Abstract
Levels of gene expression show considerable variation in eukaryotes, but no fine-scale maps have been made of the fitness consequences of such variation in controlled genetic backgrounds and environments. To address this, we assayed fitness at many levels of up- and down-regulated expression of a single essential gene, LCB2, involved in sphingolipid synthesis in budding yeast Saccharomyces cerevisiae. Reduced LCB2 expression rapidly decreases cellular fitness, yet increased expression has little effect. The wild-type expression level is therefore perched on the edge of a nonlinear fitness cliff. LCB2 is upregulated when cells are exposed to osmotic stress; consistent with this, the entire fitness curve is shifted upward to higher expression under osmotic stress, illustrating the selective force behind gene regulation. Expression levels of LCB2 are lower in wild yeast strains than in the experimental lab strain, suggesting that higher levels in the lab strain may be idiosyncratic. Reports indicate that the effect sizes of alleles contributing to variation in complex phenotypes differ among environments and genetic backgrounds; our results suggest that such differences may be explained as simple shifts in the position of nonlinear fitness curves.
Keywords: sphingolipids, gene regulatory evolution, Saccharomyces cerevisiae, fitness function, gene expression
Introduction
Some mutations have large effects on the fitness of organisms, whereas others have little or no effect on fitness (Fisher 1928; Wright 1929; Haldane 1930; Orr 1991; Deutschbauer et al. 2005). One explanation for this is that the impact of mutations on fitness (healthy growth and reproduction) is often not linearly affected by the amount that a mutation changes gene activity or expression. Although a linear relationship is often assumed, it has been demonstrated that allelic differences can have a nonlinear effect on fitness (Wright 1934; Hartl et al. 1985; Dykhuizen et al. 1987; Birchler et al. 2001; Veitia 2002; Papp et al. 2003). For example, either an increase or a decrease in the expression of a gene can create a stoichiometric imbalance among proteins in a complex, and thus either type of change may decrease fitness (Birchler et al. 2001; Veitia 2002; Papp et al. 2003). Another nonlinear fitness function describes the diminishing returns of increases in the effective activity of metabolic enzymes when alleles vary either in the protein sequence or expression level of the enzyme (Wright 1934; Hartl et al. 1985; Dykhuizen et al. 1987). Gene expression is a phenotype that shows extensive natural variation, and comparative analyses suggest that much of this variation has little physiological effect (Khaitovich et al. 2005; Yanai and Hunter 2009). The shape of the expression–fitness curve for a gene is a tool for more directly determining the causes and consequences of such variation within a species. The shape of expression–fitness curves for lac genes has been studied in bacteria, and the resulting curves are nonlinear and change in different environments (Dekel and Alon 2005; Perfeito et al. 2011). Expression–fitness curves for eukaryotic genes could similarly yield insight into the distribution of the consequences of mutations. However, no published studies have characterized fitness as a function of gene expression at high resolution in eukaryotes (Bayer 2010), which would permit a more powerful test of the hypothesis that genetic variation has a nonlinear effect on fitness.
As a first step toward this goal, we chose to quantitatively measure the expression–fitness curve, in Saccharomyces cerevisiae, of the essential and conserved metabolic gene, LCB2, which encodes a key enzyme in the sphingolipid synthesis pathway (fig. 1). This pathway synthesizes and manages a spectrum of functionally diverse products and metabolites with key structural and signaling roles that mediate responses to changing physiologic and environmental cues (Hannun and Obeid 2008). Sphingolipids are essential components of the plasma membrane, and are involved in processes including signaling, proteolysis, cytoskeletal changes, nutrient uptake, regulation of cell growth, and stress response (Nagiec et al. 1994; Cowart and Obeid 2007). Lcb2 binds in stoichiometric complex with Lcb1 to form serine palmitoyltransferase (SPT), which catalyzes the first committed step in sphingolipid synthesis (fig. 1) (Gable et al. 2002). Misregulation of sphingolipids has been shown to cause disruptions in their key structural and signaling roles and to have severe consequences for the cell (Hannun and Obeid 2008). The role of LCB2 in sphingolipid synthesis makes it an intriguing gene for studying the effects of variation in gene expression on cellular fitness. Importantly, sphingolipids and their diverse downstream products can be measured, and thereby give an additional quantitative phenotype between gene expression and fitness. In addition, LCB2 is a good choice for studying expression–fitness curves, because 1) it exhibits stoichiometric binding, such that dosage balance may affect fitness (Gable et al. 2002), 2) it shows variation in expression patterns among ecologically divergent yeast strains (Rossouw et al. 2009; Eng et al. 2010), which can shed light on the shape of the fitness curve, and 3) it is already known to affect cell growth, resulting in a severe growth defect when repressed (Mnaimneh et al. 2004). For experimental reasons, LCB2 is an appropriate candidate because it is not cell cycle regulated (Spellman et al. 1998), not periodically expressed during metabolic bursts (Tu et al. 2005), not dynamically expressed over the course of fermentative growth (Rossouw et al. 2009), and does not show cell-to-cell variation in expression (Newman et al. 2006). Therefore, we expect that any phenotypic consequences of changes in expression will be due to the level of expression, and not changes in the temporal dynamics of expression.
Fig. 1.
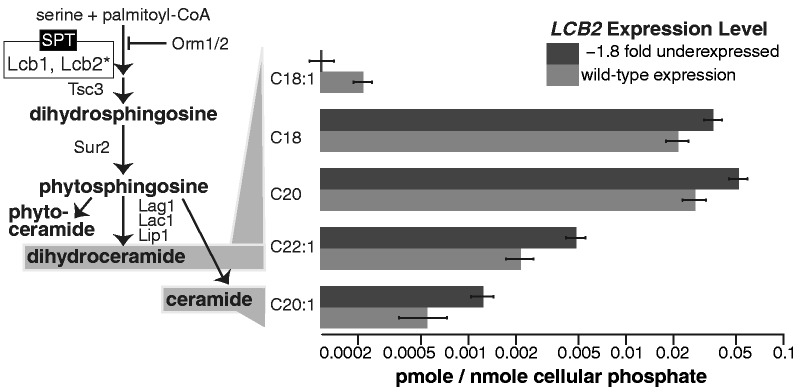
The cellular sphingolipid synthesis pathway and corresponding responses to changes in LCB2 expression. On left, the sphingolipid biosynthesis pathway in Saccharomyces cerevisiae is shown. Lcb2, the product of the titrated gene LCB2, catalyzes the first committed step in this pathway. Sphingolipid compounds were measured in response to changing levels of LCB2, and sphingolipids that displayed a significant response are shown to the right of their corresponding step in the pathway. All comparisons shown between sphingolipid levels in cells with wild-type LCB2 expression and with reduced (−2.7-fold) expression are significant according to a two tailed t test at a P value less than 0.05. Bars indicate medians and standard errors of nine biological replicates. Means, standard errors, and P values for all measured sphingolipid compounds at three LCB2 levels are provided in supplementary table S1, Supplementary Material online.
One of the challenges in assessing the fitness consequences of variation in level of gene expression is identifying a genetic system in which gene expression can be experimentally manipulated and its fitness consequences measured. One possible approach would be to mutate a gene’s promoter, but this is difficult to achieve because most promoter mutations either have no effect or lead to a coarse-grained decrease in expression (Patwardhan et al. 2009). To solve this problem, we used a chemically titratable promoter system that does not directly affect fitness to create a range of expression phenotypes for a single gene (Hughes et al. 2000; Peng et al. 2003; Mnaimneh et al. 2004). We mimicked graded allelic variation in gene expression level using the strain TetO7–LCB2, in which the native promoter of the gene LCB2 has been replaced with a doxycycline-regulated promoter (Tet-Off system) (Mnaimneh et al. 2004). We find that this titratable promoter system is an excellent tool for exploring expression–fitness curves and how they are affected by changing environments and genetic backgrounds.
Results
Fitness and Sphingolipid Flux Consequences of Variation in LCB2 Expression
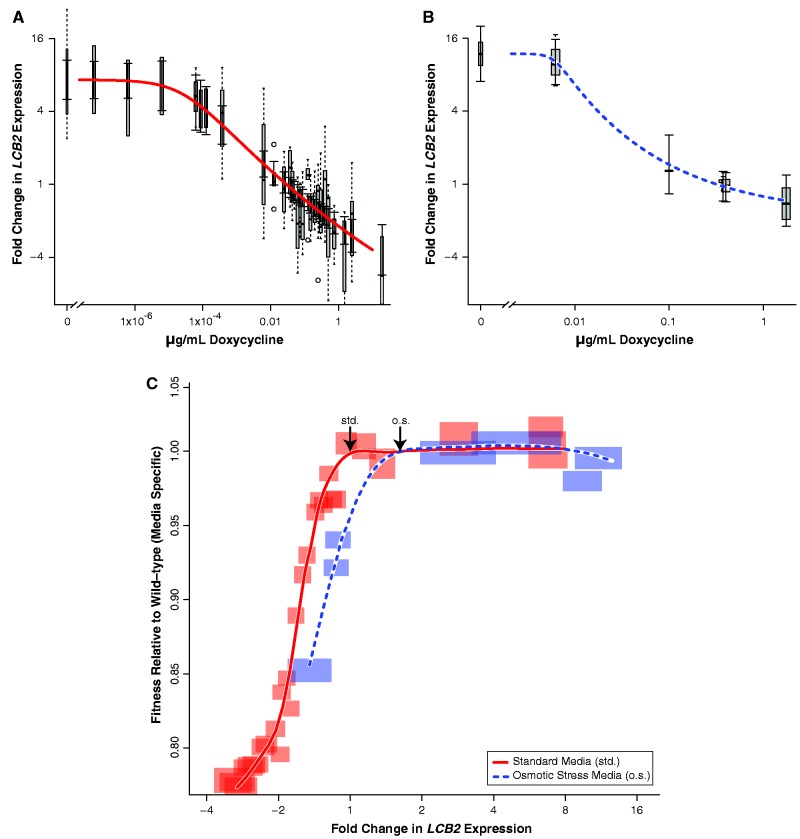
We first determined the dose-response curve between doxycycline concentration and LCB2 expression in TetO7–LCB2 (fig. 2A). Using this expression–titration function, we measured fitness at 29 expression levels under head-to-head competition in mixed culture with a fluorescent reference strain (fig. 2C). We defined fitness as the rate of cell growth and division in batch culture relative to the wild-type (parental strain R1158).
Fig. 2.
Titrated levels of gene expression reveal a fitness cliff for the gene LCB2 in Saccharomyces cerevisiae strain TetO7–LCB2. We measured the relationship between doxycycline concentration and LCB2 expression level in (A) standard media and (B) osmotic stress media. Expression levels were determined by qPCR, and each curve was fitted with a five-parameter log-logistic regression. Box and whisker plots (gray bars and dashed lines) indicate error estimates for qPCR measurements from at least three biological replicates, and capped solid lines indicate error estimates based on a regression model. (C) We assessed cellular fitness relative to levels of LCB2 expression for TetO7–LCB2 grown in two environments. The sharp fitness cost for decreases in LCB2 expression shows that the wild-type level of expression is on the edge of a fitness cliff. The shape of the fitness function in osmotic stress media (dashed line) is similar to its shape in standard media (solid line), although shifted toward higher levels of LCB2 expression in the former. Gene expression is relative to the level of expression in the parental wild-type (R1158) in standard media; arrows indicate wild-type expression levels in each environment. Fitness is normalized to the wild-type equivalent in each environment. Semitransparent boxes indicate error boundaries for fitness (standard error of at least three, and on average five, biological replicates) and for expression estimates (regression model-based error).
As expression was reduced from the wild-type level in standard media, fitness rapidly decreased, dropping 19% in conjunction with a 2-fold (50%) drop in LCB2 expression (solid red curve in fig. 2C). This represents a dramatic fitness cost—such alleles would very quickly be removed from wild populations by natural selection. Our high-resolution data reveal that the wild-type expression level is located directly at the edge of this fitness cliff, so that even a small decrease in expression has a large fitness cost. In contrast, overexpression of LCB2 up to a level 7.3-fold greater than wild-type did not markedly change fitness. The fact that the slope of the fitness curve is steep in response to decreases in LCB2 expression but flat in response to increases in expression indicates that changes in LCB2 expression have highly nonlinear fitness consequences.
We expected that changes in LCB2 gene expression would affect sphingolipid levels because of its key position at the head of the sphingolipid synthesis pathway (fig. 1). We quantitated several of the earliest sphingolipid metabolites in the de novo synthesis pathway, including dihydrosphingosine and 20 species of ceramides, which are intermediates in the formation of complex sphingolipids. In cells with a 7.3-fold increase in LCB2 expression, we did not observe changes in any sphingolipids measured (supplementary table S1, Supplementary Material online), consistent with the lack of an obvious fitness consequence (fig. 2C). However, the decrease in fitness we observed when we titrated LCB2 expression 2.7-fold (63%) below wild type was associated with changes in the levels of several ceramides. Specifically, we observed a significant decrease in the level of C18:1 dihydroceramide (two-tailed t test, P = 0.022; fig. 1), whereas four other ceramide species increased [two-tailed t test: C18 (P = 0.037), C20 (P = 0.044), and C22:1 dihydroceramide (P = 0.011), C20:1 ceramide (P = 0.038)]. Individual ceramide species influence key aspects of longevity, including cell growth, regulation, differentiation, and death (reviewed in Hannun and Obeid 2011). We saw both decreases and increases in stearoyl (C18) ceramides, which have an 18 carbon fatty acid in the ceramide or dihydroceramide moiety. Effects of stearoyl ceramides on cell growth have been observed elsewhere, for example, they are involved in the progression of squamous cell carcinomas in human head and neck tumors (Koybasi et al. 2004).
Reduction in LCB2 levels from wild-type also results in the altered transcription of many genes. According to data collected by Mnaimneh et al. (2004), a 3.8-fold (74%) reduction in LCB2 expression results in a severe growth defect and with genome-wide transcriptional changes including repression of structural ribosomal genes and induction of genes associated with cellular stress response (supplementary table S2, Supplementary Material online).
The Fitness Curve Shifts in Response to Altered Environmental Conditions
The levels and types of sphingolipids required by a cell change across different environments, for example, in response to osmotic stress (Patton et al. 1992; Hannun and Obeid 2008), and we predicted that effect of varying LCB2 levels would therefore also change among such environments. To determine the extent to which environmental change affects the LCB2 expression–fitness curve, we measured the curve when the cells were subjected to osmotic stress. We noted that in the wild-type parental strain (R1158), LCB2 expression was upregulated 1.6-fold in osmotic stress media compared with standard media (arrows in fig. 2C; fig. 3; two-tailed t test, P = 0.004). We first determined the dose-response function for the TetO7–LCB2 strain in osmotic stress media (fig. 2B). We then measured how changes in LCB2 expression affected fitness in osmotic stress media. As expression increased 7.3-fold, there was no consistent change in fitness (dashed blue line in fig. 2C). As expression decreased 2.2-fold, there was a 12.6% reduction in fitness. Interestingly, the expression–fitness curve under osmotic stress is similar in shape to the curve in standard media but shifted to a region of higher expression. This shift in the fitness curve indicates the extent of selection for environment-specific responses in gene regulation.
Fig. 3.
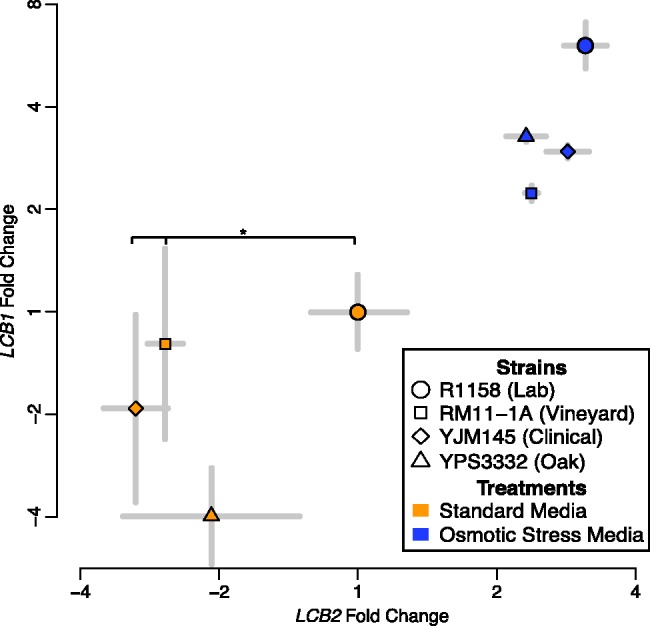
LCB2 and LCB1 gene expression levels covary among wild strains of Saccharomyces cerevisiae and between growth environments. LCB2 gene expression is upregulated in osmotic stress media (blue points) in comparison with levels of expression in standard media (orange points). LCB2 levels are significantly lower in wild strains YJM145 and RM11-1a than in the auxotrophic lab strain R1158, suggesting that the expression–fitness function has evolved in the lab strain (asterisk; two-tailed t test, P = 0.028 and P = 0.047, respectively). Fold change was ascertained by qPCR and is relative to the expression level of the same gene in the lab strain (R1158) when grown in standard media. Standard errors of three or more biological replicates are shown.
Evidence for Distinct Fitness Curves in Different Genetic Backgrounds
The shape of the fitness curve indicates the relative fitness cost of mutant alleles compared with the wild-type and accordingly predicts the expected levels of standing allelic variation in populations (Eanes 2011). Because low-expression alleles would have pronounced negative fitness consequences (fig. 2C), we would expect them to be rapidly purged from populations by natural selection. By the same token, high-expression alleles have no measurable fitness consequence and we would expect such alleles to segregate freely in populations. To test this hypothesis, we measured expression levels of LCB2 in three ecologically and genetically divergent strains of S. cerevisiae: vineyard strain RM11-1a, pathogenic strain YJM145-a, and oak strain YPS 3332. Contrary to what the LCB2 fitness curve led us to expect, two wild strains had significantly lower levels of LCB2 expression than the lab strain both in standard media and in osmotic stress media (fig. 3). The level of LCB2 expression was 1.5-fold (33%) lower in the pathogenic strain YJM145 and 1.4-fold (27%) lower in the vineyard strain RM11-1a than in the lab strain when both were grown in standard media (two-tailed t test, P = 0.028 and P = 0.047, respectively). This among-strain analysis requires that the reference gene we used for quantitative polymerase chain reaction (qPCR), ALG9, has not changed, as has been shown in previous studies across a variety of growth conditions and genetic backgrounds (Kvitek et al. 2008; Teste et al. 2009). We also checked our results using another stable qPCR reference gene according to these studies, TFC1, and found that YJM145 still had significantly different (1.5-fold lower) expression in comparison with the lab strain (two-tailed t test, P = 0.009).
If the fitness curves for LCB2 expression were the same in YJM145 and RM11-1a as in the lab strain, the wild strains would presumably have lower fitness. A more likely interpretation is that the LCB2 fitness curve itself has shifted in the lab strain due to epistasis with the genetic background and/or selection on sphingolipid levels. To the extent that different strains exhibit different expression–fitness curves, the level of permissible expression variation will be correspondingly affected. We hypothesize that there has been selection for higher levels of LCB2 expression in the lab strain. Given that sphingolipids are involved in amino acid sensing and transport (Dickson 2010), one very plausible cause for this shift may have been the deletion in the lab strain of four genes that are involved in amino acid synthesis (Brachmann et al. 1998).
Expression Responses of Stoichiometric Interactors
To perform its function, Lcb2 binds in a stoichiometric complex with Lcb1 to form SPT (fig. 1). In turn, SPT is regulated through stoichiometric interactions with a repressor complex (Orm1/2) and an activator (Tsc3) (Monaghan et al. 2002; Breslow et al. 2010). It is important to consider LCB1 expression when studying variation in LCB2 expression, because the extent to which the subunits are coregulated or one subunit is limiting determines the level of functional SPT complex. To investigate this, we measured levels of LCB1 in the lab and wild strains in both standard media and osmotic stress media. We found that LCB2 and LCB1 covary with each other across the strains and environments, consistent with this stoichiometric expectation (linear regression model, P = 0.005; fig. 3).
Discussion
The position of wild-type LCB2 gene expression at the edge of a fitness cliff, where the slope rapidly changes, supports the hypothesis that the effects of mutations can be highly nonlinear. It is important to know how variation in a simple trait, such as gene expression, may affect more complex outcomes, such as disease or fitness. If complex outcomes respond linearly to trait variation, then the contribution of variation in many simple traits (e.g., from many loci) to complex outcomes may be easily calculated and inferred (e.g., in genetic association studies). Early work in bacteria suggested that linearity might prevail (Elena and Lenski 1997). However, the lack of success in genome-wide association studies in humans and its associated missing heritability have suggested that the picture may instead be much more complicated (Manolio et al. 2009). Other work has indicated that the quantitative contributions of loci to complex-trait variation may change completely in different combinations of environments and genetic backgrounds (Gerke et al. 2010). Indeed, we found here that for LCB2 the effects of genetic variation are nonlinear and differ across environments and genetic backgrounds (Dekel and Alon 2005; Perfeito et al. 2011). Yet our continuous fitness curve (fig. 2C) also shows that missing information regarding the genetic basis of variation in complex phenotypes (Manolio et al. 2009) is not an enigma but is instead consistent with fundamental theory about the rugged landscape of outcomes that results when nonlinear outcomes from many loci recombine in a population (Wright 1932). For nonlinear curves, predicting the effects of mutation ab initio is difficult, because the magnitude of the effect depends on the specific region of the fitness curve traversed by the mutation. In particular, it is challenging to distinguish between neutral and functional variation. However, by mapping fitness curves, the results of variation in different environments and genetic backgrounds can indeed be predicted. Titration of expression alleles in different environments (as demonstrated here) and in different genetic backgrounds holds great utility for interpreting variation in expression among cells, tissues, individuals, and populations.
We observed that any decreases in the expression level of LCB2 from wild-type has a striking fitness cost (fig. 2C). Consistent with this, LCB2 has low cell-to-cell variation in gene expression (Newman et al. 2006), as one might expect for a gene perched on a fitness cliff, where any variation that results in an expression decrease would be harmful. A large cost for relatively small LCB2 dosage reductions makes sense, considering that sphingolipids require precise regulation (Hannun and Obeid 2008; Lebman and Spiegel 2008). This precise regulation of SPT (Lcb1/Lcb2) activity is achieved not by transcriptional responses but by direct interaction with a repressor, Orm1/2 (fig. 1) (Breslow et al. 2010). The Orm1/2 complex is phosphorylated in response to low levels of cellular sphingolipids, which diminishes the ability of the complex to oligomerize with and inhibit SPT; the resulting increase in SPT activity yields increased sphingolipid flux. Reduced LCB2 expression leads to changes in flux through the sphingolipid pathway and to altered levels of essential ceramides (fig. 1); reduced LCB2 expression therefore phenocopies Orm overexpression (Breslow et al. 2010). Sensitivity to decreased LCB2 expression is consistent with the hypothesis that even small changes in the regulation of sphingolipids in humans can cause diseases such as childhood asthma, Crohn’s disease, type 1 diabetes, and biliary cirrhosis. We found that LCB2 expression is on the edge of a fitness cliff, consistent with SPT having a very high level of metabolic control. Sensitivity to variation in SPT activity may be due in part to its position near the head of the sphingolipid synthesis pathway (fig. 1), where variation has been suggested to exert the strongest control (Eanes 1999; Wright and Rausher 2010). However, we note that our result of a fitness cliff for reduction of LCB2 in haploid yeast contrasts with the lack of a significant fitness cost for the hemizygous deletion of LCB2 in diploids (Deutschbauer et al. 2005). Determining whether this discrepancy is due to different effects of LCB2 reduction among haploids and diploids, or because of differences in growth conditions between our fitness assay (batch growth) and the hemizygous fitness assay (exponential growth) will require additional experimental work.
Why is overexpression of LCB2 close to neutral with respect to fitness (fig. 2C)? Increases in LCB2 transcription do not necessarily mediate a proportional increase in Lcb2 protein levels. In particular, Lcb2 is unstable unless it is associated with Lcb1 (Gable et al. 2000; Yasuda et al. 2003), so if LCB1 expression does not increase along with LCB2, any excess Lcb2 would be degraded. However, LCB1 levels appear to be upregulated in response to upregulation of LCB2 (fig. 3), and it is therefore unlikely that Lcb1 limitation is the sole factor underlying the absence of a fitness effect. Even if additional SPT is formed, this increased dose may not affect sphingolipid flux and fitness if there is enough Orm1/2 to inhibit SPT activity. Because wild-type levels of Lcb1 and Lcb2 are high enough that sphingolipid flux levels are toxic in the absence of Orm1/2 repression (Breslow et al. 2010), our findings suggest that Orm1/2 may be capable of buffering the cell against SPT levels that are greater than those found in wild-type.
It would seem advantageous, therefore, for the wild-type expression to fall within the region of high expression that offers a buffer from the genetic, environmental, or stochastic perturbations that may lower LCB2 levels. In other words, if the LCB2 expression level were higher, then mutations that decrease LCB2 expression would not result in severely compromised fitness. There are several possible explanations for the precarious position of wild-type expression of LCB2 on the edge of this fitness cliff (fig. 2C). One possibility is that there are other growth environments in which there is a cost to overexpression (Hillenmeyer et al. 2008). A second possibility is that there is a small deleterious fitness consequence for expressing LCB2 above wild-type levels that is beyond our measurement resolution. It has been estimated that an otherwise neutral 2-fold increase in gene expression will be selected against solely due to the metabolic costs of mRNA and protein production, for all genes except those with the lowest levels of expression (Wagner 2005, 2007). The critical selection coefficient in Saccharomyces, above which selection on an allele will predominate over drift, has been calculated to be very small (∼2.93 × 10−7) based on estimates of mutation rate and of a large historical population size (Wagner 2005). This is well below the resolution of our fitness measurements, so the fitness curve in figure 2C may indeed have a peak at the wild-type expression level, albeit with a shallow slope on the right-hand side. It is also possible that there is no accessible mutational path for the LCB2 promoter to achieve higher expression, or that the mutational target for LCB2 regulation is much larger than expected, and drift thus predominates (i.e., the critical selection coefficient is higher than estimated). The position on the edge of a cliff may also permit sphingolipid synthesis to be very rapidly reduced in environments where cell longevity is more important than rapid growth (Huang et al. 2012). An additional possibility, we propose, is that the lab strain has recently experienced strong selection for increased LCB2 expression associated with the loss of amino acid synthesis pathways, and given enough time, the expression level may evolve away from its precipitous position at the edge of the fitness cliff.
The cliff-like shape of the LCB2 expression–fitness curve mirrors the shape observed by varying expression levels in a model of the mitogen-activated protein kinase (MAPK) signaling cascade developed by Nijhout et al. (2003). The phenotypic output in their system is the level of active MAPK, and they examine the effect of varying expression and activity at each level of the cascade. In particular, at the highest two levels of the cascade (MAPKKK and MAPKK), the effect of variation is large at low expression levels but saturates at higher expression levels, as we observe with LCB2. The authors point out that wild-type model parameters, such as expression level, are typically unknown. However, they suggest that wild-type phenotypes are unlikely to occur deep within plateaus of a fitness curve or where the fitness curve is steeply sloped. Instead, they conclude that it is most likely that wild-type phenotypes will lie near the edges of fitness curves, as we observe here for LCB2.
Although our fitness curve measured in the lab is real, it is unclear if it reflects aspects of fitness that are important in the wild. Yeast grows exponentially when in glucose, but growth slows as glucose is used up and the population saturates, and we serially diluted the culture into fresh media every 24 h so that growth could continue. As each culture went through several cycles of lag phase, exponential growth, and saturation, our fitness measurements include both absolute growth rate and other factors such as recovery and exit from lag phase. However, it is likely that the effect of LCB2 expression on relative fitness would differ among growth conditions. For example, other components of fitness that have been investigated are sporulation efficiency in the wild (Gerke et al. 2006), exponential growth in chemostats (Dykhuizen and Hartl 1983), and long-term survival under nutrient limited conditions (Gresham et al. 2008).
The laboratory adapted strain S288C, from which TetO7–LCB2 is derived, has experienced recurrent bottlenecks and therefore shows high evolutionary rates (Gu et al. 2005). This mosaic strain lacks a key gene for flocculation (Liu et al. 1996), and TetO7–LCB2 lacks genes required for synthesis of three amino acids. Consistent with this history of bottlenecks, genetic manipulation, and elevated rates of evolution, S288C and its derivatives are outliers among S. cerevisiae for a variety of phenotypes including growth on ethanol and maltose (Warringer et al. 2011) and sporulation efficiency (Deutschbauer and Davis 2005). It is possible that our observation of LCB2 wild-type expression at the edge of a fitness cliff could similarly be an outlier. It will be important in future work to measure the LCB2 expression–fitness curves in wild strains to determine whether their wild-type expression levels are similarly positioned.
LCB2 is an essential gene with a relatively low expression level, and these factors likely contribute to the steepness of its fitness curve as expression decreases. Previous work indicates that the consequences of decreased expression for essential genes are distributed similarly to nonessential genes (Delneri et al. 2008). However, there is substantial heterogeneity among all genes in the phenotypic and fitness effects of both overexpression (Sopko et al. 2006) and reduced expression (Giaever et al. 2002; Mnaimneh et al. 2004; Deutschbauer et al. 2005; Delneri et al. 2008). There are likely to be many different shapes of expression–fitness curves, reflecting the proteome’s diversity of functions and structures and the complex web of interactions among genes and proteins.
These caveats highlight the need to expand this work to a panel of wild-strains, as well as to additional genes and environments. We expect there will be multiple classes of expression–fitness curves, and we foresee that such systematic investigations will begin to quantitatively describe how standing variation results from the evolutionary integration over environment and genetic-background specific fitness curves. Systems like the Tet-Off promoter, as we use here, allow genes to be systematically titrated, and the physiological and fitness consequences quantitatively examined. Our results show that such systems hold the possibility for producing larger scale catalogs of expression–fitness curves that would serve as a tool for interpreting natural variation in gene expression levels.
Materials and Methods
Titration of LCB2 Expression
We altered levels of LCB2 expression in S. cerevisiae strain TetO7–LCB2 (pLCB2::kanR-TetO7-TATA URA3::CMV-tTA MATa his3-1 leu2-0 met15-0), in which the native LCB2 promoter has been replaced by a tetracycline/doxycycline repressible promoter, TetO7 (Mnaimneh et al. 2004). LCB2 mRNA levels were measured at as many as 29 different concentrations of doxycycline, depending on the growth condition.
Fitness Competitions
Fitness at each level of doxycycline was measured via head-to-head competition between TetO7–LCB2 and a fluorescent reference strain, GPM1–GFP (Huh et al. 2003). Our growth protocol was as follows: TetO7–LCB2 and reference cells were grown separately in liquid media for 24 h, diluted to a density of 1.5 × 107 cells/mL, and then grown for ∼8 additional hours. The two strains were then mixed in 10:1 (TetO7–LCB2: reference) ratios and 8 × 106 cells were inoculated into 150 µL of media at the appropriate concentration of doxycycline hyclate (Calbiochem). Additional wells contained pure cultures of the reference strain or the parental strain (R1158; URA3::CMV-tTA MATa his3-1 leu2-0 met15-0). Cultures were grown in triplicate in black-wall, clear-bottom 96-well plates (Nunc) covered with foil seals (Corning) and shaken at 1,300 rpm (DTS4, Elmi) at 30°C. The standard growth environment was synthetic defined media without tryptophan (SD-Trp, Sunrise Scientific). The osmotic stress environment contained 0.3 M (instead of 0.1 M) NaCl. Competitions were diluted 1:50 into fresh media every 24 h for 4 days. The optical density A (OD units at 600 nm) and raw fluorescence B (485 nm excitation, 530 nm emission) of each microwell culture were recorded hourly (F500, Tecan). We calculated F, fluorescence per unit OD:
| (1) |
where one unit OD contains 7.39 × 107 cells/mL. Relative fitness, ω, of TetO7–LCB2 in each doxycycline concentration, C, and environment, E, was derived from the rate of change over 4 days in the proportion of TetO7–LCB2 to the reference strain in the competition culture, as measured by changes in Fcompetition, the fluorescence per unit OD of the competition culture:
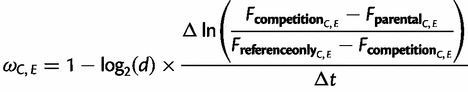 |
(2) |
where d is the dilution factor (1:50) per unit time t (1,440 min), and Fparental and Freferenceonly are the fluorescence per unit OD of parental cells only or reference cells only, respectively. We only used F when A fell between 0.57 and 0.70 (equivalent to 4.2 × 107 and 5.2 × 107 cells/ml), which allowed us to compare data among wells and days that fell in a similar optical range. Each doxycycline concentration and environment combination had, on average, five biological replicates on unique plates and days.
Measurement of LCB2 and LCB1 mRNA Levels
TetO7–LCB2 was grown to measure levels of LCB2 expression at different concentrations of doxycycline in each environment. All growth conditions were identical to the competition protocol (e.g., in microwell plates), except that TetO7–LCB2 was grown in monoculture. Subset of samples was grown both in test tubes and in microwell plates. In addition, LCB2 expression in each environment was assayed for the parental strain (R1158) and for oak (YPS 3332; MATa ho::Nat) (Murphy et al. 2006), vineyard (RM11-1a; MATa leu2Δ ura3Δ ho::Kan) (Brem et al. 2002), and pathogenic (YJM145a; MATa gal2 HO) (McCusker et al. 1994) wild strains. Growth of each treatment and strain combination was repeated at least three times on unique days and plates. RNA was extracted on the second day of growth once cultures reached an optical density of 0.6 (∼4.4 × 107 cells/ml; Zymolyase, Spin Columns, Zymo Research; Turbo DNA-free, Ambion). Expression levels were quantified using quantitative reverse transcriptase-PCR with the LCB2 primers 5′-TTGCTGTTGTTGTTGTTGCTTATCCTGCT-3′ and 5′-CGTCGTAACTGGATTTGCCGGAATTTGAT-3′ and LCB1 primers 5′-GCACACATCCCAGAGGTTTT-3′ and 5′-TGGTCCTGTATGGATCGTCA-3′ (Brilliant II SYBR 1-Step, Agilent; Mx3000P, Stratagene). ALG9 served as a reference gene for normalization of the qPCR reactions, with primers 5′-CACGGATAGTGGCTTTGGTGAACAATTAC-3′ and 5′-TATGATTATCTGGCAGCAGGAAAGAACTTGGG-3′. We normalized a subsample of reactions with an additional reference gene, TFC1, with primers: 5′-GCTGGCACTCATATCTTATCGTTTCACAATGG-3′ and 5′-GAACCTGCTGTCAATACCGCCTGGAG-3′. None of the primer sets (LCB2, ALG9, and TFC1) contained mismatches to their cognate sequences in the wild strains analyzed. Each qPCR reaction was repeated at least three times on separate plates. qPCR data were analyzed according to the 2−ΔΔCt method (Livak and Schmittgen 2001). For each environment, we fitted the relationship between doxycycline concentration and fold change with a five-parameter log-logistic function and used the function to predict final values and estimate error (R v2.14; package drc v2.2-1).
Sphingolipid Analysis
Sphingolipid metabolites at critical points in the sphingolipid synthesis pathway were extracted from monocultures of TetO7–LCB2 grown under conditions identical to those for qPCR. Nine biological replicate cultures were grown, each at three concentrations of doxycycline: 0, 0.0293, and 2.5 µg/ml, equivalent to 7.3-fold higher, the same as, and 2.7-fold lower than wild type expression, respectively. Sphingolipid levels were quantitated using LC-MS/MS by the lipidomics core facility at Medical University of South Carolina, following methods that are described in Bielawski et al. (2006).
Supplementary Material
Supplementary tables S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Walter Eanes, Geoffrey Morris, Aman Gill, Niamh O’Hara, Bin He, Jeffrey Rest; two anonymous reviewers for comments on the manuscript; and Jason O’Rawe, Geoff Bolen, and Laura Praissman for assistance in the lab. Paul Sniegowski provided strain YPS 3332. This work was supported by startup funds from Stony Brook University to J.S.R.
References
- Bayer TS. Using synthetic biology to understand the evolution of gene expression. Curr Biol. 2010;20:R772–R779. doi: 10.1016/j.cub.2010.06.049. [DOI] [PubMed] [Google Scholar]
- Bielawski J, Szulc ZM, Hannun YA, Bielawska A. Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods. 2006;39:82–91. doi: 10.1016/j.ymeth.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Bhadra U, Bhadra MP, Auger DL. Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev Biol. 2001;234:275–288. doi: 10.1006/dbio.2001.0262. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Brem RB, Yvert Gl, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, Ejsing CS, Weissman JS. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463:1048–1053. doi: 10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowart LA, Obeid LM. Yeast sphingolipids: recent developments in understanding biosynthesis, regulation, and function. Biochim Biophys Acta. 2007;1771:421–431. doi: 10.1016/j.bbalip.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekel E, Alon U. Optimality and evolutionary tuning of the expression level of a protein. Nature. 2005;436:588–592. doi: 10.1038/nature03842. [DOI] [PubMed] [Google Scholar]
- Delneri D, Hoyle DC, Gkargkas K, et al. (12 co-authors) Identification and characterization of high-flux-control genes of yeast through competition analyses in continuous cultures. Nat Genet. 2008;40:113–117. doi: 10.1038/ng.2007.49. [DOI] [PubMed] [Google Scholar]
- Deutschbauer AM, Davis RW. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat Genet. 2005;37:1333–1340. doi: 10.1038/ng1674. [DOI] [PubMed] [Google Scholar]
- Deutschbauer AM, Jaramillo DF, Proctor M, Kumm J, Hillenmeyer ME, Davis RW, Nislow C, Giaever G. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics. 2005;169:1915–1925. doi: 10.1534/genetics.104.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RC. Roles for sphingolipids in Saccharomyces cerevisiae. In: Chalfant C, Del Poeta M, editors. Sphingolipids as signaling and regulatory molecules. New York: Springer; 2010. pp. 217–231. [Google Scholar]
- Dykhuizen DE, Dean AM, Hartl DL. Metabolic flux and fitness. Genetics. 1987;115:25–31. doi: 10.1093/genetics/115.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen DE, Hartl DL. Selection in chemostats. Microbiol Rev. 1983;47:150. doi: 10.1128/mr.47.2.150-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eanes WF. Analysis of selection on enzyme polymorphisms. Annu Rev Ecol Syst. 1999;30:301–326. [Google Scholar]
- Eanes WF. Molecular population genetics and selection in the glycolytic pathway. J Exp Biol. 2011;214:165–171. doi: 10.1242/jeb.046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena SF, Lenski RE. Test of synergistic interactions among deleterious mutations in bacteria. Nature. 1997;390:395–398. doi: 10.1038/37108. [DOI] [PubMed] [Google Scholar]
- Eng KH, Kvitek DJ, Keleş S, Gasch AP. Transient genotype-by-environment interactions following environmental shock provide a source of expression variation for essential genes. Genetics. 2010;184:587–593. doi: 10.1534/genetics.109.107268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. The possible modification of the response of the wild type to recurrent mutations. Am Nat. 1928;62:115–126. [Google Scholar]
- Gable K, Han G, Monaghan E, Bacikova D, Natarajan M, Williams R, Dunn TM. Mutations in the yeast LCB1 and LCB2 genes, including those corresponding to the hereditary sensory neuropathy type I mutations, dominantly inactivate serine palmitoyltransferase. J Biol Chem. 2002;277:10194–10200. doi: 10.1074/jbc.M107873200. [DOI] [PubMed] [Google Scholar]
- Gable K, Slife H, Bacikova D, Monaghan E, Dunn TM. Tsc3p is an 80-amino acid protein associated with serine palmitoyltransferase and required for optimal enzyme activity. J Biol Chem. 2000;275:7597–7603. doi: 10.1074/jbc.275.11.7597. [DOI] [PubMed] [Google Scholar]
- Gerke J, Lorenz K, Ramnarine S, Cohen B. Gene-environment interactions at nucleotide resolution. PLoS Genet. 2010;6:e1001144. doi: 10.1371/journal.pgen.1001144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke JP, Chen CTL, Cohen BA. Natural isolates of Saccharomyces cerevisiae display complex genetic variation in sporulation efficiency. Genetics. 2006;174:985. doi: 10.1534/genetics.106.058453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, DeSevo CG, Botstein D, Dunham MJ. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 2008;4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, David L, Petrov D, Jones T, Davis RW, Steinmetz LM. Elevated evolutionary rates in the laboratory strain of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2005;102:1092–1097. doi: 10.1073/pnas.0409159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS. A note on Fisher's theory of the origin of dominance, and on a correlation between dominance and linkage. Am Nat. 1930;64:87–90. [Google Scholar]
- Hannun YA, Obeid LM. Principles of bioactive lipid signaling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- Hannun YA, Obeid LM. Many ceramides. J Biol Chem. 2011;286:27855–27862. doi: 10.1074/jbc.R111.254359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Dykhuizen DE, Dean AM. Limits of adaptation: the evolution of selective neutrality. Genetics. 1985;111:655–674. doi: 10.1093/genetics/111.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, et al. (14 co-authors) The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Liu J, Dickson RC. Down-regulating sphingolipid synthesis increases yeast lifespan. PLoS Genet. 2012;8:e1002493. doi: 10.1371/journal.pgen.1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Pääbo S, Weiss G. Toward a neutral evolutionary model of gene expression. Genetics. 2005;170:929–939. doi: 10.1534/genetics.104.037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koybasi S, Senkal CE, Sundararaj K, et al. (12 co-authors) Defects in cell growth regulation by C18:0-ceramide and longevity assurance gene 1 in human head and neck squamous cell carcinomas. J Biol Chem. 2004;279:44311–44319. doi: 10.1074/jbc.M406920200. [DOI] [PubMed] [Google Scholar]
- Kvitek DJ, Will JL, Gasch AP. Variations in stress sensitivity and genomic expression in diverse S. cerevisiae isolates. PLoS Genet. 2008;4:e1000223. doi: 10.1371/journal.pgen.1000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebman DA, Spiegel S. Thematic review series: sphingolipids. Cross-talk at the crossroads of sphingosine-1-phosphate, growth factors, and cytokine signaling. J Lipid Res. 2008;49:1388–1394. doi: 10.1194/jlr.R800008-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Styles CA, Fink GR. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, et al. (27 co-authors) Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker JH, Clemons KV, Stevens DA, Davis RW. Genetic characterization of pathogenic Saccharomyces cerevisiae isolates. Genetics. 1994;136:1261–1269. doi: 10.1093/genetics/136.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnaimneh S, Davierwala AP, Haynes J, et al. (24 co-authors) Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Monaghan E, Gable K, Dunn T. Mutations in the Lcb2p subunit of serine palmitoyltransferase eliminate the requirement for the TSC3 gene in Saccharomyces cerevisiae. Yeast. 2002;19:659–670. doi: 10.1002/yea.864. [DOI] [PubMed] [Google Scholar]
- Murphy HA, Kuehne HA, Francis CA, Sniegowski PD. Mate choice assays and mating propensity differences in natural yeast populations. Biol Lett. 2006;2:553–556. doi: 10.1098/rsbl.2006.0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagiec MM, Baltisberger JA, Wells GB, Lester RL, Dickson RC. The LCB2 gene of Saccharomyces and the related LCB1 gene encode subunits of serine palmitoyltransferase, the initial enzyme in sphingolipid synthesis. Proc Natl Acad Sci U S A. 1994;91:7899–7902. doi: 10.1073/pnas.91.17.7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JRS, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- Nijhout HF, Berg AM, Gibson WT. A mechanistic study of evolvability using the mitogen-activated protein kinase cascade. Evol Dev. 2003;5:281–294. doi: 10.1046/j.1525-142x.2003.03035.x. [DOI] [PubMed] [Google Scholar]
- Orr HA. A test of Fisher's theory of dominance. Proc Natl Acad Sci U S A. 1991;88:11413–11415. doi: 10.1073/pnas.88.24.11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Patton JL, Srinivasan B, Dickson RC, Lester RL. Phenotypes of sphingolipid-dependent strains of Saccharomyces cerevisiae. J Bacteriol. 1992;174:7180–7184. doi: 10.1128/jb.174.22.7180-7184.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan RP, Lee C, Litvin O, Young DL, Pe'er D, Shendure J. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat Biotechnol. 2009;27:1173–1175. doi: 10.1038/nbt.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W-T, Robinson MD, Mnaimneh S, Krogan NJ, Cagney G, Morris Q, Davierwala AP, Grigull J, Yang X, Zhang W. A panoramic view of yeast noncoding RNA processing. Cell. 2003;113:919–933. doi: 10.1016/s0092-8674(03)00466-5. [DOI] [PubMed] [Google Scholar]
- Perfeito L, Ghozzi S, Berg J, Schnetz K, Lässig M. Nonlinear fitness landscape of a molecular pathway. PLoS Genet. 2011;7:e1002160. doi: 10.1371/journal.pgen.1002160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossouw D, Olivares-Hernandes R, Nielsen J, Bauer FF. Comparative transcriptomic approach to investigate differences in wine yeast physiology and metabolism during fermentation. Appl Environ Microbiol. 2009;75:6600–6612. doi: 10.1128/AEM.01251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R, Huang D, Preston N, et al. (12 co-authors) Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teste M-A, Duquenne M, Francois J, Parrou J-L. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol Biol. 2009;10:99. doi: 10.1186/1471-2199-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- Veitia RA. Exploring the etiology of haploinsufficiency. BioEssays. 2002;24:175–184. doi: 10.1002/bies.10023. [DOI] [PubMed] [Google Scholar]
- Wagner A. Energy constraints on the evolution of gene expression. Mol Biol Evol. 2005;22:1365–1374. doi: 10.1093/molbev/msi126. [DOI] [PubMed] [Google Scholar]
- Wagner A. Energy costs constrain the evolution of gene expression. J Exp Zool B Mol Dev Evol. 2007;308B:322–324. doi: 10.1002/jez.b.21152. [DOI] [PubMed] [Google Scholar]
- Warringer J, Zörgö E, Cubillos FA, et al. (13 co-authors) Trait variation in yeast is defined by population history. PLoS Genet. 2011;7:e1002111. doi: 10.1371/journal.pgen.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KM, Rausher MD. The evolution of control and distribution of adaptive mutations in a metabolic pathway. Genetics. 2010;184:483–502. doi: 10.1534/genetics.109.110411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Fisher's theory of dominance. Am Nat. 1929;63:274–279. [Google Scholar]
- Wright S. The roles of mutation, inbreeding, crossbreeding, and selection in evolution. Proceedings of the 6th International Congress of Genetics, Menasha, WI. 1932;Vol. 1:356–366. [Google Scholar]
- Wright S. Physiological and evolutionary theories of dominance. Am Nat. 1934;68:24–53. [Google Scholar]
- Yanai I, Hunter CP. Comparison of diverse developmental transcriptomes reveals that coexpression of gene neighbors is not evolutionarily conserved. Genome Res. 2009;19:2214–2220. doi: 10.1101/gr.093815.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S, Nishijima M, Hanada K. Localization, topology, and function of the LCB1 subunit of serine palmitoyltransferase in mammalian cells. J Biol Chem. 2003;278:4176–4183. doi: 10.1074/jbc.M209602200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.