Abstract
Most infections result from colonization by more than one microbe. Within such polymicrobial infections, microbes often display synergistic interactions that result in increased disease severity. Although many clinical studies have documented the occurrence of synergy in polymicrobial infections, little is known about the underlying molecular mechanisms. A prominent pathogen in many polymicrobial infections is Pseudomonas aeruginosa, a Gram-negative bacterium that displays enhanced virulence during coculture with Gram-positive bacteria. In this study we discovered that during coinfection, P. aeruginosa uses peptidoglycan shed by Gram-positive bacteria as a cue to stimulate production of multiple extracellular factors that possess lytic activity against prokaryotic and eukaryotic cells. Consequently, P. aeruginosa displays enhanced virulence in a Drosophila model of infection when cocultured with Gram-positive bacteria. Inactivation of a gene (PA0601) required for peptidoglycan sensing mitigated this phenotype. Using Drosophila and murine models of infection, we also show that peptidoglycan sensing results in P. aeruginosa-mediated reduction in the Gram-positive flora in the infection site. Our data suggest that P. aeruginosa has evolved a mechanism to survey the microbial community and respond to Gram-positive produced peptidoglycan through production of antimicrobials and toxins that not only modify the composition of the community but also enhance host killing. Additionally, our results suggest that therapeutic strategies targeting Gram-positive bacteria might be a viable approach for reducing the severity of P. aeruginosa polymicrobial infections.
Keywords: Staphylococcus, quorum sensing, PQS, glucosamine
Microbes seldom exist in isolation. Instead, they inhabit complex polymicrobial communities where interactions between individuals shape the composition and biological activities of the population. Although this fact has been appreciated since the time of Pasteur (1), most laboratory studies have focused on a single bacterial species grown in isolation. Such studies are particularly prominent in the field of bacterial pathogenesis, where molecular and biochemical studies have enhanced our knowledge of monoculture infections both in vitro and in vivo. However, it is clear that the presence of other bacteria within polymicrobial infections, including commensal bacteria, can impact virulence of extracellular pathogens (2–7). Indeed, microbes within polymicrobial infections often display synergy, defined as the cooperative interaction of two or more microbes to produce a result not achieved by the individual bacterium acting alone (8). Synergistic interactions can result in enhanced pathogen persistence in the infection site, increased disease severity, and increased antimicrobial resistance (2, 6, 7, 9–14).
Owing in large part to these synergistic interactions, polymicrobial infections are a significant and growing health concern both in the United States and throughout the world. Some of the most common polymicrobial infections occurring in the United States are those affecting chronic wounds. Each year between 5 and 7 million Americans are treated for chronic wounds at an estimated annual cost of 10–20 billion dollars (15). It is expected that these costs will grow over the next 50 y as the population ages and the prevalence of risk factors such as obesity and diabetes increases. Although the importance of polymicrobial infections in clinical settings is clear, the molecular details controlling synergy have generally not been elucidated. Thus, detailed mechanistic studies elucidating polymicrobial interactions necessary for enhanced persistence in vivo are a critical step toward understanding synergy and a necessary first step toward developing therapeutics to treat multispecies infections.
The Gram-negative opportunistic pathogen Pseudomonas aeruginosa is a common member of polymicrobial infections, including chronic wounds. Studies of P. aeruginosa prevalence indicate that this bacterium is present in 15–80% of chronic wounds (16–19), depending on the type of chronic wound, sampling technique, and the methodology used for bacterial identification. Recent evidence suggests that the prevalence may be higher because P. aeruginosa occupies deeper sites within the wound, hampering identification via some conventional sampling techniques (20). In addition, chronic wounds containing P. aeruginosa are more severe and heal at a slower rate (10, 18, 21), which is hypothesized to be mediated in part by synergistic interactions with Gram-positive bacteria, including Staphylococcus aureus. Indeed, in a murine chronic wound model, P. aeruginosa–S. aureus coinfections were shown to be more severe and recalcitrant to antimicrobial treatment than P. aeruginosa monoculture infections (10). Synergistic interactions were also observed in a rat lung infection model and a Drosophila infection model where S. aureus was shown to enhance P. aeruginosa virulence (22, 23).
The goal of this study was to elucidate the mechanism of synergy during P. aeruginosa coinfection with Gram-positive bacteria. This work expands on an observation from our laboratory that P. aeruginosa enhances production of the virulence factor pyocyanin upon exposure to cell wall fragments from Gram-positive bacteria. Further work revealed that the N-acetylglucosamine (GlcNAc) portion of the cell wall polymer peptidoglycan is necessary and sufficient to enhance pyocyanin production (24). Despite the fact that GlcNAc induced pyocyanin production, it was unclear whether P. aeruginosa GlcNAc sensing impacted disease severity during polymicrobial infection. Additionally, the genetic determinants of GlcNAc sensing remained unresolved. Here we report that P. aeruginosa enhances production of multiple virulence factors during coculture with Gram-positive bacteria, and the ability to sense and respond to exogenous peptidoglycan is essential for this response. Through the isolation of a P. aeruginosa mutant that cannot respond to exogenous peptidoglycan, we also show that peptidoglycan sensing is essential for enhanced P. aeruginosa virulence during coculture with Gram-positive commensal bacteria in a Drosophila model of infection. Finally, we show that peptidoglycan sensing is important for P. aeruginosa fitness in a murine chronic wound model. On the basis of these data we propose a model in which during coinfection, P. aeruginosa senses peptidoglycan shed by Gram-positive bacteria and responds to this cue through enhanced production of virulence factors.
Results
Screen for GlcNAc-Unresponsive Mutants.
We previously reported that P. aeruginosa responds to GlcNAc and the GlcNAc-containing polymer peptidoglycan through enhanced production of the extracellular toxin pyocyanin (24). Because GlcNAc is a component of all peptidoglycan, peptidoglycan from multiple bacterial species including S. aureus induced pyocyanin production (24). This response was specific to GlcNAc: the presence of other N-acetylated sugars, including the peptidoglycan component N-acetyl muramic acid, did not induce pyocyanin (Fig. S1). The goal of this study was to first identify genes required for GlcNAc sensing and subsequently examine the importance of this pathway for P. aeruginosa virulence during coinfection with Gram-positive bacteria. To accomplish this goal, a genetic screen was performed to identify P. aeruginosa mutants unable to respond to GlcNAc and peptidoglycan. This screen took advantage of the characteristic blue-green color of pyocyanin to identify strains that produced normal levels of pyocyanin in the absence of GlcNAc but did not enhance pyocyanin production in the presence of GlcNAc. The 5,760 mutants included in the P. aeruginosa PA14 nonredundant transposon mutant library (25) were screened visually in a 96-well plate format using a defined medium in the presence and absence of GlcNAc. Fifty-nine potential GlcNAc-sensing mutants were identified in this primary screen. Rescreening these isolates using a quantitative pyocyanin spectrophotometric assay (26) resulted in 14 bona fide P. aeruginosa mutants that did not enhance pyocyanin in the presence of GlcNAc (Table S1).
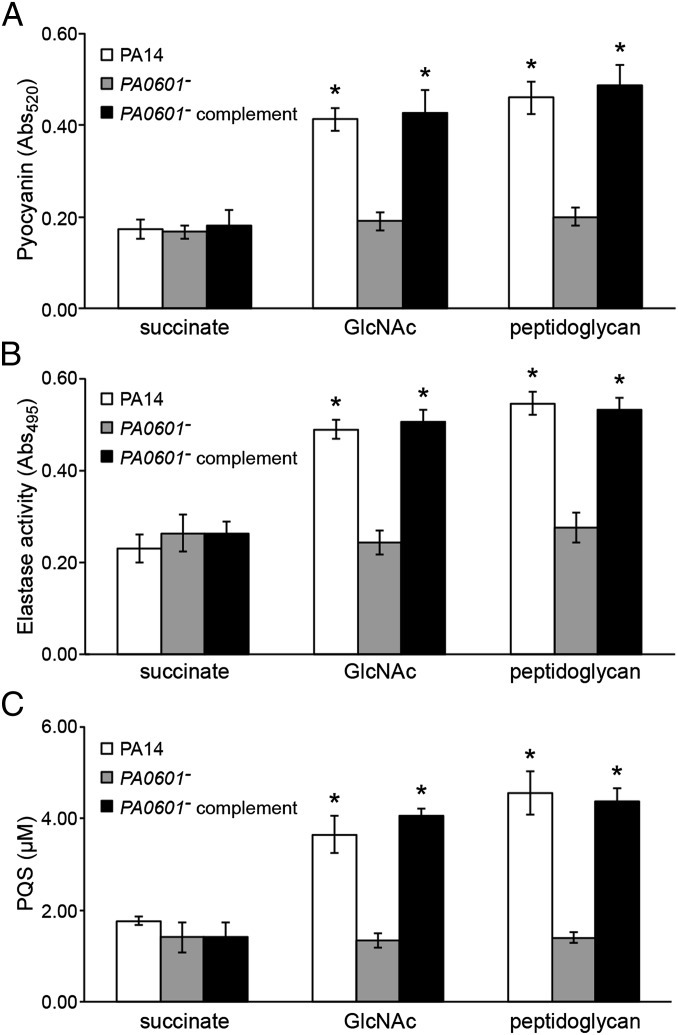
To prioritize these mutants for study, two additional criteria were used: a reduced ability to lyse Gram-positive bacteria in vitro and the ability to use GlcNAc as the sole source of energy (Table S1). The first criterion was used because peptidoglycan enhances P. aeruginosa lysis of Gram-positive bacteria (24), thus we reasoned a peptidoglycan-sensing mutant should show reduced lytic ability in vitro. The second criterion was used because our goal was to test the role of GlcNAc sensing, not GlcNAc catabolism. After these secondary screens only one mutant, containing a transposon insertion in the gene PA0601 (designated as PA14_07840 on the PA14 genome), satisfied all criteria (Table S1). PA0601 encodes a putative two-component response regulator with sequence homology to the LuxR family of transcriptional regulators [66% similarity to VsrD (27) from Burkholderia solanacearum]. Importantly, expression of PA0601 in trans restored pyocyanin production of the PA0601 mutant to WT levels in the presence of GlcNAc and peptidoglycan (Fig. 1A), indicating that the phenotype is due to inactivation of PA0601 and not polar effects on neighboring genes.
Fig. 1.
PA0601 is required for GlcNAc and peptidoglycan sensing in P. aeruginosa. (A) Pyocyanin, (B) elastase, and (C) PQS levels produced by WT P. aeruginosa (PA14), the P. aeruginosa PA0601 transposon mutant (PA0601−), and the genetically complemented PA0601 mutant (PA0601− complement) in the presence of no inducer (succinate), GlcNAc, or peptidoglycan. PA14 and PA0601− contain the empty complementation plasmid pEX1.8, whereas the PA0601− complement contains pAK601 (pEX1.8 expressing PA0601). *P < 0.05 by Student t test compared with the no-inducer (succinate) control. Error bars represent SD, n = 3.
PA0601 Is Required for Enhanced Quorum Signal Production in the Presence of Peptidoglycan.
Pyocyanin production is often coregulated with several other virulence factors in P. aeruginosa, including the extracellular protease elastase (28, 29); thus it was not surprising that addition of GlcNAc and peptidoglycan also enhanced elastase production by WT P. aeruginosa (Fig. 1B). Similar to pyocyanin, the PA0601 mutant did not display increased elastase production in the presence of GlcNAc or peptidoglycan and this phenotype was restored by expression of PA0601 in trans.
Both pyocyanin and elastase are regulated by quorum sensing (28, 29), a regulatory scheme involving the production and sensing of extracellular signaling molecules. Although P. aeruginosa uses multiple small molecule signals to regulate gene expression via quorum sensing, the signal 2-heptyl-3-hydroxy-4-quinolone (referred to as the Pseudomonas quinolone signal or PQS) controls expression of both elastase and pyocyanin (28–30). This leads to the hypothesis that P. aeruginosa enhances production of PQS in the presence of GlcNAc and peptidoglycan, resulting in increased levels of pyocyanin and elastase. In support of this hypothesis, P. aeruginosa produced approximately threefold higher levels of PQS during growth with GlcNAc and peptidoglycan, and this response was not observed in the PA0601 mutant (Fig. 1C). On the basis of these data, we conclude that GlcNAc and peptidoglycan stimulate P. aeruginosa quorum sensing, resulting in elevated levels of PQS-controlled toxins. Because PA0601 is required for this response, the PA0601 mutant provided a peptidoglycan-unresponsive strain that can be used to elucidate the importance of this response in vivo.
PA0601 Mutant Is Impaired for Virulence in a Drosophila Infection Model.
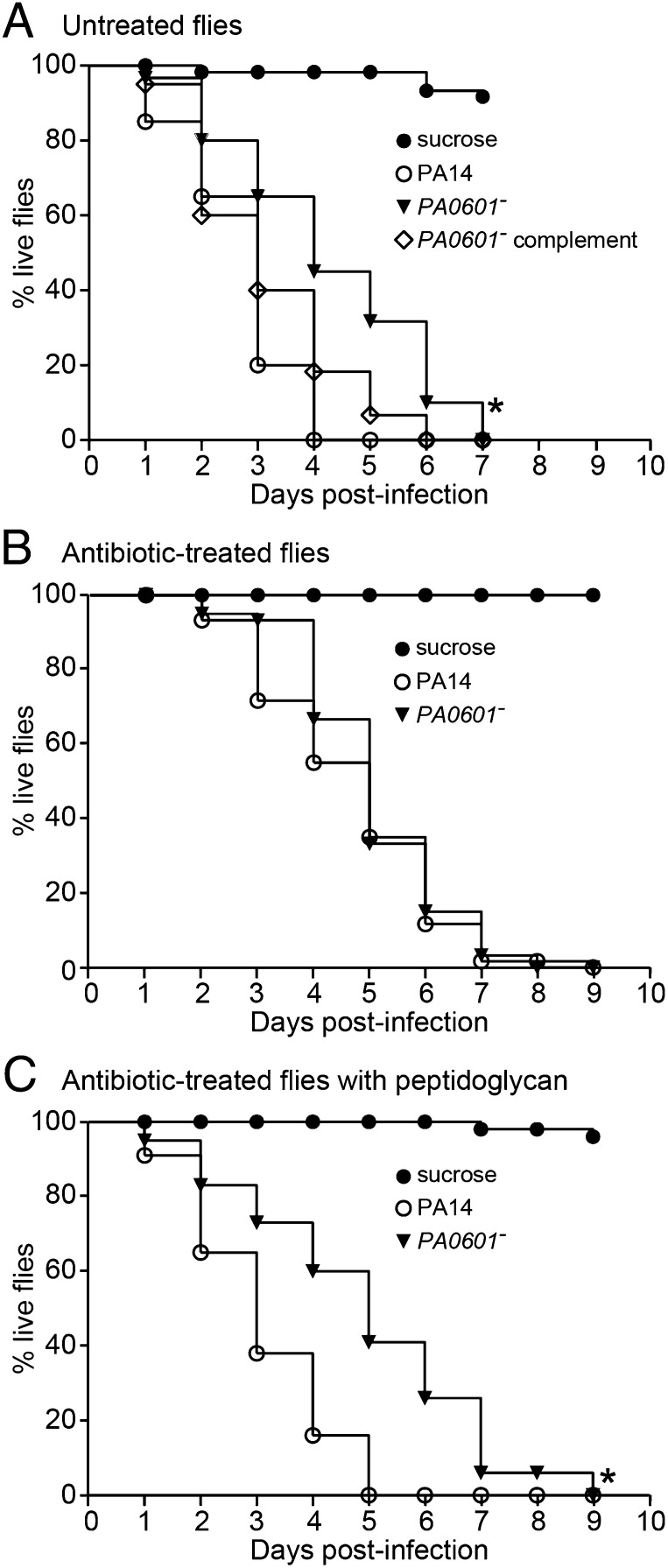
PQS-controlled factors, including elastase and pyocyanin, exert killing activity on prokaryotic and eukaryotic cells (24, 31–38), and the ability to respond to peptidoglycan during coculture growth enhances P. aeruginosa killing of Gram-positive bacteria in vitro (Fig. S2 and Table S1). On the basis of these data, we hypothesized that peptidoglycan sensing would also enhance P. aeruginosa pathogenesis during coinfection with Gram-positive bacteria. To test this hypothesis, a Drosophila model of infection was used. This is a versatile infection model previously used to study P. aeruginosa polymicrobial infections (23). In this model, flies are allowed to orally ingest P. aeruginosa, which colonizes the fly crop and ultimately causes death (23, 39). Importantly, factors such as pyocyanin and elastase required for pathogenesis in vertebrate models are also critical for killing Drosophila (40–42), thus this model is suitable for studying P. aeruginosa pathogenesis. When administered with WT P. aeruginosa, 50% of the flies were dead at 2 d (Fig. 2A); however, flies infected with the PA0601 mutant showed a significant delay in fly killing (50% dead at 4 d), and killing was restored to WT levels by expression of PA0601 in trans (Fig. 2A). To determine whether the difference in virulence was due to the reduced ability of the PA0601 mutant to colonize and persist in the fly, crops were removed and P. aeruginosa numbers measured at 2 d after infection. Despite the fact that significant killing was observed at this time point by WT P. aeruginosa but not PA0601 mutant infected flies (Fig. 2A), no difference in P. aeruginosa cell numbers was observed (Fig. S3A). These data indicate that the reduced virulence of the PA0601 mutant was not due to the inability to colonize and persist in the host.
Fig. 2.
GlcNAc/peptidoglycan sensing enhances P. aeruginosa virulence in polymicrobial infections. Kaplan-Meier survival curves of (A) antibiotic-untreated Drosophila after infection with WT P. aeruginosa (PA14), the PA0601 mutant (PA0601−), and the genetically complemented PA0601 mutant (PA0601− complement); (B) antibiotic-treated flies infected with P. aeruginosa WT or the PA0601 mutant; and (C) antibiotic-treated flies infected with WT P. aeruginosa or the PA0601 mutant in the presence and absence of peptidoglycan. Curves are representative of a minimum of two biological replicates, n = 60 for each replicate. *P < 0.0001 by the log–rank test for comparison of percentage survival after infection with PA14 compared with infection with PA0601−.
Peptidoglycan Enhances P. aeruginosa Virulence During Coculture with Gram-Positive Bacteria.
The Drosophila crop is naturally colonized with large numbers of Gram-positive bacteria (43), and the flies used in these experiments contained ∼6 × 104 Gram-positive bacteria in their crops (Fig. S3B), representing more than 60% of the total culturable bacteria. Because we were not cocolonizing the flies with Gram-positive bacteria during infection with P. aeruginosa, we hypothesized that the source of GlcNAc in these infections was peptidoglycan shed by native Gram-positive flora. To test this hypothesis, flies were fed a mixture of antibiotics designed to eliminate the native Gram-positive but not Gram-negative flora in the crop. After antibiotic treatment, no Gram-positive bacteria were detected in the Drosophila crop using conventional culturing techniques (Fig. S3B); however a considerable number of Gram-negative bacteria remained (∼3 × 103 cfu per fly). Antibiotic-treated flies showed a significant delay in killing when colonized with WT P. aeruginosa, displaying 50% killing at 4 d after infection (Fig. 2B) in comparison with 2 d after infection in antibiotic-untreated flies (Fig. 2A). No difference in killing was observed between WT P. aeruginosa and the PA0601 mutant in antibiotic-treated flies, suggesting that the native Gram-positive flora enhances pathogenesis of WT P. aeruginosa (Fig. 2B). Again, the delay in killing observed for WT P. aeruginosa in antibiotic-treated flies was not due to reduced colonization and persistence because the number of P. aeruginosa in the crop was similar in antibiotic-treated and untreated flies (Fig. S3A).
On the basis of these data, we hypothesized that delayed killing of antibiotic-treated flies by WT P. aeruginosa was due to lack of peptidoglycan in the fly crop. To test this hypothesis, antibiotic-treated flies were fed peptidoglycan along with P. aeruginosa. Peptidoglycan feeding abolished the delay in killing by WT P. aeruginosa but had no effect on killing by the P. aeruginosa PA0601 mutant (Fig. 2C). The increase in fly killing upon peptidoglycan addition was not due to increases in P. aeruginosa numbers in the fly (Fig. S3A), indicating that peptidoglycan enhances virulence factor production, not growth or persistence in vivo.
Peptidoglycan Increases pqsA Expression in Vivo.
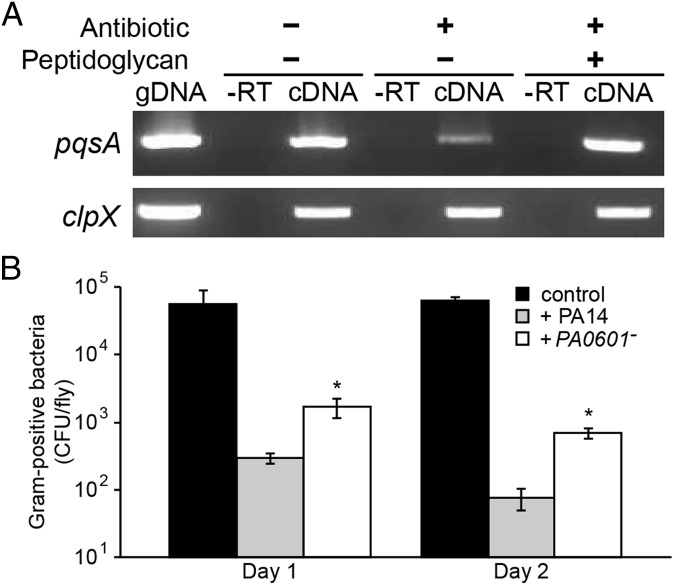
Because peptidoglycan specifically enhances production of PQS and PQS-controlled virulence factors in vitro (Fig. 1), we hypothesized that the increased virulence observed upon peptidoglycan addition was due to enhanced expression of the PQS biosynthetic genes in vivo. To test this hypothesis RNA was extracted from P. aeruginosa colonized crops, and expression of pqsA, which encodes the first gene in the PQS biosynthetic operon, was examined using reverse transcriptase PCR. Levels of the pqsA transcript were reduced approximately threefold in antibiotic-treated flies compared with untreated flies, and addition of peptidoglycan to antibiotic-treated flies restored pqsA transcript levels to those observed in antibiotic-untreated flies (Fig. 3A). It should be noted that although these results strongly implicate PQS induction as the mechanism of enhanced fly killing, they do not rule out a more general role of GlcNAc in stimulating P. aeruginosa virulence.
Fig. 3.
(A) Decreased levels of the pqsA transcript in antibiotic-treated Drosophila. Reverse transcriptase PCR was used to examine pqsA transcript levels during Drosophila infection under three conditions: (i) no antibiotic treatment of flies and no peptidoglycan added; (ii) antibiotic-treated flies with no peptidoglycan added; (iii) antibiotic-treated flies with peptidoglycan added. gDNA is the genomic DNA positive control, and −RT is the negative control using RNA as the PCR template. The constitutively expressed clpX gene was used as a control. A representative ethidium bromide stained agarose gel is shown. (B) P. aeruginosa colonization reduces the native Gram-positive flora of Drosophila. Gram-positive bacteria were enumerated by viable counts on phenylethyl alcohol agar. Shown are cfu per fly from uninfected flies (control), and flies infected with either WT P. aeruginosa (PA14) or the PA0601 mutant (PA0601−). *P < 0.01 by Student t test compared with PA14-infected flies. Error bars represent SD, n = 3.
Peptidoglycan-Sensing Enhances Killing of Commensal Gram-Positive Bacteria in Vivo.
PQS controls an array of antimicrobials, including pyocyanin and elastase, as well as the antimicrobial quinolone 4-hydroxy-2-heptylquinoline N-oxide (33). Thus, our data suggest that in addition to virulence, the ability of P. aeruginosa to sense and respond to peptidoglycan will also impact the microbial flora within an infection site. Specifically, we hypothesize that because of enhanced production of Gram-positive antimicrobial factors during coculture (Fig. 1 A and B), the number of Gram-positive bacteria will decrease in an infection site upon introduction of P. aeruginosa. In support of this hypothesis, the number of Gram-positive bacteria within the Drosophila crop decreased ∼1,000-fold when infected with WT P. aeruginosa, whereas colonization with the PA0601 mutant decreased Gram-positive bacteria significantly less (Fig. 3B). These data support a model in which during coinfection with Gram-positive commensal flora, P. aeruginosa senses peptidoglycan and responds through enhanced production of PQS-controlled factors that not only reduce commensal Gram-positive flora but also increase animal mortality. Importantly, GlcNAc transport was required for these phenotypes (Fig. S4), indicating that GlcNAc is the component of peptidoglycan sensed by P. aeruginosa in vivo.
Peptidoglycan-Sensing Is Important for P. aeruginosa Competitiveness in a Murine Chronic Wound Model.
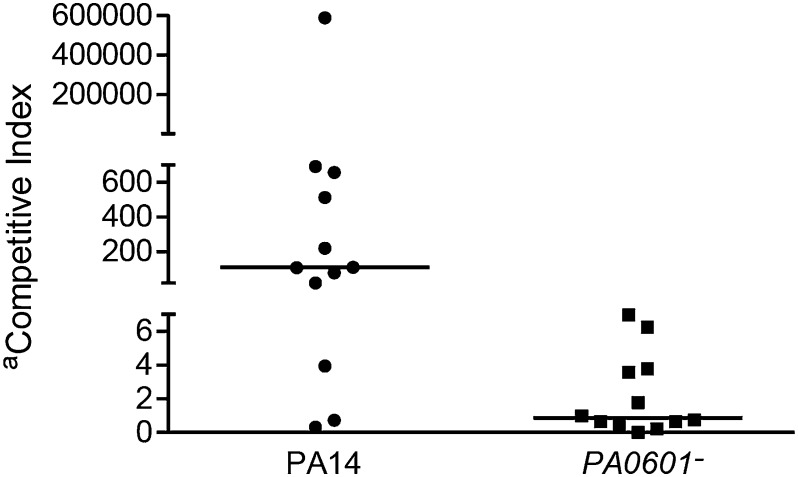
The Drosophila model provides a powerful system for probing the role of bacterial interactions in vivo, in regards to both host killing and composition of the microbial community. However, we sought to examine these interactions in a more relevant vertebrate model of polymicrobial infection. A murine chronic wound infection model was recently described that allows examination of P. aeruginosa–S. aureus interactions in vivo (44). Although this model does not provide a robust assay for virulence because it is a nonlethal model, it does allow for examination of the changes in composition of an infecting microbial community over several days. This model involves creation of a 1.5 cm × 1.5 cm surgical wound on the back of a Swiss Webster mouse that is subsequently colonized with bacteria and covered with transparent, semipermeable polyurethane dressing to protect the wound from contaminating bacteria. After 4 d, wound tissue is harvested and bacterial numbers are assessed by conventional culture techniques. Infection of the wound with monocultures of WT P. aeruginosa, the PA0601 mutant, and S. aureus showed comparable final growth yields, indicating that these strains colonized and persisted in the wounds at similar levels in isolation (Fig. S5). Coinfections of WT P. aeruginosa with S. aureus resulted in bacterial populations that were highly enriched for P. aeruginosa (Fig. 4; median 110:1 P. aeruginosa:S. aureus), despite the fact that these infections were initiated with an ∼1:1 ratio of P. aeruginosa:S. aureus. However, coculture infections of the P. aeruginosa PA0601 mutant with S. aureus were enriched for S. aureus (Fig. 4; median 0.9:1 P. aeruginosa:S. aureus). As observed in the Drosophila infections, the numbers of P. aeruginosa in WT and the PA0601 mutant cocultures did not change significantly (Fig. 4 legend), suggesting that the effect on S. aureus numbers is not due to increased persistence of WT P. aeruginosa but the ability to sense peptidoglycan and increase production of antimicrobials during coculture in vivo.
Fig. 4.
Competitive indices (CI) of WT P. aeruginosa (PA14) and the PA0601 mutant (PA0601−) in a murine wound infection model during coculture with S. aureus. Competitive index is defined as the output ratio of P. aeruginosa:S. aureus after 4 d within the chronic wound divided by the input ratio of P. aeruginosa:S. aureus used to initiate the infection at day 1. Each symbol (● or ■) represents values obtained from infection of an individual mouse. P < 0.01 by Mann-Whitney U test for PA14 CI compared with PA0601− CI, n = 12. The number of cfu/g of wound tissue for WT P. aeruginosa (average 5 × 108, SD 3 × 108) and the PA0601 mutant (average 2 × 108, SD 0.8 × 108) in coculture infections after 4 d were not significantly different, P = 0.125 via Mann-Whitney U test n = 12.
Discussion
Infections containing more than one microbe are the norm rather than the exception. The clinical significance of these infections is profound because they are often more severe and recalcitrant to therapeutic intervention than monoculture infections (2, 6, 7, 9–14). One of the most prevalent opportunistic pathogens found in polymicrobial infections is P. aeruginosa, and there is evidence that P. aeruginosa enhances virulence during coinfection with Gram-positive bacteria (10, 22, 23). Although the implications of coculture on P. aeruginosa pathogenesis are clear, mechanistic insights into this phenomenon are lacking. The goal of this manuscript was to elucidate the mechanism of enhanced P. aeruginosa virulence during coculture with Gram-positive bacteria. Our data reveal that peptidoglycan originating from commensal Gram-positive bacteria serves as a cue that enhances P. aeruginosa virulence in a Drosophila model of infection. The source of peptidoglycan in these experiments was the native Gram-positive flora in the Drosophila crop, indicating that commensal bacteria can potentiate virulence of an opportunistic pathogen (Fig. 2). Our data also show that P. aeruginosa peptidoglycan sensing impacts the composition of the microbial community, reducing the numbers of Gram-positive bacteria at the infection site in both the Drosophila model and a murine chronic wound model (Figs. 3 and 4).
Although previous work clearly shows that the GlcNAc component of peptidoglycan is necessary and sufficient for enhanced P. aeruginosa toxin production (24), there are of course other sources of GlcNAc in the animal hosts used in this study. For example, GlcNAc is a common component of chitin and mucin-like proteins in Drosophila (45–47) as well as mucin, hyaluronic acid, and serum in mammals (48–51). Indeed, chitin and mucin both stimulate pyocyanin production in P. aeruginosa (Fig. S1). However, the fact that P. aeruginosa displays reduced killing of antibiotic-treated flies (Fig. 2B) suggests that host-derived GlcNAc is not sufficient to enhance virulence in the Drosophila model. We favor peptidoglycan as the source of GlcNAc for two additional reasons: more than 25% of the cell wall is shed by Gram-positive bacteria during growth, indicating that large amounts of this polymer are shed naturally (52); and P. aeruginosa is highly lytic for Gram-positive bacteria (24, 33, 36), a process that would further release peptidoglycan fragments into the local environment. Of course other bacterial-produced GlcNAc polymers could potentially serve as sources of GlcNAc, including poly-GlcNAc exopolysaccharide capsules produced by staphylococci and streptococci (53–55).
This is not the first instance in which peptidoglycan acts as a cue to elicit specific cellular responses. For example, peptidoglycan serves as a signal for spore germination in Bacillus subtilis (56), as a signal to promote hyphal growth in Candida albicans (57), and as a ligand for nucleotide-binding oligomerization domain-containing protein 2 (NOD2), a protein receptor that plays a key role in innate immunity in many eukaryotes, including humans (58). It is likely that P. aeruginosa did not evolve the response to peptidoglycan in the context of human infection, but more likely as a means of prokaryotic surveillance (similar to NOD2 in eukaryotes) in the natural environment. The benefit of this behavior may be to eliminate competitors or provide additional nutrients for growth. This latter proposal is supported by the fact that P. aeruginosa uses lysed Gram-positive bacteria as a source of iron during coculture growth in vivo (36).
A key aspect of this work was the identification of PA0601, encoding a putative two-component response regulator critical for sensing and/or responding to GlcNAc and peptidoglycan. Importantly, inactivation of this gene did not affect production of PQS-controlled virulence factors during monoculture growth but eliminated enhanced production of these factors in the presence of GlcNAc or peptidoglycan (Fig. 1). Thus, this strain provided a means of demonstrating the critical role of GlcNAc/peptidoglycan sensing in P. aeruginosa virulence exclusively during coinfection with Gram-positive bacteria. Although this work did not elucidate the mechanism of P. aeruginosa GlcNAc sensing, it is clear that PA0601 functions upstream of PQS signaling because the PA0601 mutant did not enhance PQS production in the presence of GlcNAc (Fig. 1C). Determining whether PA0601 is a direct regulator of PQS signaling and whether this putative regulatory protein responds directly to GlcNAc is a current area of interest.
The finding that P. aeruginosa uses peptidoglycan as a cue to enhance virulence provides new insights into pathogenesis of P. aeruginosa in polymicrobial infections. These data also suggest that therapeutic strategies targeting Gram-positive bacteria may be a viable approach for treating P. aeruginosa-dominated polymicrobial infections, including chronic wounds. Finally our data also suggest that opportunistic pathogens such as P. aeruginosa have evolved strategies to respond to microbes in their natural environment, and these responses may potentiate the course of the infection in the presence of commensal bacteria.
Materials and Methods
Bacterial Strains and Media.
Bacterial strains and plasmids are listed in Table S2, and growth conditions are described in SI Materials and Methods.
DNA Manipulations.
DNA manipulations were performed using standard procedures (59). DNA sequencing was performed at the UT-Austin Institute for Cell and Molecular Biology DNA Core Facility.
Screen for GlcNAc-Unresponsive Mutants.
The 5760 transposon mutants from the PA14 nonredundant mutant library (25) were screened for their ability to respond to GlcNAc via increased pyocyanin production. Details are in SI Materials and Methods.
Gram-Positive Lysis Assays.
Quantification of Pyocyanin, PQS, and Elastase Activity.
Complementation of the PA0601 Mutant.
See SI Materials and Methods. PCR primer sequences are listed in Table S3.
P. aeruginosa–S. aureus Cocultures.
P. aeruginosa strains and S. aureus Mu50 were grown as monocultures or in coculture in a chemically defined medium (4) with glucose as a carbon source. The cfu/mL were determined by differential isolation of P. aeruginosa and S. aureus on Pseudomonas isolation agar and Baird Parker agar (Remel), respectively.
P. aeruginosa Oral Infection of Drosophila.
Drosophila melanogaster infections were performed as previously described (39, 41). Details are in SI Materials and Methods. To generate Drosophila lacking native Gram-positive flora, adult male flies were grown on yeast extract cornmeal media containing ampicillin, vancomycin, and erythromycin (each at 50 µg/mL) for 3 d, followed by growth on antibiotic-free media for 1 d. Total culturable bacteria were enumerated using viable counts on Brain Heart Infusion Agar, and Gram-positive bacteria were enumerated using phenylethyl alcohol agar (Remel). Viable counts were performed from crops excised from five flies crushed in sterile PBS.
Reverse Transcriptase PCR.
See SI Materials and Methods. PCR primer sequences are listed in Table S3.
Chronically Wounded Mouse Model.
Mice were treated humanely and in accordance with protocol #07044 approved by the Animal Care and Use Committee at Texas Tech University Health Sciences Center (Lubbock, TX). Female, Swiss Webster mice (Charles River Laboratories) weighing 20 g were administered chronic wounds as previously described (44). Briefly mice were anesthetized, shaved, and administered a dorsal, full-thickness, 1.5 × 1.5 cm surgical excision wound. The wounds were covered with a transparent, semipermeable polyurethane dressing (OPSITE; Smith & Nephew), which allowed for daily inspection of the wound, topical application of bacteria onto the wound, and protection from other contaminating bacteria. In addition, the OPSITE dressing is a mechanical barrier to wound contraction, physically holding the wound open, consequently resulting in a slow-healing wound. For monoculture infections, 103–104 CFU of P. aeruginosa or S. aureus were injected under the dressing, on top of the wound. Coculture infections administered equivalent numbers (103–104 CFU) of P. aeruginosa and S. aureus. Mice were killed at 4 d after infection and wound tissue harvested, weighed, and homogenized in sterile PBS. Bacteria were enumerated on P. aeruginosa and S. aureus selective agar to determine the cfu/g tissue.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant 5R01AI075068 (to M.W.). M.W. is a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214550110/-/DCSupplemental.
References
- 1.Pasteur L, Joubert J. Charbon et septicemie. Compt Rend Acad. 1877;85:101–105. [Google Scholar]
- 2.Armbruster CE, et al. Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. MBio. 2010;1(3):e00102–e00110. doi: 10.1128/mBio.00102-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakaletz LO. Developing animal models for polymicrobial diseases. Nat Rev Microbiol. 2004;2(7):552–568. doi: 10.1038/nrmicro928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramsey MM, Rumbaugh KP, Whiteley M. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 2011;7(3):e1002012. doi: 10.1371/journal.ppat.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramsey MM, Whiteley M. Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc Natl Acad Sci USA. 2009;106(5):1578–1583. doi: 10.1073/pnas.0809533106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weimer KE, et al. Coinfection with Haemophilus influenzae promotes pneumococcal biofilm formation during experimental otitis media and impedes the progression of pneumococcal disease. J Infect Dis. 2010;202(7):1068–1075. doi: 10.1086/656046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weimer KE, et al. Divergent mechanisms for passive pneumococcal resistance to β-lactam antibiotics in the presence of Haemophilus influenzae. J Infect Dis. 2011;203(4):549–555. doi: 10.1093/infdis/jiq087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjornson HS, et al. Association between microorganism growth at the catheter insertion site and colonization of the catheter in patients receiving total parenteral nutrition. Surgery. 1982;92(4):720–727. [PubMed] [Google Scholar]
- 9.Chen PB, Davern LB, Katz J, Eldridge JH, Michalek SM. Host responses induced by co-infection with Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans in a murine model. Oral Microbiol Immunol. 1996;11(4):274–281. doi: 10.1111/j.1399-302x.1996.tb00181.x. [DOI] [PubMed] [Google Scholar]
- 10.Dalton T, et al. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS ONE. 2011;6(11):e27317. doi: 10.1371/journal.pone.0027317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kesavalu L, Holt SC, Ebersole JL. Virulence of a polymicrobic complex, Treponema denticola and Porphyromonas gingivalis, in a murine model. Oral Microbiol Immunol. 1998;13(6):373–377. doi: 10.1111/j.1399-302x.1998.tb00694.x. [DOI] [PubMed] [Google Scholar]
- 12.Mastropaolo MD, et al. Synergy in polymicrobial infections in a mouse model of type 2 diabetes. Infect Immun. 2005;73(9):6055–6063. doi: 10.1128/IAI.73.9.6055-6063.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagashima H, Takao A, Maeda N. Abscess forming ability of streptococcus milleri group: Synergistic effect with Fusobacterium nucleatum. Microbiol Immunol. 1999;43(3):207–216. doi: 10.1111/j.1348-0421.1999.tb02395.x. [DOI] [PubMed] [Google Scholar]
- 14.Peters BM, Jabra-Rizk MA, O’May GA, Costerton JW, Shirtliff ME. Polymicrobial interactions: impact on pathogenesis and human disease. Clin Microbiol Rev. 2012;25(1):193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samson D, Lefevre F, Aronson N. Wound-healing technologies: Low-level laser and vacuum-assisted closure. Evid Rep Technol Assess (Summ) 2004;(111):1–6. [PMC free article] [PubMed] [Google Scholar]
- 16.Davies CE, et al. Use of 16S ribosomal DNA PCR and denaturing gradient gel electrophoresis for analysis of the microfloras of healing and nonhealing chronic venous leg ulcers. J Clin Microbiol. 2004;42(8):3549–3557. doi: 10.1128/JCM.42.8.3549-3557.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dowd SE, et al. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP) PLoS ONE. 2008;3(10):e3326. doi: 10.1371/journal.pone.0003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gjødsbøl K, et al. Multiple bacterial species reside in chronic wounds: A longitudinal study. Int Wound J. 2006;3(3):225–231. doi: 10.1111/j.1742-481X.2006.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirketerp-Møller K, et al. Distribution, organization, and ecology of bacteria in chronic wounds. J Clin Microbiol. 2008;46(8):2717–2722. doi: 10.1128/JCM.00501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fazli M, et al. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J Clin Microbiol. 2009;47(12):4084–4089. doi: 10.1128/JCM.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halbert AR, Stacey MC, Rohr JB, Jopp-McKay A. The effect of bacterial colonization on venous ulcer healing. Australas J Dermatol. 1992;33(2):75–80. doi: 10.1111/j.1440-0960.1992.tb00083.x. [DOI] [PubMed] [Google Scholar]
- 22.Duan K, Dammel C, Stein J, Rabin H, Surette MG. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol Microbiol. 2003;50(5):1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x. [DOI] [PubMed] [Google Scholar]
- 23.Sibley CD, et al. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 2008;4(10):e1000184. doi: 10.1371/journal.ppat.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korgaonkar AK, Whiteley M. Pseudomonas aeruginosa enhances production of an antimicrobial in response to N-acetylglucosamine and peptidoglycan. J Bacteriol. 2011;193(4):909–917. doi: 10.1128/JB.01175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liberati NT, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci USA. 2006;103(8):2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Essar DW, Eberly L, Hadero A, Crawford IP. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: Interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol. 1990;172(2):884–900. doi: 10.1128/jb.172.2.884-900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J, Carney BF, Denny TP, Weissinger AK, Schell MA. A complex network regulates expression of eps and other virulence genes of Pseudomonas solanacearum. J Bacteriol. 1995;177(5):1259–1267. doi: 10.1128/jb.177.5.1259-1267.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diggle SP, et al. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem Biol. 2007;14(1):87–96. doi: 10.1016/j.chembiol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 29.Diggle SP, et al. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol Microbiol. 2003;50(1):29–43. doi: 10.1046/j.1365-2958.2003.03672.x. [DOI] [PubMed] [Google Scholar]
- 30.Pesci EC, et al. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96(20):11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barequet IS, Ben Simon GJ, Safrin M, Ohman DE, Kessler E. Pseudomonas aeruginosa LasA protease in treatment of experimental staphylococcal keratitis. Antimicrob Agents Chemother. 2004;48(5):1681–1687. doi: 10.1128/AAC.48.5.1681-1687.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kessler E, Safrin M, Olson JC, Ohman DE. Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J Biol Chem. 1993;268(10):7503–7508. [PubMed] [Google Scholar]
- 33.Machan ZA, Taylor GW, Pitt TL, Cole PJ, Wilson R. 2-Heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J Antimicrob Chemother. 1992;30(5):615–623. doi: 10.1093/jac/30.5.615. [DOI] [PubMed] [Google Scholar]
- 34.Mahajan-Miklos S, Rahme LG, Ausubel FM. Elucidating the molecular mechanisms of bacterial virulence using non-mammalian hosts. Mol Microbiol. 2000;37(5):981–988. doi: 10.1046/j.1365-2958.2000.02056.x. [DOI] [PubMed] [Google Scholar]
- 35.Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999;96(1):47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 36.Mashburn LM, Jett AM, Akins DR, Whiteley M. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol. 2005;187(2):554–566. doi: 10.1128/JB.187.2.554-566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Norman RS, Moeller P, McDonald TJ, Morris PJ. Effect of pyocyanin on a crude-oil-degrading microbial community. Appl Environ Microbiol. 2004;70(7):4004–4011. doi: 10.1128/AEM.70.7.4004-4011.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rahme LG, et al. Plants and animals share functionally common bacterial virulence factors. Proc Natl Acad Sci USA. 2000;97(16):8815–8821. doi: 10.1073/pnas.97.16.8815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mulcahy H, Sibley CD, Surette MG, Lewenza S. Drosophila melanogaster as an animal model for the study of Pseudomonas aeruginosa biofilm infections in vivo. PLoS Pathog. 2011;7(10):e1002299. doi: 10.1371/journal.ppat.1002299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alibaud L, et al. Pseudomonas aeruginosa virulence genes identified in a Dictyostelium host model. Cell Microbiol. 2008;10(3):729–740. doi: 10.1111/j.1462-5822.2007.01080.x. [DOI] [PubMed] [Google Scholar]
- 41.Chugani SA, et al. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2001;98(5):2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang H, Duan J, Sibley CD, Surette MG, Duan K. Identification of mutants with altered phenazine production in Pseudomonas aeruginosa. J Med Microbiol. 2011;60(Pt 1):22–34. doi: 10.1099/jmm.0.022350-0. [DOI] [PubMed] [Google Scholar]
- 43.Cox CR, Gilmore MS. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect Immun. 2007;75(4):1565–1576. doi: 10.1128/IAI.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rumbaugh KP, et al. Quorum sensing and the social evolution of bacterial virulence. Curr Biol. 2009;19(4):341–345. doi: 10.1016/j.cub.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 45.Merzendorfer H, Zimoch L. Chitin metabolism in insects: Structure, function and regulation of chitin synthases and chitinases. J Exp Biol. 2003;206(Pt 24):4393–4412. doi: 10.1242/jeb.00709. [DOI] [PubMed] [Google Scholar]
- 46.Theopold U, et al. Helix pomatia lectin, an inducer of Drosophila immune response, binds to hemomucin, a novel surface mucin. J Biol Chem. 1996;271(22):12708–12715. doi: 10.1074/jbc.271.22.12708. [DOI] [PubMed] [Google Scholar]
- 47.Tian E, Ten Hagen KG. Recent insights into the biological roles of mucin-type O-glycosylation. Glycoconj J. 2009;26(3):325–334. doi: 10.1007/s10719-008-9162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dekker J, Aelmans PH, Strous GJ. The oligomeric structure of rat and human gastric mucins. Biochem J. 1991;277(Pt 2):423–427. doi: 10.1042/bj2770423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JK, Pierce M. Purification and characterization of human serum N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase. Arch Biochem Biophys. 1995;319(2):413–425. doi: 10.1006/abbi.1995.1312. [DOI] [PubMed] [Google Scholar]
- 50.Wagner S, et al. Morphological and molecular characterization of human gastric mucous cells in long-term primary culture. Pflugers Arch. 1998;436(6):871–881. doi: 10.1007/s004240050717. [DOI] [PubMed] [Google Scholar]
- 51.Weissmann B, Meyer K, Sampson P, Linker A. Isolation of oligosaccharides enzymatically produced from hyaluronic acid. J Biol Chem. 1954;208(1):417–429. [PubMed] [Google Scholar]
- 52.Blümel P, Uecker W, Giesbrecht P. Zero order kinetics of cell wall turnover in Staphylococcus aureus. Arch Microbiol. 1979;121(2):103–110. doi: 10.1007/BF00689972. [DOI] [PubMed] [Google Scholar]
- 53.Domenech M, García E, Prieto A, Moscoso M. Insight into the composition of the intercellular matrix of Streptococcus pneumoniae biofilms. Environ Microbiol. 2012 doi: 10.1111/j.1462-2920.2012.02853.x. [DOI] [PubMed] [Google Scholar]
- 54.Heilmann C, et al. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20(5):1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 55.Maira-Litrán TA, et al. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect Immun. 2002;70(8):4433–4440. doi: 10.1128/IAI.70.8.4433-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shah IM, Laaberki MH, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135(3):486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu XL, et al. Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe. 2008;4(1):28–39. doi: 10.1016/j.chom.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 58.Girardin SE, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278(11):8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 59.Ausubel F, et al. Short Protocols in Molecular Biology: A Compendium of Methods from Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1997. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.