Abstract
In higher eukaryotes most genes contain multiple introns. Introns are excised from pre-mRNAs by splicing and eventually degraded in the nucleus. It is likely that rapid intron turnover in the nucleus is important in higher eukaryotes, but this pathway is poorly understood. In order to gain insights into this pathway, we analyzed the human lariat RNA debranching enzyme1 (hDbr1) protein that catalyzes debranching of lariat-intron RNAs. Transfection experiments demonstrate that hDbr1 is localized in a nucleoplasm of HeLa cells through a bipartite type nuclear localization signal near carboxyl-terminus. The conserved GNHE motif, originally identified in protein phosphatase protein family, is critical for hDbr1 to dissolve lariat structure in vitro. Furthermore, heterokaryon experiments show that hDbr1 is a nucleocytoplasmic shuttling protein, suggesting novel role(s) of hDbr1 in the cytoplasm.
In higher eukaryotes splicing of precursor of mRNA (pre-mRNA) is a critical step for gene expression, since most genes encoded in the nucleus contain introns1,2,3. Splicing comprises two catalytic steps. As the first step, the cleavage at the 5′ splice site occurs and concurrently the 5′ end of the intron is joined to the branch nucleotide by forming a 2′–5′ phosphodiester bond. This results in the production of 5′ exon and a lariat intermediate RNA that consists of a lariat-form intron and 3′ exon. These intermediates are subject to the second step reaction in which the 3′ splice site is cleaved and two exons are ligated to produce mRNA. The excised lariat introns are supposed to be retained and degraded in the nucleus.
Splicing takes place in a large protein-RNA complex, termed spliceosome1,4. More than a hundred of protein and RNA factors were identified as components of spliceosome4. In contrast, the intron turnover pathway after splicing remained largely unclear. Only several factors for the intron turnover pathway in the nucleus have been identified almost exclusively by analyses of budding yeast mutants. Upon the completion of the splicing reaction, mRNA is released from the spliceosome, and the remaining post-splicing RNPs containing the lariat introns are disassembled by two members of the ATP-dependent DExH box RNA helicase family, Prp22 and Prp435,6. Prp22 is required for the release of the spliced RNA5, while Prp43 is necessary for the disassembly of the remaining intron-containing RNP7,8. It has been also shown that two splicing factors, Ntr1 and Ntr2, in the form of a hetero dimmer, associate with Prp43. The resultant trimeric complex is called as the ‘NTR complex’9,10. The NTR complex is functional in catalyzing the disassembly of the spliceosome9. After dissociation of U snRNPs and other splicing factors, the lariat intron is linearized by a lariat intron debranching enzyme, Dbr1, before being degraded11.
The RNA-lariat debranching enzyme 1 protein, Dbr1, specifically hydrolyzes the 2′–5′ phosphodiester bond in the intron, which converts lariat form introns into linear molecules12. The linearized intron is likely to be degraded by either XrnI, a 5′-3′ exonuclease and/or the exosome that has a 3′-5′ exonuclease activity11,13,14. Dbr1 was originally identified and cloned from budding yeast (Saccharomyces cerevisiae) by a genetic screening to identify host cellular factors involved in retrotransposon Ty1 element transposition15. The Dbr1 mutant of S. cerevisiae has reduced Ty1 transposition frequency and shows the high level accumulation of lariat introns in the cell15. This result indicates that debranching has to occur prior to intron degradation. Thus debranching is one of the key steps for intron turnover.
Dbr1 gene is not essential for cell viability in Saccharomyces cerevisiae, although Dbr1 mutant shows accumulation of lariat RNAs15. In fission yeast, Schizosaccharomyces pombe, Dbr1 is also unessential for its viability16. However, Dbr1 null mutant cells of S.pombe show severe growth defect and elongated cell shape in addition to the accumulation of RNA lariat16. One possible explanation for the difference of phenotypes between S. cerevisiae and S. pombe is the percentage of genes that contain introns. Only about 2.5% of genes contain introns in S. cerevisiae, while 40% in S. pombe17,18,19. The cDNAs of Dbr1 have been isolated from many species, such as Caenorhabditis elegans, mouse and human16,20,21. These homologues can complement S.pombe Dbr1 null mutant, indicating that the function and sequence of Dbr1 is conserved among many species. The Dbr1 cDNA was also isolated from a plant, Arabidopsis thaliana, and A. thaliana Dbr1 mutant was turned out to be embryonic lethal22. Taken together, it is likely that rapid intron turnover including debranching is important for higher eukaryotes that contain many introns.
In human, almost all genes encoded in the nucleus are separated by multiple introns that occupy, in sum, about 95% of the primary transcripts. It is therefore highly expected that the pathway for rapid intron turnover in the nucleus is critical in human. Although the homologs of the factors described above in yeast have been identified in mammals16,20,21,23,24,25,26, this pathway was not well understood. We have been analyzing this pathway in human by using in vitro splicing assaysystem. We found that introns are degraded through the formation of two complexes, IL (Intron Large) and IS (Intron Small) complexes27. IL complex is a 40S complex, and it contains U2, U5 and U6 snRNPs and hPrp19 complex proteins, while IS complex, which is a 20 S form, does not contain those snRNPs and protein factors27. We have also identified the TFIP11, a human homolog of yeast Ntr1 protein, in the IL complex and demonstrated that TFIP11 recruits the hPrp43 protein to the IL complex through interactions mediated by its N-terminal G-patch region, which is required for the transition from IL complex to IS complex27. Furthermore, we also showed that hDbr1 is accessible to IS complex, but not to IL complex27, suggesting that hDbr1 is involved in disassembly of IS complex prior to degradation of linear introns. To get further insight into the intron turnover mechanism, we decided to analyze human Dbr1 (hDbr1) protein. The hDbr1 cDNA was previously identified in the human Expressed Sequence Tag (EST) database and isolated by RT-PCR20. It was demonstrated that hDbr1 was functional in interspecies complementation experiments and that the corresponding recombinant hDbr1 protein had a debranching activity in vitro20. However the precise structural domain analysis has not been performed. Here we report the functional domain analysis of hDbr1. The hDbr1 protein is localized in the nucleoplasm of HeLa cells through a bipartite classical type nuclear localization signal (NLS) near its carboxyl-terminus. The protein phosphatase 1 like motif, point mutations of which were shown to inactivate yeast Dbr1p in vitro and in vivo28,29, is also critical for debranching activity of hDbr1 in vitro. Furthermore, in spite of its nuclear localization at steady state, hDbr1 protein has a capacity to shuttle between the nucleus and the cytoplasm. This result strongly suggests that hDbr1 has novel function(s) in the cytoplasm.
Results
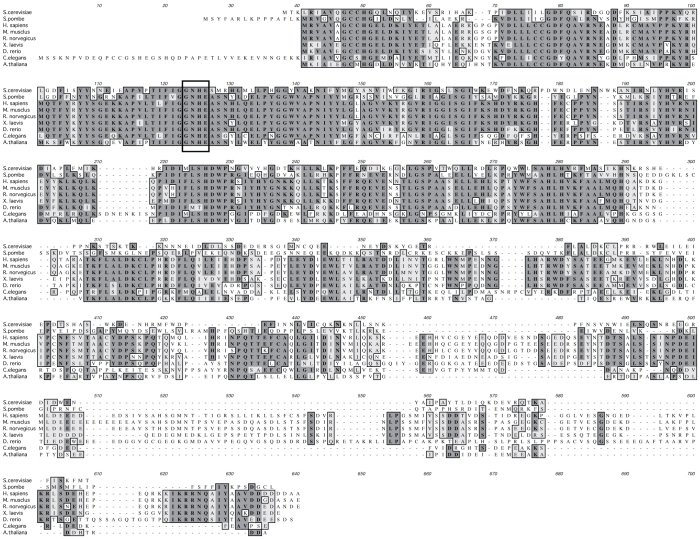
The homologues of Dbr1 protein from many species were isolated16,20,21,22 and the amino acid sequence of this protein is well conserved (Figure 1). The amino terminus of Dbr1 protein especially shows high similarity, while the carboxyl terminus is relatively diverse (Figure 1). In order to analyze the function of human Dbr1 protein, we first determined the subcellular localization of human Dbr1 protein by transfection experiments. The full-length hDbr1 protein, fused to either myc or Flag tag was transiently expressed in HeLa cells. The immunofluorescence experiments shown in Figure 2 revealed that both myc- and Flag- tagged hDbr1 proteins were localized in the nucleoplasm of HeLa cells (Figure 2, panel FITC). This localization is consistent with the function of hDbr1, because hDbr1 catalyzes debranching reaction of lariat introns, which is supposed to occur in the nucleus.
Figure 1. Amino acid sequence alignment of Dbr1 from various organisms.
Dbr1 proteins from Saccharomyces cerevisiae (NP_012773.1), Schizosaccharomyces pombe (NP_593470.2), Homo sapiens (NP_057300.2), Mus musclus (NP_113580.2), Rattus norvegicus (NP_001102907.1), Xenopus laevis (NP_001080368.1), Danio rerio (NP_955947.1), Caenorhabditis elegans (NP_491868.2) and Arabidopsis thaliana (NP_567881.1) were aligned using ClustalW alignment program (http://www.genome.jp/tools/clustalw/). Identical residues are indicated by dark shading and similar residues are shown by light shading. An open box indicates the position of a conserved GNHE motif that is also found in Protein Phosphatase 1 (PP1) as a catalytic center.
Figure 2. hDbr1 localizes to the nucleoplasm by immunofluorescence.
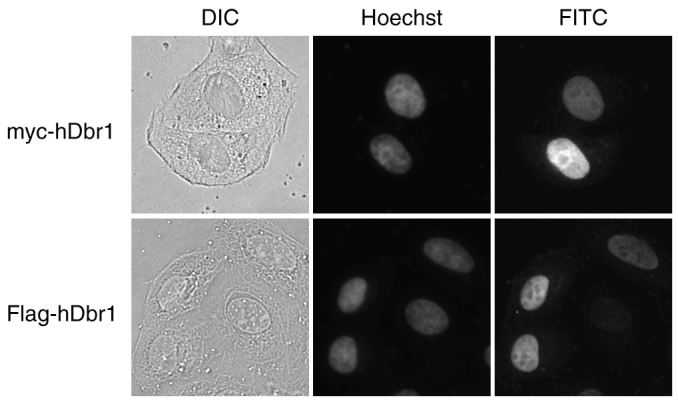
Expression vectors encoding either myc-tagged (upper panels) or Flag-tagged hDbr1 (lower panels) were transfected into HeLa cells. Twenty-four hours after transfection, the cells were stained with either anti-myc (MC045, Nacalai Tesque, Japan, panel FITC, upper right) or anti-Flag (M2, SIGMA, panel FITC, lower right). The cells were also stained with Hoechst 33342 (SIGMA, panel Hoechst, middle) to label the nuclei. Differential interference contrast (DIC) images of the cells are also shown as DIC panels at the left.
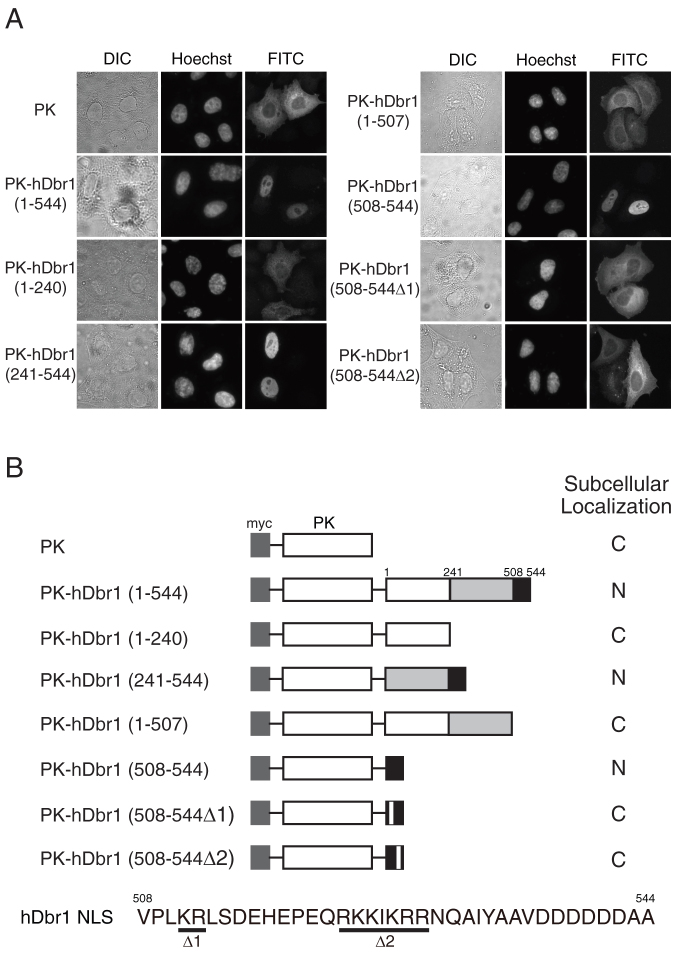
The nuclear localization of exogenously expressed hDbr1 protein predicted that hDbr1 protein has a nuclear localization signal (NLS). In order to identify the hDbr1 NLS, we fused several domains of hDbr1 to the reporter protein and tested the subcellular localization of the corresponding fusion proteins in HeLa cells. As a reporter protein, we used the myc-tagged chicken Pyruvate Kinase (myc-PK), which is exclusively localized in the cytoplasm (Figure 3A)30,31. If the domain from hDbr1 has a NLS activity, the fusion protein should be localized to the nucleus. When the full-length hDbr1 was fused to myc-PK, the chimeric protein was localized in the nucleus (Figure 3A, PK-hDbr1(1-544)). We first separated hDbr1 into two parts and fused to myc-PK. We found that the fusion of N-terminal 240 aa of hDbr1 to PK (PK-hDbr1(1-240)) produced cytoplasmic protein, while PK fused to the C-terminal 304 aa of hDbr1 protein (PK-hDbr1(241-544)) showed exclusively nuclear staining (Figure 3A). These results indicate that C-terminal region of hDbr1 confers its nuclear localization. We further deleted C-terminal region of hDbr1 and found that the fusion protein of C-terminal 37 aa of hDbr1 to PK (PK-hDbr1(508-544)) is localized in the nucleus (Figure 3A). Since PK fused to the the rest of hDbr1 showed exclusive cytoplasmic staining (Figure 3A, PK-hDbr1(1-507)), it was strongly suggested that hDbr1 has one NLS in its C-terminal 37 aa. region.
Figure 3. Identification of the nuclear localization signal (NLS) of hDbr1.
A) HeLa cells were transfected with expression vectors encoding myc-tagged PK fusion proteins. The numbers above each panel correspond to the amino acids within hDbr1 that were fused to PK. Δ1 and Δ2 indicate that the corresponding regions shown at the bottom of B by underlines were deleted. The subcellular localization of each protein was determined by indirect immunofluorescence using anti-myc monoclonal antibody, MC045. DIC images and Hoechst staining pattern of the cells are also presented as DIC and Hoechst, respectively. B) Schematic drawing of the PK fusion proteins and summary of their intracellular localization as displayed in (A). Also the bipartite NLS of hDbr1 is shown at the bottom marked with underlines. The Δ1 mutant has a deletion of amino-acid residues, KR, whereas the Δ2 mutant contains a deletion of RKKIKRR residues.
In the last 37 amino acids of hDbr1, we could find two basic residue stretches that were good candidates for classical type nuclear localization signal (Figure 3B). To test whether these regions are critical for nuclear localization or not, we expressed the mutant proteins that had deletion in either the first or the second basic stretch from myc-tagged PK with 37 amino acids of hDbr1 protein (PK-hDbr1(508–544Δ1) and PK-hDbr1(508–544Δ2), Figure 3B). As shown in Figure 3, neither of the deletion mutants were localized in the nucleus. The deletion of the last 37 amino acids from full-length hDbr1 protein fused to myc-PK resulted in the cytoplasmic localization (Figure 3A). These results indicate that hDbr1 has a bipartite type NLS that is located in the last 37 amino acid region of hDbr1.
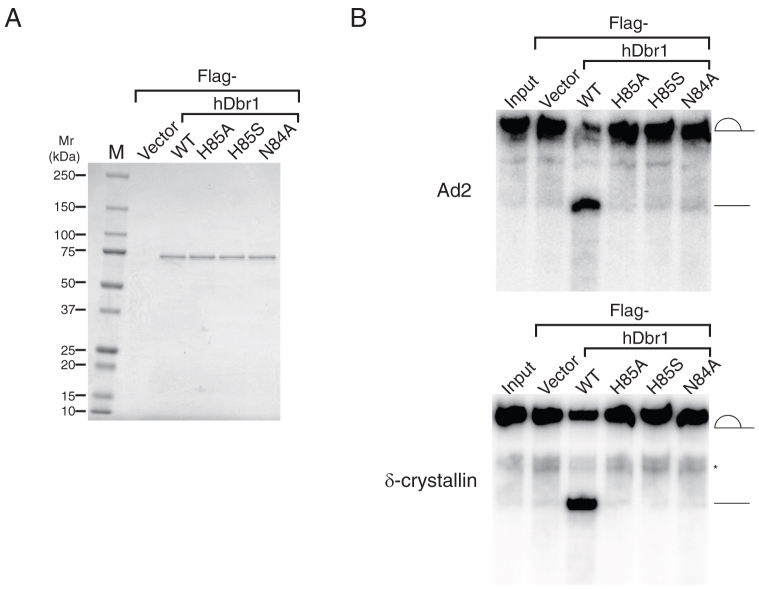
Dbr1 protein has a GNHE motif, which is one of the signatures of metallophosphoesterase superfamily protein23,32. This protein superfamily consists of λ phosphoprotein phosphatase, protein phosphatases 1 and 2, Mre11 and calcineurin and this motif is demonstrated to be important for protein phosphatase activity by binding to metal ion24,25,26. This GNHE motif is conserved among Dbr1 homologues from many species (Figure 1), suggesting that this motif is also important for Dbr1 protein function. Indeed, it is demonstrated that this GNHE motif is critical for debranching activity of yeast Dbr1p28,29. In order to test whether GNHE motif also has a vital role in debranching activity of hDbr1, we decided to prepare several mutants that have a point mutation in either 84th Asparagine (N) residue or 85th Histidine (H) residue according to the mutations used in the previous study with both protein phosohatase 126 and yeast Dbr1p28,29. These residues were substituted by either Alanine (A) or Serine (S), and the resultant mutants are called as N84A, H85S and H85A. It is already known that the corresponding mutations cause the loss of activity in the case of protein phosphatase 126, and it was also demonstrated that these residues are critical in yeast Dbr1p for both debranching activity and Ty1 transposition28,29. These mutant proteins with Flag-tag were transiently expressed to determine their subcellular localization. In HeLa cells all the mutant proteins as well as the wild type protein were localized in the nucleoplasm (Supplementary Figure 1). To obtain both wild type and mutant proteins, we transfected HEK293T cells with the plasmids carrying cDNA of either wild type or mutant proteins with Flag-tag and prepared the whole cell extracts from transfected cells33. To test the effect of these mutations on debranching activity of hDbr1, we carried out in vitro debranching assays using purified Flag-tagged proteins from the whole cell extracts by using anti-Flag M2 resin34. After elution with 3X Flag peptides, the eluates were analyzed by SDS-PAGE. As shown in Figure 4A, almost equal amount of proteins could be recovered from the extracts. These proteins were incubated with32P-labeled lariat intron RNAs derived from Adeno virus immediate early gene pre-mRNA, Ad2, which had been gel-purified from in vitro splicing reaction products. The results in Figure 4B demonstrated that the wild type hDbr1 protein could debranch the lariat intron (lane WT). However H85A, H85S and N84A mutants had a greatly reduced debranching activity (Figure 4B, lanes H85A, H85S and N84A). Essentially the same results were obtained with the lariat RNA from chicken δ-crystallin pre-mRNA (Figure 4B). The results shown in Figure 4 indicate that the GNHE motif, which is also found in protein phosphatases, is critical for debranching activity of hDbr1.
Figure 4. The conserved GNHE motif homologous to protein phosphatase 1 is essential for debranching activity of hDbr1 in vitro.
A) Equal amount (0.1 μg each) of recombinant proteins purified from either Flag-vector- or the hDbr1 cDNA-transfected HEK293T cells on a Coomassie Brilliant Blue-stained 5–20% gradient SDS-polyacrylamide gel. The plasmids used for transfection are indicated above the panel as follows; Vector : Flag-pCDNA3 vector, WT : Flag-wild type hDbr1, H85A : Flag-hDbr1 containing histidine to alanine mutation at amino-acid position 85, H85S : Flag-hDbr1 harboring histidine to serine change at amino-acid position 85, N84A : Flag-hDbr1 carrying asparagine to alanine mutation at amino-acid position 84. Input lanes contain the lariat RNAs used for the assays. B) In vitro debranching assays with purified proteins shown in A). Lariat intron RNAs derived from either Ad2 (upper panel) and δ-crystallin (lower panel) pre-mRNA were incubated with recombinant proteins and analyzed by 6% denaturing polyacrylamide gel electrophoresis. The structure of RNAs corresponding to each band is demonstrated schematically. The asterisk shows the contaminated pre-mRNAs.
Dbr1p was originally isolated as a host gene product critical for yeast retrotransposon element Ty115. During its life cycle, Ty1 RNA is synthesized in yeast nucleus and exported to the cytoplasm to serve as a template for the translation of the encoding proteins. Ty1 RNAs are incorporated into virus like particles (VLPs) with Ty1 encoding proteins including reverse transcriptase and integrase, and these particles are subsequently imported to the nucleus35,36. The synthesis of Ty1 cDNA by reverse transcription is thought to occur in VLPs in the cytoplasm. In Dbr1 mutants, the accumulation of Ty1 cDNAs is reduced, whereas the protein synthesis from Ty1 mRNA is at the wild-type level37. These results suggest that DBR1 protein is required for reverse transcription and/or the stability of Ty1 cDNA. Since Ty1 cDNA reverse transcription is supposed to occur in the cytoplasm, it was possible that DBR1 shuttles between the nucleus and the cytoplasm, although it is nuclear at the steady state.
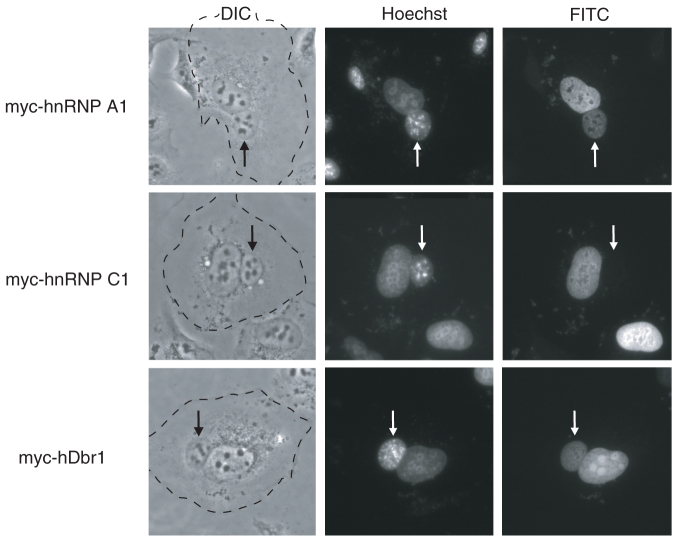
To determine if hDbr1 has a shuttling activity, we carried out a heterokaryon assay (Figure 5). Heterokaryons were formed by Polyethylen Glycol (PEG)-mediated fusions of HeLa cells that had been previously transfected with the indicated constructs and mouse NIH3T3 cells. Staining of the cells with the DNA dye, Hoechst, allows us to distinguish between the HeLa and the NIH3T3 nuclei. Fusions and subsequent incubations were carried out in the presence of cycloheximide to ensure that the observed signals result from proteins in the HeLa nucleus and not from newly synthesized proteins in the cytoplasm30,38,39,40,41. A construct containing the non-shuttling hnRNP C1 protein served as a negative control, while the rapid shuttling protein hnRNP A1 as a positive control. The results presented in Figure 5 show that hDbr1 shuttles between the nucleus and the cytoplasm, since the hDbr1 signal was clearly seen in both HeLa and NIH3T3 nuclei. This result strongly suggests that hDbr1 has novel cytoplasmic function(s).
Figure 5. The hDbr1 protein shuttles between the nucleus and the cytoplasm.
Expression vectors encoding hnRNP A1, C1, and hDbr1 with myc tag were transfected into HeLa cells. After expression of the transfected cDNAs, the cells were fused with mouse NIH3T3 cells to form heterokaryones and incubated in media containing 100 μg/ml cycloheximide for 2 hours. The cells were then fixed and stained for immunofluorescence microscopy with anti-myc tag antibody (panel FITC) to localize the proteins, and Hoechst 33342 (SIGMA, panel Hoechst), which differentiates the human and mouse nuclei within the heterokaryon. The arrows identify the mouse nuclei. The panels marked DIC show the phase-contrast image of the heterokaryons and the cytoplasmic edge is highlighted by a broken line.
Discussion
In this paper we analyzed the several domains of hDbr1 protein, which is involved in intron turnover with its debranching activity. The hDbr1 is a nuclear protein that has a bipartite classical type NLS near its Carboxyl terminus (Figure 2,3). The similar NLS-like sequence can be found in mouse, rat, Xenopus, zebra fish proteins, but not in other organisms (Figure 1). Dbr1 proteins show high similarity especially in the region of first 200 amino acids. However, carboxyl terminal half is relatively diverse among species (Figure 1). The budding yeast Dbr1p is a 406 amino acid protein, while hDbr1 is 544 amino acids long with a longer carboxyl terminus. It is reported that Green Fluorescent Protein (GFP) tagged-yeast Dbr1p is localized both in the nucleus and cytoplasm42, and its NLS has not been identified. It may have a different type NLS, rather than a classical type NLS. Further delineation of yeast Dbr1 NLS remains to be performed.
The hDbr1 protein has a conserved GNHE motif, which is a signature of metallophosphatase superfamily. This motif is known to bind to metal ion in metallophosphatases and the point mutation in asparagine (N) or histidine (H) in this motif abolishes their phosphatase activity26. The GNHE motif is also demonstrated to be important for debranching activity of yeast Dbr1p both in vitro and in vivo28,29. The motif is perfectly conserved among Dbr1 proteins from many species, suggesting that this GNHE signature is catalytically important in Dbr1 proteins from other species. As expected, the mutations of asparagine and histidine in this motif of hDbr1 abolished its debranching activity in vitro (Figure 4). These two amino-acid residues are proposed to contact to the 2′-5′ phosphodiester of the RNA by analogy to the Mre11-dAMP complex29. The yeast Dbr1 protein was demonstrated to prefer manganese to magnesium as the metal cofactor for debranching29. It is of great interest to dissolve crystal structure of Dbr1 protein with manganese and branched RNA.
The heterokaryon assay revealed that hDbr1 is a nucleocytoplasmic shuttling protein (Figure 5). In this respect, the cytoplasmic function(s) of hDbr1 is proposed as a novel feature of hDbr1. One possible function is to debranch introns in the cytoplasm. It was demonstrated that the introns in the homeobox gene Pem could be detected in the cytoplasmic fraction43. In addition, it was shown that the splice-defective splicing intermediates (lariat introns with 3′ exon) are debranched by Dbr1p and degraded by the cytoplasmic exonuclease Xrn1p and cytoplasmic exosome44. These results strongly suggest that the intron RNAs that have a lariat structure can be exported to the cytoplasm possibly by accident and hDbr1 debranches them in the cytoplasm. It is possible that hDbr1 monitors the presence of lariat-introns in the cytoplasm and rapidly debranches them to degrade. In order to test these possibilities, further experiments are required to detect lariat RNAs from the cytoplasm when hDbr1 protein level is reduced by siRNA or shRNA.
Another possible function of hDbr1 in the cytoplasm is to be involved in the cDNA synthesis by reverse transcription. The Dbr1 gene was originally isolated by a genetic screening to identify host factors required for Ty1 retrotransposon transposition15. The Dbr1 mutant strains exhibit low level of Ty1 cDNA, suggesting that Dbr1p is involved in reverse transcription of Ty1 mRNA. It was reported that Ty1 RNA forms a 2′-5′ branch structure characteristic of a lariat intron and this lariat structure is an intermediate of Ty1 cDNA synthesis45. This branch structure was suggested to facilitate the minus-strand transfer from the upstream to the downstream of Ty1 RNA during cDNA synthesis. The structural similarity of Ty1 to the animal retroviruses suggests that retrovirus genomic RNA forms a branch structure as well. It was also reported that reduction of hDbr1 mRNA level by siRNA led to the inhibition of Human Immunodeficiency Virus-1 (HIV-1) replication by reducing viral cDNA and protein production46. This paper suggests that debranching activity of hDbr1 is required for reverse transcription completion. However, it is still controversial whether Ty1 RNA has a branch structure or not, since the lariat structure of Ty1 RNA could not be detected in many different methods47,48. Therefore the molecular basis for the involvement of Dbr1p in Ty1 transposition is still unclear. Further analysis of hDbr1 including the identification of specific interactors are required to uncover the bona fide cytoplasmic function(s) of hDbr1.
Methods
Plasmid construction
Full-length hDbr1 cDNA was amplified by PCR reaction from Marathon ready HeLa cDNA library (Clontech) and cloned between BamHI and XhoI sites of myc- (myc-hDbr1) and Flag-pCDNA3 (Flag-hDbr1)41,49. For myc-chicken Pyruvate Kinase (PK)-hDbr1 fusion constructs, myc-hDbr1 was used as a series of PCR reactions. Primers were designed which included an KpnI site in the 5′ partner and an XhoI site in the 3′ partner to amplify fragments of the hDbr1 coding sequences corresponding to amino acids 1–544 (Full length), 1–240, 241–544, 241–517, 1–507 and 508–544. These individual fragments were then digested with KpnI and XhoI and subcloned into similarly digested myc-PK30. For the series of NLS deletion constructs, by using the plasmid encoding myc-PK fused with amino acids 508–544 of hDbr1 as a template, the entire region of the template plasmid except the portion that has to be deleted was amplified by PCR reaction. The hDbr1 mutant cDNAs that have a point mutation were also prepared by PCR-based mutagenesis by using Flag-hDbr1 plasmid as a template with a QuikChange site-directed mutagenesis kit (STRATAGENE) as the manufacturer recommends.
Cell culture and transfection
HeLa and HEK293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM, Nacalai tesque, Japan) supplemented with 10% fetal bovine serum (EQUITECH-BIO, Texas) and 1% penicillin-streptomycin (Gibco-BRL) in an incubator adjusted to 37°C with 5% CO2. Transfection of cultured cells was carried out by using Lipofectamine2000 (Invitrogen) as a manufacturer recommends.
Protein expression in HEK 293T cells and purification
Both wild type and mutant hDbr1 proteins with Flag-tag were transiently expressed in HEK293T cells by transfection. Whole cell lysates were prepared from those transfected cells as previously described33. Flag-tagged proteins were purified from whole cell lysates by using anti Flag-M2 resin (SIGMA) as described previously34.
Immunofluorescence
Transfected HeLa cells were fixed and stained for immunofluorescence microscopy with either anti-myc (MC045, Nakalai tesque, Japan) or anti-Flag M2 antibody (SIGMA) as described previously50. Hoechst 33342 (SIGMA) was included at 5 μg/ml during the secondary antibody incubations.
In vitro splicing and debranching assays
HeLa cell nuclear extracts were obtained from CILBIOTECH (Belgium). Lariat-intron was prepared by in vitro splicing reaction. In vitro splicing reaction was carried out as described previously33,41,51. Briefly, the splicing reaction mixture was supplied onto 6% denaturing acrylamide gel and the band corresponding to lariat-intron RNA was excised and recovered from the gel. About 0.1 pmol of the lariat-intron RNA was incubated with 50 ng (approximately 0.7 pmol) of purified Flag-tagged proteins in the debranching reaction mixture (20 mM Hepes-KOH, pH7.9, 40 mM KCl, 3 mM MgCl2, 4% Glycerol) 30 minutes at 30°C in 20 μl reaction mixture. After incubation RNA was recovered by phenol extraction and ethanol precipitation, followed by analysis on 6% denaturing polyacrylamide gel electrophoresis.
Heterokaryon assay
Heterokaryon assay was performed as described previously30,31,38,39,40,41. Briefly, HeLa cells grown in 100 mm dishes were transfected as described. Twenty-four hours later the cells were trypsinyzed and transferred to 6 hole plates containing 18 mm2 glass coverslips. Next day an equal number of NIH3T3 cells that had been cultured in the presence of 75 μg/ml cycloheximide 30 minutes were seeded on the coverslips. Cells were fused as described previously30,31,38,39,40,41 and cultured in the medium containing 100 μg/ml cycloheximide for another 2 hours prior to fixation for immunofluorescence.
Author Contributions
N.K., M.H. and M.O. designed experiments and N.K. and I.D. performed them. N.K. and M.O. analyzed the data and N.K., M.H. and M.O. wrote the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We are grateful to Dr. Gideon Dreyfuss for the plasmids. We also thank members of both Ohno laboratory and Hagiwara laboratory for discussions. This work is supported by Grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan. N.K. was supported by Program for Improvement of Research Environment for Young Researchers from Special Coordination Funds for Promoting Science and Technology (SCF) commissioned by the MEXT of Japan.
References
- Hastings M. L. & Krainer A. R. Pre-mRNA splicing in the new millennium. Curr Opin Cell Biol 13, 302–309 (2001). [DOI] [PubMed] [Google Scholar]
- Maniatis T. & Reed R. An extensive network of coupling among gene expression machines. Nature 416, 499–506 (2002). [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Kim V. N. & Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 3, 195–205 (2002). [DOI] [PubMed] [Google Scholar]
- Jurica M. S. & Moore M. J. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell 12, 5–14 (2003). [DOI] [PubMed] [Google Scholar]
- Company M., Arenas J. . & Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature 349, 487–493 (1991). [DOI] [PubMed] [Google Scholar]
- Staley J. P. & Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92, 315–326 (1998). [DOI] [PubMed] [Google Scholar]
- Martin A., Schneider S. & Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J Biol Chem 277, 17743–17750 (2002). [DOI] [PubMed] [Google Scholar]
- Arenas J. E. & Abelson J. N. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc Natl Acad Sci U S A 94, 11798–11802 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai R. T. et al. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev 19, 2991–3003 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N., Aronova A. & Schwer B. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev 21, 2312–2325 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. J. Nuclear RNA turnover. Cell 108, 431–434 (2002). [DOI] [PubMed] [Google Scholar]
- Nam K. et al. Yeast lariat debranching enzyme. Substrate and sequence specificity. J Biol Chem 269, 20613–20621 (1994). [PubMed] [Google Scholar]
- Mitchell P. & Tollervey D. mRNA turnover. Curr Opin Cell Biol 13, 320–325 (2001). [DOI] [PubMed] [Google Scholar]
- Coller J. & Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem 73, 861–890 (2004). [DOI] [PubMed] [Google Scholar]
- Chapman K. B. & Boeke J. D. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell 65, 483–492 (1991). [DOI] [PubMed] [Google Scholar]
- Nam K., Lee G., Trambley J., Devine S. E. & Boeke J. D. Severe growth defect in a Schizosaccharomyces pombe mutant defective in intron lariat degradation. Mol Cell Biol 17, 809–818 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhala G., Rosenberg G. H. & Kaufer N. F. Architectural features of pre-mRNA introns in the fission yeast Schizosaccharomyces pombe. Yeast 8, 171–182 (1992). [DOI] [PubMed] [Google Scholar]
- Rodriguez-Medina J. R. & Rymond B. C. Prevalence and distribution of introns in non-ribosomal protein genes of yeast. Mol Gen Genet 243, 532–539 (1994). [DOI] [PubMed] [Google Scholar]
- Kalogeropoulos A. Automatic intron detection in nuclear DNA sequences of Saccharomyces cerevisiae. Yeast 11, 555–565 (1995). [DOI] [PubMed] [Google Scholar]
- Kim J. W. et al. Human RNA lariat debranching enzyme cDNA complements the phenotypes of Saccharomyces cerevisiae dbr1 and Schizosaccharomyces pombe dbr1 mutants. Nucleic Acids Res 28, 3666–3673 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. C., Kim G. M., Yang J. M. & Ki J. W. Cloning, expression, and complementation test of the RNA lariat debranching enzyme cDNA from mouse. Mol Cells 11, 198–203 (2001). [PubMed] [Google Scholar]
- Wang H., Hill K. & Perry S. E. An Arabidopsis RNA lariat debranching enzyme is essential for embryogenesis. J Biol Chem 279, 1468–1473 (2004). [DOI] [PubMed] [Google Scholar]
- Zhuo S., Clemens J. C., Stone R. L. & Dixon J. E. Mutational analysis of a Ser/Thr phosphatase. Identification of residues important in phosphoesterase substrate binding and catalysis. J Biol Chem 269, 26234–26238 (1994). [PubMed] [Google Scholar]
- Egloff M. P., Cohen P. T., Reinemer P. & Barford D. Crystal structure of the catalytic subunit of human protein phosphatase 1 and its complex with tungstate. J Mol Biol 254, 942–959 (1995). [DOI] [PubMed] [Google Scholar]
- Goldberg J. et al. Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1. Nature 376, 745–753 (1995). [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhang Z., Brew K. & Lee E. Y. Mutational analysis of the catalytic subunit of muscle protein phosphatase-1. Biochemistry 35, 6276–6282 (1996). [DOI] [PubMed] [Google Scholar]
- Yoshimoto R., Kataoka N., Okawa K. & Ohno M. Isolation and characterization of post-splicing lariat-intron complexes. Nucleic Acids Res 37, 891–902 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem L. A., Boucher C. L. & Menees T. M. Relationship between RNA lariat debranching and Ty1 element retrotransposition. J Virol 77, 12795–12806 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalid M. F., Damha M. J., Shuman S. & Schwer B. Structure-function analysis of yeast RNA debranching enzyme (Dbr1), a manganese-dependent phosphodiesterase. Nucleic Acids Res 33, 6349–6360 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael W. M., Eder P. S. & Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. Embo J 16, 3587–3598 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N. et al. Specific Y14 domains mediate its nucleo-cytoplasmic shuttling and association with spliced mRNA. Scientific reports 1, 92 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V. Conserved sequence pattern in a wide variety of phosphoesterases. Protein Sci 3, 356–358 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N. & Dreyfuss G. A simple whole cell lysate system for in vitro splicing reveals a stepwise assembly of the exon-exon junction complex. J Biol Chem 279, 7009–7013 (2004). [DOI] [PubMed] [Google Scholar]
- Pellizzoni L., Baccon J., Rappsilber J., Mann M. & Dreyfuss G. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J Biol Chem 277, 7540–7545 (2002). [DOI] [PubMed] [Google Scholar]
- Roth J. F. The yeast Ty virus-like particles. Yeast 16, 785–795 (2000). [DOI] [PubMed] [Google Scholar]
- Wilhelm F. X., Wilhelm M. & Gabriel A. Reverse transcriptase and integrase of the Saccharomyces cerevisiae Ty1 element. Cytogenet Genome Res 110, 269–287 (2005). [DOI] [PubMed] [Google Scholar]
- Karst S. M., Rutz M. L. & Menees T. M. The yeast retrotransposons Ty1 and Ty3 require the RNA Lariat debranching enzyme, Dbr1p, for efficient accumulation of reverse transcripts. Biochem Biophys Res Commun 268, 112–117 (2000). [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S. & Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature 355, 730–732 (1992). [DOI] [PubMed] [Google Scholar]
- Michael W. M., Choi M. & Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell 83, 415–422 (1995). [DOI] [PubMed] [Google Scholar]
- Nakielny S. & Dreyfuss G. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J Cell Biol 134, 1365–1373 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N. et al. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell 6, 673–682 (2000). [DOI] [PubMed] [Google Scholar]
- Kumar A. et al. Subcellular localization of the yeast proteome. Genes Dev 16, 707–719 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement J. Q., Maiti S. & Wilkinson M. F. Localization and stability of introns spliced from the Pem homeobox gene. J Biol Chem 276, 16919–16930 (2001). [DOI] [PubMed] [Google Scholar]
- Hilleren P. J. & Parker R. Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol Cell 12, 1453–1465 (2003). [DOI] [PubMed] [Google Scholar]
- Cheng Z. & Menees T. M. RNA branching and debranching in the yeast retrovirus-like element Ty1. Science 303, 240–243 (2004). [DOI] [PubMed] [Google Scholar]
- Ye Y., De Leon J., Yokoyama N., Naidu Y. & Camerini D. DBR1 siRNA inhibition of HIV-1 replication. Retrovirology 2, 63 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes C. E. & Boeke J. D. An evaluation of detection methods for large lariat RNAs. Rna 11, 323–331 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratico E. D. & Silverman S. K. Ty1 reverse transcriptase does not read through the proposed 2',5'-branched retrotransposition intermediate in vitro. Rna 13, 1528–1536 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V. N., Kataoka N. & Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science 293, 1832–1836 (2001). [DOI] [PubMed] [Google Scholar]
- Kataoka N., Diem M. D., Kim V. N., Yong J. & Dreyfuss G. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon-exon junction complex. Embo J 20, 6424–6433 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L., Kataoka N., Charroux B. & Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell 95, 615–624 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information