Abstract
The binding of a ligand to orotidine 5’-monophosphate decarboxylase (OMPDC) is accompanied by a conformational change from an open, inactive conformation (Eo) to a closed, active conformation (Ec). As the substrate traverses the reaction coordinate to form the stabilized vinyl carbanion/carbene intermediate, interactions are enforced that destabilize the carboxylate group of the substrate as well as stabilize the intermediate (in the Ec•S‡ complex). Focusing on the OMPDC from Methanothermobacter thermautotrophicus, the “remote” 5’-phosphate group of the substrate activates the enzyme 2.4 × 108-fold; the activation is equivalently described by an intrinsic binding energy (IBE) of 11.4 kcal/mol. We studied residues in the activation that 1) directly contact the 5’-phosphate group; 2) participate in a hydrophobic cluster near the base of the active site loop that sequesters the bound substrate from solvent; and 3) form hydrogen-bonding interactions across the interface between the “mobile” and “fixed” half-barrel domains of the (β/α8-barrel structure. Our data support a model in which the IBE provided by the 5’-phosphate group is used to enable interactions both near the N-terminus of the active site loop and across the domain interface that stabilize both the Ec•S and Ec•S‡ complexes relative to the Eo•S complex. The conclusion that the IBE of the 5’-phosphate group provides stabilization of both the Ec•S and Ec•S‡ complexes, not just the Ec•S‡ complex, is central to understanding the structural origins of enzymatic catalysis as well as the requirements for the de novo design of enzymes that catalyze novel reactions.
INTRODUCTION
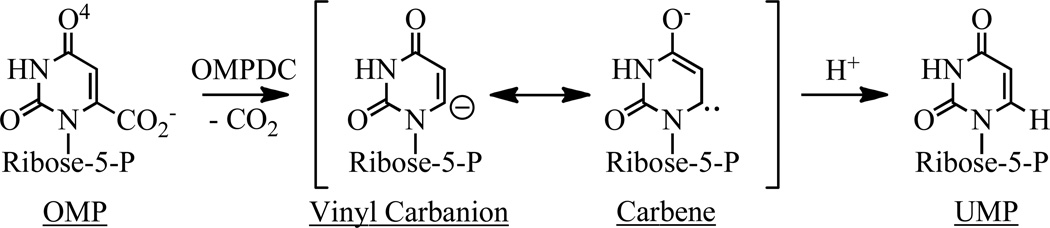
Orotidine 5’-monophosphate decarboxylase (OMPDC;1 Scheme 1) that catalyzes the final step in pyrimidine biosynthesis is a remarkable catalyst: the rate enhancement (kcat/knon = 7.1 × 1016) and the catalytic proficiency [(kcat/Km)/knon = 4.8 × 1022 M]; affinity for the transition state) are among the largest measured for enzyme-catalyzed reactions (2, 3).
Scheme 1.
We, and others (4–7), have been interested in defining the strategies used by OMPDC to reduce the value of ΔG‡ by 23 kcal/mol. The reaction coordinate involves a vinyl carbanion/carbene intermediate that is stabilized by ≥14 kcal/mol by its interactions with the active site (8–10){Tsang, 2012 #60}.
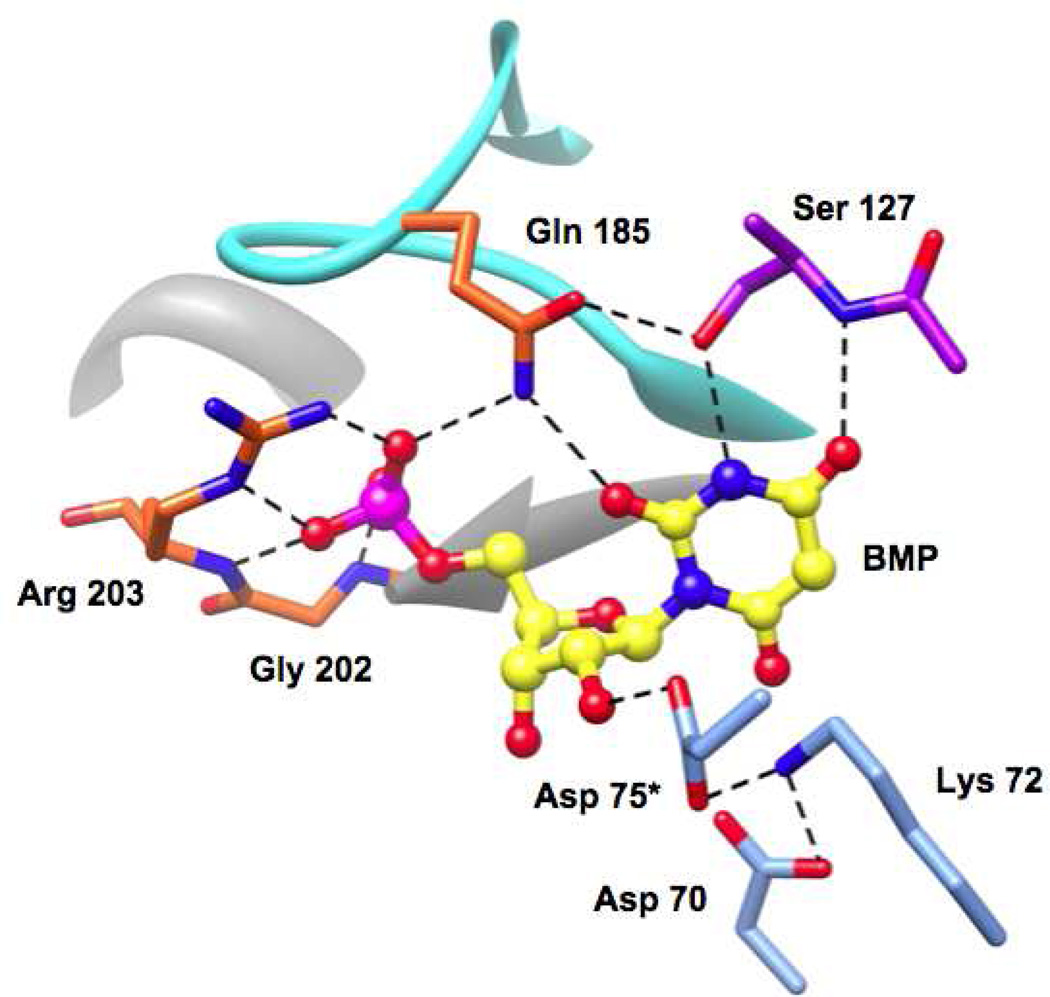
MtOMPDC, like all OMPDCs, is an obligate dimer, with the active sites located at the dimer interface. The interactions of conserved active site residues in the OMPDC from Methanothermobacter thermautotrophicus, MtOMPDC, the focus of this article, with BMP [l-(5-phospho-β-ribofuranosyl)barbituric acid], an intermediate/transition state analog (11), are shown in Figure 1. Each active site contains an essential Asp-Lys-Asp motif involving Asp 70 and Lys 72 from one polypeptide and Asp 75* from the symmetry related polypeptide as well as hydrogen-bonding interactions between the pyrimidine ring and both the OH group and the backbone amide group of Ser 127.
Figure 1.
Active site of MtOMPDC showing the interactions of active site residues with BMP, an intermediate/transition state analog.
Two structural strategies can be envisioned for destabilization of the OMP substrate as the transition state is formed: 1) electrostatic/steric destabilization of the carboxylate group by the proximal Asp (Asp 70) (12, 13); and 2) desolvation of the carboxylate group by a proximal hydrophobic cavity (formed by Ile 96, Leu 123, Val 155, and Pro 180 (14, 15). The importance of these interactions has been tested; each contributes ~4 kcal/mol to the reduction in ΔG‡.
Also, at least two structural strategies can be envisioned for stabilization of the anionic intermediate as the transition state is formed: 1) attractive electrostatic interactions with the ε-ammonium group of Lys 72 that protonates C6 of the intermediate (6); and 2) enhanced hydrogen-bonding interactions with the backbone amide group of Ser 127 (16, 17). For the latter strategy to be effective, the negative charge must be delocalized to O4 in the pyrimidine ring (as described by the carbene resonance structure; Scheme 1); calculations support the participation of this interaction, although its importance has not been tested directly (16, 17).
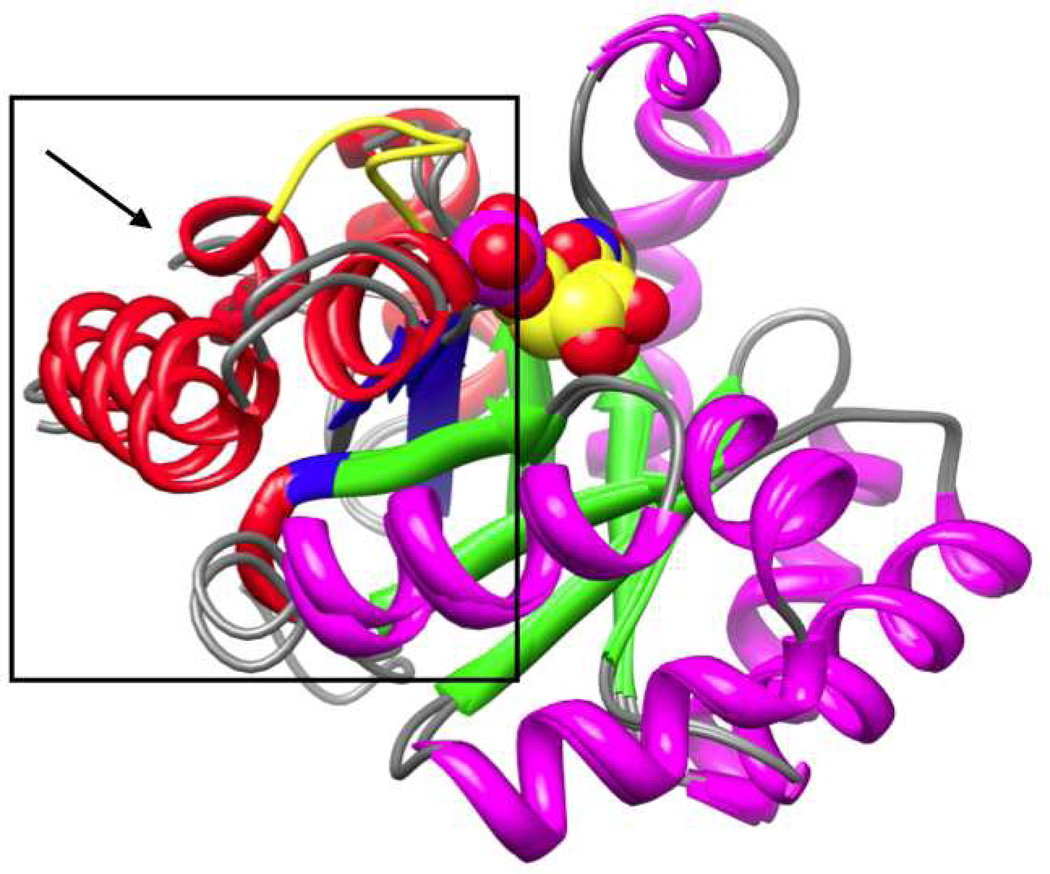
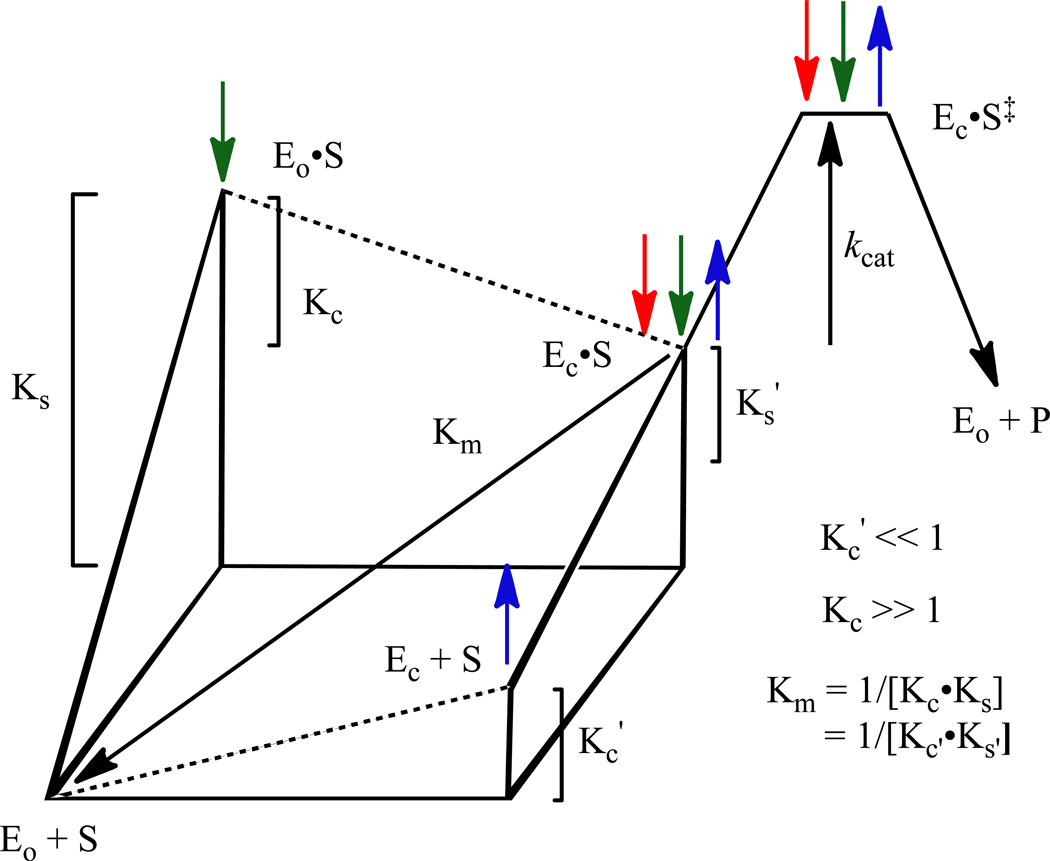
These destabilizing/stabilizing interactions are “enforced” by a closed, catalytically active conformation (Ec) that is observed crystallographically in the presence of the intermediate/transition state analogs 6-azaUMP (18) and BMP (11); in their absence, MtOMPDC exists in an open, catalytically inactive conformation (Eo) (Figure 2) (12, 13). The more favorable transition from Eo to Ec in the Eo•S and Ec•S complexes [Kc vs. Kc’ in the kinetic model shown in Scheme 2 and Figure 3 (19); vide infra] is responsible, in part, for a 2.4 × 108-fold activation of the enzyme and involves 1) closure of the nine-residue active site loop (Pro 180 to Asp 188) that sequesters the substrate from solvent, and 2) formation of a more compact conformation for the (β/α)8-barrel structure that juxtaposes the substrate with the residues involved in substrate destabilization and intermediate stabilization.
Figure 2.
Superposition of the structures of individual polypeptides of unliganded (Eo) and BMP-liganded (Ec) MtOMPDC, showing the movement (within the box; in the direction of the arrow) of the “mobile” domain (red α-helices and blue β-strands) toward the “fixed” domain (magenta α-helices and green β-strands). The ordered active site loop is colored yellow in the BMP-liganded structure.
Scheme 2.
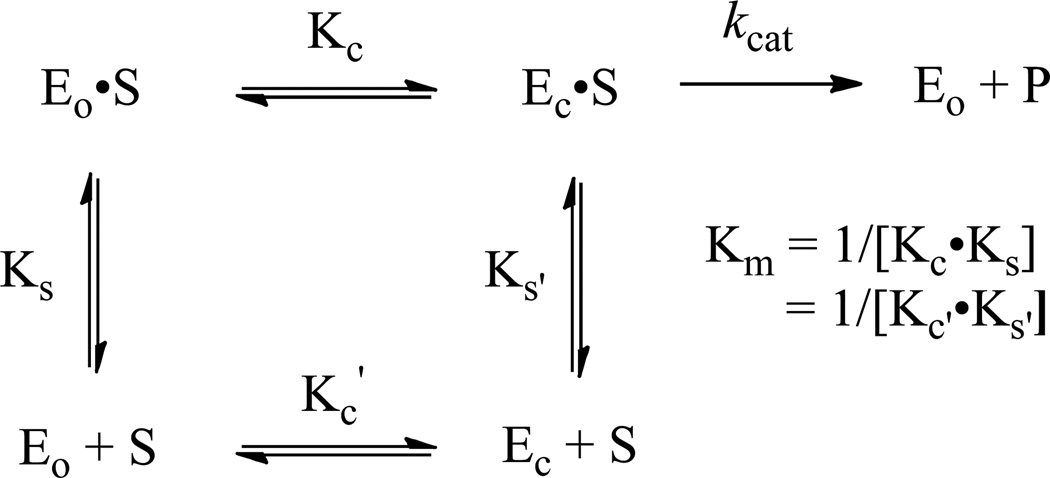
Figure 3.
Three-dimensional free energy diagram for the kinetic model depicted in Scheme 2. See the text for a detailed explanation.
Each (β/α)8-barrel in the obligate dimer is formed from two “half-barrel” domains, a “fixed” domain (including β-strands 2, 3, 4, and 5) located at the dimer interface and a “mobile” domain (including β-strands 6, 7, 8, and 1) (20); in the transition from Eo and Ecthe “mobile” domain moves toward the “fixed” domain, constricting the active site cavity that envelops the substrate (Figure 2).2 Because the active site loop sequesters the substrate from solvent in the Ec•S complex, substrate binding to Eo must precede the conformational change. Thus “induced fit”, not “conformational selection”, is responsible for altering the distribution between Eo and Ec when the substrate binds (21).3
The energy obtained from formation of the Eo•S complex from Eo + S is partitioned between 1) “gripping” the substrate [with the value of Km providing a measure of the affinity for the substrate (1/[Kc•Ks] in Scheme 2; Ks is defined as an association complex but Km is a dissociation constant)]; and 2) stabilizing both Ec•S and Ec•S‡ (vide infra) relative to Eo•S (Kc in Scheme 2). That Ec is unstable relative to Eo in the absence of substrate (Kc’) facilitates release of the UMP product; if the total energy obtained from substrate binding were used only to “grip” the substrate, release of the product from the Ec•P complex likely would be rate-determining (22). Therefore, the structural and energetic coupling between substrate binding to Eo and the subsequent transition from Eo•S to Ec•S is essential for understanding the catalytic strategy that produces the extraordinary rate enhancement.
We recently reported initial efforts to identify structural features that stabilize Ec•S (19). Those focused on a hydrophobic cluster that includes Val 182 in the active site loop as well as Ile 199, Val 201, and Ile 218 located near the C-terminal ends of the seventh and eighth β-strands in the “mobile” domain. Substitutions of these residues that are “remote” from the binding site for the 5’-phosphate group with Ala increase the value of Km but have little effect on the value of kcat. The simplest interpretation is that the hydrophobic cluster stabilizes the closed conformation of the active site loop and, therefore, contributes to the stability of Ec•S. Without this stabilization, the formation of Ec•S from Eo + S is less favorable, requiring increased concentrations of substrate to provide the energy to enable its formation (as revealed by an increase in the value of Km). However, when Ec•S is formed, it is fully competent to yield product via the vinyl anion/carbene intermediate and the transition state that precedes it on the reaction coordinate (Ec•S‡) so the value of kcat (the energy difference between Ec•S and Ec•S‡) is unchanged.
In this article we focus on residues involved in hydrogen-bonding interactions between the “mobile” and “fixed” domains observed in Ec but not in Eo (involving Thr 159, Arg 160, and Tyr 206), also “remote” from the binding site for the 5’-phosphate group, to determine whether these contribute to the stability of Ec•S. We also target the only two residues (Gln 185 and Arg 203) that provide direct side chain contacts with the 5’-phosphate group of the substrate and, therefore, reasonably would be involved in generation of the intrinsic binding energy (IBE) of the 5’-phosphate group that is used to enable stabilization of both Ec•S and Ec•S‡ relative to Eo + S (23). Finally, we examine double mutants involving one “remote” residue that contributes to the stability of Ec•S (Val 182, Thr 159, or Arg 160) and one residue that generates the IBE of the 5’-phosphate group (Arg 203). The kinetic and structural properties of the mutants are consistent with a structure-based model in which Ec•S and Ec•S‡ are stabilized equally relative to Eo•S by 1) the interaction of the 5’-phosphate group with its binding site that allows/directs reorganization of the active site loop, 2) assembly of the hydrophobic cluster near the N-terminus of the reorganized active site loop that stabilizes its closed conformation, and 3) formation of interactions across the domain interface in Ec•S and Ec•S‡ some of which (those involving Thr 159 and Arg 160) are spatially proximal to the reorganized active site loop and, therefore, are influenced by its structure. This analysis clarifies the role of the IBE of the 5’-phosphate group in reducing the value of ΔG‡ and provides the framework for a more detailed investigation of the structural features involved in both generation of the IBE and its use to facilitate the conversion of Eo•S to both Ec•S and Ec•S‡.
The conclusion that the IBE generated by interaction of the 5’-phosphate group with its binding site allows stabilization of both the Ec•S and Ec•S‡ complexes, not just the Ec•S‡ complex, relative to Eo + S by enabling formation of “remote” interactions that stabilize Ec likely can be applied to many (all?) enzymes in which conformational changes accompany substrate binding.
MATERIALS AND METHODS
Materials
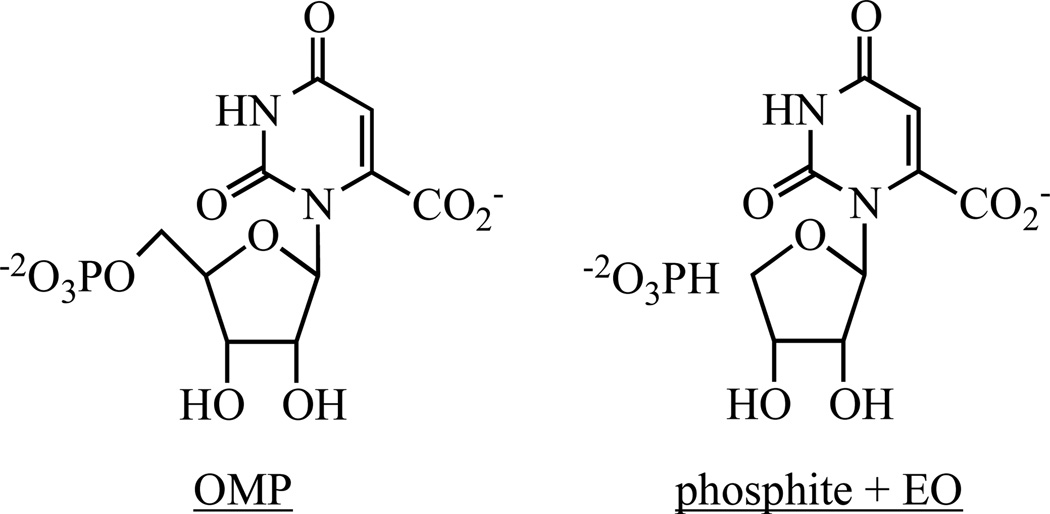
OMP, EO, FEO, and BMP were prepared by published procedures (6, 11, 13, 24–26). All solutions were prepared with Millipore Ultrapure filtered water.
Protein Expression and Purification and Site-Directed Mutagenesis
The gene encoding MtOMPDC was previously cloned in the pET-15b vector (Novagen). The site-mutants were constructed (overlap extension) and the wild type and mutant proteins were expressed and prepared following previously published procedures (13, 15, 24).
Assay for Decarboxylation of OMP
Assays were performed at 25 ° and in 10 mM MOPS, pH 7.1, containing 100 mM NaCl following published procedures (13, 15, 24).
Assay for Decarboxylation of EO
The values of kcat/Km for decarboxylation of EO were determined as previously described (15, 25).
Assay for Decarboxylation of FEO
For wild type MtOMPDC and single mutants, the values of kcat/Km for decarboxylation of FEO were determined by quantitating the first order decay of FEO at 290 nm using procedures developed for EO (15, 25).
For double and triple mutants, the values of kcat/Km for decarboxylation of FEO were determined using HPLC-based end point assays using procedures developed for EO (15, 25); the amount of FEU product was quantitated using 5-fluorouridine as the reference standard (ε = 9660 M−1 cm−1 in 0.1 N HCl).
Crystallization and Data Collection
Eight different crystal forms (Table 1) were grown by the sitting drop method at room temperature for single mutants of MtOMPDC liganded with BMP: (1) Q185A•BMP, (2) R203A•BMP, (3) T159V•BMP, (4) T159A•BMP, (5) T159S•BMP, (6) R160A•BMP, (7) Y206F•BMP, and (8) K82A•BMP. The protein solutions for all eight crystallizations contained the corresponding protein (30 mg/mL) in 20 mM Hepes, pH 7.5, 100 mM NaCl, 3 mM DTT, and 40 mM BMP. The precipitants were the following:
For the Q185•BMP complex, the precipitant contained 30% PEG 4000, 0.1 M Tris•HCl, pH 8.5, and 0.2 M lithium sulfate.
For the R203A•BMP complex, the precipitant contained 2.4 M sodium malonate, pH 7.0.
For the T159V•BMP complex, the precipitant contained 30% PEG 4000, 0.1 M sodium citrate, pH 5.6, and 0.2 M ammonium sulfate.
For the T159A•BMP complex, the precipitant contained 20% PEG 4000, 20% 2-propanol, and 0.1 M sodium citrate, pH 5.6.
For the T159S•BMP complex, the precipitant contained 20% PEG 4000, 20% 2-propanol, and 0.1 M sodium citrate, pH 5.6.
For the R160A•BMP complex, the precipitant contained 30% PEG 4000, 0.1 M sodium citrate, pH 5.6, and 0.2 M ammonium acetate.
For the Y206F•BMP complex, the precipitant contained 30% PEG 4000, 0.1 M HEPES, pH 7.5, and 0.2 M magnesium chloride.
For the K82A•BMP complex, the precipitant contained 20% PEG 8000, 0.1 M Tris•HCl, pH 8.5, and 0.2 M magnesium chloride.
Table 1.
Data collection and refinement statistics for BMP complexes of single mutants
| Q185A | R203A | T159V | T159A | T159S | R160A | Y206F | K82A | |
|---|---|---|---|---|---|---|---|---|
| Data collection | ||||||||
| Space group | P21 | P21212 | P21 | P212121 | P21 | P21 | P21 | P21 |
| No. of molecules | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| in asym. unit | ||||||||
| Cell dimensions | ||||||||
| a (Å) | 59.80 | 80.00 | 59.78 | 52.61 | 58.67 | 59.80 | 59.62 | 59.78 |
| b (Å) | 63.86 | 64.00 | 64.12 | 74.04 | 73.87 | 64.14 | 63.41 | 64.01 |
| c (Å) | 61.47 | 73.30 | 61.89 | 117.50 | 59.52 | 61.78 | 61.04 | 61.63 |
| β (°) | 115.28 | 115.46 | 119.50 | 115.67 | 115.02 | 115.52 | ||
| Resolution (Å) | 1.37 | 1.50 | 1.40 | 1.60 | 1.3 | 1.4 | 1.42 | 1.49 |
| No. of unique | 87599 | 58327 | 79912 | 54292 | 104965 | 76701 | 76776 | 68389 |
| reflections | ||||||||
| Rmerge | 0.065 | 0.082 | 0.091 | 0.076 | 0.038 | 0.083 | 0.073 | 0.043 |
| Completeness (%) | 99.7 | 95.6 | 95.8 | 88.5 | 96.8 | 91.5 | 98.1 | 99.7 |
| Refinement | ||||||||
| Resolution (Å) | 25.0–1.37 | 25.0–1.3725.0 | 25.0–1.40 | 25.0–1.60 | 25.0–1.3 | 25.0–1.40 | 25.0–1.42 | 25–1.49 |
| Rcryst | 0.165 | 0.223 | 0.188 | 0.204 | 0.165 | 0.199 | 0.172 | 0.153 |
| Rfree | 0.178 | 0.247 | 0.206 | 0.231 | 0.176 | 0.222 | 0.193 | 0.174 |
| No. atoms | ||||||||
| Protein | 3421 | 3412 | 3364 | 3322 | 3418 | 3374 | 3379 | 3416 |
| Waters | 454 | 144 | 396 | 170 | 410 | 410 | 347 | 444 |
| Bound ligands | BMP SO4 |
BMP | BMP | BMP GOL |
BMP GOL, POL |
BMP | BMP GOL |
BMP GOL |
| Ligand atoms | 54 | 44 | 44 | 50 | 82 | 44 | 50 | 50 |
| R.m.s. deviations | ||||||||
| Bond lengths (Å) | 0.006 | 0.005 | 0.006 | 0.007 | 0.006 | 0.006 | 0.006 | 0.006 |
| Bond angles (°) | 1.0 | 1.2 | 1.1 | 1.1 | 1.1 | 1.1 | 1.1 | 1.1 |
| PDB entry | 3V1P | 3LI0 | 3P60 | 3P5Y | 3P5Z | 3P61 | 3RLV | 3RLU |
Ten different crystal forms (Tables 2) were grown by sitting drop method at room temperature for double and triple mutants of MtOMPDC liganded with BMP: (1) Q185A•R203A•BMP, (2) T159V•V182A•BMP, (3) T159V•Y206F•BMP, (4) V182A•Y206F•BMP, (5) R160A•V182A•BMP, (6) R160A•Y206F•BMP, (7) R203A•V182A•BMP, (8) R203A•T159V•BMP, (9) R203A•R160A•BMP, and (10) T159V•V182A•Y206F•BMP. The protein solutions for all ten co-crystallizations contained the corresponding protein (30 mg/mL) in 20 mM Hepes, pH 7.5, 100 mM NaCl, 3 mM DTT, and 40 mM BMP. The precipitants were the following:
For the Q185A•F203A•BMP complex, the precipitant contained 2.0 M ammonium sulfate, pH 4.9.
For the T159V•V182A•BMP complex, the precipitant contained 0.4 M sodium phosphate, 1.6 M potassium phosphate, 0.1 M imidazole, pH 8.0, and 0.2 M sodium chloride.
For the T159V•Y206F•BMP complex, the precipitant contained 1.4 M sodium citrate, and 0.1 M HEPES, pH 7.5.
For the V182A•Y206F•BMP complex, the precipitant contained 0.8 M sodium phosphate, 1.2M potassium phosphate, and 0.1 M acetate, pH 4.5.
For the R160A•V182A•BMP complex, the precipitant contained 30% PEG 4000, 0.1 M Tris•HCl, pH 8.5, and 0.2 M lithium sulfate.
For the R160A•Y206F•BMP complex, the precipitant contained 30% PEG 4000, and 0.1 M Tris•HCl, pH 8.5, and 0.2 M magnesium chloride.
For the R203A•V182A•BMP complex, the precipitant contained 1.0 M sodium citrate, and 0.1 M cacodylate, pH 6.5.
For the R203A•T159V•BMP complex, the precipitant contained 0.8 M sodium phosphate, 1.2 M potassium phosphate, and 0.1 M acetate, pH 4.5.
For the R203A•R160A•BMP complex, the precipitant contained 3.5 M sodium formate, pH 7.0.
For the T159V•V182A•Y206F•BMP complex, the precipitant contained 60% tacsimate, pH 7.0.
Table 2.
Data collection and refinement statistics for BMP complexes of double and triple mutants
| Q185A/ R203A |
T159V/ V182A |
T159V/ Y206F |
V182 A/ Y206F |
R160A/ V182A |
R160A/ Y206F |
T159V/ V182A/ Y206F |
R203A/ V182A |
R203A/ T159V |
R203A/ R160A |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Data collection | ||||||||||
| Space group | P41212 | P21 | P21 | P21 | P21 | P21 | P21 | P21 | P21 | P212121 |
| No. of molecules | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| in asym. unit | ||||||||||
| Cell dimensions | ||||||||||
| a (Å) | 91.51 | 59.77 | 59.93 | 59.56 | 59.77 | 60.08 | 59.81 | 55.08 | 54.35 | 56.62 |
| b (Å) | 91.51 | 64.07 | 63.67 | 63.62 | 64.07 | 63.83 | 63.98 | 66.40 | 63.06 | 56.61 |
| c (Å) | 135.38 | 61.64 | 61.43 | 61.26 | 61.64 | 61.91 | 61.43 | 59.55 | 54.51 | 127.21 |
| β (°) | 115.46 | 115.34 | 115.00 | 115.46 | 115.53 | 115.48 | 101.85 | 98.95 | ||
| Resolution (Å) | 1.94 | 1.54 | 1.34 | 1.32 | 1.32 | 1.26 | 1.32 | 1.53 | 1.71 | 1.42 |
| No. of unique | 41820 | 55241 | 87118 | 96995 | 95690 | 111994 | 97927 | 62002 | 38041 | 77257 |
| reflections | ||||||||||
| Rmerge | 0.095 | 0.075 | 0.065 | 0.055 | 0.088 | 0.073 | 0.058 | 0.047 | 0.092 | 0.065 |
| Completeness (%) | 96.8 | 89.4 | 93.0 | 99.8 | 97.4 | 98.4 | 99.8 | 98.0 | 96.1 | 92.2 |
| Refinement | ||||||||||
| Resolution (Å) | 25–1.94 | 25–1.54 | 25–1.34 | 25.0–1.32 | 25.0–1.32 | 25.0–1.26 | 25–1.32 | 25–1.53 | 25–1.71 | 25–1.4 |
| Rcryst | 0.172 | 0.173 | 0.173 | 0.165 | 0.231 | 0.176 | 0.164 | 0.146 | 0.225 | 0.184 |
| Rfree | 0.198 | 0.206 | 0.192 | 0.178 | 0.247 | 0.196 | 0.183 | 0.172 | 0.268 | 0.211 |
| No. atoms | ||||||||||
| Protein | 3254 | 3390 | 3477 | 3434 | 3327 | 3444 | 3426 | 3499 | 3314 | 3359 |
| Waters | 191 | 367 | 406 | 342 | 553 | 464 | 373 | 74 | 225 | |
| Bound ligands | BMP SO4 |
BMP GOL |
BMP GOL |
BMP GOL |
BMP | BMP | BMP GOL |
BMP GOL |
BMP | BMP |
| Ligand atoms | 54 | 50 | 50 | 50 | 44 | 44 | 50 | 69 | 44 | 44 |
| R.m.s. deviations | ||||||||||
| Bond lengths (Å) | 0.007 | 0.007 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.008 | 0.006 |
| Bond angles (°) | 1.00 | 1.1 | 1.1 | 1.1 | 1.0 | 1.1 | 1.1 | 1.1 | 1.0 | 1.1 |
| PDB entry | 4FX8 | 3QEZ | 3QF0 | 3QMT | 3QMR | 3SJ3 | 3QMS | 4FX6 | 4FXR | 4GC4 |
Prior to data collection, the crystals of all 18 crystal forms (Tables 1 and 2) were transferred to cryoprotectant solutions composed of their mother liquids and 20% glycerol and flash-cooled in a nitrogen stream. The X-ray diffraction data sets (Tables 1, 2) were collected at the NSLS X4A beamline (Brookhaven National Laboratory) on an ADSC CCD detector and at NSLS X29A beamline on the 315q CCD detector. Diffraction intensities were integrated and scaled with programs DENZO and SCALEPACK (27). The data collection statistics are given in Tables 1, 2.
Structure Determination and Model Refinement
All 18 MtOMPDC structures (Tables 1 and 2) were solved by molecular replacement with fully automated molecular replacement pipeline BALBES (28), using only input diffraction and sequence data. Partially refined structures of all 18 MtOMPDC crystal forms (Tables 1 and 2) were the outputs from BALBES. Subsequent several iterative cycles of refinement were performed for each crystal form including: model rebuilding with COOT (29), refinement with PHENIX (30), and automatic model rebuilding with ARP (31).
For all 18 BMP-liganded structures, 1) all loops are well-defined, 2) the electron density for all BMP ligands is well-ordered, 3) no nonglycine residue lies in disallowed regions of the Ramachandran plots, 4) all crystallize as dimers, and 4) the monomers within each dimer are connected by a noncrystallographic two-fold axis.
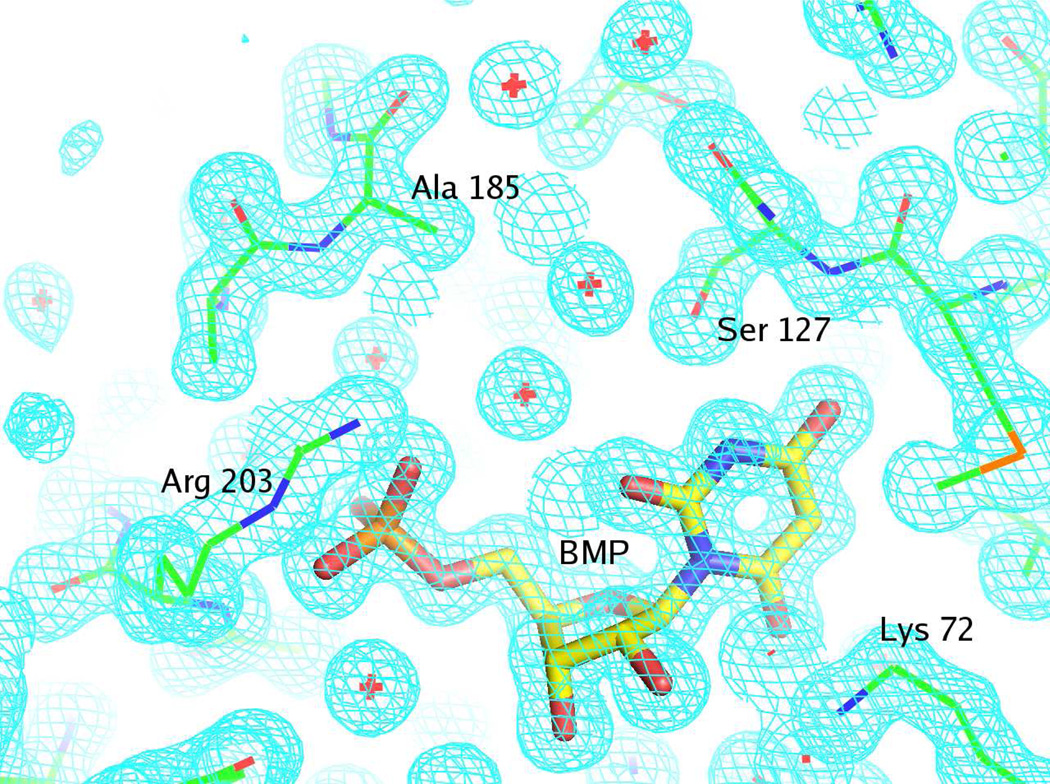
Representative electron density for the Q185A mutant liganded with BMP is shown in Figure 4.
Figure 4.
Representative electron density map for the active site of the Q185A mutant complexed with BMP and contoured at 1.5 σ. The figure was produced with PyMOL (41). The details of the interactions between BMP and the active site are described in the text.
Final crystallographic refinement statistics for all determined MtOMPDC structures are provided in the Tables 1 and 2.
RESULTS AND DISCUSSION
Intrinsic Binding Energy (IBE) of the 5’-Phosphate Group
As described by Jencks (23), the IBE of a remote substituent, e.g., the 5’-hosphate group of OMP, is used “to pay for substrate destabilization through distortion, electrostatic interactions, and desolvation, for bringing about the necessary loss of entropy by freezing the substrates in the proper position for reaction, and for binding to the transition state. The maximum binding energy is then not realized directly in the binding of the substrate, but is more completely realized in the transition state.” For MtOMPDC, the 5’-phosphate group of OMP “activates” decarboxylation 2.4 × 108-fold as measured by comparing the values of kcat/Km for OMP and EO that lacks the 5’-phosphate group [Scheme 2 (19, 26, 32)], 2.9 × 106 M−1 sec−1 and 12.4 × 10−3 M−1 sec−1, respectively (Table 3); the IBE calculated from this ratio is 11.4 kcal/mol (Table 3)].
Table 3.
Kinetic Constants for OMP at pH 7.1 and 25 °C.
| MtOMPDC |
kcat (s−1) |
Km (µM−1) |
kcatKm (M−1 s−1) |
Fold change kcat |
Fold change Km |
Fold change kcat/Km |
ΔΔG† (kcal/mol)b |
|---|---|---|---|---|---|---|---|
| Wild type | 5.3 ± 0.1 | 1.8 ± 0.5 | 2.9 × 106 | - | - | - | - |
| T159V | 2.3 ± 0.1 | 33 ± 2 | 7.0 × 104 | 2 | 18 | 41 | 2.2 |
| R160A | 2.0 ± 0.1 | 30 ± 1 | 6.7 × 104 | 3 | 17 | 43 | 2.2 |
| V182A | 3.0 ± 0.1 | 24 ± 1 | 1.3 × 105 | 2 | 13 | 22 | 1.8 |
| Y206F | 4.1 ± 0.2 | 5.5 ± 1.0 | 7.5 × 105 | 1 | 3 | 4 | 0.8 |
| T159V/V182A | 0.56 ± 0.02 | 700 ± 90 | 8.0 × 102 | 10 | 390 | 3600 | 4.8 |
| T159V/Y206F | 1.1 ± 0.1 | 300 ± 50 | 3.7 × 103 | 5 | 170 | 780 | 3.9 |
| R160A/V182A | 1.0 ± 0.1 | 600 ± 60 | 1.7 × 103 | 5 | 330 | 1700 | 4.4 |
| R160A/Y206F | 1.1 ± 0.1 | 130 ± 10 | 8.5 × 103 | 5 | 72 | 340 | 3.4 |
| V182A/Y206F | 1.5 ± 0.1 | 100 ± 15 | 1.5 × 104 | 4 | 56 | 190 | 3.1 |
| T159V/V182A/Y206F | 0.24 ± 0.01 | 1900 ± 200 | 1.3 × 102 | 22 | 1100 | 2.2 × 104 | 5.9 |
| Q185A | 1.4 ± 0.1 | 110 ± 15 | 1.3 × 104 | 4 | 61 | 220 | 3.1 |
| R203A | 1.5 ± 0.1 | 980 ± 10 | 1.5 × 103 | 4 | 540 | 1900 | 4.4 |
| Q185A/R203A | - | - | 9.6a | - | - | 3.0 × 105 | 7.4 |
| T159V/R203A | - | - | 9.9a | - | - | 3.0 × 105 | 7.4 |
| R160A/R203A | - | - | 16a | - | - | 1.8 × 105 | 7.1 |
| V182A/R203A | - | - | 32a | - | - | 9.1 × 104 | 6.7 |
Velocity is directly proportional to substrate concentration.
For kcat/Km.
The challenge has been to provide a structure-based understanding for how the IBE of a “remote” group of the substrate is both generated and used to activate the enzyme. Given the conformational change induced by ligand binding (Figure 2), a plausible explanation is that in MtOMPDC the IBE of the 5’-phosphate group not only increases the affinity of the enzyme for the substrate [Ks (and Ks’) in the kinetic model in Scheme 2/Figure 3; green arrows in Figure 3] but also increases the fraction of the enzyme that exists as Ec•S instead of Eo•S (Kc ≫ 1 in contrast to Kc’ ≪ 1 for Ec and Eo; red and blue arrows, respectively, in Figure 3) by enabling stabilizing contacts between the “mobile” and “fixed” domains.
Model to Explain the Role of the IBE of the 5’-Phosphate Group in the Activation of MtOMPDC
We previously proposed the kinetic model in Scheme 2 to understand the relationships among EoEcS, and the values of Km and kcat (19); the three-dimensional energy diagram for the model is shown in Figure 3.
In the model, Eo predominates in the absence of substrate (KC’ ≪ 1); Ec predominates in its presence (KC ≫ 1; as Ec•S). The values of Kc’ and Kc are determined by the relative stabilities of Eo and Ec in the absence and presence of substrate, respectively. The dissociation constant for S, [Eo][S]/[Ec•S], is assumed to be the value of Km1/[Kc•Ks] (= 1/[Kc’•Ks’]).3
The value of the IBE for the 5’-phosphate group is obtained from the increase in the value of kcat/Km associated with the 5’-phosphate group occupying its binding site in Ec (from the ratio of the values for OMP and EO). Because the carboxylate group of the substrate (in either OMP or EO) is distal from the binding site for the 5’-phosphate group (Figure 1), the value of kcat (the energy difference between Ec•S and Ec•S‡) can be expected to be independent of the absence/presence of the 5’-phosphate group in the absence of “long range” interactions with the carboxylate group of the substrate and/or the anionic intermediate, i.e., the same value for OMP and EO if it were possible to fully populate Ec•S when EO is the substrate. The model also assumes that interaction of the 5’-phosphate group is required to stabilize Ec•S relative to Eo•S (Kc vs. Kc’ in the unliganded enzyme), so the value of kcat/Km for EO is determined by the fraction of the unliganded enzyme that exists as Eci.e., from the value of Kc’.
The energy obtained from the 5’-phosphate group interacting with its binding site (available to OMP but not EO) can be used to 1) increase the stabilities of the Eo•S and Ec•S complexes relative to Eo + S (green arrows in Figure 3; “gripping” the substrate); and 2) increase the stability of Ec•S relative to EoS (red arrows in Figure 3; stabilizing the active conformation). Both effects stabilize the Ec•S complex relative to Eo + S, thereby decreasing the value of Km and increasing the value of kcat/Km. Thus, the activation by the 5’-phosphate group is caused by two independent effects that cannot be deconvoluted: effects on Ks and/or Kc when OMP is the substrate. If the value of kcat is independent of the interactions of the 5’-phosphate group with its binding site (evaluated by the loss of individual interactions by site-directed mutagenesis; vide infra), the activation by the 5’-phosphate group is the result of 1) an increase in the affinity of the enzyme for the OMP substrate (reflected in the value of Ks), and 2) an increase in the fraction of the enzyme that exists as Ec (as Ec•S, i.e., Kc vs. Kc’).
The stability of Ec•S relative to Eo + S (measured by the Km) is also influenced by residues “remote” from the binding site for the 5’-phosphate group. The structures of the unliganded and BMP-liganded structures of MtOMPDC allow the proposal that the hydrophobic cluster involving Val 182 in the active site loop as well as hydrogen bonds involving Thr 159, Arg 160, and Tyr 206 between the “mobile” and “fixed” domains observed in Ec but not in Eo contribute to the stability of Ec relative to Eo both in the absence and presence of ligand (Kc’ and Kcrespectively; blue arrows in Figure 3).
In our initial studies of this model, we observed that Ala substitutions for residues in the hydrophobic cluster (Val 182, Ile 199, Val 201, and Ile 218) that are “remote” from the 5’-phosphate group and its binding site increase the value of Km but do not influence the value of kcat (19). We determined the structures of the mutants in the presence of BMP and observed that they are superimposable on that of wild type with the exception of the side chains of the substituted residues. Thus, the changes in the values of Km could be associated with an incomplete “remote” hydrophobic cluster that reduces the values of both Kc’ and Kc (blue arrows in Figure 3); the unchanged values for kcat were explained by the unperturbed interactions of the active site with the OMP substrate and transition state in the Ec•S and Ec•S‡ complexes (Figure 1), so the energy difference between Ec•S and Ec•S‡ is unchanged.
The Ala substitutions in the hydrophobic core do not alter the values of the IBEs, i.e., the values of kcat/Km for OMP and EO are equivalently reduced by the substitutions (19). Because the values for EO measure the fraction of the unliganded enzyme present as Ec (the value of Kc’), the equivalent reductions demonstrate that the interaction of the 5’-phosphate group with its binding site is not the sole determinant of the stability of the Eo•S complex relative to Eo + S (Km = 1/[Kc•Ks]) and of Ec relative to Eo in the Ec•S and Eo•S complexes (Kc).
In the remainder of this manuscript and in the context of this model, we investigate the roles of the hydrogen-bonding interactions involving Thr 159, Arg 160, and Tyr 206 between the “mobile” and “fixed” domains observed in Ec but not in Eo as well as Gln 185 and Arg 203 that contribute to the binding site for the 5’-phosphate group in the activation by the 5’-phosphate group of OMP.
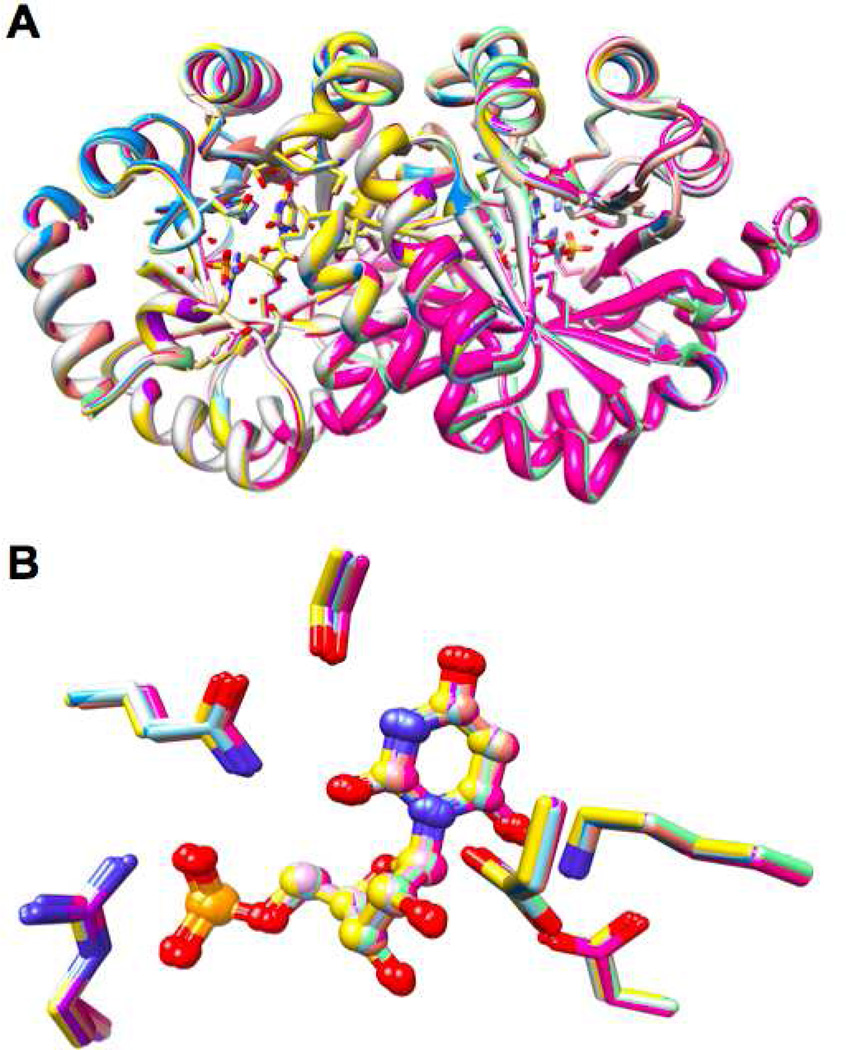
Identification of Interactions at the Domain Interface That Stabilize Ec
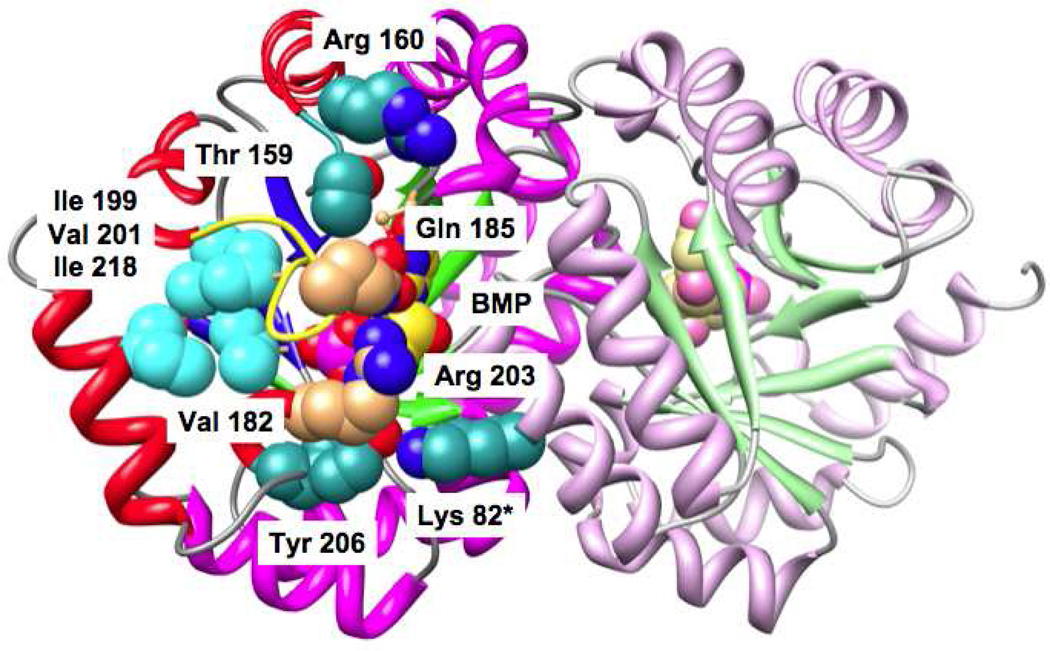
MtOMPDC undergoes a conformational change from Eo in the absence of a ligand to Ec in the presence of BMP. As discussed in the Introduction, this conformational change involves closure of the active site loop (Pro 180 to Asp 188; yellow backbone in Figure 2) as well as the movement of a “mobile” domain (β-strands 6, 7, 8, and 1; red α-helices and blue β-strands in polypeptide A in Figure 5) toward a “fixed” domain (β-strands 2, 3, 4, and 5; magenta α-helices and green β-strands), with the “fixed” domains forming the dimer interface.
Figure 5.
The structure of the dimer of MtOMPDC liganded with BMP, with the residues studied in this manuscript labeled. Ile 199, Val 201, and Ile 218 that participate in the hydrophobic cluster near the N-terminus of the active site loop (with Val 182) are also labeled (19).
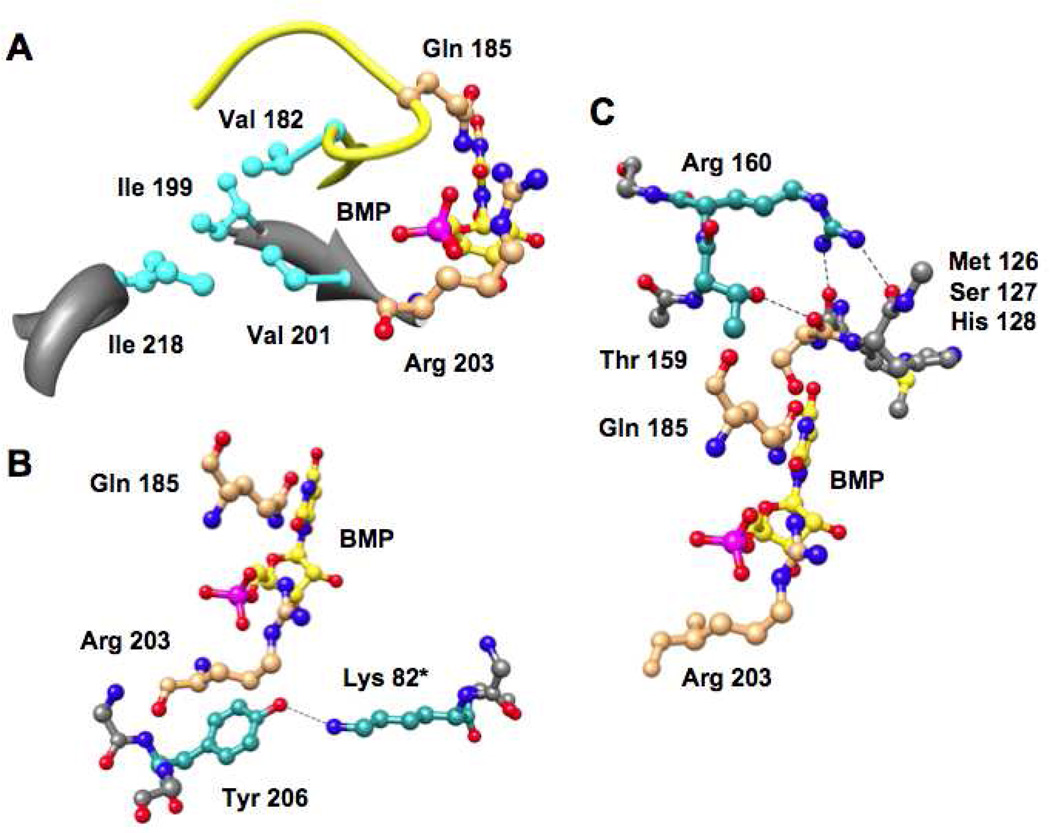
When the active site loop becomes ordered in Ec (yellow backbone in Figures 2 and 5), the hydrophobic cluster involving Val 182 in the active site loop is assembled [Val 182, Ile 199, Val 201, and Ile 218 (19); cyan residues in Figure 5], and hydrogen-bonding contacts also “remote” from the binding site for the 5’-phosphate group are established between the “mobile” and “fixed” domains (Tyr 206 with Lys 82* in the symmetry related polypeptide in the dimer, Thr 159 with the backbone carbonyl oxygen of Ser 127, and Arg 160 with the backbone carbonyl oxygens of both Met 126 and His 128; dark cyan residues in Figure 5). Enlarged views of these interactions are shown in Figure 6 (Gln 185 and Arg 203 that interact directly with the 5’-phosphate group are colored brown). Our hypothesis is that these “remote” interactions at the domain interface also contribute to the stability of Ec relative to Eo as we previously concluded for the hydrophobic cluster (19).
Figure 6.
Interactions formed by closure of the “mobile” and “fixed” domains. Panel A, the hydrophobic cluster involving Val 182 at the N-terminus of the active site loop. Panel B, the hydrogen bond between Tyr 206 and Lys 82* in the symmetry-related polypeptide in the dimer. Panel C, the hydrogen bonds involving 1) the OH group of Thr 159 and the backbone carbonyl oxygen of Ser 127 and 2) the guanidinium group of Arg 160 and the backbone carbonyl oxygens of Met 126 and His 128.
Thr 159, Arg 160, and Tyr 206 in the “mobile” domain were targeted, with the expectation that substitutions of these with Ala (Thr 159 and Arg 160) or Phe (Tyr 206) would destabilize Ec (as determined by the values of kcat/Km for EO). The substitutions were not expected to alter the IBE, as previously observed for the residues that participate in the hydrophobic cluster at the base of the active site loop (19), because they are “remote” from the binding site for the 5’-phosphate group.
Identification of Interactions That Generate the IBE of the 5’-Phosphate Group
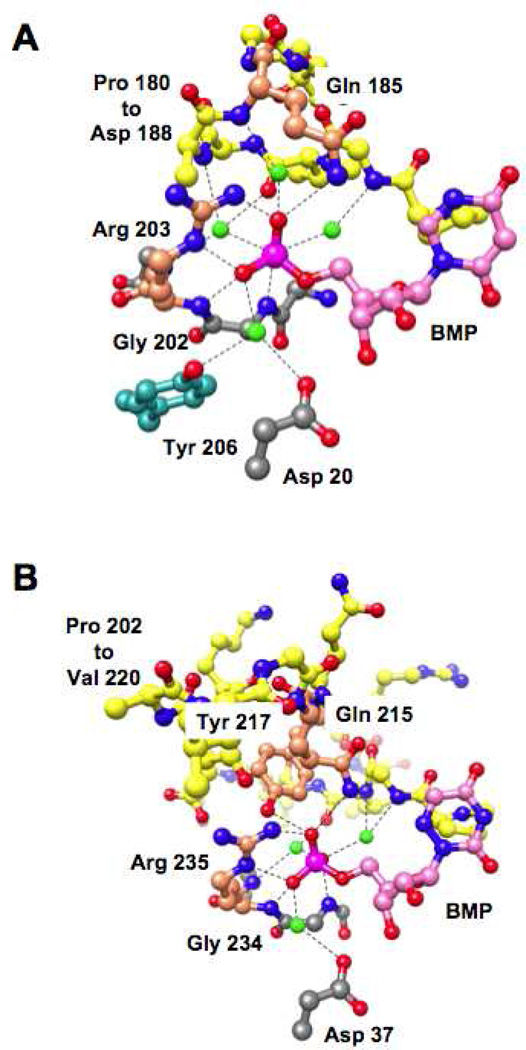
Each of the nonesterified oxygens of the 5’-phosphate group participates in three hydrogen-bonds (a total of nine). As shown in Figure 7, panel A, the 5’-phosphate group of BMP directly contacts the carboxamide nitrogen of the side chain of Gln 185 in the active site loop (via a single hydrogen bond) and the guanidinium group of Arg 203 at the end of the eighth β-strand in the “mobile” domain (via two hydrogen bonds); it also contacts the backbone amide groups of Gly 202 and Arg 203 (via a single hydrogen bond to each). The importance of the interactions with the side chains of Gln 185 and Arg 203 in determining the activation by the 5’-phosphate group can be quantitated by characterization of mutants in which the side chains are “removed”, e.g., the Q185A and R203A substitutions. However, the importance of the interactions with the backbone amide groups cannot be investigated by site-directed mutagenesis-based approaches.
Figure 7.
Hydrogen-bonding interactions of the nonesterified oxygens of the 5’-phosphate group of BMP. Panel A, the active site of MtOMPDC (details are provided in the text). Panel B, the active site of ScOMPDC (details are provided in the text). The water molecules that are involved in hydrogen bonds to the 5’-phosphate groups are colored green.
In addition, the 5’-phosphate group is hydrogen-bonded to four water molecules, with three participating in hydrogen bonds to the backbone amide groups of Gly 181, Val 182, and Gln 185 in the active site loop; the fourth water molecule is hydrogen-bonded to both Tyr 206 in the “mobile” domain and Asp 20 in the “fixed” domain. The importance of these “indirect” contacts of the 5’-phosphate group with the protein also is difficult to investigate using site-directed mutagenesis-based approaches.
Our hypothesis is that all of the contacts made by the 5’-phosphate group are used to generate the total interaction energy that is partitioned between 1) “gripping” the substrate (Ks in Scheme 2/Figure 3; green arrows); and 2) stabilizing both Ec•S and Ec•S‡ (vide infra) relative to Eo•S (Kc in Scheme 2/Figure 3; red arrows). When the active site loop assumes its closed conformation when Gln 185 interacts with the 5’-phosphate group and the hydrophobic cluster involving Val 182 is formed, hydrogen-bonding contacts involving the loop’s backbone, including the carbonyl oxygen of Pro 180, reposition the spatially proximal Thr 159 and Arg 160 in the “mobile” domain so that their side chains can form hydrogen bonds across the domain interface to the backbone carbonyl oxygens of Ser 127 as well as those of Met 126 and His 128 that flank Ser 127 in the “fixed” domain. The carboxamide oxygen of Gln 185 also forms a hydrogen bond with the OH group of Ser 127 in the “fixed” domain. Thus, we propose that IBE generated by the 5’-phosphate group is used, in part, to enable assembly of the hydrophobic cluster at the base of the active site loop as well as the interactions across the domain interface, with these providing independent contributions to the stabilization of Ec•S and Ec•S‡ relative to Eo•S (Figures 5 and 6).
Gln 185 and Arg 203 were targeted for site-directed mutagenesis, with the expectation that Ala substitutions would destabilize Ec•S and Ec•S‡ relative to Eo + S as judged by a decrease in the IBE as the result of a reduction in the value of kcat/Km for OMP but not for EO. Although the Q185A could be expected to decrease the value of Kc because of the interaction of the carboxamide group with Ser 127 in the “mobile” domain (Figure 1; as measured by the value of kcat/Km for EO), R203A is not expected to alter the value of Kc because it is “remote” from the domain interface.
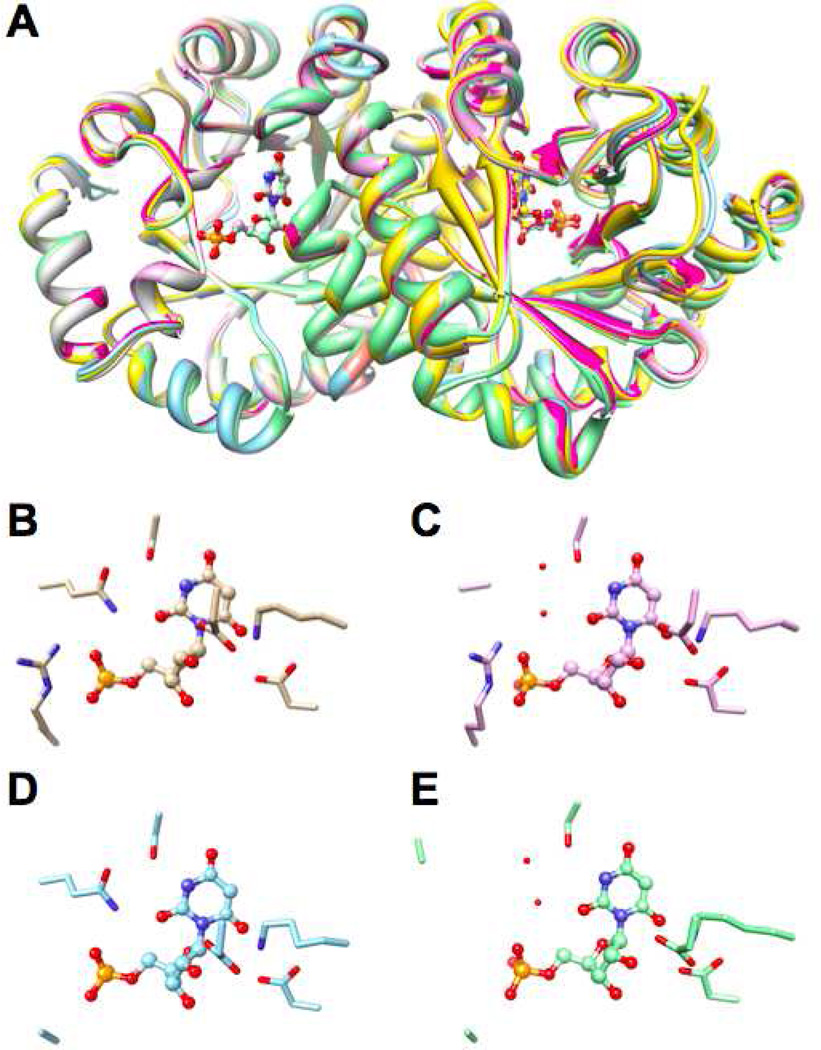
Structures of Mutants of Residues that Stabilize Ec
Five single mutants of residues that stabilize Ec [T159A, T159S, T159V, R160A, and Y206F; V182A was previously described and structurally characterized (19)] as well as six double/triple mutants that combine four of these (T159V/V182A, T159V/Y206F, R160A/V182A, R160A/Y206F, V182A/Y206F, and T159V/V182A/Y206F) were constructed and subjected to structural characterization in the presence of BMP. In these structures, the catalytic triads, Asp 70, Lys 72, and Asp 75* (from the symmetry related polypeptide in the dimer) as well as Ser 127 that hydrogen-bonds to the pyrimidine ring, superimpose well on the wild type enzyme (Figure 8). With the exception of the T159A substitution, the only structural changes are those associated with the side chain of the substituted residue; in the case of T159A, the conformation of the loop flanking the substitution (Pro 157 – Arg 163) is altered. Not surprisingly, the T159A mutant displayed significantly compromised kinetic constants (the value of kcat/Km is reduced 104-fold from that measured for wild type; data not shown) compared to all of the other mutants of residues that stabilize Ec. Our observation that the values of kcat measured for the remaining mutants are “unchanged” relative to that measured for the wild type enzyme is consistent with the observation that the structures of the active sites are not perturbed by the substitutions.
Figure 8.
Superposition of the BMP-liganded structures of wild type MtOMPDC and the K82A, T159V, R160A, Y206F, T159V/V182A, T159V/Y206F, V182A/Y206F, and T159V/V182A/Y206F mutants of residues that stabilize Ec. Panel A, the polypeptide dimers. Panel B, structures of the active sites.
Structures of Mutants of Residues that Generate the IBE of the 5’-Phosphate Group
We constructed the Q185A and R203 single substitutions as well as the Q185A/R203A double substitution to remove the direct contacts between the 5’-phosphate group and the side chains of the protein. In the structure of the Q185A/R203A double mutant, the active site loop (Pro 180 to Asp 188) is partially disordered in the A polypeptide (Ala 184 and Ala 185; with increased B-values for the remaining “visible” loop residues) and assumes an altered conformation (also with increased B-values) in the B polypeptide (Figure 9).
Figure 9.
Superposition of the BMP-liganded structures of wild type MtOMPDC and the Q185A, R203A, and Q185A/R203A mutants of residues that generate the IBE of the 5’-phosphate group. Panel A, the polypeptide dimers. Panel B, active site of wild type. Panel C, active site of the Q185A mutant. Panel D, Active site of the R203A mutant. Panel E, active site of the Q185A/R203A mutant. The red spheres represent water molecules.
In the mutant proteins, the structures of the active sites are not affected by the substitution(s); this is consistent with our observation that the values of kcat for the Q185A and R203A single mutants are “unchanged” from that measured for wild type enzyme (vide infra); the value of kcat could not be measured for the Q185A/R203A double mutant because the value of Km is significantly elevated (Ks is significant decreased) by the loss of these contacts with the 5’-phosphate group. In the Q185A and Q185A/R203A mutants, two “new” water molecules occupy the position of the carboxamide group of Gln 185 in the wild type enzyme: one is located in the same position as the carboxamide amino group and is hydrogen-bonded to one of the nonesterified phosphoryl oxygens; the second is located in the same position as the carboxamide carbonyl oxygen and is hydrogen-bonded to both the first water molecule and the OH group of Ser 127. This hydrogen-bonded “chain” maintains an indirect interaction of the 5’-phosphate group with the “fixed” domain and, therefore, is expected to contribute the stability of the domain interface although the carboxamide group is “missing”. Finally, in the R203A and Q185A/R203A mutants, a “new” water molecule located in the same position as the ε-nitrogen of the guanidinium group in the wild type protein is hydrogen-bonded to a second nonesterified oxygen.
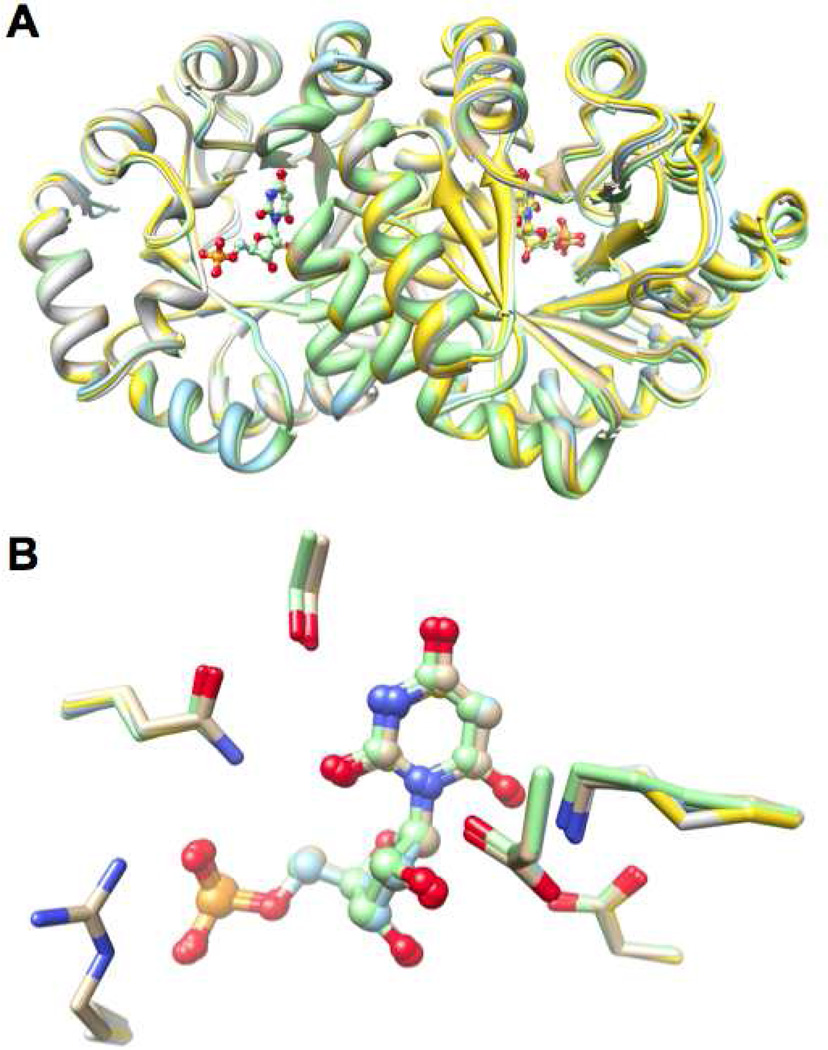
Structures of Double Mutants of Residues that Stabilize Ec and Generate the IBE of the 5’-Phosphate Group
The R203A substitution was combined separately with the spatially remote T159V, R160A, and V182A substitutions. As expected based on the structures of the single substitutions and the unchanged values for kcat (vide infra), the structures of the active sites were unchanged in the mutants (Figure 10). Also, as in the structure of R203A, in each structure a “new” water molecule is located in the same position as the ε-nitrogen of the guanidinium group in the wild type protein.
Figure 10.
Superposition of the BMP-liganded structures of wild type MtOMPDC and the T159V/R203A, R160A/R203A, and V182A/R203A double mutants of residues that stabilize Ec and generate the IBE of the 5’-phosphate group. Panel A, the polypeptide dimers. Panel B, structures of the active sites.
Kinetic Constants for Mutants of Residues that Stabilize Ec
The values of kcatKmand kcat/Km using OMP as substrate were determined for the wild type and mutants that contain “remote” substitutions for residues hypothesized to stabilize Ec relative to Eo (Table 3). The values of kcat/Km for EO and/or FEO, (kcat/Km)/KD for activation of decarboxylation of EO by phosphite (the ability of phosphite to mimic the 5’-phosphate group), and the IBEs for the 5’-phosphate group also were determined (Table 4).
Table 4.
Kinetic Constants for EO and FEO and IBEs of the 5'-Phosphate Group of OMP at pH 7.1 and 25 °C
| MtOMPDC |
kcat/Km OMPa (M−1 s−1) |
ΔΔG‡ (kcal/mol) |
kcat/Km EOb (M−1 s−1) |
ΔΔG‡ (kcal/mol) |
KcatKm FEOb (M−1 s−1) |
FEO/EO | (kcat/Km)/KD EO•HPib M−2 s−1 |
ΔΔG‡ (kcal/mol) |
5'-Phosphate IBEc kcal/mol |
|---|---|---|---|---|---|---|---|---|---|
| Wild type | 2.9 ×106 | - | 12.4 ×10−3 | - | 4.8 | 390 | 2300 | - | 11.4d |
| T159V | 7.0 ×104 | 2.2 | 0.4 ×10−3 | 2.0 | 0.083 | 210 | 35 | 2.4 | 11.2 |
| R160A | 6.7 ×104 | 2.2 | 0.2 ×10−3 | 2.4 | 0.21 | 1100 | 56 | 2.1 | 11.6 |
| V182A | 1.3 ×105 | 1.8 | 1.4 ×10−3 | 1.3 | 0.63 | 470 | 150 | 1.6 | 10.8 |
| Y206F | 7.5 ×105 | 0.8 | 6.0 ×10−3 | 0.4 | 2.2 | 370 | 280 | 1.2 | 11.0 |
| T159V/V182A | 8.0 ×102 | 4.8 | 6.2 × 10−5e | 3.1 | 0.034 | 9.6 | |||
| T159V/Y206F | 3.7 ×103 | 3.9 | 6.3 × 10−5 e | 3.1 | 0.035 | 10.5 | |||
| R160A/V182A | 1.7 ×103 | 4.4 | 4.6 × 10−5e | 3.3 | 0.025 | 10.3 | |||
| R160A/Y206F | 8.5 ×103 | 3.4 | 5.8×10−5e | 3.1 | 0.032 | 11.1 | |||
| V182A/Y206F | 1.5 ×104 | 3.1 | 5.5 × 10−4e | 1.8 | 0.3 | 10.1 | |||
| T159V/V182A/Y206F | 1.3 ×102 | 5.9 | 3.1×10−5e | 3.5 | 0.017 | 9.0 | |||
| Q185A | 1.3 ×104 | 3.1 | 2 × 10−3 | 1.0 | 0.92 | 470 | 24 | 2.7 | 9.2 |
| R203A | 1.5 ×103 | 4.4 | 5.5 ×10−3 | 0.4 | 4.7 | 860 | 2.4 | 4.0 | 7.4 |
| Q185A/R203A | 9.6 | 7.4 | 3.2 ×10−3 | 0.8 | 4.7 | ||||
| T159V/R203A | 9.9 | 7.4 | 0.2 ×10−3 | 2.4 | 6.4 | ||||
| R160A/R203A | 16 | 7.1 | 0.2 ×10−3 | 2.4 | 6.6 | ||||
| V182A/R203A | 32 | 6.7 | 1.3 ×10−3 | 1.3 | 5.9 |
From Table 3.
The estimated error is ±10%.
Calculated from the ratio of the values of kcat/Km for OMP and EO.
A lower estimate because the value of kcat/Km for OMP is partially diffusion-controlled (24).
Calculated from the value of kcat/Km for FEO by dividing the value by 550 (the average ratio of the values for kcat/Km for EO and FEO for the wild type and those mutants for which the value for EO could be reliably measured).
For the double/triple mutants, the more reactive FEO was used to estimate the values of kcat/Km for EO (25, 33, 34). For these mutants, the values of Km for OMP and, therefore, both EO and FEO are significantly elevated because the fraction of the enzyme that exists as Ec is reduced (Kc’ is reduced; blue arrows in Figure 3) because interactions that stabilize Ec are absent. Because of the increased reactivity of FEO, lower concentrations of enzyme (and/or shorter reaction times) can be used to measure the values of kcat/Kmthereby making these measurements possible. The correction factor for the double/triple mutants, 550, was obtained by averaging the values of the ratios of kcat/Km for EO and FEO for wild type MtOMPDC and the single mutants (Table 4).
For the single and most of the double mutants using OMP as substrate, the values of kcat are “unchanged” relative to that measured for wild type enzyme; in contrast, the values of Km are significantly increased. [The values for kcat for OMP are (modestly) decreased for the T159V/V182A and T159V/V182A/Y206F mutants, suggesting small changes in structure and/or dynamics.] These observations are similar to those we reported for Ala substitutions for the residues in the hydrophobic cluster at the base of the active site loop that includes Val 182 (19). The increases in the values of Km for the double/triple mutants are approximately multiplicative relative to the single mutants (additive in free energy), providing support that these “remote” residues provide “independent” contributions that stabilize Ec•S relative to Eo + S.
The values of kcat/Km for EO for the single mutants are decreased from that measured for wild type; the calculated values for the double/triple mutants are more significantly reduced, with the changes again approximately multiplicative relative to the single mutants. That the fold-decreases in the values of kcat/Km for OMP and EO are equivalent reinforces the interpretation that the targeted residues are important in determining the stability of Eci.e., the substitutions give decreased values for Kc and Kc’ (blue arrows in Figure 3).
The values of (kcat/Km)/KD for activation of the decarboxylation of EO by phosphite were determined for the single mutants; these demonstrate that phosphite retains the ability to activate the mutants. That the values of kcat/Km for OMP, kcat/Km for EO, and (kcat/Km)/KD for phosphite are equivalently decreased is consistent with the interpretation that the changes are determined by the values of Kci.e., the 5’-phosphate group of OMP and phosphite both retain the ability to increase the stability of Ec•S (and E•S‡) relative to EoS (red arrows in Figure 3)
As a result of the equivalent effects of the substitutions on the values of kcat/Km for OMP and EO, the IBEs for the 5’-phosphate group for the single and most of the double mutants are unchanged from that measured for the wild type enzyme; the same observation was made for Ala substitutions for the residues in the hydrophobic cluster (19). [For the T159V/V182A and T159V/V182A/Y206F mutants, the values of the IBEs are modestly decreased, likely the result of the same small structural changes that may affect the values of kcat.] We conclude that although the 5’-phosphate group provides the same IBE, Ec•S and E•S‡ are less stabilized for the mutants than the wild type enzyme because the interactions that stabilize the closed conformation of the active site loop (Val 182) and stabilize the domain interface (Thr 159, Arg 160, and Tyr 206) have been disrupted.
Kinetic Constants for Mutants of Residues that Generate the IBE of the 5’-Phosphate Group
Using both OMP and EO, we measured the values of kinetic constants for Q185A, R203A, and Q185A/R203A, the two single mutants and one double mutant with substitutions for the two residues with side chains that directly contact the 5’-phosphate group of the substrate (Tables 3 and 4). With OMP, the values of kcat and Km and, therefore, kcat/Km could be measured for Q185A and R203A; however, only the value kcat/Km could be measured for Q185A/R203A.
The values of kcat for the Q185A and R203A mutants are “unchanged” from that measured for wild type enzyme (Table 3), so the energy difference between the Ec•S and Ec•S‡ complexes is not affected by the decreased ability of the “remote” 5’-phosphate group to activate the enzyme. Therefore, these substitutions alter the energies of both Ec•S and Ec•S‡ relative to Eo + S, i.e., the 5’-phosphate group does not preferentially stabilize the transition state complex, Ec•S‡, but also equally stabilizes the Michaelis complex, Ec•S, because both share the Ec conformation.
The values of kcat/Km for OMP are decreased but the values for EO are “unchanged” from that measured for wild type. The decreased value for OMP is explained by the weaker affinity of enzyme for the 5’-phosphate group, i.e., a decrease in the value of Ks (green arrow in Figure 3). That the values for EO are unchanged provides evidence that Gln 185 and Arg 203 are not involved in determining the relative energies of Eo and Ec.4 Thus, we hypothesize that the IBE enables the conformational reorganization of the active site loop that allows formation of the interdomain contacts involving Thr 159, Arg 160, and Tyr 206 as well as assembly of the hydrophobic cluster involving Val 182 in the active site loop, with these determining the stability of Ec relative to Eo.
The value of the IBE of the 5’-phosphate group for the wild type enzyme is 11.4 kcal/mol (Table 4). For the Q185A and R203A mutants, the values are decreased by 2.1 and 4.0 kcal/mol, respectively; for Q185A/R203A, the value is decreased by 6.7 kcal/mol (approximately the sum of the those for the single mutants). But, despite the complete absence of the direct contacts of the 5’-phosphate group with side chains, the Q185A/R203A mutant is activated 2,900-fold by the 5’-phosphate group (the IBE is 4.7 kcal/mol). The 5’-phosphate group retains its interactions with the backbone amide groups of Gly 202 and Arg 203 as well as several ordered water molecules, including one that also hydrogen-bonds with Asp 20 in the “fixed” domain (Figure 7, panel A).
The values of (kcat/Km)/KD for activation of the decarboxylation of EO by phosphite were also determined for the single mutants. The values are decreased significantly, as expected because the values of Ks for phosphite binding should be decreased by the absence of these direct interactions.
That the side chain contacts with the 5’-phosphate group are not entirely responsible for generating the IBE of the 5’-phosphate group contrasts with results we reported for similar mutagenesis-based experiments with the OMPDC from Saccharomyces cerevisiae (ScOMPDC) (35). In ScOMPDC, the 5’-phosphate group interacts with the side chains of three residues, Gln 215 and Arg 235, structural homologues of Gln 185 and Arg 203 in MtOMPDC, as well as Tyr 217 in the longer (19-residue) active site loop (Figure 7, panel B). In ScOMPDC, the 5’-phosphate group is hydrogen-bonded to the backbone amide groups of Gly 234 and Arg 235, structural homologues of Gly 202 and Arg 203 in MtOMPDC. Also, in ScOMPDC, the 5’-phosphate group is hydrogen-bonded to three (not four) water molecules, two hydrogen-bonded to the backbone of the active site loop and the third hydrogen-bonded to Asp 37, the structural homologue of Asp 20 in MtOMPDC.
Kinetic analyses for single and multiple substitutions for the residues that contact the 5’-phosphate group (Q215A, Y217F, and R203A) revealed progressive decreases in the values of the IBE, with the values of kcat/Km for OMP only 12-fold greater than that for EO for the Q215A/Y217F/R235A triple mutant. An explanation for why the direct contacts with the side chains apparently are “fully” responsible for generating the IBE in ScOMPDC but not in MtOMPDC is not obvious. However, the active site loops differ in both length and conformation; also, structures are not available for the mutants of ScOMPDC, so unlike MtOMPDC, the effects of the substitutions on the structure of the protein and active site cannot be assessed.
Kinetic Constants for Double Mutants of Residues that Stabilize Ec and Generate the IBE of the 5’-Phosphate Group
To obtain evidence that the “remote” interactions that stabilize Ec•S and Ec•S‡ relative to Eo•S and those that generate the IBE of the 5’-phosphate group by direct contacts are independent, we constructed and characterized the R203A/T159V, R203A/R160A, and R203A/V182A double mutants. We chose the R203A instead of the Q185A substitution because Gln 185 is hydrogen-bonded not only to the 5’-phosphate group but also to the OH group of Ser 127 in the “mobile” domain, i.e., Gln 185 could contribute to both stabilization of Ec (and, therefore, the value of Kc’) as well as generation of the IBE; in fact, the Q185A substitution does cause a six-fold decrease in the value of kcat/Km for EO.4 The kinetic constants for these mutants also are included in Tables 3 and 4.
The values of kcat and Km for OMP could not be measured for these mutants: because all of the single mutants effect the stabilities of Ec•S relative to Eo + S, the values of the Km were expected to be and are, in fact, too large to be measured (Table 3). The values of kcat/Km for EO for the double mutants are essentially identical to those measured for the T159V, R160A, and V182A single mutants that alter the stability of Ec •S relative to Eoi.e., the value of kcat/Km for EO for the R203A mutant is “unchanged” (only two-fold decreased) from that measured for the wild type enzyme (Table 4). Thus, the effects of the substitutions are additive with respect to stabilization of Ec in both Ec•S and Ec•S‡. Also, the values for the IBE for the 5’-phosphate group are essentially the same as that measured for the R203A mutant, i.e., the effects of the substitutions on the IBE also are additive (Table 4).
Independent Structural Strategies to Enable and Accomplish the Stabilization of Ec
Taken together, our structural and kinetic observations provide persuasive evidence that the model presented in Scheme 2/Figure 3 provides a structure-based understanding of how the IBE of the 5’-phosphate group is both generated and used to activate the enzyme. The direct interactions of the 5’-phosphate group with the side chains of Gln 185 in the active site loop and Arg 203 (as well as the backbone amide groups of Gly 202 and Arg 203 and water molecules that interact with the active site loop; green arrows in Figure 3) organize the active site loop (red arrows) so that the hydrophobic cluster at the base of the active site loop and the “remote” interdomain interactions can be formed to stabilize Ec•S and Ec•S‡ relative to Eo•S (blue arrows).
This “cascade” of structurally expanding interactions, triggered by the binding of OMP to Eo and likely orchestrated by changes in the structure of the active site loop directed by the “remote” 5’-phosphate group, results in the remarkable 2.4 × 108-fold rate enhancement as the stabilizing/destabilizing interactions are enforced in Ec•S and Ec•S‡. Further studies of interactions of the 5’-phosphate group with its binding site, including the importance of the water-mediated contacts with the active site loop, are in progress.
Conclusions
The importance of the IBEs of “remote” substituents in achieving the transition state stabilization that are responsible for the impressive rate enhancements of enzyme-catalyzed reactions has long been recognized (23). However, a unifying structure-based description and understanding for how the IBE of remote substituents is generated and used to effect transition state stabilization has been elusive. Like triose phosphate isomerase (22), MtOMPDC provides a useful system to explore and define how the IBE of the “remote” phosphate group of the substrate is generated and used to contribute to its impressive 1017 rate acceleration: the reaction is unimolecular and requires no cofactors, the wild type enzyme and mutants can be structurally characterized, a sensitive direct spectrophotometric assay is available, and substrate analogs (EO and FEO) can be used to “isolate” the role of the activating 5’-phosphate group. Prior structural characterization established that substrate binding in MtOMPDC is accompanied by a conformational change in which a catalytically active, closed conformation (Ec) that is unfavorable in the absence of the substrate predominates when substrate is bound. We now have obtained persuasive kinetic and structural evidence that the IBE of the 5’-phosphate group provides the energy for a local reorganization of the structure of its binding site in the Eo•S complex (in particular, the ordering of the active site loop that includes Gln 185) that then enables the formation of additional “remote” interactions at the interface between the “mobile” and “fixed” domains that provide the energy to stabilize Ec in both the Ec•S and Ec•S‡ complexes. As a result of this essential conformational change in which Ecthe catalytically active conformation, is unstable relative to Eothe unliganded conformation, product release is more favorable than if the total energy obtained from substrate binding were used “simply” to bind the substrate. We expect that this strategy will be applicable to understanding the structural basis for the activation of other enzymes that undergo an essential conformational change from an open, inactive conformation (Eo) to a closed, active conformation (Ec) as the result of substrate binding.
Our studies using both the intact OMP substrate and EO/FEO that lack the 5’-phosphate group (19, 25, 26, 32) provide a kinetics/structure-based approach for understanding the role of the substrate and the IBE of its pieces in enabling the conformational changes that are required for efficient enzymatic catalysis. The dynamic characteristics of conformational changes that accompany catalysis have been studied in other enzymes, including adenylate kinase (36, 37) and dihydrofolate reductase (38–40). The principles that emerge from the kinetics-based approaches described in this manuscript and previous studies of triose phosphate isomerase (22) as well as dynamics-based approaches that employ time-resolved NMR spectroscopy, isotope exchange, and simulations together 1) provide both a description and an understanding of Nature’s structural strategies for achieving large rate accelerations, and 2) inform the (re)design of enzymic catalysts for novel reactions by revealing the requirements for substrate binding to Eo, utilization of energy provided by binding interactions with “remote” portions of the substrate to, in part, effect a necessary conformational transition from Eo•S to Ec•S that allows catalysis via Ec•S‡ and, finally, a favorable conformational relaxation to Eo to permit product dissociation.
Scheme 3.
ACKNOWLEDGEMENTS
We are grateful to the staff of NSLS beamlines X4A and X29A for their help with collecting diffraction data.
Footnotes
This research was supported by NIHGM039754 (to J. P. R.) and GM065155 (to J.A.G.). Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081). The X-ray coordinates and structure factors for the following structures have been deposited in the Protein Data Bank with the indicated accession codes: the K82A mutant of MtOMPDC in the presence of BMP (PDB accession code 3RLU), the T159A mutant of MtOMPDC in the presence of BMP (PDB accession code 3P5Y), the T159S mutant of MtOMPDC in the presence of BMP (PDB accession code 3P5Z), the T159V mutant of MtOMPDC in the presence of BMP (PDB accession code 3P60), the R160A mutant of MtOMPDC in the presence of BMP (PDB accession code 3P61), the Q185A mutant of MtOMPDC in the presence of BMP (PDB accession code 3V1P), the R203A mutant of MtOMPDC in the presence of BMP (PDB accession code 3LI0), the Y206F mutant of MtOMPDC in the presence of BMP (PDB accession code 3RLV), the T159V/V182A mutant of MtOMPDC in the presence of BMP (PDB accession code 3QEZ), the T159V/R203A mutant of MtOMPDC in the presence of BMP (PDB accession code 4FXR), the T159V/Y206F mutant of MtOMPDC in the presence of BMP (PDB accession code 3QF0), the R160A/V182A mutant of MtOMPDC in the presence of BMP (PDB accession code 3QMR), the R160A/R203A mutant of MtOMPDC in the presence of BMP (PDB accession code 4GC4), the R160A/Y206F mutant of MtOMPDC in the presence of BMP (PDB accession code 3SJ3), the V182A/R203A mutant of MtOMPDC in the presence of BMP (PDB accession code 4FX6), the V182A/Y206F mutant of MtOMPDC in the presence of BMP (PDB accession code 3QMT, and the Q185A/R203A mutant of MtOMPDC in the presence of BMP (PDB accession code 4FX8), and the T159V/V182A/Y206F mutant of MtOMPDC in the presence of BMP (PDB accession code 3QMS).
Abbreviations: BMP, l-(5-phospho-β-ribofuranosyl)barbituric acid; FOMP, 5-fluoroorotidine 5’-monophosphate; EO, 1-(β-D-erythrofuranosyl)orotic acid; FEO, 5-fluoroEO; IBE, intrinsic binding energy; OMPDC, OMP decarboxylase; MtOMPDC, OMPDC from Methanothermobacter thermautotrophicus; ScOMPDC, OMPDC from Saccharomyces cerevisiae.
All OMPDCs that have been structurally characterized in the absence and presence of a ligand (intermediate/transition state analog or UMP product) undergo a conformational change from an open, catalytically inactive conformation (Eo) in the absence of the ligand to a closed, catalytically active conformation (Ec) in the presence of the ligand. This article focuses on MtOMPDC because the wild type protein and its mutants are particularly amenable to structure determination by x-ray crystallography: to date, structures have been determined for all mutants we have constructed and kinetically characterized, including the eighteen described in this article. Although the active site residues that deliver substrate stabilization/destabilization are conserved in all OMPDCs, the lengths and sequences of the active site loops that sequester the substrate from solvent differ so the identities and locations of the residues in the loops that interact with the 5’-phosphate group of the substrate are not conserved (1). However, we hypothesize that the general structural strategy by which substrate binding favors the transition of Eo•S to Ec•S is conserved, although the details are not conserved.
The only possible pathway from Eo to Ec•S is via [Eo•S] because the closed active site loop in Ec prevents binding of S to Ec. Also, Km = 1/[Kc•Ks] because Km is defined as a dissociation complex but Ks is defined as an association constant.
For Q185A, the value of kcat/Km for EO is reduced six-fold, perhaps reflecting the hydrogen-bonding of the carboxamide group of to hydrogen-bond to the OH group of Ser 127 in the “mobile” domain. However, as described previously, the structure of the Q185A mutant reveals that the carboxamide group is replaced by two “new” water molecules that form a hydrogen-bond chain between the 5’-phosphate group and Ser 127.
REFERENCES
- 1.Toth K, Amyes TL, Wood BM, Chan KK, Gerlt JA, Richard JP. An examination of the relationship between active site loop size and thermodynamic activation parameters for orotidine 5'-monophosphate decarboxylase from mesophilic and thermophilic organisms. Biochemistry. 2009;48:8006–8013. doi: 10.1021/bi901064k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radzicka A, Wolfenden R. A proficient enzyme. Science. 1995;267:90–93. doi: 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- 3.Wolfenden R. Benchmark reaction rates, the stability of biological molecules in water, and the evolution of catalytic power in enzymes. Annu Rev Biochem. 2011;80:645–667. doi: 10.1146/annurev-biochem-060409-093051. [DOI] [PubMed] [Google Scholar]
- 4.Miller BG, Snider MJ, Short SA, Wolfenden R. Contribution of enzyme-phosphoribosyl contacts to catalysis by orotidine 5'-phosphate decarboxylase. Biochemistry. 2000;39:8113–8118. doi: 10.1021/bi000818x. [DOI] [PubMed] [Google Scholar]
- 5.Miller BG, Snider MJ, Wolfenden R, Short SA. Dissecting a charged network at the active site of orotidine-5'-phosphate decarboxylase. J Biol Chem. 2001;276:15174–15176. doi: 10.1074/jbc.M011429200. [DOI] [PubMed] [Google Scholar]
- 6.Van Vleet JL, Reinhardt LA, Miller BG, Sievers A, Cleland WW. Carbon isotope effect study on orotidine 5'-monophosphate decarboxylase: support for an anionic intermediate. Biochemistry. 2008;47:798–803. doi: 10.1021/bi701664n. [DOI] [PubMed] [Google Scholar]
- 7.Wepukhulu WO, Smiley VL, Vemulapalli B, Smiley JA, Phillips LM, Lee JK. A substantial oxygen isotope effect at O2 in the OMP decarboxylase reaction: mechanistic implications. Org Biomol Chem. 2008;6:4533–4541. doi: 10.1039/b812979g. [DOI] [PubMed] [Google Scholar]
- 8.Sievers A, Wolfenden R. Equilibrium of formation of the 6-carbanion of UMP, a potential intermediate in the action of OMP decarboxylase. J Am Chem Soc. 2002;124:13986–13987. doi: 10.1021/ja021073g. [DOI] [PubMed] [Google Scholar]
- 9.Toth K, Amyes TL, Wood BM, Chan K, Gerlt JA, Richard JP. Product deuterium isotope effect for orotidine 5'-monophosphate decarboxylase: evidence for the existence of a short-lived carbanion intermediate. J Am Chem Soc. 2007;129:12946–12947. doi: 10.1021/ja076222f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amyes TL, Wood BM, Chan K, Gerlt JA, Richard JP. Formation and stability of a vinyl carbanion at the active site of orotidine 5'-monophosphate decarboxylase: pKa of the C-6 proton of enzyme-bound UMP. J Am Chem Soc. 2008;130:1574–1575. doi: 10.1021/ja710384t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine HL, Brody RS, Westheimer FH. Inhibition of orotidine-5'- phosphate decarboxylase by 1-(5'-phospho-beta-d-ribofuranosyl)barbituric acid, 6- azauridine 5'-phosphate, and uridine 5'-phosphate. Biochemistry. 1980;19:4993–4999. doi: 10.1021/bi00563a010. [DOI] [PubMed] [Google Scholar]
- 12.Wu N, Mo Y, Gao J, Pai EF. Electrostatic stress in catalysis: structure and mechanism of the enzyme orotidine monophosphate decarboxylase. Proc Natl Acad Sci U S A. 2000;97:2017–2022. doi: 10.1073/pnas.050417797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan KK, Wood BM, Fedorov AA, Fedorov EV, Imker HJ, Amyes TL, Richard JP, Almo SC, Gerlt JA. Mechanism of the orotidine 5'- monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization. Biochemistry. 2009;48:5518–5531. doi: 10.1021/bi900623r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis CA, Jr, Wolfenden R. Orotic acid decarboxylation in water and nonpolar solvents: a potential role for desolvation in the action of OMP decarboxylase. Biochemistry. 2009;48:8738–8745. doi: 10.1021/bi901085m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iiams V, Desai BJ, Fedorov AA, Fedorov EV, Almo SC, Gerlt JA. Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site. Biochemistry. 2011;50:8497–8507. doi: 10.1021/bi2012355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JK, Houk KN. A proficient enzyme revisited: the predicted mechanism for orotidine monophosphate decarboxylase. Science. 1997;276:942–945. doi: 10.1126/science.276.5314.942. [DOI] [PubMed] [Google Scholar]
- 17.Houk KN, Lee JK, Tantillo DJ, Bahmanyar S, Hietbrink BN. Crystal structures of orotidine monophosphate decarboxylase: does the structure reveal the mechanism of nature's most proficient enzyme? Chembiochem. 2001;2:113–118. doi: 10.1002/1439-7633(20010202)2:2<113::AID-CBIC113>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 18.Handschumacher RE. Orotidylic acid decarboxylase: inhibition studies with azauridine 5'-phosphate. J Biol Chem. 1960;235:2917–2919. [PubMed] [Google Scholar]
- 19.Wood BM, Amyes TL, Fedorov AA, Fedorov EV, Shabila A, Almo SC, Richard JP, Gerlt JA. Conformational changes in orotidine 5'- monophosphate decarboxylase:"remote" residues that stabilize the active conformation. Biochemistry. 2010;49:3514–3516. doi: 10.1021/bi100443a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris P, Poulsen JC, Jensen KF, Larsen S. Substrate binding induces domain movements in orotidine 5'-monophosphate decarboxylase. J Mol Biol. 2002;318:1019–1029. doi: 10.1016/S0022-2836(02)00200-0. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan SM, Holyoak T. Enzymes with lid-gated active sites must operate by an induced fit mechanism instead of conformational selection. Proc Natl Acad Sci U S A. 2008;105:13829–13834. doi: 10.1073/pnas.0805364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richard JP. A Paradigm for Enzyme-Catalyzed Proton Transfer at Carbon: Triosephosphate Isomerase. Biochemistry. 2012;51:2652–2661. doi: 10.1021/bi300195b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jencks WP. Binding energy, specificity, and enzymic catalysis: the circe effect. Adv Enzymol Relat Areas Mol Biol. 1975;43:219–410. doi: 10.1002/9780470122884.ch4. [DOI] [PubMed] [Google Scholar]
- 24.Wood BM, Chan KK, Amyes TL, Richard JP, Gerlt JA. Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: effect of solvent viscosity on kinetic constants. Biochemistry. 2009;48:5510–5517. doi: 10.1021/bi9006226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goryanova B, Amyes TL, Richard JP. Phosphite Dianion Activation of Orotidine 5'-Phosphate Decarboxylase: Substituent Effects on Reactions of the Substrate Pieces. Biochemistry. 2012 in preparation. [Google Scholar]
- 26.Amyes TL, Richard JP, Tait JJ. Activation of orotidine 5'- monophosphate decarboxylase by phosphite dianion: the whole substrate is the sum of two parts. J Am Chem Soc. 2005;127:15708–15709. doi: 10.1021/ja055493s. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CWJ, Sweet RM, Abelson JN, Simon MI, editors. Methods in Enzymology. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 28.Long F, Vagin AA, Young P, Murshudov GN. BALBES: a molecularreplacement pipeline. Acta Crystallogr D Biol Crystallogr. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamzin VS, Wilson KS. Automated refinement for protein crystallography. Methods Enzymol. 1997;277:269–305. doi: 10.1016/s0076-6879(97)77016-2. [DOI] [PubMed] [Google Scholar]
- 32.Barnett SA, Amyes TL, Wood BM, Gerlt JA, Richard JP. Dissecting the total transition state stabilization provided by amino acid side chains at orotidine 5'-monophosphate decarboxylase: a two-part substrate approach. Biochemistry. 2008;47:7785–7787. doi: 10.1021/bi800939k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toth K, Amyes TL, Wood BM, Chan K, Gerlt JA, Richard JP. Product deuterium isotope effects for orotidine 5'-monophosphate decarboxylase: effect of changing substrate and enzyme structure on the partitioning of the vinyl carbanion reaction intermediate. J Am Chem Soc. 2010;132:7018–7024. doi: 10.1021/ja102408k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsang WY, Wood BM, Wong FM, Wu W, Gerlt JA, Amyes TL, Richard JP. Proton Transfer from C-6 of Uridine 5'-Monophosphate Catalyzed by Orotidine 5'-Monophosphate Decarboxylase: Formation and Stability of a Vinyl Carbanion Intermediate and the Effect of a 5-Fluoro Substituent. J Am Chem Soc. 2012;134:14580–14594. doi: 10.1021/ja3058474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amyes TL, Ming SA, Goldman LM, Wood BM, Desai BJ, Gerlt JA, Richard JP. Orotidine 5'-Monophosphate Decarboxylase: Transition State Stabilization from Remote Protein-Phosphodianion Interactions. Biochemistry. 2012;51:4630–4632. doi: 10.1021/bi300585e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M, Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450:913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 37.Henzler-Wildman KA, Thai V, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson MA, Petsko GA, Karplus M, Hubner CG, Kern D. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- 38.Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 39.Boehr DD, Dyson HJ, Wright PE. Conformational relaxation following hydride transfer plays a limiting role in dihydrofolate reductase catalysis. Biochemistry. 2008;47:9227–9233. doi: 10.1021/bi801102e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhabha G, Lee J, Ekiert DC, Gam J, Wilson IA, Dyson HJ, Benkovic SJ, Wright PE. A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science. 2011;332:234–238. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific LLC: San Carlos, CA; 2002. [Google Scholar]