Abstract
While the quality of the current CHARMM22/CMAP additive force field for proteins has been demonstrated in a large number of applications, limitations in the model with respect to the equilibrium between the sampling of helical and extended conformations in folding simulations have been noted. To overcome this, as well as make other improvements in the model, we present a combination of refinements that should result in enhanced accuracy in simulations of proteins. The common (non Gly, Pro) backbone CMAP potential has been refined against experimental solution NMR data for weakly structured peptides, resulting in a rebalancing of the energies of the α-helix and extended regions of the Ramachandran map, correcting the α-helical bias of CHARMM22/CMAP. The Gly and Pro CMAPs have been refitted to more accurate quantum-mechanical energy surfaces. Side-chain torsion parameters have been optimized by fitting to backbone-dependent quantum-mechanical energy surfaces, followed by additional empirical optimization targeting NMR scalar couplings for unfolded proteins. A comprehensive validation of the revised force field was then performed against data not used to guide parametrization: (i) comparison of simulations of eight proteins in their crystal environments with crystal structures; (ii) comparison with backbone scalar couplings for weakly structured peptides; (iii) comparison with NMR residual dipolar couplings and scalar couplings for both backbone and side-chains in folded proteins; (iv) equilibrium folding of mini-proteins. The results indicate that the revised CHARMM 36 parameters represent an improved model for the modeling and simulation studies of proteins, including studies of protein folding, assembly and functionally relevant conformational changes.
Keywords: Molecular dynamics simulation, NMR spectroscopy, empirical energy function, protein folding
Introduction
Accurate force field parameters are essential for the application of empirical potential energy functions to study protein structure, stability, folding and function. As a result of many years of careful refinement, current additive protein energy functions are of sufficient quality that they may be used predictively for studying protein dynamics, protein-protein interactions and in pharmacological applications.1 It is clear that the next major step in advancing protein force field accuracy requires a different representation of the molecular energy surface: in particular, the effects of charge polarization must be included, as fields induced by ions, solvent, other macromolecules and the protein itself will affect the charge distribution2–5. However, since it may be some time before such energy functions are widely available, and computationally accessible for mainstream use, it is important to ensure the highest possible accuracy of current “additive” force fields. Indeed, over the last few years more rigorous evaluation of force fields in the context of conformational sampling of polypeptides and protein folding have lead to new variants of the AMBER and CHARMM22 force fields, including AMBER ff036, ff03*7 and ff03w8, AMBER ff99SB9, ff99SBnmr110, ff99SB*7, ff99SB*-ILDN11, ff99SB*-ILDN-Q12 and CHARMM C22*13.
From the perspective of protein folding, it has been shown that in long simulations with the additive CHARMM22/CMAP14–16 force field, certain fast-folding proteins will reach the native state, starting from a completely unfolded configuration (e.g. Villin headpiece subdomain)17. However, there are some indications of significant deficiencies: examples include misfolding encountered in long simulations of the pin WW domain starting from an unfolded configuration, and differences of the Villin folding mechanism from that inferred experimentally17,18. In the case of the WW domain, free energy calculations subsequently showed that the misfolded states were in fact lower in free energy than the folded state, confirming that the energy function was at fault19. A number of studies have suggested that such discrepancies may be largely due to relatively small inaccuracies in the potential for the backbone, resulting in a net “bias” toward either structure, which may be corrected by means of a minor adjustment to the backbone potential7,20–22.
Here we report a revised set of CHARMM23 all-atom protein force field parameters (C36) that represents a significant improvement in representing the potential energy surface of proteins, within the context of the current functional form. The changes described in the present work are (i) a new backbone CMAP potential, refined against a range of data for dipeptides as well as experimental data on small peptides such as hairpins and helices and (ii) new side-chain dihedral potentials optimized against quantum mechanical energies from dipeptides and NMR data from unfolded proteins. Other differences from the previous C22/CMAP protein force field include previously published revised Lennard-Jones (LJ) parameters for aliphatic hydrogens24, improved treatment of the internal parameters for the guanidinium ion25 and new parameters for tryptophan26. The backbone and side-chain improvements have been undertaken in parallel such that the new force field is balanced with respect to the contribution of these moieties to protein structure and dynamics. Validation of the C36 force field against a wide range of test systems, including comparison with other state-of-the-art protein force fields, indicates improvements in the quality of the force field in reproducing a number of experimental observables.
Methods
1. Simulation Methods
Several simulation codes were used to perform the calculations in this work, reflecting the packages in which the CHARMM force field is currently implemented: CHARMM23 itself, NAMD227, and GROMACS 4.5.328. All molecular dynamics (MD) simulations were performed in explicit water with periodic boundary conditions, and with long range electrostatics treated using Particle-Mesh Ewald (PME). Non-bonded pair interactions were treated similarly in all packages, as described below. The water model used was TIP3P29: for simulations with CHARMM and NAMD2, a modified TIP3P including Lennard-Jones parameters for hydrogen30 was used, whilst for GROMACS, the standard TIP3P29 was used. Comparison of results with the two TIP3P variants for the Ala5 and Ac-(AAQAA)3-NH2 test cases discussed below showed negligible differences.
(i) Simulations of Ala5 and other short peptides
Ala3, Ala5, Ala7, Val3, and Gly3 were simulated in the NPT ensemble at 298K and 1 atm pressure under periodic boundary conditions. All peptides were unblocked and had protonated C-termini (experimental pH is ~2)31. Initial box sizes were 32.13 Å3, 34.56 Å3, and 38.34 Å3 for tri-, penta-, and heptapeptides, respectively. PME summation32 was used to calculate the electrostatic interactions with a real-space cutoff set to 12 Å and a 1 Å grid spacing while the LJ interactions were treated with a switching function from 10 to 12 Å. The equations of motion were integrated with a 2 fs time step while SHAKE was used to constrain covalent bonds involving non-water hydrogen bonds and SETTLE33 was used to maintain rigid water geometries. All of the peptides were simulated for 400 ns each with the new force field. In addition, Ala5 was also simulated for 200 ns with the previous C22/CMAP force field16, Amber ff99SB9, Amber ff99SB*7, OPLS-AA34, and Gromos 53a635. All of the simulations were carried out with NAMD version 2.7b2. The equilibration protocol for all of the simulations consisted of initial minimization followed by step-wise heating to 298K. Simulations of zwitterionic GPGG were run using GROMACS with a 30 Å cubic box for 100 ns at 300 K, using the same non-bonded treatment, thermostat and barostat as those for Ac-(AAQAA)3-NH2 below.
(ii) Replica exchange simulations of Ac-(AAQAA)3-NH2
The N-terminally acetylated and C-terminally amidated peptide, initially in a helical conformation, was solvated in a truncated octahedron simulation cell with a distance between nearest faces of 42 Å using GROMACS 4.5.3. The resulting system contained 1833 water molecules. It was found that a larger box size was necessary than was used in earlier work with Amber force fields7, as the coil state was more expanded. A 200 ps simulation was run at a constant pressure of 1 bar to obtain equilibrium box dimensions at 300 K. The peptide was then unfolded using a 5 ns constant volume simulation at 800 K. Starting from the final configuration from this simulation (in which all peptide bonds were trans), constant volume replica exchange MD was run using GROMACS 4.5.3, with 32 replicas spanning a temperature range from 278 to 416 K and exchange attempts every 10 ps, for a total of 150 ns per replica. Electrostatic interactions were computed with PME using a real-space cut-off of 12 Å and a 1 Å grid spacing and Lennard-Jones interactions were switched off smoothly between 10 and 12 Å. A Langevin thermostat with a friction coefficient of 1 ps−1 was used, with a time step of 2 fs to integrate the equations of motion. A residue was defined as being helical if it lay within a stretch of at least three residues in the α-helical region of the Ramachandran map, defined as |−65°−φ|< 35° and |−37° −ψ|< 30°. To define residues which lay within the wider α minimum (but not necessarily part of a helix), a larger range of angles, denoted α+, was used, i.e. −160° < φ < −120° and −120° <ψ < 50°.
(iii) Solute tempering simulation of unfolded proteins in urea
Simulations of unfolded proteins (GB1 and ubiquitin) in urea were performed at a constant pressure of 1 bar using a truncated octahedron cell with a distance between nearest faces of 70 Å using GROMACS 4.5.3. The protein was solvated in an 8 M solution of urea in TIP3P water, with the KBFF36 model being used for urea, based on the favorable results obtained in peptide simulations using this model37. Four potassium ions were added to the GB1 simulation to neutralize the overall charge. Simulations of ~100 ns duration of the unfolded proteins were found to be insufficient to get a representative sampling of side-chain rotamer distributions, particularly for the bulky aromatic side chains. Since for systems of this size (~ 24000 atoms), standard replica exchange would require a very large number of replicas, we instead used the solute tempering replica exchange proposed by Liu et al38. In this scheme, the pairwise forces are split into protein-protein, protein-water and water-water interactions, and the energy of the water-water interactions is scaled in such a way that the water-water energy vanishes from the replica exchange acceptance criterion. Specifically, the potential energy of replica m is given by 39:
| [1] |
In equation 1, Epp is the sum over all pair interactions between two atoms that are both in the protein, and, similarly, Eww and Epw are the sums over water-water and protein-water pair interactions. The particular scaling39 given in the above equation can be implemented by scaling all “solvent” (urea, water and ions) charges by (β0/βm)1/2 and angle terms (urea only) and LJ ε (urea, water and ions) by β0/βm, for replica m, where βm=1/kBTm with kB being Boltzmann’s constant and Tm the temperature of replica m. Replica 0 is the replica for which the correct equilibrium distribution is desired. Sampling was done via a general Hamiltonian replica exchange utility implemented in a modified version of GROMACS 4.5.3. Simulation parameters were the same as for Ac-(AAQAA)3-NH2, with a 1 ps interval between replica exchange attempts.
(iv) Replica exchange simulations of HEWL19
REMD simulations were also performed for the unblocked peptide HEWL19, derived from hen egg-white lysozyme with sequence KVFGRC(SMe)ELAAAMKRHGLDN. The structure and parameters for the S-methylated Cys 6 were adapted from those for methionine and are given in the Supporting Information (SI), Figure S1 and Table S1 respectively. Both termini as well as all acidic side chains were protonated, corresponding to the experimental conditions of pH 231. The peptide was solvated in a truncated octahedron simulation cell with a 42 Å distance between nearest faces, and equilibrated at a constant pressure of 1 bar for 200 ps at 300 K. Constant volume REMD was run with 32 replicas spanning the temperature range 278–472 K, for 50 ns, of which the first 10 ns were discarded in the analysis. All other simulation parameters were the same as for Ac-(AAQAA)3-NH2.
(v) Crystal Structure Simulations
Simulations of the proteins in their crystal environments (Table 1), which were used previously during optimization of the C22/CMAP force field 40, were performed using CHARMM on full unit cells with added waters and counterions to fill the vacuum space. Once the full unit cell was constructed based on the coordinates in the protein databank, a box of water with dimensions that encompassed the full unit cell was overlaid onto the crystal coordinates while preserving crystal waters, ions, and ligands. Water molecules with oxygen within 2.8 – 4.0 Å of any of the crystallographic non-hydrogen atoms were removed, as described below, as well as those occupying space beyond the full unit cell. To neutralize the total charge of each system, sodium or chloride ions were added to the system at random locations at least 3.0 Å from any crystallographic non-hydrogen atom or previously added ions and 0.5 Å from any water oxygen. Final selection of the water molecule deletion distance was performed by initially applying a 2.8 Å criteria to all systems followed by system equilibration and an NPT production run of 5 ns following which the lattice parameters were analyzed. The deletion distances were then increased and the equilibration and 5 ns production NPT simulation were repeated until the final lattice parameters were in satisfactory agreement with experimental data. The final water deletion distances and unit cell parameters from the full 40 ns production simulations are presented in Table S2 of the SI. For the minimization and MD simulations, electrostatic interactions were treated with PME using a real space cutoff of 10 Å. The LJ interactions were included with force switching from 8 Å to 10 Å, while the list of nonbonded atoms was kept for interatomic distances of up to 14 Å and updated heuristically. Each crystal system was first minimized with 100 steps of steepest-decent (SD) with non-water, non-ion crystallographic atoms held fixed followed by 200 steps of SD with harmonic positional restraints of 5 kcal/mol/Å2 on solute non-hydrogen atoms. The minimized system was then subject to an equilibration phase consisting of 100 ps of NVT simulation41 in the presence of harmonic positional restraints followed by 5 ns (100 ps for 135L and 3ICB) of fully relaxed NVT simulation with a time step of 2 fs. During the simulations all covalent bonds involving hydrogens were constrained using SHAKE42. Production phase simulations were conducted for 40 ns in the isothermal and isobaric NPT ensemble43. The only symmetry enforced was translational (i.e. periodic boundaries). Reference temperatures were set to match the crystallographic conditions (Table S2) and maintained by the Nosé-Hoover thermostat with a thermal piston mass of 1,000 kcal ps2/mol while a pressure mass of 600 amu was used with the Langevin piston. The first 5 ns of the production simulations were considered as equilibration and therefore discarded from analysis, which was performed on coordinate sets saved every 5 ps. The boundaries for α helices and β strands were obtained from a consensus of author annotations and structural assignments calculated by DSSP44 and STRIDE45 from the crystal structures.
Table 1.
Protein crystal test systems
| PDB ID | Protein | Resolution Å | Unit Cell | Comments | Reference |
|---|---|---|---|---|---|
| 135L | Lysozyme | 1.3 | Monoclinic (110.06°) | α-helix, β-sheet, αL, 298K | 86 |
| 3EBX | Erabutoxin b | 1.4 | Orthorhombic | antiparallel β-sheet, 298K | 81 |
| 1EJG | Crambin | 0.54 | Monoclinic (90.47) | α-helix, β-sheet, 100K | 82 |
| 1I27 | RAP74 Subunit | 1.02 | Orthorhombic | α-helix, β-sheet, 282K | 87 |
| 5PTI | Trypsin inhibitor | 1.00 | Orthorhombic | α-helix, β-sheet, 298K | 88 |
| 3ICB | Calcium-binding protein | 2.30 | Orthorhombic | 2 EF-hands, α-helix, 298K | 89 |
| 1BYI | Dethiobiotin Synthase | 0.97 | Monoclinic (107.24) | α-helix, parallel β-sheet, 100K | 90 |
| 1BZP | Deoxy and CO myoglobin | 1.15 | Monoclinic (105.58) | α-helix, 298K | 91 |
(vi) Simulation of a dimeric coiled-coil in solution
Simulation of a dimeric coiled-coil in solution was initiated from model 1 of the NMR structures in 1UOI.46 System preparation using CHARMM, with inputs initially generated using the CHARMM-GUI,47 involved solvating the dimer with an aqueous solution of approximately 150 mM KCl. All non-hydrogen atoms in the peptides were then harmonically restrained (5 kcal/mol/Å2) and the system subjected to a 500 step SD minimization followed by a 50 ps NPT simulation at 298 K in the presence of the restraints. The final coordinate set was used to initiate the production simulation using NAMD with the coordinates saved every 10 ps for analysis. Simulations involved a 2 fs time step, treatment of electrostatics via PME with a real space cutoff of 12 Å with smoothing of the LJ interactions via a switching function over 10 to 12 Å.
(vii) Simulations of folded proteins in solution
Simulations using Gromacs 4.5.3 were carried out for the following four folded globular proteins: bovine pancreatic trypsin inhibitor (BPTI), ubiquitin, GB3 and hen Lysozyme, starting from the published experimental structures (PDB entries 5PTI48, 1UBQ49, 1P7E50, 6LYT51 respectively). Each protein was solvated in a truncated octahedron simulation cell filled with TIP3P water, with nearest distance between images of 45 Å for all proteins except for lysozyme, for which the distance was 60 Å. Sodium and chloride ions were added as needed to yield a final salt concentrations of ~100 mM, with adjustments to ensure charge neutrality. For each protein, a 100-step SD energy minimization of the whole system was performed, followed by 200 ps of MD at a constant pressure of 1 bar and temperature of 300 K, in which harmonic positional restraints of 2.39 kcal/mol/Å2 were applied to each Cartesian component of each protein non-hydrogen atom using the minimized structure as a reference. Each protein was then simulated at a constant pressure of 1 bar and a temperature of 300 K for 200 ns. Pressure was regulated by a Parinello-Rahman barostat52 with a coupling time of 2.5 ps; otherwise all details were as described for Ac-(AAQAA)3-NH2 above.
(viii) Replica exchange folding simulation of β-hairpins
REMD simulations were performed on two well-studied β hairpins, the GB1 hairpin (residues 41–56 of protein G)53,54 and GB1m355,56 using Gromacs 4.5.3. Completely unfolded structures obtained from high temperature vacuum simulations were solvated in a 49 Å truncated octahedron box with 2225 water molecules. The REMD simulations spanned a temperature range 278 to 595 K utilizing 40 replicas, sufficient to achieve exchange probability around 0.2, with a frequency of attempted exchanges of 1 ps. An initial sampling of 500 ns per replica was obtained with a preliminary version of the force field, with a further 111 ns per replica obtained with the final parameter set, of which the first 11 ns was discarded as initial equilibration22,57; all other details were as described for Ac-(AAQAA)3-NH2 above.
(ix) Quantum mechanical calculations
Quantum mechanical calculations of the Gly and Pro dipeptides were performed to obtain 2-dimensional (2D) Ramachandran φ,ψ potential energy surfaces. With φ and ψ constrained in 15° increments the dipeptides were optimized at the MP2/aug-cc-pVDZ level of theory using Gaussian 0358,59. The optimized structures were then subjected to single point energy determinations at the RIMP2/cc-pVTZ and RIMP2/cc-pVQZ levels of theory using QCHEM.58 These energies were then used to extrapolate to the complete basis set (CBS) limit using the method of Halkier et al. 60 For the Pro dipeptide the surface was limited to φ = −180 to 30° as the remainder of the surface is energetically highly unfavorable. The resulting Gly and Pro RIMP2/CBS//MP2/aug-cc-pVDZ CMAPs were then calculated from the QM surfaces as previously described.15,16
2. Optimization methods
Backbone Optimization
Optimization of backbone parameters was done via the 2D CMAP potential VCMAP(ϕ,ψ) 15,16, for which separate parameters are used for each of (i) non-Gly, Pro residues, (ii) Gly and (iii) Pro. For non-Gly, Pro residues the starting point was the C22/CMAP potential. This CMAP was further optimized, as described further below, to obtain an acceptable match to NMR data for both a short peptide (Ala5) and a longer helix-forming peptide Ac-(AAQAA)3-NH2. The target functions for optimization were the deviations from the experimental observables, i.e. 3J scalar couplings for Ala5, and carbonyl chemical shifts for Ac-(AAQAA)3-NH2. The target function optimized was defined as:
| [2] |
In equation 2, Fobs,i is the experimental data, Fcalc,i the data calculated from the simulation, σi the error on the ith data point and N the total number of data. The error estimate was based on the uncertainty in the prediction of the experimental observables from the simulation, i.e. the estimated error in the predictions from the Karplus equation and the SPARTA+ algorithm61. The motivation for this choice is that χ2 should be ~1 when the average deviation from experiment is comparable to the error in the prediction, although it differs from the standard definition of the χ2 statistic. In most cases, the error of the experimental measurement and the statistical error from the simulation are small, relative to this prediction error. Typical values for the statistical uncertainty of backbone J-couplings from simulations are ~0.05 Hz (e.g. see Table 6) compared with 0.2–0.9 Hz for the error in the Karplus equation prediction, and for chemical shifts ~0.03 p.p.m statistical error compared with ~1 p.p.m prediction error. Values of σ used for each type of scalar coupling and chemical shift are given in Table S3 of the SI. For side-chain J-couplings, the sampling errors can be more significant, due to the slow transitions about χ torsion angles. For these, we have propagated the statistical error in the J-couplings to report an error on the estimate of χ2. For Gly and Pro, CMAPs based on the respective RIMP2/CBS//MP2/aug-cc-pVDZ energy surfaces were used without additional modification.
Table 6.
Scalar couplings of the central residue (Ala 10) of HEWL19 in different force fields. All J-coupling values in Hz.
| a expt | σ | b ff99SB | b ff99SB* | c C22/CMAP | c C36 | |
|---|---|---|---|---|---|---|
| A10 1JNCα (ψ10) | 10.58 | 0.59 | 10.84 (0.07) | 10.27 (0.03) | 9.78 (0.02) | 10.35 (0.05) |
| A10 2JNCα (ψ10) | 7.24 | 0.50 | 7.50 (0.06) | 7.06 (0.11) | 6.49 (0.06) | 7.03 (0.05) |
| A10 3JHαC (ψ10) | 1.72 | 0.38 | 2.55 (0.13) | 1.82 (0.08) | 1.18 (0.04) | 2.10 (0.10) |
| A10 3JHNC (ψ10) | 1.33 | 0.59 | 1.10 (0.06) | 0.65 (0.03) | 0.77 (0.02) | 0.88 (0.04) |
| A10 3JHNCβ (ψ10) | 2.19 | 0.39 | 2.23 (0.12) | 3.01 (0.06) | 3.73 (0.02) | 2.86 (0.06) |
| A10 3JHNHα (ψ10) | 5.10 | 0.91 | 5.72 (0.16) | 6.46 (0.12) | 4.83 (0.04) | 6.19 (0.14) |
| A10 3JHNCα (ψ10,ψ9) | 0.46 | 0.10 | 0.52 (0.01) | 0.39 (0.02) | 0.19 (0.01) | 0.37 (0.01) |
|
| ||||||
| χ2(J) [Hz2] | 1.3 | 1.0 | 4.7 | 0.6 | ||
|
| ||||||
| % α+ | - | - | 34.9 | 69.4 | 90.3 | 57.3 |
| % β | - | - | 23.5 | 12.9 | 2.66 | 14.6 |
| % ppII | - | - | 27.9 | 15.2 | 6.67 | 18.2 |
| % helix | - | - | 4.5 | 30.9 | 87.0 | 39.4 |
Experimental data from Graf et al.31;
Data from Ref7;
Data from present work. The set of Karplus parameters denoted “DFT2” in Ref20 was used. χ 2 values calculated using equation 2. Statistical errors on individual J-couplings are given in brackets next to the values, based on block error analysis92.
Sidechain χ1 and χ2 dihedral parameter optimization
Dipeptides corresponding to each residue, excluding Gly, Pro and Ala, were generated with the program CHARMM. The dipeptides contain N-acetylated and N′-methylamidated termini and are representative of the amino acid sidechains in the local environment of peptide backbone. For the initial fitting of χ parameters we used a published set of energy surfaces derived from QM calculations62 as target data. The selected QM level, RIMP2/cc-pVTZ//MP2/6-31G* (6-31+G* for the Asp and Glu dipeptides) represents a compromise between accuracy and computational accessability that has been shown to yield conformational energies comparable to CCSD(T)/complete basis sets calculations on complex molecules including those containing anomeric oxygens63. CHARMM potential energy surfaces were generated in a manner consistent with the QM target data which consisted of full 2D scans of the χ1 and χ2 dihedral angles in 15° increments for each dipeptide constrained at each of the following backbone conformations: α (−60.0°/−45.0°), β (−120.0°/120.0°), or αL (63.5°/34.8°). Initial coordinates were obtained from the QM-optimized geometries. Structures were minimized for 500 steps of conjugate-gradient (CONJ) and 500 steps of adopted basis Newton-Raphson (ABNR) with a convergence criteria of the root-mean-square (RMS) gradient < 10−5 kcal/mol/Å with harmonic dihedral restraints of 104 kcal/mol/rad on the respective dihedrals. The resulting χ1/χ2 energy surfaces were offset relative to the respective global minimum for each residue type.
A Monte Carlo simulated annealing (MCSA) automated fitting program64 was used for the optimization of sidechain dihedral parameters. This program allows simultaneous fitting of multiple dihedrals for an arbitrary number of dipeptides as required for fitting parameters for dihedrals common to more than one sidechain. During optimization of the dihedral parameters the multiplicities, n, were restricted to 1, 2 and 3, the force constant, K, was limited to K ≤ 3.0 and the phase δ limited to either 0° or 180°. Energy surfaces from all three backbone conformations were considered together as target data. The objective function of the fitting program, shown in equation 3,
| [3] |
is the RMS energy difference, RMSD, where EQM and EMM are the relative QM and MM energies, respectively, for conformation i, c is an offset to minimize the RMSD and w is a weighting factor for conformation i. In practice, the RMSD is calculated with the force constants on the dihedral parameters to be optimized set to zero such that during each iteration of the MCSA only the dihedral energies need to be evaluated and the change in the RMSD calculated in a computationally expedient fashion. During fitting we applied weighting factors of w = 5 on energy surfaces from α and β backbone conformations and w = 1 for data from αL. Each set of side-chain parameters was fitted with a minimum of 100,000 MCSA steps and an initial temperature of 1,000 K. The initial parameters for condensed phase simulation testing were obtained as the arithmetic mean of five individual MCSA runs that differed by the initial random number seed. Convergence of each run was verified by monitoring the RMSD throughout the optimization. Optimized parameters from the automated fitting program were subsequently subjected to manual adjustments focused on the relative energy of the local minima in the QM surfaces to improve the reproduction of experimental relative rotamer populations in MD simulations as described below.
For comparison of χ1 and χ2 from the crystal simulations with experimental data, crystal distributions for χ1 and χ2 were obtained from a survey of the protein data bank65 with the data converted to probability distributions using a bin size of 15°.62 The analogous probability distributions were calculated from the protein crystal simulations. To measure the agreement between the crystallographic and simulated χ distributions, the 1D overlap coefficients (OC) for the two probability distributions were calculated over the sampled grid points as previously described using equation 4.66,67
| [4] |
3. Calculation of experimental observables
Residual Dipolar Couplings
Residual dipolar couplings for the folded proteins were calculated as described previously,7 by fitting the global alignment tensor that results in the smallest RMS difference between experimental and calculated couplings.
Three-bond Scalar Couplings
Scalar couplings for backbone and sidechains were determined from the torsion angles using Karplus relationships. Alan, Glyn, Valn: both flavodoxin-based Karplus parameters68 and DFT-based Karplus parameters (DFT2) described in Ref69 were used as previously described20. Unfolded proteins in urea: the amino acid-dependent Karplus parameters of Perez et al. for sidechain dihedral χ1 were used.70 Folded proteins: the Perez parameters70 were used for all J-couplings, with the exception of 3JNCG and 3JC′CG couplings, for which the parameters of Chou and Bax were used. 71
Chemical Shifts
Chemical shifts were calculated with the SPARTA+ empirical shift prediction package61 for individual structures and ensemble-averaged over the simulation.
Results: Parameter Optimization
The main aim of the optimization was to improve the parameter set for torsion angles in the force field. This was motivated, in the case of the backbone, by the results of folding simulations that showed the backbone parameters for C22/CMAP to have an excessive helical bias18,19. In addition, since the torsion parameters had never been explicitly optimized in a side-chain specific fashion, it was felt this was an area in which improvements could be made. The backbone and side-chain optimization were initially undertaken independently, and the results then combined – it was found that there was some coupling between the backbone and side-chain parameters as described below. The final optimized C36 parameters may be obtained from the MacKerell Laboratory web page at http://mackerell.umaryland.edu/CHARMM_ff_params.html
Backbone Optimization
The CHARMM additive protein force field contains, in addition to the usual Fourier terms for the backbone torsion angles ϕ and ψ, a two dimensional cubic spline potential or CMAP, VCMAP(ϕ,ψ) 15,16. The spline is specified by energies determined on a 2D lattice with a 15° grid spacing. For internal residues, we only modify the CMAP terms for the backbone, since these effectively include any features of the energy surface that could be captured by the backbone torsion terms (some independent optimization of the backbone torsion angles for terminal Gly residues was carried out as discussed below). The previous CMAP, optimized for use with the C2214 all-atom protein force field, was based on the adiabatic QM vacuum energy surface of alanine dipeptide, with empirical modifications to correct for systematic deviations of protein crystal structure simulations from the X-ray diffraction structure. We had initially aimed to base the new CMAP on the QM dipeptide energy surface once more; however, it was found that the previous empirical corrections captured additional cooperativity that was lacking in the dipeptide-based maps (to be described in a separate publication). We therefore retained the original CMAP as a starting point for further optimization. For completeness, we summarize here the strategy used to derive the C22 CMAP16. The CMAP was initially fitted to minimize the difference between the MM energy and that obtained from the QM surface (calculated with LMP2/cc-pVQZ(-g)//MP2/6-31G*). Although this brought about some improvements, it was found that simulations of proteins in their crystal environment exhibited systematic deviations from the backbone (ϕ,ψ) angles in the experimental structures. A probable cause of this deviation is that the additive energy function cannot capture many-body effects important in folded proteins72. As a compromise, small manual adjustments of the CMAP were made in order to match the minima for regions of canonical secondary structure more closely to those in the PDB. With these changes, the average backbone torsion angles from simulations of folded proteins in their crystal environment now matched those in the experimental structures.
In the present work, the grid energies were initially optimized using an automated Monte Carlo-based protocol to improve agreement with Ala5-based NMR scalar coupling data. Iterative manual adjustments were then made in order to further improve the match with the experimental NMR data for peptides in solution, specifically the following data sets: (i) an extensive set of 3-bond scalar coupling data for Ala5 published by the group of Schwalbe31 and (ii) a set of chemical shift data reflecting helix formation in the 15-residue peptide Ac-(AAQAA)3-NH273. The purpose of the first data set is to capture “intrinsic” propensity for populating the different minima in the Ramachandran map (in the absence of any significant secondary structure), while the second data set is for a peptide with a significant helical population. In each round of optimization, a perturbation approach74 was used to estimate the correction needed to better match experimental data, following which further sampling was performed to test the new parameter set. An important principle that emerged was that one should not apply more than the minimal changes needed to match the experimental data to within the estimated error (in calculating the experimental observables from simulation), since overfitting to one data set may result in deleterious effects. For example, it is possible to match the data for Ala5 more closely by making the polyproline II region of the map even lower in energy, but we found that this has a very destabilizing effect on β-hairpins: even after formation of interstrand hydrogen bonds, the backbone remains in the polyproline II region of the Ramachandran map.
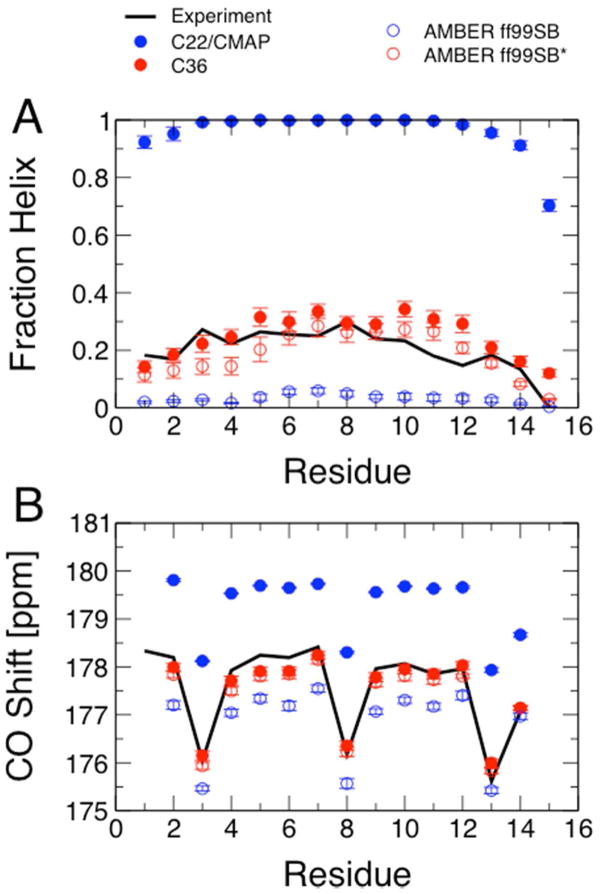
We summarize the final results of the optimization for the two peptides Ala5 and Ac-(AAQAA)3-NH2 in Table 2, together with the analogous statistics for the C22/CMAP15,16, the Amber ff99SB9 and the modified Amber ff99SB* force fields11. We see that the overall agreement with scalar couplings for Ala5 is much improved relative to C22/CMAP, and also relative to ff99SB and ff99SB*. The individual residues sample more of the “polyproline II” helix region of the Ramachandran map (52%) than any of the other force fields, and less of the enlarged α+ alpha-helical region (defined as −160° ≤ ψ ≤ −20° and −120° ≤ ψ ≤ 50°). For the longer, helix-forming peptide, Ac-(AAQAA)3-NH2 we find that the overall fraction helix at 300 K (21%) is much more reasonable than that (95%) obtained with the previous C22/CMAP, and more in line with the experimental estimate of ~19% obtained by Shalongo and Stellwagen from NMR chemical shifts in D2O73 and the similar estimate of ~22% which can be interpolated from the circular dichroism data of Scholtz et al. on the related peptide Ac-(AAQAA)3-Y-NH2 in water75,76. This agreement can also be seen at the level of individual residues, as we show in Figure 1A. Since the fraction helix from experiment is only inferred by fitting a thermodynamic model, we have also calculated carbonyl chemical shifts using the SPARTA+ chemical shift prediction program61. SPARTA+ uses a neural network trained on sets of known folded protein structures and corresponding chemical shifts in order to predict shifts from structure. The estimated error in carbonyl shift prediction is approximately 1 p.p.m. Although the training set does not include weakly structured peptides such as we consider, it does include loops in folded proteins, which are also not in canonical secondary structures. In Figure 1B we compare the predicted shifts with the carbonyl chemical shifts measured in the original experiments73, confirming the improvement afforded by the backbone optimization. A final noteworthy feature of the data for the helix-forming peptide is that the overall fraction helix is slightly higher than that in ff99SB* (21 % vs 14 %), yet the overall population of the α+ region of the Ramachandran map is lower (44 % vs 48 %). This suggests additional cooperativity as mentioned above, which will be explored further in a future publication.
Table 2.
Properties of peptides used in parameter optimization: Ala5 and Ac-(AAQAA)3-NH2. J-couplings calculated with “DFT2” set of Karplus parameters20 and chemical shifts with SPARTA+61 at 300 K. Statistical errors from a block error analysis92 are given in brackets.
| Peptide | Property | ff99SB | ff99SB* | C22/CMAP | C36 |
|---|---|---|---|---|---|
| Ala5 | |||||
| % α+ | 15.7 (1.2) | 22.5 (2.2) | 41.5 (3.2) | 13.1 (1.3) | |
| % β | 37.8 (1.4) | 34.5 (1.3) | 25.3 (2.2) | 30.8 (1.3) | |
| % ppII | 42.3 (1.9) | 39.8 (1.6) | 26.2 (1.8) | 51.9 (1.1) | |
| % α-helix | 0.0 (0.2) | 0.6 (0.3) | 3.7 (2.1) | 0.1 (0.1) | |
| χ2(J) | 1.7 | 1.7 | 2.0 | 1.2 | |
| Ac-(AAQAA)3-NH2 | |||||
|
| |||||
| % α+ | 26.9 (1.2) | 48.5 (1.3) | 98.7 (0.4) | 44.0 (1.8) | |
| % β | 32.4 (0.8) | 22.7 (0.7) | 0.46 (0.2) | 19.1 (0.6) | |
| % ppII | 31.8 (0.9) | 21.8 (0.6) | 0.41 (0.2) | 29.5 (1.2) | |
| % α-helix | 1.8 (0.3) | 14.2 (2.1) | 95.3 (0.1) | 21.0 (1.7) | |
| χ2(δC′) | 0.5 | 0.06 | 2.6 | 0.04 | |
Figure 1.
Helix formation in Ac-(AAQAA)3-NH2. (A) Fraction helix per residue estimated from experiment73 at 300 K (black lines) compared with average fraction helix calculated from REMD simulations with C22/CMAP15,16 (solid blue symbols), C36 (solid red symbols), Amber ff99SB (open blue symbols) and Amber ff99SB* (open red symbols) using the replica closest in temperature to 300 K. (B) Average carbonyl carbon chemical shifts calculated with SPARTA+61 from the simulations in (A) compared with experimental shifts.73 Color scheme as in (A).
Gly and Pro CMAPs were determined by taking the difference between the target QM and force field energies for Gly and Pro dipeptides, respectively. The new CMAPs were already found to result in good agreement with NMR data for Gly3 and GPGG, with a significant improvement for Gly, and were therefore not further adjusted. Since the Gly termini are not affected by the CMAP, we have performed a simple optimization on the N-terminal ψ and C-terminal ϕ torsion parameters. In each case, only a single parameter was optimized, namely a dihedral force constant, in such a way as to minimize the χ2 between simulated and experimental J-couplings for Gly3 (each terminus was independently optimized). The old and new parameters are listed in Table S11 in the SI.
Side-chain optimization
Dihedral parameters that modulate sidechain χ1 and χ2 conformational energies were initially optimized using QM energy surfaces as target data using a MCSA-based fitting protocol64 followed by subsequent empirical optimization targeting the rotamer populations obtained from both solution and crystal simulations, as described below. We note that an alternative method to MCSA for optimizing torsion angle coefficients is linear least squares fitting77. We have opted for MCSA as it is more generally applicable and due to the greater flexibility afforded, such as the inclusion of constraints on parameters as was used in the present study. Dihedral parameter multiplicities of up to the third term are used in conjunction with an upper boundary of 3.0 kcal/mol/deg for the force constant (K). The latter constraint reduces the likelihood of overfitting where individual parameters with large K values cancel each other, thereby maintaining parameter transferability. We also limited the phase (δ) to 0 or 180° such that the resulting parameters are not specific to chirality. In addition, during fitting only those χ1, χ2 conformations with relative energies less than 12.0 kcal/mol were included in the calculation of the RMSD (eq. 3) to avoid fitting the high-energy regions at the expense of the low energy regions that are accessible during MD simulations. Higher-energy cutoffs (20.0 kcal/mol for Arg and Lys or 25.0 kcal/mol for Asp, Glu and Hsp) were used for the charged sidechains as favorable electrostatic interactions with the backbone in selected conformations led to deep minima in the energy surface such that other local minima are 12.0 kcal/mol above the global minima. In addition to the criteria in the preceding paragraph, χ1 and χ2 parameters were grouped based on sidechain type. The groupings are shown in Table S4 of the SI. For example, initially Lys, Arg, Gln, Glu and Met have the same χ1 parameters. Fitting tests were undertaken in which selected atom types were changed to make the parameters for one or more of the sidechains unique (not shown). Based on this analysis, unique atom types were introduced at the Cβ atom of Glu, Asp, and protonated His (Hsp) thereby allowing for improvements in the quality of the fits of those sidechains as well as of the remaining sidechains in the respective groups.
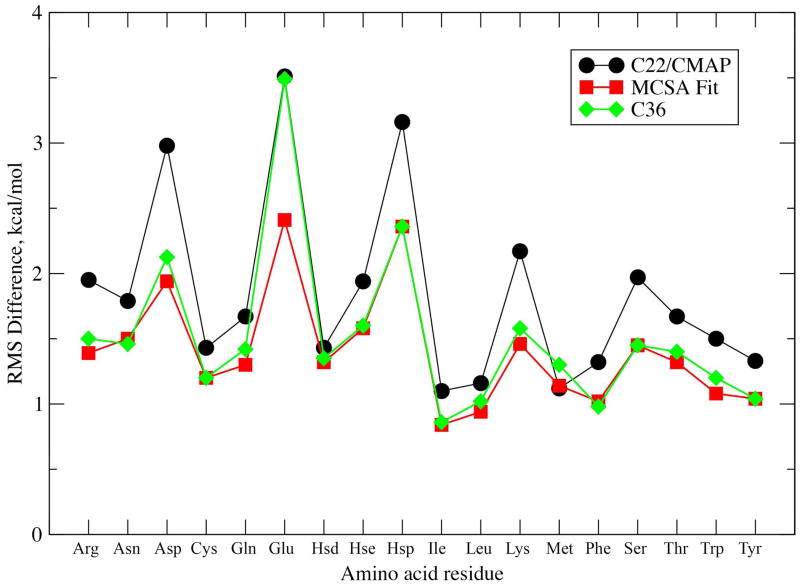
RMS differences between MM and QM energy profiles for those conformations below the relative cutoff energies are shown in Figure 2. As is evident, significant improvement was made in the overall agreement following the initial MCSA fit with respect to C22/CMAP. This is expected as the C22/CMAP χ1/χ2 parameters were typically assigned values associated with alkanes and not explicitly optimized targeting QM or other data. In Table 3 the average and RMS differences in relative energies of the expected χ1/χ2 minima, or rotamers, between MM and QM levels of theory are presented. The individual relative energies and definitions of the minima for the studied amino acids are shown in Tables S5, S6 and S7 of the SI. The results indicate that although the MCSA-fit brought the energies closer to QM as compared to C22, empirical fitting based on reproduction of the χ1/χ2 rotamer distributions from protein simulations (see below) narrowed the gap further for the α and αL backbone conformations. The RMSD values increased with the β conformation due to compromises made to better satisfy the agreement of the other secondary structures and better reproduce sampling in the protein simulations (see below). For most sidechains, the relative energies of the various rotamers are satisfactorily reproduced although the quality of the agreement varied significantly between the different residues (Tables S5, S6, and S7, SI). A significant exception occurred with Glu, which has the highest RMSD, associated, in part, with the charged nature of the residue. While MCSA fitting lead to improvement over the C22/CMAP RMSD, these parameters yielded poor agreement between crystallographic χ1/χ2 rotamer distribitutions and results from crystal MD simulations (see below). Accordingly, additional optimization was performed for the χ2 torsion of Glu via adjustments to the 1-fold torsion term to improve agreement with crystallographic survey distributions. Poor agreement (Firure 2, RMSD > 2.0 kcal/mol) is also seen for Asp and Hsp. This is due to poor reproduction of the local minima required to better reproduce the high-energy regions, a compromise that is due to inherent limitations in the form of the potential energy function that limits the ability to match the entire energy surfaces. Analysis of the RMSD for low-relative energy regions (i.e. > 2 kcal/mol above the energy minima) versus high-relative energy regions (i.e. 2 – 12 kcal/mol above the minima) show the overall improvement for the low energy regions to be minimal with the largest improvements in the higher energy regions (Table S8, SI); such changes indicate that the new model may have a larger impact on sidechain dynamics rather than on sampling of the different χ1/χ2 rotamer populations (see below). While alternate fitting protocols may alleviate the lack of significant improvements in the reproduction of the QM data in the low energy regions, as has been shown previously, rigorous reproduction of QM gas phase data does not always assure satisfactory reproduction of condensed phase properties.40
Figure 2.
RMS Differences in relative energies between MM and QM potential energy surfaces. RMS Differences were calculated for relative energies less than 12.0 kcal/mol above the global minimum for the three 1D or 2D surfaces. Higher-energy cutoffs of 20.0 kcal/mol for Arg and Lys or 25.0 kcal/mol for Asp, Glu and Hsp were used for the charged amino acids.
Table 3.
Average and RMS differences (RMSD) in kcal/mol between MM and QM calculated energies for all χ1, χ2 rotamers over all the amino acids.
| Average Difference | RMSD | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| α | β | αL | All | α | β | αL | All | |
| C22/CMAP | 1.80 | 0.74 | 3.72 | 2.09 | 3.55 | 2.46 | 4.73 | 3.69 |
| MCSA-fit | 1.68 | 0.45 | 3.64 | 1.92 | 2.55 | 1.49 | 4.64 | 3.17 |
| C36 | 1.19 | 0.22 | 2.74 | 1.39 | 2.47 | 1.78 | 3.67 | 2.75 |
Target data for further optimization of the χ1/χ2 parameters following the initial MCSA fitting was NMR J-coupling data for ubiquitin and GB1 in 8 M urea. Simulations of the two unfolded proteins were undertaken with the results showing that the initial MCSA-based parameter set did not correctly reproduce the experimental χ rotamer distributions as judged by the reproduction of NMR data (Table 4). Therefore, empirical adjustments of the χ dihedral parameters were undertaken to optimize the relative energy of the local minima in order to obtain improved sidechain sampling in the unfolded proteins while maintaining overall fit to QM data. This process primarily involved adjustments of the 1-and 2-fold dihedral terms to shift the relative energies of the minima without significantly impacting the higher energy regions of the χ1/χ2 surfaces. Following several iterations of parameter adjustments and simulations, the final parameter set was obtained. The agreement with the NMR target data on the unfolded proteins is significantly improved (Table 4) while the RMSD for the final set of parameters with respect to the QM data was generally only slightly worse than that based on the MCSA fit parameters (Figure 3). The one exception is Glu where significant disagreement with crystal data was observed requiring significant adjustment of the χ1/χ2 torsion parameters. The final set of χ dihedral parameters is anticipated to treat the higher energy regions of the sidechain conformations more accurately based on better reproduction of the QM data (Table S8) as well as of the relative sampling of the χ rotamers based on reproduction of the NMR data.
Table 4.
χ2 Values for side-chain J-couplings measured in unfolded ubiquitin and GB1, for the MCSA-derived and final sets of side-chain torsion parameters. χ2 values calculated using equation 2. Combined χ2 values for Ubiquitin and GB1 are obtained as an average over the two proteins, weighted by the number of data points for each. Overall χ2 are obtained as an unweighted average of the χ2 for the different types of residue. Statistical uncertainty from a block error analysis is given in brackets next to each number92.
| RES | MCSA-derived Parameters | Final Parameters | ||||
|---|---|---|---|---|---|---|
| Ubiquitin | GB1 | Combined | Ubiquitin | GB1 | Combined | |
| Arg | 6.1 (0.7) | N.A. | 6.1 (0.7) | 4.1 (0.6) | N.A. | 4.1 (0.6) |
| Asn | 46.7 (3.7) | 6.1 (1.3) | 16.3 (2.8) | 2.7 (1.3) | 9.7 (1.1) | 8.0 (1.2) |
| Asp | 32.4 (1.7) | 32.8 (1.9) | 32.7 (1.8) | 3.8 (0.9) | 5.5 (0.8) | 4.8 (0.9) |
| Cys | N.A. | |||||
| Gln | 11.1 (0.9) | 8.3 (1.4) | 10.3 (1.2) | 5.7 (0.6) | 3.4 (0.8) | 5.0 (0.7) |
| Glu | 21.9 (2.1) | 38.0 (1.8) | 29.4 (2.0) | 3.4 (0.5) | 5.6 (0.7) | 4.4 (0.6) |
| N.A. | N.A. | |||||
| His | 10.7 (2.3) | 10.7 (2.3) | 1.1 (1.1) | 1.1 (1.1) | ||
| Ile | 12.6 (2.4) | 7.4 (3.6) | 11.5 (3.1) | 3.6 (1.0) | 19.5 (4.9) | 7.0 (2.9) |
| Leu | 52.1 (2.4) | 64.0 (4.2) | 53.6 (3.3) | 3.4 (0.5) | 3.0 (0.8) | 3.4 (0.7) |
| Lys | 8.9 (0.6) | 13.0 (1.1) | 10.2 (0.8) | 3.7 (0.4) | 14.7 (0.8) | 7.3 (0.6) |
| Met | N.A. | |||||
| Phe | 19.4 (3.8) | 14.3 (2.1) | 16.6 (2.9) | 3.8 (0.5) | 12.9 (2.0) | 8.7 (1.3) |
| Pro | 9.5 (0.8) | N.A. | 9.5 (0.8) | 7.0 (0.7) | N.A. | 7.0 (0.7) |
| Ser | 9.9 (2.1) | N.A. | 9.9 (2.1) | 3.6 (0.9) | N.A. | 3.6 (0.9) |
| Thr | 28.5 (2.3) | 18.3 (1.9) | 22.1 (2.1) | 10.0 (1.1) | 15.3 (1.2) | 13.3 (1.2) |
| Trp | N.A. | 5.0 (3.1) | 5.0 (3.1) | N.A. | 1.3 (0.5) | 1.3 (0.5) |
| Tyr | 53.7 (0.4) | 14.3 (2.5) | 25.1 (1.5) | 4.3 (2.0) | 2.7 (0.8) | 3.1 (1.5) |
| Val | 9.1 (1.7) | 3.6 (0.8) | 6.1 (1.1) | 0.7 (0.3) | 2.2 (0.7) | 1.5 (0.5) |
|
| ||||||
| Overall | 20.8 (2.0) | 18.8 (1.9) | 17.2 (2.0) | 4.1 (0.9) | 8.0 (0.9) | 5.2 (0.9) |
N.A. indicates that no amino acids of that type are available in the respective proteins.
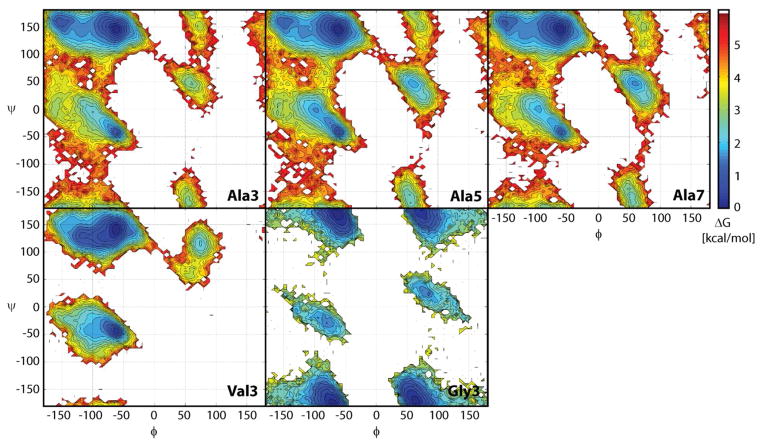
Figure 3.
Sampling of backbone φ/ψ torsion angles in the central residues of Ala3, Ala5, Ala7, Val3, and Gly3. Colors indicate relative free energies according to color bar given on the right.
Results: Validation of new parameters
Results for short peptides
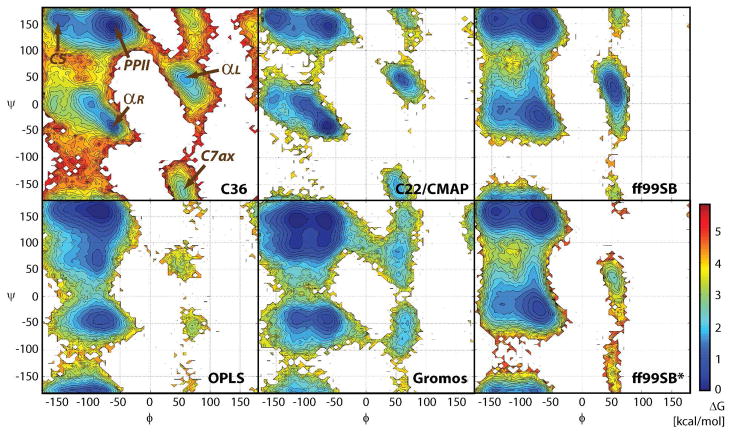
The φ/ψ-sampling for Ala3, Ala5, Ala7, Val3, and Gly3 is shown in Figure 3. In alanine- and valine-based peptides, the dominant minimum lies at PPII but additional minima at C5 and αR are only slightly higher in energy. Additional minima at αL and C7ax are about 2–3 kcal/mol higher than the PPII conformation. There is only a small difference between Ala3, Ala5, and Ala7, but the sampling for Val3 is significantly different because of the presence of the bulkier hydrophobic side chain. Gly3 exhibits symmetric sampling with similar minima at αR/αL and PPII/C7ax. In Figure 4 the φ/ψ-map with the new parameters is compared against the C22/CMAP map and maps from other popular force fields. It can be seen that the changes with respect to C22/CMAP are relatively subtle with the main differences consisting of a deeper PPII minimum, a higher and more focused αR minimum, and less sampling of the αL state. The changes go in the direction of the QM-based map16, where the PPII minimum is even deeper. Among the other force fields, the Amber ff99SB9 map is closest to the new CHARMM force field but significant differences exist. OPLS34 is qualitatively different with a pronounced minimum at C7eq and the Gromos force field35 has two similarly populated minima in the αR-basin and a low-energy transition region between αR and C7ax that is not present in any of the other force fields.
Figure 4.
Sampling of φ/ψ torsion angles in the central residues of Ala5 with different force fields: the new force field (C36), the previous C22/CMAP force field (C22)16, Amber ff99SB9, OPLS-AA,34 Gromos 53a635 and Amber ff99SB*7.
The φ/ψ-sampling was further compared with experimental J-coupling constants that are available from NMR experiments. Table 5 summarizes the results with details given in Tables S9 and S10 of the SI. The agreement is very good for the alanine-based peptides and for Gly3, and reasonable for Val3. For Ala5 we also compared with other force fields (Amber ff99SB9, OPLS/AA34, Gromos 53a678). The new C36 force field has lower χ2 values than all of the force fields and there is, in particular, significant improvement over the previous C22/CMAP force field. This improvement is largely due to decreased sampling of αR conformations and increased sampling of PPII (Figures 3 and 4). The main reason for the significant improvement over C22/CMAP is the correction of the known bias toward αR in that force field; the improvement is reflected in the J-couplings probing the ψ backbone torsion angle. However, the main reason for the improvement over other force fields (e.g. ff99SB) is due to the J-couplings probing the ϕ torsion angle (in fact most of the couplings are probing this torsion angle). This improvement largely reflects the balance between the β and ppII regions of the Ramachandran map, which are those contributing the most towards the ensemble-averaged J-couplings in most force fields. Although the χ2 value for Val is slightly worse, it should be noted that substituent effects make a significant contribution to the J-coupling and the use of an alternative set of Karplus equation parameters (Table S9 of SI) reduces many of the deviations from experiment. Note that although Gly3 data was used to derive Gly parameters for the termini (Table S11 of the SI), the internal glycine J-couplings have not been used as target data in the parametrization, with the 2D QM surface used directly to create the Gly φ/ψ CMAP term in the force field. As an independent test of terminal glycine parameters, and of the proline parameters, which were also based directly on the 2D φ/ψ QM surface, we have compared our results with data for the peptide GPGG79, obtaining much improved agreement over C22/CMAP, particularly for the C-terminal Gly in GPGG (Table S12 of the SI).
Table 5.
χ2 values from comparison of experimental NMR J-coupling constants with simulation results. All values were calculated with flavodoxin-based Karplus parameters68. For the new force field, results with DFT-based Karplus parameters (DFT2)69 are given in parentheses. For Gly3 and GPGG only DFT-based79 results are given.
| Ala3 | Ala5 | Ala7 | Val3 | Gly3 | GPGG | |
|---|---|---|---|---|---|---|
| C36 | 0.61 (1.22) | 0.74 (1.16) | 0.43 (0.68) | 2.33 (3.57) | 2.33 | 2.78 |
| C22/CMAP | 1.73 | 3.68 | 13.37 | |||
| Amber ff99SB | 1.40 | 3.04 | 2.39 | |||
| OPLS-AA | 1.35 | |||||
| Gromos 53a6 | 1.68 |
19-residue disordered fragment of Hen Lysozyme
In addition to tests on structure-forming peptides, it is important to also consider peptides that form little helix or sheet structure. As a test for such a largely disordered peptide, we chose a 19-residue fragment of hen Lysozyme, for which scalar couplings have been measured by Graf et al.31 Extensive sampling of this peptide was obtained by means of replica exchange molecular dynamics, from which the degree of structure formed was assessed by means of backbone scalar couplings. In Table 6, we report scalar couplings for the central alanine residue (full data set in Table S13 of the SI). Overall, the agreement with the couplings is much better than the previous C22/CMAP force field and slightly better than the previously optimized Amber ff99SB* force field. In particular, the agreement with scalar couplings reflecting the ψ torsion angle was the best out of the force fields considered.
Stability of dimeric coiled coil
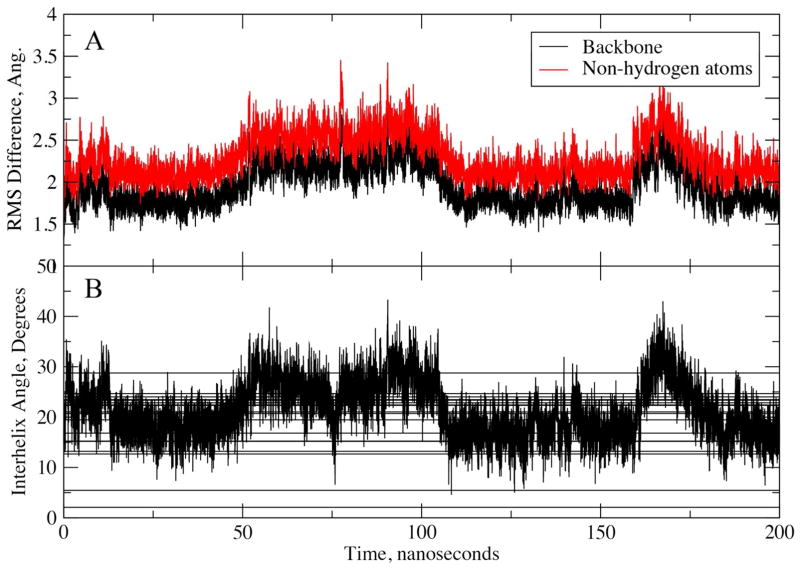
The developed parameters were also tested for their ability to reproduce the structure of a dimeric coiled-coil protein. These structures include two or more individual helices that interact primarily via hydrophobic amino acids protruding along one side of the helices. This class of peptides has been subject to a number of MD simulations, where in some cases obtaining stable native state simulations has been challenging.80 One such example is a heterodimeric parallel coiled coil (PDB identifier 1U0I)46, which was found to give poor results with a number of force fields, the closest RMS difference from the native being approximately 4 Å. Accordingly, the dimeric peptide represents a good test of the force field in the light of the previous simulation results as well as the overall structure being dominated by hydrophobic interactions between the two helices in the dimer. Figure 5A shows the RMSD as a function of simulation time for a 200 ns simulation initiated with model one from the collection of NMR structures.46 The RMSD being in the vicinity of 2 to 2.5 Å, with the C- and N-terminal residues excluded, indicates the overall structure to be stable, representing a significant improvement over previously reported results. The ability of the model to reproduce the experimental structure is futher judged by comparison of the inter-helical angles between the two helices from the simulation with those obtained from the 20 NMR models (Figure 5B). The overlap is excellent, indicating C36 to satisfactorily model the interactions between the two peptides. Additional analysis involved alignment of one of the two helices and calculation of the RMSD for that helix as well as the second helix (Figure S2 of the SI). The RMSD of the backbone atoms of the self-aligned helices are all ~1 Å indicating that the helical structure of the individual peptides was maintained: an important outcome, as the developed FF was designed to yield a lower population of helices and an additional indication that the balance of the FF between sampling of extended and helical conformations is satisfactory. Also shown are the RMSD of the second, non-aligned helix, with the difference being in the range of 2 to 6 Å with the presence of larger fluctuations showing that the helices are moving relative to each other in the simulation indicating that the force field is not overly restraining the dimer in a conformation close to that in the experimental structure. Overall, these results indicate that the FF adequately reproduces the structure of the coiled-coil dimer and thus appropriately models hydrophobic interactions and the helical character of individual helices.
Figure 5.
A) RMS difference from experimental structure and B) Inter-helical angle of the dimeric coiled-coil 1U0I as a function of simulation time. RMS differences are for the backbone (N, Cα, C, O) or non-hydrogen atoms of both helices following least squares alignment of the backbone atoms in both helices. The N- and C-terminal residues were excluded from the analysis. For the inter-helical angles vectors defining the helical axes were calculated using the non-terminal residue Cα atoms using the approach of Chothia et al.95. Horizontal lines represent the inter-helical angles from the 20 models generated in the NMR study.
Crystal simulations
To assess the quality of the new parameters on full proteins, simulations were performed on eight proteins of different morphology and size (Table 1) in their crystal environments. By performing these calculations on the full unit cells, the sampling was significantly enhanced, as the unit cells contain two to four monomers of each protein. The analysis emphasizes shifts in the φ/ψ population that may indicate systematic problems in the backbone parameters16. In addition, analysis of the χ1/χ2 distributions was undertaken. Average differences in φ/ψ obtained from the 40-ns simulations are shown in Table 7; average differences were calculated to identify local systematic deviations in the backbone conformations. The overall agreement is good with the MD simulations resulting in minimal deviation from the crystallographic backbone geometries (Table 7). While larger deviations (> 6°) occurred in selected φ/ψ torsions of β-strand regions of Erabutoxin B (PDB id: 3EBX)81 and crambin (PDB id: 1ejg),82 the overall agreement of C36 is similar to that of C22/CMAP. The larger deviations in C36 were not unexpected as the original C22/CMAP was specifically optimized to reproduce the local φ/ψ sampling. Building upon the previous CMAP in the present study appears to lead to the small, yet acceptable, degradation in the local φ/ψ sampling indicating that optimization did not lead to a bias in the sampling of φ and ψ while yielding significant improvements in the treatment of the smaller polypeptides.
Table 7.
Average differences in backbone φ and ψ torsions between MD average and crystallographic values averaged over all residues
| C22/CMAP | all | α-helix | β-strands | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| φ | SDM | ψ | SDM | φ | SDM | ψ | SDM | φ | SDM | ψ | SDM | |
| 135L | −0.67 | 0.51 | 0.03 | 0.64 | 1.57 | 0.31 | −0.49 | 0.41 | 0.03 | 0.82 | 3.18 | 0.72 |
| 1BYI | −1.57 | 0.11 | 1.08 | 0.30 | −1.17 | 0.02 | 0.27 | 0.05 | −0.75 | 0.13 | 2.94 | 0.49 |
| 1BZP | −0.59 | 0.35 | 1.44 | 0.54 | −0.35 | 0.11 | −0.36 | 0.18 | ||||
| 1EJG | −2.25 | 0.13 | 8.02 | 0.33 | −3.91 | 0.06 | 8.79 | 0.05 | 1.71 | 0.45 | −4.08 | 0.35 |
| 1I27 | −0.57 | 0.40 | −3.71 | 0.70 | −0.20 | 0.21 | −0.93 | 0.25 | −0.84 | 0.62 | −3.62 | 0.56 |
| 3EBX | 1.90 | 1.41 | 1.15 | 0.85 | 2.65 | 3.19 | 5.59 | 1.05 | ||||
| 3ICB | −3.22 | 0.29 | 2.84 | 0.84 | −5.27 | 0.38 | 6.79 | 0.92 | ||||
| 5PTI | 2.72 | 0.93 | 1.53 | 0.94 | −0.89 | 0.29 | 0.08 | 0.54 | −3.20 | 0.57 | −0.25 | 0.45 |
|
| ||||||||||||
| Avg | −0.05 | 2.69 | −2.04 | 1.67 | 1.24 | 0.85 | ||||||
|
| ||||||||||||
| C36 | all | α-helix | β-strands | |||||||||
|
| ||||||||||||
| 135L | 0.42 | 0.59 | −2.06 | 0.36 | 1.19 | 1.35 | −2.12 | 0.45 | 1.23 | 0.13 | −2.01 | 0.35 |
| 1BYI | −0.74 | 0.01 | −0.23 | 0.86 | 0.33 | 0.03 | −0.85 | 0.05 | −0.47 | 0.08 | 0.20 | 3.73 |
| 1BZP | 2.31 | 1.03 | 2.18 | 1.00 | 0.03 | 0.42 | 0.24 | 0.45 | ||||
| 1EJG | 8.29 | 0.08 | −1.38 | 0.06 | 7.42 | 0.09 | 6.51 | 0.05 | 9.96 | 0.72 | −11.57 | 0.39 |
| 1I27 | −0.25 | 0.65 | −2.24 | 0.97 | 0.74 | 0.21 | −0.30 | 0.23 | 0.85 | 1.84 | −3.33 | 1.69 |
| 3EBX | 3.45 | 3.25 | −2.68 | 2.44 | 0.99 | 7.57 | −3.93 | 5.11 | ||||
| 3ICB | −1.48 | 0.11 | 5.62 | 0.61 | −4.27 | 0.11 | 9.41 | 1.02 | ||||
| 5PTI | 6.77 | 2.28 | 1.11 | 3.26 | 0.60 | 0.34 | 2.53 | 0.52 | 4.29 | 0.45 | −2.90 | 1.29 |
|
| ||||||||||||
| Avg | 2.35 | 0.04 | 0.86 | 2.20 | 2.81 | −3.92 | ||||||
Standard deviation of the mean (SDM) calculated from averages of 5 ns intervals between 5 ns and 40ns of the simulation.
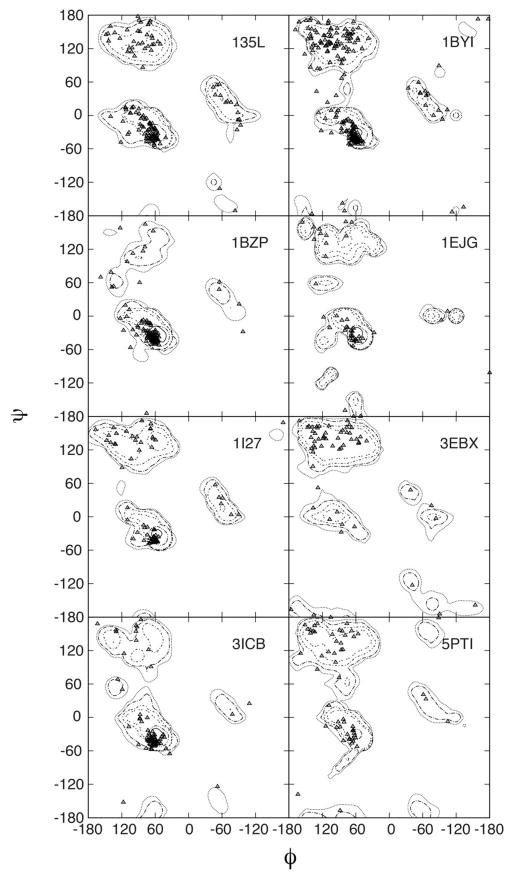
A more detailed analysis of the experimental and calculated backbone φ/ψ distributions is presented in Figure 6. Ramanchandran83 diagrams were constructed by overlaying crystallographic φ/ψ values for each of the simulated proteins with the respective probability distributions calculated from the MD simulations. Overall, φ/ψ sampling yields population distributions centered on crystallographic φ/ψ values. Worth noting is the reproduction of backbone conformations in loop regions as depicted by data points falling outside of the classic Ramachandran secondary structure regions. These results demonstrate the robustness of C36 in maintaining crystal backbone conformations.
Figure 6.
Ramanchandran plot for backbone sampling. Crystallographic φ/ψ values (triangles) are overlaid onto probability distributions obtained from the MD simulation of full unit cells. Probabilities calculated with snapshots between 5ns-40ns at 5ps intervals.
To analyze the conformational properties of the side chains in the crystal simulations χ1 and χ2 distributions for each residue type were extracted from the 5ns–40ns portion of the protein crystal simulations and compared to distributions obtained from a crystallographic survey.62 The level of agreement was quantified by calculating the overlap coefficients (Eq. 4) between simulation and protein database survey results. Simulation results for the three protonation types of histidine Hsd, Hse, and Hsp were summed and normalized together as appropriate for comparisons with the His distribution from the database survey. The results from this comparison are presented in Table 8. Overall, the crystal simulations with C36 yield excellent agreement with database distributions and demonstrate a small overall improvement over that with C22/CMAP. The χ1 torsion in Glu and that in Lys were among the torsions that have improved significantly. With Glu, improvement of sampling required sacrificing the quality of agreement with the QM data (Figure 2), as discussed above. For χ2, Asp benefitted appreciably from the new torsion parameters. However, in a number of cases there are slight degradations in C36 relative to C22/CMAP such that the overall level of agreement for the χ1/χ2 distributions is similar for the two models, with both yielding satisfactory agreement with the crystallographic survey data.
Table 8.
Overlap coefficients between χ1 and χ1 probability distributions of protein crystal simulations and crystallographic survey data.
| Residue | χ1 | Residue | χ2 | ||||
|---|---|---|---|---|---|---|---|
| C22/CMAP | C36 | diff | C22/CMAP | C36 | diff | ||
| Arg | 0.97 | 0.96 | −0.01 | Arg | 0.92 | 0.93 | 0.01 |
| Asn | 0.90 | 0.94 | 0.04 | Asn | 0.89 | 0.85 | −0.05 |
| Asp | 0.92 | 0.96 | 0.04 | Asp | 0.63 | 0.96 | 0.33 |
| Cys | 0.94 | 0.94 | 0.00 | Gln | 0.93 | 0.96 | 0.03 |
| Gln | 0.96 | 0.97 | 0.01 | Glu | 0.99 | 0.96 | −0.02 |
| Glu | 0.87 | 0.97 | 0.10 | His | 0.86 | 0.85 | −0.01 |
| His | 0.87 | 0.88 | 0.01 | Ile | 0.93 | 0.89 | −0.04 |
| Ile | 0.89 | 0.93 | 0.04 | Leu | 0.95 | 0.95 | 0.00 |
| Leu | 0.98 | 0.97 | −0.01 | Lys | 0.99 | 0.89 | −0.10 |
| Lys | 0.87 | 0.95 | 0.08 | Met | 0.93 | 0.90 | −0.04 |
| Met | 0.97 | 0.97 | 0.00 | Phe | 0.83 | 0.88 | 0.05 |
| Phe | 0.91 | 0.95 | 0.04 | Trp | 0.89 | 0.85 | −0.04 |
| Ser | 0.84 | 0.90 | 0.06 | Tyr | 0.95 | 0.97 | 0.02 |
| Thr | 0.89 | 0.92 | 0.03 | ||||
| Trp | 0.82 | 0.87 | 0.05 | ||||
| Tyr | 0.93 | 0.98 | 0.05 | ||||
| Val | 0.82 | 0.93 | 0.11 | ||||
|
| |||||||
| Average | 0.90 | 0.94 | 0.90 | 0.91 | |||
| SD | 0.05 | 0.03 | 0.09 | 0.05 | |||
Data for His is based on arithmetic average over the OC for Hsd and Hse.
Comparison with NMR data for folded proteins
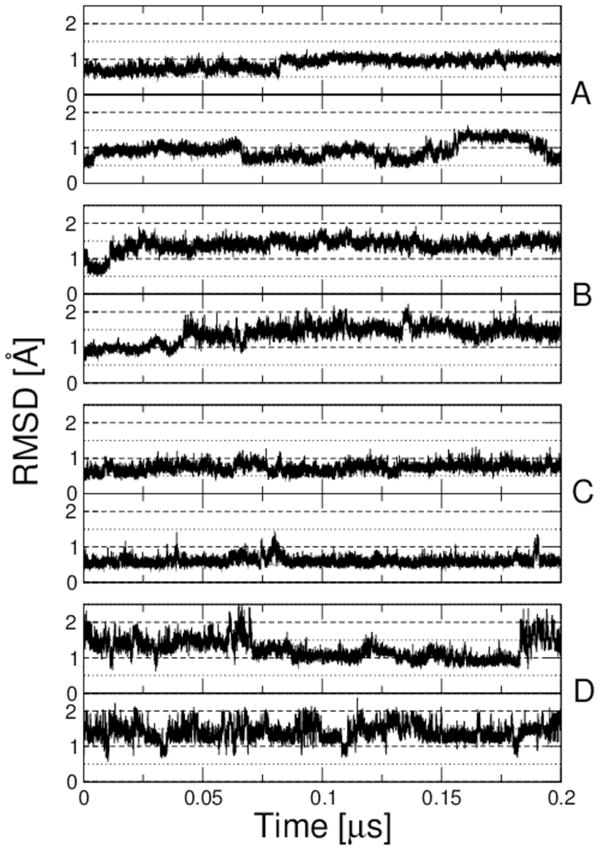
NMR data forms a valuable complement to crystal structure data for validating backbone and side-chain parameters, since it is very sensitive to the full distribution of dihedral angles, particularly the population of multiple rotamers. Accordingly, 200 ns MD simulations were performed on a set of four proteins for which extensive NMR data are available: bovine pancreatic trypsin inhibitor (BPTI), hen lysozyme, GB3 and ubiquitin. A similar data set was recently used by Lindorff-Larsen et al. in their tests of their modified Amber ff99SB force field (ff99SB-ILDN).11 In Figure 7, we show the RMSD of each protein from the experimental reference. In all cases, the backbone RMSD obtained with C36 is comparable or lower than that with C22/CMAP, with deviations generally being ~1.5 Å or less. The larger jumps observed for BPTI have been seen in other force fields and arise from transitions between multiple substates of the native state.84
Figure 7.
RMSD from experimental structure for long runs of folded proteins. (A) Ubiquitin, (B) Hen Lysozyme, (C) GB3 and (D) BPTI. Upper plots show results with C22/CMAP, lower plots with C36. Various dotted lines indicated RMSD values of 0.5, 1.0, 1.5 and 2.0 Å.
Two types of NMR data were used for validation. The first were scalar couplings reporting on the side-chain χ1 torsion angle in folded proteins; this data set was collated from published and unpublished data by Lindorff-Larsen et al. In Table 9, we present the χ2 values including the data for all four folded proteins, split by residue type; we also include results for the unfolded proteins used to guide the side-chain optimization. However, we note that the side-chain J-couplings cannot be considered as a true validation, as they were used in the optimization. The χ2 values for C36 with respect to this data would therefore be artificially lowered, relative to other force fields. Ideally, a validation would be done for unfolded proteins not included in the present study, which provide a sensitive test of torsion angle distributions. However, the only other data set we are aware of is that reported for Hen Lysozyme by Schwalbe and co-workers85 and the size of this system is not amenable to current computational resources. We find that the revised C36 side-chain parameters represent an overall improvement over C22/CMAP, with an overall χ2 of 5.2 and 8.5 for unfolded and folded proteins respectively, compared with 9.8 and 9.2 for the original C22/CMAP. The results for C36 are also comparable with the χ2 of 9.7 and 8.9 for unfolded and folded proteins obtained with Amber ff99SB-ILDN.
Table 9.
Comparison with side-chain NMR J-coupling data for folded and unfolded proteins. χ2 values representing agreement with experimental J-coupling data on unfolded proteins in urea from Vajpal et al.93 and with data for folded proteins collated in Ref11 for three force fields (Amber ff99SB-ILDN, C22/CMAP and C36). Unfolded proteins are ubiquitin and GB1 and folded proteins are ubiquitin, GB3, BPTI and hen lysozyme. The three right-most columns give the number of J-coupling measurements used to calculate each χ2 value.
| RES | Amber ff99SB-ILDN | C22/CMAP | C36 | # DATA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unf.c | Fold.a | All | Unf.c | Fold.b | All | Unf.c | Fold.b | All | Unf. | Fold. | All | |
| Arg | 2.2 | 12.7 | 7.5 | 7.9 | 7.0 | 7.4 | 4.1 | 7.7 | 5.9 | 22 | 14 | 36 |
| Asn | 7.8 | 10.6 | 9.2 | 16.1 | 8.8 | 12.5 | 8.0 | 7.5 | 7.8 | 24 | 56 | 80 |
| Asp | 13.8 | 12.8 | 13.3 | 10.7 | 17.4 | 14.0 | 4.8 | 12.5 | 8.7 | 48 | 46 | 94 |
| Cys | 9.7 | 9.7 | 8.7 | 8.7 | 8.6 | 8.6 | 0 | 46 | 46 | |||
| Gln | 2.4 | 6.8 | 4.6 | 7.1 | 9.0 | 8.1 | 5.0 | 6.2 | 5.6 | 38 | 18 | 56 |
| Glu | 6.6 | 13.2 | 9.9 | 15.5 | 11.3 | 13.4 | 4.4 | 11.3 | 7.9 | 56 | 10 | 66 |
| His | 28.1 | 12.0 | 20.0 | 8.7 | 14.8 | 11.7 | 1.1 | 10.2 | 5.6 | 5 | 6 | 11 |
| Ile | 8.8 | 7.1 | 8.0 | 28.3 | 4.8 | 16.6 | 7.0 | 5.2 | 6.1 | 14 | 44 | 58 |
| Leu | 2.3 | 12.3 | 7.3 | 7.4 | 16.3 | 11.9 | 3.4 | 10.9 | 7.1 | 50 | 26 | 76 |
| Lys | 1.9 | 9.4 | 5.6 | 3.9 | 13.1 | 8.5 | 7.3 | 11.8 | 9.5 | 56 | 30 | 86 |
| Metd | 10.3 | 10.3 | 9.1 | 9.1 | 4.7 | 4.7 | 0 | 14 | 14 | |||
| Phe | 5.0 | 6.1 | 5.5 | 2.9 | 5.0 | 3.9 | 8.7 | 5.4 | 7.1 | 22 | 32 | 54 |
| Pro | 17.4 | 7.8 | 12.6 | 5.8 | 10.6 | 8.2 | 7.0 | 18.4 | 12.7 | 9 | 4 | 13 |
| Ser | 11.1 | 8.7 | 9.9 | 6.3 | 9.3 | 7.7 | 3.6 | 12.0 | 7.8 | 12 | 14 | 26 |
| Thr | 8.0 | 3.5 | 5.8 | 10.0 | 5.1 | 7.6 | 13.3 | 4.5 | 8.9 | 43 | 82 | 125 |
| Trp | 5.3 | 6.7 | 6.0 | 5.6 | 6.4 | 6.0 | 1.3 | 6.3 | 3.8 | 5 | 10 | 15 |
| Tyr | 9.8 | 7.8 | 8.8 | 10.4 | 6.6 | 8.5 | 3.1 | 6.8 | 5.0 | 22 | 30 | 52 |
| Val | 24.8 | 3.6 | 14.2 | 10.5 | 2.8 | 6.7 | 1.5 | 2.2 | 1.9 | 22 | 74 | 96 |
|
| ||||||||||||
| ALL | 9.7 | 8.9 | 9.3 | 9.8 | 9.2 | 9.5 | 5.2 | 8.5 | 6.9 | 448 | 278 | 726 |
Data taken from 1.2 μs equilibrium simulations in11.
Data from 0.2 μs equilibrium simulations in present work.
Data from solute tempering simulations in present work.
Note that the assignments of Met52 Hβ protons in BPTI are the reverse of those reported in Ref11 (Kresten Lindorff-Larsen, private communication). Combined χ2 values for folded and unfolded are obtained as a simple average over the χ2 values for folded and unfolded. Overall χ2 are obtained as an unweighted average of the χ2 for the different types of residue. An indication of the errors on these values is given in Table 4 for C36, but is omitted here in the interests of clarity.
The second type of data considered were residual dipolar couplings (RDCs) for the folded proteins, which are very sensitive to small structural deviations. The agreement between experiment and simulation has been quantified in this case by the Q parameter conventionally used in structure refinement with RDCs, defined as:
| [5] |
where Di,obs and Di,calc represent, respectively, the ith residual dipolar coupling from experiment and as back-calculated from the simulation. The results for various alignment media and folded proteins are given in Table 10. Although RDCs have also been measured for the unfolded proteins, because of uncertainties in calculating the alignment tensor in this case, we have not attempted to compute these data from the unfolded state simulations. We find that the backbone RDCs obtained with C36 are generally slightly better than, or comparable to those obtained with C22/CMAP or Amber ff99SB. Relative to C22/CMAP, the improvement in side-chain RDCs is clear for GB3 and HEWL, and the results for ubiquitin are comparable. The results for side-chains are for a significant improvement Amber ff99SB – however, these runs did not include the ILDN side-chain correction.
Table 10.
RDC Q-factors describing agreement between RDC’s computed from simulation and experiment. Smaller values correspond to better agreement with experiment (Equation 5). Errors from a bootstrap analysis are given in brackets94.
| Data set | Amber ff99SB | C22/CMAP | C36 |
|---|---|---|---|
| Backbone: | |||
| Ubiquitin, Medium 1 | 0.29 (0.02) | 0.20 (0.01) | 0.23 (0.01) |
| Ubiquitin, Medium 2 | 0.32 (0.03) | 0.20 (0.01) | 0.23 (0.01) |
| GB3, Medium 1 | 0.14 (0.01) | 0.22 (0.02) | 0.14 (0.02) |
| GB3, Medium 2 | 0.14 (0.01) | 0.20 (0.02) | 0.15 (0.01) |
| GB3, Medium 3 | 0.19 (0.01) | 0.22 (0.02) | 0.20 (0.02) |
| GB3, Medium 4 | 0.18 (0.01) | 0.25 (0.04) | 0.19 (0.03) |
| GB3, Medium 5 | 0.17 (0.02) | 0.23 (0.02) | 0.16 (0.02) |
| HEWL, Medium 1 | 0.30 (0.03) | 0.39 (0.04) | 0.42 (0.04) |
| HEWL, Medium 2 | 0.31 (0.03) | 0.42 (0.04) | 0.42 (0.04) |
| Side-chain: | |||
| Ubiquitin, Medium 1 | 0.57 (0.02) | 0.38 (0.05) | 0.38 (0.04) |
| Ubiquitin, Medium 2 | 0.56 (0.03) | 0.32 (0.06) | 0.28 (0.05) |
| GB3 | 0.66 (0.12) | 0.59 (0.13) | 0.31 (0.06) |
| HEWL | 0.89 (0.15) | 0.44 (0.10) | 0.37 (0.09) |
We have also used the backbone RDCs to assess whether the manual adjustments to the CMAP in order to reduce systematic deviations from canonical secondary structure have any deleterious effects on residues not in elements of canonical secondary structure. For each of the proteins GB3, Ubiquitin and Hen Lysozyme, we have classified the (ϕ,ψ) angles of each residue the Ramachandran map into narrowly defined “α”, “β”, “polyproline II”, “αL” and “coil”. The definitions (given in the legend of Table 11) are chosen to be mutually exclusive and exhaustive, with “coil” comprising angles not within one of the other regions. For each of these regions, we have calculated a root-mean-square Q-factor Qk over all alignment media k for all proteins in the following way:
| [6] |
Table 11.
Breakdown of RDC Q-factors by Ramachandran angles in the native state (i.e. crystal structure). The following regions of Ramachandran space have been defined: α region: −100°≤ϕ <−30° and −67°≤ψ <−7°; β region: −180°≤ϕ<−100° and 120°≤ψ<180°; ppII region: −100°≤ϕ<−30° and 100°≤ψ<180°; αL: 30°≤ϕ<100° and 0°≤ψ<80°; coil region: all residues not in other regions. Errors from a bootstrap analysis are given in brackets94.
| Ramachandran Region | Amber ff99SB | C22/CMAP | C36 |
|---|---|---|---|
| α | 0.22 (0.02) | 0.24 (0.02) | 0.20 (0.02) |
| β | 0.18 (0.02) | 0.27 (0.05) | 0.27 (0.06) |
| Polyproline II | 0.33 (0.06) | 0.36 (0.04) | 0.22 (0.06) |
| αL | 0.50 (0.03) | 0.24 (0.05) | 0.22 (0.06) |
| Coil | 0.23 (0.03) | 0.24 (0.04) | 0.26 (0.04) |
The values of Qr.m.s. for each region and each force field are shown in Table 11. C36 performs well for both the α and β regions, and represents a significant improvement over Amber ff99SB and C22/CMAP for the polyproline II region, as might be anticipated. Interestingly, one area in which both C22/CMAP and C36 are better than Amber ff99SB is the αL region (which was not manually tweaked in the CMAP). Finally, we note that the Qr.m.s. for the coil region is not appreciably worse than for Amber ff99SB, suggesting that the tweaks did not lead to detrimental effects on residues outside canonical regions of the Ramachandran map.
Equilibrium folding simulations of β hairpins
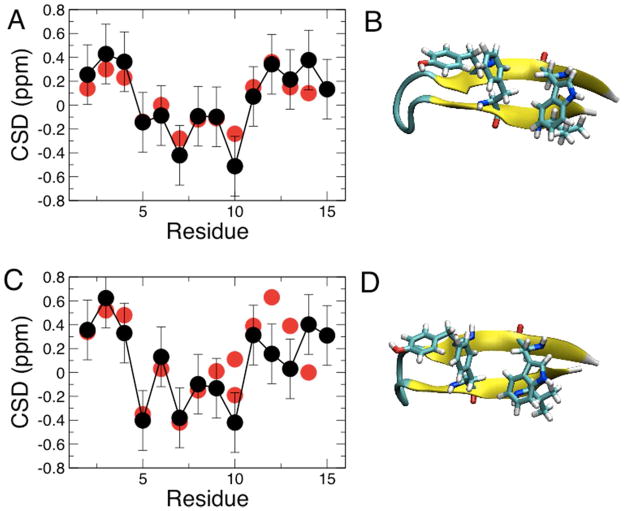
Since most of the backbone optimization focused on helix-forming peptides, we have also determined the folding equilibrium of two β-hairpins using temperature replica exchange. The two hairpins are the original native C-terminal GB1 hairpin, residues 41–56 of protein G53,54, and the GB1m3 variant 55,56, which has been optimized for greater stability. Replica exchange runs of both hairpins resulted in well-folded structures. Defining the folded state as being within 1.5 Å backbone RMSD of the hairpin in the protein G crystal structure, a folded fraction of 77 % for GB1 and 99 % for GB1m3 at 300 K is obtained, compared with experimental estimates of 60 % for GB154 and 86 % for GB1m355 (folded structures are shown in Figure 8B and D). However, these estimates are based, in the case of the simulations, on an arbitrary definition of the folded state, and, in the case of the experiment, on fitting a two-state model to a global experimental signal. It is therefore more desirable to compare an ensemble-averaged observable from the simulation with the experiment. Accordingly, ensemble-averaged NMR chemical shift deviations (CSDs) were computed over the trajectory using the SPARTA+ program, with the resulting CSDs for the Hα protons plotted in Figure 8A and C. For both peptides, the agreement with the experimental CSDs is very good, and the more positive CSD values near the termini for GB1m3 (Figure 8C) compared with GB1 (Figure 8A) are consistent with the higher fraction of folded hairpin for the former sequence.
Figure 8.
β-hairpin folding test. Hα chemical shifts from experiment (red symbols) and calculated from simulation using SPARTA+ (black symbols) for: (A) the GB1 hairpin (folded structure from simulation shown in (B)); and (C) the designed GB1m3 hairpin (structure shown in (D)). Error bars on calculated data are the typical RMSD between experiment and Hα shifts predicted by SPARTA+61. Experimental data at 280 K from Fesinmeyer et al;55 simulation data taken from 278 K replica.
Conclusions and Outlook
A revised version of the CHARMM additive protein force field (C36) is reported, the main features of which are a newly optimized backbone CMAP and sidechain torsion potentials. The model is able to correct the propensity of the backbone parameters in the previous C22/CMAP force field to overstabilize helices: as a consequence more reasonable results for the fraction helix in a helix-forming peptide are obtained, with the model still able to fold β-hairpin peptides. For the side chains, the new fit to QM data followed by empirical adjustments results in a significant improvement in agreement with side-chain scalar coupling data for the χ1 torsion for both folded and unfolded proteins, compared with C22/CMAP. In the future, it would also be interesting to compare with experimental data for the distribution of χ2, χ3 and χ4 for proteins in solution, but at the time of writing no such data is available in the literature. However, comparison with χ2 crystallographic distributions indicate C36 to satisfactorily treat this degree of freedom.
Our optimization procedure highlighted the importance of considering a range of experimental data measured in the condensed phase, in order to fine-tune the parameters initially obtained from targeting gas-phase QM calculations on model dipeptides. It is anticipated that the lessons learned will be equally useful in optimizing the next generation of polarizable force fields.
Supplementary Material
Acknowledgments
ADM acknowledges financial support from the NIH (GM 051501 and GM 072558) and the NSF (CHE 0823198) as well as computational support from the Department of Defense, the Pittsburgh Supercomputing Center and the NSF TeraGrid resources. MF acknowledges financial support from the NIH (GM 084953 and GM 092949) and the NSF (CBET 0941055) as well as access to computational resources at the High Performance Computing Center at Michigan State University. These calculations made use of the Biowulf cluster at the National Institutes of Health. RB is supported by a Royal Society University Research Fellowship.
Footnotes
Supporting Information Available. Supporting Information includes 14 tables and 2 figures including the new side-chain torsion parameters. The C36 toppar files may be obtained from the MacKerell web page at http://mackerell.umaryland.edu/CHARMM_ff_params.html and the supporting information may be obtained free of charge from the Internet at http://pubs.acs.org.
References
- 1.MacKerell AD., Jr J Comput Chem. 2004;25:1584. doi: 10.1002/jcc.20082. [DOI] [PubMed] [Google Scholar]
- 2.Stone AJ. Science. 2008;321:787. doi: 10.1126/science.1158006. [DOI] [PubMed] [Google Scholar]
- 3.Freddolino PL, Harrison CB, Liu Y, Schulten K. Nat Phys. 2010;6:751. doi: 10.1038/nphys1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warshel A, Kato M, Pisliakov AV. J Chem Theor Comput. 2007;3:2034. doi: 10.1021/ct700127w. [DOI] [PubMed] [Google Scholar]
- 5.Lopes PEM, Roux B, MacKerell AD., Jr Theor Chem Acc. 2009;124:11. doi: 10.1007/s00214-009-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman PA. J Comput Chem. 2003;24:1999. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 7.Best RB, Hummer G. J Phys Chem B. 2009;113:9004. doi: 10.1021/jp901540t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Best RB, Mittal J. J Phys Chem B. 2010;114:14916. doi: 10.1021/jp108618d. [DOI] [PubMed] [Google Scholar]
- 9.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Proteins. 2006;65:712. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li DW, Brüschweiler R. J Chem Theor Comput. 2011;7:1773. doi: 10.1021/ct200094b. [DOI] [PubMed] [Google Scholar]
- 11.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Proteins. 2010;78:1950. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Best RB, De Sancho D, Mittal J. Biophys J. 2012;102:1462. doi: 10.1016/j.bpj.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piana S, Lindorff-Larsen K, Shaw DE. Biophys J. 2011;100:L47. doi: 10.1016/j.bpj.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacKerell AD, Jr, Bashford D, Bellot M, Dunbrack JRL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Kuczera J, Yin D, Karplus M. J Phys Chem B. 2000;102:3586. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 15.MacKerell AD, Jr, Feig M, Brooks CL. J Am Chem Soc. 2004;126:698. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 16.MacKerell AD, Jr, Feig M, Brooks CL. J Comput Chem. 2004;25:1400. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 17.Freddolino PL, Schulten K. Biophys J. 2009;97:2338. doi: 10.1016/j.bpj.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freddolino PL, Liu F, Gruebele M, Schulten K. Biophys J. 2008;95:L75. doi: 10.1529/biophysj.108.131565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freddolino PL, Park S, Roux B, Schulten K. Biophys J. 2009;96:3772. doi: 10.1016/j.bpj.2009.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Best RB, Buchete NV, Hummer G. Biophys J. 2008;95:L07. doi: 10.1529/biophysj.108.132696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mittal J, Best RB. Biophys J. 2010;99:L26. doi: 10.1016/j.bpj.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Best RB, Mittal J. J Phys Chem B. 2010;114:8790. doi: 10.1021/jp102575b. [DOI] [PubMed] [Google Scholar]
- 23.Brooks BR, Brooks CL, MacKerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. J Comput Chem. 2009;30:1545. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vorobyov IV, Anisimov VM, MacKerell AD., Jr J Phys Chem B. 2005;109:18988. doi: 10.1021/jp053182y. [DOI] [PubMed] [Google Scholar]
- 25.Mason PE, Neilson GW, Enderby JE, Saboungi M-L, Dempsey CE, MacKerell AD, Jr, Brady JW. J Am Chem Soc. 2004;126:11462. doi: 10.1021/ja040034x. [DOI] [PubMed] [Google Scholar]
- 26.Macias AT, MacKerell AD., Jr J Comput Chem. 2005;26:1452. doi: 10.1002/jcc.20281. [DOI] [PubMed] [Google Scholar]
- 27.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Laxmikant K, Schulten K. J Comput Chem. 2005;26:1781. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hess B, Kutzner C, Van der Spoel D, Lindahl E. J Chem Theor Comput. 2008;4:435. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 29.Jorgensen WL, Chandrasekhar J, Madura JD. J Chem Phys. 1983;79:926. [Google Scholar]
- 30.Durell SR, Brooks BR, Ben-Naim A. J Phys Chem. 1994;98:2198. [Google Scholar]
- 31.Graf, Nguyen PH, Stock G, Schwalbe H. J Am Chem Soc. 2007;129:1179. doi: 10.1021/ja0660406. [DOI] [PubMed] [Google Scholar]
- 32.Darden T, York D, Pedersen L. J Chem Phys. 1993;103:8577. [Google Scholar]
- 33.Miyamoto S, Kollman PA. J Comp Chem. 1992;13:952. [Google Scholar]
- 34.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. J Phys Chem B. 2001;105:6474. [Google Scholar]
- 35.Oostenbrink C, Villa A, Mark AE, van Gunsteren WF. J Comput Chem. 2004;25:1656. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 36.Weerasinghe S, Smith PE. J Phys Chem B. 2003;107:3891. [Google Scholar]
- 37.Kokubo H, Pettit BM. J Phys Chem B. 2007;111:5233. doi: 10.1021/jp067659x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu P, Kim B, Friesner RA, Berne BJ. Proc Natl Acad Sci U S A. 2005;102:13749. doi: 10.1073/pnas.0506346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Camilloni C, Provasi D, Tiana G, Broglia RA. Proteins. 2007;71:1647. doi: 10.1002/prot.21852. [DOI] [PubMed] [Google Scholar]
- 40.Mackerell AD, Jr, Feig M, Brooks CL., 3rd J Comput Chem. 2004;25:1400. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 41.Nosé S. Mol Phys. 1984;52:255. [Google Scholar]
- 42.Ryckaert JP, Cicotti G, Berendsen HJC. J Comp Phys. 1977;23:327. [Google Scholar]
- 43.Feller SE, Zhang Y, Pastor RW, Brooks BR. J Chem Phys. 1995;103:4613. [Google Scholar]
- 44.Kabsch W, Sander C. Biopolymers. 1983;22:2577. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 45.Heinig M, Frischman D. Nucleic Acids Res. 2004;32:W500. doi: 10.1093/nar/gkh429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuler B, Eaton WA. Curr Opin Struct Biol. 2008;18:16. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jo S, Kim T, Iyer VG, Im W. J Comput Chem. 2008;29:1859. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 48.Wlodawer A, Walter J, Huber R, Sjolin L. J Mol Biol. 1984;180:301. doi: 10.1016/s0022-2836(84)80006-6. [DOI] [PubMed] [Google Scholar]
- 49.Vijay-Kumar S, Bugg CE, Cook WJ. J Mol Biol. 1987;194:531. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 50.Ulmer TS, Ramirez BE, Delaglio F, Bax A. J Am Chem Soc. 2003;125:9179. doi: 10.1021/ja0350684. [DOI] [PubMed] [Google Scholar]
- 51.Young ACM, Dewan JC, Nave C, Tilton RF. J Appl Cryst. 1993;26:309. [Google Scholar]
- 52.Parinello M, Rahman A. J Appl Phys. 1981;52:7182. [Google Scholar]
- 53.Blanco FJ, Rivas G, Serrano L. Nat Struct Biol. 1994;1:584. doi: 10.1038/nsb0994-584. [DOI] [PubMed] [Google Scholar]
- 54.Muñoz V, Eaton WA. Nature. 1997;390:196. doi: 10.1038/36626. [DOI] [PubMed] [Google Scholar]
- 55.Fesinmeyer RM, Hudson FM, Andersen NH. J Am Chem Soc. 2004;126:7238. doi: 10.1021/ja0379520. [DOI] [PubMed] [Google Scholar]
- 56.Du DG, Tucker MJ, Gai F. Biochemistry. 2006;45:2668. doi: 10.1021/bi052039s. [DOI] [PubMed] [Google Scholar]
- 57.Best RB, Mittal J. Proteins. 2011;79:1318. doi: 10.1002/prot.22972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shao Y, Fusti-Molnar L, Jung Y, Kussmann J, Ochsenfeld C, Brown ST, Gilbert ATBSLV, Levchenko SV, O’Neill DP, ADJR, Lochan RCWT, Beran GJO, Besley NA, Herbert JM, Lin CYVVT, Chien SH, Sodt A, Steele RP, Rassolov VA, Maslen PEKPP, Adamson RD, Austin B, Baker J, Byrd EFC, Dachsel HDRJ, Dreuw A, Dunietz BD, Dutoi AD, Furlani TRGSR, Heyden A, Hirata S, Hsu C-P, Kedziora G, Khalliulin RZKP, Lee AM, Lee MS, Liang W, Lotan I, Nair NPB, Proynov EI, Pieniazek PA, Rhee YM, Ritchie J, Rosta ESCD, Simmonett AC, Subotnik JE, Woodcock HL, III, Zhang WBAT, Chakraborty AK, Chipman DM, Keil FJ, Warshel A, Hehre WJSIHF, Kong J, Krylov AI, Gill PMW, Head-Gordon M. Phys Chem Chem Phys. 2006;8:3172. [Google Scholar]
- 59.Frisch MJTGW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery J, JA, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian, Inc; Wallingford CT: 2004. [Google Scholar]
- 60.Halkier A, Helgakerb T, Jorgensena P, Klopperb W, Kocha H, Olsenc J, Wilson AK. Chem Phys Lett. 1998;286:243. [Google Scholar]
- 61.Shen Y, Bax A. J Biomol NMR. 2010;48:13. doi: 10.1007/s10858-010-9433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu X, Shim J, Lopes PEM, MacKerell AD., Jr J Chem Inf Modell. 2012 doi: 10.1021/ci300079. In Press. [DOI] [Google Scholar]
- 63.Woodcock HL, Moran D, Pastor RW, MacKerell AD, Jr, Brooks BR. Biophys J. 2007;93:1. doi: 10.1529/biophysj.106.099986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guvench O, MacKerell AD., Jr J Mol Model. 2008;14:667. doi: 10.1007/s00894-008-0305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berman HM, Henrick K, Nakamura H. Nat Struct Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 66.Manders EMM, Verbeek FJ, Aten JA. J Microsc. 1993;169:375. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 67.Bernard D, Coop A, MacKerell AD., Jr J Med Chem. 2007;50:1799. doi: 10.1021/jm0612463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmidt JM, Blümel M, Löhr F, Rüterjans H. J Biomol NMR. 1999 doi: 10.1023/A:1008345303942. [DOI] [PubMed] [Google Scholar]
- 69.Case DA, Scheurer C, Brüschweiler R. J Am Chem Soc. 2000;122:10390. [Google Scholar]
- 70.Pérez C, Löhr F, Rüterjans H, Schmidt JM. J Am Chem Soc. 2001;123:7081. doi: 10.1021/ja003724j. [DOI] [PubMed] [Google Scholar]
- 71.Chou JJ, Case DA, Bax A. J Am Chem Soc. 2003;125:8959. doi: 10.1021/ja029972s. [DOI] [PubMed] [Google Scholar]
- 72.Bartlett GJ, Choudhary A, Raines RT, Woolfson DN. Nature Chem Biol. 2010;6:615. doi: 10.1038/nchembio.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shalongo W, Dugad L, Stellwagen E. J Am Chem Soc. 1994;116:8288. [Google Scholar]
- 74.Zwanzig RW. J Chem Phys. 1954;22:1420. [Google Scholar]
- 75.Scholtz JM, York EJ, Stewart JM, Baldwin RL. J Am Chem Soc. 1991;113:5102. [Google Scholar]
- 76.Muñoz V, Serrano L. J Mol Biol. 1995;245:275. doi: 10.1006/jmbi.1994.0023. [DOI] [PubMed] [Google Scholar]
- 77.Halgren TA, Nachbar RB. J Comput Chem. 1996;17:587. [Google Scholar]
- 78.Oosterbrink C, Villa A, Mark AE, Van Gunsteren WF. J Comput Chem. 2004;25:1656. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 79.Aliev AE, Courtier-Murias D. J Phys Chem B. 2010;114:12358. doi: 10.1021/jp101581h. [DOI] [PubMed] [Google Scholar]
- 80.Best RB, Merchant KA, Gopich IV, Schuler B, Bax A, Eaton WA. Proc Natl Acad Sci U S A. 2007;104:18964. doi: 10.1073/pnas.0709567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smith JL, Corfield PWR, Hendrickson WA, Low BW. Acta Crystallogr, Sect A: Found Crystallogr. 1988;44:357. doi: 10.1107/s0108767388000303. [DOI] [PubMed] [Google Scholar]
- 82.Jelsch C, Teeter MM, Lamzin V, Pichon-Pesme V, Blessing RH, Lecomte C. Proc Natl Acad Sci U S A. 2000;97:3171. doi: 10.1073/pnas.97.7.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ramachandran GN, Ramakrishnan C, Sasisekharan V. J Mol Biol. 1963;7 doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 84.Shaw DE, Maragakis P, Lindorff-Larsen K, Piana S, Dror RO, Eastwood MP, Bank JA, Jumper JM, Salmon JK, Shan Y, Wriggers W. Science. 2010;330:341. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 85.Hennig M, Bermel W, Dobson CM, Smith LJ, Schwalbe H. J Mol Biol. 1999;288:705. doi: 10.1006/jmbi.1999.2722. [DOI] [PubMed] [Google Scholar]
- 86.Harata K. Acta Crystallogr, Sect A: Found Crystallogr. 1993;44:357. [Google Scholar]
- 87.Kamada K, De Angelis J, Roeder RG, Burley SK. Proc Natl Acad Sci U S A. 2001;98:3115. doi: 10.1073/pnas.051631098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wlodawer A, Walter J, Huber R, Sjölin L. J Mol Biol. 1984;180:301. doi: 10.1016/s0022-2836(84)80006-6. [DOI] [PubMed] [Google Scholar]
- 89.Szebenyi DM, Moffat K. J Biol Chem. 1986;261:8761. [PubMed] [Google Scholar]
- 90.Szebenyi DM, Moffat K. J Biol Chem. 1986;261:8761. [PubMed] [Google Scholar]
- 91.Kachalova GS, Popov AN, Bartunik HD. Science. 1999;284:473. doi: 10.1126/science.284.5413.473. [DOI] [PubMed] [Google Scholar]
- 92.Flyvbjerg H, GPH J Chem Phys. 1989;91:461. [Google Scholar]
- 93.Vajpai N, Gentner M, Huang J-r, Blackledge M, Grzesiek S. J Am Chem Soc. 2010;132:3196. doi: 10.1021/ja910331t. [DOI] [PubMed] [Google Scholar]
- 94.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical recipes in C. Cambridge University Press; Cambridge, U.K: 1992. [Google Scholar]
- 95.Nettels D, Gopich IV, Hoffmann A, Schuler B. Proc Natl Acad Sci U S A. 2007;104:2655. doi: 10.1073/pnas.0611093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.