Abstract
Background
Cis-natural antisense transcripts (cis-NATs) are RNAs transcribed from the antisense strand of a gene locus, and are complementary to the RNA transcribed from the sense strand. Common techniques including microarray approach and analysis of transcriptome databases are the major ways to globally identify cis-NATs in various eukaryotic organisms. Genome-wide in silico analysis has identified a large number of cis-NATs that may generate endogenous short interfering RNAs (nat-siRNAs), which participate in important biogenesis mechanisms for transcriptional and post-transcriptional regulation in rice. However, the transcriptomes are yet to be deeply sequenced to comprehensively investigate cis-NATs.
Results
We applied high-throughput strand-specific complementary DNA sequencing technology (ssRNA-seq) to deeply sequence mRNA for assessing sense and antisense transcripts that were derived under salt, drought and cold stresses, and normal conditions, in the model plant rice (Oryza sativa). Combined with RAP-DB genome annotation (the Rice Annotation Project Database build-5 data set), 76,013 transcripts corresponding to 45,844 unique gene loci were assembled, in which 4873 gene loci were newly identified. Of 3819 putative rice cis-NATs, 2292 were detected as expressed and giving rise to small RNAs from their overlapping regions through integrated analysis of ssRNA-seq data and small RNA data. Among them, 503 cis-NATs seemed to be associated with specific conditions. The deep sequence data from isolated epidermal cells of rice seedlings further showed that 54.0% of cis-NATs were expressed simultaneously in a population of homogenous cells. Nearly 9.7% of rice transcripts were involved in one-to-one or many-to-many cis-NATs formation. Furthermore, only 17.4-34.7% of 223 many-to-many cis-NAT groups were all expressed and generated nat-siRNAs, indicating that only some cis-NAT groups may be involved in complex regulatory networks.
Conclusions
Our study profiles an abundance of cis-NATs and nat-siRNAs in rice. These data are valuable for gaining insight into the complex function of the rice transcriptome.
Keywords: Oryza sativa, Cis-NATs, Nat-siRNAs, SsRNA-seq, Transcriptome
Background
Cis-natural antisense transcripts (cis-NATs) are endogenous RNA molecules that are transcribed from the opposite DNA strand of the same genomic locus and overlap partly with sense RNA by convergent or divergent orientation. It has been clearly demonstrated that cis-NATs are an important biogenesis mechanism to generate endogenous short interfering RNAs (siRNAs) known as ‘natural antisense siRNAs’ (nat-siRNAs) [1,2]. Numerous evidence indicates that NATs use diverse transcriptional and post-transcriptional gene regulatory mechanisms to carry out different biological roles [3], including RNA interference [4], gene silencing [5-7], RNA masking-induced alternative splicing [8] and RNA editing [9].
Since a large number of natural antisense transcripts were first identified to be widespread in the human genome [10], computational analysis of data generated from large-scale sequencing projects has been widely used to globally identify cis-NATs in various eukaryotic organisms. Common techniques and databases, including in silico analyses of expressed sequence tag (EST) databases [11,12], genomic annotation of large transcript sets [13-17], large-scale sequencing of full-length complementary DNAs (cDNAs) [18-20] and tiling arrays [21-24], have been applied to identify NATs. Recently, more efforts were made to characterize nat-siRNAs and NATs at a genome-wide level: massively parallel signature sequencing (MPSS) data [25,26], combining pyrophosphate-based high-throughput sequencing and computational analyses of genomic annotation datasets [27,28] and asymmetric strand-specific analysis of gene expression (ASSAGE) [29]. It has been reported that about 5-70% in mammals and 7-9% in plants of all transcripts are overlapped as cis-NATs. These studies have demonstrated four major characteristics of cis-NATs. (i) Both cis-NAT pairs can encode proteins or be non-protein-coding transcripts. In the mammalian genome, a non-protein-coding antisense RNA partner of a protein-coding transcript is considered the most prominent form [13,15]. In Arabidopsis, ~88% of the sense-antisense transcripts (6858 of 7805) were shown to be pairs of protein-coding genes (AGI code genes) and non-protein-coding RNAs from non-AGI transcriptional units [21,24]; however, it has been reported that > 86% of rice bidirectional transcript pairs included a coding sequence in both strands [19]. (ii) The distribution of antisense transcripts in mammals was found to be non-random across the genome [29]. (iii) Expression levels of sense and antisense transcripts can be either positively or negatively correlated [15,21,30,31]. (iv) In Arabidopsis, most of the cis-NATs are arranged in convergent orientation. Several genes were found to be involved in two cis-NATs as a network: one pair is convergent, another is divergent [20,27].
However, there are still some limitations to comprehensively identifying cis-NATs. Firstly, the percentage of cis-NATs in different eukaryotic genomes, especially in plants, is estimated mainly by the alignment of full-length cDNAs, ESTs and predicted coding sequences to the genome. The transcriptomes are still not sequenced deeply enough to provide all transcripts, including low copy number and non-coding RNAs (ncRNAs). Secondly, as important evidence to determine the regulation of cis-NATs by RNA interference, nat-siRNAs are still far from saturated, despite many efforts in plants (rice and Arabidopsis) in recent years [26-28,32-35]. Thirdly, it is also essential to accurately quantify the expression levels of sense and antisense transcripts at a global level.
The advent of second-generation sequencing technology enables deep sequencing of transcripts. The paired-end tag sequencing strategy of strand-specific cDNA sequencing technology (ssRNA-seq) has the potential to globally produce abundant and novel transcripts with clear polarity and to accurately assess gene activity [36,37]. Moreover, each cell type has its unique transcriptome, so a single-cell-level description of gene expression and regulation can be instructive concerning cell populations [38,39]. The latest RNA-seq applied to single cells gives more precise transcriptome quantifications than a PCR-based amplification method [40]. Meanwhile, high-throughput sequencing techniques have made it feasible to obtain all small RNAs species, genome-wide, as it can generate hundreds of millions reads in a single sequencing run.
Here, we took advantage of ssRNA-seq technology to deeply sequence cDNAs with clear transcriptional orientations in the model plant species rice (Oryza sativa L.). All mRNAs were derived from seedlings grown under normal (seedling mixture, and only epidermal cells as well) and abiotic stressed conditions for assessing rice cis-NATs at the best possible resolution. We also deeply sequenced small RNAs to investigate nat-siRNAs from rice seedlings under normal and several stresses conditions. In addition, we tried to sequence transcriptome of rice leaf epidermal cells and evaluate the expression of cis-NATs identified in this research. We identified 2292 rice cis-NATs with both evidence of gene expression and nat-siRNAs from their overlapping regions. About 54.0% of them were shown to be simultaneously expressed in epidermal cells. Some cis-NATs gave rise to nat-siRNAs exclusively in the overlap regions, and some cis-NATs seemed to be expressed under specific abiotic stress conditions. This study was the first attempt of applying ssRNA-seq to deeply investigate novel transcripts and revealed widespread occurrence of cis-NATs in rice.
Results
ssRNA-seq and assembly of rice transcripts
In order to comprehensively identify rice cis-NATs, we applied ssRNA-seq to deeply sequence rice cDNAs for assessing transcripts with clear transcriptional orientations. Rice seedlings grown under normal and three abiotic stress conditions (salt, cold and drought treatments) were collected for preparations of mRNA and small RNAs (see Methods), and four strand-specific cDNA libraries were then constructed according to the empirical protocol [37]. In general, a modified RNA-seq method is used for incorporation of deoxy-UTP during second-strand cDNA synthesis and subsequent destruction of the uridine-containing strand in the sequencing library. This enables the identification of transcript orientation. The high-throughput sequencing of strand-specific cDNAs was performed on the Illumina GAIIX. In total, 14.7, 11.5, 14.2 and 13.2 million paired-end reads of 2 × 120 bp with high-quality, which were generated from the untreated and salt, cold and drought stress treatments libraries, respectively, matched unambiguously and uniquely to the rice reference genome [41] (Additional file 1). We estimated the accuracy of transcriptional orientation by comparing the sequencing reads with the annotated gene datasets [42]. About 89.4-95.5% of the mapped reads appeared to be aligned with correct transcriptional orientation, providing strong evidence for ssRNA-seq data in a strand-specific manner. In contrast to ssRNA-seq data, only half of the mapped reads from the previous RNA-seq data were consistent with the gene models at the same transcriptional orientations. In addition, we also deeply sequenced epidermal cells of normal rice seedlings, and about 10.4 million paired-end reads of 2 × 100 bp were generated as high-quality data.
Using the software TopHat and Cufflinks [43-46], we assembled ssRNA-seq data combined with the rice genome annotation [42] into 76,013 transcripts corresponding to 45,844 unique gene loci (including 4873 novel gene loci). Of them, 25,924 were identified as novel transcripts, which were composed of 5063 ncRNA, 16,494 CDS with protein hits and 4367 CDS without any protein hits (Additional file 2).
Identification of putative cis-NATs in rice
Based on renewed assembled transcripts, we identified putative cis-NATs which overlapped, but were opposite, from the same or adjacent gene locus. In total, 5813 pairs of rice cis-NATs (Additional file 3) were preliminarily screened out. After excluding those in which either of the pairs encoded a transposon, rRNA, tRNA, snRNA, snoRNA or miRNA, we obtained 3819 putative cis-NATs with mean overlapped length of 785 nt (Table 1). According to the directions of the involved transcripts, 2149 (56.3%) cis-NATs were categorized in enclosed, 898 (23.5%) in convergent (3′-3′ overlap), and 772 (20.2%) in divergent (5′-5′ overlap) orientations. Of rice cis-NATs, 36.1% (1378 of 3819) were pairs of protein-coding genes and non-protein-coding RNAs (Table 1). Another 33.4% (1275 of 3819) were a PFAM domain-containing transcript partner of a predicted CDS without any PFAM domain. In general, the majority of cis-NATs (3358 or 87.9%) were one-to-one type, i.e. one transcript in a cis-NAT pair had only one antisense partner. The remaining 461 cis-NATs (composed of 685 transcripts) were involved in networks of 223 cis-NAT groups.
Table 1.
Statistics of 3819 cis-NATs identified in rice
| Chr. | Transcripts | cis-NATsa | Enclosedb | 3′-3′c | 5′-5′d | CDS-pevs. ncRNA | CDS-p vs. CDS-nf | CDS-p vs. CDS-p | CDS-n vs. ncRNA | CDS-n vs. CDS-n | ncRNA vs. ncRNA |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
10,536 |
558 |
301 |
146 |
11 |
205 |
176 |
92 |
39 |
30 |
16 |
| 2 |
8,527 |
440 |
239 |
115 |
86 |
159 |
141 |
63 |
36 |
30 |
11 |
| 3 |
9,197 |
491 |
296 |
120 |
75 |
175 |
156 |
90 |
29 |
22 |
19 |
| 4 |
6,762 |
361 |
185 |
95 |
81 |
100 |
132 |
55 |
27 |
27 |
20 |
| 5 |
5,940 |
290 |
161 |
71 |
58 |
107 |
108 |
34 |
21 |
11 |
9 |
| 6 |
6166 |
299 |
170 |
69 |
60 |
111 |
100 |
38 |
23 |
13 |
14 |
| 7 |
5,862 |
278 |
163 |
63 |
52 |
90 |
96 |
30 |
24 |
19 |
19 |
| 8 |
5,290 |
239 |
138 |
51 |
50 |
103 |
74 |
27 |
19 |
10 |
6 |
| 9 |
4,256 |
189 |
100 |
34 |
55 |
70 |
71 |
10 |
16 |
10 |
12 |
| 10 |
4,144 |
221 |
128 |
41 |
52 |
85 |
74 |
24 |
19 |
10 |
9 |
| 11 |
4,861 |
231 |
139 |
47 |
45 |
92 |
75 |
25 |
23 |
7 |
9 |
| 12 |
4,472 |
222 |
129 |
46 |
47 |
81 |
72 |
24 |
20 |
15 |
10 |
| Total | 76,013 | 3,819 | 2,149 | 898 | 772 | 1,378 | 1,275 | 512 | 296 | 204 | 154 |
acis-NAT pairs without transposons.
b One transcript being entirely reverse-complementarily overlapped by the other.
c Convergent cis-NAT (with 3′-ends overlapped).
d Divergent cis-NAT (with 5′-ends overlapped).
e Coding sequence with PFAM domain-containing.
f Predicted as coding sequence but without any PFAM domain hits.
Small RNAs and nat-siRNAs in rice
To investigate the complexity of small RNAs in rice, we generated four small RNA libraries from rice seedlings under normal conditions and three abiotic stress treatments (salt, cold and drought), and deeply sequenced the libraries on the Illumina GAIIX. The small RNA libraries were made with an RNA ligation method which produces strand-specific libraries. After removing low-quality reads and those mapped to rRNA, tRNA, sn/snoRNA, mitochondria and chloroplasts, 4,843,040, 3,973,627, 2,894,255 and 5,492,145 distinct small RNAs (representing 17,632,759, 12,923,509, 8,720,251 and 20,069,157 of 48,683,191, 49,254,272, 25,705,840 and 50,192,805 raw reads, respectively) from corresponding untreated, salt, cold and drought conditions, were identified (Additional file 1). Small RNAs of 24-nt were the predominant size class (Additional file 4A). We found that the majority were located in transposon-related regions, followed by upstream, intergenic, downstream, and intron regions (Additional file 4C).
To determine the amount of nat-siRNAs, we searched in the four small RNA libraries that matched uniquely and perfectly to the overlapping regions of the 3819 cis-NATs. A total of 180,239 reads corresponding to 90,977 unique small RNAs (normal: 25,420; salt: 18,598; cold: 18,152 and drought: 28,807) were derived from overlapped regions (Additional files 1 and 5). Here, nat-siRNAs showed wide ranges in size of 18-34 nt, with 21-25 nt the most common size (Additional file 4B). The 5′-first nucleotide of nat-siRNAs was predominantly adenosine (Additional file 4F), which differed from that of the total small RNAs (Additional file 4E).
cis-NATs and nat-siRNAs with gene expression evidence under normal or abiotic stress conditions
To gain further insight into regulation of cis-NATs, we examined the expression levels of 3358 one-to-one type cis-NATs. The 2292 cis-NAT pairs were detected as expressed [i.e. Fragments Per Kilobase of exon Models (FPKM) > 0] with both sense and antisense transcripts under normal (1789 pairs), cold (1668), salt (1572) or drought (1668) conditions, and also with nat-siRNAs in an overlapping region (Table 2 and Additional file 6). We found that 166 pairs produced siRNAs exclusively and had more than five unique small RNA reads in the overlapping regions (Additional file 7). To explore whether small RNAs were more enriched in the overlapping compared to non-overlapping regions, we calculated small RNA densities of cis-NATs in these two regions. The siRNA densities of 13 cis-NATs were > 5 times that in the non-overlapping regions (Additional file 8). We further investigated cis-NATs which generated nat-siRNAs with strand bias: 25-28% of cis-NAT pairs exhibited strong strand bias in spawning nat-siRNAs with > 5-fold change in both normal and abiotic stresses (Additional file 9A). More than 75% of cis-NATs generated small RNAs from different directions with > 2-fold change (Additional file 9B). Interestingly, cis-NAT pairs, which were composed of the protein-coding gene partner of non-protein-coding RNAs, overwhelmingly produced small RNAs from the strand of protein-coding transcripts (Additional file 9).
Table 2.
Numbers of cis-NATs with expression evidence and nat-siRNAs, under four different conditions and in epidermal cells
| Both expressed ciscbsgNAT pairs | Validated in epidermal cells | Pairs with siRNAs only in overlap regionf | Pairs with siRNAs enriched in overlap regiong | |
|---|---|---|---|---|
| sda |
1,789 |
1,043 |
72 |
9 |
| STb |
1,572 |
949 |
58 |
5 |
| CDc |
1,668 |
1,003 |
66 |
5 |
| DTd |
1,668 |
986 |
75 |
5 |
| Co- |
1,072 (46.7%)e |
725 (58.6%)e |
10 (5.9%)e |
2 (15.4%)e |
| Total | 2,292 | 1,238 | 166 | 13 |
a 14-d-old-seedling.
b 14-d-old-seedling treated with 200 mM NaCl.
c 14-d-old-seedling grown under cold stress at 4°C for 24 h in darkness.
d 14-d-old-seedling treated with 20% PEG-6000.
e The percentage of co-cis-NATs in the corresponding total cis-NATs was calculated.
f Small RNAs were only enriched in the overlapping regions with more than five reads.
g The density of siRNAs in overlapping regions was > 5 times that in non-overlapping regions.
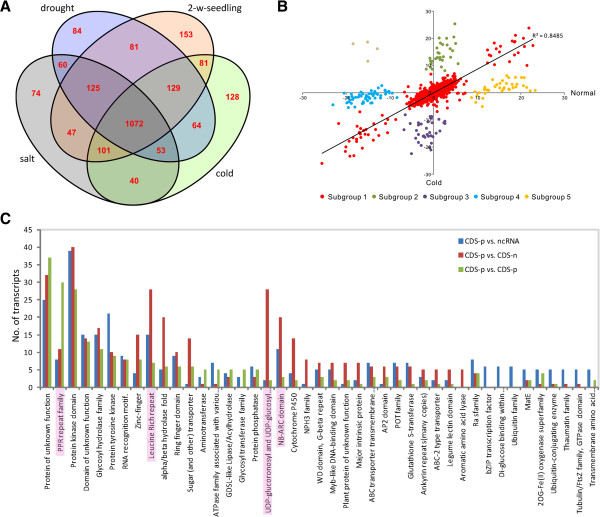
Of 2292 cis-NATs, 1072 (46.7%) cis-NATs were expressed in both normal and abiotic stresses (Figure 1A). The cis-NAT pairs could be divided into five subgroups based on a scatter plot comparing transcripts expressional ratio trends of cis-NAT pairs between normal and cold stressed conditions (Figure 1B). We also investigated the functional bias among each group of cis-NAT pairs through functional domain and expressional profiling analyses. We found that Protein kinase domains were commonly identified in cis-NAT pairs among the five subgroups. However, other functional domains and expression levels of sense/antisense transcripts were detected to be associated with different subgroups of cis-NAT pairs.
Figure 1.
Overview of 2292 one-to-one cis-NATs. (A) Venn diagram showing the shared cis-NATs that expressed under normal (control), cold, salt and drought conditions. The numbers of cis-NATs that expressed under different conditions are noted in the corresponding parentheses. (B) Scatter plot showing comparison of transcripts expression ratio trends of 1072 co-expressed cis-NAT pairs between normal and cold stress conditions. Five subgroups 1, 2, 3, 4 and 5 are indicated by red, green, purple, blue and orange points, respectively. (C) Three sets of genes are categorized on the basis of components of each cis-NAT pair. Set I (blue bar): 767 cis-NAT pairs of protein-coding genes and non-protein-coding RNAs; Set II (red bar): 818 cis-NAT pairs of protein-coding genes and predicted CDS without any PFAM domain; Set III (green bar): 377 cis-NATs composed of both protein-coding genes. Y-axis represents the number of transcripts. X-axis represents 43 protein families that were the major enriched terms in either of three sets. Four proteins of pentatricopeptide repeat (PPR) families, leucine rich repeat, NB-ARC domain and UDP-glucoronosyl and UDP-glucosyl transferase, which exhibited distinctively enriched terms with P < 0.001 in Sets II and III, are highlighted in pink in the X-axis.
Subgroup-1 cis-NATs were predominantly associated with the pentatricopeptide repeat (PPR) and Protein tyrosine kinase domains. We also found that the expression levels of the cis-NAT pairs of Subgroup-1 under stress conditions were similar to that under normal conditions. Subgroup-2 cis-NATs were identified to contain domains of leucine rich repeats and glycosyltransferase. In this subgroup, the expression levels of sense-transcripts of the cis-NAT pairs were significantly increased under stress conditions, while expression levels of antisense transcripts were greatly reduced. Thus, expression levels were higher for sense than for antisense transcripts under stress conditions. Subgroup-3 can be classified as the family of alpha/beta hydrolase fold. The expression levels of sense-transcripts of the Subgroup-3 cis-NAT pairs were significantly reduced under stress conditions, while expression levels of antisense transcripts significantly increased. Thus, expression levels were lower for sense than for antisense transcripts under stress conditions. Subgroup-4 cis-NATs can be classified as the families of the eukaryotic aspartyl protease and sugar (and other) transporter. Subgroup-5 cis-NATs were identified to be the families of glycosyl hydrolases and ubiquitin-conjugating enzyme. The remaining four cis-NAT pairs were not classified into any subgroups, as the expression levels of sense transcripts were not correlated with that of anti-sense partner under normal and abiotic conditions. We identified these four cis-NAT pairs as abnormal values (Figure 1B; indicated as the brown spots). Subgroup-1 was shown to be represented the major part. Ratio values of 913 cis-NAT pairs (Subgroup-1) between normal and cold stressed conditions were highly correlated, with R2 > 0.85. Similar results were also observed between normal and salt/drought stressed conditions with R2 > 0.93 of 917 pairs and R2 > 0.93 of 898 pairs, respectively (Additional file 10). In total, of 1072 cis-NAT pairs, 95.5% (1024) showed positive correlations between sense and antisense transcripts. One cis-NAT pair (Os05t0500000-00 vs. Os05t0500101-01) was taken as an example to compare expression changes under normal and cold stressed conditions (Figure 2). Northern blot analyses also demonstrated their differential expression levels under abiotic stresses (Figure 2C). Sense transcript Os05t0500000-00 is annotated as an UDP-glucoronosyl and an UDP-glucosyl transferase, which is an important enzyme for catalyzing transportation of sugars. Antisense transcript Os05t0500101-01 had no hits to any protein families.
Figure 2.

An example of regulatory small RNAs derived from a cis-NAT pair. (A) A cis-NAT pair was formed with genes UDP-glucoronosyl and UDP-glucosyl transferase (Os05t0500000-00) and predicted CDS without any protein hits (Os05t0500101-01). Os05t0500000-00 was re-annotated by ssRNA-seq data (short blue lines) from normal condition in the plus strand. Os05t0500101-01 was re-annotated by ssRNA-seq data (short red lines) in the minus strand. The cis-NATs spawned five unique small RNAs, mainly from the plus strand (short green lines). (B) Under cold-stress conditions, expression of this cis-NAT pair was dramatically down-regulated. (C) On the left: Northern blot analysis of the cis-NATs confirmed differential expression levels under normal and cold treatment. On the right: the figure shows the FPKM results of this cis-NAT pair. FPKM refers to Fragments Per Kilobase of exon Models.
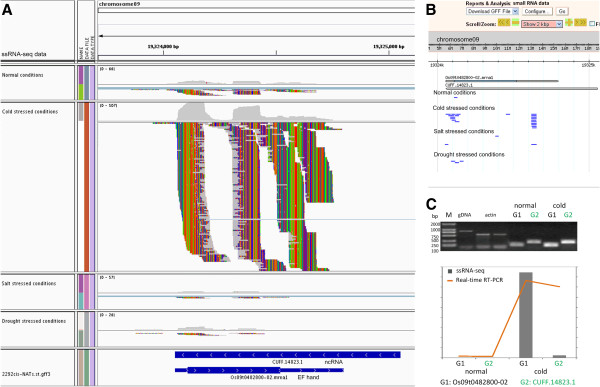
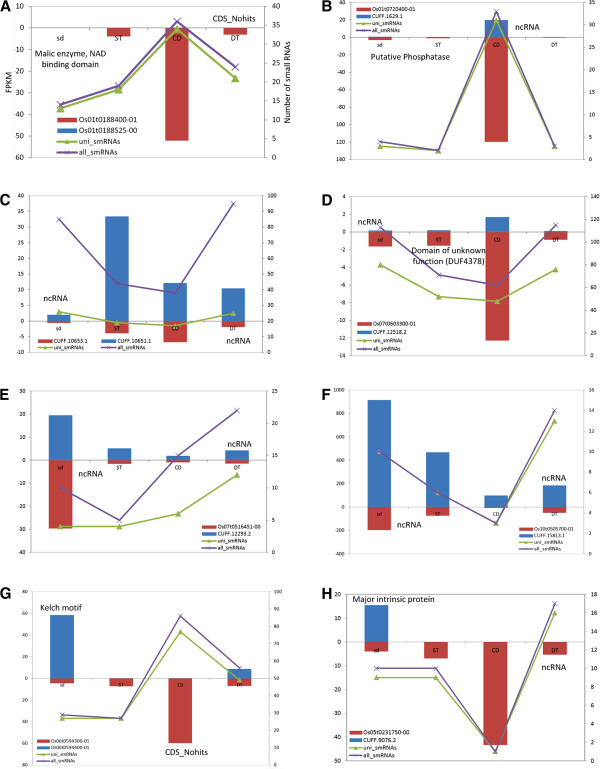
The numbers of cis-NATs expressed under different conditions were quite different (Figure 1A). The shared cis-NATs expressed under normal (control), cold, salt and drought conditions were also identified. Among them, 503 (21.9%) cis-NATs were detected preferentially under abiotic stresses compared to normal conditions. There were 84, 74 and 128 cis-NAT pairs clearly expressed under drought, salt and cold stresses, respectively. As an example, under cold treatment, sense and antisense transcripts of cis-NAT pair Os09t0482800-02 vs. CUFF.14823.1 were both much more expressed than under normal or other stressed conditions (Figure 3). Semi-quantitative RT-PCR and reverse transcription PCR (RT-PCR) were used to validate the expression of the cis-NAT pair under normal and cold conditions (Figure 3B). Functional annotation of this cis-NAT pair was composed of EF-hand protein (Os09t0482800-02) and ncRNA (CUFF.14823.1). Moreover, 1238 of 2292 (54.0%) cis-NATs were expressed in leaf epidermal cells. Of 1238 cis-NATs, 725 belonged to co-expressed pairs (Table 2). We further performed analysis of differentially expressed genes (DEGs). In total, we identified 112 cis-NATs with DEGs of at least either of each pair, which belonged to different subgroups (Additional file 11). Of them, expression levels of 69 cis-NATs (either sense or antisense transcripts) were up-regulated under abiotic stresses, while 46 cis-NATs were down-regulated under abiotic stresses. Moreover, the results showed no clear expression pattern between transcripts and nat-siRNAs (Figure 4). The number of nat-siRNAs notably increased along with higher transcriptional expression level under cold-stress conditions (Figure 4A and B); in contrast, some cis-NATs generated less nat-siRNAs, along with higher expression levels (Figure 4C and D). In addition, some cis-NATs generated nat-siRNAs with more complex expression patterns (Figure 4E-H).
Figure 3.
An example of an overwhelmingly up-regulated cis-NAT under abiotic stresses. (A) Expressions of a cis-NAT pair of an annotated gene Os09t0482800-02 and a novel ncRNA CUFF.14823.1 are dramatically up-regulated under cold-stress conditions. Expression levels of this cis-NAT pair under normal, cold, salt and drought conditions are relatively low and displayed. (B) The unique small RNAs that are generated mainly from the plus strand (short blue lines) of the cis-NAT pair of the gene Os09t0482800-02 and the ncRNA CUFF.14823.1 are shown. (C) Expressions of this cis-NAT pair were detected through semi-quantitative RT-PCR analysis. Rice actin expression was used as a control. The PCR products of the cis-NAT were amplified by 36 cycles, while actin products were amplified by 26 cycles. Real-time RT-PCR analysis of the cis-NATs was used to confirm the differential expressed genes under normal and cold conditions. Expression data from ssRNA-seq (FPKM, Fragments Per Kilobase of exon Models) are represented as gray blocks; and the data from real-time RT-PCR are indicated as orange lines.
Figure 4.
Expression patterns of eight one-to-one cis-NATs and the distribution of small RNAs located in overlapped regions of these gene pairs. (A-H) Expression patterns of eight cis-NATs are displayed. The left Y-axis of each graph represents FPKM of each transcript. Blue bars indicate FPKM of plus- transcripts (above the X-axis); red bars indicate FPKM of its partner of minus- transcripts (below the X-axis). The right Y-axis indicates the number of small RNAs located in overlapped regions. The green curves represent unique nat-siRNAs, and the purple curves represent all nat-siRNAs. FPKM refers to Fragments Per Kilobase of exon Models.
We used an enrichment analysis of PFAM protein families [47] to functionally characterize three sets that could be annotated from 2292 cis-NATs. The three sets were 767 cis-NAT pairs of protein-coding genes and non-protein-coding RNAs (Set I), 818 cis-NAT pairs of protein-coding genes and predicted CDS without any PFAM domain-containing (Set II), and 377 cis-NATs composed of both protein-coding genes (Set III), respectively. Roughly, 43 protein families were the major enriched terms in either of three sets (Figure 1C). Of them, four proteins exhibited distinctively enriched terms with P-values < 0.001. PPR families were enriched in Set III, leucine rich repeat and NB-ARC domain were significantly reduced in Set III, and UDP-glucoronosyl and UDP-glucosyl transferase were significantly enriched in Set II (Figure 1C).
Networks formed by cis-NAT groups
We further investigated networks formed by cis-NATs in rice, named ‘many-to-many’ cis-NATs, i.e. one transcript in a cis-NAT had more than one antisense partner. Networks formed by cis-NATs reflect the complexity of their post-transcriptional regulation. It has been reported that in Arabidopsis, several genes are involved in two cis-NATs, one is convergent and the other divergent [20,27]. In rice, we found 461 cis-NATs (composed of 685 transcripts) involved in 223 cis-NAT groups (Additional file 12). Of them, 209 groups belonged to one-to-two type, nine belonged to one-to-three type, four belonged to two-to-two groups, and one belonged to one-to-four group, respectively. Five types were identified in one-to-two cis-NAT groups (Additional file 12). Except 46 groups which were composed of convergent together with divergent cis-NATs, it also included 74 enclosed together with enclosed, 40 enclosed together with divergent, 48 enclosed together with convergent, and one convergent together with convergent. Interestingly, we found only 17.4-34.7% cis-NAT groups could be detected with simultaneous expression evidence and nat-siRNAs in the same sample from normal, cold, salt or drought conditions; another 29.2-48.6% cis-NAT groups were either expressed (Additional file 13). Similar to one-to-one type cis-NATs, protein kinase domain, leucine rich repeats, NB-ARC domain and the PPR repeat family were the major enriched PFAM protein domains.
Discussion
Increasing amounts of evidence indicate that antisense transcripts are frequently functional and play various biological roles using different transcriptional and post-transcriptional gene regulatory mechanisms [1,2,48-53]. Consequently, we tried to advance rice cis-NATs analysis through an integrated approach of ssRNA-seq of rice transcriptome and genome-wide in silico transcriptome data analysis.
There have been five previous studies using global investigation of rice cis-NATs. An analysis of 32,127 full-length rice cDNA sequences showed 687 bidirectional transcript pairs [19]. Comprehensive expression profiles of rice using MPSS technology identified 11,001 antisense signatures corresponding to 8023 annotated genes with highly specific expression patterns [26]. A comparative analysis of cis-NATs in eukaryotes identified 1088 rice cis-NATs referring to MSU and KOME gene datasets [17]. Combining pyrophosphate-based high-throughput sequencing of rice small RNA and computational analysis of the MSU rice gene models identified 344 cis-NATs formed by protein-coding genes [28]. Recently, 767 pairs of cis-NATs were identified using the rice genome annotation data (MSU TGAP 6.1) [35]. In this study, we identified 3819 pairs of rice cis-NATs. Of them, 2292 were identified as expressed and gave rise to small RNAs from their overlapping regions. The pairs of cis-NATs were confirmed to be formed in laser microdissection-captured rice seedling leaf epidermal cells, which were developed from a single-cell tissue. This indicated that antisense transcripts were more widespread and of more complex occurrence than previously found in the rice genome.
Moreover, qRT-PCR and Northern blot analyses confirmed the presence of cis-NATs and differential expression levels under normal and abiotic stresses. Nearly 9.7% of rice transcripts were involved in one-to-one or many-to-many cis-NATs formation. An overwhelming majority of cis-NATs (3358 of 3819, i.e. 87.9%) were of one-to-one type. Small RNAs generated from cis-NATs were broadly detected under either normal developmental conditions or stresses. Similar to the situation for mammals [15], we found that the most prominent form of antisense transcription in rice was a non-coding RNA partner of a protein-coding transcript.
We identified 4873 novel gene loci in this study. Compared with previous research, more novel transcribed activity regions with reliable transcriptional orientation were identified by ssRNA-seq. This also indicated that exploration of the rice transcriptome atlas is far from finished. Along with further progress of high-throughput sequencing technology and development of more efficient software for de novo transcriptome assembly, more accurate transcriptional units (TUs) could be defined in the future.
It should be pointed out that in our research about 4.5-10.6% of reads appeared to be aligned to antisense transcripts in error (Additional file 1). This is higher than the 3.88% rate reported by Wang et al. [54], who used a slightly modified version of this dUTP method by increasing the incubation time with UDG to enforce the complete degradation of dUTPs.
In consideration of the limited amount of RNAi-related small RNAs (mainly including microRNAs and small interfering RNAs) in public databases, we proposed the following: (i) more microRNAs and siRNAs need to be identified and (ii) more classes of small RNAs that are engaged in RNAi-related machinery or those that are not will be found in future.
Conclusions
By applying a strand-specific RNA-seq approach, we systematically identified rice cis-natural antisense transcripts and putative nat-siRNAs. Our study profiled the most abundant of transcriptional active regions and revealed widespread occurrence of cis-NATs in rice, suggesting that regulation through cis-NATs and nat-siRNAs could be a common biological phenomenon. Both ssRNA-seq data and small RNA data obtained by high-throughput sequencing technology would supply important resources for further rice transcriptome analysis. It also indicated the feasibility of global investigations of cis-NATs by ssRNA-seq in eukaryotic genomes.
Methods
Materials and growth conditions
Rice seeds (O. sativa ssp. japonica cv. Nipponbare) were used in all experiments. Seeds germinated at 28°C in darkness for 2 d were transferred to a plant growth chamber to grow for 14 d under controlled conditions (12/12 h and 30/24°C of light/dark cycles) to produce seedlings and epidermal cells. For drought and salt stress treatments, 14-d-old seedlings were incubated in solutions containing 20% PEG-6000 and 200 mM NaCl, for 4 h at 30°C, respectively. For cold treatment, seedlings at the same developmental stage were treated at 4°C for 24 h in darkness.
Strand-specific cDNA library construction and sequencing
We prepared the strand-specific cDNA libraries according to a protocol [37]. The ssRNA-seq is a simple modification of the RNA-Seq method that incorporates deoxy-UTP during second-strand cDNA synthesis and subsequent destruction of the uridine-containing strand in the sequencing library. Thus it enables identifying the orientation of transcripts. Total RNA was isolated using the TRIzol reagent (Invitrogen), then total genomic DNA was removed from tissues using DNase (New England Biolabs), which was examined by gel electrophoresis. The OligoTex mRNA midi kit (Qiagen) was used to purify poly(A) mRNA from the total RNA samples. Next, mRNAs were fragmented using the RNA fragmentation kit (Ambion). The first cDNA strand was synthesized using random hexamer primers and second-strand cDNA was synthesized where dUTP was used instead of dTTP. In this step, Actinomycin D was used to increase strand specificity by inhibiting second-strand cDNA synthesis. At 15°C 0.5 μl of actinomycin D solution (120 ng/μl), 0.5 μl of RNase OUT (40 units/μl, Invitrogen) and 0.5 μl of SuperScript III polymerase (200 units/μl,Invitrogen) were added to the reaction. Then EB (20 μl) (10mM Tris–Cl, pH 8.5, Qiagen) was added to the reaction, and the dNTPs were removed by purification of the first strand mixture on a self-made 200 μl G-50 gel filtration spin-column equilibrated with 1mM Tris–Cl, pH 7.0. After second strand synthesis and DNA fragmentation process, the sequencing libraries were further constructed by following the manufacturer’s instructions (Illumina). Fragments of 300-400 bp were recovered and purified, and then enriched by PCR for 15 cycles. Each library was loaded into one lane of the Illumina GA IIX for 2 × 120 bp pair-end sequencing at a concentration of 2 pM, except that library of normal seedlings was loaded into two lanes. Image analysis and base calling were finished using the Illumina GA processing pipeline v1.4.
Laser microdissection (LM)-captured rice seedling leaf epidermal cells and aRNA preparation
Leaves of 15-d-old seedlings of rice variety TP309 grown in a growth chamber at 12/12h and 25/22°C of day/night cycle were used. Seedling leaves were cut into pieces about 5 mm long and immediately processed by microwave-accelerated acetone fixation (BP-111-RS laboratory microwave, Microwave Research & Applications Inc.) and paraffin-embedded as described by Tang et al. [55]. Cross-sections of 10 μm thickness, parallel to leaf vascular bundles, were obtained using a Leica RM2235 rotary microtome. Paraffin-tape transfer system (Instrumedics) and Veritas Microdissection Instrument (Acturus Bioscience) were used for LM-capturing epidermal cells.
We carried out microdissection of epidermal cells, which are a group of cells (a homogenous population cells). There were about 200-500 cells per sample. The total RNAs of epidermal cells were extracted by PicoPure RNA isolation kit (Acturus, CA, USA) with the DNase (RNase-free, Qiagen) treatment. The integrity of the total RNAs was evaluated by Agilent 2100 Bioanalyzer using RNA-6000 Pico LabChips (Agilent Technologies). Because the total RNA quantity of LM-captured epidermal cells was about 10 ng, we used a TargetAmp two-round aminoallyl-aRNA (antisense RNA) amplification kit (Epicentre Biotechnologies, Madison, WI, U.S.A.) with Super-Script III and SuperScript II reverse transcriptases (Invitrogen, Carlsbad, CA U.S.A.) to amplify laser-microdissected RNA [56]. For each amplification, approximately 0.5 ng of total RNA (in a 2-μl volume) was used as starting material and, typically, 5 to 10 μg aminoallyl cRNA was recovered. For evaluating the fidelity of two-round RNA amplification, the TargetAmp one-round aminoallyl-aRNA amplification kit (Epicentre) was used as a one-round amplification control. For each one-round amplification, 400 ng of total RNA was used as starting material and, typically, more than 10 μg of aminoallyl cRNA was recovered. The quality of amplified RNA was also evaluated by Agilent 2100 Bioanalyzer, and only those with a ‘bell-shaped’ curve with peak size > 300 nucleotides were used for RNA-seq.
Small RNA sequencing
The same materials described above were used for small RNA library construction using the Illumina small RNA preparation kit (v1.5) following the manufacturer’s instructions. Small RNAs of 18-34 bp were enriched by polyacrylamide gel electrophoresis, and ligated to 5′ and 3′ adapters. The ligation product was reverse-transcribed into cDNA, which was then amplified by 15 PCR cycles and subjected to Illumina’s Solexa proprietary sequencing. Here, each library was loaded into two lanes of the Illumina GA IIX for 35-bp sequencing, except that the library of cold treatment was loaded into one lane.
Northern analysis and qRT-PCR
The PCR primers and Northern probes were designed for cis-NAT validation (Additional file 14). Northern blot analysis was carried out as described [57]. To ensure that each pair of primers had specificity of its transcript, one was selected from the overlapping region of cis-NATs, and the other from the flanking region. Total RNA was extracted using the Trizol reagent (Invitrogen) according to the manufacturer’s instructions. After treated with DNaseI (NEB), 5 μg of total RNA was used to synthesize the oligo (dT) primed first-strand cDNA using SuperScript™ II reverse transcriptase (Invitrogen). For semi-quantitative RT-PCR, rice actin expression was used as a control, cis-NAT PCR products were amplified by 36 cycles, while actin products were amplified by 26 cycles. Real-time RT-PCR was performed on the Applied Biosystems 7500 real-time PCR System. Diluted cDNA was amplified using SYBR Premix Ex Taq™ (TaKaRa). The expression levels of transcripts were normalized by endogenous eEF-1α (AK061464) transcripts. Three technical replicates were taken for each set.
Assembly of rice transcripts
All ssRNA-seq data obtained from normal and cold, salt and drought stress conditions were respectively mapped to the rice reference genome [41] using software TopHat [43]. Tolerances were set to allow at most two mismatches for paired-end reads in each alignment; and reads with multiple alignments were ignored. The corresponding outputs together with RAP-DB genome annotation data [42] were subsequently used for transcript assembly to detect known and unannotated transcripts and isoforms by another software package, Cufflinks [44,46]. Then, we integrated these transcripts together according to another command, Cuffmerge, from the Cufflinks package. Finally, referring to the renewed gene models, we calculated FPKM (Fragments Per Kilobase of exon Models) of each transcript under normal, abiotic stressed conditions, and leaf epidermal cells as well [45]. Open reading frames (ORFs) of novel transcripts were predicted using the ‘getorf’ program of EMBOSS package [58], with the longest ORF extracted for each transcript. For functional annotation, all transcripts were searched against the PFAM database [47] using HMMER v3.0 (E-value < 0.0001) [59]. The MSU Oryza Repeat Database [60] was used to determine transposable element coordinates on the rice pseudomolecules and all transcribed regions, which were annotated using RepeatMasker [61].
Identification of cis-NATs and nat-siRNAs
The renewed gene annotation results were applied as models. Those assembled genes with uncertain transcriptional orientation were filtered out first. To profile all potential cis-NAT pairs in the rice genome, transcripts that originated from the same locus but from opposite strands and with non-redundant overlapped length > 25 nt were chosen. From each group of sense-antisense pairs extracted, we selected as representative pairs those with the longest exonic overlap. We wrote perl scripts to classify cis-NAT types: convergent, divergent or enclosed. Furthermore, we examined expression levels of all assembled genes using ssRNA-seq data. Thus, we could identify expressed transcripts from putative cis-NATs. According to the number of antisense partners in each cis-NAT pair, two types of cis-NATs were defined. One was one-to-one type, i.e. one transcript in a cis-NAT pair has only one antisense partner. Another is many-to-many cis-NATs, i.e. one transcript in a cis-NAT has more than one antisense partner.
Raw data of small RNA reads from four corresponding small RNA libraries were aligned to the rice genome [41] by SMALT v0.5.7 with default parameters [62]. Mapping scores below the threshold were not reported. After removing the 5′- and 3′-adapters, only those continuously and perfectly matched reads with length of 18-34 bp were extracted for further analysis. Reads with multiple alignments or with any mismatches were completely filtered out. Next, we discarded those sequences that could be unambiguously mapped to rRNA, tRNA, sn/snoRNA, mitochondria and chloroplasts. We calculated small RNA densities according to a published method [28]. Small RNAs (at least one unique read) which fully located in overlapped regions of cis-NATs were extracted as putative nat-siRNAs. The cis-NATs were identified from the set of DEGs across normal and three abiotic-stress libraries. The analysis was carried out based on the following criteria: FPKM of transcripts were used for comparison by computing fold changes (with absolute value ≥ 2) and Fisher’s exact test (p < 0.001) according to an ‘R’ statistical package named ‘DEGseq’ [63].
In addition, R2 is the square of the Pearson product-moment correlation coefficient relating the regressor and the response variable. R2 gives some information about the goodness-of-fit of a model. In regression, R2 is a statistical measure of how well the regression line approximates the real data points. ‘R’ software was used to calculate R2[64] of cis-NATs between normal and stressed conditions.
Data release
The raw sequences were deposited in the EBI European Nucleotide Archive with accession number E-MTAB-721 (http://www.ebi.ac.uk/arrayexpress/browse.html?keywords=E-MTAB-721) and ERP001962 (http://www.ebi.ac.uk/ena/data/search?query=ERP001962). The assembled transcripts can be freely downloaded xfrom http://www.ncgr.ac.cn/scientific_data.asp.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TL and BH initiated the project; TL and BH designed the experiments; GL, YG, YZ, ZZ, WL, WT and QF carried out the experiments; TL, CZ and YZ performed the computational analyses; TL, YZ and BH participated in the experimental analyses. TL and BH coordinated the research. TL and BH wrote the paper. All authors have read and approved the manuscript for publication.
Supplementary Material
Summary of pair-end reads of ssRNA-seq and small RNAs from normal and three abiotic stress conditions.
Statistics of rice transcripts.
Summary of 5813 putative cis-NATs in rice. This table lists all cis-NATs identified from rice renewed assembled transcripts. Each column represents plus transcripts (transcriptional orientations are the same as the reference), its length, minus transcripts, length, overlapped length and detailed overlapped locations in rice genome (IRGSP v5.0) as indicated.
Overview of small RNAs in rice. (A) Distribution of the lengths of all unique small RNAs generated by high-throughput sequencing from rice under four different conditions. (B) Length distribution of nat-siRNAs. (C) Distribution of all unique small RNAs located in different sequence components. (D) Distribution of nat-siRNAs located in different sequence components. The blue, red, green and purple bars represent small RNAs or nat-siRNAs from normal, salt stressed, cold stressed and drought stressed conditions, respectively (A-D). (E) First-nucleotide distribution of all unique small RNAs under four different conditions. (F) First-nucleotide distribution of nat-siRNAs under four different conditions.
Sequences of small RNAs mapped to the overlapped regions ofcis-NATs. The 25,420, 18,598, 18,152 and 28,807 unique small RNAs from normal, salt, cold and drought conditions, respectively, are shown to be perfectly mapped to the overlapped regions of cis-NATs.
Details of 2292 expressedcis-NATs. The detailed information of 2292 cis-NAT pairs with expression evidence (i.e. FPKM > 0) of both sense and antisense transcripts and with nat-siRNAs in the overlapping region under normal (1789 pairs), cold (1668), salt (1572) and drought (1668) conditions are shown. Columns 1-4 and 9-11 represent cis-NATs type, transcripts, protein domain and the length of transcripts, respectively. Columns 5-8 and 12-15 represent FPKM of transcripts under normal, salt, cold and drought conditions, respectively. Column 16 shows the overlapped length. Columns 17-20 show the number of corresponding nat-siRNAs located in the overlapped regions. The rest of the columns are the detailed strand bias of nat-siRNAs from four different conditions.
Lists of 166cis-NATs that produced nat-siRNAs exclusively in the overlapping regions with more than five unique small RNAs from four different conditions. Of 166 cis-NATs, 72, 58, 66 and 75 cis-NATs were obtained from normal, salt, cold and drought treatments, respectively. These cis-NATs are listed separately in four tables, respectively indicated by blue, orange, red and green colour.
Lists of 13cis-NATs that produced nat-siRNAs more enriched in the overlapping regions than the non-overlapping regions. Of 13 cis-NATs, nine, six, five and five cis-NATs obtained from normal, salt, cold and drought treatments, respectively. These cis-NATs are listed separately in four tables, respectively indicated by blue, orange, red and green colour.
Strand bias ofcis-NATs that gave rise to nat-siRNAs. The strand bias is computed among different types of cis-NAT pairs under four different conditions. A total of nine types are listed. Here, plus indicates that the orientation of transcripts are the same as the reference genome, minus indicates opposite orientation of transcripts. One strand means only one strand of the cis-NATs gave rise to small RNAs. The plus/minus or minus/plus means one strand of the cis-NATs spawns at least five (part A) or two (part B) fold more small RNAs than the other.
Scatter plots of expression of 1072cis-NATs. The two scatter plots compare transcripts expression ratio trends of 1072 co-expressed cis-NAT pairs between normal and drought-stress conditions (A), and between normal and salt-stress conditions (B). The results show five subgroups: red, green, purple, blue and orange spots represent subgroups 1-5, respectively.
Lists of 112cis-NATs with DEGs. Differentially Expressed Genes (DEGs) were identified in one-to-one cis-NATs under salt, drought and cold treatments by comparing their FPKM with that under normal conditions. The gene names, FPKM, fold change (with absolute value > 2) and p-value (< 0.001) are listed.
All many-to-manycis-NAT groups. In total, 223 cis-NAT groups were identified as many-to-many type. Of them, 209 groups belonged to one-to-two type, nine belonged to one-to-three type, four belonged to two-to-two groups, and one belonged to the one-to-four group. The meaning of each column is as given in Additional file 6.
Statistics of 209 networks formed bycis-NAT groups. The numbers of one-to-two cis-NAT groups with expression evidence under normal, salt, cold and drought conditions are shown. These one-to-two cis-NAT groups were divided into five classes according to their cis-NATs types. Here, ‘All EXP’ indicates all cis-NAT groups with expression evidence (FPKM > 0) of both sense and antisense transcript, and with nat-siRNAs (number of small RNAs > 1) in overlapping region as well. ‘Either EXP’ indicates either of cis-NAT groups with expression evidence of both sense and antisense transcript, and with nat-siRNAs in the overlapping region as well.
Primers designed for real-time RT-PCR and Northern Blots in this research. (DOCX 13 kb)
Contributor Information
Tingting Lu, Email: ttlu@ncgr.ac.cn.
Chuanrang Zhu, Email: crzhu@ncgr.ac.cn.
Guojun Lu, Email: gjlv@ncgr.ac.cn.
Yunli Guo, Email: gyli@ncgr.ac.cn.
Yan Zhou, Email: yzhou@ncgr.ac.cn.
Zhiyong Zhang, Email: zhiyong@sippe.ac.cn.
Yan Zhao, Email: yzhao@ncgr.ac.cn.
Wenjun Li, Email: wjli@ncgr.ac.cn.
Ying Lu, Email: ylu@ncgr.ac.cn.
Weihua Tang, Email: whtang@sibs.ac.cn.
Qi Feng, Email: qfeng@ncgr.ac.cn.
Bin Han, Email: bhan@ncgr.ac.cn.
Acknowledgements
We thank Professor Jirong Huang and his student Fenhong Hu (Institute of Plant Physiology & Ecology, Shanghai Institutes for Biological Sciences, CAS) for helping carry out Northern Blot analysis. This work was supported by the Knowledge Innovation Program of Shanghai Institutes for Biological Sciences, CAS (2010KIP203), the National Natural Science Foundation of China (Grant 31000560) and the Chinese Academy of Sciences (KSCX2-EW-Q-1-05) to TL. The authors gratefully acknowledge the support of K.C. Wong Education Foundation, Hong Kong to TL. This work was also supported by the Chinese Academy of Sciences (KSCX2-YW-N-094), the Ministries of Science and Technology (2011CB100205), and Agriculture (2011ZX08001-004 and 2011ZX08009-002) of China, and the National Natural Science Foundation of China (30821004) to BH.
References
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M, Alandete SM, Eshed WL, Fletcher JC, McCormick S. Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev. 2011;24:1010–1021. doi: 10.1101/gad.1882810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott EM, Proudfoot NJ. Transcriptional collision between convergent genes in budding yeast. Proc Natl Acad Sci USA. 2002;99:8796–8801. doi: 10.1073/pnas.132270899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/S0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet. 2003;34:157–165. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas AJ, Zhu JK, Staskawicz BJ, Jin H. A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci USA. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings ML, Milcarek C, Martincic K, Peterson ML, Munroe SH. Expression of the thyroid hormone receptor gene, erbAalpha, in B lymphocytes: alternative mRNA processing is independent of differentiation but correlates with antisense RNA levels. Nucleic Acids Res. 1997;25:4296–4300. doi: 10.1093/nar/25.21.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters NT, Rohrbach JA, Zalewski BA, Byrkett CM, Vaughn JC. RNA editing and regulation of Drosophila 4f-rnp expression by sas-10 antisense read through mRNA transcripts. RNA. 2003;9:698–710. doi: 10.1261/rna.2120703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B, Williams G, Campbell RD, Sanderson CM. Antisense transcripts in the human genome. Trends Genet. 2002;18:63–65. doi: 10.1016/S0168-9525(02)02598-2. [DOI] [PubMed] [Google Scholar]
- Shendure J, Church GM. Computational discovery of sense-antisense transcription in the human and mouse genomes. Genome Biol. 2002;3:research0044.1–research0044.14. doi: 10.1186/gb-2002-3-9-research0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, Zhou G, Shi RZ, Rowley JD. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res. 2004;32:4812–4820. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosawa H, Yamanaka I, Osato N, Kondo S, Hayashizaki Y. RIKEN GER Group, GSL Members. Antisense transcripts with FANTOM2 clone set and their implication for gene regulation. Genome Res. 2003;13:1324–1334. doi: 10.1101/gr.982903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, Nemzer S, Pinner E, Walach S, Bernstein J, Savitsky K, Rotman G. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group). FANTOM Consortium. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu XS, Liu QR, Wei L. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res. 2006;34:3465–3475. doi: 10.1093/nar/gkl473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata K, Okada Y, Saito R, Kiyosawa H, Kanai A, Tomita M. Comparative analysis of cis-encoded antisense RNAs in eukaryotes. Gene. 2007;392:134–141. doi: 10.1016/j.gene.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, Pham P, Cheuk R, Karlin-Newmann G, Liu SX, Lam B, Sakano H, Wu T, Yu G, Miranda M, Quach HL, Tripp M, Chang CH, Lee JM, Toriumi M, Chan MM, Tang CC, Onodera CS, Deng JM, Akiyama K, Ansari Y. et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- Osato N, Yamada H, Satoh K, Ooka H, Yamamoto M, Suzuki K, Kawai J, Carninci P, Ohtomo Y, Murakami K, Matsubara K, Kikuchi S, Hayashizaki Y. Antisense transcripts with rice full-length cDNAs. Genome Biol. 2003;5:R5. doi: 10.1186/gb-2003-5-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Gaasterland T, Chua NH. Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol. 2005;6:R30. doi: 10.1186/gb-2005-6-4-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA, Okamoto M, Nambara E, Nakajima M, Kawashima M, Satou M, Kim JM, Kobayashi N, Toyoda T, Shinozaki K, Seki M. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 2008;49:1135–1149. doi: 10.1093/pcp/pcn101. [DOI] [PubMed] [Google Scholar]
- Coram TE, Settles ML, Chen X. Large-scale analysis of antisense transcription in wheat using the Affymetrix GeneChip Wheat Genome Array. BMC Genomics. 2009;10:253. doi: 10.1186/1471-2164-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriadis A, Oliver GR, Tanney A, Kendrick H, Smalley MJ, Jat P, Neville AM. Identification of differentially expressed sense and antisense transcript pairs in breast epithelial tissues. BMC Genomics. 2009;10:324. doi: 10.1186/1471-2164-10-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Tatematsu K, Matsui A, Morosawa T, Ishida J, Tanaka M, Endo TA, Mochizuki Y, Toyoda T, Kamiya Y, Shinozaki K, Nambara E, Seki M. Genome-wide analysis of endogenous abscisic acid-mediated transcription in dry and imbibed seeds of Arabidopsis using tiling arrays. Plant J. 2010;62:39–51. doi: 10.1111/j.1365-313X.2010.04135.x. [DOI] [PubMed] [Google Scholar]
- Meyer BC, Vu TH, Tej SS, Ghazal H, Matvienko M, Agrawal V, Ning J, Haudenschild CD. Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nat Biotechnol. 2004;22:1006–1011. doi: 10.1038/nbt992. [DOI] [PubMed] [Google Scholar]
- Nobuta K, Venu RC, Lu C, Beló A, Vemaraju K, Kulkarni K, Wang W, Pillay M, Green PJ, Wang GL, Meyers BC. An expression atlas of rice mRNAs and small RNAs. Nat Biotechnol. 2007;25:473–477. doi: 10.1038/nbt1291. [DOI] [PubMed] [Google Scholar]
- Jin H, Vacic V, Girke T, Lonardi S, Zhu JK. Small RNAs and the regulation of cis-natural antisense transcripts in Arabidopsis. BMC Mol Biol. 2008;9:6. doi: 10.1186/1471-2199-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Sunkar R, Jin H, Zhu JK, Zhang W. Genome-wide identification and analysis of small RNAs originated from natural antisense transcripts in Oryza sativa. Genome Res. 2009;19:70–78. doi: 10.1101/gr.084806.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henz SR, Cumbie JS, Kasschau KD, Lohmann JU, Carrington JC, Weigel D, Schmid M. Distinct expression patterns of natural antisense transcripts in Arabidopsis. Plant Physiol. 2007;144:1247–1255. doi: 10.1104/pp.107.100396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Seki M. Expression profile and 5′-terminal structure of Arabidopsis antisense transcripts expressed in seed. Plant Signal Behav. 2011;6:691–693. doi: 10.4161/psb.6.5.14976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Xia J, Lii YE, Barrera-Figueroa BE, Zhou X, Gao S, Lu L, Niu D, Chen Z, Leung C, Wong T, Zhang H, Guo J, Li Y, Liu R, Liang W, Zhu JK, Zhang W, Jin H. Genome-wide analysis of plant nat-siRNAs reveals insights into their distribution, biogenesis and function. Genome Biol. 2012;13:R20. doi: 10.1186/gb-2012-13-3-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, Soldatov A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Bodovitz S. Single cell analysis: the new frontier in ‘omocs’. Trends Biotechol. 2010;28:281–290. doi: 10.1016/j.tibtech.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Lao L, Surani A. Development and applications of single-cell transcriptom analysis. Nat Methods. 2011;8:S6–S11. doi: 10.1038/nchembio.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: Single-Cell RNA-Seq by Multiplexed Linear Amplification. Cell Rep. 2012;2:1–8. doi: 10.1016/j.celrep.2012.05.015. [DOI] [PubMed] [Google Scholar]
- IRGSPv5.0. http://rgp.dna.affrc.go.jp/IRGSP/Build5/build5.html.
- RAP-DB. http://rapdblegacy.dna.affrc.go.jp/download/index.html.
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin KE, Callen BP, Egan JB. Transcriptional interference - a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crampton N, Bonass WA, Kirkham J, Rivetti C, Thomson NH. Collision events between RNA polymerases in convergent transcription studied by atomic force microscopy. Nucleic Acids Res. 2006;34:5416–5425. doi: 10.1093/nar/gkl668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduri C. Functional insights into long antisense noncoding RNA Kcnq1ot1 mediated bidirectional silencing. RNA Biol. 2008;5:208–211. doi: 10.4161/rna.7113. [DOI] [PubMed] [Google Scholar]
- Ohhata T, Hoki Y, Sasaki H, Sado T. Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification. Development. 2008;135:227–235. doi: 10.1242/dev.008490. [DOI] [PubMed] [Google Scholar]
- Richards M, Tan SP, Chan WK, Bongso A. Reverse serial analysis of gene expression (SAGE) characterization of orphan SAGE tags from human embryonic stem cells identifies the presence of novel transcripts and antisense transcription of key pluripotency genes. Stem Cells. 2006;24:1162–1173. doi: 10.1634/stemcells.2005-0304. [DOI] [PubMed] [Google Scholar]
- Wang L, Si Y, Dedow LK, Shao Y, Liu P, Brutnell TP. A low-cost library construction protocol and data analysis pipeline for Illumina-based strand-specific multiplex RNA-seq. PLoS One. 2011;6:e26426. doi: 10.1371/journal.pone.0026426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WS, Coughlan S, Crane E, Beatty M, Duvick J. The application of laser microdissection to in planta gene expression profiling of the maize anthracnose stalk rot fungus Colletotrichum graminicola. Mol Plant Microbe Interact. 2006;19:1240–1250. doi: 10.1094/MPMI-19-1240. [DOI] [PubMed] [Google Scholar]
- Tang X, Zhang ZY, Zhang WJ, Zhao XM, Li X, Zhang D, Liu QQ, Tang WH. Global Gene Profiling of Laser-Captured Pollen Mother Cells Indicates Molecular Pathways and Gene Subfamilies Involved in Rice Meiosis. Plant Physiol. 2010;154:1855–1870. doi: 10.1104/pp.110.161661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Takano T, Akita S. Expression of α-expansin genes in young seedlings of rice (Oryza sativa L.) Planta. 2000;211:467–473. doi: 10.1007/s004250000311. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- HMMER. http://hmmer.janelia.org/
- MSU 6.0 repeat database. ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_6.0.
- RepeatMasker. http://www.repeatmasker.org.
- SMALT version 0.5.7. ftp://ftp.sanger.ac.uk/pub/users/hp3/
- Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- R software package. http://www.r-project.org/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of pair-end reads of ssRNA-seq and small RNAs from normal and three abiotic stress conditions.
Statistics of rice transcripts.
Summary of 5813 putative cis-NATs in rice. This table lists all cis-NATs identified from rice renewed assembled transcripts. Each column represents plus transcripts (transcriptional orientations are the same as the reference), its length, minus transcripts, length, overlapped length and detailed overlapped locations in rice genome (IRGSP v5.0) as indicated.
Overview of small RNAs in rice. (A) Distribution of the lengths of all unique small RNAs generated by high-throughput sequencing from rice under four different conditions. (B) Length distribution of nat-siRNAs. (C) Distribution of all unique small RNAs located in different sequence components. (D) Distribution of nat-siRNAs located in different sequence components. The blue, red, green and purple bars represent small RNAs or nat-siRNAs from normal, salt stressed, cold stressed and drought stressed conditions, respectively (A-D). (E) First-nucleotide distribution of all unique small RNAs under four different conditions. (F) First-nucleotide distribution of nat-siRNAs under four different conditions.
Sequences of small RNAs mapped to the overlapped regions ofcis-NATs. The 25,420, 18,598, 18,152 and 28,807 unique small RNAs from normal, salt, cold and drought conditions, respectively, are shown to be perfectly mapped to the overlapped regions of cis-NATs.
Details of 2292 expressedcis-NATs. The detailed information of 2292 cis-NAT pairs with expression evidence (i.e. FPKM > 0) of both sense and antisense transcripts and with nat-siRNAs in the overlapping region under normal (1789 pairs), cold (1668), salt (1572) and drought (1668) conditions are shown. Columns 1-4 and 9-11 represent cis-NATs type, transcripts, protein domain and the length of transcripts, respectively. Columns 5-8 and 12-15 represent FPKM of transcripts under normal, salt, cold and drought conditions, respectively. Column 16 shows the overlapped length. Columns 17-20 show the number of corresponding nat-siRNAs located in the overlapped regions. The rest of the columns are the detailed strand bias of nat-siRNAs from four different conditions.
Lists of 166cis-NATs that produced nat-siRNAs exclusively in the overlapping regions with more than five unique small RNAs from four different conditions. Of 166 cis-NATs, 72, 58, 66 and 75 cis-NATs were obtained from normal, salt, cold and drought treatments, respectively. These cis-NATs are listed separately in four tables, respectively indicated by blue, orange, red and green colour.
Lists of 13cis-NATs that produced nat-siRNAs more enriched in the overlapping regions than the non-overlapping regions. Of 13 cis-NATs, nine, six, five and five cis-NATs obtained from normal, salt, cold and drought treatments, respectively. These cis-NATs are listed separately in four tables, respectively indicated by blue, orange, red and green colour.
Strand bias ofcis-NATs that gave rise to nat-siRNAs. The strand bias is computed among different types of cis-NAT pairs under four different conditions. A total of nine types are listed. Here, plus indicates that the orientation of transcripts are the same as the reference genome, minus indicates opposite orientation of transcripts. One strand means only one strand of the cis-NATs gave rise to small RNAs. The plus/minus or minus/plus means one strand of the cis-NATs spawns at least five (part A) or two (part B) fold more small RNAs than the other.
Scatter plots of expression of 1072cis-NATs. The two scatter plots compare transcripts expression ratio trends of 1072 co-expressed cis-NAT pairs between normal and drought-stress conditions (A), and between normal and salt-stress conditions (B). The results show five subgroups: red, green, purple, blue and orange spots represent subgroups 1-5, respectively.
Lists of 112cis-NATs with DEGs. Differentially Expressed Genes (DEGs) were identified in one-to-one cis-NATs under salt, drought and cold treatments by comparing their FPKM with that under normal conditions. The gene names, FPKM, fold change (with absolute value > 2) and p-value (< 0.001) are listed.
All many-to-manycis-NAT groups. In total, 223 cis-NAT groups were identified as many-to-many type. Of them, 209 groups belonged to one-to-two type, nine belonged to one-to-three type, four belonged to two-to-two groups, and one belonged to the one-to-four group. The meaning of each column is as given in Additional file 6.
Statistics of 209 networks formed bycis-NAT groups. The numbers of one-to-two cis-NAT groups with expression evidence under normal, salt, cold and drought conditions are shown. These one-to-two cis-NAT groups were divided into five classes according to their cis-NATs types. Here, ‘All EXP’ indicates all cis-NAT groups with expression evidence (FPKM > 0) of both sense and antisense transcript, and with nat-siRNAs (number of small RNAs > 1) in overlapping region as well. ‘Either EXP’ indicates either of cis-NAT groups with expression evidence of both sense and antisense transcript, and with nat-siRNAs in the overlapping region as well.
Primers designed for real-time RT-PCR and Northern Blots in this research. (DOCX 13 kb)