Abstract
Dopaminergic neurons derived from pluripotent stem cells are among the best investigated products of in vitro stem cell differentiation owing to their potential use for neurorestorative therapy of Parkinson's disease. However, the classical differentiation protocols for both mouse and human pluripotent stem cells generate a limited percentage of dopaminergic neurons and yield a considerable cellular heterogeneity comprising numerous scarcely characterized cell populations. To improve pluripotent stem cell differentiation protocols for midbrain dopaminergic neurons, we established extensive and strictly quantitative gene expression profiles, including markers for pluripotent cells, neural progenitors, non-neural cells, pan-neuronal and glial cells, neurotransmitter phenotypes, midbrain and nonmidbrain populations, floor plate and basal plate populations, as well as for Hedgehog, Fgf, and Wnt signaling pathways. The profiles were applied to discrete stages of in vitro differentiation of mouse embryonic stem cells toward the dopaminergic lineage and after transplantation into the striatum of 6-hydroxy-dopamine-lesioned rats. The comparison of gene expression in vitro with stages in the developing ventral midbrain between embryonic day 11.5 and 13.5 ex vivo revealed dynamic changes in the expression of transcription factors and signaling molecules. Based on these profiles, we propose quantitative gene expression milestones that predict the efficiency of dopaminergic differentiation achieved at the end point of the protocol, already at earlier stages of differentiation.
Introduction
Pluripotent stem cell-derived midbrain dopaminergic (mDAergic) neurons carry high hopes for cell replacement strategies of Parkinson's disease (PD). In vitro differentiation of mouse embryonic stem (ES) cells into mDAergic neurons can be achieved as the end point of a multistep cell culture protocol, which is based on an in vitro recapitulation of neuronal development in response to patterning factors defining ventral midbrain (VM) identity in vivo [1]. A large number of studies have confirmed the generation of mouse and human mDAergic neurons from pluripotent cells in protocols following this rational [2–6].
The process of early midbrain development in vivo is best studied in rodent embryos, where extrinsic factors, including Sonic hedgehog (Shh) [7–9], fibroblast growth factor 8 (Fgf8) [10–15], and Wnt1/3a/5a [14,16,17], as well as intrinsic factors, including Otx2 [13,18–21], En1/2 [22–24], Lmx1a/b [9,14,25–28], and Foxa2 [9,14,28–30] have been identified to contributing to mDAergic neuronal identity. Terminal differentiation of mDAergic neurons is classically documented by the expression of neurotransmitter phenotype markers, including tyrosine hydroxylase (Th), dopamine transporter (Dat), Vmat2 [31] and additional markers, such as Girk2 and Calbindin [24].
Expression of these midbrain patterning markers and mDAergic specification markers is used to characterize populations of cells differentiating from pluripotent stem cells in cell culture. The corresponding cell culture protocols extend over several weeks and follow strict timelines based on days in vitro (DIV). The central treatment consists of a combination of Shh and Fgfs in the patterning step [1,6,32]. A frequently observed problem is that, unless specific genetic selection procedures are applied, after completion of the protocols, the proportion of cells displaying morphological and cellular properties of mDAergic neurons is generally low and typically ranges between 5% and 15% [1,3,33]. The remaining cells are a poorly characterized mixed population of stem cell derivatives.
After transplantation in animal models of PD, such mixed populations of stem cell derivatives, including mDAergic neurons, derived from both mouse and human pluripotent cells integrate and survive. The functionality of the grafts is demonstrated by the finding that they can provide behavioral recovery [2,3,33–37]. However, only a small number of Th-positive cells has been found within the grafts [3,37] and the fate of multiple other populations of grafted cells has remained largely unknown.
In view of the already successful initial strategies to generate functional mDAergic neurons for neuroregenerative approaches in vitro, the existing methods for producing pluripotent cell-derived neurons need to be further refined. A major goal is to generate a population of neurons and glia that reflects the phenotypes of cells in the developing midbrain floor plate more closely [20].
The intrinsic difficulty to steer cell–cell interactions during differentiation in vitro frequently results in sporadic differentiation giving rise to heterogeneous cell populations. To improve the in vitro differentiation protocols, 2 complementary approaches can be taken. First, it is important to improve the monitoring methods for all relevant cell populations to achieve a higher percentage of the wanted midbrain cell populations and concomitant low levels of unwanted cells, in particular, proliferative cells giving rise to tumors and non-neural populations. Second, it is equally important to establish quantitative correlations between gene expression during midbrain specification processes in vivo and in stem cell-derived populations in vitro and following transplantation.
To address these problems, we systematically investigated cell fates during differentiation by quantitative monitoring of gene expression levels. In our experiments we employed a well-established 5-stage protocol for mDAergic differentiation of mouse ES cells [1]. We hypothesized that the definition of specific milestones for in vitro differentiation based on quantitative profiling of gene expression in cultures at early and intermediate stages of differentiation would predict cell fate decisions at terminal differentiation stages. To this end, we studied midbrain- and basal plate-specific regional markers and components of the Hedgehog (Hh), Fgf and Wnt signaling pathways, as well as the pan-neural markers, in a comparative approach in vitro versus in vivo. Additionally, we have initiated the analysis of lineage marker expression in stem cell-derived populations grafted into the striatum of 6-hydroxy-dopamine (OHDA) lesioned rats as an animal model of PD.
Materials and Methods
Cell cultures
The mouse ES cell lines used were B6G-2 (passage 16–25) derived from the C57BL/6 mouse expressing enhanced green fluorescent protein (eGfp) [38] from the RIKEN Bio Research Center and E14tg2a (passage 9–15) [39]. The classical 5-stage protocol for mDAergic differentiation was followed [1], with minor modifications as described in Fig. 1.
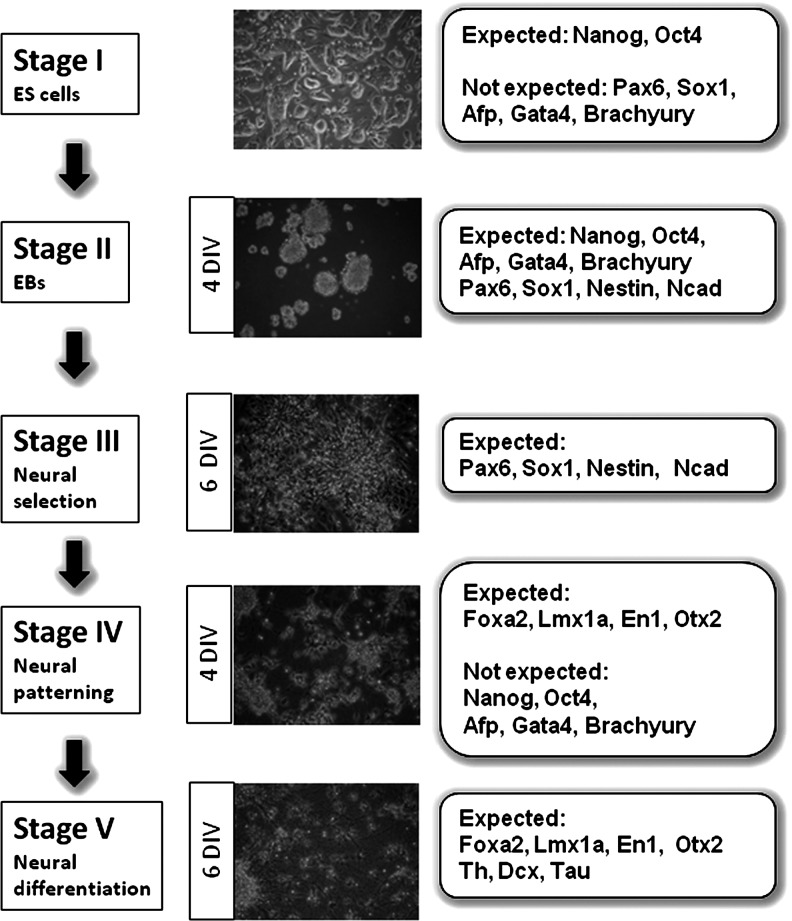
FIG. 1.
Schematic representation of the 5-stage protocol of mouse ES cell differentiation toward mDAergic neurons. Expected and not expected patterns of gene expression in each stage. ES, embryonic stem; mDAergic, midbrain dopaminergic; DIV, days in vitro.
Stage I: ES cells culture
ES cells were grown on gelatin-coated tissue culture plates in the presence of 1,400 U/mL of leukemia inhibitory factor (LIF; Millipore) in the ES cell medium consisting of the knockout Dulbecco's minimal essential medium (DMEM) supplemented with 15% knockout serum replacement, 0.1 mM MEM nonessential amino acids, 0.5 mM 2-mercaptoethanol, and 2 mM L-glutamine (all from Invitrogen Corporation).
Stage II: formation of embryoid bodies
To induce the differentiation, ES cells were dissociated with 0.05% trypsin/ethylenediaminetetraacetic acid (Sigma) and plated onto nonadherent bacterial dishes at a density of 2×104 cells/cm2 and cultured in the ES cell medium without LIF for 4 DIV.
Stage III: neural selection
Embryoid bodies (EBs) were plated onto adhesive tissue culture surface and left overnight to adhere, and then the medium was replaced with the neural selection medium, which consisted of DMEM/F12 with glutamax (Invitrogen), 1.5 mg/mL glucose, 0.24% NaHCO3, 5 μg/mL fibronectin, and 1× insulin/transferrin/sodium selenite supplement (all from Sigma). Cells were cultured for 6 DIV; meanwhile, neural progenitor cells were visible and formed neural rosettes.
Stage IV: neural patterning
Cells were dissociated with accutase (Invitrogen) and plated on polyornithin/laminin-coated tissue culture dishes or coverslips at a concentration of 2×105 cells/cm2 in the N2 medium consisting of DMEM/F12 with Glutamax (Invitrogen), 1.5 mg/mL glucose, 0.17% NaHCO3, 1% ascorbic acid (Sigma), 1% N-2 Plus Media Supplement (Invitrogen), and supplemented with 0–3 μM purmorphamine (Calbiochem), 10 ng/mL Fgf2 (Sigma), and 100 ng/mL human Fgf8 isoform b (Peprotech). The patterning treatment was kept for 4 DIV. In a preliminary experiment, we determined that Shh as in Lee et al. [1] can be replaced by the small molecule agonist, purmorphamine, at 2 μM (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/scd), in concordance with other reported studies [40–42].
Stage V: neural differentiation
Differentiation was induced by withdrawing of Fgf2, Fgf8, and purmorphamine from the N2 medium, additionally supplemented with 1% ascorbic acid (Sigma). The cells were cultured for 6 DIV.
Embryonic midbrain dissection
Embryonic mouse brain regions at E11.5, E12.5, and E13.5 were isolated from the mouse line C57BL/6-TgN(act-eGfp)OsbC14-Y01-FM131 from RIKEN Bio Research Centre [43,44] according to Dunnett and Björklund [45]. Briefly, embryos were harvested from staged pregnant mice and the brains were collected in the glucose saline medium; the midbrain was isolated and the tube was opened by cutting along its lumen. The dorsal midbrain was removed by cutting half of the wings of the opened tube, leaving a butterfly shape that represents the VM. The pieces were collected in the lysis buffer (Invitrogen) for mRNA extraction.
Quantitative real-time polymerase chain reaction
mRNA was isolated using Dynabeads® Oligo (dT)25 (Invitrogen) following the manufacturer's protocol. Briefly, mRNA was annealed to the beads by incubation in the lysis/binding buffer on a rotating mixer for 10 min at room temperature. After several washing steps, mRNA was eluted from the beads by adding 13 μL 10 mM Tris-HCl, and incubated for 3 min at 85°C. The eluted mRNA was quickly transferred to a new tube, chilled on ice, and immediately used for cDNA synthesis according to the High-Capacity cDNA Reverse Transcription Kit protocol (Applied Biosystems). The levels of cDNA were assessed by quantitative real-time polymerase chain reaction using the Fast SYBR® Green Master Mix (Applied Biosystems). Standard curves and melting curves were determined for each set of primers (Supplementary Table S1) to confirm that a single amplicon was generated. All results from 3 to 6 experiments, each experiment with 3 technical replicates were normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh) or to eGfp and expressed as ΔCt values (low ΔCt levels indicate high expression). Relative expression ratios were calculated by the ΔΔCt method [46].
Immunocytochemistry
Adherent cells on glass coverslips were fixed with 4% buffered paraformaldehyde in phosphate-buffered saline (PBS; 0.01 M, pH 7.4), for 20 min at room temperature and washed twice with PBS. Permeabilization was carried out using 0.3% Triton-X-100 in PBS (PBST) for 1 h at room temperature. After blocking (10% goat normal serum, 1% bovine serum albumin in PBST), cells were incubated overnight at 4°C with primary antibodies against Nestin, Tau [mouse immunoglobulin G (IgG), 1:200; Santa Cruz], Tau, glial fibrillary acidic protein (Gfap rabbit IgG, 1:1,000; Dako), oligodendrocyte-specific protein (Osp rabbit IgG, 1:500; Abcam), Th, GABA (mouse IgG, 1:1,000; Sigma), Glutamate (rabbit IgG, 1:10,000; Sigma), Vacht (rabbit IgG, 1:1,000; Sigma), and Otx2 (rabbit IgG, 1:1,000; Millipore).
After several rinses with PBST, cells were incubated for 1 h with 488- or 555-conjugated Alexa Fluor secondary antibodies (goat anti-mouse and goat anti-rabbit, 1:2,000; Invitrogen) and washed 3 times in PBS. The coverslips were incubated for 2 min with 4′, 6-diamidino-2-phenylindole (300 nM in PBS) for nuclear staining, rinsed in deionized water, and mounted on slides with Mowiol (Sigma). Specificity of the antibodies was tested on mouse embryonic brain samples. Negative controls, without a primary antibody, were performed in all experiments to monitor the nonspecific staining. Pictures were taken with an ApoTome Imaging System based on Axio Observer Z1 (Zeiss) using AxioVision software. The populations of Nestin-, Tau-, Otx2-, and Th-positive cells for 3 coverslips in each experimental group for at least 5 fields per coverslip were counted, using Metamorph software (Molecular Devices).
Statistics
For statistical analyses, the Statview (version 5.0.1) software was used (SAS Institute). Three to six independent biological samples for each time point contributed to the data set. Data are presented as mean±standard error of the mean. The Student's t-test was applied to compare between 2 groups. When multiple groups were compared, one way analysis of variance was employed, followed by the Student-Newman-Keul's post hoc test. Differences were considered significant when P<0.05.
In vivo transplantation
Animal model of PD
Experimental procedures were performed according to a standard protocol [4,47] in accordance with local ethical regulations (reference number do.ZI. 6545). Male Wistar rats weighing 200–250 g at the beginning of the experiment were used. Under anesthesia with isoflurane (induction 3.5%, follow up 1.5%–2%), the animals received a unilateral injection of 4 μL 6-OHDA (2 μg/μL in 0.9% NaCl with 0.1% ascorbic acid; Sigma) at a rate of 1 μL/min, into the left medial forebrain bundle at the following coordinates: AP −2.2, L +1.5, and V −8.0 (in mm, with reference to bregma and dura).
Grafting procedure
ES cell-derived population was prepared for grafting and grafted intrastriatally in previously 6-OHDA lesioned rats, using standard procedures [34]. Viability and cell number were assessed by Trypan blue dye exclusion. A total of 2 μL of cell suspension (∼4,000 cells) was injected into the striatum, in 2 deposits at the following coordinates: AP 0.48; L 2.2; V1 −5.5; V2 −4.5; tooth bar −3.3. The sham animals received the same injection only with culture medium. All animals received intraperitoneally 10 mg/kg cyclosporine-A daily; after 6 weeks, they were sacrificed under deep thiopental (120–150 mg/kg) anesthesia. The brains were removed, dissected, and cut into 400-μm slices. Fluorescent grafts as well as the sham striatum were dissected with the use of a Leica M205FA high-magnification fluorescent stereomicroscope and collected individually into the lysis buffer for mRNA extraction.
Results
Establishment of marker profiles for the 5-stage protocol of mDAergic differentiation from mouse ES cells
Molecular profiles for the mDAergic differentiation in vitro were established for the 5-stage protocol of ES cell differentiation first described by Lee et al. [1]. Individual sets of marker genes were defined for each step. The initial profiles were composed of genes expected to be expressed in a successful experiment combined with markers indicating undesirable experimental outcome (Fig. 1). These profiles contained the pluripotent markers Nanog and Oct4, the neural progenitor markers Pax6, Sox1, Ncad, and Nestin, the pan-neuronal markers Dcx and Tau, and the VM markers Otx2, En1, Foxa2, and Lmx1a. Th was chosen as the screening marker for DAergic neurons, since the most specific marker, Dat, is only expressed at very terminal stages of DAergic neuronal differentiation. As an a priori criterion for the success of a differentiation experiment a percentage of Th-positive cells >8% at the end point was defined based on previously published data [1,3,33].
This set of markers was tested in a first round of 6 independent experiments with 2 mouse ES cell lines. Cell samples were analyzed after each step of the protocol (Fig. 2). Nanog and Oct4 were highly expressed in ES cells (stage I) with ΔCt: 4.5±0.4 and 0.8±0.5 and EBs (stage II) with ΔCt: 4.8±0.3 and 1.5±0.2. Expression levels decreased after neural induction in stage III (∼6-fold and 10-fold, respectively), and further after neural patterning in stage IV (∼85-fold and 40-fold, respectively). At stage V both transcripts were below the detection limit of the method (Fig. 2A).
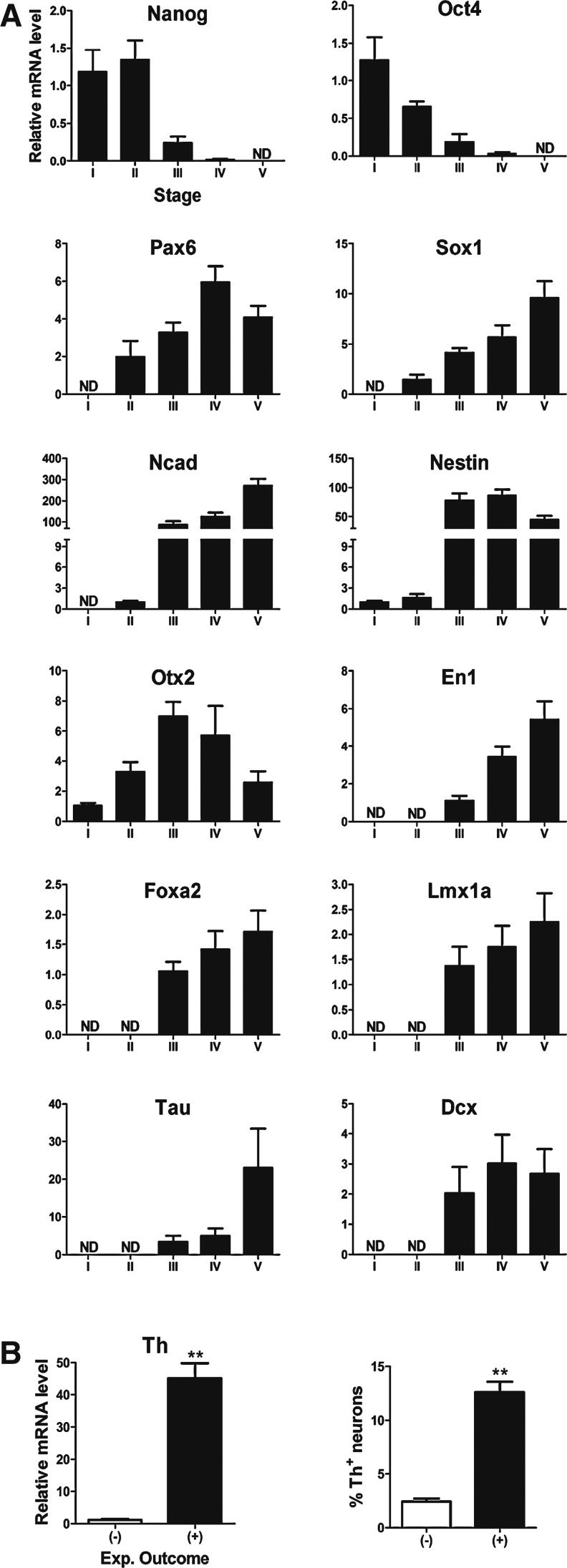
FIG. 2.
Molecular profiling of the temporal expression of expected genes in the 5-stage of mouse ES cell differentiation toward mDAergic neurons. (A) Relative mRNA expression levels for pluripotent markers, Nanog and Oct4, neural progenitor markers, Pax6, Sox1, Nestin, and Ncad, ventral midbrain (VM) markers, En1, Otx2, Foxa2, and Lmx1a as well as pan-neuronal markers, Tau and Dcx. Mean±SEM; n=6. All markers were compared to the stage, where the expression was first detected by real-time PCR. Gene expression was considered as not detected (ND) when ΔCt >14 cycles. (B) Relative mRNA levels and percentage of Th+neurons in stage V show 2 different experimental outcomes: one positive (+), where Th+ neurons were found, and another negative (−), where very few Th+ neurons were counted (see also Supplementary Table S2). Mean±SEM; n=3. **P<0.01 significantly different from (−) experimental outcome by Student t-test. Th, tyrosine hydroxylase; SEM, standard error of the mean; PCR, polymerase chain reaction.
The expression of the neural progenitor markers, Pax6, Sox1, Ncad, and Nestin, was first detected at the end point of stage II (ΔCt: 7.4±0.8, 6.3±0.6, 10.1±0.2, and 9.1±0.5, respectively), and was generally upregulated at the end point of stage III (ΔCt: 5.9±0.2, 4.3±0.2, 3.8±0.3, and 3.2±0.2, respectively). The largest increase was observed for Ncad and Nestin and their high expression levels were maintained during the consecutive stages (Fig. 2A).
The pan-neuronal markers, Dcx and Tau, were expressed in stage III with variations between experiments (ΔCt: 5.2±0.9 and 8.5±1.2, respectively). At the end point of the experiments, Dcx expression remained at the same high levels (ΔCt: 4.8±0.8), while Tau expression was ∼20-fold upregulated (ΔCt: 4.3±0.6) (Fig. 2A).
mRNAs for the mDAergic progenitor markers, En1, Foxa2, and Lmx1a, were detected at moderate levels at the end point of stage III (ΔCt: 8.4±0.3, 7.0±0.2, and 9.0±0.6) and their concentration increased in stage IV and stage V (Fig. 2A). Otx2 expression, consistent with its role in controlling the transcription in embryonic and neural stem cells [48], was already detected in stage I (ΔCt: 8.2±0.2), its expression was ∼6-fold upregulated at stage III, maintained at high levels at stage IV, and decreased at stage V (Fig. 2A).
The mRNA expression of the mDAergic marker, Th, was variable between experiments. The expression in some experiments was high in stage V (ΔCt <8), whereas in other experiments the levels were low or not detected (ΔCt >12) (Fig. 2B). This outcome was also confirmed by immunocytochemistry. The percentage of Th+neurons was >8% in the experiments with ΔCt <8. No any or very few Th+neurons were found in the experiments with low Th mRNA expression levels (Supplementary Tables S2 and S3).
We conclude that all 6 experiments, performed with 2 different cell lines, resulted in an overall neural and midbrain differentiation. However, when the a priori criterion for Th expression was applied, only 3 out of the 6 experiments were classified as positive (+) experiments, whereas the other 3 experiments were negative (−). The differences in Th expression between these groups were found statistically significant at both mRNA and protein levels (P<0.001) (Fig. 2B).
Gene expression differences between experiments with positive and negative outcome
To identify differences in gene expression between (+) and (−) experiments retrospectively, the marker profiles for the individual steps of differentiation were grouped (Fig. 3). Between these groups, significant differences were found in stage II for the neural precursor markers, Pax6, Sox1, and Nestin (∼15-fold, 5-fold, and 4-fold, respectively), and in stage III for the pan-neuronal markers, Dcx and Tau (∼15-fold and 50-fold) (Fig. 3A). In stage IV and V, the expression levels of Dcx and Tau decreased in the (−) experiments with differences being significant at the end point of stage V (Fig. 3A).
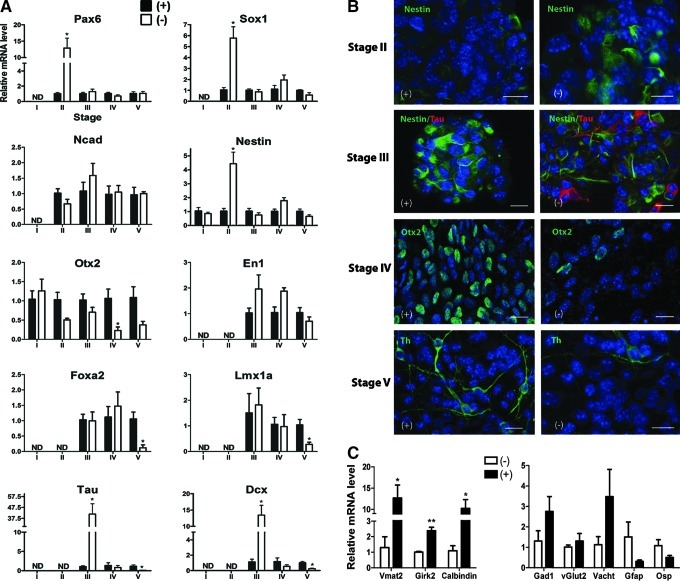
FIG. 3.
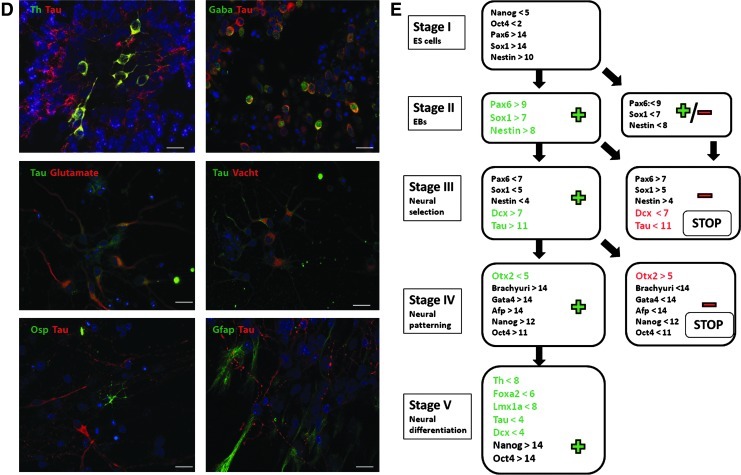
A stage-related comparison of gene expression between the experiments with (+) and (−) outcome regarding the Th expression revealed milestones with predictive potential. (A) mRNA expression levels of neural progenitor markers, Pax6, Sox1, Nestin, and Ncad, VM markers, En1, Otx2, Foxa2, and Lmx1a as well as pan-neuronal markers, Tau and Dcx, in negative (−) experiments, relative to positive (+) experiments. Mean±SEM; n=3. *P<0.05 (Student t-test). (B) Immunostaining for Nestin in stage II, Tau and Nestin in stage III, Otx2 in stage IV, and Th in stage V in 2 representative (+) and (−) experiments. (C) Relative mRNA levels in stage V of the (+) experiments as compared to the (−) ones for the dopaminergic (Vmat2, Girk2, and Calbindin), GABAergic (Gad1), glutamatergic (vGlut2), cholinergic (Vacht), and glial (Gfap and Osp) markers. Mean±SEM; n=3. *P<0.05, **P<0.01 (Student t-test). (D) Immunostaining for Tau, Th, Gfap, Osp, GABA, Glutamate, and Vacht confirmed their expression in stage V in one representative (+) experiment. (E) Schematic representation of 5-stage protocol showing the gene expression levels in each stage that predict positive (+, green) and negative (−, red) end point outcome regarding the Th expression. The milestones are represented as mean ΔCt values (related to GAPDH). When a milestone is not reached, the prediction for a (−) outcome is followed by stopping the experiment (STOP). (B, D) Nuclear staining with DAPI (blue); scale bar: 20 μm. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; DAPI, 4′, 6-diamidino-2-phenylindole. Color images available online at www.liebertpub.com/scd
Therefore, strong and significant differences between the experiments with positive and negative outcomes were observed already at stage II. Negative outcome correlates with high expression levels of Pax6, Sox1, and Nestin in stage II and Tau and Dcx in stage III. Nestin immunoreactivity was higher in the (−) group compared with the (+) group in stage II, but similar in (+) and (−) experiments from stage III onward. Tau immunoreactivity was detected at the end point of stage III in a higher proportion of cells in the (−) group compared with the (+) group (Fig. 3B and Supplementary Tables S2 and S3). These results indicate a premature, untimely neural differentiation in (−) experiments as the underlying phenomenon.
For the mDAergic progenitor markers, Otx2 gene expression was significantly lower in (−) experiments as compared to (+) experiments in stage IV. This finding was confirmed by immunocytochemistry (Fig. 3A, B and Supplementary Tables S2 and S3). No significant differences were detected at the end point of stage III and V (Otx2) as well as for En1 expression at any stage, whereas a significantly higher expression was observed in stage V for Foxa2 (∼20-folds) and Lmx1a (∼4-fold) in the (+) experiments (Fig. 3A).
Consistent with higher levels of mDAergic neurons, significantly higher expression levels of Vmat2 (∼13-fold), Girk2 (∼2-fold), and Calbindin (∼10-fold) were detected at the end point of stage V in the (+) experiments as compared to the (−) ones (Fig. 3C).
To identify other neuronal lineages than mDAergic in our cultures, GABAergic (Gad1), glutamatergic (vGlut2), and cholinergic (Vacht) marker gene expression was compared between (+) and (−) experiments at the end point of the protocol (Fig. 3C). No significant differences were found, with only a trend for higher expression for Gad1 and Vacht in (+) experiments (P=0.2). The glial markers, Osp and Gfap, were detected at high expression levels at the end of the protocol, but no significant differences between the (+) and (−) experiments were found (P=0.1 and P=0.2). The expression of neurotransmitter markers and the glial markers in stage V was confirmed by immunocytochemistry (Fig. 3D).
To test for the presence of unwanted nonectodermal populations in our cultures, we analyzed Gata4, Afp, and Brachyury expression levels as non-neural markers characteristic for endoderm or mesoderm. The specific markers for forebrain (Foxg1), diencephalon (Irx5), hindbrain (Hoxb1), and spinal cord (Hoxb9) were determined to test for unwanted central nervous system populations. Expression levels of these markers at each stage of differentiation are presented in the Supplementary Table S4. No significant differences were detected between (+) and (−) experiments (P>0.05), but a trend of increasing in non-neural (Brachyury, Gata4) differentiation was observed in the (−) experiments (Supplementary Table S4).
Based on these results, we propose quantitative milestones for marker gene expression, which can be used to monitor the positive outcome for the mDAergic neuronal differentiation in multistep differentiation protocols (Fig. 3E). For each stage-related milestone, expression values for positive markers are combined with values for markers indicating undesired or untimely cellular differentiation. In our experiments, the expression of milestone markers did not significantly differ in experiments conducted with 2 different pluripotent lines (Supplementary Fig. S2).
To validate the profiles and the milestones, 3 additional experiments were performed and monitored, based on our strategy (Supplementary Table S3). In all 3 experiments, Th expression values met the a priori criterion for a successful experiment.
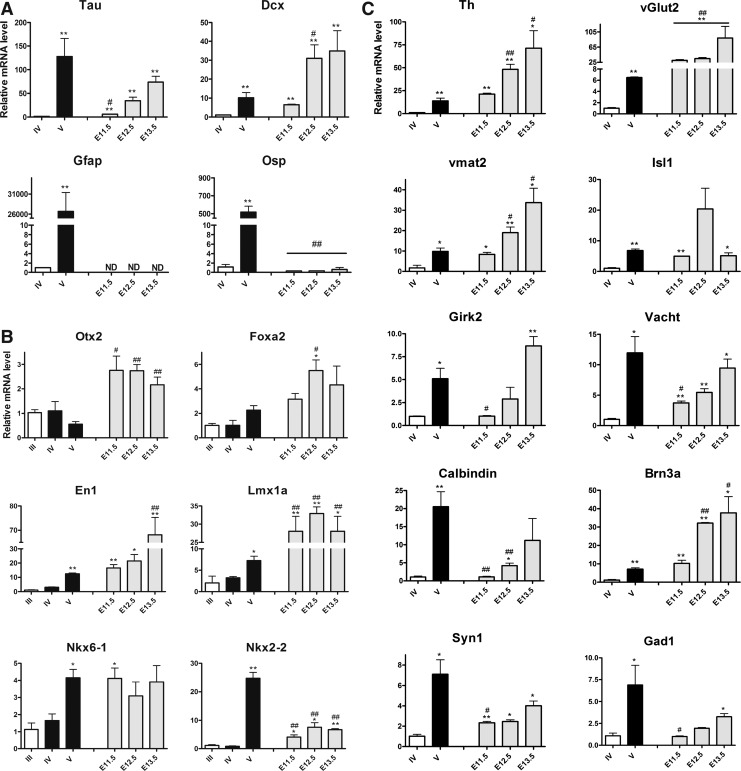
Quantitative comparison of gene expression profiles during midbrain differentiation in vitro and in vivo
In the next step, we compared the expression profiles at stage IV and V of 4 (+) experiments from the B6G-eGfp line to profiles obtained from ex vivo VM, collected at E11.5, E12.5, and E13.5. Regarding the pan-neuronal markers, the level of mRNA for Tau at stage V (ΔCt: 3.7±0.4) was similar to the levels in VM at E13.5 (ΔCt: 3.7±0.2), while the Dcx levels at stage V (ΔCt: 3.3±0.7), were matched better to those identified in VM at E11.5 (ΔCt: 4.2±0.1) (Fig. 4A). For the glial markers Gfap and Osp, which are low abundantly expressed during stage IV (ΔCt: 12.6±0.1 and 10.7±0.6, respectively), expression increased substantially during stage V (P=0.001) (Fig. 4A). These glial markers were very low abundant or not detectable in VM at all embryonic ages analyzed (Fig. 4A) indicating that gliogenesis in vitro occurs early and massively compared to in vivo.
FIG. 4.
Molecular profiling for the neural patterning and differentiation markers in vitro and in vivo. (A) Neuronal and glial differentiation. Relative mRNA expression levels of the neuronal (Tau and Dcx) and the glial (Gfap and Osp) markers after neural patterning and differentiation in all experiments presenting (+) milestones and in the mouse embryonic VM at embryonic (E) day 11.5, E12.5, and E13.5. Levels of expression are compared with stage IV and presented as mean±SEM; n=3–4. (B) VM patterning in vitro and in vivo. Relative mRNA expression levels of the midbrain (Otx2 and En1), floor plate (Foxa2 and Lmx1a), and basal plate (Nkx6-1 and Nkx2-2) markers after neural patterning and differentiation in all experiments with (+) milestones and in the mouse VM at E11.5, E12.5, and E13.5. Levels of expression are compared with stage III and presented as mean±SEM; n=3–4. (C) Neuronal specification in vitro and in vivo. Relative mRNA expression levels of the dopaminergic markers (Th, Vmat2, Girk2, and Calbindin), the glutamatergic marker (vGlut2), and the GABAergic markers (Gad1 and Brn3a) as well as the cholinergic markers (Vacht and Isl1) and the synaptic marker (Syn1) after neural differentiation in the 5-stage protocol experiments with (+) milestones and in the mouse VM at E11.5, E12.5, and E13.5. Mean±SEM; n=3–4; *P<0.05; **P<0.01 significantly different from stage IV by Student t-test. #P<0.05; ##P<0.01 significantly different from stage V by Student t-test. ND, not detected.
In respect to midbrain markers, high expression of Otx2 was detected after stage III (ΔCt: 5.3±0.2) with relatively unchanged expression in stages IV and V. Slightly higher levels were determined in VM at all 3 stages of in vivo development, but did not differ significantly from stage IV (P>0.05), and reached significance in comparison to stage V (Fig. 4B). En1 mRNA was ∼10-fold higher in stage V (ΔCt: 5.6±0.2) as related to stage IV (ΔCt: 7.4±0.2). The comparison with the VM showed no significant difference from VM E11.5 (ΔCt: 5.0±0.2, P=0.2) or E12.5 (ΔCt: 4.5±0.3, P=0.1), while the significance was reached as compared to VM E13.5 (ΔCt: 3.0±0.2) (Fig. 4B). Expression of the floor plate markers, Foxa2 and Lmx1a, increased in stage V (ΔCt: 6.0±0.3 and 7.3±0.2) as compared to stage IV (ΔCt: 7.2±0.6 and 8.6±0.2), reaching significance for Lmx1a. Foxa2 expression at stage V was not different from VM E11.5. Lmx1a expression was ∼20-fold higher in vivo for all ages than in vitro at stage V (Fig. 4B).
To additionally test for alternative midbrain patterning and specification fates, the dorsal marker, Pax7, and the basal plate markers, Nkx6-1, Nkx2-2, Brn3a, and Isl1 were added to the profile [49–51]. Pax7 was very low expressed in stage V (ΔCt: 12.5±0.4) or was not detected, as it was in the embryonic VM (ΔCt >14). The basal plate markers, Nkx6-1 and Nkx2-2, were found to be expressed in vitro, starting with stage III (ΔCt: 7.1±0.5 and 11.3±0.4, respectively), and a significant upregulation was detected in stage V for Nkx6-1 (∼3-fold) and Nkx2-2 (∼22-fold). Similar levels of Nkx6-1 expression in stage V in vitro compared with VM E11.5, E12.5, and E13.5 were identified (P=0.9, P=0.3, and P=0.8, respectively), whereas Nkx2-2 expression was ∼20-fold higher at stage V compared to E11.5, 12.5, and 13.5 (Fig. 4B).
These data indicate that in vitro patterning induces not only a floor plate phenotype expressing Foxa2 and Lmx1a, but also basal plate-like cell identity characterized by expression of Nkx6-1 and Nkx2-2. Although some VM markers (Foxa2, En1, and Nkx6-1) had similar expression as age-matched embryonic VM tissue, Lmx1a expression was lower, whereas Nkx2-2 expression was higher in vitro than in vivo.
The level of expression of the mature mDAergic markers was very low in stage IV: Th (ΔCt: 11.5±0.4), Vmat2 (ΔCt: 10.7±0.9), Girk2 (ΔCt: 11.3±0.1), and Calbindin (ΔCt: 9.3±0.3). Their expression significantly increased in stage V (Th ∼20-fold, Vmat2 ∼10-fold, Girk2 ∼4-fold, Calbindin ∼20-fold) as compared with stage IV, reaching similar levels as in mouse VM: Th and Vmat2 expressions were not different from VM E11.5 (ΔCt: 7.1±0.1, P=0.1 and 7.6±0.2, P=0.5), Girk2 expression was comparable to VM E13.5 (ΔCt: 8.2±0.2, P=0.07), and Calbindin expression was also similar to VM E13.5 (ΔCt: 5.8±0.7, P=0.1) (Fig. 4C). The late mDAergic marker, Dat, was still barely detectable in stage V (ΔCt: 12.9±0.9).
Regarding alternative neuronal markers, vGlut2 showed low expression in stage IV (ΔCt: 9.5±0.3), its expression was significantly upregulated in stage V (∼5-fold), but its levels in vitro were at least ∼20-fold lower compared to all 3 VM stages (Fig. 4C). For Gad1, levels increased ∼6-fold in stage V as compared to stage IV (ΔCt: 5.6±0.4) reaching expression levels not different with VM E13.5 (ΔCt: 3.9±0.1) (P=0.2) (Fig. 4C). For the specific GABAergic marker, Brn3a, the low expression at stage IV (ΔCt: 11.6±0.5) increased during stage V (∼6-fold, P=0.002) to reach levels similar to VM E11.5 (ΔCt: 8.2±0.2), but significantly lower than VM E12.5 (P<0.001) or VM E13.5 (P=0.02). Vacht showed low expression in stage IV (ΔCt: 11.5±0.3), while in stage V, levels were significantly upregulated (∼10-fold) and comparable to in vivo VM E12.5 (ΔCt: 10.7±0.2) and E13.5 (ΔCt: 9.9±0.2) (Fig. 4C). A similar observation was made for cholinergic marker, Isl, showing similar expression levels for stage V, E11.5 (ΔCt: 7.3±0.1) and E13.5 (ΔCt: 7.2±0.2) (Fig. 4C). The synaptic marker, Syn1, was significantly upregulated in stage V as compared to stage IV (∼6-fold) at even higher levels than determined in VM E13.5 (ΔCt: 5.3±0.2) (Fig. 4C).
When the expression of the extended set of midbrain markers was applied to the groups of positive (+) and negative (−) experiments, Brn3a expression was found to be 3-fold higher in the (+) experiment as compared to the (−) experiments (Supplementary Fig. S3), while for Nkx2-2, Nkx6-1, and Isl1 the differences were not significant (P=0.08, 0.09 and 0.2, respectively).
We conclude that neural differentiation and specification in the successful in vitro experiments not only lead to generation of mDAergic floor plate-derived neurons expressing Th, Vmat2, Girk2, and Calbindin, but also to the formation of GABAergic, glutamatergic, and cholinergic basal plate neurons, expressing Gad1, vGlut2, Vacht, Isl1, and Brn3a. However, although the neuronal markers, Tau and Dcx, were expressed at similar levels in cultures and age-matched embryonic VM, an overshooting high rate of gliogenesis was observed in vitro compared to in vivo, identified by the high expression values of Gfap and Osp.
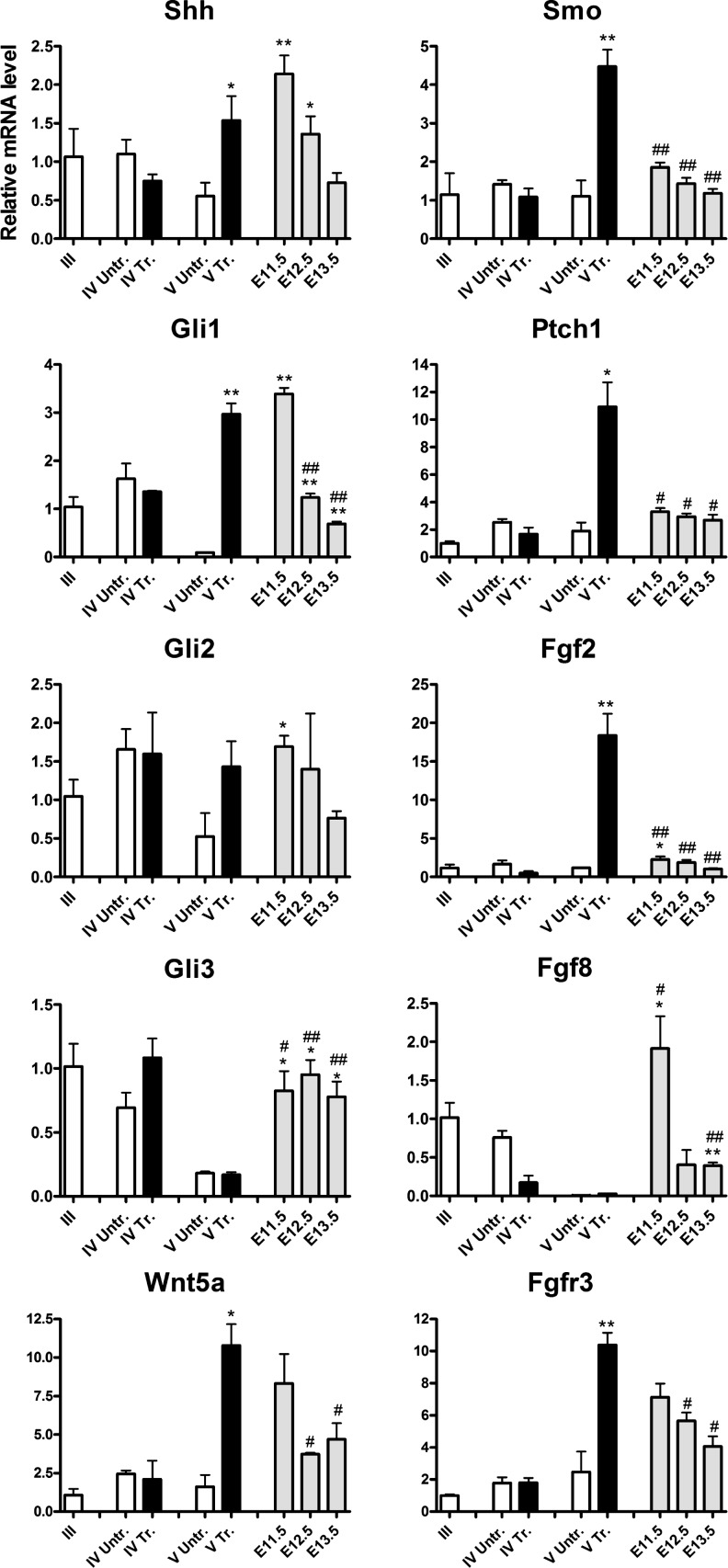
Profiles for signaling pathway components reveal their modulation in vitro and in the developing VM
Next, we analyzed the expression of genes encoding components of signaling pathways, including the Hh pathway (Shh, Gli1, Gli2, Gli3, Ptch1, and Smo), Fgf signaling (Fgf8, Fgf2, and Fgfr3), and Wnt signaling (Wnt1, Wnt3a, and Wnt5a). Again the expression in cell culture was compared to the expression in the developing VM at E11.5, E12.5, and E13.5 (Fig. 5). The mRNA expression was analyzed in cultures treated in stage IV with a combination of agonists for these pathways (purmorphamine+Fgf2+Fgf8) and compared to untreated (control) cultures. Shh was abundantly expressed before treatment at stage III (ΔCt: 5.3±0.5). This expression remained similar in the untreated cultures during the subsequent stages. Treatment with patterning factors increased expression levels of endogenous Shh significantly in stage V to reach levels comparable with the VM E11.5 (ΔCt: 4.2±0.1, P=0.2) and E12.5 (ΔCt: 4.9±0.2, P=0.7). Gli1 expression after stage III (ΔCt: 7.4±0.3) decreased in the untreated cultures (ΔCt: 10.9±0.1 in stage V). Agonist treatment increased Gli1 expression in stage V, similar levels were present in VM E11.5 (ΔCt: 5.6±0.1). Gli2 and Gli3 expression levels were low in the control conditions (ΔCt: 9.8±0.9 and 12.2±0.1, respectively), and their expression was not influenced significantly by the treatment (Fig. 5). Transcripts of the Smo and Ptch1 Hh receptor genes were present at similar levels in untreated cultures at stages III (ΔCt: 5.1±0.8 and 5.5±0.2), IV (ΔCt: 4.6±0.1 and 4.2±0.1), and V (ΔCt: 5.1±0.5 and 4.7±0.4) and embryonic VM at all stages. Remarkably, their levels increased in stage V after treatment, ∼3-fold for Smo (P=0.009) and ∼10-fold for Ptch1 (P=0.03), overcoming their levels in vivo (Fig. 5).
FIG. 5.
Molecular profiling for the Hh, Fgf, and Wnt pathways in vitro versus in vivo. Relative mRNA expression levels of the Hh pathway (Shh, Gli1, Gli2, and Gli3) and receptors (Smo and Ptch1), Fgf signaling (Fgf2, Fgf8, and Fgfr3) and Wnt signaling (Wnt1 and Wnt5a) in protocol of mouse ES differentiation toward dopaminergic neurons in treated (Tr., purmorphamine+Fgf2+Fgf8) and untreated (Untr.) experimental conditions of mouse ES differentiation and in the mouse VM at embryonic (E) day 11.5, E12.5, and E13.5. Mean±SEM; n=3–4; *P<0.05; **P<0.01 significantly different from stage V untreated by Student t-test. #P<0.05; ##P<0.01 significantly different from stage V treated by Student t-test. Hh, Hedgehog.
Endogenous Fgf8 expression was high in stage III (ΔCt: 6.3±0.3) and levels decreased subsequently in both treated and untreated conditions. In contrast, Fgf2 expression was low in stage III (ΔCt: 10.8±0.5) and stage IV (ΔCt: 10.2±0.4). A ∼20-fold increase was observed in the treated cultures in stage V (P=0.004), reaching levels higher than in embryonic VM (P<0.01) (Fig. 5). The patterning treatment caused also a significant increase (∼10-fold, P=0.006) in the expression of Fgfr3 in stage V, with expression higher, but approaching the level in VM E11.5 (ΔCt: 3.8±0.2) (Fig. 5).
Wnt1 was moderately expressed after stage III (ΔCt: 9.4±0.5) and its expression decreased in both control and treated cultures at later stages. Wnt3a was barely detectable both in vitro and in vivo (data not shown). In contrast, Wnt5a was significantly upregulated after patterning treatment in stage V (∼10-fold, P=0.02) to similar levels as in VM E11.5 (ΔCt: 4.6±0.3, P=0.4) (Fig. 5).
We conclude that mouse ES cell-derived neural populations respond to Hh and Fgf pathway stimulation with increasing the levels of components of the corresponding signaling pathways. The regulated transcripts reach similar levels (for Shh, Gli1, Fgfr3, and Wnt5a) or even higher levels (for Fgf2, Smo, and Ptch1) as detected in the developing VM.
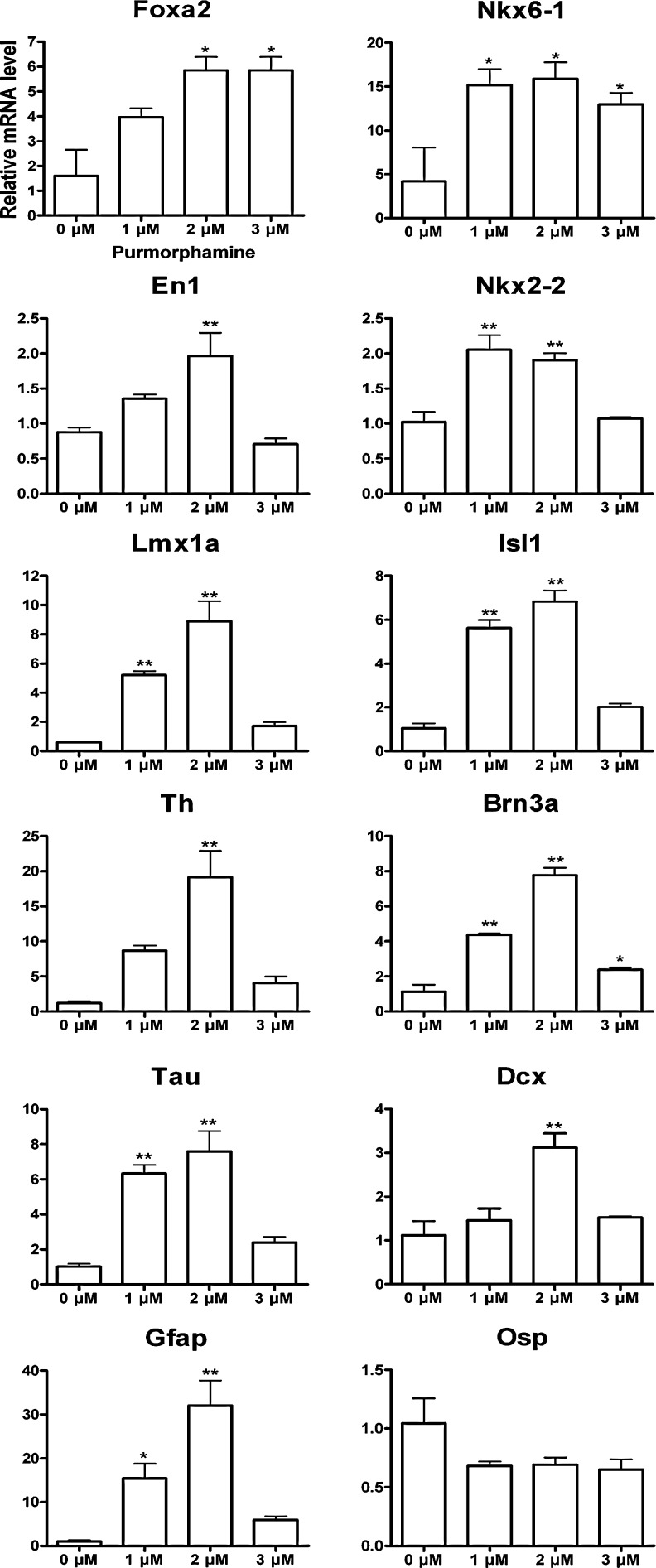
Monitoring of the effect of Hh signaling on in vitro cell fates
To test whether the gradient in Hh modulation during stage IV exerts an influence on in vitro cell heterogeneity and midbrain patterning, the Hh agonist purmorphamine at 3 different doses (1, 2, and 3 μM) combined with Fgf2 (10 ng/mL) and Fgf8 (100 ng/mL) were applied after reaching the milestones at stage III. Expression of the neuronal markers, Tau and Dcx, and the glial markers, Gfap and Osp, were compared between purmorphamine treated and untreated cultures at stage V together with VM, floor plate (Th, En1, Lmx1a, and Foxa2), and basal plate markers (Nkx6-1, Nkx2-2, Isl1, and Brn3a) (Fig. 6). A dose-dependent increase in the expression of Tau, Dcx, Gfap and the floor plate markers Th, En1, and Lmx1a was observed, peaking at 2 μM with a decrease at 3 μM. For Foxa2, no difference was observed between 2 and 3 μM. Compared to controls, expression levels increased for the basal plate transcription factors, Nkx6-1 and Nkx2-2, at 1, 2, and 3 μM for Nkx6-1 (∼10-fold, P=0.03) and at 1 and 2 μM for Nkx2-2 (P<0.001). A peak at 2 μM was seen for the neuronal basal plate markers, Isl1 and Brn3a. Osp expression was unaffected by the treatment.
FIG. 6.
Molecular profiling for the VM patterning, neuronal and glial specification related to Hh modulation in vitro. Relative mRNA expression levels of the floor plate markers Th, Foxa2, En1, and Lmx1a, and the basal plate markers Nkx6-1, Nkx2-2, Isl1, and Brn3a, the neuronal markers Tau and Dcx and the glial markers Gfap and Osp at the end point of the 5-stage protocol treated with purmorphamine concentrations of 1, 2, and 3 μM, as compared to untreated controls (0 μM). Mean±SEM; n=3; *P<0.05; **P<0.01 significantly different from 0 μM purmorphamine by Student-Newman-Keul's post hoc test.
Therefore, a dose of 2 μM purmorphamine was optimal for stimulating neuronal and floor plate mDAergic differentiation, whereas 1 μM of purmorphamine was sufficient to stimulate expression of the basal plate markers. Expression of glial markers could not effectively be reduced by varying Hh signaling.
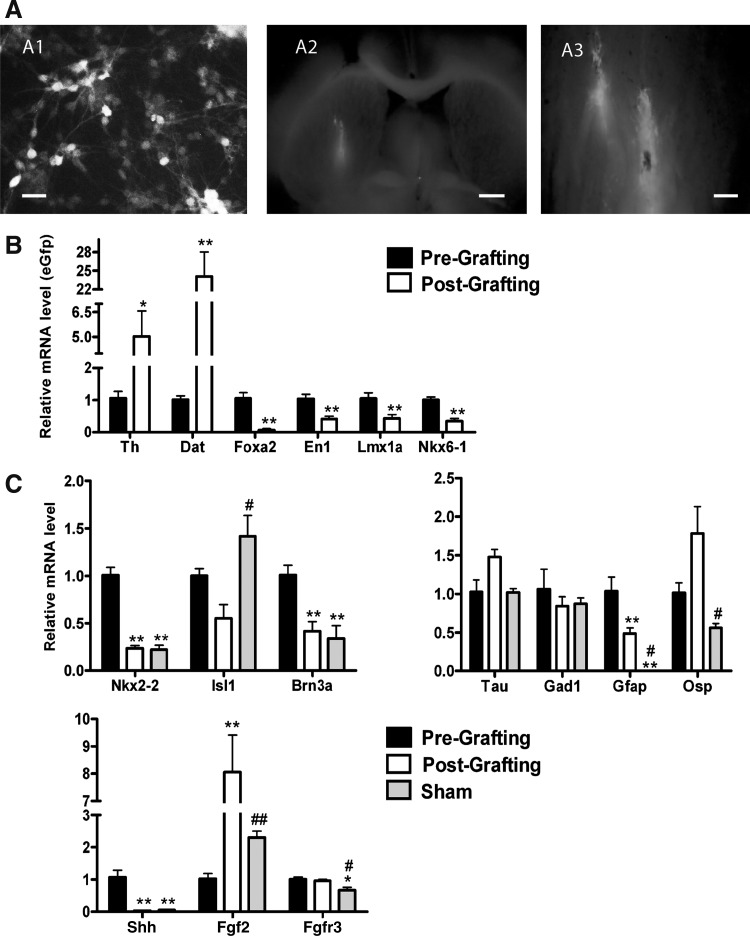
Expression profiles are useful for the investigation of cell fates in stem cell-derived grafts in 6-OHDA models of PD
Next, we applied the established profiles to grafted stem cell derivatives, following their in vivo differentiation. ES cells expressing eGfp constitutively were differentiated to neural cells in vitro (Fig. 7A1) and were grafted into the striatum of 6-OHDA lesioned rats. Gfp expressing grafts were dissected from the host tissue 6 weeks after grafting. In 6 out of 12 transplanted rats, eGfp expression was readily detectable in the grafts. Tumor formation was not observed. The graft core was integrated within the striatum and surrounded by neuronal fibers extending into the host tissue (Fig. 7A2, A3). Then, we analyzed the expression levels of marker genes in the grafts and compared them with the population of pregrafted cells and the striatum of sham-operated animals. Values were obtained for the VM developmental markers, Foxa2, En1, Lmx1a, Nkx6-1, Nkx2-2, Isl1, and Brn3a. In addition, markers for various neuronal lineages (Th, Dat, Girk2, Calbindin, Vmat2, Gad1, and vGlut2) and glial cells (Gfap, Osp) were analyzed (Fig. 7B, C). ∼5-fold higher transcript levels for Th were detected in grafts compared to the pregrafted population (P=0.04). Dat expression levels postgrafting increased ∼22-fold (P=0.005). Midbrain progenitor markers, Foxa2, En1, Lmx1a, Nkx6-1, and Nkx2-2 were downregulated during the differentiation in vivo. Together, these indicate a robust further maturation and mDAergic differentiation of the grafted population (Fig. 7B, C). In contrast to the mDAergic neuronal markers, the expression of the basal plate neuronal markers was decreased postgrafting. Isl1 expression was reduced after transplantation and Vacht expression was not detectable in the grafts. The mRNA levels of Brn3a were reduced postgrafting to levels similar to those found in the striatum of the sham-operated animals. No difference was observed pre- and postgrafting for Gad1 (Fig. 7C). vGlut2 was very low expressed (ΔCt >11) or not detectable in the transplants. The glial marker, Gfap, was ∼2-fold less abundant in the grafts and Osp levels remained unchanged compared to the pregrafted population (Fig. 7C). These results indicate that the VM floor plate mDAergic neurons were selectively supported by the host environment in comparison to the basal plate-derived neuronal populations.
FIG. 7.
Molecular profiling for the neural, midbrain, and signaling pathway markers in the cells grafted in an animal model of Parkinson's disease. (A) Fluorescent cells detected before transplantation (A1) and in the striatum (A2, A3) of a 6-hydroxy-dopamine-lesioned rat model. Scale bars: 20 μm (A1), 1 mm (A2), 200 μm (A3). (B) Relative mRNA expression levels of Th, Dat, Foxa2, En1, Lmx1a, and Nkx6-1 of the mouse eGfp-ES cell-derived grafts 6 weeks after transplantation, as compared to pregrafted cells at the end point of the 5-stage protocol. Levels of expression are normalized to eGfp and presented as mean±SEM; n=6 at 6 weeks after grafting; *P<0.05; **P<0.01 significantly different from pregrafting by Student t-test. (C) Relative mRNA expression levels of Nkx2-2, Isl1, and Brn3a as well as Tau, Gad1, Gfap, Osp and Shh, Fgf2 and Fgfr3. Levels of expression are normalized to Gapdh, compared to the levels in the striatum of sham animals and presented as mean±SEM; n=6; *P<0.05; **P<0.01 significantly different from pregrafting by Student-Newman-Keul's post hoc test. #P<0.05, ##P<0.01 significantly different from postgrafting by Student-Newman-Keul's post hoc test. eGfp, enhanced green fluorescent protein.
To investigate the molecular mechanisms involved in the integration and survival of the transplanted neural populations, the expression of signaling molecules Shh, Fgf2, Fgfr3, and Wnt5a were analyzed in grafted tissues, since we had observed significant changes for these genes during in vitro patterning (Fig. 7C). The analysis of the grafts revealed a strong loss of Shh transcripts (∼30-fold, P<0.001) after transplantation. Similar low levels were detected in the striatum of sham animals (ΔCt: 9.8±0.1). Wnt5a levels were slightly reduced after transplantation, but remained detectable at moderate to high levels (ΔCt: 4.6±0.2). No difference was determined for Fgfr3 expression between pre- and postgrafting, whereas expression of Fgf2 was ∼7-fold higher after transplantation and exceeded the levels in the striatum of sham-operated animals (P=0.002).
Concluding, the postgrafted cells express significant levels of signaling molecules, in particular, Fgf2 and Wnt5a, which are likely to support the specification of grafted cells toward the mDAergic phenotype compared to alternative fates.
Discussion
The exciting potential of stem cells for cellular models and therapeutics for PD [52] is an essential motivation for research on the developmental keys that govern patterning and specification of the VM. Recently, the generation of induced pluripotent stem cells from PD patient fibroblasts and their differentiation into mDAergic neurons has been reported, a finding that further spurs on hopes in patient-derived neurons [53]. However, major challenges still remain to be addressed: for replacement strategies the undesirable heterogeneity of cell populations obtained in vitro, the variable survival of mDAergic neurons within the grafts, and the risk of tumor formation.
One way to address these problems is to establish a strict quantitative comparison of critical steps of in vitro differentiation with in vivo development. To this end, we have established expression profiles of specific marker genes. These profiles were put to a test in a well-established 5-stage protocol of neural differentiation of mouse ES cells. This particular protocol lends itself well for this kind of study, since the prototypical difficulties of the in vitro neural differentiation methodology are displayed, in particular, a high variability of outcome and heterogeneity of the generated populations. Our profiles include positive markers, genes which are expected to be expressed at specific stages in a successful experiment, as well as negative markers. Examples of negative indicators include the non-neural markers, Afp, Gata4, and Brachyury. Other markers in our profiles are dynamic that is, they are expected to be expressed at specific stages, but not at others. For example, the pluripotency markers, Nanog and Oct4, should be expressed at stage I, downregulated during neural induction, and absent at the end point of the protocol. The neural progenitor markers, Pax6, Sox1, Nestin, and Ncad, should be gradually upregulated during neural induction (stages II and III) reaching high levels during the neural patterning (stage IV) as a consequence of an efficient selection of neural progenitors.
Successful experiments in our study were defined combining quantitative expression values for the Th mRNA level with the percentages of Th immunoreactive neurons. Applying these criteria, we grouped experiments regarding their positive and negative outcome and compared the differences in the proposed profiles at each stage (Fig. 3). This strategy allowed us to establish milestones with predictive value. It became evident that the patterning treatment must be started before the expression of Dcx and Tau reaches specific values at the end point of stage III, when neural progenitor markers, Pax6, Sox1, and Nestin, need to be expressed at high levels indicating efficient neural induction. The same neural progenitor markers expressed earlier in stage II are correlatives for negative outcome and indicate premature neural induction typically resulting in undesirable high expression of Tau in stage III. These results can henceforth be used to replace the classical way to follow a strict timing of experiments based on days in vitro paying tribute to the difficulty to precisely time differentiation processes.
Neurogenesis and gliogenesis in vitro versus in vivo
We have referenced the progress of the cells in vitro against parallel ex vivo mouse embryonic VM samples from relevant embryonic stages E11.5–E13.5, when patterning and specification occur in vivo. This comparison points to substantial differences in the timing of gliogenesis versus neurogenesis in vivo and in vitro. While for neurogenesis, we found that the expression levels of neural progenitor and neuronal markers in stage V reached similar levels as detected in VM, for gliogenesis, the glial markers were highly expressed, comparing to their very low levels in VM (Fig. 4). Therefore, it appears important to control premature and overshooting gliogenesis in cell cultures.
VM patterning in vitro versus in vivo
Expression levels of the midbrain floor plate markers, En1 and Foxa2, were comparable between in vivo VM (E11.5–E13.5) and cell cultures, whether Lmx1a levels were low in vitro compared to in vivo. It has been reported that recombinant overexpression of Lmx1a promotes in vitro formation of mDAergic neurons [23,54,55]. In view of our finding, a high level of recombinant Lmx1a might be viewed as a correction of an Lmx1a deficit in vitro compared to developing progenitors in vivo.
In our study, we have complemented mDAergic markers with markers for alternative progenitor and neuronal midbrain related populations. These markers were selected based on recent developmental studies regarding molecular mechanism of patterning and specification in the midbrain. In addition to substantia nigra, the VM contains other specific regions, including the oculomotor complex and the red nucleus, comprising specific glutamatergic, GABAergic, and cholinergic neurons. During development, these populations are formed from different progenitor domains in the basal plate, expressing specific markers, including Nkx6-1/2 [50,51] and Nkx2-2 [17–19]. Nkx6-1 was expressed in our cultures at similar levels as in developing VM, while Nkx2-2 expression in stage V was high compared to in vivo levels (Fig. 4B). Markers characteristic of basal plate-derived neuronal populations were found in vitro as well as in embryonic VM at similar (vGlut2 and Isl1) or different (Vacht, Brn3a, and Gad1) levels (Fig. 4C). Nkx6-1 was shown to suppress floor plate marker expression, especially Foxa2 [9]. Hence, it would be important to decrease its expression in vitro, to facilitate mDAergic differentiation.
Interestingly, Otx2 was expressed differentially in stage IV between the experiments with (+) and (−) outcome. Its expression level constitutes a valuable milestone at this stage. This result, combined with recent studies showing the dynamics of Otx2 at the VM levels, indicates that Otx2 should be expressed at high levels in vitro to confer neurogenic activity to the floor plate cells and determine a mDAergic fate [9]. This could possibly be achieved by blocking the Fgf pathway at an early step of in vitro neural differentiation, since this treatment was shown to increase the expression of Otx2 and mDAergic specification [56].
Molecular profiling of the Hh, Fgf, and Wnt pathways revealed novel dynamics in their in vitro and in vivo expression
Previous studies have demonstrated that Hh, Fgf, and Wnt pathways are essential and sufficient for induction of mDAergic neurons [12,17,57–60]. Shh is a major factor for dorsal–ventral patterning of the neural tube, including the midbrain [17,60]. Here, we addressed the direct stimulation of the Hh pathways by the Smo agonist purmorphamine [40–42]. We found that levels of the Shh ligand and Gli1 were specifically increased, while no effect was observed for Gli2 and Gli3 during in vitro patterning. In addition, Smo and Ptch1 receptor transcript levels were also highly enriched by the purmorphamine treatment. Compared with the developing midbrain, Shh and Gli1 expression levels in vitro reached similar levels as in VM E11.5, while Smo and Ptch1 expression levels surpassed the levels found in VM between E11.5 and E13.5 (Fig. 5).
Fgf signaling controls cell proliferation and differentiation of specific neuronal cell types during midbrain development. Fgf8 overexpression can induce the development of ectopic midbrain structures [61]. In a rat midbrain explant culture model, Fgf8 promoted the development of mDAergic neurons, whereas inhibition of Ffg8 signaling had an inhibitory effect [12]. Fgf2 is involved in the development, maintenance, and survival of many cells in the nervous system and, as one of the most potent survival factors, exerts neurotrophic activity on mDAergic neurons in vitro and in vivo [62,63]. In our experiments, purmorphamine and Fgfs treatment did not affect endogenous expression of Fgf8, but exerted a strong stimulatory effect on Fgfr3, in agreement with previous studies [64,65], providing evidence for dynamics of Fgfrs in midbrain development in vivo and in vitro.
Wnt family members are important regulators of mDAergic development [16,17,58,59,66]. A severe mid-hindbrain phenotype was described for the Wnt1 KO [67]. In addition, Wnt3a stimulated proliferation of precursor cells and Wnt5a increased the maturation and differentiation of mDAergic precursors into neurons [16,66]. In our study, Hh and Fgf pathway stimulation prolonged expression of Wnt1 in vitro and stimulated the expression of Wnt5a, in agreement with recently published studies proving an interaction between Wnt signaling and Hh to regulate the generation of mDAergic neurons [57,59].
The set of experiments in which we have studied the effect of different doses of Hh agonist, purmorphamine, revealed that 1 μM was sufficient to induce a peak expression of the basal plate markers, Nkx6-1 and Nkx2-2, whereas higher concentrations (2 μM) stimulated expression of floor plate progenitor and neuronal markers. This result is in concordance with studies indicating a gradient role of Shh in VM specification in vivo [68,69]. No reduction was observed at high doses of purmorphamine for markers of unwanted cells, including the glial cells (Fig. 6).
In summary, our in vitro versus in vivo comparison provided proof that VM patterning and specification have occurred in vitro generating similar populations as present in vivo. However, our expanded profile pinpoints quantitative differences by showing that the selective generation and maintenance of the floor plate populations occur in vitro only if the related signaling is available at a proper time for neurogenesis and at a proper concentration.
Cell fate analysis of ES cell-derived populations after transplantation in a rodent model of PD
Extending previous studies of mDAergic cells within transplants into the striatum of 6-OHDA lesioned rats, we have analyzed the expression not only of mDAergic markers, but also of an array of floor plate and basal plate markers, as well as components of signaling pathways. We found that mainly Th and Dat were increased in the grafts, indicating that mDAergic cells further maturated after transplantation. At the same time, the progenitor markers, Foxa2, En1, and Lmx1a, were decreased, as well as the basal plate markers, Nkx2-2 and Brn3a, and the astrocytic marker, Gfap. No expression was detected for graft-derived Vacht and vGlut2 after transplantation, whereas Gad1 and Osp levels remained unaltered. Therefore, it appears that a highly selective specification or selection process occurs in the grafts leading to decreasing expression of markers for progenitor cells, basal plate, and astrocytes, whereas for the GABAergic and oligodendrocyte markers the levels did not change before and after transplantation.
On the one hand, these results can be explained by the lack of extrinsic signals provided by the host striatum that support the survival and differentiation of the midbrain basal plate-like cells, and by the presence of extrinsic signals supporting the survival and differentiation of the midbrain floor plate-like cells on the other hand. We addressed this question by testing the expression of signaling pathway molecules previously shown to be upregulated during in vitro differentiation and in the developing embryonic VM. We found that high levels of Th and Dat correlated with high Fgf2 and Wnt5a levels, while the Shh level decreased after transplantation.
Conclusion
Taking all together, our new profiling methodology correlated with the quantitative ex vivo analysis of the developing brain, open new opportunities to explore and improve cell culture protocols for midbrain fates differentiation of pluripotent stem cells and to study cell fate decision and signaling in different stem cell-derived neuronal grafts.
Supplementary Material
Acknowledgments
This work was supported by Integrierte Forschungs- und Therapiezentrum (IFTZ) grant 2008; the Innsbruck Medical University and SPIN FWF W1206-B05, Austria. The authors acknowledge Dr. Rana E.L. Rawas and Dr. Galina Apostolova for the useful advises and for critically reading the manuscript, Mag. Eva-Maria Müller for English corrections and Tanja Massimo for technical support with mES cell cultures.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Lee SH. Lumelsky N. Studer L. Auerbach JM. McKay RD. Efficient generation of midbrain and hindbrain neurons from mouse embryonic stem cells. Nat Biotechnol. 2000;18:675–679. doi: 10.1038/76536. [DOI] [PubMed] [Google Scholar]
- 2.Barberi T. Klivenyi P. Calingasan NY. Lee H. Kawamata H, et al. Neural subtype specification of fertilization and nuclear transfer embryonic stem cells and application in parkinsonian mice. Nat Biotechnol. 2003;21:1200–1207. doi: 10.1038/nbt870. [DOI] [PubMed] [Google Scholar]
- 3.Kim JH. Auerbach JM. Rodriguez-Gómez JA. Velasco I. Gavin D, et al. Dopamine neurons derived from embryonic stem cells function in an animal model of Parkinson's disease. Nature. 2002;418:50–56. doi: 10.1038/nature00900. [DOI] [PubMed] [Google Scholar]
- 4.Cho MS. Lee Y-E. Kim JY. Chung S. Cho YH, et al. Highly efficient and large-scale generation of functional dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:3392–3397. doi: 10.1073/pnas.0712359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan Y. Yang D. Zarnowska ED. Du Z. Werbel B, et al. Directed differentiation of dopaminergic neuronal subtypes from human embryonic stem cells. Stem cells. 2005;23:781–790. doi: 10.1634/stemcells.2004-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gale E. Li M. Midbrain dopaminergic neuron fate specification: of mice and embryonic stem cells. Mol Brain. 2008;1:8. doi: 10.1186/1756-6606-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smidt MP. Burbach JPH. How to make a mesodiencephalic dopaminergic neuron. Nat Rev Neurosci. 2007;8:21–32. doi: 10.1038/nrn2039. [DOI] [PubMed] [Google Scholar]
- 8.Abeliovich A. Hammond R. Midbrain dopamine neuron differentiation: factors and fates. Dev Biol. 2007;304:447–454. doi: 10.1016/j.ydbio.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 9.Nakatani T. Kumai M. Mizuhara E. Minaki Y. Ono Y. Lmx1a and Lmx1b cooperate with Foxa2 to coordinate the specification of dopaminergic neurons and control of floor plate cell differentiation in the developing mesencephalon. Dev Biol. 2010;339:101–113. doi: 10.1016/j.ydbio.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 10.Wurst W. Bally-Cuif L. Neural plate patterning: upstream and downstream of the isthmic organizer. Nat Rev Neurosci. 2001;2:99–108. doi: 10.1038/35053516. [DOI] [PubMed] [Google Scholar]
- 11.Wang MZ. Jin P. Bumcrot DA. Marigo V. McMahon AP, et al. Induction of dopaminergic neuron phenotype in the midbrain by Sonic hedgehog protein. Nat Med. 1995;1:1184–1188. doi: 10.1038/nm1195-1184. [DOI] [PubMed] [Google Scholar]
- 12.Ye W. Shimamura K. Rubenstein JL. Hynes MA. Rosenthal A. FGF and Shh signals control dopaminergic and serotonergic cell fate in the anterior neural plate. Cell. 1998;93:755–766. doi: 10.1016/s0092-8674(00)81437-3. [DOI] [PubMed] [Google Scholar]
- 13.Joyner AL. Liu A. Millet S. Otx2, Gbx2 and Fgf8 interact to position and maintain a mid-hindbrain organizer. Curr Opin Cell Biol. 2000;12:736–741. doi: 10.1016/s0955-0674(00)00161-7. [DOI] [PubMed] [Google Scholar]
- 14.Chung S. Leung A. Han B-S. Chang M-Y. Moon J-I, et al. Wnt1-lmx1a forms a novel autoregulatory loop and controls midbrain dopaminergic differentiation synergistically with the SHH-FoxA2 pathway. Cell. 2009;5:646–658. doi: 10.1016/j.stem.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olander S. Nordström U. Patthey C. Edlund T. Convergent Wnt and FGF signaling at the gastrula stage induce the formation of the isthmic organizer. Mech Dev. 2006;123:166–176. doi: 10.1016/j.mod.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Castelo-Branco G. Wagner J. Rodriguez FJ. Kele J. Sousa K, et al. Differential regulation of midbrain dopaminergic neuron development by Wnt-1, Wnt-3a, and Wnt-5a. Proc Natl Acad Sci U S A. 2003;100:12747–12752. doi: 10.1073/pnas.1534900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prakash N. Wurst W. Genetic networks controlling the development of midbrain dopaminergic neurons. J Physiol. 2006;575:403–410. doi: 10.1113/jphysiol.2006.113464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puelles E. Annino A. Tuorto F. Usiello A. Acampora D, et al. Otx2 regulates the extent, identity and fate of neuronal progenitor domains in the ventral midbrain. Development. 2004;131:2037–2048. doi: 10.1242/dev.01107. [DOI] [PubMed] [Google Scholar]
- 19.Vernay B. Koch M. Vaccarino F. Briscoe J. Simeone A, et al. Otx2 regulates subtype specification and neurogenesis in the midbrain. J Neurosci. 2005;25:4856–4867. doi: 10.1523/JNEUROSCI.5158-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ono Y. Nakatani T. Sakamoto Y. Mizuhara E. Minaki Y, et al. Differences in neurogenic potential in floor plate cells along an anteroposterior location: midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development. 2007;134:3213–3225. doi: 10.1242/dev.02879. [DOI] [PubMed] [Google Scholar]
- 21.Omodei D. Acampora D. Mancuso P. Prakash N. Di Giovannantonio LG, et al. Anterior-posterior graded response to Otx2 controls proliferation and differentiation of dopaminergic progenitors in the ventral mesencephalon. Development. 2008;135:3459–3470. doi: 10.1242/dev.027003. [DOI] [PubMed] [Google Scholar]
- 22.Simon HH. Saueressig H. Wurst W. Goulding MD. O'Leary DD. Fate of midbrain dopaminergic neurons controlled by the engrailed genes. J Neurosci. 2001;21:3126–3134. doi: 10.1523/JNEUROSCI.21-09-03126.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albéri L. Sgadò P. Simon HH. Engrailed genes are cell-autonomously required to prevent apoptosis in mesencephalic dopaminergic neurons. Development. 2004;131:3229–3236. doi: 10.1242/dev.01128. [DOI] [PubMed] [Google Scholar]
- 24.Sgadò P. Albéri L. Gherbassi D. Galasso SL. Ramakers GMJ, et al. Slow progressive degeneration of nigral dopaminergic neurons in postnatal Engrailed mutant mice. Proc Natl Acad Sci U S A. 2006;103:15242–15247. doi: 10.1073/pnas.0602116103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersson E. Tryggvason U. Deng Q. Friling S. Alekseenko Z, et al. Identification of intrinsic determinants of midbrain dopamine neurons. Cell. 2006;124:393–405. doi: 10.1016/j.cell.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 26.Smidt MP. Asbreuk CH. Cox JJ. Chen H. Johnson RL, et al. A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat Neurosci. 2000;3:337–341. doi: 10.1038/73902. [DOI] [PubMed] [Google Scholar]
- 27.Yan CH. Levesque M. Claxton S. Johnson RL. Ang S-L. Lmx1a and lmx1b function cooperatively to regulate proliferation, specification, and differentiation of midbrain dopaminergic progenitors. J Neurosci. 2011;31:12413–12425. doi: 10.1523/JNEUROSCI.1077-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin W. Metzakopian E. Mavromatakis YE. Gao N. Balaskas N, et al. Foxa1 and Foxa2 function both upstream of and cooperatively with Lmx1a and Lmx1b in a feed forward loop promoting mesodiencephalic dopaminergic neuron development. Dev Biol. 2009;333:386–396. doi: 10.1016/j.ydbio.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 29.Kittappa R. Chang WW. Awatramani RB. McKay RDG. The foxa2 gene controls the birth and spontaneous degeneration of dopamine neurons in old age. PLoS Biol. 2007;5:e325. doi: 10.1371/journal.pbio.0050325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee H-S. Bae E-J. Yi S-H. Shim J-W. Jo A-Y, et al. Foxa2 and Nurr1 synergistically yield A9 nigral dopamine neurons exhibiting improved differentiation, function, and cell survival. Stem cells. 2010;28:501–512. doi: 10.1002/stem.294. [DOI] [PubMed] [Google Scholar]
- 31.Di Porzio U. Zuddas A. Cosenza-Murphy DB. Barker JL. Early appearance of tyrosine hydroxylase immunoreactive cells in the mesencephalon of mouse embryos. Int J Dev Neurosci. 1990;8:523–532. doi: 10.1016/0736-5748(90)90044-3. [DOI] [PubMed] [Google Scholar]
- 32.Kawasaki H. Mizuseki K. Nishikawa S. Kaneko S. Kuwana Y, et al. Induction of midbrain dopaminergic neurons from ES cells by stromal cell-derived inducing activity. Neuron. 2000;28:31–40. doi: 10.1016/s0896-6273(00)00083-0. [DOI] [PubMed] [Google Scholar]
- 33.Rodríguez-Gómez JA. Lu J-Q. Velasco I. Rivera S. Zoghbi SS, et al. Persistent dopamine functions of neurons derived from embryonic stem cells in a rodent model of Parkinson disease. Stem cells. 2007;25:918–928. doi: 10.1634/stemcells.2006-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bjorklund LM. Sánchez-Pernaute R. Chung S. Andersson T. Chen IYC, et al. Embryonic stem cells develop into functional dopaminergic neurons after transplantation in a Parkinson rat model. Proc Natl Acad Sci U S A. 2002;99:2344–2349. doi: 10.1073/pnas.022438099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D-W. Chung S. Hwang M. Ferree A. Tsai H-C, et al. Stromal cell-derived inducing activity, Nurr1, and signaling molecules synergistically induce dopaminergic neurons from mouse embryonic stem cells. Stem cells. 2006;24:557–567. doi: 10.1634/stemcells.2005-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung S. Moon J-I. Leung A. Aldrich D. Lukianov S, et al. ES cell-derived renewable and functional midbrain dopaminergic progenitors. Proc Natl Acad Sci U S A. 2011;108:9703–9708. doi: 10.1073/pnas.1016443108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai J. Yang M. Poremsky E. Kidd S. Schneider JS, et al. Dopaminergic neurons derived from human induced pluripotent stem cells survive and integrate into 6-OHDA-lesioned rats. Stem Cells Dev. 2010;19:1017–1023. doi: 10.1089/scd.2009.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimizukawa R. Sakata A. Hirose M. Takahashi A. Iseki H, et al. Establishment of a new embryonic stem cell line derived from C57BL/6 mouse expressing EGFP ubiquitously. Genesis. 2005;42:47–52. doi: 10.1002/gene.20122. [DOI] [PubMed] [Google Scholar]
- 39.Doetschman T. Gregg RG. Maeda N. Hooper ML. Melton DW, et al. Targeted correction of a mutant HPRT gene in mouse embryonic stem-cells. Nature. 1987;330:576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- 40.El-Akabawy G. Medina L. Jeffries A. Price J. Modo M. Purmorphamine increases DARPP-32 differentiation in human striatal neural stem cells through the hedgehog pathway. Stem Cells Dev. 2011;20:1873–1887. doi: 10.1089/scd.2010.0282. [DOI] [PubMed] [Google Scholar]
- 41.Sinha S. Chen JK. Purmorphamine activates the Hedgehog pathway by targeting smoothened. Nat Chem Biol. 2006;2:29–30. doi: 10.1038/nchembio753. [DOI] [PubMed] [Google Scholar]
- 42.Nat R. Salti A. Suciu L. Ström S. Dechant G. Pharmacological modulation of the hedgehog pathway differentially affects dorsal/ventral patterning in mouse and human embryonic stem cell models of telencephalic development. Stem Cells Dev. 2012;21:1016–1046. doi: 10.1089/scd.2011.0271. [DOI] [PubMed] [Google Scholar]
- 43.Okabe M. Ikawa M. Kominami K. Nakanishi T. Nishimune Y. “Green mice” as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 44.Hadjantonakis AK. Gertsenstein M. Ikawa M. Okabe M. Nagy A. Generating green fluorescent mice by germline transmission of green fluorescent ES cells. Mech Dev. 1998;76:79–90. doi: 10.1016/s0925-4773(98)00093-8. [DOI] [PubMed] [Google Scholar]
- 45.Dunnett SB. Björklund A. Dissecting Embryonic Neural Tissues for Transplantation. Neuromethods: Cell and Tissue Transplantation in the CNS. Vol. 36. Humana Press; Totowa: 2000. pp. 3–25. [Google Scholar]
- 46.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puschban Z. Scherfler C. Granata R. Laboyrie P. Quinn NP. Autoradiographic study of striatal dopamine re-uptake sites and dopamine D1 and D2 receptors in a 6-hydroxydopamine and quinolinic acid double-lesion rat model of striatonigral degeneration (multiple system atrophy) and effects of embryonic ventral mesenc. Neuroscience. 2000;95:377–388. doi: 10.1016/s0306-4522(99)00457-1. i. [DOI] [PubMed] [Google Scholar]
- 48.Hailesellasse Sene K. Porter CJ. Palidwor G. Perez-Iratxeta C. Muro EM, et al. Gene function in early mouse embryonic stem cell differentiation. BMC Genomics. 2007;8:85. doi: 10.1186/1471-2164-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fedtsova N. Turner EE. Signals from the ventral midline and isthmus regulate the development of Brn3.0-expressing neurons in the midbrain. Mech Dev. 2001;105:129–144. doi: 10.1016/s0925-4773(01)00399-9. [DOI] [PubMed] [Google Scholar]
- 50.Moreno-Bravo JA. Perez-Balaguer A. Martinez S. Puelles E. Dynamic expression patterns of Nkx6.1 and Nkx6.2 in the developing mes-diencephalic basal plate. Dev Dyn. 2010;239:2094–2101. doi: 10.1002/dvdy.22327. [DOI] [PubMed] [Google Scholar]
- 51.Prakash N. Puelles E. Freude K. Trümbach D. Omodei D, et al. Nkx6-1 controls the identity and fate of red nucleus and oculomotor neurons in the mouse midbrain. Development. 2009;136:2545–2555. doi: 10.1242/dev.031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKay R. Kittappa R. Will stem cell biology generate new therapies for Parkinson's disease? Neuron. 2008;58:659–661. doi: 10.1016/j.neuron.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 53.Soldner F. Hockemeyer D. Beard C. Gao Q. Bell GW, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Friling S. Andersson E. Thompson LH. Jönsson ME. Hebsgaard JB, et al. Efficient production of mesencephalic dopamine neurons by Lmx1a expression in embryonic stem cells. Proc Natl Acad Sci U S A. 2009;106:7613–7618. doi: 10.1073/pnas.0902396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sánchez-Danés A. Consiglio A. Richaud Y. Rodríguez-Pizà I. Dehay B, et al. Efficient generation of A9 midbrain dopaminergic neurons by lentiviral delivery of LMX1A in human embryonic stem cells and induced pluripotent stem cells. Hum Gene Ther. 2012;23:56–69. doi: 10.1089/hum.2011.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jaeger I. Arber C. Risner-Janiczek JR. Kuechler J. Pritzsche D, et al. Temporally controlled modulation of FGF/ERK signaling directs midbrain dopaminergic neural progenitor fate in mouse and human pluripotent stem cells. Development. 2011;138:4363–4374. doi: 10.1242/dev.066746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Joksimovic M. Anderegg A. Roy A. Campochiaro L. Yun B, et al. Spatiotemporally separable Shh domains in the midbrain define distinct dopaminergic progenitor pools. Proc Natl Acad Sci U S A. 2009;106:19185–19190. doi: 10.1073/pnas.0904285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Joksimovic M. Yun BA. Kittappa R. Anderegg AM. Chang WW, et al. Wnt antagonism of Shh facilitates midbrain floor plate neurogenesis. Nat Neurosci. 2009;12:125–131. doi: 10.1038/nn.2243. [DOI] [PubMed] [Google Scholar]
- 59.Tang M. Villaescusa JC. Luo SX. Guitarte C. Lei S, et al. Interactions of Wnt/beta-catenin signaling and sonic hedgehog regulate the neurogenesis of ventral midbrain dopamine neurons. J Neurosci. 2010;30:9280–9291. doi: 10.1523/JNEUROSCI.0860-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bayly RD. Ngo M. Aglyamova GV. Agarwala S. Regulation of ventral midbrain patterning by Hedgehog signaling. Development. 2007;134:2115–2124. doi: 10.1242/dev.02850. [DOI] [PubMed] [Google Scholar]
- 61.Nakamura H. Katahira T. Matsunaga E. Sato T. Isthmus organizer for midbrain and hindbrain development. Brain Res Rev. 2005;49:120–126. doi: 10.1016/j.brainresrev.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 62.Ferrari G. Minozzi MC. Toffano G. Leon A. Skaper SD. Basic fibroblast growth factor promotes the survival and development of mesencephalic neurons in culture. Dev Biol. 1989;133:140–147. doi: 10.1016/0012-1606(89)90305-9. [DOI] [PubMed] [Google Scholar]
- 63.Engele J. Bohn MC. The neurotrophic effects of fibroblast growth factors on dopaminergic neurons in vitro are mediated by mesencephalic glia. J Neurosci. 1991;11:3070–3078. doi: 10.1523/JNEUROSCI.11-10-03070.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Timmer M. Cesnulevicius K. Winkler C. Kolb J. Lipokatic-Takacs E, et al. Fibroblast growth factor (FGF)-2 and FGF receptor 3 are required for the development of the substantia nigra, and FGF-2 plays a crucial role for the rescue of dopaminergic neurons after 6-hydroxydopamine lesion. J Neurosci. 2007;27:459–471. doi: 10.1523/JNEUROSCI.4493-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saarimäki-Vire J. Peltopuro P. Lahti L. Naserke T. Blak A, et al. Fibroblast growth factor receptors cooperate to regulate neural progenitor properties in the developing midbrain and hindbrain. J Neurosci. 2007;27:8581–8592. doi: 10.1523/JNEUROSCI.0192-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schulte G. Bryja V. Rawal N. Castelo-Branco G. Sousa KM, et al. Purified Wnt-5a increases differentiation of midbrain dopaminergic cells and dishevelled phosphorylation. J Neurochem. 2005;92:1550–1553. doi: 10.1111/j.1471-4159.2004.03022.x. [DOI] [PubMed] [Google Scholar]
- 67.McMahon AP. Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62:1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- 68.Ribes V. Balaskas N. Sasai N. Cruz C. Dessaud E, et al. Distinct Sonic Hedgehog signaling dynamics specify floor plate and ventral neuronal progenitors in the vertebrate neural tube 2. Genes Dev. 2010;24:1186–1200. doi: 10.1101/gad.559910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dessaud E. Yang LL. Hill K. Cox B. Ulloa F, et al. Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature. 2007;450:717–720. doi: 10.1038/nature06347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.