Abstract
Chromatin regulation is a fundamental mechanism underlying stem cell pluripotency, differentiation, and the establishment of cell type-specific gene expression profiles. To examine the role of chromatin regulation in stem cells in vivo, we study regeneration in the freshwater planarian Schmidtea mediterranea. These animals possess a high concentration of pluripotent stem cells, which are capable of restoring any damaged or lost tissues after injury or amputation. Here, we identify the S. mediterranea homologs of the SET1/MLL family of histone methyltransferases and COMPASS and COMPASS-like complex proteins and investigate their role in stem cell function during regeneration. We identified six S. mediterranea homologs of the SET1/MLL family (set1, mll1/2, trr-1, trr-2, mll5–1 and mll5–2), characterized their patterns of expression in the animal, and examined their function by RNAi. All members of this family are expressed in the stem cell population and differentiated tissues. We show that set1, mll1/2, trr-1, and mll5–2 are required for regeneration and that set1, trr-1 and mll5–2 play roles in the regulation of mitosis. Most notably, knockdown of the planarian set1 homolog leads to stem cell depletion. A subset of planarian homologs of COMPASS and COMPASS-like complex proteins are also expressed in stem cells and implicated in regeneration, but the knockdown phenotypes suggest that some complex members also function in other aspects of planarian biology. This work characterizes the function of the SET1/MLL family in the context of planarian regeneration and provides insight into the role of these enzymes in adult stem cell regulation in vivo.
Keywords: stem cells, regeneration, neoblasts, planarian, SET1, MLL, COMPASS, histone methyltransferase, H3K4
Introduction
Coordinated changes in gene expression underlie the transition from a pluripotent stem cell state through lineage commitment and differentiation into a vast array of cell types. Covalent modifications of histones, including methylation, acetylation, ubiquitination, phosphorylation, and other types of marks1 establish and propagate gene expression programs over multiple cellular generations. The SET1/MLL family of histone methyltransferases regulates gene expression by methylating lysine 4 of histone H3, which is associated with an active chromatin state.2,3 Members of this family include Set1 in yeast,4 SET-2 and SET-16 in Caenorhabditis elegans,5 dSET1, Trithorax (TRX) and Trithorax-related (TRR) in Drosophila melanogaster,6 and SET1A, SET1B and MLL1-MLL5 in humans.3,7 SET1/MLL family members (excluding MLL5) act as the catalytic subunit in COMPASS (complex proteins associated with Set1) and COMPASS-like complexes with other proteins that are essential for their methyltransferase activity.2,6,8-12 These other complex members aid in complex assembly, modulate the methyltransferase activity of the SET domain-containing member, and can aid in recruitment to targets.2,13 In species with more than one SET1/MLL family member, the proteins play non-redundant roles in regulating diverse sets of genes,2,3 and their targets include key developmental regulators such as Hox genes.2
Besides playing critical roles in epigenetic regulation of gene expression during development, SET1/MLL proteins have been implicated in human diseases, including cancer. Chromosomal translocations that fuse the N-terminus of MLL to other proteins lead to myeloid and lymphoblastic leukemia.14 Many common MLL fusion partners are members of a Super Elongation Complex that controls transcription elongation,15 and misregulation of elongation at normal MLL targets such as the Hox genes may be an underlying cause of disease pathogenesis.15 MLL translocations can transform hematopoietic cells at different stages of development, including hematopoietic stem cells and committed progenitor cells such as common myeloid progenitors and granulocyte macrophage progenitors, into leukemia stem cells, restoring capacity for self-renewal in more differentiated cell types.14,16,17 A deeper understanding of the role of the SET1/MLL family in epigenetic programming of gene expression in normal stem cells could help elucidate how these genes contribute to cancer stem cell formation.14
The planarian flatworm Schmidtea mediterranea is an excellent model system for studying stem cells in vivo. These animals are capable of regenerating any tissues lost to injury and constantly replace cells of all types through normal homeostatic turnover.18,19 Their regenerative ability is owed to a population of adult stem cells, called neoblasts, which make up ~20–35% of the total cell number in the worm18,20 and are maintained throughout their lifetime. Recent studies have demonstrated that a subset of the neoblast population is truly pluripotent.21 Furthermore, comparisons of the gene expression profile of neoblasts to that of mammalian embryonic stem cells identified conserved pluripotency factors22-24 and illustrate that the insights gained into stem cell biology in planarians are applicable to other species. Through studies using this model, we can potentially gain a better understanding of the epigenetic changes underlying stem cell pluripotency and fate specification during normal cell turnover or in regeneration.
In this study, we identify the S. mediterranea homologs of SET1/MLL family proteins and COMPASS and COMPASS-like complex proteins and characterize their role in stem cell regulation. We found six planarian members of the SET1/MLL family and determined their evolutionary relationship to members of the family from other species, characterized their patterns of expression, and examined their function by RNA interference (RNAi). All members of this family are expressed in stem cells and differentiated tissues, and most are required for normal regeneration. The family also plays a role in maintaining the proper number of mitotic cells, and knockdown of Smed-set1 leads to a loss of stem cells. Most of the planarian homologs of COMPASS and COMPASS-like complex proteins are also expressed in stem cells and required for stem cell function, but the knockdown phenotypes for some members of the complex suggest that they also function in other aspects of planarian biology. This work characterizes the SET1/MLL family and provides insights into the role of specific histone methyltransferases in adult stem cell regulation during tissue regeneration in vivo.
Results
SET1/MLL family proteins in S. mediterranea
We identified six SET1/MLL family proteins in S. mediterranea by performing standalone tblastn searches with the SET domain portion of members of this family from humans, mice, worms, flies and yeast against the S. mediterranea genome and transcriptomes (see Materials and Methods). We performed phylogenetic analysis to determine the relationship of the S. mediterranea proteins within the family (Fig. 1) and named the newly identified proteins based on homology. The predicted protein product of each gene contains a SET [Su(var)3–9, enhancer-of-zeste, Trithorax] domain, which is the methyltransferase catalytic domain.25
Figure 1. Phylogenetic tree of SET1/MLL family proteins. Relationships of planarian SET1/MLL proteins (highlighted in red) to those of other species based on Neighbor-joining analysis of their SET domains. Numbers represent the percentage of Bootstrap replicates that include that node. The predicted domain structure of each protein is diagramed to the right. The members of the larger family fall into four sub-groups indicated by brackets. Sequences were selected from Homo sapiens (HUMAN), Mus musculus (MOUSE), Caenorhabditis elegans (CAEEL), Schizosaccharomyces pombe (SCHPO), Saccharomyces cerevisiae (YEAST), Drosophila melanogaster (DROME) and Schmidtea mediterranea (SMED).
Other domains are also shared between the planarian SET1/MLL proteins and their homologs from other species. SMED-SET1 contains an N-terminal RNA recognition motif, as do many other members of the SET1 sub-family. SMED-MLL1/2, which groups with Drosophila Trithorax (TRX) and vertebrate MLL1 and MLL2, is most similar to MLL2 in its domain structure, having the CXXC-type and PHD-type zinc fingers and FYR-N-terminal domain common to this sub-family but lacking the nuclear hormone receptor binding domain of TRX and the bromodomain of MLL1. SMED-TRR-1 and SMED-TRR-2 both fall within the TRR/MLL3/MLL4 sub-family, with SMED-TRR-1 grouping closely with Drosophila Trithorax-related (TRR) and SMED-TRR-2 relating to the sub-family more generally. Each of these planarian proteins contains FYR N- and C-terminal domains in close proximity to each other near the SET domain, a feature common to all members of this sub-family. The N-terminal halves of MLL3 and MLL4, which contain various combinations of PHD-type, RING-type and DHHC-type zinc fingers, are represented in Drosophila by a separate protein called LPT (lost plant homeodomains of TRR)6 that acts in the same complex as TRR so that together they fulfill the roles of MLL3 or MLL4. Interestingly, this function seems to be split between LPT and TRR in S. mediterranea as in Drosophila; SMED-TRR-1 and SMED-TRR-2 lack zinc fingers, and a potential LPT homolog encoded on a separate genomic contig (v31.007060) is expressed based on transcriptome data (isotig2294926 and BPKG19822). The MLL5 subgroup is the most divergent and shares homology with yeast Set3/4 and Ash1 families27 in addition to its relatedness to the rest of the SET1/MLL group.28 SMED-MLL5-1 and SMED-MLL5-2 cluster with vertebrate MLL5 and contain no other domains besides the SET domain, which in this sub-family is found closer to the N-terminus of the protein rather than at the C-terminus.
Planarian set1/mll genes are expressed in stem cells and differentiated tissues
To determine set1/mll gene expression patterns in planarians, we performed whole mount in situ hybridization. All six genes are detected throughout the animal with stronger expression in the mesenchyme, outlining the cephalic ganglia, and in the intestine (Fig. 2). In addition, Smed-mll1/2 and Smed-mll5–2 show strong epidermal expression. We treated worms with 60 Gy of γ-irradiation three days prior to fixation to destroy the stem cells and compared them to untreated controls. Mesenchymal expression of all six transcripts was reduced following irradiation, indicating that they are expressed in stem cells. Quantitative real-time polymerase chain reaction (qPCR) measuring the amount of each mRNA in samples from control and irradiated animals confirmed that there is a decrease in transcript levels following irradiation (Fig. S1). These results are consistent with recently published transcriptome expression studies,22-24 which classify these genes as expressed both in the X1 irradiation sensitive stem cell population and in differentiated tissues (Table S1). All six genes are strongly expressed in the blastema during regeneration with “peak” expression on specific days post-amputation (Fig. S2), suggesting possible roles of these genes in differentiation.
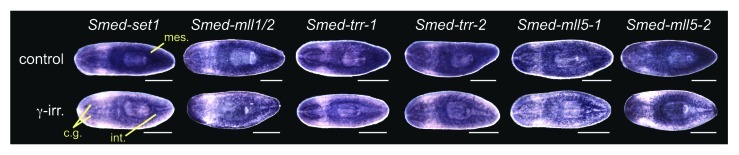
Figure 2. Expression of planarian set1/mll family genes. Whole mount in situ hybridization showing the mRNA expression patterns of genes in the S. mediterranea set1/mll family. The lower worm of each pair was treated with 60 Gy of γ−irradiation three days prior to fixation. mes, mesenchyme; cg, cephalic ganglia; int, intestine. Animals are shown ventral side up with anterior to the left. Scale bars = 0.5 mm.
Knockdown of set1/mll family genes results in defects in regeneration and homeostasis
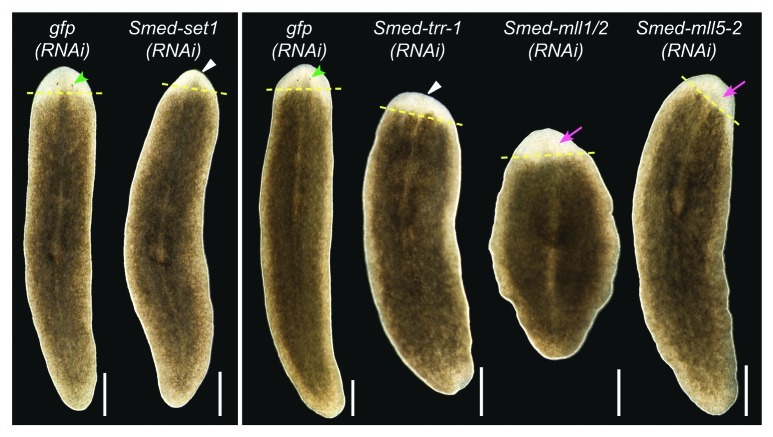
Regeneration in planarians serves as a readout of stem cell function because it requires the stem cells to proliferate and differentiate in response to injury to replace the missing tissues. To assay the function of set1/mll family members in planarian regeneration, we fed animals bacterially expressed dsRNA targeting each gene29 then amputated the animals anterior to the pharynx and observed them during regeneration. Control animals that were fed gfp dsRNA formed a regeneration blastema and fully regenerated all missing structures within 10 days (Fig. 3). Knockdown of Smed-set1 or Smed-trr-1 led to reduced blastema size and a delay in formation of or failure to regenerate visible photoreceptors (Table S2). Smed-mll1/2(RNAi) and Smed-mll5–2(RNAi) animals formed normally sized blastemas but displayed delayed or reduced photoreceptor regeneration.
Figure 3.set1/mll family knockdown phenotypes. Planarians were fed bacterially expressed double-stranded RNA targeting each set1/mll gene and then transected anterior to the pharynx and observed during regeneration. gfp(RNAi) serves as a negative control. Animals in the left panel were fed three times over one and a half weeks and imaged after six days of regeneration. The animals in the right panel were fed six times over three weeks and imaged after seven days of regeneration. Yellow dashed lines mark the plane of amputation. White triangles indicate reduced blastema growth, green arrowheads indicate normal photoreceptors, and magenta arrows indicate lack of or underdeveloped photoreceptors. Anterior is up. Scale bars = 0.5 mm.
In addition to regeneration specific phenotypes, we also observed other defects in both amputated and intact animals treated with dsRNA targeting Smed-set1 or Smed-mll1/2. For Smed-set1, these included ventral curling, lesions, lysis and death in amputated animals after three dsRNA feedings and in intact animals after four or more feedings. These phenotypes are associated with loss of stem cells, suggesting that Smed-set1 may be required for stem cell maintenance or proliferation. Following regeneration, Smed-mll1/2 knockdown animals did not readily move or stretch out to their full length and instead remained in one place with a shortened and ruffled appearance. We also observed areas of edema (tissue with a swollen, lighter appearance) in a few (n = 3/29) uninjured Smed-mll1/2(RNAi) animals after nine dsRNA feedings, indicating a potential defect in the protonephridia, the planarian excretory system. Planarians use ventral cilia for movement,30 and the protonephridia contain ciliated cells.31 Thus, the loss of mobility and edema could be associated with defects in cilia function. Staining of regenerating Smed-mll1/2(RNAi) animals with anti-acetylated tubulin showed that the density of the cilia was considerably reduced compared to gfp(RNAi) controls, both in the blastema and uninjured areas (Fig. S3A). We also stained regenerating Smed-mll1/2(RNAi) animals by in situ hybridization to EGFR5 and inx10, which label ciliated sections of the protonephridia,31 and observed a marked reduction of staining for these markers in the blastema (Fig. S3B). Thus, our data suggest that Smed-mll1/2 is implicated in the differentiation or maintenance of the ventral cilia and might have a role in protonephridia development among other processes.
No phenotypes were observed following knockdown of Smed-trr-2 or Smed-mll5-1 using our standard scheme of up to six dsRNA feedings. When we extended the knockdown to nine feedings, intact Smed-trr-2(RNAi) animals displayed head regression and lysis in one out of three independent experiments. We never observed a phenotype in Smed-mll5-1(RNAi), even after extending the dsRNA treatment to nine feedings. Preliminary qPCR measurements suggest knockdown of Smed-mll5-1 mRNA is incomplete (data not shown) and that analysis of this gene may require longer feeding schemes or improving the design of our RNAi constructs. In addition, we performed simultaneous knockdown of each of these genes with their closest homolog (Smed-trr-1 with Smed-trr-2, and Smed-mll5-1 with Smed-mll5-2). We observed an enhancement of the Smed-trr-1 phenotype when Smed-trr-2 was also inhibited. Smed-trr-1 RNAi by itself led to reduced regeneration, but the animals were able to form a small blastema and regenerate photoreceptors; however, in the double knockdown condition, there was little to no blastema formation, and the majority of the animals curled ventrally and died by lysis (Table S2). This strengthening of the stem cell loss phenotypes in the double knockdown suggests that there may be some redundancy in the function of Smed-trr-1 and Smed-trr-2 in stem cell maintenance. We did not observe an enhancement of the Smed-mll5-2 phenotype when we simultaneously knocked down Smed-mll5-1.
set1/mll genes regulate stem cell proliferation
To examine the role of these genes in controlling proliferation of planarian stem cells, we stained animals treated with dsRNA against set1/mll genes with anti-phospho-Histone H3 (PH3). We found that the number of mitotic cells more than doubled following knockdown of Smed-set1, from 34 ± 5 cells/mm2 in gfp(RNAi) controls to 74 ± 8 cells/mm2 in Smed-set1(RNAi) (Fig. 4A and B). We also observed a statistically significant increase in the number of PH3+ cells in Smed-mll5-2(RNAi), to 64 ± 9 cells/mm2 from 32 ± 2 in controls. Knockdown of Smed-trr-1 had the opposite effect; the number of mitotic cells decreased to 16 ± 2 cells/mm2. We conclude that Smed-mll5-2 negatively regulates stem cell mitosis and that Smed-trr-1 promotes it. The ventral curling, head regression, and lysis phenotypes associated with Smed-set1(RNAi) are stereotypical signs of stem cell loss. Therefore, although we observed an increase in PH3+ cells following knockdown of Smed-set1, we hypothesized that these animals had a reduced number of stem cells overall.
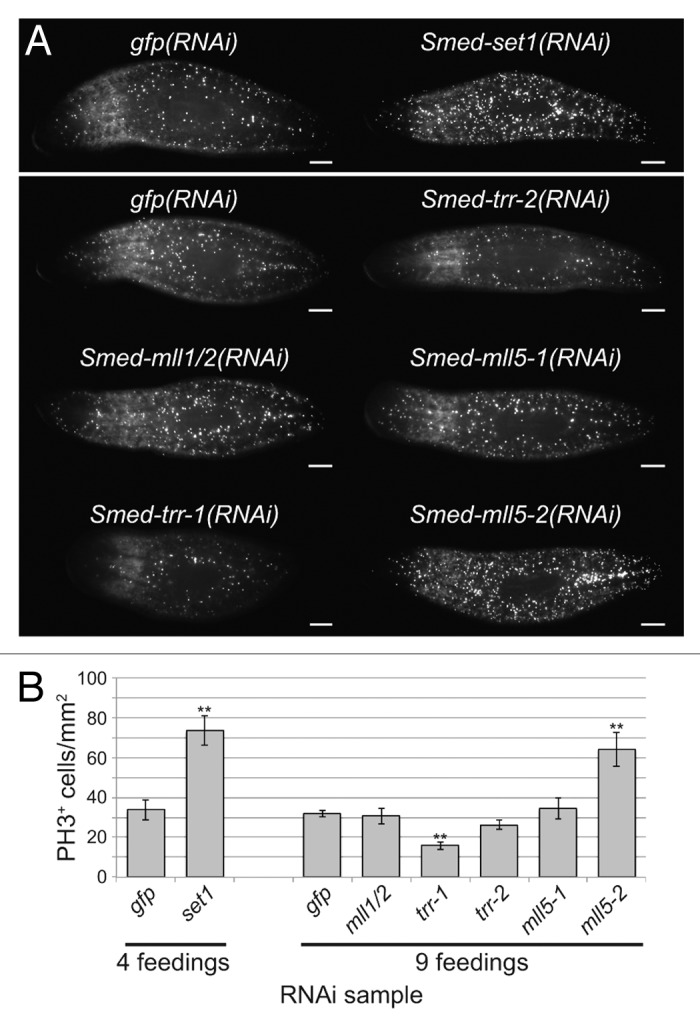
Figure 4. Effect of set1/mll knockdown on the mitotic stem cell population. (A) Anti-phospho-Histone H3 (Ser10) staining following RNAi against set1/mll family genes. Animals in the upper panel received four RNAi feedings over the course of two weeks, and worms in the lower panel received nine feedings over 4.5 weeks. Samples were fixed five days after the final feeding. Scale bars = 0.5 mm. (B) Quantitation of the number of phospho-Histone H3-positive cells in each group shown in A. Error bars represent standard error of the mean. Results significantly different from gfp(RNAi) control are marked by ** (p value < 0.01, Student’s t-test).
Smed-set1 RNAi leads to loss of the stem cell population
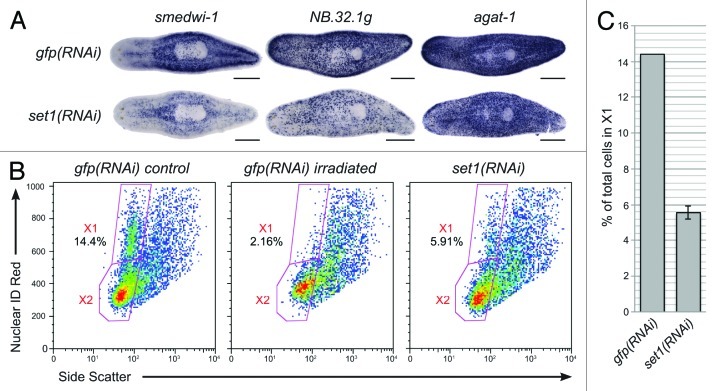
To test the hypothesis that Smed-set1 is required for stem cell maintenance, we used additional markers to label the stem cells and their descendants following Smed-set1 RNAi. gfp(RNAi) and Smed-set1(RNAi) worms that were fed bacterially expressed dsRNA four times over two weeks were fixed and processed for whole mount in situ hybridization to markers of the stem cells (Smedwi-1) or their descendants (NB.32.1g and agat-1) (Fig. 5A).32 Compared with controls, Smed-set1(RNAi) worms had reduced staining for each of the three markers, suggesting that Smed-set1 is required for maintenance of the stem cell pool. We used flow cytometry to verify that there was a loss of the cycling stem cell population and not simply a reduction in marker expression. Planarians that were dissociated following Smed-set1 RNAi showed a decrease in the percentage of cells in the X1 population (the cycling stem cells)33 from a mean of 14.4% in the gfp(RNAi) controls to 5.6% in Smed-set1(RNAi) animals (Fig. 5B and C). We conclude that Smed-set1 is indispensable for stem cell maintenance.
Figure 5. RNAi of Smed-set1 results in loss of stem cells. (A) In situ hybridization to markers of stem cell and progeny cell populations following knockdown of Smed-set1. Worms were fed dsRNA against Smed-set1 or gfp (negative control) four times over two weeks and then fixed and stained for markers of stem cells and their descendants. Scale bars = 0.5 mm. (B) Flow cytometric analysis of Smed-set1(RNAi) worms. Worms from the same RNAi knockdown as in A were dissociated and analyzed by flow cytometry. Some gfp(RNAi) worms were treated with 100 Gy γ-irradiation four days prior to dissociation to aid in identification of the cycling cells. (C) Summary of change in the percentage of cells in X1 from two replicates of the flow cytometric analysis shown in B. Error bars represent standard error of the mean. The difference between the two groups is statistically significant (p value < 0.01, Student’s t-test).
Identification and functional analysis of planarian COMPASS and COMPASS-like complex members
SET1/MLL family members interact with other proteins in COMPASS and COMPASS-like complexes that are essential for their activity. Core components ASH2, DPY30, RBBP5 and WDR5 are common to all COMPASS and COMPASS-like complexes (Fig. 6A).13 The human COMPASS complex, which associates with SET1A and SET1B, additionally includes CXXC1, WDR82 and HCF1.9,11 MLL1 and MLL2 each enter into COMPASS-like complexes that include HCF1 and Menin,12 while MLL3 and MLL4 interact with PTIP, UTX, PA1 and NCOA6 as complex specific subunits10 in addition to the core components. SET1/MLL family proteins in other species act in analogous complexes.6,8
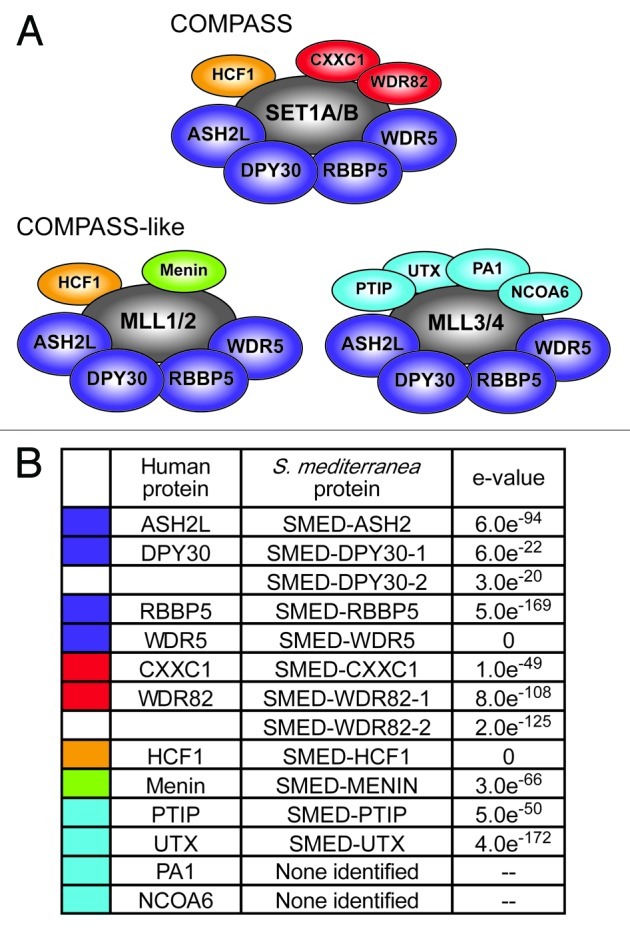
Figure 6. Identification of planarian COMPASS proteins. (A) Human COMPASS and COMPASS-like complexes. SET1/MLL family members (shown in gray) act in complex with other proteins. Core subunits common to all COMPASS and COMPASS-like complexes are shown in dark blue. Complex-specific subunits are color-coded by complex: red, SET1/COMPASS complex; orange, SET1/COMPASS and MLL1/2 COMPASS-like complexes; green, MLL1/2 COMPASS-like complex; light blue, MLL3/4 COMPASS-like complex. (B) S. mediterranea homologs of COMPASS proteins. Color-coding matches that in A. e-values are for reciprocal protein blast of the predicted full-length planarian protein against the top human hit in the NCBI non-redundant protein sequence database.
To further investigate the activity of the SET1/MLL family in planarians, we sought to identify and characterize the COMPASS complex members that may work with them. We identified the planarian homologs of COMPASS and COMPASS-like proteins by tblastn with the yeast and human proteins against the S. mediterranea genome and transcriptomes. Figure 6B lists these homologs and the e-values from reciprocal blastp of the planarian proteins against their corresponding human proteins. There was one planarian protein for each human COMPASS and COMPASS-like protein, with the exception of WDR82 and DPY30, which each had two potential planarian homologs, and PA1 and NCOA6, which had no hits meeting our criteria.
We subsequently examined the planarian COMPASS and COMPASS-like genes by whole mount in situ hybridization and found that expression of many of the COMPASS genes resembled that of SET1/MLL family members (Fig. 7). Core complex members Smed-ash2, Smed-dpy30-2, Smed-rbbp5 and Smed-wdr5 are widely expressed throughout the animal, and treatment with γ-irradiation led to a reduction in mesenchymal staining, indicating that these genes are expressed in the stem cells. Mesenchymal expression of Smed-dpy30-1 and SET1/COMPASS-specific subunits Smed-cxxc1, Smed-wdr82-1, and Smed-wdr82-2 was also slightly reduced following γ-irradiation; however, strong intestinal expression in the irradiated animals made this reduction less striking. As with SET1/MLL family members, our observation that each of these genes is expressed both in stem cells and in differentiated tissues is consistent with recent transcriptome data (summarized in Table S1).22-24 Smed-hcf1, the final SET1/COMPASS member is similarly expressed in stem cells, raising the possibility of a complete COMPASS complex interacting with Smed-set1 in those cells. All of the core complex members and COMPASS-specific subunits are also expressed in the intestine, and different subsets are found in the cephalic ganglia and epidermis. With the exception of the dpy30 homologs, each of these genes is upregulated in the blastema during the earliest days of regeneration (day 1–3; Fig. S4). Staining patterns that did not overlap with those seen for Smed-set1 include stronger expression around the anterior periphery of the head for Smed-dpy30-1 and Smed-dpy30-2 and the appearance of dark puncta in the mesenchyme around the pharynx with Smed-wdr82-1 and Smed-wdr82-2.
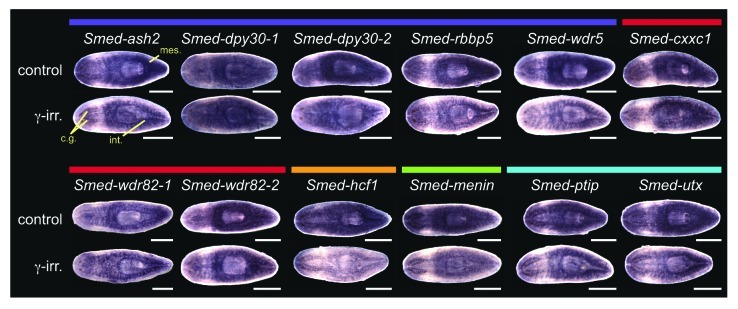
Figure 7. Expression of planarian COMPASS genes. Whole mount in situ hybridization to homologs of COMPASS and COMPASS-like complex members. Colored bars above the gene names indicate complex membership as in Figure 6. The lower worm of each pair was treated with 60 Gy γ-irradiation three days prior to fixation. Mes, mesenchyme; cg, cephalic ganglia; int, intestine. Animals are shown ventral side up with the anterior to the left. Scale bars = 0.5 mm.
Smed-menin, which based on homology is predicted to participate in a Smed-mll1/2-containing COMPASS-like complex, is expressed in stem cells, as well as in the intestine and cephalic ganglia. Unlike Smed-mll1/2, however, Smed-menin expression was not detected in the epidermis. The staining patterns of Smed-ptip and Smed-utx closely resemble those of Smed-trr-1 and Smed-trr-2, with fairly strong expression in the cephalic ganglia and intestine in addition to the stem cells. This similarity of expression suggests that these proteins may interact in a COMPASS-like complex analogous to those of Drosophila TRR and its mammalian homologs MLL3 and MLL4. Smed-menin, Smed-ptip and Smed-utx show high expression in the blastema early during regeneration and all are strongly expressed by day 3 of regeneration (Fig. S4).
To test the hypothesis that COMPASS proteins cooperate with SET1/MLL family members in planarians, we examined their function by RNAi. Worms were fed dsRNA targeting each gene of interest six times over three weeks and then amputated anterior to the pharynx and observed over ten days of regeneration. Knockdown of core complex members Smed-ash2 and Smed-rbbp5, as well as SET1/COMPASS members Smed-wdr82-2 and Smed-hcf1 led to reduced blastema formation and failure to regenerate visible photoreceptors (Fig. 8). Animals undergoing knockdown of any of those four genes eventually began to curl ventrally and died by lysing, indicative of a loss of stem cells. These phenotypes match those of Smed-set1, suggesting that these COMPASS members act together with Smed-set1.
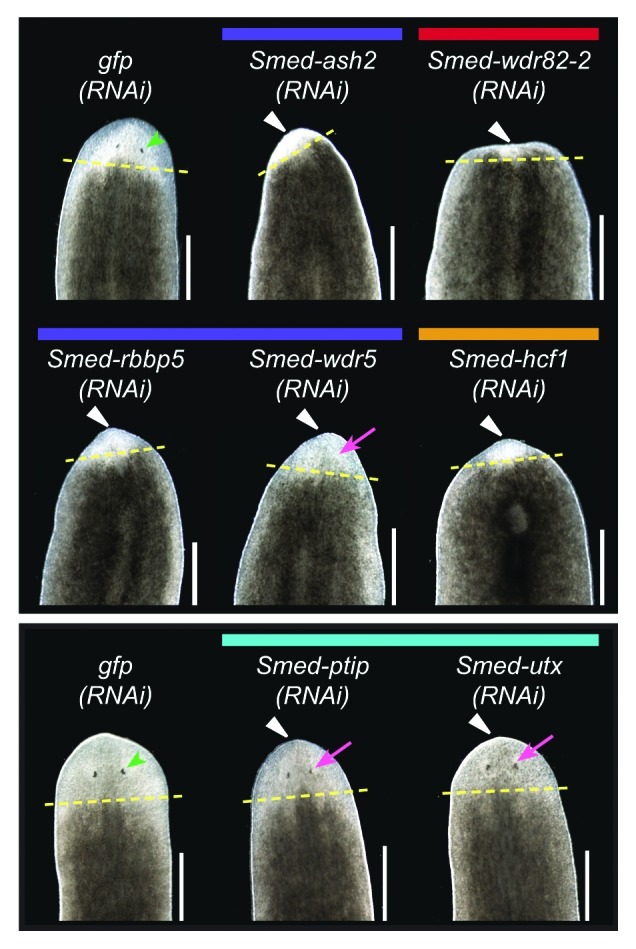
Figure 8. RNAi knockdown of COMPASS complex genes. Planarians were fed dsRNA targeting the indicated gene six times over three weeks and then amputated pre-pharyngeally to assay regeneration. gfp(RNAi) served as a negative control. The worms in the upper and lower panels were imaged after six or ten days of regeneration, respectively. Yellow dashed lines mark the plane of amputation. White triangles indicate reduced blastema formation, green arrowheads indicate normal photoreceptors, and magenta arrows indicate underdeveloped photoreceptors. Scale bars = 0.5 mm.
We did not observe loss of stem cell phenotypes when knocking down core complex components Smed-dpy30-1, Smed-dpy30-2 and Smed-wdr5 or the COMPASS members Smed-wdr82-1 and Smed-cxxc1, but we did find other interesting defects. Smed-wdr5 RNAi led to reduced blastema formation, and while these worms showed some signs of photoreceptor regeneration, the photoreceptors appeared underdeveloped (smaller with lighter pigment) relative to controls throughout the regeneration time course (Fig. 8). Smed-cxxc1 or Smed-dpy30-2 RNAi led to a delay in photoreceptor development; however, all Smed-cxxc1(RNAi) and many Smed-dpy30-2(RNAi) animals fully regenerated and were not substantially different than controls by day 10. Smed-dpy30-1(RNAi) worms regenerated normally, but both the head and tail fragments displayed abnormal inching movements. Staining with anti-acetylated tubulin did not reveal an obvious difference in the abundance of ventral cilia (not shown). It remains to be tested if the inching phenotype is due to cilia regulation or function. Finally, loss of Smed-wdr82-1 did not lead to any discernable phenotype.
Knockdown of Smed-menin, which we predicted might recapitulate the reduced regeneration or cilia defects we observed with Smed-mll1/2 RNAi, did not result in any obvious external phenotype. Smed-ptip(RNAi) and Smed-utx(RNAi) animals had reduced blastema formation and smaller, fainter photoreceptors relative to controls. Smed-utx additionally showed abnormal “freckly” pigmentation in the blastema. These results suggest that while Smed-ptip and Smed-utx may work with Smed-trr-1 to achieve normal blastema growth, Smed-utx may act outside this complex as well to control pigment production.
Discussion
Epigenetic control of gene expression is essential during development, for stem cell and somatic cell regulation, and has been implicated in the etiology of multiple diseases. Thus, understanding the precise epigenetic configuration of different cell types has attracted intense research efforts culminating in genome-wide studies in model organisms such as C. elegans and Drosophila and in humans.34,35 The role of epigenetic regulation in regeneration has been examined in species that undergo limited regeneration of particular appendages or organs through dedifferentiation and transdifferentiation.36,37 Planarians have extensive stem cell-based regeneration capabilities and represent an excellent model to investigate epigenetic regulation during tissue replacement.23,38-41
In this study, we have identified and characterized the SET1/MLL family and COMPASS complex homologs in S. mediterranea. We found at least one homolog in each of the four SET1/MLL subfamilies and one-to-one homologs for most of the members of COMPASS and COMPASS-like complexes. The conservation of these genes across diverse branches of the phylogeny underscores the importance of epigenetic regulation in eukaryotic development and allows us to apply knowledge gained from studies in model systems to make predictions about the function of these genes in other species, including humans.
Planarian SET1/COMPASS is required for stem cell maintenance
SET1 is conserved from yeast to humans and serves as the main H3K4 methyltransferase in all species tested,4,11,42-44 acting broadly to promote active gene expression. WDR5, RBBP5, ASH2L, and DPY30 are core members common to all COMPASS and COMPASS-like complexes.13 Knockdown of Smed-set1 (Fig. 3) or the homologs of any of the core COMPASS members except DPY30 (Fig. 8) leads to reduced blastema formation and failure to fully regenerate missing structures such as photoreceptors. WDR5, RBBP5 and ASH2L are integral structural components of the complex and necessary for full enzymatic activity of their SET-domain containing partners.13,45-47 These three members plus the C-terminal fragment of MLL1 are sufficient to reconstitute a complex with in vitro methyltransferase activity similar to that of the MLL1 holocomplex.46
DPY30 increases the rate of catalysis and stimulates the methyltransferase activity of SET1/MLL family members48,49 but plays a less integral role than the other three core components described above.48,50 This may explain why the phenotypes we observed for the two potential DPY30 homologs we identified did not match those of the other core members; the inching behavior of Smed-dpy30-1(RNAi) and slight delay in regeneration of Smed-dpy30-2(RNAi) suggest that, as in other species, planarian DPY30 homologs may be less critical for H3K4 methylation than other complex members and may act in other processes outside their role in COMPASS.
The SET1/COMPASS complex also includes proteins that are not shared among all of the related complexes; WDR82 and CXXC1 are found only in complex with SET1, and HCF1 is found with SET1, MLL1 or MLL2.9,11 We observed defects in blastema formation following knockdown of Smed-wdr82-2 and Smed-hcf1 (Fig. 8) and a delay in regeneration in Smed-cxxc1(RNAi). In other species, WDR82, CXXC1, and HCF1 are involved in recruitment of the SET1/COMPASS complex to its targets through a variety of mechanisms.9,51-53 They are themselves brought to specific regions of the genome through interactions with other chromatin features, including mono-ubiquitinated histone H2B54 and non-methylated CpG dimers,52 as co-activators of transcription factors,53 or by direct interaction with RNA polymerase II.51 The phenotypes of these complex specific subunits in planarians support our conclusion that the SET1/COMPASS complex is required for processes underlying stem cell regulation and regeneration, and their mode of action may be through recruitment of the rest of the complex to specific targets.
Planarian SET1/COMPASS also plays a role in stem cell maintenance. RNAi against Smed-set1 or COMPASS genes Smed-ash2, Smed-rbbp5, Smed-wdr82-2 or Smed-hcf1 leads to loss of stem cell phenotypes such as curling and lysis, and reduced stem cell number was confirmed for Smed-set1(RNAi) by staining with smedwi-1 (Fig. 5). Ang et al.55 recently demonstrated that COMPASS and COMPASS-like complexes regulate self-renewal of mammalian embryonic stem cells through interactions between WDR5 and pluripotency factor OCT4. This interaction recruits the histone methyltransferase complexes to OCT4 targets, where they act as transcriptional co-activators. Although a definitive homolog of OCT4 has not yet been clearly identified in planarians, homologs of OCT4 regulators and direct targets show enriched expression in planarian stem cells compared with differentiated tissues.23 Thus, the requirement of SET1/COMPASS for planarian stem cell maintenance may be through a conserved mechanism.
In Smed-set1(RNAi), we observed an increase in PH3+ mitotic cells at the same time point that staining with a stem cell marker and flow cytometry showed a decrease in the total number of stem cells (Figs. 4 and 5). This may indicate a defect in mitotic progression that causes the residual stem cells to become “stuck” in mitosis before their eventual death, as observed in planarians following RNAi against Smed-lissencephaly-1.56 Intriguingly, mammalian WDR82 acts separately from its role in COMPASS to promote exit from mitosis into interphase as a member of the PP1 phosphatase complex.57 Mutations affecting the PP1 complex cause cells to arrest at mitotic exit due to uneven chromatin decondensation and subsequently die by apoptosis.57 Smed-wdr82-2 may play a similar role during mitosis in planarians, and disruption of the COMPASS complex in Smed-set1(RNAi) may indirectly interfere with this function. Alternatively, the increase in mitosis coupled with failed stem cell self-renewal may be analogous to that seen in the adult hematopoietic stem cells (HSCs) of Mll1 knockout mice; inducible inactivation of Mll1 in bone marrow causes HSCs to ectopically enter the cell cycle, which depletes the pool of quiescent stem cells as these dividing cells proceed to differentiate.58 It is possible this function of regulating cell cycle entry falls on the SET1/COMPASS complex in planarians, rather than on Smed-mll1/2.
A planarian MLL1/2 homolog is involved in cilia development or function
Members of the TRX/MLL1/MLL2 subfamily activate expression of a smaller collection of genes than SET12 and serve non-redundant, essential functions during development. The best-known targets of these proteins are the Hox genes, transcription factors important for anterior-posterior patterning, segment identity, and hematopoiesis.2 Members of this subfamily participate in a COMPASS-like complex that includes Menin in addition to the other proteins shared with COMPASS.12
We identified Smed-mll1/2 as the only member of this subfamily (Fig. 1) and found that it is required for maintenance of ciliated cells of the ventral epidermis and protonephridia regeneration. Based on the conserved role of TRX, MLL1 and MLL2, we hypothesize that Smed-mll1/2 may regulate Hox genes, which have been partially characterized in other planarian species.59-61 It will also be interesting to examine if Smed-mll1/2 targets genes required for ciliogenesis such as kif27, fused, iguana, Smed-ift88 and Smed-kif3a,30,62 or those needed for regeneration of the protonephridia, which include Smed-EGFR-5, Six1/2-2, POU2/3, hunchback, Eya and Sall.31,63
The Smed-trr-1 COMPASS-like complex is required for regeneration
Smed-trr-1 and Smed-trr-2 fall within a subgroup of the SET1/MLL family that includes Drosophila TRR, mammalian MLL3 and MLL4 and C. elegans SET-16 (Fig. 1). These proteins in other species function as co-activators of nuclear hormone receptor signaling, immunoglobulin class switch recombination, and adipogenesis, among other processes, through their role in H3K4 methylation.2 In planarians, knockdown of Smed-trr-1 caused reduced blastema formation and a reduction in the number of mitotic cells (Figs. 3 and 4), which suggests a role for Smed-trr-1 in cell proliferation.
We also identified S. mediterranea homologs of PTIP and UTX, which are subunits unique to MLL3/MLL4 COMPASS-like complexes,10 but did not find homologs of PA1 and NCOA6 (Fig. 6B). Smed-ptip(RNAi) and Smed-utx(RNAi) displayed blastema growth defects and compromised photoreceptor development similar to Smed-trr-1(RNAi) (Fig. 8), suggesting that these three proteins may work together. The freckly blastema pigmentation in Smed-utx(RNAi) (Fig. 8) that was not seen with knockdown of any of the other putative complex members may be due to its participation in other processes. UTX in flies and humans is a H3K27 specific demethylase that functions in the interplay between Trithorax and Polycomb group proteins, coordinating removal of the repressive H3K27 mark with the addition of the activating H3K4 mark.64 The fact that Smed-utx likely does not directly participate in H3K4 methylation may explain why its knockdown phenotype is different than that of the other complex members.
A homolog of MLL5 regulates mitosis in planarians
MLL5 is the most divergent member of the SET1/MLL family in mammals, differing in sequence and mode of action. Unlike other members of the family, MLL5 does not participate in a COMPASS-like complex, but instead interacts with other proteins.65 We identified two potential MLL5 homologs in S. mediterranea (Fig. 1). While knockdown of Smed-mll5-1 did not lead to any overt external morphological phenotype, knockdown of Smed-mll5-2 caused impaired photoreceptor regeneration (Fig. 3) and an increase in the number of dividing stem cells (Fig. 4). This latter phenotype is consistent with reports that mammalian MLL5 is involved in cell cycle regulation and maintenance of quiescent hematopoietic stem cells. Both ectopic overexpression and siRNA knockdown of MLL5 in human cells lead to cell cycle arrest and growth defects,66,67 a result that has been explained by the finding that MLL5 acts at multiple points in the cell cycle with opposing effects.66 MLL5 was flagged in a screen for genes associated with quiescence68 and is needed to prevent ectopic expression of S-phase genes during G0.69 Furthermore, MLL5 knockout mice have impaired hematopoietic stem cell function, with reduced numbers and excessive cycling of HSCs and progenitors, suggesting a failure in maintenance of the quiescent stem cell pool.70 The increased number of mitotic stem cells in Smed-mll5-2(RNAi) suggests that MLL5 may play a conserved role in negatively regulating the cell cycle and maintaining stem cell quiescence in planarians.
Planarians as a model to examine epigenetic regulation of stem cells during regeneration
Studies previous to the work we have reported in this paper have begun to examine the role of epigenetic regulation of stem cells in planarians.23,38-41 Önal et al.23 found that chromatin proteins, including the gene we have named Smed-mll1/2 and several members of COMPASS (Smed-ash2, Smed-rbbp5, Smed-wdr5 and Smed-hcf1) are upregulated in the X1 stem cell population in S. mediterranea, and Rossi et al.41 observed a similar enrichment of chromatin remodelers on their list of Dugesia japonica stem cell genes. Our work, which is focused on the SET1/MLL family, also demonstrates that these genes are expressed in stem cells and play distinct roles in their regulation during regeneration. In future studies, it would be interesting to characterize other families of histone modifiers whose action is related to that of SET1/MLL. For example, members of the Polycomb group complex, which catalyzes repressive H3K27 methylation at many targets shared with SET1/MLL, have been shown to play important roles in planarian stem cells.23,40
Interestingly, flatworms appear to lack DNA methylation altogether (reviewed in ref. 71), and it is possible that modification of histones is the predominant epigenetic process in planarians. Given the experimental accessibility of planarians and the ability to isolate their stem cells or immediate progeny, there is great potential for further investigation aimed at identifying targets of histone methyltransferase complexes. Targets could be identified by chromatin immunoprecipitation combined with high-throughput sequencing or by using RNA-seq to assay changes in expression following RNAi knockdown of any of these family members. These studies will help us to gain insights into conserved epigenetic mechanisms regulating stem cells in tissue regeneration.
Materials and Methods
Planarians
Asexual Schmidtea mediterranea (CIW4) were maintained in 1X Montjuïc salts and fed pureed calf liver as previously described.72 Worms used for in situ hybridization were 1–2 mm in length, and those used for RNAi were 3–5 mm at the time of the first feeding. All animals were starved for one week prior to experiments.
Identification of SET1/MLL family and COMPASS complex proteins
Planarian proteins in the SET1/MLL family were identified by tblastn (e-value ≤ 1 × 10−4) against the S. mediterranea genome,73 EST database74 and high throughput sequencing transcriptomes22,26,75 using the SET domain portions of all known Homo sapiens [SET1A (O15047), SET1B (Q9UPS6), MLL1 (Q03164), MLL2 (Q9UMN6), MLL3 (Q8NEZ4), MLL4 (O14686), MLL5 (Q8IZD2)], Mus musculus [SET1B (Q8CFT2), MLL1 (P55200), MLL2 (O08550), MLL3 (Q8BRH4), MLL4 (Q6PDK2), MLL5 (Q3UG20)], Drosophila melanogaster [DSET1 (Q5LJZ2), TRX (P20659), TRR (Q8IRW8)] Caenorhabditis elegans [SET2 (Q18221), SET16 (G5EGI1)], Schizosaccharomyces pombe [Set1 (Q9Y7R4)], and Saccharomyces cerevisae [SET1 (P38827)] members of this family. Membership in the gene family was confirmed by reciprocal blastx of the predicted planarian protein against the NCBI non-redundant protein sequence database (nr) looking for members of this family as the top hits, and names were assigned based on homology from examination of the phylogenetic tree. Gene models for the planarian sequences were built based on bioinformatic predictions and available EST and transcriptome data. Protein domains were identified using PROSITE.76
Homologs of COMPASS and COMPASS-like complex members were identified by tblastn of the full-length proteins from H. sapiens, S. pombe, S. cerevisiae and/or D. melanogaster against the S. mediterranea databases described above. The GenBank accession numbers of the query proteins are: ASH2 (O60070, Q9UBL3, P43132), DPY30 (Q9C005, Q03323, O74861), RBBP5 (Q15291, P39706, O42858), WDR5 (P61964, P38123, O43017), CXXC1 (Q9P0U4, Q03012, O74508), WDR82 (Q6UXN9, P36104, O60137), HCF1 (P51610), Menin (O00255, Q7KXY9), PTIP (Q6ZW49, Q9VUB7), UTX (O15550, Q9VL07), PA1 (Q9BTK6, Q9VYZ8), NCOA6 (Q14686, A8DYV5). The planarian protein with the highest similarity (cut-off e-value < 1 × 10−4) matching the query protein as its top hit in reciprocal blast searches was assigned as the intended homolog.
Phylogenetic analysis
The sequence of the SET domain was isolated from each full-length predicted protein using PROSITE76 and aligned by ClustalW within MEGA576 using default settings. A phylogenetic tree was built in MEGA5 using the Neighbor-joining method and based on the Jones-Taylor-Thornton model with uniform mutation rates and pairwise deletion of gaps. The tree was tested by Bootstrap resampling with 1,000 replications and displayed rooted at the midpoint.
Cloning
Constructs available in a collection of sequenced cDNA clones74 were obtained from glycerol stocks. Other genes were directionally cloned into pJC53.277 using gene specific primers. Primer sequences and GenBank accession numbers for each clone are listed in Table S3.
In situ hybridization
DNA templates for Digoxigenin-labeled riboprobe synthesis were amplified by PCR with T3 and T7 primers and in vitro transcribed with T3 polymerase (Promega). Whole mount in situ hybridization was performed as previously described.78 After formaldehyde fixation, all subsequent steps, including anti-Dig AP antibody (Roche, 1:2,000) incubation and MABT washes, were performed in an Intavis InsituPro VS liquid handling robot. Signal was developed by incubation with NBT/BCIP substrate in AP buffer, and animals were post-fixed and placed in 50–80% glycerol for imaging. For irradiation experiments, worms were treated with 60 Gy γ-irradiation in a JL Shepherd Mark I Cesium-137 irradiator and fixed three days later. Control and irradiated samples were processed side-by-side and signals developed for the same length of time.
Immunostaining
To stain planarians with anti-phospho-Histone H3 (Ser 10) (1:1,000, D2C8, Cell Signaling), worms were treated with 2% HCl for 5 min. on ice, fixed with Carnoy’s solution (6:3:1 ethanol, chloroform, and acetic acid) for 2 h at 4°C, and then incubated in methanol for 1 h at 4°C. Further processing and development were performed as previously described.56 Staining with anti-acetylated tubulin (1:1,000, clone 6-11B-1, Sigma) was performed in the same manner except that the HCl treatment was shortened to 30 sec.
RNAi
Planarians were fed bacterially expressed dsRNA as previously described.79 Inserts in pBluescript II SK(+) were shuttled to pJC53.277 by ligation independent cloning,80 using LIC-BSinsF and R primers to amplify the inserts from pBluescript II SK(+) and LIC-pJC532vecF and R primers to amplify the vector (see Table S3 for primer sequences). dsRNA expression was induced from this vector in HT115(DE3) E. coli strain with IPTG. Bacterial pellets were mixed with liver puree, water, and red food coloring for feeding to planarians, and feedings were spaced 3–4 d apart. The number of feedings was adjusted for each gene as required to produce a phenotype. Bacterially expressed gfp dsRNA was used as a negative control. In experiments to assay regeneration ability, worms were amputated pre-pharyngeally one day after the final feeding. For simultaneous knockdown of two genes, bacterial pellets were prepared by mixing equal amounts of the two cultures to the same total volume as in single knockdown experiments. Each gene was also tested in combination with gfp to control for the effect of dilution. At least 10 animals were included in each knockdown group. The RNAi phenotypes observed and their penetrance are summarized in Table S2.
Imaging
Live animals and those stained by in situ hybridization were imaged using a Leica DFC290 camera mounted on a Leica M205C microscope. Image adjustments were made in Adobe Photoshop to normalize levels, color, and brightness; control and γ-irradiated worms were treated identically for all imaging and adjustments. Fluorescent samples were imaged with an Axiocam MRm camera mounted on a Zeiss SteREO Lumar.V12 stereomicroscope, or a Zeiss Axio Observer.Z1 inverted microscope using an ApoTome for optical sectioning.
Cell counting
Phospho-Histone H3 positive cells were counted by hand in ImageJ 4 software81 (n = 5–10 animals per group). Cell counts were divided by the area of the animal in mm2 to normalize for size.
Flow cytometric analyses of the stem cell population
Planarians (n = 6–10) were placed into “C” tubes (Miltenyi Biotec) and rinsed with calcium-magnesium-free media (CMF).82 6 mL CMF + 30 μg/mL Trypsin Inhibitor (Sigma) was added, tubes were placed on a GentleMACS tissue dissociator (Miltenyi Biotec), and program “A.01” (25 sec blending program) was run once. Samples were then incubated for 10 min on a nutator at 4°C. Fourteen mL cold CMF was added, and tubes were gently vortexed and then centrifuged at RT for 5 min at 300 × g. After aspiration, 6 mL CMF + 0.25% (w/v) Trypsin (Sigma) + 5 μg/mL DNaseI (Sigma) was added, and program “A.01” was run again on the GentleMACS. Samples were incubated for 10 min on a nutator at 4°C, followed by program “A.01” twice. Fourteen mL of CMF was added, and tubes were gently vortexed or inverted and centrifuged as before. Samples were aspirated, resuspended in 2 mL CMF + 0.5 μg/mL DNaseI, and filtered through a 50 μM nylon mesh. A portion of the cell suspension was incubated with 1:2,000 Nuclear-ID Red Cell Cycle Dye (Enzo Life Sciences) for 30 min. in the dark. Before acquisition, 2 μg/ml Propidium Iodide was added to a separate tube of cells to assess general viability at the time of acquisition. Flow cytometry data were acquired with a BD FACSCanto flow cytometer equipped with 488- and 633-nm lasers (BD Biosciences) and analyzed using FlowJo (Tree Star, Inc.). Samples were initially gated to exclude non-nucleated cells.
Supplementary Material
Acknowledgments
We would like to thank Jared Hoffman and Thomas Phan who helped with the initial characterization of SET domain genes; Jim Collins for the pJC53.2 vector and Phil Newmark for cDNA clones; and Bret Pearson for sharing RNA-seq data ahead of publication. FACS analysis was performed in the SDSU Flow Cytometry Core Facility. M.W.C. gratefully acknowledges support from the San Diego Chapter of the ARCS Foundation. This work was supported by National Institutes of Health IRACDA Postdoctoral Fellowship GM68524-08 to AH and California Institute for Regenerative Medicine Grant RN2-00940-1 to RMZ.
Glossary
Abbreviations:
- COMPASS
complex proteins associated with Set1
- dsRNA
double-stranded RNA
- H3K4me3
histone H3 lysine 4 trimethyl
- H3K27me3
histone H3 lysine 27 trimethyl
- MLL
mixed-lineage leukemia
- PH3
phospho-histone H3
- SET
Su(var)3-9, enhancer-of-zeste, Trithorax
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/epigenetics/article/23211
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/23211
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansari KI, Mandal SS. Mixed lineage leukemia: roles in gene expression, hormone signaling and mRNA processing. FEBS J. 2010;277:1790–804. doi: 10.1111/j.1742-4658.2010.07606.x. [DOI] [PubMed] [Google Scholar]
- 4.Nislow C, Ray E, Pillus L. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol Biol Cell. 1997;8:2421–36. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wenzel D, Palladino F, Jedrusik-Bode M. Epigenetics in C. elegans: facts and challenges. Genesis. 2011;49:647–61. doi: 10.1002/dvg.20762. [DOI] [PubMed] [Google Scholar]
- 6.Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, Washburn MP, et al. The COMPASS family of H3K4 methylases in Drosophila. Mol Cell Biol. 2011;31:4310–8. doi: 10.1128/MCB.06092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol. 2010;339:240–9. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, et al. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci U S A. 2001;98:12902–7. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem. 2005;280:41725–31. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- 10.Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282:20395–406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, et al. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28:7337–44. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–49. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ernst P, Vakoc CR. WRAD: enabler of the SET1-family of H3K4 methyltransferases. Brief Funct Genomics. 2012;11:217–26. doi: 10.1093/bfgp/els017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–33. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 15.Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25:661–72. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faber J, Armstrong SA. Mixed lineage leukemia translocations and a leukemia stem cell program. Cancer Res. 2007;67:8425–8. doi: 10.1158/0008-5472.CAN-07-0972. [DOI] [PubMed] [Google Scholar]
- 17.Cozzio A, Passegué E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–35. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elliott SA, Sánchez Alvarado A. The history and enduring contributions of planarians to the study of animal regeneration. Wiley Interdisciplinary Reviews: Developmental Biology. 2012 doi: 10.1002/wdev.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reddien PW. Constitutive gene expression and the specification of tissue identity in adult planarian biology. Trends Genet. 2011;27:277–85. doi: 10.1016/j.tig.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baguñà J. The planarian neoblast: the rambling history of its origin and some current black boxes. Int J Dev Biol. 2012;56:19–37. doi: 10.1387/ijdb.113463jb. [DOI] [PubMed] [Google Scholar]
- 21.Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 2011;332:811–6. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labbé RM, Irimia M, Currie KW, Lin A, Zhu SJ, Brown DDR, et al. A comparative transcriptomic analysis reveals conserved features of stem cell pluripotency in planarians and mammals. Stem Cells. 2012;30:1734–45. doi: 10.1002/stem.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Önal P, Grün D, Adamidi C, Rybak A, Solana J, Mastrobuoni G, et al. Gene expression of pluripotency determinants is conserved between mammalian and planarian stem cells. EMBO J. 2012;31:2755–69. doi: 10.1038/emboj.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Resch AM, Palakodeti D, Lu YC, Horowitz M, Graveley BR. Transcriptome analysis reveals strain-specific and conserved stemness genes in Schmidtea mediterranea. PLoS One. 2012;7:e34447. doi: 10.1371/journal.pone.0034447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qian C, Zhou MM. SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cell Mol Life Sci. 2006;63:2755–63. doi: 10.1007/s00018-006-6274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adamidi C, Wang Y, Gruen D, Mastrobuoni G, You X, Tolle D, et al. De novo assembly and validation of planaria transcriptome by massive parallel sequencing and shotgun proteomics. Genome Res. 2011;21:1193–200. doi: 10.1101/gr.113779.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glaser S, Schaft J, Lubitz S, Vintersten K, van der Hoeven F, Tufteland KR, et al. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development. 2006;133:1423–32. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- 28.Emerling BM, Bonifas J, Kratz CP, Donovan S, Taylor BR, Green ED, et al. MLL5, a homolog of Drosophila trithorax located within a segment of chromosome band 7q22 implicated in myeloid leukemia. Oncogene. 2002;21:4849–54. doi: 10.1038/sj.onc.1205615. [DOI] [PubMed] [Google Scholar]
- 29.Newmark PA, Reddien PW, Cebrià F, Sánchez Alvarado A. Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc Natl Acad Sci U S A. 2003;100(Suppl 1):11861–5. doi: 10.1073/pnas.1834205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rompolas P, Patel-King RS, King SM. Chapter 4 - Schmidtea mediterranea: A Model System for Analysis of Motile Cilia. In: Stephen MK, Gregory JP, eds. Methods in Cell Biology: Academic Press, 2009:81-98. [DOI] [PubMed] [Google Scholar]
- 31.Rink JC, Vu HT, Sánchez Alvarado A. The maintenance and regeneration of the planarian excretory system are regulated by EGFR signaling. Development. 2011;138:3769–80. doi: 10.1242/dev.066852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eisenhoffer GT, Kang H, Sánchez Alvarado A. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell. 2008;3:327–39. doi: 10.1016/j.stem.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayashi T, Asami M, Higuchi S, Shibata N, Agata K. Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev Growth Differ. 2006;48:371–80. doi: 10.1111/j.1440-169X.2006.00876.x. [DOI] [PubMed] [Google Scholar]
- 34.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, et al. modENCODE Consortium Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–97. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrero MJ, Izpisua Belmonte JC. Regenerating the epigenome. EMBO Rep. 2011;12:208–15. doi: 10.1038/embor.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katsuyama T, Paro R. Epigenetic reprogramming during tissue regeneration. FEBS Lett. 2011;585:1617–24. doi: 10.1016/j.febslet.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 38.Bonuccelli L, Rossi L, Lena A, Scarcelli V, Rainaldi G, Evangelista M, et al. An RbAp48-like gene regulates adult stem cells in planarians. J Cell Sci. 2010;123:690–8. doi: 10.1242/jcs.053900. [DOI] [PubMed] [Google Scholar]
- 39.Scimone ML, Meisel J, Reddien PW. The Mi-2-like Smed-CHD4 gene is required for stem cell differentiation in the planarian Schmidtea mediterranea. Development. 2010;137:1231–41. doi: 10.1242/dev.042051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner DE, Ho JJ, Reddien PW. Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell Stem Cell. 2012;10:299–311. doi: 10.1016/j.stem.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossi L, Salvetti A, Marincola FM, Lena A, Deri P, Mannini L, et al. Deciphering the molecular machinery of stem cells: a look at the neoblast gene expression profile. Genome Biol. 2007;8:R62. doi: 10.1186/gb-2007-8-4-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hallson G, Hollebakken RE, Li T, Syrzycka M, Kim I, Cotsworth S, et al. dSet1 is the main H3K4 di- and tri-methyltransferase throughout Drosophila development. Genetics. 2012;190:91–100. doi: 10.1534/genetics.111.135863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J. 2011;30:2817–28. doi: 10.1038/emboj.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao Y, Bedet C, Robert VJ, Simonet T, Dunkelbarger S, Rakotomalala C, et al. Caenorhabditis elegans chromatin-associated proteins SET-2 and ASH-2 are differentially required for histone H3 Lys 4 methylation in embryos and adult germ cells. Proc Natl Acad Sci U S A. 2011;108:8305–10. doi: 10.1073/pnas.1019290108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–72. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 46.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nature Structural &. Mol Biol. 2006;13:713–9. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 47.Steward MM, Lee J-S, O'Donovan A, Wyatt M, Bernstein BE, Shilatifard A. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nature Structural &. Mol Biol. 2006;13:852–4. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 48.Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, Roeder RG. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell. 2011;144:513–25. doi: 10.1016/j.cell.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel A, Dharmarajan V, Vought VE, Cosgrove MS. On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. J Biol Chem. 2009;284:24242–56. doi: 10.1074/jbc.M109.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dehé PM, Dichtl B, Schaft D, Roguev A, Pamblanco M, Lebrun R, et al. Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J Biol Chem. 2006;281:35404–12. doi: 10.1074/jbc.M603099200. [DOI] [PubMed] [Google Scholar]
- 51.Lee JH, Skalnik DG. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol. 2008;28:609–18. doi: 10.1128/MCB.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clouaire T, Webb S, Skene P, Illingworth R, Kerr A, Andrews R, et al. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev. 2012;26:1714–28. doi: 10.1101/gad.194209.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Narayanan A, Ruyechan WT, Kristie TM. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc Natl Acad Sci U S A. 2007;104:10835–40. doi: 10.1073/pnas.0704351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, et al. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–96. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 55.Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–97. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cowles MW, Hubert A, Zayas RMA. A Lissencephaly-1 homologue is essential for mitotic progression in the planarian Schmidtea mediterranea. Dev Dyn. 2012;241:901–10. doi: 10.1002/dvdy.23775. [DOI] [PubMed] [Google Scholar]
- 57.Lee JH, You J, Dobrota E, Skalnik DG. Identification and characterization of a novel human PP1 phosphatase complex. J Biol Chem. 2010;285:24466–76. doi: 10.1074/jbc.M110.109801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1:324–37. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saló E, Tauler J, Jimenez E, Ramón Bayascas J, Gonzalez-Linares J, Garcia-Fernàndez J, et al. Hox and ParaHox Genes in Flatworms: Characterization and Expression. Am Zool. 2001;41:652–63. doi: 10.1668/0003-1569(2001)041[0652:HAPGIF]2.0.CO;2. [DOI] [Google Scholar]
- 60.Bayascas JR, Castillo E, Muñoz-Mármol AM, Saló E. Planarian Hox genes: novel patterns of expression during regeneration. Development. 1997;124:141–8. doi: 10.1242/dev.124.1.141. [DOI] [PubMed] [Google Scholar]
- 61.Orii H, Kato K, Umesono Y, Sakurai T, Agata K, Watanabe K. The planarian HOM/HOX homeobox genes (Plox) expressed along the anteroposterior axis. Dev Biol. 1999;210:456–68. doi: 10.1006/dbio.1999.9275. [DOI] [PubMed] [Google Scholar]
- 62.Rink JC, Gurley KA, Elliott SA, Sánchez Alvarado A. Planarian Hh signaling regulates regeneration polarity and links Hh pathway evolution to cilia. Science. 2009;326:1406–10. doi: 10.1126/science.1178712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scimone ML, Srivastava M, Bell GW, Reddien PW. A regulatory program for excretory system regeneration in planarians. Development. 2011;138:4387–98. doi: 10.1242/dev.068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–4. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 65.Fujiki R, Chikanishi T, Hashiba W, Ito H, Takada I, Roeder RG, et al. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 2009;459:455–9. doi: 10.1038/nature07954. [DOI] [PubMed] [Google Scholar]
- 66.Cheng F, Liu J, Zhou SH, Wang XN, Chew JF, Deng LW. RNA interference against mixed lineage leukemia 5 resulted in cell cycle arrest. Int J Biochem Cell Biol. 2008;40:2472–81. doi: 10.1016/j.biocel.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 67.Deng LW, Chiu I, Strominger JL. MLL 5 protein forms intranuclear foci, and overexpression inhibits cell cycle progression. Proc Natl Acad Sci U S A. 2004;101:757–62. doi: 10.1073/pnas.2036345100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sambasivan R, Pavlath GK, Dhawan J. A gene-trap strategy identifies quiescence-induced genes in synchronized myoblasts. J Biosci. 2008;33:27–44. doi: 10.1007/s12038-008-0019-6. [DOI] [PubMed] [Google Scholar]
- 69.Sebastian S, Sreenivas P, Sambasivan R, Cheedipudi S, Kandalla P, Pavlath GK, et al. MLL5, a trithorax homolog, indirectly regulates H3K4 methylation, represses cyclin A2 expression, and promotes myogenic differentiation. Proc Natl Acad Sci U S A. 2009;106:4719–24. doi: 10.1073/pnas.0807136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Madan V, Madan B, Brykczynska U, Zilbermann F, Hogeveen K, Döhner K, et al. Impaired function of primitive hematopoietic cells in mice lacking the Mixed-Lineage-Leukemia homolog MLL5. Blood. 2009;113:1444–54. doi: 10.1182/blood-2008-02-142638. [DOI] [PubMed] [Google Scholar]
- 71.Regev A, Lamb MJ, Jablonka E. The Role of DNA Methylation in Invertebrates: Developmental Regulation or Genome Defense? Mol Biol Evol. 1998;15:880. doi: 10.1093/oxfordjournals.molbev.a025992. [DOI] [Google Scholar]
- 72.Cebrià F, Newmark PA. Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development. 2005;132:3691–703. doi: 10.1242/dev.01941. [DOI] [PubMed] [Google Scholar]
- 73.Robb SMC, Ross E, Sánchez Alvarado A. SmedGD: the Schmidtea mediterranea genome database. Nucleic Acids Res. 2008;36(Database issue):D599–606. doi: 10.1093/nar/gkm684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zayas RM, Hernández A, Habermann B, Wang Y, Stary JM, Newmark PA. The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: analysis of ESTs from the hermaphroditic strain. Proc Natl Acad Sci U S A. 2005;102:18491–6. doi: 10.1073/pnas.0509507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sandmann T, Vogg MC, Owlarn S, Boutros M, Bartscherer K. The head-regeneration transcriptome of the planarian Schmidtea mediterranea. Genome Biol. 2011;12:R76. doi: 10.1186/gb-2011-12-8-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34(Web Server issue):W362-5. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Collins JJ, 3rd, Hou X, Romanova EV, Lambrus BG, Miller CM, Saberi A, et al. Genome-wide analyses reveal a role for peptide hormones in planarian germline development. PLoS Biol. 2010;8:e1000509. doi: 10.1371/journal.pbio.1000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Sánchez Alvarado A. Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev Dyn. 2009;238:443–50. doi: 10.1002/dvdy.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gurley KA, Rink JC, Sánchez Alvarado A. β-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 2008;319:323–7. doi: 10.1126/science.1150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res. 1990;18:6069–74. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abramoff MD, Magalhães PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 82.Kang H, Sánchez Alvarado A. Flow cytometry methods for the study of cell-cycle parameters of planarian stem cells. Dev Dyn. 2009;238:1111–7. doi: 10.1002/dvdy.21928. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.