Abstract
A comprehensive study of the phylogeography and population genetics of the largest wild artiodactyl in the arid and cold-temperate South American environments, the guanaco (Lama guanicoe) was conducted. Patterns of molecular genetic structure were described using 514 bp of mtDNA sequence and 14 biparentally-inherited microsatellite markers from 314 samples. These individuals originated from 17 localities throughout the current distribution across Peru, Bolivia, Argentina and Chile. This confirmed well-defined genetic differentiation and subspecies designation of populations geographically separated to the northwest (L. g. cacsilensis) and southeast (L. g. guanicoe) of the central Andes plateau. However, these populations are not completely isolated, as shown by admixture prevalent throughout a limited contact zone, and a strong signal of expansion from north to south in the beginning of the Holocene. Microsatellite analyses differentiated 3 northwestern and 4-5 southeastern populations, suggesting patterns of genetic contact among these populations. Possible genetic refuges were identified, as were source-sink patterns of gene flow at historical and recent time scales. Conservation and management of guanaco should be implemented with an understanding of these local population dynamics while also considering the preservation of broader adaptive variation and evolutionary processes.
Keywords: Camelids, microsatellites, d-loop, contact zone, Holocene, Patagonia
Introduction
The guanaco (Lama guanicoe) is the most ecologically-important and abundant native ungulate of the arid and cold-temperate region in the southern cone of South America, occupying diverse habitats from sea level to over 4,000 meters (Franklin 1982). West of the Andes, guanacos are widely distributed from northern Peru to Tierra del Fuego in Argentina and the Navarino Islands in Chile. To the east, they extend southward from the Bolivian and Paraguayan Chaco across the pampas throughout most of Argentina (Torres 1985; González et al. 2006). The current distribution of guanacos corresponds to less than 30% of the range at the time of the arrival of the Europeans to South America (Raedeke, 1979). Currently fewer than 5% of the guanaco population occupies less than 30% of their range, compared with when Europeans arrived to South America in the XVII century (Raedeke, 1979;). Uncontrolled hunting, habitat reduction and deterioration, and displacement by introduced livestock have caused local and regional extinctions resulting ain a reduced and fragmented population (Torres 1992). In spite of the ecological importance of the guanaco, it's past and more recent evolutionary history and broad phylogenetic biogeographic patterns are not well-described.
The genus Lama descended from the feeder-browser llama-like Hemiauchenia that originated in North America and subsequently migrated to South America around 2.5-3.0 million years ago across the Isthmus of Panama as part of the great American interchange (Webb 1974). This migration was probably facilitated by a sub-tropical, relatively arid savannah-like corridor from the Gulf of Mexico, across the Mexican plateau, and through the entire isthmus (Webb 1978, 1991). Through South America, this corridor divided into two main routes: a “high road” along the western slopes of the Andes and pacific coast, and a more eastern “low road” beginning in the northwestern low-lands of Colombia through the Amazonian and Chaco regions (Webb 1978). Although it was initially postulated that modern camelids evolving from Hemiauchenia arose in the Andes (Webb 1974; Webb & Stehli 1995), most Plio-Pleistocene camelid fossils have been found in non-Andean locations (Ochsenius 1995; Menegaz et al. 1989; Scherer 2009), suggesting that the early South American camelid radiation may have occurred elsewhere, such as the pampas of Argentina, and that descendants later colonized Andean and subtropical regions (Menegaz et al. 1989; Scherer 2009).
Paleontological and zooarcheological evidence suggest that guanacos expanded from north to south. The earliest fossils, from the Pliocene and late Pleistocene, were found in the Pampas of Buenos Aires and the “mesopotamic” region of Argentina, in northern Uruguay and northeastern and southern Brazil (Scherer 2009). Subsequently, L. guanicoe is found along the Reconquista River, east of the Pampean region, in strata from the middle Pleistocene (< 0.78 Ma and > 0.126 Ma) (Menegaz et al. 1989; Cajal et al. 2010). In the late Pleistocene, guanaco have been found in Paso Otero (Necochea County) in the La Chumbiada (35 000 radiocarbon years BP, Cajal et al. 2010) and Guerrero Members (ca. 24 000 and ca. 10 000 radiocarbon years BP, Tonni et al. 2003) of the Luján Formation. L. guanicoe was found in four sites in the Santa Cruz Province in Patagonia: La María (10 967 ± 55 radiocarbon years BP, Paunero, 2002), Los Toldos (12 600 ± 650 radiocarbon years BP; Cardich et al. 1973; Cardich 1987), El Ceibo (ca. 11 000 radiocarbon years BP; Cardich 1987) and Piedra Museo (units 6 to 4, dated between 12 890 ± 90 and 9230 ± 105 radiocarbon years BP; Miotti et al. 2003). This radiocarbon date is similar to other taxon-dates obtained from L. guanicoe bones from the Cueva del Medio, 10,450 ± 100, 10,710 ± 190, and 10,850 ± 130 radiocarbon years before present (Nami & Nakamura 1995; Massone & Prieto 2004). In Tierra del Fuego L. guanicoe dates to 10,130 and 11,880 AMS years before present (Massone & Prieto 2004). Finally, guanaco bones associated with signs of human settlements from the Holocene around 6,000 years ago have been found along the Beagle Channel, between Tierra del Fuego and Navarino Island (Tivoli & Zangrando 2011), suggesting a more recent colonization of guanaco to these islands. Nevertheless, fossil records are abundant in the Pampas from the late Pleistocene, but are scarce in Patagonia, making it difficult to document their arrival time. However, fossils are not unequivocal and are affected by degradation, taphonomic processes, and ease of discovery (Borrero 2005; Fernandez 2008; Borrero 2008), making local geographic findings difficult to interpret on a broad scale.
Traditionally, four subspecies of modern guanacos have been recognized (Wheeler 1995) based on their distribution and phenotype (body and skull size, and coloration): Lama guanicoe guanicoe Müller 1776, L. g. huanacus Molina 1782, L. g. cacsilensis Lönnberg 1913, and L. g. voglii Krumbiegel 1944. However, Franklin (1982, 1983) speculated that there were only two guanaco subspecies, separated by the salt plains of southern Bolivia and the crests of the Andean chain: the smaller, lighter-colored L. g. cacsilensis, restricted to the north-western slopes of the Andes between 8° and 41° S and the larger and darker L. g. guanicoe located on the south-eastern side of the Andes between 18 and 55° S (Raedeke 1979; González et al. 2006). This hypothesis was partially confirmed from an analysis of sequence variation in two mitochondrial genes (Marin et al. 2008), but the geographic limits for each taxon could not be clearly defined. Additionally, substructure within these subspecies has not been assessed, although analyses of microsatellite variation have suggested that low-levels of genetic structure may occur between neighbouring populations, especially where these are isolated such as in Tierra del Fuego (Sarno et al. 2001; Maté et al. 2005).
Here we present a comprehensive assessment of the molecular diversity of guanaco based on samples collected throughout its distribution range and analyze variation in14 microsatellite loci and sequence variation in mtDNA control region sequence. We present evidence of range-wide phylogeographic structure linked with guanaco evolutionary history to 1) define patterns of molecular genetic structure among guanacos, 2) link patterns of genetic variation with phylogeographic history and barriers to gene flow, and 3) describe and contrast the evolutionary history and patterns of gene flow among these populations.
Materials and methods
Sample collection and DNA extraction
Material for DNA analysis was collected from guanacos throughout their range. DNA was extracted from blood samples from 88 wild-caught adults following chemical immobilization (Sarno et al. 1996) at five localities, from muscle or skin samples from 55 dead animals from 6 localities, and from 150 fresh faecal samples from different dung piles at 8 localities (Table 1, Fig. 1A). DNA was also obtained opportunistically from liver samples of 26 adult males in Valle Chacabuco (Valchac Ltd.), Chile, under a sustainable-use program authorized by the Chilean Government. Locations of sample sites and the geographic position of individuals collected at each site are given in Figure 1 and Table 1. All samples were stored at –70° C in the Laboratorio de Genómica y Biodiversidad, Departamento de Ciencias Básicas, Facultad de Ciencias, Universidad del Bio-Bío, Chillán, Chile or at CONOPA in Lima, Peru. We followed guidelines of the American Society of Mammalogists during the collection and handling of animals (Gannon et al. 2007). Total genomic DNA was extracted from blood and bone marrow using the Wizard Genomic DNA Purification Kit (Promega, Madison, Wisconsin). DNA from liver and muscle samples was purified using proteinase K digestion and a standard phenol-chloroform protocol (Sambrook et al. 1989). DNA from faeces was extracted using the QIAamp DNA Stool Mini Kit (QIAGEN, Valencia, California) in a separate non-genetic-oriented laboratory.
Table 1.
Summary of the Lama guanicoe samples used in the analyses, including localities (ordered roughly from north to south), type of sample (B indicates blood; F, faecal; M, muscle; S, skin and L, liver), number of samples used from each locality for each genetic marker. Demarcation of regions was assisted by the empirical results of spatial analyses.
| Region Population | Localities, Country (Abbreviation) | Sample type | Samples mtDNA (N = 306) | Samples Microsatellites (N = 314) |
|---|---|---|---|---|
| Northwestern Region | ||||
| Hiper-Arid Peru | Huallhua, Peru (HU) | F | 10 | 18 |
| Chilean Pre-altiplano | Putre, Chile (PU) | B, F | 20 | 21 |
| Arid Chile and Chilean Peri-Puna | Pan de Azucar National Park, Chile (PA) | F | 11 | 12 |
| Llanos de Challe National Park, Chile (LC) | B, M, F | 28 | 21 | |
| Illapel, Chile (IL) | B, M | 20 | 19 | |
| Southeastern Region | ||||
| Bolivian Chaco | Kaa-Iya National Park, Bolivia (KA) | F | 20 | 17 |
| Northern and central Patagonia | La Payunia Reserve, Argentina (LP) | B | 10 | 10 |
| Loma Blanca, Argentina (LB) | F | 20 | 20 | |
| Telsen, Argentina (TE) | S | 13 | 15 | |
| Península Valdés, Argentina (PV) | S | 21 | 15 | |
| Camarones, Argentina (CA) | S | 13 | 15 | |
| Western Patagonia | Valle Chacabuco, Chile (VC) | L | 20 | 26 |
| Southern Patagonia | Bosques Petrificados, Argentina (BP) | F | 20 | 20 |
| Monte Leon, Argentina (ML) | F, M | 20 | 22 | |
| Torres del Paine National Park, Chile (TP) | B | 20 | 23 | |
| Pali-Ayke National Park, Chile (PK) | F | 20 | 20 | |
| Fuegian zone | Tierra del Fuego, Chile (TF) | B | 20 | 20 |
Fig. 1.
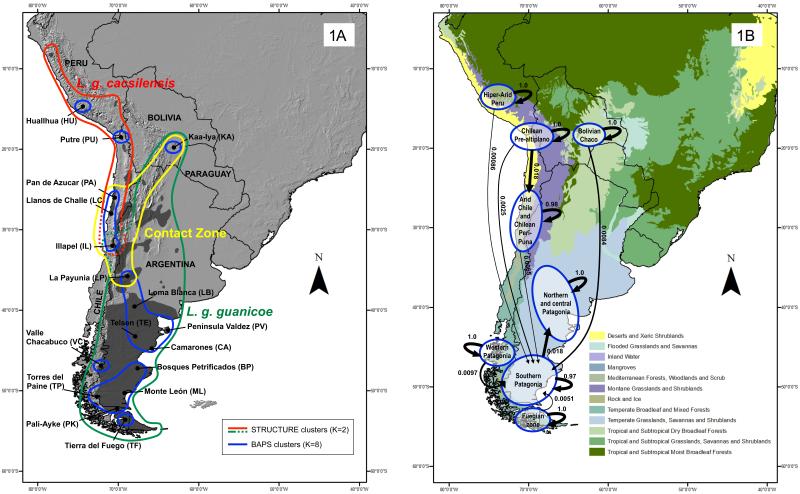
Geographic distribution of Lama guanicoe in South America (in dark-grey based on González et al. 2006) showing genetic divisions by subspecies determined by mitochondrial BAPS (red and green continuous lines) and STRUCTURE K=2 (red and green-backed line) analyses, contact zone (yellow line) obtained by microsatellite assignations (STRUCTURE, see Table 3), and populations (blue lines) identified by microsatellite BAPS (1A). Gene flow among populations identified by microsatellite BAPS analyses. The thickness of the arrow indicates the magnitude of gene flow (1B).
Microsatellites markers
Fourteen autosomal dinucleotide microsatellite loci, designated YWLL08, YWLL29, YWLL36, YWLL38, YWLL40, YWLL43, YWLL46 (Lang et al. 1996), LCA5, LCA19, LCA22, LCA23 (Penedo et al. 1998), LCA65 (Penedo et al. 1999) and LGU49, LGU68 (Sarno et al. 2000) were analyzed. Amplification was carried out in a 10 μL reaction volume, containing 50 - 100 ng of template DNA, 1.5 - 2.0 mm MgCl2, 0.325 μm of each primer, 0.2 mm dNTP, 1X polymerase chain reaction (PCR) buffer (QIAGEN) and 0.4 U Taq polymerase (QIAGEN). All PCR amplifications were performed in a PE9700 (Perkin Elmer Applied Biosystems) thermal cycler with cycling conditions of: initial denaturation at 95 °C for 15 min, followed by 40 cycles of 95 °C for 30 s, 52–57 °C for 90 s and 72 °C for 60 s, and a final extension of 72 °C for 30 min. Amplification and genotyping of DNA from faecal samples was repeated 2 or 3 times. One primer of each pair was labelled with a fluorescent dye on the 5′-end, and fragments analysed on an ABI-3100 sequencer (Perkin Elmer Applied Biosystems). Data collection, sizing of bands and analyses were carried out using GeneScan software (Applied Biosystems).
We identified multiple faecal samples that came from the same individual by searching for matching microsatellite genotypes using the Excel Microsatellite Toolkit (Park 2001) and eliminated samples from the study if they showed more than 85% overlap. We also evaluated the existence of null alleles using the program Micro-Checker version 2.2.3 (van Oosterhout et al. 2004). The software FSTAT 2.9.4 (Goudet 2005) was used to estimate allele frequency, observed heterozygosity (HO), and expected heterozygosity (HE). The inbreeding coefficient FIS, which estimates the heterozygote deficit or excess within populations, was estimated following Weir & Cockerham (1984) using FSTAT 2.9.4.
Mitochondrial DNA
The left domain of the mitochondrial control region (514 bp) was amplified using the camelid and guanaco specific primers LthrArtio (5’-GGT CCT GTA AGC CGA AAA AGG A-3’), H15998V (5’-CCA GCT TCA ATT GAT TTG ACT GCG-3’), HLoop550G: 5′ ATG GAC TGA ATA GCA CCT TAT G 3′ (Marin et al. 2007). Amplification was performed in a 50 μl reaction volume with ~ 30 ng genomic DNA, 1x PCR buffer (8 mM Tris-HCl (pH 8.4); 20 mM KCl (InvitrogenGibco, Life Technologies), 2 mM MgCl2, 25 μM each of dNTP, 0.5 μM each primer and 0.1U/μ Taq polymerase (InvitrogenGibco, Life Technologies®). All PCR amplifications were performed in a PE9700 (Perkin Elmer Applied Biosystems) thermal cycler with cycling conditions as follows: initial denaturation at 95 °C for 10 min, followed by 30-35 cycles of 94°C for 45 s, 62°C for 45 s and 72°C for 60 s, and a final extension of 72 °C for 5 min. PCR products were purified using the GeneClean Turbo for PCR Kit (Bio101) following the manufacturer's instructions. Products were sequenced in forward and reverse directions using BigDye chemistry on an ABI Prism 377 or 3100 semi-automated DNA analyzer. The reactions were carried out in a 10-μL volume containing approximately 100 ng of purified DNA, 1 μL of either forward or reverse primer and 2 μL of BigDye Terminator Kit version 3.1 (PerkinElmer). Sequence reactions were visualized using an ABI-3100 sequencer (Perkin Elmer Applied Biosystems).
Sequences were aligned using Clustal_X (Thompson et al. 1997) and were checked by eye. The number of segregating sites (S) and haplotypes (nh), haplotype diversity (h) (Nei 1987), nucleotide diversity (π) and average number of nucleotide differences between pairs of sequences (k) were estimated using ARLEQUIN 3.5.1.2 (Excoffier & Lischer 2010). These estimates were calculated for each locality independently and for the two broad regions (as detected by STRUCTURE, see Results).
Intraspecific mtDNA genealogies were inferred using the method of statistical parsimony (Templeton et al. 1992) implemented in the program TCS v1.21 (Clement et al. 2000). TCS estimates phylogenetic relationships when there are low levels of divergence and provides a 95% plausible set for all haplotype connections. Three polymorphic sites were deleted to reduce homoplasy among haplotypes for a better interpretation of the network.
Estimation and delimitation of genetic units
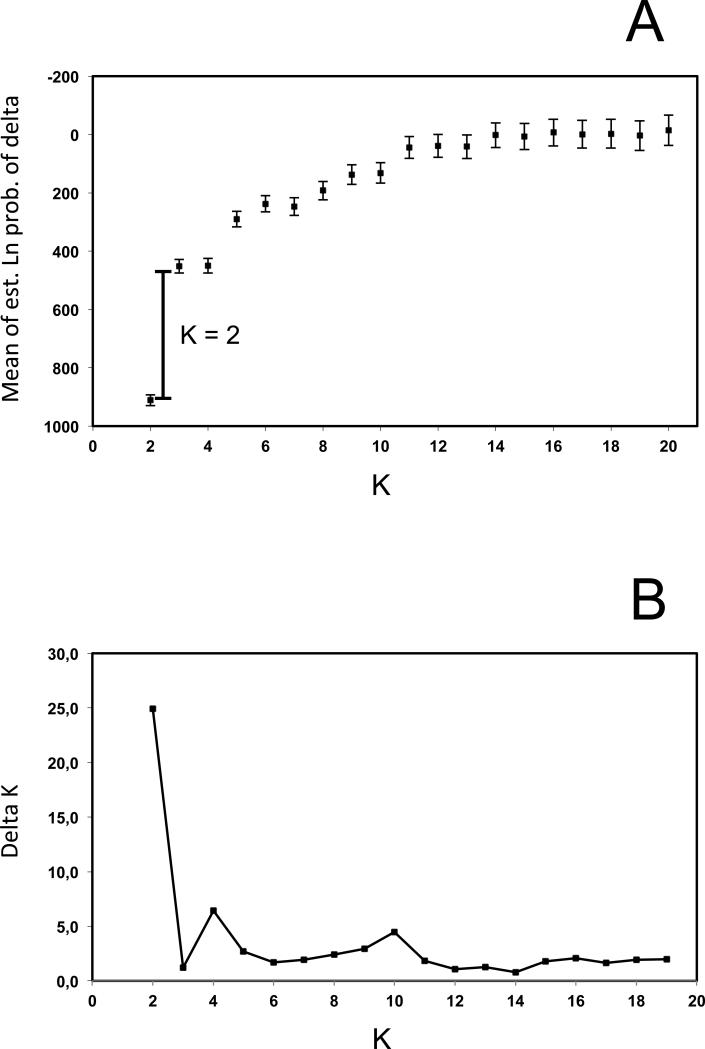
The genetic structure of Lama guanicoe populations was investigated with a Bayesian clustering algorithm implemented in STRUCTURE version 2.3.3 (Pritchard et al. 2000) using the total microsatellite genotypes data set (n = 314) assuming that sampled individuals belong to an unknown number of K genetically distinct clusters. For each K (from 1 to 20), we performed 20 runs and calculated the mean posterior probability of the data [‘log probability of data’, L(K)]. We determined the most probable number of populations, K, by evaluating the significance of the posterior probabilities (Pritchard et al. 2007) (Figure 2A), and by using the method described by Evanno et al. (2005) that examines ΔK, an ad hoc quantity related to the change in posterior probability between runs of different K (Figure 2B). We ran the program using the admixture model and correlated allele frequencies option for 500,000 iterations after a burn-in period of 30,000 iterations. These parameters are considered most appropriate for detecting structure among populations that are likely to be similar due to migration or shared ancestry (Falush et al. 2003; Pritchard et al. 2007). In our data set, the greatest amount of variation was explained by separating two genetic groups (K = 2) (see Results). To detect potential substructure in each group, we assigned individuals based on their highest q-value – an estimate of the proportion of an individual's genome attributed to each of the identified genetic clusters – and then analyzed each group separately.
Fig. 2.
Support for defining of the number of guanaco population based on the microsatellite data set. (A) Mean L(K) ± SD over 20 runs as a function of K. (B) ΔK (Delta K = mean(|L’’(K)|) /sd(L(K))) following Evanno et al (2005) as a function of K.
We assessed genetic differentiation using estimates of FST (Weir & Cockerham 1984) from the microsatellite data using ARLEQUIN 3.5.1.2 (Excoffier & Lischer 2010) with 10,000 permutations for each of the eight populations and two regions. For comparison, genetic differentiation between each pair of populations was also estimated using Jost's D in GENODIVE version 2.0b22 (Meirmans & van Tienderen, 2004), a method which is independent of the amount of within-population diversity (Jost 2008).
The spatial method, implemented in the program BAPS 5.2 (Corander et al. 2008), incorporates coordinate data to assign a non-uniform prior such that neighbouring individuals are more likely to be assigned to the same genetic cluster. We performed spatial mixture clustering of individuals for 10 runs with the maximum number of populations (Kmax) set to 20. Results from each run were stored and merged by the program, and the optimal K value was selected as the partition with the maximum likelihood (L(K)) and highest posterior probability (p). The assignments from the mixture analysis were then used to perform admixture analysis with the recommended parameter values, including 100 iterations for individuals, 20 iterations for reference individuals, and 200 reference individuals from each population. Similar to our STRUCTURE analyses, we assigned individuals to the group with the highest Q-value, flagging potentially admixed individuals (Q < 0.75).
To investigate hierarchical levels of population structure of mitochondrial sequences, an analysis of molecular variance (AMOVA) was performed using ARLEQUIN 3.5.1.2 (Excoffier & Lischer 2010), considering genetic distances between haplotypes and their frequencies among the defined populations, and with statistical significance determined by 10000 permutations. This analysis was based on the geographical division of the population as determinate by STRUCTURE analyses.
Historical demography and Migration
Deviations from a neutral Wright-Fisher model were estimated with Tajima's D and Fu's Fs statistics (Tajima 1989; Fu 1997). A mismatch analysis was performed using ARLEQUIN 3.5.1.2 (Excoffier & Lischer 2010) to test for evidence of population expansion in the groups identified. ARLEQUIN uses a parametric bootstrap approach (10000 simulations) to compute 95% confidence intervals for τ (Tau), the time since the expansion in units of mutational time (τ = 2μt, where μ is the mutational rate and t is the generation time in years), θ0, the initial population size (θ0 = 2μN0) and θ1, the final population size (θ1 = 2μN1). ARLEQUIN also computes the raggedness statistic (Harpending 1994) and the sum-of-squared deviations (SSD), which test the fit of the observed data to the expected population growth model (Schneider & Excoffier 1999). To estimate t, we assumed a mutation rate of 13%/Myr (similar to used in water buffalo, Lau et al. 1998; hartebeest, Flagstad et al. 2001; African buffalo, van Hooft et al. 2002; roan antelope, Alpers et al. 2004; giraffe, Brown et al. 2007 and other artiodactyls), with an average generation time of 3 years (Gonzalez et al. 2006).
To identify demographic scenarios that explain the current diversity patterns in the two regions, we used an Approximate Bayesian Computation approach (Beaumont 2010) implemented in the popABC program (Lopes et al. 2009) using mitochondrial sequences and microsatellite loci together for each of the two STRUCTURE clusters (Northwest and Southeast, see Results). We adopted an “Isolation-Migration” model (Nielsen & Wakeley 2001) that assumes the two populations diverged from a common ancestral population and that these populations may have been connected by migration since separation. The regression step was performed in R using scripts by M. Beaumont (http://code.google.com/p/popabc/model_choice.r) and additional scripts from the popABC package that were modified to fit our analyses. Priors of population parameters were: uniform distributions bound between minimum and maximum values (see supplementary Table 6, Supplementary Material online) with or without gene flow.
Similarly, we also used LAMARC v. 2.1.8 (Kuhner 2006) to estimate ML migration rates between two sub-regions of L. guanicoe. This approach, based on coalescence using MCMC searches, takes both history and asymmetrical gene flow into account, unlike classical migration–drift equilibrium approaches, and allows simultaneous estimation of population growth or decline (Beerli & Felsenstein 2001). We ran 10 short chains (using 500 of 10,000 sampled trees) and 2 long chains (using 10,000 of 200,000 sampled trees), assuming two clusters with possible population growth and bi-directional migration. LAMARC searches were run with a starting value of θ and with the Mixed K-Allele/Stepwise model (MixedKS) that considers both mutation to adjacent states (like the Stepwise model) and mutation to arbitrary states (like the K-Allele model).
Results
Genetic diversity
Of the 320 replicated microsatellite genotypes, we found four pairs of faecal samples that were identified as being from the same individual and therefore were removed from the analysis. We detected 209 alleles at 14 loci in 314 individuals. The number of alleles per locus ranged from 9 to 30, and 43 alleles were unique to a single population. Within populations, we found no significant departures from H-W equilibrium (P > 0.0011, the Bonferoni-adjusted critical value), no evidence for linkage among the five loci (P > 0.0005, the adjusted critical value, for each pair of loci across all populations), and low frequencies of null alleles per locus (fo < 0.08). We found consistently high levels of genetic diversity (mean expected heterozygosity ranged from 0.56 to 0.84) and high values for allelic richness (mean RA ranged from 4.41 to 9.33) (Table 2). Thirty-nine variable positions (7.6%), 34 transitions, 7 transversions and one insertion from 514 nucleotides and 35 haplotypes were identified in 306 sequences of the 5’ end of the control region. Haplotype (h) and nucleotide diversity (π) is detailed in Table 2 and the distribution of haplotypes within 17 localities is given in Table 5. Sequences were deposited in GenBank with accession numbers JX678291 - JX678596.
Table 2.
Genetic diversity indices from 14 microsatellite loci and mtDNA Control Region sequences by localities (defined in Table 1 and Fig 1A).
| Region Localities | Microsatellites |
mtDNA |
|||||
|---|---|---|---|---|---|---|---|
| A ± SD | Ho ± SD | He ± SD | n | np | h ± SD | π | |
| Northwest | 14.27 ± 4.85 | 0.70 ± 0.096 | 0.85 ± 0.05 | 17 | 15 | 0.87 ± 0.01 | 0.0079 ± 0.0044 |
| HU | 5.83 ± 1.95 | 0.72 ± 0.79 | 0.69 ± 0.13 | 5 | 5 | 0.64 ± 0.15 | 0.0100 ± 0.0060 |
| PU | 8.17 ± 2.66 | 0.76 ± 0.13 | 0.79 ± 0.09 | 5 | 4 | 0.76 ± 0.06 | 0.0074 ± 0.0044 |
| PA | 8.42 ± 2.40 | 0.76 ± 0.16 | 0.84 ± 0.06 | 3 | 0 | 0.54 ± 0.07 | 0.0087 ± 0.0052 |
| LC | 9.33 ± 2.89 | 0.60 ± 0.14 | 0.80 ± 0.08 | 5 | 1 | 0.67 ± 0.05 | 0.0053 ± 0.0032 |
| IL | 7.33 ± 3.08 | 0.68 ± 0.23 | 0.75 ± 0.11 | 6 | 3 | 0.66 ± 0.06 | 0.0018 ± 0.0014 |
| Southeast | 15.53 ± 5.50 | 0.70 ± 0.081 | 0.79 ± 0.09 | 20 | 18 | 0.33 ± 0.04 | 0.0008 ± 0.0008 |
| KA | 6.58 ± 2.35 | 0.58 ± 0.17 | 0.72 ± 0.13 | 2 | 0 | 0.00 ± 0.00 | 0.0000 ± 0.0000 |
| LP | 5.92 ± 2.97 | 0.64 ± 0.25 | 0.67 ± 0.18 | 5 | 2 | 0.53 ± 0.18 | 0.0024 ± 0.0018 |
| LB | 7.83 ± 3.51 | 0.89 ± 0.10 | 0.76 ± 0.11 | 4 | 1 | 0.18 ± 0.10 | 0.0003 ± 0.0005 |
| TE | 7.33 ± 3.28 | 0.73 ± 0.20 | 0.75 ± 0.13 | 5 | 2 | 0.29 ± 0.15 | 0.0009 ± 0.0009 |
| PV | 6.33 ± 3.31 | 0.76 ± 0.25 | 0.70 ± 0.14 | 4 | 2 | 0.40 ± 0.11 | 0.0008 ± 0.0008 |
| CA | 6.58 ± 2.42 | 0.64 ± 0.26 | 0.73 ± 0.12 | 4 | 2 | 0.15 ± 0.12 | 0.0003 ± 0.0005 |
| VC | 6.33 ± 2.49 | 0.63 ± 0.16 | 0.71 ± 0.10 | 10 | 6 | 0.64 ± 0.11 | 0.0015 ± 0.0013 |
| BP | 7.58 ± 3.29 | 0.80 ± 0.16 | 0.75 ± 0.09 | 4 | 2 | 0.28 ± 0.12 | 0.0006 ± 0.0007 |
| ML | 7.16 ± 2.91 | 0.69 ± 0.14 | 0.74 ± 0.11 | 6 | 2 | 0.41 ± 0.11 | 0.0008 ± 0.0009 |
| TP | 6.75 ± 3.27 | 0.62 ± 0.18 | 0.70 ± 0.12 | 5 | 2 | 0.42 ± 0.12 | 0.0018 ± 0.0014 |
| PK | 6.75 ± 2.76 | 0.69 ± 0.13 | 0.71 ± 0.10 | 6 | 1 | 0.35 ± 0.12 | 0.0007 ± 0.0008 |
| TF | 4.41 ± 1.97 | 0.57 ± 0.20 | 0.56 ± 0.18 | 4 | 2 | 0.18 ± 0.10 | 0.0003 ± 0.0005 |
A. mean number of alleles per locus; He, mean expected heterozygosity; Ho. mean observed heterozygosity; n. number of haplotypes; np. number of private haplotypes; h. haplotype diversity; π. nucleotide diversity; p. number of polymorphic sites. Standard error values are in parentheses.
Table 5.
Distribution of the 35 control region haplotypes observed in 307 guanacos from 17 localities. The vertical numbers indicate the position of polymorphic sites relative to haplotype 1. Localities are divided in Northern and Southern region based on the results of the AMOVA analysis.
| Haplotypes | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 4 | 5 | Region | Localities |
N | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 2 | 2 | 3 | 3 | 3 | 3 | 3 | 3 | 4 | 4 | 6 | 6 | 7 | 7 | 8 | 8 | 9 | 2 | 7 | 8 | 9 | 9 | 1 | 1 | 1 | 3 | 4 | 5 | 7 | 8 | 8 | 2 | 5 | 3 | 0 | |||||||||||||||||||||
| 1 | 5 | 2 | 5 | 6 | 0 | 4 | 5 | 6 | 8 | 9 | 0 | 4 | 5 | 9 | 0 | 7 | 2 | 5 | 1 | 6 | 5 | 1 | 1 | 6 | 2 | 8 | 9 | 5 | 3 | 9 | 1 | 4 | 7 | 1 | 6 | 1 | 8 | HU | PU | PA | LC | KA | IL | LP | LB | TE | PV | CA | VC | BP | ML | TP | PK | TF | |||
| 1 | A | G | – | G | T | T | T | A | A | G | T | T | G | T | A | C | A | C | A | C | A | T | T | C | C | C | A | T | C | A | G | T | C | C | C | C | A | G | NORTHWESTRN | 1 | 1 | ||||||||||||||||
| 2 | . | . | – | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | 2 | 2 | |||||||||||||||||
| 3 | . | . | – | . | C | C | C | . | . | . | C | C | . | . | G | . | . | . | . | T | . | . | . | . | T | . | . | . | . | . | . | . | T | . | . | . | . | . | 5 | 5 | |||||||||||||||||
| 4 | . | . | – | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | 1 | 1 | |||||||||||||||||
| 5 | . | . | – | . | C | C | C | . | . | . | C | C | . | . | G | . | . | . | . | T | . | . | . | . | T | . | . | . | . | . | . | . | T | . | . | . | . | C | 1 | 1 | |||||||||||||||||
| 6 | . | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | T | . | . | . | . | T | T | G | . | . | . | . | . | . | . | . | . | . | . | 1 | 1 | |||||||||||||||||
| 7 | . | . | – | . | . | . | C | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | 4 | 4 | |||||||||||||||||
| 8 | . | . | – | . | . | . | C | . | . | A | . | . | . | . | . | . | . | . | . | . | . | C | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | 5 | 5 | |||||||||||||||||
| 9 | . | . | – | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | C | . | . | A | . | . | . | . | . | . | . | 8 | 5 | 13 | ||||||||||||||||
| 10 | . | . | T | . | . | . | C | G | G | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | 6 | 6 | 14 | |||||||||||||||
| 11 | . | . | – | A | . | . | C | . | . | . | C | . | T | . | . | . | . | . | . | T | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | SOUTHEASTRN | 1 | 1 | ||||||||||||||||
| 12 | . | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | T | . | . | . | . | T | . | G | . | . | . | . | . | . | . | . | . | . | . | 8 | 8 | |||||||||||||||||
| 13 | . | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | T | . | . | . | . | T | T | . | . | . | . | . | . | . | . | . | . | G | . | 1 | 1 | |||||||||||||||||
| 14 | . | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | T | . | . | . | . | T | . | . | . | . | . | . | . | T | . | . | . | . | . | 1 | 1 | |||||||||||||||||
| 15 | . | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | T | . | . | . | . | T | T | . | . | . | . | . | . | . | . | . | . | . | . | 9 | 9 | |||||||||||||||||
| 16 | . | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | T | . | T | . | . | . | . | T | . | . | . | T | . | . | . | . | . | . | . | . | . | 1 | 1 | 2 | ||||||||||||||||
| 17 | . | . | – | . | . | . | C | . | . | . | C | . | . | C | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | T | . | . | . | . | . | . | . | 1 | 1 | |||||||||||||||||
| 18 | . | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | T | . | . | . | . | T | . | . | C | . | . | . | . | . | . | . | . | . | . | 2 | 2 | |||||||||||||||||
| 19 | . | . | – | . | . | . | C | . | . | . | C | . | A | . | . | . | . | . | . | T | . | . | . | . | T | . | . | . | T | . | . | . | . | . | . | . | . | . | 1 | 1 | |||||||||||||||||
| 20 | . | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | G | T | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 1 | |||||||||||||||||
| 21 | . | . | – | . | . | . | C | . | . | . | C | . | . | . | . | T | . | . | . | T | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | 4 | 4 | |||||||||||||||||
| 22 | . | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | T | . | . | C | . | T | . | . | . | T | . | . | . | . | . | . | . | . | . | 1 | 1 | |||||||||||||||||
| 23 | C | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | T | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 1 | |||||||||||||||||
| 24 | T | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | T | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 1 | |||||||||||||||||
| 25 | . | A | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | T | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | 2 | |||||||||||||||||
| 26 | . | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | T | . | C | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 1 | |||||||||||||||||
| 27 | . | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | T | . | . | . | . | T | . | . | . | . | . | . | . | . | G | . | . | . | . | 1 | 1 | |||||||||||||||||
| 28 | . | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | G | . | . | T | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 1 | 2 | ||||||||||||||||
| 29 | . | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | T | . | . | . | . | T | . | . | . | . | G | . | . | . | . | . | . | . | . | 3 | 3 | |||||||||||||||||
| 30 | . | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | T | . | . | . | T | T | . | . | . | . | . | . | . | . | . | T | . | . | . | 2 | 2 | |||||||||||||||||
| 31 | . | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | T | G | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 1 | 1 | 4 | 3 | 10 | |||||||||||||
| 32 | . | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | T | . | . | . | T | T | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 1 | |||||||||||||||||
| 33 | . | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | T | . | . | . | . | T | . | . | . | . | . | . | C | . | . | . | . | . | . | 1 | 2 | 2 | 5 | |||||||||||||||
| 34 | . | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | T | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 1 | |||||||||||||||||
| 35 | . | . | – | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | T | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | 13 | 20 | 8 | 7 | 18 | 11 | 16 | 12 | 12 | 17 | 15 | 15 | 16 | 17 | 197 | ||||
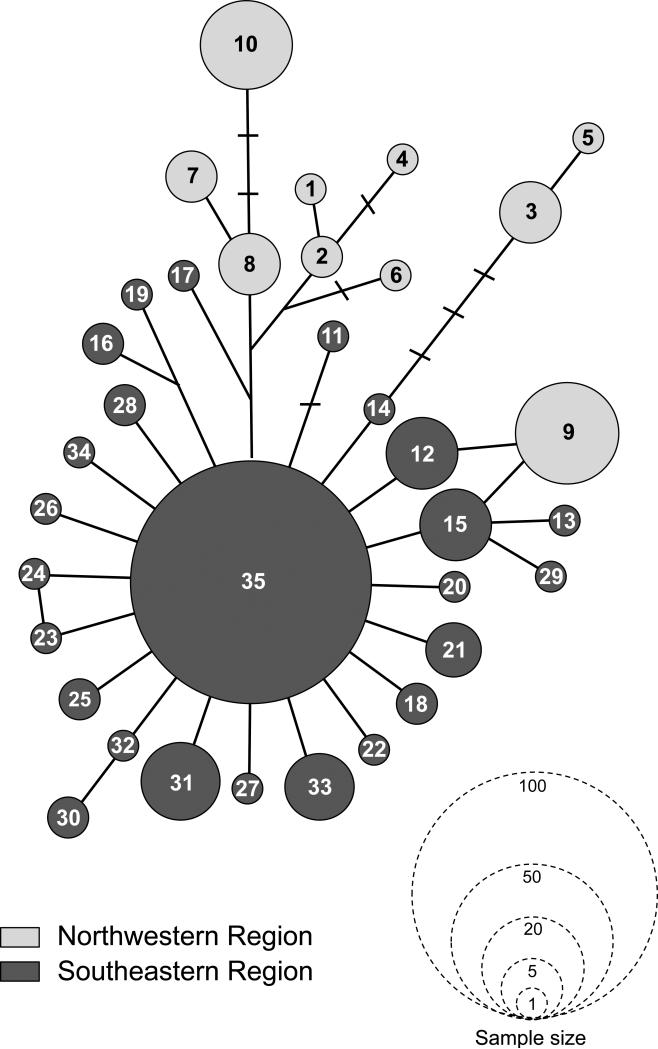
The 35 Control Region sequences are connected through a network with a maximum of 12 mutational steps (Fig. 4). Although the network did not completely delineate a genetic partition corresponding with a geographic separation between the northwest and southeast, there is a clear trend that the most-divergent haplotypes are found in the northwest. A single dominant haplotype (H35) was observed exclusively in the mitochondrial southern cluster, from which related sequences were separated by one or two mutational steps. Up to six mutational steps separated the haplotypes of Northwest population from Southeast population. Haplotype 35 was shared among localities throughout the southern group, including LC and IL, two locations within the contact zone (Fig. 1A).
Fig. 4.
Minimum spanning network representing the relationships among 35 control region haplotypes (see Table 5). Circle sizes correspond to haplotype frequencies.
Genetic units
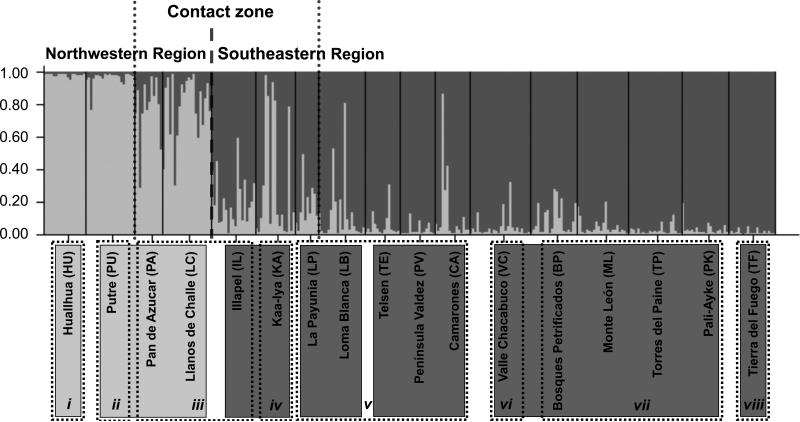
Results of initial STRUCTURE analysis with all samples indicated strongest support for K = 2 (Northwest vs. Southeast localities; Fig. 3), based on ΔK and the assignment plots, though the L(K) values increased asymptotically with higher K (Fig. 2). Subsequent analysis of only the Northwest group provided evidence of additional substructure (K = 2) between Hyper-Arid Peru (HU) and Chilean Pre-altiplano and arid Chile (PU+PA+LC). The Southeast group also showed evidence of substructure (K = 5), delimiting the Bolivian Chaco and Chilean Peri-Puna (KA+IL), Northern Patagonia (LP+LB), Central Patagonia, (TE+PV+CA), Southern and western Patagonia (VC+BP+ML+TP+PK), and the Fuegian zone (TF) (data not shown).
Fig. 3.
Plot of posterior probability of assignment for 319 guanacos (vertical lines) to two genetic clusters based on Bayesian analysis of variation at 14 microsatellite loci. Individuals are grouped by locality, and localities are indicated along the horizontal axis. Light grey = Genetic Cluster 1: Northwest group; dark grey = Genetic Cluster 2: Southeast group. The tables contain the locations, in light grey and dark grey, show the 7 clusters obtained from STRUCTURE analyses (citation). The numbers in the tables indicates BAPS grouping for comparisons; where i: Hyper-Arid Peru, ii: Chilean Pre-altiplano; iii: Arid Chile and Chilean Peri-Puna, iv: Bolivian Chaco, v: Northern and central Patagonia, vi: Western Patagonia, vii: Southern Patagonia, and viii: Fuegian zone.
All runs at K = 2 produced identical clustering solutions. Of 314 individuals, 271 had a clear predominant heritage (i.e. Q > 75%). Of the 91 individuals sampled in the Northwest region, 51 had a consistent genetic heritage, 26 were of mixed southeastern and northwestern origin and 14 had clear Southeastern genetic heritages. The 14 of Southeastern origin were all sampled in the northwestern localities of PA and LC (Fig. 3) and also LC had the dominant southwestern mtDNA haplotype (Haplotype 35, Fig. 4). Of the 223 individuals sampled in the Southeast region, 206 had consistent genetic heritages and 15 were of mixed origin, and 2 were assigned to the Northwest cluster. Overall, most of the individuals of mixed or incongruent heritage were sampled in localities that were in relatively close geographical proximity to the putative boundary between the Northwest and Southeast (Fig. 3, Table 3).
Table 3.
Percentage of individuals assigned to the Northwest or Southeast genetic clusters with nuclear (Q > 75%, STRUCTURE analysis). The number of individuals from each locality is in parentheses. Localities containing animals from of both clusters, considered hybrids localities (Q > 25%), are shaded and in bold (localities are defined in Table 1 and Fig 1A).
| Localities | Microsatellites |
||
|---|---|---|---|
| Northwestern | Southeastern | Hybrids | |
| HU | 100.0 (18) | 0.0 (0) | 0.0 (0) |
| PU | 95.2 (20) | 0.0 (0) | 4.8 (1) |
| PA | 33.3 (4) | 8.3 (1) | 58.4 (7) |
| LC | 42.8 (9) | 4.8 (1) | 52.4 (11) |
| IL | 0.0 (0) | 63.2 (12) | 36.8 (7) |
| KA | 11.8 (2) | 64.7 (11) | 23.5 (4) |
| LP | 0.0 (0) | 60.0 (6) | 40.0 (4) |
| LB | 0.0 (0) | 90.0 (18) | 10.0 (2) |
| TE | 0.0 (0) | 93.3 (14) | 6.7 (1) |
| PV | 0.0 (0) | 100.0 (15) | 0.0 (0) |
| CA | 0.0 (0) | 86.7 (13) | 13.3 (2) |
| VC | 0.0 (0) | 96.2 (25) | 3.8 (1) |
| BP | 0.0 (0) | 95.0 (19) | 5.0 (1) |
| ML | 0.0 (0) | 100.0 (22) | 0.0 (0) |
| TP | 0.0 (0) | 100.0 (23) | 0.0 (0) |
| PK | 0.0 (0) | 100.0 (20) | 0.0 (0) |
| TF | 0.0 (0) | 100.0 (20) | 0.0 (0) |
Results of the BAPS analyses were concordant with the hierarchical STRUCTURE analysis. The optimal solution was obtained with K = 8 using all samples (L(K) = -18299.36) and consisted of (blue line in Figure 1A and dashed black line in Figure 3): i) Hyper-Arid Peru: HU; ii) Chilean Pre-altiplano: PU; iii) Arid Chile and Chilean Peri-Puna: PA, LC and IL; iv) Bolivian Chaco: KA; v) Northern and central Patagonia: LP, LB, TE, PV and CA; vi) Western Patagonia: VC; vii) Southern Patagonia: BP, ML, TP and PK; and viii) Fuegian zone: TF. The BAPS analysis also infers a network based on relative gene flow among clusters as indicated by weighted arrows reflecting the relative proportion of ancestry from the source cluster found among the individuals assigned to the target cluster. For example, our results suggest that the southern Patagonia population has had historic gene flow from 5 of the 7 other clusters, and that southern Patagonia is the main source of gene flow to northern and central Patagonia (Fig. 1B).
Most of the groups were significantly differentiated from one another based on pair-wise FST and D values using microsatellites data and concordant results were achieved whether BAPS vs. STRUCTURE or eight vs. two populations were considered (Table 4). Pairwise-population differentiation estimated as FST values were modest (FST ≈ 0.06 - 0.424), with the Hyper-Arid Peru and Fuegian Zone (Tierra del Fuego) clusters being most distinct from the other populations (FST = 0.424). However, pairwise population differentiation estimated as D (Jost 2008) was more pronounced (D ≈ 0.142 - 0.738) and the Hyper-Arid Peru and Bolivian Chaco clusters were most differentiated from the other populations (D = 0.738).
Table 4.
Pairwise population differentiation based on allelic frequencies of 14 polymorphic microsatellites estimated as Jost D values (below the diagonal) and Slatkin linearized FST values (above the diagonal) for each of the eight (top) and two (bottom) values used in microsatellite population scenarios. All comparisons were statistically significant (P < 0.005) based on 10,000 permutations.
| Region (K=2) and Population (K=8) | N | Northwestern |
Southeastern |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Hiper-Arid Peru | Chilean Pre-altiplano | Arid Chile and Chilean Peri-Puna | Bolivian Chaco | Northern and central Patagonia | Western Patagonia | Southern Patagonia | Fuegian zone | ||
| Hyper-Arid Peru | 18 | - | 0.184 | 0.199 | 0.295 | 0.208 | 0.265 | 0.249 | 0.424 |
| Chilean Pre-altiplano | 21 | 0.552 | - | 0.072 | 0.123 | 0.111 | 0.165 | 0.115 | 0.278 |
| Arid Chile and Chilean Peri-Puna | 52 | 0.662 | 0.322 | - | 0.060 | 0.058 | 0.086 | 0.051 | 0.191 |
| Bolivian Chaco | 17 | 0.739 | 0.395 | 0.232 | - | 0.087 | 0.119 | 0.080 | 0.265 |
| Northern and central Patagonia | 75 | 0.599 | 0.413 | 0.249 | 0.286 | - | 0.081 | 0.061 | 0.143 |
| Western Patagonia | 26 | 0.626 | 0.505 | 0.283 | 0.317 | 0.245 | - | 0.053 | 0.242 |
| Southern Patagonia | 85 | 0.682 | 0.408 | 0.196 | 0.264 | 0.221 | 0.143 | - | 0.145 |
| Fuegian zone | 20 | 0.724 | 0.610 | 0.492 | 0.500 | 0.333 | 0.457 | 0.339 | - |
| Northwest | 91 | - | 0.042 | ||||||
| Southeast | 223 | 0.200 | - | ||||||
Similarly, the mitochondrial DNA data showed a hierarchical AMOVA pattern with significant structure (ΦCT = 0.404, P = 0.00136) when the sample sites were divided into the two groups: (i) HU, PU, PA and (ii) LC, IL and all sites of the Southeastern Region. This result supports the differentiation between the Northwest and Southeast, but further reduces the number of locations assigned to the Northwest Region.
Historical demography and Migration
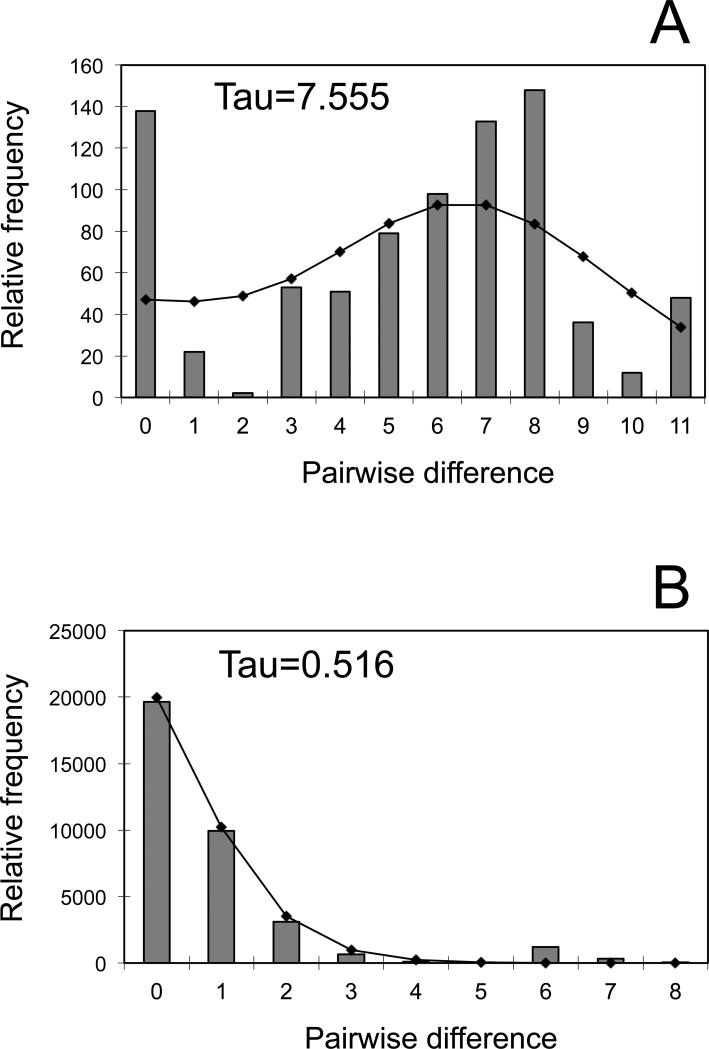
Tajima's D and Fu's F values were negative and statistically significant for the Southeast region (Fs = -3.032, P<0.02; D = -1.832, P<0.05) but not significant for the Northwest region (Fs = -0.628, P>0.10; D = -0.242, P>0.10), hence, we inferred a scenario of expansion for the Southeast region and non-expansion for the Northwest region. Furthermore, Fu's test, which is considered a powerful test to detect past population expansion (Fu 1997; Ramos-Onsins & Rozas 2002), was negative and significant for the Southeast region, indicating an excess of lower frequency haplotypes compared with predicted values under the Wright-Fisher model. Mismatch distributions were consistent with a model of a rapidly expanding population for the Southeast region (τ = 0.516, θ = 0.255) (Fig. 5B), but not the Northwest region (τ = 7.555, θ = 0.004) (Fig. 5A). Based on the expansion parameter (τ), sudden growth of the South-eastern population would have occurred around 11,583 years ago (tτ Low bound= 0 and tτ Up bound= 50,508).
Fig. 5.
Mismatch distribution of pairwise nucleotide differences among control region sequences of northwestern guanacos (A), and southeastern guanacos (B). Gray histograms represent the observed differences and the thin lines represent the ideal distribution as predicted by the model.
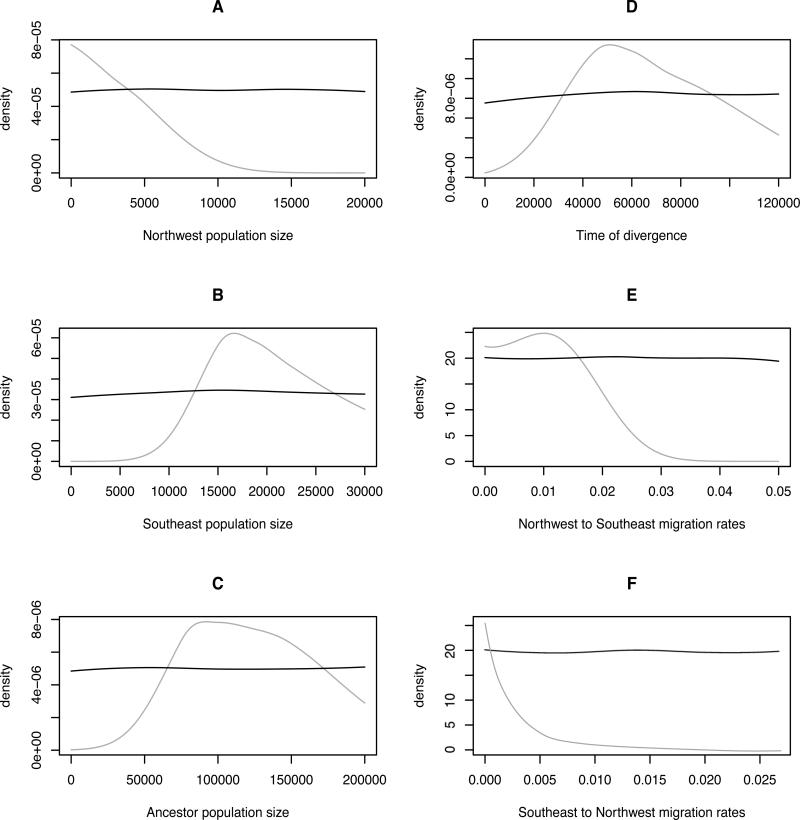
Although STRUCTURE is very efficient at assigning individuals to genetic clusters, it fails to detect historical gene flow events (Pritchard et al. 2000). Therefore, the programs LAMARC and popABC were used to investigate relative patterns of population size and gene flow between the Northwest and Southeast regions. Rates of gene flow and effective population size were jointly estimated with popABC using both mitochondrial and nuclear markers. Estimated posterior distributions for all parameters are shown in Fig 6. Effective population size (Ne) of the Southeast population was estimated to be around 17,000 individuals (Fig. 6B), although the estimated posterior distribution was substantially different between the Southeast and Northwest (Fig. 6A). Prior to the divergence of the two populations around 50,000 years ago (Fig. 6D), the models predict an estimated population of between 8,000 and 12,000 individuals (Fig. 6C). The effective migration rate between the Northwest and Southeast populations was low, but slightly higher from north to south (Fig. 6E and 6F).
Fig 6.
(A) Plot of prior (black) and posterior (gray) distributions of Northwest effective population sizes, (B) Southeast and (C) ancestral of Lama guanicoe. (D) Prior (black) and posterior (gray) distributions of divergence time (in years) between Northwest and Southeast clusters. (E) Northwest to Southeast migration rates and (F) Southeast to Northwest migration rates.
Results from multiple Maximum Likelihood LAMARC runs were mostly consistent with popABC results, with largely concordant confidence intervals. Diversity in the Northwest (Theta=0.3355) was higher than in the Southeast region (Theta=0.1534). Migration rates, expressed as the number of migrants per generation, were higher from the Northwest to the Southeast (M12=8.5279) compared with from the Southeast to the Northwest (M21=5.3502).
Discussion
Genetic structure
Microsatellite and mtDNA sequence variation revealed two relatively distinct genetic groups of guanaco that were segregated geographically by the Central Andes Plateau. In the North-western region guanacos range over arid ecosystems such as desert and xeric shrub lands that include the Sechura and Atacama deserts between Peru and northern Chile. Southern guanacos inhabit a more diverse array of ecosystems distributed from Bolivia southward, including tropical and subtropical dry broadleaf forests represented by Chaco savannas, temperate grasslands, savannas and shrub lands that include Argentinean Monte, Patagonian steppe, Patagonian grasslands, parts of the Southern Andean steppe in the montane grasslands and shrub lands ecosystem, and portions of temperate broadleaf and mixed forest represented by the sub-polar Nothofagus forest (Dinerstein et al.1995; González et al.2006). The Central Andes Plateau formed by the “altiplano and puna” appears to have been the most important geographic barrier, significantly reducing gene flow between guanaco populations on either side of this portion of the Andes, which is 1800 km long and 300-400 km wide. Paleontological evidence indicates that guanacos occupied the regions surrounding the altiplano since at least 0.78 million years ago (MYA) (see review in Cajal et al. 2010) and their distribution has certainly been influenced by repeated periods of glacial build-up and melting even as recently as the Last Glacial Maximum near 25,000 years ago (MacQuarrie et al. 2005, Hoffstetter 1986, Rabassa et al 2011). During glacial maxima glaciers were found as low as 3,800 meters above sea level (masl), over 1000 m below the current glacial line. Further, during inter-glacial periods large lakes (Hoffstetter 1986) may have also been physical barriers to guanaco displacement.
The climatic conditions and vegetative cover in the Andean plateau during the Holocene likely contributed greatly to this hypothesized longitudinal ecological barrier limiting guanaco dispersal. A warmer and drier climate was established around 7,000-8,000 years ago with a brief interlude of increased humidity around 4,000-5,000 years ago (Baied & Wheeler 1993), which would have favoured microthermic vegetation adapted to arid conditions and high solar radiation, by developing hard silica cuticles and other water and energy saving strategies (Menegaz et al. 1989; Adler et al. 2004). This vegetation would not have been very palatable for the guanaco, in spite of its generalist foraging strategy. In contrast, the vicuña (Vicugna vicugna) was better adapted to these extreme environments with its smaller body-size, lower energy requirements, hair that was better adapted to low temperatures and intense solar radiation, and continuous tooth growth better adapted to abrasive vegetation (Franklin 1983; Lucherini 1996; Arazmendia et al. 2006; Weinstock et al. 2009; Borgnia et al. 2010; Mosca & Piug 2010). Currently there is a relatively narrow portion of the Altiplano and Puna where guanacos and vicuñas are both found, but where they segregate spatially in different habitats. This pattern suggests that direct competition for food may occur periodically (Lucherini 1996; Lucherini & Birocho 1997; Cajal et al. 2010) and that competition for limited resources with vicuña in the Altiplano and the Puna may have also helped limit movement of guanacos across this region.
Although the altiplano is not a biogeographical barrier for all species, the environmental conditions found in the area have helped isolate and differentiate some species, such as the Sigmodont rodents. For example, Phyllotis xantophygus chilensis lives in the Puna and Phyllotis limatus inhabits lowlands/pre-Puna area, where climate and the composition of vegetation differ because of the altitudinal effect (Steppan 1998; Palma et al. 2005). In contrast, Abrothryx olivaceus is characterized by having one of the largest ranges in Chile, encompassing most of the north, the central valley and the southern tip of the country, whereas A. andinus is restricted to high elevations in the pre-Puna and Altiplano regions of the Andes (Palma et al. 2005). Although these small-sized mammals have smaller home-ranges and generational time than guanacos, they are indicative of the strength of selection and isolation pressures on this region's biota.
Although geographically fairly well defined by several analyses, the two groups of guanaco are clearly not completely isolated as there were several examples of populations with individuals of both north-western and south-eastern heritage, as well as individuals of mixed genetic heritage. However, our results suggest that gene flow between groups is asymmetric and is mostly from the Northwest to Southeast, as shown by the LAMARC and popABC analyses. Prevalent migration from Northwest to Southeast agrees with the expansion history of guanaco. However, the Northwest population has more unique alleles than the Southeast population, allowing for easier detection of migrants using assignment tests and this difference in level of genetic diversity has potentially resulted in an underestimate of the migration rate into the Northwest. Nevertheless, migration rates in both directions are low and the previous conclusion that these populations should be considered demographically independent is supported by the migration rates estimated from our popABC analysis. A good example was the population from Illapel (IL), which is located in the Chilean mountains at the south of the altiplano-puna zone. Throughout this contact zone, evidence of admixture was prevalent, but dissipated in populations further north and south based on microsatellite assignation. For example, in the populations of Pan de Azucar (PA), Llanos de Challe (LC), Illapel (IL) and La Payunia (LP) just south of the Atacama Desert and the Altiplano, 36 to 58% of the individuals were of mixed origin. Levels of admixture decreased in localities farther away, as was observed in Kaa-Iya (KA). This contact region is consistent with the absence of complete reciprocal monophyly between L. g. cacsilensis and L. g. guanicoe using mtDNA control region and CytB haplotypes in a smaller sampling of guanacos (González et al. 2006; Marín et al. 2007; Marín et al. 2008). Several mutually nonexclusive factors may have contributed to the fairly large numbers of individuals of northwest genetic heritage in the south-eastern populations of La Payunia (LP), Loma Blanca (LB) and Camarones (CA). This includes closer geographic and ecological affinity among these localities than is evident today because of the extirpation of intermediate populations, long range dispersal patterns, or significant anthropic translocation by Amerindians or early Europeans of individuals from the Andes to the coast (Politis et al. 2011).
There was significant structure of microsatellite variation at the regional level, suggesting only moderate levels of genetic connectivity. Although slightly fewer populations were recognized when the spatial component was included in genetic variation (BAPS) relative to the non-spatial Bayesian analysis (STRUCTURE), the results were largely comparable. In the north-western cluster there were 2-3 populations, one encompassing Huallhua (HU) (Peru) and possibly unsampled areas further to the north and one to two populations between Putre (PU), and Llanos de Challe (LC) and Illapel (IL) in Chile. Among south-eastern guanacos there is a less-well-defined gradient of related populations roughly consisting of: those from the central Andean corridor from the Bolivan Chaco and peri-puna in Chile to the northern and/or central Patagonia, southern and/or western Patagonia, and the Fuegian zone. This pattern corresponds with the phylogeographic units observed in Patagonian sigmodontine rodents (Lessa et al. 2010) and likely reflect incipient regional barriers to gene flow as well as behavioural phylopatry exhibited by guanacos (Ortega & Franklin 1995; Young & Franklin 2004). This is consistent with other molecular studies done at a regional spatial scale where populations from the Argentine provinces of Chubut and northern Santa Cruz southward to southern Patagonia had little genetic differentiation and low genetic structure (Maté et al. 2005) while remote isolated populations separated by geographical barriers such as Torres del Paine National Park and Tierra del Fuego had increased genetic uniqueness (Sarno et al. 2001). Broad geographic differences do not correlate well with guanaco population structure, suggesting that more studies are needed at the landscape scale level to better understand the processes influencing guanaco population dynamics.
Locally, patterns of microsatellite genetic variation suggest that the relatively low levels of gene flow among populations has occurred mostly from north to south, and that guanacos likely occupied several glacial refuges during the recent ice age (Fig. 1B). The southern Patagonian population shows evidence of gene flow from most of the other areas, including the distant populations of Bolivia and Peru, suggesting that gene flow and interconnectivity among guanacos during the Holocene was very high. However, gene flow among populations is uneven, and generally in only one direction. Western Patagonia and the Fuegian populations also contributed to the genetic variation in Southern Patagonia, suggesting that these regions may have been reservoirs of genetic variation and a manifestation of ice age refuges. The apparent gene flow from Southern Patagonia towards north and central Patagonia probably reflects more-recent animal movements and gene flow, perhaps associated with habitat degradation, persecution and an associated “source/sink” phenomenon. Further north, there is less evidence of connectivity between geographically proximate populations, such as between Hyper-Arid Peru and Chilean pre-altiplano, and between these populations and the Bolivian Chaco. Populations in this region have probably been stable for longer periods of time (Marín et al. 2008) due to more enduring geographic barriers to gene flow caused by the Andean altiplano.
Genetic interchange among populations is also evident from the significant number of individuals with discordant mtDNA and microsatellites patterns and/or incongruent (<75%) population assignment with STRUCTURE and/or BAPS. This asymmetric pattern may reflect sexual differences in dispersal through generations. Although males and females in family groups are territorial during the reproductive period and show phylopatry since they use the same territory for several years (Bank et al. 2003; Young & Franklin 2004), adults males coming from non-territorial male groups are more likely to disperse longer distances looking for new territories (Franklin 1983; Young & Franklin 2004). These differences in dispersal between sexes may be facilitated at the population-level by long-range movements of groups looking for forage and better weather conditions in the autumn-winter. Although most guanaco populations are largely sedentary with small annual home ranges (Franklin 1982; Marino & Baldi 2008), facultative migration has also been documented in the mountains of central Chile/Argentina and parts of Patagonia (Ortega & Franklin 1995; González et al. 2008; Moraga pers. comm.). These patterns would help contact among neighbouring populations and facilitate genetic interchange.
Population expansion
The mismatch distribution of Southeast guanacos was close to the theoretically expected unimodal distributions for population expansion, while Tajima's D and Fu's F values were significantly negative. The combination of these two observations suggests a recent expansion ≈11,500 years ago, at the start of the Holocene deglaciation process in southern South America (McCulloch et al 2000). The uniformity of mitochondrial haplotypes across a large geographic range, with only one common haplotype that are shared in guanacos from the Bolivian Chaco to the Fuegian zone, provided further support for a past demographic expansion. This pattern is expected when there are high levels of gene flow or small effective size in populations that have not been affected by long-term biogeographic barriers (Avise 2000) and is common in populations where numbers increased rapidly as they expanded their ranges. This pattern would result in genealogical trees with shallow branches, many of which are dispersed widely across the species current ranges, as we observed with southern guanacos. It is also possible that the shallow divergence levels reflect the historical mixing of populations resulting from climatic and landscape changes that affected Patagonia throughout the Quaternary.
During the late Quaternary it is likely that the southward expansion of guanacos on the eastern side of the Andean plateau in Peru and northern Chile (Scherer 2009) was influenced and limited by the Atacama Desert, which has persisted, except for brief interludes, for over 200 million years (Clarke 2006; Dunai et al. 2005; Hartley & Chong, 2002). The severe climate of the Atacama continues to impede guanaco movements in the region, limiting dispersion southwards to more humid habitats. Similarly, the Atacama desert separates the two vicuña (Vicugna vicugna) subspecies, V. v. vicugna to the arid area of the altiplano and puna under the Dry Diagonal, and V. v. mensalis to the more-humid altiplano at the north (Marín et al. 2007). Other mammalian genera have similar distributional patterns, with sister species occupying different sides of this xeric diagonal. These include north-south disjunct distributions between Abrocoma cinerea and Abrocoma bennetti (Abrocomidae), Chinchilla brevicaudata and Chinchilla lanigera (Chinchillidae)(Hershkovitz 1972), and Phyllotis magister and Phyllotis darwini (Cricetidae)(Steppan 1998). Although these small- and medium-sized mammals have smaller home-ranges than guanacos, they are indicative of the strength of different selection and isolation pressures that influenced this regions biota. The small and restricted North-western guanaco cluster of mixed genetic heritage, which is seemingly confined to the Sechura and Atacama Deserts, may have been established and reconnected sporadically with populations to the south and east during relatively brief periods of increased moisture, such as those that occurred 16.2-10.5 MYA and between 8-3 MYA when the amount of vegetation east of the desert increased (Betancourt et al. 2000).
The western slopes of the Andes, perhaps to the northwest of their current distributions, likely served as refugia for guanacos during the Pleistocene glacial periods, as evidenced by patterns of genetic diversity, the network of mitochondrial haplotypes and the mismatch distributions. Current populations in Peru and northern Chile have the highest mtDNA and microsatellite diversity and multiple private and derived mtDNA haplotypes, indicating that guanaco populations have existed in these areas for extended time periods and with little suggestion of population expansions or extended contractions. A similar multimodal mismatch distribution pattern was observed in populations described as L. g. cacsilensis (Marín et al. 2008). Glacial or ecological refugia perhaps also existed east of the Andean plateau, but may not have been sampled because past extinctions/extirpations and because current guanaco populations in this region are severely reduced and isolated. Habitat changes from the LGM to the beginning of the Holocene, when open savannah habitats and grasslands were replaced by dense savannah and evergreen Amazon forests (de Vivo & Carmignotto 2004), likely led to the local extinction of guanacos from north-eastern and southern Brazil, Uruguay and the Pampeana region of Argentina towards the end of the Pleistocene (Couto 1983; Guérin & Faure 1999; Scherer 2009). Moreover, in some areas of the Pampas, guanacos inhabited drier and cooler conditions until the late Holocene (between ca. AD 800-1200), when local extinctions occurred following environmental changes produced by the Medieval Thermal Maximum (Politis & Pedrotta, 2006; Politis et al. 2011).
The evolution and expansion of guanacos in southern South American was probably also affected by the changing availability of ecological niches left following the extinction of Megatherium, mylodones, gliptodontes, Hippidion, Lama gracilis, vicuñas, and other herbivores, through the end of the Pleistocene (Markraft 1985; Barnosky & Lindsey 2010; Weinstock et al. 2009). By the end of Quaternary, guanacos had increased their effective population size sharply and had likely colonized much of southern South America by the Holocene. They are theorized to have already colonized and been isolated on the island of Tierra del Fuego by the formation of the Straits of Magellan around that time (Sarno et al. 2001).
Patterns of local extinction and colonization are common in Patagonian terrestrial mammals during the repeated glacial advances and retreats, with some species surviving glacial periods in one or a few southern refugia while others inhabited and subsequently recolonized from northern areas (Sersic et al. 2011, Pardiñas et al. 2011). It is therefore reasonable that some guanacos may have survived in Patagonian refugia, perhaps in Valle Chacabuco (VC), where estimates of mtDNA variation are the highest (Table 2). The Late Holocene was a period of low humidity or dry climate, which was favourable for guanaco populations (Stine & Stine 1990). Similar patterns have been proposed for austral huemul deer (Marín et al. 2012) and several rodent species (Lessa et al. 2010; Pardiñas et al. 2011). However, timing of the demographic expansion in rodent generally exceeds 21000 years, to be older than the LGM (Lessa et al. 2010).
Finally our data also reflect the precipitous reduction in population sizes that occurred as intensive sheep ranching spread throughout the region during the last centuries and guanacos were harvested for their pelts (Dennler de La Tour 1954; Mares & Ojeda 1984), likely resulting in the loss of rare haplotypes. Chile exported around 35,000 pelts during the 20th century (Iriarte & Jaksic 1986), but far fewer than Argentina, which exported around 223,000 units between 1976 and 1979 (Mares & Ojeda 1984).
Taxonomic, conservation and management implications
Given the dynamic history of the guanaco during the last 20,000 years, we suggest that both clusters should be considered to be incipient subspecies, with a broad contact area with individuals with different degrees of genetic heritage from each subspecies. The recognition of two subspecies is supported by the presence of unique derived haplotypes and significant differences in microsatellite structure in each. L. g. cacsilensis would encompass populations from Peru to northern Chile, including guanacos from Llanos de Challe (LC) and Illapel (IL) and L. g. guanicoe would include all areas further south and east. These two groups could also be considered as different Evolutionary Significant Units (ESU) as defined by Moritz (1984). Currently the guanaco is classified as a species of “Least Concern” by the IUCN (Baldi et al. 2008), and the recognition of two different subspecies would facilitate more precise assessments of population threats and management needs for both. This would also be the first step for defining management units and to better understanding whether the observed genetic structure is due in part to climatic, topographic/geographic, or ecological factors (PasbØll et al. 2006).
Overall, our analysis has provided estimates of demographic parameters that fit well with our current understanding of guanaco demographic history and population status. Our results confirm the recent splitting of the Northwest and Southeast populations, the low migration rates between the two populations and the low effective population size of the Northwest guanacos. The results provide new insights that are important to guanaco conservation management. Although the Northwest population appears to have been relatively stable during the last 20,000 years, and second, our estimates of effective population size suggest the census size of this population is possibly larger than is currently believed. These findings reinforce the importance of incorporating better monitoring and management of the Northwest population into the guanaco recovery plan and the maintenance of healthy guanaco populations throughout its distribution.
Conservation and management actions should be implemented at a local scale, taking into consideration the preservation of adaptive variation and evolutionary process throughout the entire range of the species, while correcting or mitigating adverse effects of human activities (Crandall et al. 2000; Frankham et al. 2003). Therefore it is important to: 1) protect populations with large amounts of genetic diversity, 2) promote the connectivity among localities within the same populations detected in the STRUCTURE/BAPS analyses by improving corridors for dispersal, 3) translocate individuals if necessary among localities with animals of similar genetic heritage to limit extreme effects of inbreeding (Franklin and Grigione, 2005), and 4) carefully monitor populations being legally harvested (Marín et al. 2009).
Several guanaco populations need close demographic and genetic monitoring, including 1) Peruvian populations, which are heavily impacted by hunting and habitat deterioration, 2) the small fragmented populations in Bolivia and Paraguay, 3) the isolated and fragmented populations of northern Chile and Argentina, and 4) the large and legally harvested, but genetically less diverse and more isolated population of Tierra del Fuego (Zapata et al. 2009). For naturally isolated populations or for population fragmented by human activities, the introduced guanacos on the Falkland Islands may serve as an example of founder effects and bottlenecks on the genetic and reproductive features of this species (Franklin & Grigione 2005).
Supplementary Material
Acknowledgements
Financial support from the following organizations is gratefully acknowledged: CONICYT, Chile (Beca de Apoyo a Tesis Doctoral); FONDECYT, Chile Grant 101105 and Postdoctoral Grant 3050046; Darwin Initiative for the Survival of Species (UK), The British Embassy (Lima), NERC (UK) grant GST/02/828, Darwin Initiative grant 12/022, European Commission INCO-DC ICA4-2000-10229, MACS. This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH) and the National Cancer Institute (NCI) (US). This work was written by Benito A. González during his PhD studies in the Programa de Doctorado en Ciencias Silvoagropecuarias y Veterinarias of the Universidad de Chile supported by CONICYT scholarship. In Peru we thank to Maria Luisa del Rio (MINAM), Carlos Loret de Mola (CONAM), Gustavo Suarez de Freitas and Antonio Morizaki (INRENA), Wilder Trejo, Daniel Rivera, Daniel Arestegui, Leonidas Rodriguez, Carlos Flores, and Dirky Arias at CONACS, Carlos Ponce (Conservation International). In Chile we thank the Servicio Agrícola y Ganadero, SAG (Permit 447, 2002), the Corporación Nacional Forestal, CONAF (permit 6/02, 2002), for granting other collection permits and help in collecting samples. We especially thanks C. Bonacic (Pontificia Universidad Católica de Chile), O. Skewes (Universidad de Concepción), F. de Smet (Valchac Ltd.), F. Novoa (Centro de Ecología Aplicada Ltda), and Minera Los Pelambres, R. Baldi and V. Burgi (CENPAT, Argentina), E. Cuellar (WCS-Bolivia), A. Duarte (Zoológico de Mendoza, Argentina) for sharing samples; Homero Pezoa, Nicolás Aravena, for support in the laboratory and Pablo Orozco-teWengel (Cardiff University), for support in the software analyses. Special thanks are due to Jane C. Wheeler (CONOPA) for facilitating samples and collaboration, and Michael Bruford (Cardiff University) and Angel Spotorno (Universidad de Chile) for useful information, discussions, advice and support. Samples were transported under CITES authorization numbers 6282, 4222, 6007, 5971, 5177, 5178, 23355, 22967 and 22920.
Footnotes
Data accessibility
DNA sequences: Genbank accessions numbers JX678291 - JX678596. Individual-by-individual Genbank accession numbers, microsatellite data, haplotypes sequences and DNA alignment files: DRYAD entry doi:10.5061/dryad.hm550.
Supporting information
Additional supporting information may be found in the online version of this article.
References
- Adler PB, Milchunas DG, Lauenroth Wk, Sala Oe, Burke IC. Functional traits of graminoids in semi-arid steppes: A test of grazing histories. Journal of Applied Ecology. 2004;41:653–663. [Google Scholar]
- Alpers DL, Van Vuuren BJ, Arctander P, Robinson TJ. Population genetics of the roan antelope (Hippotragus equinus) with suggestions for conservation. Molecular Ecology. 2004;13:1771–1784. doi: 10.1111/j.1365-294X.2004.02204.x. [DOI] [PubMed] [Google Scholar]
- Arzamendia Y, Cassini MH, Vilá VL. Habitat use by vicuña Vicugna vicugna in Laguna Pozuelos Reserve, Jujuy, Argentina. Oryx. 2006;40:1–6. [Google Scholar]
- Avise JC. Phylogeography: The History and Formation of Species. Harvard University Press; Cambridge, Massachusetts: 2000. [Google Scholar]
- Baied CA, Wheeler JC. Evolution of High Andean Puna Ecosystems: Environment, climate, and culture change over the last 12,000 years in the Central Andes. Mountain Research and Development. 1993;13:145–156. [Google Scholar]
- Baldi R, Lichtenstein G, González B, Funes M, Cuéllar E, Villalba L, Hoces D, Puig S. Lama guanicoe. IUCN 2010. IUCN Red List of Threatened Species. Version 2010.1. 2008 www.iucnredlist.org http://www.iucnredlist.org/apps/redlist/details/11186/0.
- Bank MS, Sarno RJ, Franklin WL. Spatial distribution of guanaco mating sites in southern Chile: conservation implications. Biological Conservation. 2003;112:427–434. [Google Scholar]
- Barnosky AD, Lindsey EL. Timing of Quaternary megafaunal extinction in South America in relation to human arrival and climate change. Quaternary International. 2010;217:10–29. [Google Scholar]
- Beaumont MA. Approximate Bayesian Computation in evolution and ecology. Annual Review of Ecology, Evolution, and Systematics. 2010;41:379–406. [Google Scholar]
- Beerli P, Felsenstein J. Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetic. 1999;152:763–773. doi: 10.1093/genetics/152.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt JI, Latorre C, Rech JA, Quade J, Rylander KA. 22.000-Year record of monsoonal precipitation from northern Chile's Atacama Desert. Science. 2000;289:1542–1546. doi: 10.1126/science.289.5484.1542. [DOI] [PubMed] [Google Scholar]
- Borgnia M, Vilá BL, Cassini MC. Foraging ecology of Vicuña, Vicugna vicugna, in dry Puna of Argentina. Small Ruminant Research. 2010;88:44–53. [Google Scholar]
- Borrero LA. Taphonomy of Late Pleistocene faunas at Fuego–Patagonia. Journal of South American Earth Sciences. 2005;20:115–120. [Google Scholar]
- Borrero LA. Extinction of Pleistocene megamammals in South America: The lost evidence. Quaternary International. 2008;185:69–74. [Google Scholar]
- Brown DM, Brenneman RA, Koepfli KP, et al. Extensive population genetic structure in the giraffe. BMC Biology. 2007;5:57. doi: 10.1186/1741-7007-5-57. doi: 10.1186/1741-7007-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajal J, Eduardo P Tonni, Viviana Tartarini. The extinction of some South American Camelids: The case of Lama (Vicugna) Gracilis. Mastozoología Neotropical. 2010;17:129–134. [Google Scholar]
- Cardich AI, Hajduk A. Secuencia Arqueológica y Cronológica Radiocarbónica de la cueva 3 de los Toldos (Santa Cruz. Argentina). Relaciones sociedad Argentina de Antropología. 1973;7:85–123. [Google Scholar]
- Cardich A. Arqueología de los Toldos y el Ceibo (Provincia de Santa Cruz. Argentina). Investigaciones Paleoindias al sur de la línea Ecuatorial. Estudios Atacameños. 1987;8:98–117. [Google Scholar]
- Clarke JDA. Antiquity of Aridity in the Chilean Atacama Desert. Geomorphology. 2006;73:101–114. [Google Scholar]
- Clement M, Posada D, Crandall K. TCS: A computer program to estimate gene genealogies. Molecular Ecology. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Corander J, Sirén J, Arjas E. Spatial modelling of genetic population structure. Computational Statistics. 2008;23:111–129. [Google Scholar]
- Couto CP. Fossil Mammals from the cenozoic of acre. Brasil. vii - Miscellanea. Iheringia Série Geologia Porto Alegre. 1983;8:101–120. [Google Scholar]
- Crandall KA, Bininda-Emonds ORP, Mace GM, Wayne RK. Considering evolutionary processes in conservation biology. Trends an Ecology and Evolution. 2000;15:290–295. doi: 10.1016/s0169-5347(00)01876-0. [DOI] [PubMed] [Google Scholar]
- Dennler de La Tour G. The guanaco. Oryx. 1954;2:273–279. [Google Scholar]
- de Vivo M, Carmignotto P. Holocene vegetation change and the mammal faunas of South America and Africa. Journal of Biogeography. 2004;31:943–957. [Google Scholar]
- Dinerstein E, Olson DM, Graham DJ, Webster AL, Primm SA, Bookbinder MP, Ledec G. A Conservation Assessment of the Terrestrial Ecoregions of Latin America and the Caribbean. The World Bank; Washington, DC: 1995. [Google Scholar]
- Dunai TJ, González López GA, Juez-Larréet J. Oligocene-Miocene age of aridity in the Atacama desert revealed by exposure dating of erosion-sensitive landforms. Geology. 2005;33:321–324. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software Structure: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under linux and windows. Molecular Ecology Resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández PM. Taphonomy and zooarchaeology in the Neotropics: A view fromnorthwestern Patagonian forest and steppe. Quaternary International. 2008;180:63–74. [Google Scholar]
- Flagstad Ø , Syvertsen PO, Stenseth NC, Jakobsen KS. Environmental change and rates of evolution: the phylogeographic pattern within the hartebeest complex as related to climatic variation. Proceedings of the Royal Society of London, B. 2001;268:667–677. doi: 10.1098/rspb.2000.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R, Ballou JD, David A. Briscoe introduction to conservation Genetics. Cambridge University Press; 2003. [Google Scholar]
- Franklin W. Biology Ecology and Relationship to Man of the South American Camelids. In: Mares MA, Genoways HH, editors. Mamalian Biology in South America. Vol. 6. Pymatuning Laboratory of Ecology, University of Pittsburgh; Pittsburgh: 1982. pp. 457–489. [Google Scholar]
- Franklin WL. Contrasting socioecologies of South American's wild camelids: the vicuña and guanaco. In: Eisenberg JF, Kleiman DG, editors. Advances in the study of mammalian behavior. American Society of Mammalogists; 1983. pp. 573–629. Special Publication No. 7. [Google Scholar]
- Franklin WL, Grigione MM. The enigma of guanacos in the Falklands Islands: the legacy of John Hamilton. Journal of Biogeography. 2005;32:661–675. [Google Scholar]
- Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon WL, Sises RS, The Animal Care And Use Committee Of The American Society Of Mammalogists Guidelines of the American Society of Mammalogists for the use of wild mammals in research. Journal of Mammalogy. 2007;88:809–823. doi: 10.1093/jmammal/gyw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González BA, Palma RE, Zapata B, Marín JC. Taxonomic and Biogeographic Status of Guanaco Lama Guanicoe (Artiodactyla. Camelidae). Mammal Review. 2006;36:157–178. [Google Scholar]
- González BA, Novoa F, Saffer K. Desplazamiento altitudinal y rango de hogar de guanacos (Lama guanicoe) mediante seguimiento con collares satelitales. In: Novoa FF, Contreras M, editors. Biodiversidad de fauna en Minera Los Pelambres. Ediciones del Centro de Ecología Aplicada Ltda; Chile: 2008. pp. 57–77.pp. 312 [Google Scholar]
- Goudet J. FSTAT Version 2.9.4.: A Program to Estimate and Test Population Genetics Parameters. 2005 Available from URL: http://www2.unil.ch/popgen/softwares/fstat.htm.
- Guérin C, Faure M. Palaeolama (Hemiauchenia) Niedae Nov.sp. nouveau camelidae du nordeste brasilien. Et sa place parmi les lamini D'Amérique du sud. Geobios. 1999;32:625–659. [Google Scholar]
- Harpending HC. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Human Biology. 1994;66:591–600. [PubMed] [Google Scholar]
- Hartley A, J Chong G. Late Pliocene age for the Atacama Desert: Implications for the desertification of western South America. Geology. 2002;30:43–46. [Google Scholar]
- Hershkovitz P. The recent mammals of the neotropical region: A Zoogeographic and 430 Ecological Review. In: Keast A, Erk FC, Glass B, editors. Evolution, Mammals and the 431 southern continents. New York State University Press; New York: 1972. pp. 311–421. [Google Scholar]
- Hoffstetter R. High Andean mammalian faunas during the Plio-Pleistocene. In: Vulleumier F, Monasterio M, editors. High Altitude Tropical Biogeography. Oxford University Press; Oxford: 1986. pp. 218–245. [Google Scholar]
- Iriarte JA, Jaksic FM. The fur trade in Chile: an overview of seventy-five years of export data (1910–1984). Biological Conservation. 1986;38:243–253. [Google Scholar]
- Jost L. GST and its relatives do not measure differentiation. Molecular Ecology. 2008;17:4015–4026. doi: 10.1111/j.1365-294x.2008.03887.x. [DOI] [PubMed] [Google Scholar]
- Krumbiegel I. Die neuweltlichen Tylopoden. Zoologischer Anzeiger. 1944;145:45–70. [Google Scholar]
- Kuhner MK. lamarc 2.0: maximum likelihood and Bayesian estimation of population parameters. Bioinformatics. 2006;22:768–770. doi: 10.1093/bioinformatics/btk051. [DOI] [PubMed] [Google Scholar]
- Lang KDM, Wang Y, Plante Y. Fifteen polymorphic dinucleotide microsatellites in llamas and alpacas. Animal Genetics. 1996;27:293. doi: 10.1111/j.1365-2052.1996.tb00502.x. [DOI] [PubMed] [Google Scholar]
- Lau CH, Drinkwater RD, Yusoff K, et al. Genetic diversity of Asian water buffalo (Bubalus bubalis): mitochondrial DNA D-loop and cytochrome b sequence variation. Animal Genetics. 1998;29:253–264. doi: 10.1046/j.1365-2052.1998.00309.x. [DOI] [PubMed] [Google Scholar]
- Lessa EP, D'Elía G, Pardiñas UFJ. Genetic footprints of late Quaternary climate change in the diversity of Patagonian-Fueguian rodents. Molecular Ecology. 2010;19:3031–3037. doi: 10.1111/j.1365-294X.2010.04734.x. [DOI] [PubMed] [Google Scholar]
- Lönnberg E. Notes on guanacos. Arkiv für Zoologi. 1913;8:1–8. [Google Scholar]
- Lopes JS, Balding D, Beaumont MA. popABC: a program to infer historical demographic parameters. Bioinformatics. 2009;25:2747–2749. doi: 10.1093/bioinformatics/btp487. [DOI] [PubMed] [Google Scholar]
- Lucherini M. Group size, spatial segregation and activity of wild sympatric vicuñas Vicugna vicugna and guanacos Lama guanicioë. Small Ruminant Research. 1996;20:193–198. [Google Scholar]
- Lucherini M, Birocho DE. Lack of agresión and avoidance between vicuña and guanaco herds grazing in the same Andean habitat. Studies of Neotropical Fauna & Environment. 1997;32:72–75. [Google Scholar]
- MacQuarrie N, Horton BK, Zandt J, Beck S, Decelles PG. Lithospheric Evolution of the Andean fold–thrust belt. Bolivia and the origin of the central Andean Plateau. Ectonophysics. 2005;399:15–37. [Google Scholar]
- Mares MA, Ojeda RA. Faunal commercialization and conservation in South America. BioScience. 1984;34:580–4. [Google Scholar]
- Marin JC, Casey CS, Kadwell M, Yaya K, Hoces D, Olazabal J, Rosadio R, Rodriguez J, Spotorno A, Bruford MW, Wheeler JC. Mitochondrial phylogeography and demographic history of the vicuna: implications for conservation. Heredity. 2007;99:70–80. doi: 10.1038/sj.hdy.6800966. [DOI] [PubMed] [Google Scholar]
- Marín JC, Zapata B, González BA, Bonacic C, Wheeler JC, Casey C, Bruford M, Palma E, Poulin E, Alliende MA, Spotorno AE. Sistemática. Taxonomía y domesticación de alpacas y llamas: Nueva evidencia cromosómica y molecular. Revista Chilena de historia Natural. 2007;80:121–140. [Google Scholar]
- Marín JC, Spotorno AE, González BA, Bonacic C, Wheeler JC, Casey CS, Bruford MW, Palma RE, Poulin E. Mitochondrial DNA variation and systematics of the guanaco (Lama Guanicoe. Artiodactyla: Camelidae). Journal of Mammalogy. 2008;89:269–281. [Google Scholar]
- Marín JC, Corti P, Saucedo C, González BA. application of DNA forensic techniques for identifying poached guanacos (lama guanicoe) in chilean patagonia. Journal of Forensic Sciences. 2009;54:1073–1076. doi: 10.1111/j.1556-4029.2009.01087.x. [DOI] [PubMed] [Google Scholar]
- Marín JC, Varas V, Vila AR, López R, Corti P. Refugia in Patagonian fjords and eastern Andes during the Last Glacial Maximum revealed by huemul (Hippocamelus bisulcus) phylogeographic patterns and genetic diversity. Submitted Journal of Biogeography. 2012 [Google Scholar]
- Marino A, Baldi R. Vigilance Patterns of Territorial Guanacos (Lama guanicoe): The Role of Reproductive Interests and Predation Risk. Ethology. 2008;114:413–423. [Google Scholar]
- Markgraf V. Late Pleistocene faunal extinctions in southern Patagonia. Science. 1985;228:1110–1112. doi: 10.1126/science.228.4703.1110. [DOI] [PubMed] [Google Scholar]
- Massone M, Prieto A. Evaluación de la modalidad cultural Fell 1 en Magallanes. Chungara, Revista de Antropología Chilena. 2004:303–315. Volumen Especial. pp. [Google Scholar]
- Maté ML, Bustamante A, Giovambattista G, De Lamo D, Von Thüngen J, Zambelli A, Vidal-Rioja L. Genetic diversity and differentiation of guanaco populations from Argentina inferred from Microsatellite data. Animal Genetics. 2005;36:316–321. doi: 10.1111/j.1365-2052.2005.01307.x. [DOI] [PubMed] [Google Scholar]
- McCulloch RD, Bentley MJ, Purves RS, et al. Climatic inferences from glacial and palaeoecological evidence at the last glacial termination, southern South America. Journal of Quaternary Science. 2000;15:409–417. [Google Scholar]
- Menegaz A N, Goin F J, Ortiz Jaureguizar E. Análisis Morfológico y Morfométrico multivariado de los representantes fósiles y vivientes del género Lama (Artiodactyla. Camelidae). Sus implicancias sistemáticas. Biogeográficas. Ecológicas y Biocronológicas. Ameghiniana. 1989;26:153–172. [Google Scholar]
- Meirmans PG, Van Tienderen PH. GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Molecular Ecology Notes. 2004;4:792–794. [Google Scholar]
- Miotti L, Salemme M, Rabassa J. Radiocarbon chronology at Piedra Museo Locality. In: Miotti L, Salemme M, Flegenheimer N, editors. Where the South Winds Blow. Centre for the Study of First Americans and Texas A and M University Press; 2003. pp. 99–104. [Google Scholar]
- Molina GI. Saggio Sulla Storia Naturale del Chili. Tomasso d'Aquino; Bologna, Italy: 1782. [Google Scholar]
- Moritz C. Defining “Evolutionarily Significant Units” for conservation. Trends in Ecology and Evolution. 1994;9:373–375. doi: 10.1016/0169-5347(94)90057-4. [DOI] [PubMed] [Google Scholar]
- Mosca Torres ME, Puig S. Seasonal diet of vicuñas in the Los Andes protected area (Salta, Argentina): Are they optimal foragers? Journal of Arid Environments. 2010;74:450–457. [Google Scholar]
- Müller PLS. Erste Classe, Säugende Thiere. Des Ritters Carl von Linné vollständiges Naturalsystem nach der zwölften Lateinischen Ausgabe. 1776:1773–1776. pp. 1-62 + 3 pls., Suppl. 384 pp, Register, 36 unnumbered pp. + 536 pp.
- Nami HG, Nakamura T. Cronología Radiocarbónica con ams sobre muestras de hueso procedentes del sitio cueva del medio (Última Esperanza. Chile). Anales del Instituto de la Patagonia. 1995;23:125–134. [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. Columbia University Press; New York: 1987. [Google Scholar]
- Nielsen R, Wakeley J. Distinguishing migration from isolation: a Markov Chain Monte Carlo approach. Genetics. 2001;158:885–896. doi: 10.1093/genetics/158.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenius . Late Pleistocene Paleoecology of the South American Aridity: A case of continental dichotomy and the search of a new paradigm in Paleoclimatology. In: Argollo Jaime, Mourguiart Philippe., editors. Climas Cuaternarios en America del Sur. Orstom - Institut Francais de recherche scientifique pour le développement en coopération; La Paz. Bolivia: 1995. pp. 3–27. [Google Scholar]
- Ortega IM, Franklin W. Social organization, distribution and movements of a migratory guanaco population in the Chilean Patagonia. Revista Chilena de Historia Natural. 1995;68:498–500. [Google Scholar]
- Palma RE, Marquet PA, Boric-Bargetto D. Inter- and intraspecific phylogeography of small mammals in the Atacama Desert and adjacent areas of northern Chile. Journal of Biogeography. 2005;32:1931–1941. [Google Scholar]
- Pardiñas UFJ, Teta P, D'Elía G, Lessa EP. The evolutionary history of sigmodontine rodents in Patagonia and tierra del Fuego. Biological Journal of the Linnean Society. 2011;103:495–513. [Google Scholar]
- Park SDE. PhD thesis. University of Dublin; 2001. Trypanotolerance in West African cattle and the population genetic effects of selection. [Google Scholar]
- PasbØll PJ, Bérubé M, Allendorf FW. Identification of management units using population genetic data. Trends in Ecology and Evolution. 2006;22:11–16. doi: 10.1016/j.tree.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Paunero RS. The presence of a Pleistocenic colonizing culture in La Maria archaeological locality: Casa del minero 1. In: Miotti L, Salemme M, Flegenheimer N, editors. Ancient evidences for paleo South Americans: From where the south winds blow. Texas A & M University press; 2002. pp. 127–132. [Google Scholar]
- Penedo MCT, Casetano AR, Cordova KI. Microsatellite markers for South American camelids. Animal Genetics. 1998;29:411–412. [PubMed] [Google Scholar]
- Penedo MCT, Caetano AR, Cordova K. Eight microsatellite markers for South American camelids. Animal Genetics. 1999;30:166–167. doi: 10.1046/j.1365-2052.1999.00382-8.x. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Wen X, Falush D. Documentation for structure software: version 2.2. University of Chicago; Chicago: 2007. pp. 1–36. [Google Scholar]
- Politis GG, Pedrotta V. Recursos faunísticos y estrategias de subsistencia en el este de la región pampeana durante el holoceno tardío: El caso del Guanaco (Lama Guanicoe). Relaciones de la Sociedad Argentina de Antropología. 2006;31:301–336. [Google Scholar]
- Politis GG, Prates L, Merino ML, Tognelli MF. Distribution parameters of guanaco (Lama guanicoe), pampas deer (Ozotoceros bezoarticus) and marsh Deer (Blastocerus dichotomus) in central Argentina: archaeological and paleoenvironmental implications. Journal of Archaeological Science. 2011;38:1405–1416. [Google Scholar]
- Rabassa J, Coronato A, Martinez O. Late Cenozoic glaciations in Patagonia and Tierra del Fuego: an updated review. Biological Journal of the Linnean Society. 2011;103:316–335. [Google Scholar]
- Raedeke K. Doctoral Dissertation. College of Forest Resources University of Washington; 1979. Population dynamics and socioecology of the guanaco (Lama guanicoe) of Magallanes, Chile. [Google Scholar]
- Ramos-Onsins SE, Rozas J. Statistical properties of new neutrality tests against population growth. Molecular Biology and Evolution. 2002;19:2092–2100. doi: 10.1093/oxfordjournals.molbev.a004034. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning - A Laboratory Manual. Second Edition Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sarno R, Hunter R, Franklin W. Immobilization of guanacos by use of tiletamine/zolazepam. Journal of American Veterinary Medicine Association. 1996;208:408–409. [PubMed] [Google Scholar]
- Sarno RJ, David VA, Franklin WL, O'Brien SJ, Johnson WE. Development of microsatellite markers in the guanaco, Lama guanicoe: utility for South American Camelids. Molecular Ecology. 2000;9:1922–1924. doi: 10.1046/j.1365-294x.2000.01077-3.x. [DOI] [PubMed] [Google Scholar]
- Sarno R, Franklin W, O'brien S, Johnson W. Patterns of mtdna and microsatellite variation in an Island and Mainland Population of Guanacos in Southern Chile. Animal Conservation. 2001;4:93–101. [Google Scholar]
- Scherer CS. Phd Dissertation. Universidade federal do rio grande do sul. Instituto de Geociências; Brazil: 2009. Os Camelidae Lamini (Mammalia. Artiodactyla) Do Pleistoceno da América do Sul: Aspectos Taxonômicos e Filogenéticos. [Google Scholar]
- Schneider S, Excoffier L. Estimation of demographic para- meters from the distribution of pairwise differences when the mutation rates vary among sites: Application to human mitochondrial DNA. Genetics. 1999;152:1079–1089. doi: 10.1093/genetics/152.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sersic A, Cosacov A, Cocucci AA, et al. Emerging phylogeographic patterns of plants and terrestrial vertebrates from Patagonia. Biological Journal of the Linnean Society. 2011;103:475–494. [Google Scholar]
- Steppan SJ. Phylogenetic relationships and species limits within Phyllotis (Rodentia: Sigmodontinae) concordance between mtDNA sequence and morphology. Journal of Mammalogy. 1998;79:573–593. [Google Scholar]
- Stine S, Stine M. A record from Lake Cardiel of climate change in southern South America. Nature. 1990;345:705–708. [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivoli AM, Zangrando AF. Subsistence variations and landscape use among maritime hunter-gatherers. A zooarchaeological analysis from the Beagle Channel (Tierra del Fuego. Argentina). Journal of Archaeological Science. 2011;38:1148–1146. [Google Scholar]
- Tonni EP, Huarte R, Carbonari JE, Figini A. New Radiocarbon chronology for the Guerrero member of the Luján Formation (Buenos Aires. Argentina): Palaeoclimatic Significance. Quaternary International. 2003;109-110:45–48. [Google Scholar]
- Torres H. Distribution and Conservation of the Guanaco. Special report 2. iucn/ssc South American Camelids Specialist Group; Gland. Switzerland: 1985. [Google Scholar]
- Torres H. South American Camelids: An action plan for their conservation. iucn/ssc South American Camelids Specialist Group; Gland. Switzerland: 1992. [Google Scholar]
- Van Hooft WF, Groen AF, Prins HHT. Phylogeography of the African buffalo based on mitochondrial and Y-chromosomal loci: Pleistocene origin and population expansion of the Cape buffalo subspecies. Molecular Ecology. 2002;11:267–279. doi: 10.1046/j.1365-294x.2002.01429.x. [DOI] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- Webb SD. Pleistocene Llamas of Florida with a brief review of the Lamini. In: Webb s. d., editor. Pleistocene Mammals of Florida. University Of Florida Press; Gainsville: 1974. pp. 170–213. [Google Scholar]
- Webb SD. A History of Savanna vertebrates in the new world. Part II: South America and Great interchange. Annual Review of Ecology and Systematics. 1978;9:393–426. [Google Scholar]
- Webb SD. Ecogeography and the Great American Interchange. Paleobiology. 1991;17:266–280. [Google Scholar]
- Webb SD, Stehli FG. Bulletin of the Florida Museum of Natural History. 19. Vol. 37. Gainesville; 1995. Selenodont Artiodactyla (Camelidae and Cervidae) from the leisey shell pits. Hills borough county. Florida. pp. 621–643. [Google Scholar]
- Weinstock J, Shapiro B, Prieto A, Marín JC, González BA, Gilbert MT, Willerslev E. The Late Pleistocene distribution of vicuñas (Vicugna vicugna) and the “extinction” of the gracile llama (“Lama gracilis”): new molecular data. Quaternary Science Reviews. 2009;28:1369–1373. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-Statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wheeler J. Evolution and present situation of the South-American Camelidae. Biological Journal of the Linnean Society. 1995;52:271–295. [Google Scholar]
- Young JK, Franklin WL. Territorial fidelity of males guanacos in the Patagonia of southern Chile. Journal of Mammalogy. 2004;85:72–78. [Google Scholar]
- Zapata B, González BA, Marin JC, Cabello JL, Johnson W, Skewes O. Finding of polydactyly in a free-ranging guanaco (Lama guanicoe). Small Ruminant Research. 2008;76:220–222. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.