Abstract
Sirtuins are a family of NAD+-dependent enzymes that was proposed to control organismal life span about a decade ago. While such role of sirtuins is now debated, mounting evidence involves these enzymes in numerous physiological processes and disease conditions, including metabolism, nutritional behavior, circadian rhythm, but also inflammation and cancer. SIRT1, SIRT2, SIRT3, SIRT6, and SIRT7 have all been linked to carcinogenesis either as tumor suppressor or as cancer promoting proteins. Here, we review the biological rationale for the search of sirtuin inhibitors and activators for treating cancer and the experimental approaches to their identification.
Keywords: Sirtuin modulators, cancer, drug design, epigenetics, drug discovery.
SIRTUINS: AT THE CROSSROAD OF METABOLISM, CANCER, AND INFLAMMATION
Sirtuins are enzymes that require nicotinamide adenine dinucleotide (NAD+) to catalyze their reactions [1-4]. The latter include activities as mono-ADP-ribosyltransferases [5] or as deacetylases. In mammals, seven sirtuins have been identified, of which two are predominantly nuclear, SIRT6 and SIRT7, two are nuclear and cytosolic, SIRT1 and SIRT2, and three are mitochondrial, SIRT3-5. Sirtuins have been ascribed roles in numerous physiological and disease conditions, including aging, metabolism, circadian clock regulation, nutritional behavior, but also cancer and inflammation [4, 6]. Due to their broad involvement in key biological functions, sirtuins are considered appealing targets for the development of pharmaceuticals. In this review, we will focus on the role of sirtuins in cancer, we will discuss the rationale behind the development of sirtuin-targeting agents for the treatment of human malignancies, and we will review the approaches through which modulators of sirtuin activity are currently investigated.
SIRTUINS AND CANCER
The sirtuins for which a role in cancer has been proposed include SIRT1, SIRT2, SIRT3, SIRT6 and SIRT7. In genetic mouse models, SIRT1, SIRT2, and SIRT3 were shown to act as tumor suppressors [7-10]. In the case of SIRT1, its activity as a tumor suppressor has been ascribed to its capacity to deacetylate and consequently inhibit the RelA/p65 subunit of NF-kappaB [11], a transcription factor with antiapoptotic and pro-inflammatory activity. Moreover, studies show that SIRT1 also deacetylates β-catenin and thereby suppresses its ability to activate transcription and drive cell proliferation [12]. Disruption of SIRT2, which is a tubulin deacetylase [13], in the mouse was found to increase the levels of mitotic regulators, such as Aurora-A and -B, aneuploidy, and mitotic cell death [8]. SIRT2-deficient mice developed gender-specific tumorigenesis, with females developing mammary tumors and males developing hepatocellular carcinoma. Moreover, human breast cancers and hepatocellular carcinomas were reported to exhibit reduced SIRT2 levels as compared with normal tissues. SIRT3 was proposed to oppose cancer development through its role in mitochondrial metabolism, reactive oxygen species production and genome stability [9, 10]. Its deficiency was shown to favor cell transformation in response to oncogenic Ras or Myc and to lead to HIF-1α stabilization with consequent induction of the Warburg effect. SIRT6 has also been suggested to act as a tumor suppressor since its acute overexpression in cancer cell lines of different histology was found to induce apoptosis [14]. Interestingly, this biological activity of SIRT6 appears to be linked to its mono-ADP-ribosyltransferase but not to its deacetylase activity. Finally, SIRT6 anticancer activity could also be ascribed to its capacity to negatively regulate NF-κB and HIF-1α [15, 16], the latter being a transcription factor that promotes the expression of glycolytic and pro-angiogenic genes.
The apparent consistency of this picture is complicated by evidence that these same sirtuins may actually favor certain aspects of neoplastic growth and that, at least in certain instances, their inhibition may be preferable to their activation. Numerous reports indicate that SIRT1 also has cancer-promoting functions which include deacetylation and inactivation of p53 [17, 18] and of proapoptotic FOXO transcription factors [19], deacetylation of Ku-70 with consequent sequestration of Bax away from mitochondria [20], as well as inhibition of senescence and of apoptosis in c-Myc- and in PML-driven cancers [21, 22]. Accordingly, many studies attribute direct anticancer activity to SIRT1 inhibitors or show how such compounds sensitize cancer cells to anticancer agents or to oxidative stress [23-29]. Similar results were also reported for SIRT2. SIRT2 downregulation or chemical inhibition were found to induce anti-cancer effects that include p53 accumulation, cell cycle arrest and apoptosis [30-32]. In a recent study, the antileukemia effects of SIRT2 inhibition (with AC93253) were accompanied by Akt acetylation, de-phosphorylation, and consequent inhibition [33]. Interestingly, evidence exists that SIRT2 inhibition could be counterproductive in certain instances, such as upon treatment with microtubules inhibitors [34]. Namely, SIRT2 downregulation appears to make cancer cells resistant to these agents by prolonging chronic mitotic arrest. Also SIRT3’s role as a tumor suppressor does not seem to extend to all types of cancer. In particular, in oral squamous cell carcinomas, SIRT3 was found to be overexpressed and its downregulation had antiproliferative activity and sensitized carcinoma cells to radiation and to cisplatin [35]. Finally, in the case of SIRT6, studies show that this enzyme is crucial for telomere maintenance, DNA repair and genome stability [36, 37]. Thus, SIRT6 inhibitors could conceivably be used to sensitize cancer cells to chemotherapeutics or radiotherapy [38]. Moreover, SIRT6 promotes TNF synthesis by a mechanism that appears to entail the enhancement of the efficiency with which Tnf mRNA is translated [39, 40]. TNF plays a central role in some of the systemic manifestations of cancer, such as fever and cachexia, and its role in shaping the tumor microenvironment is acknowledged [41]. SIRT6 inhibition could theoretically serve the purpose of reducing the levels of this unwanted mediator of cancer-induced inflammation and thereby interfere with invalidating systemic manifestations of disease.
Finally, a recent study suggests that SIRT7 may promote tumorigenesis by deacetylating lysine 18 of histone H3, thereby repressing genes with tumor suppressor function [42]. SIRT7 was found to be stabilized at target promoters by interaction with the ETS family transcription factor ELK4. According to these authors, H3K18 deacetylation by SIRT7 would be necessary to maintain essential features of cancer cells, such as anchorage-independent cell growth and loss of contact inhibition. Indeed, in line with this model, SIRT7 depletion reduced the tumorigenicity of human cancer xenografts in mice.
Overall, while it is eventually becoming clear that sirtuins play important roles in cancer pathophysiology, the final judgment as to whether a defined sirtuin should be blocked or rather activated in order to achieve a therapeutic benefit may vary depending on the type of cancer, its molecular features, stage of disease, and clinical manifestations. Either way, a crucial step in the pursuit of sirtuin-targeting approaches is the identification of compounds that could be used to specifically modulate sirtuin activity in vivo.
SIRTUIN STRUCTURE
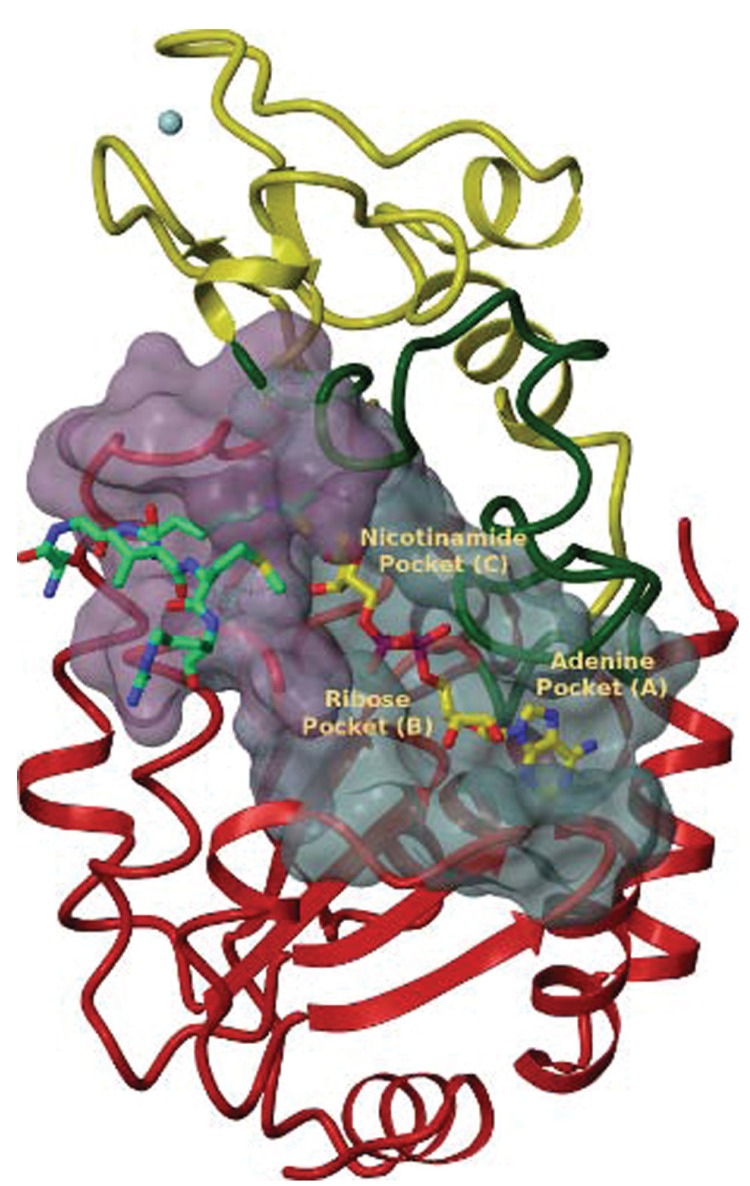
Sirtuins contain a conserved catalytic core with good structural superposition among the various isoforms. The main sirtuin structure consists of a large Rossman-fold domain (a structural motif that is typical of NAD+ binding proteins), a small zinc-binding domain, and a number of flexible loops that bring together the two domains to form an extended cleft accommodating both cofactor and substrate (Fig. 1). The large Rossman-fold domain is characterized by the presence of six parallel β-strands surrounded by a different number of α-helices on either side, depending on the type of sirtuin. This domain shows the typical features of NAD+ binding sites, such as a Gly-X-Gly motif, that is important for phosphate binding, in addition to specific features that are deputed to the recognition of nicotinamide or ribose groups. The adenine base binds the C-terminal half of this domain while nicotinamide enters the N-terminal part which is typical in inverted Rossman-fold domains.
Fig. (1).
X-ray structure of human SIRT3 in complex with ADPR bound to Acetyl-coenzyme A synthetase 2 peptide containing a thioacetyl lysine (PDB code 3GLT). Different domains of the protein are color-coded (red for the Rossman-fold domain, yellow for zinc-binding domain, green for the cofactor-binding loop. The molecular structure of substrate peptide (green) and cofactor (yellow) are reported. The surfaces of substrate-binding site (purple) and NAD+-binding site (light blue) are also highlighted. (The color version of the figure is available in the electronic copy of the article).
The small zinc-binding domain comprises three antiparallel β sheets, a variable α helical region and a Zn2+ cation coordinated by four conserved cysteine residues. The zinc ion does not directly participate in the catalytic mechanism of sirtuins, like in the case of class I/II HDACs [43], but its presence is required to ensure enzyme functionality [44]. This domain shows large variability in three-dimensional arrangements among different sirtuins and its position relatively to the large domain is also dependent on the presence of substrate and NAD+. This variability may have a potential role in substrate specificity and enzyme localization, highlighting this domain as an attractive potential binding site for selective sirtuin inhibitors.
The so-called cofactor binding loop is the largest loop that connects the large and the small domains of sirtuins and it is one of the most flexible regions of the enzyme. This loop forms part of the cofactor binding site and its conformation is largely influenced by the presence of NAD+ or other reaction intermediates. This loop can take a disordered conformation when NAD+ is not bound, while two different ordered conformations, open or closed, are induced by NAD+ and by the reaction product 2-O-acetyl-ADP-ribose (OAADPr), respectively.
The NAD+ binding site, consisting in the large domain and in the loop region which is mainly represented by the cofactor binding loop, can be divided in an adenine binding pocket (pocket A), a nicotinamide ribose binding pocket (pocket B) and a nicotinamide binding pocket (pocket C). The cofactor molecule undergoes several interactions within the binding site and some of these are conserved among different sirtuins. The adenine base, for instance, is positioned in a way that allows the establishment of several van der Waals interactions including two conserved glycines and several hydrogen bonds, e.g. those formed by the amide nitrogen with a glutamate or serine residue, while the hydroxyls of the adenine ribose mainly interact with asparagine residues. Phosphate and nicotinamide ribose moieties undergo a complex interaction network that, in both cases, is affected by the conformation of the cofactor binding loop. Similarly, conformational changes occur also in the nicotinamide binding pocket. In fact, crystallographic structures available in the Protein Data Bank show that the nicotinamide moiety of NAD+ is often bound outside of the C pocket in a conformation that is not compatible with acetyl-lysine binding and deacetylation reaction, the so-called non-productive conformation. Vice versa, a productive conformation of NAD+ requires the presence of an acetyl-lysine containing peptide so that the nicotinamide moiety can bind the C pocket that undergo interactions with various conserved residues, including the hydrophobic invariant motif Gly-Ala-Gly.
The substrate peptide binding site is located in a cleft between large and small domains. Peptide binding induces a conformational change where the two domains merge together in order to form the binding site and the acetyl-lysine binding tunnel. The substrate peptide backbone forms several hydrogen bond interactions with two flanking strands in the enzyme, known as β staple, while the acetyl-lysine side chain makes several van der Waals interactions with conserved hydrophobic residues and an hydrogen bond between the N-ε atom and a backbone carbonyl of conserved valine residue.
Three-dimensional structures of SIRT 2, 3 5 and 6 have been resolved by X-ray crystallography and it is worth to note that the recently obtained crystal structure of human SIRT6 [45] revealed distinctive features of this enzyme, such as a more open conformation due to a displacement of the small domain and the lack of the cofactor binding loop which is replaced by a stable single helix. The combination of structural, kinetic and biochemical studies highlight SIRT6 as the unique Sirtuin able to bind NAD+ in the absence of acetyl-lysine substrate opening to the possibility that it evolved away from NAD+-dependent deacetylase function toward a NAD+ metabolite sensor function.
An extended analysis on sirtuin structures and the current availability of crystallographic data has been recently published by Sanders et al. and Andreoli et al. [46, 47].
SIRTUIN INHIBITORS
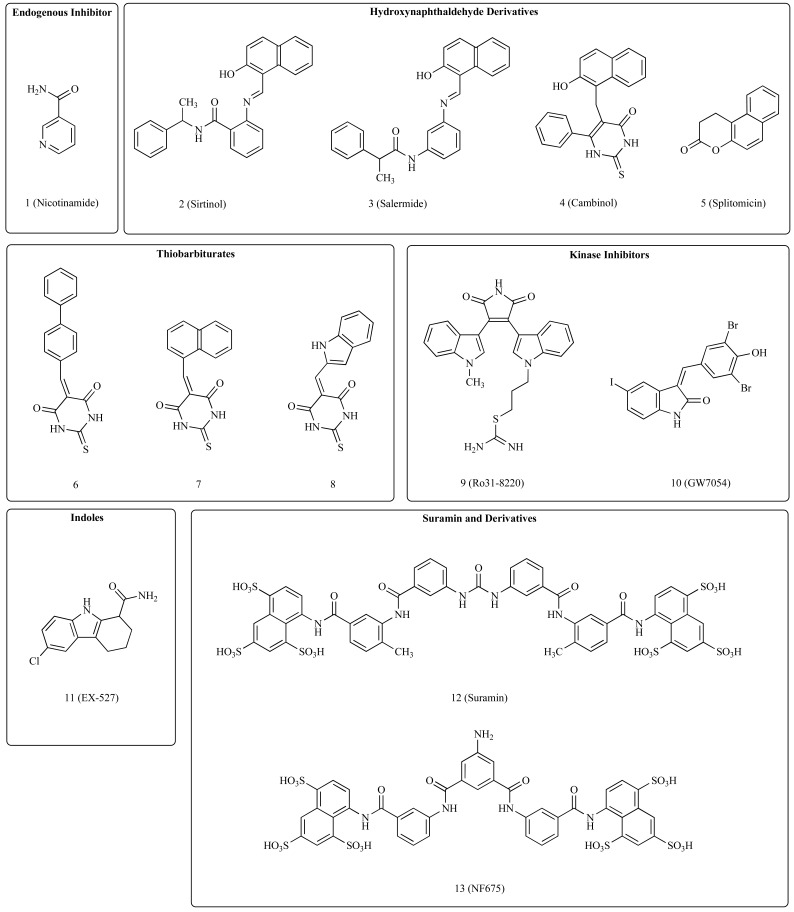
Although sirtuins emerged in the last years as therapeutic targets for small molecule-based interventions, a relatively small number of highly active molecules have been developed so far. The available inhibitors belong to several structural classes that reflect both different drug discovery strategies and the complexity of the catalytic machinery of these enzymes. The simplest approach used to identify novel active compounds was the evaluation of substrate and/or product mimetics. Among the first compounds studied were the endogenous inhibitor nicotinamide (1, (Fig. 2)), which is a product of the deacetylation reaction, and its derivatives. Because of its simple chemical structure, nicotinamide is a micromolar non-competitive inhibitor of SIRT1 and SIRT2 [48] that interact with the pocket C in the binding site. Similarly, slightly modified NAD+ molecules such as carbamido-NAD+, compete with NAD+ for the cofactor binding site. SIRT1 inhibition with nicotinamide was reported to have anticancer activity in B-cell chronic lymphocytic leukemia and in prostate cancer [27, 49]. However, nicotinamide and the NAD+ analogs have limited therapeutic potential as demonstrated by unsuccessful searches for more potent inhibitors of this class [50, 51]. Several active molecules were designed starting from substrate peptides, such as small fragments of p53 protein, that were modified in order to block or reduce the catalytic activity. Thioacetyllysine derived inhibitors and other modifications of acetylated lysine residue have been demonstrated to be potent sirtuin inhibitors [52-59]. A recent study highlighted SIRT5 preference to catalyze the hydrolysis of malonyl and succinyl group from the lysine residue rather than acetyl group, demonstrating that this preference could be used to the design of selective inhibitors [60].
Fig. (2).
Sirtuin inhibitors.
A phenotypic screen of a small compound library led to the discovery of sirtinol (2, Fig. 2), a hydroxynaphthaldehyde derivative that is active on different sirtuin isoforms [61] and has cytotoxic activity against cancer cells of different origin, including breast, lung, prostate and leukemia cells [25, 30, 49, 62]. A number of subsequent structure-activity relationship studies were carried out in search of derivatives of this molecule with improved activity and/or properties. Some examples are salermide (3, Fig. 2) [63], cambinol (4, Fig. 2) [23] and splitomicin (5, Fig. 2) [64]. With the exception of splitomicin, which is not active on human sirtuins [65], also these compounds were reported to have anticancer properties [23, 30, 63, 66]. A series of thiobarbiturates derivatives that are similar to cambinol and inhibit SIRT1 and SIRT2 at micromolar concentrations, was identified through a structure-based virtual screening approach followed by binding energy estimation and biological testing (6, 7 and 8, Fig. 2) [67]. A large high-throughput screening effort led to the discovery of a series of indole compounds as interesting inhibitors of SIRT1, including one of the most potent and selective compounds known so far, EX-527 (11, Fig. 2). This compound inhibits SIRT1 at nanomolar concentrations and shows remarkable selectivity over SIRT2 and SIRT3, as well as good pharmacokinetic properties [68]. We found that EX-527 cooperates with HDAC inhibitors to induce apoptosis in leukemia cells [25]. However, due to its selectively for SIRT1 and to its poor activity on other sirtuins, the anticancer activity of this inhibitor as a single agent appears to be weak [30].
Starting from the rationale that sirtuins and protein kinases contain an adenosine binding site, a series of known kinase inhibitors was tested against SIRT2 revealing bisindolylmaleinimides and indolinone as interesting scaffolds that are able to exert biological activity. The most interesting compounds, Ro31-8220 (9, Fig. 2) and GW5074 (10, Fig. 2) showed SIRT1/SIRT2 inhibitory properties in the low micromolar range [69].
Suramin (12, Fig. 2), an adenosine receptor antagonist, was discovered as a potent inhibitor of SIRT1/SIRT2 while searching for sirtuin activators [70]. This molecule inhibits SIRT1/SIRT2 at nanomolar concentrations and appears to be selective for SIRT1. Suramin has anticancer activity and has been studied in clinical trials [71]. However, it is still unclear to which extent such activity is due to sirtuin inhibition or, instead, to other modes of action, such as adenosine receptor activation or ceramide accumulation [72]. The optimization of the suramin scaffold led to NF675 (13, Fig. 2) which is the most potent and selective molecules in this series, showing a 20-fold selectivity ratio for SIRT1 over SIRT2 [73]. The binding mode of suramin was investigated by co-crystallization with human SIRT5 [74]. This structure highlights that suramin binds SIRT5 by occupying the nicotinamide ribose pocket (B-pocket), the nicotinamide pocket (C-pocket) and part of the substrate-binding site. Although this compound shows interesting potency and selectivity profile, its modest drug-likeness, especially given its high molecular weight and its anionic nature, limits the therapeutic applications.
Several other compounds with various structural cores were reported to inhibit sirtuins at micromolar concentrations, such as tenovins [24], AGK2 [75], 1,4-dihydropyridines [76], bisnaphtalimidopropyl derivatives [77], AC-93253 [32], and a series of natural products such as aristoforin [78], amurensin G [79], polyphenols [80] and tanikolide [81]. Among these, tenovin-1 and tenovin-6 were reported to have strong anticancer activity that is associated to p53 activation [24].
Several reviews concerning sirtuin inhibitors have been published [82-87].
IN SILICO APPROACH TO THE DESIGN OF SIRTUIN INHIBITORS
Computer-based drug design techniques are frequently used in the drug discovery pipeline to identify and/or optimize hit and lead molecules with the desired bioactivity profile. When the three-dimensional structure of the target protein is known, structure-based methods are often the first choice since they allow estimating directly the molecular interactions between a ligand and a receptor. Nowadays, even inexpensive computing machines, e.g. personal computer or small hardware infrastructures, can process large database of molecules that can be virtually screened in order to select novel candidate compounds for subsequent experimental tests. In the case of sirtuin enzymes, a variety of crystallographic structures derived from different organisms have been published in the last decade. The structures of human SIRT2, SIRT3, SIRT5 and SIRT6 are available in the Protein Data Bank, in unbound form or in complex with NAD+, substrates, or different reaction intermediates. Only one structure reports SIRT5 in complex with the known inhibitor suramin [74]. This amount of structural information was used in several computational studies aimed to discover new sirtuin inhibitors [48, 65, 67, 75, 88-93]. While most of these studies made use of virtual screening techniques as an effective method to screen commercial compound libraries to select compounds for in vitro testing, molecular modeling efforts were also directed to elucidate binding modes of known inhibitors and to rationalize the structure-activity relationship of identified molecules. In particular, modeling techniques such as prediction of the ligand binding affinity [67] or generation of three-dimensional models of different receptor conformations [75] were used. Overall, molecular docking results suggest that sirtuin inhibitors can bind different pockets depending on the sirtuin and on the specific ligand scaffold.
Despite the amount of knowledge that has been gathered in last years on sirtuin structures and on new small-molecule inhibitors, the drug discovery process driven by computational approaches is far from being saturated. For instance, compound libraries used in virtual screening experiment were so far rather limited. Considering the capability of virtual screening algorithms and the increasing power of computing infrastructures, it is nowadays feasible to virtually screen million-size databases of candidates molecules, even by using several freely accessible databases available to the scientific community [94, 95]. In this context, it is expected that additional virtual screening studies will be directed to sirtuins in search of more potent and diverse classes of sirtuin inhibitors. In addition it is expected that further structural insights, i.e. the elucidation of new crystallographic structures or the co-crystal structure of sirtuins with new ligands, will play an essential role to allow a more effective use of computational approaches, not only for improving the knowledge of binding processes, but also to assist the lead optimization process.
SIRTUINS ACTIVATORS
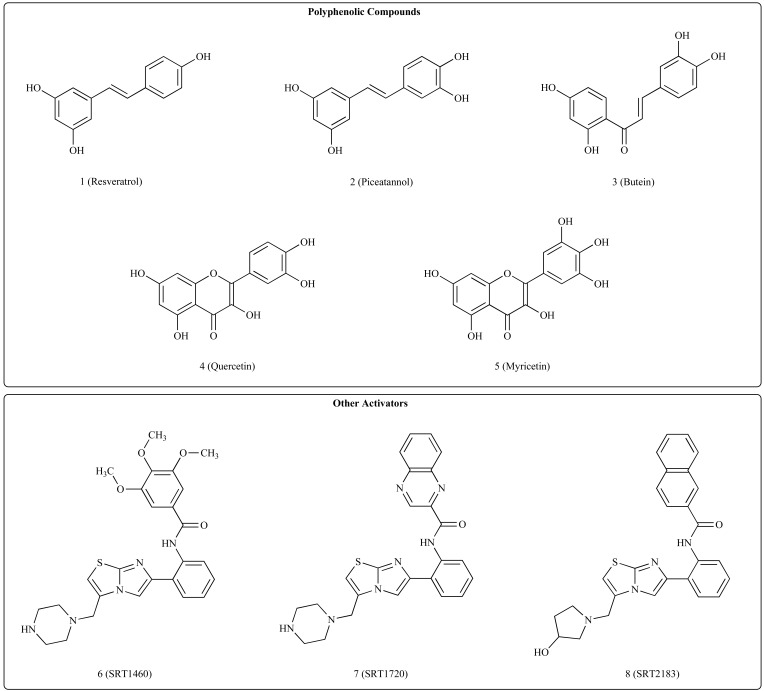
Because sirtuins have been involved in several physiopathological conditions that include aging, metabolism and nutritional behavior [96], a major interest has been to define possible pharmacological actions that could activate sirtuin activity. The first small molecules that were reported as activators of SIRT1 were polyphenolic compounds such as resveratrol, piceatannol, butein, quercetin, and myricetin (respectively compounds 1-5 in (Fig. 3)) [70]. More potent and chemically diverse activators were subsequently reported, known as SRT1460, SRT1720 and SRT2183 (compounds 6-8 in (Fig. 3)) [97], while a recent study reported allosteric modulation of SIRT1 by nonpolyphenolic compounds [98]. Most of these compounds were reported to have anticancer activity [99, 100]. However, it should be noted that their mechanism of action is still unclear [88, 101] and a matter of controversy is the fact that sirtuin activation could frequently only be demonstrated with fluorescently tagged substrates [102-105]. Such activation might be ascribed to other in vivo and in vitro effects that are not mediated by SIRT1. Alternatively, increased SIRT1 activity in response to these compounds may be indirect, i.e. reflect the activity of these agents on proteins that are SIRT1 interactors in the cell. Recent works by Park et al. [106] and Price et al. [107, 108] investigate further the role of resveratrol. In the first study authors show that resveratrol indirectly activates Sirt1 in vivo due to its effect on cAMP signaling, in particular activating the cAMP-Epac1-AMPK-Sirt1 pathway; it is also speculated that other known putative Sirt1 activators such as SRT1720, SRT2183, and SRT1460, because of their similarity in metabolic effects, should act in a similar way of resveratrol. The second work establises a connection between SIRT1 and resveratrol by providing evidence that increased mitochondrial biogenesis and function, AMPK activation, and increased NAD+ levels in skeletal muscles are obtained when mice are treated with a moderate dose of resveratrol.
Fig. (3).
Sirtuins activators.
A comprehensive review on small molecules that activate sirtuins has been recently written by Alcaín and Villalba [109].
BIOCHEMICAL APPROACHES TO THE IDENTIFICATION OF SIRTUIN INHIBITORS
The identification of modulators of sirtuin activity requires efficient assays, possibly suitable for high throughput screening of a high number of compounds, for instance, but not solely, identified through in silico approaches. Here we will describe the most common assays recently used to screen set of molecules as possible inhibitors of human sirtuins.
Homogeneous fluorescent deacetylase assay -
Recombinant sirtuins are incubated in the presence of the fluorescent histone deacetylase substrate peptide ZMAL (a fluorescent acetyl lysine derivative) and NAD+. At the end of the incubation, a buffer containing trypsin and nicotinamide (to inhibit sirtuin activity) is added. ZMAL deacetylation forms the metabolite, ZML, which is a substrate for trypsin, while ZMAL is not. In the latter reaction, the fluorescent 7-aminomethylcoumarin (AMC) is generated, and its fluorescence can be detected in a plate reader. In case the compounds to be screened as possible sirtuin inhibitors are fluorescent, or when a plate reader is not available, a protocol for detection and quantification by HPLC can be used [110]. The homogenous fluorescent assay was used to screen as possible sirtuin inhibitors different families of compounds: adenosine mimetics, inhibitors of kinases or phosphatases [69]; suramin analogs [73]; splitomicin derivatives [65]; thiobarbiturates [67]; novel 3-arylideneindolin-2-ones [111].
SIRT1 Fluorescence Polarization (FP) Assay -
The SIRT1 FP assay, published in 2007 [97], is a coupled enzyme assay, as the one described above, where the deacetylation reaction is followed by a trypsin cleavage. More in detail, in the SIRT1 FP assay, SIRT1 activity is monitored using a specific peptide derived from the sequence of p53. The peptide is N-terminally linked to biotin and C-terminally modified with a fluorescent tag. The first reaction (catalyzed by SIRT1) determines lysine deacetylation. In the second reaction the peptide is cleaved by trypsin at the newly exposed lysine residue. The reaction is stopped by addition of nicotinamide and trypsin, and streptavidin is then added to accentuate the mass differences between substrate and product: fluorescent polarization is determined using excitation and emission wavelengths of 650 nm and 680 nm, respectively. This approach was used to screen large number of compounds and allowed the identification of SIRT1 activators.
Sirtuin fluorimetric activity assay -
The most commonly used method to screen molecules as possible sirtuin inhibitors are sirtuin fluorimetric activity assays with different substrates depending on the sirtuin whose deacetylase activity is assayed. These kits are based on similar principles as the homogeneous fluorescent deacetylase assay described above, as well as in the method described in [112]. In the so-called Fluor de Lys® (Fluorescent deacetylation of lysine) assay system, the deacetylation of the substrate by the sirtuin, sensitizes the substrate so that, in a second step, treatment with the appropriate developer produces a fluorophore. The fluorescence of the wells is then measured using a fluorometric reader and the percentage of inhibition exerted by each compound is calculated. The most recent sirtuin inhibitors identified by these assay kits include: indoles [68]; a non-peptide molecule containing an N-thioacetyl lysine [59]; four novel scaffolds [113]; tri- and tetrecyclic pyrimidinediones [114]; molecules containing N(epsilon)-modified lysine [58]; tenovins [24]; pseudopeptidic compounds [88]; JGB1741, a small molecule developed on sirtinol structure [115] and other sirtinol analogues [116]; cambinol analogs [117]; 2-anilinobenzamide derivatives [118]. It has been suggested that the aminomethylcoumarin moiety of the fluorogenic substrates used in the SIRT1 Fluor de Lys® assay can affect the binding of other small molecules to the enzyme [102, 103]. Therefore, confirmation of results with other assays might be recommended.
Charcoal Binding Assays -
In this assay, recombinant sirtuins are incubated with the appropriate [3H]-acetylated peptide substrate (depending on the tested sirtuin) and NAD+. In this setting, sirtuin activity produces [3H]-labeled OAADPr, nicotinamide and deacetylated peptide. Activated charcoal is then added to terminate the reaction: both OAADPr and the acetylated peptide bind to the activated charcoal, but under the conditions described in [50, 119], the [3H]-labeled acetate is hydrolyzed from OAADPr and remains in the supernatant, which is collected and radioactivity is quantified by liquid scintillation counting. Thus, this method measures the amount of [3H]-labeled OAADPr in the form of free acetate. This assay has been efficiently used to screen nicotinamide analogs and NAD+ metabolites as inhibitors of SIRT2 [50, 51].
Microfluidic mobility shift assay -
The microfluidic mobility shift assay technology separates substrates and products of enzymatic reactions: the differences in electrophoretic mobility depend on the charge and on the mass of the molecule. Quantitation of remaining substrate and newly generated product is achieved by measuring the fluorescence intensities of both product and substrate peaks. The mobility shift assay described in [120] effectively separate SIRT1 substrate fluorescein-labeled peptides containing a neutral acetylated lysine residue from product peptides containing a deacetylated lysine bearing an additional positive formal charge. Although this assay could be amenable to high throughput screenings, it appears more suitable for hit confirmation and mechanistic studies.
Bioluminescence assay -
This assay utilizes label-free peptide substrate and is based upon quantitation of remaining NAD+, after a deacetylation reaction catalyzed by sirtuins. The NAD+ bioluminescence assay couples NAD+ consumption to the bacterial luciferase-catalyzed oxidation of decanal. The method combines the quantitative reduction of remaining NAD+ to NADH with the measurement of NADH using Photobacterium fischeri luciferase and NAD(P)H: FMN-oxidoreductase in the presence of long-chain aldehyde. This method is applicable to assay of sirtuins with any peptide substrate and is highly amenable to high throughput screenings [120].
Radiochemical assay of Nicotinamide Release -
The activity of recombinant sirtuins can be measured with an assay using an appropriate peptide containing an acetylated lysine (substrate of the investigated sirtuin) and [carbonyl-14C]-NAD+. Among the possible methods to detect the release of [14C]nvestigated sirtuin) and]nicotinamide from [carbonyl-14C]-NAD+, the one developed by McDonagh and coll. uses commercially available materials and allows for high-throughput screenings. Incubations can be performed in 96-well plates and the deacetylase reaction is stopped by the addition of a boronic acid resin, which binds unreacted NAD+: by means of a filter plate on a vacuum manifold, the unbound fraction, containing released nicotinamide, can be collected and the radioactivity content can be detected in a scintillation counter [121]. By the use of this method, the identification of indoles as sirtuins inhibitor was confirmed [68].
Sirtuin enzyme-coupled assay for nicotinamide detection -
Although the above described method to detect nicotinamide release is suitable for high throughput screenings, the use of radioactive compounds raises problems of safety and waste disposal. In 2009, an enzyme-coupled assay for detection of nicotinamide, produced by different enzymes, including sirtuins, was developed [122]. With this spectrophotometric assay, nicotinamide formation is continuously measured using a coupled enzyme system with nicotinamidase and glutamate dehydrogenase. Nicotinamidase hydrolyzes nicotinamide to nicotinic acid and ammonia. Glutamate dehydrogenase then converts ammonia, α-ketoglutarate, and NAD(P)H to glutamate and NAD(P). NAD(P)H oxidation/consumption is measured spectrophotometrically at 340 nm. As far as we know, this method has not been used for large screenings of compounds as possible sirtuin inhibitors. However, the coupled assay could be adapted to a high-throughput format, and a continuous assay should avoid many of the artifacts associated with fluorescent and end-point assays.
Capillary electrophoresis-based sirtuin assay using non-peptide substrates -
Recently, non-peptide substrates for SIRT1 have been synthesized and two capillary electrophoresis assays have been developed and validated to monitor the deacetylation process of the substrates and to screen the activity of sirtuin inhibitors [123]. Small peptides containing a fluorescence label at the N-terminus had been previously established as substrates for sirtuins in CE-based assays [124, 125].
Following high throughput screenings based on the evaluation of enzymatic activity of recombinant sirtuins, the identification of possible sirtuin inhibitors is often confirmed with assays in which cells are exposed to the compounds, putative sirtuin inhibitors. Here we review the most commonly performed “functional” tests.
Western blot for identification of acetylated proteins -
This is by far the most used analysis: cells are exposed to different putative sirtuin inhibitors and total protein extracts are subjected to SDS-PAGE and Western blot. The membranes are then probed with antibody specific for acetylated lysine in proteins that are substrate of sirtuin activity (usually p53, histone 3, γ-tubulin): thus, different levels of acetylated proteins are quantified. An incomplete list of studies employing Western blot analyses to evaluate sirtuin inhibition includes: [23, 59, 65, 88, 111, 114, 115, 117, 126, 127].
Cell-based ELISA-type assay -
Huber and coll. exploited this method, developed to identify and evaluate histone acetyltransferase inhibitors [128], to confirm novel 3-Arylideneindolin-2-ones as SIRT2 inhibitors [111]. In this assay, well-suitable for high throughput screenings, cells are seeded into 96-well microtiter plates and treated with the different compounds. Cells are then fixed and permeabilized, incubated with a blocking solution and subsequently exposed to an antibody against acetylated α-tubulin and to a secondary anti-IgG mouse antibody properly labeled. The Enhancement solution is then added to each well and fluorescence intensity is measured [111]. Similarly, acetylated α-tubulin and α-tubulin are visualized by indirect immunofluorescence in [23].
CONCLUSION
The hunt for compounds that act as modulators of sirtuin activity is open and is eventually leading to the discovery of new (or old) molecules that could one day be used as therapeutics. The preclinical data that are currently available suggest numerous possible applications of this type of molecules for the treatment of human diseases and, possibly, for their prevention. However, a note of caution should be raised in that the true significance of sirtuin activity inside the cells is still elusive. So are also the biological significance of the protein modifications that sirtuins perform and the interplay between sirtuins and other regulatory proteins, such as macrodomain proteins, that act in concert with them to regulate intracellular ADP-ribose levels [129, 130]. The answers to these questions will come with time and will help design rational applications of sirtuin-targeting agents in humans.
ACKNOWLEDGEMENTS
This work was supported in part by the Associazione Italiana per la Ricerca sul Cancro (AIRC Start-up grant 6108 to AN and IB, and AIRC Emilia Romagna Start-up grant 6266 to MDP and ADR), by the European Seventh Framework Program (project number 256986, PANACREAS, to AN and SB), by the University of Genova.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
REFERENCES
- 1. Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260:273–9. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 2. Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999; 99:735–45. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- 3. Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000; 403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 4. Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009; 460:587–91. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dani N, Barbosa AJM, Del Rio A, Di Girolamo M. ADPribosylated proteins as old and new drug targets for anticancer therapy: the example of ARF6. Curr Pharm Des. 2013;19(4):624–33. [PubMed] [Google Scholar]
- 6. Libert S, Pointer K, Bell EL, et al. SIRT1 activates MAO-A in the brain to mediate anxiety and exploratory drive. Cell. 2011;147:1459–72. doi: 10.1016/j.cell.2011.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang RH, Sengupta K, Li C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008; 14:312–23. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim HS, Vassilopoulos A, Wang RH, et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell. 2011; 20:487–99. doi: 10.1016/j.ccr.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim HS, Patel K, Muldoon-Jacobs K, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010; 17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Finley LW, Carracedo A, Lee J, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19:416–28. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yeung F, Hoberg JE, Ramsey CS, et al. Modulation of NF-kappaBdependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004; 23:2369–80. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Firestein R, Blander G, Michan S, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PloS One. 2008; 3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003; 11:437–44. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 14. Van Meter M, Mao Z, Gorbunova V, Seluanov A. SIRT6 overexpression induces massive apoptosis in cancer cells but not in normal cells. Cell Cycle. 2011; 10:3153–8. doi: 10.4161/cc.10.18.17435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kawahara TL, Michishita E, Adler AS, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009; 136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhong L, D'Urso A, Toiber D, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010; 140:280–93. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luo J, Nikolaev AY, Imai S, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001; 107:137–48. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 18. Vaziri H, Dessain SK, Ng Eaton E, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001; 107:149–59. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 19. Brunet A, Sweeney LB, Sturgill JF, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004; 303:2011–5. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 20. Cohen HY, Miller C, Bitterman KJ, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004; 305:390–2. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 21. Langley E, Pearson M, Faretta M, et al. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002; 21:2383–96. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Menssen A, Hydbring P, Kapelle K, et al. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc Natl Acad Sci USA. 2012; 109:E187–96. doi: 10.1073/pnas.1105304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heltweg B, Gatbonton T, Schuler AD, et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006; 66:4368–77. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- 24. Lain S, Hollick JJ, Campbell J, et al. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell. 2008; 13:454–63. doi: 10.1016/j.ccr.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cea M, Soncini D, Fruscione F, et al. Synergistic interactions between HDAC and sirtuin inhibitors in human leukemia cells. PloS One. 2011; 6:e22739. doi: 10.1371/journal.pone.0022739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang B, Hasan MK, Alvarado E, Yuan H, Wu H, Chen WY. NAMPT overexpression in prostate cancer and its contribution to tumor cell survival and stress response. Oncogene. 2011; 30:907–21. doi: 10.1038/onc.2010.468. [DOI] [PubMed] [Google Scholar]
- 27. Audrito V, Vaisitti T, Rossi D, et al. Nicotinamide blocks proliferation and induces apoptosis of chronic lymphocytic leukemia cells through activation of the p53/miR-34a/SIRT1 tumor suppressor network. Cancer Res. 2011; 71:4473–83. doi: 10.1158/0008-5472.CAN-10-4452. [DOI] [PubMed] [Google Scholar]
- 28. Yuan H, Wang Z, Li L, et al. Activation of stress response gene SIRT1 by BCR-ABL promotes leukemogenesis. Blood. 2012;119:1904–14. doi: 10.1182/blood-2011-06-361691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li L, Wang L, Li L, et al. Activation of p53 by SIRT1 Inhibition Enhances Elimination of CML Leukemia Stem Cells in Combination with Imatinib. Cancer Cell. 2012; 21:266–81. doi: 10.1016/j.ccr.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peck B, Chen CY, Ho KK, et al. SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol Cancer Ther. 2010; 9:844–55. doi: 10.1158/1535-7163.MCT-09-0971. [DOI] [PubMed] [Google Scholar]
- 31. Li Y, Matsumori H, Nakayama Y, et al. SIRT2 down-regulation in HeLa can induce p53 accumulation via p38 MAPK activationdependent p300 decrease, eventually leading to apoptosis. Genes Cells. 2011; 16:34–45. doi: 10.1111/j.1365-2443.2010.01460.x. [DOI] [PubMed] [Google Scholar]
- 32. Zhang Y, Au Q, Zhang M, Barber JR, Ng SC, Zhang B. Identification of a small molecule SIRT2 inhibitor with selective tumor cytotoxicity. Biochem Biophys Res Commun. 2009;386:729–33. doi: 10.1016/j.bbrc.2009.06.113. [DOI] [PubMed] [Google Scholar]
- 33. Dan L, Klimenkova O, Klimiankou M, et al. The role of Sirtuin 2 activation by nicotinamide phosphoribosyltransferase in the aberrant proliferation and survival of myeloid leukemia cells. Haematologica. 2011; 97:551–9. doi: 10.3324/haematol.2011.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Inoue T, Nakayama Y, Yamada H, et al. SIRT2 downregulation confers resistance to microtubule inhibitors by prolonging chronic mitotic arrest. Cell cycle. 2009; 8:1279–91. doi: 10.4161/cc.8.8.8245. [DOI] [PubMed] [Google Scholar]
- 35. Alhazzazi TY, Kamarajan P, Joo N, et al. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer. 2011;117:1670–8. doi: 10.1002/cncr.25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mostoslavsky R, Chua KF, Lombard DB, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006; 124:315–29. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 37. Jia G, Su L, Singhal S, Liu X. Emerging roles of SIRT6 on telomere maintenance, DNA repair, metabolism and mammalian aging. Mol Cell Biochem. 2012; 364:345–50. doi: 10.1007/s11010-012-1236-8. [DOI] [PubMed] [Google Scholar]
- 38. McCord RA, Michishita E, Hong T, et al. SIRT6 stabilizes DNAdependent protein kinase at chromatin for DNA double-strand break repair. Aging (Albany NY) 2009; 1:109–21. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Van Gool F, Galli M, Gueydan C, et al. Intracellular NAD levels regulate tumor necrosis factor protein synthesis in a sirtuindependent manner. Nat Med. 2009; 15:206–10. doi: 10.1038/nm.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Truyers C, Kellen E, Arbyn M, et al. The use of human tissue in epidemiological research, ethical and legal considerations in two biobanks in Belgium. Med Health Care Philos. 2010; 13:169–75. doi: 10.1007/s11019-009-9230-y. [DOI] [PubMed] [Google Scholar]
- 41. Allavena P, Germano G, Marchesi F, Mantovani A. Chemokines in cancer related inflammation. Exp Cell Res. 2011; 317:664–73. doi: 10.1016/j.yexcr.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 42. Barber MF, Michishita-Kioi E, Xi Y, et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature . 2012; 487:114–8. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cea M, Cagnetta A, Gobbi M, et al. New Insights into the Treatment of Multiple Myeloma with Histone Deacetylase Inhibitors. Curr Pharm Des. 2013;19(4): 734–44. [PMC free article] [PubMed] [Google Scholar]
- 44. Min J, Landry J, Sternglanz R, Xu RM. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001; 105:269–79. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- 45. Pan PW, Feldman JL, Devries MK, Dong A, Edwards AM, Denu JM. Structure and biochemical functions of SIRT6. J Biol Chem . 2011;286(16):14575–87. doi: 10.1074/jbc.M111.218990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sanders BD, Jackson B, Marmorstein R. Structural basis for sirtuin function: what we know and what we don't. Biochim Biophys Acta . 2010; 1804:1604–16. doi: 10.1016/j.bbapap.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Andreoli F, Barbosa AJM, Parenti MD, Del Rio A. Modulation of epigenetic targets for anticancer therapy: clinicopathological relevance, structural data and drug discovery perspectives. Curr Pharm Des. 2013;19(4): 578–613. doi: 10.2174/138161213804581918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tervo AJ, Kyrylenko S, Niskanen P, et al. An in silico approach to discovering novel inhibitors of human sirtuin type 2. J Med Chem . 2004; 47:6292–8. doi: 10.1021/jm049933m. [DOI] [PubMed] [Google Scholar]
- 49. Jung-Hynes B, Nihal M, Zhong W, Ahmad N. Role of sirtuin histone deacetylase SIRT1 in prostate cancer. A target for prostate cancer management via its inhibition? J Biol Chem. 2009;284:3823–32. doi: 10.1074/jbc.M807869200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jackson MD, Schmidt MT, Oppenheimer NJ, Denu JM. Mechanism of nicotinamide inhibition and transglycosidation by Sir2 histone/protein deacetylases. J Biol Chem. 2003; 278:50985–98. doi: 10.1074/jbc.M306552200. [DOI] [PubMed] [Google Scholar]
- 51. Schmidt MT, Smith BC, Jackson MD, Denu JM. Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J Biol Chem. 2004; 279:40122–9. doi: 10.1074/jbc.M407484200. [DOI] [PubMed] [Google Scholar]
- 52. Fatkins DG, Monnot AD, Zheng W. Nepsilon-thioacetyl-lysine: a multi-facet functional probe for enzymatic protein lysine Nepsilondeacetylation. Bioorg Med Chem Lett. 2006; 16:3651–6. doi: 10.1016/j.bmcl.2006.04.075. [DOI] [PubMed] [Google Scholar]
- 53. Jamonnak N, Fatkins DG, Wei L, Zheng W. N(epsilon)-methanesulfonyl-lysine as a non-hydrolyzable functional surrogate for N(epsilon)-acetyl-lysine. Org Biomol Chem. 2007; 5:892–6. doi: 10.1039/b617185k. [DOI] [PubMed] [Google Scholar]
- 54. Jamonnak N, Hirsch BM, Pang Y, Zheng W. Substrate specificity of SIRT1-catalyzed lysine Nepsilon-deacetylation reaction probed with the side chain modified Nepsilon-acetyl-lysine analogs. Bioorg Chem. 2010; 38:17–25. doi: 10.1016/j.bioorg.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 55. Smith BC, Denu JM. Acetyl-lysine analog peptides as mechanistic probes of protein deacetylases. The J Biol Chem. 2007; 282:37256–65. doi: 10.1074/jbc.M707878200. [DOI] [PubMed] [Google Scholar]
- 56. Smith BC, Settles B, Hallows WC, Craven MW, Denu JM. SIRT3 substrate specificity determined by peptide arrays and machine learning. ACS Chem Biol. 2011; 6:146–57. doi: 10.1021/cb100218d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kiviranta PH, Suuronen T, Wallen EA, et al. N(epsilon)-thioacetyllysine-containing tri-, tetra-, and pentapeptides as SIRT1 and SIRT2 inhibitors. J Med Chem. 2009; 52:2153–6. doi: 10.1021/jm801401k. [DOI] [PubMed] [Google Scholar]
- 58. Huhtiniemi T, Suuronen T, Lahtela-Kakkonen M, et al. N(epsilon)-Modified lysine containing inhibitors for SIRT1 and SIRT2. Bioorg Med Chem. 2010; 18:5616–25. doi: 10.1016/j.bmc.2010.06.035. [DOI] [PubMed] [Google Scholar]
- 59. Suzuki T, Asaba T, Imai E, Tsumoto H, Nakagawa H, Miyata N. Identification of a cell-active non-peptide sirtuin inhibitor containing N-thioacetyl lysine. Bioorg Med Chem Lett. 2009; 19:5670–2. doi: 10.1016/j.bmcl.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 60. He B, Du J, Lin H. Thiosuccinyl peptides as sirt5-specific inhibitors. J Am Chem Soc. 2012; 134:1922–5. doi: 10.1021/ja2090417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Grozinger CM, Chao ED, Blackwell HE, Moazed D, Schreiber SL. Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J Biol Chem. 2001; 276:38837–43. doi: 10.1074/jbc.M106779200. [DOI] [PubMed] [Google Scholar]
- 62. Ota H, Tokunaga E, Chang K, et al. Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MAPK signaling in human cancer cells. Oncogene. 2006; 25:176–85. doi: 10.1038/sj.onc.1209049. [DOI] [PubMed] [Google Scholar]
- 63. Lara E, Mai A, Calvanese V, et al. Salermide, a Sirtuin inhibitor with a strong cancer-specific proapoptotic effect. Oncogene. 2009;28:781–91. doi: 10.1038/onc.2008.436. [DOI] [PubMed] [Google Scholar]
- 64. Bedalov A, Gatbonton T, Irvine WP, Gottschling DE, Simon JA. Identification of a small molecule inhibitor of Sir2p. Proc Natl Acad Sci USA. 2001; 98:15113–8. doi: 10.1073/pnas.261574398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Neugebauer RC, Uchiechowska U, Meier R, et al. Structureactivity studies on splitomicin derivatives as sirtuin inhibitors and computational prediction of binding mode. J Med Chem. 2008;51:1203–13. doi: 10.1021/jm700972e. [DOI] [PubMed] [Google Scholar]
- 66. Marshall GM, Liu PY, Gherardi S, et al. SIRT1 promotes N-Myc oncogenesis through a positive feedback loop involving the effects of MKP3 and ERK on N-Myc protein stability. PLoS Genet. 2011;7:e1002135. doi: 10.1371/journal.pgen.1002135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Uciechowska U, Schemies J, Neugebauer RC, et al. Thiobarbiturates as sirtuin inhibitors: virtual screening, free-energy calculations, and biological testing. ChemMedChem. 2008; 3:1965–76. doi: 10.1002/cmdc.200800104. [DOI] [PubMed] [Google Scholar]
- 68. Napper AD, Hixon J, McDonagh T, et al. Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J Med Chem. 2005; 48:8045–54. doi: 10.1021/jm050522v. [DOI] [PubMed] [Google Scholar]
- 69. Trapp J, Jochum A, Meier R, et al. Adenosine mimetics as inhibitors of NAD+-dependent histone deacetylases, from kinase to sirtuin inhibition. J Med Chem. 2006; 49:7307–16. doi: 10.1021/jm060118b. [DOI] [PubMed] [Google Scholar]
- 70. Howitz KT, Bitterman KJ, Cohen HY, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003; 425:191–6. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 71. McGeary RP, Bennett AJ, Tran QB, Cosgrove KL, Ross BP. Suramin: clinical uses and structure-activity relationships. Mini Rev Med Chem. 2008; 8:1384–94. doi: 10.2174/138955708786369573. [DOI] [PubMed] [Google Scholar]
- 72. Gill JS, Windebank AJ. Suramin induced ceramide accumulation leads to apoptotic cell death in dorsal root ganglion neurons. Cell Death Differ. 1998; 5:876–83. doi: 10.1038/sj.cdd.4400410. [DOI] [PubMed] [Google Scholar]
- 73. Trapp J, Meier R, Hongwiset D, Kassack MU, Sippl W, Jung M. Structure-activity studies on suramin analogues as inhibitors of NAD+-dependent histone deacetylases (sirtuins) ChemMedChem. 2007; 2:1419–31. doi: 10.1002/cmdc.200700003. [DOI] [PubMed] [Google Scholar]
- 74. Schuetz A, Min J, Antoshenko T, et al. Structural basis of inhibition of the human NAD+-dependent deacetylase SIRT5 by suramin. Structure. 2007; 15:377–89. doi: 10.1016/j.str.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 75. Outeiro TF, Kontopoulos E, Altmann SM, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science. 2007; 317:516–9. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 76. Mai A, Valente S, Meade S, et al. Study of 1,4-dihydropyridine structural scaffold: discovery of novel sirtuin activators and inhibitors. J Med Chem. 2009; 52:5496–504. doi: 10.1021/jm9008289. [DOI] [PubMed] [Google Scholar]
- 77. Tavares J, Ouaissi A, Kong Thoo Lin P, et al. Bisnaphthalimidopropyl derivatives as inhibitors of Leishmania SIR2 related protein 1. ChemMedChem. 2010; 5:140–7. doi: 10.1002/cmdc.200900367. [DOI] [PubMed] [Google Scholar]
- 78. Gey C, Kyrylenko S, Hennig L, et al. Phloroglucinol derivatives guttiferone G, aristoforin, and hyperforin: inhibitors of human sirtuins SIRT1 and SIRT2. Angew Chem Int Ed Engl. 2007;46:5219–22. doi: 10.1002/anie.200605207. [DOI] [PubMed] [Google Scholar]
- 79. Oh WK, Cho KB, Hien TT, et al. Amurensin G, a potent natural SIRT1 inhibitor, rescues doxorubicin responsiveness via downregulation of multidrug resistance 1. Mol Pharmacol. 2010; 78:855–64. doi: 10.1124/mol.110.065961. [DOI] [PubMed] [Google Scholar]
- 80. Kahyo T, Ichikawa S, Hatanaka T, Yamada MK, Setou M. A novel chalcone polyphenol inhibits the deacetylase activity of SIRT1 and cell growth in HEK293T cells. J Pharmacol Sci. 2008; 108:364–71. doi: 10.1254/jphs.08203fp. [DOI] [PubMed] [Google Scholar]
- 81. Gutierrez M, Andrianasolo EH, Shin WK, et al. Structural and synthetic investigations of tanikolide dimer, a SIRT2 selective inhibitor, and tanikolide seco-acid from the Madagascar marine cyanobacterium Lyngbya majuscula. J Org Chem. 2009; 74:5267–75. doi: 10.1021/jo900578j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Neugebauer RC, Sippl W, Jung M. Inhibitors of NAD+ dependent histone deacetylases (sirtuins) Curr Pharm Des. 2008; 14:562–73. doi: 10.2174/138161208783885380. [DOI] [PubMed] [Google Scholar]
- 83. Cen Y. Sirtuins inhibitors: the approach to affinity and selectivity. Biochim Biophys Acta. 2010; 1804:1635–44. doi: 10.1016/j.bbapap.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 84. Lawson M, Uciechowska U, Schemies J, Rumpf T, Jung M, Sippl W. Inhibitors to understand molecular mechanisms of NAD(+)- dependent deacetylases (sirtuins) Biochim Biophys Acta. 2010;1799:726–39. doi: 10.1016/j.bbagrm.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 85. Chen L. Medicinal chemistry of sirtuin inhibitors. Curr Med Chem. 2011; 18:1936–46. doi: 10.2174/092986711795590057. [DOI] [PubMed] [Google Scholar]
- 86. Stunkel W, Campbell RM. Sirtuin 1 (SIRT1): the misunderstood HDAC. J Biomol Screen. 2011; 16:1153–69. doi: 10.1177/1087057111422103. [DOI] [PubMed] [Google Scholar]
- 87. Moniot S, Weyand M, Steegborn C. Structures, substrates, and regulators of Mammalian sirtuins - opportunities and challenges for drug development. Front Pharmacol. 2012; 3:16. doi: 10.3389/fphar.2012.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Huhtiniemi T, Salo HS, Suuronen T, et al. Structure-based design of pseudopeptidic inhibitors for SIRT1 and SIRT2. J Med Chem . 2011; 54:6456–68. doi: 10.1021/jm200590k. [DOI] [PubMed] [Google Scholar]
- 89. Schlicker C, Boanca G, Lakshminarasimhan M, Steegborn C. Structure-based development of novel sirtuin inhibitors. Aging (Albany NY) 2011; 3:852–72. doi: 10.18632/aging.100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sivaraman P, Mattegunta S, Subbaraju GV, Satyanarayana C, Padmanabhan B. Design of a novel nucleoside analog as potent inhibitor of the NAD dependent deacetylase, SIRT2. Syst Synth Biol. 2010; 4:257–63. doi: 10.1007/s11693-011-9069-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Huhtiniemi T, Suuronen T, Rinne VM, et al. Oxadiazolecarbonylaminothioureas as SIRT1 and SIRT2 inhibitors. J Med Chem. 2008; 51:4377–80. doi: 10.1021/jm800639h. [DOI] [PubMed] [Google Scholar]
- 92. Kiviranta PH, Leppanen J, Kyrylenko S, et al. N,N'-Bisbenzylidenebenzene-1,4-diamines and N,N'-Bisbenzylidenenaphthalene- 1,4-diamines as Sirtuin Type 2 (SIRT2) Inhibitors. J Med Chem. 2006; 49:7907–11. doi: 10.1021/jm060566j. [DOI] [PubMed] [Google Scholar]
- 93. Tervo AJ, Suuronen T, Kyrylenko S, et al. Discovering inhibitors of human sirtuin type 2: novel structural scaffolds. J Med Chem . 2006; 49: 7239–41. doi: 10.1021/jm060686r. [DOI] [PubMed] [Google Scholar]
- 94. Barbosa AJ, Del Rio A. Freely accessible databases of commercial compounds for high-throughput virtual screenings. Curr Top Med Chem. 2012; 12:886–77. doi: 10.2174/156802612800166710. [DOI] [PubMed] [Google Scholar]
- 95. Del Rio A, Barbosa AJ, Caporuscio F, Mangiatordi GF. CoCoCo: a free suite of multiconformational chemical databases for highthroughput virtual screening purposes. Mol Biosyst. 2010; 6:2122–8. doi: 10.1039/c0mb00039f. [DOI] [PubMed] [Google Scholar]
- 96. Mak L, Liggi S, Tan L, et al. Anti-cancer drug development: Computational strategies to identify and target proteins involved in cancer metabolism. Curr Pharm Des. 2013;19(4): 532–77. [PubMed] [Google Scholar]
- 97. Milne JC, Lambert PD, Schenk S, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007; 450:712–6. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Dai H, Kustigian L, Carney D, et al. SIRT1 activation by small molecules: kinetic and biophysical evidence for direct interaction of enzyme and activator. J Biol Chem. 2010; 285:32695–703. doi: 10.1074/jbc.M110.133892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Gupta SC, Kim JH, Prasad S, Aggarwal BB. Regulation of survival, proliferation, invasion, angiogenesis, and metastasis of tumor cells through modulation of inflammatory pathways by nutraceuticals. Cancer Metastasis Rev. 2010; 29:405–34. doi: 10.1007/s10555-010-9235-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chauhan D, Bandi M, Singh AV, et al. Preclinical evaluation of a novel SIRT1 modulator SRT1720 in multiple myeloma cells. Br J Haematol. 2011; 155:588–98. doi: 10.1111/j.1365-2141.2011.08888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Huber JL, McBurney MW, Distefano PS, McDonagh T. SIRT1-independent mechanisms of the putative sirtuin enzyme activators SRT1720 and SRT2183. Future Med Chem. 2010;2:1751–9. doi: 10.4155/fmc.10.257. [DOI] [PubMed] [Google Scholar]
- 102. Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem. 2005; 280:17187–95. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 103. Kaeberlein M, McDonagh T, Heltweg B, et al. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005; 280:17038–45. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 104. Pacholec M, Bleasdale JE, Chrunyk B, et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem. 2010; 285:8340–51. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Beher D, Wu J, Cumine S, et al. Resveratrol is not a direct activator of SIRT1 enzyme activity. Chem Biol Drug Des. 2009;74:619–24. doi: 10.1111/j.1747-0285.2009.00901.x. [DOI] [PubMed] [Google Scholar]
- 106. Park SJ, Ahmad F, Philp A, et al. Resveratrol ameliorates agingrelated metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012; 148:421–33. doi: 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Price NL, Gomes AP, Ling AJY, et al. SIRT1 Is Required for AMPK Activation and the Beneficial Effects of Resveratrol on Mitochondrial Function. Cell Metab. 2012; 15:675–90. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Denu JM. Fortifying the Link between SIRT1, Resveratrol, and Mitochondrial Function. Cell Metab. 2012; 15:566–7. doi: 10.1016/j.cmet.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Alcain FJ, Villalba JM. Sirtuin activators. Expert Opin Ther Pat. 2009; 19:403–14. doi: 10.1517/13543770902762893. [DOI] [PubMed] [Google Scholar]
- 110. Heltweg B, Trapp J, Jung M. In vitro assays for the determination of histone deacetylase activity. Methods. 2005; 36:332–7. doi: 10.1016/j.ymeth.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 111. Huber K, Schemies J, Uciechowska U, et al. Novel 3-arylideneindolin-2-ones as inhibitors of NAD+ -dependent histone deacetylases (sirtuins) J Med Chem. 2010; 53:1383–6. doi: 10.1021/jm901055u. [DOI] [PubMed] [Google Scholar]
- 112. Marcotte PA, Richardson PL, Guo J, et al. Fluorescence assay of SIRT protein deacetylases using an acetylated peptide substrate and a secondary trypsin reaction. Anal Biochem. 2004; 332:90–9. doi: 10.1016/j.ab.2004.05.039. [DOI] [PubMed] [Google Scholar]
- 113. Sanders BD, Jackson B, Brent M, et al. Identification and characterization of novel sirtuin inhibitor scaffolds. Bioorg Med Chem. 2009; 17:7031–41. doi: 10.1016/j.bmc.2009.07.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Rotili D, Mai A. Targeting Histone Demethylases: A New Avenue for the Fight against Cancer. Genes Cancer. 2011; 2:663–79. doi: 10.1177/1947601911417976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kalle AM, Mallika A, Badiger J, Alinakhi Talukdar P. Sachchidanand. Inhibition of SIRT1 by a small molecule induces apoptosis in breast cancer cells. Biochem Biophys Res Commun. 2010; 401:13–9. doi: 10.1016/j.bbrc.2010.08.118. [DOI] [PubMed] [Google Scholar]
- 116. Mai A, Massa S, Lavu S, et al. Design, synthesis, and biological evaluation of sirtinol analogues as class III histone/protein deacetylase (Sirtuin) inhibitors. J Med Chem. 2005; 48:7789–95. doi: 10.1021/jm050100l. [DOI] [PubMed] [Google Scholar]
- 117. Medda F, Russell RJ, Higgins M, et al. Novel cambinol analogs as sirtuin inhibitors: synthesis, biological evaluation, and rationalization of activity. J Med Chem. 2009; 52:2673–82. doi: 10.1021/jm8014298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Suzuki T, Imai K, Imai E, et al. Design, synthesis, enzyme inhibition, and tumor cell growth inhibition of 2-anilinobenzamide derivatives as SIRT1 inhibitors. Bioorg Med Chem. 2009; 17:5900–5. doi: 10.1016/j.bmc.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 119. Borra MT, Denu JM. Quantitative assays for characterization of the Sir2 family of NAD(+)-dependent deacetylases. Methods Enzymol . 2004; 376:171–87. doi: 10.1016/S0076-6879(03)76011-X. [DOI] [PubMed] [Google Scholar]
- 120. Liu Y, Gerber R, Wu J, Tsuruda T, McCarter JD. High-throughput assays for sirtuin enzymes: a microfluidic mobility shift assay and a bioluminescence assay. Anal Biochem. 2008; 378:53–9. doi: 10.1016/j.ab.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 121. McDonagh T, Hixon J, DiStefano PS, Curtis R, Napper AD. Microplate filtration assay for nicotinamide release from NAD using a boronic acid resin. Methods. 2005; 36:346–50. doi: 10.1016/j.ymeth.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 122. Smith BC, Hallows WC, Denu JM. A continuous microplate assay for sirtuins and nicotinamide-producing enzymes. Anal Biochem . 2009; 394:101–9. doi: 10.1016/j.ab.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Fan Y, Hense M, Ludewig R, Weisgerber C, Scriba GK. Capillary electrophoresis-based sirtuin assay using non-peptide substrates. J Pharm Biomed Anal. 2011; 54:772–8. doi: 10.1016/j.jpba.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 124. Fan Y, Ludewig R, Imhof D, Scriba GK. Development of a capillary electrophoresis-based assay of sirtuin enzymes. Electrophoresis. 2008; 29:3717–23. doi: 10.1002/elps.200800361. [DOI] [PubMed] [Google Scholar]
- 125. Fan Y, Ludewig R, Scriba GK. 9-Fluorenylmethoxycarbonyllabeled peptides as substrates in a capillary electrophoresis-based assay for sirtuin enzymes. Anal Biochem. 2009; 387:243–8. doi: 10.1016/j.ab.2009.01.038. [DOI] [PubMed] [Google Scholar]
- 126. Solomon JM, Pasupuleti R, Xu L, et al. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol Cell Biol. 2006; 26:28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Suzuki T, Imai K, Nakagawa H, Miyata N. 2-Anilinobenzamides as SIRT inhibitors. ChemMedChem. 2006; 1:1059–62. doi: 10.1002/cmdc.200600162. [DOI] [PubMed] [Google Scholar]
- 128. Wynne Aherne G, Rowlands MG, Stimson L, Workman P. Assays for the identification and evaluation of histone acetyltransferase inhibitors. Methods. 2002; 26:245–53. doi: 10.1016/S1046-2023(02)00028-2. [DOI] [PubMed] [Google Scholar]
- 129. Peterson FC, Chen D, Lytle BL, et al. Orphan macrodomain protein (human C6orf130) is an O-acyl-ADP-ribose deacylase: solution structure and catalytic properties. J Biol Chem. 2011;286:35955–65. doi: 10.1074/jbc.M111.276238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Chen D, Vollmar M, Rossi MN, et al. Identification of macrodomain proteins as novel O-acetyl-ADP-ribose deacetylases. J Biol Chem. 2011;286:13261–71. doi: 10.1074/jbc.M110.206771. [DOI] [PMC free article] [PubMed] [Google Scholar]