Abstract
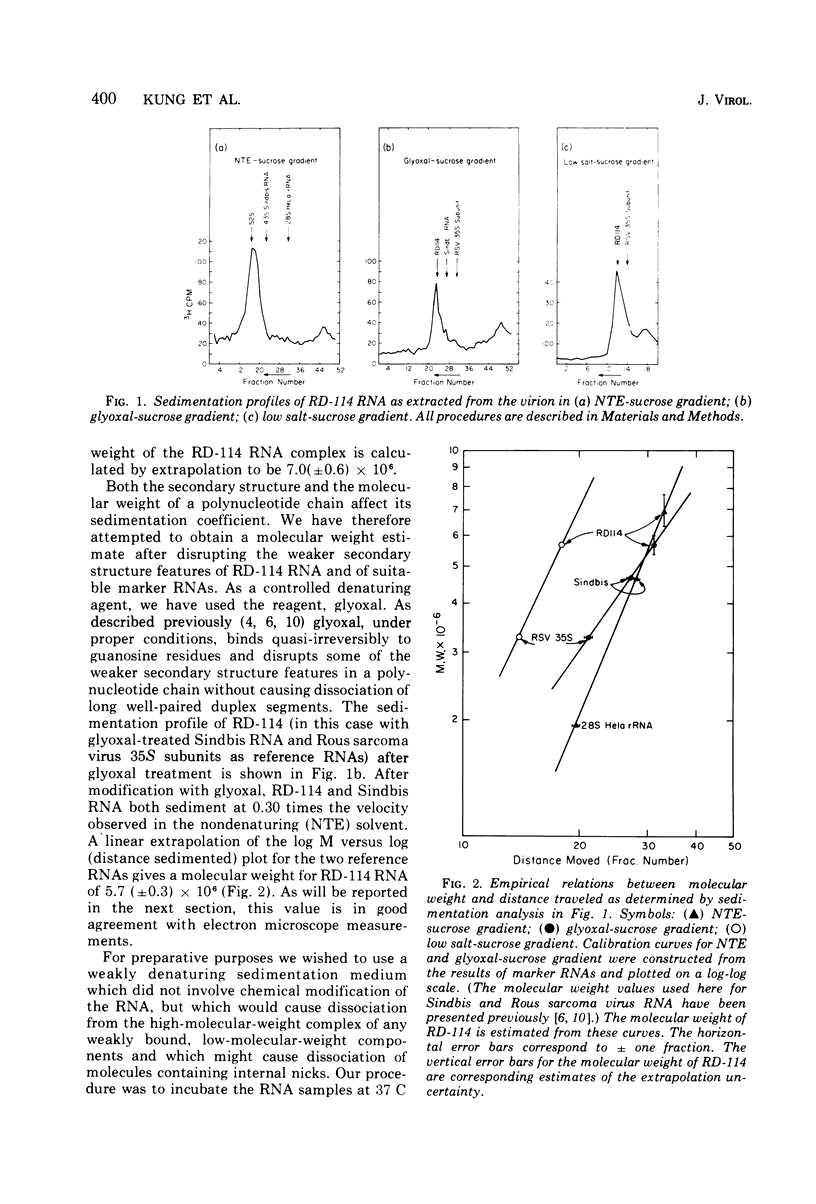
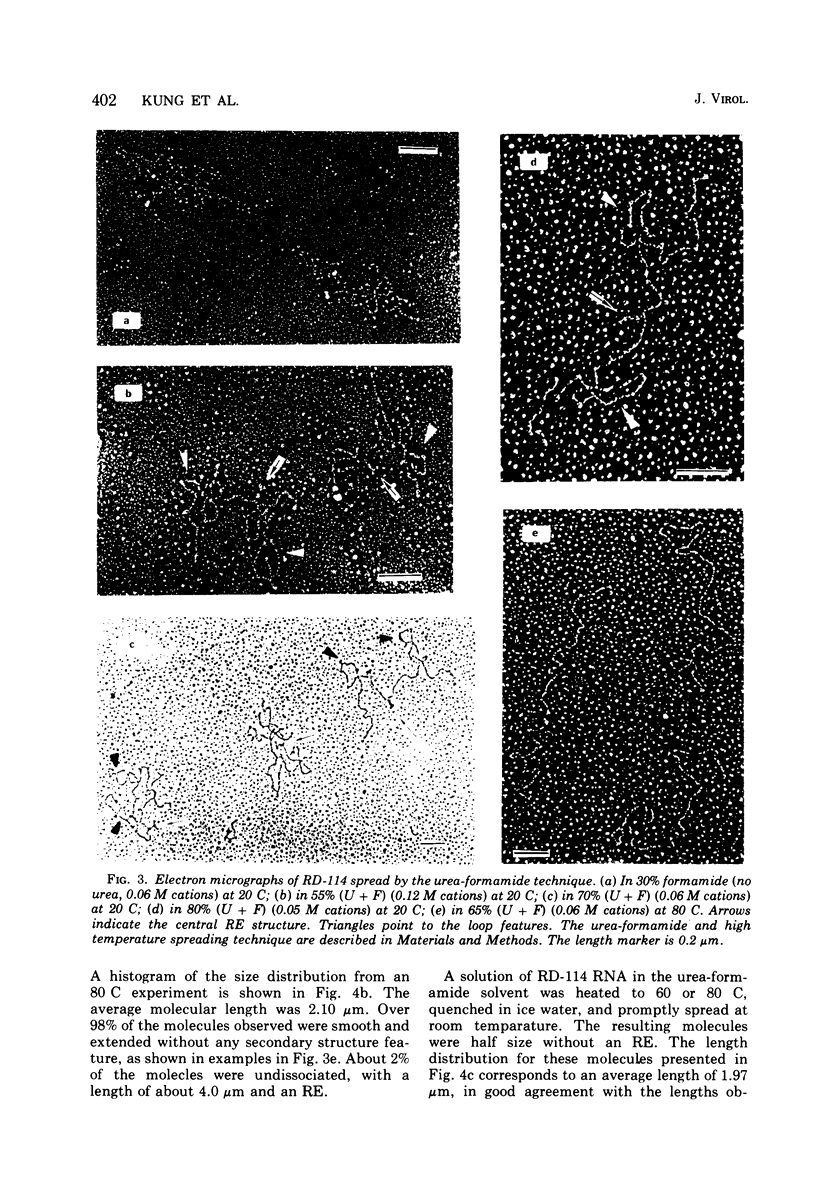
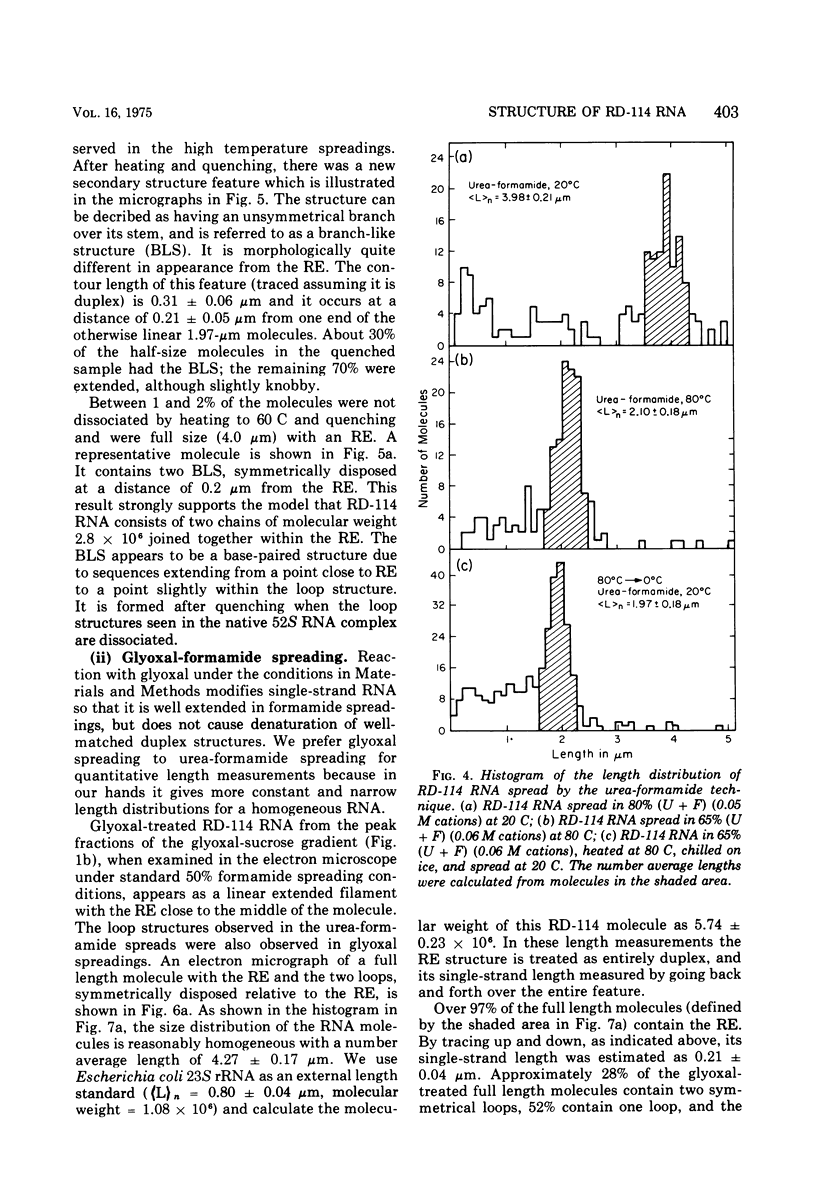
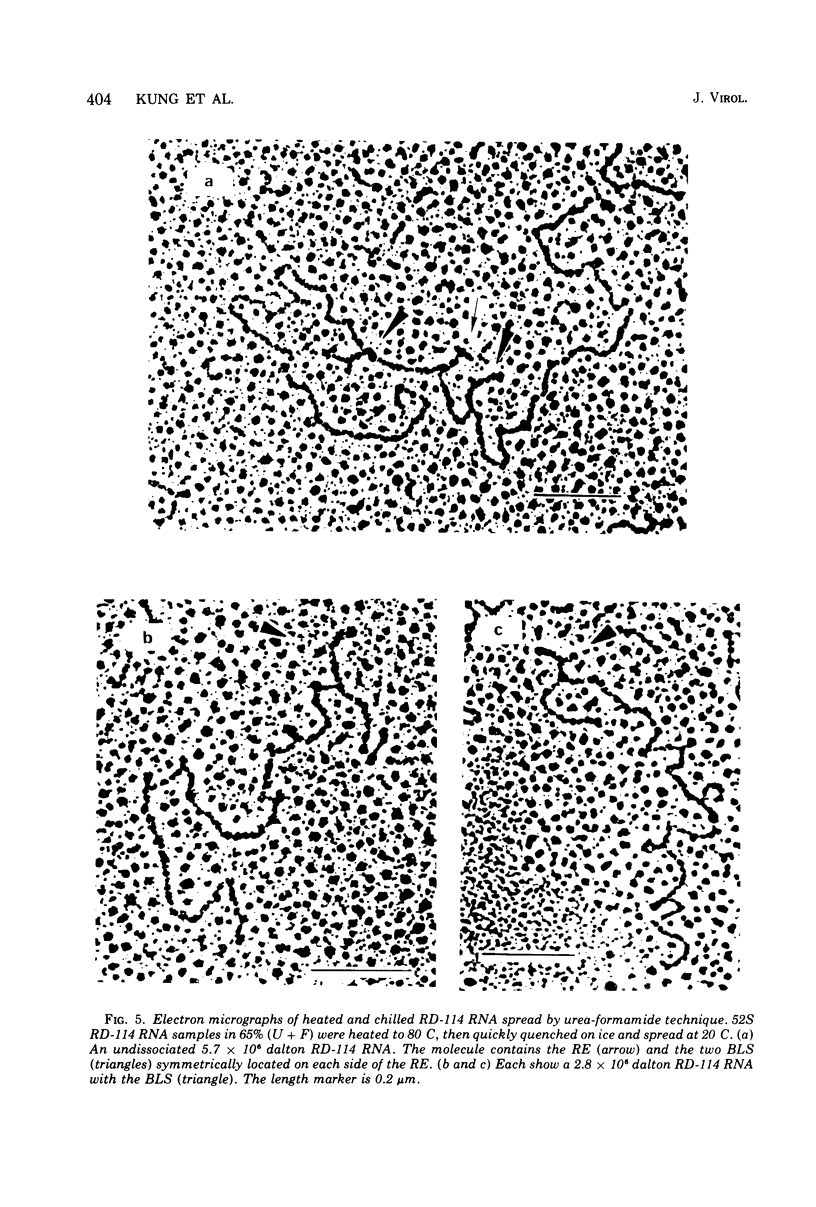
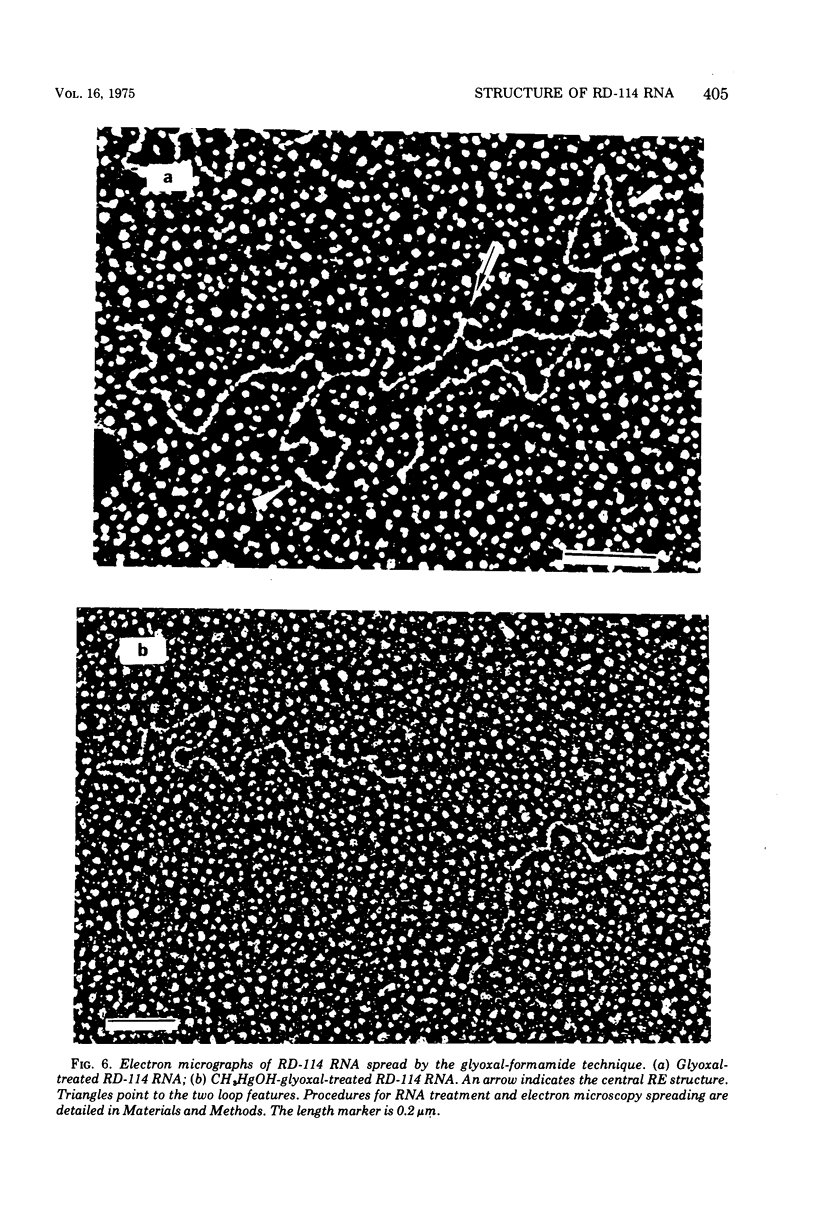
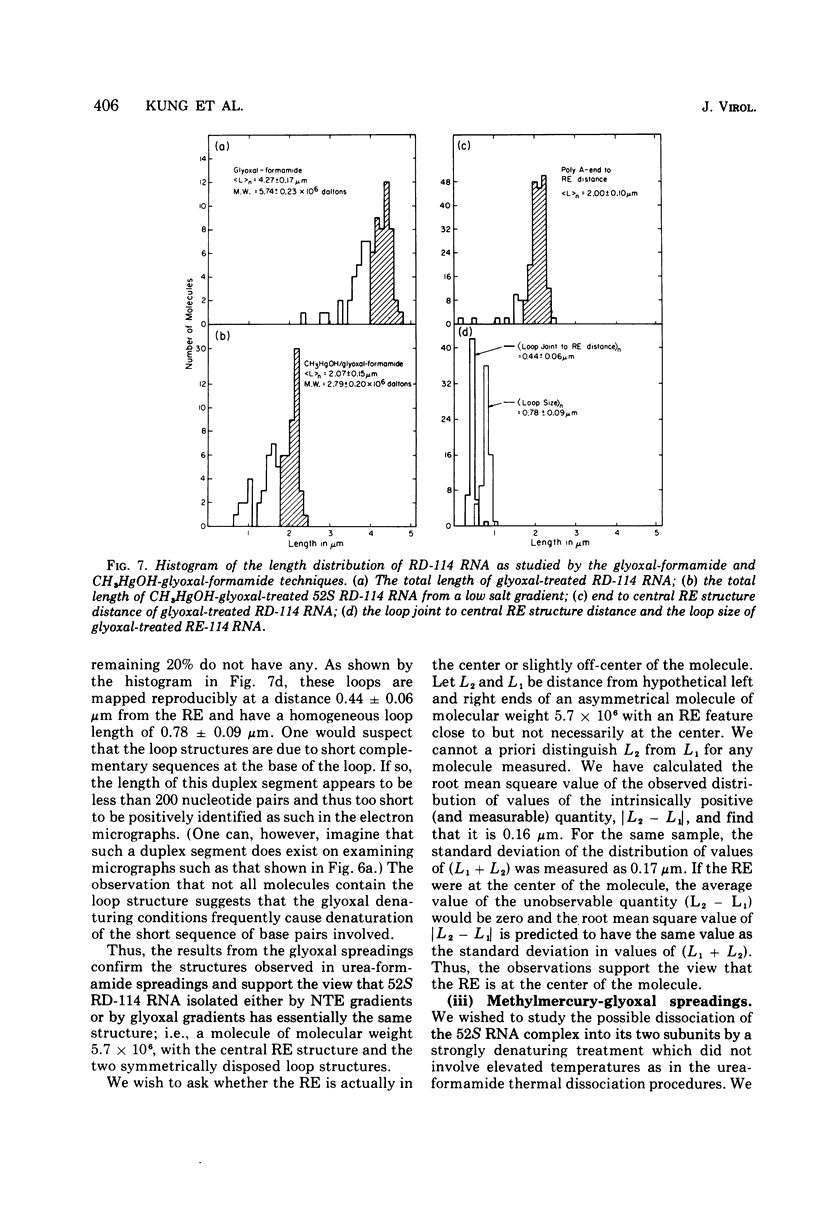
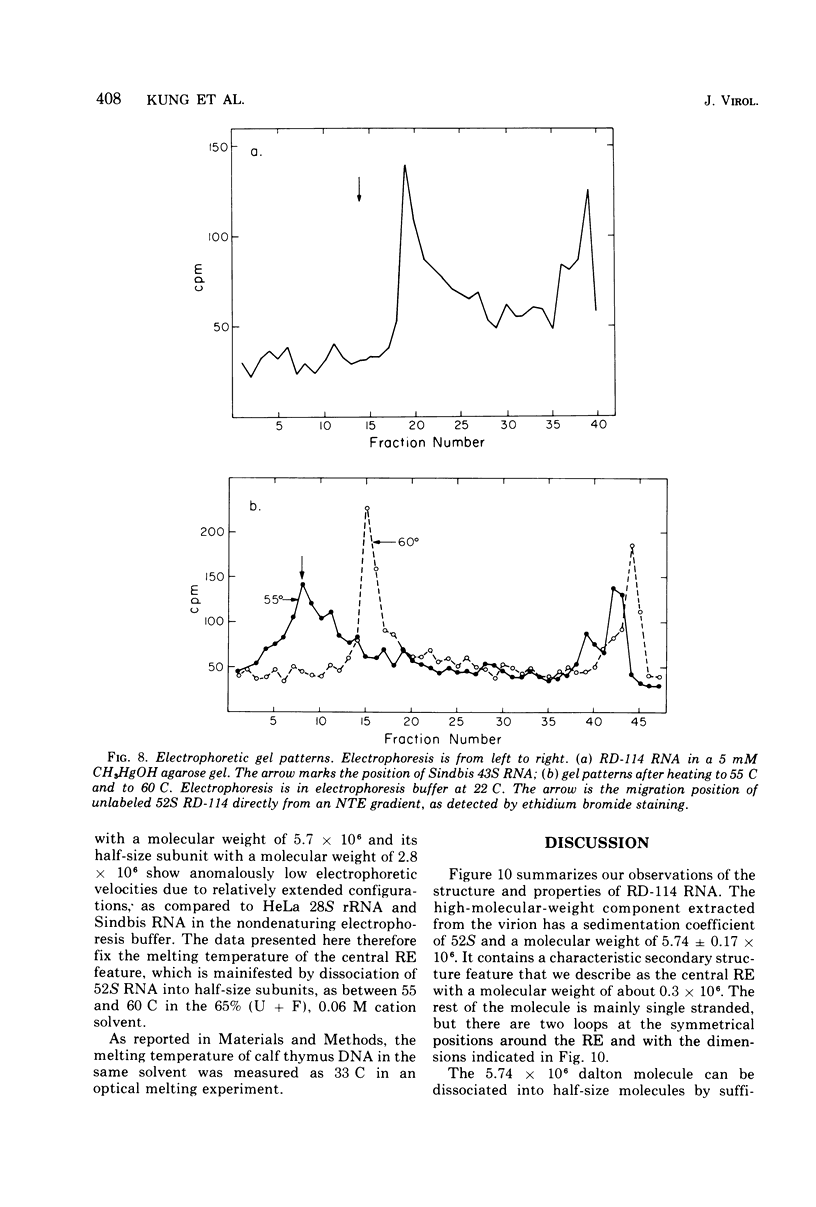
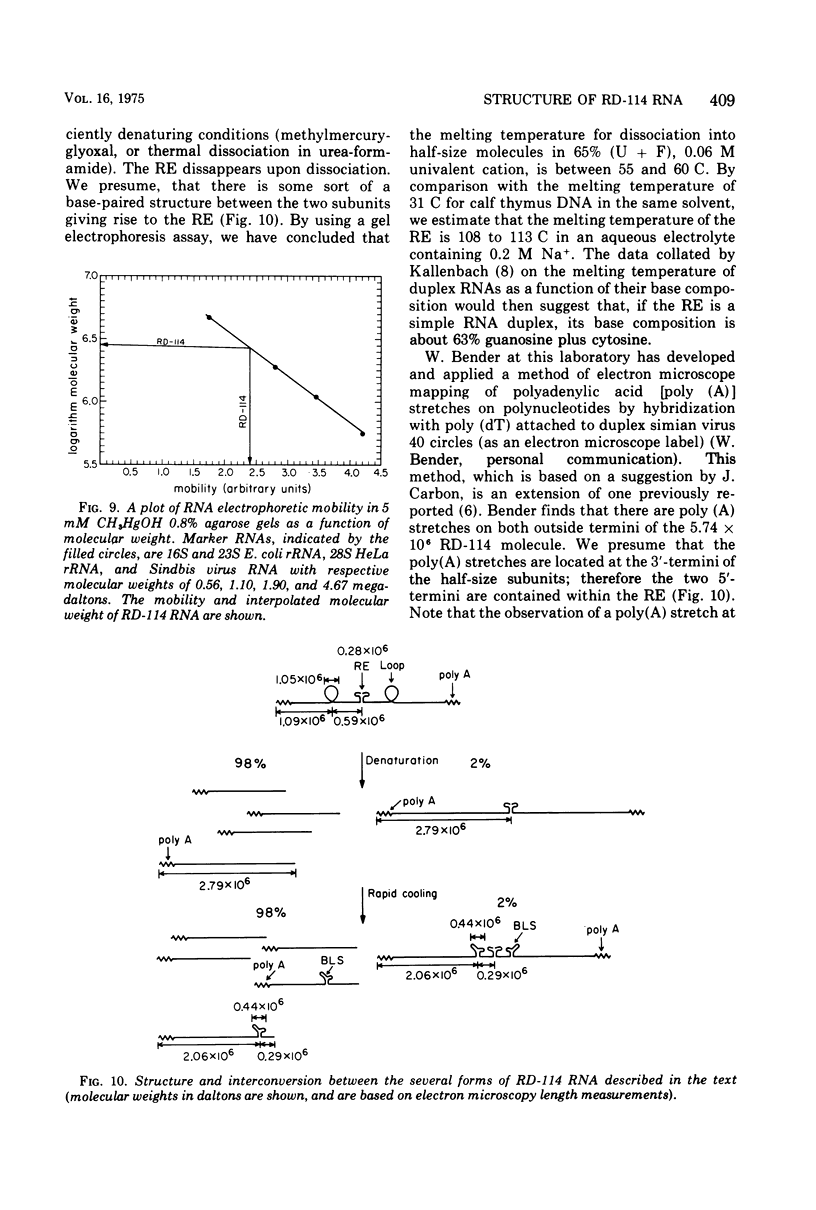
The properties and subunit composition of the RNA extracted from RD-114 virions have been studied. The RNA extracted from the virion has a sedimentation coefficient of 52S in a nondenaturing aqueous electrolyte. The estimated molecular weight by sedimentation in nondenaturing and weakly denaturing media is in the range 5.7 X 10(6) to 7.0 X 10(6). By electron microscopy, under moderately denaturing conditions, the 52S molecule is seen to be an extended single strand with a contour length of about 4.0 mum corresponding to a molecular weight of 5.74 X 10(6). It contains two characteristic secondary structure features: (i) a central Y- or T-shaped structure (the rabbit ears) with a molecular weight of 0.3 X 10(6), (ii) two symmetreically disposed loops on each side of and at equal distance from the center. The 52S molecule consists of two half-size molecules, with molecular weight 2.8 X 10(6), joined together within the central rabbit ears feature. Melting of the rabbit ears with concomitant dissociation of the 52S molecule into subunits, has been caused by either one of two strongly denaturing treatments: incubation in a mixture of CH3HgOH and glyoxal at room temperature, or thermal dissociation in a urea-formamide solvent. When half-size molecules are quenched from denaturing temperatures, a new off-center secondary structure feature termed the branch-like structure is seen. The dissociation behavior of the 52S complex and the molecular weight of the subunits have been confirmed by gel electrophoresis studies. The loop structures melt at fairly low temperatures; the dissociation of the 52S molecule into its two subunits occurs at a higher temperature corresponding to a base composition of about 63% guanosine plus cytosine. Polyadenylic acid mapping by electron microscopy shows that the 52S molecule contains two polyadenylic acid segments, one at each end. It thus appears that 52S RD-114 RNA consists of two 2.8 X 10(6) dalton subunits, each with a characteristic secondary structure loop, and joined at the 5' ends to form the rabbit ears secondary structure feature. The observations are consistent with but do not require the conclusion that the two 2.8 X 10(6) dalton subunits of 52S RD-114 RNA are identical.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- East J. L., Knesek J. E., Allen P. T., Dmochowski L. Structural characteristics and nucleotide sequence analysis of genomic RNA from RD-114 virus and feline RNA tumor viruses. J Virol. 1973 Nov;12(5):1085–1091. doi: 10.1128/jvi.12.5.1085-1091.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filbert J. E., McAllister R. M., Nicolson M. O., Gilden R. V., Lennette E. H. RD-114 virus infectivity assay by measurements of DNA polymerase activity and virus group specific antigen. Proc Soc Exp Biol Med. 1974 Feb;145(2):366–370. doi: 10.3181/00379727-145-37812. [DOI] [PubMed] [Google Scholar]
- Forsheit A. B., Davidson N., Brown D. D. An electron microscope heteroduplex study of the ribosomal DNAs of Xenopus laevis and Xenopus mulleri. J Mol Biol. 1974 Dec 5;90(2):301–314. doi: 10.1016/0022-2836(74)90375-1. [DOI] [PubMed] [Google Scholar]
- Gruenwedel D. W., Davidson N. Complexing and denaturation of DNA by methylmercuric hydroxide. I. Spectrophotometric studies. J Mol Biol. 1966 Oct 28;21(1):129–144. doi: 10.1016/0022-2836(66)90084-2. [DOI] [PubMed] [Google Scholar]
- Hsu M. T., Kung H. J., Davidson N. An electron microscope study of Sindbis virus RNA. Cold Spring Harb Symp Quant Biol. 1974;38:943–950. doi: 10.1101/sqb.1974.038.01.096. [DOI] [PubMed] [Google Scholar]
- Inman R. B., Schnös M. Partial denaturation of thymine- and 5-bromouracil-containing lambda DNA in alkali. J Mol Biol. 1970 Apr 14;49(1):93–98. doi: 10.1016/0022-2836(70)90378-5. [DOI] [PubMed] [Google Scholar]
- Kallenbach N. R. Theory of thermal transitions in low molecular weight RNA chains. J Mol Biol. 1968 Nov 14;37(3):445–466. doi: 10.1016/0022-2836(68)90114-9. [DOI] [PubMed] [Google Scholar]
- Kung H. J., Bailey J. M., Davidson N., Nicolson M. O., McALLISTER R. M. Structure and molecular length of the large subunits of RD-114 viral RNA. J Virol. 1974 Jul;14(1):170–173. doi: 10.1128/jvi.14.1.170-173.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung H. J., Bailey J. M., Davidson N., Vogt P. K., Nicolson M. O., McAllister R. M. Electron microscope studies of tumor virus RNA. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):827–834. doi: 10.1101/sqb.1974.039.01.096. [DOI] [PubMed] [Google Scholar]
- Robberson D., Aloni Y., Attardi G., Davidson N. Expression of the mitochondrial genome in HeLa cells. VI. Size determination of mitochondrial ribosomal RNA by electron microscopy. J Mol Biol. 1971 Sep 28;60(3):473–484. doi: 10.1016/0022-2836(71)90182-3. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Trávnícek M., Ríman J. Subunits of oncornavirus high-molecular-weight RNA. I. Stepwise conversion of 60 S AMV (avian myeloblastosis virus) RNA to subunits. Biochem Biophys Res Commun. 1973 Jul 2;53(1):217–223. doi: 10.1016/0006-291x(73)91422-8. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B., Kelley D. E., Perry R. P. Secondary structure maps of ribosomal RNA. II. Processing of mouse L-cell ribosomal RNA and variations in the processing pathway. J Mol Biol. 1974 Oct 25;89(2):397–407. doi: 10.1016/0022-2836(74)90527-0. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. Secondary structure maps of RNA: processing of HeLa ribosomal RNA. Proc Natl Acad Sci U S A. 1973 Oct;70(10):2827–2831. doi: 10.1073/pnas.70.10.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]





