Abstract
Background
Tribe Fabeae comprises about 380 legume species, including some of the most ancient and important crops like lentil, pea, and broad bean. Breeding efforts in legume crops rely on a detailed knowledge of closest wild relatives and geographic origin. Relationships within the tribe, however, are incompletely known and previous molecular results conflicted with the traditional morphology-based classification. Here we analyse the systematics, biogeography, and character evolution in the tribe based on plastid and nuclear DNA sequences.
Results
Phylogenetic analyses including c. 70% of the species in the tribe show that the genera Vicia and Lathyrus in their current circumscription are not monophyletic: Pisum and Vavilovia are nested in Lathyrus, the genus Lens is nested in Vicia. A small, well-supported clade including Vicia hirsuta, V. sylvatica, and some Mediterranean endemics, is the sister group to all remaining species in the tribe. Fabeae originated in the East Mediterranean region in the Miocene (23–16 million years ago (Ma)) and spread at least 39 times into Eurasia, seven times to the Americas, twice to tropical Africa and four times to Macaronesia. Broad bean (V. faba) and its sister V. paucijuga originated in Asia and might be sister to V. oroboides. Lentil (Lens culinaris ssp. culinaris) is of Mediterranean origin and together with eight very close relatives forms a clade that is nested in the core Vicia, where it evolved c. 14 Ma. The Pisum clade is nested in Lathyrus in a grade with the Mediterranean L. gloeosperma, L. neurolobus, and L. nissolia. The extinct Azorean endemic V. dennesiana belongs in section Cracca and is nested among Mediterranean species. According to our ancestral character state reconstruction results, ancestors of Fabeae had a basic chromosome number of 2n=14, an annual life form, and evenly hairy, dorsiventrally compressed styles.
Conclusions
Fabeae evolved in the Eastern Mediterranean in the middle Miocene and spread from there across Eurasia, into Tropical Africa, and at least seven times to the Americas. The middle-Atlantic islands were colonized four times but apparently did not serve as stepping-stones for Atlantic crossings. Long-distance dispersal events are relatively common in Fabeae (seven per ten million years). Current generic and infrageneric circumscriptions in Fabeae do not reflect monophyletic groups and should be revised. Suggestions for generic level delimitation are offered.
Keywords: Lathyrus, Legumes, Lentil, Long-distance dispersal, Macaronesia, Pea, Pisum, Vicia
Background
Legumes (Fabaceae/Leguminosae) are grown on 12-15% of the world’s arable surface and account for c. 27% of the world’s primary crop production [1]. The tribe Fabeae Rchb. (not Vicieae (Bronn) DC., nom. illeg.) in particular, a group of five genera and c. 380 species ([2,3], our Additional file 1: Annex 1, Figure 1 for some examples) contains several important crop species. These include the pea (Pisum sativum L., Figure 1F), lentil (Lens culinaris Medik.), broad bean (Vicia faba L.), and bitter vetch (Vicia ervilia Willd.) that, together with wheat, barley and flax are considered the founder crops of Neolithic agriculture in the Fertile Crescent of Western Asia [4-6]. In addition to crops of worldwide importance, the tribe also contains minor crop species (e.g., Lathyrus sativus L., L. sphaericus Retz.), forage or green manure species (e.g., Vicia sativa L., V. villosa L.) and popular ornamentals (e.g. L. odoratus L.).
Figure 1.
Examples of morphological diversity in Fabeae.A) Vicia cirrhosa, Gran Canaria, Canary Islands, Spain; B) Vicia bithynica, Pico, Azores, Portugal; C) Vicia lutea, Gran Canaria, Canary Islands, Spain; D) Lathyrus angulatus, Gran Canaria, Canary Islands, Spain; E) Lathyrus tingitanus, Santa Maria, Azores, Portugal; F) Pisum sativum ssp. sativum, Washington D.C., USA (cultivated); G) Vicia tenuissima, Flores, Azores, Portugal; H) Vicia hirsuta, Cambridge, USA (invasive); I) Vicia sylvatica, Tegernsee, Bavaria, Germany (scale bar: 5 mm; all pictures © H. Schaefer).
Fabeae have an almost worldwide distribution: of the five genera in the tribe, Vicia L. (c. 216 species) and Lathyrus L. (c. 150 species) are both most diverse in the Eastern Mediterranean but extend throughout Europe, and both have species in Asia, Northern to Tropical Africa, and North and South America [7,8]. Vicia has also colonized Hawaii in the Pacific [9] and the middle-Atlantic archipelagos of the Canaries, Madeira and Azores. The remaining three genera are small and more localized: Lens Medik., with 4–8 species, and Pisum L. with perhaps two species and several subspecies are both most diverse in the Eastern Mediterranean region; the genus Vavilovia Al.Fed. with one or two species is native to the West Asian mountains [2,10]. Fabeae are absent from most of the lowland Tropics and have been introduced to Australia and Polynesia only very recently with European settlers.
While our knowledge of the systematics and biogeography of the Fabaceae is increasing steadily, there are still remarkable gaps. For the tribe Fabeae, the last detailed and comprehensive morphology-based systematic revision was produced by Alefeld [11], who divided the group in two tribes ‘Viciidae’ and ‘Orobidae’ with, collectively, 25 genera. The currently used infrageneric classifications in Vicia and Lathyrus largely follow the systems suggested by Kupicha [7,8]. For Lathyrus, Kupicha accepted 13 sections [8], which have subsequently been modified by Asmussen & Liston [12] and more recently by Kenicer et al. [13], who accepted eleven sections. In Vicia, Kupicha [7] introduced two subgenera, Vicia and Vicilla, and 22 sections with an additional four sections later introduced by Maxted [14]. One species was placed in a separate genus Anatropostylia[15] but has also been treated as a monotypic section Anatropostylia of Vicia[16].
Recent molecular phylogenetic studies have focussed on sections or genera. Thus, studies on Vicia included up to 55 species [17-23]; phylogenetic studies on the genus Lathyrus included up to 53 species [12,13,24-26]; several studies analysed the genera Lens[27-29] and Pisum[30-33]; finally, Oskoueiyan et al. [10] analysed the phylogenetic position of Vavilovia. Full chloroplast genomes have recently become available for Lathyrus sativus and Pisum sativum[34]. In contrast to this growing body of analyses at the generic and sectional level, there are few studies dealing with the entire tribe. The only exceptions are family-wide or multi-tribe legume phylogenetic studies, which included a limited sample of species across Fabeae (e.g., [35-39]). Those studies suggested that Fabeae is monophyletic and sister group to the clover genus Trifolium L. Even though sampling of Fabeae in those family-wide studies has been limited, they suggest that plastid DNA sequence data do not agree with morphology-based genus circumscriptions in the tribe and that Vicia might be paraphyletic [36]. Altogether, these studies highlight the need of a comprehensive tribal-level study of Fabeae.
Several hypotheses have been proposed to explain the worldwide distribution of Fabeae but there has been no comprehensive biogeographic analysis of the entire tribe. Simola [40] suggested a South American origin of Lathyrus followed by a dispersal event to Africa and then into the Mediterranean. In contrast, Kupicha [8] suggested that Lathyrus and Vicia originated at high latitudes in the Old World, and might have migrated later to the Mediterranean and to North America via Greenland or from Asia via Beringia to Alaska. From North America, the lineages could have spread into South America in the late Tertiary. A North American origin of the South American Notolathyrus group was also suggested by Burkart [41] and Asmussen & Liston [12]. Based on DNA sequence data, Kenicer et al. [13] suggested an eastern Mediterranean origin for Lathyrus followed by range expansion into northern Eurasia. The Beringian land bridge would then have allowed migration into North America. According to Kenicer et al. [13], the most likely origin of the South American Lathyrus species is directly from Eurasia via long-distance dispersal (sea currents). In this context the question arises whether those lineages used stepping-stone islands in the middle-Atlantic as suggested by Axelrod [42] and more recently by Fernández-Palacios et al. [43].
The stepping-stone hypothesis is supported by the existence of forty-five species of Fabeae on the volcanic oceanic Atlantic islands of which nine are currently considered as endemic to one or more of the islands. On Iceland in the extreme North, five species are possibly native (Lathyrus japonicus Willd., L. palustris L., L. pratensis L., Vicia cracca L. and V. sepium L.) and four occur as casuals (V. angustifolia L., V. hirsuta (L.) Gray, V. sativa, V. villosa) [44]. Further to the South, in the Azores archipelago, a total of 14 Vicia, 10 Lathyrus, and Lens culinaris have been reported [45,46]. All are considered introduced [45], with the exception of Vicia dennesiana H. C. Watson (Figure 2), which is endemic to the Azorean island of São Miguel but has not been seen since the mid 19th century [45,47,48]. For Madeira, 15 Vicia, 11 Lathyrus, and Lens culinaris have been reported [49]. Of those, three species are endemic to Madeira (V. capreolata Lowe, V. costae A. Hansen and V. ferreirensis Goyder from Porto Santo), 15 others considered native, and nine are probably introduced [49]. In the Canary Islands, 22 Vicia, 12 Lathyrus, two Lens, and one Pisum species have been found [50]. Five species are currently considered to be endemic to the Canaries: Vicia chaetocalyx Webb & Berthel., V. cirrhosa C. Sm. ex Webb & Berthel. (Figure 1A), V. filicaulis Webb & Berthel., V. nataliae U. Reifenb. & A.Reifenb., and V. scandens R.P.Murray. Of the remainder 13 are considered at least possibly native, and 19 are very likely recent introductions [50]. No native Fabeae have been reported from the Selvagens or, further south, from Cape Verdes, St. Helena, or Tristan da Cunha.
Figure 2.
Vicia dennesiana H. C. Watson. The extinct Azorean endemic Vicia dennesiana, here shown to be nested in the Mediterranean section Cracca. This drawing originally published in Curtis’s Botanical Magazine in 1887, ser. 3, vol. 43, was made of a plant cultivated at Royal Botanical Gardens Kew from seeds of plants cultivated in the private garden of H. C. Watson, who obtained his seeds from T. C. Hunt, then British consul on São Miguel Island, Azores [47]. The Kew plants were subsequently lost in a late May frost [48] and the species was not rediscovered ever since.
The aims of this study were to examine the evolution, biogeography and classification of Fabeae. Newly generated plastid and nuclear DNA sequence data (rbcL, matK, trnL-trnF, trnS-trnG, psbA-trnH, and ITS (internal transcribed spacer) region) for Fabeae species from the middle-Atlantic islands, North and South America, Africa, and Eurasia are analysed in conjunction with previously published data to establish a phylogeny of the tribe that includes c. 70% of the accepted species (often more than one accession per taxon), with representatives sampled from all sections and all geographic regions in which the tribe occurs. This phylogenetic framework is used to (i) test current morphology-based generic and infrageneric circumscriptions and (ii) analyse the evolution of characters used for generic and infrageneric delimitation, notably stylar and life form characters [7,8]. Using a Bayesian relaxed molecular clock approach and ancestral range reconstruction, we further test the hypothesis of a Eurasian origin followed by dispersal to the Americas (a) across the Atlantic or (b) via land bridges or ‘stepping stone’ islands.
Results
Phylogenetic analyses
Maximum likelihood (ML) and Bayesian analyses of chloroplast (rbcL, matK, trnL/trnL-trnF, trnS-trnG, psbA-trnH) and nuclear ribosomal ITS sequence data for 262 of the c. 380 species currently accepted in the tribe (Additional file 2: Table S1, Additional file 1: Annex 1) yield a generally well-resolved and statistically supported phylogeny although some nodes of interest are only weakly or not supported (see Figure 3 for ML phylogeny of combined data, and Additional file 3: Figure S1-S7 Additional file 4: Figure S2, Additional file 5: Figure S3, Additional file 6: Figure S4, Additional file 7: Figure S5, Additional file 8: Figure S6, Additional file 9: Figure S7 for phylogenies from individual markers; Additional file 10: Figure S8 for Bayesian phylogeny from combined data). The nuclear ribosomal ITS (Additional file 3: Figure S1) and combined plastid (S7) phylogenies are congruent (based on comparison of clades with BS ≥ 65%). We thus find no evidence for a strong effect of hybridization in the evolution of the Fabeae and analyses of a combined matrix is justified.
Figure 3.
Maximum likelihood phylogeny of the Fabeae. Best maximum likelihood tree based on the combined chloroplast and ITS dataset for 470 ingroup accessions (262 species) plus seven outgroups (6200 aligned nucleotides plus 83 gap characters). Likelihood bootstrap values ≥ 50% are given at the nodes. The five currently accepted genera in the tribe plus the proposed Ervum and Ervilia and the sections [7,8,14] are indicated.
Our results confirm the monophyly of Lens and Pisum (99 and 100% BS respectively, Figure 3; 1.0 PP, Figure S8) and the paraphyly of Vicia. In contrast to previous studies, we find that Lathyrus is not monophyletic and includes Pisum and Vavilovia (Figure 3; Additional file 10: Figure S8). Currently recognized subgenera in Vicia and sections with more than one species in both Vicia and Lathyrus are mostly not recovered as monophyletic groups (see Table 1). Of the 13 sections in Lathyrus, sections Neurolobus, Nissolia, Orobon, Orobastrum, and Viciopsis are monotypic. Our sampling strategy allowed us to test the monophyly of seven of the eight sections of Lathyrus that comprise multiple species. Only Notolathyrus and Pratensis are found to be monophyletic (100 and 88% BS respectively, Figure 3; 1.0 PP, Additional file 10: Figure S8). We had material of only one of the two species in section Aphaca but since the two species are distinguishable only through slight differences in style shape and fruit morphology, section Aphaca is very likely also monophyletic. Sections Orobon and Orobastrum are both nested in section Lathyrus. Section Linearicarpus is polyphyletic: L. hygrophilus Taub. and L. inconspicuus L. group together (76% BS, Figure 3; 1.0 PP, Additional file 10: Figure S8), whereas L. angulatus L. (Figure 1D) belongs to section Lathyrus (70% BS, Figure 3; 1.0 PP, Additional file 10: Figure S8) and L. sphaericus groups with section Lathyrostylis (99% BS, Figure 3; 1.0 PP, Additional file 10: Figure S8). Lathyrus gloeosperma Warb. & Eig from Kupicha’s section Clymenum groups with L. nissolia L. in the ML reconstruction (but <50% BS, Figure 3). Lathyrus saxatilis (Vent.) Vis. (representing Kupicha’s monotypic section Viciopsis) is placed in the core Vicia and may be sister to a clade that includes V. dumetorum L. and V. americana Muhl. ex Willd. although its closest relatives in Vicia are unclear due to low support values (Figure 3; Additional file 10: Figure S8).
Table 1.
Overview of morphology-based sections in Fabeae compared to molecular results.
| Genus | Subgenus | Section | Geographic range | No. of accepted species | No. of sequenced species | Monophyletic? |
|---|---|---|---|---|---|---|
|
Lathyrus |
|
Aphaca |
Eurasia, Mediterranean region |
2 |
1 |
? |
| |
|
Clymenum |
Mediterranean region |
4 |
3 |
No |
| |
|
Lathyrus |
Eurasia |
31 |
15 |
No |
| |
|
Lathyrostylis |
Eurasia |
16 |
6 |
No |
| |
|
Linearicarpus |
Eurasia, Mediterranean region |
7 |
3 |
No |
| |
|
Neurolobus |
Crete |
1 |
1 |
NA |
| |
|
Nissolia |
Eurasia, Mediterranean region |
1 |
1 |
NA |
| |
|
Notolathyrus |
S America, Southern N America |
22 |
14 |
Yes |
| |
|
Orobastrum |
Eurasia, Mediterranean region |
1 |
1 |
NA |
| |
|
Orobon |
Caucasus Mts. |
1 |
1 |
NA |
| |
|
Orobus |
Eurasia, Mediterranean region, N & C America |
44 |
36 |
No |
| |
|
Pratensis |
Eurasia, Mediterranean region |
6 |
2 |
Yes |
| |
|
Viciopsis |
S Europe, Turkey, NW Africa |
1 |
1 |
NA |
|
Lens |
|
|
Mediterranean region, Asia Minor |
5 |
5 |
Yes |
|
Pisum |
|
|
Mediterranean region, Asia Minor |
2 |
2 |
Yes |
|
Vavilovia |
|
|
Caucasus Mts. |
2 |
2 |
Yes |
|
Vicia |
Vicia |
Atossa |
Eurasia, Mediterranean region |
4 |
2 |
No |
| |
|
Faba |
Eurasia, Mediterranean region |
2 |
2 |
Yes |
| |
|
Bithynicae |
Mediterranean region |
1 |
1 |
NA |
| |
|
Wiggersia |
Eurasia, Mediterranean region |
2 |
2 |
No |
| |
|
Microcarinae |
Mediterranean region |
1 |
1 |
NA |
| |
|
Narbonensis |
Mediterranean region, Asia Minor |
7 |
7 |
Yes |
| |
|
Hypechusa |
Eurasia, Mediterranean region |
13 |
11 |
No |
| |
|
Peregrinae |
Mediterranean region, Asia Minor |
3 |
3 |
Yes |
| |
|
Vicia |
Eurasia, Mediterranean region |
5 |
5 |
No |
| |
Vicilla |
Americanae |
East Asia, N America |
2 |
2 |
Yes |
| |
|
Anatropostylia |
Asia |
1 |
0 |
NA |
| |
|
Australes |
Mexico, C & S America |
17 |
6 |
No |
| |
|
Cassubicae |
Eurasia, Mediterranean region, Pacific coast of N & S America |
7 |
7 |
No |
| |
|
Cracca |
Eurasia, Mediterranean region, N America |
45 |
29 |
No |
| |
|
Ervilia |
Mediterranean region, Asia Minor |
2 |
1 |
? |
| |
|
Ervoides |
Mediterranean region, Asia Minor |
1 |
1 |
NA |
| |
|
Ervum |
Eurasia, Mediterranean region |
3 |
3 |
Yes |
| |
|
Lentopsis |
Turkey |
1 |
1 |
NA |
| |
|
Mediocinctae |
Southern USA |
1 |
1 |
NA |
| |
|
Panduratae |
Mediterranean region, Asia Minor |
3 |
2 |
No |
| |
|
Pedunculatae |
S Europe, Asia Minor, NW Africa |
3 |
2 |
No |
| |
|
Perditae |
Azores |
1 |
1 |
NA |
| |
|
Subvillosae |
Asia |
1 |
1 |
NA |
| |
|
Trigonellopsis |
Mediterranean region, Asia Minor |
3 |
2 |
Yes |
| |
|
Variegatae |
S Europe, Caucasus Mts., Asia |
3 |
1 |
? |
| |
|
Vicilla |
Eurasia, Mediterranen region |
15 |
13 |
No |
| Volutae | Southeastern Europe, Caucasus Mts. | 1 | 1 | NA |
Within Vicia s.str., most of the species of Kupicha’s subgenus Vicia group together, with the notable exception of Vicia narbonensis L. and close relatives (section Narbonensis sensu Maxted) from the Eastern Mediterranean and Asia Minor. The second subgenus, Vicilla, is clearly polyphyletic (see below; Figure 3). Nine of the 27 sections currently recognized in Vicia are monotypic: sections Anatropostylia, Bithynicae, Ervoides, Lentopsis, Mediocinctae, Microcarinae, Perditae, Subvillosae, and Volutae (Table 1). Our sampling strategy allowed us to test the monophyly of 16 of the 18 remaining sections. Of these, sections Americanae, Ervum, Faba, Narbonensis, Peregrinae, and Trigonellopsis are found to be monophyletic. Section Cracca is not monophyletic because (i) sections Lentopsis, Panduratae, Perditae, Variegatae, and Volutae are nested in a clade otherwise comprising species from this section and (ii) Vicia chaetocalyx and V. hirsuta that were previously assigned to this section are placed outside the Cracca clade (Figure 3). With regards to section Vicilla, V. americana, V. bungei Ohwi, V. crocea (Desf.) B. Fedtsch., V. dumetorum, V. gigantea, and V. nigricans are not resolved with other members of the section. These species are resolved as paraphyletic with respect to sections Cassubicae and Amurense. Section Australes is shown to be paraphyletic with respect to section Mediocinctae and section Pedunculatae includes section Americanae and V. dumetorum (Figure 3). Section Vicia is also not monophyletic because sections Atossa, Hypechusa, Peregrinae, and Faba are nested within it. Vicia section Ervum is sister to all Lathyrus species (except L. saxatilis) plus Pisum and Vavilovia (98% BS, Figure 3; 1.0 PP, Additional file 10: Figure S8). Vicia sections Ervilia, Ervoides, and Trigonellopsis group together (98% BS, Figure 3; 0.99 PP, Additional file 10: Figure S8) and form a clade that is sister to all other Fabeae (95% BS, Figure 3; 1.0 PP, Additional file 10: Figure S8).
At the species level, our material of the Macaronesian endemic Vicia chaetocalyx is nested in a highly supported clade of V. lutea L. accessions (99% BS, Figure 3). There is no resolution in the V. cirrhosa group from the Canaries and Madeira in any of the analyses (Figure 3, Additional file 3: Figure S1-S7, Additional file 4: Figure S2, Additional file 5: Figure S3, Additional file 6: Figure S4, Additional file 7: Figure S5, Additional file 8: Figure S6, Additional file 9: Figure S7). In contrast, the V. sativa group is genetically quite heterogeneous, especially in nuclear ITS sequences (Additional file 3: Figure S1) and while V. sativa s.l. is monophyletic, the different accessions of V. angustifolia and V. sativa s.str. do not group together. In the V. tetrasperma lineage, V. tenuissima Schinz & Thell. and V. pubescens (DC.) Link are genetically distinct from V. tetrasperma (L.) Schreb. s. str. both in ITS (Additional file 3: Figure S1) and combined plastid sequences (Additional file 9: Figure S7). Nuclear ITS sequence differences also support the separation of North and South American populations of V. nigricans s. l. as V. gigantea Hook. and V. nigricans Hook. & Arn. s. str. respectively (Additional file 3: Figure S1). Heterogeneity at the genetic level is also evident for the Lathyrus aphaca L. clade, a morphologically variable taxon widespread from the Macaronesian islands to the Himalayas (Figure 3; Additional file 3: Figure S1; Additional file 9: Figure S7). In contrast, Lathyrus undulatus Boiss. and L. miniatus Steven do not appear to be genetically distinct from L. rotundifolius Willd. s. str. and the Romanian endemic L. hallersteinii Baumg. is nested in a highly supported clade of L. pratensis accessions (99% BS, Figure 3).
Molecular dating and ancestral range reconstruction
The results of our Bayesian molecular clock and ancestral range analyses suggest a Mediterranean origin and a crown age of c. 23–16 Myr for Fabeae, with a best estimate for the associated substitution rate of 0.0045 substitutions per site per Myr (subst/site/My) (Figure 4; Additional file 11: Figure S9). We reconstruct a minimum of three dispersal events to the middle-Atlantic islands (four if V. chaetocalyx is accepted) and seven to the Americas. None of the New World lineages originates from the Atlantic islands (Figures 4, 5). The oldest Macaronesian lineage, the Vicia scandens clade, split from a Mediterranean ancestor 4.9-2.4 Ma, 0.0019 subst/site/My (Additional file 11: Figure S9) and colonized the Canary Islands and Madeira plus Porto Santo (crown age 0.9-0.15 Ma, 0.0026 subst/site/My). The second dispersal led to the establishment of the Vicia dennesiana lineage in the Azores. The age estimate for this event is not very robust due to the low resolution in the V. cracca clade, where V. dennesiana is placed but it occurred probably more recently than the Canary island lineage, between 2.9-1.1 Ma, 0.0028 subst/site/My (Figure 4, Additional file 11: Figure S9). The remaining colonization events are even more recent: the Madeiran species or form Vicia pectinata Lowe split from the Mediterranean Vicia sativa 0.8-0.1 Ma (0.002 subst/site/My), whereas the Canary island Vicia chaetocalyx is nested in a clade of the mainly Mediterranean Vicia lutea (Figure 1C) and might have arrived only with the first human settlers.
Figure 4.
Biogeographic history of the Fabeae. Reconstruction of the biogeographical history of the tribe Fabeae using BEAST and the approach of Lemey et al. [79]. Posterior probability values for the range reconstructions are given at the nodes. Asterisks indicate LDD events. Geographical regions are colour-coded: Mediterranean - purple, Central and Western Europe - orange, Asia - red, Hawaii - green, Macaronesia - turquoise, Africa - dark blue, North America - light blue, South America - yellow. The vertical light green bar marks the first opening of the Bering Strait c. 5.4-5.5 Ma [57].
Figure 5.

Overview map of the biogeographic history of Fabeae. World map showing the reconstructed biogeographical scenario for the Fabeae: origin 23–16 Ma in the Mediterranean, then at least 24 times range expansion into Asia and 15 times into Central and Western Europe. Furthermore, at least two range expansions or dispersal events into tropical Africa. South America was colonized twice via LDD from the Mediterranean and once via range expansion from North America. North America was colonized once via LDD from Europe/Mediterranean, three times from Asia and perhaps three times through range expansion of South American lineages. Macaronesia was reached four times by LDD from the Mediterranean with one colonisation of the Azores and three of the Canaries/Madeira.
We reconstruct seven migration events to the New World: (1) the Vicia nigricans clade split from the Eurasian Vicia lineages 13.7-8.5 Ma (0.0014 subst/site/My). This split is reconstructed as a long distance dispersal (LDD) event from the Mediterranean region to South America with high support (0.97 PP, Figure 4) but the Vicia nigricans clade might also be sister to the Caucasian V. crocea (Additional file 11: Figure S9), which would indicate dispersal from Asia via Beringia. The ancestor of the Hawaiian endemic Vicia menziesii Spreng. then dispersed from Western America to Hawaii 2.5-0.1 Ma, 0.0014 subst/site/My (Additional file 11: Figure S9); (2) the North American Vicia ludoviciana Nutt. clade split from a Mediterranean ancestral lineage 8.9-6.5 Ma and dispersed across the Atlantic, possibly floating in surface water (Figure 4; Additional file 11: Figure S9); (3) the mainly South American Vicia section Australes (incl. V. leucophaea Greene) split from a Mediterranean ancestral lineage 8.5-4.8 Ma (0.0016 subst/site/My) and is again most likely a case of LDD across the Atlantic (Figure 4; Additional file 11: Figure S9); (4) the South American section Notolathyrus split from a Mediterranean ancestral lineage 8.6-6.1 Ma (0.0018 subst/site/My), representing the third LDD event in the same time window of 9–5 Ma (Figure 4; Additional file 11: Figure S9); (5) the large clade of North American species in Lathyrus section Orobus split from an Asian ancestral lineage 2.9-1.8 Ma, 0.0017 subst/site/My (Figure 4; Additional file 11: Figure S9); (6) colonization of the North American coasts by the Lathyrus littoralis/L. japonicus lineage started c. 3–1.7 Ma, 0.0017 subst/site/My (Figure 4; Additional file 11: Figure S9); (7) and finally the split between the North American Vicia americana and its Asian sister V. bungei is dated to 2.3-0.2 Ma, 0.002 subst/site/My (Additional file 11: Figure S9).
Origin of the crop species in Fabeae
The pea, Pisum sativum s. l. (including P. elatius M.Bieb. and P. humile Mill.) is sister to the Eastern Mediterranean Pisum fulvum Sibth. & Sm. (100% BS, Figure 3; 1.0 PP, Additional file 10: Figure S8) and both are sister to Vavilovia formosa (Stev.) Fed. (≡ Pisum formosum (Stev.) Alef.) and V. aucheri (Jaub. & Spach) Fed. (≡ Pisum aucheri Jaub. & Spach) from the Caucasus Mountains (98% BS, Figure 3; 1.0 PP, Additional file 10: Figure S8). The crown age of the Pisum clade is estimated to 2.3-0.8 Ma (Additional file 11: Figure S9); the divergence between Pisum and Vavilovia dates back to 9.8-4.8 Ma (Additional file 11: Figure S9). Lentil, Lens culinaris ssp. culinaris, is nested among a group of very closely related taxa. Their mostly allopatric distribution, morphological similarity and very recent divergence estimates suggest that they might be best treated as subspecies or varieties of Lens culinaris. All together are sister to the Mediterranean Lens nigricans (M.Bieb.) Godr. (99% BS, Figure 3; 1.0 PP, Additional file 10: Figure S8). The split between the L. culinaris group and L. nigricans occurred 4.9-1.9 Ma, whereas the stem lineage of Lens might have split from its Vicia ancestors 14.9-12.6 Ma. Faba bean or broad bean, Vicia faba is sister to the Himalayan V. paucijuga B. Fedtsch., which is often treated as a subspecies of V. faba. Both are nested in section Vicia (incl. sect. Atossa, Bithynicae, Hypechusa, Microcarinae, Peregrina, and Wiggersia) with an estimated stem age of 8–3.8 Ma but this is not robust due to the lack of a well supported placement of the lineage in any of our phylogeny estimates (Figure 3; Additional file 10: Figure S8, Additional file 12: Figure S10). The ancient crop species Vicia ervilia is placed in the Ervilia clade that is here recovered as the sister group to all other Fabeae. It split from its closest relatives in the V. sylvatica L. clade 11.2-5.0 Ma (Additional file 11: Figure S9). Vicia sativa belongs in a group of closely related and insufficiently studied Mediterranean taxa. In our ML and Bayesian analyses, the V. sativa group (including V. pectinata, V. angustifolia, V. incisa M.Bieb., and V. amphicarpa Dorthes) is consistently reconstructed as sister to V. pyrenaica Pourr. (but <50% BS, Figure 3; <0.8 PP, Figure S8) from which it split 5.6-2.2 Ma. The economically most important species in the genus Lathyrus, the ornamental sweet pea, L. odoratus, from southern Italy and Sicily, is sister to the wider Mediterranean L. hirsutus L. (94% BS, Figure 3; 1.0 PP, Figure S8), from which it diverged 2.5-0.4 Ma (Figure S9). Finally, the minor crop L. sphaericus is sister to the Mediterranean L. bauhinii Genty clade (99% BS, Figure 3; 1.0 PP, Additional file 10: Figure S8), from which it diverged 7.6-4.5 Ma (Additional file 11: Figure S9).
Character evolution
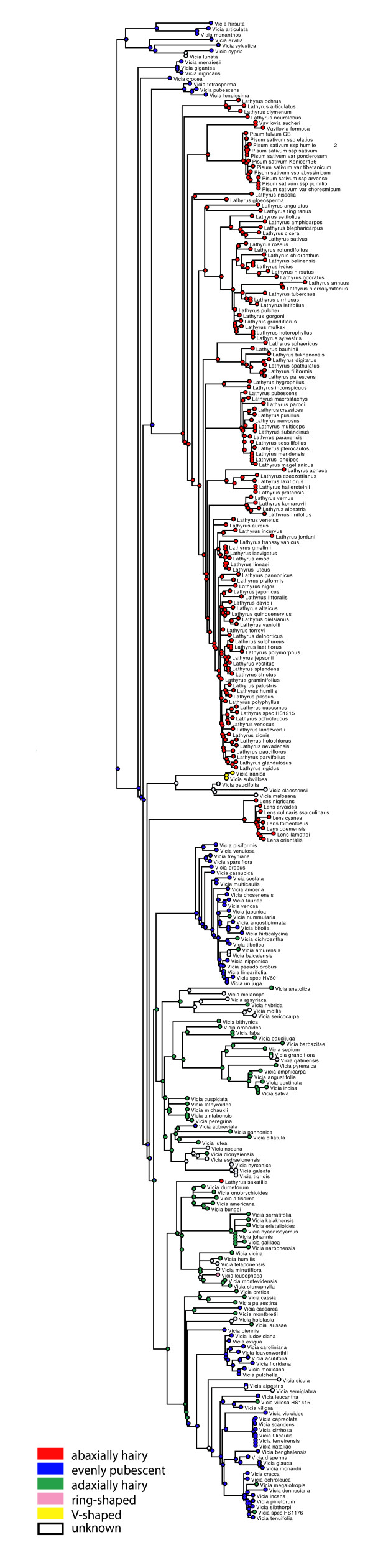
Annual life form is reconstructed as ancestral in the tribe and a perennial life form evolved at least 20 times independently followed by several reversals to annual life form (Figure 6). The ML ancestral character reconstruction indicates that a basic chromosome number of 2n=14 is most likely for the origin of the tribe (Figure 7). From that, reductions to 2n=12 or 2=10 and duplications to 2n=24, 2n=28, and 2=42 have occurred. Major clades in Fabeae seem to be characterized by common stylar pubescence patterns (Figure 8). From an originally evenly hairy style (still prominent in the Ervum, Ervilia, and early-branching Vicia clades), the Lathyrus-Pisum-Vavilovia clade evolved an adaxially hairy style, whereas some of the more derived Vicia clades are dominated by an abaxially hairy style. There are, however, many exceptions: Lathyrus saxatilis, for example, with an adaxially hairy style is deeply nested in Vicia. The Asian Vicia subvillosa (Ledeb.) Boiss. has a unique type of style with V-shaped hairy areas. It is unknown whether this character is shared with its African sister species V. malosana (Baker) Baker f., V. paucifolia Baker, and V. claessensii De Wild. Another unique pattern is found in V. leucophaea, which has a densely hairy ring on its style that seems to have evolved from an abaxially hairy style (Figure 8). Style shape, another traditional character for Fabeae classification, varies little within the tribe: most species have a dorsiventrally compressed style, which is also reconstructed as most likely ancestral condition in the tribe (Figure 9). Exceptions are most of the Vicia species in section Cracca, which have a laterally compressed style. A terete style (circular in cross section) has evolved six to seven times independently in the tribe: in the Mediterranean V. pubescens, the Eurasian V. hirsuta and Vicia crocea, the North American V. leucophaea, and the East Asian V. amurensis Oett., V. tibetica Prain ex C.E.E. Fisch., V. nummularia Hand.-Mazz., and V. dichroantha Diels. Furthermore, Pisum and Vavilovia are characterised by a longitudinally folded style otherwise unknown in the tribe.
Figure 6.
Life form evolution in Fabeae. Evolution of life form in Fabeae reconstructed under the Markov k-state one-parameter model on the best maximum likelihood tree using Mesquite ver. 2.75 [82]. Colour code: blue - perennial, red - annual (rarely biannual Vicia biennis).
Figure 7.
Evolution of chromosome numbers in Fabeae. Evolution of chromosome number in Fabeae reconstructed under the Markov k-state one-parameter model on the best maximum likelihood tree using Mesquite ver. 2.75 [82]. Colour code: yellow - 2n=10, blue - 2n=12, red - 2n=14, purple 2n=24, green - 2n=28, orange 2n=42.
Figure 8.
Evolution of stylar pubescence in Fabeae. Evolution of stylar pubescence in Fabeae reconstructed under the Markov k-state one-parameter model on the best maximum likelihood tree using Mesquite ver. 2.75 [82]. Colour code: red - abaxially hairy, blue - evenly hairy, green - adaxially hairy, pink - densely hairy ring, yellow - V-shaped hairy zone.
Figure 9.
Evolution of stylar shape in Fabeae. Evolution of stylar shape in Fabeae reconstructed under the Markov k-state one-parameter model on the best maximum likelihood tree using Mesquite ver. 2.75 [82]. Colour code: red - laterally compressed, yellow - terete, green - folded longitudinally, blue - dorsiventrally compressed.
Discussion
Taxonomic implications
The molecular data show that most of the currently recognized genera and subgenera, as well as many of the traditional sections are not monophyletic. Pisum and Vavilovia are sister groups and nested in Lathyrus, which together with Lens is nested in Vicia. This grouping is also supported from the (few) currently available whole plastid genomes for Fabeae: Lathyrus sativus and Pisum sativum share four gene losses (infA, rps16, rpl22, and rpl23) and the loss of the first intron of clpP and the cis-intron of rps12 [34]. Pisum and Lathyrus furthermore share the phytoalexin pisatin, which was not found in Vicia and Lens[51]. The two latter genera both had the phytoalexin wyerone in all analysed species except in Vicia articulata and V. ervilia[52], which we find to be sister to all Vicieae. Thus the presence of wyerone could be a synapomorphy for Vicia and Lens.
Apart from Alefeld’s concept of splitting the tribe into a multitude of tiny genera [11], the most radical solution to obtain a more natural classification of Fabeae would be the transfer of all species of the four currently accepted smaller genera to a broadly circumscribed genus Vicia. This, however, would require more than 100 new combinations. An alternative (and perhaps preferable) solution would be to transfer Pisum and Vavilovia to a then monophyletic Lathyrus. Vicia section Ervum (the clade comprising V. tetrasperma, V. tenuissima (Figure 1G), and V. pubescens) could be raised to genus-level; the name Ervum L. with the lectotype E. tetraspermum L. is available. A monophyletic Vicia could then be obtained by transferring Lens and Lathyrus saxatilis to Vicia. The clade comprising Vicia cypria Kotschy, V. lunata (Boiss. & Bal.) Boiss., V. ervilia, V. hirsuta (Figure 1H), V. sylvatica (Figure 1I), V. monanthos (L.) Desf. and V. articulata Hornem. (mainly Kupicha’s sections Ervilia, Ervoides, and Trigonellopsis [7] could be split from Vicia s. str. and raised to genus-level. Among the available names, Ervilia Link seems the most appropriate. Of the species not sequenced, V. koeieana Rech.f. (= Anatropostylia koeieana (Rech.f.) Kupicha) and V. quadrijuga P.H.Davis very likely belong here too, so that a genus Ervilia would probably comprise at least nine species.
Most of the sections also need to be revised if they are supposed to reflect monophyletic groupings: section Cracca should include the former sections Lentopsis, Panduratae, Perditae, Variegatae, and Volutae. Section Vicilla should be recircumscribed to include sections Amurense and Cassubicae. Section Mediocinctae could be lumped with section Australes. Section Pedunculatae should be merged with section Americanae. Section Vicia could be defined in a broader sense to accommodate sections Atossa, Bithynicae, Faba (sensu Maxted [14]), Hypechusa, Microcarinae, Peregrinae, and Wiggersia. In Lathyrus, the clade including L. alpestris (Waldst. & Kit.) Kit. ex Reichb. and L. vernus (L.) Bernh. could be split from section Orobus. Sections Orobon and Orobastrum could be lumped with section Lathyrus. The circumscriptions of sections Linearicarpus, Lathyrus, and Lathyrostylis all need to be slightly modified in order to obtain monophyletic groups. Newly discovered Fabeae clades that might warrant description of additional sections include (i) the sister group of section Subvillosae, comprising the African V. claessensii, V. malosana, and V. paucifolia (delimitation of the rarely collected species in this clade needs further studies); (ii) the Pacific coast clade including V. nigricans, V. gigantea, and V. menziesii; (iii) Vicia crocea, which might best be placed in a monotypic section even though additional work might reveal a sister group relationship to the V. nigricans clade.
The results of our phylogenetic analyses show that Lathyrus saxatilis is currently included in the wrong genus: the two accessions we have are both placed in the core Vicia in our phylogeny and even though our data are not sufficient to pinpoint its exact position (grouping with V. dumetorum and V. americana in the Bayesian but not in the ML phylogeny) it should certainly be treated as Vicia saxatilis (Vent.) Tropea. The taxon was described as Orobus saxatilis by Ventenat but interestingly, it was already transferred to Vicia in the early 20th century (see Kupicha [8] for details). Kupicha discusses the morphological affinity to Vicia (especially Vicia sections Vicia and Hypechusa) but although expressing some doubts, the dorsally compressed and adaxially pubescent style led her to include it in Lathyrus in a new monospecific section Viciopsis [8].
Regarding the Azorean endemic V. dennesiana (Figure 2), we reject Kupicha’s concept of a monotypic section Perditae and find that the species belongs in the Mediterranean clade of section Cracca, where it also fits well based on stylar morphology [7]. The available herbarium specimens for this presumably extinct taxon show a striking overall morphological similarity to the V. nigricans group of the American Pacific coasts and Hawaii [9] but our partial DNA sequence data (nuclear ITS and fragments of plastid matK from two specimens) reject a close relationship. Kupicha [7] suggested that this species might be a biogeographic link explaining some of the amphi-Atlantic distribution patterns in Vicia but our analyses strongly suggest a single LDD event out of the Mediterranean to the Azores.
Our phylogeny estimates cast some doubt on species concepts applied in the Canary islands and Madeira: the Canarian endemic Vicia chaetocalyx is nested in Macaronesian and Mediterranean V. lutea accessions and it seems clear that the sequenced material should be assigned to the latter species because they are morphologically almost identical. However, our material came from a recently described population in Gran Canaria [52] and not from the original type material collected by Webb and Berthelot and now in the Florence herbarium, which should be used for further genetic study. A similar problem was reported in a recent study on an endemic bird species from the Cape Verde archipelago [53]. The four Canary and Madeira endemics in the V. cirrhosa group show no genetic differences in any of the sequenced regions. Since morphological differences are also minor, a re-evaluation of the group might be necessary (and possibly lumping of all four taxa into V. cirrhosa). In contrast, we find a high genetic variability among the relatively few samples included from the V. sativa group, where additional Macaronesian endemics might have been overlooked. Lowe’s Vicia pectinata for example, has been synonymized with the widespread V. sativa but we find three substitutions in the ITS region, which seem to be unique to Lowe’s taxon. Together with some morphological differences and the obvious isolation of the population on an oceanic island, there might be enough evidence to reinstate V. pectinata. However, a more detailed study of the Vicia sativa group in Macaronesian with several samples per island would be required to properly address this question.
Biogeographic history of the tribe
The origin of Fabeae is estimated to lie in the Middle Miocene (23–16 Ma). This result, however, could be expected since we constrained the Vicia s.l. crown to 17.5 Ma under a normal prior distribution with standard deviation of 1.9 million years. LDD events (here defined as dispersal across oceans) are relatively common in Fabeae. We reconstruct 12 LDD events over the period of c. 18 million years, which translates into a rate of c. 7 successful LDD events per 10 million years. The same rate was found for Cucurbitaceae [54] but sampling density in that study was lower (c. 25% of the accepted species), so the cucurbit rate is very likely an underestimate.
Migration via Greenland and the North Atlantic land bridge is not an option in Fabeae since the entire tribe is too young (climate cooling made use of that connection very likely impossible some 40 Ma [55]). Migration across the Beringian land bridge is likely for at least one lineage: the ancestors of the Pacific coast species Vicia nigricans, V. gigantea, and the Hawaiian V. menziesii. However, since the lineage later managed to colonize the isolated Hawaiian archipelago, LDD across the Pacific from Asia to the American Pacific coast is also an option or, less parsimonious, LDD from the Mediterranean to the American continent followed by spread to the West coast and extinction in the East. The next three Fabeae lineages, Vicia section Australes, Lathyrus section Notolathyrus, and the V. ludoviciana clade, all reached the American continent in the same time window (9–5 Ma) via LDD from the Mediterranean region across the Atlantic. This suggests favourable conditions for legume seed dispersal in the late Miocene, a ‘window of opportunity’ in the sense of Carine [56]. With the opening of the Bering Strait, c. 5.5-5.4 Ma [57], it became temporarily impossible to migrate to the Americas over land but later reconnections might have allowed terrestrial migration of cold-adapted lineages. For the ancestors of Lathyrus japonicus/L. littoralis, L. palustris, the large New World clade in Lathyrus section Orobus, and Vicia americana we can thus not decide between terrestrial migration and LDD. More recently, there might have been some transport across the region by Inuit, who use the roasted seeds to prepare drinks [58]. For L. japonicus and L. palustris the most likely direction is a spread from Asia to North America. In contrast, the V. americana lineage might have come the opposite way from North America (V. americana s.str., diploid) to Asia (V. bungei, tetraploid to hexaploid). Both V. americana and V. bungei are sister to the European Vicia onobrychioides L. and V. dumetorum and the ancestral lineage of the latter species might have spread into North America across the North Atlantic.
The now extinct Azorean endemic Vicia dennesiana might have reached the Azorean archipelago in the North Atlantic via LDD c. 2 Ma and the Canary islands and Madeira endemics of the Vicia scandens clade colonized those archipelagos via LDD about 4 Ma. We can therefore reject the hypothesis of ancient stepping stone dispersal via the Atlantic islands because all the island lineages are relatively recent arrivals compared to the age of Macaronesia (c. 60 million years [43]) and none of the American lineages is nested in an Atlantic island clade. In some cases our age estimates might also be biased old because of extinction events. The Vicia dennesiana and V. scandens lineages could represent the last survivors of previously more species-rich Eurosiberian clades in Macaronesian refugia and might be even younger than estimated if the Northwest African species of section Cracca were included, for which we failed to obtain material.
Evolution of life form and floral morphology in Fabeae
Ancestral character reconstruction shows that life form, stylar characters, and chromosome number are of limited value for systematic grouping of Fabeae. Style shape, a traditional character for Fabeae classification [7,8], varies little within the tribe: most species have a dorsiventrally compressed style, which is also reconstructed as most likely ancestral condition in the tribe (Figure 9). The finding that an annual life form is most likely the ancestral state in the tribe is unexpected and rejects earlier hypotheses of multiple evolution of annual life form as an adaptation to range expansion into xeric habitats [59]. It fits, however, with the result of the biogeographic analysis locating the region of origin in the Mediterranean and not in the temperate or boreal forests, where a perennial life form would have been an advantage. A basic chromosome number of 2n=14 has already been suggested by other authors (e.g. [60]), an ancestral, evenly hairy, dorsiventrally compressed style fits with recent results of small-scale character reconstructions [23].
Origin of legume crops
Our analyses shed light on a group of generally overlooked Mediterranean Lathyrus species that apart from the Caucasian Vavilovia species might be very interesting for breeding of new Pisum varieties due to benefitial traits including drought tolerance and perennial life form: L. gloeosperma, L. neurolobus Boiss. & Heldr., and L. nissolia all have affinities to the Pisum plus Vavilovia clade and might be included in breeding programs. The placement of V. faba far from the other members of section Faba sensu Kupicha [7] and somewhere among V. oroboides Wulfen, V. bithynica (L.) L., and the V. sativa-V. sepium clade (Figure 3; Additional file 10: Figure S8; Additional file 12: Figure S10) is in agreement with earlier morphological, cytological and DNA studies [14,17,18,61]. Vicia faba differs in chromosome number from most of the suggested relatives found in our analyses. Vicia bithynica, V. oroboides, and V. sepium all have 2n=14 chromosomes, whereas V. faba (and V. sativa s.l.) has 2n=12 (Additional file 2: Table S1; Figure 7), which will make crossings for plant breeding purposes difficult. The Lens clade has its closest relatives among early branching Vicia clades followed by c. 10 million years of independent evolution. While this could be a big obstacle for breeding efforts, the species of the African V. paucifolia complex and the Asian V. subvillosa might be worth testing if initial screenings reveal benefitial traits. For the V. sativa complex, a sister group relationship to V. pyrenaica seems very likely and the species should be explored. The high sequence diversity detected among our accessions of V. sativa s.l. corresponds to earlier findings of karyological and AFLP studies [62,63] and a detailed revision of the group is recommended in order to detect the most useful growth forms.
Conclusions
Based on nuclear and chloroplast phylogenies, we conclude that tribe Fabeae evolved in the Eastern Mediterranean in the middle Miocene. Ancestral Fabeae probably were annual plants with a chromosome number of 2n=14, and evenly hairy, dorsiventrally compressed style. From the Mediterranean, the tribe expanded its range at least 15 times into Central and Western Europe and 24 times to Asia, twice to Tropical Africa, and at least seven times across the Atlantic/Pacific to the Americas. The middle-Atlantic islands were colonized four times but did not serve as stepping-stones for lineages colonizing the New World. Our biogeographic analyses show that long distance dispersal events are relatively common in Fabeae (one successful event per 1.5 million years). Current generic and infrageneric circumscriptions in Fabeae do not reflect monophyletic groups and should be revised.
Methods
Sampling and DNA extraction
Leaf samples of 262 species of Fabeae were collected in the field and dried in silica gel or taken from herbarium vouchers, 125 of them never sequenced before (Additional file 2: Table S1). We tried to cover all described sections and all biogeographical regions of the Fabeae worldwide distribution. The morphologically most divergent species among the not sequenced taxa is V. koeieana, native to the Eastern Mediterranean/Asia Minor. Based on overall morphology, the species is suspected to be close to V. cypria, which we have included. Total genomic DNA was isolated from c. 100 mg dry leaf material with commercial plant DNA extraction kits (DNeasy, Qiagen; NucleoSpin, Machery-Nagel), following the manufacturers’ manuals. For the polymerase chain reaction (PCR) we used standard protocols with primer annealing at 48°C for rbcL, 49°C for matK and psbA-trnH, and 52–55°C for the trnL, trnS-trnG, and ITS1-5.8S-ITS2 regions. Reaction products were purified using ExoSAP-IT (USB Corporation, Cleveland, OH, USA) and cycle sequencing was performed with BigDye Terminator v3.1 cycle sequencing kits on an ABI 3730 sequencer (Applied Biosystems, Foster City, CA, USA) or sent to Functional Biosciences, Inc. (Madison, WI, USA) for sequencing. To amplify rbcL, we used the primers rbcL-1F, rbcL-1460R and the internal primers rbcL-600F and rbcL-724R [64], for matK, we used matK-F1, matK-1100F and matK-1932R [36,37], and for psbA-trnH we used the primer pair psbA (5′- GTT ATG CAT GAA CGT AAT GCT C) and trnH (5′- CGC GCA TGG TGG ATT CAC AAA TC) by Sang et al. [65]. For trnL/trnL-trnF, trnS-trnG, and the ITS region, we designed new primer pairs: trnL-V (5′-GCC TTG GTA TGG AAA CTT ACC A-3′) and trnF-V (′5-CGA CCA TTC TTG ACG CAC-3′), trnS-V (5′-GAT ACS TCG SAT AAA CAA AAA GAA C-3′) and trnG-V (5′-CAT GTT TCG TAA AGG GCC CCC TAA TG-3′), and ITS-VF (5′-TCG ATG CCT TAC ATG CAG TG-3′) and ITS-VR (5′-TAG AAA CGC ATA TGG GTA AAA GAG-3′).
Sequence alignment and phylogenetic analyses
Nine hundred and two sequences were generated for this study and combined with all available sequences from previous studies (mainly [13,23]). Additional file 2: Table S1 lists the relevant taxonomic names with authors, plant sources, and GenBank accession numbers. Sequences from other authors were carefully selected and compared to all available sequence data to avoid including misidentified material. All doubtful sequences were left aside.
Sequences were edited with Sequencher (4.9; Gene Codes, Ann Arbor, Michigan, USA) and Geneious pro v.5.0.4 [66], and aligned using MAFFT [67]; the final alignments were checked visually in MacClade v.4.08 [68]. The aligned nuclear ITS matrix comprised 394 ingroup accessions of 260 species with 668 nucleotides (1.4% gaps and 8.6% missing data); the rbcL matrix comprised 79 ingroup accessions of 72 species with 1352 nucleotides and 14% missing data; the matK matrix comprised 163 ingroup accessions of 118 species with 1587 nucleotides (0.2% gaps and 28% missing data); for the three spacer regions, we included 191 ingroup accession (146 species) and 733 nucleotides for trnL-trnF, 171 ingroup accessions (125 species) with 1174 nucleotides for trnS-trnG, and 150 ingroup accessions (111 species) with 688 aligned nucleotides for psbA-trnH. The percentages of gaps or missing data were 3% gaps/20% missing data for trnL-trnF, 3% gaps/40% missing data for trnS-trnG, and 15% gaps/29% missing data for psbA-trnH. Twenty-one unambiguous gaps in the ITS matrix, 51 in the trnS-trnG matrix, and ten in the psbA-trnH matrix were coded as binary characters based on Simmons & Ochoterena [69]. We furthermore combined all plastid regions into one alignment (341 ingroup accessions, 218 species, 5501 nucleotides, 1% gaps/60% missing data) and then combined the plastid and the nuclear ITS datasets into one final matrix with 470 ingroup accessions (262 species), 6200 aligned nucleotides, 1% gaps and 66% missing data. Finally, to obtain a condensed input dataset for the BEAST analyses (see below), we built a combined plastid and ITS matrix with multiple accessions per species merged into a single consensus sequence using the “merge sequence” function in MacClade 4.08 [68], which resulted in 262 ingroup taxa (6274 nucleotides, 1% gaps, 65% missing data).
Maximum likelihood (ML [70]) tree searches and ML bootstrap searches [71] were performed using RAxML 7.0.3 [72]. Based on the Akaike Information Criterion [73] as implemented in jModeltest [74] we selected the GTR + Γ model (six general time-reversible substitution rates, assuming gamma rate heterogeneity), with model parameters estimated over the duration of specified runs. Analyses in RAxML were run on all seven sequence data sets and both with the combined un-partitioned data and with a model that partitioned the plastid data from the ITS data.
Bayesian MCMC inference [75] used the GTR + Γ model (with the default four rate categories) plus a proportion of invariable sites and relied on MrBayes 3.1.2 [76]. Markov chain Monte Carlo (MCMC) runs started from independent random trees, were repeated twice, and extended for five million generations, with trees sampled every 100th generation. We used the default priors in MrBayes, namely a flat Dirichlet prior for the relative nucleotide frequencies and rate parameters, a discrete uniform prior for topologies, and an exponential distribution (mean 1.0) for the gamma-shape parameter and branch lengths. Convergence was assessed using Tracer v. 1.5 [77]. The data matrices and trees have been deposited in TreeBASE (http://www.treebase.org/) study number S13228.
Molecular dating analysis
To translate genetic distances into absolute times, we used Bayesian time estimation with an uncorrelated-rates model as implemented in BEAST v. 1.7.1 [78]. Since we have been unable to trace any fossils for the tribe, we use four secondary calibration points based on [38] and a normal prior distribution. Specifically, we constrained the root node (the split Ononis - rest to 24.7 and standard deviation (SD) of 2.3 Ma, the Vicia sensu lato crown age was set to 17.5, SD=1.9, the Vicia sensu stricto crown clade to 11.8, SD=1.7 Ma, and the clade comprising Lathyrus sativus and L. latifolius to 6.3, SD=1.3 Ma. BEAST analyses used the GTR + Γ model with six rate categories. Metropolis coupled Monte Carlo Markov chains were run for 20 million generations, sampling every 1000th generation. Of the 20,001 posterior trees, we excluded the first 2500 as burnin based on convergence assessment using Tracer v. 1.5 [77].
Biogeographic analyses
To reconstruct the biogeographical history of Fabeae, we coded species’ geographic ranges as an unordered multi-state character, using the following eight character states: (i) Central and Western Europe, (ii) Mediterranean region, (iii) Macaronesian islands, (iv) Asia, (v) Tropical Africa, (vi) North America, (vii) South America, and (viii) Hawaii. We then performed Bayesian ancestral area reconstructions [79] using BEAST v. 1.7.1 and a CTMC model with a gamma prior (shape = 1) and an exponential prior (mean = 1). Otherwise we used the same calibration points and MCMC settings as in the previously described BEAST molecular dating runs.
Ancestral state reconstruction
To analyse evolution of life form, chromosome number, and stylar morphology, we used the ML tree built from the condensed dataset for the 262 species (Additional file 12: Figure S10). We mapped the evolution of stylar shape, stylar hair patterns, chromosome number and annual versus perennial habit onto the phylogeny by defining four, five, six, and two unordered states respectively (see Additional file 2: Table S1). Information on morphological traits came from personal study of herbarium material and from the detailed lists in [7,8,23,80,81]. To infer ancestral states, we used maximum likelihood as implemented in Mesquite ver. 2.75 [82] (http://mesquiteproject.org/mesquite/mesquite.html). All analyses were carried out on the preferred highest likelihood tree, with branch lengths set to the maximum likelihood values obtained under the GTR + G model (above). Likelihood analyses in Mesquite used the Markov k-state one-parameter model, which is a generalization of the Jukes–Cantor model [83] and assumes a single rate for all transitions between character states. We let Mesquite estimate the transition parameters of the model, based on the tip trait states in the 262-taxon tree and its branch lengths.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
All authors collected data, HS analyzed data, all authors discussed results and wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Annex 1. Accepted species of the tribe Fabeae. Overview table for Fabeae with genus, subgenus, and section, distribution and synonyms for each accepted species. Data compiled from [3,7,8] and herbarium label information.
Table S1. Material used for phylogenetic analyses. Data for sequences used in the phylogenetic analyses: name, origin, Genbank accession number and voucher specimen for all sequenced taxa and for the sequences retrieved from Genbank.
Figure S1. Best ITS maximum likelihood phylogeny of the Fabeae. Best maximum likelihood tree based on the ITS dataset for 394 ingroup accessions (260 species) plus five outgroup species (668 aligned nucleotides). Likelihood bootstrap values ≥ 50% are given at the nodes.
Figure S2. Best rbcL maximum likelihood phylogeny of the Fabeae. Best maximum likelihood tree based on the rbcL dataset for 79 ingroup accessions (72 species) plus five outgroup species (1352 aligned nucleotides). Likelihood bootstrap values ≥ 50% are given at the nodes.
Figure S3. Best matK maximum likelihood phylogeny of the Fabeae. Best maximum likelihood tree based on the matK dataset for 163 ingroup accessions (118 species) plus five outgroup species (1587 aligned nucleotides). Likelihood bootstrap values ≥ 50% are given at the nodes.
Figure S4. Best trnS-trnG maximum likelihood phylogeny of the Fabeae. Best maximum likelihood tree based on the trnS-trnG dataset for 171 ingroup accessions (125 species) plus three outgroup species (1174 aligned nucleotides). Likelihood bootstrap values ≥ 50% are given at the nodes.
Figure S5. Best trnL/trnL-trnF maximum likelihood phylogeny of the Fabeae. Best maximum likelihood tree based on the trnL/trnL-trnF dataset for 191 ingroup accessions (146 species) plus four outgroup species (733 aligned nucleotides). Likelihood bootstrap values ≥ 50% are given at the nodes.
Figure S6. Best psbA-trnH maximum likelihood phylogeny of the Fabeae. Best maximum likelihood tree based on the psbA-trnH dataset for 150 ingroup accessions (111 species) (688 aligned nucleotides). Likelihood bootstrap values ≥ 50% are given at the nodes.
Figure S7. Best plastid marker maximum likelihood phylogeny of the Fabeae. Best maximum likelihood tree based on the combined plastid marker dataset for 353 ingroup accessions (230 species) plus seven outgroup species (5501 aligned nucleotides). Likelihood bootstrap values ≥ 50% are given at the nodes.
Figure S8. Bayesian consensus phylogeny for the combined dataset. Bayesian phylogeny reconstruction using MrBayes on a combined sequence alignment. When multiple accessions were present in the full dataset, we merged them into one consensus sequence per species resulting in 262 ingroup species (6274 nucleotides). Bayesian posterior probability values ≥ 0.8 are given at the nodes. Major clades (currently accepted genera plus Ervum and Ervilia) colour-coded according to the system in Figure 3.
Figure S9. Chronogram of the Fabeae. Dated phylogeny of the Fabeae estimated using BEAST on a combined sequence alignment. When multiple accessions were present in the full dataset, we merged them into one consensus sequence per species resulting in 262 ingroup species (6274 nucleotides). The age estimates with 95% confidence intervals (green bars) are given at the nodes.
Figure S10. Consensus ML phylogeny of the Fabeae. Best maximum likelihood tree based on a combined plastid and ITS matrix with multiple accessions per species merged into a single consensus sequence resulting in 262 ingroup species (6274 nucleotides).
Contributor Information
Hanno Schaefer, Email: hanno.schaefer@tum.de.
Paulina Hechenleitner, Email: phechenleitner@rbge.org.uk.
Arnoldo Santos-Guerra, Email: asantos@icia.es.
Miguel Menezes de Sequeira, Email: sequeira@uma.pt.
R Toby Pennington, Email: pennington@rbge.ac.uk.
Gregory Kenicer, Email: g.kenicer@rbge.ac.uk.
Mark A Carine, Email: m.carine@nhm.ac.uk.
Acknowledgements
We thank F. G. Dunkel (Karlstadt), Sylvia Norton (UK Plant heritage national collection holder), G. Lewis (RBG Kew), S. Harris & S. Marner (OXF) and the keepers of the following herbaria BM, E, GH, M, and U for material. We are grateful to P. Smykal (Olomouc) and two anonymous reviewers for comments on earlier drafts of the manuscript.
References
- Graham PH, Vance CP. Legumes: importance and constraints to greater use. Plant Physiol. 2003;131:872–877. doi: 10.1104/pp.017004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock JM, Maxted N. In: Legumes of the world: 505–510. Lewis G, Schrire B, Mackinder B, Lock M, editor. Kew, The Royal Botanic Gardens.; 2005. Tribe Fabeae. [Google Scholar]
- ILDIS. International Legume Database and information Service. 2010. Available at http://www.ildis.org/
- Zohary D. The mode of domestication of the founder crops of southwest Asian agriculture. UCL Press, London; 1996. (Harris DR (ed) The origins and spread of agriculture and pastoralism in Eurasia). [Google Scholar]
- Zohary D, Hopf M. Domestication of Pulses in the Old World - Legumes were companions of wheat and barley when agriculture began in the Near East. Science. 1973;182:887–894. doi: 10.1126/science.182.4115.887. [DOI] [PubMed] [Google Scholar]
- Zohary D, Hopf M. Domestication of plants in the Old world. 3. Oxford University Press, New York; 2002. [Google Scholar]
- Kupicha FC. The infrageneric structure of Vicia. Notes Roy Bot Gard Edinburgh. 1976;34:287–326. [Google Scholar]
- Kupicha FC. The infrageneric structure of Lathyrus. Notes Roy Bot Gard Edinburgh. 1983;41:209–244. [Google Scholar]
- Lassetter JS, Gunn CR. Vicia menziesii Sprengel (Fabaceae) rediscovered: its taxonomic relationships. Pac Sci. 1979;33:85–101. [Google Scholar]
- Oskoueiyan R, Kazempour Osaloo S, Maassoumi AA, Nejadsattari T, Mozaffarian V. Phylogenetic status of Vavilovia formosa (Fabaceae-Fabeae) based on nrDNA ITS and cpDNA sequences. Biochem Syst Ecol. 2010;38:313–319. doi: 10.1016/j.bse.2010.01.011. [DOI] [Google Scholar]
- Alefeld F. Über Vicieen. Bonplandia. 1861;9:66–72. 99–105, 116–131, 139–153. [Google Scholar]
- Asmussen CB, Liston A. Chloroplast DNA characters, phylogeny, and classification of Lathyrus (Fabaceae) Amer J Bot. 1998;85:387–401. doi: 10.2307/2446332. [DOI] [PubMed] [Google Scholar]
- Kenicer GJ, Kajita T, Pennington RT, Murata J. Systematics and biogeography of Lathyrus (Leguminosae) based on internal transcribed spacer and cpDNA sequence data. Amer J Bot. 2005;92:1199–1209. doi: 10.3732/ajb.92.7.1199. [DOI] [PubMed] [Google Scholar]
- Maxted N. An Ecogeographical Study of Vicia subgenus Vicia. International Plant Genetic Resources Institute, Rome; 1995. (Systematic and Ecogeographic Studies on Crop Genepools8). [Google Scholar]
- Kupicha FC. Studies in the Vicieae I: the new genus Anatropostylia. Notes Roy Bot Gard Edinburgh. 1973;32:247–250. [Google Scholar]
- Gunn CR, Kluve J. Androecium and pistil characters for tribe Vicieae (Fabaceae) Taxon. 1976;25:563–575. doi: 10.2307/1220110. [DOI] [Google Scholar]
- van de WTG V, Duncan N, Ramsay G, Phillips M, Powell W, Waugh R. Taxonomic relationships between V. faba and its relations based on nuclear and mitochondrial RFLPs and PCR analysis. Theor Appl Genet. 1993;86:71–80. doi: 10.1007/BF00223810. [DOI] [PubMed] [Google Scholar]
- Fennell SR, Powell W, Wright F, Ramsay G, Wangh R. Phylogenetic relationships between Vicia faba (Fabaceae) and related species inferred from chloroplast trnL sequences. Plant Syst Evol. 1998;212:247–259. doi: 10.1007/BF01089741. [DOI] [Google Scholar]
- Potokina E, Tomooka N, Vaughan DA, Alexandrova T, Xu R. Phylogeny of Vicia subgenus Vicia (Fabaceae) based on analyis of RAPDs and RFLP of PCR-amplified chloroplast genes. Genet Res Crop Evol. 1999;46:149–161. doi: 10.1023/A:1008640322198. [DOI] [Google Scholar]
- Jaaska V. Isozyme variation and phylogenetic relationships in Vicia subgenus Cracca (Fabaceae) Ann Bot. 2005;96:1085–1096. doi: 10.1093/aob/mci260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B, Seok D, Endo Y, Ohashi H. Phylogenetic significance of stylar features in genusVicia(Leguminosae): an analysis with molecular phylogeny. J Plant Res. 2006;119:513–523. doi: 10.1007/s10265-006-0015-6. [DOI] [PubMed] [Google Scholar]
- Endo Y, Choi BH, Ohashi H, Delgado-Salinas A. Phylogenetic relationships of New World Vicia (Leguminosae) inferred from nrDNA Internal Transcribed Spacer sequences and floral characters. Syst Bot. 2008;33:356–363. doi: 10.1600/036364408784571536. [DOI] [Google Scholar]
- Endo Y, Choi B, Kakinuma D, Kenicer G, Zhu X-Y, Ohashi H. Molecular phylogeny of Vicia sect. Amurense (Leguminosae) J Jpn Bot. 2010;85:337–349. [Google Scholar]
- Croft AM, Pang PCK, Taylor PWJ. Molecular analysis of Lathyrus sativus L. (grasspea) and related Lathyrus species. Euphytica. 1999;107:167–176. doi: 10.1023/A:1003520721375. [DOI] [Google Scholar]
- Badr A, El Shazly H, El Rabey H, Watson LE. Systematic relationships in Lathyrus sect. Lathyrus (Fabaceae) based on amplified fragment length polymorphism (AFLP) data. Canad J Bot. 2002;80:962–969. doi: 10.1139/b02-084. [DOI] [Google Scholar]
- Ben Brahim N, Salhi A, Chtourou N, Combes D, Marrakchi M. Isozymic polymporphism and phylogeny of 10 Lathyrus species. Gen Res Crop Evol. 2002;49:427–436. [Google Scholar]
- Mayer MS, Soltis PS. Chloroplast DNA phylogeny of Lens (Leguminosae): origin and diversity of the cultivated lentil. Theor Appl Genet. 1994;87:773–781. doi: 10.1007/BF00221128. [DOI] [PubMed] [Google Scholar]
- Mayer MS, Bagga SK. The phylogeny of Lens (Leguminosae): new insight from ITS sequence analysis. Plant Syst Evol. 2002;232:145–154. doi: 10.1007/s006060200038. [DOI] [Google Scholar]
- Sonnante GI, Galasso I, Pignone D. ITS sequence analysis and phylogenetic inference in the genus Lens Mill. Ann Bot. 2003;91:49–54. doi: 10.1093/aob/mcg007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polans NO, Saar DE. ITS sequence variation in wild species and cultivars of pea. Pisum Genetics. 2002;34:9–13. [Google Scholar]
- Kosterin OE, Bogdanova VS. Relationship of wild and cultivated forms of Pisum L. as inferred from an analysis of three markers, of the plastid, mitochondrial and nuclear genomes. Genet Res Crop Evol. 2008;55:735–755. doi: 10.1007/s10722-007-9281-y. [DOI] [Google Scholar]
- Kosterin OE, Zaytseva OO, Bogdanova VS, Ambrose M. New data on three molecular markers from different cellular genomes in Mediterranean accessions reveal new insights into phylogeography of Pisum sativum L. subsp. elatius (Bieb.) Schmalh. Genet Res Crop Evol. 2010;57:733–739. doi: 10.1007/s10722-009-9511-6. [DOI] [Google Scholar]
- Smykal P, Kenicer G, Flavell AJ, Corander J, Kosterin O, Redden RJ, Ford R, Coyne CJ, Maxted N, Ambrose MJ, Ellis NTH. Phylogeny, phylogeography and genetic diversity of the Pisum genus. Plant Genetic Resources: Characterization and Utilization. 2011;9:4–18. doi: 10.1017/S147926211000033X. [DOI] [Google Scholar]
- Magee AM, Aspinall S, Rice DW, Cusack BP, Semon M, Perry AS, Stefanovic S, Milbourne D, Barth S, Palmer JD, Gray JC, Kavanagh TA, Wolfe KH. Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 2010;20:1700–1710. doi: 10.1101/gr.111955.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski MF, Sanderson MJ, Steele KP, Liston A. In: Advances in Legume Systematics 9: 277–298. Herendeen PS, Bruneau A, editor. Kew, Royal Botanic Gardens; 2000. Molecular phylogeny of the “Temperate Herbaceous Tribes” of papilionoid legumes: a supertree approach. [Google Scholar]
- Steele KP, Wojciechowski MF. In: Advances in Legume Systematics 10. Klitgaard BB, Bruneau A, editor. The Royal Botanic Gardens, Kew; 2003. Phylogenetic analyses of tribes Trifolieae and Vicieae, based on sequences of the plastid gene matK (Papilionoideae: Leguminosae) pp. 355–370. [Google Scholar]
- Wojciechowski MF, Lavin M, Sanderson MJ. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Amer J Bot. 2004;91:1845–1861. doi: 10.3732/ajb.91.11.1846. [DOI] [PubMed] [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. Syst Biol. 2005;54:575–594. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- Simon MF, Grether R, de Queiroz LP, Skema C, Pennington RT, Hughes CE. Recent assembly of the Cerrado, a neotropical plant diversity hotspot, by in situ evolution of adaptations to fire. PNAS. 2009;106:20359–20364. doi: 10.1073/pnas.0903410106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simola LK. Comparative studies on number of leaflets, venation and epidermal structure in the genus Lathyrus. Canad J Bot. 1968;46:71–84. doi: 10.1139/b68-014. [DOI] [Google Scholar]
- Burkart A. Revision de las especias de Lathyrus de la Republica Argentina. Revista de la Faculdad de Agronomia y Veterinaria. 1935;8:1–149. [Google Scholar]
- Axelrod DI. Evolution and biogeography of Madrean-Tethyan sclerophyll vegetation. Ann Mo Bot Gard. 1975;62:280–334. doi: 10.2307/2395199. [DOI] [Google Scholar]
- Fernández-Palacios JM, de Nascimento L, Otto R, Delgado JD, García-del-Rey E, Arévalo JR, Whittaker RJ. A reconstruction of Palaeo-Macaronesia, with particular reference to the long-term biogeography of the Atlantic island laurel forests. J Biogeo. 2011;38:226–246. doi: 10.1111/j.1365-2699.2010.02427.x. [DOI] [Google Scholar]
- Stefánsson S. Flóra Íslands. 3 ed. Prentverk Odds Björnssonar, Akureyri; 1948. [Google Scholar]
- Schaefer H. Chorology and diversity of the Azorean flora. J. Cramer, Stuttgart (Germany); 2003. (Dissertationes Botanicae 374). [Google Scholar]
- Silva L, Moura M, Schaefer H, Rumsey F, Dias EF. In: A list of the terrestrial and marine biota from the Azores. Borges PAV, Costa A, Cunha R, Gabriel R, Gonçalves V, Martins AF, Melo I, Parente M, Raposeiro P, Rodrigues P, Santos RS, Silva L, Vieira P, Vieira V, editor. Princípia, Cascais (Portugal); 2010. Vascular plants (Tracheobionta) pp. 117–146. [Google Scholar]
- Watson HC. In: Natural history of the Azores or Western Islands. Cane Godman F, editor. J. Van Voorst, London; 1870. Botany of the Azores; pp. 113–288. [Google Scholar]
- Hooker JD. Vicia dennesiana. Curtis’s Bot Mag. 1887;43:6967. [Google Scholar]
- Jardim R, Menezes De Sequeira M. In: A list of the terrestrial fungi, flora and fauna of Madeira and Selvagens archipelagos. Borges PAV, Abreu C, Aguiar AMF, Carvalho P, Jardim R, Melo I, Oliveira P, Sérgio C, Serrano ARM, Vieira P, editor. Direcção Regional do Ambiente da Madeira and Universidade dos Açores, Funchal and Angra do Heroísmo, Portugal; 2008. Lista das plantas vasculares (Pteridophyta and Spermatophyta) p. 440. Pp. 179–208. [Google Scholar]
- Izquierdo I, Martín JL, Zurita N, Arechavaleta M. Lista de especies silvestres de Canarias (hongos, plantas, y animales terrestres) Gobierno de Canarias, La Laguna (Spain); 2004. [Google Scholar]
- Robeson DJ, Harborne JB. A chemical dichotomy in Phytoalexin induction within the tribe Vicieae of the Leguminosae. Phytochemistry. 1980;19:2359–2365. doi: 10.1016/S0031-9422(00)91027-6. [DOI] [Google Scholar]
- Dunkel F-G. Vicia chaetocalyx Webb & Berthel. on Gran Canaria - A Canarian endemic plant rediscovered. Bot Macaronesica. 2008;27:107–113. [Google Scholar]
- Johnson JA, Watson RT, Mindell DP. Prioritizing species conservation: does the Cape Verde kite exist? Proc Roy Soc B. 2005;272:1365–1371. doi: 10.1098/rspb.2005.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer H, Heibl C, Renner SS. Gourds afloat: a dated phylogeny reveals an Asian origin of the gourd family (Cucurbitaceae) and numerous oversea dispersal events. Proc Roy Soc B. 2009;276:843–851. doi: 10.1098/rspb.2008.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffney BH. The Eocene North Atlantic land bridge: its importance in Tertiary and modern phylogeography of the Northern Hemisphere. J Arnold Arb. 1985;66:243–273. [Google Scholar]
- Carine MA. Spatio-temporal relationships of the Macaronesian endemic flora: a relictual series or window of opportunity? Taxon. 2005;54:895–903. doi: 10.2307/25065476. [DOI] [Google Scholar]
- Gladenkov AY, Oleinik AE, Marincovich L, Barinov KB. A refined age for the earliest opening of Bering Strait. Palaeogeography Palaeoclimatology Palaeoecology. 2002;183:321–328. doi: 10.1016/S0031-0182(02)00249-3. [DOI] [Google Scholar]
- Anderson JP. Plants used by the Eskimo of the northern Bering Sea and arctic regions of Alaska. Amer J Bot. 1939;26:714–716. doi: 10.2307/2437021. [DOI] [Google Scholar]
- Hanelt P, Mettin D. Biosystematics of the genus Vicia L. Annu Rev Ecol Syst. 1989;20:199–223. [Google Scholar]
- Cubero JI. In: Genetic resources and their exploitation - chickpeas, faba beans and lentils. Witcombe JR, Erskine W, editor. Junk Publishers, Hague, Martinus Nijhoff/Dr W; 1984. Taxonomy, distribution and evolution of the faba bean and its wild relatives; pp. 131–144. [Google Scholar]
- Jaaska V. Isoenzyme diversity and phylogenetic affinities in Vicia subgenus Vicia (Fabaceae) Gen Res Crop Evol. 1997;44:557–574. doi: 10.1023/A:1008630003045. [DOI] [Google Scholar]
- Zohary D, Plitmann U. Chromosome polymorphism, hybridization and colonization in the Vicia sativa group (Fabaceae) Plant Syst Evol. 1979;131:143–156. doi: 10.1007/BF00984128. [DOI] [Google Scholar]
- Potokina E, Blattner FR, Alexandrova T, Bachmann K. AFLP diversity in the common vetch (Vicia sativa L.) on the world scale. Theor Appl Genet. 2002;105:58–67. doi: 10.1007/s00122-002-0866-8. [DOI] [PubMed] [Google Scholar]
- Olmstead RG, Michaels HJ, Scott KM, Palmer JD. Monophyly of the Asteridae and identification of their major lineages inferred from DNA sequences of rbcL. Ann Mo Bot Gard. 1992;79:249–265. doi: 10.2307/2399768. [DOI] [Google Scholar]
- Sang T, Crawford DJ, Stuessy TF. Chloroplast DNA phylogeny, reticulate evolution and biogeography of Paeonia (Paeoniaceae) Am J Bot. 1997;84:1120–1136. doi: 10.2307/2446155. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, Kearse M, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A. Geneious, version 5.0.4. 2010. http://www.geneious.com.
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT vs. 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison DR, Maddison WP. MacClade 4.08. Sinauer Associates, Sunderland, MA; 2005. [Google Scholar]
- Simmons MP, Ochoterena H. Gaps as characters in sequence-based phylogenetic analyses. Syst Biol. 2000;49:369–381. [PubMed] [Google Scholar]
- Felsenstein J. Maximum likelihood and minimum-steps methods for estimating evolutionary trees from data on discrete characters. Syst Zool. 1973;22:240–249. doi: 10.2307/2412304. [DOI] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web-servers. Syst Biol. 2008;75:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Akaike H. A new look at the statistical model identification. IEEE Trans. Automat Control. 1974;19:716–723. doi: 10.1109/TAC.1974.1100705. [DOI] [Google Scholar]
- Posada D. jModelTest: Phylogenetic model averaging. Molec Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Yang Z, Rannala B. Bayesian phylogenetic inference using DNA sequences: a Markov Chain Monte Carlo method. Molec Biol Evol. 1997;14:717–724. doi: 10.1093/oxfordjournals.molbev.a025811. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer 1.5.0. MCMC Trace Analysis Tool. 2009. Available at: http://beast.bio.ed.ac.uk/Tracer.
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Comput Biol. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y, Ohashi H. The morphology of styles and stigmas in Vicia (Leguminosae), and its systematic implications. J Plant Res. 1995;108:17–24. doi: 10.1007/BF02344301. [DOI] [Google Scholar]
- Endo Y, Ohashi H. The morphology and anatomy of styles in Vicieae (Leguminosae) J Jpn Bot. 1997;72:9–18. [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 2.75. 2011. Available from: http://mesquiteproject.org.
- Lewis PO. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst Biol. 2001;50:913–925. doi: 10.1080/106351501753462876. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Annex 1. Accepted species of the tribe Fabeae. Overview table for Fabeae with genus, subgenus, and section, distribution and synonyms for each accepted species. Data compiled from [3,7,8] and herbarium label information.
Table S1. Material used for phylogenetic analyses. Data for sequences used in the phylogenetic analyses: name, origin, Genbank accession number and voucher specimen for all sequenced taxa and for the sequences retrieved from Genbank.
Figure S1. Best ITS maximum likelihood phylogeny of the Fabeae. Best maximum likelihood tree based on the ITS dataset for 394 ingroup accessions (260 species) plus five outgroup species (668 aligned nucleotides). Likelihood bootstrap values ≥ 50% are given at the nodes.
Figure S2. Best rbcL maximum likelihood phylogeny of the Fabeae. Best maximum likelihood tree based on the rbcL dataset for 79 ingroup accessions (72 species) plus five outgroup species (1352 aligned nucleotides). Likelihood bootstrap values ≥ 50% are given at the nodes.
Figure S3. Best matK maximum likelihood phylogeny of the Fabeae. Best maximum likelihood tree based on the matK dataset for 163 ingroup accessions (118 species) plus five outgroup species (1587 aligned nucleotides). Likelihood bootstrap values ≥ 50% are given at the nodes.
Figure S4. Best trnS-trnG maximum likelihood phylogeny of the Fabeae. Best maximum likelihood tree based on the trnS-trnG dataset for 171 ingroup accessions (125 species) plus three outgroup species (1174 aligned nucleotides). Likelihood bootstrap values ≥ 50% are given at the nodes.
Figure S5. Best trnL/trnL-trnF maximum likelihood phylogeny of the Fabeae. Best maximum likelihood tree based on the trnL/trnL-trnF dataset for 191 ingroup accessions (146 species) plus four outgroup species (733 aligned nucleotides). Likelihood bootstrap values ≥ 50% are given at the nodes.
Figure S6. Best psbA-trnH maximum likelihood phylogeny of the Fabeae. Best maximum likelihood tree based on the psbA-trnH dataset for 150 ingroup accessions (111 species) (688 aligned nucleotides). Likelihood bootstrap values ≥ 50% are given at the nodes.
Figure S7. Best plastid marker maximum likelihood phylogeny of the Fabeae. Best maximum likelihood tree based on the combined plastid marker dataset for 353 ingroup accessions (230 species) plus seven outgroup species (5501 aligned nucleotides). Likelihood bootstrap values ≥ 50% are given at the nodes.
Figure S8. Bayesian consensus phylogeny for the combined dataset. Bayesian phylogeny reconstruction using MrBayes on a combined sequence alignment. When multiple accessions were present in the full dataset, we merged them into one consensus sequence per species resulting in 262 ingroup species (6274 nucleotides). Bayesian posterior probability values ≥ 0.8 are given at the nodes. Major clades (currently accepted genera plus Ervum and Ervilia) colour-coded according to the system in Figure 3.
Figure S9. Chronogram of the Fabeae. Dated phylogeny of the Fabeae estimated using BEAST on a combined sequence alignment. When multiple accessions were present in the full dataset, we merged them into one consensus sequence per species resulting in 262 ingroup species (6274 nucleotides). The age estimates with 95% confidence intervals (green bars) are given at the nodes.
Figure S10. Consensus ML phylogeny of the Fabeae. Best maximum likelihood tree based on a combined plastid and ITS matrix with multiple accessions per species merged into a single consensus sequence resulting in 262 ingroup species (6274 nucleotides).