Summary
Two-Partner Secretion (TPS) systems use β-barrel proteins of the Omp85-TpsB superfamily to transport large exoproteins across the outer membranes of Gram-negative bacteria. The Bordetella FHA/FhaC proteins are prototypical of TPS systems in which the exoprotein contains a large C-terminal prodomain that is removed during translocation. Although it is known that the FhaB prodomain is required for FHA function in vivo, its role in FHA maturation has remained mysterious. We show here that the FhaB prodomain is required for the extracellularly-located mature C-terminal domain (MCD) of FHA to achieve its proper conformation. We show that the C-terminus of the prodomain is retained intracellularly and that sequences within the N-terminus of the prodomain are required for this intracellular localization. We also identify sequences at the C-terminus of the MCD that are required for release of mature FHA from the cell surface. Our data support a model in which the intracellularly-located prodomain affects the final conformation of the extracellularly-located MCD, which, we hypothesize, triggers cleavage and degradation of the prodomain.
Introduction
Two Partner Secretion (TPS) systems are widespread amongst Gram-negative bacteria. They use β-barrel, channel-forming proteins (TpsB family members) to translocate large, predominantly β-helical, exoproteins (TpsA family members) across the outer membrane to the cell surface (Mazar and Cotter, 2007; Jacob-Dubuisson et al., 2009). TpsA family members include adhesins, such as filamentous hemagglutinin (FHA) of Bordetella species and the HMW proteins of Haemophilus influenzae; cytolysins, such as ShlA of Serratia marcescens; and contact dependent growth inhibition (CDI) mediators, such as CdiA of Escherichia coli (Jacob-Dubuisson et al., 2004; Aoki et al., 2005). TpsB proteins are members of the Omp85-TpsB superfamily, which includes BamA, the main component of the Bam complex that inserts β-barrel proteins into the outer membranes of Gram-negative bacteria (Genevrois et al., 2003; Voulhoux et al., 2003; Wu et al., 2005; Hagan et al., 2011), and also Tob55/Sam50 and Toc75, which insert proteins into the outer membranes of mitochondria and chloroplasts, respectively (Reumann and Keegstra, 1999; Kozjak et al., 2003). Used by bacteria and organelles of fungi, plants, and animals, protein translocation by the Omp85-TpsB superfamily is the most widely distributed outer membrane protein secretion mechanism known (Yen et al., 2002).
Omp85-TpsB family members are composed of soluble N-terminal regions containing one to five polypeptide transport associated (POTRA) domains and C-terminal regions of about 350 aa that form a transmembrane β-barrel. The Bordetella pertussis TpsB family member FhaC has been crystallized and its structure solved with 3.15 Å resolution (Clantin et al., 2007). Sequence comparisons of bacterial TpsB proteins indicate that they can be separated into at least four distinct subfamilies, with FhaC, HMW1B (which exports H. influenzae HMW1), ShlB (which exports S. marcescens ShlA), and CdiB (which exports E. coli CdiA) representing the best characterized member of each subfamily.
TpsA family members are less well conserved. Their amino acid similarity is greatest within an ~250 aa region at the N-terminus called the “TPS domain” (Jacob-Dubuisson et al., 2004). The TPS domains of Bordetella FHA and H. influenzae HMW1 are required for secretion and each has been shown to be sufficient, as a polypeptide fragment, for translocation by their cognate TpsB protein when produced in E. coli (Renauld-Mongenie et al., 1996; Grass and St Geme, 2000). The TPS domain of FHA has been shown to interact with the POTRA domains of FhaC and to do so in an unfolded state (Hodak et al., 2006). Although they share less amino acid sequence similarity in regions C-terminal to the TPS domains, all TpsA proteins are predicted to contain a substantial amount of β-helical character, the folding of which has been hypothesized to provide the energy for translocation across the outer membrane (Thanassi et al., 2005). Comparison of the amino acid sequences of TPS domains separates TpsA proteins into the same four subfamilies identified by comparison of TpsB family members, and the different subfamilies appear to correlate with different TpsA biogenesis processes. For example, HMW1 contains a N-terminal “pro-piece” that is removed during outer membrane translocation and mature HMW1 is anchored to the cell surface by its C-terminus (Grass and St Geme, 2000; Buscher et al., 2006). FHA, by contrast, is synthesized with a C-terminal prodomain that is ultimately removed and mature FHA is anchored to the cell via its N-terminus (Renauld-Mongenie et al., 1996; Mazar and Cotter, 2006). In the case of CdiA, there appears to be no pro-peptide processing (other than removal of the signal sequence) and given the C-terminal location of the toxic domain, it is assumed that the protein is anchored via its N-terminus (Aoki et al., 2010; Hayes et al., 2010). Similarly, ShlA also does not have an evident pro-peptide, and a C-terminal pore-forming effector domain suggests that ShlA is surface-associated by its N-terminus (Hertle, 2005).
FHA is one of the primary virulence factors produced by B. pertussis, the causative agent of whooping cough, and the closely related, broad host-range pathogen, B. bronchiseptica. FHA mediates adherence to epithelial cells and macrophages in vitro and is required for tracheal colonization in vivo (Urisu et al., 1986; Locht et al., 1993; Cotter et al., 1998; Inatsuka et al., 2005; Julio et al., 2009). FHA also modulates the inflammatory response to infection, contributing to persistence in the lower respiratory tract (Inatsuka et al., 2005; Henderson et al., 2012), and FHA is one of the primary components of acellular pertussis vaccines (Sato and Sato, 1999). As one of the best-studied TPS pathways, FHA/FhaC is one of the primary models used for understanding the TPS mechanism.
The current model for FHA secretion is shown in Supplemental Figure 1A. FHA is initially synthesized as an ~375 kDa preproprotein called FhaB. FhaB contains a 71 aa signal peptide, which includes an N-terminal extension of 21 residues, that targets the protein to the general secretory machinery in a SecAB-dependent, post-translational manner (Lambert-Buisine et al., 1998; Chevalier et al., 2004; Desvaux et al., 2007). The N-terminal TPS domain associates with the periplasmic POTRA domains of FhaC to initiate outer membrane translocation (Sanchez-Pulido et al., 2003; Hodak et al., 2006). Translocation through FhaC is thought to initiate with an extended hairpin forming through the FhaC channel, then the C-terminus of the TPS domain folds into a β-helix as it emerges on the cell surface. As more residues reach the surface, they continue to adopt a right-handed β-helical fold, starting from the cell surface and folding outward to a length of ~35 nm (Makhov et al., 1994; Kajava et al., 2001; Clantin et al., 2004). C-terminal to the β-helical shaft is a domain of poor structural definition that becomes the mature C-terminal domain (MCD) of FHA, which is critical for FHA function in vivo and in vitro (Mazar and Cotter, 2006; Julio et al., 2009). Cleavage of FhaB occurs C-terminal to the MCD, yielding the ~250 kDa mature FHA protein. The autotransporter SphB1 is required for cleavage of FhaB somewhere within a PLFETRIKFID sequence (residues 2472–2482 of B. bronchiseptica and 2362–2372 of B. pertussis) and at least one site ~100 aa N-terminal to this sequence, although SphB1 has not been shown to cleave FhaB directly (Coutte et al., 2001). One or more unidentified proteases are also involved in FHA maturation, as FhaB is processed to a slightly larger protein, referred to as FHA* in B. pertussis or FHA’ in B. bronchiseptica, in sphB1 deletion strains (Coutte et al., 2001; Mazar and Cotter, 2006). The C-terminus of FHA’ in B. bronchiseptica strain RB50 is located between residues 2485 and 2545 (Mazar and Cotter, 2006). Although mature FHA can be detected on the cell surface with its C-terminus facing outward, a substantial amount is released into the extracellular environment. Neither the mechanism controlling surface retention versus release nor the biological relevance of FHA release is known.
The C-terminal ~1200 aa prodomain of FhaB is not detectable after SphB1-dependent or SphB1-independent cleavage and is thought to be rapidly degraded (Delisse-Gathoye et al., 1990; Mazar and Cotter, 2006). Although models for FhaB secretion and maturation have suggested that the prodomain remains in an intracellular compartment (Renauld-Mongenie et al., 1996; Mazar and Cotter, 2007), its fate is unknown and it is possible that it functions from the cell surface (Supplemental Figure 1B and C, respectively). B. bronchiseptica and B. pertussis strains in which FhaB is synthesized with truncated prodomains produce FHA proteins that appear to be translocated to the cell surface, processed in an SphB1-dependent manner, and released into the extracellular environment, yet these strains are defective for adherence to epithelial cells and macrophages in vitro and, in the case of B. bronchiseptica, for tracheal colonization in vivo (Mazar and Cotter, 2006). The goal of this work was to determine the role of the FhaB prodomain in maturation and function of FHA.
Results
Bioinformatic analysis of the FhaB prodomain
A PSI-BLAST search using the entire B. bronchiseptica FhaB prodomain as query revealed substantial similarity over the entire length of the prodomain (E values ≤ e−30) with other FHA-related proteins in Bordetella species as well as a few related proteins in Pasteurella multocida, Haemophilus ducreyi, Citrobacter rodentium, and Erwinia billingiae. Strikingly, however, hundreds of predicted TpsA proteins, many of which are known to belong to the FHA subfamily, were identified with significant similarity (alignment scores between 80 and 200) to residues 2772–2601 of the FhaB prodomain, which we are now calling the prodomain N-terminus (PNT). ClustalW analysis revealed ~50% similarity overall amongst the proteins identified in our analysis (Supplemental Figure 2). No similarity with other proteins above the threshold level set by PSI-BLAST was identified for regions of the FhaB prodomain C-terminal to the PNT. We did, however, notice a proline rich region near the C-terminus of the prodomain by simple visual examination of the amino acid sequence. This proline rich region has been noted previously (Locht et al., 1992) and, curiously, a putative FHA ortholog in Bordetella avium contains a region with even more prolines near its C-terminus. There are no reports of functional characterization for any sequences with which similarity with the prodomain was identified.
The FhaB prodomain is required for the MCD to attain its proper conformation
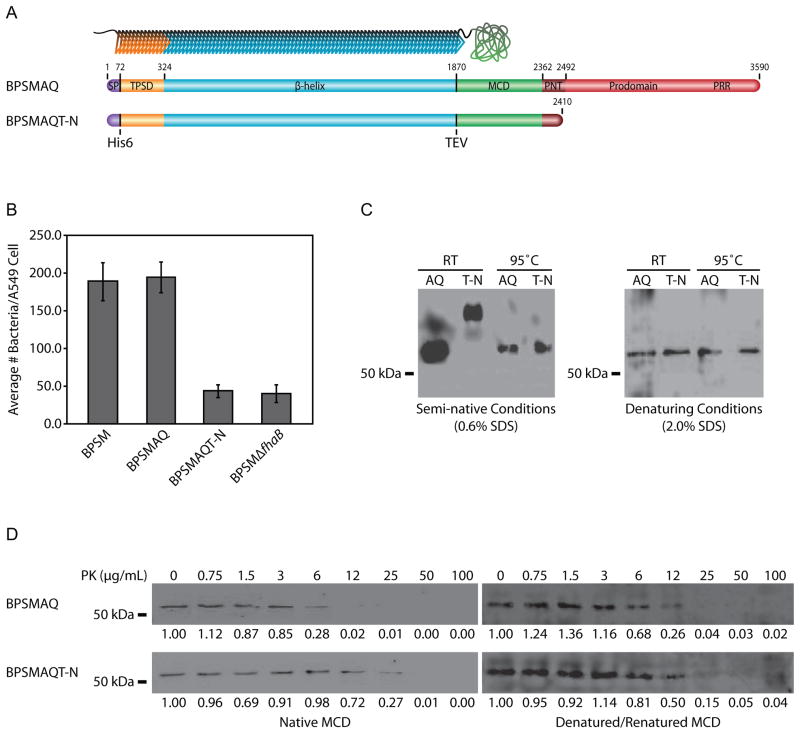
Because the prodomain is required for FHA function in vivo (Mazar and Cotter, 2006) and there is evidence that the MCD is an important functional domain (Julio et al., 2009; Henderson et al., 2012), we hypothesized that the FhaB prodomain is required for the MCD to attain its proper conformation. To test this hypothesis, two B. pertussis strains were constructed that produce FhaB proteins containing hexahistidine (His6) insertions immediately C-terminal to Q72 (the N-terminal amino acid of mature FHA) and tobacco etch virus (TEV) protease cleavage sites at the junction between the β-helical shaft and the MCD. One strain (BPSMAQ) produced full-length FhaB and the other (BPSMAQT-N) contained a stop codon in fhaB 106 codons 3’ to the region encoding the primary SphB1-dependent cleavage site (Figure 1A–note that residue numbering of the schematic corresponds to B. pertussis FhaB, not B. bronchiseptica FhaB). We used B. pertussis strain BPSM for these experiments because it releases substantially more FHA into culture supernatants than B. bronchiseptica strain RB50, facilitating our ability to recover a sufficient amount of protein with minimal manipulation (Jacob-Dubuisson et al., 2000; Mazar and Cotter, 2006). The presence of the His6 tag and the TEV protease cleavage site did not affect the ability of otherwise wildtype FHA to function as an adhesin in vitro (Figure 1B). Bacteria were grown under fhaB-inducing conditions, and culture supernatants were collected under native conditions and incubated with nickel-chelating resin, which was then washed and incubated with AcTEV protease to release the MCD from FHA. The MCD polypeptides were resolved on a semi-native polyacrylamide gel (0.6% SDS). Samples were then transferred to a nitrocellulose membrane and probed with anti-MCD antibody. The MCD polypeptides recovered from the BPSMAQ and BPSMAQT-N strains under native conditions migrated with different mobilities on the semi-native gel, but they migrated with the same mobility after heat denaturation (Figure 1C). The MCD polypeptides migrated with the same mobility regardless of pre-heating when separated by standard, denaturing, SDS-PAGE (2.0% SDS). These results suggest that the presence of the prodomain in the nascent FhaB polypeptide affects the conformation of the MCD in mature FHA.
Figure 1.
The FhaB prodomain affects the conformation of the MCD. (A) At top is an illustration of mature FHA with an N-teriminal hexahistadine tag and a TEV protease cleavage site upstream of the MCD. Below are schematics of the B. pertussis FhaB proteins as initially synthesized. From left, the domains illustrated are the signal peptide (purple), the TPS domain (orange), the β-helical shaft (light blue), the MCD (green), the PNT (dark red), and the remainder of the prodomain (red). The hexahistadine tag is located between the signal peptide and the TPS domain. The TEV protease cleavage site is located between the β-helix and the MCD. The approximate site of SphB1-dependent cleavage is noted. The proline-rich region (PRR) of the prodomain is also noted. Residue numbering is based on the B. pertussis FhaB protein. (B) The strains were tested to determine if the hexahistadine tag and TEV protease site altered FHA mediated adherence to A549 human lung epithelial cells. BPSMAQ adhered in a manner indistinguishable from wildtype B. pertussis, while BPSMAQT-N adhered as inefficiently as an fhaB deletion strain. (C) Anti-MCD immunoblot of purified MCD polypeptides. Semi-native gel analysis indicated altered mobility of the MCD derived from the prodomain-truncated (T-N) FhaB compared to the MCD derived from full-length (AQ) FhaB. When denatured by either heat or chemical denaturation, the MCDs of both proteins migrate identically. (D) Western blot of MCD polypeptides after proteinase-K treatment. Purified MCDs were tested for sensitivity to proteinase-K. Samples were incubated with increasing concentrations of proteinase-K, separated by SDS-PAGE, and detected with anti-MCD antibody. Additionally, samples were denatured/renatured prior to analysis of proteinase- K sensitivity. Densitometery values proportionate to the undigested sample of each Western blot are listed below the blots.
We also treated the native MCD polypeptides with increasing concentrations of proteinase-K and resolved them by SDS-PAGE. While the MCD from BPSMAQ (which produces full-length FhaB) was completely digested by 12 μg/mL of proteinase-K, 50 μg/mL of proteinase-K was required to completely degrade the MCD from BPSMAQT-N (Figure 1D). As a control, the MCDs from both strains were boiled to denature the proteins, then allowed to renature, and were then incubated with proteinase-K. After being subjected to denaturing and renaturing conditions in vitro, both MCD polypeptides were completely degraded by 25 μg/mL proteinase-K, suggesting that the conformations of the in vitro renatured proteins were the same (but different from those of the naturally folded polypeptides, which were digested by either 25 or 50 μg/mL proteinase-K). Together, these data indicate that the conformation of the MCD that forms after translocation to the bacterial surface differs depending on whether the initially synthesized FhaB protein contains the C-terminal prodomain.
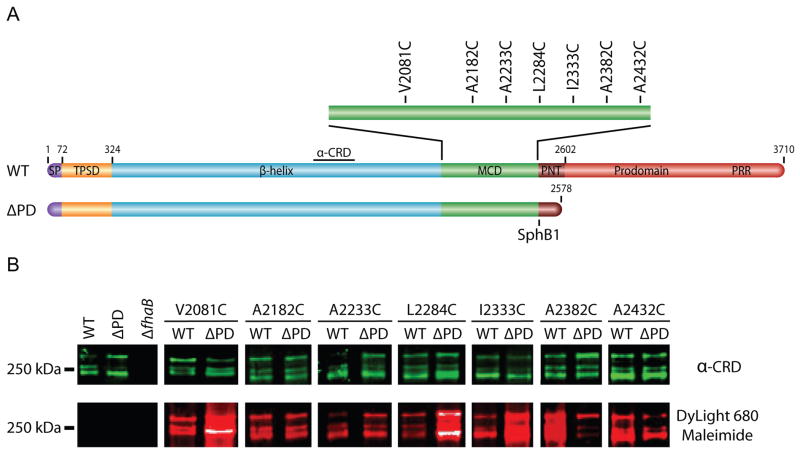
As an additional approach to probe the conformation of the MCD, we performed cysteine-accessibility experiments, taking advantage of the fact that FHA contains no cysteine residues. We constructed a series of B. bronchiseptica strains – in both wildtype (WT) and prodomain-truncated (ΔPD) backgrounds – in which a single codon in the MCD-encoding region of fhaB was changed to a cysteine codon (Figure 2A), with substitutions regularly spaced ~50 aa apart. (We used B. bronchiseptica for these experiments so that the mutant strains could subsequently be compared with wildtype bacteria for their ability to establish respiratory infections in natural-host animal models.) The strains shown in Figure 2 are those that grew and adhered to L2 rat lung epithelial cells in a manner indistinguishable from wildtype B. bronchiseptica (data not shown) and they released FHA as efficiently as wildtype bacteria. Culture supernatants were concentrated under native conditions and incubated with DyLight 680 Maleimide, which forms a covalent bond with the sulfhydryl group on cysteine residues. Samples were then separated by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with an antibody that recognizes a central region of FHA (the carbohydrate recognition domain (CRD)) to determine the relative amounts of FHA contained in each sample. The anti-CRD antibody was detected with anti-chicken antibody conjugated to a near-infrared dye that fluoresces at 800 nm, shown in green in Figure 2B. The fluorophore of the DyLight 680 Maleimide fluoresces at 680 nm, shown in red in Figure 1D. For the A2382C substitution, and to a lesser extent, the A2432C substitution, FHA produced in the wildtype strain displayed greater DyLight-dependent fluorescence than FHA produced in the ΔPD strain, indicating that these cysteines were more accessible in the FHA produced by the wildtype strain than in the FHA produced by the ΔPD strain (Figure 2B). Conversely, for the V2081C, L2284C and I2233C substitutions, FHA produced by the wildtype strain displayed weaker fluorescence than FHA produced by the ΔPD strain. These data indicate that the accessibility of the cysteine residues differed depending on whether FhaB contained an intact prodomain and therefore provide further support for the conclusion that the prodomain affects the conformation of the MCD.
Figure 2.
The FhaB prodomain affects the surface-accessibility of MCD residues. (A) Schematic of B. bronchiseptica FhaB cysteine-substitution proteins. Single cysteine residues were substituted into wildtype (WT) and prodomain-truncated (ΔPD) proteins. (B) Cysteine-accessibility immunoblot of FHA recovered under native conditions from culture supernatants. The Western blots were immuno-stained with an anti-CRD antibody and displayed four isoforms of mature, released FHA. Native FHA was incubated with DyLight 680 Maleimide prior to SDS-PAGE.
The C-terminus of the prodomain remains in an intracellular compartment
We envisaged three mechanisms by which the FhaB prodomain could affect the conformation of the MCD on mature FHA: 1) the prodomain could function as a scaffold or ‘foldase’ for the MCD, analogous to the prodomains of many secreted proteases of Gram-positive bacteria (Shinde and Thomas, 2011); 2) the prodomain could remain in an intracellular compartment, functioning as a tether to constrain the possible conformations that the MCD can sample before attaining a stably folded state; 3) the prodomain could control the rate of translocation of the MCD to the cell surface and that controlled translocation could be required for proper folding (e.g., perhaps the N-terminal region needs to fold first, followed by the C-terminal region). In the first mechanism, the entire prodomain would be translocated to the cell surface before subsequent maturation (assuming that a fully folded MCD cannot be translocated through FhaC) while in the second and third mechanisms, the C-terminus of the prodomain would remain in an intracellular compartment. We therefore sought to determine the location of the prodomain during maturation of FHA. We were specifically interested in determining the location of the C-terminus of the prodomain before cleavage of FhaB to form FHA.
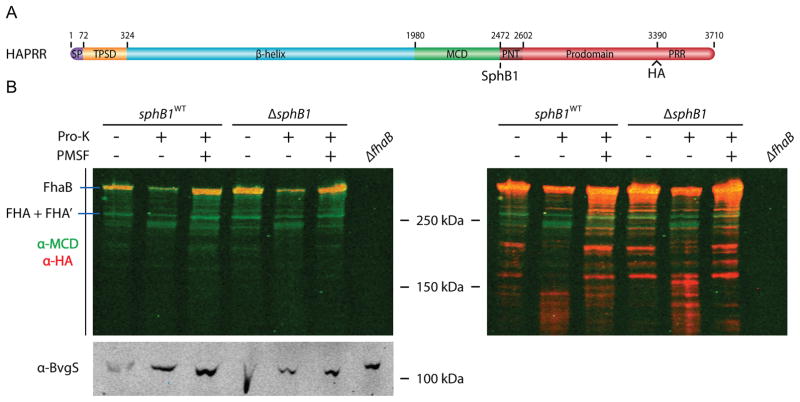
We first constructed a B. bronchiseptica strain (CTHA) producing FhaB with a hemagglutinin (HA) epitope seven residues N-terminal to the C-terminus of FhaB (Supplemental Figure 3A). Immunoblot analysis indicated that the HA epitope disrupted the stability of full-length FhaB (no full-length FhaB protein was detected in this strain by either the anti-MCD or anti-HA antibodies (Supplemental Figure 3B)), rendering the strain unsuitable for localization studies. We therefore constructed a B. bronchiseptica strain (HAPRR) producing FhaB with an HA epitope after residue 3265, just N-terminal to the proline-rich region in the FhaB prodomain (Supplemental Figure 3A). This epitope did not affect secretion, maturation, or release of FhaB, as the proportion of FhaB and FHA polypeptides was indistinguishable between this strain and wildtype bacteria (Supplemental Figure 3B). Moreover, preliminary data suggest this strain is indistinguishable from wildtype B. bronchiseptica in its ability to cause respiratory infection in mice (our unpublished results).
To determine if the prodomain remains in an intracellular location, the HAPRR strain was grown under FHA-inducing conditions, cells were harvested by centrifugation and suspended in buffer with 1 μg/mL proteinase-K to cleave surface-exposed proteins, washed to remove extracellular protein fragments, then whole-cell lysates (WCLs) were prepared and analyzed by immunoblot (Figure 3B. Note that the left and right panels in Figure 3B are the same immunoblot. The intensity of the red channel was increased in the panel on the right to facilitate visualization of polypeptides detected by the anti-HA antibody. Similar manipulation of the green channel did not reveal any additional bands). In the untreated sample, full-length, unprocessed FhaB (~375 kDa) was detected by both anti-MCD and anti-HA antibodies and mature FHA (FHA and FHA’, ~250 kDa) was detected by only the anti-MCD antibody. Some polypeptides of ~200 kDa were also detected by the anti-HA antibody (evident in the right panel), suggesting they result from proteolysis near the N-terminus of FhaB. Since FHA is anchored to the cell surface by its N-terminus, we hypothesize that these polypeptides result from proteolysis during sample preparation. Consistent with previous results indicating that the prodomain is degraded rapidly after FhaB processing (Delisse-Gathoye et al., 1990; Mazar and Cotter, 2006), no polypeptide of ~130 kDa was recognized by the anti-HA antibody in the untreated samples. Proteinase-K treatment caused a decrease in the amount of full-length FhaB (and mature FHA) and PMSF inhibited this proteolysis, indicating that this proteolysis was proteinase-K-dependent. Concomitant with the proteinase-K-dependent decrease in FhaB was the appearance of polypeptides of ~230 kDa that were recognized by only the anti-MCD antibody (green channel) and polypeptides of ~120–140 kDa that were recognized by only the anti-HA antibody (Figure 3B, red channel in the right panel). Proteinase-K activity was confined to the extracellular space, as indicated by the integrity of BvgS, a sensor kinase that spans the cytoplasmic membrane and contains a large periplasmic domain. The ~230 kDa polypeptide that predominates after proteinase-K treatment is most likely the N-terminus and β-helical shaft region of FHA, consistent with β-helical proteins’ resistance to proteinase-K (Ieva and Bernstein, 2009; Junker et al., 2009). The appearance of cell-associated HA-containing polypeptides of ~120–140 kDa after proteinase-K treatment, but not before proteinase-K treatment, indicates that they arose from polypeptides in which the C-terminus of the prodomain was intracellular while the central region of FhaB (most of the β-helical shaft, the MCD, and perhaps the N-terminus of the prodomain) was extracellular. In the ΔsphB1 strain, more polypeptides in the range of ~120–160 kDa were recognized by the anti-HA antibody in the proteinase-K treated sample than in the proteinase-K treated sample of the strain that is wildtype for sphB1 (Figure 3B, right). These data suggest that the C-terminus of the prodomain remains in an intracellular compartment during outer membrane translocation of the β-helix and MCD regions, but these data cannot distinguish between periplasmic versus cytoplasmic localization of the prodomain C-terminus.
Figure 3.
The C-terminus of the prodomain resides intracellularly during FHA maturation. (A) Schematic of B. bronchiseptica HA-tagged FhaB protein used in proteinase-K digestion experiments. The site of the HA tag is noted. (B) Left, anti-MCD and anti-HA immunoblot of proteinase-K treated whole-cells, lysed to evaluate cellular protein content. PMSF was added prior to proteinase-K incubation as indicated. The panel on the right is an analysis of the immunoblot on left, with the intensity of the 700 nm (anti-HA) channel increased for visualization of proteinase-K dependent intracellular prodomain. Bottom, anti-BvgS immunoblot of whole-cell lysates.
The region surrounding FhaB maturation sites is required for the C-terminus of the prodomain to remain in an intracellular compartment and for mature FHA to be released from the cell surface
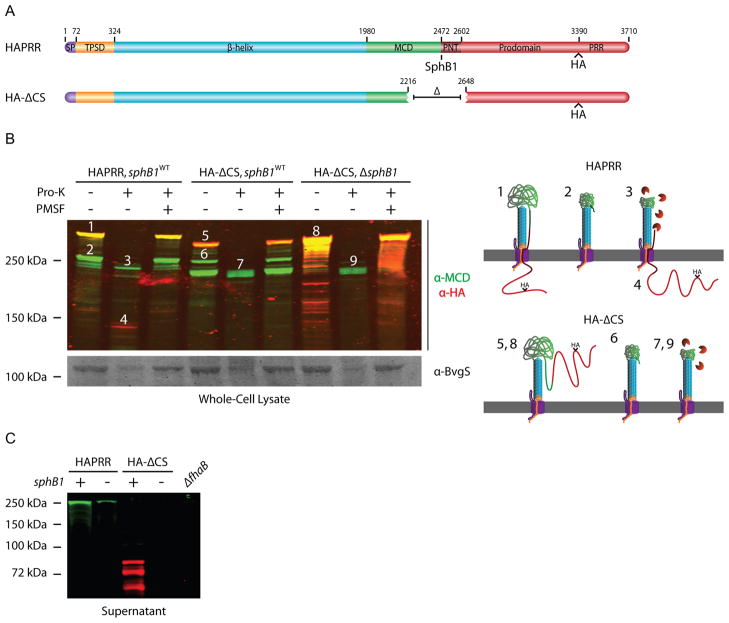
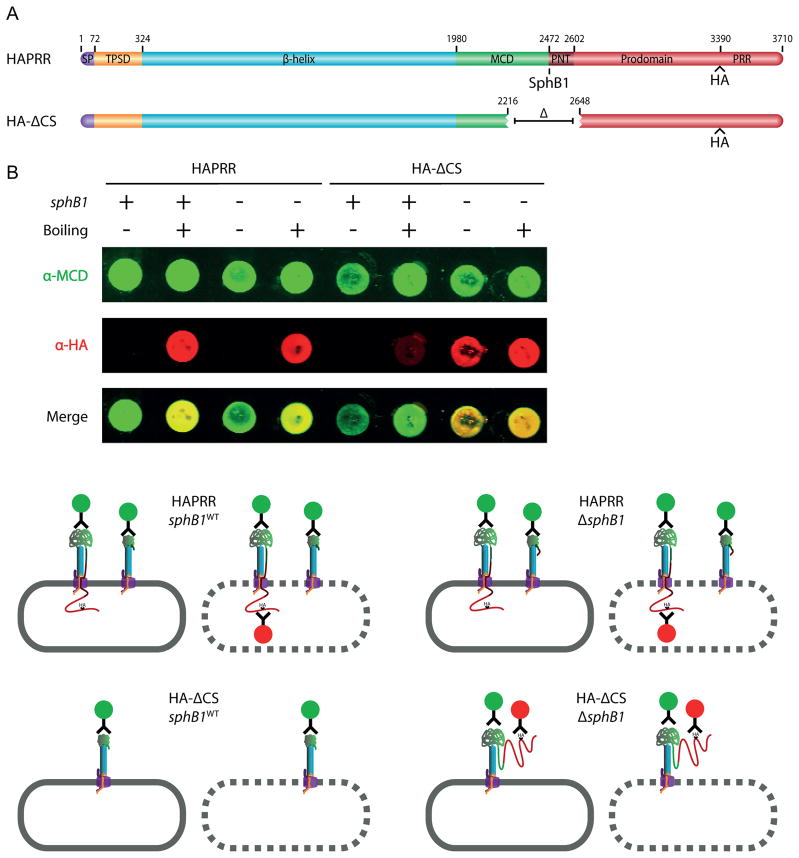
Although the data shown in Figure 3 suggest an intracellular location for the FhaB prodomain, the experiment is complicated by the fact that FhaB is processed by both SphB1 and one or more unidentified proteases, making it difficult to ‘capture’ the pre-cleavage FhaB secretion intermediate. In an attempt to eliminate both SphB1-dependent and SphB1-independent processing, we constructed a strain (HA-ΔCS) producing an FhaB protein with a 431 aa deletion encompassing the known SphB1-dependent cleavage sites and the region that becomes the C-terminus of FHA’, which is produced by cleavage/degradation by the unidentified protease(s). The FhaB produced by this strain contains an HA epitope N-terminal to the PRR, the same as in the HAPRR strain (Figure 4A). Unprocessed FhaB in the HA-ΔCS strain was smaller than FhaB in wildtype bacteria (consistent with the size of the deletion) and was detected with both anti-MCD and anti-HA antibodies (Figure 4B, band 5. Illustrations to the right of Figure 4B illustrate how we envision FhaB/FHA exist on the cell surface with and without proteinase-K treatment. Band numbers on the Western blot correspond to illustration numbers). Unexpectedly, FhaB was still processed in this strain, resulting in the formation of polypeptides of ~250 and ~220 kDa (Figure 4B, bands 6 (green)) that were detected only by the anti-MCD antibody and therefore represent the N-terminal ~2300–2500 aa of FhaB. Processing to form these polypeptides did not occur in a strain that was isogenic except for deletion of sphB1 (referred to as HA-ΔCS,ΔsphB1), indicating that the processing was SphB1-dependent (Figure 4B). (Note, however, that this strain contained an increased amount of FhaB in WCL and many breakdown products were present that were recognized by the anti-HA antibody and therefore resulted from degradation from the N-terminus of FhaB, presumably during sample preparation.) Proteinase-K treatment of the HA-ΔCS strain resulted in cleavage of FhaB and FHA (Figure 4B, band 7), but no polypeptides recognized by the anti-HA antibody were generated that could be detected in WCL. This result suggested that, in contrast to the case with wildtype bacteria, the C-terminus of the prodomain was not retained intracellularly in the HA-ΔCS strain. Comparison of proteins present in culture supernatants showed that wildtype and ΔsphB1 bacteria released FHA and FHA’, respectively, and did not release HA-containing polypeptides into the extracellular environment (Figure 4C). By contrast, the HA-ΔCS strain released a large amount of HA-containing polypeptides of about 50–80 kDa, but released no mature FHA into the culture supernatant. (Proteins were separated using an 8% polyacrylamide gel for the immunoblot shown in Figure 4C, which allows lower molecular weight proteins to be visualized, but limits resolution of higher molecular weight proteins.) The prodomain, therefore, appears to not be retained intracellularly in the HA-ΔCS strain and not only are prodomain polypeptides released into the culture supernatant, they are relatively stable. In the HA-ΔCS,ΔsphB1 strain, FhaB remained predominantly unprocessed, as WCL contained mostly full-length FhaB and several breakdown products, but no polypeptides corresponding to FHA. Proteinase-K treatment of the HA-ΔCS,ΔsphB1 strain resulted in the formation of a predominant polypeptide of about 220 kDa that was recognized by the anti-MCD antibody but not the anti-HA antibody (Figure 4B, band 9), therefore corresponding to the β-helical shaft and part of the MCD. No HA-containing polypeptides were released into the culture supernatant of the HA-ΔCS,ΔsphB1 strain, consistent with the conclusion that processing of FhaB does not occur in this strain and that the HA-containing polypeptides present in the supernatants of the HA-ΔCS strain were extracellularly located prodomain fragments released after SphB1-dependent processing. Together, these data indicate that sequences within the region deleted in the HA-ΔCS strain are required for retention of the C-terminal region of the prodomain in an intracellular compartment and are also required for release of mature FHA into the extracellular environment. Although some degradation of BvgS by proteinase-K was evident in this experiment, indicating that some amount of proteinase-K entered the cells, the presence of large amounts of HA-stained polypeptides of ~75 kDa in supernatants of the HA-ΔCS strain, but no HA-containing polypeptides in supernatants of wildtype B. bronchiseptica, strongly suggests that the prodomain is retained in an intracellular compartment in wildtype bacteria but not in HA-ΔCS bacteria. Complete elimination of HA-containing polypeptides in the HA-ΔCS,ΔsphB1 strain after proteinase-K treatment also supports this conclusion.
Figure 4.
The region surrounding the native SphB1-dependent cleavage site regulates FHA release and keeps the C-terminus of the prodomain intracellular. (A) Schematic of proteins used in proteinase-K digestion experiments. (B) Top, anti-MCD and anti-HA immunoblot of whole-cell lysate of the cleavage-site deletion strain. Cells were incubated with proteinase-K as indicated. PMSF was added prior to proteinase-K incubation as indicated. Numbers above protein bands correspond to illustrations on the right side of the figure. Illustrations 1–4 represent products of the HAPRR strain, while illustrations 5–9 represent products of the HA-ΔCS strain. These illustrations show how we envision FhaB/FhaC exist at the outer membrane (OM, gray), with the area above the OM representing the extracellular space and the region below the OM representing the intracellular space. Proteinase-K is colored orange. Illustrations are not drawn to scale. Bottom, anti-BvgS immunoblot of whole-cell lysates. (C) Anti-MCD and anti-HA immunoblot of concentrated supernatants. Samples were run on an 8% polyacrylamide gel to resolve the released forms of FHA and prodomain.
As an alternate approach, we performed dot blots of the HAPRR and HA-ΔCS strains (Figure 5). Whole cells in PBS were applied to a nitrocellulose membrane for detection of surface antigens and boiled lysates were applied to the membrane for detection of total cellular protein content. Whole cells of the HAPRR strains (sphB1WT and ΔsphB1 backgrounds) were stained by only the anti-MCD antibody (Figure 5B; note that the schematics below the dot blots indicate only the various polypeptide species, not their relative abundances). Boiling the samples however, allowed detection by both the anti-MCD and anti-HA antibodies, indicating that the C-terminus of the prodomain in the HAPRR strains was located intracellularly. In the HA-ΔCS strain (sphB1WT), there was no surface detection of HA epitopes, consistent with the previous result indicating SphB1-dependent cleavage and release of a surface-localized prodomain in this strain. Boiling the sample resulted in weak detection of the HA tag, which we hypothesize represents FhaB molecules prior to translocation across the outer membrane. In the HA-ΔCS,ΔsphB1 strain, detection of both surface proteins and cell lysate revealed MCD and HA epitopes, consistent with idea that the prodomain crossed the outer membrane, but was not cleaved in the absence of SphB1. (Note: The western blot in Fig. 4B indicates that some full-length, surface-exposed FhaB is present in the HA- CS,sphB1WT strain (band 5). Although very faint staining of this strain by the anti-HA antibody was apparent in some dot blot experiments, it is not apparent in the one shown in Fig. 5B. Lack of significant staining by dot blot compared to western blot may be due to differences in sample preparation and differences in sensitivity of the two assays.) As a control, a B. bronchiseptica strain without an HA tag in FhaB (RBX11) was used in an identical experiment. Although both whole cells and boiled lysates were recognized by the anti-MCD antibody, neither were recognized by the anti-HA antibody (Supplemental Figure 4B), demonstrating that there was no nonspecific fluorescence in our samples probed with the anti-HA antibody. Taken together, these data indicate that the C-terminus of the prodomain is retained in an intracellular compartment (either the periplasm or the cytoplasm) during translocation and maturation of FHA and that the prodomain is degraded rapidly after SphB1-dependent and/or SphB1-independent processing when retained intracellularly. Sequences within the region deleted in the HA-ΔCS strain are required for the C-terminus of the prodomain to remain intracellular and for mature FHA to be released from the cell surface.
Figure 5.
The prodomain C-terminus of full-length FhaB remains intracellular. (A) Schematic of proteins used in dot blot experiments. (B) At the top, anti-MCD and anti-HA immuno-dot-blot of HAPRR and HA-ΔCS strains. Normalized amounts of whole cells and boiled lysates were applied. Below, illustrations of immuno-stained samples. The rounded gray rectangle represents the B. bronchiseptica outer membrane, with a solid edge representing the outer membrane of an intact cell and a dashed edge representing an outer membrane disrupted by boiling. MCD staining is represented by green-circled antibodies and HA staining is represented by red-circled antibodies.
The C-terminus of the MCD controls FHA release competency while the N- terminus of the prodomain controls intracellular prodomain retention
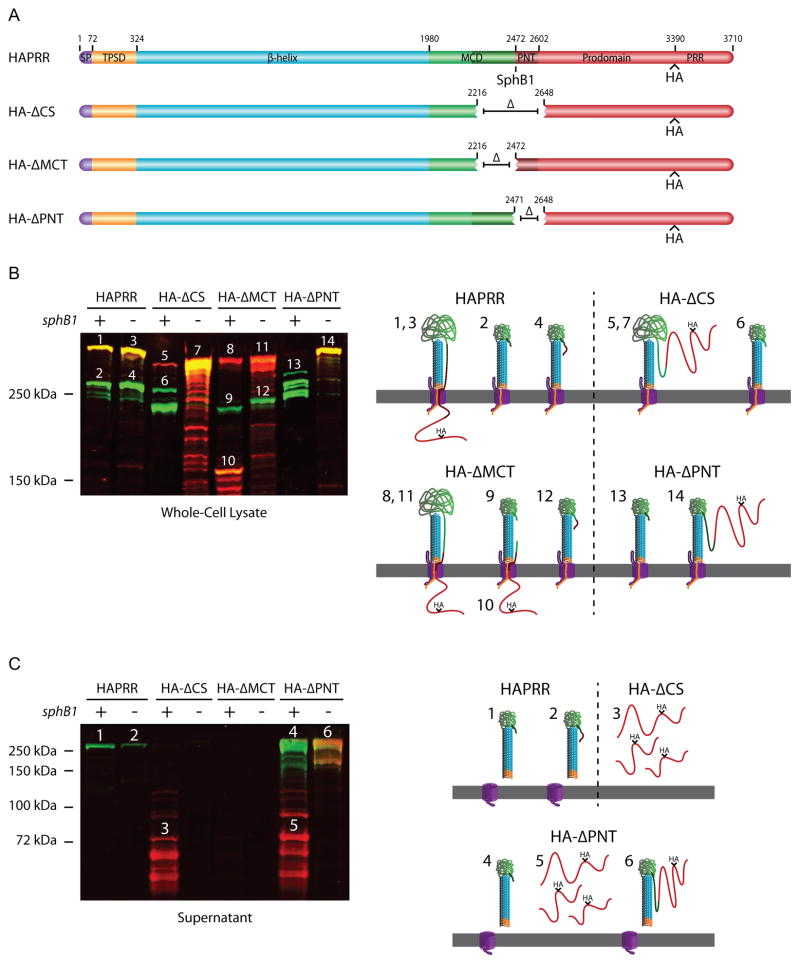
To investigate which sequences within the region deleted in the HA-ΔCS strain were responsible for retention of the prodomain in an intracellular compartment and for release of mature FHA, we constructed strains producing FhaB proteins with either a 255 aa deletion of the MCD C-terminus (the MCT, shaded dark green in Figure 6) or a 176 aa deletion of the prodomain N-terminus (the PNT, shaded dark red). These strains, which contain the sequence encoding an HA epitope just 5’ to the PRR-encoding region, were named HA-ΔMCT and HA-ΔPNT, respectively. The ΔPNT deletion encompasses the primary SphB1-dependent cleavage site (the sequence of which is PLFETRIKFID).
Figure 6.
The MCT controls FHA release and the PNT keeps the prodomain C-terminus intracellular. (A) Schematic of FhaB proteins used in experiment. The MCT is represented as dark green. (B) Anti-MCD and anti-HA immunoblot of whole-cell lysates. Numbers above protein bands correspond to illustrations on the right side of the figure. Illustrations 1–4 represent products of the HAPRR strain, 5–7 represent products of the HA-ΔCS strain, 8–12 represent products of the HA-ΔMCT strain, and 13 and 14 represent products of the HA-ΔPNT strain. Layout of these illustrations is as described in Figure 4B. (C) Anti-MCD and anti-HA immunoblot of concentrated supernatants. Numbers above protein bands correspond to illustrations on the right side of the figure. Illustrations 1 and 2 represent products of the HAPRR strain, 3 represents the product of the HA-ΔCS strain, and 4–6 represent products of the HA-ΔPNT strain. Layout of these illustrations is as described in Figure 4B. Deletion of the MCT prevents FHA release, while deletion of the PNT promotes release. Additionally, deletion of the PNT results in translocation of the prodomain into the extracellular space where it can be separated from FHA in an SphB1-dependent manner.
Immunoblot analysis (resolved by 5% polyacrylamide SDS-PAGE) showed that both unprocessed and processed FhaB/FHA polypeptides were detectable in WCL of the HA-ΔMCT strain (Figure 6B, bands 8 and 9, respectively. Bands of the Western blot and illustration are labeled as in Figure 4B). In addition, a polypeptide of ~150 kDa that was recognized by both anti-MCD and anti-HA antibodies (Figure 6B, band 10), as well as a few slightly smaller polypeptides, were detected. Recognition of the ~150 kDa polypeptide by both antibodies indicates that it includes the prodomain and at least part of the MCD. These polypeptides were not detected in WCL of the HA-ΔMCT strain that also contained the ΔsphB1 deletion, demonstrating that they resulted from SphB1-dependent cleavage. The fact that analogous polypeptides are not present in the HAPRR strain suggests that SphB1-independent cleavage/degradation is inefficient in the HA-ΔMCT strain, since SphB1-independent cleavage would have separated the MCD part of the polypeptide from the C-terminus. The presence of these polypeptides in WCL, but not in supernatants (Figure 6C; proteins in culture supernatants were resolved using an 8% polyacrylamide gel, allowing lower molecular weight proteins to be visualized at the expense of separating higher molecular weight proteins), also indicates that the C-terminus of the prodomain is present in an intracellular compartment in this strain as these polypeptides would have been released into the supernatant fraction if they had been located extracellularly. In WCL of the HA-ΔMCT strain that also contains the ΔsphB1 deletion, more unprocessed FhaB protein was detected, as well as a polypeptide of about 230 kDa, which is presumably a product of cleavage by the unidentified protease(s) (Figure 6B, bands 11 and 12, respectively). No polypeptides of any size were detected by either antibody in supernatant fractions of the HA-ΔMCT strain, regardless of whether sphB1 was wildtype or deleted, indicating that the MCT is required for FHA release. The MCT, therefore, is required for release of FHA, but not for intracellular retention of the prodomain.
In the HA-ΔPNT strain, FhaB was extensively processed, resulting in the formation of several polypeptides of around 250 kDa (Figure 6B, bands 13). The processing was SphB1-dependent as only full-length FhaB, which was recognized by both the anti-MCD and anti-HA antibodies, was present in WCL of the HA-ΔPNT strain that also contained the ΔsphB1 deletion (Figure 6B, band 14). The HA-ΔPNT strain released a large amount of ~250 kDa polypeptides that were recognized by only the anti-MCD antibody and lower molecular weight (~50–100 kDa) polypeptides that were recognized only by the anti-HA antibody, into the culture supernatant (Figure 6C, bands 4 and 5, respectively). The HA-ΔPNT strain with the ΔsphB1 deletion released a large amount of full-length protein that was recognized by both antibodies (Figure 6C, band 6). These results indicate that sequences within the PNT are required for retention of the C-terminus of the prodomain in an intracellular compartment and are not required for release of FhaB/FHA from the cell surface. In fact, the HA-ΔPNT strain releases more FhaB/FHA from the surface than the wildtype strain, suggesting that the PNT influences FHA release in a negative manner.
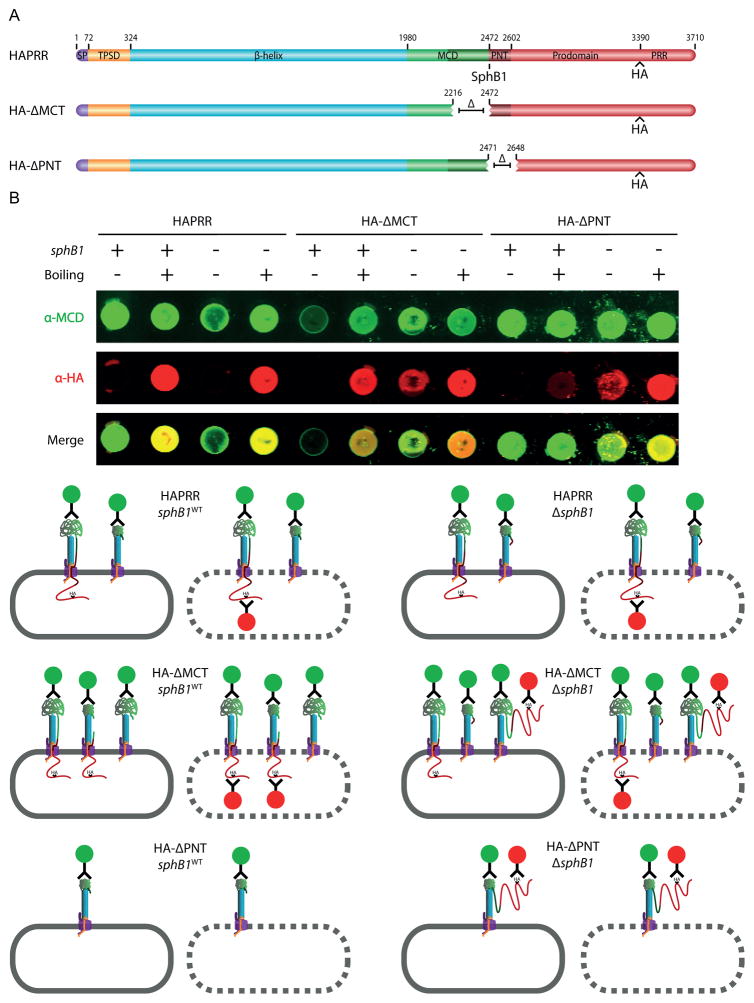
To further investigate the location of the prodomain in the HA-ΔMCT and HA-ΔPNT strains, we performed dot blots (Figure 7). In the HA-ΔMCT strain (sphB1WT), only the MCD, and not the HA epitope, was detected on the surface of whole cells (Figure 7B; note that as with Fig. 5B, the schematics below the dot blots indicate only the various polypeptide species, not their relative abundances). Lysing these cells resulted in detection of the HA tag, indicative of an intracellular prodomain C-terminus. Whole cells of the ΔMCTΔsphB1 strain were detected at a low level by the anti-HA antibody, suggesting that some prodomain C-terminus was surface-localized. This result suggests that lack of the MCT can overcome, to some extent, the ability of the PNT to block translocation of the prodomain through FhaC to the cell surface. If so, then in the ΔMCT strain that is wildtype for sphB1, the small amount of prodomain that is translocated to the surface would be cleaved in an SphB1-dependent manner and released into the supernatant. Indeed, a closer analysis of concentrated supernatants revealed that the ΔMCT strain released cleaved prodomain polypeptides, albeit much less efficiently than the ΔCS and ΔPNT strains (Supplemental Figure 5B). Boiling the ΔMCTΔsphB1 strain increased detection by the anti-HA antibody, indicating that although some prodomain crossed the outer membrane, most of it was retained intracellularly.
Figure 7.
The prodomain of the HA-ΔMCT strain is mostly intracellularly-localized while that of the HA-ΔPNT strain is extracellularly-localized. (A) Schematic of proteins used in dot blot experiments. (B) At the top, anti-MCD and anti-HA immuno-dot-blot of HAPRR, HA-ΔMCT and HA-ΔPNT strains. Normalized amounts of whole cells and boiled lysates were applied. Below, illustrations of immuno-stained samples. Layout of these illustrations is as described in Figure 5B.
In the HA-ΔPNT strain (sphB1WT), no HA epitopes were detected on the surface of whole-cells (Figure 7B), consistent with the previous observation of extensive SphB1-dependent cleavage of surface-localized prodomain (which was also observed in the HA-ΔCS strain). Boiling these cells resulted in weak detection of the HA tag, which, as with the HA-ΔCS strain, likely represents FhaB prior to translocation across the outer membrane. In the HA-ΔPNT,ΔsphB1 strain, both the anti-MCD and the anti-HA antibodies were able to detect surface protein, consistent with the prodomain crossing the outer membrane but not being cleaved in the absence of SphB1. Collectively, these data implicate to the PNT as the subdomain responsible for intracellular localization of the prodomain C-terminus.
Discussion
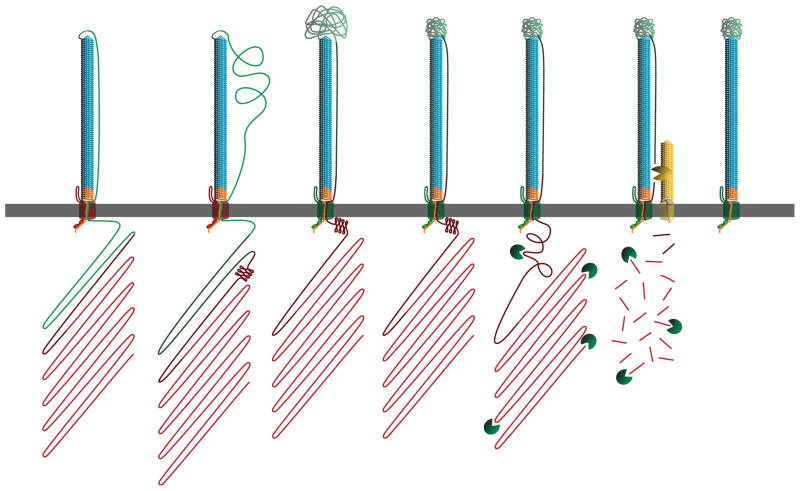
The experiments described in this work yielded two primary findings: 1) the FhaB prodomain remains in an intracellular compartment during FHA biogenesis and 2) the prodomain affects the conformation of the FHA MCD. Our results also show that sequences within the C-terminal half of the MCD (the MCT) are required for FHA to be released from the cell surface and that sequences at the N-terminus of the prodomain (the PNT) are required for the C-terminus of the prodomain to be retained intracellularly. Based on these results, we propose a revised model for secretion by the FhaB-subfamily of TPS systems. The revisions relate primarily to events occurring after the β-helical shaft has been exported to the cell surface (Figure 8). The model states that as the MCT is secreted, FhaC changes from a release-incompetent state to a release-competent state (the shift in FhaC release competency is indicated in Figure 8 by a change in FhaC color from red to green). Subsequent FhaB transit through FhaC is halted by the PNT, which we postulate folds into a conformation that is incompatible with translocation. According to this model, the prodomain functions as a tether that constrains the conformations that the extracellularly-located MCD may sample while folding into a functional structure. Alternatively, the prodomain may control the rate of translocation of the MCD, with rate-limited translocation being required for the MCD to fold properly. We hypothesize that upon achieving a functional conformation, the MCD signals maturation-cleavage readiness, resulting in FhaB cleavage by the unidentified protease(s). The prodomain is rapidly degraded and additional SphB1-dependent processing occurs. Fully matured FHA is then released into the extracellular environment by an, as yet, unknown mechanism.
Figure 8.
Revised model for FHA biogenesis. Our model continues from Supplemental Figure 1A, with the β-helix surface-localized and the MCD and prodomain still inside the cell. During the secretion process, the PNT adopts a conformation that prevents prodomain transit through FhaC. Next, the MCT of FhaB translocates to the extracellular space, priming FhaC for release of FHA, as indicated by the shift in FhaC color from red to green. The translocation-impaired PNT confers tension across the extracellular space to the MCD, restricting the conformations the MCD may sample while folding. FhaB undergoes maturation into FHA upon completion of MCD folding, with FHA maturation being accomplished by SphB1-independent cleavage, degradation of the prodomain and SphB1-dependent cleavage. At this point, FHA is fully mature and can remain either surface-associated or be released into the supernatant.
We reported previously that the prodomain is required for FHA function in vivo and we also provided evidence that the MCD is an important functional domain, if not the functional domain, of FHA during infection (Mazar and Cotter, 2006; Julio et al., 2009). In this study we used semi-native PAGE, proteinase-K digestion, and cysteine residue accessibility experiments to determine that the FhaB prodomain affects the conformation of the MCD. These results indicate that the prodomain functions as an intramolecular chaperone that controls the folding of the MCD. Proteinase-K digestion and dot blot experiments demonstrated that the prodomain remains intracellular while the MCD is extracellular, but they did not reveal whether the C-terminus of the prodomain remains in the cytoplasm or the periplasm. Regardless of its exact intracellular location, the FhaB intramolecular chaperone domain (the prodomain) and the functional domain it controls (the MCD) are separated by at least one membrane. Although intramolecular chaperones that affect the conformation of functional domains have been described, they typically co-localize with their functional domains, acting as molecular scaffolds that guide the folding of the active domain of the mature protein via direct protein-protein interactions. The best-characterized example of this family of intramolecular chaperones is subtilisin E of B. subtilis (Shinde and Thomas, 2011). The FhaB prodomain appears to function in an entirely different manner since it does not co-localize with the conformationally mature MCD. The simplest model is that the prodomain, via its intracellular location, functions as a tether that constrains the folding space of the MCD such that its functional conformation is the preferred fold amongst those available for sampling. Although a more complex scenario, it is also possible that the FhaB prodomain, either directly or indirectly from inside the cell, influences the translocation rate of the MCD such that the MCD is translocated across the outer membrane in a manner compatible with its proper folding on the cell surface. To our knowledge, neither of these mechanisms for the control of protein folding has been described previously. However, a similar concept has been described in the case of the E. coli autotransporter EspP. In this case, it is hypothesized that, prior to cleavage by leader peptidase, a cytoplasmically-located extended signal peptide alters the conformation of the periplasmically-located β-domain and, consequently, its ability to integrate into the outer membrane (Szabady et al., 2005).
Our experiments identified the PNT as the subdomain responsible for intracellular retention of the prodomain. One mechanism by which the PNT may mediate intracellular prodomain retention is by folding into a conformation incompatible with translocation through FhaC (Figure 8). Alternatively, the PNT may interact with another molecule or molecular complex that prevents export through FhaC. Whatever the mechanism, because this subdomain is conserved amongst TpsA proteins of the FHA subfamily, intracellular retention of C-terminal prodomains and the mechanism by which PNTs function may also be conserved.
As suggested by our model, we do not believe that the only role for the prodomain is to function as a tether (if the tether hypothesis is even correct). We showed previously that a B. bronchiseptica strain that produces an FhaB protein lacking only the C-terminal 339 aa (and hence containing an intact PNT) is defective for tracheal colonization in a rats (Mazar and Cotter, 2006) and we showed in this work that insertion of an HA epitope near the extreme C-terminus of FhaB resulted in an inability to detect full-length FhaB, suggesting the importance of this region for protein stability and/or resistance to protease digestion. These data provide evidence that sequences C-terminal to the PNT are required for prodomain function. We and others have also noted the presence of a proline rich region near the C-terminus of the prodomain, but the role of this region is currently unknown. We hypothesize that proper folding of the MCD on the cell surface is somehow sensed by the prodomain to trigger continued FHA maturation (i.e., to stimulate cleavage and degradation of the prodomain and then release of mature FHA). This would occur regardless of whether the prodomain functions as a tether or a translocation rate limiter and we hypothesize that regions of the prodomain C-terminal to the PNT are important for these steps in FHA biogenesis. If the prodomain functions as a limiter of MCD translocation rate, then sequences C-terminal to the PNT may also be involved in somehow controlling the rate of translocation through FhaC, possibly by transiently influencing the conformation of the MCD in the periplasm. Our future studies will be aimed at testing these hypotheses.
Although SphB1-independent proteolysis of FhaB to yield FHA* and FHA’ was apparent in previous studies (Coutte et al., 2001; Mazar and Cotter, 2006), this cleavage was considered ‘aberrant’ and occurring primarily in the absence of SphB1 (Coutte et al., 2001). In this work, we observed two circumstances in which prodomain polypeptides were generated that were not cleaved by the unidentified protease. In each case, the prodomain polypeptides were stable. The first instance was after proteinase-K digestion of the HAPRR strain (sphB1WT and ΔsphB1). The second was in the ΔMCT strain, in which FhaB underwent SphB1-dependent cleavage, but not SphB1-independent proteolysis. The fact that stable prodomain polypeptides were present in both of these cases, despite the fact that they were localized intracellularly and hence presumably accessible to the unidentified protease(s), suggests that the unidentified protease(s) is responsible for degradation of the prodomain and that it does so only in response to the normal sequence of events that occurs during FhaB translocation and maturation. We speculate, therefore, that cleavage/degradation by the unidentified protease(s) is an important regulated event during FHA biogenesis and we are currently working to identify the protease(s), determine its spectrum of activity, and understand how it is regulated.
While our results suggest a role for the unidentified protease(s), they failed to reveal an important role for SphB1 with regard to FHA biogenesis. SphB1-dependent cleavage of FhaB occurs at multiple locations and with varying efficiencies in wildtype B. bronchiseptica and B. pertussis. Target promiscuity is especially evident in the HA-ΔCS strain, in which, despite a sizeable deletion surrounding the PLFETRIKFID region, SphB1-dependent cleavage of FhaB occurred at alternate sites. Furthermore, the FhaB prodomains in the HA-ΔCS and HA-ΔPNT strains reached the extracellular space, where they were cleaved in an SphB1-dependent manner, suggesting that SphB1-dependent cleavage occurs regardless of prodomain location. Although ΔsphB1 mutants of B. pertussis and B. bronchiseptica are severely defective in mouse models of infection ((Coutte et al., 2003) and our unpublished data), this virulence defect cannot be attributed solely to altered processing of FhaB because SphB1-dependent cleavage of other Bordetella proteins occurs (our unpublished data). It is also possible that SphB1 interacts with host cells in a manner independent of its affects on other bacterial proteins. Understanding the roles SphB1 plays in Bordetella biology, therefore, remains a challenging and currently unattained goal.
When grown in broth, B. pertussis and B. bronchiseptica release a substantial amount of FHA into the culture medium. As FHA contributes to adherence of the bacteria to eukaryotic cells in vitro more than any other Bordetella virulence factor (Relman et al., 1990; Cotter et al., 1998; Mattoo et al., 2000), the fact that FHA is so efficiently released from the cell surface has been perplexing. Recent data have indicated that FHA also functions to modulate the inflammatory response (Inatsuka et al., 2005; Julio et al., 2009; Henderson et al., 2012). Although in vitro studies suggest that the released form of FHA can affect host cell signaling (McGuirk and Mills, 2000; Abramson et al., 2008; Dieterich and Relman, 2011), whether the released form of FHA plays an important role during infection is unknown. The major obstacle to addressing this question is our current inability to construct a strain that is defective only for FHA release. Previous studies with B. pertussis showed that although short FHA molecules corresponding to the N-terminal ~30 and ~80 kDa of the protein (the TPS domain and part of the β-helical shaft) were efficiently released into culture supernatants, longer molecules that included most of the β-helical shaft or all of the β-helical shaft plus the N-terminal half of the MCD were poorly released (Renauld-Mongenie et al., 1996). If the initially synthesized FhaB polypeptide contained most of the prodomain, however, mature FHA was efficiently released (Renauld-Mongenie et al., 1996). These data suggest that sequences within the C-terminal half of the β-helical shaft render the system incompetent for release and that sequences C-terminal to that region restore release competency. Our experiments now reveal a critical role for the MCT in allowing FHA release, suggesting that sequences within this region somehow cause the FhaB/FhaC complex to switch from a release-incompetent to release-competent state. How this switch might occur mechanistically is completely unknown, but we are optimistic that further investigation will reveal an understanding to a level that will allow the construction of strains that fail to release otherwise functional FHA proteins so that the role of FHA release during infection can be investigated.
Although our results have provided insight into the mechanism of FHA secretion and maturation, they also raise the question of why FHA maturation and release is so complex, and they remind us that data obtained by studying bacteria in culture in the laboratory must be interpreted with caution. During infection, Bordetella spp produce FHA while residing on the mucosal surface of the respiratory tract. We and others have assumed that the “mature” ~250 kDa FHA protein is the functional form during infection. However, it is possible that precleaved FhaB (currently considered a secretion intermediate) performs an important biological role. Our data suggest that the prodomain confers tension across the outer membrane, restraining mobility of the MCD, and we propose that once the MCD has folded, conformational information may also be sent back across the membrane to trigger cleavage and degradation of the prodomain by the unidentified protease(s). The potential for the MCD to communicate signals across the outer membrane to the prodomain raises the intriguing possibility that those signals could be generated in response to events that occur during infection, such as binding to specific host cell receptors. Moreover, the possibility exists that those signals could affect processes in the bacterial cell in addition to those related to FHA maturation, such as, perhaps, signal transduction and gene expression changes. Our future studies will be aimed at testing these hypotheses.
Experimental Procedures
Bioinformatics
Proteins containing regions of similarity to the FhaB PNT were identified with a PSI-BLAST search. ClustalW alignment was used to identify similar and identical residues amongst the resulting TpsA proteins. WebLogo 3 was used for presentation of the ClustalW alignment (Crooks et al., 2004).
Adherence assay
Adherence to rat lung epithelial cells (L2) was performed as described (Cotter et al., 1998).
Semi-native polyacrylamide gel analysis of FHA MCD
25 mL overnight cultures of BPSMAQ and BPSMAQT-N were grown to OD600 of 3–5. Cultures were spun down at 11,000 rpm for 10 minutes at 4°C. Supernatants were collected and sterilized using a Millex-GV syringe driven filter units by Millipore (0.22 μm PVDF). 2 ml of ProBond slurry were added to filter-sterilized supernatants and the mixes were incubated end-over-end for one hour at room temperature. The slurry-supernatant mixes were spun at 3,500 rpm for five minutes and supernatant set aside. The pelleted resin was washed twice in Native Binding Buffer and twice in Native Washing Buffer. The pelleted resin was then washed twice in 250 μL of Native Elution Buffer (500 mM Imidazole), with each wash and the resin being saved for analysis. Samples can be stored overnight at 4°C. Samples were resolved on 6% SDS-PAGE gel (10 μL loaded).
The gel was stained by coomassie blue to assess elution yields. The elution with the highest FHA yield was dialyzed in 1 L sterile PBS pH 7.4 overnight at 4°C to remove excess Imidazole. 20x TEV buffer, 1 μL (10 units) AcTEV protease and 1 mM DTT (final) were added to dialyzed samples. Samples were incubated at 30°C for six hours. 250 μL of ProBond resin were added to the samples and samples were incubated end-over-end overnight at 4°C to remove N-terminal portion of TEV protease-cleaved FHA from the supernatant, leaving behind cleaved MCD in the flow-through. 15 μL of the flow-through sample was added to 15 μL of 2x Native Sample Buffer. Samples were incubated at room temperature or 95°C for 5 minutes. Samples were resolved on 0.6% and 2.0% SDS-PAGE gels.
Proteinase-K digest of purified MCD
For digest of native MCD, 100 ng of protein was digested with various concentrations of proteinase-K (from 0 to 100 μg/mL) for 15 minutes at RT. 2x sample buffer was then added to each digest. After boiling, samples were separated using SDS-PAGE, transferred to nitrocellulose, and probed with the anti-MCD antibody. For digest of denatured/renatured MCD, 1 μg of protein was incubated in 50 μL 6 M Guanidinium for 30 minutes at 50°C, then raised to 1 mL with PBS and concentrated using 10 kDa MW cut-off centrifugation filters. The sample was then resuspended in PBS and incubated for 30’ at 37°C, vortexing consistently. Samples were then split, loading 1/10 of each sample into individual tubes and digested with various concentrations of proteinase-K (from 0 to 100 μg/mL) for 15 minutes at RT. 2x sample buffer was then added to each digest. After boiling, samples were separated using SDS-PAGE, transferred to nitrocellulose, and probed with the anti-MCD antibody. Densitometry analyses were performed using ImageJ (Schneider et al., 2012).
Accessibility of cysteine substitutions in the MCD
Overnight cultures of RBX11, RBX11-T-N, their respective cysteine substitutions and RBX20 were grown overnight. Cultures were spun down at 14,000 rpm for three minutes. Supernatants were collected and sterilized with a 0.22 μm filter. Filter-sterilized supernatants were the concentrated using a Microcon Ultracel YM-100, spun at 14,000 for 15 minutes until all supernatant had been concentrated. Columns were washed in 500 μL sterile PBS, pH 7.4. 100 μL of PBS was added to the columns; the columns were then inverted in the epindorf tube and spun to elute concentrated and washed supernatant protein. Samples were brought to 10 mM DTT and incubated at room temperature for 30 minutes. 2 μL of 10 mM DyLight 680 Maleimide (1 mg in 100 μL DMF) was added to each sample to bring them to 0.2 mM DyLight. Samples were incubated for two hours at room temperature. 4:1–2x Sample Buffer: 1M DTT was added to samples, which were resolved by SDS-PAGE gel, transferred to nitrocellulose membrane and analyzed by western blot.
Immunoblotting
Whole cell lysates and concentrated supernatants were prepared from Bordetella cultures as described (Mazar and Cotter, 2006). Samples were resolved by 5.0% and 8.0% polyacrylamide gels (2.0% SDS) as indicated in results section. For anti-MCD immunodetection, IRDye 800CW Goat anti-Rabbit IgG (H + L) was used (Li-cor, diluted 1:25,000). For anti-HA immunodetection, IRDye 680LT Goat anti-Mouse IgG (H + L) was used (Li-Cor, diluted 1:15,000). For anti-BvgS immunodetection, IRDye 800CW Goat anti-Mouse IgG (H + L) was used (Li-Cor, diluted 1:25,0000). Membranes imaged on a Li-Cor Odyssey infrared imaging system.
Proteinase-K digestion of FhaB
B. bronchiseptica cultures were grown to an OD600 of 1.5 to 3.0. Three samples per strain of 2 OD units were spun at 5000 rpm for ten minutes at 4°C to pellet bacteria. Supernatants were aspirated and pellets were suspended to 200 μL cold HEPES buffer, pH 7.4. Samples were spun at 5000 rpm for five minutes at 4°C and supernatants were aspirated. 200 μL of mock reaction buffer (HEPES, pH 7.4), 1.0 μg/mL proteinase-K (Sigma-Aldrich) in HEPES, or 1.0 μg/mL proteinase-K with 1 mM PMSF (MP Biomedicals) in HEPES were added to each of the three samples per strain. Reactions proceeded for 30 minutes at 16°C. 2 μL of PMSF (1 mM final) were added to proteinase-K digested samples to neutralize digestion. All samples were spun at maximum speed for one minute at room temperature and supernatants were aspirated. Whole-cell lysates were prepared, resolved and immunoblotted as described.
Dot blot analysis
B. bronchispetica cultures were grown to an OD600 of 2.0 to 4.0. Volumes equivalent to 2 OD units were spun at max speed for one minute to pellet bacteria. Supernatants were aspirated and pellets were washed in PBS. Samples were spun again at max speed for one minute and the wash was aspirated. Samples were suspended in 1 mL of PBS and split into 500 μL aliquots. Bacteria in one tube were boiled for five minutes to lyse cells. Intact and lysed cells in were spotted (50 μL) onto a nitrocellulose membrane using a Whatman® Minifold® I 96 well slot-blot array system. The membrane was blocked in 5% milk-PBS for one hour. Immunodetection by anti-MCD and anti-HA primary antibodies and goat anti-rabbit and goat anti-mouse secondary antibodies proceeded as described (here and (Mazar and Cotter, 2006)).
Supplementary Material
Acknowledgments
We thank members of our laboratory for many insightful discussions. We thank Scott Stibitz for the gift of anti-BvgS antibody. This work was supported by funds from the NIH (AI43986 and AI094991 to P.A.C).
References
- Abramson T, Kedem H, Relman DA. Modulation of the NF-kappaB pathway by Bordetella pertussis filamentous hemagglutinin. PLoS ONE. 2008;3:e3825. doi: 10.1371/journal.pone.0003825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, Braaten BA, Jones AM, Webb JS, Hayes CS, Cotter PA, et al. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature. 2010;468:439–442. doi: 10.1038/nature09490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA. Contact-dependent inhibition of growth in Escherichia coli. Science. 2005;309:1245–1248. doi: 10.1126/science.1115109. [DOI] [PubMed] [Google Scholar]
- Buscher AZ, Grass S, Heuser J, Roth R, St Geme JW. Surface anchoring of a bacterial adhesin secreted by the two-partner secretion pathway. Mol Microbiol. 2006;61:470–483. doi: 10.1111/j.1365-2958.2006.05236.x. [DOI] [PubMed] [Google Scholar]
- Chevalier N, Moser M, Koch HG, Schimz KL, Willery E, Locht C, Jacob-Dubuisson F, Muller M. Membrane targeting of a bacterial virulence factor harbouring an extended signal peptide. J Mol Microbiol Biotechnol. 2004;8:7–18. doi: 10.1159/000082076. [DOI] [PubMed] [Google Scholar]
- Clantin B, Delattre AS, Rucktooa P, Saint N, Meli AC, Locht C, Jacob-Dubuisson F, Villeret V. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science. 2007;317:957–961. doi: 10.1126/science.1143860. [DOI] [PubMed] [Google Scholar]
- Clantin B, Hodak H, Willery E, Locht C, Jacob-Dubuisson F, Villeret V. The crystal structure of filamentous hemagglutinin secretion domain and its implications for the two-partner secretion pathway. Proc Natl Acad Sci USA. 2004;101:6194–6199. doi: 10.1073/pnas.0400291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PA, Yuk MH, Mattoo S, Akerley BJ, Boschwitz J, Relman DA, Miller JF. Filamentous hemagglutinin of Bordetella bronchiseptica is required for efficient establishment of tracheal colonization. Infect Immun. 1998;66:5921–5929. doi: 10.1128/iai.66.12.5921-5929.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutte L, Alonso S, Reveneau N, Willery E, Quatannens B, Locht C, Jacob-Dubuisson F. Role of adhesin release for mucosal colonization by a bacterial pathogen. J Exp Med. 2003;197:735–742. doi: 10.1084/jem.20021153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutte L, Antoine R, Drobecq H, Locht C, Jacob-Dubuisson F. Subtilisin-like autotransporter serves as maturation protease in a bacterial secretion pathway. EMBO J. 2001;20:5040–5048. doi: 10.1093/emboj/20.18.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delisse-Gathoye AM, Locht C, Jacob F, Raaschou-Nielsen M, Heron I, Ruelle JL, de Wilde M, Cabezon T. Cloning, partial sequence, expression, and antigenic analysis of the filamentous hemagglutinin gene of Bordetella pertussis. Infect Immun. 1990;58:2895–2905. doi: 10.1128/iai.58.9.2895-2905.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvaux M, Scott-Tucker A, Turner SM, Cooper LM, Huber D, Nataro JP, Henderson IR. A conserved extended signal peptide region directs posttranslational protein translocation via a novel mechanism. Microbiology. 2007;153:59–70. doi: 10.1099/mic.0.29091-0. [DOI] [PubMed] [Google Scholar]
- Dieterich C, Relman DA. Modulation of the host interferon response and ISGylation pathway by B. pertussis filamentous hemagglutinin. PLoS ONE. 2011;6:e27535. doi: 10.1371/journal.pone.0027535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevrois S, Steeghs L, Roholl P, Letesson JJ, van der Ley P. The Omp85 protein of Neisseria meningitidis is required for lipid export to the outer membrane. EMBO J. 2003;22:1780–1789. doi: 10.1093/emboj/cdg174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass S, St Geme JW. Maturation and secretion of the non-typable Haemophilus influenzae HMW1 adhesin: roles of the N-terminal and C-terminal domains. Mol Microbiol. 2000;36:55–67. doi: 10.1046/j.1365-2958.2000.01812.x. [DOI] [PubMed] [Google Scholar]
- Hagan CL, Silhavy TJ, Kahne D. β-Barrel membrane protein assembly by the Bam complex. Annu Rev Biochem. 2011;80:189–210. doi: 10.1146/annurev-biochem-061408-144611. [DOI] [PubMed] [Google Scholar]
- Hayes CS, Aoki SK, Low DA. Bacterial contact-dependent delivery systems. Annu Rev Genet. 2010;44:71–90. doi: 10.1146/annurev.genet.42.110807.091449. [DOI] [PubMed] [Google Scholar]
- Henderson MW, Inatsuka CS, Sheets AJ, Williams CL, Benaron DJ, Donato GM, Gray MC, Hewlett EL, Cotter PA. Contribution of Bordetella filamentous hemagglutinin and adenylate cyclase toxin to suppression and evasion of interleukin-17-mediated inflammation. Infect Immun. 2012;80:2061–2075. doi: 10.1128/IAI.00148-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertle R. The family of Serratia type pore forming toxins. Curr Protein Pept Sci. 2005;6:313–325. doi: 10.2174/1389203054546370. [DOI] [PubMed] [Google Scholar]
- Hodak H, Clantin B, Willery E, Villeret V, Locht C, Jacob-Dubuisson F. Secretion signal of the filamentous haemagglutinin, a model two-partner secretion substrate. Mol Microbiol. 2006;61:368–382. doi: 10.1111/j.1365-2958.2006.05242.x. [DOI] [PubMed] [Google Scholar]
- Ieva R, Bernstein HD. Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc Natl Acad Sci USA. 2009;106:19120–19125. doi: 10.1073/pnas.0907912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inatsuka CS, Julio SM, Cotter PA. Bordetella filamentous hemagglutinin plays a critical role in immunomodulation, suggesting a mechanism for host specificity. Proc Natl Acad Sci USA. 2005;102:18578–18583. doi: 10.1073/pnas.0507910102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob-Dubuisson F, Fernandez R, Coutte L. Protein secretion through autotransporter and two-partner pathways. Biochim Biophys Acta. 2004;1694:235–257. doi: 10.1016/j.bbamcr.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Jacob-Dubuisson F, Kehoe B, Willery E, Reveneau N, Locht C, Relman DA. Molecular characterization of Bordetella bronchiseptica filamentous haemagglutinin and its secretion machinery. Microbiology. 2000;146:1211–1221. doi: 10.1099/00221287-146-5-1211. [DOI] [PubMed] [Google Scholar]
- Jacob-Dubuisson F, Villeret V, Clantin B, Delattre AS, Saint N. First structural insights into the TpsB/Omp85 superfamily. Biol Chem. 2009;390:675–684. doi: 10.1515/BC.2009.099. [DOI] [PubMed] [Google Scholar]
- Julio SM, Inatsuka CS, Mazar J, Dieterich C, Relman DA, Cotter PA. Natural-host animal models indicate functional interchangeability between the filamentous haemagglutinins of Bordetella pertussis and Bordetella bronchiseptica and reveal a role for the mature C-terminal domain, but not the RGD motif, during infection. Mol Microbiol. 2009;71:1574–1590. doi: 10.1111/j.1365-2958.2009.06623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker M, Besingi RN, Clark PL. Vectorial transport and folding of an autotransporter virulence protein during outer membrane secretion. Mol Microbiol. 2009;71:1323–1332. doi: 10.1111/j.1365-2958.2009.06607.x. [DOI] [PubMed] [Google Scholar]
- Kajava AV, Cheng N, Cleaver R, Kessel M, Simon MN, Willery E, Jacob-Dubuisson F, Locht C, Steven AC. Beta-helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol Microbiol. 2001;42:279–292. doi: 10.1046/j.1365-2958.2001.02598.x. [DOI] [PubMed] [Google Scholar]
- Kozjak V, Wiedemann N, Milenkovic D, Lohaus C, Meyer HE, Guiard B, Meisinger C, Pfanner N. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J Biol Chem. 2003;278:48520–48523. doi: 10.1074/jbc.C300442200. [DOI] [PubMed] [Google Scholar]
- Lambert-Buisine C, Willery E, Locht C, Jacob-Dubuisson F. N-terminal characterization of the Bordetella pertussis filamentous haemagglutinin. Mol Microbiol. 1998;28:1283–1293. doi: 10.1046/j.1365-2958.1998.00892.x. [DOI] [PubMed] [Google Scholar]
- Locht C, Bertin P, Menozzi FD, Renauld G. The filamentous haemagglutinin, a multifaceted adhesion produced by virulent Bordetella spp. Mol Microbiol. 1993;9:653–660. doi: 10.1111/j.1365-2958.1993.tb01725.x. [DOI] [PubMed] [Google Scholar]
- Locht C, Geoffroy MC, Renauld G. Common accessory genes for the Bordetella pertussis filamentous hemagglutinin and fimbriae share sequence similarities with the papC and papD gene families. EMBO J. 1992;11:3175–3183. doi: 10.1002/j.1460-2075.1992.tb05394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhov AM, Hannah JH, Brennan MJ, Trus BL, Kocsis E, Conway JF, Wingfield PT, Simon MN, Steven AC. Filamentous hemagglutinin of Bordetella pertussis. A bacterial adhesin formed as a 50-nm monomeric rigid rod based on a 19-residue repeat motif rich in beta strands and turns. J Mol Biol. 1994;241:110–124. doi: 10.1006/jmbi.1994.1478. [DOI] [PubMed] [Google Scholar]
- Mattoo S, Miller JF, Cotter PA. Role of Bordetella bronchiseptica fimbriae in tracheal colonization and development of a humoral immune response. Infect Immun. 2000;68:2024–2033. doi: 10.1128/iai.68.4.2024-2033.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazar J, Cotter PA. Topology and maturation of filamentous haemagglutinin suggest a new model for two-partner secretion. Mol Microbiol. 2006;62:641–654. doi: 10.1111/j.1365-2958.2006.05392.x. [DOI] [PubMed] [Google Scholar]
- Mazar J, Cotter PA. New insight into the molecular mechanisms of two-partner secretion. Trends Microbiol. 2007;15:508–515. doi: 10.1016/j.tim.2007.10.005. [DOI] [PubMed] [Google Scholar]
- McGuirk P, Mills KH. Direct anti-inflammatory effect of a bacterial virulence factor: IL-10-dependent suppression of IL-12 production by filamentous hemagglutinin from Bordetella pertussis. Eur J Immunol. 2000;30:415–422. doi: 10.1002/1521-4141(200002)30:2<415::AID-IMMU415>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Relman D, Tuomanen E, Falkow S, Golenbock DT, Saukkonen K, Wright SD. Recognition of a bacterial adhesion by an integrin: macrophage CR3 (alpha M beta 2, CD11b/CD18) binds filamentous hemagglutinin of Bordetella pertussis. Cell. 1990;61:1375–1382. doi: 10.1016/0092-8674(90)90701-f. [DOI] [PubMed] [Google Scholar]
- Renauld-Mongenie G, Cornette J, Mielcarek N, Menozzi FD, Locht C. Distinct roles of the N-terminal and C-terminal precursor domains in the biogenesis of the Bordetella pertussis filamentous hemagglutinin. J Bacteriol. 1996;178:1053–1060. doi: 10.1128/jb.178.4.1053-1060.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Keegstra K. The endosymbiotic origin of the protein import machinery of chloroplastic envelope membranes. Trends Plant Sci. 1999;4:302–307. doi: 10.1016/s1360-1385(99)01449-1. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Devos D, Genevrois S, Vicente M, Valencia A. POTRA: a conserved domain in the FtsQ family and a class of beta-barrel outer membrane proteins. Trends Biochem Sci. 2003;28:523–526. doi: 10.1016/j.tibs.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Sato Y, Sato H. Development of acellular pertussis vaccines. Biologicals. 1999;27:61–69. doi: 10.1006/biol.1999.0181. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde U, Thomas G. Insights from bacterial subtilases into the mechanisms of intramolecular chaperone-mediated activation of furin. Methods Mol Biol. 2011;768:59–106. doi: 10.1007/978-1-61779-204-5_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabady RL, Peterson JH, Skillman KM, Bernstein HD. An unusual signal peptide facilitates late steps in the biogenesis of a bacterial autotransporter. Proc Natl Acad Sci USA. 2005;102:221–226. doi: 10.1073/pnas.0406055102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanassi DG, Stathopoulos C, Karkal A, Li H. Protein secretion in the absence of ATP: the autotransporter, two-partner secretion and chaperone/usher pathways of gram-negative bacteria. Mol Membr Biol. 2005;22:63–72. doi: 10.1080/09687860500063290. [DOI] [PubMed] [Google Scholar]
- Urisu A, Cowell JL, Manclark CR. Filamentous hemagglutinin has a major role in mediating adherence of Bordetella pertussis to human WiDr cells. Infect Immun. 1986;52:695–701. doi: 10.1128/iai.52.3.695-701.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Yen MR, Peabody CR, Partovi SM, Zhai Y, Tseng YH, Saier MH. Protein-translocating outer membrane porins of Gram-negative bacteria. Biochim Biophys Acta. 2002;1562:6–31. doi: 10.1016/s0005-2736(02)00359-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.