Abstract
Melanomas resist conventional chemotherapeutics in part through intrinsic disrespect of apoptotic checkpoint activation. In this study, using an unbiased genome-wide RNAi screen we identified RhoJ and its effector Pak1, as key modulators of melanoma cell sensitivity to DNA damage. We find that RhoJ activates Pak1 in response to drug-induced DNA damage, which then uncouples ATR from its downstream effectors, ultimately resulting in a blunted DNA damage response (DDR). In addition, ATR suppression leads to the decreased phosphorylation of ATF2, and consequent increased expression of the melanocyte survival gene Sox10 resulting in a higher DDR threshold required to engage melanoma cell death. In the setting of normal melanocyte behavior, this regulatory relationship may facilitate appropriate epidermal melanization in response to UV-induced DNA damage. However, pathological pathway activation during oncogenic transformation produces a tumor that is intrinsically resistant to chemotherapy and has the propensity to accumulate additional mutations. These findings identify DNA damage agents and pharmacological inhibitors of RhoJ/PAK1 as novel synergistic agents that can be used to treat melanomas that are resistant to conventional chemotherapies.
Introduction
Melanoma, an aggressive and fatal malignancy resistant to current therapies, has increased in incidence and mortality over the last several decades (1). The DNA alkylating agents dacarbazine or temozolomide are still utilized to treat metastatic melanoma, but only 16% of patients respond with no improvement in overall survival (2). Melanomas are resistant to a spectrum of chemotherapies, including cisplatin, paclitaxel, docetaxel, combination chemotherapy, immunotherapy, and even newly developed BRAF inhibitors (3) (4). Melanoma cells acquire the ability to invade adjacent tissues and resist chemotherapy early during their evolution (4), further underscoring the importance of developing more effective treatments for this tumor.
Melanoma cells upregulate multiple pathways that allow them to be intrinsically resistant to apoptosis (4)- this includes the activation of anti-apoptotic factors (IAP family, FLIP), the downregulation of pro-apoptotic genes (APAF-1, BAD, BIM), and the activation of prosurvival pathways (NF-kB, AKT). Despite the fact that only 10–20% of melanomas contain p53 mutations, most melanoma tumors contain additional mechanisms to suppress the function of p53, a central regulator of chemoresponsiveness (5). Transcriptional (6) and enzymatic regulators of melanogenesis (7) also modulate melanoma chemoresponsiveness. While extensive studies have identified multiple pathways that control melanoma chemoresistance, this information has not yet led to the development of more effective regimens to treat melanoma.
Synthetic lethal functional genomics screening is an emerging strategy to identify drug targets for the rational design of synergistic agents with selective toxicity towards cancer cells (8, 9). A gene and a drug have a synthetic lethal relationship if mutation or depletion of that gene sensitizes cells to sub-lethal concentrations of a drug (8, 9). Synthetic-lethal screening has been used to identify genes that regulate lung cancer, cervical cancer, and breast cancer chemoresistance (9–11). In this study, we utilize a systems-level screening approach to identify regulators of melanoma chemoresistance with the goal of discovering pathways that could be the molecular targets of new synergistic chemotherapy regimens. This screen identified RhoJ, a CDC42 homologue that regulates endothelial cell migration and angiogenesis (12), as a novel regulator of melanoma cell chemoresponsiveness. We find that RhoJ activates Pak1 in response to DNA damage, which then suppresses ATR’s ability to activate its downstream effectors Chk1 and ATF2. ATR suppression ultimately results in decreased DNA damage-induced apoptosis and the increased expression of prosurvival genes. Taken together, these studies uncover a new signaling pathway that coordinately regulates survival and chemoresistance.
Materials and Methods
Cell culture and reagents
MNT-1 cells were a gift of M. Marks (University of Pennylvania) and were cultured as described (13). C8161 melanoma cells were obtained from Frank Meyskens (University of California, Irvine). SK-Mel-28 melanoma cells were obtained from the ATCC. SK-Mel-28 and C8161 melanoma cells were cultured in RPMI medium with 10% fetal bovine serum. Darkly pigmented normal human melanocytes were purchased from Cascade Biologics and cultured as recommended by the manufacturer. The genome wide siRNA library used in these studies and the transfection protocols were previously described (13). Dacarbazine and cisplatin were purchased from Sigma and IPA-3 was purchased from Tocris Biosciences. Cell line verification was performed by Powerplex genotyping before use.
Antibodies
The antibodies for cleaved PARP, β-Actin, P-Ser345-Chk1, Chk1, P-Ser20-p53, p73, Phospho-Ser343-NBS1, NBS1, Tubulin, ATF2, PLK1, PAK1 and Phospho-PAK1,3 (Ser199/204)/PAK2 (Ser192/197) were purchased from Cell Signaling Technology. RhoJ (monoclonal from Abnova and polyclonal from Sigma), p53 (rabbit polyclonal, Santa Cruz Biotechnology), Sox10 (goat polyclonal, Santa Cruz Inc.), Phospho-Ser490/498-ATF2 (Thermo Scientific), and P-Thr68-Chk2, Claspin (Abcam) were purchased as indicated.
High Throughput Transfection Protocol
High throughput transfection was performed as described (13). MNT-1 melanoma cells in plates were incubated for 72 hours at 37°C/5% CO2 and light activated dacarbazine (14) was added to make a final concentration of 0.1 mg/ml. After an additional 48 hours of drug incubation, a Cell-Titer-Glo Reagent (CTG) (Promega) was delivered to each well and the luminescence values for each well was determined. All SMART-pooled siRNAs used in this study were purchased from Dharmacon. Inc.
Clonogenicity Assays
SK-MEL-28 melanoma cells were transfected with 50 nM pooled siRNAs. 48 hours after transfection, cells were incubated in the presence and absence of 10 µM cisplatin. 72 hours after cisplatin treatment, treated cells were washed with PBS and incubated in fresh media for an additional five days to allow colony regrowth. Relative cell numbers were quantified using a sulforhodamine B assay (Sigma).
Flow Cytometry
For the cell cycle analysis, melanoma cells were trypsinized and washed with PBS, and then were fixed in 70% ethanol. Cells were washed with PBS after fixation, and were stained in a PBS solution containing 40µg/ml Propidium Iodide, 0.1mg/ml RNaseA, 0.1% Triton-X 100. For the apoptosis assays, cells were trypsined, washed with PBS, and stained with Alexa Fluor 488-Annexin V/ Propidium Iodide according to the manufacturer’s protocol (V13245 Invitrogen). After 15 mins staining, cells were subjected to flow cytometry analysis (BD FACSCalibur) and the resulting data were analyzed using FlowJo. We used cells that were not treated with cisplatin to establish gating parameters. Cells in the early stage apoptosis were defined as the PI-negative Annexin V-positive population, while late stage apoptotic cells were defined as the PI-positive Annexin V-positive population.
IPA-3 Cisplatin sensitization studies
SK-MEL-28 Melanoma cells or melanocytes were plated on 96-well plates. 24 hours after plating, cisplatin and IPA-3 was added as indicated. 48 hours after drug treatment, a Cell-Titler-Glo kit (Promega) was utilized to quantify the number of surviving cells. The relative percent of surviving cells in IPA-3 treated and vehicle treated samples was measured to calculate the synergy between IPA-3 and cisplatin.
Results
Genome wide siRNA Screen to Identify Candidate Genes that Modulate Melanoma Chemoresponsiveness
We employed a previously described (9) Dharmacon siRNA library of 84,508 siRNAs corresponding to four unique siRNA duplexes, targeting each of the 21,127 unique human genes arrayed in a one-gene/one-well format (4 siRNAs towards each gene) on 96 well microtiter plates to identify genes that selectively supported melanoma cell survival in the presence of dacarbazine (DTIC). MNT-1 cells, a cell line with intermediate resistance to a previously utilized in vitro genotoxic stress (light-activated dacarbazine) (14), were transfected with 35 nM target siRNA using published protocols (13) and incubated in the presence and absence of an empirically determined sublethal dose of dacarbazine (Figure 1A). An ATP-dependent luminescence cell viability assay (CellTiter-Glo) was used to quantify the impact of each individual siRNA or siRNA + dacarbazine on cell viability. To identify genes that selectively support cell survival in the presence of dacarbazine, we calculated normalized luminescence values from dacarbazine treated and untreated wells for each siRNA and used these values to generate cell viability ratios (Table S1). SiRNAs with low viability ratios (lower cell numbers in siRNA-transfected, dacarbazine-treated wells than in siRNA-transfected, carrier treated wells) correspond to dacarbazine synthetic lethal genes. In addition to identifying 140 candidate genes that modulate melanoma chemoresponsiveness (Figure S1A, red dots), we identified an even larger number of siRNAs that appeared to selectively promote cell survival in the presence of dacarbazine (Z-score greater than 4). Closer analysis of these high Z-score siRNAs revealed that they all potently induced cell death in the absence of dacarbazine (Figure S1B), while the addition of dacarbazine appeared to promote the survival of a fraction of the siRNA transfected cells. Interestingly, this set of genes was enriched in kinases including BRAF (Figure S1C), the target of new melanoma therapies (15). While BRAF siRNAs potently inhibited ATP accumulation in MNT-1 cells (Figure S1B, red bar), the addition of either dacarbazine or cisplatin slightly inhibited the impact of BRAF depletion on ATP accumulation (Figure S1D). Recent clinical trials have demonstrated that the combination of dacarbazine and BRAF inhibitors offer no improvement in survival when compared to patients treated with dacarbazine alone (43), suggesting that DNA damage agents and kinase inhibitors are not necessarily synergistic in the clinical setting. These findings reinforce the importance of establishing a mechanistic rationale for combination chemotherapies prior to the initiation of clinical trials.
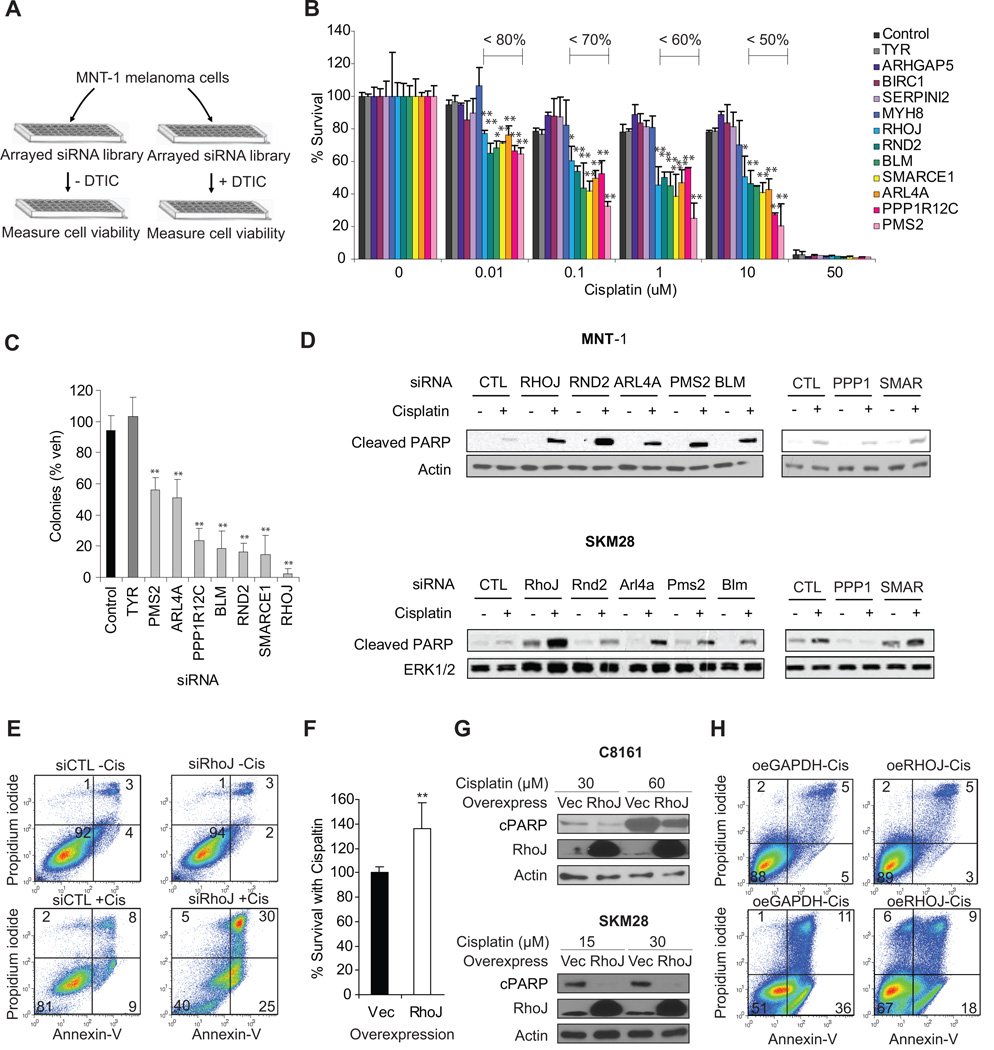
Figure 1. Genome-wide siRNA screen identifies core regulators of melanoma chemoresistance.
A. Identification of regulators of dacarbazine resistance in MNT-1 cells MNT-1 melanoma cells were transfected with a genome-wide siRNA library. 72 hours post transfection, duplicate plates were incubated in the presence and absence of a sublethal dose of dacarbazine for an additional 48 hours. Cell titer glo values were determined for each well and normalized to internal reference samples on each plate, followed by normalization to the experimental mean for each well calculated from the full data set. Similarly adjusted luminescence values from drug treated samples were generated and used to calculate a normalized ratio (drug treated/untreated). B. Identification of Gene Targets that Potently Sensitize Melanoma Cells to Cisplatin. SKM28 melanoma cells were transfected with the indicated siRNA and incubated with cisplatin as indicated. Relative cell number was determined using a Cell TiterGlo assay. The fraction of surviving cells is indicated to highlight siRNAs that significantly sensitize cells to cisplatin. **, p< 0.01 comparing to siControl, determined by Student’s t-t-test. C. Rho GTPases sensitize Melanoma Cells to Chemotherapy Induced Apoptosis. SKM28 and MNT-1 melanoma cells were transfected with 50 nM target siRNAs and incubated in the presence and absence of the 30µM cisplatin. 24 hours after drug treatment, lysates were prepared and subjected to immunoblotting with cleaved PARP and actin loading control antibodies. Representative blot is shown. D. Clonogenicity assays identify genes that regulate cell survival in the presence of cisplatin. SKM28 melanoma cells were transfected with the indicated siRNAs and incubated with 10 µM cisplatin for 72 hours. Wells were then washed and media was repleted. Relative cell number in cisplatin treated and untreated samples were quantified using a sulforhodamine B assay and used to calculate the ratio of surviving cells. **, p< 0.01 comparing to siControl, determined by Student’s t-t-test. E. RhoJ Depletion Sensitizes Melanoma Cells To Cisplatin-induced Apoptosis. MNT-1 cells were transfected with the indicated siRNAs for 48 hours and incubated in the presence and absence of 30 µM cisplatin for 48 hours. Apoptotic cells were defined as Annexin V-positive, while necrotic cells were defined as PI-positive Annexin V-negative population by flow cytometry. F. Overexpression of RhoJ promotes melanoma chemoresistance. The relative sensitivity of vector infected or RhoJ overexpressing C8161 cells to 30 µM cisplatin for 48hrs was measured using a Cell-titer-glo ATP accumulation assay. **, t-test p-value<0.01 versus vector. Bar represents mean ± STD (left panel). G. RhoJ Overexpression Inhibits Cisplatin-induced PARP cleavage. C8161 and SK-MEL-28 RhoJ overexpressing or control vector infected cells were incubated in the presence of the indicated doses of cisplatin for 24hrs. The relative accumulation of cleaved PARP was measured by immunoblotting. H. RhoJ overexpression inhibits Cisplatin induced Apoptosis. C8161 RhoJ overexpressing or control vector infected cells were incubated in the presence of 60 µM cisplatin for 24 hours. Apoptotic cells were defined as Annexin V-positive, while necrotic cells were defined as PI-positive Annexin V-negative by flow cytometry.
Identification of Synthetic Lethal Genes That Sensitize Melanoma Cells to Chemotherapy-Induced Apoptosis
Dacarbazine induces the methylation of guanine bases, resulting in base mispairings which are ultimately repaired by the DNA mismatch repair system (16). A dacarbazine-based synthetic lethal screening approach would be predicted to identify candidate genes that control mismatch repair while also identifying other candidate genes that suppress the cellular response to DNA damage. In order to identify the subset of candidates that suppress the DNA damage response, we next sought to identify siRNAs that sensitized melanoma cells to both dacarbazine and cisplatin, an agent that induces the formation of more bulky DNA adducts which can be repaired by several different DNA repair mechanisms (16). Initial studies eliminated false-positives present in our candidate list (Figure S2A) and utilized a pool deconvolution approach to confirm that the observed dacarbazine sensitization phenotypes for our top 12 hits were not a result of RNAi off-target effects using well established criteria (Fig S2B) (9, 13, 17). The top 12 siRNAs identified in the screen also modulated dacarbazine resistance in another melanoma cell line (SK-MEL-28 melanoma cells) (Figure S2C). In addition, we validated that the siRNAs used in the study effectively inhibited the expression of their target genes (Fig S4A). Dose response studies revealed that seven of the 12 identified siRNAs potently sensitize SK-MEL-28 melanoma cells to cisplatin at low doses (RND2, RHOJ, SMARCE1, PPP1R12C, BLM, PMS2, and ARL4A) (Figure 1B). As predicted, two regulators of DNA damage resistance identified in the screen were DNA repair genes (PMS2, BLM) known to repair damage induced by cisplatin (18, 19) (Table 1). Two other genes (SMARCE1, PPP1R12C) regulate essential processes that are required for DNA replication: chromatin remodeling (20) and mitosis (21), respectively. Three of the seven validated genes were Ras superfamily GTPases (RhoJ, Arl4a, Rnd2), implicating a novel role for Ras superfamily GTPases in the DNA damage response. Colony formation assays revealed that depletion of RHOJ, RND2, BLM, PPP1R12C, and SMARCE1 had profound effects on the proliferation/survival of cisplatin treated SK-MEL-28 cells (Figure 1B). Once we had identified the subset of candidate genes that potently regulate cell proliferation/survival, we next measured the accumulation of apoptosis markers (cleaved PARP) in siRNA-transfected, drug treated cells to identify which of these siRNAs sensitize melanoma cells to cisplatin-induced apoptosis. While low dose cisplatin treatment induced baseline levels of PARP cleavage in MNT-1 and SK-Mel-28 cells treated with control siRNAs (Figure 1C), low dose cisplatin induced significantly more PARP cleavage in MNT-1 and SK-Mel-28 cells treated with RHOJ, RND2, ARL4A, PMS2, and BLM siRNAs (Figure 1C). Of interest, one of the genes that sensitized cells to cisplatin in both cell lines was ARL4A, an Arf-like GTPase that is genetically amplified in melanoma (22). The gene that most potently sensitized cells to cisplatin-induced death was RHOJ. Retrospective analysis of previously published microarray datasets revealed that RhoJ is overexpressed in metastatic melanoma (Table S2) (23), implicating a role for RhoJ in modulating chemoresponsiveness in human tumors. RhoJ siRNA efficiently inhibited RhoJ expression at both the RNA and protein level (Fig S4B). Moreover, RhoJ depletion potently sensitized MNT-1 cells (Figure 1E) and SK-Mel-28 cells (Figure S3A, S3B) to cisplatin-induced apoptosis as measured by the accumulation of Annexin-V positive cells. In contrast, RhoJ overexpression enhanced melanoma chemoresistance (Figure 1F), supressed cisplatin-induced PARP cleavage (Figure 1G), and also inhibited cisplatin-induced apoptosis (Figure 1H). Taken together, these results identify RhoJ as an supressor of DNA-damage induced apoptosis.
Table 1. Ras Family GTPases Potently Sensitize Melanoma Cells to Cisplatin.
GO annotation data and Pubmed searches were used to identify critical cancer phenotypes regulated by targets identified in this screen.
| Gene | EMBL-EBI Family | GO Biological Processes | Go molecular function | Comments |
|---|---|---|---|---|
| RHOJ | Ras GTPase; Small GTPase, Rho type; Ras | actin cytoskeleton organization; regulation of cell shape; regulation of small GTPase mediated signal transduction | GTP binding; GTPase activity | Lower RHOJ expression correlates with sensitivity to epigenetic therapy for sarcoma |
| RND2 | Ras GTPase; Small GTPase, Rho type; Ras | small GTPase mediated signal transduction | GTP binding; GTPase activity | Activates RhoA |
| ARL4A | Ras GTPase; ADP-ribosylation factor; ARF/SAR superfamily | small GTPase mediated signal transduction | GTP binding; protein binding | Genetically amplified in melanoma |
| PPP1R12C | Protein phosphatase 1, regulatory subunit 12A/B/C, eukaryote | not available | not available | Regulates mitosis |
| PMS2 | DNA mismatch repair protein | mismatch repair; reciprocal meiotic recombination; somatic hypermutation of immunoglobulin genes | ATP binding; ATPase activity; endonuclease activity | Rapair for damages caused by Cisplatin; direct p53 effector; interact with MLH1 |
| BLM | DNA helicase, ATP-dependent, RecQ type | double-strand break repair via homologous recombination; G2 phase of mitotic cell cycle; G2/M transition DNA damage checkpoint | ATP binding; bubble DNA binding; DNA strand annealing activity | BLM deficient cells are more sensitive to cisplatin; interact with MLH1 and p53 |
| SMARCE1 | Other | chromatin modification; negative regulation of transcription, DNA-dependent; nervous system development | chromatin binding; DNA binding; N-acetyltransferase activity | Regulates chromatin remodeling during replication |
RhoJ Regulates Melanoma Chemoresistance by Uncoupling ATR From Chk1
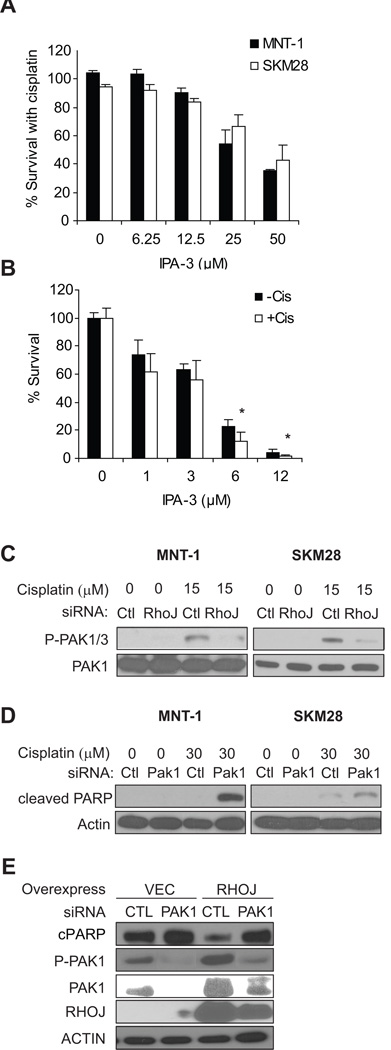
While published studies have revealed that Rho and Cdc42 signaling pathways regulate melanoma invasion (24), these GTPases were not known to regulate chemoresistance. To determine how RhoJ regulates melanoma chemoresistance, we sought to identify downstream pathways activated by RhoJ and determined how these pathways modulate the DNA damage response. RhoJ is a CDC42 homologue, a class of GTPases that can bind and activate group I Pak kinases that contain Pak autoinhibitory domains (25). We utilized a group I Pak inhibitor (IPA-3) to determine whether Pak inhibition sensitized melanoma cells to cisplatin-induced apoptosis. IPA-3 and cisplatin acted synergistically to inhibit ATP accumulation in melanoma cells (Fig 2A). On the other hand, melanocytes were very sensitive to low dose IPA-3 (Fig 2B) and addition of cisplatin only slightly enhanced the inhibitory effect of IPA-3 on ATP accumulation (Fig 2B). Next, we asked whether group I Pak kinases (Pak1, Pak2, or Pak3) are activated by RhoJ. Using an antibody that can recognize phospho-Ser199/204 Pak1/3 or Ser192/197Pak2, we observed that cisplatin treatment induced the accumulation of a band that corresponds to p-Pak1/Pak3 in both MNT-1 and SK-Mel-28 melanoma cells (Figure 2C). Cisplatin did not induce the accumulation of a band that corresponds to phospho-Pak2, which migrates at a different molecular weight, indicating that cisplatin activates either Pak1/Pak3. Using antibodies specific for Pak1 and Pak3, we demonstrated that Pak3 expression is undetectable in the cells studied (Fig S4C), suggesting that cisplatin activates Pak1. Importantly, cisplatin induced the accumulation of phospho Pak1 in a RhoJ-dependent manner (Fig 2C) while Pak1 depletion sensitized both SK-MEL-28 and MNT-1 melanoma cells to cisplatin-induced PARP cleavage (Fig 2D). These results are consistent with published studies which have demonstrated that RhoJ can activate Pak1 (26) and indicate that RhoJ modulates melanoma chemoresponsiveness by activating Pak1. If this is true, then depletion of Pak1 in RhoJ overexpressing cells should sensitize these cells to cisplatin-induced apoptosis. While RhoJ overexpression induced the phosphorylation of Pak1 and suppressed cisplatin-induced PARP cleavage (Fig 2E), depletion of Pak1 in the context of RhoJ overexpression inhibited phospho-Pak1 acumulation and restored the level of PARP cleavage observed in vector expressing cells (Fig 2E), consistent with the contention that Pak1 is activated by RhoJ to deflect DNA damage induced death. While we cannot exclude that RhoJ can activate other group I Paks in other melanoma cell lines when they are expressed, our results indicate that RhoJ activates Pak1 to modulate melanoma chemoresponsiveness in the cell lines studied.
Figure 2. RhoJ Regulates the Activation of Group I Pak kinases.
A. Group I Pak inhibitors sensitize melanoma cells to cisplatin. SK-MEL-28 cells were treated with 30 µM cisplatin and the indicated dose of IPA-3 for 48 hours. The ratio of surviving cells in cisplatin+IPA-3 treated wells was normalized to wells treated with cisplatin alone. B. Group I Pak inhibitors slightly sensitizes melanocytes to cisplatin. Deeply pigmented human epidermal melanocytes are treated with indicated doses of the Pak1 inhibitor IPA-3 in the presence or absence of 20µM cisplatin for 48hrs. The ratios of Cell-Titer-Glo values were normalized to the value in vehicle-treated cells. *, p-value from Student’s t-test < 0.05 compared to IPA-3 alone in the same dose. C. RhoJ Activates Pak1/Pak3 in Response to DNA Damage. SK-MEL-28 and MNT-1 melanoma cells were transfected with the indicated siRNAs and incubated with or without cisplatin for 24hrs. The accumulation of phosphorylated Pak1, Pak2, or Pak3 was determined. The accumulation of phosphorylated Pak2, which migrates at a lower molecular weight than Pak1/Pak3, was not observed. D. Pak1 depletion sensitizes melanoma cells to cisplatin induced PARP cleavage. MNT-1 or SK-MEL-28 cells were transfected with Pak1 siRNAs. 48 hours after transfection cells were incubated with the indicated doses of cisplatin for 20 hours. Cell lysates were prepared and subjected to immunoblotting with the indicated antibodies. E. Pak1 depletion sensitizes RhoJ overexpressing cells to cisplatin-induced PARP cleavage. Vector or RhoJ-overexpressing C8161 melanoma cells are transfected with control or Pak1 siRNAs for 48hrs, and were incubated with 30 µM cisplatin for 24hrs. The relative levels of cleaved PARP, P-Ser199-Pak1, Pak1, RhoJ and Actin were measured.
Once we had identified a potential downstream mediator of RhoJ signaling, we next sought examine if RhoJ/Pak1 modulates the DNA damage response. Our initial screen revealed that RhoJ depletion sensitized cells to dacarbazine, an agent that generates DNA mismatches that are known to activate the ATR kinase {Smits, 2010 #10926}. Once activated, ATR phosphorylates multiple different targets, including H2AX and Chk1, which can then mark the sites of stalled replication forks or induce cell cycle arrest/apoptosis, respectively (16). There is some inherent redundancy in this system, as ATM, a kinase that is induced upon double stranded break formation, can also phosphorylate H2AX when ATR is not active (27). Cisplatin induced similar amounts of p-H2Ax accumulation in RhoJ depleted or control siRNA treated cells (Fig S5A). RhoJ depletion itself did not induce DNA damage and cisplatin induced a similar amount of DNA damage in RhoJ depleted or control siRNA transfected cells (Fig S5B). Taken together, these observations indicate that RhoJ does not inhibit DNA damage from occurring.
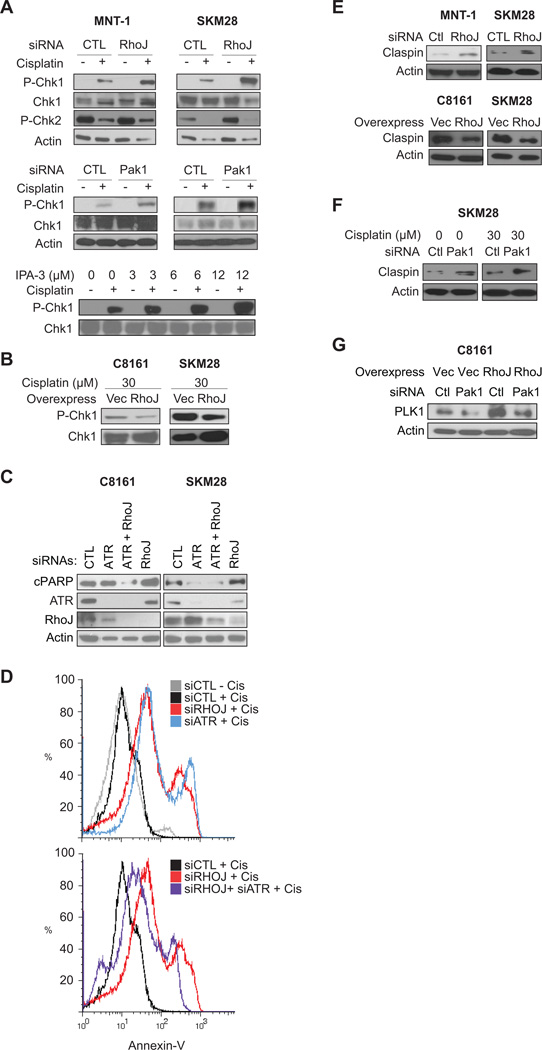
Once we had determined that RhoJ does not prevent DNA damage, we sought to determine whether RhoJ/Pak1 modulates the cellular response to DNA damage. Upon DNA damage, ATR localizes to chromatin where it is activated (28). ATR then phosphorylates Chk1 on Ser345, which can initiate cell cycle arrest or induce apoptosis (28). RhoJ depletion when coupled with cisplatin treatment induced the selective accumulation of Chk1 phosphorylated at its ATR dependent sites (Figure 3A), indicating that RhoJ modulates ATR activity. Surprisingly, pChk2, a kinase that is phophorylated specifically by ATM (29), accumulates in untreated cells but not in cisplatin treated cells (Fig 3A), suggesting that RhoJ modulates ATR and not ATM activity. Pak1 depletion or IPA-3 treatment when coupled with cisplatin treatment also induced the accumulation of pChk1 (Fig 3A), indicating that RhoJ/Pak1 normally suppress Chk1 activation. Finally RhoJ overexpression suppressed cisplatin-induced accumulation of pSer345 Chk1 (Figure 3B), demonstrating that RhoJ and Pak1 inhibits ATR’s ability to phosphorylate Chk1 (28).
Figure 3. RhoJ and Pak kinases Suppress ATR Activation.
A. Depletion of RhoJ/Pak1 or Group I Pak inhibition Enhances Chk1 Activation. MNT-1 or SK-MEL-28 melanoma cells were transfected with the indicated siRNAs for 48hrs and incubated in the presence of 30 µM cisplatin (RhoJ depleted cells) or 15 µM cisplatin (Pak1 depleted cells) for 24hrs. Phospho-ser 345 Chk1 (ATR dependent site), Chk1, Phospho-Thr 68 Chk2 (ATM dependent site) and actin accumulation was measured by immunoblotting. SK-MEL-28 cells were also treated with the indicated dose of the group I Pak inhibitor IPA-3 and cisplatin and phospho-ser 345 Chk1 accumulation was measured. B. RhoJ Suppresses Chk-1 activation upon cisplatin treatment. C8161 and SK-MEL-28 cells overexpressing RhoJ or infected with the control vector were incubated in the presence of the indicated doses of cisplatin for 24hrs. The relative accumulation of pChk1 was measured by immunoblotting. C. ATR Depletion Mitigates the Effects of RhoJ Depletion on Cisplatin induced PARP cleavage. C8161 and SK-MEL-28 melanoma cells were treated with indicated siRNA for 48hrs and then incubated with 30µM cisplatin for 24hrs. Relative accumulation of cleaved PARP was measured. D. ATR Depletion Mitigates the Effects of RhoJ Depletion on Cisplatin induced Apoptosis. MNT-1 melanoma cells were transfected with the indicated siRNAs for 48 hours. Subsequently cells were incubated with 30 µM cisplatin for 48 hours. Apoptosis was quantified by measuring Annexin V staining. E. RhoJ Modulates Claspin accumulation. MNT-1/ SK-MEL-28 melanoma cells were transfected with the indicated siRNAs for 72hrs and the relative accumulation of Claspin was measured by immunoblotting. C8161 and SK-MEL-28 melanoma cells overexpressing RhoJ expressed lower levels of Claspin when compared to cells that express a control vector (right panel). F. RhoJ Modulates Claspin Accumulation via a mechanism involving Pak1. SK-MEL-28 cells transfected with control or Pak1 siRNAs for 48hrs, then treated with or without 30 µM cisplatin for 24hrs. The relative accumulation of claspin was measured by by immunoblotting. G. Pak1 depletion restores RhoJ overexpressing-induced Plk1 accumulation. Vector or RhoJ overexpressing C8161 cells treated with control or Pak1 siRNA for 72hrs and analyzed by immunoblotting.
Once we had demonstrated that RhoJ suppresses ATR activation, we next asked whether RhoJ regulates cisplatin-induced apoptosis in an ATR-dependent manner. While RhoJ depletion when coupled with cisplatin induced the accumulation of cleaved PARP, co-depletion of ATR and RhoJ suppressed cisplatin-induced cleaved PARP accumulation in both p53 wild type (C8161) and p53 mutant (SK-Mel-28) cells (Fig 3C). Similarly, depletion of RhoJ or ATR alone when coupled with cisplatin treatment induced the accumulation of Annexin-V positive cells (Figure 3D, top panel), while co-depletion of ATR and RhoJ suppressed the cisplatin-induced accumulation of Annexin-V positive cells (Fig 3D, bottom panel). Depletion of Chk1 alone was highly toxic, so we were unable to test whether RhoJ modulates chemoresistance in a Chk1 dependent manner (data not shown). Nonetheless, our observations indicate that RhoJ suppresses the DNA damage response by an ATR-dependent mechanism.
Next, we sought to determine how RhoJ/Pak1 uncouples ATR from Chk1. Claspin, a scaffold that couples ATR to its downstream effector Chk1 (30), can be phosphorylated by the Pak1 target Plk1 (31). Phosphorylated Claspin is subsequently degraded by the ubiquitin-proteasome pathway (30). We found that RhoJ overexpression suppressed Claspin accumulation while RhoJ depletion stimulated Claspin accumulation (Figure 3E), suggesting that RhoJ modulates ATR signaling by modulating Claspin accumulation. Similarly, Pak1 depletion modulated Claspin acumulation both in the absence and presence of cisplatin (Fig 3F). RhoJ overexpression induced the accumulation of Plk1, the kinase that phosphorylates Claspin and induces its degradation. Pak1 depletion suppressed the accumulation of Plk1 induced by RhoJ overexpression (Fig 3G), consistent with a role for Pak1 in activating Plk1. Taken together, our results indicate that RhoJ modulates the DNA damage response by suppressing the ability of ATR to phosphorylate its downstream effectors, including Chk1.
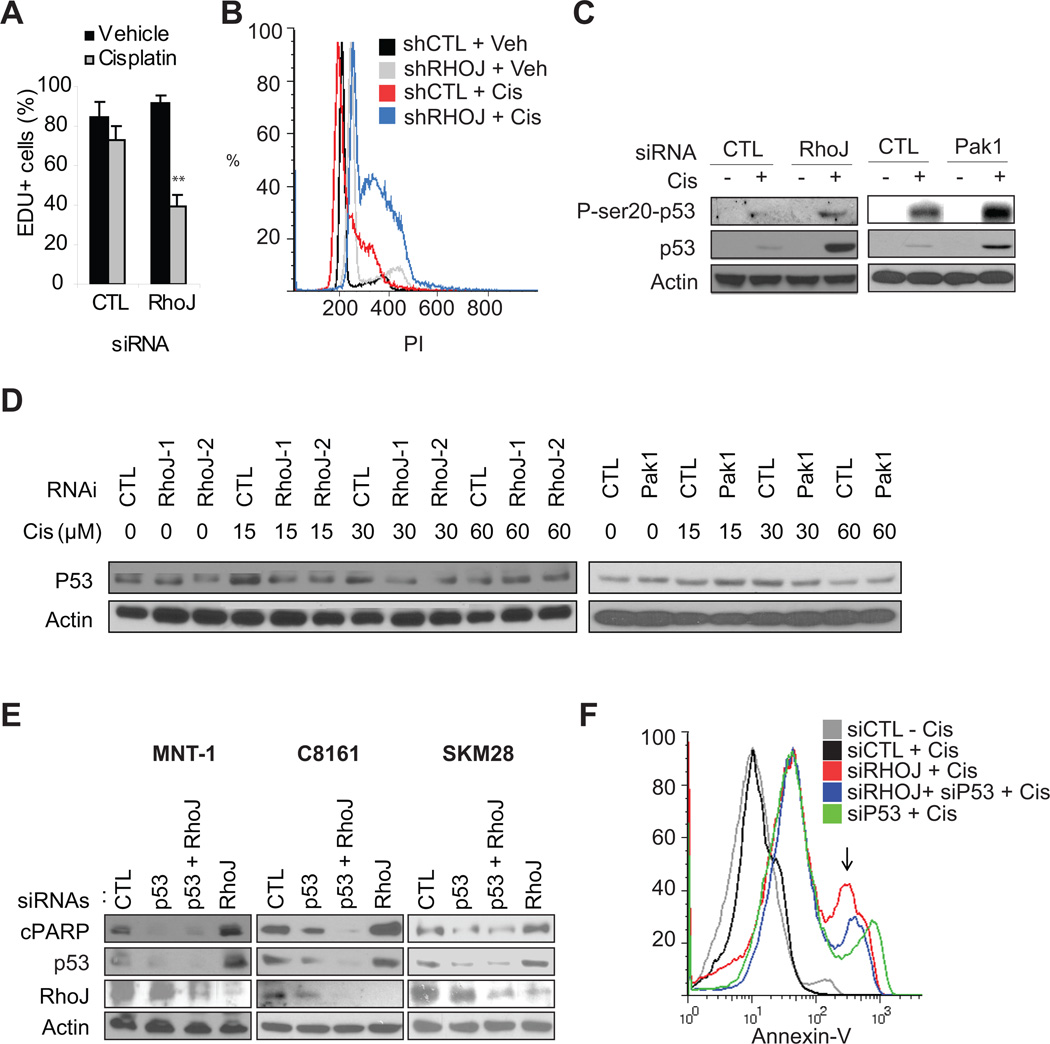
Published studies have indicated that Chk1 primarily functions to induce cell cycle arrest (32), although it can also initiate apoptosis by phosphorylating p53 (33). Depletion of RhoJ suppressed proliferation (Figure 4A, representative images Figure S5D) and enhanced S-phase arrest (Figure 4B) in the presence but not in the absence of cisplatin in both p53 wild type and mutant cells, indicating that RhoJ suppressed Chk1 dependent cell cycle arrest. As Chk1 can also induce apoptosis by activating p53, we next sought to determine whether RhoJ regulates cisplatin-induced apoptosis in a p53 dependent manner. Pak1 and RhoJ depletion induced the selective accumulation of p53 and p53 phosphorylated at Ser20 (a Chk1 dependent phosphorylation site (33)) in p53 wild type cells (Figure 4C). In contrast, Pak1 or RhoJ depletion did not induce the acumulation of p53 (Figure 4D) or pSer20-p53 (data not shown) in p53 mutant SK-Mel-28 melanoma cells. Co-depletion of p53 and RhoJ suppressed cisplatin-induced PARP cleavage (Fig 4E) and cisplatin-induced Annexin-V accumulation in p53 wild type cells (Fig 4F). While co-depletion of p53 and RhoJ suppressed cisplatin-induced PARP cleavage in p53 mutant cells (SK-MEL-28) (Fig 4E), it did not inhibit Annexin-V accumulation in these cells (Fig S3C). Taken together these results suggest that RhoJ regulates cisplatin-induced apoptosis in a p53 dependent manner in wild type cells. In p53 mutant cells, RhoJ’s ability to modulate chemoresponsiveness is only partially dependent on p53, consistent with published studies indicating that mutant p53 can be partially functional (34).
Figure 4. RhoJ Inhibits Drug-Induced Cell Cycle Arrest and Apoptosis.
A. RhoJ Depletion Synergizes with Chemotherapy to Inhibit Proliferation. SK-MEL-28 cells were transfected with 50nM siRNAs and incubated with 1µM cisplatin or vehicle for 48 hours. EdU incoporation was quantified. Representative images are contained in Figure S4D. B. RhoJ Depletion Synergizes with Cisplatin to Induce S phase arrest. C8161 melanoma cells expressing control or RhoJ shRNAs were incubated in the presence and absence of 15µM cisplatin for 24hrs and subjected to FACS analysis as described. Note the accumulation of cells in the peak between G1 (200) and G2 (400). C. RhoJ/Pak1 Modulates Cisplatin-induced p53 accumulation in p53 wild type cells. MNT-1 cells were transfected with the indicated siRNAs and incubated with 30µM cisplatin for 24hr. The accumulation of pSer20-p53 (Chk1 dependent phosphorylation site) and p53 was measured. D. RhoJ/Pak1 does not modulate Cisplatin-induced p53 accumulation in p53 mutant cells. SK-MEL-28 cells expressing the indicated shRNA (left panel) or transfected with the indicated siRNAs (right panel) were incubated with the indicated doses of cisplatin for 24 hours. As pSer20-p53 could not be detected in these cells (data not shown), the accumulation of p53 was measured. E. p53 Depletion Mitigates the Effects of RhoJ Depletion on Cisplatin induced PARP cleavage in p53 wild type cells. P53 wild type (C8161, MNT-1) and p53 mutant (SK-MEL-28) melanoma cells were incubated with the indicated siRNAs for 48 hours followed by 30 µM cisplatin for 24 hours. Relative accumulation of cleaved PARP was measured. F. p53 Depletion partially mitigates the effects of RhoJ Depletion on Cisplatin induced Apoptosis. MNT-1 melanoma cells were transfected with the indicated siRNAs for 48 hours. Subsequently cells were incubated with 30 mM cisplatin for 48 hours. Apoptosis was quantified by measuring the intensity of Annexin V staining.
Next, we sought to better understand how RhoJ regulates melanoma chemoresponsiveness in p53 mutant cells. In the absence of functional p53, Chk1 can modulate apoptosis by phosphorylating the p53 homologue p73 (35). Unfortunately, co-depletion of p73 and RhoJ did not inhibit cisplatin-induced Annexin V accumulation (Fig S3C). Still other studies have identified NBS1 as a protein that is a downstream target of ATR (36) that can modulate DNA-damage induced apoptosis (37). Co-depletion of NBS1 and RhoJ did suppress cisplatin-induced apoptosis in p53 mutant cells (Fig S3C). Moreover, cisplatin treatment induced the the phosphorylation of NBS1 at its ATR specific phosphorylation site only in RhoJ siRNA treated cells (Fig S3D), indicating that RhoJ normally suppresses cisplatin-induced NBS1 phosphorylation. Taken together, these results suggest that RhoJ regulates melanoma chemoresistance by suppressing the ability of ATR to phoshporylate its downstream effectors, resulting in decreased activation of Chk1 and the decreased phosphorylation of Chk1 targets including p53. In p53 mutant cells, RhoJ can suppress cisplatin induced apoptosis both by inhibiting the activation of hypofunctional p53 and by suppressing the ability of ATR to phosphorylate other effectors, such as NBS1. Interestingly, other targets identified in our screen did not modulate Chk1 activation but did modulate p53 accumulation (Figure S5C), reinforcing the concept that p53 is a central regulator of melanoma chemoresponsiveness.
RhoJ Modulates Melanoma Chemoresistance by Modulating the Expression of Sox10
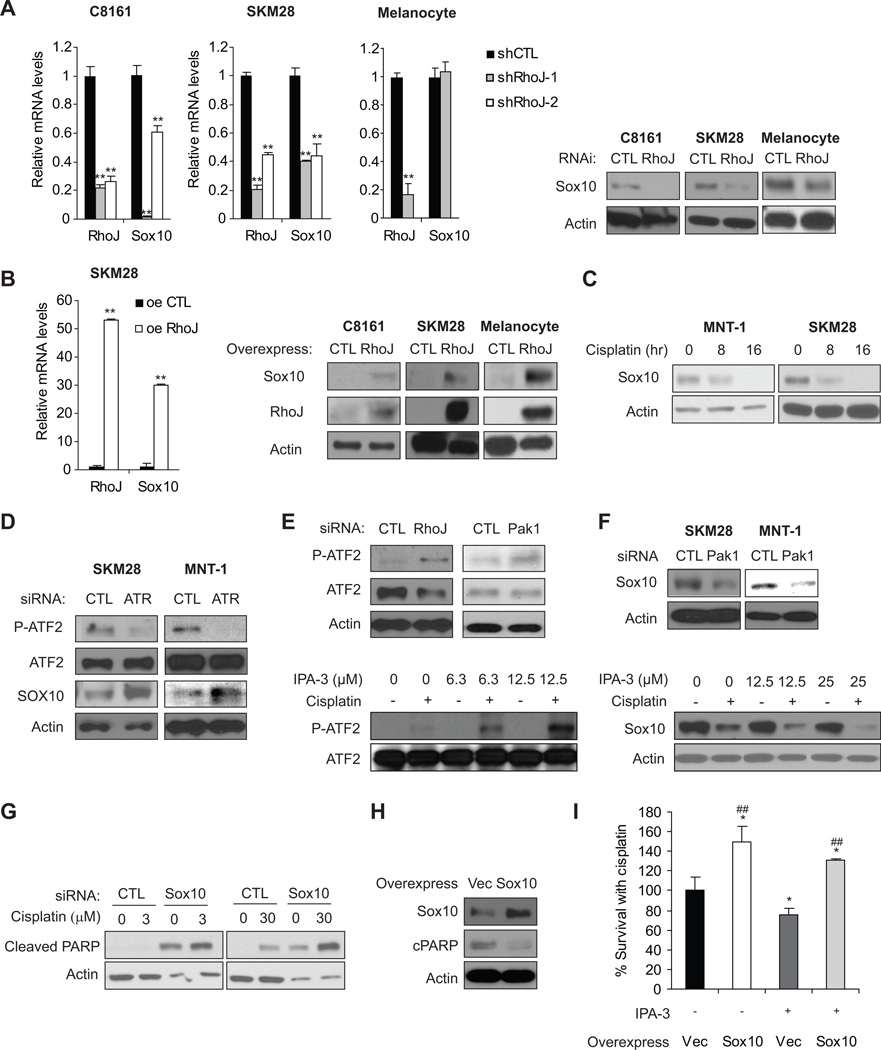
To gain a better appreciation of molecular pathways regulated by RhoJ, we next sought to identify RhoJ-regulated genes and pathways using a microarray-based approach (see dataset, Table S3). Gene set enrichment analysis (38) revealed that regulators of neural development were significantly up or downregulated in cells expressing RhoJ shRNA when compared to cells expressing control shRNAs (Figure S6A). In particular, chronic RhoJ depletion potently inhibited the expression of Sox10 (>100 fold) as well as Sox10 target genes (Figure S6B). Chronic RhoJ depletion was sufficient to inhibit both Sox10 mRNA/protein accumulation in C8161 and SK-MEL-28 melanoma cells (Figure 5A). While RhoJ depletion did not inhibit the accumulation of Sox10 mRNA in melanocytes, it did inhibit the accumulation of Sox10 protein in these cells (Figure 5A). RhoJ overexpression induced the accumulation of Sox10 mRNA and protein in both SK-MEL-28 cells and normal melanocytes (Figure 5B), indicating that RhoJ regulates the expression of Sox10.
Figure 5. RhoJ Inhibits Sox10 Expression in Melanoma Cells.
A. RhoJ is Required for Sox10 Expression in Melanoma Cells but not Melanocytes. The relative mRNA expression of RhoJ and SOX10 was measured by RT-qPCR in C8161, SK-MEL-28 melanoma cells and melanocytes expressing control or two different RhoJ shRNAs (left panel). **, p< 0.01 comparing to shCTL, determined by Student’s t-t-test.Sox10 protein accumulation in RhoJ deficient melanocytes and melanoma cells was measured via immunoblotting (right panel). B. RhoJ Modulates the Expression of Sox10 in Melanoma cells and Melanocytes. Relative mRNA expression of RhoJ and Sox10 in RhoJ overexpressing SK-MEL-28 melanoma cells was quantified using RT-qPCR (left panel). **, p< 0.01 comparing to shCTL, determined by Student’s t-t-test. Lysates from C8161, SK-MEL-28 melanoma cells and melanocytes overexpresing RhoJ were subjected to western blotting with Sox10 and actin antibodies to measure the relative accumulation of RhoJ (right panel). C. Sox10 expression is modulated by the DNA damage response. MNT-1 and SK-MEL-28 cells were treated with 30µM cisplatin at the indicated time points. The relative amount of Sox10 protein was measured via immunoblotting. D. ATR modulates ATF2 phosphorylation. SK-MEL-28 and MNT-1 cells were transfected with control or ATR siRNAs followed by incubation with 30 µM cisplatin for 24hrs. The accumulation of phospho-Thr 490/498 ATF2, a downstream target of ATR, and Sox10 was measured by immunoblotting. E. RhoJ/Pak1 Depletion or Group I Pak inhibition Modulates ATF2 phosphorylation. SK-MEL-28 cells were either transfected with the indicated siRNAs followed by incubation with 30 µM cisplatin for 24hrs or were incubated in the presence or absence 30 µM cisplatin combined with indicated doses of IPA-3 for 24hrs. The accumulation of phospho-Thr-490/498 ATF2 and ATF2 was measured via immunoblotting. F. Pak1 depletion or Inhibition Modulates Sox10 accumulation. SK-MEL-28 cells were transfected with the indicated siRNAs followed by incubation with 30 µM cisplatin for 24hrs or were incubated in the presence or absence of 30 µM cisplatin and the indicated dose of IPA-3 for 24 hours. Relative accumulation of Sox10 and Actin was measured by western blotting. G. Sox10 Depletion Sensitizes Melanoma Cells to Chemotherapy Induced Apoptosis. MNT-1 cells were treated with control or Sox10 siRNA and the indicated dose of cisplatin for 24hrs. Relative accumulation of cleaved PARP and actin was measured via immunoblotting. H. Sox10-overexpression melanoma cells are more resistant to cisplatin-induced cell death. SK-MEL-28 cells overexpressing Sox10 or vector were treated with 30 µM cisplatin for 24hrs before immunolblotting analysis. I. Sox10 Overexpression Partially Mitigates the Effect of Pak Inhibitors on Cisplatin-induced Apoptosis. SK-MEL-28 cells overexpressing Sox10 or infected with virus encoding the empty vector were treated with or without 3 µM IPA-3 in the presence of 30 µM cisplatin for 72 hrs. Cell survival was quantified using a Cell-titer-Glo assay. *, p≤0.05 by Student’s t-test; ##, p≤0.01 using a Student’s t-test.
Sox10, a known oncogene in melanomas, regulates the survival of neural crest precursor cells prior to lineage commitment in the developing embryo (39), and is a reliable marker for melanoma tumor initiating cells (40). Recent studies have determined that Sox10 is required for melanoma formation in both mice and humans and determined that depletion of Sox10 alone is sufficient to induce apoptosis in melanoma cells (41). ATF2, a transcription factor that is activated by ATM/ATR signaling, has recently been shown to suppress Sox10 expression in melanoma cells (42). Depletion of ATR inhibited the phosphorylation of ATF2 at its ATR/ATM target sites while also inducing the accumulation of Sox10 (Figure 5C), consistent with a role for ATR in modulating ATF2 function and repressing Sox10 expression. In contrast, RhoJ or Pak1 depletion or Pak1 inhibition resulted in the accumulation of ATF2 phosphorylated at its ATR target sites (Fig 5D). Pak1 depletion or inhibition ultimately resulted in the decreased accumulation of Sox10 (Fig 5E). Sox10 depletion inhibited melanoma cell survival (Figure S6C) and sensitized melanoma cells to cisplatin-induced PARP cleavage (Figure 5F). In addition, cisplatin treatment inhibited Sox10 accumulation (Figure 5G), indicating that ATR activation can suppress Sox10 expression. Intriguingly, Sox10 overexpression inhibited cisplatin-induced apoptosis in p53 mutant cells (Fig 5H) and Sox10 overexpression partially mitigated the impact of Pak inhibitors on cisplatin sensitivity (Fig 5I). These results indicate that Sox10 expression also promotes melanoma chemoresistance. In summary, our results indicate that RhoJ activates Pak1 in response to DNA damage. Activated Pak1 then suppresses Claspin accumulation, which uncouples ATR from its downstream effectors Chk1 and ATF2. This ultimately results in the functional inactivation of p53 and the increased expression of prosurvival genes (Figure 6).
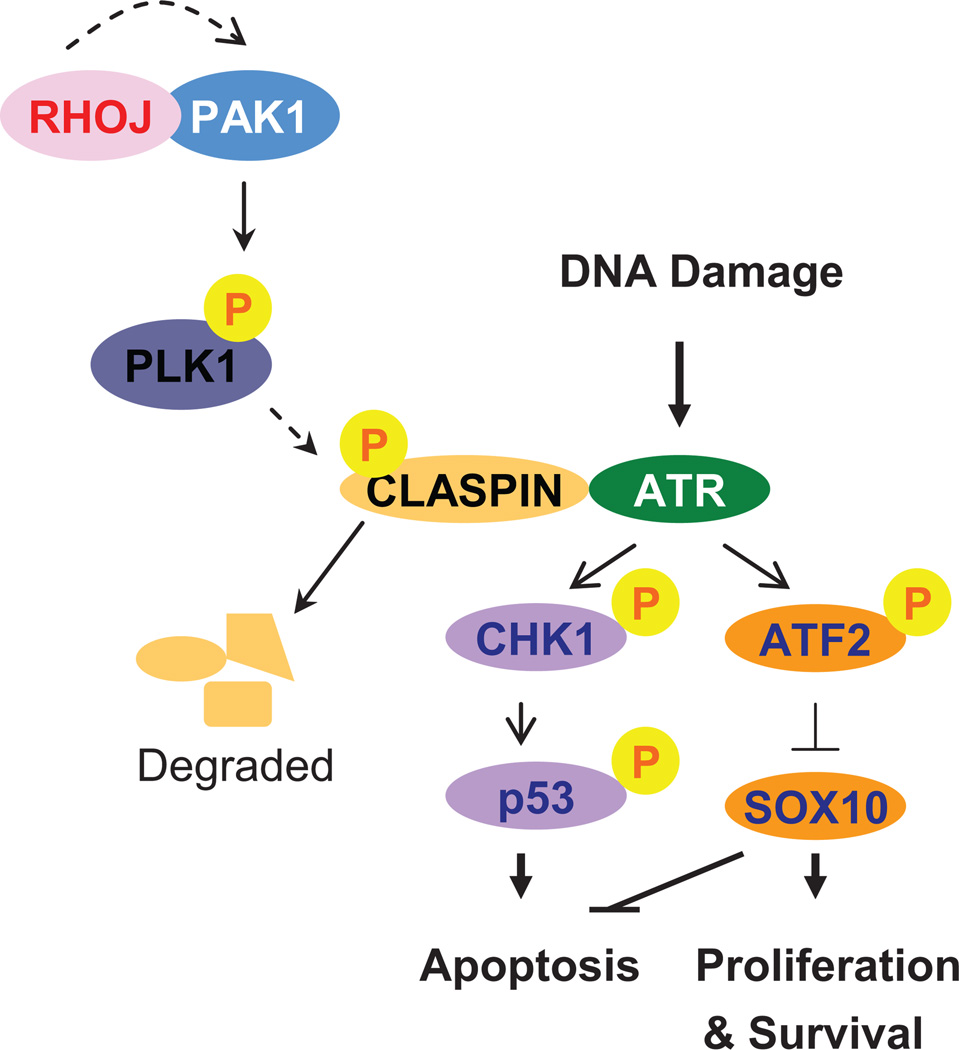
Figure 6. RhoJ/Pak1 Regulate Melanoma Chemoresistance by Suppressing Cellular Mechanisms that Sense DNA Damage.
Our studies revealed that RhoJ/ Pak kinases activate Plk1, resulting in the phosphorylation and degradation of Claspin. In the absence of Claspin, ATR is not able to activate its downstream effectors Chk1 and ATF2. In p53 wild type cells, Chk1 is no longer able to phosphorylate p53 and induce apoptosis while ATF2 is no longer able to regulate the expression of Sox10, which also suppresses apoptosis and stimulates cell proliferation.
Discussion
In this study, we have utilized an unbiased genome-wide functional genomics approach to identify RhoJ and its downstream kinase Pak1 as novel regulators of melanoma cell’s response to DNA damage. Cisplatin activates Pak1 in a RhoJ-dependent fashion, indicating that RhoJ activates Pak1 in order to modulate melanoma chemoresistance. While RhoJ does not inhibit cisplatin-induced DNA damage, it does suppress the phosphorylation of the ATR downstream effectors Chk1 and ATF2. In p53 wild type cells, RhoJ-induced ATR suppression leads to a functional inactivation of p53. In p53 mutant cells, RhoJ-induced ATR suppression results in the decreased phosphorylation of the ATR target NBS1 and the increased expression of Sox10, which can also suppress drug-induced apoptosis. Importantly, parallel studies have determined that Pak1 depletion or Pak kinase inhibition recapitulated all of the phenotypes observed upon RhoJ depletion, suggesting that RhoJ regulates all of these phenotypes in part by activating Pak1. Taken together, these studies identify the suppression of DNA damage sensing, as opposed to the commonly accepted mechanism of apoptosis execution, as a linchpin of chemoresistance in melanoma and nominate RhoJ and Pak1 as optimal therapeutic targets for the rational design of novel synergistic chemotherapy regimens.
Melanocytes in the epidermis produce melanin in response to UVB irradiation and transfer this pigment to keratinocytes, protecting them from UV-induced DNA damage (44). In order to accomplish this, melanocytes must possess mechanisms to resist DNA damage-induced apoptosis and also facilitate the expression of genes that regulate melanin production and survival (45). Of interest, our results suggest that RhoJ and Pak1 limit the response to DNA damage while concomitantly facilitating the expression of central regulators of melanogenesis. RhoJ/Pak1 suppress ATR activation, which can also be induced by UV, allowing the melanocyte to tolerate higher levels of DNA damage than other epidermal cells. ATR suppression in turn leads to the increased expression of Sox10, which can in turn regulate MITF, a transcription factor which controls the expression of multiple genes required for melanin production (46). Elevated expression of Sox10 is observed in melanoma tumors, and Sox10 is required for melanoma formation (41). Similarly, RhoJ is overexpressed in advanced melanomas as compared to primary melanomas (Table S3). In addition, melanoma tumors that occur in UV exposed skin are known to accumulate large numbers of mutations (47), suggesting they have an increased DNA damage tolerance. These observations suggest that, in the normal setting, RhoJ/Pak1 activity allows melanocytes to tolerate of limited amounts of DNA damage in order to facilitate the production of melanin in response to UV stress. When this pathway is activated in melanoma, the resulting tumor cells are profoundly resistant to DNA damage agents and have an increased DNA damage tolerance as evidenced by their accumulation of multiple mutations. Taken in context, our results indicate that melanoma cells are intrinsically chemoresistant because they activate cell-autonomous, lineage-selective pathways that blunt the responsiveness of the DNA-damage surveillance machinery, thus allowing them to persist in the face of high levels of DNA damage. Development of agents that selectively disable RhoJ/PAK1 pathway activity may therefore represent an opportunity to re-sensitize melanoma tumors to conventional chemotherapeutic agents.
Supplementary Material
Acknowledgments
We thank Stacie Loftus and Bill Pavan for their suggestions, and Keith Hoek for his assistance with the RhoJ tumor expression data, and Kyoko Yokomori and Roberta Baronio for their advice regarding the comet assay and p53 sequencing.
Grant Support
This work was supported by the NIH (1K08AR056001 to AKG and CA71443 to MAW), the UC Cancer Research Coordinating Committee, the American Cancer Society (121540-RSG-11-128-01-CSM to AKG), Outrun the Sun, Inc., and the Robert E. Welch Foundation (I-1414 to MAW). The microscopy studies were supported by a P41-RR01192. This work was also partially supported by UL1 RR031985 from the National Center for Research Resources (NCRR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: siRNA, RT PCR
The authors report no conflicts of interest
References
- 1.Tsao H, Atkins MB, Sober AJ. Management of cutaneous melanoma. N Engl J Med. 2004;351:998–1012. doi: 10.1056/NEJMra041245. [DOI] [PubMed] [Google Scholar]
- 2.Bradbury PA, Middleton MR. DNA repair pathways in drug resistance in melanoma. Anticancer Drugs. 2004;15:421–426. doi: 10.1097/01.cad.0000127665.74096.93. [DOI] [PubMed] [Google Scholar]
- 3.Solit DB, Rosen N. Resistance to BRAF inhibition in melanomas. N Engl J Med. 2011;364:772–774. doi: 10.1056/NEJMcibr1013704. [DOI] [PubMed] [Google Scholar]
- 4.Soengas MS, Lowe SW. Apoptosis and melanoma chemoresistance. Oncogene. 2003;22:3138–3151. doi: 10.1038/sj.onc.1206454. [DOI] [PubMed] [Google Scholar]
- 5.Lee JT, Herlyn M. MEK'ing the most of p53 reactivation therapy in melanoma. J Invest Dermatol. 2012;132:263–265. doi: 10.1038/jid.2011.362. [DOI] [PubMed] [Google Scholar]
- 6.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 7.Pak BJ, Lee J, Thai BL, Fuchs SY, Shaked Y, Ronai Z, et al. Radiation resistance of human melanoma analysed by retroviral insertional mutagenesis reveals a possible role for dopachrome tautomerase. Oncogene. 2004;23:30–38. doi: 10.1038/sj.onc.1207007. [DOI] [PubMed] [Google Scholar]
- 8.Mizuarai S, Irie H, Schmatz DM, Kotani H. Integrated genomic and pharmacological approaches to identify synthetic lethal genes as cancer therapeutic targets. Curr Mol Med. 2008;8:774–783. doi: 10.2174/156652408786733676. [DOI] [PubMed] [Google Scholar]
- 9.Whitehurst AW, Bodemann BO, Cardenas J, Ferguson D, Girard L, Peyton M, et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007;446:815–819. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- 10.Bartz SR, Zhang Z, Burchard J, Imakura M, Martin M, Palmieri A, et al. siRNA Screens Reveal Enhanced Cisplatin Cytotoxicity in Tumor Cells Having Both BRCA Network and TP53 Disruptions. Mol Cell Biol. 2006 doi: 10.1128/MCB.01229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 12.Leszczynska K, Kaur S, Wilson E, Bicknell R, Heath VL. The role of RhoJ in endothelial cell biology and angiogenesis. Biochem Soc Trans. 2011;39:1606–1611. doi: 10.1042/BST20110702. [DOI] [PubMed] [Google Scholar]
- 13.Ganesan AK, Ho H, Bodemann B, Petersen S, Aruri J, Koshy S, et al. Genome-Wide siRNA-Based Functional Genomics of Pigmentation Identifies Novel Genes and Pathways That Impact Melanogenesis in Human Cells. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000298. e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lev DC, Ruiz M, Mills L, McGary EC, Price JE, Bar-Eli M. Dacarbazine causes transcriptional up-regulation of interleukin 8 and vascular endothelial growth factor in melanoma cells: a possible escape mechanism from chemotherapy. Mol Cancer Ther. 2003;2:753–763. [PubMed] [Google Scholar]
- 15.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roos WP, Kaina B. DNA damage-induced apoptosis: From specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Whither RNAi? Nat Cell Biol. 2003;5:489–490. doi: 10.1038/ncb0603-490. [DOI] [PubMed] [Google Scholar]
- 18.Shimodaira H, Yoshioka-Yamashita A, Kolodner RD, Wang JY. Interaction of mismatch repair protein PMS2 and the p53-related transcription factor p73 in apoptosis response to cisplatin. Proc Natl Acad Sci U S A. 2003;100:2420–2425. doi: 10.1073/pnas.0438031100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slupianek A, Gurdek E, Koptyra M, Nowicki MO, Siddiqui KM, Groden J, et al. BLM helicase is activated in BCR/ABL leukemia cells to modulate responses to cisplatin. Oncogene. 2005;24:3914–3922. doi: 10.1038/sj.onc.1208545. [DOI] [PubMed] [Google Scholar]
- 20.Kazantseva A, Sepp M, Kazantseva J, Sadam H, Pruunsild P, Timmusk T, et al. N-terminally truncated BAF57 isoforms contribute to the diversity of SWI/SNF complexes in neurons. J Neurochem. 2009;109:807–818. doi: 10.1111/j.1471-4159.2009.06005.x. [DOI] [PubMed] [Google Scholar]
- 21.Banko MR, Allen JJ, Schaffer BE, Wilker EW, Tsou P, White JL, et al. Chemical genetic screen for AMPKalpha2 substrates uncovers a network of proteins involved in mitosis. Mol Cell. 2011;44:878–892. doi: 10.1016/j.molcel.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68:664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calvo F, Sanz-Moreno V, Agudo-Ibanez L, Liu S, Ren S, Morris C, et al. RasGRF suppresses Cdc42-mediated tumour cell movement, cytoskeletal dynamics and transformation. Nat Cell Biol. 2011;13:819–826. doi: 10.1038/ncb2271. [DOI] [PubMed] [Google Scholar]
- 25.Dummler B, Ohshiro K, Kumar R, Field J. Pak protein kinases and their role in cancer. Cancer Metastasis Rev. 2009;28:51–63. doi: 10.1007/s10555-008-9168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vignal E, De Toledo M, Comunale F, Ladopoulou A, Gauthier-Rouviére C, Blangy A, et al. Characterization of TCL, a new GTPase of the rho family related to TC10 andCcdc42. J Biol Chem. 2000;275:36457–36464. doi: 10.1074/jbc.M003487200. [DOI] [PubMed] [Google Scholar]
- 27.Chanoux RA, Yin B, Urtishak KA, Asare A, Bassing CH, Brown EJ. ATR and H2AX cooperate in maintaining genome stability under replication stress. J Biol Chem. 2009;284:5994–6003. doi: 10.1074/jbc.M806739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smits VA, Warmerdam DO, Martin Y, Freire R. Mechanisms of ATR-mediated checkpoint signalling. Front Biosci. 2010;15:840–853. doi: 10.2741/3649. [DOI] [PubMed] [Google Scholar]
- 29.Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- 30.Peschiaroli A, Dorrello NV, Guardavaccaro D, Venere M, Halazonetis T, Sherman NE, et al. SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol Cell. 2006;23:319–329. doi: 10.1016/j.molcel.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 31.Maroto B, Ye MB, von Lohneysen K, Schnelzer A, Knaus UG. P21-activated kinase is required for mitotic progression and regulates Plk1. Oncogene. 2008;27:4900–4908. doi: 10.1038/onc.2008.131. [DOI] [PubMed] [Google Scholar]
- 32.Sorensen CS, Syljuasen RG. Safeguarding genome integrity: the checkpoint kinases ATR, CHK1 and WEE1 restrain CDK activity during normal DNA replication. Nucleic Acids Res. 2012;40:477–486. doi: 10.1093/nar/gkr697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rong JJ, Hu R, Song XM, Ha J, Lu N, Qi Q, et al. Gambogic acid triggers DNA damage signaling that induces p53/p21(Waf1/CIP1) activation through the ATR-Chk1 pathway. Cancer Lett. 2010;296:55–64. doi: 10.1016/j.canlet.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 34.Jordan JJ, Inga A, Conway K, Ha J, Lu N, Qi Q, et al. Altered-function p53 missense mutations identified in breast cancers can have subtle effects on transactivation. Mol Cancer Res. 2010;8:701–716. doi: 10.1158/1541-7786.MCR-09-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez S, Prives C, Cordon-Cardo C. p73alpha regulation by Chk1 in response to DNA damage. Mol Cell Biol. 2003;23:8161–8171. doi: 10.1128/MCB.23.22.8161-8171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Olson E, Nievera CJ, Lee AY, Chen L, Wu X. The Mre11-Rad50-Nbs1 complex acts both upstream and downstream of ataxia telangiectasia mutated and Rad3-related protein (ATR) to regulate the S-phase checkpoint following UV treatment. J Biol Chem. 2007;282:22939–22952. doi: 10.1074/jbc.M702162200. [DOI] [PubMed] [Google Scholar]
- 37.So EY, Ausman M, Saeki T, Ouchi T. Phosphorylation of SMC1 by ATR is required for desferrioxamine (DFO)-induced apoptosis. Cell Death Dis. 2011;2:e128. doi: 10.1038/cddis.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elkon R, Linhart C, Sharan R, Shamir R, Shiloh Y. Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res. 2003;13:773–780. doi: 10.1101/gr.947203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris ML, Baxter LL, Loftus SK, Pavan WJ. Sox proteins in melanocyte development and melanoma. Pigment Cell Melanoma Res. 2010;23:496–513. doi: 10.1111/j.1755-148X.2010.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Civenni G, Walter A, Kobert N, Mihic-Probst D, Zipser M, Belloni B, et al. Human CD271-positive melanoma stem cells associated with metastasis establish tumor heterogeneity and long-term growth. Cancer Res. 2011;71:3098–3109. doi: 10.1158/0008-5472.CAN-10-3997. [DOI] [PubMed] [Google Scholar]
- 41.Shakhova O, Zingg D, Schaefer SM, Hari L, Civenni G, Blunschi J, et al. Sox10 promotes the formation and maintenance of giant congenital naevi and melanoma. Nat Cell Biol. 2012 doi: 10.1038/ncb2535. [DOI] [PubMed] [Google Scholar]
- 42.Shah M, Bhoumik A, Goel V, Dewing A, Breitwieser W, Kluger H, et al. A role for ATF2 in regulating MITF and melanoma development. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001258. e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Meara E, Cruet-Hennequart S, Carty MP. Analysis of protein phosphorylation in cisplatin-treated human cells following annexin V-based separation and multi-antibody screening. Cancer Genomics Proteomics. 2010;7:279–286. [PubMed] [Google Scholar]
- 44.Goding CR. Melanocytes: the new Black. Int J Biochem Cell Biol. 2007;39:275–279. doi: 10.1016/j.biocel.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 45.Abdel-Malek ZA, Kadekaro AL, Swope VB. Stepping up melanocytes to the challenge of UV exposure. Pigment Cell Melanoma Res. 2010;23:171–186. doi: 10.1111/j.1755-148X.2010.00679.x. [DOI] [PubMed] [Google Scholar]
- 46.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 47.Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.