Abstract
Oncogenic mutations in PIK3CA, which encodes the phosphoinositide-3-kinase (PI3K) catalytic subunit p110α, occur in ∼25% of human breast cancers. In this study, we report the development of a knock-in mouse model for breast cancer where the endogenous Pik3ca allele was modified to allow tissue-specific conditional expression of a frequently found Pik3caH1047R (Pik3cae20H1047R) mutant allele. We found that activation of the latent Pik3caH1047R allele resulted in breast tumors with multiple histological types. Whole-exome analysis of the Pik3caH1047R-driven mammary tumors identified multiple mutations, including Trp53 mutations that appeared spontaneously during the development of adenocarinoma and spindle cell tumors. Further, we used this model to test the efficacy of GDC-0941, a PI3K inhibitor, in clinical development, and showed that the tumors respond to PI3K inhibition.
Keywords: Pik3ca, H1047R, knock-in, mammary gland, Trp53, exome sequencing
Introduction
The class-IA phosphoinositide-3-kinase (PI3K) catalytic subunits, p110α, p110β and p110δ, occur as an obligate heterodimer in complex with p85α-, p55α-, p50α-, p85β- or p55γ-regulatory subunits.1, 2, 3 In normal cells, PI3Ks are maintained in a basal inactive state and become active following growth factor stimulation and cell-surface-receptor engagement. Activated PI3Ks phosphorylate and convert phosphatidylinositol-4,5-bisphosphate (PIP2) to phosphatidylinositol-3,4,5-trisphosphate (PIP3). Elevated cellular PIP3 levels promote the activation of PI3K effectors, which in turn regulate a number of cellular processes, including cell growth, proliferation and survival.1, 2, 3
Frequent somatic mutations in PIK3CA, the gene that encodes p110α, have been reported in colorectal, breast and liver cancers.4, 5, 6 The majority of the reported p110α mutations occur in three hotspots—two (E542K and E545K) in the helical domain and one (H1047R) in the kinase domain.7, 8 These hotspot mutations result in an oncogenic p110α capable of constitutively activating downstream signaling.8 Further, the p110α mutants when expressed in cells can lead to cellular transformation and support tumor formation in xenograft models.4, 9, 10, 11, 12
Genetically engineered mouse models (GEMMs) of human cancer serve as an important resource for understanding tumor initiation and progression.13 GEMMs have also been used to test therapeutics and develop treatment strategies.14 Studies aimed at understanding oncogenic transformation by mutant Pik3ca have primarily used cell-based systems, xenograft models and conditional transgenic models.4, 10, 11, 12, 15, 16, 17, 18 Given the high frequency of PIK3CA mutations and ongoing efforts to develop small-molecule inhibitors, a knock-in mouse model that can express endogenous levels of mutant Pik3ca from its native promoter and genomic architecture would be a valuable tool to study tumor initiation, progression and treatment. Also, such a model would lend itself to identification of lesions that cooperate with mutant Pik3ca during tumor development. With this in mind, we generated a GEMM where we have modified the endogenous Pik3ca allele by placing a dormant copy of an oncogenic mutant Pik3ca exon 20, encoding H1047R, adjacent to the wild-type coding exon 20. Prior to activation of the dormant mutant allele, the engineered mice expressed the modified wild-type Pik3ca (Pik3cae20mwt) allele. However, upon Cre-mediated recombination, the mutant Pik3cae20H1047R allele was conditionally activated in the mouse mammary glands, leading to its expression and tumorigenesis.
Although next-generation sequencing has enabled basepair-level characterization of human tumors as they evolve and progress,19, 20 mouse models of cancer provide a defined experimentally tractable system that allows systematic sampling and characterization of tumors as they evolve. In an effort to characterize the tumors at the sequence level, we performed whole-exome capture and sequencing of tumors, and identified the emergence of spontaneous Trp53 mutations as a potential cooperating event in spindle cell tumor and adenocarcinoma formation. We further report additional somatic mutations and copy-number aberrations in breast tumors from this model. In addition to molecular characterization, we tested the ability of a PI3K small-molecule inhibitor for efficacy and show that the tumors respond to inhibitor treatment.
Results
Engineering conditionally activatable Pik3caH1047R mice
To study the role of Pik3ca mutations in tumor initiation, development and progression, we have engineered a mouse capable of conditionally expressing the mutant Pik3ca H1047R allele driven by its native promoter. The engineered mouse, Pik3cae20mwt, has a wild-type Pik3ca exon 20 that was modified to contain flanking loxP sites. The modified wild-type exon is followed by a transcriptional stop cassette and a copy of Pik3ca exon 20 encoding an H1047R mutation (Figures 1a–d). A targeting vector (Figure 1b) was used to modify the endogenous Pik3ca locus in mouse embryonic stem (ES) cells. Two independent ES cell clones containing the appropriate modification, identified by Southern blotting (Figures 1f and g), were used to generate chimeric mice that showed germline transmission of the Pik3cae20mwt allele. Intercrosses involving heterozygous Pik3cae20mwt/+ mice resulted in progenies with the appropriate genotypes at expected Mendelian ratios (Supplementary Table 1). Unlike the embryonic lethality observed in Pik3ca−/−-null mice,21 prior to Cre-mediated recombination, we found that the homozygous Pik3cae20mwt/e20mwt animals were born at the expected Mendelian frequency (Supplementary Table 1), indicating that the modified Pik3cae20mwt functioned analogous to the wild-type Pik3ca allele.
Figure 1.
Generation of Pik3cae20H1047R conditionally activatable knock-in allele. (a–d) Genomic locus encoding Pik3ca locus (a); targeting construct (b); targeted allele (c); targeted locus in the ES after removal of the neomycin cassette (d); and Pik3cae20H1047R allele following Cre-mediate activation (e) are shown. (f, g) Southern blotting using a 5′ probe (f) and a 3′ probe (g) was used to identify the appropriately targeted knock-in (ki) and wild-type (wt) allele. (h–i) Sanger sequencing of the cDNA obtained from Pik3cae20H1047R/+ mammary gland (h) and kidney (i) following Cre-mediated activation confirms the expression of the Pik3cae20H1047R allele in the mammary gland.
Mammary gland-specific expression of Pik3cae20H1047R allele
As PIK3CA is mutated in over 25% of human breast cancers9 we tested the role of Pik3caH1047R in breast tumorigenesis by breeding the Pik3cae20mwt mouse to an MMTV-Cre transgenic mouse.22, 23 The MMTV-Cre strain expresses P1 Cre recombinase under the control of a mammary gland-permissive MMTV-LTR promoter, allowing recombination and expression of Pik3cae20H1047R. We confirmed the expression of the Pik3cae20H1047R mutant allele and the wild-type Pik3ca allele by sequencing the cDNA corresponding to the Pik3ca mRNA extracted from the mammary glands of the Pik3cae20H1047R/+ mouse (Figure 1h). In contrast, RNA extracted from kidney showed the expression of only wild-type Pik3ca from the Pik3cae20mwt allele (Figure 1i), confirming mammary-specific expression of the mutant Pik3cae20H1047R allele.
Expression of Pik3cae20H1047R leads to enhanced mammary branch morphogenesis
Previous studies have shown that expression of oncogenes or loss of tumor suppressors, such as Pten, affects branching and morphogenesis of mouse mammary glands.17, 24, 25 Hence, we studied the effect of expression of Pik3cae20H1047R on mammary gland development using whole-mount staining. At 12 weeks, Pik3cae20H1047R/+ mutant mammary glands showed precocious lobulo-alveolar development and hyper-branched ductal structures compared to control mammary glands (Supplementary Figures 1a–d). We found that there was a ∼2-fold increase in ductal branch points in mutant animals (n=6) as compared with the control group (MMTV-Cre (n=3), P=0.003; Pik3cae20mwt/+ (n=4), P=0.001; Supplementary Figure 1e). By 50 weeks, the Pik3cae20H1047R/+ mutant mammary glands showed a feathery, hyper-branched morphology compared with the control mammary glands (Supplementary Figures 1f and g). Further, histological sections of the Pik3cae20H1047R/+ mutant mammary glands showed evidence of tumor nodules at 50 weeks of age (Supplementary Figures 1h and i). This is similar to the mammary branching morphogenesis defects reported in previous studies involving Pten conditional null mice and other mammary-specific transgenic models with PI3K pathway activation.17, 24
Pik3cae20H1047R expression promotes mammary tumorigenesis
Female Pik3cae20H1047R/+, Pik3cae20mwt/+ and MMTV-Cre mice were aged to see if they develop mammary tumors. We found that the Pik3cae20H1047R/+ animals developed mammary gland tumors (Figures 2a–c), with a median tumor-free survival of 478.5 days. The majority of the animals developed tumors in the thoracic mammary glands (#1, 2, 3, 6 7 and 8), whereas some developed tumors in the inguino-abdominal mammary glands (#4, 5, 9 and 10), and a few developed tumors in both thoracic and inguino-abdominal mammary glands. In contrast to Pik3cae20H1047R/+, the control Pik3cae20mwt/+ and MMTV-Cre animals remained tumor-free for the duration of the study. The mammary gland tumor phenotype with a similar latency was also observed in Pik3cae20H1047R/+ mice derived from a second independent targeted ES clone (line 2; Figure 2c), further confirming the specificity of the Pik3cae20H1047R expression-driven tumor phenotype.
Figure 2.
Pik3cae20H1047R/+ mice develop mammary tumors. (a, b) Mouse bearing tumor in the abdominal mammary (a) and thoracic mammary (b) glands. The white arrowheads indicate the location of the tumors. (c) Kaplan–Meier plot depicting tumor-free survival of two independent Pik3cae20H1047R lines following MMTV-Cre-mediated activation of the latent Pik3cae20mwt allele. Animals bearing mammary tumors (86.4% of line 1 and 84.2% of line 2 Pik3cae20H1047R/+ female mice had developed tumors) at the end point are shown in the plot. Two control groups, MMTV-Cre mice and Pik3cae20mwt, were tumor-free for the duration of the study. (d–g) Histology of Pik3cae20H1047R/+ mammary tumors. Analysis of H&E-stained tumors sections show the presence of multiple histological types, which include fibroadenoma (d), adenosquamous carcinoma (e), adenocarcinoma (f) and spindle cell tumor (g). (h) Western blot of Pik3cae20H1047R/+ mammary tumors representative of the histological types shows elevated pAKT levels (lanes 3–10) and pS6 (lanes 3–10) in all tumor types compared with control mammary glands (lanes 1–2). Tumors 9 and 10 were derived from passaging a primary spindle tumor in the SCID mice mammary fat pad.
MMTV-LTR promoter drives gene expression in both virgin and lactating mammary glands.22 Given this, we compared tumor incidence in nulliparous and multiparous mice, and found that latency was significantly shortened from 492 to 465 days (P=0.0002) in multiparous Pik3cae20H1047R/+ mice as compared with nulliparous Pik3cae20H1047R/+ animals (Supplementary Figure 2), suggesting that pregnancy-related mammary gland changes potentially contribute to accelerated tumorigenesis. Alternatively, during pregnancy, an increase in progenitor cells undergoing Cre-mediated Pik3ca activation may contribute to the accelerated tumorigenesis observed.26
Although the MMTV-LTR promoter directs Cre expression starting at day 6 after birth in mammary epithelial cells primarily, it is also known to drive expression in salivary glands and male seminal vesicles.23 In performing histological analysis of salivary glands from mice bearing mammary tumors and seminal vesicles from males aged between 35 and 41 weeks, we found no evidence of tumors (Supplementary Figures 3a and b). However, morphologically the seminal vesicles were larger in Pik3cae20H1047R/+ mice as compared with the control animals. Histological analysis of seminal vesicle cross-sections indicated an increase in seminal vesicle fluid secretion leading to ductal distention and enlargement (Supplementary Figures 3c and d). However, this did not affect the breeding and fertility of Pik3cae20H1047R/+ males.
Histology and signaling activation in Pik3cae20H1047R-driven tumors
We performed histological analysis of Pik3cae20H1047R/+ tumors to understand its pathological features and found that the majority of the tumors showed features consistent with fibroadenoma (76.9% (20/26)) (Figure 2d), whereas a few showed histopathological features of adenocarcinoma (15.4% (4/26)) (Figures 2e and f) or spindle cell neoplasia (7.7% (2/26)) (Figure 2g).
To further characterize tumors, we tested the PI3K pathway activation status of the histologically distinct tumor types. In all tumors types, we observed elevated pAKT and pS6-kinase (Figure 2h), both downstream from PI3K, indicative of constitutive activation of the pathway in these tumors.
In an effort to further understand the mammary tumor types observed, we studied the expression of basal cytokeratin 5 (CK5), luminal cytokeratin 18 (CK18), estrogen receptor (ER), progesterone receptor (PR) and a mesenchymal marker, vimentin, in them (Figure 3). While we found fibroadenomas and adenocarcinomas to be CK5+, CK18+, ER+ and PR+, the spindle cell tumors were ER−/PR− (Figure 3o) and CK5− (data not shown). Interestingly, this is reminiscent of the ER− and PR− status reported for human mammary spindle cell tumors.27, 28 In addition, the spindle cell tumors were CK18+/vimentin+ (Figures 3n and p), indicating that the cells are undergoing epithelial–mesenchymal transition (EMT).
Figure 3.
Immunofluorescence staining of Pik3cae20H1047R mammary tumors. (a–p) Serial sections of a fibroadenoma (a–d), two adenocarcinomas (e–h and i–l) and a spindle cell tumor (m–p) stained with H&E (a, e, i, m), anti-CK18 and anti-CK5 (b, f, j), anti-ERα (c, g, k), anti-ERα and anti-PR (o), anti-PR (d, h, l), and anti-CK18 and anti-vimentin (n, p) antibodies. The section in panel p was obtained from a third-passage spindle cell tumor explant.
Genomic analysis of Pik3cae20H1047R-driven mammary tumors
Prior to performing genomic analysis of Pik3caH1047R mammary tumors, given the long latency for tumor formation, we first tested whether tumor explants derived from mammary tumors would grow as xenografts in SCID mice. The majority of fibroadenomas, adenocarcinomas and spindle cell tumors that we tested formed tumors. Interestingly, the histology of some xenograft tumors derived from fibroadenoma explants showed features of adenocarcinoma, indicating that fibroadenomas are likely benign and that only a subset of cells with tumorigenic potential within the original explants contributed to the development of tumors upon passage (Supplementary Figures 4a and b). However, the spindle cell tumor explants maintained their histological features during passage (Supplementary Figures 4c and d).
We performed microarray-based gene expression analyses of different mammary tumor subtypes and control mammary glands to further understand their molecular characteristics. Hierarchical clustering of expression data using markers for epithelial cells, stem cells and EMT29, 30 showed that primary fibroadenomas, adenocarcinomas and tumors derived from passaging the explants from these tumor types clustered together, indicating a common origin and a shared set of gene expression changes (Figure 4a). In contrast to the spindle cell type, both primary and passaged tumors, clustered together showing features of EMT, suggests a distinct cellular origin or is reflective of a distinct set of cooperating mutations in this tumor type (Figure 4a).
Figure 4.
Expression profile and genomic aberrations in Pik3cae20H1047R-driven mammary tumors. (a) Heatmap derived by hierarchical clustering of the histologically distinct mammary tumor types using the expression level for the set of genes indicated. Mammary tumors of different histological types show distinct expression profiles. Spindle cell tumors are Claudin-low and show elevated expression of EMT genes including Snail, Twist and Zeb1/2. Samples shown are spindle cell tumors (1–4; 2–4 are serially passaged tumors derived from tumor 1), control mammary gland (Mg) (5–10), fibroadenoma (11–15) and adenocarinoma (16–20). Tumor samples 17 and 18 were derived by serial passage of tumor 16. Similarly, tumor sample 19 was derived from passaging tumor 11. Samples 5, 7 and 8 were mammary glands from control groups. Samples 6, 9 and 10 were matched mammary glands from animals bearing tumors 12, 13 and 14, respectively. (b) Spontaneous Trp53 mutations in adenocarcinoma and spindle cell tumors. The genomic region, the Trp53 exons, sequencing reads (black lines) from the region of interest with mutation (red dots) are shown. The Sanger sequencing trace file depicting the A135V mutation is also shown. The three Trp53 mutations identified mapped on to its domain architecture are shown. PR, proline-rich domain; Reg, carboxy-terminal regulatory domain; TA, transactivation domain; Tet, tetramerization domain; DBD, DNA-binding domain.
Whole-exome capture and sequencing have been applied in identifying somatic mutations in the coding regions of genomes.31 In order to understand the emergence of secondary mutations in the various mammary tumor types observed in our model, we performed whole-exome capture and sequencing of a select set of samples (Supplementary Table 2). Besides the presence of the engineered Pik3ca H1047R mutation, the tumors also contained several additional somatic changes (Supplementary Table 3 and Supplementary Figure 5). We found that the fibroadenomas had a low level of somatic mutations (between 2 and 13), followed by adenocarcinomas (between 4 and 61 mutations) and spindle cell tumors (between 44 and 88 mutations). Notably, we found the Trp53 I192N mutation in an adenocarcinoma and an R245H mutation in a spindle cell tumor. The R245H mutation is equivalent of R248H TP53 hotspot mutation found in human cancers.32 Additionally, to understand the prevalence of the Trp53 mutation in the oncogenic Pik3ca-driven mouse tumors, we sequenced the coding exons of Trp53 in an independent spindle cell tumor and identified an A135V mutation (Figure 4b, and Supplementary Tables 3 and 5). All Trp53 mutations identified in this study occur in the DNA-binding domain and are known to impair Trp53 function and have been reported in human cancers.32, 33, 34 Further, the Trp53 mutation and the majority of the mutations identified in the primary spindle cell tumor were also found in the explant-derived tumors following passage, indicating that the primary spindle tumor was likely genetically less heterogeneous (Supplementary Figure 5). As observed in the spindle cell tumors, the majority of the mutations present in the primary adenocarcinoma were also present in the tumors derived following transplantation and passaging (Supplementary Figure 5). Analysis of the effect of protein-altering somatic changes using SIFT35 and/or PolyPhen36 identified 39 mutations, including those in Plk1, Tssk2 and Smg1 kinases, to have a functional effect suggesting that some of these may act as drivers (Supplementary Table 3 and Supplementary Figure 5). Interestingly, SMG1 mutations have been reported previously in human breast cancer.37
In addition to whole-exome mutational analysis, we profiled the tumors on CGH arrays to understand chromosomal copy-number alteration. We found that both fibroadenomas and adenocarcinomas showed few regions of copy-number aberrations (Supplementary Table 4). In contrast to fibroadenomas and adenocarcinomas, we found several regions with copy-number alterations in spindle cell tumor samples when compared with normal mammary glands (Supplementary Table 4 and Supplementary Figures 6s and 7a). Spindle cell tumors are also characterized by two regions of chromosomal loss that fall within chromosomes 11 and 12, both of which contain several coding genes. In particular, deletion in chromosome 11 resulted in loss of Nf1 (Supplementary Figure 7) and is consistent with its downregulated expression (log2 ratio of −2.09; P=8.338e−05; Figure 4a), suggesting that the Ras–MAPK pathway might also be engaged in these tumors.
Pik3cae20H1047R/+ mammary tumors respond to PI3K inhibitor
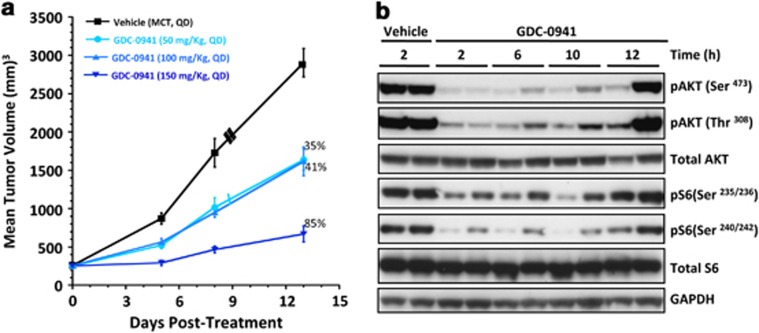
Engineered mouse models provide an ideal platform for candidate drug testing in a preclinical setting.14 As the mice in this study develop mammary tumors after a long latency, we used our explant-derived tumor model to test its utility in assessing drug efficacy. Explants derived from spindle cell tumors with Trp53 mutation were implanted subcutaneously near the mammary fat pad and treated with a PI3K inhibitor, GDC-0941,38, 39 once the tumors had reached a volume of 200–300 mm3. Tumor-bearing mice were treated in four groups of nine animals each with 50, 100 and 150 mg/kg GDC-0941 or a vehicle. We found that animals treated with GDC-0941 showed the highest tumor growth inhibition (TGI) of 85% at 150 mg/kg, although TGI was also observed at the other tested doses compared with vehicle treatment (Figure 5a), indicating that these tumors were still dependent on the mutant Pik3ca signaling for their growth. Further, log-rank tests of these data regarding time to progression showed that all the drug treatment groups showed a statistically significant difference when compared with the vehicle-treated animals at all the doses tested (P<0.0001 at 150 mg/kg; Supplementary Figure 8). Consistent with the TGI observed, tumors from mice treated with GDC-0941 showed a significant decrease in pAKT and pS6 at least up to 10 h following treatment (Figure 5b), confirming the contribution of PI3K inhibition in tumor growth inhibition.
Figure 5.
In vivo antitumor activity of PI3K inhibitor. (a) Pik3ca e20H1047R mammary tumor explants grown as xenografts respond to treatment with GDC-0941. 35, 41 and 85% on the graph represent %TGI with respect to control (n=9) for each of the treatment dose (n=9 for each dose). (b) Tumor from animals treated with GDC-0941 show evidence of PI3K pathway inhibition.
Discussion
GEMMs of cancer are invaluable for understanding tumor initiation, progression and therapy evaluation.40 In addition, application of next-generation sequencing technologies to these models can provide valuable information on the molecular aberrations that arise during the evolution of tumors. Here we describe a conditionally activatable Pik3caH1047R mouse model of breast cancer and show that these mice develop mammary tumors of multiple histological types, including fibroadenomas, adenocarcinomas and spindle cell neoplasia. We found that the fibroadenomas and adenocarcinomas were positive for hormone receptors and cytokeratins (CK5+/CK18+), indicating they are epithelial and myoepithelial in origin. By contrast, the spindle cell tumors, whereas being positive for cytokeratin 18, were distinct in that they were ER/PR-negative but vimentin-positive. This is reminiscent of human mammary spindle cell tumors.27, 28
PIK3CA mutations are observed in multiple human breast cancer subtypes, including of hormone receptor-positive (HR+), Her2+ triple-negative (TN) and metaplastic types.41 This is consistent with the multiple histological types observed in our breast cancer model and other published transgenic Pik3caH1047R breast tumor models.16, 17, 18 However, the latency of breast tumor development in our model is longer compared with these studies where the Pik3caH1047R mutant cDNA was expressed under the control of either an ROSA26 locus promoter (median survival ∼140 days in line A; >500 days in line NLST)16 or a chicken β-actin promoter (average latency 214±22.6 days)17, 24, 25 following activation of its expression using MMTV-Cre or induction of expression from a tetracycline-inducible promoter (average latency 210 days).16, 17, 18 This likely reflects the fact that our knock-in model expresses the mutant Pik3ca allele from its endogenous promoter present in its native genomic configuration and hence more closely mimics the PIK3CAH1047R-driven tumorigenesis observed in human tumors.
The advent of next-generation sequencing technology has allowed comprehensive characterization of cancers at the sequence level.42 In applying next-generation sequencing technology, we have characterized tumors derived from our mouse model to understand additional hits required for breast tumor development following expression of the Pik3caH1047R mutant allele. Among several candidate secondary hits that may cooperate with Pik3caH1047R, we report the occurrence of Trp53 mutations in two spindle cell tumors and one adenocarcinoma sample. All three Trp53 mutations, R245H, A135V and I192N (equivalent to human TP53 mutations R248H, A138V and I195N), observed in this study occur in human cancers.32, 33, 34 TP53 and PIK3CA mutations co-occur in human breast cancer.43 Recently, loss of Trp53 has been shown to accelerate tumorigenesis in an oncogenic Pik3ca transgenic mouse breast tumor model.16 The spontaneous emergence of Trp53 mutations in our tumor model mimics the cooperative TP53 lesions observed in human breast cancer. Hence, our mouse model will be ideal to study therapeutic intervention strategies for Pik3ca-driven mammary tumors. In line with this, we demonstrate the utility of our model in testing the efficacy of GDC-0941, a PI3K inhibitor, in a subset of mammary tumor with EMT features. A previous report studied the efficacy of NVP-BEZ235, a dual pan-PI3K/mTOR inhibitor, in a Pik3caH1047R transgenic lung tumor model.15 However, given that PIK3CA mutations are more frequent in human breast cancers, our model can serve as a genetically defined system to test the efficacy of PI3K inhibitors in breast cancer and potentially help to develop treatment regimens for clinical drug testing. Also, the model can be used to understand the development of resistance to targeted therapeutics. In this context, it remains to be seen whether the c-Myc-mediated mechanism of resistance, recently reported in a transgenic oncogenic Pik3ca mammary tumor model, has a role in our model system.16, 17, 18
Besides its utility in studying mammary tumorigenesis, this model can be used to study other cancers where PIK3CA mutations have been observed, by activating the mutant Pik3ca allele using an appropriate tissue-specific Cre line. In conjunction with next-generation sequencing technologies, our model can be used to discover and understand additional driver mutations that cooperate with mutant Pik3ca in other tumor types. Given that PI3K pathway activation has been implicated in resistance to therapy,44 the Pik3caH1047R mice can also be used to model drug resistance and study therapeutic strategies to overcome resistance.
Materials and methods
Generation of conditionally activatable Pik3caH1047R mice
We used a targeting vector template (Supplementary Figure 9) to assemble the final targeting vector. Using mouse genomic DNA from C57B/6N ES cells as template, a 2.045-kb fragment encompassing Pik3ca exons 18 and 19 (left arm), a 588-bp region containing Pik3ca wild-type exon 20 (mid-arm), and a 3.137-kb genomic DNA containing exon 20 and an additional 3′ intronic sequence (right arm) were PCR-amplified and used for constructing the vector. Prior to cloning into the template targeting vector, codon 1047 encoded by exon 20 in the left arm was mutated to code for histidine (R1047H) using standard mutagenesis techniques. In cloning the mid-arm into the targeting template vector, the wild-type exon 20 region was modified by addition of a human growth hormone (hgh) polyA signal present in the vector. A neomycin (neo) expression cassette and a 4 × transcriptional stop cassette45 were part of the vector used to assemble the targeting construct. In the final configuration, the neomycin cassette was flanked by ‘frt' sites to facilitate its excision following modification of the ES cells. Also, the modified wild-type exon was flanked by ‘loxP' sites to facilitate its removal, upon which the mutant exon 20 can be activated (Figure 1). Accuracy of the targeting vector was verified by Sanger sequencing. The targeting construct was electroporated into C57B/6N ES cells and selected for neomycin resistance. Appropriately targeted ES clones were identified by 5′ and 3′ Southern blotting. The 5′ probe recognized a 6.6-kb wild-type fragment and a 9.1-kb fragment in the targeted ES cell (Figures 1c and f). Similarly, the 3′ probe recognized a 6.9-kb wild-type fragment and a 4.8-kb fragment in the ES cells where the Pik3ca allele was modified (Figures 1c and g). Following removal of the neo cassette and confirmation of the architecture of the modified genomic region encoding Pik3ca by PCR and sequencing, the ES clones were injected into blastocysts to generate chimeric mice. A 290-bp PCR product, as opposed to a 201-bp fragment in wild-type mice, generated using primers 20F (AGCCAGCAGACAATAATTCTTAGCACA) and 20R (CAGTGCTTGAACATTGGAGGTCAGT) was used to follow germline transmission and also genotype-modified, Pik3ca allele-bearing mice. To activate the latent Pik3caH1047R mutant allele in mammary glands, we bred this mouse with an MMTV-Cre line (Jackson Labs, Bar Harbor, ME, USA; # 003551; line A). Mice that showed any body condition score<2,46 a hunched posture and >20% loss of body weight were killed as per the Institutional Animal Care and Use Committee (IACUC) guidelines. Our endpoint study included mice with tumors that reached ∼2500 mm3 and also those with palpable tumors that were killed in accordance with IACUC recommendations. All mice were maintained in our animal facility as per the IACUC guidelines.
Histological, immunohistochemical and whole-mount analysis
Five-micrometer, formalin-fixed, paraffin-embedded specimens was used for routine hematoxylin and eosin (H&E) staining and histology evaluation. For whole-mount analysis, the mammary glands were dissected, spread and fixed with Carnoy's fixative for 2–4 h. Tissues were hydrated and stained in Carmine alum overnight.
Immunofluorescence staining was performed using 10-μm sections Tissue-Tek OCT (Sakura Finetek, Torrance, CA, USA)-embedded frozen samples. The sectioned samples were fixed in 4% paraformaldehyde for 10 min and then blocked for 30 min with PBT (PBS with 0.1% Triton) containing 1% bovine serum albumin. The blocked sections were then stained with an appropriate primary antibody diluted in PBT with 0.1% bovine serum albumin overnight at 4 °C in a humidified chamber. The slides were washed three times in PBT and then incubated with appropriate secondary antibodies for 60 min at room temperature in a humidified chamber. Unbound secondary antibody was removed by washing with PBT. The Prolong Gold anti-fade reagent (Molecular Probes, Eugene, OR, USA) was used to mount the slides. Primary antibodies against cytokeratin 5 (ab53121), cytokeratin 18 (ab668), PR (ab2764) and vimentin (ab45939) were obtained from Abcam (Cambridge, MA, USA). The ERα (Ab-21) was purchased from Thermo Fisher Scientific (Waltham, MA, USA). Appropriate secondary antibodies conjugated with Alexa 488 or 647 (Molecular Probes) were used for detecting the bound primary antibody.
Western blot analysis
Frozen tumors were weighed and lysed with a pestle PP (Scienceware, Pequannock, NJ, USA) in cell extraction buffer (Invitrogen, Carlsbad, CA, USA) supplemented with protease inhibitors (Roche Applied Science, Indianapolis, IN, USA) 1 mM phenylmethysulfonyl fluoride, and phosphatase inhibitor cocktails 1 and 2 (Sigma, St Louis, MO, USA). Protein concentrations were determined using the Pierce BCA Protein Assay kit (Rockford, IL, USA). For western blots, equal amounts of protein were separated by electrophoresis using NuPage Bis–Tris 4–12% gradient gels (Invitrogen) and then transferred onto nitrocellulose pore membranes using the iBlot system (Invitrogen). The primary antibodies, pAkt (Ser473), pAkt (Thr308), total Akt, pS6 (Ser235/236), pS6 (Ser240/242) and total S6, were obtained from Cell Signaling Technology (Danvers, MA, USA). Anti-actin antibody was purchased from Sigma-Aldrich. Chemiluminescence western blotting detection (Amersham Biosciences, Pittsburgh, PA, USA) coupled with appropriate secondary, horseradish peroxidase-conjugated secondary IgG antibodies was used to detect the primary, antibody-bound proteins.
DNA/RNA preparation
DNA from tumors and normal tissues was prepared using the QiagenAllPrep DNA/RNA kit (Qiagen, Valencia, CA, USA). RNA samples were prepared using the RNeasy Mini kit (Qiagen).
Expression analysis
The quantity and quality of total RNA samples were determined, respectively, using an ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). A Cy dye-labeled cRNA preparation and array hybridization were prepared as per the manufacturer's (Agilent Technologies) instructions. Briefly, total RNA sample was converted to double-stranded cDNA and then to Cy dye-labeled cRNA using an Agilent Quick Amp Labeling kit. The labeled cRNA was purified using an RNeasy mini kit (Qiagen). cRNA yield and Cy dye incorporation were determined using the ND-1000 spectrophotometer (Thermo Scientific). A 750-ng weight of the labeled cRNA was fragmented and hybridized to Agilent Whole Mouse Genome 4 × 44 K arrays. All samples were labeled with Cy5 dye and hybridized against a Cy3 dye-labeled universal mouse reference (Stratagene, La Jolla, CA, USA). Samples were hybridized for 17 h at 65 °C with constant agitation on a Pro Hybridization station HS 4800 (Tecan US, Durham, NC, USA). Following hybridization, the arrays were washed, dried and scanned using an Agilent scanner. The Agilent Feature Extraction software 10.7 was used to analyze the acquired array images. The data were processed using methods implemented in the limma R package.47 First, data were background-corrected, after which a within-array loess fit normalization and a between-array quintile normalization were applied.48 Differentially expressed gene signatures were obtained using limma's linear model fit and probes with a false discovery rate-adjusted P-value of 0.05 or lower, and a log2 fold change of at least 2, were deemed differentially expressed. Hierarchical clustering of samples and EMT-related genes was computed on the mean centered gene expression profiles using the Euclidian distance metric and complete linkage clustering.
CGH arrays
Mammary tumor samples from Pik3cae20H1047R/+ mice were profiled on Agilent CGH arrays according to the manufacturer's protocol (Agilent Technologies). Briefly, 500 ng DNA of both tumor and a reference sample (C57BL/6J mouse; The Jackson Laboratory, ME, USA) were digested with AluI and RsaI (Promega, Madison, WI, USA) and subsequently purified using the QIAprep Spin Miniprep kit (Qiagen). Digested samples were labeled with Cy5-dUTP (test sample) or Cy3-UTP (reference sample) using the Genomic DNA Labeling Kit Plus (Agilent Technologies). Test samples were pooled with the reference and subsequently purified using MicroCon YM-30 (Millipore, Billerica, MA, USA). Labeled probes were mixed with Cot-1 DNA (Invitrogen), 10 × blocking agent and 2 × Hi-RPM hybridization buffer (Agilent Technologies), and hybridized to an Agilent 244K Mouse Genome CGH microarray. The samples were hybridized for 24 h at 67 °C with constant agitation using a Pro Hybridization station HS 4800 (Tecan US). The arrays were washed, dried and scanned using the Agilent scanner according to the manufacturer's protocol (Agilent Technologies). Individual log2 ratios of background-subtracted signal intensities were obtained from the Agilent Feature Extraction software version 10.7. The log2 ratios were centered to a median of zero and the resulting log2 ratio values for each probe were segmented using GLAD.49 All genes within the genomic bounds of a given GLAD-derived segment were assigned the mean copy-number value of the probes within that segment. Copy-number values ⩾0.3 log2 ratio units represented gain and values <=−0.3 log2 ratio units represented loss.
Exome capture and sequencing
Targeted sequence capture was performed using 3 μg of genomic DNA and the SureSelectXT Mouse All Exon kit according to the manufacturer's protocol (Agilent Technologies). The Mouse All Exon kit is designed to capture 49.6 Mb of a targeted region and covers 221 784 exons within 24 306 genes. The pre-capture library was amplified using four PCR cycles, whereas 12 PCR cycles were used in post-capture amplification. Fragment size distribution of post-capture amplified libraries was determined using Bioanalyzer 2100 using a DNA high-sensitivity chip (Agilent Technologies). Concentration of the libraries was measured by the Kapa library quantification kit (Kapa Biosystems, Woburn, MA, USA). Each library was sequenced on HiSeq 2000 to generate 2 × 75 bp reads per the manufacturer's instructions (Illumina, San Diego, CA, USA). The sequencing reads were mapped to the UCSC mouse genome (NCBI37/mm9) using the BWA software50 set to default parameters. Local realignment, duplicate marking and raw variant calling were performed as described previously.51 Single-nucleotide polymorphisms represented in dbSNP Build 13152 were used to remove known germline variations. In addition, variants that were present in both tumor and normal samples were removed as germline variations. The remaining variations present in the tumor sample, but absent in the matched normal, were predicted to be somatic. The predicted somatic variations were additionally filtered to include only positions with a minimum of 10 × coverage in both the tumor and matched normal, as well as an observed variant allele frequency of <1% in the matched normal. The effect of protein somatic mutation on gene function was predicted using SIFT35 and PolyPhen36 (Supplementary Table 3).
Sanger sequencing
Spindle cell tumor from animal #113 (Supplementary Table 2) was analyzed for Trp53 mutations using DNA derived from formalin-fixed, paraffin-embedded material. We used a pair of nested primers (outer and inner) to PCR-amplify the Trp53 exons (Supplementary Table 5). The inner primers were m13-tagged to facilitate sequencing. The primers used for amplification are provided in Supplementary Table 5. PCR was performed using Platinum Taq kit (Invitrogen) as per the manufacturer's protocols. The following parameters were used for PCR amplification: 94 °C for 2 min, followed 30 cycles of denaturation at 94 °C for 30 s, annealing at 56 °C for 30 s and extension at 72 °C for 1 min. Samples, prior to analysis, were held at 4 °C after a final extension at 72 °C for 8 min. PCR products were purified using the ExoSAP-IT kit (Affymetrix, Santa Clara, CA, USA) and sequenced using ABI3730xl per the manufacturer's instructions (Life Technologies, Foster City, CA, USA). Sequencing reads were analyzed using Sequencher (GeneCodes, Ann Arbor, MI, USA).
Drug efficacy study
Mammary tumor pieces were implanted subcutaneously near the mammary fat pad of 8- to 10-week-old female C.B-17 SCID beige mice weighing between 20 and 25 g (Charles River Laboratories, Wilmington, MA, USA). Tumors were monitored until they reached a mean tumor volume of 200–300 mm3, and animals were randomly distributed into four groups of nine animals each before initiation of dosing. Mice were orally gavaged daily for 13 days with 50, 100 and 150 mg/kg GDC-0941 dissolved in 0.5% methylcellulose with 0.2% Tween 80 (MCT). Tumor sizes and mouse body weights were recorded twice weekly over the course of the study. Tumor volumes were measured in two dimensions using a caliper and calculated using the following formula: Tumor size (mm3)=(Longer measurement × Shorter measurement2) × 0.5. Percent weight change was calculated using the following formula: Group percent weight change=((New weight−Initial weight)/Initial weight) × 100. Mean tumor volumes (±s.e.m.) and percent weight change were calculated and plotted using Kaleidagraph (version 4.03; Synergy Software, Reading, PA, USA). Mice with tumor volumes ⩾2000 mm3 or with losses in body weight of ⩾20% from initial body weight were promptly killed based on the IACUC guideline. To analyze the repeated measurement of tumor volumes from the same animals over time, a mixed modeling approach was used.53 This approach addresses both repeated measurements and modest dropouts owing to any non-treatment-related death of animals before study end. Cubic regression splines were used to fit a non-linear profile to the time courses of log2 tumor volume at each dose level. These non-linear profiles were then related to dose within the mixed model. Tumor growth inhibition as a percentage of vehicle (%TGI) was calculated as the percentage of the area under the fitted curve (AUC) for the respective dose group per day in relation to the vehicle, using the formula: %TGI=100 × (1−AUCDose/AUCVeh). To get uncertainty intervals for %TGI, the fitted curve and the fitted covariance matrix were used to generate a random sample as an approximation to the distribution of %TGI. The random sample is composed of 1000 simulated realizations of the fitted-mixed model, where %TGI has been re-calculated for each realization. Our reported uncertainty interval is values for which 95% of the time the re-calculated values of %TGI will fall in this region given the fitted model. The 2.5 and 97.5 percentiles of the simulated distribution were used as the upper and lower uncertainty intervals. Plots were generated using R (version 2.8.1; R Development Core Team 2008, R Foundation for Statistical Computing, Vienna, Austria) and Excel. Data were analyzed using R and the mixed models were fitted within R using the nlme package.53
Acknowledgments
We thank the Genentech animal facility, DNA sequencing, microarray and oligo synthesis group for help with the study. We also thank to Chie Sakanaka, Mallika Singh, Melissa Juttila and Bob Liu for helpful discussions during the course of the project.
All the authors are employees of Genentech and hold Roche stocks.
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- Hawkins PT, Anderson KE, Davidson K, Stephens LR. Signalling through Class I PI3Ks in mammalian cells. Biochem Soc Trans. 2006;34:647–662. doi: 10.1042/BST0340647. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Ali K, Bilancio A, Geering B, Foukas LC. Signalling by PI3K isoforms: insights from gene-targeted mice. Trends Biochem Sci. 2005;30:194–204. doi: 10.1016/j.tibs.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3:772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- Lee JW, Soung YH, Kim SY, Lee HW, Park WS, Nam SW, et al. PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene. 2004;24:1477–1480. doi: 10.1038/sj.onc.1208304. [DOI] [PubMed] [Google Scholar]
- Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005;5:921–929. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci USA. 2008;105:2652–2657. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27:5486–5496. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakoff SJ. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- Bader AG. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci USA. 2006;103:1475–1479. doi: 10.1073/pnas.0510857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S, Bergholz U, Jücker M, McCubrey JA, Trümper L, Stocking C, et al. Mutations in the catalytic subunit of class IA PI3K confer leukemogenic potential to hematopoietic cells. Oncogene. 2008;27:4096–4106. doi: 10.1038/onc.2008.40. [DOI] [PubMed] [Google Scholar]
- Walrath JC, Hawes JJ, Van Dyke T, Reilly KM. Genetically engineered mouse models in cancer research. Adv Cancer Res. 2010;106:113–164. doi: 10.1016/S0065-230X(10)06004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Lima A, Molina R, Hamilton P, Clermont AC, Devasthali V, et al. Assessing therapeutic responses in Kras mutant cancers using genetically engineered mouse models. Nat Biotechnol. 2010;28:585–593. doi: 10.1038/nbt.1640. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JR, Xu K, Liu JC, Agamez NMR, Loch AJ, Wong RG, et al. Cooperation between Pik3ca and p53 mutations in mouse mammary tumor formation. Cancer Res. 2011;71:2706–2717. doi: 10.1158/0008-5472.CAN-10-0738. [DOI] [PubMed] [Google Scholar]
- Meyer DS, Brinkhaus H, Muller U, Muller M, Cardiff RD, Bentires-Alj M. Luminal expression of PIK3CA mutant H1047R in the mammary gland induces heterogeneous tumors. Cancer Res. 2011;71:4344–4351. doi: 10.1158/0008-5472.CAN-10-3827. [DOI] [PubMed] [Google Scholar]
- Liu P, Cheng H, Santiago S, Raeder M, Zhang F, Isabella A, et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat Med. 2011;17:1116–1120. doi: 10.1038/nm.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi L, Okabe I, Bernard DJ, Wynshaw-Boris A, Nussbaum RL. Proliferative defect and embryonic lethality in mice homozygous for a deletion in the p110alpha subunit of phosphoinositide 3-kinase. J Biol Chem. 1999;274:10963–10968. doi: 10.1074/jbc.274.16.10963. [DOI] [PubMed] [Google Scholar]
- Wagner KU, Wall RJ, St-Onge L, Gruss P, Wynshaw-Boris A, Garrett L, et al. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25:4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KU, McAllister K, Ward T, Davis B, Wiseman R, Hennighausen L. Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Res. 2001;10:545–553. doi: 10.1023/a:1013063514007. [DOI] [PubMed] [Google Scholar]
- Li G, Robinson GW, Lesche R, Martinez-Diaz H, Jiang Z, Rozengurt N, et al. Conditional loss of PTEN leads to precocious development and neoplasia in the mammary gland. Development. 2002;129:4159–4170. doi: 10.1242/dev.129.17.4159. [DOI] [PubMed] [Google Scholar]
- Fishler T, Li YY, Wang RH, Kim HS, Sengupta K, Vassilopoulos A, et al. Genetic instability and mammary tumor formation in mice carrying mammary-specific disruption of Chk1 and p53. Oncogene. 2010;29:4007–4017. doi: 10.1038/onc.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiede B, Kang Y. From milk to malignancy: the role of mammary stem cells in development, pregnancy and breast cancer. Cell Res. 2011;21:245–257. doi: 10.1038/cr.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H. Spindle cell carcinoma of the breast: a case series of a rare histological subtype. Eur J Surg Oncol. 2003;29:600–603. doi: 10.1016/s0748-7983(03)00107-0. [DOI] [PubMed] [Google Scholar]
- Carter MR, Hornick JL, Lester S, Fletcher CD. Spindle cell (sarcomatoid) carcinoma of the breast: a clinicopathologic and immunohistochemical analysis of 29 cases. Am J Surg Pathol. 2006;30:300–309. doi: 10.1097/01.pas.0000184809.27735.a1. [DOI] [PubMed] [Google Scholar]
- Weigelt B, Reis-Filho JS. Histological and molecular types of breast cancer: is there a unifying taxonomy. Nat Rev Clin Oncol. 2009;6:718–730. doi: 10.1038/nrclinonc.2009.166. [DOI] [PubMed] [Google Scholar]
- Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA. 2010;107:15449–15454. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amikura T, Sekine M, Hirai Y, Fujimoto S, Hatae M, Kobayashi I, et al. Mutational analysis of TP53 and p21 in familial and sporadic ovarian cancer in Japan. Gynecol Oncol. 2006;100:365–371. doi: 10.1016/j.ygyno.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Grochova D, Vankova J, Damborsky J, Ravcukova B, Smarda J, Vojtesek B, et al. Analysis of transactivation capability and conformation of p53 temperature-dependent mutants and their reactivation by amifostine in yeast. Oncogene. 2007;27:1243–1252. doi: 10.1038/sj.onc.1210748. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C, et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–592. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- Folkes A, Shuttleworth S, Chuckowree I, Oxenford S, Wan NC, Castanedo G, et al. Preparation of thienopyrimidines and furopyrimidines as lipid kinase inhibitors for treating cancer and other diseases PCT Int Appl 2007206CODEN: PIXXD2 WO 2007127175 A2 20071108.
- Salphati L, Wong H, Belvin M, Bradford D, Edgar KA, Prior WW, et al. Pharmacokinetic-pharmacodynamic modeling of tumor growth inhibition and biomarker modulation by the novel phosphatidylinositol 3-kinase inhibitor GDC-0941. Drug Metab Dispos. 2010;38:1436–1442. doi: 10.1124/dmd.110.032912. [DOI] [PubMed] [Google Scholar]
- Tuveson D, Hanahan D. Translational medicine: cancer lessons from mice to humans. Nature. 2011;471:316–317. doi: 10.1038/471316a. [DOI] [PubMed] [Google Scholar]
- Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyault S, Drouet Y, Navarro C, Bachelot T, Lasset C, Treilleux I, et al. Mutational characterization of individual breast tumors: TP53 and PI3K pathway genes are frequently and distinctively mutated in different subtypes. Breast Cancer Res Treat. 2011;132:29–39. doi: 10.1007/s10549-011-1518-y. [DOI] [PubMed] [Google Scholar]
- Hynes NE, Dey JH. PI3K inhibition overcomes trastuzumab resistance: blockade of ErbB2/ErbB3 is not always enough. Cancer Cell. 2009;15:353–355. doi: 10.1016/j.ccr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman-Cullere MH, Foltz CJ. Body condition scoring: a rapid and accurate method for assessing health status in mice. Lab Anim Sci. 1999;49:319–323. [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Hupe P, Stransky N, Thiery JP, Radvanyi F, Barillot E. Analysis of array CGH data: from signal ratio to gain and loss of DNA regions. Bioinformatics. 2004;20:3413–3422. doi: 10.1093/bioinformatics/bth418. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D. nlme: linear and nonlinear mixed effects models. R Package Version. 2008;3:1–89. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.