Abstract
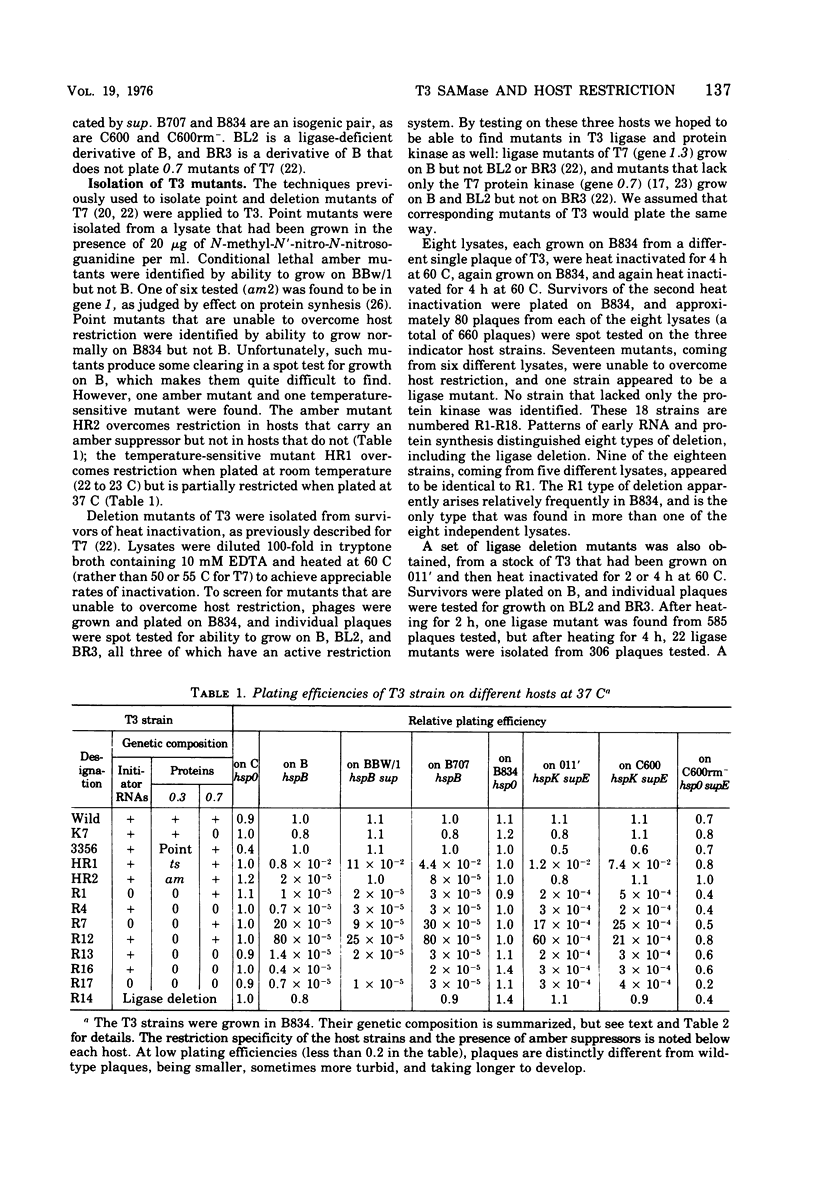
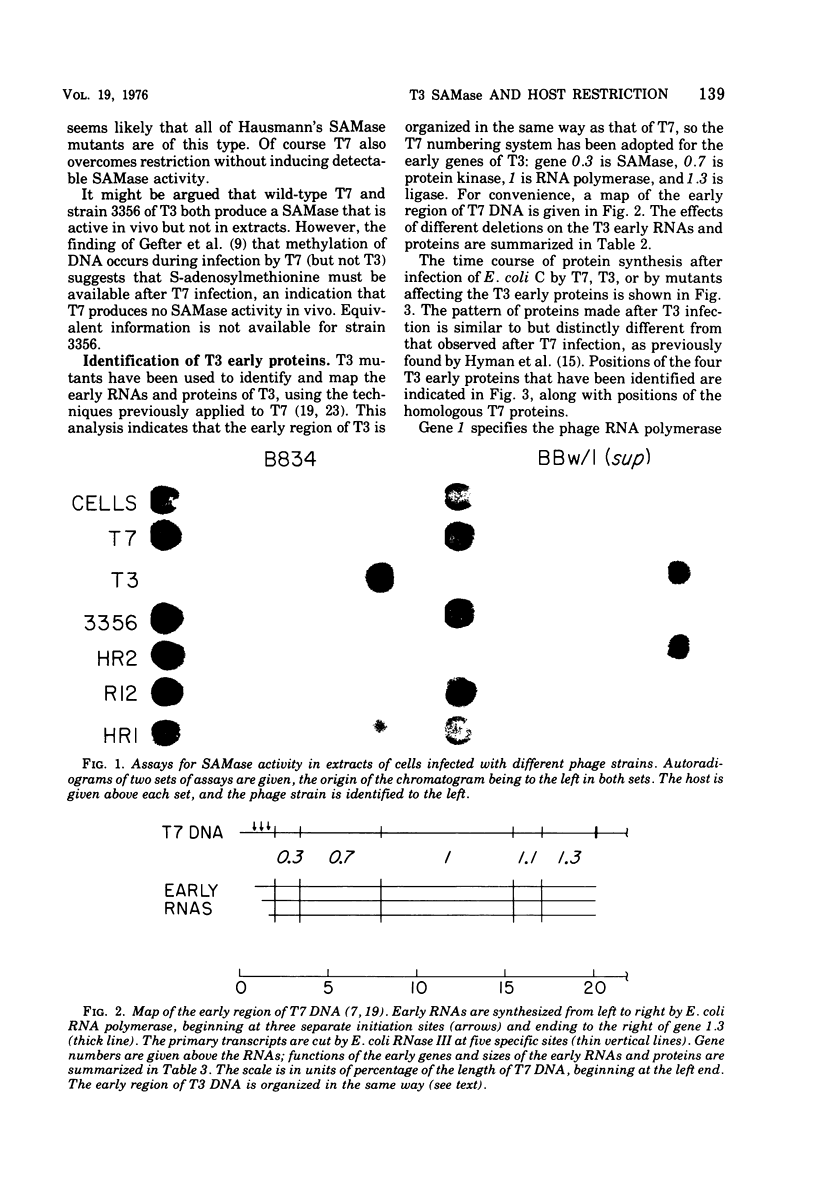
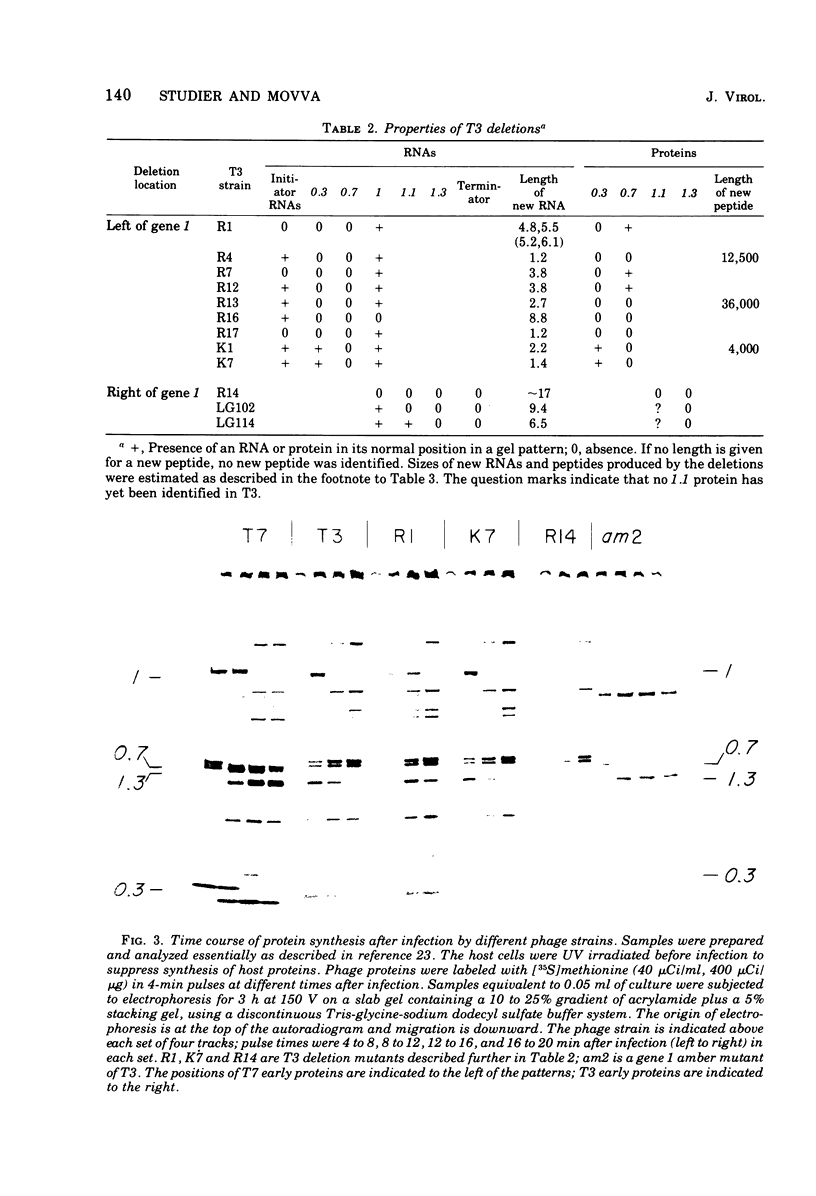
Deletion and point mutants of T3 have been isolated and used to show that the early region of T3 DNA is organized in the same way as that of T7 DNA. Homologous early RNAs and proteins of the two phages have been identified by electrophoresis on polyacrylamide gels in the presence of sodium dodecyl sulfate. Both phages have five early mRNA's, numbered 0.3, 0.7, 1,1.1 and 1.3 from left to right, although no T3 protein that corresponds to the 1.1 protein of T7 has yet been identified. In general, corresponding early RNAs and proteins of the two phages migrate differently on gels, indicating that they differ in molecular weight and/or conformation. In both T7 and T3, gene 0.3 is responsible for overcoming the DNA restriction system of the host, gene 0.7 specifies a protein kinase, gene 1 specifies a phage-specific RNA polymerase, and gene 1.3 specifies a polynucleotide ligase. The 0.3 protein of T3 is responsible for the S-adenosylmethionine cleaving activity (SAMase) induced after T3 (but not T7) infection. However, cleaving of S-adenosylmethionine does not appear to be the primary mechanism by which T3 overcomes host restriction, since at least one mutant of T3 has lost the SAMase activity without losing the ability to overcome host restriction.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arber W., Linn S. DNA modification and restriction. Annu Rev Biochem. 1969;38:467–500. doi: 10.1146/annurev.bi.38.070169.002343. [DOI] [PubMed] [Google Scholar]
- Beier H., Hausmann R. Genetic map of bacteriophage T3. J Virol. 1973 Aug;12(2):417–419. doi: 10.1128/jvi.12.2.417-419.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin M., McGrath J., Waskell L. New RNA polymerase from Escherichia coli infected with bacteriophage T7. Nature. 1970 Oct 17;228(5268):227–231. doi: 10.1038/228227a0. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Hyman R. W. A study in evolution: the DNA base sequence homology between coliphages T7 and T3. J Mol Biol. 1971 Dec 14;62(2):287–301. doi: 10.1016/0022-2836(71)90428-1. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Bautz F. A., Bautz E. K. Different template specificities of phage T3 and T7 RNA polymerases. Nat New Biol. 1971 Mar 17;230(11):94–96. doi: 10.1038/newbio230094a0. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Effect of RNAase III, cleavage on translation of bacteriophage T7 messenger RNAs. J Mol Biol. 1975 Dec 15;99(3):487–499. doi: 10.1016/s0022-2836(75)80140-9. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. T7 early RNAs are generated by site-specific cleavages. Proc Natl Acad Sci U S A. 1973 May;70(5):1559–1563. doi: 10.1073/pnas.70.5.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefter M., Hausmann R., Gold M., Hurwitz J. The enzymatic methylation of ribonucleic acid and deoxyribonucleic acid. X. Bacteriophage T3-induced S-adenosylmethionine cleavage. J Biol Chem. 1966 May 10;241(9):1995–2006. [PubMed] [Google Scholar]
- Golomb M., Chamberlin M. A preliminary map of the major transcription units read by T7 RNA polymerase on the T7 and T3 bacteriophage chromosomes. Proc Natl Acad Sci U S A. 1974 Mar;71(3):760–764. doi: 10.1073/pnas.71.3.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann R., Gomez B. Amber mutants of bacteriophages T3 and T7 defective in phage-directed deoxyribonucleic acid synthesis. J Virol. 1967 Aug;1(4):779–792. doi: 10.1128/jvi.1.4.779-792.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann R. Synthesis of an S-adenosylmethionine-cleaving enzyme in T3-infected Escherichia coli and its disturbance by co-infection with enzymatically incompetent bacteriophage. J Virol. 1967 Feb;1(1):57–63. doi: 10.1128/jvi.1.1.57-63.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrlich P., Schweiger M. T3 and T7 bacteriophage deoxyribonucleic acid-directed enzyme synthesis in vitro. J Virol. 1970 Dec;6(6):750–753. doi: 10.1128/jvi.6.6.750-753.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman R. W., Brunovskis I., Summers W. C. A biochemical comparison of the related bacteriophages T7, phiI, phiII, W31, H, and T3. Virology. 1974 Jan;57(1):189–206. doi: 10.1016/0042-6822(74)90120-2. [DOI] [PubMed] [Google Scholar]
- Meselson M., Yuan R., Heywood J. Restriction and modification of DNA. Annu Rev Biochem. 1972;41:447–466. doi: 10.1146/annurev.bi.41.070172.002311. [DOI] [PubMed] [Google Scholar]
- Rahmsdorf H. J., Pai S. H., Ponta H., Herrlich P., Roskoski R., Jr, Schweiger M., Studier F. W. Protein kinase induction in Escherichia coli by bacteriophage T7. Proc Natl Acad Sci U S A. 1974 Feb;71(2):586–589. doi: 10.1073/pnas.71.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro A. L., Viñuela E., Maizel J. V., Jr Molecular weight estimation of polypeptide chains by electrophoresis in SDS-polyacrylamide gels. Biochem Biophys Res Commun. 1967 Sep 7;28(5):815–820. doi: 10.1016/0006-291x(67)90391-9. [DOI] [PubMed] [Google Scholar]
- Simon M. N., Studier F. W. Physical mapping of the early region of bacteriophage T7 DNA. J Mol Biol. 1973 Sep 15;79(2):249–265. doi: 10.1016/0022-2836(73)90004-1. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Analysis of bacteriophage T7 early RNAs and proteins on slab gels. J Mol Biol. 1973 Sep 15;79(2):237–248. doi: 10.1016/0022-2836(73)90003-x. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Bacteriophage T7. Science. 1972 Apr 28;176(4033):367–376. doi: 10.1126/science.176.4033.367. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Gene 0.3 of bacteriophage T7 acts to overcome the DNA restriction system of the host. J Mol Biol. 1975 May 15;94(2):283–295. doi: 10.1016/0022-2836(75)90083-2. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Genetic analysis of non-essential bacteriophage T7 genes. J Mol Biol. 1973 Sep 15;79(2):227–236. doi: 10.1016/0022-2836(73)90002-8. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Genetic mapping of a mutation that causes ribonucleases III deficiency in Escherichia coli. J Bacteriol. 1975 Oct;124(1):307–316. doi: 10.1128/jb.124.1.307-316.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Maizel J. V., Jr T7-directed protein synthesis. Virology. 1969 Nov;39(3):575–586. doi: 10.1016/0042-6822(69)90105-6. [DOI] [PubMed] [Google Scholar]
- Studier F. W. The genetics and physiology of bacteriophage T7. Virology. 1969 Nov;39(3):562–574. doi: 10.1016/0042-6822(69)90104-4. [DOI] [PubMed] [Google Scholar]