). In the table S6, the list of target mRNAs of each differentially expressed miRNA has been expanded, including the most relevant obesity-related genes putatively regulated by each miRNA.
Table 3. Short selection of target mRNAs of the microRNAs with differences in expression between responders and non-responders to the energy-restricted.
| miRNA | Sequence | miRanda algorithm | TargetScan algorithm | |
| (source mirbase.org ) | MicroCosm | microRNA | TargetScan | |
| hsa-mir-935 | GGCGGGGGCGCGGGCGGCAGUGGCGGGAGCGGCCCCUCGGCCAUCCUCCGUCUGCCCAGUUACCGCUUCCGCUACCGCCGCCGCUCCCGCU | HDAC1, ADRA1A, LPL, RBP5 | HIF1A | ADRB1 |
| hsa-mir-223 | CCUGGCCUCCUGCAGUGCCACGCUCCGUGUAUUUGACAAGCUGAGUUGGACACUCCAUGUGGUAGAGUGUCAGUUUGUCAAAUACCCCAAGUGCGGCACAUGCUUACCAG | IGFL2c, DNMT1, HDAC2, HDAC8, SIRT5 | LIPG | IGF1R, HDAC4 |
| hsa-mir-224 | GGGCUUUCAAGUCACUAGUGGUUCCGUUUAGUAGAUGAUUGUGCAUUGUUUCAAAAUGGUGCCCUAGUGACUACAAAGCCC | MC3R, SREBF1 | ACAT1 | IRS2 |
| hsa-mir-376b | CAGUCCUUCUUUGGUAUUUAAAACGUGGAUAUUCCUUCUAUGUUUACGUGAUUCCUGGUUAAUCAUAGAGGAAAAUCCAUGUUUUCAGUAUCAAAUGCUG | FABP4, CPT2 | LIPH, PPARG, G6PC2, HDAC9, NR3C1 | HDAC9, IGF1R |
| hsa-mir-433 | CCGGGGAGAAGUACGGUGAGCCUGUCAUUAUUCAGAGAGGCUAGAUCCUCUGUGUUGAGAAGGAUCAUGAUGGGCUCCUCGGUGUUCUCCAGG | HDAC6, ADRA1A, IGFBP1, ACOX2, LPL, LIPE | GCLC, FRZB, | ADRA1A, LEPR |
| has-mir-154 | GUGGUACUUGAAGAUAGGUUAUCCGUGUUGCCUUCGCUUUAUUUGUGACGAAUCAUACACGGUUGACCUAUUUUUCAGUACCAA | ADRA1A, IGF1 | ||
| hsa-mir-27b | ACCUCUCUAACAAGGUGCAGAGCUUAGCUGAUUGGUGAACAGUGAUUGGUUUCCGCUUUGUUCACAGUGGCUAAGUUCUGCACCUGAAGAGAAGGUG | PPARG, INSR, MC4R, SIRT5 | SCD5, ACAT1, | PPARG, INSR, IRS1, CNR1, PPARGC1B, IGF1 |
| hsa-mir-409 | UGGUACUCGGGGAGAGGUUACCCGAGCAACUUUGCAUCUGGACGACGAAUGUUGCUCGGUGAACCCCUUUUCGGUAUCA | FZD3, PPARGC1A, APOF, AGPAT9 | ||
| has-mir-183 | CCGCAGAGUGUGACUCCUGUUCUGUGUAUGGCACUGGUAGAAUUCACUGUGAACAGUCUCAGUCAGUGAAUUACCGAAGGGCCAUAAACAGAGCAGAGACAGAUCCACGA | HDAC10, HDAC8, HDAC6, CRTC2, DGKH, GK5, PLB1, CPT2 | ACAD8 | PEX19 |
| hsa-mir-542-3p | UGUGACAGAUUGAUAACUGAAA | IGFBP1, IGFBP6, FABP3, PLA2G4B, LIPA, APOL5, ELOVL3, SLC27A3, PPARD, TNF | FABP3 | GDPD4, PLA2R1, |
| hsa-mir-4772-3p | CCUGCAACUUUGCCUGAUCAGA | ACSL1, PDK3, PPARGC1A, ERLIN2, IGF2BP1, IGF2, LRP8, CREBBP | ||
ACAD: acyl-CoA dehydrogenase family; ACAT: acetyl-coa acetyltransferase; ACOX: acyl-coenzyme A oxidase, peroxisomal; ACSL: acyl-CoA synthetase long-chain family member; ADRA1A: alpha-1A adrenergic receptor; ADRB1: beta-1 adrenergic receptor; AGPAT: 1-acylglycerol-3-phosphate o-acyltransferase; APO: apolipoprotein; CREBBP: CREB binding proteína; CNR: cannabinoid receptor; CPT: Carnitine O-palmitoyltransferase, mitochondrial precursor; CRTC: transducer of regulated cAMP response element-binding protein (CREB); DGKH : diacylglycerol kinase eta; DNMT: DNA (cytosine-5)-methyltransferase; ELOVL: elongation of very long chain fatty acids protein; ERLIN: ER lipid raft associated; FABP: fatty acid-binding protein, adipocyte; FRZB: frizzled-related protein; FZD: frizzled, drosophila, homolog of; GCLC: glutamate-cysteine ligase, catalytic subunit; GDPD: glycerophosphodiester phosphodiesterase domain containing; GIPR: gastric inhibitory polypeptide receptor; GK: glycerol kinase; G6PC: glucose-6-phosphatase, catalytic; HDAC: histone deacetylase; HIF1A: hypoxia-inducible factor; IGF1: insulin-like growth factor 1; IGF1R: insulin-like growth factor 1 receptor; IGFBP: insulin-like growth factor-binding protein precursor; IGFL2c: insulin growth factor-like family member 2 precursor; INSR: insulin receptor; IRS: insulin receptor substrate; LEPR: leptin receptor; LIPA: lysosomal acid lipase/cholesteryl ester hydrolase precursor; LIPE: hormone-sensitive lipase; LIPG: factor 1, alpha subunit; lipase, endothelial; LIPH: lipase h; LRP: Low density lipoprotein receptor-related protein, apolipoprotein e receptor;LPL: Lipoprotein lipase precursor; MCR: Melanocortin receptor; NR3C1: glucocorticoid receptor; PDK: pyruvate dehydrogenase kinase, isozyme; PEX: peroxisomal biogenesis factor; PLA2G4B: cytosolic phospholipase A2 beta; PLA2R1: phospholipase A2 receptor 1; PLB: phospholipase B; PPARD: Peroxisome proliferator-activated receptor delta; PPARG: Peroxisome proliferator-activated receptor gamma; PPARGC: peroxisome proliferator-activated receptor gamma, coactivator; RBP: retinol-binding protein, cellular; SLC27A: long-chain fatty acid transport protein; SCD: stearoyl-coa desaturase; SIRT5: NAD-dependent deacetylase sirtuin-5; SREBF1: sterol regulatory element-binding protein 1; TNF: tumor necrosis factor precursor.
Chromosomal Localization of miRNAs
Sequenced miRNAs with a significant fold change of 2 or greater (in absolute values) have been mapped by chromosome and shown as supplementary data (tables S2 and S3) accompanied by the differences between responders and non-responders. Most of the miRNAs, as determined by the total number of reads mapped per chromosome, are located in the chromosomes 14 and X. When the miRNAs were clustered based on chromosome location and, in the case of chromosomes X and 14, on inter-miRNA distance <100000 (according to miRBase), three clusters located in chromosomes 9, X and 18 were significantly up-regulated. Another three clusters were significantly down-regulated, two of them in chromosome X and one in chromosome 7.
Associations/correlations between miRNAs and Anthropometric Measurements or Indexes
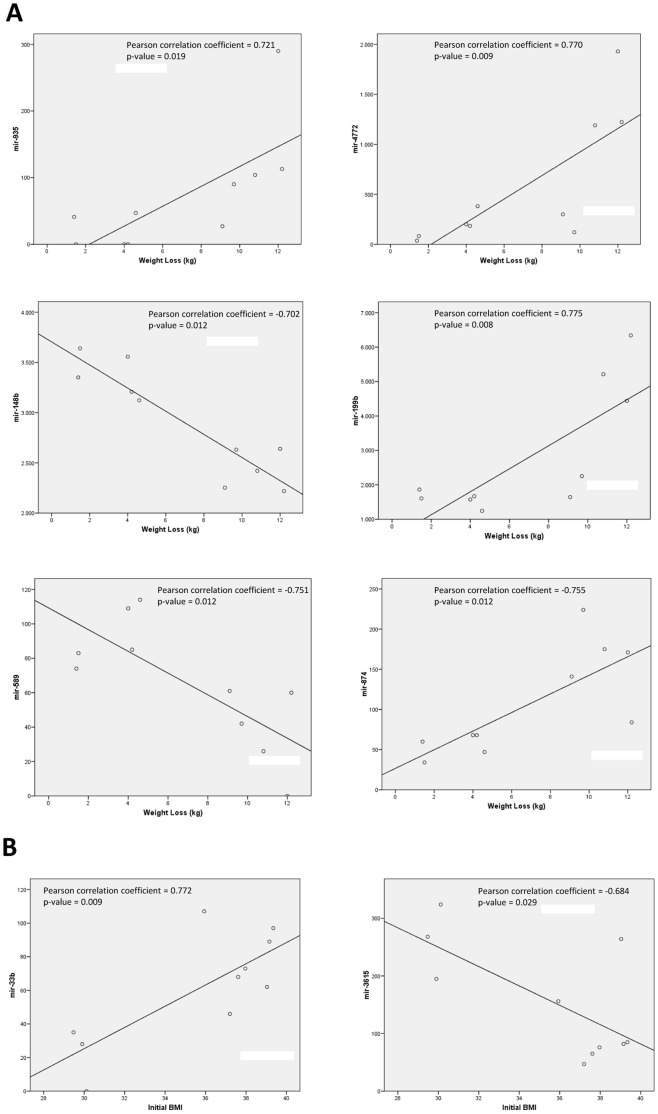
Both mir-935 and mir-4772 showed close associations with the magnitude of weight loss (figure 3a, with Pearson correlation coefficients of 0.721 and 0.770, respectively). Interestingly, similar results were observed with other miRNAs presenting low P-values in the non-adjusted test, such as mir-874 (P = 0.014), mir-199b (P = 0.020), mir-766 (P = 0. 052), mir-589 (P = 0.076), and mir-148b (P = 0.389). Other miRNAs differentially expressed between responders and non-responders shown in figure 1, as is the case of mir-223 and mir-224, evidenced a trend towards significance in their correlation with the magnitude of weight loss (Pearson correlation coefficients of 0.597 and 0.599 and P-values of 0.069 and 0.067, respectively).
Figure 3. a. Associations between the expression of some miRNAs and body weight change during the energy restriction period.
miRNA expression as number of transcripts sequenced. Pearson product-moment correlation coefficient has been applied. b. Associations between the expression of some miRNAs and the baseline body mass index (BMI) of the subjects. miRNA expression as number of transcripts sequenced. Pearson product-moment correlation coefficient has been applied.
There were no baseline differences between the participants in the study regarding their anthropometric and biochemical measurements (table 1). However, we wanted to know if the expression of some miRNAs in PBMCs were associated with initial BMI, the most used parameter of adiposity (figure 3b). Moderate associations were found between the expression of some miRNAs and this phenotypic marker of adiposity, with mir-3615 and mir-33b being the more relevant. These miRNAs associated to initial body weight could be candidates for being implicated in obesity onset and complications, although it should be elucidated in later studies.
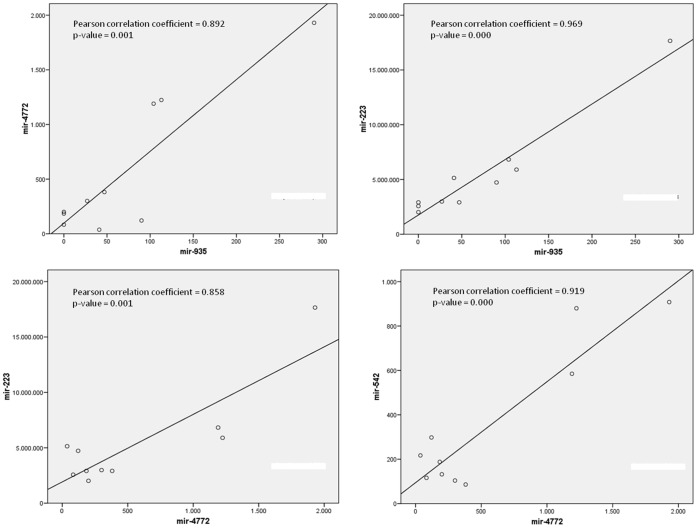
It is noteworthy that the expression of some of the analysed miRNAs was highly correlated among them, as observed when comparing the miRNAs that were more associated with weight loss, such as mir-935, mir-4772, mir-223, mir-409 and mir-27b (figure 4).
Figure 4. Associations between the expression of different miRNAs that are involved in the response to the dietary treatment.
Data represent the number of transcripts sequenced. Pearson product-moment correlation coefficient has been applied.
Discussion
The current research is pioneer in the use of high-throughput sequencing in the search for expression biomarkers in obesity, by determining the whole miRNA transcriptome in human PBMCs. The current article employed a similar approach that Ahn et al. [39] when determined the miRNA transcriptome in the newborn mouse ovaries. In our case, we studied the miRNA transcriptome in peripheral blood cells of ten obese women that followed a weight-loss program, with 5 of them categorized as high responders and the other 5 as non-responders, according to Campion et al. [40]. This kind of cells has been selected because they are widely and successfully used in the search for non-invasive biomarkers as surrogates of tissues that are not easily accessible, such as adipose tissue [41]. We found 5 miRNAs whose expression was different at baseline between both groups (with an adjusted p-value lower than 0.1). Among them, only mir-223 (fold change of 2.5) and mir-224 (fold change of −4.1) had been previously related to obesity. Thus, mir-223 has been reported to regulate the expression of many genes, as for example GLUT4 [42], and is overexpressed in epididymal adipocytes of obese mice [43] and down-regulated in plasma of diabetic patients [40]. On the other hand, mir-224 is the only differentially expressed miRNA that has been previously related to adiposity, being up-regulated in 3T3-L1 mature adipocytes 9 days post induction when compared with preadipocytes [44].
Regarding the expression of the other 3 miRNAs shown in figure 1 (mir-935, mir-4772 and mir-376b), this study is apparently the first that finds differences in PBMC cells and that relates their expression with the susceptibility to lose weight. More studies (in vitro and in animals) are necessary to confirm a role of these 3 transcripts in obesity and energy metabolism.
Some of the miRNAs reported in table 2 have been previously associated with different diseases, especially cancer. Among them, there are three miRNAs (mir-183, mir-433 and mir-27b) that seem to play an important role in the regulation of gene expression in metabolic disorders. Thus, regarding mir-183 (fold change of 9.0), the incubation of 3T3-L1 adipocytes with insulin inhibits its expression [45]. mir-433 (fold change of −9,6) is activated by the estrogen receptor ESRRG, in which several SNPs have been associated with higher risk of type 2 diabetes by GWAS [46] and that acts as a co-activator of the demethylating enzyme Dnmt1 [47]. Although most articles studying this specific miRNA are focused on cancer [48], it is also differentially expressed in liver of patients with non-alcoholic fatty liver disease [49]. Finally, mir-27b (fold change of −3,4), which is an oxidative stress-responsive miRNA that suppresses lipopolysaccharide-induced activation of NFκB [50], is up-regulated in epididymal adipocytes of obese mice [43] and impairs human adipocyte differentiation by targeting PPARγ [51].
The moderate correlations observed between the expression of the different miRNAs in PBMCs and the BMI at baseline (figure 3b) suggest that small differences in adiposity do not appears to strongly affect miRNA expression in this kind of cells. However, the magnitude of weight loss strongly correlated with the expression of several miRNAs at baseline (figure 3a), suggesting a role of these miRNAs in body weight regulation, energy homeostasis and response to the diet. The significant correlations found between the expression of different miRNAs (figure 4) suggest that, at least in PBMCs, their expression could be regulated by common factors and even that they might share similar regulatory functions.
Some of the potential targets of the miRNAs identified in the present study are represented in table 3. Those that have been extensively related to obesity, energy homeostasis and insulin signalling, as well as those that can act at an epigenetic level, have been reflected in the table. This is the case of genes like peroxisome proliferator-activated receptor gamma (PPARG), glucocorticoid receptor (NR3C1), hypoxia-inducible factor 1, alpha subunit (HIF1A), gastric inhibitory polypeptide receptor (GIPR), histone deacetylases (HDAC) and methyltransferases (DNMT). None of the miRNAs shown in table 3 have been previously studied in relation to the expression of these genes in models of obesity or weight regulation. So that, in vitro studies with siRNAs or addition of miRNAs to the culture medium of 3T3-L1 cells, primary adipocytes, monocytes, macrophages or other key cell types in the development of obesity, are needed to elucidate the real importance of these miRNAs on the regulation of the expression of these genes. However, previous studies have revealed an interaction between some miRNAs, none of them differentially expressed in our study, and the expression of HIF1A (miR-210 [52] and miR-101 [53], among many others), PPARG (such as miR-130 [54]), or NR3C1 (including miR-18 and 124a, [55]), suggesting that miRNAs may play an important role in the regulation of these genes and the related pathways. It is noteworthy that some of the analyzed miRNAs have histone deacetylases and methyltransferases as predicted targets, suggesting an epigenetic loop between miRNAs and histone modifications in gene expression regulation.
Some of the highly abundant miRNA transcripts observed in the present study coincide with those reported by Vaz et al. [10] in a previous analysis of microRNA transcriptome in PBMCs by deep sequencing, as for example mir-29a, mir-21, mir-191 or let-7g. However, the disparities could be partially attributed to the difference in technology (SOLiD v4 versus SOLEXA).
This is the first work that determines the whole miRNA transcriptome by using high-throughput sequencing in the search for biomarkers related to obesity and body weight regulation. The results of this study stress the importance of miRNA expression in blood cells as biomarkers of weight loss and response to the diet (prognostic biomarkers), which could be used in the design of personalized nutrition. Also, they suggest that the expression of different miRNAs in blood cells might be implicated in the regulation of energy homeostasis and body weight. However, it should be emphasized that some of the miRNA expression differences observed between responders and non-responders could be due to (or influenced by) the diet at baseline or other factors, such as physical activity, genetic background, dietary habits or immune function, that should be analyzed in further studies.
To conclude, the present study proposes the basal expression levels of different miRNAs in blood cells, particularly mir-935, mir-4772, mir-223, mir-224 and mir-376b, as prognostic biomarkers of response to the diet. This research represents a milestone in the design of personalized nutrition by taking into account miRNA expression in blood cells, and opens the door to the study of different miRNAs as novel biomarkers of weight loss.
Supporting Information
All the miRNA transcripts sequenced and the differences between responders and non-responders (as fold change), categorized by p-value adjusted for multiple comparisons.
(DOC)
miRNA clusters (according to miRBase) upregulated in peripheral blood cells of non-responders to the low-calorie diet when compared to the responders, categorized by chromosomal location.
(DOC)
miRNA clusters (according to miRBase) downregulated in peripheral blood cells of non-responders to the low-calorie diet when compared to the responders, categorized by chromosomal location.
(DOC)
The most expressed miRNA transcripts in PBMC cells, based on read counts, and the chromosome where they are located.
(DOC)
List of predicted novel miRNAs (according to miRBase) found in peripheral blood mononuclear cells.
(DOC)
Target mRNAs of the microRNAs with differences in expression between responders and non-responders to the energy-restricted.
(DOC)
Acknowledgments
The technical assistance of Ana Lorente in RNA isolation and miRNA quality assessment is gratefully acknowledged. We thank Juan Carlos Triviño and Sheila Zúñiga for the bioinformatic analysis, and to the Unidad de Nuevas Tecnologías of Sistemas Genómicos (Victoria Fernández-Pedrosa, Carmen Collado, María José García and Raquel Rodríguez) for the library generation, PCR emulsion and SOLiD 4.0 sequencing. We also acknowledge the work of Salome Perez (nurse), Maria Hernandez (dietitian) and Blanca Martínez de Morentin (physician) in sample collection.
Funding Statement
This work has been supported by the Linea Especial LE/97 of the University of Navarra, the RETICS and CIBERobn Schemes funded by Instituto de Salud Carlos III, the Government of the Basque Country (IT-386-10 and IT-572-13) and the University of the Basque Country UPV/EHU (ELDUNANOTEK UFI11/32). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Alegría-Torres JA, Baccarelli A, Bollati V (2011) Epigenetics and lifestyle. Epigenomics 3: 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carthew RW, Sontheimer EJ (2009) Origins and Mechanisms of miRNAs and siRNAs. Cell 136: 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ali AS, Ali S, Ahmad A, Bao B, Philip PA, et al. (2011) Expression of microRNAs: potential molecular link between obesity, diabetes and cancer. Obes Rev 12: 1050–1062. [DOI] [PubMed] [Google Scholar]
- 4. McGregor RA, Choi MS (2011) microRNAs in the regulation of adipogenesis and obesity. Curr Mol Med 11: 304–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xie H, Lim B, Lodish HF (2009) MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes 58: 1050–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ortega FJ, Moreno-Navarrete JM, Pardo G, Sabater M, Hummel M, et al. (2010) MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoS One 5: e9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nadler ST, Stoehr JP, Schueler KL, Tanimoto G, Yandell BS, et al. (2000) The expression of adipogenic genes is decreased in obesity and diabetes mellitus. Proc Natl Acad Sci U S A 97: 11371–11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bengestrate L, Virtue S, Campbell M, Vidal-Puig A, Hadaschik D, et al. (2011) Genome-wide profiling of microRNAs in adipose mesenchymal stem cell differentiation and mouse models of obesity. PLoS One 6: e21305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klöting N, Berthold S, Kovacs P, Schön MR, Fasshauer M, et al. (2009) MicroRNA expression in human omental and subcutaneous adipose tissue. PLoS One 4: e4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vaz C, Ahmad HM, Sharma P, Gupta R, Kumar L, et al. (2010) Analysis of microRNA transcriptome by deep sequencing of small RNA libraries of peripheral blood. BMC Genomics 11: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sarzynski MA, Jacobson P, Rankinen T, Carlsson B, Sjöström L, et al. (2011) Associations of markers in 11 obesity candidate genes with maximal weight loss and weight regain in the SOS bariatric surgery cases. Int J Obes (Lond) 35: 676–683. [DOI] [PubMed] [Google Scholar]
- 12. Mutch DM, Temanni MR, Henegar C, Combes F, Pelloux V, et al. (2007) Adipose gene expression prior to weight loss can differentiate and weakly predict dietary responders. PLoS One 2: e1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Campión J, Milagro F, Martínez JA (2010) Epigenetics and obesity. Prog Mol Biol Transl Sci 94: 291–347. [DOI] [PubMed] [Google Scholar]
- 14. Milagro FI, Campión J, García-Díaz DF, Goyenechea E, Paternain L, et al. (2009) High fat diet-induced obesity modifies the methylation pattern of leptin promoter in rats. J Physiol Biochem 65: 1–9. [DOI] [PubMed] [Google Scholar]
- 15. Plagemann A, Harder T, Brunn M, Harder A, Roepke K, et al. (2009) Hypothalamic proopiomelanocortin promoter methylation becomes altered by early overfeeding: an epigenetic model of obesity and the metabolic syndrome. J Physiol 587: 4963–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Widiker S, Karst S, Wagener A, Brockmann GA (2010) High-fat diet leads to a decreased methylation of the Mc4r gene in the obese BFMI and the lean B6 mouse lines. J Appl Genet 51: 193–197. [DOI] [PubMed] [Google Scholar]
- 17. Lomba A, Martínez JA, García-Díaz DF, Paternain L, Marti A, et al. (2010) Weight gain induced by an isocaloric pair-fed high fat diet: a nutriepigenetic study on FASN and NDUFB6 gene promoters. Mol Genet Metab 101: 273–278. [DOI] [PubMed] [Google Scholar]
- 18. Stepanow S, Reichwald K, Huse K, Gausmann U, Nebel A, et al. (2011) Allele-specific, age-dependent and BMI-associated DNA methylation of human MCHR1. PLoS One 6: e17711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang X, Zhu H, Snieder H, Su S, Munn D, et al. (2010) Obesity related methylation changes in DNA of peripheral blood leukocytes. BMC Med 8: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Campión J, Milagro FI, Goyenechea E, Martínez JA (2009) TNF-alpha promoter methylation as a predictive biomarker for weight-loss response. Obesity (Silver Spring) 17: 1293–1297. [DOI] [PubMed] [Google Scholar]
- 21. Bouchard L, Rabasa-Lhoret R, Faraj M, Lavoie ME, Mill J, et al. (2010) Differential epigenomic and transcriptomic responses in subcutaneous adipose tissue between low and high responders to caloric restriction. Am J Clin Nutr 91: 309–320. [DOI] [PubMed] [Google Scholar]
- 22. Milagro FI, Campión J, Cordero P, Goyenechea E, Gómez-Uriz AM, et al. (2011) A dual epigenomic approach for the search of obesity biomarkers: DNA methylation in relation to diet-induced weight loss. FASEB J 25: 1378–1389. [DOI] [PubMed] [Google Scholar]
- 23. Cordero P, Campion J, Milagro FI, Goyenechea E, Steemburgo T, et al. (2011) Leptin and TNF-alpha promoter methylation levels measured by MSP could predict the response to a low-calorie diet. J Physiol Biochem 67: 463–470. [DOI] [PubMed] [Google Scholar]
- 24. Parra P, Serra F, Palou A (2010) Expression of adipose microRNAs is sensitive to dietary conjugated linoleic acid treatment in mice. PLoS One 5: e13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakanishi N, Nakagawa Y, Tokushige N, Aoki N, Matsuzaka T, et al. (2009) The up-regulation of microRNA-335 is associated with lipid metabolism in liver and white adipose tissue of genetically obese mice. Biochem Biophys Res Commun 385: 492–496. [DOI] [PubMed] [Google Scholar]
- 26. Zhao E, Keller MP, Rabaglia ME, Oler AT, Stapleton DS, et al. (2009) Obesity and genetics regulate microRNAs in islets, liver, and adipose of diabetic mice. Mamm Genome 20: 476–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park JH, Ahn J, Kim S, Kwon DY, Ha TY (2011) Murine hepatic miRNAs expression and regulation of gene expression in diet-induced obese mice. Mol Cells 31: 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heneghan HM, Miller N, McAnena OJ, O’Brien T, Kerin MJ (2011) Differential miRNA expression in omental adipose tissue and in the circulation of obese patients identifies novel metabolic biomarkers. J Clin Endocrinol Metab 96: E846–850. [DOI] [PubMed] [Google Scholar]
- 29. Pogribny IP, Starlard-Davenport A, Tryndyak VP, Han T, Ross SA, et al. (2010) Difference in expression of hepatic microRNAs miR-29c, miR-34a, miR-155, and miR-200b is associated with strain-specific susceptibility to dietary nonalcoholic steatohepatitis in mice. Lab Invest 90: 1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martínez-González MA, López-Fontana C, Varo JJ, Sánchez-Villegas A, Martinez JA (2005) Validation of the Spanish version of the physical activity questionnaire used in the Nurses’ Health Study and the Health Professionals’ Follow-up Study. Public Health Nutr 8: 920–927. [DOI] [PubMed] [Google Scholar]
- 31. Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, et al. (2008) Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 26: 407–415. [DOI] [PubMed] [Google Scholar]
- 34. Livak K, Schmittgen T (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 35. Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Betel D, Wilson M, Gabow A, Marks DS, Sander C (2008) The microRNA.org resource: targets and expression. Nucleic Acids Res 36: D149–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- 39. Ahn HW, Morin RD, Zhao H, Harris RA, Coarfa C, et al. (2010) MicroRNA transcriptome in the newborn mouse ovaries determined by massive parallel sequencing. Mol Hum Reprod 16: 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, et al. (2010) Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res 107: 810–817. [DOI] [PubMed] [Google Scholar]
- 41. Crujeiras AB, Parra D, Milagro FI, Goyenechea E, Larrarte E, et al. (2008) Differential expression of oxidative stress and inflammation related genes in peripheral blood mononuclear cells in response to a low-calorie diet: a nutrigenomics study. OMICS 12: 251–261. [DOI] [PubMed] [Google Scholar]
- 42. Lu H, Buchan RJ, Cook SA (2010) MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc Res 86: 410–420. [DOI] [PubMed] [Google Scholar]
- 43. Xie H, Sun L, Lodish HF (2009) Targeting microRNAs in obesity. Expert Opin Ther Targets 13: 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kajimoto K, Naraba H, Iwai N (2006) MicroRNA and 3T3-L1 pre-adipocyte differentiation. RNA 12: 1626–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chang CL, Au LC, Huang SW, Fai Kwok C, Ho LT, et al. (2011) Insulin up-regulates heme oxygenase-1 expression in 3T3-L1 adipocytes via PI3-kinase- and PKC-dependent pathways and heme oxygenase-1-associated microRNA downregulation. Endocrinology 152: 384–393. [DOI] [PubMed] [Google Scholar]
- 46. Rampersaud E, Damcott CM, Fu M, Shen H, McArdle P, et al. (2007) Identification of novel candidate genes for type 2 diabetes from a genome-wide association scan in the Old Order Amish: evidence for replication from diabetes-related quantitative traits and from independent populations. Diabetes 56: 3053–3062. [DOI] [PubMed] [Google Scholar]
- 47. Zhang Y, Wang L (2011) Nuclear receptor SHP inhibition of Dnmt1 expression via ERRγ. FEBS Lett 585: 1269–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, et al. (2010) Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol 11: 136–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Estep M, Armistead D, Hossain N, Elarainy H, Goodman Z, et al. (2010) Differential expression of miRNAs in the visceral adipose tissue of patients with non-alcoholic fatty liver disease. Aliment Pharmacol Ther 32: 487–497. [DOI] [PubMed] [Google Scholar]
- 50. Thulasingam S, Massilamany C, Gangaplara A, Dai H, Yarbaeva S, et al. (2011) miR-27b*, an oxidative stress-responsive microRNA modulates nuclear factor-kB pathway in RAW 264.7 cells. Mol Cell Biochem 352: 181–188. [DOI] [PubMed] [Google Scholar]
- 51. Karbiener M, Fischer C, Nowitsch S, Opriessnig P, Papak C, et al. (2009) microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochem Biophys Res Commun 390: 247–251. [DOI] [PubMed] [Google Scholar]
- 52. Nakada C, Tsukamoto Y, Matsuura K, Nguyen TL, Hijiya N, et al. (2011) Overexpression of miR-210, a downstream target of HIF1α, causes centrosome amplification in renal carcinoma cells. J Pathol 224: 280–288. [DOI] [PubMed] [Google Scholar]
- 53. Cao P, Deng Z, Wan M, Huang W, Cramer SD, et al. (2010) MicroRNA-101 negatively regulates Ezh2 and its expression is modulated by androgen receptor and HIF-1alpha/HIF-1beta. Mol Cancer 9: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee EK, Lee MJ, Abdelmohsen K, Kim W, Kim MM, et al. (2011) miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol 31: 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vreugdenhil E, Verissimo CS, Mariman R, Kamphorst JT, Barbosa JS, et al. (2009) MicroRNA 18 and 124a down-regulate the glucocorticoid receptor: implications for glucocorticoid responsiveness in the brain. Endocrinology 150: 2220–2228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All the miRNA transcripts sequenced and the differences between responders and non-responders (as fold change), categorized by p-value adjusted for multiple comparisons.
(DOC)
miRNA clusters (according to miRBase) upregulated in peripheral blood cells of non-responders to the low-calorie diet when compared to the responders, categorized by chromosomal location.
(DOC)
miRNA clusters (according to miRBase) downregulated in peripheral blood cells of non-responders to the low-calorie diet when compared to the responders, categorized by chromosomal location.
(DOC)
The most expressed miRNA transcripts in PBMC cells, based on read counts, and the chromosome where they are located.
(DOC)
List of predicted novel miRNAs (according to miRBase) found in peripheral blood mononuclear cells.
(DOC)
Target mRNAs of the microRNAs with differences in expression between responders and non-responders to the energy-restricted.
(DOC)