Abstract
Yeast Pah1p phosphatidate phosphatase (PAP) catalyzes the penultimate step in the synthesis of triacylglycerol. PAP plays a crucial role in lipid homeostasis by controlling the relative proportions of its substrate phosphatidate and its product diacylglycerol. The cellular amounts of these lipid intermediates influence the synthesis of triacylglycerol and the pathways by which membrane phospholipids are synthesized. Physiological functions affected by PAP activity include phospholipid synthesis gene expression, nuclear/endoplasmic reticulum membrane growth, lipid droplet formation, and vacuole homeostasis and fusion. Yeast lacking Pah1p PAP activity are acutely sensitive to fatty acid-induced toxicity and exhibit respiratory deficiency. PAP is distinguished in its cellular location, catalytic mechanism, and physiological functions from Dpp1p and Lpp1p lipid phosphate phosphatases that utilize a variety of substrates that include phosphatidate. Phosphorylation/dephosphorylation is a major mechanism by which Pah1p PAP activity is regulated. Pah1p is phosphorylated by cytosolic-associated Pho85p-Pho80p, Cdc28p-cyclin B, and protein kinase A and is dephosphorylated by the endoplasmic reticulum-associated Nem1p-Spo7p phosphatase. The dephosphorylation of Pah1p stimulates PAP activity and facilitates the association with the membrane/phosphatidate allowing for its reaction and triacylglycerol synthesis.
Keywords: Lipid synthesis, Triacylglycerol, Phosphatidate, Diacylglycerol, phosphatidate phosphatase, Lipid phosphate phosphatase, Yeast
1. Introduction
With the increasing prevalence of obesity and its associated diseases, much interest has been placed on the study of mechanisms that control lipid homeostasis. The triacylglycerol (TAG) molecule constitutes the most calorie-dense form of cellular energy storage, allowing organisms to withstand prolonged periods of nutrient deprivation [1]. Moreover, stores of TAG may provide a source of fatty acids and diacylglycerol (DAG) for membrane biosynthesis during cellular growth. This dual function of TAG as a reservoir for energy substrates and membrane lipid precursors makes it a central player in lipid homeostasis [2]. The regulation of TAG synthesis and storage is crucial in human health because both an excess and a defect in fat storage results in lipid-associated disorders such as obesity, lipodystrophy, insulin resistance, diabetes, hypertension, cardiovascular disease, and cancer [1].
Phosphatidate phosphatase (PAP), the enzyme involved in the penultimate step in TAG synthesis, catalyzes the dephosphorylation of phosphatidate (PA) to yield DAG and Pi [3] (Fig. 1). In de novo lipid synthesis in the yeast Saccharomyces cerevisiae, the DAG generated in the reaction is used for the synthesis of TAG as well as the phospholipids phosphatidylcholine (PC) and phosphatidylethanolamine (PE) via the Kennedy pathway [4–7], while the reaction substrate PA serves as a precursor for all major phospholipids via the CDP-DAG pathway [4–6] (Fig. 1). In mammalian cells, however, the phospholipids phosphatidylserine, PE and PC are derived from DAG [8]. In addition, both the substrate and the product of this reaction have lipid signaling functions. PA is implicated in transcription, activation of cell growth, membrane proliferation, secretion, and vesicular trafficking, while DAG is primarily involved in the activation of protein kinase C in higher eukaryotes [9–18]. By the nature of the reaction, PAP activity controls the cellular concentrations of these two important lipid mediators, playing a role in lipid signaling. Thus, the regulation of PAP activity may govern whether cells make storage lipids or membrane phospholipids, determine the pathways by which these lipids are synthesized, and control the cellular levels of important signaling lipids. Genetic and biochemical studies in yeast and mammalian cells have revealed PAP as a major regulator of lipid metabolism and cell physiology [1,7,19–31].
Fig. 1.
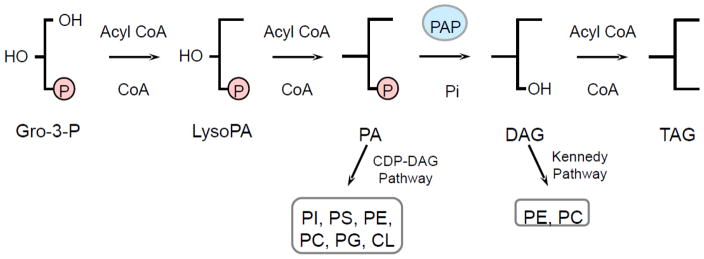
Lipid synthesis in yeast. The pathways shown for the synthesis of TAG and phospholipids include the relevant steps discussed in this review. PAP (highlighted in blue) catalyzes the dephosphorylation of PA to form DAG in the penultimate step in TAG synthesis. The PAP substrate PA and product DAG are utilized for the synthesis of phospholipids via the CDP-DAG pathway and Kennedy pathway, respectively. Gro-3-P, glycerol-3-phosphate; LysoPA, lysophosphatidate; PA, phosphatidate; DAG, diacylglycerol; TAG, triacylglycerol; CDP-DAG, CDP-diacylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; CL, cardiolipin.
2. Discovery of PAP activities and their involvement in lipid metabolism and signaling
The PAP reaction was first described in animal tissues by Smith et al. in 1957 [3], providing a link between the neutral and phospholipid synthesis pathways in mammalian cells [6,32]. Subsequent studies demonstrated that this enzymatic reaction requires Mg2+ ions, and that the majority of activity resides in the soluble fraction of cell lysates [33], in contrast to other enzymes in the TAG and phospholipid synthesis pathways, which are integral membrane proteins [34–38]. Furthermore, PAP activity was found to translocate from the cytosol to the membrane fraction of cells treated with fatty acids [4,39,40]. Due to the instability of the mammalian enzyme, the isolation of PAP remained elusive for more than three decades.
In 1984, Hosaka and Yamashita identified PAP activity in the cytosolic and membrane fractions of the yeast S. cerevisiae [33,41]. This simple eukaryote synthesizes lipids by pathways common to those of more complex organisms and is an attractive model system due to its molecular and genetic tractability [42–44]. The use of this model organism allowed for the purification of a 91-kDa PAP enzyme from the total membrane fraction in 1989 [45]. Additionally, 104-kDa, 75-kDa, and 45-kDa forms of PAP were isolated from microsomes, cytosol, and mitochondria, respectively [33,46]. The 91-kDa enzyme was later shown to be a degradation product of the 104-kDa form [46], while precursor-product relationships do not exist between the 75-kDa, 45-kDa, and 104-kDa proteins [46]. Characterization studies indicated that all forms of the PAP enzyme require Mg2+ ions for catalysis, and they are highly specific for PA as a substrate [33,45,46]. Unfortunately, none of the schemes used to purify these enzymes resulted in sufficient amounts of protein for sequencing by classical Edman degradation analysis, and so the gene(s) encoding them could not been identified.
In subsequent studies, Wu and coworkers isolated a 34-kDa protein from yeast microsomes that exhibited PAP activity [47]. Characterization of the enzyme revealed that it catalyzes the removal of the β-phosphate from diacylglycerol pyrophosphate (DGPP) to form PA, and subsequently dephosphorylates PA to produce DAG; thus, the enzyme was termed DGPP phosphatase [47]. In contrast to the enzymes discussed above, this phosphatase enzyme does not require Mg2+ ions for catalysis [47]. Sufficient enzyme was purified to obtain amino acid sequence information, which was matched to a gene in the S. cerevisiae database and designated DPP1 (for diacylglycerol pyrophosphate phosphatase) [48]. Dpp1p1 is 289 amino acids in length and has a molecular mass of 33.5 kDa [48], which is in close agreement with the size of the purified DGPP phosphatase [47]. The identification of a conserved phosphatase sequence motif contained within several lipid phosphatases [49] revealed DGPP phosphatase shares sequence homology with a mammalian Mg2+-independent PAP (designated PAP2) believed to be involved in lipid signaling [50–53]. Shortly after this discovery, the LPP1 (for lipid phosphate phosphatase) gene was identified based on protein sequence homology with Dpp1p [54]. Lpp1p is 274 amino acids in length and has a predicted molecular mass of 31.6 kDa [54]. Studies with the dpp1Δ lpp1Δ double mutant show that the DPP1 and LPP1 genes encode essentially all Mg2+-independent PAP activity in yeast, with DPP1 being the major contributor [48,54,55]. Detailed characterization studies showed that the PAP activity of these enzymes is distinct from the conventional Mg2+-dependent PAP enzymes, whose gene(s) had yet to be identified. In particular, the broad substrate specificity (discussed in later sections) of the Mg2+-independent Dpp1p and Lpp1p enzymes has resulted in their designation as lipid phosphate phosphatase (LPP) enzymes [56].
In a fortunate twist of scientific fate, a preparation of the 91-kDa enzyme was recovered from frozen storage, analyzed for enzymatic activity, and sequenced by mass spectrometry [21], a much more sensitive method than Edman degradation. The deduced protein sequence matched that of the deduced product of the SMP2 gene, which had been implicated in plasmid maintenance and respiration [57]. The molecular function of Smp2p, however, had yet to be established. Overexpression of SMP2 was reported to complement the aberrant nuclear membrane expansion phenotype of nem1Δ and spo7Δ mutants lacking the endoplasmic reticulum (ER)-associated Nem1p-Spo7p phosphatase complex [20,58]. Upon identification of the molecular function as a PAP enzyme, the SMP2 gene was renamed PAH1 (for phosphatidic acid phosphohydrolase) [21]. Pah1p is 862 amino acids in length and has a predicted molecular mass of 95-kDa; however, when expressed in S. cerevisiae it migrates as a 124-kDa protein upon SDS-PAGE [21]. While phosphorylation of Pah1p results in a shift in electrophoretic mobility to a position of a slightly higher molecular mass [20,22,25,27], Pah1p expressed in E. coli migrates as a 114-kDa protein product upon SDS-PAGE analysis [21]. Therefore, the discrepancy in the predicted vs. observed size of Pah1p cannot be attributed solely to modification by phosphorylation.
The identification and characterization of yeast Pah1p revealed its homology to the mammalian proteins known as lipins, encoded by the murine Lpin1, 2, and 3 genes [21,59]. Pah1p and mouse lipin-1 share structural similarity in two conserved regions: the NLIP and CLIP domains, found at the N-terminus and C-terminus, respectively [59]. Lpin1 was identified as the gene whose mutation is responsible for the transient fatty liver dystrophy (fld) phenotype of mice [59,60]. A loss of lipin-1 was shown to prevent normal adipose tissue development, resulting in lipodystrophy and insulin resistance, while an excess of lipin-1 promotes obesity and insulin sensitivity [59,61]. However, the molecular function of this protein was not known at the time. In view of the observation that Pah1p PAP activity is dependent on a haloacid dehalogenase (HAD)-like domain possessing the DXDX(T/V) catalytic motif [21,23], which is contained in the CLIP domain, lipin-1 was characterized as a PAP enzyme [21,62]. Further enzymological studies confirmed that all isoforms of lipin-1 (α, β, and γ splice variants), as well as lipin-2 and -3, also exhibit PAP activity [62,63]. LPIN1 mutations in humans are associated with metabolic syndrome, type 2 diabetes, and recurrent acute myoglobinuria in children, while mutations in LPIN2 result in anemia and inflammatory disorders associated with Majeed syndrome [1,30,64]. Little is known about the consequences of LPIN3 mutations. Mammalian lipin-1 and -2 were shown to complement phenotypes exhibited by yeast pah1Δ mutant cells [65], indicating the functions of PAP enzymes are evolutionarily conserved. Indeed, the discovery of yeast Pah1p led to the identification of genes encoding PAP enzymes in humans [21,63], mice [59,62], flies [66,67], worms [68], and plants [69,70]. All PAP enzymes have the HAD-like domain that contains a DXDX(T/V) catalytic motif and the NLIP domain of unknown function [21,23,59,71]. The reader is directed to recent reviews that summarize our current understanding of mammalian lipins [19,31,32,72,73].
3. Biochemical, enzymological and structural properties of PAP enzymes
Since the initial characterization of the PAP reaction in 1957 [3], both Mg2+-dependent and -independent enzymes have been identified in yeast [21,48,54]. Besides the difference in their cofactor requirement, these enzymes are distinguished by several other properties (Figs 2 and 3).
Fig. 2.
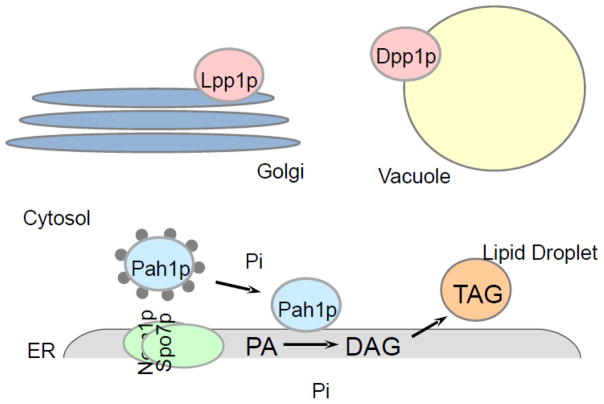
PAP and LPP enzymes are localized in different cellular compartments. The cartoon shows the integral membrane enzymes Dpp1p and Lpp1p in the vacuole and Golgi, respectively. Pah1p in the cytosol is phosphorylated on multiple sites (symbols decorating enzyme). At the ER membrane, Pah1p is dephosphorylated by the Nem1p-Spo7p phosphatase complex, which allows for its interaction with the membrane where its substrate PA resides. The dephosphorylated form of Pah1p catalyzes the dephosphorylation of PA to generate DAG for the synthesis of TAG that is stored in lipid droplets.
Fig. 3.
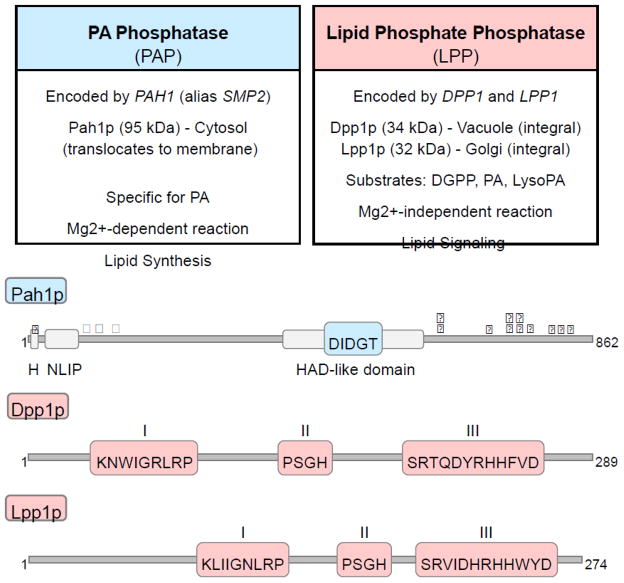
Distinguishing characteristics of PAP and LPP enzymes. The basic characteristics of the yeast PAP (Pah1p) and LPP (Dpp1p and Lpp1p) enzymes, including their catalytic motifs and phosphorylation sites, are summarized in the figure. H, amphipathic helix; HAD, haloacid dehalogenase;
 , approximate positions of the sites (Ser110, Ser114, Ser168, Ser602, Thr723, Ser744, and Ser748) phosphorylated by Pho85p-Pho80p; ★, approximate positions of the sites (Ser602, Thr723, and Ser744) phosphorylated by Cdc28p-cyclin B;
, approximate positions of the sites (Ser110, Ser114, Ser168, Ser602, Thr723, Ser744, and Ser748) phosphorylated by Pho85p-Pho80p; ★, approximate positions of the sites (Ser602, Thr723, and Ser744) phosphorylated by Cdc28p-cyclin B;
 , approximate positions of the sites (Ser10, Ser677, Ser773, Ser774, and Ser788) phosphorylated by protein kinase A. The diagrams of each protein are not drawn to scale.
, approximate positions of the sites (Ser10, Ser677, Ser773, Ser774, and Ser788) phosphorylated by protein kinase A. The diagrams of each protein are not drawn to scale.
Dpp1p and Lpp1p are relatively small integral membrane proteins confined to the vacuole and Golgi membranes, respectively (Fig. 2). Both proteins possess six transmembrane domains distributed over their polypeptide sequences [48,54]. In contrast, Pah1p is a much larger protein that contains no transmembrane domains in its sequence; it is primarily found in the cytosol, but must translocate to the membrane to access its substrate PA for catalysis [21] (Fig. 2). In addition, Pah1p has also been found to localize to the nucleus where studies have indicated it interacts with the promoter of phospholipid synthesis genes [19,20]. Thus, the spatial differences in the subcellular localization of the PAP and LPP enzymes point to distinct cellular functions.
While mammalian Mg2+-dependent PAP and Mg2+-independent LPP enzymes are also differentiated based on their sensitivity to the thioreactive compound N-ethylmaleimide (NEM) [4,50], this distinction is not applicable to their yeast counterparts. In fact, Dpp1p activity is insensitive to NEM [47], whereas the activity of Lpp1p is potently inhibited by this compound [74]. Moreover, NEM has no effect on the PAP activity of Pah1p [21]. Similarly, the synthetic compound propranolol inhibits both Pah1p [46] as well as Lpp1p [74], even though it is thought to hinder activity by interacting with the Mg2+ binding site of enzymes [75]. Therefore, inhibition by NEM or propranolol should not be used to distinguish yeast PAP and LPP enzymes.
Dpp1p and Lpp1p do not require divalent cations for activity [47,54,74], while maximum activity of Pah1p is dependent on Mg2+ ions [21]. This distinction in cofactor requirement can be explained by the differences in the catalytic motifs that govern the activity of each class of enzymes (Fig. 3). The PAP activity of Pah1p is governed by a DXDX(T/V) motif found in members of a superfamily of Mg2+-dependent phosphatase enzymes [76,77] that include mammalian lipin PAP enzymes [1,21]. Consistent with other Mg2+-independent LPP enzymes [49,78,79], Dpp1p and Lpp1p contain a three-domain lipid phosphatase motif comprised of the consensus sequences KXXXXXXRP (I), PSGH (II) and SRXXXXXHXXXD (III) [49], which confers these proteins their enzymatic activity [80].
Finally, the PAP and LPP enzymes in yeast differ with respect to their substrate specificities. Dpp1p and Lpp1p utilize a variety of lipid phosphate substrates, including PA, DGPP, lysoPA, sphingoid base phosphates, and isoprenoid phosphates [47,48,54,55,74,81]; however, only DGPP and PA have been shown to be substrates in vivo [54]. In addition, the enzymological properties of Lpp1p differ significantly from those of Dpp1p. While Dpp1p can utilize PA in the absence of DGPP, it has a 10-fold higher specificity for DGPP [47]. Conversely, PA is the preferred substrate for Lpp1p, followed by DGPP and lysoPA [54,74]. Moreover, the affinity of Lpp1p for PA, DGPP, and lysoPA as substrates is greater than the affinity of Dpp1p for these substrates [74]. In contrast, Pah1p is specific for PA [33,45–47]. Additionally, both the substrates and products of the reactions catalyzed by the LPP enzymes are important signaling molecules, suggesting that these enzymes are involved in lipid signaling, and are not responsible for the de novo synthesis of phospholipids and TAG that occurs in the ER [6,48,54]. In fact, dpp1Δ lpp1Δ mutations do not affect lipid synthesis, whereas, the pah1Δ mutation affects both the synthesis of TAG as well as the synthesis of phospholipids [21,26]. Thus, the synthesis of the DAG required for phospholipid and TAG synthesis in S. cerevisiae is attributed to the Pah1p enzyme [21,26,82].
4. Pah1p PAP is a key player in de novo lipid synthesis
The essential role of PAP in de novo lipid metabolism has been established through studies using the yeast pah1Δ mutant [20,22,23,55]. Consistent with a lack of enzyme activity, this mutant shows elevated levels of PA and decreased levels of DAG and TAG [21,23,26]. Additionally, the amounts of phospholipids, fatty acids, and sterol esters are also elevated in response to the pah1Δ mutation [21,23,26], indicating that PAP regulates overall lipid synthesis. The effects on TAG (> 90 % decrease) are most pronounced in the stationary phase of growth [21,23], where the synthesis of TAG is predominant over the synthesis of membrane phospholipids [83].
In addition to the alteration in lipid metabolism, phenotypes of the pah1Δ mutant include slow growth [20], aberrant expansion of the nuclear/ER membrane [20], respiratory deficiency [21], defects in lipid droplet formation [28] and morphology [84], vacuole homeostasis and fusion [29], fatty-acid induced lipotoxicity [26], and a growth sensitivity to elevated temperature [20,21]. Catalytically inactive mutant forms of PAP, with mutations in either a conserved glycine (e.g., G80R) at the N-terminus or DIDGT catalytic motif residues (D398E or D400E) exhibit the same phenotypes associated with the pah1Δ mutation, indicating these effects are specifically linked to the loss of PAP activity [23,26,28,29].
A contributing factor for the increased amounts of phospholipids and fatty acids in the pah1Δ mutant is the derepression of UASINO-containing lipid synthesis genes in response to elevated PA levels [20,21,23,85–89]. The UASINO element contains the binding site for the Ino2p-Ino4p complex that stimulates expression of lipid synthesis genes [85]. The PA-mediated regulation of these genes is controlled by the transcriptional repressor Opi1p [90], whose function is determined by its localization. In its inactive state, Opi1p is tethered to the nuclear/ER membrane through interactions with Scs2p and PA [91,92]. A reduction in PA levels destabilizes this interaction, allowing for the translocation of Opi1p into the nucleus, where it suppresses transcription of UASINO-containing genes by binding to the Ino2p subunit of the Ino2p-Ino4p activator complex [92,93]. Additionally, the decreased capacity of pah1Δ mutant cells to incorporate fatty acids into TAG may contribute to the observed alterations in phospholipids, fatty acids, and sterol esters. This misregulation of lipid metabolism may also be a factor for the susceptibility of the pah1Δ mutant to fatty acid-induced toxicity [26].
The yeast DGK1-encoded DAG kinase (Dgk1p) has recently been identified as an enzyme whose function counterbalances that of the Pah1p PAP [94] (Fig. 4). This nuclear/ER integral membrane enzyme is unique in that, in contrast to other DAG kinases in bacteria, plants, and animals [13,95–98], it utilizes CTP rather than ATP as the phosphate donor in the phosphorylation reaction [94] (Fig. 4). Cells bearing a dgk1Δ mutation do not exhibit any remarkable phenotypes under standard growth conditions [94]. However, like the pah1Δ mutation, overexpression of Dgk1p causes the accumulation of PA at the nuclear/ER membrane and the derepression of UASINO-containing genes [94], providing evidence that Dgk1p activity antagonizes that of the Pah1p enzyme by regulating the cellular levels of PA (Fig. 4).
Fig. 4.
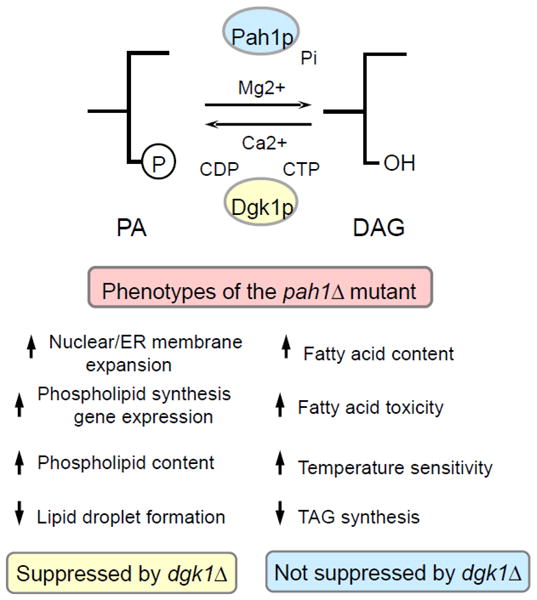
Summary of pah1Δ mutant phenotypes. The reactions catalyzed by Pah1p PAP and Dgk1p DAG kinase control the balance of PA and DAG. The pah1Δ phenotypes suppressed and not suppressed by the dgk1Δ mutation are listed in the figure. Phosphatidate Phosphatase, a Key Regulator of Lipid Homeostasis Florencia Pascual and George M. Carman*
The aberrant membrane expansion phenotype of the pah1Δ mutant has been attributed to the abnormal increase in phospholipid synthesis associated with this mutation [20]; however, recent reports have shown that increased expression of phospholipid biosynthetic genes alone is not sufficient for nuclear/ER membrane expansion [22,94]. In addition, DGK1 overexpression results in anomalous nuclear/ER membrane morphology [94], while the introduction of the dgk1Δ mutation to the pah1Δ background restores PA levels, the suppression of UASINO-containing genes, and a normal nuclear/ER membrane structure [94]. Thus, increased phospholipid synthesis coupled to increased PA levels result in the aberrant nuclear/ER morphology displayed by the pah1Δ mutant. Among the phenotypes associated with the pah1Δ mutant, the defect in lipid droplet formation that results from loss of Pah1p function can also be complemented by the dgk1Δ mutation [26,28], indicating that elevated PA levels might be the basis for this phenotype. In support of this hypothesis, a recent study has implicated PA as an important regulator of lipid droplet morphology [84]. The defect in lipid droplet formation and structure associated with the pah1Δ mutation has been attributed to the decreased DAG levels caused by loss of PAP activity [28]. Thus, this phenotype might result from a combination of both an elevated PA content and reduced DAG levels.
In contrast with the pah1Δ mutant phenotypes described above, the defect in TAG synthesis [94], increase in fatty acid content [94], fatty acid-induced toxicity [26], and temperature sensitivity2 cannot be suppressed by loss of Dgk1p function. These observations seem to indicate that these phenotypes are related to depletion of DAG rather than increase in PA content. Another phenotype exhibited by the pah1Δ mutant is the defect in vacuole homeostasis and fusion [29]. While this phenotype is due to the loss of Pah1p PAP activity [29], it is unknown whether it is based on alterations in PA and/or DAG. The analysis of vacuole morphology in the pah1Δ dgk1Δ double mutant might shed light on the mechanism underlying this phenotype.
5. PAP activity is regulated at several levels
The involvement of PAP enzymes in lipid homeostasis and lipid-associated disorders in animals has prompted interest in understanding how their expression and activity are regulated. In addition to the pah1Δ mutant phenotypes discussed previously, overexpression of a phosphorylation-deficient form of Pah1p (7A, discussed in later sections) in S. cerevisiae is also detrimental to cell growth [22,25,94]. The cause of this phenotype appears to be consistent with a reduction in PA and accumulation of DAG to a toxic level. Thus, a balance in PA and DAG levels mediated by Pah1p PAP activity must be achieved to maintain lipid homeostasis and normal cell physiology. In view of the variety of cellular processes in which PA and DAG play a role, the regulatory mechanisms governing PAP activity are complex, and occur at many levels, affecting Pah1p transcription, posttranslational modification, subcellular localization, and biochemical properties.
5.1. Regulation of PAP expression
Lpin1/2-encoded PAP enzymes are regulated at the transcriptional level in response to various conditions. For instance, several transcription factors have been shown to interact with the Lpin1 promoter, including the glucocorticoid receptor (GR) [99–101], sterol response element binding protein-1 (SREBP-1) [102], and cAMP response element binding protein (CREBP) [103]. In contrast, Lpin2 is known to be induced by fasting [100,104], but this regulation is not affected by cAMP signaling or glucocorticoids [100]. Compared with its mammalian orthologs, relatively little is known about the transcriptional regulation of the yeast Pah1p PAP enzyme. Microarray data has indicated that PAH1 expression is induced upon transition from glucose-based fermentative growth to glycerol- and ethanol-based respiratory growth [105]. Furthermore, despite the observed increase in Mg2+-dependent PAP activity in stationary phase cells [41], reports suggest that PAH1 is repressed during the diauxic shift [106]. Additional studies are needed to determine the effect of growth phase on PAH1 expression.
Recent work by Soto-Cardalda and colleagues has shown the expression of PAH1-encoded PAP activity is affected by intracellular levels of zinc [107]. This essential nutrient in S. cerevisiae and higher eukaryotes [108,109] serves as a cofactor for numerous enzymes, and is a structural component of many proteins [108,110,111]. Tight control of intracellular zinc levels must therefore be exerted through the action of zinc transporters located in the plasma, vacuole, ER, and mitochondrial membranes [111–120]. A deficiency in zinc causes an induction in the expression of many of these transporters, which is accompanied by changes in membrane phospholipid composition that result from the transcriptional regulation of various phospholipid synthesis genes [107,109,121–126]. PAH1 expression is induced in response to zinc depletion in a Zap1p-dependent manner through its interaction with zinc-responsive upstream activating (UASZRE) sequences in the PAH1 promoter [107]. This induction correlates with an increase in Pah1p PAP activity and elevated TAG levels [107].
5.2. Biochemical regulation of PAP
5.2.1. Regulation by lipids and nucleotides
Pah1p PAP activity is stimulated by the phospholipids CDP-DAG, PI and CL, which act as mixed competitive activators of PAP activity by decreasing the Km for PA [127]. On the other hand, PAP activity is inhibited by the sphingoid bases sphingosine, phytosphingosine, and sphinganine [127] in a parabolic competitive mechanism, by which more than one inhibitor molecule contributes to the exclusion of PA from the enzyme.
The nucleotides ATP and CTP, precursors of phospholipid synthesis [42], inhibit Pah1p PAP activity by a complex mechanism which affects both the Vmax and Km for PA and might also involve chelation of the Mg2+ cofactor [128]. Moreover, cellular ATP and CTP levels correlate with synthesis of phospholipids and TAG [128]: high ATP levels favor increased PA content and phospholipid synthesis, while low levels result in reduced PA content and increased TAG synthesis; high CTP levels increase PA content and thus result in derepression of UASINO-containing genes [129].
5.2.2. Regulation by phosphorylation/dephosphorylation
At 3,910 molecules per cell [130], Pah1p is a relatively abundant enzyme in S. cerevisiae [38]. While Pah1p is found mostly in the cytosol, its substrate PA resides in the nuclear/ER membrane, and therefore translocation of the enzyme is vital for in vivo function [21,24,25,27]. Recent studies have demonstrated phosphorylation/dephosphorylation of Pah1p governs it subcellular localization, thus serving as the major regulator of PAP activity [22,24,25,27]. Phosphorylated Pah1p resides in the cytosol, while its dephosphorylated form is associated with the membrane [25] (Fig. 2). Recruitment of the phosphorylated enzyme to the nuclear/ER membrane is dependent on the Nem1p-Spo7p protein phosphatase complex, which dephosphorylates PAP and thus allows for its association with the membrane through a process mediated by a short N-terminal amphipathic helix [20,24,25,58] (Fig. 2).
The nuclear/ER membrane-associated Nem1p-Spo7p complex was identified in studies that showed it is essential for the formation of a spherical nucleus [20]. Further work revealed the protein phosphatase activity of the complex is dependent on the catalytic motif DXDX(T/V) in Nem1p, as well as binding of the regulatory subunit Spo7p to Nem1p [20]. A defect in either Nem1p or Spo7p results in the same aberrant nuclear/ER membrane expansion phenotype exhibited by the pah1Δ mutant, indicating both subunits of the phosphatase complex are required for PAP function in vivo, and confirming that Pah1p is dephosphorylated exclusively by Nem1p-Spo7p [20,58]. Overexpression of the Nem1p-Spo7p complex is lethal only in the presence of its substrate Pah1p [20], suggesting that dephosphorylation of Pah1p by Nem1p-Spo7p is a key modulator of PAP function. Moreover, the expression level of Nem1p is 10-fold lower than that of Pah1p [130], supporting this theory. Thus, under normal conditions, the level of membrane-associated Pah1p may be controlled by the amount of the Nem1p-Spo7p complex on the membrane, which would result in low PAP activity.
Large-scale analysis of the yeast proteome has identified Pah1p as a phosphoprotein with target sites for several protein kinases, including those encoded by PHO85 [131,132] and CDC28 [133]. In S. cerevisiae, Cdc28p is essential and sufficient for cell cycle progression, while the non-essential Pho85p supports many additional functions [134]. Cross-talk between these two kinases allows for regulation of cell morphology, gene expression, macromolecular metabolism, and signaling in response to environmental stimuli [134–137]. Mass spectrometry and immunoblot analyses of yeast purified Pah1p have identified 14 sites of phosphorylation, seven of which (Ser110, Ser114, Ser168, Ser602, Thr723, Ser744, and Ser748) are contained within the minimal (Ser/Thr)-Pro motif that is a target for cell cycle-regulated protein kinases [22]. In fact, all seven sites are phosphorylated by Pho85p-Pho80p [27], and three of the sites (Ser602, Thr723, and Ser744) are also targets of Cdc28p-cyclin B [25] (Fig. 3). The phosphorylation efficiency of the three common sites is much greater for Pho85p-Pho80p when compared with Cdc28p-cyclin B [27]. Phosphorylation by Pho85p-Pho80p results in a 6-fold reduction in the catalytic efficiency (Vmax/Km) of PAP [27], whereas activity is not affected by phosphorylation via Cdc28p-cyclin B [25]. That these kinases phosphorylate Pah1p in vivo is supported by the analysis of Pah1p phosphorylation in mutants lacking Pho85p or functional Cdc28p [20,27].
Pah1p is also a substrate for protein kinase A [138,139], the principal mediator of signals transmitted through the RAS/cAMP pathway in S. cerevisiae [140,141]. Protein kinase A phosphorylates Pah1p on Ser10, Ser677, Ser773, Ser774, and Ser788 with specificity similar to that shown for Pho85p-Pho80p and Cdc28p-cyclin B [139]. The protein kinase A-mediated phosphorylation of Pah1p inhibits its PAP activity by decreasing catalytic efficiency (1.8-fold), but to a lesser extent as that observed for Pho85p-Pho80p [27,139]. The inhibitory effect of protein kinase A on PAP activity is primarily conferred by phosphorylation at Ser10 [139].
Analysis of phosphorylation-deficient forms of Pah1p has provided insight into the biochemical and physiological roles of phosphorylation by Pho85p-Pho80p, Cdc28p-cyclin B, and protein kinase A [22,25,27,139]. The purified 7A mutant enzyme, where all seven (Ser/Thr)-Pro sites are mutated to nonphosphorylatable alanine, exhibits elevated PAP activity and increased interaction with phospholipid vesicles [22,25]. In vivo, expression of the 7A mutant enzyme complements the pah1Δ nem1Δ double mutant phenotypes that include temperature sensitivity, nuclear/ER membrane expansion, and derepression of phospholipid synthesis genes [22,25]. Moreover, the 7A mutations facilitate the translocation of Pah1p from the cytosol to the membrane, and in a nem1Δ mutant background, cause an increase in the synthesis of TAG [24,25]. Cells lacking the Nem1p-Spo7p complex exhibit reduced TAG due to loss of PAP function, thus indicating that lack of phosphorylation of the seven sites renders Pah1p capable of bypassing the Nem1p-Spo7p requirement for in vivo function [25]. Simultaneous mutation of the three Cdc28p-cyclin B phosphorylation sites (3A) only partially mimics the physiological consequences of the 7A mutations [25].
Analysis of the S10A and S10D mutations (mimicking dephosphorylation and phosphorylation, respectively, by protein kinase A), alone or in combination with the 7A mutations, indicate that phosphorylation at Ser10 inhibits its association with membranes, PAP activity, and TAG synthesis [139]. In fact, the S10A mutation enhances the physiological effects caused by the 7A mutations, whereas the S10D mutation attenuates the effects of the 7A mutations [139]. Thus, the protein kinase A-mediated phosphorylation of Ser10 functions in conjunction with the phosphorylations mediated by Pho85p-Pho80p and Cdc28p-cyclin B, and that Ser10 should be dephosphorylated for proper PAP function [139].
In addition to Pho85p-Pho80p, Cdc28p-cyclin B, and protein kinase A, Pah1p is phosphorylated by protein kinase C and casein kinase II [138]. These protein kinases are also known to regulate phospholipid synthesis in yeast [142]. Phosphorylation of Pah1p by protein kinase C and casein kinase II decreases its interaction with model membranes [138], suggesting an inhibitory effect of phosphorylation on PAP activity in vivo. Further studies identifying the protein kinase C and casein kinase II phosphorylation sites and characterizing their effects on PAP activity are yet to be performed.
Like the yeast Pah1p PAP, the phosphorylation of mammalian lipin-1 and -2 affects their subcellular localization, thereby indirectly inhibiting PAP activity [143]. For instance, the phosphorylation of lipin-1 in response to insulin and amino acids in rat and mouse adipocytes is mTOR-dependent and promotes cytosolic versus membrane localization [144], thus hindering PAP in vivo function by limiting access of the enzyme to its substrate. In addition, reduced PAP activity during mitosis has been linked to lipin-1 and -2 phosphorylation [65], confirming an evolutionarily conserved role of phosphorylation as a modulator of PAP activity. Like yeast Pah1p, lipin-1 is specifically dephosphorylated by the Nem1p human ortholog C-terminal domain nuclear envelope phosphatase 1 (CTDNEP1, formerly called dullard) [144,145]. In addition, recent work indicates CTDNEP1 can dephosphorylate lipin-1α, -1β, and -2 only in the presence of envelope phosphatase 1-regulatory subunit 1 (NEP1-R1), the metazoan ortholog of Spo7p [145].
6. Regulation by protein abundance
Stationary phase is the stage of growth where the synthesis of TAG predominates over the synthesis of phospholipids [83], and this change in lipid metabolism correlates with an increase in PAP activity [41,83]. That the pah1Δ mutation results in elevated phospholipids and a dramatic decrease in TAG in stationary phase [21,26] supports the notion that Pah1p PAP is a major contributor to the regulation of lipid synthesis in response to growth phase. Thus in exponential phase, where membrane phospholipid synthesis is essential, PAP function should be attenuated to allow partitioning of PA into phospholipids. On the other hand, the increased synthesis of TAG that occurs when cells progress into stationary phase should require stimulation of PAP function for channeling PA into the storage lipid TAG. As discussed above, phosphorylation/dephosphorylation is a major modulator of PAP function. We speculate that attenuation of Pah1p function in exponential phase is mediated by phosphorylation of the enzyme, whereas in cells entering stationary phase PAP activity is stimulated by dephosphorylation.
Ironically, phosphorylation deficiency caused by the 7A mutations or by the loss of Pho85p-Pho80p phosphorylation of Pah1p in pho85Δ mutant cells cause dramatic reductions (50–60 %) in Pah1p abundance [25,27]. In addition, the abundance of the enzyme decreases in cells progressing from the early- to late-exponential phase, but this effect is attenuated in nem1Δ mutant cells lacking the Nem1p-Spo7p phosphatase complex [27]. These observations indicate that phosphorylation stabilizes Pah1p, whereas dephosphorylation causes loss of abundance. We know that controlling excess PAP activity is important because the overexpression of the 7A mutant form of Pah1p [22,25,94], as well as the overexpression of Nem1p-Spo7p [20], is deleterious to growth [20,22,25,94]. Thus, the paradoxical effects of phosphorylation/dephosphorylation on Pah1p function and enzyme abundance appear to be a mechanism by which cells control the levels of PA and DAG to maintain homeostasis of lipids. It remains to be determined whether Pah1p abundance is regulated by means of programmed proteolysis.
7. Perspectives/Future developments
With the identification and characterization of the yeast PAH1 gene and its protein product, monumental advances have been made in establishing the role of PAP in lipid homeostasis. The importance of understanding the mechanisms that regulate PAP activity is underscored by the involvement of this enzyme in lipid-based disorders in human physiology. Genetic and biochemical studies with yeast Pah1p PAP have provided insights into the basic biochemical properties of the enzyme and how its activity is controlled by effector molecules, subcellular localization, and posttranslational modifications.
Phosphorylation is clearly a key modulator of PAP function. This process is mediated by multiple kinases, the action of which will likely result in different physiological outcomes. The effect of phosphorylation by the cell-cycle regulated kinases Pho85p-Pho80p and Cdc28p-cyclin B and by protein kinase A has been examined. Although other kinases have been shown to phosphorylate Pah1p, their individual effects on PAP activity have yet to be elucidated. While each of these kinases can phosphorylate Pah1p in the absence of prior phosphorylations by other kinases, it is not known whether phosphorylation at a particular site by one kinase might stimulate or inhibit the phosphorylation of other sites by the same kinase, or by different kinases. Thus, studies examining the interdependencies of the various phosphorylations will advance our understanding of the mechanisms that fine-tune PAP function.
Dephosphorylation of Pah1p by the Nem1p-Spo7p complex governs its subcellular localization, thereby affecting PAP physiological activity. Additional knowledge of the factors regulating Nem1p-Spo7p function will further our understanding of the mechanism by which Pah1p translocates to the membrane and thus becomes activated in vivo. In this regard, structural information of Pah1p will also shed light on the mechanisms underlying its interaction with the membrane.
Research Highlights.
Phosphatidate phosphatase (PAP) catalyzes the penultimate step in triacylglycerol synthesis.
PAP plays a crucial role in lipid homeostasis and cell physiology.
Multiple protein kinases phosphorylate PAP.
Nem1p-Spo7p dephosphorylates PAP.
Phosphorylation/dephosphorylation regulates PAP function.
Acknowledgments
We acknowledge Gil-Soo Han and Symeon Siniossoglou for the critical reading of the manuscript. This work was supported in part by United States Public Health Service Grants GM-28140 and GM-50679 from the National Institutes of Health.
Abbreviations
- TAG
triacylglycerol
- DAG
diacylglycerol
- PA
phosphatidate
- PAP
PA phosphatase
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- PS
phosphatidylserine
- CDP-DAG
CDP-diacylglycerol
- CL
cardiolipin
- DGPP
diacylglycerol pyrophosphate
- ER
endoplasmic reticulum
- NEM
N-ethylmaleimide
- UASINO
inositol-responsive upstream activating sequence
Footnotes
The protein product of a yeast gene is designated by the gene acronym followed by the letter p (e.g., Dpp1p, Lpp1p, and Pah1p).
Unpublished observations from the laboratories of S. Siniossoglou and G. M. Carman.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reue K, Brindley DN. Multiple roles for lipins/phosphatidate phosphatase enzymes in lipid metabolism. J Lipid Res. 2008;49:2493–2503. doi: 10.1194/jlr.R800019-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kohlwein SD. Triacylglycerol homeostasis: insights from yeast. J Biol Chem. 2010;285:15663–15667. doi: 10.1074/jbc.R110.118356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith SW, Weiss SB, Kennedy EP. The enzymatic dephosphorylation of phosphatidic acids. J Biol Chem. 1957;228:915–922. [PubMed] [Google Scholar]
- 4.Brindley DN. Intracellular translocation of phosphatidate phosphohydrolase and its possible role in the control of glycerolipid synthesis. Prog Lipid Res. 1984;23:115–133. doi: 10.1016/0163-7827(84)90001-8. [DOI] [PubMed] [Google Scholar]
- 5.Nanjundan M, Possmayer F. Pulmonary phosphatidic acid phosphatase and lipid phosphate phosphohydrolase. Am J Physiol Lung Cell Mol Physiol. 2003;284:L1–23. doi: 10.1152/ajplung.00029.2002. [DOI] [PubMed] [Google Scholar]
- 6.Carman GM, Han GS. Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem Sci. 2006;31:694–699. doi: 10.1016/j.tibs.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brindley DN, Pilquil C, Sariahmetoglu M, Reue K. Phosphatidate degradation: phosphatidate phosphatases (lipins) and lipid phosphate phosphatases. Biochim Biophys Acta. 2009;1791:956–961. doi: 10.1016/j.bbalip.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vance DE. In: Glycerolipid Biosynthesis in Eukaryotes. Vance DE, Vance J, editors. Amsterdam: 1996. pp. 153–181. [Google Scholar]
- 9.Bishop WR, Ganong BR, Bell RM. Attenuation of sn-1,2-diacylglycerol second messengers by diacylglycerol kinase. J Biol Chem. 1986;261:6993–7000. [PubMed] [Google Scholar]
- 10.Kearns BG, McGee TP, Mayinger P, Gedvilaite A, Phillips SE, Kagiwada S, Bankaitis VA. Essential role for diacylglycerol in protein transport from the yeast Golgi complex. Nature. 1997;387:101–105. doi: 10.1038/387101a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waggoner DW, Xu J, Singh I, Jasinska R, Zhang QX, Brindley DN. Structural organization of mammalian lipid phosphate phosphatases: implications for signal transduction. Biochim Biophys Acta. 1999;1439:299–316. doi: 10.1016/s1388-1981(99)00102-x. [DOI] [PubMed] [Google Scholar]
- 12.Sciorra VA, Morris AJ. Roles for lipid phosphate phosphatases in regulation of cellular signaling. Biochim Biophys Acta. 2002;1582:45–51. doi: 10.1016/s1388-1981(02)00136-1. [DOI] [PubMed] [Google Scholar]
- 13.Testerink C, Munnik T. Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci. 2005;10:368–375. doi: 10.1016/j.tplants.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Devaiah SP, Zhang W, Welti R. Signaling functions of phosphatidic acid. Prog Lipid Res. 2006;45:250–278. doi: 10.1016/j.plipres.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Brindley DN. Lipid phosphate phosphatases and related proteins: signaling functions in development, cell division, and cancer. J Cell Biochem. 2004;92:900–912. doi: 10.1002/jcb.20126. [DOI] [PubMed] [Google Scholar]
- 16.Howe AG, McMaster CR. Regulation of phosphatidylcholine homeostasis by Sec14. Can J Physiol Pharmacol. 2006;84:29–38. doi: 10.1139/Y05-138. [DOI] [PubMed] [Google Scholar]
- 17.Foster DA. Regulation of mTOR by phosphatidic acid? Cancer Res. 2007;67:1–4. doi: 10.1158/0008-5472.CAN-06-3016. [DOI] [PubMed] [Google Scholar]
- 18.Carrasco S, Merida I. Diacylglycerol, when simplicity becomes complex. Trends Biochem Sci. 2007;32:27–36. doi: 10.1016/j.tibs.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Siniossoglou S. Phospholipid metabolism and nuclear function: roles of the lipin family of phosphatidic acid phosphatases. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbalip.2012.09.014. [DOI] [PubMed] [Google Scholar]
- 20.Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 2005;24:1931–1941. doi: 10.1038/sj.emboj.7600672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han G-S, Wu W-I, Carman GM. The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem. 2006;281:9210–9218. doi: 10.1074/jbc.M600425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Hara L, Han GS, Peak-Chew S, Grimsey N, Carman GM, Siniossoglou S. Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J Biol Chem. 2006;281:34537–34548. doi: 10.1074/jbc.M606654200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han GS, Siniossoglou S, Carman GM. The cellular functions of the yeast lipin homolog Pah1p are dependent on its phosphatidate phosphatase activity. J Biol Chem. 2007;282:37026–37035. doi: 10.1074/jbc.M705777200. [DOI] [PubMed] [Google Scholar]
- 24.Karanasios E, Han G-S, Xu Z, Carman GM, Siniossoglou S. A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc Natl Acad Sci U S A. 2010;107:17539–17544. doi: 10.1073/pnas.1007974107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi H-S, Su W-M, Morgan JM, Han G-S, Xu Z, Karanasios E, Siniossoglou S, Carman GM. Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae. Identification of Ser602, Thr723, and Ser744 as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J Biol Chem. 2011;286:1486–1498. doi: 10.1074/jbc.M110.155598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fakas S, Qiu Y, Dixon JL, Han GS, Ruggles KV, Garbarino J, Sturley SL, Carman GM. Phosphatidate phosphatase activity plays a key role in protection against fatty acid-induced toxicity in yeast. J Biol Chem. 2011;286:29074–29085. doi: 10.1074/jbc.M111.258798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi H-S, Su WM, Han GS, Plote D, Xu Z, Carman GM. Pho85p-Pho80p phosphorylation of yeast Pah1p phosphatidate phosphatase regulates its activity, location, abundance, and function in lipid metabolism. J Biol Chem. 2012;287:11290–11301. doi: 10.1074/jbc.M112.346023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adeyo O, Horn PJ, Lee S, Binns DD, Chandrahas A, Chapman KD, Goodman JM. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J Cell Biol. 2011;192:1043–1055. doi: 10.1083/jcb.201010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasser T, Qiu QS, Karunakaran S, Padolina M, Reyes A, Flood B, Smith S, Gonzales C, Fratti RA. The yeast lipin 1 orthologue Pah1p regulates vacuole homeostasis and membrane fusion. J Biol Chem. 2011;287:2221–2236. doi: 10.1074/jbc.M111.317420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reue K, Dwyer JR. Lipin proteins and metabolic homeostasis. J Lipid Res. 2009;50(Suppl):S109–S114. doi: 10.1194/jlr.R800052-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Csaki LS, Reue K. Lipins: multifunctional lipid metabolism proteins. Annu Rev Nutr. 2010;30:257–272. doi: 10.1146/annurev.nutr.012809.104729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris TE, Finck BN. Dual function lipin proteins and glycerolipid metabolism. Trends Endocrinol Metab. 2011 doi: 10.1016/j.tem.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hosaka K, Yamashita S. Partial purification and properties of phosphatidate phosphatase in Saccharomyces cerevisiae. Biochim Biophys Acta. 1984;796:102–109. [PubMed] [Google Scholar]
- 34.Habeler G, Natter K, Thallinger GG, Crawford ME, Kohlwein SD, Trajanoski Z. YPL.db: the Yeast Protein Localization database. Nucleic Acids Res. 2002;30:80–83. doi: 10.1093/nar/30.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar A, Agarwal S, Heyman JA, Matson S, Heidtman M, Piccirillo S, Umansky L, Drawid A, Jansen R, Liu Y, Cheung KH, Miller P, Gerstein M, Roeder GS, Snyder M. Subcellular localization of the yeast proteome. Genes Dev. 2002;16:707–719. doi: 10.1101/gad.970902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 37.Natter K, Leitner P, Faschinger A, Wolinski H, McCraith S, Fields S, Kohlwein SD. The spatial organization of lipid synthesis in the yeast Saccharomyces cerevisiae derived from large scale green fluorescent protein tagging and high resolution microscopy. Mol Cell Proteomics. 2005;4:662–672. doi: 10.1074/mcp.M400123-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Henry SA, Kohlwein S, Carman GM. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 2012;190:317–349. doi: 10.1534/genetics.111.130286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hubscher G, Brindley DN, Smith ME, Sedgwick B. Stimulation of biosynthesis of glyceride. Nature. 1967;216:449–453. doi: 10.1038/216449a0. [DOI] [PubMed] [Google Scholar]
- 40.Martin-Sanz P, Hopewell R, Brindley DN. Long-chain fatty acids and their acyl-CoA esters cause the translocation of phosphatidate phosphatase from the cytolic to the microsomal fraction of rat liver. FEBS LETTERS. 1984;175:284–288. doi: 10.1016/0014-5793(84)80752-8. [DOI] [PubMed] [Google Scholar]
- 41.Hosaka K, Yamashita S. Regulatory role of phosphatidate phosphatase in triacylglycerol synthesis of Saccharomyces cerevisiae. Biochim Biophys Acta. 1984;796:110–117. [PubMed] [Google Scholar]
- 42.Carman GM, Henry SA. Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog Lipid Res. 1999;38:361–399. doi: 10.1016/s0163-7827(99)00010-7. [DOI] [PubMed] [Google Scholar]
- 43.Gaspar ML, Aregullin MA, Jesch SA, Nunez LR, Villa-Garcia M, Henry SA. The emergence of yeast lipidomics. Biochim Biophys Acta. 2007;1771:241–254. doi: 10.1016/j.bbalip.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 44.Carman GM, Han G-S. Regulation of phospholipid synthesis in yeast. J Lipid Res. 2009;50:S69–S73. doi: 10.1194/jlr.R800043-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Y-P, Carman GM. Purification and characterization of phosphatidate phosphatase from Saccharomyces cerevisiae. J Biol Chem. 1989;264:8641–8645. [PubMed] [Google Scholar]
- 46.Morlock KR, McLaughlin JJ, Lin Y-P, Carman GM. Phosphatidate phosphatase from Saccharomyces cerevisiae. Isolation of 45-kDa and 104-kDa forms of the enzyme that are differentially regulated by inositol. J Biol Chem. 1991;266:3586–3593. [PubMed] [Google Scholar]
- 47.Wu W-I, Liu Y, Riedel B, Wissing JB, Fischl AS, Carman GM. Purification and characterization of diacylglycerol pyrophosphate phosphatase from Saccharomyces cerevisiae. J Biol Chem. 1996;271:1868–1876. doi: 10.1074/jbc.271.4.1868. [DOI] [PubMed] [Google Scholar]
- 48.Toke DA, Bennett WL, Dillon DA, Chen X, Oshiro J, Ostrander DB, Wu W-I, Cremesti A, Voelker DR, Fischl AS, Carman GM. Isolation and characterization of the Saccharomyces cerevisiae DPP1 gene encoding for diacylglycerol pyrophosphate phosphatase. J Biol Chem. 1998;273:3278–3284. doi: 10.1074/jbc.273.6.3278. [DOI] [PubMed] [Google Scholar]
- 49.Stukey J, Carman GM. Identification of a novel phosphatase sequence motif. Protein Science. 1997;6:469–472. doi: 10.1002/pro.5560060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jamal Z, Martin A, Gomez-Munoz A, Brindley DN. Plasma membrane fractions from rat liver contain a phosphatidate phosphohydrolase distinct from that in the endoplasmic reticulum and cytosol. J Biol Chem. 1991;266:2988–2996. [PubMed] [Google Scholar]
- 51.Balsinde J, Dennis EA. Bromoenol lactone inhibits magnesium-dependent phosphatidate phosphohydrolase and blocks triacylglycerol biosynthesis in mouse P388D1 macrophages. J Biol Chem. 1996;271:31937–31941. doi: 10.1074/jbc.271.50.31937. [DOI] [PubMed] [Google Scholar]
- 52.Kocsis MG, Weselake RJ. Phosphatidate phosphatases of mammals, yeast, and higher plants. Lipids. 1996;31:785–802. doi: 10.1007/BF02522974. [DOI] [PubMed] [Google Scholar]
- 53.Brindley DN, Waggoner DW. Phosphatidate phosphohydrolase and signal transduction. Chem Phys Lipids. 1996;80:45–57. doi: 10.1016/0009-3084(96)02545-5. [DOI] [PubMed] [Google Scholar]
- 54.Toke DA, Bennett WL, Oshiro J, Wu WI, Voelker DR, Carman GM. Isolation and characterization of the Saccharomyces cerevisiae LPP1 gene encoding a Mg2+-independent phosphatidate phosphatase. J Biol Chem. 1999;273:14331–14338. doi: 10.1074/jbc.273.23.14331. [DOI] [PubMed] [Google Scholar]
- 55.Faulkner AJ, Chen X, Rush J, Horazdovsky B, Waechter CJ, Carman GM, Sternweis PC. The LPP1 and DPP1 gene products account for most of the isoprenoid phosphatase activities in Saccharomyces cerevisiae. J Biol Chem. 1999;274:14831–14837. doi: 10.1074/jbc.274.21.14831. [DOI] [PubMed] [Google Scholar]
- 56.Brindley DN, Waggoner DW. Mammalian lipid phosphate phosphohydrolases. J Biol Chem. 1998;273:24281–24284. doi: 10.1074/jbc.273.38.24281. [DOI] [PubMed] [Google Scholar]
- 57.Irie K, Takase M, Araki H, Oshima Y. A gene, SMP2, involved in plasmid maintenance and respiration in Saccharomyces cerevisiae encodes a highly charged protein. Mol Gen Genet. 1993;236:283–288. doi: 10.1007/BF00277124. [DOI] [PubMed] [Google Scholar]
- 58.Siniossoglou S, Santos-Rosa H, Rappsilber J, Mann M, Hurt E. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 1998;17:6449–6464. doi: 10.1093/emboj/17.22.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Péterfy M, Phan J, Xu P, Reue K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat Genet. 2001;27:121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- 60.Langner CA, Birkenmeier EH, Ben-Zeev O, Schotz MC, Sweet HO, Davisson MT, Gordon JI. The fatty liver dystrophy (fld) mutation. A new mutant mouse with a developmental abnormality in triglyceride metabolism and associated tissue-specific defects in lipoprotein lipase and hepatic lipase activities. J Biol Chem. 1989;264:7994–8003. [PubMed] [Google Scholar]
- 61.Phan J, Reue K. Lipin, a lipodystrophy and obesity gene. Cell Metab. 2005;1:73–83. doi: 10.1016/j.cmet.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 62.Donkor J, Sariahmetoglu M, Dewald J, Brindley DN, Reue K. Three Mammalian Lipins Act as Phosphatidate Phosphatases with Distinct Tissue Expression Patterns. J Biol Chem. 2007;282:3450–3457. doi: 10.1074/jbc.M610745200. [DOI] [PubMed] [Google Scholar]
- 63.Han G-S, Carman GM. Characterization of the human LPIN1-encoded phosphatidate phosphatase isoforms. J Biol Chem. 2010;285:14628–14638. doi: 10.1074/jbc.M110.117747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reue K, Donkor J. Genetic factors in type 2 diabetes: all in the (lipin) family. Diabetes. 2007;56:2842–2843. doi: 10.2337/db07-1288. [DOI] [PubMed] [Google Scholar]
- 65.Grimsey N, Han G-S, O’Hara L, Rochford JJ, Carman GM, Siniossoglou S. Temporal and spatial regulation of the phosphatidate phosphatases lipin 1 and 2. J Biol Chem. 2008:29166–29174. doi: 10.1074/jbc.M804278200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Valente V, Maia RM, Vianna MC, Paco-Larson ML. Drosophila melanogaster lipins are tissue-regulated and developmentally regulated and present specific subcellular distributions. FEBS J. 2010;277:4775–4788. doi: 10.1111/j.1742-4658.2010.07883.x. [DOI] [PubMed] [Google Scholar]
- 67.Ugrankar R, Liu Y, Provaznik J, Schmitt S, Lehmann M. Lipin is a Central Regulator of Adipose Tissue Development and Function in Drosophila. Mol Cell Biol. 2011;31:1646–1656. doi: 10.1128/MCB.01335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Golden A, Liu J, Cohen-Fix O. Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J Cell Sci. 2009;122:1970–1978. doi: 10.1242/jcs.044743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nakamura Y, Koizumi R, Shui G, Shimojima M, Wenk MR, Ito T, Ohta H. Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc Natl Acad Sci U S A. 2009;106:20978–20983. doi: 10.1073/pnas.0907173106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eastmond PJ, Quettier AL, Kroon JT, Craddock C, Adams N, Slabas AR. Phosphatidic acid phosphohydrolase 1 and 2 regulate phospholipid synthesis at the endoplasmic reticulum in Arabidopsis. Plant Cell. 2010;22:2796–2811. doi: 10.1105/tpc.109.071423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carman GM, Han G-S. Phosphatidic acid phosphatase, a key enzyme in the regulation of lipid synthesis. J Biol Chem. 2009;284:2593–2597. doi: 10.1074/jbc.R800059200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Siniossoglou S. Lipins, lipids and nuclear envelope structure. Traffic. 2009;10:1181–1187. doi: 10.1111/j.1600-0854.2009.00923.x. [DOI] [PubMed] [Google Scholar]
- 73.Bou KM, Blais A, Figeys D, Yao Z. Lipin - The bridge between hepatic glycerolipid biosynthesis and lipoprotein metabolism. Biochim Biophys Acta. 2010;1801:1249–1259. doi: 10.1016/j.bbalip.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 74.Furneisen JM, Carman GM. Enzymological properties of the LPP1-encoded lipid phosphatase from Saccharomyces cerevisiae. Biochim Biophys Acta. 2000;1484:71–82. doi: 10.1016/s1388-1981(99)00202-4. [DOI] [PubMed] [Google Scholar]
- 75.Abdel-Latif AA, Smith JP. Studies on the effects of Mg2+ ion and propranolol on iris muscle phosphatidate phosphatase. Can J Biochem Cell Biol. 1984;62:170–177. doi: 10.1139/o84-024. [DOI] [PubMed] [Google Scholar]
- 76.Collet JF, Stroobant V, Pirard M, Delpierre G, Van Schaftingen E. A new class of phosphotransferases phosphorylated on an aspartate residue in an amino-terminal DXDX(T/V) motif. J Biol Chem. 1998;273:14107–14112. doi: 10.1074/jbc.273.23.14107. [DOI] [PubMed] [Google Scholar]
- 77.Collet JF, Stroobant V, Van Schaftingen E. Mechanistic studies of phosphoserine phosphatase, an enzyme related to P-type ATPases. J Biol Chem. 1999;274:33985–33990. doi: 10.1074/jbc.274.48.33985. [DOI] [PubMed] [Google Scholar]
- 78.Hemrika W, Renirie R, Dekker HL, Barnett P, Wever R. From phosphatases to vanadium peroxidases: a similar architecture of the active site. Proc Nat Acad Sci USA. 1997;94:2145–2149. doi: 10.1073/pnas.94.6.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Neuwald AF. An unexpected structural relationship between integral membrane phosphatases and soluble haloperoxidases. Protein Science. 1997;6:1764–1767. doi: 10.1002/pro.5560060817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Toke DA, McClintick ML, Carman GM. Mutagenesis of the phosphatase sequence motif in diacylglycerol pyrophosphate phosphatase from Saccharomyces cerevisiae. Biochemistry. 1999;38:14606–14613. doi: 10.1021/bi991472x. [DOI] [PubMed] [Google Scholar]
- 81.Dillon DA, Chen X, Zeimetz GM, Wu W-I, Waggoner DW, Dewald J, Brindley DN, Carman GM. Mammalian Mg2+-independent phosphatidate phosphatase (PAP2) displays diacylglycerol pyrophosphate phosphatase activity. J Biol Chem. 1997;272:10361–10366. doi: 10.1074/jbc.272.16.10361. [DOI] [PubMed] [Google Scholar]
- 82.Sorger D, Daum G. Triacylglycerol biosynthesis in yeast. Appl Microbiol Biotechnol. 2003;61:289–299. doi: 10.1007/s00253-002-1212-4. [DOI] [PubMed] [Google Scholar]
- 83.Taylor FR, Parks LW. Triacylglycerol metabolism in Saccharomyces cerevisiae relation to phospholipid synthesis. Biochim Biophys Acta. 1979;575:204–214. doi: 10.1016/0005-2760(79)90022-5. [DOI] [PubMed] [Google Scholar]
- 84.Fei W, Shui G, Zhang Y, Krahmer N, Ferguson C, Kapterian TS, Lin RC, Dawes IW, Brown AJ, Li P, Huang X, Parton RG, Wenk MR, Walther TC, Yang H. A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 2011;7:e1002201. doi: 10.1371/journal.pgen.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carman GM, Henry SA. Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J Biol Chem. 2007;282:37293–37297. doi: 10.1074/jbc.R700038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schuller HJ, Hahn A, Troster F, Schutz A, Schweizer E. Coordinate genetic control of yeast fatty acid synthase genes FAS1 and FAS2 by an upstream activation site common to genes involved in membrane lipid biosynthesis. EMBO J. 1992;11:107–114. doi: 10.1002/j.1460-2075.1992.tb05033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chirala SS. Coordinated regulation and inositol-mediated and fatty acid-mediated repression of fatty acid synthase genes in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1992;89:10232–10236. doi: 10.1073/pnas.89.21.10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hasslacher M, Ivessa AS, Paltauf F, Kohlwein SD. Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J Biol Chem. 1993;268:10946–10952. [PubMed] [Google Scholar]
- 89.Chirala SS, Zhong Q, Huang W, Al-Feel W. Analysis of FAS3/ACC regulatory region of Saccharomyces cerevisiae: identification of a functional UASINO and sequences responsible for fatty acid mediated repression. Nucleic Acids Res. 1994;22:412–418. doi: 10.1093/nar/22.3.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.White MJ, Hirsch JP, Henry SA. The OPI1 gene of Saccharomyces cerevisiae, a negative regulator of phospholipid biosynthesis, encodes a protein containing polyglutamine tracts and a leucine zipper. J Biol Chem. 1991;266:863–872. [PubMed] [Google Scholar]
- 91.Loewen CJR, Roy A, Levine TP. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Loewen CJR, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, Levine TP. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science. 2004;304:1644–1647. doi: 10.1126/science.1096083. [DOI] [PubMed] [Google Scholar]
- 93.Ambroziak J, Henry SA. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J Biol Chem. 1994;269:15344–15349. [PubMed] [Google Scholar]
- 94.Han G-S, O’Hara L, Carman GM, Siniossoglou S. An unconventional diacylglycerol kinase that regulates phospholipid synthesis and nuclear membrane growth. J Biol Chem. 2008;283:20433–20442. doi: 10.1074/jbc.M802903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Raetz CRH, Dowhan W. Biosynthesis and function of phospholipids in Escherichia coli. J Biol Chem. 1990;265:1235–1238. [PubMed] [Google Scholar]
- 96.Sakane F, Imai S, Kai M, Yasuda S, Kanoh H. Diacylglycerol kinases: why so many of them? Biochim Biophys Acta. 2007;1771:793–806. doi: 10.1016/j.bbalip.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 97.Merida I, vila-Flores A, Merino E. Diacylglycerol kinases: at the hub of cell signalling. Biochem J. 2008;409:1–18. doi: 10.1042/BJ20071040. [DOI] [PubMed] [Google Scholar]
- 98.Topham MK, Prescott SM. Mammalian diacylglycerol kinases, a family of lipid kinases with signaling functions. J Biol Chem. 1999;274:11447–11450. doi: 10.1074/jbc.274.17.11447. [DOI] [PubMed] [Google Scholar]
- 99.Péterfy M, Phan J, Reue K. Alternatively spliced lipin isoforms exhibit distinct expression pattern, subcellular localization, and role in adipogenesis. J Biol Chem. 2005;280:32883–32889. doi: 10.1074/jbc.M503885200. [DOI] [PubMed] [Google Scholar]
- 100.Manmontri B, Sariahmetoglu M, Donkor J, Bou KM, Sundaram M, Yao Z, Reue K, Lehner R, Brindley DN. Glucocorticoids and cAMP selectively increase hepatic lipin-1 expression and insulin acts antagonistically. J Lipid Res. 2008;49:1056–1067. doi: 10.1194/jlr.M800013-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang P, O’Loughlin L, Brindley DN, Reue K. Regulation of lipin-1 gene expression by glucocorticoids during adipogenesis. J Lipid Res. 2008;49:1519–1528. doi: 10.1194/jlr.M800061-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ishimoto K, Nakamura H, Tachibana K, Yamasaki D, Ota A, Hirano K, Tanaka T, Hamakubo T, Sakai J, Kodama T, Doi T. Sterol-mediated Regulation of Human Lipin 1 Gene Expression in Hepatoblastoma Cells. J Biol Chem. 2009;284:22195–22205. doi: 10.1074/jbc.M109.028753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ryu D, Oh KJ, Jo HY, Hedrick S, Kim YN, Hwang YJ, Park TS, Han JS, Choi CS, Montminy M, Koo SH. TORC2 regulates hepatic insulin signaling via a mammalian phosphatidic acid phosphatase, LIPIN1. Cell Metab. 2009;9:240–251. doi: 10.1016/j.cmet.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 104.Gropler MC, Harris TE, Hall AM, Wolins NE, Gross RW, Han X, Chen Z, Finck BN. Lipin 2 Is a Liver-enriched Phosphatidate Phosphohydrolase Enzyme That Is Dynamically Regulated by Fasting and Obesity in Mice. J Biol Chem. 2009;284:6763–6772. doi: 10.1074/jbc.M807882200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Roberts GG, Hudson AP. Transcriptome profiling of Saccharomyces cerevisiae during a transition from fermentative to glycerol-based respiratory growth reveals extensive metabolic and structural remodeling. Mol Genet Genomics. 2006;276:170–186. doi: 10.1007/s00438-006-0133-9. [DOI] [PubMed] [Google Scholar]
- 106.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 107.Soto-Cardalda A, Fakas S, Pascual F, Choi HS, Carman GM. Phosphatidate phosphatase plays role in zinc-mediated regulation of phospholipid synthesis in yeast. J Biol Chem. 2011;287:968–977. doi: 10.1074/jbc.M111.313130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vallee BL, Falchuk KH. The biochemical basis of zinc physiology. Physiol Rev. 1993;73:79–118. doi: 10.1152/physrev.1993.73.1.79. [DOI] [PubMed] [Google Scholar]
- 109.Eide DJ. Homeostatic and adaptive responses to zinc deficiency in Saccharomyces cerevisiae. J Biol Chem. 2009;284:18565–18569. doi: 10.1074/jbc.R900014200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schwabe JW, Klug A. Zinc mining for protein domains [news; comment] Nat Struct Biol. 1994;1:345–349. doi: 10.1038/nsb0694-345. [DOI] [PubMed] [Google Scholar]
- 111.Ellis CD, Wang F, MacDiarmid CW, Clark S, Lyons T, Eide DJ. Zinc and the Msc2 zinc transporter protein are required for endoplasmic reticulum function. J Cell Biol. 2004;166:325–335. doi: 10.1083/jcb.200401157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.MacDiarmid CW, Gaither LA, Eide D. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 2000;19:2845–2855. doi: 10.1093/emboj/19.12.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhao H, Eide D. The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J Biol Chem. 1996;271:23203–23210. doi: 10.1074/jbc.271.38.23203. [DOI] [PubMed] [Google Scholar]
- 114.Zhao H, Eide D. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc Natl Acad Sci U S A. 1996;93:2454–2458. doi: 10.1073/pnas.93.6.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Waters BM, Eide DJ. Combinatorial control of yeast FET4 gene expression by iron, zinc, and oxygen. J Biol Chem. 2002;277:33749–33757. doi: 10.1074/jbc.M206214200. [DOI] [PubMed] [Google Scholar]
- 116.MacDiarmid CW, Milanick MA, Eide DJ. Induction of the ZRC1 Metal Tolerance Gene in Zinc-limited Yeast Confers Resistance to Zinc Shock. J Biol Chem. 2003;278:15065–15072. doi: 10.1074/jbc.M300568200. [DOI] [PubMed] [Google Scholar]
- 117.Miyabe S, Izawa S, Inoue Y. The Zrc1 is involved in zinc transport system between vacuole and cytosol in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2001;282:79–83. doi: 10.1006/bbrc.2001.4522. [DOI] [PubMed] [Google Scholar]
- 118.Devirgiliis C, Murgia C, Danscher G, Perozzi G. Exchangeable zinc ions transiently accumulate in a vesicular compartment in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2004;323:58–64. doi: 10.1016/j.bbrc.2004.08.051. [DOI] [PubMed] [Google Scholar]
- 119.Ellis CD, MacDiarmid CW, Eide DJ. Heteromeric protein complexes mediate zinc transport into the secretory pathway of eukaryotic cells. J Biol Chem. 2005;280:28811–28818. doi: 10.1074/jbc.M505500200. [DOI] [PubMed] [Google Scholar]
- 120.Muhlenhoff U, Stadler JA, Richhardt N, Seubert A, Eickhorst T, Schweyen RJ, Lill R, Wiesenberger G. A specific role of the yeast mitochondrial carriers MRS3/4p in mitochondrial iron acquisition under iron-limiting conditions. J Biol Chem. 2003;278:40612–40620. doi: 10.1074/jbc.M307847200. [DOI] [PubMed] [Google Scholar]
- 121.Han G-S, Johnston CN, Chen X, Athenstaedt K, Daum G, Carman GM. Regulation of the Saccharomyces cerevisiae DPP1-encoded diacylglycerol pyrophosphate phosphatase by zinc. J Biol Chem. 2001;276:10126–10133. doi: 10.1074/jbc.M011421200. [DOI] [PubMed] [Google Scholar]
- 122.Han S-H, Han G-S, Iwanyshyn WM, Carman GM. Regulation of the PIS1-encoded phosphatidylinositol synthase in Saccharomyces cerevisiae by zinc. J Biol Chem. 2005;280:29017–29024. doi: 10.1074/jbc.M505881200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kersting MC, Carman GM. Regulation of the Saccharomyces cerevisiae EKI1-encoded ethanolamine kinase by zinc depletion. J Biol Chem. 2006;281:13110–13116. doi: 10.1074/jbc.M601612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Soto A, Carman GM. Regulation of the Saccharomyces cerevisiae CKI1-encoded choline kinase by zinc depletion. J Biol Chem. 2008;283:10079–10088. doi: 10.1074/jbc.M800502200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Eide DJ. Multiple Regulatory Mechanisms Maintain Zinc Homeostasis in Saccharomyces cerevisiae. J Nutr. 2003;133:1532S–1535S. doi: 10.1093/jn/133.5.1532S. [DOI] [PubMed] [Google Scholar]
- 126.Carman GM, Han GS. Regulation of phospholipid synthesis in Saccharomyces cerevisiae by zinc depletion. Biochim Biophys Acta. 2007;1771:322–330. doi: 10.1016/j.bbalip.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wu W-I, Lin Y-P, Wang E, Merrill AH, Jr, Carman GM. Regulation of phosphatidate phosphatase activity from the yeast Saccharomyces cerevisiae by sphingoid bases. J Biol Chem. 1993;268:13830–13837. [PubMed] [Google Scholar]
- 128.Wu W-I, Carman GM. Regulation of phosphatidate phosphatase activity from the yeast Saccharomyces cerevisiae by nucleotides. J Biol Chem. 1994;269:29495–29501. [PubMed] [Google Scholar]
- 129.Ostrander DB, O’Brien DJ, Gorman JA, Carman GM. Effect of CTP synthetase regulation by CTP on phospholipid synthesis in Saccharomyces cerevisiae. J Biol Chem. 1998;273:18992–19001. doi: 10.1074/jbc.273.30.18992. [DOI] [PubMed] [Google Scholar]
- 130.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 131.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, McCartney RR, Schmidt MC, Rachidi N, Lee SJ, Mah AS, Meng L, Stark MJ, Stern DF, De Virgilio C, Tyers M, Andrews B, Gerstein M, Schweitzer B, Predki PF, Snyder M. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 132.Dephoure N, Howson RW, Blethrow JD, Shokat KM, O’Shea EK. Combining chemical genetics and proteomics to identify protein kinase substrates. Proc Natl Acad Sci U S A. 2005;102:17940–17945. doi: 10.1073/pnas.0509080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 134.Enserink JM, Kolodner RD. An overview of Cdk1-controlled targets and processes. Cell Div. 2010;5:11. doi: 10.1186/1747-1028-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Moffat J, Huang D, Andrews B. Functions of Pho85 cyclin-dependent kinases in budding yeast. Prog Cell Cycle Res. 2000;4:97–106. doi: 10.1007/978-1-4615-4253-7_9. [DOI] [PubMed] [Google Scholar]
- 136.Carroll AS, O’Shea EK. Pho85 and signaling environmental conditions. Trends Biochem Sci. 2002;27:87–93. doi: 10.1016/s0968-0004(01)02040-0. [DOI] [PubMed] [Google Scholar]
- 137.Huang D, Friesen H, Andrews B. Pho85, a multifunctional cyclin-dependent protein kinase in budding yeast. Mol Microbiol. 2007;66:303–314. doi: 10.1111/j.1365-2958.2007.05914.x. [DOI] [PubMed] [Google Scholar]
- 138.Xu Z, Su WM, Carman GM. Fluorescence spectroscopy measures yeast PAH1-encoded phosphatidate phosphatase interaction with liposome membranes. J Lipid Res. 2011;53:522–528. doi: 10.1194/jlr.M022798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Su W-M, Han G-S, Casciano J, Carman GM. Protein kinase A-mediated phosphorylation of Pah1p phosphatidate phosphatase functions in conjunction with the Pho85p-Pho80p and Cdc28p-cyclin B kinases to regulate lipid synthesis in yeast. J Biol Chem. 2012;287 doi: 10.1074/jbc.M112.402339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Broach JR, Deschenes RJ. The function of RAS genes in Saccharomyces cerevisiae. Adv Cancer Res. 1990;54:79–139. doi: 10.1016/s0065-230x(08)60809-x. [DOI] [PubMed] [Google Scholar]
- 141.Thevelein JM. Signal Transduction in Yeast. Yeast. 1994;10:1753–1790. doi: 10.1002/yea.320101308. [DOI] [PubMed] [Google Scholar]
- 142.Carman GM, Han G-S. Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Ann Rev Biochem. 2011;80:859–883. doi: 10.1146/annurev-biochem-060409-092229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Harris TE, Huffman TA, Chi A, Shabanowitz J, Hunt DF, Kumar A, Lawrence JC., Jr Insulin Controls Subcellular Localization and Multisite Phosphorylation of the Phosphatidic Acid Phosphatase, Lipin 1. J Biol Chem. 2007;282:277–286. doi: 10.1074/jbc.M609537200. [DOI] [PubMed] [Google Scholar]
- 144.Kim Y, Gentry MS, Harris TE, Wiley SE, Lawrence JC, Jr, Dixon JE. A conserved phosphatase cascade that regulates nuclear membrane biogenesis. Proc Natl Acad Sci U S A. 2007;104:6596–6601. doi: 10.1073/pnas.0702099104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Han S, Bahmanyar S, Zhang P, Grishin N, Oegema K, Crooke R, Graham M, Reue K, Dixon JE, Goodman JM. Nuclear Envelope Phosphatase 1-Regulatory Subunit 1 (Formerly TMEM188) Is the Metazoan Spo7p Ortholog and Functions in the Lipin Activation Pathway. J Biol Chem. 2012;287:3123–3137. doi: 10.1074/jbc.M111.324350. [DOI] [PMC free article] [PubMed] [Google Scholar]