Abstract
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a glycolytic enzyme that also functions in transcriptional regulation, oxidative stress, vesicular trafficking, and apoptosis. Because GAPDH is required for cellular heme insertion into inducible nitric oxide synthase (Chakravarti et al, PNAS 2010, 107(42):18004-9), we extensively characterized the heme binding properties of GAPDH. Substoichiometric amounts of ferric heme bound to GAPDH (1 heme per GAPDH tetramer) to form a low-spin complex with UV-visible maxima at 362, 418 and 537 nm, and when reduced to ferrous gave maxima at 424, 527 and 559 nm. Ferric heme association and dissociation rate constants at 10 °C were kon =17,800 M−1s−1 and koff1 = 7.0 × 10−3 s−1; koff2 = 3.3 × 10−4 s−1 respectively, giving approximate affinities of 19–390 nM. Ferrous heme bound more poorly to GAPDH and dissociated with a koff = 4.2 × 10−3 s−1. Magnetic circular dichroism (MCD), resonance Raman (rR) and EPR spectroscopic data on the ferric, ferrous, and ferrous-CO complexes of GAPDH showed that the heme is bis-ligated with His as the proximal ligand. The distal ligand in ferric complex was not displaced by CN− or N3− but in ferrous complex was displaceable by CO at a rate of 1.75 s−1 (for [CO]>0.2 mM). Studies with heme analogs revealed selectivity toward the coordinating metal and porphyrin ring structure. GAPDH-heme was isolated from bacteria induced to express rabbit GAPDH in the presence of δ-amino levulinic acid. Our finding of heme binding to GAPDH expands the protein’s potential roles. The strength, selectivity, reversibility, and redox sensitivity of heme binding to GAPDH is consistent with it performing heme sensing or heme chaperone-like functions in cells.
Keywords: Heme binding protein, glyceraldehyde-3-phosphate dehydrogenase, spectroscopy, heme transfer, kinetics, affinity
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH, EC 1.2.1.12) catalyzes the conversion of glyceraldehyde-3-phosphate into glycerate-1,3-bisphosphate. Besides its housekeeping role in the glycolytic pathway (4), eukaryotic GAPDHs participate in an array of cellular functions including transcription, oxidative stress, apoptosis and autophagy (5, 6). A great deal of this functional diversity is attained via changes in subcellular localization and/or posttranslational modifications. GAPDHs are widely distributed in Nature, ranging from bacteria and Archaea to animals, including humans (7). Prokaryotic GAPDHs are similarly versatile; some of their functions include binding to mammalian proteins, receptor-mediated acquisition of transferrin-bound iron, ADP ribosylation and prokaryotic-eukaryotic cell-cell communication (8).
Since its original isolation from rabbit muscle in 1964 by W. Ferdinand (9), research on the functional versatility of GAPDH has mounted steadily. A perhaps underappreciated feature of GAPDH concerns its reported yet unexplored interactions with heme (10–12) and the recently proposed role of heme in the regulation of the protein’s expression in red blood cells (13). Our laboratory has recently shown that GAPDH is required for heme transfer into the apo-form of inducible NO synthase and that this process is regulated by the protein’s ability to undergo S-nitrosylation (14).
The superfamily of canonical1 heme proteins includes globins, cytochromes, cytochrome P450 monooxygenases, nitric oxide synthases, heme oxygenases, guanylate cyclases, catalases and peroxidases. These proteins accomplish a wide range of functions, including oxygen transport, electron transfer, gaseous-molecule sensing and heme degradation. The heme binding site in each member of this superfamily is highly conserved across species. The recent years have witnessed the identification of a number of non-canonical heme-binding proteins. These proteins present non-standard heme binding sites (15, 16), usually bind the cofactor in a reversible fashion, and in some cases their function requires the switching of the axial ligands coordinating the heme moiety (17–19). Non-canonical heme binding proteins participate in diverse functions including heme trafficking, intracellular signaling and the regulation of gene expression.
The major experimental setback in identifying the proteins involved in heme metabolism has been the inability to isolate cellular synthesis and degradation of heme from the intracellular trafficking events (20). We proposed that GAPDH may participate in the regulation of heme metabolism by controlling heme insertion into apo-proteins (14). However, the biophysical characteristics of heme binding to GAPDH remain unexplored. Here, we set out to investigate the details of the novel interaction between GAPDH and Fe(III)-protoporphyrin IX. We found that GAPDH binds heme to form a complex with spectral features that are reminiscent of certain b-type cytochromes and other heme-binding proteins featuring bis-His axial ligation. Binding of heme by GAPDH is a reversible and redox-sensitive process, and in general is reminiscent of non-canonical heme-binding proteins involved in heme sensing and transport.
MATERIALS AND METHODS
Reagents
Rabbit muscle GAPDH (rGAPDH) and human erythrocyte GAPDH (hGAPDH) were purchased from Sigma (St. Louis, MO). Hemin chloride was purchased from MP Biomedicals (Solon, OH). CO gas was obtained from Praxair, Inc. (Danbury, CT). EPPS was purchased from Fisher Scientific (Pittsburgh, PA). DTT was purchased from RPI Corp. (Mount Prospect, IL). All other reagents were purchased from Sigma (St. Louis, MO). Apo-myoglobin (expression plasmid kindly provided by Dr. John Olson) was produced in-house according to a published procedure (21).
Protein expression and purification
The wild type rabbit GAPDH gene was synthesized by Genscript™ with codon optimization for optimal expression in Escherichia coli, according to the cDNA sequence reported in the NCBI database (GI:160332366; http://www.uniprot.org/uniprot/P46406). Wild type and mutant rabbit GAPDH (rrGAPDH) coding sequences were subcloned in the pET15b vector using the NdeI and XhoI restriction sites. The resulting rrGAPDH constructs containing a His6 tag attached to their N-termini were overexpressed in E. coli strain BL21 (DE3). All the proteins were expressed and purified according to standard procedures. Briefly, 25 mL of an overnight Luria broth-culture of E. coli carrying the construct of interest was added to 0.5 L of terrific broth supplemented with 125 µg/mL ampicillin. Cells were induced with 1 mM IPTG after the optical density reached 0.8–1.0, followed by additional growth for 48 h at room temperature. When appropriate, TB cultures were supplemented with 0.5 mM δ-amino levulinic acid (heme biosynthesis precursor) at the time of induction in order to isolate heme-bound rabbit, human or S. suis GAPDHs. Purification of wild type and rrGAPDH variants was performed by lysis and sonication followed by Ni-NTA affinity chromatography according to the manufacturer’s instructions (Amersham Biosciences). Digestion with thrombin yielded 85% His6-free rrGAPDH. Purity of the final preparations (90–95% rrGAPDH) was assessed by SDS-PAGE. The UV-visible and kinetic properties (kon and koff for heme) of commercial and recombinant rabbit GAPDHs were identical, hence, commercial rGAPDH was utilized for all studies not requiring in vivo expression and reconstitution with heme.
Human recombinant GAPDH (hrGAPDH) was expressed as a GST-fusion protein according to a published procedure (22). When appropriate, cultures were supplied with 0.5 mM δ-amino levulinic acid (heme biosynthesis precursor) at the time of induction in order to isolate heme-bound hrGAPDH. All the heme-binding studies presented herein were performed with pure hrGAPDH obtained upon digestion of GST-hrGAPDH with thrombin followed by purification on a GST-resin according to the manufacturer’s instructions (Novagen).
Streptococcus suis
GAPDH-6 His was expressed according to a published procedure (23). Purification of the 6 His-tagged protein was carried out using Ni-NTA affinity chromatography as described for rrGAPDH.
Analytical Gel Filtration
The oligomeric state and the stability of heme binding to GAPDH were assessed by analytical gel filtration. Protein samples (~300 µM) in EPPS buffer (40 mM, pH 7.6, 150 mM NaCl) were incubated with 30 µM hemin for 1 h at 4 °C. Samples of apo-GAPDH or GAPDH-heme complexes were injected on a Superdex 200 resin pre-equilibrated with EPPS buffer (40 mM, pH 7.6, 150 mM NaCl).
Activity assays
GAPDH activity was measured by the method of Ferdinand (9). The assay solution contained triethanolamine (40 mM), Na2HPO4 (50 mM) and EDTA 0.2 mM (pH 8.9), 2 mM NAD and 2 mM D-glyceraldehyde 3-phosphate solution. The reaction was started by the addition of ~ 1 µg enzyme. The reaction was followed at 340 nm (formation of NADH, ε340 = 6.22 mM−1cm−1).
Heme staining assay
Heme retention by wild type and GAPDH variants was assessed by in-gel heme staining according to the modified procedure of Guo et al. (24). Apo or heme-loaded GAPDH (~ 30 µg) were examined for their ability to retain heme under non-reducing 10% SDS-PAGE conditions.
Redox titrations
Spectroelectrochemical titrations were carried out in a glove-box (Belle Technology, Dorset, UK) under N2 atmosphere as previously described (25). Briefly, GAPDH-heme samples were made anaerobic by gel filtration in a Sephadex G-25 column (PD 10, GE Healthcare) equilibrated with anaerobic buffer (100 mM phosphate buffer, pH 7.0, 125 mM NaCl). Protein samples were diluted to a 3.5-ml final volume (final concentration ≈ 10 µM) and the following electron mediator dyes (0.5–1 µM) were used: potassium ferricyanide (Em = +450 mV), quinhydrone (Em = +280 mV), dichlorophenolindophenol (Em = +217 mV), phenazine methosulfate (Em = +80 mV), 1,4-naphtoquinone (Em = +60 mV), 2-hydroxy-1,4-napthoquinone (Em = −137 mV) anthraquinone-2-sulfonate (Em = −225 mV), phenosafranine (Em = −252 mV), benzyl viologen (Em = −358 mV) and methyl viologen (Em = −450 mV). The titration was carried out at 15 °C by bolus additions of a potassium ferricyanide (oxidative titrations) or sodium dithionite (reductive titration). Absorption spectra were recorded with a Cary 50 spectrophotometer equipped with a dip-probe detector, and the potentials were measured with an Accumet AB15 pH meter (Fisher Scientific) using a silver/silver chloride microelectrode saturated with 4 M KCl.
UV-visible spectroscopy
UV-visible spectra of GAPDH, hemin (or hemin analogues), and GAPDH-hemin complexes were generated from freshly prepared samples of GAPDH and hemin. Hemin (or hemin analogue) stock solutions (2.5 mM) were prepared by dissolving solid hemin in 0.1M NaOH and 5% dimethylsulfoxide. The later was included to prevent heme dimer formation (17, 26). The concentration of hemin was calculated based on ε385 = 58.4 mM−1cm−1 (27) or by means of the pyridine hemochrome method (28, 29). The concentration of heme-free GAPDH was determined using an absorption coefficient of 30.4 mM−1cm−1 at 280 nm. Typically, ferrous GAPDH-heme complexes were generated by addition of a slight excess of dithionite. Ferrous-CO complexes were generated by direct bubbling of CO into a pre-reduced GAPDH-heme complex.
Ethoxylation of His residues with diethylpyrocarbonate (DEPC)
Two hundred microliters of 100 µM rGAPDH or preformed rGAPDH-heme (100 µM rGAPDH/20µM heme) were incubated with 15 mM DEPC (44 µL of a 68 mM freshly prepared stock solution of DEPC) for 2 °C on ice. The reactions were performed in 40 mM EPPS, pH 7.6 supplemented with 150 mM NaCl and 10% glycerol. Three rounds of buffer exchange were then applied using 0.5 mL microcentrifuge spin columns with a MWCO of 10 kDa. The concentrated samples (~ 100 µL) were diluted to 500 µL with the same buffer (final concentration of protein ~ 40 µM) and the spectra recorded before and after the addition of 8 µM freshly prepared hemin (1.5 µL of a 2.4 mM stock).
Resonance Raman Spectroscopy
Samples (50 µL) of ferric, ferrous or ferrous-GAPDH-CO solutions at 70–150 µM were prepared in gas-tight quartz EPR tubes and disposed in a homemade spinning cell, at room temperature, to avoid local heating and to prevent photodissociation and degradation. Raman excitation at 413.1 nm was obtained with a laser power around 10 mW. Resonance Raman spectra were recorded using a modified single-stage spectrometer (Jobin-Yvon T64000, HORIBA Jobin Yvon S.A.S., Chilly Mazarin, France) equipped with a liquid N2-cooled back-thinned CCD detector. Stray scattered light was rejected using a holographic notch filter (Kaiser Optical Systems, Ann Arbor, MI). Spectra correspond to the average of three different 1-hour accumulations. Neutral density filters were used for the ferrous-CO complexes to decrease laser power (below 5 milliwatts at the sample) and avoid photodissociation and photo-oxidation. Spectral accuracy was estimated to be ±1 cm−1. Spectral resolution was about 3 cm−1. Baseline correction was performed using GRAMS 32 software (Galactic Industries, Salem, NH).
EPR spectroscopy
X-band (9.4 GHz) EPR spectra were recorded on a Bruker ELEXSYS 500 spectrometer equipped with a standard TE cavity (Bruker) and an Oxford Instruments continuous flow liquid helium cryostat interfaced with a temperature control system. Simulations were performed using the Easyspin software package (30).
Magnetic Circular Dichroism
MCD spectra were measured with a magnetic field strength of 1.41 T by using a JASCO J815 spectropolarimeter. This instrument was equipped with a JASCO MCD-1B electromagnet and interfaced with a Silicon Solutions PC through a JASCO IF-815-2 interface unit. Data acquisition and manipulation using Cary or Jasco software has been previously described (31). Apo-rGAPDH was loaded with Fe(III) hemin to form the ferric complex. To ensure homogeneity, ferricyanide was used to fully oxidize the heme center, followed by desalting chromotography. The resulting spectra were compared to data from other proteins and the most accurate match was with that of a bis-His (imidazole) coordinated heme center. Histidine is a common protein backbone heme axial ligand, so the data from Fe(III) rGAPDH was compared to two well-characterized ferric bis-His systems: cytochrome b5 and the bis-imidazole-ligated H93G sperm whale myoglobin (Mb) mutant. Native ferric cytochrome b5 has a six coordinate, low-spin bis-His heme structure and the H93G Mb system has two open axial pockets created by a mutation of the proximal ligand that allows for two exogenous ligands to bind to the heme and, in this case, imidazole (Im) is used to create the same bis-His-type environment. All spectral measurements for rGAPDH-heme were carried out with a 0.2 cm quartz cuvette at 4 °C. Complete reduction of the heme iron was accomplished by adding a few microliters of concentrated sodium dithionite solution (25 mg/mL of H2O) with a microliter syringe. Ferrous–CO adducts were prepared by bubbling CO gas into the ferrous rGAPDH-heme samples. UV−visible absorption spectra were recorded with a Cary 400 spectrophotometer interfaced to a PC before and after the MCD measurements to verify sample integrity.
Binding affinities
Spectrophotometric titrations were carried out by step-wise addition of 0–35 µM hemin to a sample of GAPDH (and to the protein-free buffer in the reference cuvette). Formation of the GAPDH-heme complex was characterized by a red-shift in wavelength from 385 to 415 nm. Binding affinities (Kd) and stoichiometry were determined by plotting ΔA (415–700 nm) against the concentration of added hemin. The data were fit to the following equation:
| (Equation 1) |
, which accounts for the concentration of protein present in the assay (32). [hemin] is the concentration of hemin added in the sample and reference cuvettes, ε415 is the absorption coefficient of the GAPDH-bound hemin (ε415 = 80 mM−1cm−1) and Amax is the absorbance of the maximal binding.
Kinetics of heme binding to GAPDH, kon
Heme binding to rGAPDH was examined by stopped-flow at 10 °C, using a Hi-Tech SF61-DX2 stopped-flow instrument (Hi-Tech Scientific, Salisbury, UK) coupled to a diode array detector. Briefly, 5 µM freshly prepared hemin was mixed with varying concentrations of rGAPDH (10–80 µM) in EPPS buffer (100 mM, pH 7.6) supplemented with 10% glycerol and 150 mM NaCl. Observed rate constants, kobs, were calculated by fitting the data to a single exponential equation.
Kinetics of heme transfer from GAPDH to apo-myoglobin, koff
Ferric heme transfer from rGAPDH-heme complexes (10 µM GAPDH/ 2 µM hemin) to apo-myoglobin (50 µM) was measured by UV-vis spectrophotometry following the formation of holo-myoglobin at 411 nm. GAPDH-heme complexes (molar ratio 5:1) was mixed with 5-fold excess apo-myoglobin and the reaction was allowed to proceed for 2 hours. UV-vis spectra were recorded every 2 min. Transfer of ferrous-heme to apo-myoglobin was performed under the same conditions, except that protein samples and buffers were made anaerobic prior to the experiment. The reactions were carried out in a rubber-septum capped anaerobic cuvette. koff rates were calculated by plotting the absorbance difference at 408 nm (absorption maximum for myoglobin) and 416 nm (absorption maximum for GAPDH-heme) versus time.
Kinetics of CO binding to Fe(II)-GAPDH
Rabbit GAPDH-heme complexes (5:1) were prepared by mixing freshly prepared GAPDH (40 µM) and hemin solutions (10 µM) in EPPS buffer (100 mM, pH 7.6) supplemented with 10% glycerol and 150 mM NaCl. The samples were made anaerobic and ferrous GAPDH-heme complexes (Soret band appearing at 425 nm) were generated by adding a slight excess of dithionite. The samples were transferred to an anaerobic stopped-flow device using a gastight syringe and mixed with anaerobic buffer containing different concentrations of CO. Formation of the Fe(II)-CO-GAPDH complex was followed at 420 nm. Observed rate constants, kobs1 and kobs2, were obtained by fitting the data to a 2-exponential sequential model as the traces displayed a biphasic behavior.
Kinetics of NO binding to Fe(II)-GAPDH
Rabbit GAPDH-heme complexes (5:1) were prepared by mixing freshly prepared GAPDH (final concentration 100 µM) and hemin solutions (final concentration 20 µM) in EPPS buffer (100 mM, pH 7.6) supplemented with 10% glycerol and 150 mM NaCl. The samples were made anaerobic and ferrous GAPDH-heme were generated by adding a slight excess of dithionite. The samples were transferred to an anaerobic stopped-flow device using a gastight syringe and mixed with anaerobic buffer containing different concentrations of NO. Formation of the Fe(II)-NO-GAPDH complex was followed at 424 nm (disappearance of ferrous rGAPDH-heme) and at 385 nm (formation of a Fe(II)-NO rGAPDH complex). Observed rate constants, kobs1 and kobs2, were obtained by fitting the data to a 2-exponential sequential model as the traces displayed a biphasic behavior.
RESULTS
Heme binding to rGAPDH
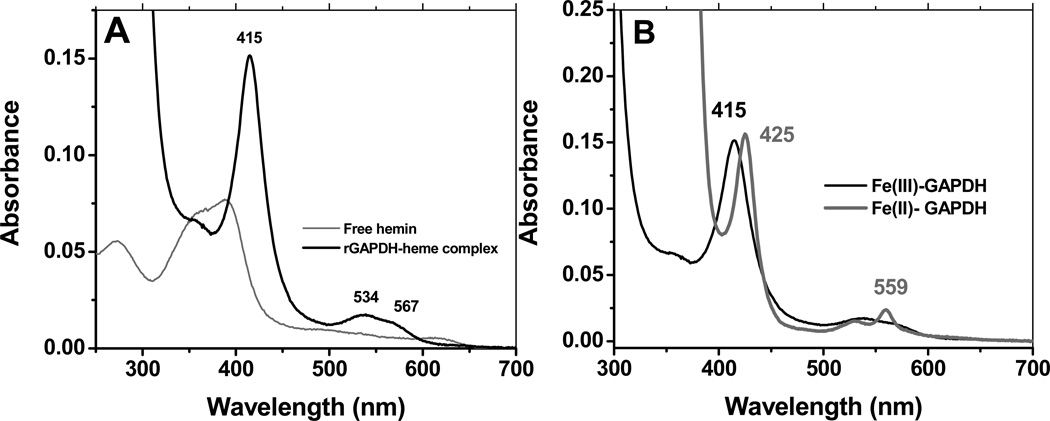
We first examined the interaction of free hemin with rGAPDH by UV-visible spectroscopy. At a ratio of 1 heme to 1 GAPDH tetramer, rGAPDH bound hemin to form a low-spin, six-coordinated complex that is red-shifted with respect to free hemin (Soret band at 385 nm) and possesses a substantially higher molar absorptivity (80 mM−1cm−1 versus 58.4 mM−1cm−1 for free hemin (33)), as determined using the pyridine hemochrome assay (28, 29) (Fig. 1 panel A). The ferric rGAPDH-heme complex presents a Soret band with peak wavelength maximum at 415 nm, indicative of axial coordination to the heme. Reduction of the rGAPDH-heme complex with sodium dithionite generated a species with a Soret band at 425 nm and a sharp α band at 559 nm (Fig 1, panel B), while reaction of rGAPDH-heme with pyridine and dithionite led to formation of the characteristic band at 556 nm (data not shown).
Figure 1.
UV-vis spectrum of rGAPDH-heme complex at pH 7.4. Panel A: UV-vis spectra of free hemin and hemin-bound to rGAPDH. The ferric rGAPDH-heme complex is in a low-spin configuration. Panel B: UV-vis wavelength maxima of ferric and ferrous rGAPDH-heme complexes. Ferrous GAPDH displays absorption maxima at 425, 527 and 559 nm.
Analysis of a Cys-null mutant of rGAPDH (all four Cys replaced by Ser) showed no differences in the spectra compared to wild type rGAPDH, indicating that Cys residues are not directly coordinated to the heme (data not shown). Heme binding affinity (Kd for Cys-null rGAPDH 21 nM) and gel filtration experiments confirmed that the Cys-null mutant does not differ from wild type GAPDH in its ability to bind heme (see sections below). His-Fe-Met coordinated proteins display an additional band to the red of the Q bands (34–36) that is absent in the rGAPDH-heme spectra. Finally, the UV-visible spectra of His-Fe-Tyr proteins usually present a Soret band around 406–411 nm in the ferric form (37, 38), which is significantly blue-shifted with respect to the Soret of ferric rGAPDH-heme (416–420 nm, depending on the pH). These data suggest ferric GAPDH complex maybe bis-His ligated. We are currently performing mutagenesis to replace each His to Ala or Gly in order to identify the residues involved in heme coordination.
Stability of the rGAPDH-heme complex
Gel filtration of pre-formed rGAPDH-heme was done to examine the stability of the complex and the oligomeric state of the protein bound to heme. The results showed that rGAPDH-heme is maintained as a tetramer with a molecular weight of ~ 130 kDa (Fig S1, panel A). UV-visible analysis of the eluted fractions indicates that the properties of post-column rGAPDH-heme are identical to those of the complex prior to gel filtration (Fig S1, panel B). This suggests that binding of heme to rGAPDH does not alter the oligomeric state of the protein, and that rGAPDH-heme is relatively stable under these experimental conditions. We found that rGAPDH-heme can be stored at 4 °C for up to one month while preserving its spectral properties. The heme bound to rGAPDH could also be detected using an in-gel heme staining method following SDS-PAGE of the complex (Fig S2, Supporting Information).
Effect of heme binding on GAPDH secondary structure and dehydrogenase activity
Far UV-circular dichroism was employed to determine whether heme binding to rGAPDH induced major changes on the protein’s secondary structure. The CD spectrum of native rGAPDH agrees well with previously published results (Fig. S3) (39). A comparison of the spectra of apo-rGAPDH and rGAPDH-heme (Fig. S3) indicated that heme binding does not substantially alter the secondary structure of the protein. Moreover, we found that heme binding does not affect the dehydrogenase activity of rGAPDH or hGAPDH (data not shown), suggesting that heme binding does not perturb the enzyme’s active site.
Binding of rGAPDH to porphyrin analogues
We utilized Fe(III)-protoporphyrin IX analogues to investigate the specificity of heme binding in rGAPDH. A comparison of the UV-visible properties of free and rGAPDH-bound heme analogues is provided in Table 1. We found that Fe(III)-mesoheme, Fe(III)-deuteroheme and Co(III)-protoporphyrin IX each bound GAPDH to form a red-shifted (with respect to free ligand) complex as we observed for the naturally occurring heme. In contrast, negligible binding was indicated with Fe(III)-2,4-diacetylprotoporphyrin IX, Zn-protorpohyrin IX and the metal-free protoporphyrin IX. The results suggest: a) a coordinating metal center is required for proper binding of heme to rGAPDH; b) the axial ligand/s display a preference for Fe(III) and Co(III) metal centers versus Zn, and c) The presence of bulky peripheral groups (as in 2,4-diacetylporph) impedes porphyrin binding to rGAPDH, implying the existence of a sterically selective heme binding site.
Table 1.
Binding of rabbit GAPDH to several protoporphyrin IX analogs at pH 7.4 (Tris, 100 mM, 10% glycerol), room temperature.
| Wavelength maxima (nm) | Comments | |
|---|---|---|
| Free porphyrin | ||
| Protoporphyrin IX (Porph IX) | 347, 471, 539, 591, 642 | |
| Fe(III)-protoporph IX | 352, 392, 497, 613 | CT |
| Fe(III)-mesoporph IX | 351, 389, 490, 604 | CT |
| Fe(III)-deuteroporph IX | 342, 386, 490, 602 | CT |
| Fe(III)-2,4-diacetyldeuteroporph IX | 342, 403, 426 (s), 625 | CT |
| Co(III)-protoporph IX | 351, 417, 531, 565 | LS |
| Zn(II)-protoporph IX | 347 (s), 393, 550, 589 | LS |
| GAPDH-porphyrin complex | ||
| Protoporph IX | 347, 471, 539, 591, 642 | No binding |
| Fe(III)-protoporph IX | 366, 412, 536 | LS, red-shift |
| Fe(III)-mesoporph IX | 353, 403, 533 | LS, red-shift |
| Fe(III)-deuteroporph IX | 344, 404, 531 | LS, red-shift |
| Fe(III)-2,4-diacetyldeuteroporph IX | 347, 404, 428, 443 (s) | LS, partial binding, red-shift |
| Co(III)-protoporph IX | 352, 426, 536, 568 | LS, red-shift |
| Zn(II)-protoporph IX | 343 (s), 393, 420, 549, 588 | LS, partial binding, red-shift |
Redox potentiometry
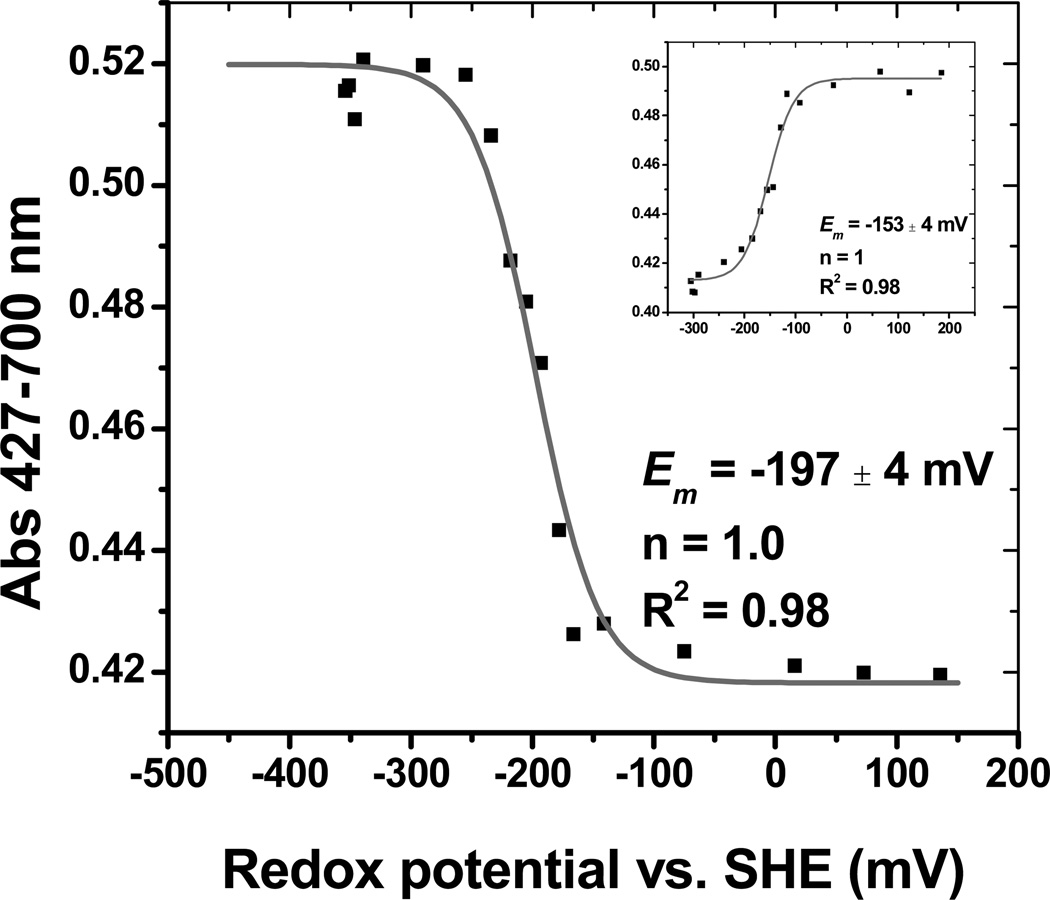
We examined the redox behavior of pre-formed GAPDH-heme. Stepwise oxidation of ferrous GAPDH-heme with potassium ferricyanide led to the ferric complex. The accompanying spectral change could be fit to the Nernst equation (Fig. 2), to yield a calculated midpoint potential of Em = −197 ± 9 mV (one-electron transfer, n=1). Titration of the ferric complex with sodium dithionite displayed marked hysteresis, resulting in deviations of about + 40–60 mV from the midpoint value determined via the oxidative titration (Fig. 2, inset). The reasons for this behavior are currently unknown.
Figure 2.
Redox potentiometry of rGAPDH-heme. Oxidative titration of ferrous-GAPDH-heme followed at 427 nm (disappearance of ferrous to form ferric GAPDH-heme). The GAPDH-heme complex exhibits Nernst behavior, and could be best fit to a one-electron transfer model with a midpoint potential of −197 ± 4 mV versus SHE. The inset shows the reductive titration of ferric-GAPDH-heme. The midpoint potential was −153 ± 4 mV.
Stoichiometry and Binding affinity of rGAPDH for heme
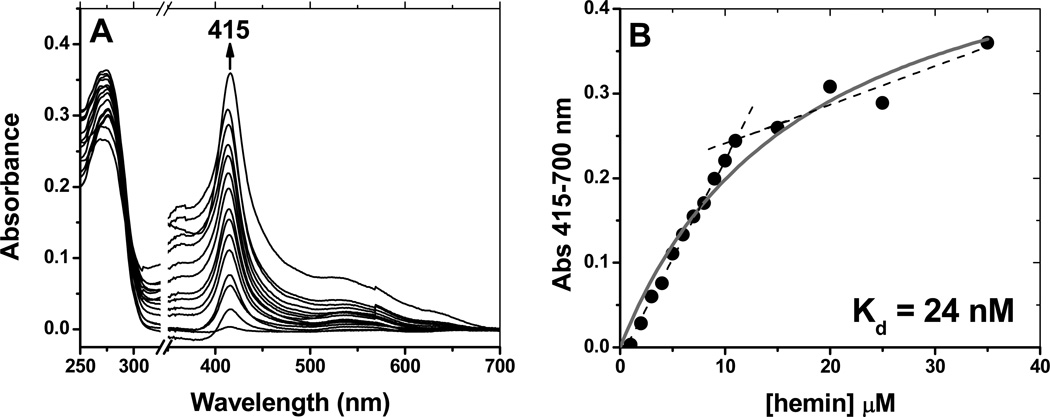
UV-visible spectroscopy showed that rGAPDH binding to heme can involve more than one binding mode. A complex with the aforementioned characteristics forms at high rGAPDH to heme ratios. However, increasing the heme ratio beyond 1 heme per 1 GAPDH tetramer produced a distinct complex with features undistinguishable from those of free hemin in aqueous solution (Fig. S4, Supporting Information). Nonetheless, the heme moiety was retained under these conditions, as evidenced by desalting experiments. Briefly, G-25 gel filtration of the GAPDH-heme complex obtained upon completion of the titration shown in Fig. S4 resulted in negligible heme-loss from GAPDH. This suggests that the free-hemin-like species detected by UV-visible spectroscopy is associated with the protein. Because under physiological conditions the concentration of GAPDH exceeds that of free heme by several orders of magnitude ([free heme]~0.03–0.1 µM (40)), we focused on characterizing the binding parameters of the complex that forms under conditions of [rGAPDH]>[heme]. Direct titration of rGAPDH with increasing concentrations of hemin led to the formation of the ferric-rGAPDH complex, with a wavelength peak at 416 nm (Fig. 3 panel A). At [rGAPDHtetramer]/[hemin] < 1, a second complex formed with wavelength maximum at ~ 400 nm. A fit of the data to the equation given in Methods yielded the line shown in Fig. 3 panel B, and gave a calculated dissociation constant (Kd) of 24 nM. A deflection occurs in the line at a stoichiometric value of 1:1 (1 heme per GAPDHmonomer). Beyond this point, non-specific binding takes place to form a complex with spectroscopic features that do not differ markedly from those of free hemin. We attempted to crystallize rGAPDH-heme without success. This is possibly due to the existence of multiple heme binding modes.
Figure 3.
Binding affinity of rGAPDH for hemin. Spectrophotometric titrations were performed by stepwise addition of hemin (0–35 µM) to a 10 µM solution of rGAPDH in buffer (100 mM Tris, pH 7.4, 10% glycerol). A reference was made by stepwise addition of hemin (0–35 µM) to the same buffer. Panel A: The titration profile (shown as differential spectra, GAPDH-heme minus free hemin) displays a maximum change at 415 nm. The break (covering 325 to 350 nm) is positioned at 20% of the axis length. Panel B: Plot of ΔA at 415 nm versus the concentration of hemin. A deflection point is observed at [hemin] ~ 10 µM, indicating that beyond 1:1 ratios of rGAPDH to hemin a second, less specific binding occurs.
Heme binding kinetics, kon and koff
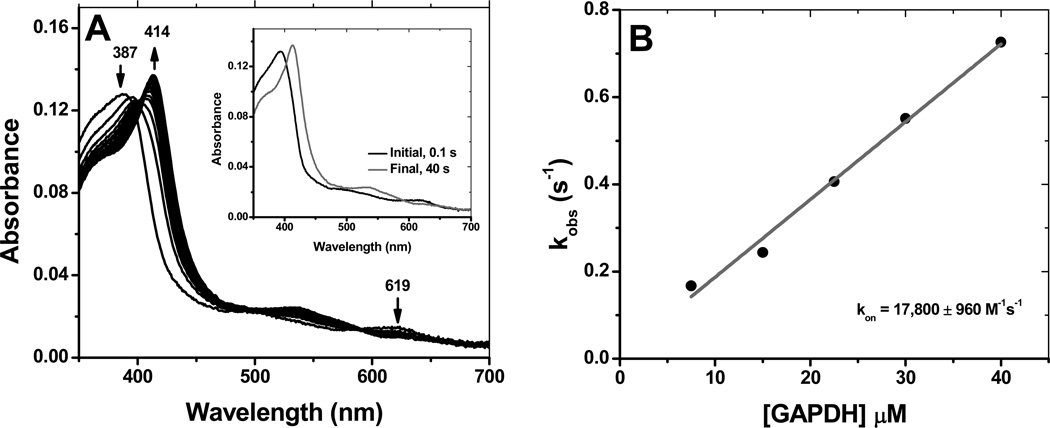
Binding of heme to rGAPDH was studied by stopped-flow spectroscopy. The reaction proceeds as a single step process (Fig 4, panel A) involving two species with isosbestic points occurring at 400 and 590 nm. The spectral data could be fit to a single exponential equation, and the corresponding kobs values were plotted against the concentration of hemin to give a second-order rate constant, kon of 17,800 M−1s−1 at 10 °C (Fig 4, panel B). This value is within the range reported for other non-canonical heme binding proteins (Table 2).
Figure 4.
Kinetics of heme binding to GAPDH. Heme binding to GAPDH was assessed by stopped-flow. The reaction proceeded cleanly to form ferric GAPDH-heme in a single step process (Panel A). The initial and final spectra given in the inset were collected at 0.1 s and 40 s, respectively. Plot of the observed rate constants for different doses of GAPDH yielded a binding constant kon of 17,800 M−1s−1 at 10 °C (Panel B).
Table 2.
Kinetic parameters of heme binding proteins with various functions.
| Protein | kon (M−1s−1) | koff (s−1) | Kd (koff/kon) (M) |
Kd (direct) (M) |
Function | Reference |
|---|---|---|---|---|---|---|
| Wild type rGAPDH | 1.78 × 104 | 3.3 × 10−4 7.0 × 10−3 |
19 × 10−9 39 × 10−8 |
24 × 10−9 | Unknown | This work |
| Nuclear hormone receptor, Rev-erb β (oxidized) | ND | ND | ND | 1.17 × 10−7 | Gene repression; circadian cycle, inflammation | (14) |
| Nuclear hormone receptor, Rev-erb β (reduced) | ND | ND | ND | 23 × 10−9 | Gene repression; circadian cycle, inflammation | (14) |
| Sperm whale Mb | 7.6 × 107 | 8.4 × 10−7 | 1.1 ×10−14 | ND | O2 transport | (56) |
| HRI (heme regulated inhibitor), eIF2α | 1.1 × 107 | 1.5 × 10−3 | 1.4 × 10−10 | ND | Regulation of protein synthesis by iron | (60) |
| BSA | 5.0 × 107 | 1.1 × 10−2 | 2.2 × 10−10 | ND | Fatty acid transport; heme metabolism | (56) |
| Apo-DHR51-LBD (D. melanogaster, homologous of human nuclear receptor Reverb α and β | ND | ND | ND | 4.3 × 10−7 | Nuclear receptor | (81) |
| CcmE | ND | ND | ND | 2 × 10−7 (high affinity) 10 × 10−6 (low affinity) |
Heme chaperone; cytochrome c biogenesis in Gram-negative bacteria | (82) |
| HasAp | 1.6 × 107 | ND | 35 ×10−6 | ND | Heme acquisition from Hb in P. aeruginosa | (34) |
| HtsA | 8 × 107 | ND | 8 ×10−6 | ND | Heme acquisition by S. pyogenes | (83) |
| Shp | 1.6 × 106 | ND | 22 × 10−6 | ND | Cell surface hemin transfer protein in S. pyogenes | (84) |
| HemQ | ND | ND | ND | 30–40 | Heme synthesis by M. tuberculosis | (71) |
| HTP (PhuT) | ND | 3.8 × 10−4 | ND | 0.0012 | Heme acquisition by P. aeruginosa | (21) |
| PhuS | 1.8 × 105 | 3.6 × 10−2 | 0.2 ×10−6 | ND | Heme acquisition by P. aeruginosa | (57) |
| pa-HO | 1.1 × 105 | 6.6 × 10−2 | 0.6 × 10−6 | ND | Heme degradation by P. aeruginosa | (57) |
| Rv0203 | 1.3 × 108 | 8.2 ×10−2 | 6.2 × 10−10 | ND | Heme binding protein M.tuberculosis | (85) |
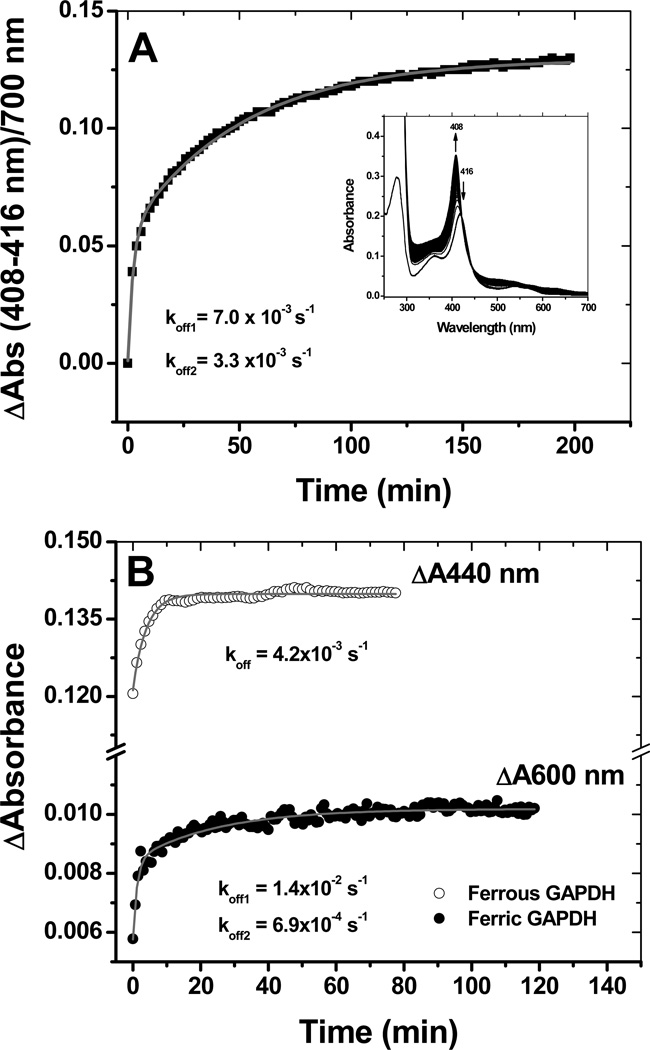
The reversibility of heme binding to rGAPDH was examined using sperm whale apo-myoglobin as described in the literature (41). Heme binding to rGAPDH was a reversible process. The transfer of ferric heme displayed a biphasic behavior (Fig. 5, panel A) with koff rates of 7.0 ×10−3 s−1 and 3.3 ×10−4 s−1 (at 10 °C). These values lie within the range reported for other non-canonical heme binding proteins (Table 2). Transfer of reduced heme from rGAPDH to apomyoglobin (Fig. S5) occurred with a koff = 4.2 × 10−3 s−1 at 25 °C, whereas the transfer of ferric heme at the same temperature yielded koff = of 1.4 ×10−2 s−1 and 6.9 ×10−3 s−1 (Fig. 5 panel B). rGAPDH bound very poorly to pre-reduced heme (Fig. S6, Supporting Information), with only 18% of the total available reduced heme forming a complex with rGAPDH after 10 min. This suggests that the protein has greater affinity toward ferric versus ferrous heme.
Figure 5.
Heme transfer from GAPDH to apomyoglobin. Heme binding to GAPDH is a reversible process. Incubation of ferric GAPDH-heme with a 5-fold excess of apo-myoglobin resulted in the stoichiometric transfer of heme with rates of 7.0 ×10−3 s−1 and 3.3 ×10−4 s−1 at 10 °C (bi-exponential fit) (Panel A). Time courses for ferric and ferrous heme transfer to apomyoglobin at 25 °C (Panel B). Heme transfer from ferrous GAPDH to apo-myoglobin was derived from the absorption changes at 440 nm (maximum absorption for ferrous Mb) and was best fit to a single exponential equation. In panel B, heme transfer from ferric GAPDH to apo-myoglobin was derived from the absorption changes at 600 nm (charge transfer band of holo-Mb) to give koff rates of 1.4 ×10−2 s−1 and 6.9 ×10−3 s−1.
Reaction of ferric and ferrous-rGAPDH-heme with small ligands
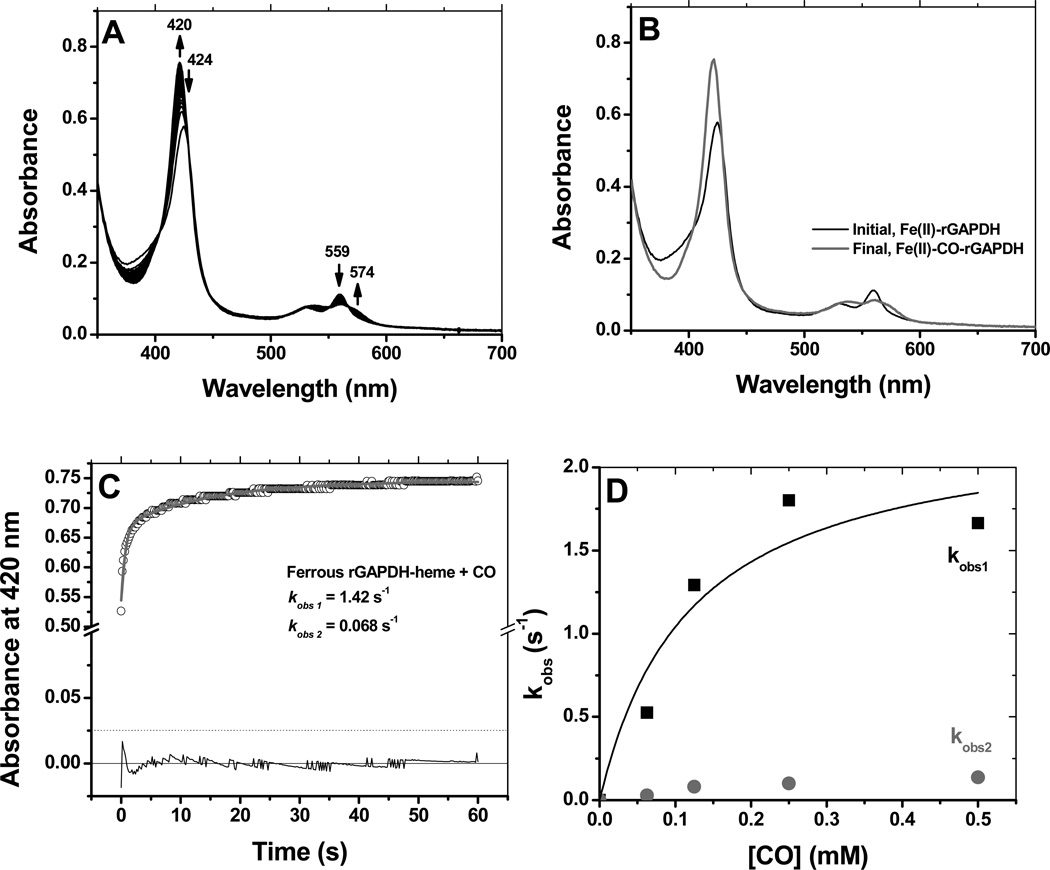
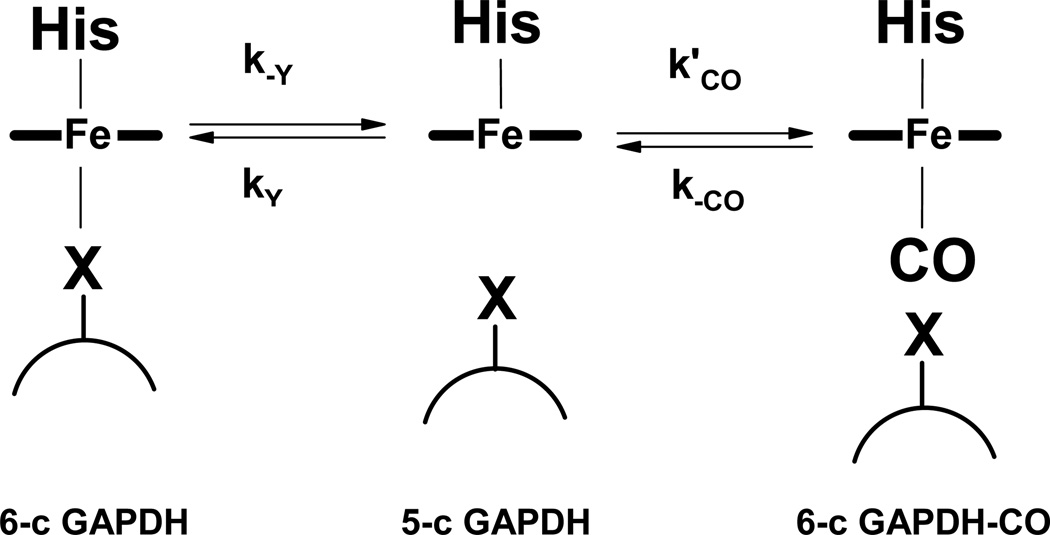
Ferric rGAPDH-heme was probed with the classical small ligand molecules utilized to characterize axial substitution in hemoproteins (42, 43). No spectral shift was observed with 100 mM cyanide, 100 mM thiocyanate or 250 mM imidazole, indicating that these ligands did not bind. Ferrous rGAPDH-heme reacted relatively slowly with CO to form a complex with wavelength maxima appearing at 420, 564 and 574 (Fig 6, panels A and B). The reaction profile displayed a biphasic behavior. Global analysis of the data using a 2-exponential sequential model yielded kobs1 = 1.5 s−1 and kobs2 = 0.068 s−1 for CO-saturated buffer (1 mM final concentration) (Fig. 6, panel C). The successive pseudo-first order constants differed by more than an order of magnitude. The fast phase was [CO]-dependent in a non-linear manner and showed saturation behavior (Fig. 6, panel D). This is the trend expected if association of CO becomes limited by the dissociation of a pre-existing endogenous axial ligand. A general mechanism for this reaction is provided in Scheme 1. Thus, CO binding to ferrous GAPDH can be expressed as:
| (Equation 2) |
where Y is the distal axial ligand that undergoes displacement by CO. The fitting of Equation 2 to the data for the fast phase (kobs1) gives an axial ligand dissociation rate constant k−Y of ~ 2 s−1 and a ligand rebinding rate ratio (curvature) kY/k’CO ~0.1 mM. A similar rebinding rate ratio was reported for the displacement of Cys52 by CO in CBS (106 µM) (44).
Figure 6.
Reaction of ferrous GAPDH with CO. The reaction of ferrous GAPDH with CO was examined by stopped-flow kinetics at 10 °C. Panels A and B: reaction profile and initial and final species detected by global analysis, respectively. Panel C: Time course at 420 nm for the formation of ferrous-CO under conditions of CO-saturation (final concentration 0.5 mM). Panel D: Dependence of kobs (average of two independent experiments) with the dose of CO. The observed rate varied with the dose of CO in a non-linear manner, and displayed saturation behavior.
Scheme 1.
General mechanism for distal ligand displacement by CO in GAPDH-heme.
The slow phase of the CO binding had much smaller amplitude than the fast phase and did not display a clear CO concentration dependence with the concentration of CO. We attribute this second phase to a minor subpopulation of ferrous GAPDH-heme with more stable axial ligand association. An additional, spectroscopically different rGAPDH-heme-CO forms when the reaction is run at [rGAPDHtetramer]/[heme]< than 1:1. This complex presents a Soret band at 410 nm, and its rate of formation was 4-fold faster than that of rGAPDH-heme-CO at 420 nm (Fig. S7, Supporting Information).
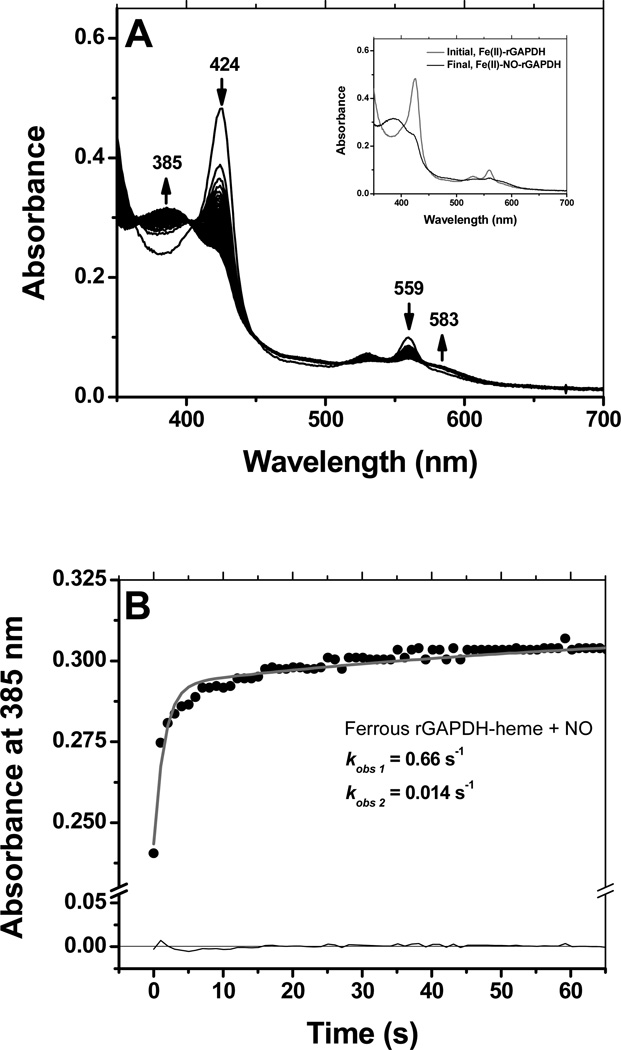
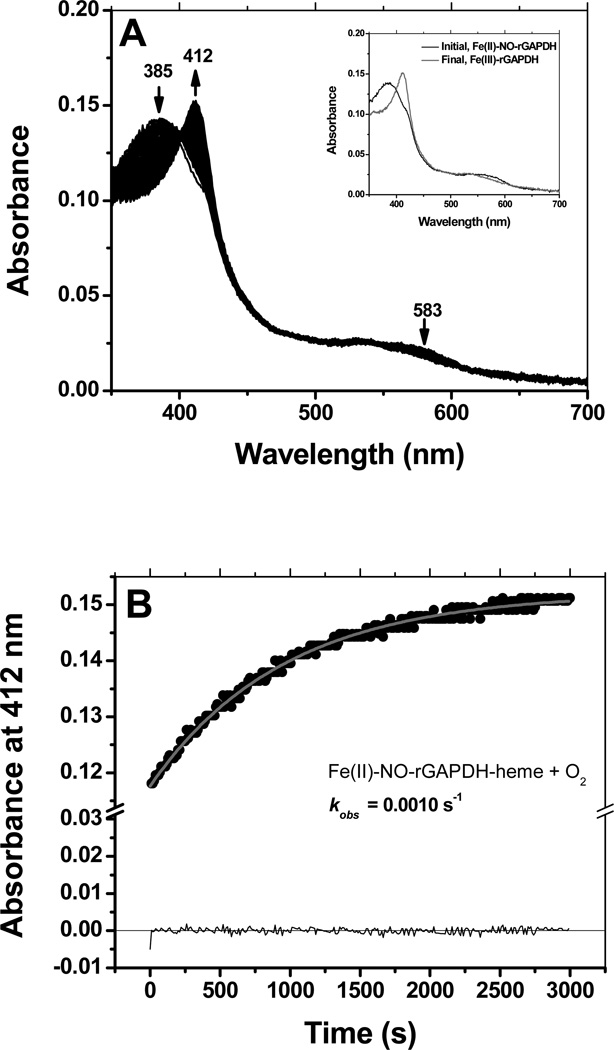
Ferrous rGAPDH-heme also reacted with NO to form a stable, long-lived pentacoordinated complex with a Soret band appearing at 385 nm (Fig. 7, panels A and its inset). Similarly to what was observed for CO, binding of NO could be best fit to a two-exponential model, with kobs values kobs1 =0.66 s−1 and kobs 2 =0.014 s−1 under conditions of NO-saturation (Fig. 7, panel B). Reaction of pre-formed Fe(II)-NO-rGAPDH with dioxygen was slow and led to the formation of ferric GAPDH-heme as the final product (Fig. 8, panel A and its inset). Under conditions of O2 saturation, the observed rate constant for the reaction of Fe(II)-NO rGAPDH with dioxygen to form ferric GAPDH-heme was 0.001 s−1 (Fig. 8, panel B).
Figure 7.
Reaction of ferrous GAPDH with NO. The reaction of ferrous GAPDH with NO was monitored by stopped-flow kinetics at 10 °C. Reaction of ferrous GAPDH with NO led to the formation of a stable, 5-coordinate complex featuring a broad Soret band at 385 nm (Panels A) The insert shows the spectra of the initial and final species. As observed for CO binding, the reaction with NO displayed a biphasic behavior, thus the reaction time course could be best fit to a sequential 2-exponential model (Panel B).
Figure 8.
Reaction of ferrous-GAPDH-NO with dioxygen. The ferrous-GAPDH-NO complex reacted slowly with dioxygen to form ferric GAPDH-heme (Panels A and B). An estimated kobs of 0.0010 s−1 was determined at 10 °C, in oxygen-saturated buffer.
Magnetic Circular Dichroism
UV-visible absorption and magnetic circular dichroism (MCD) spectroscopies were employed to help establish the coordination structure of heme bound to GAPDH as well as the changes in ligation due to heme oxidation/reduction and the addition of small exogenous ligand molecules.
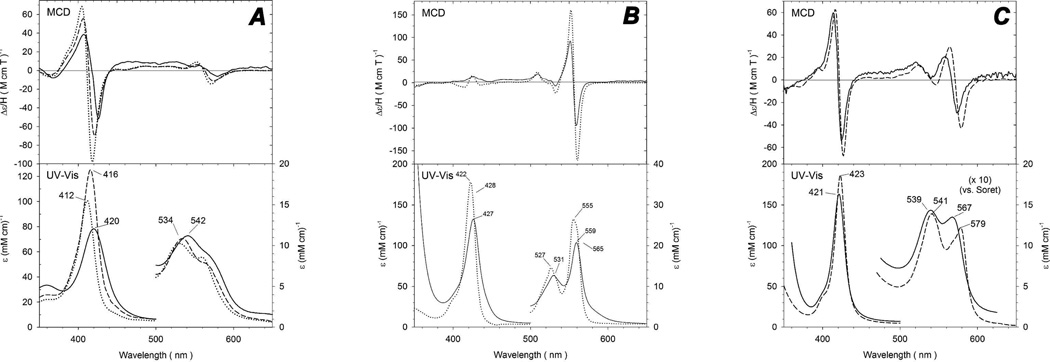
The spectral comparison of ferric rGAPDH to two bis-His examples, cytochrome b5 and H93G (bis-Im) sperm whale Mb, is shown in Figure 9, panel A (1, 2). The spectra of ferric rGAPDH are in good agreement with those of both bis-His examples even though the MCD features in the Soret region are slightly red-shifted (4–6 nm) and are the least intense of the three systems. The MCD bands in the visible region of all three derivatives are also similar. The major UV-Vis absorption peaks of ferric rGAPDH are slightly red-shifted when compared to those of cytochrome b5 and H93G (bis-Im) Mb and the Soret peak intensity is the lowest of the three. Nonetheless, the UV-Vis absorption and MCD spectra of ferric rGADPH are generally similar to those of the two ferric bis-His-ligated complexes. This suggests that ferric rGAPDH has a similar bis-His coordination structure.
Figure 9.
MCD and UV-vis spectra of GAPDH-heme and model systems cytochrome b5 and sperm whale Mb. Panel A: spectra of ferric GAPDH-heme (solid line) at pH 7.8, cytochrome b5 (dotted line) and (Bis-Im) H93G Mb (dashed line). Spectra of ferric cytochrome b5 are replotted from reference (1) and spectra of ferric (Bis-Im) H93G Mb are replotted from reference (2). Panel B: Spectra of ferrous GAPDH-heme (solid line) and cytochrome b5 (dotted line). Spectra of ferric cytochrome b5 are replotted from reference (1). Panel C: Spectra of ferrous-CO complexes of GAPDH (solid line) and H93G-Im Mb (dashed line). Spectra of ferrous-CO of H93G-Im Mb are replotted from reference (3).
In order to study ferrous rGAPDH, the protein was reduced using a minimal amount of sodium dithionite. This reduction was judged to be complete when the UV-Vis absorption spectra had shifted from 420 nm for the Fe(III) heme to 427 nm for the Fe(II) system. The ferrous rGAPDH were once again compared to cytochrome b5. The overlaid spectra are shown in Figure 9, panel B. The UV-vis and MCD spectra of ferrous rGAPDH resemble those of the ferrous cytochrome b5 bis-His system (1). In the visible region, the peak-to-trough MCD intensity of the spectrum of ferrous rGAPDH compares especially well with that of cytochrome b5. The UV-vis absorption spectrum of ferrous rGAPDH is also similar to that of the bis-His-ligated complexes. The Soret absorption peak of ferrous rGAPDH is once again slightly red-shifted compared to that of ferrous cytochrome b5. The UV-vis absorption spectra of the ferrous proteins in the visible region feature sharp, defined alpha and beta bands for each protein. The position of the beta band for rGAPDH at 531 nm is identical to that of cytochrome b5. The more pronounced alpha bands of the two systems are very similar in intensity as well as in relative position with the alpha band of ferrous rGAPDH (559 nm) located slightly to the red of that of ferrous cytochrome b5 (555 nm). The close spectral similarities between ferrous cytochrome b5 and rGAPDH again favor bis-His ligation in the latter.
Ferrous-CO rGADPH was also studied with UV-Vis and MCD spectroscopy. The spectra of Fe(II)-CO rGADPH and of sperm whale Mb are shown in Figure 9, panel C. The spectra of ferrous-CO rGAPDH have very similar characteristics to the His-Fe-CO present in H93G Mb (3). When analyzing the UV-Vis data, the spectra of ferrous-CO rGAPDH are blue shifted when compared with those of the ferrous-CO complexes of native Mb and the Mb H93G mutant (data not shown), however, the visible region shows that the rGAPDH has fully converted to a Fe(II)-CO species due to the change in relative intensities for the alpha and beta bands. The most noticeable spectral difference for the ferrous-CO complex is seen with the rGAPDH alpha band, which is blue-shifted by 12 nm when compared to that of native Fe(II)-CO Mb. Nevertheless, the relative intensity and shape of the alpha bands are comparable throughout, consistent with His-Fe-CO ligation.
Resonance Raman analysis of ferric, ferrous, and ferrous-CO rGAPDH-heme complexes
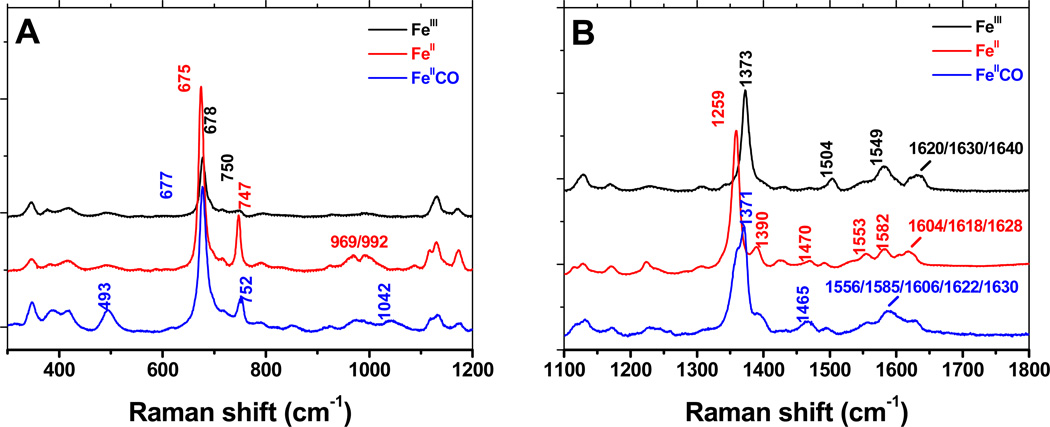
We utilized resonance Raman to further examine the electronic environment of the heme bound to rGAPDH. Figure 10 shows the rR spectra of ferric, ferrous and ferrous-CO rGAPDH-heme in the low and high frequency regions (panels A and B, respectively).
Figure 10.
Resonance Raman spectra of ferric, ferrous and ferrous-GAPDH-CO complexes. High frequency region of the spectrum of the GAPDH-heme complex was obtained with laser excitation at 406 nm. The observed bands are consistent with a low-spin, 6-coordinate complex, with at least one His as the axial ligand.
The ferric GAPDH complex is clearly a low-spin 6-coordinated (LS-6c) species. This is particularly indicated by the frequency of the ν3 band at 1504 cm−1. The frequencies of the ν2, ν3, ν4 and ν10 modes are compatible with bis-His coordinated heme. The bands at 1630 and 1640 cm−1 are reminiscent of bis-His globins (45–47) and other bis-His complexes (48, 49), respectively; however, this assignment is somewhat speculative due to the superimposition of several modes (ν10 and vinyl stretching modes) of possibly different species.
The oxidation state, π* electron density band v4, occurs at 1373 cm−1 in ferric rGAPDH-heme and at 1359 cm−1 in reduced rGAPDH-heme, confirming the ferric state of the native GAPDH-heme complex and its complete reduction to ferrous upon addition of dithionite. The value of the ν4 frequency is a good indication that the proximal ligand is a histidine residue. Additionally, the presence of different v3 bands at 1470 cm−1 (high-spin, five-coordinate, HS-5c) and 1492 cm−1 (low-spin, six-coordinated, LS-6c) indicate the presence of multiple species, which might arise from photolysis, thermal equilibrium or heterogeneity of heme-protein complexes. The nature of this equilibrium is currently under investigation. A summary of all the rR stretching values is provided in Table 3. These spectral features are consistent with previously characterized bis-His coordinated hemes (50, 51).
Table 3.
Resonance Raman shifts for the ferric, ferrous and ferrous-CO complexes of the rGAPDH-heme complex
| Low frequency region of the spectrum of rGAPDH-heme complex | ||||||||||||||||||||||
| FeIII | WT | 346 | 382 | 419 | 494 | 546/559/587 | 678 | 692/715 | 750 | 1055 | 1121 | 1132 | 1171 | |||||||||
| FeII | WT | 346 | 381 | 417 | 493 | - | 675 | 697/717 | 747 | - | 1117 | 1130 | 1174 | |||||||||
| FeIICO | WT | 347 | 385 | 418 | 493 | - | 677 | 698/719 | 752 | 1042 | 1117 | 1133 | 1172 | |||||||||
| High frequency region of the spectrum of rGAPDH-heme complex | ||||||||||||||||||||||
| FeIII | WT | 1373 | 1397 | 1469 | 1489 | 1504 | 1549 | 1582 | 1600 | 1620 | 1630 | 1639 | ||||||||||
| FeII | WT | 1359 | 1390 | 1470 | 1491 | - | 1553 | 1582 | 1604 | 1618 | 1628 | - | ||||||||||
| FeIICO | WT | 1371 | 1393 | 1465 | 1496 | - | 1556 | 1585 | 1606 | 1622 | 1630 | - | ||||||||||
We recorded the resonance Raman spectrum of the FeII-CO complexes for wild-type rGAPDH and a series of mutants. All spectra of the Fe-CO complex seem also to correspond to a mixture of various FeII and FeII-CO species (ν4 at 1359/1372 cm−1), indicating photo-dissociation of CO upon irradiation. Further evidence of weak distal axial ligation was provided by the concomitant presence of HS-5c FeII and LS-6c FeII-CO complexes (ν3 at 1465/1496 cm−1) under these conditions. The observed frequencies match the ones reported for 6c-LS (FeII-CO) bis-His of neuroglobin (1374/1497 cm−1)(45). The analysis of the frequencies of the Fe-CO and C-O stretching modes gives an insight on the proximal ligation. Indeed carbon monoxide is a σ-donor and a π-acceptor ligand when bound to heme. The back donation from the iron d π* orbital to the empty anti-bonding π* CO orbital results in a strengthening of the Fe-CO bond and a weakening of the C-O bond (52–55). Thus, changes in the electrostatic and polar properties of the heme distal pocket will modify this back donation and lead to an inverse linear correlation between the νFe-CO and νC-O frequencies. However, changes in the electron donating properties of the proximal ligand induce a σ-competition between the sigma donor orbitals of CO and of the proximal ligand for the iron orbital, leading to a modification of the νFe-CO frequency and a shift of the correlation curve. As a result, heme-CO complexes of proteins with similar proximal ligands align on the same νFe-CO/νC-O correlation curve (52–54). The frequency of the ν(Fe-CO) mode is observed at 493 cm−1 for wild-type and all mutants, while the v(C-O) stretch appears at 1965-7 cm−1 for most of the samples and is poorly enhanced for the other ones (including the wild-type, data not shown), which is reminiscent of Fe-CO complexes described for other His-ligated proteins (50, 51). Plotting these values on the ν(C-O)/ν(Fe-CO) correlation graph (54) shows that the GAPDH-heme falls right on the correlation line for His-ligated heme (52–54).
However, bis-His ligated proteins often display (at least) two different Fe-CO conformations (46) characterized by two different ν(Fe-C) frequencies around 493 and 523 cm−1. Removal of the distal His by mutagenesis was shown to lead to the disappearance of the 523 cm−1 band, which suggested that this conformer is characterized by an H-bond (or electrostatic) interaction between the CO ligand and the distal histidine that remains in the heme pocket (46). In our case, the presence of a single band at 493 cm−1 (as observed in GAPDH-heme) supports a His-free distal environment that could be attributed either to the displacement of the distal His (far) away from the heme or to a weak distal ligand other than histidine.
Electronic Paramagnetic Resonance of the ferric rGAPDH-heme complex
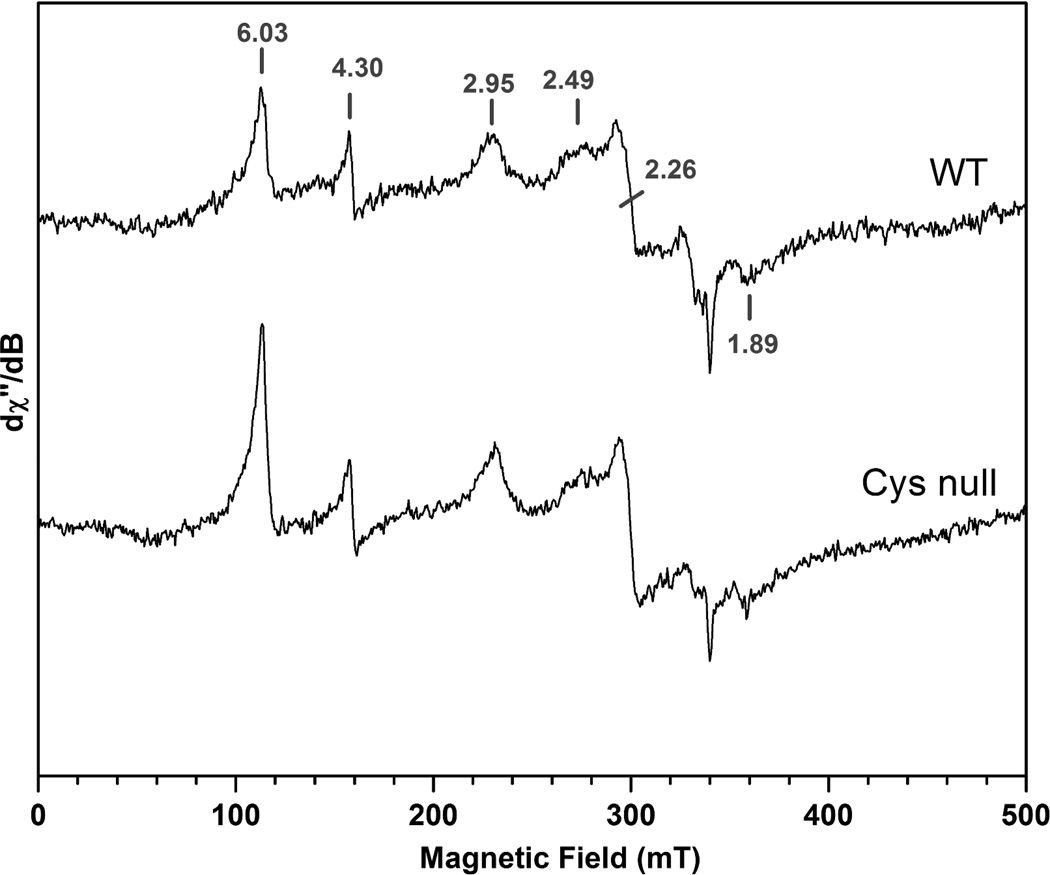
To further examine the electronic environment of the heme bound to rGAPDH, we collected X-band EPR measurements on the ferric heme complexes of rGAPDH and the Cys-null mutant. In the high-field region, the spectrum of the wild type complex presents resonances at g values of 2.95, 2.49, 2.26 and 1.89, consistent with two low-spin, six-coordinate binding modes with principal values (2.95, 2.26, 1.89) and tentatively (2.49, 2.26, 1.89) (Fig. 11). The signature g value of the first species with gmax at 2.95 is consistent with bis-His ligation, whereas the resonance at 2.49 could be tentatively assigned to a His-Fe-Cys binding mode. However, the EPR spectrum of the Cys-null mutant did not differ markedly from that of wild type rGAPDH (Fig. 11), indicating that Cys is not involved in the ligation of heme in either complex. In addition, signals in the low field region with features at g=4.30 (non-specific iron) and g=6.03 (high-spin ferric heme, likely from uncoordinated hemin) are also observed by EPR.
Figure 11.
X-Band EPR spectra of ferric heme-wild type and -Cys-null rGAPDH. The data suggest that two heme subpopulations are present under these conditions, one of which is consistent with bis-His axial ligation (g = 2.95, 2.26 and 1.89). The nature of the species displaying a g value of 2.49 is currently unknown. The spectra were recorded at 10 K using 2 mT field modulation amplitude and 0.5 mW microwave power; microwave frequency: 9.4956 GHz.
Chemical characterization
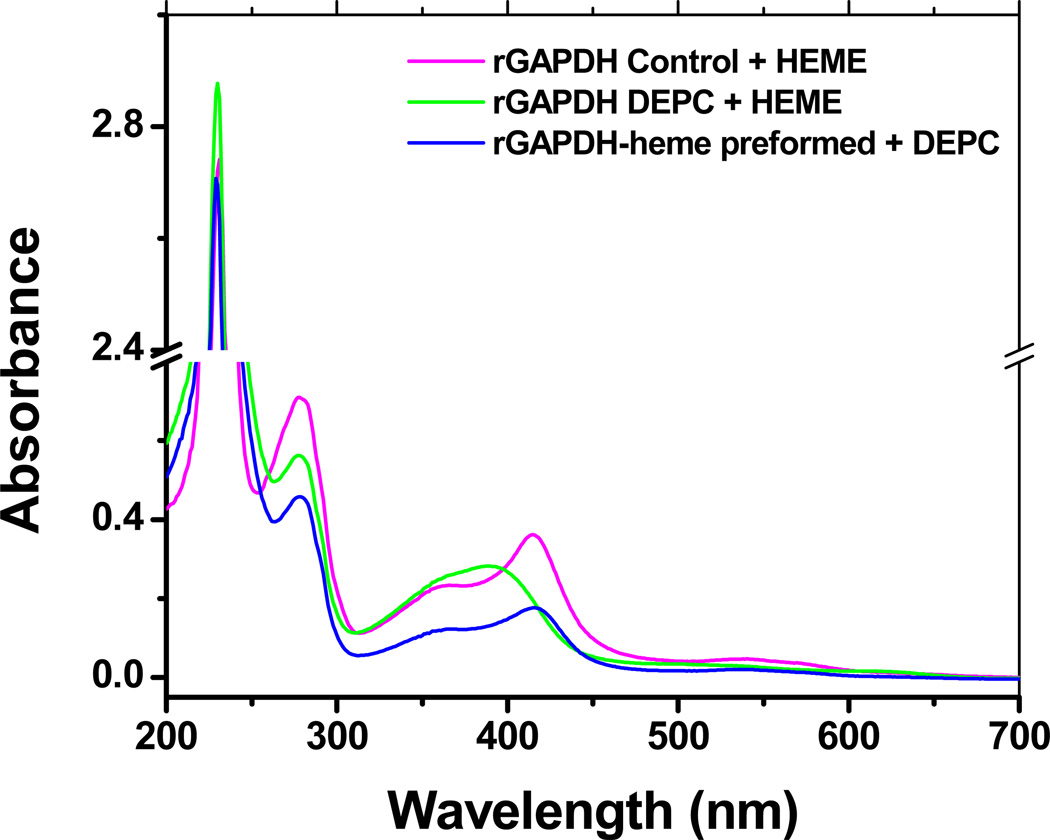
Treatment of rGAPDH with DEPC (an agent that modifies His residues by ethoxylation) hampered heme binding completely, whereas the addition of DEPC to preformed rGAPDH-heme had no effect on its spectral properties (Fig. 12). These results suggest that heme coordination by rGAPDH is accomplished via one or two His residues, and that once the complex forms, at least one axial His is no longer modifiable by DEPC.
Figure 12.
Modification of His residues with DEPC. Untreated (control), heme-free, and pre-formed GAPDH-heme complex were treated with 15 mM DEPC for 2 h on ice. The samples were buffer-exchanged to remove excess DEPC and tested for their ability to bind hemin. DEPC-treated GAPDH was unable to bind heme. Treatment of preformed GAPDH-heme with DEPC does not alter the UV-vis spectral properties of the complex.
Isolation of heme-bound rGAPDH and heme binding to GAPDHs from other species
Because rGAPDH is not a canonical heme binding protein (it does not possess a defined heme binding site) we set out to examine: a) whether rGAPDH-heme could be isolated in vivo and b) whether heme binding is common among GAPDH proteins from different species. E. coli overexpressing rabbit, human or Streptococcus suis GAPDHs were grown in the presence or absence of the heme precursor δ-amino levulinic acid (δ-ALA). The proteins were purified and examined for heme content by UV-visible spectroscopy. Figure S8 (Supporting Information) shows the UV-visible spectra of human (panel A) or Streptococcus suis (panel B) GAPDHs from cultures grown in the absence or presence of δ-ALA. We found that under conditions where heme synthesis is stimulated a measurable fraction of the intracellular GAPDH appears to be heme-bound (2–8 %).
DISCUSSION
Reports that GAPDH participates in hemeprotein maturation (14) and binds to hemin-agarose affinity resins (10, 11) prompted us to investigate the GAPDH-heme interaction in detail. We found that GAPDH from three different species (H. sapiens, O. cuniculus and S. suis; amino acid sequence alignment is provided in Fig. S9) all contained some bound heme when they were expressed and purified from bacterial cells, and could also bind sub-stoichiometric amounts of heme provided to their purified apo-forms, to generate a low-spin six-coordinate complex with spectral features very similar to hemoproteins featuring bis-His axial coordination (36, 56, 57). For example, the UV-vis and MCD spectra of rGAPDH-heme matched the spectra properties of two well-known bis-His coordinated systems (cyt b5 and sperm whale Mb (1–3)) regarding the position and intensity of the Soret and α/β bands for the ferric, ferrous, and ferrous-CO states.
Similarly, the resonance Raman fingerprints of the FeIII, FeII and FeIICO species of the rGAPDH-heme complex, along with the νFe-CO/νC-O correlation, strongly support a His-ligation of the heme with some characteristics reminiscent of bis-His complex. The spectral properties of a Cys-null GAPDH mutant ruled out alternative Cys axial coordination to the heme, and chemically modifying the His residues of GAPDH prevented heme from binding. The selectivity that GAPDH displayed toward the porphyrin metal center (good binding with Fe or Co but not with Zn) was also consistent with bis-His axial ligation. Finally, the midpoint potential of the GAPDH-heme complex (−197 mV) is comparable to other bis-His coordinated proteins (43, 58, 59), which generally have lower midpoint potentials than hemoproteins containing other axial ligands, such as members of the cytochrome c family, cellobiose dehydrogenase, and Mb with H2O/His as axial ligands (34, 58, 60, 61).
Interestingly, X-band EPR results suggested the presence of two different heme-bound rGAPDH populations; one that was consistent with bis-His coordination with g values 2.95, 2.26 and 1.89, and a second population of unknown nature with a prominent gmax value at 2.49. Some uncoordinated hemin is also observed. A similar X-band EPR pattern was reported for two other proteins, namely the bacterial iron response regulator (Irr) (62) and the nuclear hormone receptor Rev-erbβ (17). In these cases, the existence of two distinct heme-bound populations was attributed to heterogeneity of the heme binding modes and redox-dependent switching of the axial ligands, respectively. According to the data gathered by other spectroscopic techniques used in this study the two GAPDH-heme populations observed by EPR are probably due to heterogeneity in the conformation or location of the heme binding sites.
The ferric rGAPDH-heme complex was stable over several days at 4 °C and only partly dissociated during gel filtration or while running the GAPDH-heme complex on non-reducing SDS-PAGE gels (24, 63). Our titration and kinetic studies confirmed that rGAPDH binds heme avidly, with a Kd of 24 nM and a kon of 1.75 × 104 M−1s−1. Based on the Kd value and the estimated intracellular levels of free hemin (0.03–0.1 µM (40)), and GAPDH (1–100 µM (64)) a fraction of GAPDH could indeed exist in heme-bound form, which we confirmed in our bacterial expression studies. Despite the stability of the ferric rGAPDH-heme complex, its heme moiety could be transferred to apomyoglobin with koff1 = 7.0 ×10−3 s−1 and koff2=3.3 ×10−4 s−1 at 10 °C. Heme dissociation from GAPDH was slower than that reported for BSA-heme (10−2 s−1, (65)) or for the cytoplasmic heme binding protein PhuS (3.6 ×10−2 s−1, (66)) but faster than the heme loss rates of most globins (10−4–10−6 s−1, (41, 67)) and nitrobindin (3.6 ×10−5 s−1, (68)) (Table 2). The ability of rGAPDH to bind heme in a reversible manner with intermediate affinity makes it plausible to act as a heme sensor or transient heme carrier in vivo. Indeed, similar koff rates for hemin were reported for eIF2α kinase (1.5 ×10−3 s−1), which exploits heme-sensing to control protein synthesis (69).
Treatment of apo-rGAPDH with DEPC, an agent that specifically blocks His residues (and to a much lesser extent Tyr and Lys residues) (70–73) abolished the protein’s ability to bind heme. However, DEPC treatment of a preformed rGAPDH-heme complex did not alter the spectral properties of the complex, suggesting the axial His residue(s) was unable to react. This is consistent with cyanide, azide, or imidazole being unable to bind to ferric rGAPDH-heme, and suggests a relatively stable bis-axial coordination to ferric heme. Our CO binding studies suggest that the bis-axial coordination may be weakened in the ferrous state. Still, CO binding was relatively slow and appeared to require displacement of a coordinating ligand. A similar behavior with CO has been reported for HtsA, a His/Met coordinated-hemeprotein (74), cystathionine-b-synthase (44) and CooA (75). Overall, our data are consistent with a relatively stable ferric heme bis-coordinated complex, that when reduced allows one of the axial ligands to become displaced by CO according to the mechanism depicted in Scheme 1. The ferrous GAPDH-heme species also bound NO to form a complex whose spectral features and slow reaction with O2 (0.001 s−1) identified it to be a 5-coordinate ferrous heme-NO complex (76). NO binding was slow and biphasic like we observed for CO, consistent with NO binding also requiring displacement of an axial ligand. Indeed, the upper rates of CO and NO binding likely approximate the dissociation rate of an axial ligand in the ferrous GAPDH-heme complex. We observed that the reaction of dioxygen with preformed GAPDH-heme nitrosyl resulted in the enzyme returning to its characteristic ferric-heme bound state, without the formation of any detectable intermediates (His-Fe-NO, His-Fe-O2, etc). Thus, oxidation of GAPDH-heme nitrosyl seems to occur via a mechanism in which re-ligation by the axial residues does not generate detectable intermediates.
Interestingly, pre-formed ferrous heme bound very poorly to GAPDH. In addition, transfer of ferrous heme from GAPDH to apomyoglobin was slightly faster than for ferric heme. Hargrove et al. showed that the major factors contributing to the rate of heme loss from a protein are solvent exposure and the number and types of protein-heme contacts (41, 67). If heme reduction weakens the bis-axial ligation, this could help explain the faster koff of the ferrous heme from GAPDH. Because the Em value for GAPDH-heme is within the reach of most cellular reductants (77), a redox-based change in heme affinity, mediated through the bis-axial coordination, could potentially facilitate heme release from GAPDH in the intracellular milieu.
When heme to GAPDH ratios exceeded 1 heme per tetramer, an additional complex formed whose properties resembled a penta-coordinate species. A similar phenomenon has been reported for prostaglandin G/H synthase (78), histidine rich protein 2 (79), the bacterial iron response regulator (Irr) (62) and the heme synthesis protein HemQ (80). Although this type of GAPDH-heme complex is unlikely to exist under physiological conditions where the concentration of GAPDH exceeds that of free heme, it was still of interest to examine its properties. The penta-coordinated species formed a Fe(II)-CO complex whose Soret absorption maximum was blue-shifted (410 nm versus 420 nm for the six-coordinate, low spin rGAPDH-heme that forms at heme to GAPDH ratios ≤ 1: 4), and whose formation rate was about 4 times faster (kobs 5-coord, 410 nm = 4.3 s−1 at saturating [CO] versus kobs, 6-coord, 420 nm = 1.2 s−1 at saturating [CO]), all consistent with there being a weaker axial ligand in place of His for the bound heme in this case.
A thorough examination of the heme binding capacity of various proteins not reported to bind heme showed that they associated with the cofactor with similar strength as described for Per2 and nPas2, two circadian rhythm proteins proposed to utilize heme to attain their regulatory functions (81). Extensive mutagenesis of the PAS domain thought to be involved in heme binding did not abolish the protein’s ability to bind heme. This led Airola et al to conclude that the interaction between heme and Per2 is likely nonspecific, mainly occurring via hydrophobic interactions and exposed ligands on the surface of the protein (81). According to Hayasaka et al, (71) the fact that extensive mutagenesis failed to eliminate the heme binding ability of Per2 and nPas2 is not surprising as the structures of heme responsive heme-sensor proteins exhibit an enhanced flexibility, which allows for facile ligand switching in the immediate neighborhood of the heme (71). Further, for proteins whose function requires relay of the heme onto other targets or quick binding reversibility, the existence of a heme binding site near the surface is a valuable feature rather than a sign of non-specificity as suggested by Airola et al (81).
The function of heme-bound rGAPDH is currently unknown. One major factor affecting heme binding is its nonspecific partitioning into hydrophobic regions of the protein (82), which also dictates the tendency of heme to adsorb to a variety of proteins (65, 83–87). In contrast, specific binding of heme is governed by bond formation with side chains of amino acids positioned near the porphyrin ring (88, 89). However, the presence of a canonical, well-defined heme binding site does not guarantee stable or constitutive heme binding to a protein. For example, HemQ, an essential protein for the biosynthesis of heme in Gram-positive bacteria possesses a canonical heme binding site similar to that present in chlorite dismutase (80), yet the recombinant protein is isolated with only small and variable amounts of heme. Reconstitution of the holo-protein requires the addition of exogenous hemin during the workup procedure (80). The authors reported poor heme binding and noted that HemQ is either capable of binding more than one heme per subunit or that the multimeric protein does not have uniform heme binding affinity (80). Recently, Hamza et al. made the distinction that a dedicated heme binding protein can be defined as such only if its binding is specific for heme, i.e. if the protein does not bind to metal-free protoporphyrin IX (90).
Based on our results, it is reasonable to propose that the specificity and reversibility of heme binding to GAPDH may serve at least three possible functions: a) it may exploit binding specificity to perform heme sensing functions, b) it may prevent free heme from binding to other proteins while in transit to its destination, and c) it may act as a transient trap to diminish the intracellular reactivity and potential toxicity of free heme. Indeed, the relative stability of the ferric GAPDH-heme complex and its poor reactivity toward small ligands may have evolved to limit the reactivity of the free heme pool during intracellular trafficking events. GAPDH may thus operate as a chaperone to control the serendipitous reactivity and toxicity of heme prior to its downstream delivery. On the other hand, the relative instability of ferrous GAPDH-heme complex and its capacity to bind NO may function as a signal to trigger downstream processes. Along these lines, a previous report by our laboratory indicated that the proposed interaction of GAPDH with heme in cultured cells was weakened by treatment with a NO donor, and this was associated with an inhibition of heme insertion into apo-NOS (14). We found evidence that this process was mediated by S-nitrosylation of GAPDH (14). In lieu of our current findings, we can also consider that NO might influence GAPDH behavior during cellular heme insertion by coordinating to the GAPDH-heme. Our study provides a platform to test this possibility.
Supplementary Material
ACKNOWLEDGMENT
We would like to thank Rajesh Vempati for helping with heme transfer experiments and the preparation of apomyoglobin.
Funding Sources
This work was supported by funding from the National Institutes of Health (HL76491, GM51491 and CA53914 to D.J.S.) and the American Heart Association (11POST650034 to L.H.). Work completed in J.H.D. Laboratory was also supported by the NIH (GM26730).
ABBREVIATIONS
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- EPR
electronic paramagnetic resonance
- MCD
magnetic circular dichroism
- DTT
dithiothreitol
- IPTG
isopropyl thiogalactoside
- LS
low-spin
- HS
high-spin
- 6c
six-coordinate
- 5c
five-coordinate
Footnotes
The term “canonical” refers to proteins with a defined heme binding site which is highly conserved across species.
ASSOCIATED CONTENT
Supporting Information. Pyridine hemochrome UV-visible spectra, in-gel heme staining, heme spectrophotometric titrations, reaction of GAPDH with ferrous heme, reaction of ferrous-GAPDH with CO and UV-visible spectra of heme-bound native human and S. suis GAPDH. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Reference
- 1.Nobeli I, Favia AD, Thornton JM. Protein promiscuity and its implications for biotechnology. Nat Biotechnol. 2009;27:157–167. doi: 10.1038/nbt1519. [DOI] [PubMed] [Google Scholar]
- 2.Pond AE, Roach MP, Thomas MR, Boxer SG, Dawson JH. The H93G myoglobin cavity mutant as a versatile template for modeling heme proteins: ferrous, ferric, and ferryl mixed-ligand complexes with imidazole in the cavity. Inorg Chem. 2000;39:6061–6066. doi: 10.1021/ic0007198. [DOI] [PubMed] [Google Scholar]
- 3.Perera R, Sono M, Sigman JA, Pfister TD, Lu Y, Dawson JH. Neutral thiol as a proximal ligand to ferrous heme iron: implications for heme proteins that lose cysteine thiolate ligation on reduction. Proc Natl Acad Sci U S A. 2003;100:3641–3646. doi: 10.1073/pnas.0737142100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barber RD, Harmer DW, Coleman RA, Clark BJ. GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol Genomics. 2005;21:389–395. doi: 10.1152/physiolgenomics.00025.2005. [DOI] [PubMed] [Google Scholar]
- 5.Sirover MA. On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochim Biophys Acta. 2011;1810:741–751. doi: 10.1016/j.bbagen.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 6.Sirover MA. Mini review. Emerging new functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. Life Sci. 1996;58:2271–2277. doi: 10.1016/0024-3205(96)00123-3. [DOI] [PubMed] [Google Scholar]
- 7.Ronimus RS, Morgan HW. Distribution and phylogenies of enzymes of the Embden-Meyerhof-Parnas pathway from archaea and hyperthermophilic bacteria support a gluconeogenic origin of metabolism. Archaea. 2003;1:199–221. doi: 10.1155/2003/162593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Modun B, Morrissey J, Williams P. The staphylococcal transferrin receptor: a glycolytic enzyme with novel functions. Trends Microbiol. 2000;8:231–237. doi: 10.1016/s0966-842x(00)01728-5. [DOI] [PubMed] [Google Scholar]
- 9.Ferdinand W. The Isolation and Specific Activity of Rabbit-Muscle Glyceraldehyde Phosphate Dehydrogenase. Biochem J. 1964;92:578–585. doi: 10.1042/bj0920578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campanale N, Nickel C, Daubenberger CA, Wehlan DA, Gorman JJ, Klonis N, Becker K, Tilley L. Identification and characterization of heme-interacting proteins in the malaria parasite, Plasmodium falciparum. J Biol Chem. 2003;278:27354–27361. doi: 10.1074/jbc.M303634200. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Wang X, Zhao K, Zhou Z, Zhao C, Yan R, Lin L, Lei T, Yin J, Wang R, Sun Z, Xu Z, Bao J, Zhang X, Feng X, Liu S. A novel approach for identifying the heme-binding proteins from mouse tissues. Genomics Proteomics Bioinformatics. 2003;1:78–86. doi: 10.1016/S1672-0229(03)01011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Famin O, Ginsburg H. The treatment of Plasmodium falciparum-infected erythrocytes with chloroquine leads to accumulation of ferriprotoporphyrin IX bound to particular parasite proteins and to the inhibition of the parasite's 6-phosphogluconate dehydrogenase. Parasite. 2003;10:39–50. doi: 10.1051/parasite/2003101p39. [DOI] [PubMed] [Google Scholar]
- 13.Omodeo Sale F, Vanzulli E, Caielli S, Taramelli D. Regulation of human erythrocyte glyceraldehyde-3-phosphate dehydrogenase by ferriprotoporphyrin IX. FEBS Lett. 2005;579:5095–5099. doi: 10.1016/j.febslet.2005.07.081. [DOI] [PubMed] [Google Scholar]
- 14.Chakravarti R, Aulak KS, Fox PL, Stuehr DJ. GAPDH regulates cellular heme insertion into inducible nitric oxide synthase. Proc Natl Acad Sci U S A. 2010;107:18004–18009. doi: 10.1073/pnas.1008133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li T, Bonkovsky HL, Guo JT. Structural analysis of heme proteins: implications for design and prediction. BMC Struct Biol. 2011;11:13. doi: 10.1186/1472-6807-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossmann MG, Argos P. The taxonomy of binding sites in proteins. Mol Cell Biochem. 1978;21:161–182. doi: 10.1007/BF00240135. [DOI] [PubMed] [Google Scholar]
- 17.Gupta N, Ragsdale SW. Thiol-disulfide redox dependence of heme binding and heme ligand switching in nuclear hormone receptor rev-erb{beta} J Biol Chem. 2011;286:4392–4403. doi: 10.1074/jbc.M110.193466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ragsdale SW, Yi L. Thiol/Disulfide redox switches in the regulation of heme binding to proteins. Antioxid Redox Signal. 2011;14:1039–1047. doi: 10.1089/ars.2010.3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yi L, Morgan JT, Ragsdale SW. Identification of a thiol/disulfide redox switch in the human BK channel that controls its affinity for heme and CO. J Biol Chem. 2010;285:20117–20127. doi: 10.1074/jbc.M110.116483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamza I. Intracellular trafficking of porphyrins. ACS Chem Biol. 2006;1:627–629. doi: 10.1021/cb600442b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aasa R, Vanngard T, Dunford HB. EPR studies on compound I of horseradish peroxidase. Biochim Biophys Acta. 1975;391:259–264. doi: 10.1016/0005-2744(75)90249-1. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Snyder SH. Nitric oxide stimulates auto-ADP-ribosylation of glyceraldehyde-3-phosphate dehydrogenase. Proc Natl Acad Sci U S A. 1992;89:9382–9385. doi: 10.1073/pnas.89.20.9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brassard J, Gottschalk M, Quessy S. Cloning and purification of the Streptococcus suis serotype 2 glyceraldehyde-3-phosphate dehydrogenase and its involvement as an adhesin. Vet Microbiol. 2004;102:87–94. doi: 10.1016/j.vetmic.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Tong Y, Guo M. Cloning and characterization of a novel periplasmic heme-transport protein from the human pathogen Pseudomonas aeruginosa. J Biol Inorg Chem. 2007;12:735–750. doi: 10.1007/s00775-007-0226-x. [DOI] [PubMed] [Google Scholar]
- 25.Hannibal L, Somasundaram R, Tejero J, Wilson A, Stuehr DJ. Influence of heme-thiolate in shaping the catalytic properties of a bacterial nitric-oxide synthase. J Biol Chem. 2011;286:39224–39235. doi: 10.1074/jbc.M111.286351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuzelova K, Mrhalova M, Hrkal Z. Kinetics of heme interaction with heme-binding proteins: the effect of heme aggregation state. Biochim Biophys Acta. 1997;1336:497–501. doi: 10.1016/s0304-4165(97)00062-7. [DOI] [PubMed] [Google Scholar]
- 27.Jarolim P, Lahav M, Liu SC, Palek J. Effect of hemoglobin oxidation products on the stability of red cell membrane skeletons and the associations of skeletal proteins: correlation with a release of hemin. Blood. 1990;76:2125–2131. [PubMed] [Google Scholar]
- 28.Paul KG, Theorell H, Akeson A. The molar light absorption of pyridine ferroprotoporphyrin (Pyridine hemochromogen) Acta Chem Scand. 1953;7:1284–1287. [Google Scholar]
- 29.DiNello RK, Dolphin DH. Substituted hemins as probes for structure-function relationships in horseradish peroxidase. J Biol Chem. 1981;256:6903–6912. [PubMed] [Google Scholar]
- 30.Stoll S, Schweiger A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J Magn Reson. 2006;178:42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 31.Ambrosi E, Capaldi S, Bovi M, Saccomani G, Perduca M, Monaco HL. Structural changes in the BH3 domain of SOUL protein upon interaction with the anti-apoptotic protein Bcl-xL. Biochem J. 2011;438:291–301. doi: 10.1042/BJ20110257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lechardeur D, Cesselin B, Liebl U, Vos MH, Fernandez A, Brun C, Gruss A, Gaudu P. Discovery of an intracellular heme-binding protein, HrtR, that controls heme-efflux by the conserved HrtB HrtA transporter in Lactococcus lactis. J Biol Chem. 2011 doi: 10.1074/jbc.M111.297531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirschner-Zilber I, Rabizadeh E, Shaklai N. The interaction of hemin and bilirubin with the human red cell membrane. Biochim Biophys Acta. 1982;690:20–30. doi: 10.1016/0005-2736(82)90234-6. [DOI] [PubMed] [Google Scholar]
- 34.Rotsaert FA, Hallberg BM, de Vries S, Moenne-Loccoz P, Divne C, Renganathan V, Gold MH. Biophysical and structural analysis of a novel heme B iron ligation in the flavocytochrome cellobiose dehydrogenase. J Biol Chem. 2003;278:33224–33231. doi: 10.1074/jbc.M302653200. [DOI] [PubMed] [Google Scholar]
- 35.Sook BR, Block DR, Sumithran S, Montanez GE, Rodgers KR, Dawson JH, Eichenbaum Z, Dixon DW. Characterization of SiaA, a streptococcal heme-binding protein associated with a heme ABC transport system. Biochemistry. 2008;47:2678–2688. doi: 10.1021/bi701604y. [DOI] [PubMed] [Google Scholar]
- 36.Wojtowicz H, Wojaczynski J, Olczak M, Kroliczewski J, Latos-Grazynski L, Olczak T. Heme environment in HmuY, the heme-binding protein of Porphyromonas gingivalis. Biochem Biophys Res Commun. 2009;383:178–182. doi: 10.1016/j.bbrc.2009.03.148. [DOI] [PubMed] [Google Scholar]
- 37.Yukl ET, Jepkorir G, Alontaga AY, Pautsch L, Rodriguez JC, Rivera M, Moenne-Loccoz P. Kinetic and spectroscopic studies of hemin acquisition in the hemophore HasAp from Pseudomonas aeruginosa. Biochemistry. 2010;49:6646–6654. doi: 10.1021/bi100692f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Izadi-Pruneyre N, Huche F, Lukat-Rodgers GS, Lecroisey A, Gilli R, Rodgers KR, Wandersman C, Delepelaire P. The heme transfer from the soluble HasA hemophore to its membrane-bound receptor HasR is driven by protein-protein interaction from a high to a lower affinity binding site. J Biol Chem. 2006;281:25541–25550. doi: 10.1074/jbc.M603698200. [DOI] [PubMed] [Google Scholar]
- 39.Erales J, Lignon S, Gontero B. CP12 from Chlamydomonas reinhardtii, a permanent specific "chaperone-like" protein of glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 2009;284:12735–12744. doi: 10.1074/jbc.M808254200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sassa S. Why heme needs to be degraded to iron, biliverdin IX alpha, and carbon monoxide? Antioxid Redox Signal. 2004;6:819–824. doi: 10.1089/ars.2004.6.819. [DOI] [PubMed] [Google Scholar]
- 41.Hargrove MS, Whitaker T, Olson JS, Vali RJ, Mathews AJ. Quaternary structure regulates hemin dissociation from human hemoglobin. J Biol Chem. 1997;272:17385–17389. doi: 10.1074/jbc.272.28.17385. [DOI] [PubMed] [Google Scholar]
- 42.Winkler WC, Gonzalez G, Wittenberg JB, Hille R, Dakappagari N, Jacob A, Gonzalez LA, Gilles-Gonzalez MA. Nonsteric factors dominate binding of nitric oxide, azide, imidazole, cyanide, and fluoride to the rhizobial heme-based oxygen sensor Fix L. Chem Biol. 1996;3:841–850. doi: 10.1016/s1074-5521(96)90070-8. [DOI] [PubMed] [Google Scholar]
- 43.Yoon J, Herzik MA, Jr, Winter MB, Tran R, Olea C, Jr, Marletta MA. Structure and properties of a bis-histidyl ligated globin from Caenorhabditis elegans. Biochemistry. 2010;49:5662–5670. doi: 10.1021/bi100710a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puranik M, Weeks CL, Lahaye D, Kabil O, Taoka S, Nielsen SB, Groves JT, Banerjee R, Spiro TG. Dynamics of carbon monoxide binding to cystathionine beta-synthase. J Biol Chem. 2006;281:13433–13438. doi: 10.1074/jbc.M600246200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Couture M, Burmester T, Hankeln T, Rousseau DL. The heme environment of mouse neuroglobin. Evidence for the presence of two conformations of the heme pocket. J Biol Chem. 2001;276:36377–36382. doi: 10.1074/jbc.M103907200. [DOI] [PubMed] [Google Scholar]
- 46.Uno T, Ryu D, Tsutsumi H, Tomisugi Y, Ishikawa Y, Wilkinson AJ, Sato H, Hayashi T. Residues in the distal heme pocket of neuroglobin. Implications for the multiple ligand binding steps. J Biol Chem. 2004;279:5886–5893. doi: 10.1074/jbc.M311748200. [DOI] [PubMed] [Google Scholar]
- 47.Pesce A, Thijs L, Nardini M, Desmet F, Sisinni L, Gourlay L, Bolli A, Coletta M, Van Doorslaer S, Wan X, Alam M, Ascenzi P, Moens L, Bolognesi M, Dewilde S. HisE11 and HisF8 provide bis-histidyl heme hexa-coordination in the globin domain of Geobacter sulfurreducens globin-coupled sensor. J Mol Biol. 2009;386:246–260. doi: 10.1016/j.jmb.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 48.Desbois A, Lutz M. Redox control of proton transfers in membrane b-type cytochromes: an absorption and resonance Raman study on bis (imidazole) and bis (imidazolate) model complexes of iron-protoporphyrin. Eur Biophys J. 1992;20:321–335. [Google Scholar]
- 49.Takeuchi F, Hori H, Obayashi E, Shiro Y, Tsubaki M. Properties of two distinct heme centers of cytochrome b561 from bovine chromaffin vesicles studied by EPR, resonance Raman, and ascorbate reduction assay. J Biochem. 2004;135:53–64. doi: 10.1093/jb/mvh006. [DOI] [PubMed] [Google Scholar]
- 50.Lukat-Rodgers GS, Rodgers KR, Caillet-Saguy C, Izadi-Pruneyre N, Lecroisey A. Novel heme ligand displacement by CO in the soluble hemophore HasA and its proximal ligand mutants: implications for heme uptake and release. Biochemistry. 2008;47:2087–2098. doi: 10.1021/bi7019518. [DOI] [PubMed] [Google Scholar]
- 51.Auclair K, Huang HW, Moenne-Loccoz P, Ortiz de Montellano PR. Cloning and expression of a heme binding protein from the genome of Saccharomyces cerevisiae. Protein Expr Purif. 2003;28:340–349. doi: 10.1016/s1046-5928(02)00699-x. [DOI] [PubMed] [Google Scholar]
- 52.Lou BS, Snyder JK, Marshall P, Wang JS, Wu G, Kulmacz RJ, Tsai AL, Wang J. Resonance Raman studies indicate a unique heme active site in prostaglandin H synthase. Biochemistry. 2000;39:12424–12434. doi: 10.1021/bi001257c. [DOI] [PubMed] [Google Scholar]
- 53.Spiro TG. Biological Applications of Raman Spectroscopy. John Wiley & Sons Canada Limited; 1988. [Google Scholar]
- 54.Vogel KM, Kozlowski PM, Zgierski MZ, Spiro TG. Role of the axial ligand in heme-CO backbonding; DFT analysis of vibrational data. Inorg Chim Acta. 2000;297:11–17. [Google Scholar]
- 55.Spiro TG, Wasbotten IH. CO as a vibrational probe of heme protein active sites. J Inorg Biochem. 2005;99:34–44. doi: 10.1016/j.jinorgbio.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 56.Argos P, Mathews FS. The structure of ferrocytochrome b5 at 2.8 A resolution. J Biol Chem. 1975;250:747–751. [PubMed] [Google Scholar]
- 57.Babcock GT, Widger WR, Cramer WA, Oertling WA, Metz JG. Axial ligands of chloroplast cytochrome b-559: identification and requirement for a heme-cross-linked polypeptide structure. Biochemistry. 1985;24:3638–3645. doi: 10.1021/bi00335a036. [DOI] [PubMed] [Google Scholar]
- 58.Dou Y, Admiraal SJ, Ikeda-Saito M, Krzywda S, Wilkinson AJ, Li T, Olson JS, Prince RC, Pickering IJ, George GN. Alteration of axial coordination by protein engineering in myoglobin. Bisimidazole ligation in the His64-->Val/Val68-->His double mutant. J Biol Chem. 1995;270:15993–16001. doi: 10.1074/jbc.270.27.15993. [DOI] [PubMed] [Google Scholar]
- 59.Halder P, Trent JT, 3rd, Hargrove MS. Influence of the protein matrix on intramolecular histidine ligation in ferric and ferrous hexacoordinate hemoglobins. Proteins. 2007;66:172–182. doi: 10.1002/prot.21210. [DOI] [PubMed] [Google Scholar]
- 60.Miller GT, Zhang B, Hardman JK, Timkovich R. Converting a c-type to a b-type cytochrome: Met61 to His61 mutant of Pseudomonas cytochrome c-551. Biochemistry. 2000;39:9010–9017. doi: 10.1021/bi000232a. [DOI] [PubMed] [Google Scholar]
- 61.Raphael AL, Gray HB. Axial ligand replacement in horse heart cytochrome c by semisynthesis. Proteins. 1989;6:338–340. doi: 10.1002/prot.340060316. [DOI] [PubMed] [Google Scholar]
- 62.Ishikawa H, Nakagaki M, Bamba A, Uchida T, Hori H, O'Brian MR, Iwai K, Ishimori K. Unusual heme binding in the bacterial iron response regulator protein: spectral characterization of heme binding to the heme regulatory motif. Biochemistry. 2011;50:1016–1022. doi: 10.1021/bi101895r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang F, Xia X, Lei HY, Wang ED. Hemin binds to human cytoplasmic arginyl-tRNA synthetase and inhibits its catalytic activity. J Biol Chem. 2010;285:39437–39446. doi: 10.1074/jbc.M110.159913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seidler NW. Basic Biology of GAPDH. Adv Exp Med Biol. 2013;985:1–36. doi: 10.1007/978-94-007-4716-6_1. [DOI] [PubMed] [Google Scholar]
- 65.Hargrove MS, Barrick D, Olson JS. The association rate constant for heme binding to globin is independent of protein structure. Biochemistry. 1996;35:11293–11299. doi: 10.1021/bi960371l. [DOI] [PubMed] [Google Scholar]
- 66.Bhakta MN, Wilks A. The mechanism of heme transfer from the cytoplasmic heme binding protein PhuS to the delta-regioselective heme oxygenase of Pseudomonas aeruginosa. Biochemistry. 2006;45:11642–11649. doi: 10.1021/bi060980l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hargrove MS, Wilkinson AJ, Olson JS. Structural factors governing hemin dissociation from metmyoglobin. Biochemistry. 1996;35:11300–11309. doi: 10.1021/bi960372d. [DOI] [PubMed] [Google Scholar]
- 68.Bianchetti CM, Blouin GC, Bitto E, Olson JS, Phillips GN., Jr The structure and NO binding properties of the nitrophorin-like heme-binding protein from Arabidopsis thaliana gene locus At1g79260.1. Proteins. 2010;78:917–931. doi: 10.1002/prot.22617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miksanova M, Igarashi J, Minami M, Sagami I, Yamauchi S, Kurokawa H, Shimizu T. Characterization of heme-regulated eIF2alpha kinase: roles of the N-terminal domain in the oligomeric state, heme binding, catalysis, and inhibition. Biochemistry. 2006;45:9894–9905. doi: 10.1021/bi060556k. [DOI] [PubMed] [Google Scholar]
- 70.Dage JL, Sun H, Halsall HB. Determination of diethylpyrocarbonate-modified amino acid residues in alpha 1-acid glycoprotein by high-performance liquid chromatography electrospray ionization-mass spectrometry and matrix-assisted laser desorption/ionization time-of-flight-mass spectrometry. Anal Biochem. 1998;257:176–185. doi: 10.1006/abio.1997.2552. [DOI] [PubMed] [Google Scholar]
- 71.Hayasaka K, Kitanishi K, Igarashi J, Shimizu T. Heme-binding characteristics of the isolated PAS-B domain of mouse Per2, a transcriptional regulatory factor associated with circadian rhythms. Biochim Biophys Acta. 2011;1814:326–333. doi: 10.1016/j.bbapap.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 72.Konkle ME, Elsenheimer KN, Hakala K, Robicheaux JC, Weintraub ST, Hunsicker-Wang LM. Chemical modification of the Rieske protein from Thermus thermophilus using diethyl pyrocarbonate modifies ligating histidine 154 and reduces the [2FE-2S] cluster. Biochemistry. 2010;49:7272–7281. doi: 10.1021/bi1007904. [DOI] [PubMed] [Google Scholar]
- 73.Tawfik DS. Ethoxyformylation of Histidine. In: Walker JM, editor. The Protein Protocols Handbook. 2nd Edition. Chapter 65. 2002. pp. 473–474. [Google Scholar]
- 74.Ran Y, Liu M, Zhu H, Nygaard TK, Brown DE, Fabian M, Dooley DM, Lei B. Spectroscopic identification of heme axial ligands in HtsA that are involved in heme acquisition by Streptococcus pyogenes. Biochemistry. 2010;49:2834–2842. doi: 10.1021/bi901987h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Puranik M, Nielsen SB, Youn H, Hvitved AN, Bourassa JL, Case MA, Tengroth C, Balakrishnan G, Thorsteinsson MV, Groves JT, McLendon GL, Roberts GP, Olson JS, Spiro TG. Dynamics of carbon monoxide binding to CooA. J Biol Chem. 2004;279:21096–21108. doi: 10.1074/jbc.M400613200. [DOI] [PubMed] [Google Scholar]
- 76.Moller JK, Skibsted LH. Mechanism of nitrosylmyoglobin autoxidation: temperature and oxygen pressure effects on the two consecutive reactions. Chemistry. 2004;10:2291–2300. doi: 10.1002/chem.200305368. [DOI] [PubMed] [Google Scholar]
- 77.Pierre JL, Fontecave M, Crichton RR. Chemistry for an essential biological process: the reduction of ferric iron. Biometals. 2002;15:341–346. doi: 10.1023/a:1020259021641. [DOI] [PubMed] [Google Scholar]
- 78.Gaspard S, Chottard G, Mahy JP, Mansuy D. Study of the coordination chemistry of prostaglandin G/H synthase by resonance Raman spectroscopy. Eur J Biochem. 1996;238:529–537. doi: 10.1111/j.1432-1033.1996.0529z.x. [DOI] [PubMed] [Google Scholar]
- 79.Choi CY, Cerda JF, Chu HA, Babcock GT, Marletta MA. Spectroscopic characterization of the heme-binding sites in Plasmodium falciparum histidine-rich protein 2. Biochemistry. 1999;38:16916–16924. doi: 10.1021/bi991665k. [DOI] [PubMed] [Google Scholar]
- 80.Dailey TA, Boynton TO, Albetel AN, Gerdes S, Johnson MK, Dailey HA. Discovery and Characterization of HemQ: an essential heme biosynthetic pathway component. J Biol Chem. 2010;285:25978–25986. doi: 10.1074/jbc.M110.142604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Airola MV, Du J, Dawson JH, Crane BR. Heme binding to the Mammalian circadian clock protein period 2 is nonspecific. Biochemistry. 2010;49:4327–4338. doi: 10.1021/bi901945w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spolaore B, De Filippis V, Fontana A. Heme binding by the N-terminal fragment 1–44 of human growth hormone. Biochemistry. 2005;44:16079–16089. doi: 10.1021/bi051374d. [DOI] [PubMed] [Google Scholar]
- 83.Hrkal Z, Vodrazka Z, Kalousek I. Transfer of heme from ferrihemoglobin and ferrihemoglobin isolated chains to hemopexin. Eur J Biochem. 1974;43:73–78. doi: 10.1111/j.1432-1033.1974.tb03386.x. [DOI] [PubMed] [Google Scholar]
- 84.Leclerc E, Leclerc L, Poyart C, Marden MC. Interaction of heme with amphiphilic peptides: use of hemin-CN to probe the interaction of calmodulin with its target peptides. Arch Biochem Biophys. 1993;306:158–162. doi: 10.1006/abbi.1993.1494. [DOI] [PubMed] [Google Scholar]
- 85.Marden MC, Dufour E, Christova P, Huang Y, Leclerc-L'Hostis E, Haertle T. Binding of heme-CO to bovine and porcine beta-lactoglobulins. Arch Biochem Biophys. 1994;311:258–262. doi: 10.1006/abbi.1994.1235. [DOI] [PubMed] [Google Scholar]
- 86.Wardell M, Wang Z, Ho JX, Robert J, Ruker F, Ruble J, Carter DC. The atomic structure of human methemalbumin at 1.9 A. Biochem Biophys Res Commun. 2002;291:813–819. doi: 10.1006/bbrc.2002.6540. [DOI] [PubMed] [Google Scholar]
- 87.Zhang L, Guarente L. Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J. 1995;14:313–320. doi: 10.1002/j.1460-2075.1995.tb07005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Poulos TL. The role of the proximal ligand in heme enzymes. Journal of Biological Inorganic Chemistry. 1996;1:356–359. [Google Scholar]
- 89.Reedy CJ, Gibney BR. Heme protein assemblies. Chem Rev. 2004;104:617–649. doi: 10.1021/cr0206115. [DOI] [PubMed] [Google Scholar]
- 90.Schultz IJ, Chen C, Paw BH, Hamza I. Iron and porphyrin trafficking in heme biogenesis. J Biol Chem. 2010;285:26753–26759. doi: 10.1074/jbc.R110.119503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.