Abstract
Persistent signaling by the oncogenic epidermal growth factor receptor (EGFR) is a major source of cancer resistance to EGFR targeting. We established that inactivation of two sterol biosynthesis pathway genes, SC4MOL (sterol C4-methyl oxidase-like) and its partner NSDHL (NADP-dependent steroid dehydrogenase-like), sensitized tumor cells to EGFR inhibitors. Bioinformatics modeling of interactions for the sterol pathway genes in eukaryotes allowed us to hypothesize, and then extensively validate an unexpected role for SC4MOL and NSDHL in controlling the signaling, vesicular trafficking and degradation of EGFR and its dimerization partners, ERBB2 and ERBB3. Metabolic block upstream of SC4MOL with ketoconazole or CYP51A1 siRNA rescued cancer cell viability and EGFR degradation. Inactivation of SC4MOL markedly sensitized A431 xenografts to cetuximab, a therapeutic anti-EGFR antibody. Analysis of Nsdhl-deficient Bpa1H/+ mice confirmed dramatic and selective loss of internalized PDGFR in fibroblasts, and reduced activation of EGFR and its effectors in regions of skin lacking NSDHL.
Introduction
The epidermal growth factor receptor (EGFR) provides essential growth and pro-survival signals to epithelial cells, and is often targeted therapeutically in epithelial malignancies. Unfortunately, EGFR-antagonizing treatment strategies are often limited in their efficacy due to acquired or primary resistance in tumors. Receptor switching, or compensatory activation of EGFR co-receptors such as the receptor tyrosine kinases (RTKs) ERBB2 (1), ERBB3 (2), and IGF-1R (3) are examples of acquired resistance, while pre-existing somatic mutations activating downstream effectors of the EGFR pathway, such as RAS and BRAF, commonly confer primary resistance to EGFR antagonists in pancreatic or colon cancers (4). Persistent EGFR signaling is a basis for resistance to EGFR antagonists, as observed in PTEN deficient tumors (5), or when EGFR internalization and degradation are reduced (6). In many cases, the source of therapeutic resistance remains unknown, limiting clinical efforts to select patients for EGFR-targeting therapeutics or to generally enhance the potency of such therapeutics in all patients.
In a network-guided siRNA-based screen (7), we recently identified SC4MOL (sterol C4-methyl oxidase-like) among genes whose silencing significantly sensitizes refractory tumor cells to EGFR inhibitors. The EGFR-focused library was designed on the basis of systems biology prediction that genes engaged in synthetic lethal relationships involve those closely linked in signaling processes (8 , 9). Among the validated hits that increased cell killing by common EGFR antagonists, SC4MOL was consistently one of the most effective modulators. SC4MOL, an intermediate enzyme in the cholesterol biosynthetic pathway, was included in the library based on its representation in a high confidence GEO transcriptional profiling dataset (GSE6521) as a transcript that rapidly undergoes significant expression change in response to stimulation or inhibition of EGFR. Active sterol biosynthesis remains an essential metabolic component of cancer, and changes in the function of this pathway are thought to contribute to resistance to some forms of cancer treatment (10). However, to date, no direct functional connections between SC4MOL or other sterol pathway proteins and EGFR signaling have been reported.
In this study, we first systematically explore the proteins of the distal cholesterol biosynthesis pathway to establish that depletion or loss of SC4MOL and its partner, NSDHL (NADP-dependent steroid dehydrogenase-like), specifically synergize with common inhibitors of EGFR. We then model from the interaction networks of the evolutionarily conserved orthologs of these proteins in yeast to determine that SC4MOL and NSDHL directly regulate intracellular vesicular trafficking of EGFR, influencing its rate of degradation. These results reveal a previously unidentified means of regulating RTK expression and activity, and suggest a more complex action for these cholesterol biosynthetic pathway enzymes than has been formerly appreciated.
Results
Probing the sterol biosynthesis pathway for regulation of response to EGFR inhibitors
To address the mechanism of SC4MOL regulation of response to the EGFR inhibitors erlotinib and cetuximab, we first considered whether this effect was general to proteins operating in the cholesterol biosynthesis pathway (Fig. 1A) or more specific. SC4MOL is highly conserved throughout evolution, as are many genes operating upstream and downstream in the sterol synthesis pathway (11). Three human catalytic enzymes, SC4MOL, NSDHL, and HSD17B7, and a gene with unknown function, C14ORF1 are orthologous to a complex of yeast C4-sterol demethylation genes that define the "ergosome" (ERG25/SC4MOL, ERG26/NSDHL, ERG27/HSD17B7, and ERG28/C14ORF1) (12).
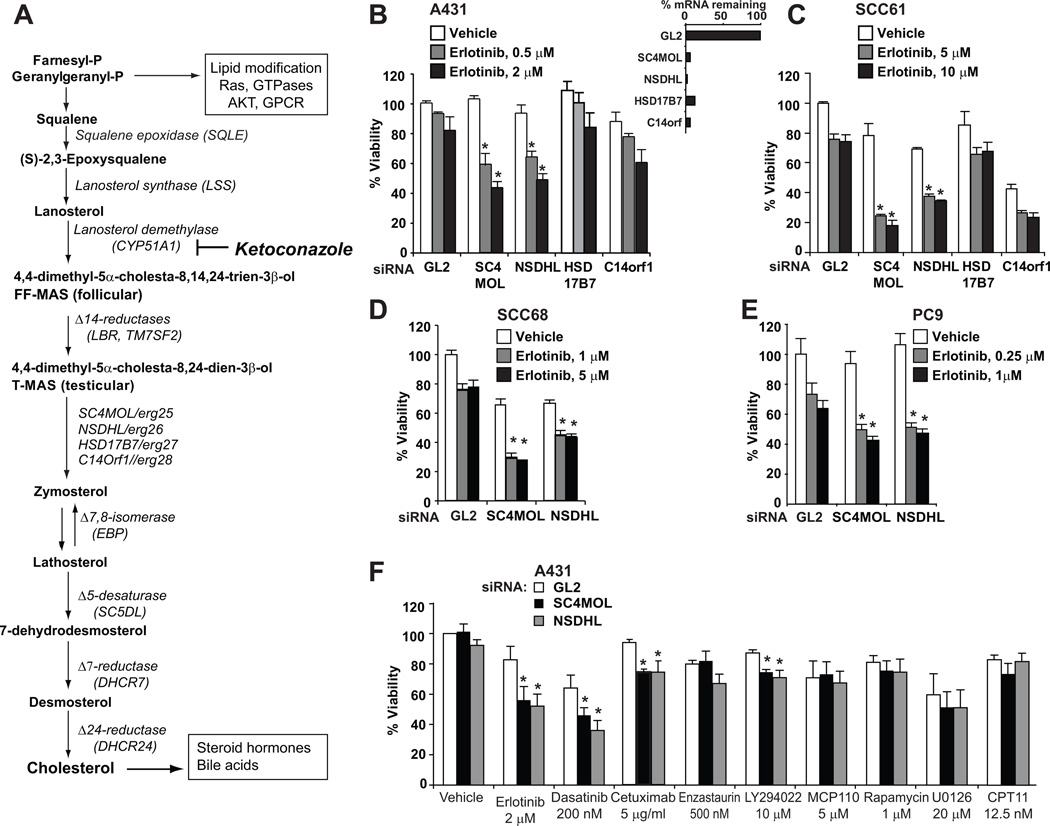
Figure 1. Effects of sterol pathway genes silencing on cancer cell drug sensitivity.
A Schema of the cholesterol pathway. Relevant enzymes are italicized, and biological functions are shown in boxed text. B–F Silencing of SC4MOL and NSDHL increased cytotoxicity of erlotinib. A431 B, F), SCC61 (C), PC9 (D) and SCC68 (E) cells were made deficient in the indicated sterol pathway genes and, 24 hours later, were either mock treated or exposed to erlotinib for 72 hours. F Specificity of activity of SC4MOL and NSDHL silencing. A431 cells made deficient in SC4MOL and NSDHL were treated with indicated inhibitors at concentrations producing 20–30% decrement in viability. In (B–F), viability data from three independent experiments were normalized to mock-treated GL2-control transfected cells. Each column represents averaged results; bars standard deviations. *, p <0.001.
Depletion of SC4MOL and NSDHL by multiple siRNA or shRNA (Fig. 1B Fig. S1A–C), but not of 6 other proteins (SQLE, LSS, CYP51A1, TM7SF2, LBR, HSD17B7, and C14ORF1) operating further upstream or downstream in the pathway (Fig. S1D–F), sensitized A431 cells to the EGFR kinase inhibitor erlotinib, suggesting a specific block focused at the C4-demethylation step in the pathway. Similar results were obtained in the head and neck squamous carcinoma cell lines SCC61 (Fig. 1C) and SCC68 (Fig. 1D) expressing moderate levels of EGFR (Fig. S2A), and in the lung adenocarcinoma cell line PC9 (Fig. 1E), which expresses a mutated form of EGFR, ΔE746–A750 (13), indicating the findings were not specific to A431 cells. Sensitization was also observed with two short hairpin RNA (shRNA) constructs targeting SC4MOL (Fig. S1C), and was associated with marked enhancement of apoptosis (Fig. S2B, S2C). In contrast to sensitization, inactivation of SC4MOL and NSDHL did not affect intrinsic cell growth of the EGFR-high A431 cells or untransformed MCF12F mammary cells, slightly reduced growth of EGFR-intermediate SCC61 and SCC68 cells, and was extremely deleterious to the EGFR-low head and carcinoma cell line, FaDu (making it difficult to assess sensitization in the last line) (Figs. 1B–D , S2D).
Targeting SC4MOL and NSDHL specifically sensitized cells to erlotinib (targeting EGFR), cetuximab (targeting EGFR), dasatinib (targeting SRC, EGFR (14), and other RTKs), minimally sensitized to LY294022 (targeting PI3K), and did not sensitize to enzastaurin (targeting PKC), MCP110 (targeting RAS/RAF interactions (15)), rapamycin (targeting mTOR), U0126 (targeting MEK1/2), or CPT11 (a DNA-damaging agent), supporting the selectivity for EGFR in cancer cell lines with activated EGFR signaling (Fig. 1F).
Congruence between sterol metabolite profile and sensitization to EGFR-targeting drugs
We next determined whether production of specific sterol metabolites correlated with sensitization to EGFR inhibitors, and was sufficient to explain the observed sensitization. SiRNA targeting SC4MOL (siSC4MOL) elevated expression of the SC4MOL substrates 4-mono- and 4,4-dimethylzymosterol (T-MAS) (Fig. 2A , S3A), and reduced downstream enzymatic products such as lathosterol. As a contrasting control, depletion of the upstream enzyme, CYP51A1 (Fig. 2A second row), specifically increased its substrate, dihydrolanosterol. Studies in yeast have previously shown that either chemical or genetic inhibition of the CYP51A1 ortholog, ERG11, rescues lethal mutants in the SC4MOL ortholog, ERG25 (16). Here, the CYP51A1 inhibitor ketoconazole reversed the accumulation of C4-methylsterol substrates in SC4MOL-silenced cells (Fig. 2B), and eliminated the SC4MOL-dependent sensitization of A431 cells to erlotinib (Fig. 2C). Similar results were obtained using siRNA to deplete CYP51A1 (Fig. 2D). As a control, we confirmed siRNA-depleted SC4MOL levels remained low in ketoconazole-treated cells (Fig. S3B), excluding indirect action. Surprisingly, although NSDHL was efficiently depleted by siRNA (Fig. S3B) this did not produce accumulation of 4-methylsterols, substrates of functional SC4MOL. This may be due to the detection limit of the GC-MS technique in identifying low levels of carboxylated derivatives of T-MAS, or to the enhanced ability of tumor cells to dispose of this metabolite.
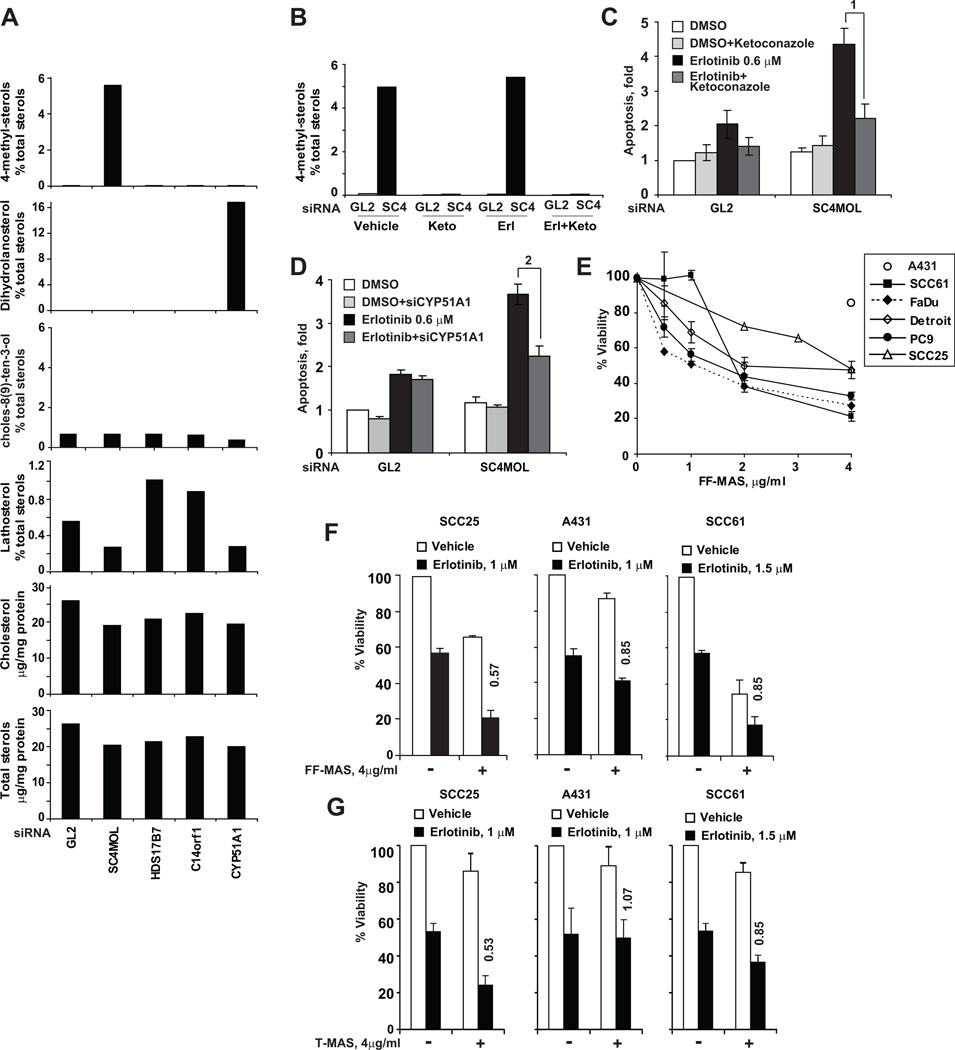
Figure 2. Concordant effects of siRNA silencing on the cellular composition of sterols.
A Sterol composition of cellular pellets was assessed by gas chromatography and mass spectroscopy following extraction of lipids. The area under the peak of each sterol species was expressed as proportion of total cellular sterols, or as per milligram of total protein (for total cholesterol and total sterols). B Ketoconazole treatment for 48 hours prevented accumulation of C4-methylsterols in A431 cells made deficient in SC4MOL with siRNA. In samples parallel to (C), C4-methylated sterols were GC-MS measured in total lipid extracts of GL2 and SC4MOL (SC4) silenced A431 cells. C Effects of CYP51A1 inhibitor ketoconazole (6 µg/ml) on apoptosis. A431 cells were made SC4MOL deficient by siRNA transfection and, in 24 hours, treated with vehicle or erlotinib for 72 hours followed by Annexin V surface labeling. Similar results were obtained with co-transfection with 15 nM CYP51A1 siRNA (D). In all samples, the total concentration of siRNA was maintained at 30 nM. p-values: (1) 0.001 (2) 0.001. (E) Effects of FF-MAS on viability of cancer cell lines. (F–G) C4-methylated sterol metabolites, FF-MAS (F) and T-MAS (G) sensitize cancer cell lines to erlotinib. Columns represent averaged results of three independent experiments; bars standard deviations. Numbers above columns indicate relative increase in erlotinib cytotoxicity, the sensitization index (SI).
Metabolite profiling also showed a small but appreciable reduction of cholesterol in SC4MOL-silenced cells (Fig. 2A) or NSDHL-deficient Bpa1H fibroblasts (Fig. S3C). However, supplementation of media with cholesterol or an upstream metabolite in the pathway such as lanosterol did not have any effect on viability or sensitivity to EGFR inhibitors (Fig. S3D–G) suggesting specific effects at the level of the C4 demethylation complex. In contrast, addition of T-MAS or, most notably, its immediate precursor, FF-MAS, to the culture medium reduced cancer cell viability (Fig. 2E) and increased cancer cells sensitivity to erlotinib (Fig. 2F, G). Taken in sum, these data support the interpretation that sensitization to erlotinib is associated with perturbation of pools of a sterol intermediate metabolite proximally upstream of SC4MOL in the metabolic pathway. The negative effect of accumulation of this substrate can be rescued by a upstream blockade, while gross changes in the abundance of more distal upstream or downstream sterols (lanosterol, cholesterol) per se are not sufficient to explain the observed effects on EGFR.
Network modeling suggests a role for SC4MOL and NSDHL in trafficking of EGFR
No previous studies have suggested a mechanism for how the SC4MOL protein might influence sensitization to EGFR inhibitors. Among all sterol metabolizing enzymes and their corresponding substrates, ERG1, ERG7, ERG11, ERG24, ERG25, ERG26, ERG27 were conserved between Saccharomyces cerevisiae and humans, such that proteins with high levels of sequence homology performed comparable functions in sterol biosynthesis (Fig. 1 S4A, S4B and Table S1). The majority of ERG genes downstream of zymosterol (ERG6, ERG2, ERG3, ERG5 and ERG4) showed little or no sequence homology with human genes (KEGG pathways (17)), but instead proteins with unrelated sequence performed comparable enzymatic activities. As a source of insight, we systematically analyzed the yeast orthologs in this highly conserved metabolic pathway. For this, we used the yeast sterol pathway proteins as seeds to mine data from large-scale yeast genetic arrays (18), affinity purification and mass spectroscopy resolution of protein complexes (19–21) and protein complementation screens (22), to gain further insight into their function (Figure S4, Table S1 and supplemental Cytoscape file).
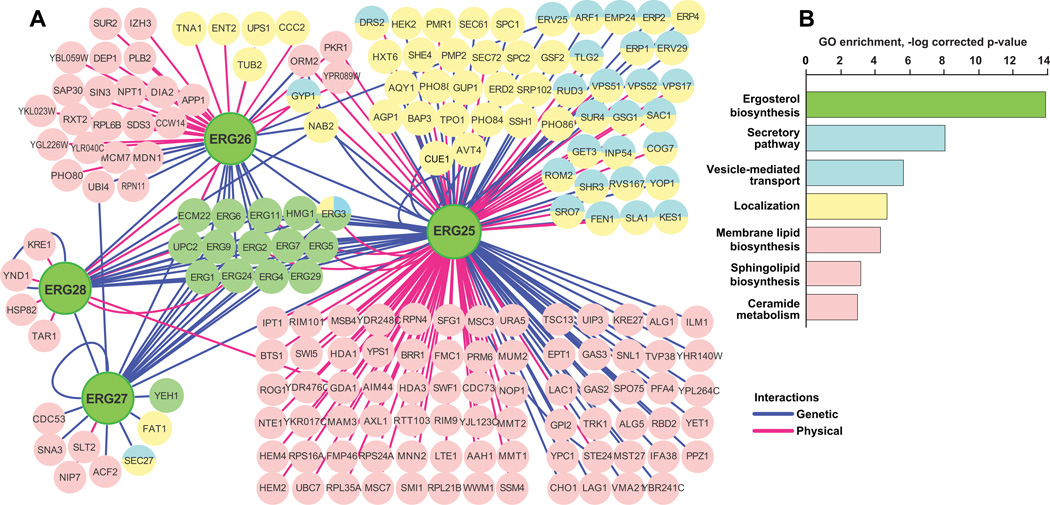
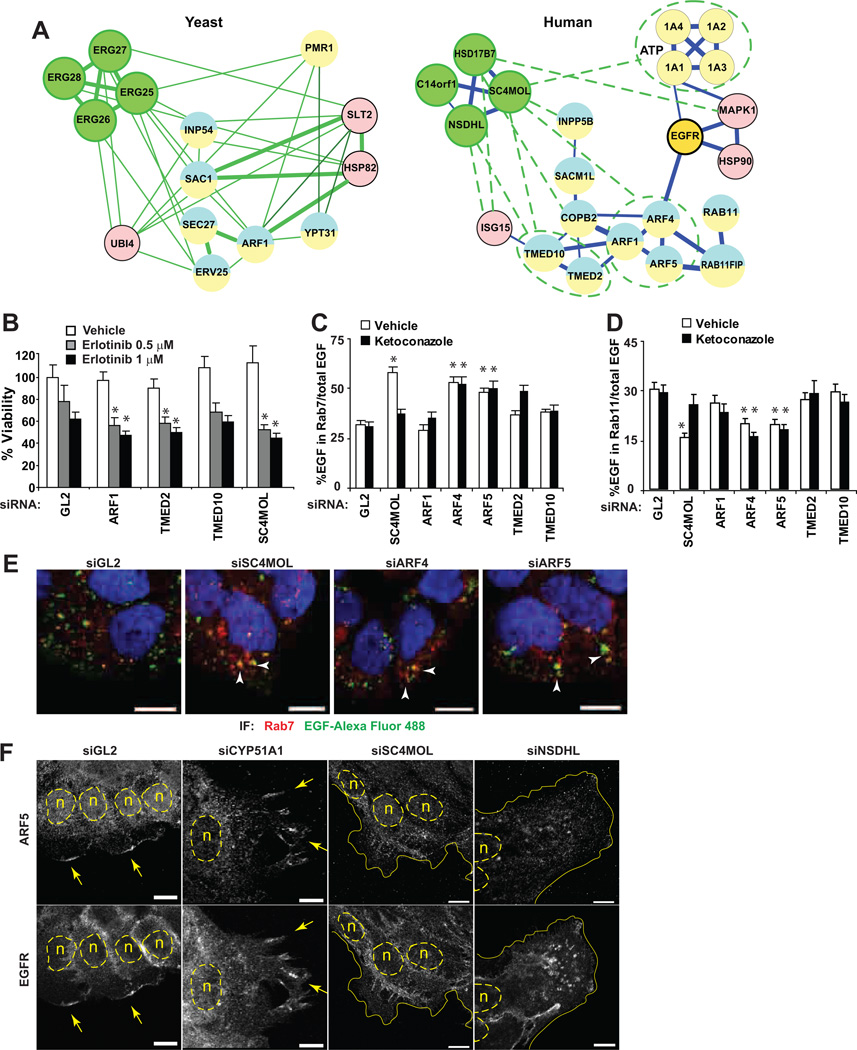
The in silico network generated for ERG25, ERG26, ERG27, ERG28 proteins (Fig. 3A S4) revealed, as expected, many interactions reflecting their participation in the linear ergosterol biosynthesis pathway (green circles in Fig. 3A) as well as additional interactions with genes annotated for roles in lipid synthesis and metabolism. Unexpectedly, multiple genetic and protein-protein interactions were also detected between ERG25 and proteins with Gene Ontology (GO) annotations indicating direct involvement in vesicular transport, secretory pathway and cellular localization: of 178 ERG25-interacting proteins, 53 had such GO annotations, representing a highly significant enrichment (e.g. vesicle-mediated transport, p=1.4*10−8) (Fig. 3B). ERG11, which rescues ERG25 mutations, also had many interactions and a significant enrichment for such GO annotations. In contrast, ERG27 and ERG28 which did not affect response to EGFR-targeting agents, interacted with only 8 and 7 non-sterol pathway genes, respectively, and fewer genes overall (Fig. S4B). ERG26 had an intermediate number of interactors (n=46), and no significant GO enrichment. However, genetic and biochemical studies in yeast (12) have noted a close physical and functional interaction between ERG25 and ERG26, suggesting NSDHL might be acting through SC4MOL to influence transport processes. Resistance to cetuximab in the clinic has been strongly linked to defects in internalization and degradation of EGFR (5); we therefore tested the idea that SC4MOL and associated proteins may regulate EGFR trafficking.
Figure 3. Interactions of the ERG25-ERG28 genes in S.cerevisiae.
A) Sterol pathway genes interact genetically (blue lines) or physically in protein complexes (red lines) with multiple genes regulating cellular localization (yellow) and vesicular transport (blue) of macromolecules. B Gene Ontology function (GO function Id) classification of genes interacting with ERG25-ERG28: ergosterol biosynthesis (6696), secretory pathway (45045), vesicle-mediated transport (16192), localization (51179), membrane lipid biosynthesis (46467), sphingolipid biosynthesis (30148), ceramide metabolism (6672). Y-axis represents negative logarithm of p-value indicating significance for enrichment considering the entire yeast genome.
Depletion of SC4MOL or NSDHL increases vesicular trafficking of EGFR to the lysosome
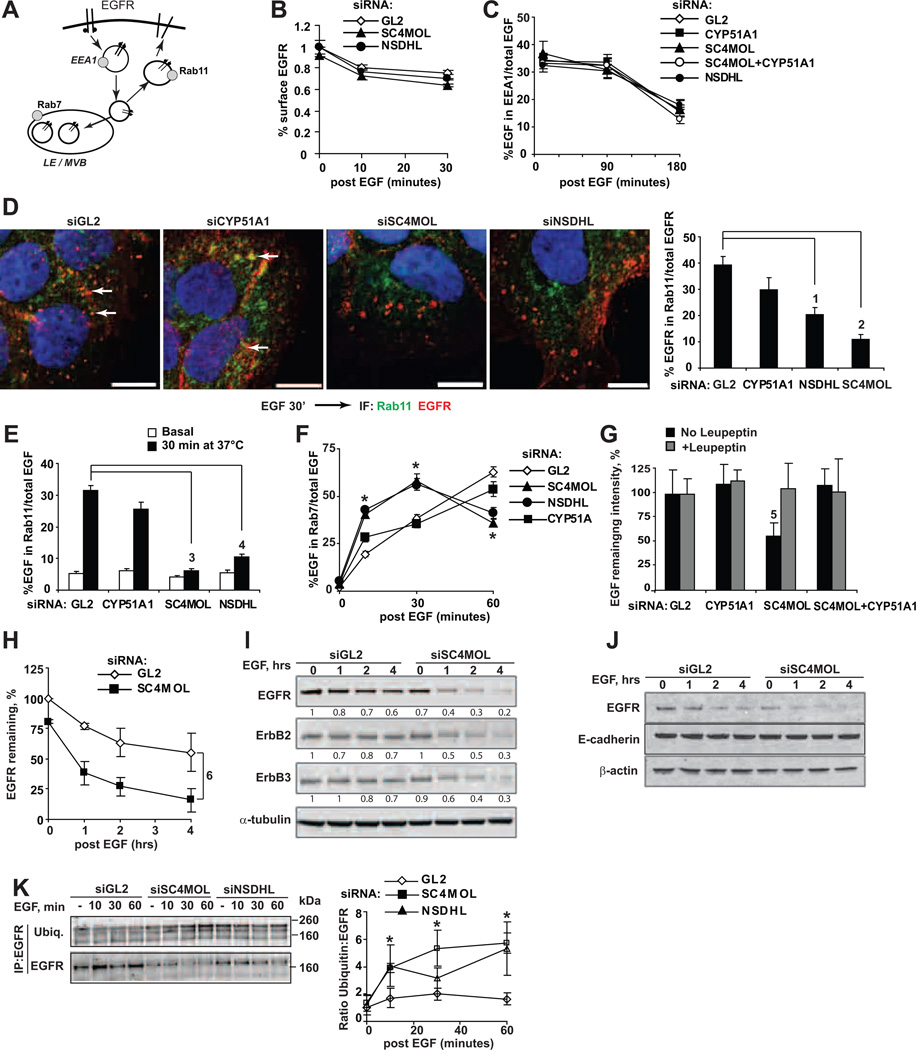
Following activation by EGF binding, or binding by antibodies such as cetuximab, EGFR is internalized from the plasma membrane, moves through a series of sorting endosomes, and then either recycles to the cell surface or moves through the late endosome/multivesicular body (MVB) to the lysosome for destruction (Fig. 4A) (23). After first excluding an effect of SC4MOL-deficiency on the rates of EGFR synthesis (Fig. S5A), we then systematically probed EGFR trafficking following siRNA depletion of SC4MOL. Levels of cell surface EGFR declined at a similar rate following EGF stimulation (Fig. 4B), and I125-EGF internalization kinetics were comparable in control or SC4MOL-depleted cells (Fig. S5B). We also did not observe differences in the transit of labeled EGF through the early endosomal compartment, as quantified (24) based on co-localization with early endosomal antigen 1 (EEA1, Fig. 4C S6A).
Figure 4. Altered endosomal trafficking of EGFR in SC4MOL and NSDHL deficient cells.
(A) Simplified schema of EGFR endocytosis and markers of compartments. EEA1 early endosomal antigen 1; LE late endosomes; MVB multivesicular bodies. (B,C) Kinetics of EGF-induced EGFR internalization in siRNA-depleted cells as measured by the surface EGFR (B), or by co-localization of EGF-Alexa Fluor 488 with EEA1 (C). (D) Silencing of SC4MOL and NSDHL prevented association of internalized EGFR (red) with RAB11-positive recycling endosomes (green) in A431 cells. Right averaged co-localization results from 2 repeats. (E) Co-localization of labeled EGF and RAB11. (F) Depletion of SC4MOL and NSDHL increased association of EGF with RAB7 endosomes (note depletion of EGF at 60 minutes). (G) EGF loss at 1 hour in SC4MOL-silenced cells was prevented by leupeptin or CYP51A1 silencing. (H) EGFR degradation in SC4MOL deficient cells following treatment with EGF. Symbols averaged Western blot densities from three independent experiments; bars StDev. I Accelerated degradation of ErbB1-3 proteins but not of E-cadherin (J) in SC4MOL deficient cells; (I, J) representative images of three independent Western blots showing identical results. Numbers below bands are their normalized densities. K Ubiquitin conjugated EGFR following immunoprecipitation with cetuximab. Right, averaged results from three independent experiments; bars SEM. p-values: (1) 3*10−4(2) 4*10−7(3) 1.4*10−27(4) 3.7*10−20(5) 0.0011 or less for all comparisons; p-values for slopes (6) 0.005. *, p <0.001 vs. GL2 at all points; NS, non-significant. Horizontal bars in all images, 10 µm.(12)
In contrast, depletion of either SC4MOL or NSDHL significantly reduced the amount of fluorescently labeled EGFR (Fig. 4D) or EGF (Fig. 4E S5C, S6B) associated with RAB11-positive recycling endosomes after 30 minutes of EGF stimulation. Conversely, SC4MOL and NSDHL-deficient cells showed an accelerated concentration of internalized fluorescently labeled EGF in RAB7-positive (10 minutes, Fig. 4F S6C) or EGFR in LAMP1-positive late endosomes (Fig. S6D), followed by marked loss of EGF in these compartments at later time-points (Fig. 4G S6D, S6E).
Indeed, although basal levels of surface EGFR were comparable, depletion of SC4MOL dramatically reduced total cellular EGFR levels within an hour of EGF treatment (Fig. 4H and 4I), with over 80% loss of EGFR within 4 hours of EGF treatment. Interestingly, ERBB2 and ERBB3 capable of binding the EGFR (but not EGF) in the membranes, also underwent similar rapid elimination in SC4MOL-targeted cells following EGF-induced EGFR internalization to endosomes (Fig. 4I), while an unrelated surface protein, E-cadherin, did not (Fig. 4J). Similar findings were observed with direct labeling of EGFR in SC4MOL-deficient cells treated with EGF for 30 and 60 minutes, which showed substantial depletion of EGFR in SC4MOL-deficient cells (Fig. S6D). Ubiquitin-conjugated immunoprecipitated forms of EGFR accumulated with accelerated kinetics in SC4MOL or NSDHL-deficient cells, compatible with early arrival at the late endosomes (Fig. 4K and S7A–C). Finally, treatment with the anti-malarial primaquine, which inhibits lysosomal acidification, or the CYP51A1 inhibitor ketoconazole, rescued EGFR levels in SC4MOL and NSDHL-depleted cells, while the proteasomal inhibitor bortezomib did not (Fig. S7D and S7E). In keeping with the possibility of rapid lysosomal clearance of EGF-EGFR complexes in SC4MOL-depleted cells, the Cys/Ser/Thr peptidase inhibitor leupeptin (25) also eliminated loss of fluorescently-labeled EGF (compared to GL2 and CYP51A1 in Fig. 4G and S6E).
Depletion of SC4MOL suppresses EGFR-dependent signaling, causes EGFR loss, and induces in vivo sensitization to cetuximab
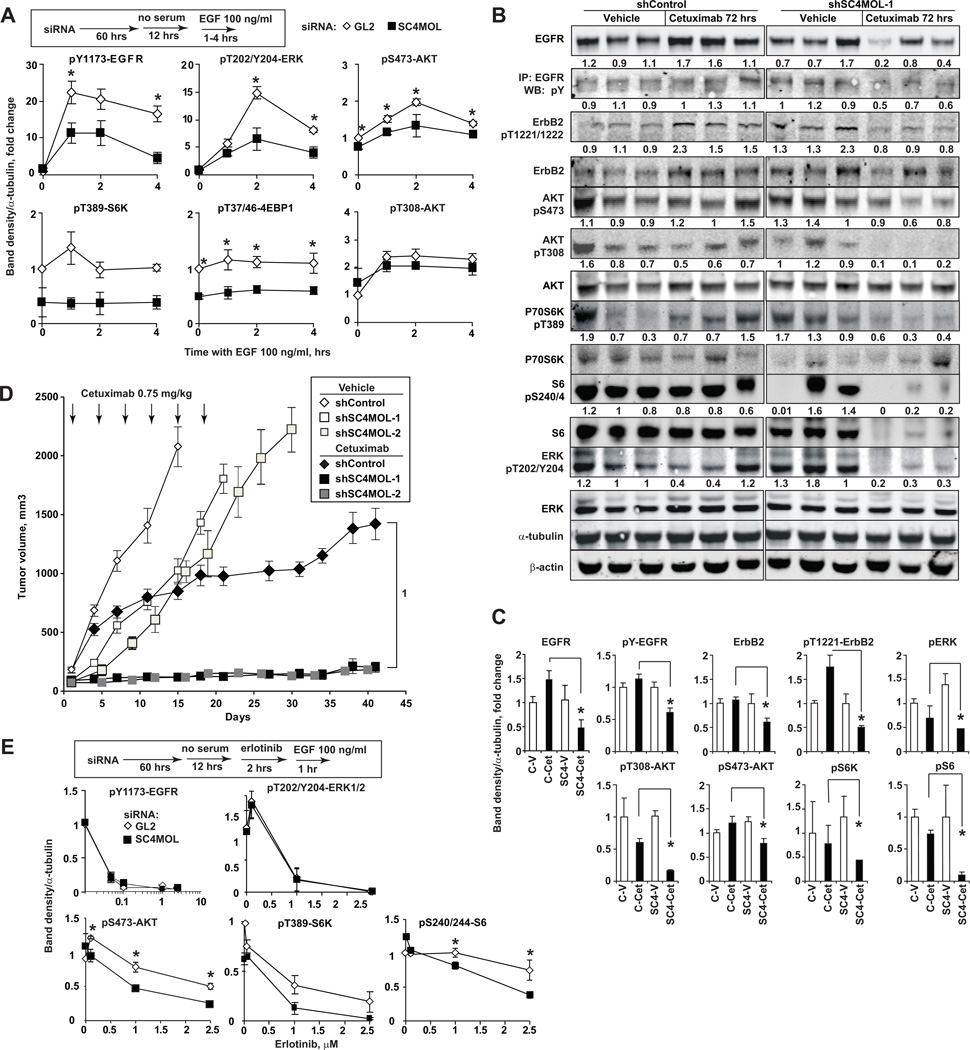
Trafficking of EGFR to late endosomes and lysosomes has been shown to rapidly terminate its signaling (26). We investigated EGFR signaling in cells with depleted SC4MOL, based on the observation of aberrant EGFR trafficking in these cells (Fig. 4). Indeed, SC4MOL siRNA-treated SCC61 cells pulsed with EGF showed reduced total and phosphorylated EGFR as well as reduced phosphorylation of its key downstream effectors, ERK, AKT (phospho-S473), S6K, 4EBP1 and S6 ribosomal protein (Fig. 5A S8A).
Figure 5. Silencing of SC4MOL suppresses EGFR signaling and sensitizes A431 xenografts to cetuximab.
(A) Silencing SC4MOL in SCC61 cells suppresses phosphorylation of EGFR downstream effectors following EGF stimulation for indicated time intervals. (B) Effects of a single dose of cetuximab on A431 xenografts at 72 hours. Numbers below bands are tubulin-normalized densities relative to vehicle-treated shControl grafts. (C) Summary of results quantified from (B); *, p<0.05. D Tumor volumes of xenografts of shRNA-modified A431 cells treated with cetuximab (closed symbols) or vehicle (open symbols); p-value (1) 2.2*10−16. (E) SCC61 cells depleted of SC4MOL are sensitized to erlotinib-induced suppression of AKT-mTOR pathway signaling. Graphs represent averaged results of 3 experiments; bars, SEM.
We used shRNA to deplete SC4MOL in A431, and obtained two independent lines for xenograft analysis, and a non-targeting control-depleted line (Fig. S2B). Lysates of xenografted A431 tumors obtained 72 hours after a single i.p. dose of cetuximab delivered on day 8 post-implantation showed reduction of total EGFR, and marked suppression of phosphorylation of EGFR, its heterodimerizing partner ERBB2, and the downstream signaling effectors AKT, S6K, S6 and ERK (Fig. 5B, C) in SC4MOL-deficient versus control xenografts. If depletion of SC4MOL sensitizes cells to EGFR inhibitors by promoting EGFR degradation, it should be particularly effective in sensitizing tumors to cetuximab in vivo, given the action of this antibody in promoting EGFR lysosomal degradation (27). Growth of SC4MOL-depleted tumors was moderately suppressed compared to control-depleted A431 cells (p=2.2*10−16) (Fig. 5D). Cetuximab treatment of control-depleted tumors beginning at day 8 after injection of mice with tumor cells transiently reduced tumor growth, with resuming after ~ 2 weeks (Fig. 5D). In striking contrast, similar cetuximab treatment of SC4MOL-depleted tumors, almost completely prevented growth of SC4MOL-deficient xenografts (Fig. 5D) and 2–3 of 10 animals in each shRNA group showed no evidence of tumor at 7 weeks. Similarly to the conserved ERG11/CYP51A1 and ERG25/SC4MOL epistasis (16) in vitro (Fig. 2B–D), SC4MOL-deficient A431 xenografts resisted cetuximab in mice given fluconazole, a CYP51A1 inhibitor, in drinking water (Fig. S8B). The effect of SC4MOL silencing on EGFR signaling was further evaluated in the context of EGFR kinase inhibitor, erlotinib (Fig. 5E), in cells treated with EGF to induce receptor internalization (28). The SC4MOL-depleted cells showed higher sensitivity to erlotinib-induced suppression of AKT, S6 kinase and S6 phosphorylation, while levels of pY1173-EGFR and pERK were equally suppressed by erlotinib in SC4MOL deficient cells and controls (Fig. 5E).
Network-predicted SC4MOL interactions control EGFR endocytic traffic
Given the clear success of the yeast protein interaction network in suggesting a role for SC4MOL and NSDHL in EGFR protein trafficking (Fig. 3A S4), we hypothesized that these data could also nominate SC4MOL effectors relevant to this process, given the growing appreciation of the existence of interologs (evolutionarily conserved interactions) among highly conserved proteins and protein modules (29). We therefore identified human orthologs for those yeast proteins shown in Fig. 3A possessing GO annotations relevant to vesicular trafficking (Fig. 6A and Table S1). Suggestively, our analysis predicted that ERG25/SC4MOL and ERG26/NSDHL might interact directly with multiple components of the mammalian exocytic machinery, including COPI, the p24 cargo receptors TMED2 and TMED10, or the ARF GTPases (18 30 31). This was particularly interesting, as we had identified ARF4 and ARF5 as hits in the initial screen yielding SC4MOL as a sensitizer to EGFR-targeting drugs (7), and as these proteins are known to form a complex that regulates the trafficking of EGFR out of the Golgi and late endosomes via a RAB11-mediated recycling pathway (32).
Figure 6. Network-predicted SC4MOL interactions control EGFR endocytic traffic.
(A) Network of yeast and human interologs for ERG25/SC4MOL and EGFR interacting proteins. Interactions inferred from yeast are shown as green dashed lines. Circles are nodes represented by individual proteins or paralogs. Blue lines depict mammalian interactions where thickness is proportionate to the confidence score of an interaction. (B) Silencing of ERG25/SC4MOL interacting proteins increased cytotoxicity of erlotinib. (C, D) Depletion of SC4MOL, and its interacting ARF4 and ARF5, promotes EGF localization to late RAB7 endosomes (C, and representative images in (E), arrowheads) and suppresses EGF entry into RAB11 recycling compartments (D). Ketoconazole pre-treatment selectively rescued EGF trafficking defect in SC4MOL silenced cells (C D, closed columns). (F) ARF5 and EGFR are expressed at the growing edge of A431 cells (arrows). Silencing of SC4MOL and NSDHL reduced EGFR and ARF5 co-localization. n nuclei marked with dashed lines, solid lines cell borders. *, p<0.001 in all graphs; horizontal bars in all images, 10 µm.
Systematically probing the ARF sub-network, we found that siRNA depletion of several components of this network increased sensitivity of the A431 cancer cell line to EGFR inhibition (Fig. 6B and in (7)). Further, silencing of ARF4 and ARF5 increased accumulation of labeled EGF in RAB7-positive endosomes and reduced entry of EGF into RAB11 compartments (Fig. 6C–E), paralleling the SC4MOL depletion phenotype. While ketoconazole, an inhibitor of CYP51, rescued the effects of SC4MOL depletion (i.e., through elimination of metabolite accumulation), it did not reverse the EGF trafficking defects due to ARF4 and ARF5 silencing in keeping with our model in Fig. 6A. We also found that EGFR and ARF5 co-localized in the leading cellular contact-free edge of control-silenced A431 cells (Fig. 6F GL2 and CYP51A1), consistent with the critical role of ARF4 and ARF5-dependent in trans-Golgi trafficking and exocytosis (33). This co-localization was completely blocked by silencing of SC4MOL or NSDHL (Fig. 6F). These data are compatible with the idea of an important contribution of SC4MOL and NSDHL to EGFR endocytic traffic within ARF4 and ARF5-positive endosomes.
Genetic loss of Nsdhl reduces EGFR expression and signaling in vivo
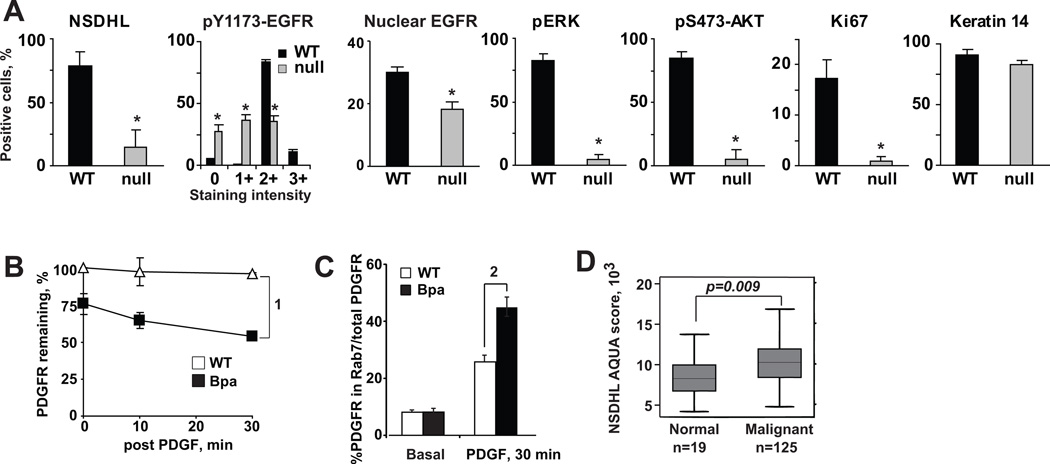
Finally, to determine if our data offer new insights into the physiological control of EGFR activity in vivo, we took advantage of the bare patches (Bpa1H mouse model, which contains a nonsense mutation (K103X) in the X-linked Nsdhl gene (34). This mutation results in a null allele with a truncated protein that is degraded. This genetic lesion in heterozygous Bpa1H/+ females produces mosaic NSDHL-null regions of the skin with characteristic patchy, scaly eruptions by postnatal day 5 that subsequently resolve, leaving linear stripes following lines of X-inactivation. Strikingly, these NSDHL-null areas of the skin (between the dashed lines in Fig. S9A) differed from the wild type areas in the patterns of EGFR expression, activation, and signaling. Total EGFR staining (Fig. 7A S9B and S9C) was predominantly membranous in the NSDHL-null areas. The nuclear EGFR was significantly reduced in the mutated patches of the skin (Fig. 7A and S9C). The reduced intracellular EGFR staining was accompanied by a marked loss of phosphorylated (activated) Y1173-EGFR (Fig. 7A S9D), T202/Y204-ERK1/2 (Fig. 7A S9E) and S473-AKT (Fig. 7A S9F) signal in the NSDHL-null areas. EGFR signaling is of critical importance for keratinocyte proliferation and differentiation (35). The NSDHL-null areas of the skin also exhibited significantly reduced proliferation, as determined by Ki-67 (Fig. 7A S9G) expression and in vivo BrdU incorporation (Fig. S9H). Global effects of loss of NSDHL protein expression were not observed, as expression of keratin-14 was normal (Fig. 7A S9I). However, the Nsdhl-mutated areas consistently showed parakeratosis accompanied by reduced expression of loricrin, a marker of terminally differentiated keratinocytes (Fig. S9J).
Figure 7. Sterol pathway genes expression regulates keratinocyte EGFR signaling in vivo.
A) Quantified results from immunohistochemical evaluation of skin tissues from mosaic female Bpa1H/+ pups (day P3); WT wild type areas, null denotes areas lacking NSDHL expression; *, t-test p<0.001. B PDGFR degradation following treatment with PDGF, 25 ng/ml, for indicated times. Symbols averaged Western blot densities from three independent experiments; bars StDev; (1) p=0.005 comparing slopes. (C) Increased association of PDGFR with RAB7 endosomes in NSDHL deficient Bpa1H murine fibroblasts; (2) p=0.0006. (D) Elevated NSDHL protein expression in head and neck squamous carcinoma.
Using NSDHL-deficient mouse embryonal fibroblasts derived from these mice, we next assessed whether a ligand-induced trafficking defect was observed for a RTK commonly expressed in fibroblasts, PDGFR. Semi-quantitative analysis of PDGFR levels in these fibroblasts treated with PDGF showed accelerated ligand-induced PDGFR loss in NSDHL-deficient Bpa1H cells, but not in controls; the former was associated with concentration of PDGFR in Rab7 endosomes (Fig. 7B, C).
Finally, while no suitable immunohistochemistry antibody reagents were available to analyze SC4MOL, there was increased expression of NSDHL protein in the proliferating basal layer of normal oral keratinocytes (Fig. S10A), thus suggesting critical requirement of the sterol pathway enzymes for the EGFR-dependent cellular growth. We further performed quantitative measurements of NSDHL using AQUA (36) and found NSDHL expression was more abundant in squamous cell head and neck carcinomas compared to normal epithelium (Fig. 7D S10B, S10C) although not affecting the survival of squamous head and neck carcinoma patients (Fig. S10D).
Discussion
We here use a network modeling-guided strategy to show that depletion of SC4MOL and NSDHL, best known as components of a C4-demethylation complex in the distal cholesterol pathway, potently sensitizes tumor cells to EGFR inhibitors, or could be directly cytotoxic in cancer cells with low EGFR. Our data suggest that the mechanistic basis for this effect is a requirement for SC4MOL and NSDHL for effective endosomal trafficking of EGFR, thus limiting the EGFR stability and the availability of an activated receptor pool. Mechanistically, the activity of SC4MOL and NSDHL mediates the production of specific sterol metabolites, and may also involve proteins regulating the endocytic recycling of receptors (such as ARF1, ARF4 and ARF5), as inferred from the yeast interaction data.
Many studies have implicated function of the sterol synthesis pathway in tumor growth and response to treatment. For example, the sterol composition of the membrane has been shown to regulate EGFR signaling (26) and sensitivity of head and neck cancer cells to apoptosis (37). Inhibition of lipogenesis has been an effective anti-tumor strategy in glioblastoma models associated with hyperactive EGFRvIII (38). Suggestively, we found increased expression of NSDHL in head and carcinoma tissues (Fig. 7D) in agreement with reports on accelerated sterol biosynthesis in cancer which associates with drug resistance and poor survival in breast and non-small cell lung cancer (39 40). However, in contrast to upstream pathway components such as farnesyltransferases, and downstream components directly regulating cholesterol production, SC4MOL and NSDHL have not been previously analyzed in the context of cancer signaling.
Our data imply a particularly important and previously unappreciated functional role for SC4MOL and NSDHL in vesicular trafficking. We observed a significant bias in routing the EGFR-containing endosomes towards RAB7 late endocytic compartments as opposed to RAB11 recycling compartments (Fig. 5 and S6). Such alteration may promote rapid EGFR dephosphorylation via membrane contact with PTP1B (41) or other EGFR phosphatases. Studies of NSDHL have also indicated this protein traffics through the Golgi, and then associates with ER membranes and lipid droplets (42). Lipid droplets are ER-derived structures that serve to compartmentalize lipids and an array of enzymes, kinases and other proteins. Lipid droplets have attracted increasing attention because of evidence that they mediate trafficking of signaling proteins to multiple organelles, and have roles in cell signaling and secretion during inflammation and cancer (43). A recent proteomic study also noted enrichment of NSDHL at lysosomes during camptothecin-induced apoptosis, implying mobilization of NSDHL-containing vesicles to this structure during induction of cell death (44). Work reported here for the first time suggests SC4MOL- and NSDHL-dependent trafficking, potentially in association with targeted delivery of specific metabolites, actively contributes to cancer pathology and drug resistance.
The C4-methylsterols, FF-MAS and T-MAS, are the substrates for the evolutionarily conserved LBR/TM7SF2/ERG24 and SC4MOL/ERG25 enzymes, respectively. Strikingly, we discovered new signaling inhibitory properties of these sterol metabolites, which have previously been studied primarily in the context of gonadal physiology (45 46). FF-MAS (Follicular Fluid Meiosis-Activating Sterol) and T-MAS (Testis Meiosis-Activating Sterol) have significant biological activities in regulating cumulus expansion and oocyte maturation. Some studies of reproductive biology (47 48) indicate that this action likely involves regulation of EGFR signaling, which is known to be important during ovulation and cumulus dispersal (49). Our data for the first time demonstrate activity for these proteins in resistance to erlotinib in cancer cells: we hypothesize that addition of large pools of external FF-MAS and T-MAS disrupts the vesicular targeting role of small endogenous pools of these metabolites.
While 4-methylcholest-8(9)-en-3β-ol; 4,4'-dimethylcholest-8(9)-en-3β-ol intermediaries were detected in SC4MOL-silenced tumor cells and in Bpa1H fibroblasts with germline nonsense mutation in exon 5 of Nsdhl (34), we did not detect these intermediates following depletion of NSDHL with siRNA (Fig. 2A) in carcinoma cell lines, a discrepancy likely due to the fact that these methylsterols are direct substrates of SC4MOL, not NSDHL, and potentially to the low residual levels of NSDHL protein due to incomplete knockdown. We thoroughly validated the idea that both SC4MOL and NSDHL regulate trafficking comparably (Fig. 4 S6). By application of C4-methylsterols in vitro, and by pharmacologic inactivation of the upstream CYP51A1 with ketoconazole or by siRNA knockdown (Fig. 2 S3), we demonstrated a direct effect of SC4MOL and NSDHL on drug sensitivity. Our data also emphasize interactions between ERG25/SC4MOL and multiple proteins associated with endosomal compartments (Fig. 3). Taken in sum, these results support the interpretation that grossly regulating abundance of a sterol intermediate substrate, or depletion of cholesterol, the end-product of the pathway, per se is not sufficient to explain the observed sensitization and trafficking effects. One plausible explanation is that the localized insertion of sterol intermediates into specific vesicular compartments facilitates targeting to specific destinations, as in the pathway recently defined in Arabidopsis (50).
Perturbations of sterol metabolism in humans and in animal models cause skin manifestations associated with accumulation of sterol metabolites but the mechanism was largely unclear. Although no SC4MOL mouse model exists, a patient with SC4MOL deficiency has been described presenting with marked diffuse psoriasiform skin rash (51). Mutations in NSDHL are associated with human CHILD syndrome (congenital hemidysplasia with ichthyosiform nevus and limb defects), an X-linked, male lethal disorder (52). Similar to the human NSDHL-deficiency syndrome (53) or the X-linked dominant Conradi-Hünermann-Happle syndrome (54), skin manifestations in the respective Bpa1H and tattered murine models of these sterol pathway disorders include atrophic and pigmentary lesions, striated hyperkeratosis, coarse lusterless hair and alopecia. These skin changes are similar to those seen in mice and humans with the EGFR pathway perturbations (35). Our studies indicate that these phenotypes may indeed be connected, as EGFR signaling is markedly downregulated in skin keratinocytes with loss of Nsdhl (Fig. 6).
Targeting EGFR in carcinomas has produced tangible therapeutic gains in the treatment of colorectal, breast, head and neck, and non-small cell lung cancers. However, the fact that only a small subset of tumors are controlled by EGFR-directed therapies makes it critical to identify biomarkers for patient response, or targets for drugs that can be combined in treatment strategies that boost the efficacy of EGFR inhibitors. In this context, blockade of sterol metabolism at the level of SC4MOL or NSDHL provides a potential unique point to influence resistance to EGFR-targeted inhibitors via regulation of the EGFR intracellular traffic and signaling activity. Moreover, the fact that depletion of SC4MOL and NSDHL also targets dimerization partners of EGFR, such as ERBB2 and ERBB3, and potentially other proteins which are commonly upregulated to compensate for resistance to EGFR inhibitors, makes this a particularly attractive strategy, as receptor targeting strategies at the level of EGFR endocytic trafficking have begun to show promising activity (55).
Finally, the approach we develop here illustrates the power of integrating systems biological analysis of protein networks into efforts to elucidate the function of relatively obscure proteins. Our work has moved from initial generation of an EGFR-focused network to guide siRNA screening, based on the hypothesis that proteins linked to EGFR by some criteria – physical, genetic, or expression change – would be more likely to modulate EGFR function (7). Having identified SC4MOL, we then modeled from orthologs and a network of their protein interactions in yeast to generate a new hypothesis regarding protein function. Having validated the functional prediction of a role in vesicular trafficking, we were then able to rapidly model interologs relevant to protein action, identifying relevant human protein partners that closed the circle by establishing a proximal link to EGFR trafficking. The success of this rapid oscillation between hypothesis-generation and hypothesis testing illustrates the powerful resources becoming available from “omics” projects, and should support more rapid development of these projects to the clinic in the future.
Methods
Cell lines, Compounds, Antibodies
The A431, FaDu, Detroit and MCF12F cells were obtained from the ATCC (USA) and maintained at the Fox Chase Cancer Center Cell Culture Facility. The identity of A431 cell line was confirmed by single tandem repeat DNA profiling (Biosynthesis, Louisville, TX). The SCC61, SCC68, SCC25 cells were kindly provided by Dr. Tanguy Y. Seiwert (University of Chicago, Chicago, IL, USA). PC9 cells were kindly provided by Dr. William Pao (Vanderbilt-Ingram Cancer Center, Nashville, TN). All cell lines were Mycoplasma-free and maintained in DMEM supplemented with 10% v/v fetal bovine serum and L-glutamine without antibiotics. Mouse embryonic fibroblasts were obtained from Bpa1H mice as described (56) and propagated in DMEM supplemented with 10% v/v fetal bovine serum and L-glutamine without antibiotics. Cetuximab and erlotinib were obtained from the Fox Chase Cancer Center pharmacy; CPT-11, cholesterol and lanosterol were purchased from Sigma-Aldrich (USA); FF-MAS (14 –demethyl-14-dehydrolanosterol, cat#700077) and T-MAS (4,4-dimethylzymosterol/4,4-dimethyl-cholest-8(9),24-dien-3ß-ol, cat#700073) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL, USA). All antibodies used in Western blot experiments were purchased from Cell Signaling (Danvers, MA, USA). Rabbit polyclonal antibody to SC4MOL was raised using the peptide antigen TEYFNIPYDWERMPRW-amide (amino acids 120–135) and affinity purified (21st Century Biochemicals, Marlboro, MA, USA). Anti-NSDHL rabbit polyclonal antiserum was generated using the peptide antigen DEAVERTVQSFHHLRKDK (amino acid residues 345–362) of mouse NSDHL.
siRNA transfections and in vitro viability assays
siRNA targeting human sterol biosynthesis genes and controls were obtained from Qiagen (Valencia, CA). Cells were transfected in triplicates with siRNA at 10 nM concentrations mixed with HiPerfect transfection reagent (Qiagen, Valencia, CA) on a 96-well plate according to the manufacturer’s reverse transfection protocol. In 24 hrs after plating, cells were treated with erlotinib, cetuximab, CPT11, or vehicle (0.02% DMSO). The viability was measured in 96 hrs using CellTiter Blue viability assay (Promega, Madison, WI). We used a sensitization index (SI) to determine if gene silencing enhances drug cytotoxicity (7). Viability was determined for each target gene and normalized to the averaged GL2 viability on each plate. Sensitization index (SI) was calculated for each individual well as SI=(Xdrug/GL2drug)/(XDMSO/GL2DMSO), where X was viability in wells transfected with targeting duplexes and GL2 was the averaged viability of 3 wells with non-targeting negative control siRNA on the same plate. The SI=1 indicates no interaction, and SI<1 indicates potentiation of the drug cytotoxicity. In viability assays with sterol metabolites, the latter were added with the drugs. Corresponding dilutions of ethanol and DMSO were added as vehicle, and did not affect viability compared to untreated cells.
Quantitative RT-PCR
For evaluation of the target genes knockdown, cells were reverse transfected in 6-well plates, and total RNA was extracted using RNeasy Minikit (Qiagen, Valencia, CA) 48 hrs after transfection. Quantitative RT-PCR reactions were performed using TaqMan probes and primers designed by the manufacturer, using an ABI PRISM 7700 detection system (Applied Biosystems, Foster City, CA). The results were analyzed using the comparative Ct method to establish relative expression curves.
Apoptosis and pathway analysis
Apoptosis was measured using the Annexin V assay (Guava Technologies, Hayward, CA). Annexin V-positive A431 cells were counted using Guava flow cytometry 72 hours post transfection, 48 hours after treatment. To measure the effect of siRNAs on the activity of EGFR effectors, cells were transfected with siRNA, and the culture media replaced with glutamine-supplemented serum-free DMEM at 24 hrs post-transfection. After overnight incubation, cells were either left untreated, or stimulated with EGF at 15 ng/ml for 15 minutes. Cell extracts were prepared using M-PER™ mammalian protein extraction buffer (Thermo Scientific, Rockford, IL) supplemented with the Halt™ phosphatase inhibitor cocktail (Thermo Scientific, Rockford, IL) and the Complete Mini™ protease inhibitor cocktail (Roche Diagnostics Gmbh, Manheim, Germany). Extracts were centrifuged at 15,000g for 10 min at 4°C. Western analysis was performed using antibodies to phosphorylated and total EGFR, AKT, ERK1/2, and to beta-actin (Cell Signaling, Beverly, MA).
Analysis of sterol composition
Sterol contents of A431 cells cultured in 1% FBS/DMEM were measured using selected ion monitoring gas liquid chromatography/mass spectrometry (GC/MS) as described (57). Briefly, cells were transfected with 30 nM siRNA and, in 72 hrs, detached from the plastic by trypsin. Two aliquots per sample were stored at −80°C until analysis for total protein and lipids.
Immunofluorescent microscopy
A431 and SCC61 cells were seeded on glass slides in the presence of 30 nM siRNA. For EGF internalization studies, cells were starved overnight in serum-free DMEM/L-Glutamine and, at 72 hrs post transfection, were labeled on ice (basal) with 100 ng/ml of EGF Alexa Fluor 488 for 1 hr as previously described (24). Then, the medium was changed to pre-warmed DMEM supplemented with L-Glutamine and transferred to 37°C incubator for 10 or 30 minutes and fixed with freshly prepared 4% paraformaldehyde (Electron Microscopy Sciences, Ft. Washington, PA). For later time-points, the internalization proceeded for 30 minutes, after which cells were washed twice in DMEM/glutamine and chased for up to 3 hrs in the presence of unlabeled EGF at 100 ng/ml to avoid binding of labeled EGF recycled to the cell surface. After two washes in ice-cold phosphate buffer saline (PBS), cells were fixed in freshly prepared 4% paraformaldehyde/PBS at ambient temperature for 10 minutes followed by two washes in PBS and stored at 4°C or labeled immediately. In some EGF internalization experiments, cells were pre-treated with leupeptin at 21 µM at 37°C for 1 hr. In these experiments, the same concentrations of inhibitors were maintained during incubations with EGF (25).
For RAB7 (Cell Signaling, Beverly, MA) and RAB11 (Invitrogen, Carlsbad, CA) antibody immunolabeling, cells were incubated in PBS with 5% normal bovine serum and 0.3% Triton X-100 followed by overnight incubation with primary antibodies diluted 1:100 in 1% BSA/0.3% Triton X-100/PBS and appropriate secondary antibody conjugated with Alexa Fluor 488 and Alexa Fluor 568 (Invitrogen, Carlsbad, CA). For experiments with ARF1, ARF4, ARF5, LAMP1 and EEA1 (Abcam, Cambridge, MA) all incubation were in the presence of 0.05% Triton X-100. All immunofluorescent imaging experiments were repeated at least twice. In multiple spots (4–5) of each slide, 10 or more fully adherent and viable (judged by labeled EGF internalization) cells were analyzed for each condition. Optical sections through the middle of the cells were acquired on Nikon C1 Spectral Confocal microscope with x60 oil objective. Analysis for marker co-localization was done in MetaMorph (Universal Imaging/Molecular Devices) software using built-in co-localization tool. The threshold of background was applied uniformly to single channel images per individual cells so that the intensity of endosomal marker did not include the diffuse background staining. The extent of co-localization was calculated as the spatial overlap between two channels expressed as the ratio of the integrated EGF or EGFR fluorescence within the indicated endosomal compartment marker over the total EGF or EGFR intensity.
Immortalized sorted Bpa1H or NSDHL-sufficient fibroblasts(56) were treated with recombinant human PDGF-BB (Invitrogen, Frederick, MD, USA) at 25 ng/ml for indicated duration of time, followed by either fixation with 4% paraformaldehyde for microscopy or lysis for PDGFR expression analysis by Western blot.
EGFR degradation and EGFR immunoprecipitaion
SCC61 cells (in six-well plate) were transfected with 30nM siRNA for 72 hours (as control non-targeting firefly luciferase GL2 siRNA was used) and then either mock treated or induced with EGF at 100 ng/ml for various time periods as indicated. At the end of incubation, cells were transferred to ice and washed twice with ice-cold PBS. Total cellular lysates were obtained by lysis in RIPA (Sigma-Aldrich, USA) supplemented with the Halt™ phosphatase inhibitor cocktail (Thermo Scientific, Rockford, IL) and the Complete Mini™ protease inhibitor cocktail (Roche Diagnostics Gmbh, Mannheim, Germany) for 10 minutes. Soluble fractions were analyzed by Western blotting, EGFR bands density was normalized to α-tubulin and mock-treated GL2. For EGFR immunoprecipitation experiments, the conditions were as above, except 10 µM N-ethylmaleimide was used in all buffers. For each condition, 500–750 micrograms of total protein was incubated with 5 micrograms of cetuximab overnight and immunoprecipitated with 40 microliters of pre-washed Protein-G conjugated beads slurry (Thermo Scientific, Rockford, IL). Beads were centrifuged at 5,000 rpm for 1 min at 4°C on a benchtop minicentrifuge, and proteins were eluted from the beads by boiling for 5 minutes in 5x Loading Buffer (Thermo Scientific, Rockford, IL). Western blot membranes were probed simultaneously with mouse antibody to EGFR and rabbit anti-ubiquitin antibody (PA3-16717, Thermo Scientific, USA) and developed using infrared Odyssey imager. All Western blot protein expression evaluations were repeated at least twice.
Tumor xenografts
Six to 8 week old ICR-C.B17-scid males were injected subcutaneously to the flanks with 3 million of A431 cells carrying stably integrated lentiviral shRNA constructs targeting SC4MOL or non-silencing-GIPZ lentiviral shRNAmir control (V2LHS_84827, V2LHS_84829, Thermo Scientific, USA). Tumors were measured in all animals on day 8 (range 70–130 mm3). Animals (n=10 per group) were randomized to receive cetuximab 0.75 mg/kg or saline intraperitoneally twice a week for 3 weeks. Tumor volumes were assessed twice a week as (length×width2)/2 (7). Treatment cohorts were followed for at least 7 weeks, or until average tumor volumes exceeded 2000 mm3ulcerated, or animals demonstrated distress or weight loss >10% as per the local IACUC guidelines. For Western blot and tissue analyses, xenografts were treated on day 8 post implantation with a single dose of cetuximab 0.75 mg/kg i.p. or saline and collected on day 11. Tumor tissues (3 animals per group) were immediately homogenized in RIPA buffer on ice, cleared by centrifugation and analyzed by Western blot.
Mouse skin tissues collection and immunohistochemistry
Following sacrifice at postnatal day 3, skin over the dorsum of the back was removed from wild type and Bpa1H/+ littermates. At this age, it is possible to discern affected females based on the appearance of hyperkeratotic patches of skin. Observed phenotypes were confirmed by genotyping of tail tips following sacrifice.Isolated skin was fixed in Bouin’s fixative and stained with NSDHL antibody at 1:500 dilution. For studies using BrdU, pups received a single intraperitoneal injection of 100 µg/gm body weight BrdU and were sacrificed 2 hr later. Samples were fixed as above and IHC performed. Heat was used for antigen retrieval (95°C for 20 min) in solution (10 mM sodium citrate, 0.05% Tween-20, pH 6). Slides were incubated with indicated primary antibodies (Cell Signaling, Danvers, MA) diluted as per the manufacturer’s instructions. Primary antibody binding was amplified using a Vectastain Elite ABC kit (Vector Laboratories, Burlingame, CA), including biotinylated anti-rabbit secondary antibody. Antibody binding was visualized using the Liquid DAB+ Substrate Chromogen System (Dako, Carpinteria, CA). Samples were counterstained for 1 min with hematoxylin. Immunostained slides were scanned by Aperio ScanScope CS scanner (Aperio, Vista, CA), and selected regions of interest (12–15 spots of NSDHL-null and normal skin from 4 skin sections each) were outlined manually by a pathologist (A.K-Z. and K.Q.C.). Expression levels of NSDHL, pY1173-EGFR, pERK, pS473-AKT and Keratin 14 were measured using the Positive Pixel Count V9 algorithm, and nuclear EGFR and Ki-67 with the Nuclear V9 algorithm using ImageScope software.
For Automated Quantitative Analysis (AQUA) of NSDHL expression, antibody (Proteintech Group, Chicago, IL) titer and dynamic range has been established on tissue microarray including cancer and normal (36). The AQUA automated image acquisition and analysis was done on Olympus BX51 microscope and analyzed with the AQUA software. Areas of tumor were distinguished from stromal elements by creating an epithelial tumor mask from the cytokeratin signal.
Network assembly and analysis
Publicly available databases (BioGrid (58), SBD (59), STRING (60)) were searched for the yeast ergosterol biosynthesis pathway genes which were used as search seeds (Fig S4). Orthologs for yeast genes were identified in higher eukaryotes using PubMed and P-POD (Princeton Protein Orthology Database (61), using Naïve Ensemble search tool). Subsequent analysis was aimed to augment information on protein-protein interactions (PPIs) in human cells, PPIs between homologous genes in model organisms, database or pathway links, and text mining (co-appearance of gene names in PubMed). Data regarding experimentally proven interactions in human and model organisms were merged in Cytoscape (Supplemental file and Table S1). To provide additional context in some analyses (Fig. 4), STRING-extracted information from pathway databases and text-mining data were merged and displayed using Cytoscape as indicated in figure legends. BINGO plugin to the Cytoscape was used to identify GO functions overrepresented in the ERG25-ERG28 interactions network in Fig.3; the nodes were color-labeled using GOlorize plugin to the Cytoscape.
Statistical analysis
For drug sensitivity testing and Western blot enumeration of protein expression, we used generalized linear models assuming gamma family and log link. For analysis of immunofluorescent imaging, Student’s two-sided t-test was used after visual assessment for normality of data distribution.
Supplementary Material
Acknowledgments
We are grateful to Calvin Shaller for technical assistance with xenograft and I125-EGF experiments, Igor Gorin for technical assistance with Western blots, Fox Chase Genomics Facility (Dr. Emmanuelle Nicolas) and to Dr. Jonathan Chernoff for critical comments on the work.
Grant Support
This work was supported by NIH core grant CA-06927, by the Pew Charitable Fund, and by a generous gift from Mrs. Concetta Greenberg to Fox Chase Cancer Center. Some of the authors were supported by NIH R01 CA-63366 and R01 CA113342 (EAG); and by Tobacco Settlement funding from the State of Pennsylvania (EAG and IA); by NIH R01 CA50633, U54 CA149147, and by the Jeannik Littlefield Award from the American Association of Cancer Research (LW); by NIH R01 HD38572 (GEH); by NIH K22 CA160725, R21 CA164205 and a career development award from Genentech (IA).
Footnotes
The authors disclose no potential conflicts of interest.
Author contributions: A.S., A.G., L.G., H.L., I.G.S., L.M.W., L.E.K., R.I.K., G.E.H., E.A.G. and I.A. designed research; A.S., A.G., L.G., H.Z., L.E.K., D.C., G.E.H., A.K.S., K.Q.C., D.R., H.L., D.Y., R.M, B.B. and I.G.S. performed research; Y.Z., R.M., B.B., D.Y., G.A., G.E.H. and D.C. contributed new reagents/analytical tools; A.S., A.G., L.K., D-H. Y., A. K-S., R.I.K., I.G.S., H.L., B.L.E. and I.A. analyzed data; A.S., A.G., E.A.G. and I.A. wrote the paper.
Significance. This work identifies a critical role for SC4MOL and NSDHL in regulation of EGFR signaling and endocytic trafficking, and suggests novel strategies to increase the potency of EGFR antagonists in tumors.
References
- 1.Erjala K, Sundvall M, Junttila TT, Zhang N, Savisalo M, Mali P, et al. Signaling via ErbB2 and ErbB3 associates with resistance and epidermal growth factor receptor (EGFR) amplification with sensitivity to EGFR inhibitor gefitinib in head and neck squamous cell carcinoma cells. Clin Cancer Res. 2006;12:4103–4111. doi: 10.1158/1078-0432.CCR-05-2404. [DOI] [PubMed] [Google Scholar]
- 2.Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guix M, Faber AC, Wang SE, Olivares MG, Song Y, Qu S, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest. 2008;118:2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, et al. Wild-Type BRAF Is Required for Response to Panitumumab or Cetuximab in Metastatic Colorectal Cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 5.Vivanco I, Rohle D, Versele M, Iwanami A, Kuga D, Oldrini B, et al. The phosphatase and tensin homolog regulates epidermal growth factor receptor (EGFR) inhibitor response by targeting EGFR for degradation. Proc Natl Acad Sci U S A. 2010;107:6459–6464. doi: 10.1073/pnas.0911188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Argiris A, Duffy AG, Kummar S, Simone NL, Arai Y, Kim SW, et al. Early tumor progression associated with enhanced EGFR signaling with bortezomib, cetuximab, and radiotherapy for head and neck cancer. Clin Cancer Res. 2011;17:5755–5764. doi: 10.1158/1078-0432.CCR-11-0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Astsaturov I, Ratushny V, Sukhanova A, Einarson MB, Bagnyukova T, Zhou Y, et al. Synthetic Lethal Screen of an EGFR-Centered Network to Improve Targeted Therapies. Sci Signal. 2010;3:ra67. doi: 10.1126/scisignal.2001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krause SA, Gray JV. The functional relationships underlying a synthetic genetic network. Commun Integr Biol. 2009;2:4–6. doi: 10.4161/cib.2.1.7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong AH, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- 10.Maiti B, Kundranda MN, Spiro TP, Daw HA. The association of metabolic syndrome with triple-negative breast cancer. Breast Cancer Res Treat. 2010;121:479–483. doi: 10.1007/s10549-009-0591-y. [DOI] [PubMed] [Google Scholar]
- 11.Vinci G, Xia X, Veitia RA. Preservation of genes involved in sterol metabolism in cholesterol auxotrophs: facts and hypotheses. PLoS One. 2008;3:e2883. doi: 10.1371/journal.pone.0002883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mo C, Valachovic M, Randall SK, Nickels JT, Bard M. Protein-protein interactions among C-4 demethylation enzymes involved in yeast sterol biosynthesis. Proc Natl Acad Sci U S A. 2002;99:9739–9744. doi: 10.1073/pnas.112202799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ono M, Hirata A, Kometani T, Miyagawa M, Ueda S, Kinoshita H, et al. Sensitivity to gefitinib (Iressa, ZD1839) in non-small cell lung cancer cell lines correlates with dependence on the epidermal growth factor (EGF) receptor/extracellular signal-regulated kinase 1/2 and EGF receptor/Akt pathway for proliferation. Mol Cancer Ther. 2004;3:465–472. [PubMed] [Google Scholar]
- 14.Li J, Rix U, Fang B, Bai Y, Edwards A, Colinge J, et al. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat Chem Biol. 2010;6:291–299. doi: 10.1038/nchembio.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato-Stankiewicz J, Hakimi I, Zhi G, Zhang J, Serebriiskii I, Guo L, et al. Inhibitors of Ras/Raf-1 interaction identified by two-hybrid screening revert Ras-dependent transformation phenotypes in human cancer cells. Proc Natl Acad Sci U S A. 2002;99:14398–14403. doi: 10.1073/pnas.222222699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gachotte D, Pierson CA, Lees ND, Barbuch R, Koegel C, Bard M. A yeast sterol auxotroph (erg25) is rescued by addition of azole antifungals and reduced levels of heme. Proc Natl Acad Sci U S A. 1997;94:11173–11178. doi: 10.1073/pnas.94.21.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. http://www.genome.jp
- 18.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 20.Kaake RM, Milenkovic T, Przulj N, Kaiser P, Huang L. Characterization of cell cycle specific protein interaction networks of the yeast 26S proteasome complex by the QTAX strategy. J Proteome Res. 2010;9:2016–2029. doi: 10.1021/pr1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 22.Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, et al. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 23.Scita G, Di Fiore PP. The endocytic matrix. Nature. 2010;463:464–473. doi: 10.1038/nature08910. [DOI] [PubMed] [Google Scholar]
- 24.Roberts M, Barry S, Woods A, van der Sluijs P, Norman J. PDGF-regulated rab4-dependent recycling of alphavbeta3 integrin from early endosomes is necessary for cell adhesion and spreading. Curr Biol. 2001;11:1392–1402. doi: 10.1016/s0960-9822(01)00442-0. [DOI] [PubMed] [Google Scholar]
- 25.Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell. 2006;21:737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 26.Sigismund S, Argenzio E, Tosoni D, Cavallaro E, Polo S, Di Fiore PP. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev Cell. 2008;15:209–219. doi: 10.1016/j.devcel.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Guo WJ, Zhang XW, Cai X, Tian S, Li J. Cetuximab enhances the activities of irinotecan on gastric cancer cell lines through downregulating the EGFR pathway upregulated by irinotecan. Cancer Chemother Pharmacol. 2011;68:871–878. doi: 10.1007/s00280-011-1559-2. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Pennock S, Chen X, Wang Z. Internalization of inactive EGF receptor into endosomes and the subsequent activation of endosome-associated EGF receptors. Epidermal growth factor. Sci STKE. 2002;2002:pl17. doi: 10.1126/stke.2002.161.pl17. [DOI] [PubMed] [Google Scholar]
- 29.Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins SR, et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science. 2008;322:405–410. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chun J, Shapovalova Z, Dejgaard SY, Presley JF, Melancon P. Characterization of class I and II ADP-ribosylation factors (Arfs) in live cells: GDP-bound class II Arfs associate with the ER-Golgi intermediate compartment independently of GBF1. Mol Biol Cell. 2008;19:3488–3500. doi: 10.1091/mbc.E08-04-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deretic D, Williams AH, Ransom N, Morel V, Hargrave PA, Arendt A. Rhodopsin C terminus, the site of mutations causing retinal disease, regulates trafficking by binding to ADP-ribosylation factor 4 (ARF4) Proc Natl Acad Sci U S A. 2005;102:3301–3306. doi: 10.1073/pnas.0500095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazelova J, Astuto-Gribble L, Inoue H, Tam BM, Schonteich E, Prekeris R, et al. Ciliary targeting motif VxPx directs assembly of a trafficking module through Arf4. EMBO J. 2009;28:183–192. doi: 10.1038/emboj.2008.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadakata T, Sekine Y, Oka M, Itakura M, Takahashi M, Furuichi T. Calcium-dependent activator protein for secretion 2 interacts with the class II ARF small GTPases and regulates dense-core vesicle trafficking. FEBS J. 2011;279:384–394. doi: 10.1111/j.1742-4658.2011.08431.x. [DOI] [PubMed] [Google Scholar]
- 34.Liu XY, Dangel AW, Kelley RI, Zhao W, Denny P, Botcherby M, et al. The gene mutated in bare patches and striated mice encodes a novel 3beta-hydroxysteroid dehydrogenase. Nat Genet. 1999;22:182–187. doi: 10.1038/9700. [DOI] [PubMed] [Google Scholar]
- 35.Schneider MR, Werner S, Paus R, Wolf E. Beyond wavy hairs: the epidermal growth factor receptor and its ligands in skin biology and pathology. Am J Pathol. 2008;173:14–24. doi: 10.2353/ajpath.2008.070942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Psyrri A, Egleston B, Weinberger P, Yu Z, Kowalski D, Sasaki C, et al. Correlates and determinants of nuclear epidermal growth factor receptor content in an oropharyngeal cancer tissue microarray. Cancer Epidemiol Biomarkers Prev. 2008;17:1486–1492. doi: 10.1158/1055-9965.EPI-07-2684. [DOI] [PubMed] [Google Scholar]
- 37.Bionda C, Athias A, Poncet D, Alphonse G, Guezguez A, Gambert P, et al. Differential regulation of cell death in head and neck cell carcinoma through alteration of cholesterol levels in lipid rafts microdomains. Biochem Pharmacol. 2008;75:761–772. doi: 10.1016/j.bcp.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 38.Guo D, Hildebrandt IJ, Prins RM, Soto H, Mazzotta MM, Dang J, et al. The AMPK agonist AICAR inhibits the growth of EGFRvIII-expressing glioblastomas by inhibiting lipogenesis. Proc Natl Acad Sci U S A. 2009;106:12932–12937. doi: 10.1073/pnas.0906606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pitroda SP, Khodarev NN, Beckett MA, Kufe DW, Weichselbaum RR. MUC1-induced alterations in a lipid metabolic gene network predict response of human breast cancers to tamoxifen treatment. Proc Natl Acad Sci U S A. 2009;106:5837–5841. doi: 10.1073/pnas.0812029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khodarev NN, Pitroda SP, Beckett MA, MacDermed DM, Huang L, Kufe DW, et al. MUC1-induced transcriptional programs associated with tumorigenesis predict outcome in breast and lung cancer. Cancer Res. 2009;69:2833–2837. doi: 10.1158/0008-5472.CAN-08-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eden ER, White IJ, Tsapara A, Futter CE. Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nat Cell Biol. 2010;12:267–272. doi: 10.1038/ncb2026. [DOI] [PubMed] [Google Scholar]
- 42.Caldas H, Herman GE. NSDHL, an enzyme involved in cholesterol biosynthesis, traffics through the Golgi and accumulates on ER membranes and on the surface of lipid droplets. Hum Mol Genet. 2003;12:2981–2991. doi: 10.1093/hmg/ddg321. [DOI] [PubMed] [Google Scholar]
- 43.Beller M, Thiel K, Thul PJ, Jackle H. Lipid droplets: a dynamic organelle moves into focus. FEBS Lett. 2010;584:2176–2182. doi: 10.1016/j.febslet.2010.03.022. [DOI] [PubMed] [Google Scholar]
- 44.Parent N, Winstall E, Beauchemin M, Paquet C, Poirier GG, Bertrand R. Proteomic analysis of enriched lysosomes at early phase of camptothecin-induced apoptosis in human U-937 cells. J Proteomics. 2009;72:960–973. doi: 10.1016/j.jprot.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Byskov AG, Yding Andersen C, Hossaini A, Guoliang X. Cumulus cells of oocyte-cumulus complexes secrete a meiosis-activating substance when stimulated with FSH. Mol Reprod Dev. 1997;46:296–305. doi: 10.1002/(SICI)1098-2795(199703)46:3<296::AID-MRD8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 46.Byskov AG, Andersen CY, Nordholm L, Thogersen H, Xia G, Wassmann O, et al. Chemical structure of sterols that activate oocyte meiosis. Nature. 1995;374:559–562. doi: 10.1038/374559a0. [DOI] [PubMed] [Google Scholar]
- 47.Coticchio G, Rossi G, Borini A, Grondahl C, Macchiarelli G, Flamigni C, et al. Mouse oocyte meiotic resumption and polar body extrusion in vitro are differentially influenced by FSH, epidermal growth factor and meiosis-activating sterol. Hum Reprod. 2004;19:2913–2918. doi: 10.1093/humrep/deh514. [DOI] [PubMed] [Google Scholar]
- 48.Faerge I, Terry B, Kalous J, Wahl P, Lessl M, Ottesen JL, et al. Resumption of meiosis induced by meiosis-activating sterol has a different signal transduction pathway than spontaneous resumption of meiosis in denuded mouse oocytes cultured in vitro. Biol Reprod. 2001;65:1751–1758. doi: 10.1095/biolreprod65.6.1751. [DOI] [PubMed] [Google Scholar]
- 49.Park JY, Su YQ, Ariga M, Law E, Jin SL, Conti M. EGF-like growth factors as mediators of LH action in the ovulatory follicle. Science. 2004;303:682–684. doi: 10.1126/science.1092463. [DOI] [PubMed] [Google Scholar]
- 50.Markham JE, Molino D, Gissot L, Bellec Y, Hematy K, Marion J, et al. Sphingolipids containing very-long-chain Fatty acids define a secretory pathway for specific polar plasma membrane protein targeting in Arabidopsis. Plant Cell. 2011;23:2362–2378. doi: 10.1105/tpc.110.080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He M, Kratz LE, Michel JJ, Vallejo AN, Ferris L, Kelley RI, et al. Mutations in the human SC4MOL gene encoding a methyl sterol oxidase cause psoriasiform dermatitis, microcephaly, and developmental delay. J Clin Invest. 2010;121:976–984. doi: 10.1172/JCI42650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Porter FD, Herman GE. Malformation syndromes caused by disorders of cholesterol synthesis. J Lipid Res. 2010;52:6–34. doi: 10.1194/jlr.R009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hummel M, Cunningham D, Mullett CJ, Kelley RI, Herman GE. Left-sided CHILD syndrome caused by a nonsense mutation in the NSDHL gene. Am J Med Genet A. 2003;122A:246–251. doi: 10.1002/ajmg.a.20248. [DOI] [PubMed] [Google Scholar]
- 54.Derry JM, Gormally E, Means GD, Zhao W, Meindl A, Kelley RI, et al. Mutations in a delta 8-delta 7 sterol isomerase in the tattered mouse and X-linked dominant chondrodysplasia punctata. jderry@immunex.com. Nat Genet. 1999;22:286–290. doi: 10.1038/10350. [DOI] [PubMed] [Google Scholar]
- 55.Spangler JB, Neil JR, Abramovitch S, Yarden Y, White FM, Lauffenburger DA, et al. Combination antibody treatment down-regulates epidermal growth factor receptor by inhibiting endosomal recycling. Proc Natl Acad Sci U S A. 2010;107:13252–13257. doi: 10.1073/pnas.0913476107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cunningham D, Swartzlander D, Liyanarachchi S, Davuluri RV, Herman GE. Changes in gene expression associated with loss of function of the NSDHL sterol dehydrogenase in mouse embryonic fibroblasts. J Lipid Res. 2005;46:1150–1162. doi: 10.1194/jlr.M400462-JLR200. [DOI] [PubMed] [Google Scholar]
- 57.Kelley RI. Diagnosis of Smith-Lemli-Opitz syndrome by gas chromatography/mass spectrometry of 7-dehydrocholesterol in plasma, amniotic fluid and cultured skin fibroblasts. Clin Chim Acta. 1995;236:45–58. doi: 10.1016/0009-8981(95)06038-4. [DOI] [PubMed] [Google Scholar]
- 58. http://thebiogrid.org
- 59. http://www.yeastgenome.org
- 60. http://string-db.org
- 61. http://ppod.princeton.edu/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.