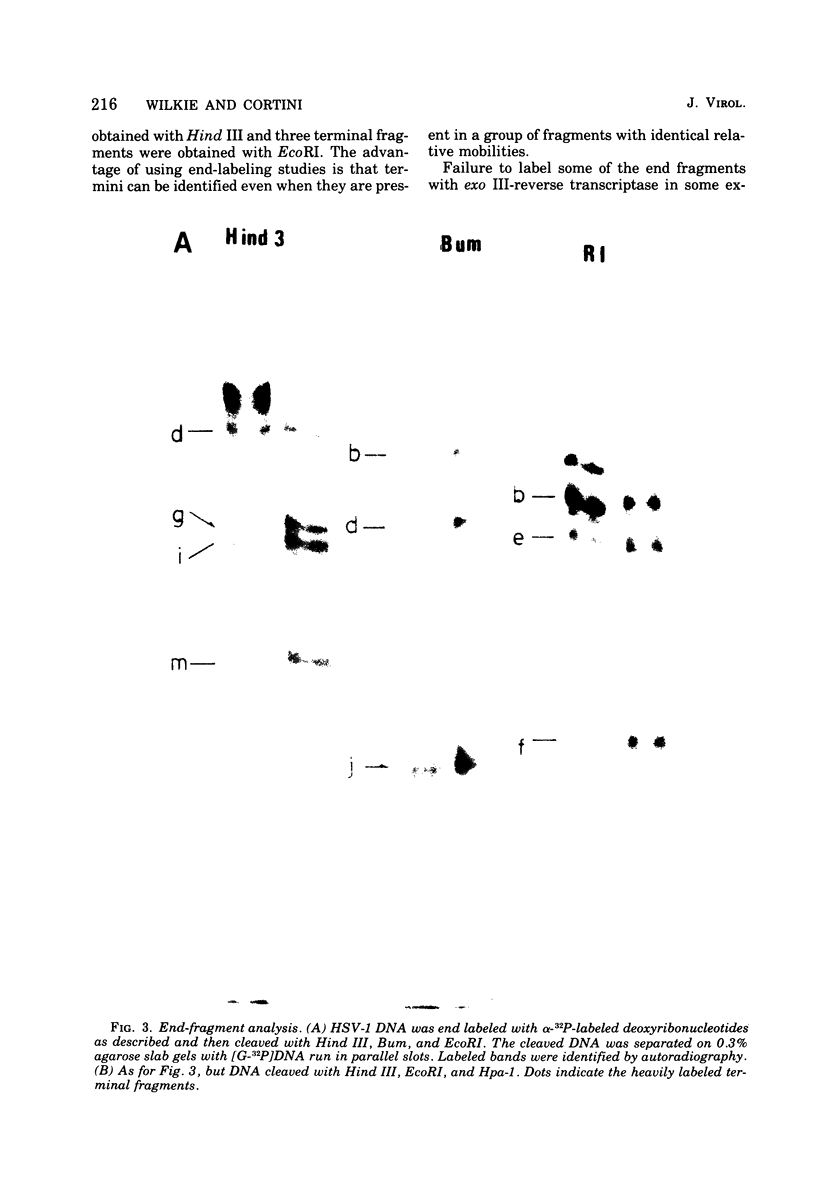
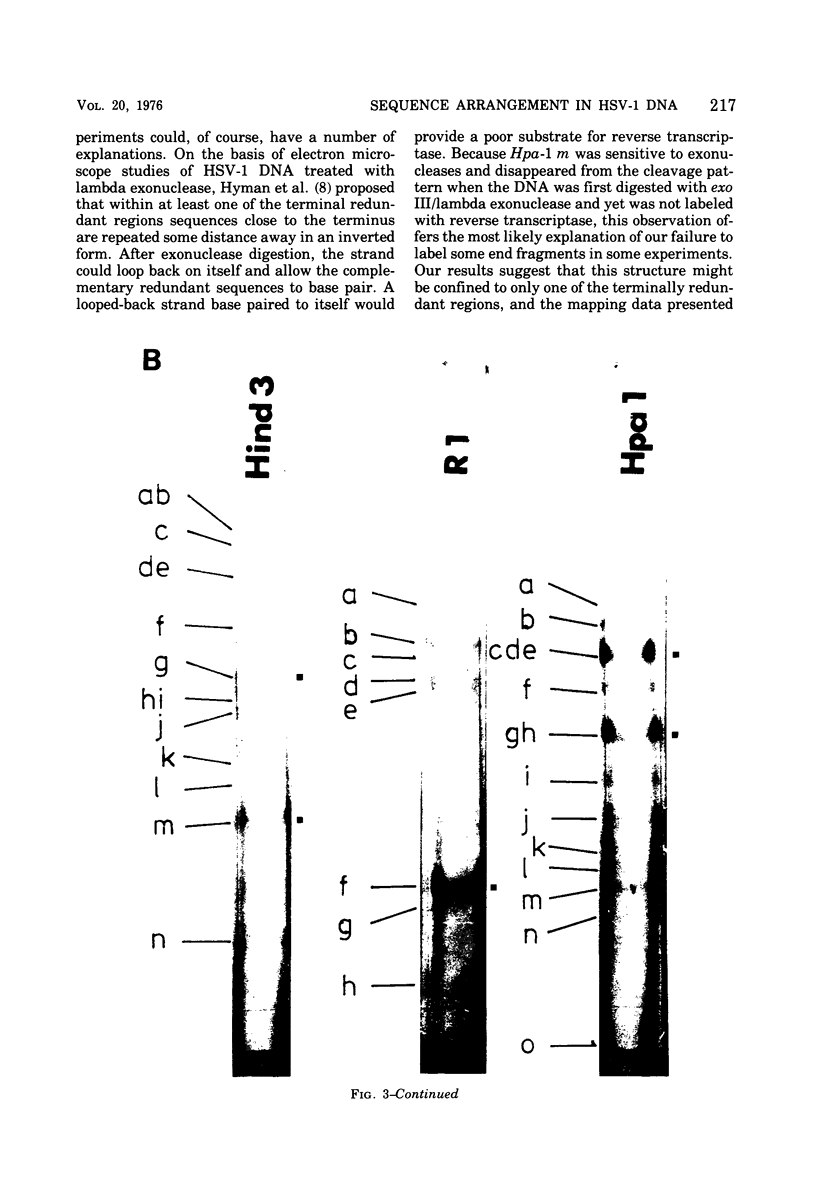
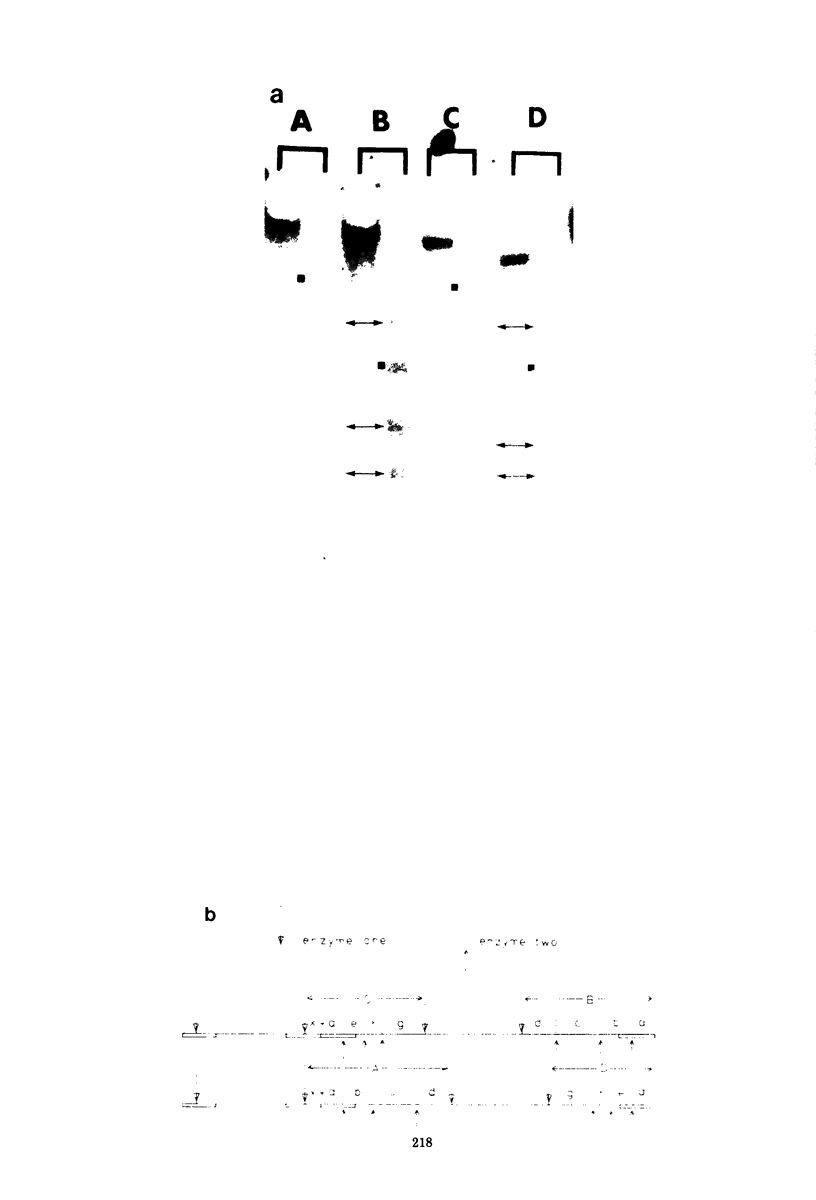
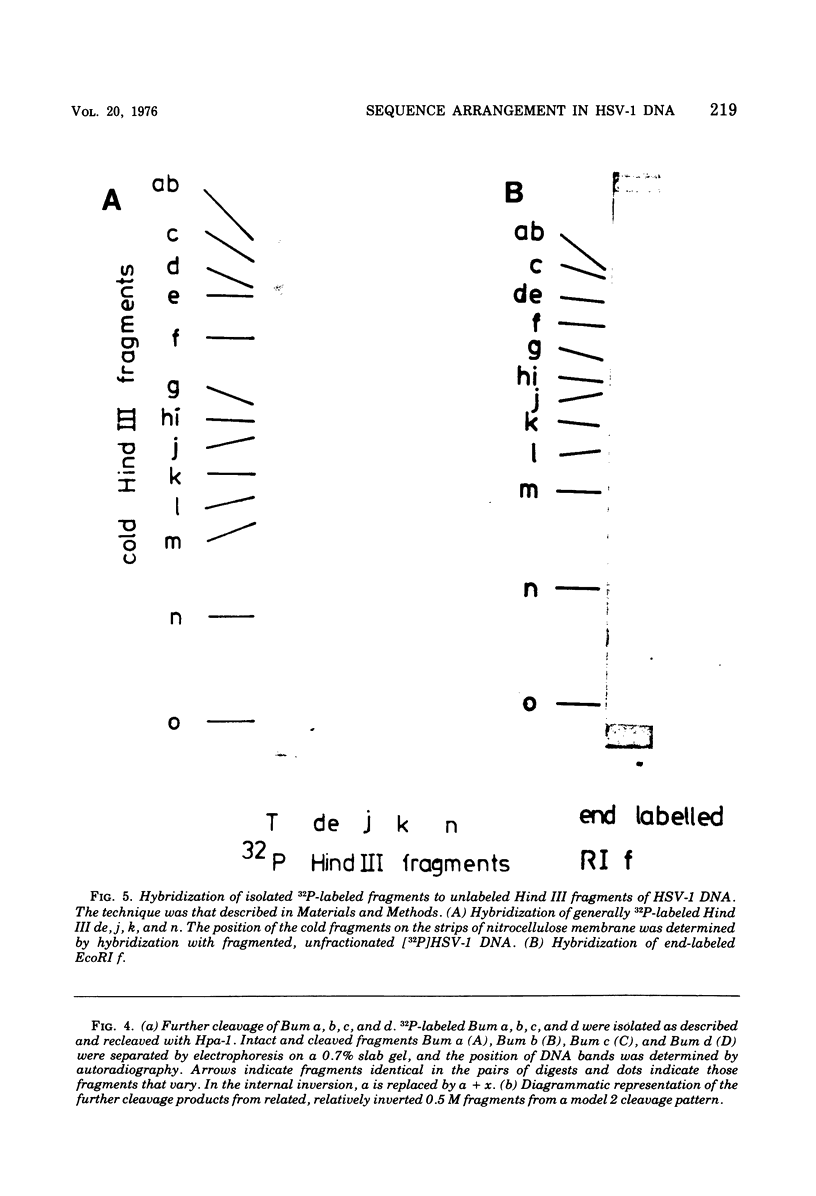
Abstract
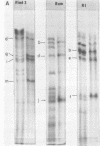
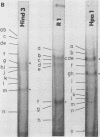
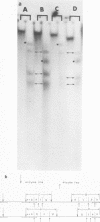
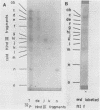
It has been proposed by Sheldrick and Berthelot (1974) that the terminal sequences of herpes simplex virus type 1 (HSV-1) DNA are repeated in an internal inverted form and that the inverted redundant sequences delimit and separate two unique sequences, S and L. In this study the sequence arrangement in HSV-1 DNA has been investigated with restriction endonuclease cleavage, end-labeling studies, and molecular hybridization experiments. The terminal fragments in digests with restriction endonucleases Hind III, Hpa-1, EcoRI and Bum were identified and shown to be consistent with the Sheldrick and Berthelot model. Inverted fragments which contain unique sequences as well as redundant sequences, and which the model predicts, were identified by DNA-DNA hybridization studies. Further cleavage of Bum fragments with Hpa-1 also revealed inversions of the terminal sequences that contained unique sequences. The results obtained showed that the unique sequences S and L are relatively inverted in different DNA molecules in the population, resulting in the presence of four related genomes with rearranged sequences in apparently equal amounts. The redundant sequences bounding S do not share complete sequence homology with those bounding L, but hybridization studies are presented which show that the terminal 0.3% of the genome is repeated in every redundant sequence.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arrand J. R., Keller W., Roberts R. J. Extent of terminal repetition in adenovirus 2 DNA. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):401–407. doi: 10.1101/sqb.1974.039.01.052. [DOI] [PubMed] [Google Scholar]
- Clements J. B., Cortini R., Wilkie N. M. Analysis of herpesvirus DNA substructure by means of restriction endonucleases. J Gen Virol. 1976 Feb;30(2):243–256. doi: 10.1099/0022-1317-30-2-243. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Grafstrom R. H., Alwine J. C., Steinhart W. L., Hill C. W., Hyman R. W. The terminal repetition of herpes simplex virus DNA. Virology. 1975 Sep;67(1):144–157. doi: 10.1016/0042-6822(75)90412-2. [DOI] [PubMed] [Google Scholar]
- Grafstrom R. H., Alwine J. C., Steinhart W. L., Hill C. W. Terminal repetitions in herpes simplex virus type 1 DNA. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):679–681. doi: 10.1101/sqb.1974.039.01.081. [DOI] [PubMed] [Google Scholar]
- Hayward G. S., Frenkel N., Roizman B. Anatomy of herpes simplex virus DNA: strain differences and heterogeneity in the locations of restriction endonuclease cleavage sites. Proc Natl Acad Sci U S A. 1975 May;72(5):1768–1772. doi: 10.1073/pnas.72.5.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman R. W., Burke S., Kudler L. A nearby inverted repeat of the terminal sequence of herpes simplex virus DNA. Biochem Biophys Res Commun. 1976 Jan 26;68(2):609–615. doi: 10.1016/0006-291x(76)91189-x. [DOI] [PubMed] [Google Scholar]
- Kieff E. D., Bachenheimer S. L., Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J Virol. 1971 Aug;8(2):125–132. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B., Kozak M., Honess R. W., Hayward G. Regulation of herpesvirus macromolecular synthesis: evidence for multilevel regulation of herpes simplex 1 RNA and protein synthesis. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):687–701. doi: 10.1101/sqb.1974.039.01.083. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Laithier M., Lando D., Ryhiner M. L. Infectious DNA from herpes simplex virus: infectivity of double-stranded and single-stranded molecules. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3621–3625. doi: 10.1073/pnas.70.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skare J., Summers W. P., Summers W. C. Structure and function of herpesvirus genomes. I. comparison of five HSV-1 and two HSV-2 strains by cleavage their DNA with eco R I restriction endonuclease. J Virol. 1975 Apr;15(4):726–732. doi: 10.1128/jvi.15.4.726-732.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Wilkie N. M., Clements J. B., Macnab J. C., Subak-Sharpe J. H. The structure and biological properties of herpes simplex virus DNA. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):657–666. doi: 10.1101/sqb.1974.039.01.079. [DOI] [PubMed] [Google Scholar]
- Wilkie N. M. Physical maps for Herpes simplex virus type 1 DNA for restriction endonucleases Hind III, Hpa-1, and X. bad. J Virol. 1976 Oct;20(1):222–233. doi: 10.1128/jvi.20.1.222-233.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie N. M. The synthesis and substructure of herpesvirus DNA: the distribution of alkali-labile single strand interruptions in HSV-1 DNA. J Gen Virol. 1973 Dec;21(3):453–467. doi: 10.1099/0022-1317-21-3-453. [DOI] [PubMed] [Google Scholar]