Summary
The UCS (UNC-45/CRO1/She4) chaperones play an evolutionarily conserved role in promoting myosin-dependent processes, including cytokinesis, endocytosis, RNA transport, and muscle development. To investigate the protein machinery orchestrating myosin folding and assembly, we performed a comprehensive analysis of Caenorhabditis elegans UNC-45. Our structural and biochemical data demonstrate that UNC-45 forms linear protein chains that offer multiple binding sites for cooperating chaperones and client proteins. Accordingly, Hsp70 and Hsp90, which bind to the TPR domain of UNC-45, could act in concert and with defined periodicity on captured myosin molecules. In vivo analyses reveal the elongated canyon of the UCS domain as a myosin-binding site and show that multimeric UNC-45 chains support organization of sarcomeric repeats. In fact, expression of transgenes blocking UNC-45 chain formation induces dominant-negative defects in the sarcomere structure and function of wild-type worms. Together, these findings uncover a filament assembly factor that directly couples myosin folding with myofilament formation.
Graphical Abstract
Highlights
► UNC-45 self-assembles a docking platform for multiple chaperone and client proteins ► Hsp70/90 and myosin bind to specific sites on the TPR and UCS domains of UNC-45 ► The UNC-45 multimer offers the proper spacing to couple myosin folding and assembly ► In vivo, UNC-45 chains support the formation of fully functional sarcomeric repeats
The myosin chaperone UNC-45 polymerizes into a linear chain that presents ordered binding sites for the myosin substrate and the chaperones Hsp70 and Hsp90, coupling myosin folding with assembly of a functional muscle myofilament.
Introduction
Muscle development and function rely on the correct assembly of structural and motor proteins into a series of contractile units called the sarcomeres. Its main components, thin (actin) and thick (myosin) filaments, are organized in a precisely ordered, quasi-crystalline protein framework that is critical to link the formation of actin-myosin crossbridges with filament gliding and muscle contraction (Gautel, 2011; Sweeney and Houdusse, 2010). Although the basic components and the overall architecture of the sarcomere have been studied in detail, little is known about its highly complex assembly process (Sanger et al., 2005). In particular, the mechanism of myosin incorporation into thick filaments is poorly understood. So far, it has been shown that folding of the myosin motor domain involves the assistance of the general chaperones Hsp70 and Hsp90 (Du et al., 2008; Etard et al., 2007; Gaiser et al., 2011; Hawkins et al., 2008; Srikakulam and Winkelmann, 2004) and of UCS-domain-containing proteins that function as myosin-specific chaperones (Barral et al., 2002; Hutagalung et al., 2002; Kachur and Pilgrim, 2008; Lord and Pollard, 2004; Wesche et al., 2003; Yu and Bernstein, 2003). One founding member of the UCS family is the Caenorhabditis elegans UNC-45 protein, which is highly conserved in metazoans (Barral et al., 1998; Epstein and Thomson, 1974; Venolia et al., 1999). In Drosophila, zebrafish, and Xenopus, UNC-45 proteins are important for skeletal and cardiac muscle function and for muscle maintenance during stress (Bernick et al., 2010; Etard et al., 2007, 2008; Geach and Zimmerman, 2010; Melkani et al., 2011; Wohlgemuth et al., 2007). Moreover, UNC-45 homologs exist in vertebrates, indicating a conserved requirement for the myosin-specific chaperone. Mice and humans have the two isoforms UNC45a and UNC45b either expressed in multiple tissues or restricted to myogenic processes in skeletal and cardiac muscles, respectively (Price et al., 2002). In line with this tissue specificity, abnormal UNC45b function is associated with severe muscle defects resulting in skeletal and cardiac myopathies (Janiesch et al., 2007; Walker, 2001). To exert its cellular function, UNC-45 employs an N-terminal tetratricopeptide repeat (TPR) domain implicated in binding the Hsp70/Hsp90 partner chaperones (Barral et al., 2002; Srikakulam et al., 2008), a central armadillo repeat (ARM) domain of unknown function and a C-terminal UCS domain that is critical to interact with the motor domain of myosin (Barral et al., 1998, 2002; Venolia et al., 1999). Mutations affecting the UCS domain of C. elegans UNC-45 result in paralyzed worms with reduced amounts of thick filaments and severe myofibril disorganization (Barral et al., 1998; Epstein and Thomson, 1974; Hoppe et al., 2004; Venolia and Waterston, 1990), indicating that UNC-45 is important for myosin maturation and sarcomere organization (Ao and Pilgrim, 2000; Kachur and Pilgrim, 2008). Indeed, UNC-45 does not only target substrate to Hsp90, but also functions itself as a chaperone promoting the folding and assembly of myosin molecules (Gaiser et al., 2011; Melkani et al., 2010).
Despite recent structural insight into the architecture of the central and UCS domain (Lee et al., 2011; Shi and Blobel, 2010), the molecular mechanisms of UNC-45 in promoting thick filament assembly and its interactions with partner chaperones are not understood. To delineate these functions, we combined biochemical and structural analyses with in vivo studies of the C. elegans UNC-45 protein. Intriguingly, our work reveals a chaperone chain in which UNC-45 tandem modules assemble a multisite docking platform that enforces collaboration with Hsp70 and Hsp90 in a precisely defined pattern to assemble myosin filaments.
Results
Crystal Structure of the UNC-45 Protomer
To obtain insight into the mechanistic role of UNC-45, we crystallized the full-length protein from C. elegans (107 kDa) and determined its crystal structure at 2.9 Å resolution (Table S1 available online). Structure comparison with related UCS proteins and analysis of local flexibility within the UNC-45 protomer reveals a four-domain architecture comprising an N-terminal TPR domain (TPR repeats 1–3), a central domain (ARM repeats 1–5), a neck domain (ARM repeats 6–9) and a C-terminal UCS domain (ARM repeats 10–17) (Figure 1A and Figure S1A). The resultant overall structure resembles a proteinous mouth that is constituted by a lower jaw (central domain and attached TPR domain), an upper jaw (UCS domain), and the neck region connecting the functional elements. In the slightly open UNC-45 fold, which is 65 Å high, 85 Å long, and 45 Å wide, the UCS domain protrudes in a 20° angle from the central domain and is situated above the interface of central and TPR domains (Figure 1B).
Figure 1.
Structure of UNC-45
(A) Ribbon presentation of the UNC-45 protomer showing its domain architecture. The numbering of the TPR and ARM repeats is given with helix H3 of each ARM repeat being highlighted. The cocrystallized Hsp90 peptide is shown in stick mode (lilac). Dashed lines represent flexible loops comprising residues 508–524 and 608–617.
(B) Orthogonal view of the UNC-45 protomer highlighting the two distinct faces of the UCS domain.
(C) Sequence alignment of the UNC-45 proteins from C. elegans (Ce; Q09981), Drosophila melanogaster (Dm; Q9VHW4), Homo sapiens (Hs; E1P642), and Danio rerio (Dr; Q6DGE9). The nomenclature of the TPR and ARM helices is given. With the exception of the UCS signature motif (loop L602–630) colored in magenta, the colors of the secondary structure elements reflect the domain colors. Dotted lines represent flexible loops that are not defined by electron density. Positions of UNC-45 temperature-sensitive (ts) mutations are indicated by asterisks. Functionally important residues are marked with a triangle (dark gray, oligomer interface inner core; light gray, oligomer interface outer residues; green, binding to Hsp90 C terminus; blue, TPR central interface). Red residues are strictly conserved in all four sequences; boxed residues are highly conserved. The overall sequence identity between the CeUNC-45 and the other listed UNC-45 proteins is about 30% (48% similarity).
Figure S1.
Domain Organization and Interfaces of CeUNC-45, Related to Figure 1
(A) Ribbon model and folding topology of UNC-45 illustrating its domain architecture and the used helix nomenclature (TPR: green; central: orange; neck: yellow; UCS: gray). Each TPR motif is made of two α helices A and B, whereas the ARM repeats are composed of three helices H1, H2 and H3. The helices H2 and H3 (shown as circles) of neighboring repeats are arranged in parallel constituting the core of the individual domains, whereas the relatively short H1 helices (shown as bars, not labeled), which are perpendicularly arranged to H2 and H3, orient adjacent ARM repeats to each other. Disordered loops are indicated by a dotted line.
(B) The central domain serves as a scaffolding unit that arranges TPR and UCS domain. The two close-up views illustrate the corresponding UNC-45 domain interfaces (polar interactions represented as dotted lines). Upper panel: Interface of central and UCS domain. Lower panel: Interface of central and TPR domain highlighting several short-distanced salt-bridges formed between Asp62-Lys269, Arg53-Asp279, Lys72-Glu281 and Glu29-Lys277.
(C) Sequence alignment of the UCS myosin-binding domain of CeUNC-45, Saccharomyces cerevisiae (Sc) She4, Schizosaccharomyces pombe (Sp) Rng3, Podospora anserina (Pa) CRO1, the founding members of the UCS protein family. Helices identified in the CeUNC-45 structure are indicated. Conserved residues are highlighted (red, strictly conserved; boxed, highly conserved) and reveal the extremely high conservation of the functionally important 13H3 helix pointing to a common mechanism in myosin binding. The signature motif in the UCS domain is colored in magenta. The sequence identity (similarity) of the UCS domain between CeUNC-45 and She4, Rng3 and CRO1 is 15% (26%), 21% (34%) and 23% (36%), respectively.
The TPR domain of UNC-45 is assembled by three TPR motifs, each containing two antiparallel α helices A and B. The helices are packed in a curved, right-handed superhelix featuring a shallow groove at its concave side that is critical to recognize and bind specific peptide ligands and to tether partner chaperones (Scheufler et al., 2000). The TPR3 motif is followed by a 54 Å long, kinked α helix (residues 115–150) that contributes to both TPR and central domains, thus constraining their relative orientation. Analysis of disorder B factors indicates that the central domain is the most rigid part of UNC-45 (average B factor of 70 Å2) functioning as a molecular scaffold to maintain the orientation of the TPR domain (102 Å2) and the UCS domain (147 Å2), the most flexible entity.
The central domain is a flat, rectangular protein ribbon that is structured by helices H1, H2, and H3 of ARM repeats 1–5. Owing to specific H1 adaptations, which impede the spiraling arrangement of adjacent H2 and H3 helices, the consecutive ARM repeats constitute a slightly curved H2/H3 helix bilayer rather than a superhelix. The edge of the bilayer that is covered by the H1 helices is used to hold the TPR domain in place. The major part of this interface is contributed by helix H1 of ARM repeat 4 (helix 4H1) and its extended N-terminal loop, which intercalate into the cleft formed by the TPR1 and TPR2 motifs and mediate several short-distanced salt bridges (Figure S1B). Because most residues of this interface are highly conserved (Figure 1C), the precise positioning of the TPR to the central domain should be generally important for the function of UNC-45 proteins. Interactions with the second functional domain, the UCS domain, are mediated by residues located on the H3 side of the central domain. Here, helices 4H3 and 5H3 form a flat binding surface to accommodate the loops protruding from ARM repeats 10–12. The relatively small size of this interface (339 Å2) and the prevalence of unspecific van der Waals interactions (Figure S1B) indicate that the UCS domain is less fixed than the TPR domain and though preoriented by leaning against the central domain may reposition when interacting with target proteins.
UCS and central domain are connected by the neck domain. Insertion of the highly flexible loop 508–524 and deletion of helices 8H1 and 9H1 disrupt the interfaces between ARM repeats 8–10, thereby unlocking the individual motifs from each other. As a result, the helix pairs of the neck domain adopt a superturn structure that accounts for a 180° bend in the UNC-45 fold directing the consecutive UCS domain back toward the N-terminal TPR domain, thus completing the UNC-45 mouth. The UCS domain itself forms an almost regular right-handed superhelix. The parallel packed helices of neighboring repeats assemble a spiraling scaffold with a long shallow groove that is lined by the H3 helices (Figure 1A). Due to the irregular packing of the H2/H3 helix pairs of ARM repeats 12–14, the UCS superhelix is interrupted at ARM repeat 13. Finally, the UCS domain encompasses an extended loop (residues 602–630) that is inserted after helix 10H3 and represents one of the most conserved signature motifs of UCS proteins (Figure S1C).
Hsp70 and Hsp90 Bind Differently to the TPR Domain of UNC-45
Hsp70 and Hsp90 chaperones exert their protein folding function by collaborating with distinct cofactors that are required for substrate targeting. The physical link between chaperone and cochaperone is commonly mediated by binding of the C terminus of Hsp70/Hsp90 to the TPR domain of the targeting factor (Scheufler et al., 2000; Young et al., 2003). In the case of UNC-45, the interactions with Hsp70 and Hsp90 have not been addressed in detail, and thus the composition and organization of the chaperone complex that promotes myosin folding and assembly are still unclear. To characterize the interplay between the three chaperones, we performed a detailed isothermal titration calorimetry (ITC) analysis (Figure 2A). For Hsp90 (DAF-21 in C. elegans), the ITC data revealed a tight interaction with UNC-45, thus confirming previous reports (Barral et al., 2002; Srikakulam et al., 2008). Notably, the apparent affinity for the C-terminal peptide (EDASRMEEVD, dissociation constant KD ∼14 μM) was slightly higher than for full-length Hsp90 (KD ∼31 μM), suggesting that the peptide-TPR interaction represents the main contact between the two chaperones. ITC measurements with the C-terminal peptide AGGPTIEEVD of Hsp70 (HSP-1 in C. elegans) revealed a weaker but robust affinity toward UNC-45 (KD ∼120 μM). Owing to the low yield of recombinant Hsp70, we monitored complex formation with UNC-45 by size exclusion chromatography (SEC). To this end, we compared the elution profiles of the isolated chaperones and complexes thereof. Importantly, UNC-45 coelutes with both of its partner chaperones and is thus capable of forming complexes with Hsp70 and Hsp90 (Figure 2B). When equal amounts of Hsp70 and Hsp90 are incubated with UNC-45, the Hsp90 chaperone is preferentially bound (Figure S2A). In contrast, incubation of UNC-45 with an excess of the Hsp70 C-terminal peptide abolished interaction with Hsp90, showing that both chaperones compete for the same binding site on the TPR domain (Figure S2A). To further exclude that complexes with Hsp70 and Hsp90 result from unspecific chaperone-substrate interactions, we introduced site-specific mutations in the TPR domain of UNC-45 (K82E) and in the C termini of Hsp70 and Hsp90 (deletion of the five terminal residues). SEC and ITC binding studies revealed that the introduced mutations abolished complex formation (Figures 2A and S2B). We thus conclude that the TPR domain of UNC-45 is the main docking site for Hsp70 or Hsp90, allowing formation of defined complexes with either of the two general chaperones.
Figure 2.
Interaction of UNC-45 with Its Partner Chaperones Hsp70 and Hsp90
(A) Representative ITC data recorded upon mixing wild-type and K82E UNC-45 with the Hsp70/90 C-terminal peptides. Raw data (top) and binding isotherm derived from the integrated heat (bottom) are shown. Calculated KD values are given.
(B) SEC/SDS-PAGE analysis reveals complex formation of UNC-45 with Hsp90 (left) and Hsp70 (right).
(C) Binding mode of the Hsp90 C terminus (lilac) to the TPR domain (green). Interacting residues of the TPR domain are labeled.
(D) Overlay of the Hsp70 (blue) and Hsp90 (lilac) peptide structures bound to the TPR domain (green). Close-up view of the Hsp90 peptide (top), Hsp70 peptide (bottom), and interacting TPR domain residues overlaid with the 2Fo – Fc omit electron density (Hsp90: 2.9 Å resolution, contoured at 1.1σ; Hsp70: 3.6 Å, contoured at 1.1σ). Interactions with Hsp90 that are not formed with Hsp70 are highlighted with an asterisk.
See also Figure S2.
Figure S2.
Details of Hsp70 and Hsp90 Binding to UNC-45, Related to Figure 2
(A) Analytical SEC and SDS-PAGE of UNC-45, Hsp70 and Hsp90. Left panel: UNC-45 and equal concentrations of Hsp70 and Hsp90 full-length proteins. Right panel: UNC-45 and Hsp90 in the presence of a decapeptide representing the C terminus of Hsp70.
(B) Analytical SEC and SDS-PAGE of UNC-45 and its partner chaperones Hsp70 or Hsp90 lacking their C-terminal motives IEEVD and MEEVD, respectively.
(C) Molecular surface of UNC-45 with mapped conservation pattern (red, highly conserved; yellow, less conserved) and bound Hsp90 peptide (lilac). The close-up view of the TPR domain shows the strictly conserved binding site for the Hsp90 peptide.
(D) UNC-45 was co-crystallized with a decapeptide representing either the Hsp70 or the Hsp90 C terminus. Left panel: Stereo view of the TPR domain (green) and the co-crystallized Hsp70 (blue) and Hsp90 (lilac) peptides shown in stick mode. The final model is overlaid with the respective 2Fo-Fc omit electron density calculated without ligand (complex with Hsp90: 2.9 Å resolution, contoured at 1.1σ; Hsp70: 3.6 Å resolution, contoured at 1.1σ). Interacting residues are labeled. To describe the specific contacts with UNC-45, we will focus on the interaction with the Hsp90 peptide, which is numbered in descending order with the C-terminal Asp0 preceded by Val-1, Glu-2, Glu-3, Met-4, Arg-5, Ser-6 and Ala-7. The Hsp90 C terminus is tethered in a bent conformation to the TPR domain undergoing multiple main-chain and side-chain interactions with highly conserved UNC-45 residues. Contacting residues of the TPR domain are highlighted in green and are labeled. Contacts to the backbone are mediated by Lys82 and Arg86 that bind to the carbonyl oxygen of Glu-2, by Asp114 that interacts with the amine nitrogen of Met-4 and finally by Arg12, Asn16 and Asn52 that compose a positively charged pocket to accommodate the carboxy-terminus of Hsp90. Notably, the geometry of the three backbone anchor points favors binding of the Hsp90 peptide in bent conformation. The observed turn structure is further stabilized by an internal hydrogen bond between the side chains of Glu-2 and Ser-6. All remaining side-chains of the Hsp90 peptide are engaged in interactions with highly conserved TPR residues ensuring the tight and specific binding of the Hsp90 C-terminal peptide. The carboxylate group of Asp0 is tightly bound by Arg51 and Lys82, the Val-1 side chain is accommodated in a hydrophobic pocket lined by residues Val19, Asn52 and Met55, Glu-3 forms a short-distanced salt-bridge with the amino group of Lys59 and the side-chain of Met-4 protrudes into a deep hydrophobic pocket bordered by Val81, Lys82, Phe85 and Ile117. Right panel: Eight residues of the Hsp90 peptide ligand (ASRMEEVD lilac) and ten residues of the Hsp70 peptide ligand (AGGPTIEEVD blue) are well-defined by electron density revealing their precise binding mode. Additional flexible residues at the C-termini of Hsp70 (∼20 amino acids) and Hsp90 (∼5 amino acids) are shown schematically as dotted lines.
To address the individual binding modes of Hsp70 and Hsp90, we cocrystallized UNC-45 with oligopeptides reflecting the two C termini, respectively. In the 2.9 Å electron density map of the UNC-45/Hsp90 complex, the eight C-terminal residues of Hsp90 could be unambiguously modeled into the TPR binding pocket. Accordingly, the Hsp90 C terminus ASRMEEVD is bound in a bent conformation undergoing multiple main-chain and side-chain interactions with highly conserved UNC-45 residues (Figure 2C, detailed description in Figure S2). In the crystal structure of the UNC-45/Hsp70 complex determined at 3.6 Å resolution, the peptide ligand AGGPTIEEVD is bound in a similarly bent conformation; however, the entire peptide is shifted ∼1.5 Å away from the center of the domain (Figure 2D). Consequently, the tip of the Hsp70 peptide turn is differently accommodated in the TPR cleft (Figure 2D). Several interactions undergone with Hsp90 are broken in the UNC-45/Hsp70 complex structure, including the hydrogen bond with Asp114, the salt bridge with Lys59, and the hydrophobic contacts with Phe85 (Figure 2D). Together, these differences should account for the preferential binding of Hsp90, as directly seen in our ITC binding studies. In addition to confirming the observed binding preferences, the structural data also visualize that both Hsp70 and Hsp90 chaperones are bound in a specific manner and are thus likely candidates to assist UNC-45 in myosin folding and assembly.
Interaction between UCS Domain and Muscle Myosin
Previous reports revealed that the UCS domain is critical to bind myosin (Barral et al., 2002; Lord and Pollard, 2004; Toi et al., 2003). To identify the corresponding interaction site, we carried out homology searches with DALI and observed that the UCS domain of UNC-45 is most similar to β-catenin (Figure S3A). Notably, most β-catenins have a conserved protein-binding groove that is undisclosed in several protein-ligand complex structures. Transferring the catenin-bound ligands to the superimposed UNC-45 allowed us to visualize the potential binding site for myosin, which is a lengthy canyon formed by highly conserved residues spanning the entire UCS domain in a screw-like manner (Figures 3A and S3BC). The bottom of the 70 Å long groove is formed by the H3 helices of ARM repeats 10–17 and is well designed to accommodate polypeptide chains in extended conformation.
Figure S3.
Potential Myosin-Binding Site of the UCS Domain, Related to Figure 3
(A) Closest structural homologs, as revealed by a DALI search, are listed for the UCS domain.
(B) Left panel: Structural alignment of the UCS domain of UNC-45 (gray) with the ARM domain of β-catenin (violet, PDB code: 1i7x). Right panel: To identify the myosin binding site of UNC-45, peptide ligands (green) were extracted from aligned β-catenin crystal structures and mapped on the UCS domain. H3 helices of the UCS domain are highlighted with helix 13H3 and the UCS loop shown in magenta.
(C) Left panel: Molecular surface of the UCS domain with mapped electrostatic potential and aligned β-catenin peptide ligand. Right panel: Molecular surface of the UCS domain with mapped conservation pattern and aligned β-catenin peptide ligand. The illustration highlights the hydrophobic surface and the conservation of the UCS canyon.
Figure 3.
UNC-45 Interaction with Myosin
(A) Ribbon model of the UCS domain highlighting mutations (magenta) used to address the myosin binding of UNC-45. To illustrate the position of substrate, a peptide ligand (green) of a superimposed β-catenin molecule was transferred to the UNC-45 fold. The zoom-up windows provide a detailed view on the Y750W and N758Y mutations, the functionally related residues in β-catenin (PDB code 1i7w, with bound E-cadherin peptide shown as stick model) and the Δ602-630 deletion expected to interrupt the UCS canyon.
(B) To estimate the rescue of the uncoordinated (unc) phenotype, young adult unc-45(m94) worms expressing the indicated UNC-45 variants were grown at 25°C and body bends counted (two independent transgenic lines per mutation, SE of the mean indicated).
(C) Immunostaining of unc-45(m94) mutant worms grown at 25°C expressing Y750W, N758Y, or Δ602–630 UNC-45.
(D) Protein-protein interactions undergone by wild-type and mutant UNC-45 (FLAG tag indicated by asterisk). The UNC-54 myosin and Hsp90 were coimmunoprecipitated from cell lysates of the indicated unc-45(m94) mutant worms expressing different UNC-45 variants grown at 25°C. Protein interactions were evaluated by western blot analysis using UNC-54-, Hsp90-, and UNC-45-specific antibodies.
To characterize the myosin-binding canyon, we carried out systematic peptide binding studies using ITC and peptide spot analyses. In contrast to the yeast UCS protein She4 (Shi and Blobel, 2010; Toi et al., 2003), UNC-45 from C. elegans did not interact with myosin-derived peptides in these assays (data not shown). We thus pursued an in vivo approach to test the relevance of the predicted myosin-binding canyon. Notably, the in vivo analysis of UNC-45 is not trivial because loss of UNC-45 immediately leads to cytokinesis defects and embryonic lethality (Kachur et al., 2004; Kachur and Pilgrim, 2008; Venolia and Waterston, 1990). We thus expressed selected UNC-45 mutants in worms harboring the unc-45 temperature-sensitive (ts) allele m94 (E781K) and analyzed to which degree muscle function of the ts mutant was restored. To this end, we generated specific mutations that distorted the UCS canyon to different degrees (detailed description in the Supplemental Information). Based on the structural comparison with β-catenin, we targeted the central passage of the canyon that is composed by helix 13H3, the most conserved stretch of the UCS domain (Figure S1C). As outlined in Figure 3A, the N758Y mutation was predicted to directly abolish the interaction with substrate, whereas the Y750W mutation was designed to modify the outer rim of the substrate-binding cleft. To further explore the myosin-binding site, we deleted the UCS signature motif 602–630 that composes an elongated, highly conserved loop next to the UCS channel. Accordingly, the Δ602–630 mutation should discontinue the substrate-binding cleft and abrogate the interaction with myosin (Figure 3A). The N758Y, Y750W, and Δ602–630 mutants were transgenically expressed and assayed for their ability to rescue the motility of unc-45(m94) worms and to bind to the myosin heavy chain UNC-54. In contrast to the Y750W mutant, which is still capable to bind myosin and restore muscle function, the N758Y and Δ602–630 mutants neither rescue the defect in sarcomere organization nor bind to UNC-54 in immunoprecipitation experiments (Figures 3B–3D and Table S2). Because all mutants interact with Hsp90 similar to wild-type, the myosin-binding mutations do not affect the structural integrity of UNC-45 (Figure 3D). Together, these findings demonstrate that the UCS canyon composes the major myosin-binding site of UNC-45 and most likely accommodates an unfolded portion of the myosin substrate, as also suggested previously (Srikakulam et al., 2008).
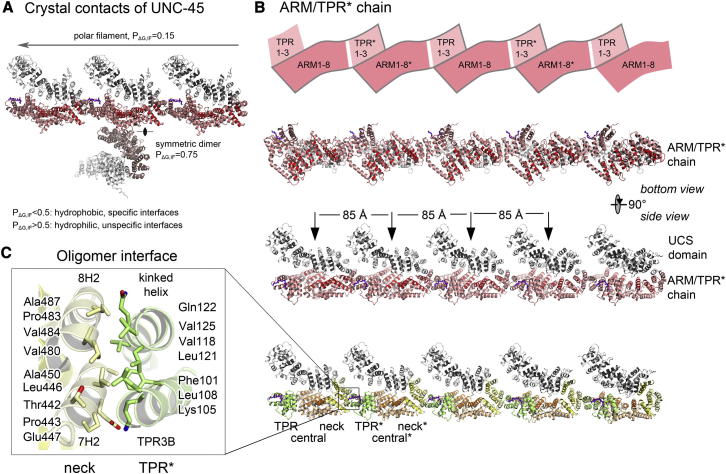
UNC-45 Crystallizes as a Polar Protein Filament
In the crystal lattice of UNC-45, two prominent crystal contacts can be discerned that lead to either a symmetric dimer, which is stabilized by juxtaposed central domains, or a continuous protein filament resulting from lateral associations of adjacent subunits (Figure 4A). To estimate the likelihood of whether the crystallographic interfaces mediate oligomerization in solution, we performed a bioinformatic analysis with PISA (Protein Surfaces, Interfaces and Assemblies). Probability measures PΔG,IF of specific interfaces were derived from the gain in solvation energy upon complexation, with PΔG,IF > 0.5 pointing to hydrophilic/unspecific and PΔG,IF < 0.5 to hydrophobic/specific interfaces. In contrast to the dimer interface that appears to be a crystal artifact (PΔG,IF = 0.75), the extremely low PΔG,IF value of the UNC-45 filament interface (0.15) lies in the range of probabilities derived from typical protein interfaces (0.1–0.4). Moreover, other structurally characterized protein filaments exhibit similar PΔG,IF values, some of which indicate an even smaller polymerization probability than UNC-45 (Table S3). We thus suppose that UNC-45 may form similar linear multimers in solution, although the relatively small size of the interface (788 Å2) suggests that UNC-45 chains are only transiently formed or may need further factors to be stabilized.
Figure 4.
Structural Organization of the UNC-45 Multimer
(A) Crystal contacts observed in the unit cell of C. elegans UNC-45. Respective PΔG,IF values reflect the likelihood that the resultant dimer or filament represents a crystallographic artifact.
(B) Architecture of the UNC-45 oligomer. The ARM/TPR∗ building block is schematically shown as enclosed box in the top panel (molecular neighbors distinguished by asterisks). The ribbon presentations in the bottom panels provide an orthogonal view on the UNC-45 chain and illustrate that TPR, central, and neck domain form the backbone of the assembly, whereas the UCS domain projects away offering myosin-binding sites in a regular array (indicated by arrows).
(C) Close-up view of the oligomer interface showing the composite four-helix bundle. Involved helices and residues are labeled.
Concerning its multimeric architecture, the UNC-45 chain represents a unique filamentous structure resulting from the mouth-to-neck assembly of single subunits (Figure 4B). In the crystal lattice, adjacent subunits in the UNC-45 chain are held together by a composite four-helix bundle to which neighboring molecules contribute two helices each (Figure 4C). With the exception of the Glu447-Lys105∗ salt bridge (the asterisk marks the partner molecule), the filament interface is exclusively formed by hydrophobic residues arranged in a zipper-like manner in the center of the four-helix bundle. The backbone of the resultant UNC-45 filament is thus composed by the tethered superhelices ARM1-8 and TPR1-3∗, whereas the UCS domain, which is proposed to bind myosin, extends in a characteristic manner away from the filament axis and does not participate in filament assembly (Figure 4B). Similarly, the side-by-side packing of the ARM/TPR∗ units does not influence the peptide-binding site of the TPR domain. This structural organization would ensure that the connected TPR and UCS domains retain their functional independence in the oligomeric state, allowing the simultaneous docking of multiple client proteins and cooperating chaperones to the UNC-45 chain. As the hydrophobicity of the multimer interface is largely conserved (Figure 1C), we presume that the observed chaperone chain represents a common feature of the UNC-45 family. Deviations from this organization, as implicated by the crystal structures of UNC-45 from Drosophila melanogaster (Lee et al., 2011) and yeast She4 (Shi and Blobel, 2010) (Figure S4), may reflect different functional states or activities, respectively. For example, the DmUNC-45 was observed in monomeric form with an entirely flexible TPR domain that could not be localized in the electron density, whereas the dimeric yeast She4 that lacks a TPR domain is involved in promoting myosin-actin interactions rather than thick filament assembly (Toi et al., 2003).
Figure S4.
Crystal Packing of CeUNC-45, DmUNC-45, and She4, Related to Figure 4
(A) Structural alignments of CeUNC-45 (gray) with DmUNC-45 (PDB code: 3now, magenta, top) and She4 (PDB code: 3opb, blue, bottom panel). The 602-630 loop of CeUNC-45 extends as unfolded segment into the protein periphery, whereas in the crystal structure of DmUNC-45, this loop folds back on the UCS domain filling partly the extended central canyon (shown in orange). We presume that the different domain organization and oligomeric states observed in the crystal structures of DmUNC-45 and She4 are due to different crystal contacts and the introduction of multiple mutations for protein crystallization, respectively.
(B) CeUNC-45 was crystallized in space group P6122 having a single molecule in the asymmetric unit (unit cell dimensions are indicated). Six UNC-45 dimers related by a crystallographic 2-fold axis are sitting in a 61-screw axis on top of each other thus yielding the long c-axis of 815 Å. Each of the 12 molecules undergoes longitudinal interactions with their crystallographic neighbors (shown for top molecule) resulting in polar UNC-45 chains that extend with a 85 Å periodicity throughout the crystal, perpendicularly to the long crystallographic c-axis. In contrast to the filament interface, the crystallographic dimer is largely stabilized by polar interactions as shown in the close-up window.
(C) DmUNC-45 was crystallized in space group P23 with a single subunit in the asymmetric unit. Owing to crystal symmetry, twelve molecules associate via their UCS domains forming a hollow sphere that is surrounded by six couples of central domains. Side-by-side packing of the DmUNC-45 spheres generates a wide-meshed crystal lattice with huge holes, into which the flexible TPR domains (indicated by green spheres) protrude. Whether the en-bloc flexibility of the TPR domain is functionally relevant or influenced by crystal packing remains to be shown.
(D) She4, a yeast UCS protein that is proposed to promote actin-myosin interactions (Shi and Blobel, 2010), was crystallized upon introducing 13 point mutations and deleting loop 343-358 (indicated by red circles). In the crystal lattice, two She4 molecules associate to generate an S-shaped dimer that is stabilized by the N-terminal helix binding to the neck domain (ARM repeats 8-9) of a molecular neighbor. It still needs to be experimentally shown that the crystallographic dimer is functionally relevant.
UNC-45 Forms Short Chains in Solution
To directly monitor the predicted UNC-45 multimer in solution, we performed a SEC analysis varying protein concentration and buffer conditions. As we could not detect higher-order oligomers in these experiments, we next developed a specific assay to target transient interactions that may stabilize the UNC-45 chain. To this end, we applied a targeted photocrosslinking approach using the UV-inducible amino acid p-benzoyl-L-phenylalanine (pBpa), which can be expressed at any specific position in the protein. To explore chain formation, we exchanged a glutamine close to the supposed filament interface by pBpa (Q452B) and compared the crosslinking capability of the wild-type Q452B with several variants affecting the structure of the donor (ARM) and acceptor (TPR∗) sites of the UNC-45 chain. For this purpose, we used the ΔTPR (Δ1–134), L121W, and V480R/V484R mutations as steric and electrostatic blocks, respectively, preventing UNC-45 chain formation.
The crosslinking experiments revealed that the wild-type protein can assemble multimers of different size, with the largest ones comprising at least five subunits. In contrast, mutating the filament interface abolished the formation of defined higher-order oligomers (Figures 5A and 5B). Consistent with the proposed filament structure, this experiment highlights the importance of residues Leu121, Val480, and Val484 in mediating UNC-45 multimerization in solution. Moreover, the photocrosslinking approach revealed that chain formation depends strongly on UNC-45 concentration, with the longer chains occurring only at elevated protein concentration. For example, in solution, UNC-45 tetramers started to accumulate upon incubating UNC-45 at 50 μM, but not at lower concentrations (Figure S5). To ultimately confirm that the crystallographic filament interface is relevant in vitro, we analyzed the SDS-PAGE bands of the different UNC-45 multimers by mass spectrometry (MS). Recorded MS spectra revealed that the UV-activated pBpa452 crosslinks to the Lys131/Ile132/Lys133 sequence (Figure 5C). Moreover, the pBpa452-131/132/133 crosslink was specifically observed in all wild-type oligomers (marked with an arrow, Figure 5A), whereas it was not detected in the wild-type monomer sample nor in the second dimer fraction resulting from unspecific crosslinking (marked with a circle, Figure 5A). Strikingly, in the structure of the UNC-45 filament, Lys131 and Ile132 are the most adjacent residues of Gln452∗ being situated 3–5 Å next to its terminal carboxamide group, thus explaining the precisely defined crosslinking pattern of UNC-45 multimers (Figure 5D). Taken together, the MS analysis of photocrosslinked UNC-45 oligomers unequivocally demonstrates that UNC-45 forms short protein chains in solution comprising two to at least five subunits, which associate in the same manner as observed in the crystal lattice.
Figure 5.
In Vitro Validation of UNC-45 Chains
(A) SDS-PAGE analysis of the photocrosslinking reaction upon pBpa452 (Q452B, indicated by “X”) activation. Samples of wild-typex, L121Wx, V480R/V484Rx, and ΔTPRx UNC-45 were exposed for 15 and 30 min to UV illumination. Schematic representations of the introduced mutations and resultant crosslinks are indicated. Specifically linked oligomers (as deduced by MS) are marked by arrows, whereas dimers resulting from unspecific crosslinking are labeled with a circle.
(B) SEC elution profile and corresponding SDS-PAGE of the wild-typex sample illustrating formation of 2–5 meric UNC-45 chains.
(C) MS2 spectrum of the crosslinked peptide identified in the marked Q452B oligomers. The inset shows the observed product ions mapped onto the sequence of the crosslinked peptide, the isotopic distribution of the crosslinked peptide, its mass-to-charge ratio (m/z), its charge (3+), and its monoisotopic mass value (m). Δmass, difference between the expected and measured masses; R, resolution of the measurement; B, pBpa; M∗, oxidized methionine.
(D) In the crystal structure of the UNC-45 chain, the identified crosslinked residues Lys131-Ile132 are juxtaposed to the pBpa452 of the molecular neighbor. Because these residues were exclusively identified in the UNC-45 2 mer, 3 mer, 4 mer, and 5 mer, the MS data are consistent with the illustrated protein chains. The close-up window illustrates the molecular details of the TPR-ARM∗ oligomer interface highlighting the mutated (black) and crosslinked (magenta) residues.
See also Figure S5.
Figure S5.
Concentration Dependency of UNC-45 Oligomer Formation, Related to Figure 5
(A) SDS-PAGE analysis of the photo-crosslinking reaction upon pBpa452 activation. Samples of wild-type UNC-45 Q452 pBpa exposed to UV illumination for 15 and 30 min at three different concentrations are shown. The unspecific dimer is marked with a circle.
(B) Quantification of the crosslinked fraction at 30 min. Percentage of dimer (lilac), trimer (red) and tetramer (green) are shown for the different concentrations. 100% crosslink at 100 μM concentration is used as reference.
Formation of UNC-45 Tandem Modules Promotes Organization of Muscle Sarcomeres
To evaluate the relevance of the UNC-45 multimer in supporting myofilament formation in vivo, we analyzed the effect of expressing the ΔTPR, L121W, and V480R/V484R interface mutations in C. elegans body wall muscles (Figure 6A). Given their similar potential in destabilizing UNC-45 chains in vitro, we expected that the three transgenes would exhibit similar phenotypes in vivo. Surprisingly, however, the V480R/V484R mutation showed the strongest effects and did not restore locomotion and sarcomere integrity of unc-45(m94) worms, as indicated by the low number of body bends and the completely disorganized sarcomere structure, respectively (Figures 6B and 6C). To address the underlying reason for the enhanced phenotype, we performed extensive pull-down experiments monitoring the interactions with the UNC-45 partner proteins. As detailed in the Supplemental Information, these experiments clearly showed that the V480R/V484R mutant hinders binding of the myosin substrate in addition to blocking chain formation (Figures 6C–6E and S6). As the L121W and ΔTPR mutations did not abrogate myosin binding, they should reflect the consequence of impaired chain formation in the unc-45(m94) background more precisely. In contrast to wild-type UNC-45, the TPR mutations could not recover full motility of m94 worms, as indicated by a 30%–50% reduced activity in body-bend assays. Moreover, muscle sarcomeres could only be in part restored and exhibited local myosin assembly defects, as well as a reduced overall size (Figures 6B and 6C and Table S2). Most notably, sarcomeres restored by L121W and ΔTPR UNC-45 had fewer myosin-containing A bands, i.e., four to six instead of seven to eight bands seen in worms expressing wild-type UNC-45 (Figures 6E and S6A and Table S4). Because this effect is not correlated with Hsp90 function (Gaiser et al., 2011), the dis- and reorganization of muscle sarcomeres should directly result from the impaired formation of UNC-45 chains.
Figure 6.
Functional Analysis of UNC-45 Oligomerization Mutants In Vivo
(A) Ribbon plot providing the close-up view of the oligomer interface with the mutated residues Val480, Val484, and Leu121 shown as orange spheres and the ΔTPR enclosed by a box.
(B–E) To estimate the rescue of the uncoordinated (unc) phenotype, young adult unc-45(m94) worms expressing the indicated UNC-45 variants were grown at 25°C. Immunostaining of UNC-45 and UNC-54 (myosin), body bend counts, the ability of UNC-45-FLAG variants to coIP UNC-54 (myosin), and sarcomeric A band quantification are shown, respectively.
(F–H) The dominant-negative effect of expressing different UNC-45-FLAG versions in unc-119/unc-45(wt) worms was assayed by body bend counts, A band staining, and quantification, respectively. The number of A bands (labeled with anti-MHC A) is given per body wall muscle cell (indicated by dotted line), with all analyzed cells located in the same area between pharynx and vulva.
All values are mean ± SEM. See also Figure S6, Table S2, and Table S4.
Figure S6.
Analysis of UNC-45 Oligomer Formation Mutants, Related to Figure 6
(A– C) (A) Immunostaining of MHC A in unc-45(m94) worms expressing either wild-type UNC-45-FLAG or the indicated UNC-45-FLAG oligomer formation mutants (FLAG-tag indicated by asterisk). Single cells are outlined by a dotted line. Total UNC-54 levels in unc-45(m94) worms (B) and unc-119/unc-45(wt) worms (C) expressing different UNC-45-FLAG versions were determined by anti-UNC-54 Western Blot analysis. An anti-tubulin Western Blot is shown as loading control.
(D) UNC-54 myosin was coimmunoprecipitated with the indicated UNC-45-FLAG versions from unc-119/unc-45(wt) worms expressing the indicated UNC-45-FLAG variants. Proteins were detected by Western Blot analysis.
(E) Molecular surface representation of UNC-45 (gray) and an aligned β-catenin peptide ligand (green) in stick mode. The site-specific oligomer formation mutants L121W and V480R/V484R are highlighted in orange.
To further substantiate the biological relevance of UNC-45 oligomerization for muscle function, we tested whether the interface mutations compromised sarcomere organization when expressed in wild-type worms. We observed that transgenes encoding the two site-specific chain mutations L121W and V480R/V484R caused a small but definite reduction in motility, whereas the ΔTPR transgene caused the most pronounced locomotion defect, as would have been predicted from our structural model (Figure 6F). Moreover, and consistent with the unc-45(m94) rescue experiments, the reduced motility was linked to a smaller number of A bands in the muscle sarcomere (Figure 6F–6H and Table S4). Most importantly, this dominant-negative phenotype was only observed when expressing UNC-45 interface mutants, but not with the myosin-binding mutants N758Y and Δ602-630 (Figure 6F). These data strongly support the UNC-45 chain model, as the interface mutants could directly interfere with forming UNC-45 scaffolding complexes, whereas the myosin binding capacity of wild-type UNC-45 cannot be compromised by the expressed myosin-binding mutants.
Extended Results.
Choice of UNC-45 Residues for Addressing Myosin Binding In Vivo
Owing to the pronounced homology to β-catenin, the UCS domain of UNC-45 is predicted to bind part of the myosin substrate in an extended conformation. As observed in β-catenin crystal structures (for example Huber and Weis, 2001), a series of histidine and asparagine residues that originate from adjacent H3 ARM helices anchor the peptide backbone of the substrate by several short-distanced hydrogen bonds. Notably, these main-chain binding sites are largely conserved among catenin and other ARM proteins (Conti et al., 1998; Huber and Weis, 2001). Our structural analysis suggests that this binding mode is also maintained in UNC-45 and relies on helix 13H3 (residues 748-761). This helix, which is highly conserved in the UCS protein family (Fig.S1C), composes the floor of the central passage of the UCS domain and is thus predicted to bind the core of potential substrate ligands. In analogy to β-catenin, the strictly conserved Asn758 should be critical for tethering the main-chain of the captured substrate, whereas the nearby Tyr750 contributes to the hydrophobic character of the outer rim of the UCS canyon (Figure 3A). Of note, UCS proteins express different hydrophobic residues at position 750 preferentially tyrosine and phenylalanine (Figure 1C, Fig.S1C). To test the catenin-based binding model, we replaced Asn758 of UNC-45 with a tyrosine residue that should abrogate the hydrogen-bonding to the substrate’s peptide backbone and block the central passage of the UCS canyon. In parallel, we introduced a conservative mutation at position 750 by replacing tyrosine with tryptophan, which can in silico obtain a similar arrangement within the UCS canyon (Figure 3A). In addition, we introduced a distinct steric block in the UCS canyon by deleting loop 602-630, the most characteristic signature motif of UCS proteins that is absent in other ARM proteins. Structural alignment with β-catenin complex structures suggests that this loop, which connects the two rims of the UCS canyon, may embrace the substrate protein. Therefore, deletion of residues 602-630 would shrink the opening of the UCS passage such that the binding site is interrupted (Figure 3A). In addition, the Δ602-630 mutant lacks highly conserved residues, which border the UCS canyon and may be directly involved in substrate binding. The in vivo analysis of the Y750W, N758Y and Δ602-630 mutations confirmed the postulated binding site and revealed that the UCS canyon observed in the C. elegans crystal structure is essential to interact with myosin.
Assessing the In Vivo Side Effects of the UNC-45 Interface Mutations
To address the formation of UNC-45 chains, we generated three distinct mutants affecting the two faces of the UNC-45 multimerization site formed by helices of the TPR domain (motif-1) and the neck domain (motif-2). Two mutants comprise site-specific exchanges in motif-1 (L121W) and motif-2 (V480R/V484R), respectively, whereas the ΔTPR mutant lacks the entire motif-1. Though all three interface mutants blocked chain formation in vitro, the structural data suggest that the ΔTPR mutant should show the strongest effect in impairing UNC-45 oligomer function in vivo. It is thus surprising that the double arginine mutant V480R/V484R exhibits the most pronounced defects and could not rescue the temperature-dependent locomotion phenotype of unc-45(m94) ts mutants, whereas expression of the two TPR transgenes (L121W and ΔTPR) rescued motility at least partially.
To better understand the observed in vivo phenotypes, we tested whether the interface mutations disturb, in addition, the interaction with partner proteins. Indeed, systematic UNC-45 pull-down experiments, which were carried out at different temperatures and using cell lysates from unc-45(m94) and wild-type worms revealed that the distinct mutations affect the binding of the myosin substrate to different degrees. Most strikingly, the V480R/V484R mutation impaired myosin binding under all conditions though the total myosin levels in the cells were not affected (Figure 6D, Fig.S6BCD). Inspection of the crystal structure provides a possible explanation for this effect. The two residues Val480 and Val484 are located in close proximity to the opening of the myosin binding canyon (Fig.S6E) and thus inserting two bulky and positively charged arginine residues at this position may impede substrate binding. In contrast to the V480R/V484R mutant, the L121W and ΔTPR mutants bound larger amounts of cellular myosin than the wild-type UNC-45. Because the corresponding TPR mutations are located far remote of the identified myosin binding site (Fig.S6E), an indirect effect should account for this observation. Notably, our in vivo analyses revealed that worms expressing the TPR mutations have only half of the number of A-bands than wild-type worms, however, total myosin levels remained the same (Fig.S6C). We thus presume that a substantial fraction of the cellular myosin is not assembled in thick filaments, as part of the A-bands, and is therefore less stable. Though we could not directly compare levels of aggregated and filamentous myosin (both species contribute to the insoluble fraction of muscle cell lysates), our interpretation is corroborated by recent publications reporting the reduced stability and aggregation of unassembled muscle myosin (Kurapati et al., 2012; Martinsson et al., 2000; Tajsharghi and Oldfors, 2012; Tajsharghi et al., 2005). Accordingly, the TPR mutants appear to retain the capability to interact with the myosin substrate, however in addition pull down unassembled, presumably aggregated myosin molecules.
In sum, the myosin coIP experiments suggest that the V480R/V484R mutation causes a profound side-effect in interfering with substrate binding. Therefore, its strong in vivo phenotype seems to result from two additive effects that include the weakened interaction with myosin on top of the impaired formation of UNC-45 chaperone chains. In contrast, the two TPR mutants retained their ability to interact with myosin and should thus reflect the biological function of UNC-45 multimers more precisely.
Discussion
Although the function of UNC-45 has been extensively studied, mechanistic insight of how UNC-45 promotes the assembly of myosin thick filaments and cooperates with the Hsp70 and Hsp90 chaperones is still elusive (Myhre and Pilgrim, 2012). In contrast to recent structural studies that revealed the folds of the central and UCS domains of yeast She4 (Shi and Blobel, 2010) and Drosophila UNC-45 (Lee et al., 2011), our structural, biochemical, and in vivo data of C. elegans UNC-45 uncover the atomic architecture of a filament assembly factor that itself forms a continuous, polar protein chain. Owing to its role in chaperoning and organizing myosin thick filaments, the UNC-45 multimer exhibits characteristic features that distinguish it from the few other protein filaments—typically cytoskeletal filaments—for which high-resolution structural data are available (Table S3). The UNC-45 oligomer has a relatively small interface that allows the rapid (dis)assembly of multimers comprising two to five subunits. Moreover, the chaperone chain is organized as a bipartite structure with a backbone formed by a series of ARM/TPR∗ superhelices and attached functional protrusions mediating interactions with client proteins and partner chaperones.
Our data suggest that the UNC-45 multimer establishes a multisite docking platform licensing the Hsp70 and Hsp90 chaperones to act in a periodic pattern on the unfolded substrate. This UNC-45 manufacturing system is ideally suited for the assembly of a substrate protein like myosin that itself forms a polar filament. It allows a protein with a single Hsp70/Hsp90 binding motif to simultaneously recruit multiple chaperones and foster their side-by-side collaboration (Figure 7A). Given the different nature of the cooperating chaperones, Hsp70 and Hsp90 may either occupy alternating positions on the UNC-45 chain or bind sequentially to carry out early and late folding steps, respectively. Moreover, the UNC-45 tandem modules offer the proper spacing of about 170 Å to work on dimeric myosin heads that protrude from the coiled-coil backbone of the thick filament with an overall periodicity of 145 Å (Craig and Woodhead, 2006) (Figure 7B). The slightly different spacing of UNC-45 and myosin might be overcome by the transient nature of UNC-45 multimers. According to our model, UNC-45 chains are not infinite in length but are, rather, organized as a series of short assembly lines that scaffold the dimeric myosin heads over short distances along the myofilament. Indeed, formation of stable UNC-45 filaments would cause severe problems in vivo, as UNC-45 is implicated not only in muscle development, but also in maintaining myosin integrity during stress situations (Etard et al., 2008). Stably formed UNC-45 chains would hinder the relocation of the myosin chaperone within sarcomeric substructures and thus limit the functionality of UNC-45 during muscle development and maintenance.
Figure 7.
Model of UNC-45-Promoted Myosin Assembly
(A) UNC-45 composes a myosin folding complex. (Top) UNC-45 (different domains indicated), its oligomers, Hsp70 (blue), and Hsp90 (lilac) represent the basic building blocks for the myosin multichaperone complex. (Bottom) UNC-45 chains compose a molecular scaffold that allows the simultaneous binding of Hsp70, Hsp90, and myosin, thus mediating the interactions between unfolded myosin and its cognate chaperones. Different lengths of the Hsp70/90 C-terminal linkers and resultant activity radii when bound to UNC-45 are indicated.
(B) UNC-45 enforces the folding of myosin in regular spacing. In the relaxed state of the muscle, dimeric myosin heads project with a periodicity of 145 Å from the coiled-coil backbone of the thick filament. The observed periodicity between UNC-45 tandem modules (170 Å) fits well to the observed spacing between protomers of adjacent myosin pairs (110 to 220 Å, derived from PDB code 3dtp), as schematically shown in the enlarged window (myosin in green with orange circles symbolizing equal segments in the asymmetric dimer). Owing to its patterning function, UNC-45 could directly link myosin assembly with myosin folding.
Owing to its unique structural features, we propose that UNC-45 chains support the precise in-register arrangement of myosin head domains, which is a prerequisite for proper thick filament and sarcomere assembly. This previously undescribed molecular patterning function should mainly depend on the multimerization properties of UNC-45 and should thus underlie its autonomous role in sarcomere organization (Gaiser et al., 2011). The biological significance of the polar chaperone chain described here is also supported by the fact that all organisms containing sarcomeric structures express UCS proteins harboring a TPR domain, which is an essential structural component for UNC-45 polymerization. As the residues contributing to the crystallographic filament interface are moderately conserved within this UCS protein family, we presume that UNC-45 chain formation might be a common principle important for myosin assembly and muscle formation. Direct experimental evidence for this function is provided by the dominant-negative effect of expressing UNC-45 interface mutations in wild-type worms. As predicted by our model, the sarcomere assembly defects could result from integrating interface mutants into cellular UNC-45 scaffolds, leading to the premature termination of myosin assembly chains. As the myosin-binding mutants did not exert this effect, substrate binding and chain formation seem to be two separate activities of UNC-45. Indeed, chain formation appears to be a highly dynamic process that depends on the amount of UNC-45 (Figure S5), which should be carefully controlled in the cell. Consistently, mutations in human p97, a ubiquitin-selective ATPase critical for the degradation of UNC45b, are directly linked to inclusion body myopathies (Janiesch et al., 2007). The deleterious effect of elevated UNC-45 levels on sarcomere and thick filament integrity is further evidenced by UNC-45 overexpression studies in worms (Hoppe et al., 2004; Landsverk et al., 2007) and by the fact that human UNC45b is one of the most upregulated proteins in heart muscle tissues of humans who died of ischemic heart failure (Stanley et al., 2007, Circulation, abstract). It will therefore be important to test which cellular factors and modifications influence UNC-45 chain formation and substrate recruitment. The present data provide the structural and mechanistic basis to address these points and get a better understanding of how myosin folding and thick filament assembly are coordinated in the cell under both normal and pathological conditions.
Experimental Procedures
Strains, Expression Constructs, and Generation of Transgenic Animals
The C. elegans Bristol N2 was used as wild-type strain. Mutant worms used in this study are unc-119(ed4)III, unc-45(e286)III, and unc-45(m94)III. The C terminally FLAG-tagged UNC-45 and mutants thereof were cloned under the muscle-specific unc-54 promoter into pPD88.27 containing a selection marker. Subsequently, the constructs were bombarded into unc-119(ed4) worms. Coimmunoprecipitation and motility assays, A band quantification, fluorescence microscopy, and immunostainings were performed following standard procedures, as described in the Extended Experimental Procedures.
Extended Experimental Procedures.
Strains, Expression Constructs, and Generation of Transgenic Animals
Worms were handled according to standard procedures (Brenner, 1974). The C. elegans Bristol N2 was used as wild-type strain. Mutant worms used in this study are unc-119(ed4)III, and unc-45(m94)III. The C-terminally FLAG tagged UNC-45 and ΔTPR (1-131) were constructed by PCR amplification and cloned into pPD88.27, under the muscle specific promoter unc-54, containing the unc-119(+) marker for selection of transgenic worms. Using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies) the following mutations were generated: L121W, V480R/V484R, Y750W, N758Y and Δ602-630. These constructs were bombarded into unc-119(ed4) worms as described previously (Praitis et al., 2001).
Antibody Generation, Fluorescence Microscopy, and Immunostaining
Purified UNC-45 His6-tag-fusion protein was used to generate two anti-UNC-45 polyclonal antisera in rabbits (BioGenes). The antiserum was purified by affinity-chromatography and tested for specificity by immunodetecion of UNC-45 using C. elegans total worm extract and recombinant UNC-45. Fluorescence images were recorded with an Axio-Imager M1 microscope equipped with an AxioCam MRm camera (Carl Zeiss) and processed with the corresponding analysis software (AxioVision 4.7, Carl Zeiss). Immunofluorescence studies were performed as previously described (Ben-Zvi et al., 2009). Worms were stained with anti-MHC A, anti-UNC-45 and anti-UNC-54. Secondary antibodies were DyLight 488 or DyLight 594 (Thermo Fisher Scientific). Animals were mounted in Dapi Fluoromount G medium (SouthernBiotech).
Coimmunoprecipitation Studies
Sonicated lysates from wild-type or mutant worms were resuspended in IP buffer (50 mM Tris pH 8.0, 100 mM NaCl, 0.5 mM EDTA, 4% Glycerol and 1% Triton X-100, 4 mM PMSF and protease inhibitor mix from Roche). For coimmunoprecipitations, worms expressing transgenic, FLAG-tagged UNC-45 were resuspended in lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 2 mM PMSF and protease inhibitor mix; Roche) and incubated with anti-Flag M2 Affinity Gel (Sigma-Aldrich) overnight at 4°C or 2 hr at room temperature. Immunoprecipitates were washed five times in lysis buffer with an increasing NaCl concentration, from 150 to 300 mM, and eluted with SDS–PAGE sample buffer. Western blotting was performed using antibodies against UNC-45, DAF-21, UNC-54 and Tubulin (Sigma). Proteins were detected by immunoblotting using an ECL kit (GE Healthcare).
Motility Assay
Body-bend assays were performed in a M9 buffer at 15°C, 20°C or 25°C. Individual young adult worms grown at indicated temperatures were placed in M9 buffer, and body bends were counted during one minute.
Quantification of A Bands Assembly
Sarcomere assembly was monitored by labeling A-bands with anti-MHC A. The number of A-bands per body-wall muscle cell was counted in the same area, located between the pharynx and vulva.
Cloning, Protein Expression, and Purification
UNC-45, Hsp70 (HSP-1 in C.elegans) and Hsp90 (DAF-21 in C. elegans) were amplified from C. elegans cDNA and cloned into pET21a and pET28a to generate the C-terminally and N-terminally His6-tagged proteins, respectively. The expression vectors for Strep-tagged, ΔTPR(1-134) and Δ602-630 UNC-45 were cloned by using accordingly designed DNA primers, whereas the site-specific amber stop codon at amino acid position 452, the K82E, L121W, V480R/V484R, Y750W and N758Y mutations were introduced with the QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene). Overexpression was carried out in BL21-DE3-RIL (native proteins) and B834-DE3 cells (selenomethionine substituted protein), which were incubated, after induction with 100 μM IPTG, for 12 hr at 18°C (UNC-45 variants) or 5 hr at 25°C (Hsp70, Hsp90). Subsequently, cells were harvested by centrifugation and lysed by sonication conducted in 50 mM Na2HPO4 pH 8.0, 300 mM NaCl (His6-tagged proteins) or 20 mM Tris pH 8.0, 150 mM NaCl (buffer A, Strep-tagged proteins). The tagged proteins were first affinity purified by submitting the cleared bacterial lysates on HisTrap (GE Healthcare) or Streptactin colums (IBA), respectively and by separating bound proteins in a step-wise gradient using either 150 mM imidazole or 2.5 mM d-desthiobiotin in the final elution step. After exchanging the buffer to 20 mM Tris pH 8.0 (buffer B), the pooled fractions were loaded on a Resource Q anion exchange column (GE Healthcare). Target proteins were separated by applying a linear gradient (0 to 500 mM NaCl in buffer B) and pooled according to SDS-PAGE gels. Finally, all proteins were subjected to size-exclusion chromatography using a Superdex 200 column (GE Healthcare) equilibrated with buffer A.
To express pBpa452 variants, UNC-45 plasmids containing the site-specific amber stop codon were co-transformed with the plasmid containing the orthogonal tRNA pair for the incorporation of pBpa (pDULE-pBpa) into BL21-DE3 cells (Chin et al., 2002). Cells were grown in LB medium supplemented with pBpa (Bachem) and the expression induced by 100 μM IPTG for 16 hr at 20°C. The His6-tagged proteins were purified by HisTrap and SEC chromatography as described above using 20 mM Na2HPO4 pH 8.0 and 150 mM NaCl as final SEC buffer.
Crystallization and Data Collection
UNC-45 crystals were grown by the sitting-drop vapor diffusion method at 19°C in 24-well plates. 2 μl protein (200 μM) were mixed with 0.4 μl additive (0.5 M NaF) and 0.5 μl reservoir solution (0.1 M HEPES pH 7.0, 10% PEG 8000, 12% ethylene glycol) yielding hexagonal plate-like crystals after two days. To co-crystallize UNC-45 with the C-terminal Hsp70/Hsp90 peptide, UNC-45 was pre-incubated with the peptide AGGPTIEEVD/EDASRMEEVD (final concentration 500 μM) for 30 min at 19°C before setting up crystallization trials. Mounted in 90°-bent loops, crystals were shortly incubated in the crystallization solution containing 25% ethylene glycol as cryoprotectant and flash-frozen in liquid nitrogen. An anomalous diffraction data set UNC-45SAD and the UNC-45Hsp70 data set (Table S1) were collected at the Swiss Light Source (SLS, beam line X06DA), whereas the UNC-45Hsp90 high resolution diffraction data were collected at the European Synchrotron Radiation Facility (ESRF, beamline ID-23-1). The UNC-45SAD data used for phasing were integrated with DENZO (Otwinowski and Minor, 1997) and scaled with SCALEPACK (Otwinowski and Minor, 1997), whereas the UNC-45Hsp70 and UNC-45Hsp90 data were processed with the XDS software package (Kabsch, 2010). The unit cell of the UNC-45 crystals resembles an extremely long rod having dimensions of a = b = 86 Å, c = 815 Å, α = β = 90°, γ = 120° and a large solvent content of 72%. The crystal parameters are summarized in Table S1.
Structure Determination and Refinement
Though the 107 kDa protein readily crystallized, structure solution was hindered by the large dimension of the crystallographic unit cell having a 815 Å long c-axis. The usage of bent loops to properly orient the plate-like crystals was critical for structure solution. To obtain the experimental phases, we collected SAD data of selenomethionine containing UNC-45 crystals at the f”-peak of the Se-edge. Essential to localize 18 out of 24 selenium sites with SHELXD (Sheldrick, 2008) was the collection of diffraction data with high redundancy. After phasing with SHARP (Bricogne et al., 2003; statistics in Table S1) and solvent flattening with SOLOMON (Abrahams and Leslie, 1996), we obtained an experimental density of excellent quality that revealed the complete peptide backbone of the UNC-45 protomer (1 molecule/asymmetric unit). Despite the rather low resolution of the electron density map (3.8 Å), well-defined bulky residues and the localized methionine residues enabled us to assign about 50% of all side chains. The partial UNC-45 model was then further refined with the UNC-45 data set to 2.9 Å resolution. Refinement and model rebuilding proceeded smoothly via rigid body, positional and later B-factor optimization using the programs CNS (Brünger et al., 1998), PHENIX (Adams et al., 2002 and Jones et al., 1991). In later refinement stages, clear electron density developed for 8 of the 10 residues of the co-crystallized Hsp90 peptide, which were included in the model. Finally, the structure was checked using simulated annealing composite omit maps. Some protein segments including residues 1-4, 508-524, 608–617 and 931-961 were hardly visible in these maps and were therefore omitted from the final model. Owing to the inherent flexibility of the UCS domain the refinement converged at a R-factor of 23.8% (Rfree 25.6%) and had a few UCS residues in the not-allowed region of the Ramachandran plot (Table S1). The UNC-45Hsp70 structure was solved by molecular replacement which was carried out with PHASER (McCoy et al., 2007) using full-length UNC-45 as search model. Refinement of the partial model using CNS (Brünger et al., 1998) yielded an improved electron density map that clearly showed all 10 residues of the co-crystallized Hsp70 peptide. The final structure was refined at 3.6 Å resolution to an R-factor of 23.4% (Rfree 28.4%). Ramachandran statistics were computed with PROCHECK (Laskowski et al., 1993), crystal contacts analyzed with PISA (Krissinel and Henrick, 2007), structural homologs identified with DALI (Holm et al., 2006; Holm and Rosenström, 2010) and molecular illustrations prepared with PyMOL (DeLano, 2002). Sequence alignments were performed with ClustalW (Chenna et al., 2003) and visualized with ESPript (Gouet et al., 1999).
Photocrosslinking
Proteins (100 μM, if not specifically indicated) containing the UV-inducible crosslinking amino acid pBpa were incubated in 20 mM NaH2PO4 pH 8.0, 150 mM NaCl on ice and irradiated with UV light (λ = 365 nm) using a BLAK-Ray UVP, Model B 100 AP UV lamp. After 15 or 30 min, the crosslinked products were separated by analytical SEC (Superose 6 PC 3.2/30 column, GE Healthcare) and SDS-PAGE for subsequent MS analysis. Quantification of the bands was carried out with ImageJ (Abramoff et al., 2004).
Mass Spectrometry and Automated Mapping of Crosslinks
Protein bands were excised and digested in-gel sequentially with trypsin (Promega) and Glu-C (Worthington) as described elsewhere with minor modifications (Wilm et al., 1996). The digested samples were resuspended in 10 μl 0.1% formic acid. For the MS/MS identification of peptide crosslinks, 3 to 5 μl of the digest were injected in an Ultimate 3000 HPLC system (LC Packings Dionex) and the effluent directly electrosprayed into the LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific). “Crossfinder” was used to automatically map crosslinks as described previously (Forne et al., 2012).
Analytical Size Exclusion Chromatography
Analytical SEC to monitor the complex formation of UNC-45 with Hsp70/Hsp90 was performed at 4°C on an Ettan LC system (GE Healthcare). The proteins were incubated at 50 μM for 30 min at 4°C in 20 mM Tris pH 8.0 (supplemented with 1 mM Hsp70 C-terminal peptide for the indicated runs) and subsequently injected on a Superose 6 PC 3.2/30 column equilibrated with the same buffer. For the Hsp70/Hsp90 competition runs 50 μM UNC-45 was incubated with 100 μM Hsp70 and Hsp90.
Isothermal Titration Calorimetry
The binding of UNC-45 to full-length Hsp90/DAF-21 and synthesized oligopeptides (EDASRMEEVD, Hsp90/DAF-21 C terminus; AGGPTIEEVD, Hsp70/HSP-1 C terminus) were characterized by ITC (VP-ITC, Microcal). All experiments were conducted in overflow mode at 20°C with proteins and ligands equilibrated in buffer A. 1.8 ml protein solution was placed in the temperature-controlled sample cell and titrated with the ligand loaded in the 300 μl mixing syringe. After an initial injection of 2 μl, injections of 10 μl ligand were dispensed into the sample cell using a 120 s equilibration time between injections with a total of 29 injections. Control experiments were carried out in order to measure and correct the heat of dilution upon buffer addition. Data were plotted and analyzed using a single site binding model with the MicroCal Origin software. All peptides were synthesized in-house on a Syro Peptide Synthesizer (Multisyntech) and HPLC purified.
Cloning, Protein Expression, and Purification
Detailed information on the construct design, cloning, and overexpression of UNC-45, UNC-45 variants, Hsp70, and Hsp90 is provided in the Extended Experimental Procedures. All proteins were purified by a three-step procedure combining affinity, ion exchange, and size exclusion chromatography.
Crystallization and Data Collection
UNC-45 crystals were grown by the sitting-drop vapor diffusion method (for details, see Extended Experimental Procedures). Mounted in 90° bent loops, anomalous diffraction data (UNC-45SAD) and data of the UNC-45Hsp70 complex were collected at the Swiss Light Source (SLS, beam line X06DA) and the high-resolution diffraction data of UNC-45Hsp90 at the European Synchrotron Radiation Facility (ESRF, beamline ID-23-1).
Structure Determination and Refinement
Usage of bent loops to properly orient the plate-like crystals (815 Å long crystallographic c axis) and collection of anomalous data with high redundancy were critical to solve the structure of CeUNC-45. The solvent-flattened SAD electron density map was of excellent quality and revealed the complete peptide backbone of the protomer. Despite the rather low resolution of the electron density map (3.8 Å), well-defined bulky residues and the localized methionine residues enabled us to assign ∼50% of all side chains. The partial UNC-45 model was further refined with the UNC-45Hsp90 data set to 2.9 Å resolution. Refinement and model rebuilding proceeded smoothly via rigid body, positional, and later B factor optimization. In later refinement stages, clear electron density developed for 8 of the 10 residues of the cocrystallized Hsp90 peptide. Finally, the structure was checked using simulated annealing composite omit maps. Some residues (1–4, 508–524, 608–617, and 931–961) were hardly visible in these maps and were therefore omitted from the final model. The refinement converged at an R factor of 23.8% (Rfree 25.6%). The UNC-45Hsp70 structure was solved by molecular replacement using full-length UNC-45 as search model. After rebuilding and refinement of the UNC-45 protomer, the omit density occurring for the Hsp70 peptide allowed the modeling of all 10 residues. The final structure was refined at 3.6 Å resolution to an R factor of 23.4% (Rfree 28.4%).
Photocrosslinking
After 15 or 30 min irradiation with UV light (λ = 365 nm), the crosslinked products were separated by analytical SEC and SDS-PAGE for subsequent MS analysis.
Mass Spectrometry and Automated Mapping of Crosslinks
Protein bands were excised and digested in-gel sequentially with trypsin (Promega) and Glu-C (Worthington), as described elsewhere with minor modifications (Wilm et al., 1996). For the MS/MS identification of peptide crosslinks, digests were injected in an Ultimate 3000 HPLC system (LC Packings Dionex) and the effluent directly electrosprayed into the LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific). “Crossfinder” was used to automatically map crosslinks, as described previously (Forne et al., 2012).
Analytical Size Exclusion Chromatography
Analytical size exclusion chromatography to monitor the complex formation of UNC-45 with Hsp70/Hsp90 was performed on an Ettan LC system (GE Healthcare). After 30 min of preincubation, protein mixtures were applied to a Superose 6 PC 3.2/30 column.
Isothermal Titration Calorimetry
The binding of UNC-45 to full-length Hsp90 and C-terminal peptides of Hsp70 and Hsp90 (AGGPTIEEVD and EDASRMEEVD, respectively) was characterized by isothermal titration calorimetry (VP-ITC, Microcal) applying standard procedures.
Acknowledgments
We are grateful to the beamline staff at the SLS and ESRF, in particular to Andrew McCarthy and Max Nanao, for their assistance during data collection; Mathias Madalinski for peptide synthesis; Axel Imhof and Peter Becker for providing access to the mass spectrometry infrastructure; the Caenorhabditis Genetics Center (funded by the NIH National Center for Research Resources) for strains; Richard I. Morimoto and Patricija van Oosten-Hawle for providing the Hsp90 antibody; and Peter G. Schultz for providing the pDULE-pBpa plasmid. With regards to author contributions, L.G. and T.C. performed the structural studies; L.G. and D.H. performed the protein biochemistry; T.L. prepared the UNC-45 antibody; W.P. performed C. elegans experiments; and I.F. and F.M.-P. performed MS analysis. T.C. outlined the structural and mechanistic part of the work and T.H. the C. elegans part. All authors contributed to writing the manuscript. This work was supported by grants from the European Community Network of Excellence RUBICON (LSHC-CT-2005-018683 to T.H.), the Deutsche Forschungsgemeinschaft (CECAD, FOR885, SFB635, HO2541/1-1, and HO2541/4-1 to T.H.), and the Austrian Science Fund (FWF P22570-B09 to D.H. and L.G.). The IMP is funded by Boehringer Ingelheim.
Published: January 17, 2013
Footnotes
Supplemental Information includes Extended Results, Extended Experimental Procedures, six figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2012.12.025.
Contributor Information
Thorsten Hoppe, Email: thorsten.hoppe@uni-koeln.de.
Tim Clausen, Email: clausen@imp.univie.ac.at.
Accession Numbers
Coordinates have been deposited to the Protein Data Bank under accession codes 4i2z (UNC-45Hsp90) and 4i2w (UNC-45Hsp70).
Supplemental Information
References
- Ao W., Pilgrim D. Caenorhabditis elegans UNC-45 is a component of muscle thick filaments and colocalizes with myosin heavy chain B, but not myosin heavy chain A. J. Cell Biol. 2000;148:375–384. doi: 10.1083/jcb.148.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral J.M., Bauer C.C., Ortiz I., Epstein H.F. Unc-45 mutations in Caenorhabditis elegans implicate a CRO1/She4p-like domain in myosin assembly. J. Cell Biol. 1998;143:1215–1225. doi: 10.1083/jcb.143.5.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral J.M., Hutagalung A.H., Brinker A., Hartl F.U., Epstein H.F. Role of the myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science. 2002;295:669–671. doi: 10.1126/science.1066648. [DOI] [PubMed] [Google Scholar]
- Bernick E.P., Zhang P.J., Du S. Knockdown and overexpression of Unc-45b result in defective myofibril organization in skeletal muscles of zebrafish embryos. BMC Cell Biol. 2010;11:70. doi: 10.1186/1471-2121-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R., Woodhead J.L. Structure and function of myosin filaments. Curr. Opin. Struct. Biol. 2006;16:204–212. doi: 10.1016/j.sbi.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Du S.J., Li H., Bian Y., Zhong Y. Heat-shock protein 90alpha1 is required for organized myofibril assembly in skeletal muscles of zebrafish embryos. Proc. Natl. Acad. Sci. USA. 2008;105:554–559. doi: 10.1073/pnas.0707330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein H.F., Thomson J.N. Temperature-sensitive mutation affecting myofilament assembly in Caenorhabditis elegans. Nature. 1974;250:579–580. doi: 10.1038/250579a0. [DOI] [PubMed] [Google Scholar]
- Etard C., Behra M., Fischer N., Hutcheson D., Geisler R., Strähle U. The UCS factor Steif/Unc-45b interacts with the heat shock protein Hsp90a during myofibrillogenesis. Dev. Biol. 2007;308:133–143. doi: 10.1016/j.ydbio.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Etard C., Roostalu U., Strähle U. Shuttling of the chaperones Unc45b and Hsp90a between the A band and the Z line of the myofibril. J. Cell Biol. 2008;180:1163–1175. doi: 10.1083/jcb.200709128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forne I., Ludwigsen J., Imhof A., Becker P.B., Mueller-Planitz F. Probing the conformation of the ISWI ATPase domain with genetically encoded photoreactive crosslinkers and mass spectrometry. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.012088. M111.012088. Published online December 13, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiser A.M., Kaiser C.J., Haslbeck V., Richter K. Downregulation of the Hsp90 system causes defects in muscle cells of Caenorhabditis elegans. PLoS ONE. 2011;6:e25485. doi: 10.1371/journal.pone.0025485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautel M. The sarcomeric cytoskeleton: who picks up the strain? Curr. Opin. Cell Biol. 2011;23:39–46. doi: 10.1016/j.ceb.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Geach T.J., Zimmerman L.B. Paralysis and delayed Z-disc formation in the Xenopus tropicalis unc45b mutant dicky ticker. BMC Dev. Biol. 2010;10:75. doi: 10.1186/1471-213X-10-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins T.A., Haramis A.P., Etard C., Prodromou C., Vaughan C.K., Ashworth R., Ray S., Behra M., Holder N., Talbot W.S. The ATPase-dependent chaperoning activity of Hsp90a regulates thick filament formation and integration during skeletal muscle myofibrillogenesis. Development. 2008;135:1147–1156. doi: 10.1242/dev.018150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe T., Cassata G., Barral J.M., Springer W., Hutagalung A.H., Epstein H.F., Baumeister R. Regulation of the myosin-directed chaperone UNC-45 by a novel E3/E4-multiubiquitylation complex in C. elegans. Cell. 2004;118:337–349. doi: 10.1016/j.cell.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Hutagalung A.H., Landsverk M.L., Price M.G., Epstein H.F. The UCS family of myosin chaperones. J. Cell Sci. 2002;115:3983–3990. doi: 10.1242/jcs.00107. [DOI] [PubMed] [Google Scholar]
- Janiesch P.C., Kim J., Mouysset J., Barikbin R., Lochmüller H., Cassata G., Krause S., Hoppe T. The ubiquitin-selective chaperone CDC-48/p97 links myosin assembly to human myopathy. Nat. Cell Biol. 2007;9:379–390. doi: 10.1038/ncb1554. [DOI] [PubMed] [Google Scholar]
- Kachur T.M., Pilgrim D.B. Myosin assembly, maintenance and degradation in muscle: Role of the chaperone UNC-45 in myosin thick filament dynamics. Int. J. Mol. Sci. 2008;9:1863–1875. doi: 10.3390/ijms9091863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachur T., Ao W., Berger J., Pilgrim D. Maternal UNC-45 is involved in cytokinesis and colocalizes with non-muscle myosin in the early Caenorhabditis elegans embryo. J. Cell Sci. 2004;117:5313–5321. doi: 10.1242/jcs.01389. [DOI] [PubMed] [Google Scholar]
- Landsverk M.L., Li S., Hutagalung A.H., Najafov A., Hoppe T., Barral J.M., Epstein H.F. The UNC-45 chaperone mediates sarcomere assembly through myosin degradation in Caenorhabditis elegans. J. Cell Biol. 2007;177:205–210. doi: 10.1083/jcb.200607084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.F., Hauenstein A.V., Fleming J.K., Gasper W.C., Engelke V., Sankaran B., Bernstein S.I., Huxford T. X-ray crystal structure of the UCS domain-containing UNC-45 myosin chaperone from Drosophila melanogaster. Structure. 2011;19:397–408. doi: 10.1016/j.str.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord M., Pollard T.D. UCS protein Rng3p activates actin filament gliding by fission yeast myosin-II. J. Cell Biol. 2004;167:315–325. doi: 10.1083/jcb.200404045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkani G.C., Lee C.F., Cammarato A., Bernstein S.I. Drosophila UNC-45 prevents heat-induced aggregation of skeletal muscle myosin and facilitates refolding of citrate synthase. Biochem. Biophys. Res. Commun. 2010;396:317–322. doi: 10.1016/j.bbrc.2010.04.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkani G.C., Bodmer R., Ocorr K., Bernstein S.I. The UNC-45 chaperone is critical for establishing myosin-based myofibrillar organization and cardiac contractility in the Drosophila heart model. PLoS ONE. 2011;6:e22579. doi: 10.1371/journal.pone.0022579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myhre J.L., Pilgrim D.B. At the Start of the Sarcomere: A Previously Unrecognized Role for Myosin Chaperones and Associated Proteins during Early Myofibrillogenesis. Biochem. Res. Int. 2012;2012:712315. doi: 10.1155/2012/712315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M.G., Landsverk M.L., Barral J.M., Epstein H.F. Two mammalian UNC-45 isoforms are related to distinct cytoskeletal and muscle-specific functions. J. Cell Sci. 2002;115:4013–4023. doi: 10.1242/jcs.00108. [DOI] [PubMed] [Google Scholar]
- Sanger J.W., Kang S., Siebrands C.C., Freeman N., Du A., Wang J., Stout A.L., Sanger J.M. How to build a myofibril. J. Muscle Res. Cell Motil. 2005;26:343–354. doi: 10.1007/s10974-005-9016-7. [DOI] [PubMed] [Google Scholar]
- Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F.U., Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- Shi H., Blobel G. UNC-45/CRO1/She4p (UCS) protein forms elongated dimer and joins two myosin heads near their actin binding region. Proc. Natl. Acad. Sci. USA. 2010;107:21382–21387. doi: 10.1073/pnas.1013038107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikakulam R., Winkelmann D.A. Chaperone-mediated folding and assembly of myosin in striated muscle. J. Cell Sci. 2004;117:641–652. doi: 10.1242/jcs.00899. [DOI] [PubMed] [Google Scholar]
- Srikakulam R., Liu L., Winkelmann D.A. Unc45b forms a cytosolic complex with Hsp90 and targets the unfolded myosin motor domain. PLoS ONE. 2008;3:e2137. doi: 10.1371/journal.pone.0002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley B.A., Shaw J., Arab S., Liu P., Kirshenbaum L.A., Van Eyk J.E. UNC-45B exhibits increased abundance in heart failure patients and functions as a novel dual regulator of myosin heavy chain transcription and assembly. Circulation. 2007;116:170. 170. [Google Scholar]
- Sweeney H.L., Houdusse A. Structural and functional insights into the Myosin motor mechanism. Annu. Rev. Biophys. 2010;39:539–557. doi: 10.1146/annurev.biophys.050708.133751. [DOI] [PubMed] [Google Scholar]
- Toi H., Fujimura-Kamada K., Irie K., Takai Y., Todo S., Tanaka K. She4p/Dim1p interacts with the motor domain of unconventional myosins in the budding yeast, Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:2237–2249. doi: 10.1091/mbc.E02-09-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venolia L., Waterston R.H. The unc-45 gene of Caenorhabditis elegans is an essential muscle-affecting gene with maternal expression. Genetics. 1990;126:345–353. doi: 10.1093/genetics/126.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venolia L., Ao W., Kim S., Kim C., Pilgrim D. unc-45 gene of Caenorhabditis elegans encodes a muscle-specific tetratricopeptide repeat-containing protein. Cell Motil. Cytoskeleton. 1999;42:163–177. doi: 10.1002/(SICI)1097-0169(1999)42:3<163::AID-CM1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Walker M.G. Pharmaceutical target identification by gene expression analysis. Mini Rev. Med. Chem. 2001;1:197–205. doi: 10.2174/1389557013407034. [DOI] [PubMed] [Google Scholar]
- Wesche S., Arnold M., Jansen R.P. The UCS domain protein She4p binds to myosin motor domains and is essential for class I and class V myosin function. Curr. Biol. 2003;13:715–724. doi: 10.1016/s0960-9822(03)00264-1. [DOI] [PubMed] [Google Scholar]
- Wilm M., Shevchenko A., Houthaeve T., Breit S., Schweigerer L., Fotsis T., Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- Wohlgemuth S.L., Crawford B.D., Pilgrim D.B. The myosin co-chaperone UNC-45 is required for skeletal and cardiac muscle function in zebrafish. Dev. Biol. 2007;303:483–492. doi: 10.1016/j.ydbio.2006.11.027. [DOI] [PubMed] [Google Scholar]
- Young J.C., Barral J.M., Ulrich Hartl F. More than folding: localized functions of cytosolic chaperones. Trends Biochem. Sci. 2003;28:541–547. doi: 10.1016/j.tibs.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Yu Q., Bernstein S.I. UCS proteins: managing the myosin motor. Curr. Biol. 2003;13:R525–R527. doi: 10.1016/s0960-9822(03)00447-0. [DOI] [PubMed] [Google Scholar]
Supplemental References
- Abrahams, J.P., and Leslie, A.G. (1996). Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D Biol. Crystallogr. 52, 30–42. [DOI] [PubMed]
- Abramoff, M.D., Magalhães, P.J., and Ram, S.J. (2004). Image processing with ImageJ. Biophotonics Intl. 11, 36–42.
- Adams, P.D., Grosse-Kunstleve, R.W., Hung, L.W., Ioerger, T.R., McCoy, A.J., Moriarty, N.W., Read, R.J., Sacchettini, J.C., Sauter, N.K., and Terwilliger, T.C. (2002). PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954. [DOI] [PubMed]
- Ben-Zvi, A., Miller, E.A., and Morimoto, R.I. (2009). Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc. Natl. Acad. Sci. USA 106, 14914–14919. [DOI] [PMC free article] [PubMed]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed]
- Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M., and Paciorek, W. (2003). Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D Biol. Crystallogr. 59, 2023–2030. [DOI] [PubMed]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. (1998). Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921. [DOI] [PubMed]
- Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T.J., Higgins, D.G., and Thompson, J.D. (2003). Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500. [DOI] [PMC free article] [PubMed]
- Chin, J.W., Martin, A.B., King, D.S., Wang, L., and Schultz, P.G. (2002). Addition of a photocrosslinking amino acid to the genetic code of Escherichiacoli. Proc. Natl. Acad. Sci. USA 99, 11020–11024. [DOI] [PMC free article] [PubMed]
- Conti, E., Uy, M., Leighton, L., Blobel, G., and Kuriyan, J. (1998). Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 94, 193–204. [DOI] [PubMed]
- DeLano, W.L. (2002). The Pymol Molecular Graphics System (San Carlos, CA: DeLano Scientific).
- Gigant, B., Curmi, P.A., Martin-Barbey, C., Charbaut, E., Lachkar, S., Lebeau, L., Siavoshian, S., Sobel, A., and Knossow, M. (2000). The 4 A X-ray structure of a tubulin:stathmin-like domain complex. Cell 102, 809–816. [DOI] [PubMed]
- Gouet, P., Courcelle, E., Stuart, D.I., and Métoz, F. (1999). ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308. [DOI] [PubMed]
- Guarné, A., Brendler, T., Zhao, Q., Ghirlando, R., Austin, S., and Yang, W. (2005). Crystal structure of a SeqA-N filament: implications for DNA replication and chromosome organization. EMBO J. 24, 1502–1511. [DOI] [PMC free article] [PubMed]
- Holm, L., and Rosenström, P. (2010). Dali server: conservation mapping in 3D. Nucleic Acids Res. 38(Web Server issue), W545–W549. [DOI] [PMC free article] [PubMed]
- Holm, L., Kääriäinen, S., Wilton, C., and Plewczynski, D. (2006). Using Dali for structural comparison of proteins. Curr. Protoc. Bioinformatics Chapter 5, Unit 5.5. [DOI] [PubMed]
- Huber, A.H., and Weis, W.I. (2001). The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell 105, 391–402. [DOI] [PubMed]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. (1991). Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119. [DOI] [PubMed]
- Kabsch, W. (2010). Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132. [DOI] [PMC free article] [PubMed]
- Krissinel, E., and Henrick, K. (2007). Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Kurapati, R., McKenna, C., Lindqvist, J., Williams, D., Simon, M., LeProust, E., Baker, J., Cheeseman, M., Carroll, N., Denny, P., et al. (2012). Myofibrillar myopathy caused by a mutation in the motor domain of mouse MyHC IIb. Hum. Mol. Genet. 21, 1706–1724. [DOI] [PubMed]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. (1993). PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291.
- Martinsson, T., Oldfors, A., Darin, N., Berg, K., Tajsharghi, H., Kyllerman, M., and Wahlstrom, J. (2000). Autosomal dominant myopathy: missense mutation (Glu-706 —> Lys) in the myosin heavy chain IIa gene. Proc. Natl. Acad. Sci. USA 97, 14614–14619. [DOI] [PMC free article] [PubMed]
- McCoy, A.J., Grosse-Kunstleve, R.W., Adams, P.D., Winn, M.D., Storoni, L.C., and Read, R.J. (2007). Phaser crystallographic software. J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Narita, A., Oda, T., and Maéda, Y. (2011). Structural basis for the slow dynamics of the actin filament pointed end. EMBO J. 30, 1230–1237. [DOI] [PMC free article] [PubMed]
- Oliva, M.A., Cordell, S.C., and Löwe, J. (2004). Structural insights into FtsZ protofilament formation. Nat. Struct. Mol. Biol. 11, 1243–1250. [DOI] [PubMed]
- Otwinowski, Z., and Minor, W. (1997). Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology: Macromolecular Crystallography, part A 276, 307–326. [DOI] [PubMed]
- Praitis, V., Casey, E., Collar, D., and Austin, J. (2001). Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157, 1217–1226. [DOI] [PMC free article] [PubMed]
- Sheldrick, G.M. (2008). A short history of SHELX. Acta Crystallogr. A 64, 112–122. [DOI] [PubMed]
- Sirajuddin, M., Farkasovsky, M., Hauer, F., Kühlmann, D., Macara, I.G., Weyand, M., Stark, H., and Wittinghofer, A. (2007). Structural insight into filament formation by mammalian septins. Nature 449, 311–315. [DOI] [PubMed]
- Tajsharghi, H., and Oldfors, A. (2012). Myosinopathies: pathology and mechanisms. Acta Neuropathol. Published online August 5, 2012. http://dx.doi.org/10.1007/s00401-012-1024-2. [DOI] [PMC free article] [PubMed]
- Tajsharghi, H., Pilon, M., and Oldfors, A. (2005). A Caenorhabditis elegans model of the myosin heavy chain IIa E706K [corrected] mutation. Ann. Neurol. 58, 442–448. [DOI] [PubMed]
- van den Ent, F., Amos, L.A., and Löwe, J. (2001). Prokaryotic origin of the actin cytoskeleton. Nature 413, 39–44. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.