Abstract
Airway occlusion in obstructive airway diseases is caused in part by the overproduction of secretory mucin glycoproteins through the up-regulation of mucin (MUC) genes by inflammatory mediators. Some pharmacological agents, including the glucocorticoid dexamethasone (Dex), repress mucin concentrations in lung epithelial cancer cells. Here, we show that Dex reduces the expression of MUC5AC, a major airway mucin gene, in primary differentiated normal human bronchial epithelial (NHBE) cells in a dose-dependent and time-dependent manner, and that the Dex-induced repression is mediated by the glucocorticoid receptor (GR) and two glucocorticoid response elements (GREs) in the MUC5AC promoter. The pre-exposure of cells to RU486, a GR antagonist, and mutations in either the GRE3 or GRE5 cis-sites abolished the Dex-induced repression. Chromatin immunoprecipitation (ChIP) assays showed a rapid temporal recruitment of GR to the GRE3 and GRE5 cis-elements in the MUC5AC promoter in NHBE and in A549 cells. Immunofluorescence showed nuclear colocalization of GR and histone deacetylase–2 (HDAC2) in MUC5AC-expressing NHBE cells. ChIP also showed a rapid temporal recruitment of HDAC2 to the GRE3 and GRE5 cis-elements in the MUC5AC promoter in both cell types. The knockdown of HDAC2 by HDAC2-specific short interfering RNA prevented the Dex-induced repression of MUC5AC in NHBE and A549 cells. These data demonstrate that GR and HDAC2 are recruited to the GRE3 and GRE5 cis-sites in the MUC5AC promoter and mediate the Dex-induced cis repression of MUC5AC gene expression. A better understanding of the mechanisms whereby glucocorticoids repress MUC5AC gene expression may be useful in formulating therapeutic interventions in chronic lung diseases.
Keywords: MUC5AC, HDAC2, dexamethasone, gene repression, glucocorticoid receptor
Clinical Relevance
Mucin overproduction contributes to morbidity and mortality in chronic airway diseases. Mechanisms whereby pharmacological agents such as glucocorticoids repress mucin gene expression are not well-studied. Here we show that the glucocorticoid receptor and histone deacetylase–2 mediate the dexamethasone-induced repression of MUC5AC, a major airway mucin gene. Understanding how glucocorticoids repress the expression of mucin genes may be important for formulating therapeutic interventions in chronic lung diseases.
Mucus covers and protects the epithelium in the mammalian respiratory, gastrointestinal, and reproductive tracts, and contributes to the mucosal defense barrier (1, 2). In the respiratory tract, secreted mucin glycoproteins (mucins), the major macromolecular components of lung mucus, are part of the innate immune defense system and mucociliary escalator that protect the airways against airborne challenges (3). Mucin production is increased in chronic lung diseases and contributes to the occlusion of the conducting airways by mucus, thereby significantly affecting disease morbidity and mortality in patients with asthma, cystic fibrosis, bronchopulmonary dysplasia, and chronic obstructive pulmonary diseases (4).
Mucin (MUC) genes encode the protein backbone of human mucins, and exhibit a selective tissue and cell specificity that is frequently altered in inflammatory diseases and in cancer (5, 6). Two polymeric mucins (MUC5AC and MUC5B) are normally expressed and secreted in the lungs by secretory cells in the conducting airway epithelium and submucosal glands, respectively. Increased production of MUC5AC mRNA and protein occurs in the airway epithelium and secretions of patients with asthma (7–10), and MUC5B concentrations are increased in the secretions of patients with chronic obstructive diseases (11). Studies from several laboratories have shown that secretory mucin gene expression is up-regulated in lung epithelial cells in vitro by inflammatory/immune response mediators that are activated in the respiratory tract in response to airborne challenges and mechanisms whereby mediators up-regulating mucin gene expression have been identified (6, 12, 13).
In contrast, mechanisms whereby secretory mucin genes are down-regulated have not been identified, although a few pharmacological agents have been reported to reduce concentrations of secretory mucins, as will be presented. These include glucocorticoids, which are used clinically to treat lung inflammation. The glucocorticoid regulation of gene expression occurs via the glucocorticoid receptor (GR), and the mechanisms are varied and complex (14). Classically, ligand-activated GR binds to glucocorticoid responsive element (GRE) cis-sites in the 5′-upstream flanking sequences (e.g., the promoter region) to cis-activate or cis-repress target genes (15, 16), but the mechanisms whereby glucocorticoids cis-repress gene expression have been minimally studied (16). Alternatively, ligand-activated GR can regulate gene expression by binding to inflammatory transcription factors such as NF-κB or adaptor protein 1 (AP1) to trans-repress the expression of genes that lack functional GRE cis-elements in their promoter region (17, 18). Ligand-activated GR also recruit various histone acetylases and coactivators or histone deacetylases (HDACs) and corepressors to the promoters of GR-targeted genes to mediate gene regulation (19–21).
The glucocorticoid dexamethasone (Dex) decreases MUC2 and MUC5AC mRNA abundance in the H292 lung cancer cell line (22), as well as MUC5AC mRNA abundance in A549 lung cancer cells (23, 24) and primary rat tracheal surface cells (23). Recently, we reported that Dex activated GR binding to two putative GRE cis-sequences (GRE3 and GRE5) in the MUC5AC promoter, resulting in the cis-repression of MUC5AC gene expression in the A549 lung epithelial cancer cell line (24). However, mucin gene regulation in cancer-derived or immortalized cell lines may not reflect normal physiology (25). Here, we report on functional studies in primary differentiated normal human bronchial epithelial (NHBE) cells, compared with A549 cells. This in vitro NHBE lung model system morphologically mimics the human conducting airway epithelium (26, 27), and is typically used to verify the biological significance of findings in lung cell lines. The data show a role for GR and HDAC2 in the Dex-induced repression of MUC5AC in both model systems.
Materials And Methods
Information on cell culture, Dex exposure, reporter plasmids, transfection, RNA isolation, real-time PCR analysis, Western blot analyses, and inhibitor experiments is provided in the online supplement.
Immunofluorescent Staining
Differentiated NHBE cells were fixed in 75% ethanol, blocked with 2% BSA/0.1% Triton-100, and labeled with rabbit anti-GR (1:50; Santa Cruz Biotechnology, Santa Cruz, CA), goat anti-HDAC2 (1:50; Santa Cruz Biotechnology), or mouse anti-MUC5AC (Clone 45M1, 1:50; NeoMarkers, Fremont, CA) antibodies at 4°C overnight. Alexa Fluor 566 donkey anti-rabbit IgG (1:100; Life Technologies, Carlsbad, CA), Alexa Fluor 647 donkey anti-goat IgG (1:125; Life Technologies), and Alexa Fluor 488 donkey anti-mouse IgG (1:100; Life Technologies) were used as secondary antibodies. Cells were imaged using a Zeiss LSM 510 confocal microscope (Carl Zeiss MicroImaging, Thornwood, NY), with excitation wavelengths of 488 nm, 561 nm, and 633 nm. Confocal images were acquired by sequential excitation through a ×63 objective (numerical aperture = 1.4). Postacquisition processing was performed using Adobe Photoshop CS3. In alternative experiments, differentiated NHBE cells on 24-well plates were fixed in 2% paraformaldehyde for 30 minutes and labeled with rabbit anti-GR (1:50; Santa Cruz Biotechnology) or goat anti-HDAC2 (1:50; Abcam, Cambridge, MA) at 4°C overnight. Texas Red–donkey anti-rabbit IgG (1:250; Jackson ImmunoResearch, West Grove, PA) and FITC–donkey anti-mouse IgG (1:200; Jackson ImmunoResearch) were used as secondary antibodies. Cells were counterstained to reveal cell nuclei with 10-μg/ml 4′-6-diamidino-2-phenylindol (Sigma Chemical Co., St. Louis, MO) for 5 minutes, and were analyzed using an Apotome microscope (Carl Zeiss MicroImaging) with a ×40 objective.
Chromatin Immunoprecipitation Assays
The protocol for cross-linking and isolating chromatin was performed using the chromatin immunoprecipitation (ChIP) assay kit according to the manufacturer’s protocol (Upstate, Charlottesville, VA). Two hundred and fifty miocroliters were taken for the input assay before adding antibodies for immunoprecipitation. Immunoprecipitation was performed using a total of 10 μg of anti-GR antibodies (PAI-511A and PAI-512; Affinity Bioreagents, Rockford, IL), anti-HDAC2 antibody (Cell Signaling, Danvers, MA), or control IgG antibody (Santa Cruz Biotechnology) overnight at 4°C. DNA associated with immunocomplexes was amplified by quantitative PCR, using SYBR-green (BioRad, Irvine, CA) and MUC5AC-specific primer pairs: GRE1/2 forward 5′-AGTGCTCAGAACAGCCTTGAG-3′ and reverse 5′-ATGGGAGGAATGGCAGGA-3′; GRE3 forward 5′-CCTTCAGGCCAAAGACTCAC-3′ and reverse 5′-GGTCTCTGGCCACCAAGAT-3′; GRE4 forward 5′-GTGGCCAGAGACCATCAAGT-3′ and reverse 5′-ATAGAACCCCTCCCTCACCA-3′; and GRE5 forward 5′-GAATGGCAGGAAAGGGAAAG-3′ and reverse 5′-GTTCCTGGTGCCCAGAAGT-3′. Values for immunoprecipitated DNA were normalized to input MUC5AC DNA. The results are expressed as mean fold change over baseline concentrations. Each sample was analyzed in triplicate. Each experiment was performed on at least three separate occasions.
HDAC2 Small Interfering RNA
NHBE and A549 cells were transfected with 10 nM of control, single (HDAC2-2), or multiple (HDAC2-1,3,4) small interfering RNAs (siRNAs; Qiagen, Valencia, CA), using HiPerFect transfection reagent (Qiagen) in accordance with the manufacturer’s protocol.
Statistical Analyses
Comparisons of the means between two groups were performed using the Student t test. Comparisons for more than two groups were performed using ANOVA (GraphPad Prism Software, San Diego, CA). Statistical significance was set at P ≤ 0.05.
Results
Dex Decreases MUC5AC mRNA and Protein Expression in NHBE Cells in a Time-Dependent and Dose-Dependent Manner
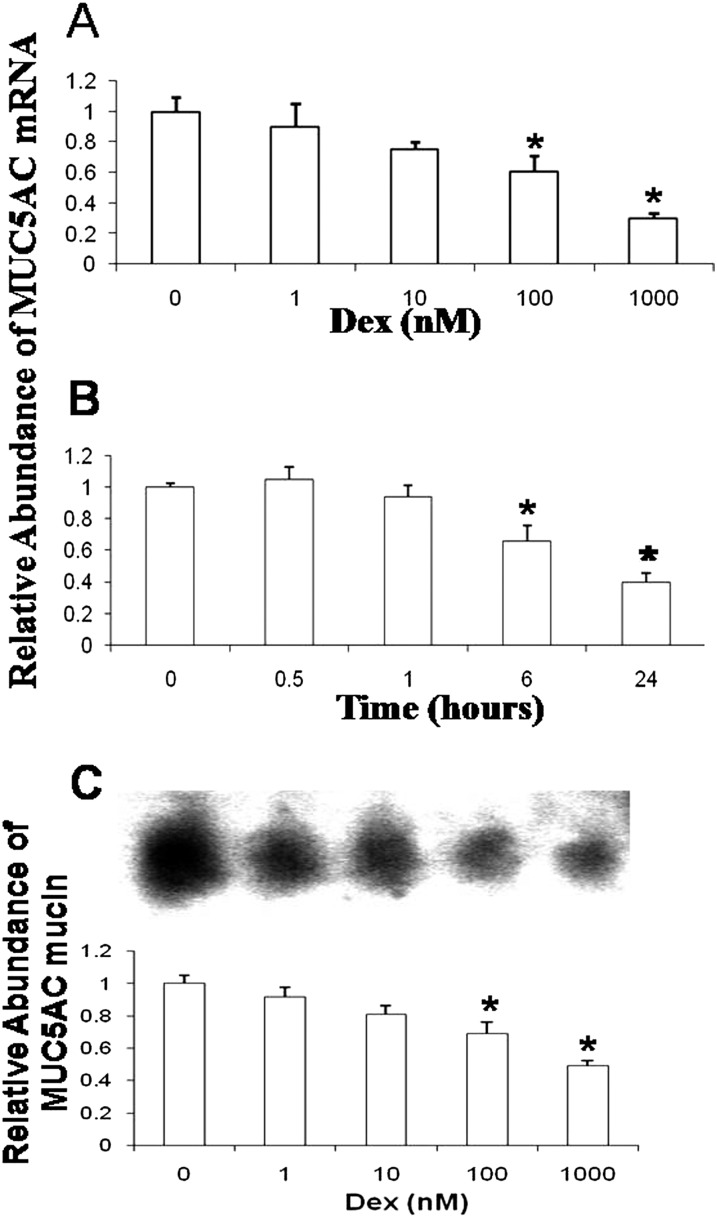
Previously, we reported that Dex decreases MUC5AC mRNA expression in the A549 lung cancer cell line, as well as in NHBE cells from one individual (24). Here, we evaluated the temporal effects, as well as the dose effects, of Dex on MUC5AC mRNA and protein expression in NHBE cells from additional individuals. In a dose–response test, 100 and 1,000 nM Dex resulted in a significant decrease of MUC5AC mRNA at 24 hours, indicating that Dex reduced the abundance of MUC5AC mRNA in NHBE cells in a dose-dependent manner. (Figure 1A). Temporal analyses showed that 1,000 nM Dex resulted in a significant decrease in the expression levels of MUC5AC mRNA at 6 hours, and a greater decrease at 24 hours (Figure 1B). This temporal pattern showed that the Dex-induced repression in NHBE cells was a delayed response, similar to that observed for MUC5AC mRNA in A549 lung cancer cells (24). To determine whether the Dex-induced repression of MUC5AC mRNA translated into reduced concentrations of mature MUC5AC mucin in NHBE cell secretions, the effects of Dex on MUC5AC mucin in apical secretions were examined. Western blot analysis showed a concentration-dependent decrease of MUC5AC (Figure 1C) mucin concentrations. Taken together, these data demonstrated that Dex decreased MUC5AC mRNA and protein abundance in vitro within 24 hours and at concentrations within a range that approximate glucocorticoid concentrations (10–1,000 nM) in airway epithelium after oral delivery (28).
Figure 1.
Dexamethasone (Dex) decreases mucin 5AC (MUC5AC) mRNA and protein concentrations in primary differentiated normal human bronchial epithelial (NHBE) cells in a dose-dependent and time-dependent manner. (A) Cells were exposed to Dex (1, 10, 100, or 1,000 nM) or vehicle for 24 hours. (B) Cells were incubated with 1,000 nM Dex or vehicle for 0, 0.5, 1, 6, and 24 hours. In A and B, MUC5AC and actin mRNA concentrations were quantified by quantitative RT-PCR, and then normalized to actin and carrier control. The results of three independent experiments with triplicate PCR analyses from two individuals are expressed as means ± standard error (SE). (C) Cells were exposed to Dex (1, 10, 100, or 1,000 nM) or vehicle for 24 hours. Equal volumes of apical secretions were analyzed by Western blotting on 1% agarose gels. An example of a typical blot is shown at the top. Mucin concentrations were evaluated by densitometry and normalized to control (unexposed) conditions. The results of experiments with duplicate analyses using NHBE cells from two individuals are expressed as means ± SE. Statistically significant differences are indicated by asterisks. *P < 0.05.
GR Is Required for the Dex-Induced Repression of MUC5AC Gene Expression
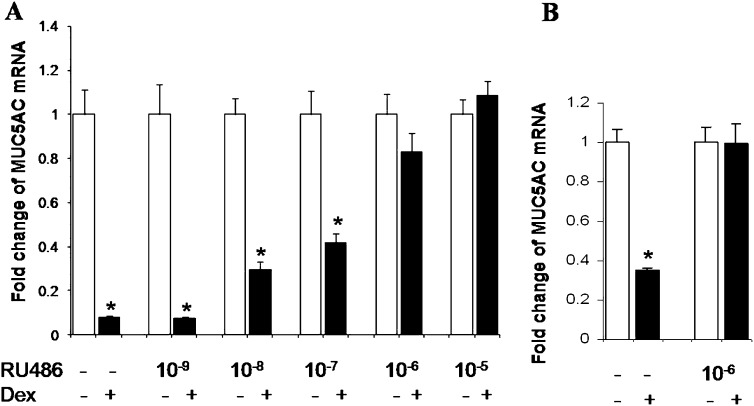
Ligand-activated GR is required for the transcriptional regulation of glucocorticoid targeted genes (29). To establish whether the Dex-induced decrease of MUC5AC mRNA was dependent on the presence of the Dex-activated GR, both A549 and NHBE cells were pre-exposed to RU486, a GR antagonist, and then to Dex. In A549 cells, increasing concentrations of RU486 (10−9 to 10−6) decreased the ability of Dex to reduce MUC5AC mRNA concentrations (Figure 2A). The Dex-induced repression of the MUC5AC gene was completely abrogated in A549 cells at 10 μM RU486 (Figure 2A) and at 1 μM RU486 in NHBE cells (Figure 2B). These concentrations are within the RU486 range that abolishes the impact of Dex on the expression of known GR-targeted genes (30, 31). These results show that GR is required for the Dex-mediated repression of MUC5AC gene expression in airway epithelial cells.
Figure 2.
The GR antagonist reduces Dex-induced repression of MUC5AC mRNA. A549 cells (A) and NHBE cells (B) were pre-exposed to RU486 (1–10,000 nM) or medium for 1 hour, and then exposed to Dex (1,000 nM) Dex for 24 hours. Cells were harvested, and MUC5AC and actin mRNA expressions were determined by quantitative RT-PCR. Values were normalized to actin mRNA. Results are expressed as the mean fold change above baseline concentrations. Each sample was analyzed in triplicate; each experiment was performed on three separate occasions. Statistically significant differences indicated by asterisks. *P < 0.05.
GRE-3 and GRE-5 cis-Sites Mediate the Dex-Induced Repression of MUC5AC
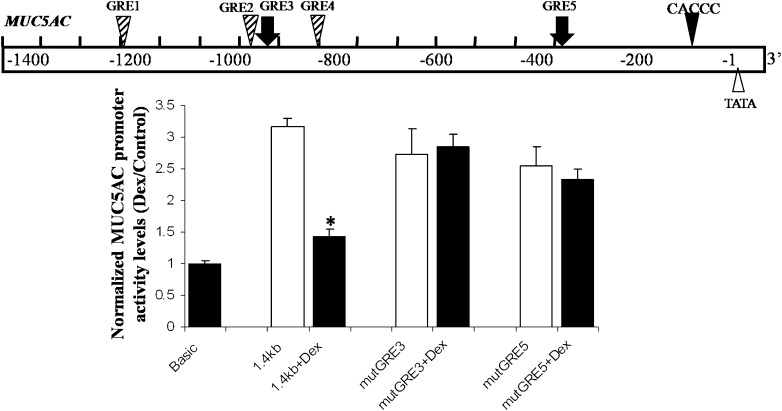
GRE3 and GRE5 cis-sites in the MUC5AC promoter (Figure 3, top) are implicated in the Dex-induced transcriptional repression of the MUC5AC gene through promoter analyses and electrophoretic mobility shift assays (EMSAs) in A549 cells (24). To assess functionally the effects of Dex on MUC5AC gene expression in NHBE cells, we established conditions that permitted the efficient transfection of plasmids into these cells. The transfection efficiency was evaluated using a pUL37×1-mEGFP fusion protein reporter construct, because the HCMV UL37×1 gene product concentrates the fluorescent signal to the secretory apparatus and mitochondria (32). Immunofluorescence data showed that the transfection efficiency was 25% in NHBE cells (see Figure E1 in the online supplement).
Figure 3.
GRE3 and GRE5 cis-sites in the MUC5AC promoter region are functionally required for Dex-induced repression in NHBE cells. Top: Potential GRE cis-sites in the 5′ flanking upstream sequences of the MUC5AC promoter. The GRE3 and GRE5 cis-sites, shown to bind GR by electrophoretic mobility shift assays (EMSAs) and wild-type and mutant (mut) MUC5AC:GRE probes after the exposure of A549 cells to Dex (24), are indicated by solid arrows. The wild-type and mutated sequences in the GRE3 and GRE5 sites are shown in Table 1. Site-directed mutagenesis was performed to alter specific base pairs individually in each sequence of the promoter constructs. NHBE cells were transected with each promoter construct (as described in the online supplement’s Materials and Methods), and cells were exposed to Dex or vehicle for 48 hours. The promoter activity of the MUC5AC promoter–Luc reporter constructs is shown relative to that of the promoter activity of the basic Luc plasmid. The results are expressed as the mean ± SE for triplicate samples from three independent experiments. *P < 0.05.
Analysis of the MUC5AC–Luc promoter in NHBE cells from three individuals showed some variability in promoter activity levels between cell cultures, as expected with primary cells from different individuals, but was maximal at 0.165–0.33 μg DNA per well in a 12-well plate (data not shown). Using this method, we produced a similar response pattern of MUC5AC promoter activity in response to Dex in NHBE cells and the A549 cancer cell line (Figure E2). These data demonstrate that NHBE cells can be relatively efficiently transfected, and that Dex transcriptionally represses MUC5AC promoter activity in NHBE cells as well as in A549 cells.
Site-directed mutagenesis was performed independently on both the GRE3 and GRE5 cis-sequences in the MUC5AC promoter (Table 1), and the transfection of wild-type and mutated MUC5AC promoter reporter constructs was performed in differentiated NHBE cells. Transfection with the mutant promoter constructs exerted a minimal effect on MUC5AC basal promoter activity. Dex induced the repression of MUC5AC wild-type promoter activity, but was unable to repress MUC5AC promoter-driven expression in NHBE cells transfected with promoter constructs mutated at either the GRE3 or GRE5 cis-sites (Figure 3, bottom). These data indicate that both the GRE3 and GRE5 cis-sequences in the MUC5AC promoter play functional roles in the Dex-induced cis-repression of MUC5AC gene expression in primary NHBE cells.
TABLE 1.
GRE3 AND GRE5 SEQUENCES IN MUC5AC PROMOTER CONSTRUCTS
| Sequence | Nucleotide Numbers | |
| GRE3 | ccTGTCCA GAG GGTACTga | −930 to −912 |
| mutGRE3* | ccTCACCA GAG GCAACTga | |
| GRE5 | ctgggcTGGGCC CCC TGTCCTgctg | −369 to −351 |
| mutGRE5* | ctgggcTCCGAC CCC TCACCTgctg |
Definition of abbreviations: GRE, glucocorticoid response element; MUC5AC, mucin 5AC gene; mut, mutated.
Sequences in boldface were mutated. Sequences adjacent to the GRE sites are shown as lowercase letters.
Expression of HDACs in NHBE Cells
In addition to the GR, the down-regulation of gene expression by glucocorticoids can involve HDACs. Because no information, to the best of our knowledge, has been reported on HDAC expression in differentiated NHBE cells, we examined their nuclear abundance, as well as that of the GR, at baseline, and monitored their temporal response after exposure to Dex by Western blot analysis. HDAC1, HDAC2, HDAC3, and HDAC5 were detectable in NHBE nuclear lysates before Dex exposure, and their nuclear abundance increased after exposure to Dex, with each HDAC exhibiting a specific temporal pattern. We also observed that the nuclear abundance of GR in quiescent NHBE cells was barely detectable, but markedly increased at 0.5 and 1 hour after Dex exposure (Figure E3). Although all HDACs evaluated were identified in NHBE cells, as well as in A549 cells (data not shown), initial studies focused on HDAC2, which was shown to be recruited by the GR to trans-repress inflammatory genes in A549 cells (33).
Localization of GR and HDAC2 in NHBE Cells
The localization of the GR and HDAC2 after Dex exposure to NHBE cells during a 24-hour period was evaluated by immunofluorescence. The data showed a nuclear localization of HDAC2 in NHBE cells under both control and Dex-exposed conditions, as expected. After Dex exposure, the nuclear translocation of the GR and colocalization with HDAC2 was observed at 0.5, 1, and 6 hours, but not at 18 or 24 hours. Localization and colocalization data for the 1-hour time point are shown in Figure E4. The nuclear colocalization of GR and HDAC2 was observed in a majority of cells in the epithelium, which include goblet, ciliated, and basal cells.
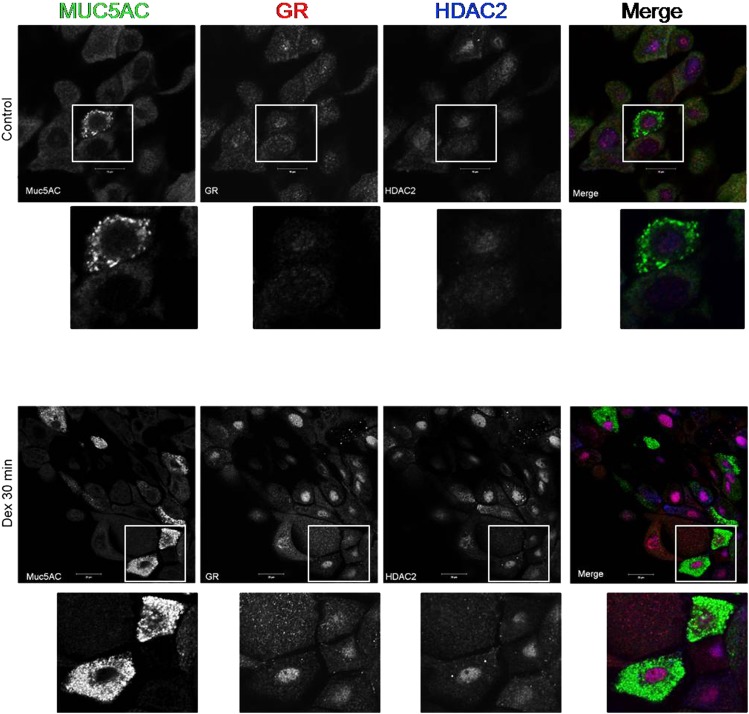
To determine whether the GR and HDAC2 were colocalized to MUC5AC-expressing goblet cells, immunofluorescence studies were performed. The colocalization of GR and HDAC2 was clearly observed in MUC5AC-expressing cells (shown in Figure 4, at the 30-min time point), suggesting that the GR and HDAC2 may comprise part of a complex that mediates the Dex-induced repression of the MUC5AC gene.
Figure 4.
Dex and histone deacetylase (HDAC) colocalize in MUC5AC-positive NHBE cells after Dex repression. NHBE cells were methanol-fixed and imaged using confocal microscopy (Zeiss LSM510). Cells were probed with anti-MUC5AC (green), anti-GR (red), and anti-HDAC2 (blue) antibodies and their corresponding secondary antibodies. Sequentially acquired channels of a single optical section of NHBE cells after exposure to vehicle (top) or Dex (1,000 nM; bottom) for 30 minutes are shown in individual (black and white) panels. Merged (color) images are shown at far right. Areas in white boxes are displayed as enlargements under each image. At far right bottom, the nuclear colocalization of GR and HDAC2 signals presents as violet or purple after Dex exposure, and was observed in the majority of cells in the heterogeneous NHBE system, including the goblet cells that express MUC5AC.
Dex Recruits the GR and HDAC2 to GRE cis-Elements in the MUC5AC Promoter
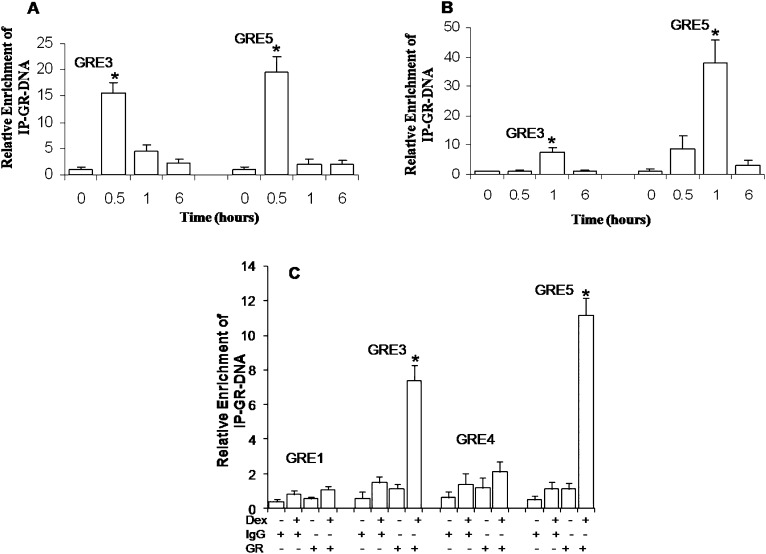
EMSAs had previously demonstrated that the GR binds to the GRE3 and GRE5 cis-sites in the MUC5AC promoter in A549 cells (24). However, EMSA shows that transcription factors in nuclear extracts can bind to synthetic oligonucleotides of a defined sequence, whereas ChIP assays demonstrate whether or not the actual binding of transcription factors occurs at specific cis-sequences in situ in target gene promoters (34). Thus, we used ChIP assays to evaluate GR binding to specific GRE cis-sites in the MUC5AC promoter at 0, 0.5, 1, and 6 hours after Dex exposure. Our analysis showed the recruitment of the GR to the GRE3 and GRE5 cis-sites in the MUC5AC promoter, with maximal binding of the GR at both GRE cis-sites at 30 minutes in A549 cells (Figure 5A). A similar pattern was observed in NHBE cells, where GR recruitment to the GRE3 and GRE5 cis-sites was maximal 1 hour after Dex exposure (Figure 5B). In addition, ChIP analyses were also performed to evaluate GR binding at GRE cis-sites in the MUC5AC promoter that did not exhibit binding in EMSA studies (24). Similar to previous data (Figure 5A), the GR bound specifically to GRE3 and GRE5, but not GRE1 or GRE4, cis-sites in the MUC5AC promoter (Figure 5C). Binding at GRE2 alone could not be evaluated via ChIP, because primer pairs that distinguish between GRE2 and GRE3 cis-sites could not be designed. However, EMSA and promoter studies in A549 cells (24) showed that GRE3, but not GRE2, is a functional GRE cis-site in the MUC5AC promoter.
Figure 5.
Chromatin immunoprecipitation (ChIP) analysis demonstrated the temporal recruitment of the GR to the GRE3 and GRE5 cis-sites in the MUC5AC promoter in lung epithelial cells. (A and B) Cells were exposed to Dex for 0, 0.5, 1, and 6 hours. The GR associated with chromatin in nuclear lysates was immunoprecipitated (IP) using anti-GR antibodies. The associated DNA was amplified and quantified by quantitative PCR with primer pairs specific for GRE3 or GRE5. Values were normalized by input DNA. The results from three experiments with triplicate analyses are expressed as the mean fold change above baseline concentrations. *P < 0.05. (A) A549 cells, 100 nM Dex. (B) NHBE cells, 1,000 nM Dex. Data represent the results of experiments using NHBE cells from two individuals. (C) A549 cells were exposed to 100 nM Dex for 0.5 hour. ChIP analyses were performed, using primer pairs that included the GRE1/2, GRE3, GRE4, and GRE5 cis-sites after immunoprecipitation with IgG-specific or GR-specific antibodies, as indicated on the y axis. The results from three experiments are expressed as the mean fold change above baseline concentrations. *P < 0.05.
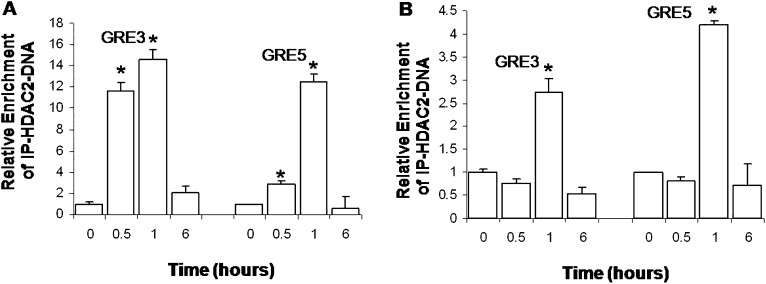
ChIP analyses were also performed to determine whether HDAC2 was recruited to the GRE3 and GRE5 cis-sites in the MUC5AC promoter. In A549 cells, HDAC2 was present at both cis-sites in the MUC5AC promoter at 0.5 and 1 hour, and was maximal at 1 hour after Dex exposure (Figure 6A). In NHBE cells, HDAC2 recruitment was maximal at 1 hour at both the GRE3 and GRE5 sites after Dex exposure (Figure 6B). These results, and particularly the kinetics of HDAC2 binding to the GRE3 and GRE5 sites in A549 cells, are consistent with the recruitment of HDAC2 to the MUC5AC cis-regulatory elements by activated GR after Dex treatment.
Figure 6.
ChIP analysis demonstrated that HDAC2 is recruited temporally to the GRE3 and GRE5 cis-sites in the MUC5AC promoter in lung epithelial cells. Cells were exposed to Dex for 0, 0.5, 1, and 6 hours. (A) A549 cells, 100 nM Dex. (B) NHBE cells, 1,000 nM Dex. The binding of HDAC2 was determined by ChIP assays, using anti-HDAC2 antibodies for immunoprecipitation. The associated DNA was amplified and quantified by quantitative PCR with primer pairs specific for MUC5AC:GRE3 and MUC5AC:GRE5. Results are expressed as the mean fold change above basal concentrations. Values are normalized by actin and input DNA. The results shown represent data from two separate experiments. *P < 0.05.
HDAC2 Is Required for the Dex-Induced Repression of the MUC5AC Gene
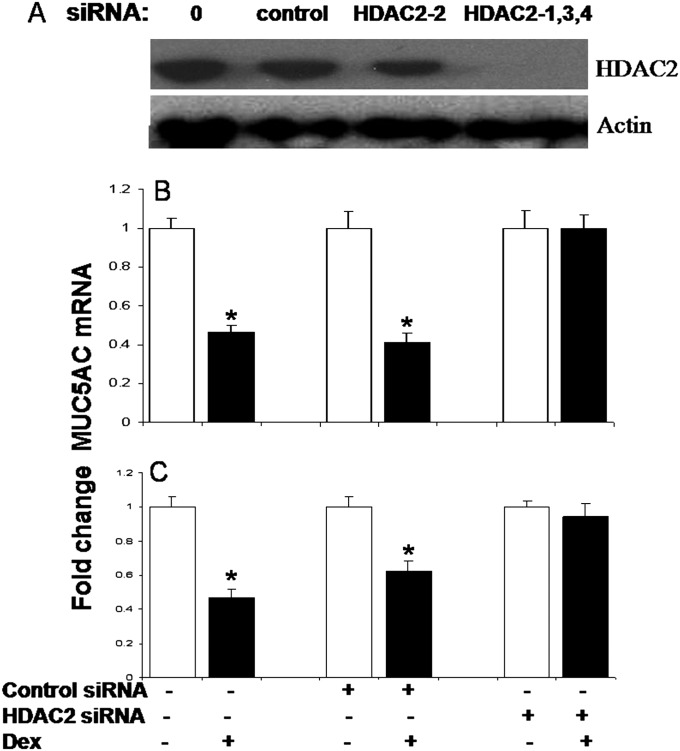
The requirement for HDAC2 in repressing MUC5AC gene expression was verified using siRNA specific to HDAC2. The maximal inhibition (70%) of HDAC2 protein expression in A549 cells was achieved using multiple HDAC2 siRNAs, rather than a single HDAC2 siRNA (Figure 7A). Dex was unable to repress MUC5AC expression when HDAC2 expression was knocked down in either A549 (Figure 7B) or NHBE (Figure 7C) cells. These data demonstrate that HDAC2 mediates the Dex-induced repression of MUC5AC gene expression.
Figure 7.
HDAC2 knockdown abolished the Dex-induced reduction of MUC5AC mRNA. (A) HDAC2 protein expression was evaluated by Western blot analyses. Maximal inhibition (70%) in A549 cells was achieved using multiple (HDAC2-1,3,4) short interfering RNAs (siRNAs), rather than a single (HDAC2-2) siRNA. HDAC2 expression was knocked down with HDAC2-1,3,4 siRNA (10 nM) in A549 (B) or NHBE (C) cells. MUC5AC and actin mRNA concentrations were quantified by quantitative RT-PCR. MUC5AC concentrations were normalized to actin and carrier control. The results are expressed as the mean fold change above baseline concentrations. Each sample was analyzed in triplicate; each experiment was performed on three separate occasions. Statistically significant differences are indicated by asterisks. *P < 0.05.
Discussion
Glucocorticoids modulate the pathological inflammatory drive that is central to chronic diseases, including lung diseases (35), and typically reduce lung mucin concentrations in vivo (36), which is not unexpected because mucin overproduction in the airways is a major consequence of lung inflammation (6). However, Dex also reduces the expression of polymeric genes (MUC5AC and MUC2) in lung epithelial cells in vitro in the absence of an inflammatory stimulus (22–24). Earlier studies from this laboratory showed that Dex cis-represses MUC5AC gene expression, and identified GRE3 and GRE5 as functional targets of activated GR in A549 lung cancer cells (24). The data reported here indicate that Dex likewise cis-represses MUC5AC expression in primary differentiated NHBE cells, and demonstrates for the first time, to the best of our knowledge, that HDAC2 plays a role in glucocorticoid-mediated cis-repression, a markedly understudied area (16). This is in contrast to trans-repression of genes where ligand-activated GR binds to inflammatory transcription factors of anti-inflammatory genes to prevent their up-regulation (16, 18).
In contrast to GR-targeted genes that are regulated by trans-repression, fewer genes are cis-repressed, as reviewed previously (16). These genes include pro-opiomelanocortin (37), collagen (38), vasoactive intestinal peptide receptor (39), osteocalcin (40), IL-1β (41), and keratins (42). More recently, glutathione S-transferase (43), mFasL (44), FASL (45), and MUC5AC (24) have been added to the list of genes that are cis-repressed by glucocorticoids.
Mechanistic studies on the role of glucocorticoids in lung inflammation have used the A549 lung cancer cell line and focused on the Dex induction of trans-repression and chromatin remodeling in target genes (46). Our initial studies on the Dex-induced repression of MUC5AC were performed in A549 cells (24), but gene regulation in cancer-derived or immortalized cell lines may not fully reflect normal physiology (25).
Thus, functional analyses in this study were performed in both primary differentiated HBE and A549 cells. The data show that the GR is required for the Dex-induced repression of MUC5AC in both types of lung cells. Promoter analyses using wild-type and mutant constructs showed that GRE3 and GRE5 cis-sites were required for the Dex-induced repression of MUC5AC in primary differentiated NHBE cells. ChIP analyses showed that the GR rapidly and simultaneously binds to the GRE3 and GRE5, but not the GRE1 or GRE4, cis-sites in the MUC5AC promoter, with similar temporal patterns in both cell types. The data provide direct evidence that the GR and GRE3 and GRE5 cis-sites in the MUC5AC promoter play functional roles in the Dex-induced repression of the MUC5AC gene in both NHBE cells and A549 lung cells.
Chromatin remodeling is tightly linked to the regulation of gene expression via histone acetylation and deacetylation, which alter the ability of transcription factors to access DNA sites. Glucocorticoids alter chromatin structure in target genes (19–21). Preliminary studies involved experiments with tricostatin A (TSA), an antifungal antibiotic that selectively inhibits mammalian HDACs by interfering with the removal of acetyl groups from histones (47). The TSA inhibitor experiments were not conclusive (data not shown), but suggested that one or more HDACs might mediate the Dex-induced repression of the MUC5AC gene. We focused on HDAC2, which had been shown to be expressed in A549 cells and recruited by the GR to trans-repress inflammatory genes (33, 35, 46). Our data showed that HDAC2 is also expressed in NHBE cells, and that it responds temporally to Dex. ChIP data also support the chromatin remodeling of MUC5AC, because HDAC2 rapidly and simultaneously binds to both the GRE3 and GRE5 cis-sites in the MUC5AC promoter with similar, but not identical, temporal patterns in primary and cancer lung cells. Confocal analyses show that the GR, which translocates to the nucleus of MUC5AC-expressing goblet cells in differentiated NHBE cells 30 minutes after Dex exposure, colocalizes with HDAC2. Intriguingly, even with an abundance of nuclear GR and HDAC2 proteins at this time point and colocalization in goblet cells, neither the GR nor HDAC2 was detectably enriched, according to ChIP analyses, above baseline concentrations at the MUC5AC:GRE3 and MUC5AC:GRE5 cis-sites at 30 minutes in NHBE cells, although the GR and HDAC2 are clearly present at these promoter sites at 1 hour after Dex exposure. This suggests an ordered loading of GR and HDAC2 onto Dex-targeted genes in goblet cells, and indicates that the MUC5AC gene is not immediately targeted earliest by incoming GR and HDAC2. siRNA experiments support the requirement for HDAC2 in the Dex-induced repression of MUC5AC in both A549 and NHBE cells. Taken together, these finding suggest the requirement of subsequent steps beyond mere nuclear localization in coordinating specific promoter associations of transcription-regulating complexes in cis-regulation. In addition, our findings demonstrate that HDAC2, which is implicated in the trans-repression of GR-targeted genes (46), is likewise used for the cis-repression of the MUC5AC gene.
The GR recruits a variety of coactivators and corepressors to the basal transcription machinery, to activate the chromatin remodeling of target genes (20, 48). Our results suggest that GR recruitment to the GRE3 and GRE5 cis-sites in the MUC5AC promoter dynamically increases HDAC2 recruitment to these sites. This, in turn, is likely to affect the chromatin remodeling and gene repression of MUC5AC in lung epithelial cells, possibly by promoting the deacetylation of histones at the MUC5AC promoter. Predictably, HDAC2 also recruits corepressors to the GRE3 or GRE5 cis-sites in the MUC5AC promoter. Corepressor complexes associated with HDACs act via the formation of large multiprotein complexes, including the NuRD and SIN3 complexes (49). Studies to identify the corepressors associated with the GR and HDAC2 at the GRE3 and GRE5 cis-sites in the MUC5AC promoter are underway, and should prove informative in elucidating the mechanisms whereby Dex induces the repression of the MUC5AC gene under baseline conditions. Moreover, studies of Dex-induced responses at the MUC5AC promoter in lung epithelial cells exposed to inflammatory mediators will be required for a better understanding of the pharmacological role of Dex in the treatment of airway diseases.
Supplementary Material
Acknowledgments
The authors thank Michael D. Smith, PhD, Diego Preciado, MD, PhD, and Lindsay Garvin, PhD candidate, for critical readings of the manuscript.
Footnotes
This work was supported by National Institutes of Health grant HL33052 (M.C.R.), and received bridge support from the Children’s National Medical Center and the Cystic Fibrosis Foundation.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2012-0009OC on July 12, 2012
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Rose MC. Mucins: structure, function, and role in pulmonary diseases. Am J Physiol Lung Cell Mol Physiol 1992;263:L413–L429 [DOI] [PubMed] [Google Scholar]
- 2.Corfield AP, Shukla AK. Mucins: vital components of the mucosal defensive barrier. Genom Proteom Technol 2003;3:20–22 [Google Scholar]
- 3.Knowles MR, Boucher RC. Mucus clearance as a primary innate defense mechanism for mammalian airways. J Clin Invest 2002;109:571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogers DF. Mucoactive agents for airway mucus hypersecretory diseases. Respir Care 2007;52:1176–1193 [PubMed] [Google Scholar]
- 5.Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer 2004;4:45–60 [DOI] [PubMed] [Google Scholar]
- 6.Rose MC, Voynow JA. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol Rev 2006;86:245–278 [DOI] [PubMed] [Google Scholar]
- 7.Ordonez CL, Khashayar R, Wong HH, Ferrando R, Wu R, Hyde DM, Hotchkiss JA, Zhang Y, Novikov A, Doglanov G, et al. Mild and moderate asthma is associated with airway goblet cell hyperplasia and abnormalities in mucin gene expression. Am J Respir Crit Care Med 2001;163:517–523 [DOI] [PubMed] [Google Scholar]
- 8.Hoshino M, Morita S, Iwashita H, Sagiya Y, Nairn AC, Naganishi A, Ashida Y, Nishimura O, Fujisawa Y, Fujino M. Increased expression of the human Ca2+-activated Cl− channel 1 (CaCC1) gene in the asthmatic airway. Am J Respir Crit Care Med 2002;165:1132–1136 [DOI] [PubMed] [Google Scholar]
- 9.Groneberg DA, Eynott PR, Lim S, Oates T, Wu R, Carlstedt I, Roberts P, McCann B, Nicholson AG, Harrison BD, et al. Expression of respiratory mucins in fatal status asthmaticus and mild asthma. Histopathology 2002;40:367–373 [DOI] [PubMed] [Google Scholar]
- 10.Hallstrand TS, Debley JS, Farin FM, Henderson WR., Jr Role of MUC5AC in the pathogenesis of exercise-induced bronchoconstriction. J Allergy Clin Immunol 2007;119:1092–1098 [Published erratum appears in J Allergy Clin Immunol 2007;120:1102.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirkham S, Kolsum U, Rousseau K, Singh D, Vestbo J, Thornton DJ. MUC5B is the major mucin in the gel phase of sputum in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2008;178:1033–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basbaum C, Lemjabbar H, Longphre M, Li D, Gensch E, McNamara N. Control of mucin transcription by diverse injury-induced signaling pathways. Am J Respir Crit Care Med 1999;160:544–548 [DOI] [PubMed] [Google Scholar]
- 13.Thai P, Loukoianov A, Waschi S, Wu R. Regulation of airway mucin gene expression. Annu Rev Physiol 2008;70:405–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou J, Cidlowski JA. The human glucocorticoid receptor: one gene, multiple proteins and diverse responses. Steroids 2005;70:407–417 [DOI] [PubMed] [Google Scholar]
- 15.Beato M. Gene regulation by steroid hormones. Cell 1989;56:335–344 [DOI] [PubMed] [Google Scholar]
- 16.Dostert A, Heinzel T. Negative glucocorticoid receptor response elements and their role in glucocorticoid action. Curr Pharm Des 2004;10:2807–2816 [DOI] [PubMed] [Google Scholar]
- 17.McKay LI, Cidlowski JA. Molecular control of immune/inflammatory responses: interactions between nuclear factor–kappa B and steroid receptor–signaling pathways. Endocr Rev 1999;20:435–459 [DOI] [PubMed] [Google Scholar]
- 18.Pelaia G, Vatrella A, Cuda G, Maselli R, Marsico SA. Molecular mechanisms of corticosteroid actions in chronic inflammatory airway diseases. Life Sci 2003;72:1549–1561 [DOI] [PubMed] [Google Scholar]
- 19.Sheldon LA, Becker M, Smith CL. Steroid hormone receptor–mediated histone deacetylation and transcription at the mouse mammary tumor virus promoter. J Biol Chem 2001;276:32423–32426 [DOI] [PubMed] [Google Scholar]
- 20.Hager GL, Elbi C, Johnson TA, Voss T, Nagaich AK, Schiltz RL, Qiu Y, John S. Chromatin dynamics and the evolution of alternate promoter states. Chrom Res 2006;14:107–116 [DOI] [PubMed] [Google Scholar]
- 21.Johnson TA, Elbi C, Parekh BS, Hager GL, John S. Chromatin remodeling complexes interact dynamically with a glucocorticoid receptor–regulated promoter. Mol Biol Cell 2008;19:3308–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kai H, Yoshitake K, Hisatsune A, Kido T, Isohama Y, Takahama K, Miayata T. Dexamethasone suppresses mucus production and MUC2 and MUC5AC gene expression by NCI-H292 cells. Am J Physiol 1996;271:L484–L488 [DOI] [PubMed] [Google Scholar]
- 23.Lu W, Lillehoj E, Kim KC. Effects of dexamethasone on MUC5AC mucin production by primary airway goblet cells. Am J Physiol Lung Cell Mol Physiol 2005;288:L52–L60 [DOI] [PubMed] [Google Scholar]
- 24.Chen YA, Nickola TJ, DiFronzo N, Colberg-Poley AM, Rose MC. Dexmethasone-mediated repression of MUC5AC mucin gene expression in human lung epithelial cells. Am J Respir Cell Mol Biol 2006;34:1–1016354749 [Google Scholar]
- 25.Hattrup CL, Gendler SJ. Structure and function of the cell surface (tethered) mucins. Annu Rev Physiol 2008;70:431–457 [DOI] [PubMed] [Google Scholar]
- 26.Wu R, Zhao YH, Chang MMJ. Growth and differentiation of conducting airway epithelial cells in culture. Eur Respir J 1997;10:2398–2403 [DOI] [PubMed] [Google Scholar]
- 27.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. Well-differentiated human airway epithelial cell cultures. In: Picot J, editor. Human cell culture protocol, 2nd ed. Totowa, NJ: Humana Press, Inc.; 2004. pp. 183–206. [DOI] [PubMed]
- 28.Dorscheid DR, Wojcik KR, Sun S, Marroquin B, White SR. Apoptosis of airway epithelial cells induced by corticosteroids. Am J Respir Crit Care Med 2001;164:1939–1947 [DOI] [PubMed] [Google Scholar]
- 29.Beato M, Chalepakis G, Schauer M, Slater EP. DNA regulatory elements for steroid hormones. J Steroid Biochem 1989;32:737–748 [DOI] [PubMed] [Google Scholar]
- 30.Donnelly LE, Newton R, Kennedy GE, Fenwick PS, Leung RH, Ito K, Russell RE, Barnes PJ. Anti-inflammatory effects of resveratrol in lung epithelial cells: molecular mechanisms. Am J Physiol Lung Cell Mol Physiol 2004;287:L774–L783 [DOI] [PubMed] [Google Scholar]
- 31.Kim YS, Park JS, Jee YK, Lee KY. Dexamethasone inhibits TRAIL- and anti-cancer drugs–induced cell death in A549 cells through inducing NFkB-independent CIAP2 expresssion. Cancer Res Treat 2004;36:330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharon-Friling R, Goodhouse J, Colberg-Poley AM, Shenk T. Human cytomegalovirus PUL37x1 induces the release of endoplasmic reticulum calcium stores. Proc Natl Acad Sci USA 2006;103:19117–19122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barnes PJ. Transcription factors in airway diseases. Lab Invest 2006;86:867–872 [DOI] [PubMed] [Google Scholar]
- 34.Melcher K. New chemical crosslinking methods for the identification of transient protein–protein interactions with multiprotein complexes. Curr Protein Pept Sci 2004;5:287–296 [DOI] [PubMed] [Google Scholar]
- 35.Adcock IM. Glucocorticoids: new mechanisms and future agents. Curr Allergy Asthma Rep 2003;3:249–257 [DOI] [PubMed] [Google Scholar]
- 36.Wojtczak HA, Kerby GS, Wagener JS, Copenhaver SC, Gotlin RW, Riches DWH, Accurso FJ. Beclomethasone diproprionate reduced airway inflammation without adrenal suppression in young children with cystic fibrosis: a pilot study. Pediatr Pulmonol 2001;32:293–302 [DOI] [PubMed] [Google Scholar]
- 37.Charron J, Drouin J. Glucocorticoid inhibition of transcription from episomal proopiomelanocortin gene promoter. Proc Natl Acad Sci USA 1986;83:8903–8907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiner FR, Czaja MJ, Jefferson DM, Giambrone MA, Tur-Kaspa R, Reid LM, Zern MA. The effects of dexamethasone on in vitro collagen gene expression. J Biol Chem 1987;262:6955–6958 [PubMed] [Google Scholar]
- 39.Pei L. Identification of a negative glucocorticoid response element in the rat Type 1 vasoactive intestinal polypeptide receptor gene. J Biol Chem 1996;271:20879–20884 [PubMed] [Google Scholar]
- 40.Meyer T, Carlsted-Duke J, Starr DB. A weak TATA box is a prerequisite for glucocorticoid-dependent repression of the osteocalcin gene. J Biol Chem 1997;272:30709. [DOI] [PubMed] [Google Scholar]
- 41.Zhang G, Zhang L, Duff GW. A negative regulatory region containing a glucocorticosteroid response element (NGRE) in the human interleukin-1beta gene. DNA Cell Biol 1997;16:145–152 [DOI] [PubMed] [Google Scholar]
- 42.Radoja N, Diaz DV, Minars TJ, Freedberg IM, Blumenberg M, Tomic-Canic M. Specific organization of the negative response elements for retinoic acid and thyroid hormone receptors in keratin gene family. J Invest Dermatol 1997;109:566–572 [DOI] [PubMed] [Google Scholar]
- 43.Ki SH, Cho IJ, Choi DW, Kim SG. Glucocorticoid receptor (GR)–associated SMRT binding to C/EBPbeta TAD and Nrf2 Neh4/5: role of SMRT recruited to GR in GSTA2 gene repression. Mol Cell Biol 2005;25:4150–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baumann S, Dostert A, Novac N, Bauer A, Schmid W, Fas SC, Krueger A, Heinzel T, Kirchhoff S, Schutz G, et al. Glucocorticoids inhibit activation-induced cell death (AICD) via direct DNA-dependent repression of the CD95 ligand gene by a glucocorticoid receptor dimer. Blood 2005;106:617–625 [DOI] [PubMed] [Google Scholar]
- 45.Novac N, Baus D, Dostert A, Heinzel T. Competition between glucocorticoid receptor and NFkappaB for control of the human FasL promoter. FASEB J 2006;20:1074–1081 [DOI] [PubMed] [Google Scholar]
- 46.Kagoshima M, Wilcke T, Ito K, Tsaprouni L, Barnes PJ, Punchard N, Adcock I. Glucocorticoid-mediated transrepression is regulated by histone acetylation and DNA methylation. Eur J Pharm 2001;429:327–334 [DOI] [PubMed] [Google Scholar]
- 47.Vanhaecke T, Papeleu P, Elaut G, Rogiers V. Trichostatin A–like hydroxamate histone deacetylase inhibitors as therapeutic agents: toxicological point of view. Curr Med Chem 2004;11:1629–1643 [DOI] [PubMed] [Google Scholar]
- 48.Belandia B, Parker MG. Nuclear receptors: a rendezvous for chromatin remodeling factors. Cell 2003;114:277–280 [DOI] [PubMed] [Google Scholar]
- 49.Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet 2000;16:351–356 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.