Abstract
Background
The phytohormone ethylene is involved in a wide range of developmental processes and in mediating plant responses to biotic and abiotic stresses. Ethylene signalling acts via a linear transduction pathway leading to the activation of Ethylene Response Factor genes (ERF) which represent one of the largest gene families of plant transcription factors. How an apparently simple signalling pathway can account for the complex and widely diverse plant responses to ethylene remains yet an unanswered question. Building on the recent release of the complete tomato genome sequence, the present study aims at gaining better insight on distinctive features among ERF proteins.
Results
A set of 28 cDNA clones encoding ERFs in the tomato (Solanum lycopersicon) were isolated and shown to fall into nine distinct subclasses characterised by specific conserved motifs most of which with unknown function. In addition of being able to regulate the transcriptional activity of GCC-box containing promoters, tomato ERFs are also shown to be active on promoters lacking this canonical ethylene-responsive-element. Moreover, the data reveal that ERF affinity to the GCC-box depends on the nucleotide environment surrounding this cis-acting element. Site-directed mutagenesis revealed that the nature of the flanking nucleotides can either enhance or reduce the binding affinity, thus conferring the binding specificity of various ERFs to target promoters.
Based on their expression pattern, ERF genes can be clustered in two main clades given their preferential expression in reproductive or vegetative tissues. The regulation of several tomato ERF genes by both ethylene and auxin, suggests their potential contribution to the convergence mechanism between the signalling pathways of the two hormones.
Conclusions
The data reveal that regions flanking the core GCC-box sequence are part of the discrimination mechanism by which ERFs selectively bind to their target promoters. ERF tissue-specific expression combined to their responsiveness to both ethylene and auxin bring some insight on the complexity and fine regulation mechanisms involving these transcriptional mediators. All together the data support the hypothesis that ERFs are the main component enabling ethylene to regulate a wide range of physiological processes in a highly specific and coordinated manner.
Keywords: Ethylene, ERF, Transcriptional regulation, cis-regulatory elements, Tomato
Background
The gaseous plant hormone ethylene is reported to play an active role in a wide range of developmental processes, including germination, flower and leaf senescence, fruit ripening, leaf abscission, root nodulation, programmed cell death, and responses to abiotic stresses and pathogen attacks [1-3]. Components of ethylene signalling have been extensively studied mainly in Arabidopsis model plant [4] revealing a linear transduction pathway that leads to the activation of transcriptional regulators belonging to the Ethylene Response Factor (ERF) type. These last components of the ethylene signalling pathway are responsible for modulating the transcription of ethylene-regulated genes. Whereas the apparent simplicity of the ethylene transduction pathway cannot account for the wide diversity of plant responses to ethylene, the molecular mechanisms that enable this hormone to drive different physiological and developmental processes in an appropriate way are yet to be elucidated. Being encoded by one of the largest family of plant transcription factors, ERF proteins are the most suited step of ethylene signalling where the diversity and specificity of ethylene responses may originate.
ERFs are trans-acting factors unique to plants shown to bind specifically to GCC box cis-acting elements found in the promoter regions of ethylene-responsive genes [5,6]. The ERF family is part of the AP2-containing domain superfamily which also contains the AP2 and RAV families of transcription factors [7]. The AP2 family is characterized by the presence of two copies of the AP2 domain initially described in the Arabidopsis homeotic gene APETALA2[8]. The RAV family contains two DNA binding domains, an AP2-like domain that binds the CAACA motif and a B3-like domain that binds the CACCTG motif [9]. The ERF type family is further divided into two major subfamilies, the ERF and the CBF/DREB (C-repeat binding factor/dehydration responsive element binding factor) subfamilies of transcription factors [10] both containing a single AP2 domain. DREB subfamily is characterized by the presence of a valine and glutamic acid respectively at position 14 and 19 in the AP2 domain, whereas alanine and aspartic acid are conserved in the corresponding positions for ERFs [10]. It has been demonstrated that the amino acid residues involved in DNA binding are not conserved between AP2 and ERFs [11,12]. ERF type proteins have been first isolated in the context of plant responses to biotic stress while DREB are associated with abiotic stress. ERFs have been shown to bind the GCC-box found in ethylene-responsive genes and DREBs to the DRE cis-regulatory element. In Arabidopsis, ERFs account for up to 53 % of the total ERF/DREB proteins [13]. In poplar and rice ERF proteins represent 54 % of the ERF/DREB family [13,14] whereas it represents 51 % in soybean [15]. Comparative structural analysis of Arabidopsis and rice ERFs, have been performed in silico using either the entire protein sequence for phylogenetic analysis [16,17], or the conserved AP2 domain [10,13] to infer relationship between ERF family members. In Arabidopsis the ERF subfamily contains 65 members and is divided into 5 subclasses based on the conservation of the AP2 domain [13]. The ERF domain, first identified as a conserved motif of 59 amino acids in four DNA-binding proteins from Nicotiana tabacum[5] is characterized by 3 β-sheets and 1 α-helix and allows binding of the ERFs to the GCC-box [5,11]. In addition to the requirement for a perfectly conserved GCC motif, it has been suggested that the binding affinity may also depend on the nucleotide environment of the GCC box as well as on the nature of some amino acid residues within the ERF binding domain [17]. Moreover, in line with the data reporting that Pti4, an ERF type transcription factor, is able to bind promoters lacking a GCC box cis-element [18], recent studies demonstrated that ERFs can also bind different cis-elements such as VWRE (Vascular Wounding Responsive Element) [19].
ERFs are ubiquitous in plant kingdom and their functional implications have been studied in various plant species and in a wide range of processes including hormonal signal transduction [5], response to biotic [20,21] and abiotic stresses [22-24], regulation of metabolic pathways [25-28] and developmental processes [29-31]. Expression studies indicated that some ERFs are regulated by abiotic stresses such as wounding and salt stress [17,32,33] and more recently, it was demonstrated that ERFs are also involved in seed germination [3]. A number of studies demonstrated that in addition to regulating the expression of ethylene-responsive genes harbouring the GCC-box, ERFs can also regulate jasmonic acid and salicylic acid-responsive genes [20,34,35]. The presence of distinctive structural features among ERF classes suggests that different members of this family may display different functionalities and binding activities. The differential binding activities of ERFs might represent the mean by which ethylene signalling targets a specific set of genes thus providing the basis of the observed tissue and developmental specificity of plant responses to the hormone. However, structural characterization of ERF proteins has been restricted to in silico analysis and so far the structure/function relationship has been only superficially addressed [36].
The present study shows that the ERF gene family in the tomato is organised into 9 subclasses defined by distinct structural features. Based on functional analysis of 28 tomato ERFs and through testing their ability to activate or repress transcriptional activity of target genes, the data suggest that functional activity is conserved among ERF proteins sharing the same structural features. Moreover, data demonstrate that flanking regions of the core GCC-box sequence are part of the discrimination mechanism by which ERFs selectively bind to their target promoters. The data also show that ERF genes display tissue-specific patterns of transcript accumulation and uncover their regulation by auxin.
Results
Isolation and phylogenetic analysis of tomato ERFs
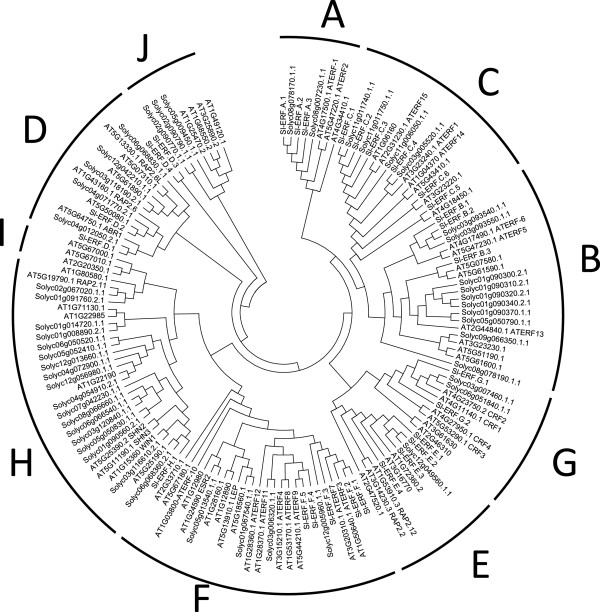
Tomato ERF cDNA clones were initially identified by TBLASTN search [37] in the tomato unigene database (http://solgenomics.net/) using a consensus sequence within the AP2/ERF domain as a query sequence. Forty nine unigenes were found from which AP2, RAV and DREB sequences were discarded based on their distinctive features regarding the number of AP2 domains, the presence of a B3-like domain and the presence of conserved amino acid residues, respectively. Using RACE-PCR extension approach, complete CDSs were obtained for 28 ERF unigenes that are representative of the main ERF sub-groups. Subsequently, building on the annotated whole tomato genome sequence recently released [38], a genome wide in silico screening allowed the identification of up to 146 genes encoding putative AP2-containing proteins distributed into 77 ERFs, 48 DREBs, 18 AP2 and 3 RAVs (Table 1). Since Arabidopsis ERFs have been classified so far using the AP2/ERF domain exclusively [10], we constructed the ERF phylogenetic trees using either the whole protein sequences or only the AP2/ERF domain. Due to the weak homology among ERF proteins outside the AP2/ERF domain, identical classification patterns were obtained with the two clustering methods (Figure 1). However, while 10 subclasses (A to J) define the ERF family in Arabidopsis, only nine are represented in tomato, which lacks subclass I (Figure 1, Table 2). The distribution of the tomato ERF proteins into nine individual subclasses is further supported by the presence within the AP2 domain of distinctive motifs and amino acid signatures specific for each subclass previously described in Arabidopsis[13] . In the absence of a consensual nomenclature and due to a lack of rational naming of ERF genes across plant species, we attempted to rename the tomato genes by giving a letter (A to J) to discriminate between different subclasses and a number to distinguish between members within the same subclass (Table 3). While complying with the most complete classification available in Arabidopsis [13] the proposed nomenclature better clarifies the correspondence between ERF subclasses in different species. The correspondence between the classification adopted here and those previously proposed for tomato ERFs [17,39] and for Arabidopsis ERFs [10,13] is given in Table 2. In this newly proposed classification class XI, defined by Sharma et al. 2010, splits into two subclasses I and J as proposed by Nakano (2006) [13] clearly demonstrating that Arabidopsis subclass I has no representatives in the tomato. Of particular note, members of subclass I harbour imperfect AP2 domain and comparatively to other subclasses is under-represented in Arabidopsis while it is missing in the tomato. Like in Arabidopsis, the overwhelming majority of tomato ERF genes (59 out of 77 genes) are intronless (Table 4). However, while Arabidopsis ERFs can bear at most a single intron, among the 18 tomato intronic genes, 5 have two introns. ERF intronic genes are found in four subclasses in Arabidopsis while they are spread across 6 subclasses in the tomato (Table 4). Although tomato ERF genes could be localized on 12 chromosomes, they present an uneven distribution with Chromosome 1 and 3 bearing 13 and 11 genes, respectively (Table 5). Fine mapping of ERFs on the tomato genome revealed that these genes could be distributed individually or in clusters. In particular, chromosome1 contains a cluster of 5 ERF genes (Solyc01g09300, Solyc01g09310, Solyc01g09320, Solyc01g09340, Solyc01g09370) that likely result from tandem duplication events (Table 5) as suggested by their position within sub-group B in the neighbourhood phylogenetic tree (Figure 1). The correspondence between the nomenclature proposed here for tomato Sl-ERFs and that issued by the ITAG 2.3 nomenclature (38) is provided in Additional file 1.
Table 1.
Summary of the tomato AP2/ERF superfamily
| Classification | Number |
|---|---|
| AP2 family |
18 |
| DREB family |
48 |
| ERF family |
77 |
| RAV family |
3 |
| Total | 146 |
Figure 1.
Phylogenetic tree of Arabidopsis and Tomato ERFs. Different subclasses are named by letters (A to J). Tomato genes for which the corresponding cDNA has been successfully isolated and that were subjected to functional analysis in this paper are named using the Sl-ERF nomenclature (Additional file 1) while other tomato ERFs are named using International Tomato Annotation Genome (ITAG 2.3) nomenclature. Phylogenetic trees were constructed with the whole protein sequences using neighbour joining method.
Table 2.
The distribution of members of the ERF gene family among subclasses constructed in the present study and in previous classifications
Table 3.
Correspondence between the proposed nomenclature of tomato ERFs and previous nomenclatures
| New name | Previous name | Reference |
|---|---|---|
| Sl-ERF.A.1 |
- |
|
| Sl-ERF.A.2 |
LeERF1 |
[17] |
| Sl-ERF.A.3 |
Pti4 |
[40] |
| Sl-ERF.B.1 |
- |
|
| Sl-ERF.B.2 |
- |
|
| Sl-ERF.B.3 |
LeERF4 |
[17] |
| Sl-ERF.C.1 |
JERF2/TERF1 |
[41] |
| Sl-ERF.C.2 |
- |
|
| Sl-ERF.C.3 |
- |
|
| Sl-ERF.C.4 |
TSRF1 |
[42] |
| Sl-ERF.C.5 |
Pti5 |
[40] |
| Sl-ERF.C.6 |
ERF5 |
[43] |
| Sl-ERF.D.1 |
- |
|
| Sl-ERF.D.2 |
- |
|
| Sl-ERF.D.3 |
- |
|
| Sl-ERF.D.4 |
- |
|
| Sl-ERF.E.1 |
LeERF2 |
[17] |
| Sl-ERF.E.2 |
JERF1 |
[42] |
| Sl-ERF.E.3 |
JERF3 |
[44] |
| Sl-ERF.E.4 |
- |
|
| Sl-ERF.F.1 |
- |
|
| Sl-ERF.F.2 |
- |
|
| Sl-ERF.F.3 |
- |
|
| Sl-ERF.F.4 |
- |
|
| Sl-ERF.F.5 |
LeERF3 |
[17] |
| Sl-ERF.G.1 |
- |
|
| Sl-ERF.G.2 |
Pti6 |
[40] |
| Sl-ERF.H.1 | - |
Reference to the first paper describing the gene is given in the last column.
Table 4.
Presence of intron on tomato ERFs
| ERF gene | Class | Intron numbers |
|---|---|---|
| Solyc01g008890 |
H |
2 |
| Solyc01g065980 |
E |
1 |
| Solyc03g116610 |
H |
1 |
| Solyc03g118190 |
D |
1 |
| Solyc03g123500 |
C |
2 |
| Solyc04g012050 |
D |
1 |
| Solyc04g051360 |
D |
1 |
| Solyc04g054910 |
H |
2 |
| Solyc04g071770 |
D |
1 |
| Solyc06g063070 |
A |
2 |
| Solyc06g065820 |
H |
1 |
| Solyc06g068360 |
H |
1 |
| Solyc06g068830 |
D |
2 |
| Solyc09g075420 |
E |
1 |
| Solyc12g042210 |
D |
1 |
| Solyc12g049560 |
E |
1 |
| Solyc12g056590 |
D |
1 |
| Solyc12g056980 | H | 1 |
Table 5.
Distribution of ERFs on tomato chromosomes
| Chromosomes genes per cluster | Number of ERFs genes | Clusters of ERFs/ERFs |
|---|---|---|
| Ch01 |
13 |
1/5 |
| Ch02 |
5 |
2/2 |
| Ch03 |
11 |
1/3 |
| Ch04 |
6 |
|
| Ch05 |
9 |
1/3 |
| Ch06 |
8 |
|
| Ch07 |
4 |
|
| Ch08 |
5 |
1/3 |
| Ch09 |
4 |
1/2 |
| Ch10 |
3 |
|
| Ch11 |
3 |
1/2 |
| Ch12 |
6 |
|
| Total | 77 | 8 clusters |
Subcellular localization of tomato ERFs
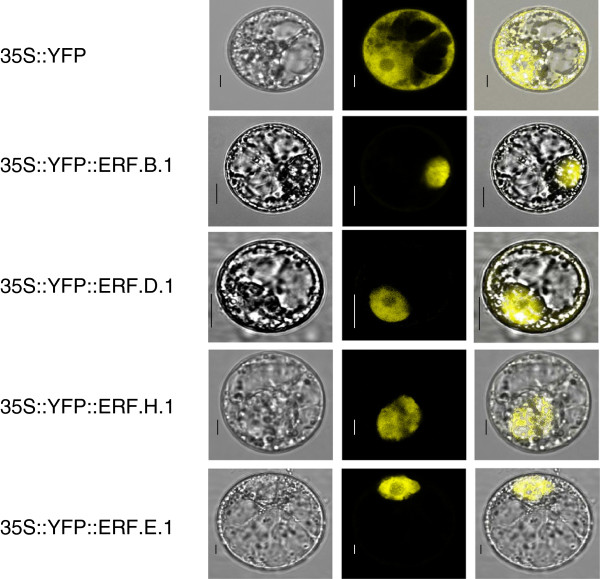
All ERF proteins display at least one canonical nuclear localization signal with the exception of ERF.B.1, D.1 and H.1 that lack any predictable nuclear targeting motif (Additional file 2). Since nuclear import of transcription factors is instrumental to their transcriptional activity, we investigated the subcellular localization of these three tomato ERFs by generating N-terminal fusions with the YFP expressed under the 35S promoter (35S:: YFP-ERF). Transient expression in tobacco BY-2 protoplasts coupled to confocal microscopy analysis clearly indicated that ERF B.1, D.1 and H.1 are exclusively targeted to the nuclear compartment similar to ERF.E.1 which contains a consensus nuclear localization signal (Figure 2).
Figure 2.
Subcellular localization of Sl-ERFs. ERF.B.1, ERF.D.1, ERF.E.1 and ERF.H.1 proteins were fused to the YFP (Yellow Fluorescent Protein) in the N-terminal region and the chimerical proteins were transiently expressed in BY-2 tobacco protoplasts under the control of the 35S promoter. Subcellular localization was then analyzed by confocal laser scanning microscopy. The merged pictures of the yellow fluorescence channel (middle panels) and the corresponding bright field (left panels) are shown (right panels). Control cells expressing YFP alone are shown in the top panel. The scale bar indicates 10 μm.
Relationship between structural features and functional activity of ERFs
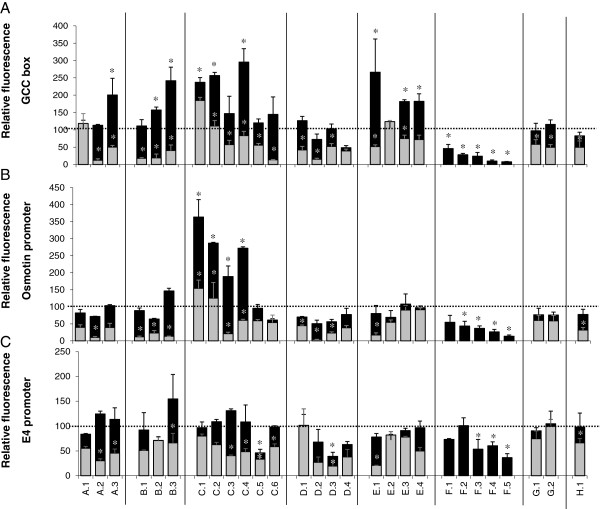
To address whether specific structural features affect the ability of ERFs from different subclasses to drive the transcriptional activity of target promoters, a transient expression assay in a single cell system was used. Tobacco BY-2 protoplasts were co-transformed with effectors constructs carrying the ERF coding sequences driven by the 35S constitutive promoter and reporter constructs consisting of the GFP coding sequence driven either by a GCC-rich synthetic promoter or a native ethylene-responsive promoter. To discriminate between the situation where the absence of activity is due to the lack of binding to the target promoter from those where the ERF binds but remains neutral on the promoter activity, we used chimerical constructs as effectors consisting of ERF coding sequences fused to the SRDX repressor motif [45]. Since the dominant repression activity of the chimerical construct is mediated by epigenetics mechanisms [46], the absence of repression with any of the ERF-SRDX constructs can be interpreted as resulting from the incapacity of the ERF to recognise the target promoter. Figure 3A shows that ERF proteins can not only act as activators or repressors, but can also be neutral on the ethylene-responsive promoters. Experiments carried out with the SRDX fusion demonstrate that all ERFs have the ability to bind the GCC box containing promoter except ERF.A.1 and ERF.E.2. Members of subclass C display the strongest activation activity whereas ERFs from subclass F show the highest repression activity (Figure 3A). ERFs from subclass A, B and E are weak activators on the GCC box whereas those from class G and H are neutral (Figure 3A).
Figure 3.
ERF-mediated transcription from native and synthetic promoters. Transient expression in single cell system has been used to assess the transcriptional activity of ERF proteins from different subclasses. The fluorescence of the reporter gene was measured by flow cytometry upon co-transfection with a reporter construct (GCC:: GFP or Sl-Osmotin Promoter:: GFP or Sl-E4promoter:: GFP) and an effector construct (35S::Sl-ERF or 35S::Sl-ERF-SRDX). The basal fluorescence obtained in the assay transfected with the reporter construct and an empty effector construct was taken as reference (100 % relative fluorescence). (A) ERF activity on synthetic promoter containing 4 direct repeats of the GCC-box. (B) ERF activity on osmotin native promoter containing the canonical GCC motif. (C) ERF activity on E4 native promoter lacking the GCC motif. The results are mean of 3 independent biological replicates. Analysis of variance (ANOVA) was performed to determine the effect of the subclass on ERF activity (p<0.05). The Mann–Whitney test indicates significant result (p<0.05). Black and gray Bars indicate relative fluorescence obtained with native or repression version of each ERF, respectively. Bars indicate SE of the mean.
To test whether the regulation of the ethylene-responsive promoters by ERF proteins is strictly dependent on the presence of a canonical GCC motif, tomato osmotin and E4 native promoters containing or lacking a conserved GCC motif, respectively, were fused to the GFP coding sequence and used as reporter constructs. The data obtained with the fused SRDX repressor motif suggest that up to 16 ERFs out of the 28 tested in this study (Figure 3B) are capable of binding the osmotin promoter (p<0.05). In line with the results obtained with the synthetic GCC promoter, the osmotin promoter is strongly activated by ERFs from subclass C and repressed by ERFs from subclass F. By contrast, the transcriptional regulation of the native osmotin promoter by several ERFs from other subclasses did not match their behaviour with the synthetic promoters. Notably, the native osmotin promoter contains a number of cis-elements that are likely to drive the transcriptional activity in a more complex way than that displayed by the synthetic promoter containing simply the GCC-box. Overall, ERFs showing the strongest effect on the synthetic GCC promoter also display the most significant effect on the osmotin complex promoter. E4 promoter is a well described ethylene-inducible promoter lacking the conserved GCC motif that represent a potential target for ERFs. The data presented in Figure 3C indicate that ERF.C.5, ERF.D.3, ERF.F.3, ERF.F.4 and ERF.F.5, are capable to bind the E4 promoter and to repress its activity, suggesting that some ERFs can be active on ethylene-responsive promoters lacking the canonical GCC box cis-element.
The data reveal that ERF proteins are more active on the synthetic GCC promoter than on native complex promoters. The activity of the ERFs on the GCC box (Figure 3A) indicate that members of the same sub-class tend to modulate the activity of the target promoters in the same way (ANOVA p<10-5). Half of the ERFs tested (14 out of 28) have significant effect on the synthetic promoter while only 8 ERFs are active on the native osmotin GCC-containing promoter and 4 on the native E4 lacking a conserved GCC motif.
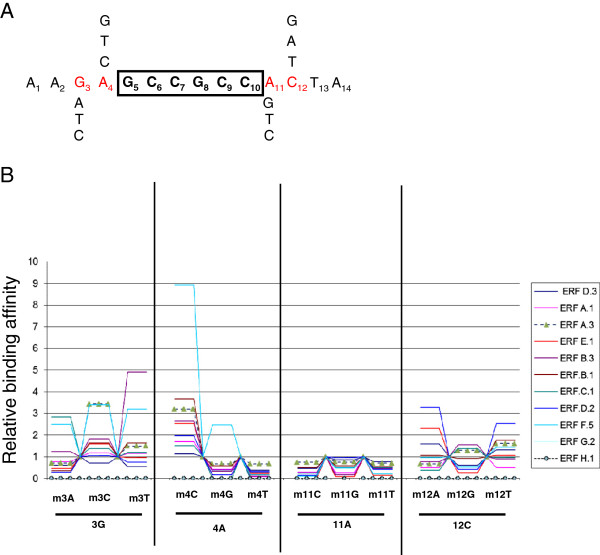
The impact of the GCC box flanking regions on the binding affinity of ERFs
We have previously demonstrated [17] that ERF proteins can display differential affinity to various promoters containing the highly conserved GCC-box. Hence, the hypothesis that the nucleotide environment of the GCC box may impact the binding affinity of the ERFs was tested. A total of 11 ERFs representing all subfamily types were challenged with the native Sl-Chitinase GCC box (5′-A1A2G3A4GCCGCCA11C12T13A14- 3′) or with mutated versions of this motif in gel shift assay experiments (Figure 4A). The four nucleotides flanking the GCC motif (G3, A4, A11 and C12) were mutated and the binding of the ERFs was tested by EMSA and assessed by phosphoimaging. Mutation of A at position 4 (A4) into T or G dramatically decreased the affinity to the GCC box of all ERFs except that of ERF.F.5 (Figure 4B) while the substitution of A4 by a C increases the affinity of all tested ERFs. Strikingly, mutation of A11 to any of the three other nucleotides resulted in a dramatic loss of affinity of ERFs to the GCC box. These data suggest that nucleotides upstream (position 4) and downstream (position 11) to the GCC box impact the binding affinity in the same way for all the ERFs tested. By contrast, substitution of G3 or C12 by another nucleotide resulted in different effect on the binding affinity depending on the ERF tested (figure 4B).
Figure 4.
Binding affinity of ERFs to the GCC-box is impacted by the nucleotide composition of the flanking regions. (A) To assess the role of the nucleotide composition of the close environment of the GCC box, nucleotides flanking the chitinase GCC box were mutated. Different mutated GCC boxes were used as probe in gel shift assay to test the binding affinity of ERFs. (B) Binding affinity of ERFs to the mutated probes. Relative affinity is calculated with non mutated Sl-Chitinase (Solyc10g055810.1) as reference. The data are mean of 3 independent replicates. Analysis of variance with the R package reveals that the flanking region of the GCC box is significantly involved in the affinity of the binding (p<0.05).
Expression pattern of ERFs
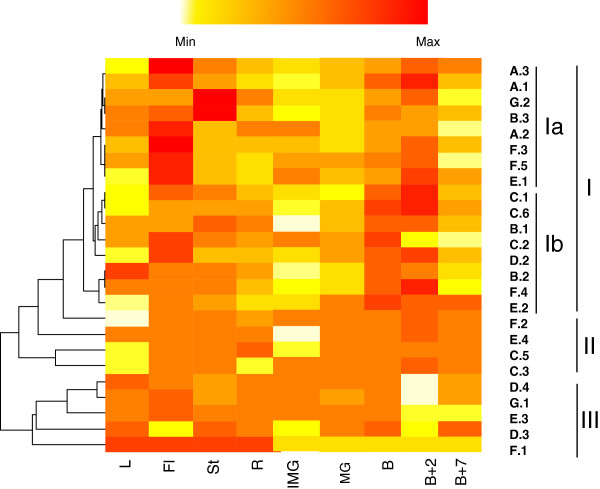
To assist with the elucidation of the physiological function of tomato ERFs, we sought to gain more insight on the spatio-temporal expression at the transcriptional level of each member of the gene family. Transcript accumulation was assessed by qRT-PCR for 25 ERFs whereas for the remaining three, transcripts could not be detected in the nine different plant tissues tested. The heatmap representation of the global expression pattern allowed the clustering of tomato ERFs in three main clades (Figure 5), with clade I (16 ERFs) corresponding to genes preferentially expressed in reproductive tissues, clade II (4 ERFs) corresponding to genes with an ubiquitous expression without tissue preference, and clade III (5 ERFs) mainly active in vegetative tissues. Subclade Ia displays a distinctive expression pattern associated with the breaker +2 and flower stages, while subclade Ib contains genes whose expression is more pronounced at the breaker + 2 and breaker stages. These results suggest that some ERFs operate specifically in vegetative tissues (clade III) or reproductive organs (clade I), whereas few ERFs seem to be ubiquitous and might be involved in the development of various organs (clade II). Interestingly some ERFs are strongly expressed in B+2 and Flowers stage (clade Ia) suggesting a role in fruit set and during ripening while others seem to be active more specifically at the onset of ripening (Breaker and Breaker + 2, clade Ib). These data also point out to the absence of a clear relationship between the structurally defined ERF subclasses and their tissue type expression.
Figure 5.
Heatmap representation of the expression of ERF genes in different tomato tissues. The data obtained by quantitative RT-PCR correspond to the levels of ERF transcripts in total RNA samples extracted from Roots (R), Leaves (L), Stem (St), Flower (Fl), Early Immature Green (IMG), Mature Green (MG), Breaker (B), Breaker + 2 days (B+2), Breaker + 7 days (B+7). The data presented correspond to 3 independent biological repetitions. Red and white colours correspond to high and weak expression of the ERF genes, respectively. Heat map was generated using R software.
Ethylene and auxin regulation of ERF genes
It has been reported that beside their regulation by ethylene, ERF genes can also be induced by other hormones among which auxin [47]. To test the responsiveness of tomato ERF genes to ethylene and auxin, transcripts accumulation in seedlings treated with ethylene or auxin for 5 and 3 hours, respectively, was assessed by qRT-PCR. E4 and SAUR, known as ethylene and auxin-responsive genes, respectively, were used as controls to validate the hormone treatment. Transcript accumulation levels (Figure 6) indicate that 13 tomato ERFs are significantly induced (p<0.05) by ethylene and 4 significantly (p<0.05) down-regulated by this hormone. Interestingly, the expression of up to 12 ERFs is also positively regulated by auxin. Among these, 4 ERF genes (ERF.B.1, Sl-ERF.E.1, Sl-ERF.F.4, and Sl-ERF.H.1) are up-regulated by both hormones whereas ERF.F.5 undergo opposite regulation by auxin and ethylene. More surprisingly, 6 ERF genes (ERF.A.1, ERF.A.3, ERF.C.4, ERF.C.6, ERF.D.4 and ERF.F.1) are significantly up-regulated by auxin while they are not responsive to ethylene.
Figure 6.
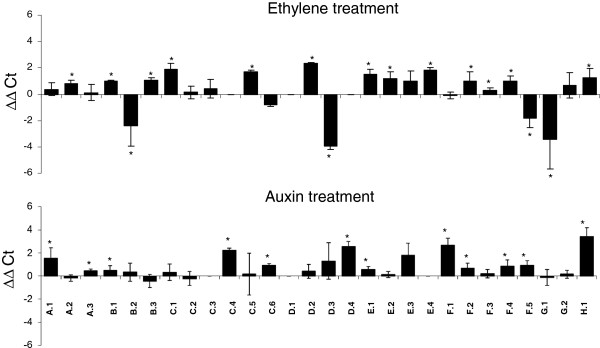
Expression of ERFs in response to ethylene and auxin treatments. Quantitative RT-PCR of ERF transcripts in total RNA samples extracted from 5-days dark growing seedlings treated for 5 hours with ethylene or with IAA for 3 hours. ΔΔCt refers to the fold of difference in ERF expression relative to the untreated seedlings. Stars indicate a statistical significance (p<0.05) using Mann-Withney test.
Discussion
A wide range of developmental processes are known to rely at least partly on ethylene action, yet, the molecular mechanisms underlying the diversity of plant responses to this hormone are still awaiting some in depth characterization. The present study brings new insight on structural features that impact the binding affinity of Ethylene Response Factors (ERFs) to ethylene responsive promoters. Upstream of ERFs, the ethylene signal propagates via a linear transduction pathway which can hardly account for the wide range of ethylene responses. In its downstream part, ethylene signalling leads to the activation of ERFs that are encoded by one of the largest multigene family of transcription factors [7], and these transcriptional mediators may therefore represent one of the main step where the diversity and selectivity of ethylene responses can take place. The structural and functional characterisation of tomato ERFs carried out here provides some clues to the means by which ethylene selects target genes that are required to bring into play the appropriate physiological responses in a tissue-specific and developmentally defined manner. Taking advantage of the recent release of the complete annotated tomato genome sequence [38], a genome wide in silico screening identified 146 genes encoding putative AP2-containing proteins distributed into 77 ERFs, 48 DREBs, 18 AP2 and 3 RAVs (Table 1). Subsequently, the functional analysis concentrated on 28 members of the tomato ERF family encompassing 8 of the 9 subclasses of the gene family.
The data indicate that all ERF proteins display a clear nuclear localization, even though three members of the family lack any predictable nuclear targeting motif suggesting that their import to the nucleus may occur by a non-conventional localization signal or they might be conveyed to this compartment via an interaction with a yet unknown nuclear localized partner.
The large diversity among ERFs is well illustrated by the presence of distinctive motifs that are sufficient to discriminate between different subclasses. Recent studies already suggested a link between the structural classification of ERFs and their physiological function [13], yet, this hypothesis has not been supported by strong experimental data at the level of whole ERF family. It was reported that within class E, the conserved specific N-terminal domain, initially described in tomato ERFs [17], is dedicated to an oxygen dependent sequence of post-translational modifications that leads to degradation of the protein under aerobic conditions. In hypoxia condition, the RAP2.12 ERF protein is relocated from plasma membrane to nucleus to activate gene expression associated with hypoxia acclimation [48]. These results suggest that members of the subclass E are involved in hypoxia response.
Transient expression assays indicated that the activity of ERFs on synthetic GCC box promoter correlates well with their structural classification. That is, ERFs from subclass C are strong activators and those belonging to subclasses A, B and E are weak activators, whereas members of subclass F are clear repressors. By contrast, ERFs from subclasses G and H display no activity when challenged with the GCC box. Noteworthy, ERFs show weaker activity on the native complex osmotin promoter than on the synthetic promoter. This may be due to the high basal expression displayed by the native osmotin promoter in the absence of ERF effectors, which likely minimizes the induction effect observed in the presence of ERFs (Additional file 3). Moreover, the experimental data show that ERFs can regulate transcription from both types of ethylene–responsive promoters either containing (osmotin promoter) or lacking (E4 promoter) the GCC-box thus revealing their capacity to bind cis-elements without the canonical GCC motif. While it cannot be ruled out that ERFs may induce indirectly the E4 promoter through the activation of primary target genes encoding transcriptional proteins active on the E4 promoter, our data suggest that ERFs can interact with other cis-acting elements beside the canonical GCC motif. This is in agreement with recent studies demonstrating that, in addition to the GCC box, ERFs can bind cis-acting elements such as VWRE and GCC-like [19,49-51]. The clustering of the ERF proteins according to their activity on the GCC box, indicates that members of the same subclass tend to modulate the activity of the target promoters in the same way (Figure 3A). While the data show that the repressor and activator activities of ERFs correlate well with their structural classification, the recognition of the GCC-box by a particular ERF seems to depend on the promoter used and might be impacted by the nucleotide environment surrounding the cis-acting element.
Indeed, gel shift assay experiments show that the environment of the GCC box greatly impacts the affinity of ERF proteins to the conserved cis-element. In particular, the data (Figure 4) clearly demonstrate that the nature of the nucleotides at position N3 and N12 can have different and sometime opposite effect on different ERFs, thus suggesting that these bases contribute to the binding specificity of various ERFs to target promoters. Taking together these results suggest that the two nucleotides directly flanking the GCC box at position N4 and N11 are essential for the binding of any ERF to the GCC-box and, therefore, their nature determines whether or not the cis-element is functional for the binding to an ERF. By contrast, the nature of nucleotides at position N3 and N12 can either enhance or reduce the binding affinity, which supports their contribution in determining the binding specificity of various ERFs to target promoters. Previous work carried out with native promoters already stressed the putative role of the GCC box flanking regions in impacting the binding activity of the ERFs [17]. The same study also postulated that variation in amino acid composition within the binding domain may also impact the binding affinity of ERFs to their target promoters [17]. It was also postulated that the conserved motifs lying outside the DNA binding domain may also contribute to the differential affinity displayed by ERF proteins towards the GCC box-containing promoters. Recent study demonstrated that the trans-activating activity of some ERFs was localized in the acidic domain [52]. In keeping with this observation, ERF.D.1 and ERF.D.2 are lacking the acidic activator domain and accordingly are unable to activate the synthetic GCC box promoter. On the other hand, subclasses A, B, C and E ERFs harbour one or more acidic domains, however, not all of them display transcriptional activation of the synthetic promoter. Members of class F have an EAR repressor domain in the C-terminal region of the protein and accordingly they are all repressors of the activity of the GCC box-containing promoters.
Unravelling the expression pattern of ERF genes provides important clues towards the elucidation of their physiological function since physiological effects of transcriptional mediators not only depend on their activation or repression function, but also on their specific pattern of expression. While consistent with previous description concerning the expression of some ERF genes like Pti4[53] and ERF1-4[17], the data distinguish different ERF groups depending on whether they are preferentially expressed in vegetative, or reproductive tissues or whether they show ubiquitous expression with no tissue preference. Noteworthy, structural subclasses did not display tissue or organ specialisation as exemplified by the fruit development process throughout which ERFs from structurally distinct subclasses are expressed from flower anthesis to fruit ripening. This could be in line with the hypothesis that different responses to ethylene taking place in different tissues and at different developmental stages are mediated by different ERF proteins. Tissue and abiotic stress expression pattern of tomato ERFs has been already reported [39], however, the hormone-dependent expression remained unknown. As expected, a high number of ERF genes are ethylene-responsive with 13 ERFs being up-regulated and 4 down-regulated upon ethylene treatment. Less expected, our study revealed that up to 12 tomato ERF genes were regulated by auxin among which 6 undergo double regulation by ethylene and auxin while the remaining 6 others are exclusively auxin-responsive. It was already reported that, beside ethylene, ERFs can be regulated by salicylic acid and jasmonic acid [20,35,54], but the auxin regulation of such a high number of ERF genes has not been reported. The auxin regulation of the ERFs shown to be active on ethylene responsive promoters, suggests their potential contribution to the cross-talk between the two hormones. The phytohormones auxin and ethylene are essential regulators of plant development and it is well documented that ethylene and auxin regulate common physiological aspects such as hook formation [55,56], root hair differentiation [57], root elongation [58], root growth [59], and hypocotyl phototropism [60]. The mechanisms underlying the interactions between the two hormones are becoming better understood even though only few molecular players of this cross-talk have been identified so far. Besides acting independently on the same target genes, ethylene and auxin can mutually regulate each other’s biosynthesis and response pathways. In support to this idea, we previously reported that the down-regulation of Sl-IAA3, encoding a tomato transcriptional regulator from the Aux/IAA type, results in phenotypic responses related to classical auxin and ethylene-regulated processes [61,62]. Therefore, uncovering the auxin responsiveness of some members of the ERF gene family, may define new actors potentially involved in the cross-talk between the two hormones. Global transcriptomic analyses of tomato lines under-expressing Sl-IAA9, another tomato Aux/IAA gene [63,64], revealed the altered expression of a high number of ethylene-related genes, further supporting the idea of an active link between auxin and ethylene signalling during the flower to fruit transition. In keeping with the complex role of ERFs in mediating various hormone responses, ERF.A.1 and ERF.D.4 that are highly expressed in mature flowers, display strong up-regulation by auxin but not by ethylene. On the other hand, ERF.A.2 and ERF.C.1 are up-regulated by ethylene but not by auxin, and they are strongly expressed in the flower. Interestingly, many ERFs are expressed in flower and at early ripening stages (B and B+2), suggesting their putative auxin-dependent role in fruit set and fruit ripening process.
Conclusion
The present study provides some molecular clues on how ERFs can contribute to the specificity and selectivity of ethylene responses through (i) the differential expression of the gene family members, (ii) the ability to negatively or positively impact transcriptional activity and, (iii) the capacity to select with specificity their target genes based on the nucleotide environment of the GCC-box. Considering the diversity of their transcriptional activity and expression patterns, ERFs possess the necessary features for channelling ethylene signalling to a selected set of genes required for the appropriate developmental responses or the desired responses to environmental cues. The insight gained in this study opens new prospects towards assigning a specific role for individual ERFs in controlling developmental processes and for identifying the direct target genes for each member of the ERF family.
Methods
Plants growing
Tomato (Solanum lycopersicum cv MicroTom) plants were grown in climatic chamber. The conditions are the following: 14-h-day/10-h-night cycle, 25/20°C day/night temperature, 80 % hygrometry, 250 μmol.m-2.s-1 irradiance.
Cloning of over expressing and repressing ERF constructs
The full length CDS were obtained by RACE using BD SMARTTM RACE cDNA Amplification kit (Clontech, http://www.clontech.com) and the complete ERF CDS were, cloned in over-expression vector, with or without SRDX fusion in C-terminal [45] (Additional files 4, 5).
Ethylene and auxin treatments
Five day-old dark-grown seedlings were treated with air or ethylene gas (50 μL/L) for 5 hours and RNA was extracted from the corresponding tissues. Auxin treatments were performed on 7 day-old light-grown seedlings by soaking (3 hours) in auxin-containing (20 μM) or auxin-free MS solution. Three independent biological repeats were performed for each experiment.
Analysis of ERF gene expression
RNA samples were obtained from different plant tissues: Immature Green fruit (17 days post anthesis), Mature Green fruit (1 day before Breaker), Breaker fruit, Breaker + 2 days, Breaker + 7 days, leaf, flowers, roots and stem. Real-time quantitative PCR was performed according to Pirrello et al. 2006 (Additional file 6). Heat map representation was performed using centring and normalized ΔCt value, with R software to visualize clustering.
Phylogenetic analysis
In order to identify Sl-ERF genes in the tomato genome, the AP2/ERF domain of a tomato Ethylene Response Factor sequence (GenBank number NP_001234308) was used as BLAST query sequence against the tomato ITAG2.30(Sl2.40) protein database . The same database was also screened by an HMM analysis using a typical AP2/ERF domain PF00847 as query. One hundred forty six genes were identified as possibly encoding proteins containing the AP2/ERF domain (Table 1). The evolutionary history was inferred using the Neighbour-Joining method [65]. The optimal tree with the sum of branch length = 13.53632771 is shown. The phylogenetic tree was linearized assuming equal evolutionary rates in all lineages [66]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method [67] and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). There were a total of 63 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 [68]. Conserved motifs were determined using MEME version 3.5.5 [69].
Transcriptional activity tests by transient expression in a single cell system
A synthetic reporter construct (4XGCC-GFP) was generated by fusing a synthetic promoter containing 4 GCC-box repeats upstream of the minimal −42 to +8 TATA box from the 35S promoter of Cauliflower mosaic virus to the coding region of the Green Fluorescent Protein (GFP). Reporter constructs were also generated with native promoters, E4 (S44898) and Sl-Osmotine (C08HBa0235H18.1), fused to GFP. Effectors constructs were generated by fusing the 35S promoter to the CDS of the ERF genes. For transient assays, tobacco (Nicotiana tabacum) BY-2 protoplasts were co-transformed with reporter and effector constructs [61]. Transformation assays were performed in three independent replicates. After 16 h, GFP expression was analysed and quantified by flow cytometry (FACS Calibur II instrument, BD Biosciences) on a flow cytometry platform (IRF31). Data were analysed using Cell Quest software. For each sample, 100–1000 protoplasts were gated on forward light scatter and the GFP fluorescence per population of cells corresponds to the average fluorescence intensity of the cell population after subtraction of autofluorescence determined with non-transformed BY-2 protoplasts. The data were normalized using an experiment with protoplasts transformed with the reporter vector in combination with the vector used as the effector plasmid but lacking the Sl-ERF coding sequence.
Subcellular localization
YFP N-terminal fusions were obtained with ERF.B1, D.1, H.1 and E.1 and used for tobacco protoplast BY-2 transfection according to Chaabouni et al. 2009 [61]. The subcellular location of the fluorescence was determined after 20 hours using confocal microscopy.
Electro mobility shift assay
Proteins used for the gel shift assay were produced in vitro using a “TNT®T7 Quick kit for PCR DNA” (Promega, http://www.promega.com). Primers used for amplification of ERFs were design according to manufacture recommendation and include Kozak T7 sequences. Gel retardation experiments were performed with the GCC-box probe mutated in the flanking nucleotides according to the procedure described in Tournier et al. 2003 [17] (Additional file 7). Scanning and radioactivity quantification were achieved by a “Phosphoimaging Fujifilm Bas 5000” and the software “Image Gauge” (Fuji Film, http://www.fujifilm.com).
Sequences
Coding DNA sequences of the 28 studied ERFs are provided in Additional file 8.
Authors’ contribution
JP and BCNP participated in the design of the study, performed the experiments, and contributed to the writing of the manuscript. WZ contributed to the cloning of ERF genes and to qRT-PCR experiments. KC participated in the design of the project. IM carried out the transcriptional activity experiments in the single cell system. MZ contributed to the in silico analysis of the ERF gene families. AL and JCP participated in the ethylene and auxin treatment experiments. MOT contributed to the design of the SRDX-repressor motif experiments. FR contributed to the design of the study and of the experiments, to bioinformatics analysis and to drafting the manuscript. MB conceived the study, and participated in its design and coordination and contributed to the writing of the final manuscript. All authors read and approved the final manuscript.
Supplementary Material
Correspondance between Sl-ERF nomenclature and ITAG 2.3 nomenclature.
Schematic diagram of protein structure of the isolated 28 tomato ERFs defining the 8 subclass. Each colored box represents the AP2/ERF domain and conserved motifs, as indicated below the diagram. The position of the motif is indicating by the number on the top of the diagram. The name of motif by Nakano is given inside the box [13]
Comparative basal activity of the synthetic promoter (4 X GCC) and the native promoters, fused to the GFP in the absence of added ERF effectors.
Primers used for RACE amplification.
Primers used for CDS amplification. Each pair of primers encompasses the complete CDS from the ATG to the STOP codon. Adaptors attB1 and attB2, have been added at the 5′ end of sense and antisense primers, respectively.
Primers used for qRT-PCR. Primers designed with PRIMER EXPRESS software in sequence specific regions for each ERF. Specificity was further checked using BLAST against all tomato unigenes (Tomato unigene database).
Primers used for the synthesis of radioactive GCC box probes utilized in EMSA experiments.
Coding sequences of the 28 studied ERF.
Contributor Information
Julien Pirrello, Email: julien.pirrello@cirad.fr.
BC Narasimha Prasad, Email: bcnp5@yahoo.com.
Wangshu Zhang, Email: zjuzws@hotmail.com.
Kunsong Chen, Email: akun@zju.edu.cn.
Isabelle Mila, Email: mila@ensat.fr.
Mohamed Zouine, Email: mohamed.zouine@ensat.fr.
Alain Latché, Email: latche@ensat.fr.
Jean Claude Pech, Email: pech@ensat.fr.
Masaru Ohme-Takagi, Email: m-takagi@aist.go.jp.
Farid Regad, Email: regad@ensat.fr.
Mondher Bouzayen, Email: bouzayen@ensat.fr.
Acknowledgement
This work forms part of the requirement for the degree of Ph.D. for JP. This research was supported by the European Integrated Project EU-SOL (FOOD-CT-2006-016214), by IFCPAR-CEFIPRA Indo-French program (Grant 3303–02) and by funds from the “Laboratoire d'Excellence” (LABEX) entitled TULIP (ANR −10-LABX-41). BCNP was recipient of postdoctoral fellowship from the IFCPAR-CEFIPRA. The work benefited from the networking activities within the European COST Action FA1106. Microscopy and cytometry analyses were performed at RIO Imaging Platform under the technical assistance of A. Jauneau and J.C Lepert.
References
- Bleecker AB, Kende H. Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol. 2000;16:1–18. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- Johnson PR, Ecker JR. The ethylene gas signal transduction pathway: a molecular perspective. Annu Rev Genet. 1998;32:227–254. doi: 10.1146/annurev.genet.32.1.227. [DOI] [PubMed] [Google Scholar]
- Pirrello J, Jaimes-Miranda F, Sanchez-Ballesta MT, Tournier B, Khalil-Ahmad Q, Regad F, Latché A, Pech JC, Bouzayen M. Sl-ERF2, a tomato ethylene response factor involved in ethylene response and seed germination. Plant Cell Physiol. 2006;47:1195–1205. doi: 10.1093/pcp/pcj084. [DOI] [PubMed] [Google Scholar]
- Benavente LM, Alonso JM. Molecular mechanisms of ethylene signaling in Arabidopsis. Mol Biosyst. 2006;2:165–173. doi: 10.1039/b513874d. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 1995;7:173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell. 1994;6:1211–1225. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Ohmiya K, Hattori T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 1999;27:470–478. doi: 10.1093/nar/27.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M. A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 1998;17:5484–5496. doi: 10.1093/emboj/17.18.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nole-Wilson S, Krizek BA. DNA binding properties of the Arabidopsis floral development protein AINTEGUMENTA. Nucleic Acids Res. 2000;28:4076–4082. doi: 10.1093/nar/28.21.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang J, Cai B, Peng R, Zhu B, Jin X, Xue Y, Gao F, Fu X, Tian Y, Zhao W, Qiao Y, Zhang Z, Xiong A, Yao Q. Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem Biophys Res Commun. 2008;371:468–474. doi: 10.1016/j.bbrc.2008.04.087. [DOI] [PubMed] [Google Scholar]
- Zhang G, Chen M, Chen X, Xu Z, Guan S, Li L, Li A, Guo J, Mao L, Ma Y. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.) J Exp Bot. 2008;59:4095–4107. doi: 10.1093/jxb/ern248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell. 2000;12:393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier B, Sanchez-Ballesta MT, Jones B, Pesquet E, Regad F, Latché A, Pech J, Bouzayen M. New members of the tomato ERF family show specific expression pattern and diverse DNA-binding capacity to the GCC box element. FEBS Lett. 2003;550:149–154. doi: 10.1016/s0014-5793(03)00757-9. [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Tuori RP, D'Ascenzo MD, Fobert PR, Despres C, Martin GB. The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell. 2003;15:3033–3050. doi: 10.1105/tpc.017574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Mitsuhara I, Seo S, Ito H, Matsui H, Ohashi Y. Two novel AP2/ERF domain proteins interact with cis-element VWRE for wound-induced expression of the Tobacco tpoxN1 gene. Plant J. 2007;50:1079–1092. doi: 10.1111/j.1365-313X.2007.03111.x. [DOI] [PubMed] [Google Scholar]
- Gu YQ, Yang C, Thara VK, Zhou J, Martin GB. Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell. 2000;12:771–786. doi: 10.1105/tpc.12.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Suzuki K, Shinshi H. Elicitor-responsive, ethylene-independent activation of GCC box-mediated transcription that is regulated by both protein phosphorylation and dephosphorylation in cultured tobacco cells. Plant J. 1999;20:571–579. doi: 10.1046/j.1365-313x.1999.00634.x. [DOI] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003;33:751–763. doi: 10.1046/j.1365-313x.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger E, Gilmour S, Thomashow M. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell. 2004;16:2463–2480. doi: 10.1105/tpc.104.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P, Poindexter P, Osborne E, Jiang C, Riechmann JL. WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc Natl Acad Sci USA. 2004;101:4706–4711. doi: 10.1073/pnas.0305574101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits L, Memelink J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science. 2000;289:295–297. doi: 10.1126/science.289.5477.295. [DOI] [PubMed] [Google Scholar]
- Zhang J, Broeckling CD, Blancaflor EB, Sledge MK, Sumner LW, Wang Z. Overexpression of WXP1, a putative medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (medicago sativa) Plant J. 2005;42:689–707. doi: 10.1111/j.1365-313X.2005.02405.x. [DOI] [PubMed] [Google Scholar]
- Banno H, Ikeda Y, Niu QW, Chua NH. Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell. 2001;13:2609–2618. doi: 10.1105/tpc.010234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Muszynski M, Kellogg E, Hake S, Schmidt RJ. The control of spikelet meristem identity by the branched silkless1 gene in maize. Science. 2002;298:1238–1241. doi: 10.1126/science.1076920. [DOI] [PubMed] [Google Scholar]
- van der Graaff E, Dulk-Ras AD, Hooykaas PJ, Keller B. Activation tagging of the LEAFY PETIOLE gene affects leaf petiole development in Arabidopsis thaliana. Development. 2000;127:4971–4980. doi: 10.1242/dev.127.22.4971. [DOI] [PubMed] [Google Scholar]
- Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang H, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA, Budworth PR, Tao Y, Xie Z, Chen X, Lam S, Kreps JA, Harper JF, Si-Ammour A, Mauch-Mani B, Heinlein M, Kobayashi K, Hohn T, Dangl JL, Wang X, Zhu T. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell. 2002;14:559–574. doi: 10.1105/tpc.010410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Park C, Lee S, Ham B, Shin R, Paek K. Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2–type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell. 2001;13:1035–1046. doi: 10.1105/tpc.13.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RL, Kazan K, McGrath KC, Maclean DJ, Manners JM. A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 2003;132:1020–1032. doi: 10.1104/pp.102.017814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell. 2003;15:165–178. doi: 10.1105/tpc.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehner N, Hartmann L, Ehlert A, Böttner S, Oñate-Sánchez L, Dröge-Laser W. High-throughput protoplast transactivation (PTA) system for the analysis of Arabidopsis transcription factor function. Plant J. 2011;68:560–569. doi: 10.1111/j.1365-313X.2011.04704.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Sato S. The Tomato Genome Consortium, et al.: The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma MK, Kumar R, Solanke AU, Sharma R, Tyagi AK, Sharma AK. Identification, phylogeny, and transcript profiling of ERF family genes during development and abiotic stress treatments in tomato. Mol Genet Genomics. 2010;284:455–475. doi: 10.1007/s00438-010-0580-1. [DOI] [PubMed] [Google Scholar]
- Zhou J, Tang X, Martin GB. The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J. 1997;16:3207–3218. doi: 10.1093/emboj/16.11.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Zhang Z, Zhang X, Zhang H, Huang D, Huang R. Tomato TERF1 modulates ethylene response and enhances osmotic stress tolerance by activating expression of downstream genes. FEBS Lett. 2004;573:110–116. doi: 10.1016/j.febslet.2004.07.064. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhang D, Chen J, Yang Y, Huang Z, Huang D, Wang X, Huang R. Tomato stress-responsive factor TSRF1 interacts with ethylene responsive element GCC box and regulates pathogen resistance to Ralstonia solanacearum. Plant Mol Biol. 2004;55:825–834. doi: 10.1007/s11103-004-2140-8. [DOI] [PubMed] [Google Scholar]
- Pan Y, Seymour GB, Lu C, Hu Z, Chen X, Chen G. An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep. 2012;31:349–360. doi: 10.1007/s00299-011-1170-3. [DOI] [PubMed] [Google Scholar]
- Wang H, Huang Z, Chen Q, Zhang Z, Zhang H, Wu Y, Huang D, Huang R. Ectopic overexpression of tomato JERF3 in tobacco activates downstream gene expression and enhances salt tolerance. Plant Mol Biol. 2004;55:183–192. doi: 10.1007/s11103-004-0113-6. [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003;34:733–739. doi: 10.1046/j.1365-313x.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- Kagale S, Rozwadowski K. EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics. 2011;6:141–146. doi: 10.4161/epi.6.2.13627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterson N, Reuber TL. Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr Opin Plant Biol. 2004;7:465–471. doi: 10.1016/j.pbi.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Licausi F, Kosmacz M, Weits DA, Giuntoli B, Giorgi FM, Voesenek LACJ, Perata P, van Dongen JT. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature. 2011;479:419–422. doi: 10.1038/nature10536. [DOI] [PubMed] [Google Scholar]
- Andriankaja A, Boisson-Dernier A, Frances L, Sauviac L, Jauneau A, Barker DG, de Carvalho-Niebel F. AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell. 2007;19:2866–2885. doi: 10.1105/tpc.107.052944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Chen X, Ren H, Zhang Z, Zhang H, Wang J, Wang X, Huang R. ERF protein JERF1 that transcriptionally modulates the expression of abscisic acid biosynthesis-related gene enhances the tolerance under salinity and cold in tobacco. Planta. 2007;226:815–825. doi: 10.1007/s00425-007-0528-9. [DOI] [PubMed] [Google Scholar]
- Xu Z, Xia L, Chen M, Cheng X, Zhang R, Li L, Zhao Y, Lu Y, Ni Z, Liu L, Qiu Z, Ma Y. Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol Biol. 2007;65:719–732. doi: 10.1007/s11103-007-9237-9. [DOI] [PubMed] [Google Scholar]
- Jung J, Won SY, Suh SC, Kim H, Wing R, Jeong Y, Hwang I, Kim M. The barley ERF-type transcription factor HvRAF confers enhanced pathogen resistance and salt tolerance in Arabidopsis. Planta. 2007;225:575–588. doi: 10.1007/s00425-006-0373-2. [DOI] [PubMed] [Google Scholar]
- Chen G, Hu Z, Grierson D. Differential regulation of tomato ethylene responsive factor LeERF3b, a putative repressor, and the activator Pti4 in ripening mutants and in response to environmental stresses. J Plant Physiol. 2008;165:662–670. doi: 10.1016/j.jplph.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Hirota A, Kato T, Fukaki H, Aida M, Tasaka M. The auxin-regulated AP2/EREBP gene PUCHI is required for morphogenesis in the early lateral root primordium of Arabidopsis. Plant Cell. 2007;19:2156–2168. doi: 10.1105/tpc.107.050674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman A, Black R, Ecker JR. HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell. 1996;85:183–194. doi: 10.1016/s0092-8674(00)81095-8. [DOI] [PubMed] [Google Scholar]
- Raz V, Ecker JR. Regulation of differential growth in the apical hook of Arabidopsis. Development. 1999;126:3661–3668. doi: 10.1242/dev.126.16.3661. [DOI] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol. 1994;106:1335–1346. doi: 10.1104/pp.106.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts RJ, Cernac A, Estelle M. Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 1998;16:553–560. doi: 10.1046/j.1365-313x.1998.00321.x. [DOI] [PubMed] [Google Scholar]
- Rahman A, Amakawa T, Goto N, Tsurumi S. Auxin is a positive regulator for ethylene-mediated response in the growth of Arabidopsis roots. Plant Cell Physiol. 2001;42:301–307. doi: 10.1093/pcp/pce035. [DOI] [PubMed] [Google Scholar]
- Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, Watahiki MK, Yamamoto K, Liscum E. The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell. 2000;12:757–770. doi: 10.1105/tpc.12.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaabouni S, Jones B, Delalande C, Wang H, Li Z, Mila I, Frasse P, Latché A, Pech J, Bouzayen M. Sl-IAA3, a tomato Aux/IAA at the crossroads of auxin and ethylene signalling involved in differential growth. J Exp Bot. 2009;60:1349–1362. doi: 10.1093/jxb/erp009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaabouni S, Latché A, Pech J, Bouzayen M. Tomato Aux/IAA3 and HOOKLESS are important actors of the interplay between auxin and ethylene during apical hook formation. Plant Signal Behav. 2009;4:559–560. doi: 10.4161/psb.4.6.8748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Schauer N, Usadel B, Frasse P, Zouine M, Hernould M, Latché A, Pech J, Fernie AR, Bouzayen M. Regulatory features underlying pollination-dependent and -independent tomato fruit Set revealed by transcript and primary metabolite profiling. Plant Cell. 2009;21:1428–1452. doi: 10.1105/tpc.108.060830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, Chaabouni S, Latché A, Pech J, Bouzayen M. The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell. 2005;17:2676–2692. doi: 10.1105/tpc.105.033415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Takezaki N, Figueroa F, Zaleska-Rutczynska Z, Takahata N, Klein J. The phylogenetic relationship of tetrapod, coelacanth, and lungfish revealed by the sequences of forty-four nuclear genes. Mol Biol Evol. 2004;21:1512–1524. doi: 10.1093/molbev/msh150. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L, In Evolving Genes and Proteins. In: Evolutionary divergence and convergence in proteins. V.and Vogel, H.J, editor. New York; 1965. pp. 97–166. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correspondance between Sl-ERF nomenclature and ITAG 2.3 nomenclature.
Schematic diagram of protein structure of the isolated 28 tomato ERFs defining the 8 subclass. Each colored box represents the AP2/ERF domain and conserved motifs, as indicated below the diagram. The position of the motif is indicating by the number on the top of the diagram. The name of motif by Nakano is given inside the box [13]
Comparative basal activity of the synthetic promoter (4 X GCC) and the native promoters, fused to the GFP in the absence of added ERF effectors.
Primers used for RACE amplification.
Primers used for CDS amplification. Each pair of primers encompasses the complete CDS from the ATG to the STOP codon. Adaptors attB1 and attB2, have been added at the 5′ end of sense and antisense primers, respectively.
Primers used for qRT-PCR. Primers designed with PRIMER EXPRESS software in sequence specific regions for each ERF. Specificity was further checked using BLAST against all tomato unigenes (Tomato unigene database).
Primers used for the synthesis of radioactive GCC box probes utilized in EMSA experiments.
Coding sequences of the 28 studied ERF.