Abstract
The fruit fly Drosophila melanogaster has emerged as a useful model for cardiac diseases, both developmental abnormalities and adult functional impairment. Using the tools of both classical and molecular genetics, the study of the developing fly heart has been instrumental in identifying the major signaling events of cardiac field formation, cardiomyocyte specification, and the formation of the functioning heart tube. The larval stage of fly cardiac development has become an important model system for testing isolated preparations of living hearts for the effects of biological and pharmacological compounds on cardiac activity. Meanwhile, the recent development of effective techniques to study adult cardiac performance in the fly has opened new uses for the Drosophila model system. The fly system is now being used to study long-term alterations in adult performance caused by factors such as diet, exercise, and normal aging. The fly is a unique and valuable system for the study of such complex, long-term interactions, as it is the only invertebrate genetic model system with a working heart developmentally homologous to the vertebrate heart. Thus, the fly model combines the advantages of invertebrate genetics (such as large populations, facile molecular genetic techniques, and short lifespan) with physiological measurement techniques that allow meaningful comparisons with data from vertebrate model systems. As such, the fly model is well situated to make important contributions to the understanding of complicated interactions between environmental factors and genetics in the long-term regulation of cardiac performance.
I. Introduction
In the late twentieth century, the rapid expansion of the field of developmental genetics has been one of the greatest triumphs of the biological sciences. Invertebrate genetic model organisms have been used to make startling progress in understanding the steps of patterning, cell specification, and organogenesis during normal development. More recently, invertebrate model systems have been employed as effective models for adult-onset and age-related diseases.
Since the fruit fly Drosophila melanogaster is the only major invertebrate model system that contains a working organ with developmental and functional homologies to the vertebrate heart, flies have been the only model system where the advantages of invertebrate genetics have been utilized to model cardiac development and disease.
The Drosophila heart undergoes developmental steps reminiscent of the early stages of vertebrate heart development. Following subdivision of the cardiac mesoderm from the broader visceral mesoderm, the cardiac field forms from two bilaterally symmetric, linear strands of cells. These two rows of presumptive cardiac cells then migrate to the midline, where they adhere to each other to form a tube with a hollow lumen. Meanwhile, cardiomyocytes are gradually specified by multiple signaling inputs that activate cell-autonomous expression of conserved transcription factors.1,2
Using molecular genetics, lineage tracing, and confocal imaging, the genetic pathways involved in fly cardiac specification have been identified and, in many cases, orthologs have been shown to be involved similarly in early vertebrate heart development.3 In parallel, disruptions in these vertebrate orthologs have been associated with developmental disorders.4,5
The accessibility and simplicity of the Drosophila larval heart have made it a useful model for testing effects of natural or synthetic compounds that regulate cardiac performance. By introducing such factors into partially dissected open preparations, the direct effects of various neurotransmitters on cardiac rate and rhythm have been assayed. The transparency of the larva has facilitated optical measurements of rate and rhythm in intact animals under both normal and pathological conditions, while electrical field measurements have proved effective in describing defects in electrical conductance.6
Excitingly, the use of Drosophila genetics has begun to make a significant impact in the field of adult cardiac performance. As a short-lived model system with well-described characteristics of aging, flies represent an excellent opportunity to model long-term changes in cardiac function that can be traced throughout life, at both the population level and the individual level. Several techniques have been developed to facilitate this process, including generation of M-mode traces from intact or semi-intact preparations, external electrical pacing, and external field recording. Already, flies have been used to model the effects of nutrition, genetic disease, exercise-training, age-related pathology, and normal aging.7
This review will discuss the uses of the Drosophila model system for the study of cardiac developmental and adult disease. We will address, in turn, what has already been learned from the study of embryonic cardiac development, larval cardiac function, and the study of aging adult flies.
II. Embryonic Cardiac Development
Using traditional forward genetics supplemented with candidate gene approaches, a detailed portrait of the specification and organogenesis of the heart has emerged in flies. In this section, we will describe the development of the fly heart in four stages: cardiac field formation, cardiomyocyte specification, division of cardiomyocytes into subtypes, and migration/adhesion of the heart tube. In conclusion, we will outline the relationship between the genetic factors involved in these events in the fly and its vertebrate counterparts.
III. Cardiac Field
The field of presumptive cardiac cells is distinguished from other mesodermal-derived tissue types ultimately by the action of two conserved extracellular signaling molecules, the Wnt family homolog wingless (wg) and the BMP family homolog decepentaplegic (dpp) (Table I). wg is expressed in the embryo in a series of stripes which mark, and, indeed, help to define, the segments of the embryonic mesoderm along the anterior–posterior axis.8–11 Meanwhile, dpp is an important component that signals position to the mesoderm along the dorsal–ventral axis.12,13 Thus, these two signaling molecules act along perpendicular axes during embryogenesis.
TABLE I.
Drosophila Homologs of Genes Involved in Cardiac Development
| Vertebrate/animal homolog | Drosophila Homolog | Reference |
|---|---|---|
| Wnt family | wingless (wg) | 8–11 |
| BMP family | decepentaplegic (dpp) | 12,13 |
| Nkx2.5 | tinman (tin) | 14–16 |
| NK family | bagpipe | 17 |
| FGF signaling | heartless (htl) | 18–22 |
| FOG family | u-shaped (ush) | 23,24 |
| COUP-TF | seven-up (svp) | 25–27 |
| GATA4 | pannier (pnr) | 28 |
| T-box (Tbx5) | nmr | 16 |
| Lbx1 | ladybird | 29,30 |
| Sarco-endoplasmic reticulum calcium ATPase (SERCA) | SERCA | 31 |
| Dystrophin | dystrophin (dys) | 32,33 |
| Sarcoglycan | d-sarcoglycan | 34–36 |
| Myosin transducer complex | Myosin heavy chain (Mhc) | 37–41 |
| KCNQ1 | KCNQ | 42–45 |
| Evx2 | even-skipped (eve) | 46 |
| ErbB0 | EGF receptor | 47–49 |
| Opa1 | dopa1 | 50,51 |
| Superoxide dismutase 2 | Sod2 | 52,53 |
| Sestrin | sesn | 54 |
| 4eBP | 4eBP | 55 |
| Fatty acid transporter | Fatp | 56,57 |
| PGC1-α | spargel | 58,59 |
The points of intersection between these two expression patterns are instructive and required for specification of cardiac cell types. Cardiac cell fate requires the combined exposure to these two signaling molecules not once but twice, at two separate stages of embryogenesis.60 Mesoderm-specific context is provided to interpret this signal by the Nkx2.5 homolog tinman (tin).14,15 Combined misexpression of wg, dpp, and tin is sufficient to induce cardiac-specific gene expression ectopically.60 Interestingly, visceral mesoderm is distinguished from dorsal mesodermal derivatives, such as heart, by the activity of another NK family homeobox protein, Bagpipe, which acts to provide context to more ventral mesodermal cells exposed to the combined signals of Wg and Dpp.17
This strategy of integrating broad extracellular signals with mesoderm-specific homeobox genes to specify cardiogenic mesoderm is broadly conserved in vertebrates as well. BMP family members are essential for induction of cardiac mesoderm in the mouse,61 chick,62 and frog,63 while SMAD-family transcription factors that act downstream of Bmp signaling are autonomously required for cardiogenesis in mice.64
Moreover, as in flies, BMP signaling cooperates with cell-autonomous activity of the tin homolog Nkx2.5 to specify cardiomyocyte lineage in mouse. Nkx2.5 mutant mice have morphological defects resulting from lack of proper specification of the cardiac tube,65 and Nkx2.5 expression is dependent on the activity of BMP-dependent SMAD transcription factors.66
The relationship between Wnt signaling and cardiac induction is more complicated in vertebrates than flies. In frog and chick embryos, it is necessary to inhibit Wnt signaling induced by ligands secreted from the neural tube to allow cardiac field formation.67 This inhibition is accomplished by the action of two proteins from the organizer region, Dickkopf-1 and Crescent.68,69 Dickkopf-1 requires the activity of the homeodomain transcription factor Hex in this context.70
However, work in conditional mouse knockouts has made it clear that at other stages, Wnt signaling is essential as a positive regulator of cardiomyocyte differentiation,71,72 and becomes essential again as a positive regulator of cardiomyocyte proliferation postnatally.73,74 A biphasic role, depending on timing, also exists in zebrafish, where Wnt signaling prior to gastrulation promotes cardiac differentiation, while signaling after gastrulation inhibits cardiac differentiation.75 Further complicating the picture, signaling through noncanonical Wnt pathways is also required for cardiac specification, and this noncanonical signaling may itself act to dampen canonical signaling.76
In addition, proliferation of cardiac progenitors in both flies and vertebrates requires secreted FGF factors. In chick and zebrafish embryos, inhibition of FGF signaling reduces the size of the cardiac field.18–20 In flies, the FGF Receptor, heartless (htl), as its name would suggest, is required for the formation of a functional heart tube.21 Cardiac defects in heartless mutants may, however, be attributable primarily to a failure in the migration step of the cardiac mesoderm to the dorsal midline.22
Despite the greater complexity found in the vertebrate system, it seems clear that the signals involved in the early stages of fly cardiac development are well conserved in vertebrate models. Since these genes are also involved in many congenital cardiac abnormalities, a combination of research in fly and vertebrate models will continue to be of interest in elucidating these relationships in increasing detail.
IV. Cardiomyocyte Differentiation
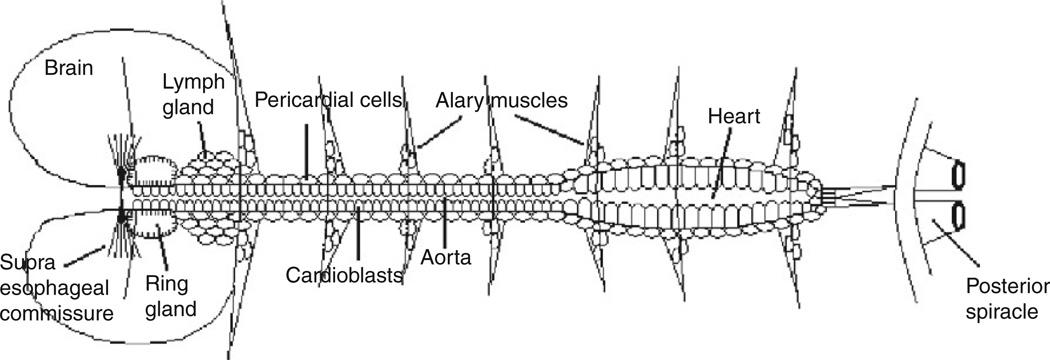
Presumptive heart cells in the fly are subdivided by the coordinate action of several conserved transcription factors.77,78 The heart becomes subdivided into a posterior pumping heart proper and a more anterior portion termed the aorta as a result of positional information provided by homeobox transcription factors (Fig. 1).54,79 Later, during pupal metamorphosis, homeobox genes act in response to hormonal signals to control remodeling of cardiomyocytes into their adult form.80 Within the cardiomyocytes forming the heart tube proper, several specific pairs of cells are fated to alter their shape81 and form the valve-like ostia that allow inflow into the tube prior to pumping.82
Fig. 1.
Diagram of embryonic heart (adapted from Ref. 12).
After the early role of tin in specifying cardiac mesoderm, it is required again during the differentiation of myocardial cells, where it is necessary to define contractile cardiomyocytes and restrict formation of ostia to their proper location.83 tin expression itself is maintained through the action of the T-box transcription factors mid and H15 (aka nmr1–2),84 and mid, H15 and tin act coordinately to promote cardiomyocyte differentiation.85,86
Also acting coordinately as part of this regulatory network is the GATA factor encoding gene pannier (pnr), which acts in response to ectodermal dpp signaling to maintain tin expression and acts both coordinately and separately to activate target genes, including tin itself.87–90 Interestingly, the pnr antagonist u-shaped (ush), a homolog of vertebrate FOG genes, antagonizes cardiogenesis early on,23 but later acts to promote cardiogenesis by maintaining tin expression.90
Another important factor acting in conjunction with tin and pnr to promote cardiomyocyte differentiation is the T-box containing Dorsocross (Doc) family of transcription factors. While the three members of this family appear to have partially redundant function, a triple knockout results in failure of cardiomyocyte specification.91 The combined activity of these genes activates genes directly involved in morphological specification, such as the Drosophila homolog of the vertebrate Hand genes.92
Although the combined activity of tin, pnr, and Doc1–3 is required for cardiomyocyte specification, Doc genes then resolve themselves into a mutually exclusive expression pattern with tin, caused by mutual repression, in which tin is expressed in four out of six cells in each segment, and Doc genes are expressed in the remaining two. The two cells where Doc is expressed become ostia in the posterior heart tube, while the four cells where tin is expressed become contractile cardiomyocytes.83
Cardiomyocyte differentiation proceeds under the direction of differentiation genes, which, in some cases are direct tin targets, such as b-3 tubulin,93 dSur,94,95 or dHand96 and in other cases are targets of transcription factors activated by tin, such as dMef2.97 A key transcription factor in the cells fated to become ostia is the COUP-TF homolog seven-up (svp).26 svp acts to repress tin expression in these cells and maintains expression of wg.25,27
Strikingly, many of the relevant transcription factors to Drosophila cardiac specification have vertebrate orthologs that are also involved in cardiac development and specification, and many of these have been associated with congenital heart diseases.4 The vertebrate homolog of tin, Nkx2.5, has been linked to atrial and ventricular septal defects,16 as well as tetralogy of Fallot.98 Also linked to septal defects are the pnr homolog GATA428 and the nmr family homolog Tbx5.16
Even the combined coordinate and mutually repressive network between transcription factors elucidated in flies shows evidence of being reflected in vertebrate cardiogenesis as well. Genetic interactions have been observed between GATA4 and Tbx5 in mice,99 and Nkx2.5 along with Nkx2.7 is necessary to downregulate Tbx factors later in cardiac development in zebrafish.100 In general, there appears to be a greater degree of redundancy in vertebrate species, for example. GATA4 and GATA5 are both necessary to carry on the activities controlled by pnr in flies,101 but the simplicity of the fly model will continue to be used as an important advantage to the identification of important regulatory families.
V. Migration/Adhesion
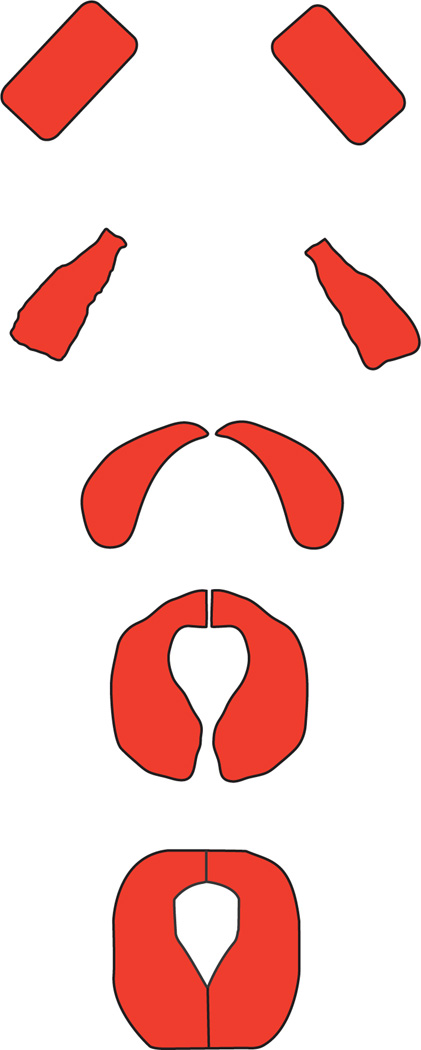
The formation of the final embryonic structure occurs in three steps. First, the bilaterally symmetric rows of cells undergo morphological changes to their membrane structure and align with each other. Then, the two rows of presumptive cardiomyocytes migrate to the dorsal midline. Lastly, the two rows adhere to each other and form a lumen between them (Fig. 2).
Fig. 2.
Diagram of migration and alignment of embryonic cardioblasts (adapted from Ref. 2).
Prior to migration, the two rows of cardiac cells undergo a mesenchymal to epithelial transition, during which they establish contacts between themselves and form a continuous monolayer.8,12 During this process, they begin to express various membrane markers and determinants of polarity.102 This process is dependent initially on cell adhesion molecules, including the Ig-family protein Faint sausage (Fas)103 and the transmembrane receptor protein Toll,104 while maintenance of the ultrastructure becomes dependent on conserved extracellular matrix proteins, including laminins,105,106 integrins,107 and cadherins later.103 In addition, the heterotrimeric G-protein encoding gene, brokenheart (bkh), is required in a cell-autonomous fashion to establish apical–basal polarity, and bkh embryos have polarity defects in tube formation resulting in dysfunctional hearts.108
Once the two rows of cells are properly aligned, they must move together along their respective rows toward the dorsal midline where they will adhere to each other. As the cells migrate, they remain in contact with the “leading edge” of the overlying ectodermal sheet, which is simultaneously migrating toward the midline in the process of dorsal closure. A Type IV-collagen-like protein, Pericardin, mediates the coordinated movements of the two germ layers in this process and is essential for the migration of cardiac precursors to the midline.109
Signaling between the secreted protein Slit and its receptors Robo and Robo2 is essential for maintaining cell polarity during migration and for adhesion when the two bilateral rows of heart cells fuse at the midline.110 In wild-type development, Slit accumulates at the points of connection between bilateral heart precursors and signals to Robo, which is localized at the apical surfaces of the cell pairs, although Robo2 can substitute for Robo in robo mutant flies.110 While fusion of the bilateral heart precursors is occurring, Slit localization is absolutely required for proper localization of membrane proteins that govern the switch from basal–lateral to apical–lateral polarity necessary for initiation of proper cardiac function.111 As a result, mutations in slit produce alignment defects reminiscent of those seen in flies carrying mutations in genes encoding the membrane proteins themselves, such as discs large and the E-cadherin-encoding shotgun.2,112 Using live imaging, it has been demonstrated that the Slit/Robo interaction also plays an additional role to regulate the cell shape changes necessary for the heart cells to contact each other dorsally and ventrally to form a lumen.2
Recently, the morphogenesis of the anterior region of the heart near the border between the heart and aorta has been more fully described29 and termed the outflow tract. Excitingly, this outflow tract has a distinctive derivation from the pharyngeal mesoderm, suggesting a direct developmental homology with the vertebrate outflow tract. In both vertebrates and flies, outflow tract is derived from cells in the pharyngeal mesoderm which express either the transcription factor ladybird in flies or the homologous transcription factor Lbx1 in vertebrates.29,30 Interestingly, the migration and assembly of these cells from the pharyngeal mesoderm to form the outflow tract also require the interaction between Slit and Robo.113
Additional conserved factors have been identified in recent years that are responsible for maintenance of adhesion and/or polarity after the initial formation of the heart tube. In genetic screens for a phenotype in which myocardial cells lose their connections with the surrounding pericardial cells, several new players were identified. One such factor was the HMG-coA reductase, along with downstream effectors of the mevalonate pathway.114 These factors act to regulate heterotrimeric G-proteins, which are also necessary for adhesion between myocardial and pericardial cells.108,114 These proteins act, in part, by regulating protein components of the septate junction, including Neurexin-IV, Sinuous, and Coracle.115
In this context, studies of Drosophila cardiac development have led to productive use of the fly heart not only as a model for early stages of vertebrate heart development, but also as a model for tubulogenesis in general.116
Indeed, rather than thinking about conservation of genetic functions at the level of individual genes, it is becoming more common to think in terms of conservation of gene regulatory networks governing development of orthologous structures. As such, these networks involving interrelated genes involved in the formation and maintenance of similar structures, such as the heart, not only act as conserved regulators of the development of homologous structures but also provide a buffered system for the development and structure of conserved organ systems to be tweaked during speciation without losing the essential function of the organ. Bioinformatic evidence suggests that conservation of gene expression in homologous organ systems is relatively high across species,117 and the gene network governing heart development continues to be elucidated in comparative experiments between flies and vertebrates.77
VI. Larval Heart Function
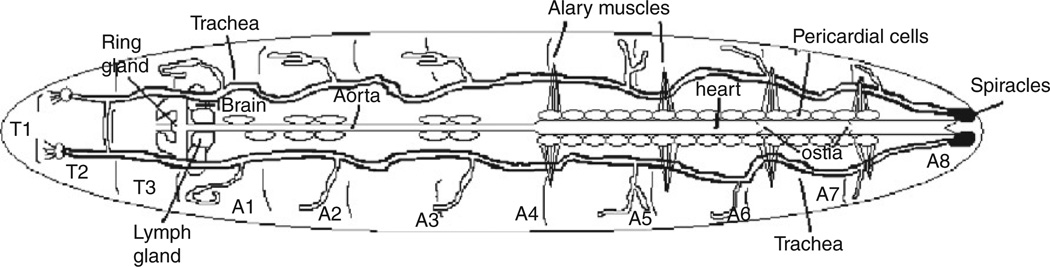
During the larval stages of Drosophila development, a tremendous degree of growth occurs, with a tiny embryo-sized first instar larva gradually reaching the size required for pupation and metamorphosis. The heart must increase its size proportionately during this process, but does so not by cell division, but rather by cell growth.6 Thus, the final third instar larval heart is structurally similar to the embryonic heart, but much larger and more suitable for physiological assays. In addition, the larva is essentially transparent, making it an ideal stage for visualization of cardiac function in intact animals (Fig. 3).82
Fig. 3.
Diagram of larval heart (adapted from Ref. 82).
Another significant advantage of the larva as a model system is that it has a myogenic pacemaker that continues to promote regular contractile activity without neuronal input.118 The larval fly heart is, however, innervated119 and neuronal input regulates rate and rhythm of cardiac contractions.120,121 Thus, partially dissected preps can be utilized to expose the heart to physiological medium and keep it beating regularly for extended periods of time. Using several different preparations and methods, the intact or semidissected larva has been used to test the effects of numerous biologically active compounds, both synthetic and naturally derived (e.g., hormones and neurotransmitters).
In this section, we will discuss the various methods developed to exploit the Drosophila larva as a model system, and summarize the findings with regard to cardiac susceptibility to various external bioactive inputs.
A. Methods
A breakthrough in automation and processing of measurements in intact larvae came with the adaption of photodiode-based measurements of late-stage, immobile larvae at the larva–pupa transition.122 Measurements of transmitted light through the larva change rhythmically in consonance with the heart movements as the heart changes both its shape and the shape of the connected viscera, then returns to its previous shape in a cyclical fashion. The darkening and then lightening of the transparent larva thus produces a cycling trace of changing light exposure in a photodiode that receives input through the microscope field of view. This trace is driven by and mimics exactly the movements of the heart over the same period. Rate and rhythmicity can then be ascertained by mathematical application to the traces. Variations on this method have subsequently been employed in which pixel-tracing camera technology has replaced the photodiode, but acquisition and processing of the data are otherwise similar.123
The application of this technique to intact animals has made it suitable for genetic applications, and several candidate mutants have been analyzed in the fly model using this system.122,124,125 In addition, a large-scale genetic screen has been conducted in which a collection of insertional mutants were screened for mutations that significantly accelerated or decelerated larval heartbeat, identifying several novel candidates for future study in the fly model.123
In addition to intact visualization, the larva is also the developmental stage most easily amenable to semi-intact dissection. Perhaps, the most common and fruitful usage of the Drosophila larva has been with preparations dissected in physiological media and then treated with various biological or pharmacological compounds. The accessibility of the larva to such treatments in combination with the many genetic tools available in the fly system has made this model a useful one for identifying and describing minimum components necessary for generation and propagation of cardioactive electrical signals, as well as for examining how neuronal inputs to cardiac activity are governed in a simple model system.
One method, which is suitable both for measurements of physiological function and as a preparation to perfuse the heart with pharmacological agents, is to perform a partial dissection, then pull a portion of the heart into a micropipette tip, which can then be employed to record spontaneous field potentials.126
Within the last 2 years, several novel ideas have emerged for preparation and measurement of larval heart activity, including two novel preparations for viewing and making recordings from intact larvae. One, known as the “ant farm” method, utilizes plastic spacers to imbed the larva in a small region where it is constrained from moving but unstressed and with both an air source and a thin layer of food available to the larva, thus avoiding potential changes to heart activity due to starvation or oxygen stress responses.127 This method is well suited for examination of the response to genetic interventions, without potential complications from dissection, and can be employed for relatively lengthy recording times. Two different groups have introduced an additional method involving restraint by glue, which introduces some caveats with respect to the restraint method, but makes the animal easily available to the introduction of electrodes for field recording or pacing stimulus.127,128 These preparations have been used in direct visual counting of heart rate127 and also used in automated optical counting protocols.129,130
The introduction of the novel automated optical protocols represents a substantial advance building upon previous optical methods, expanding their possibilities. Importantly, these methods allow data to be quantitated not just at the level of patterns of cardiac activity over time, as in prior indirect measures, but allow direct data acquisition of several parameters of individual beats on a continuing basis.129 Such parameters include systolic and diastolic volume, relaxation, and contraction velocity. Once these data have been acquired, they can be used in turn to derive other measurements such as fractional shortening. Interestingly, these methods have also been applied successfully to analyze cardiac movements in other systems, including vertebrate models such as the zebrafish.117,129,130
Another recent innovation with great potential, although requiring highly specialized equipment, is the combined use of optical coherence tomography (OCT) with laser scanning fluorescence microscopy. This technique requires the use of two dedicated imaging systems in combination, but does provide cross-sectional visualization in perpendicular planes of living animals.131
Section VII will describe how these various methods, past and present, have been employed to examine genetic, pharmacological, hormonal, and peptidergic regulation of cardiac rate and rhythm.
VII. Pacemaker Regulation
In the mid-1990s, early versions of these methods began to be employed to establish that the larval heart is moyogenic. Using partially dissected preps, treated with pharmacological compounds that block specific subsets of ion channels, it was established that calcium and potassium currents are essential for larval pacemaking, but that sodium channels are dispensable.132 At the same time, these channel-blocking experiments lent molecular support to the earlier contention that the larval heartbeat is fully myogenic.118,122,132
Mutations in genes encoding components of ion channels closely support pharmacological results, as mutants deficient in potassium channel function, such as slowpoke, shaker, and ether-a-go-go, all have deficiencies in rhythmic control of heartbeat, while disruption of calcium or chloride channels has little effect.125 The role of intracellular calcium storage and release in regulating contractile activity also appears to be conserved, as mutations in the ryanodine receptor ortholog in flies reduce contractile activity in cardiac muscle.133
Despite the myogenic nature of the insect pacemaker, it has been clearly established that regulation of the rate and rhythmicity of contractions is influenced by a variety of biological compounds. For example, injections of the insulin-signaling antagonist Adipokinetic Hormone into late larvae/prepupae had a cardioacceleratory effect.134
Using photodiodes to record heart movements, it was demonstrated that temperature-sensitive mutations in the gene no action potential (nap) caused arrhythmias which disappeared when animals were returned to the permissive temperature.122 Lending further credence to the idea that neuronal regulation modulates the activity of the myogenic pacemaker, several neurotransmitters were demonstrated to accelerate heart rate without adverse effects on rhythmicity, including serotonin, octopamine, norepinephrine, dopamine, and acetylcholine.124 Conversely, mutations affecting secretion or synthesis of these neurotransmitters tended to decelerate the heart.124 Other reports have generated partially contradictory results135 but some of these discrepancies may be explained by dose-dependent differences, as serotonin, for example, has more recently been shown to be cardioacceleratory or cardioinhibitory depending on dose of exposure.136 Additionally, several FMRFamide-related peptides, including Dromyosuppressin, were demonstrated to decelerate the larval heart.137
The Crustacean Cardioacceleratory Peptide was also found to accelerate heart rate in Drosophila larvae.138 Interestingly, release of Crustacean Cardioacceleratory Peptide is activated by Ecdysis Triggering Hormone just prior to eclosion and may be responsible for the temporary elevation in heartbeat that presages the emergence of the adult fly.139,140 Other neuropeptides involved in the molting or adult emergence process are also cardioactive, including the pyrokinin-like peptide encoded by the hugin gene.141
Regulation by neuronally produced factors is necessary not only to adjust heart rate to changing conditions but also to maintain proper heart rate under normal resting conditions, as demonstrated by examining the heart rate in flies with a temperature-sensitive mutation in the dynamin-encoding gene shibire.142 These mutants are deficient in the ability to recover endocytotic vesicles at the restrictive temperature143 and are useful for temporary induction of phenotypes dependent on neurotransmitter communication. Both optical recording and electrocardiograms were employed on intact late larvae/prepupae to establish that blocking the functions of multiple neurotransmitters by inactivating shibire leads to defects in regulation of heart rate even under resting, unstressed conditions.144
An extensive series of tests of various exogenous factors in combination with genetics has led to significant progress in understanding the makeup of the various currents associated with the larval pacemaker. Multiple classes of G-proteins have been implicated as targets of pharmacological interventions, while cGMP and Protein Kinase G seem to be critical components of changing pacemaker responses to varying conditions.145 Interestingly, some of the same genetic factors identified as key regulators in the involuntary response of heart rate to conditional change have also previously been shown to be important factors in behavioral regulation of activity levels and foraging behaviors, such as the foraging gene,146,147 and the peptide encoded by the flatline gene. flatline, despite extensive sequence homology to Manduca allostatin, does not appear to act as a Juvenile Hormone antagonist in Drosophila, but instead has a potent myotropic activity in both cardiac and visceral muscle.148
More recently, a combination of genetic approaches and electrophysiology has identified a two-pore domain potassium channel Ork1 that is essential for the regulation of heart rate and rhythm.149 Reduction of Ork1 expression throughout the animal or specifically in the heart tube leads to a proportional increase in heart rate while overexpression of Ork1 blocks heart beat entirely and action potentials are unrecordable when Ork1 is overexpressed.149 Significantly, action potentials in wild-type hearts were recorded throughout the length of the heart, supporting the idea that Drosophila cardiomyocytes are more homogeneous than vertebrate cardiomyocytes. The highly conserved Sarco-Endoplasmic Reticulum Calcium ATPase (SERCA) protein has also recently been studied using the fly larva as a conserved readout. Flies carrying mutations in the SERCA gene have disrupted heart rate and extended periods of heart stoppage.31
The combination of evolving techniques for precise cardiac physiology in insect models with the sequenced genome in flies should result in continued importance of the fly model in addressing problems such as how currents are generated and regulated to precisely govern pacemaking. The Drosophila larva is well positioned to be an important model system for use in testing various combinations of genetic and pharmacological factors contributing to cardiac disease.
VIII. Adult Functional Models
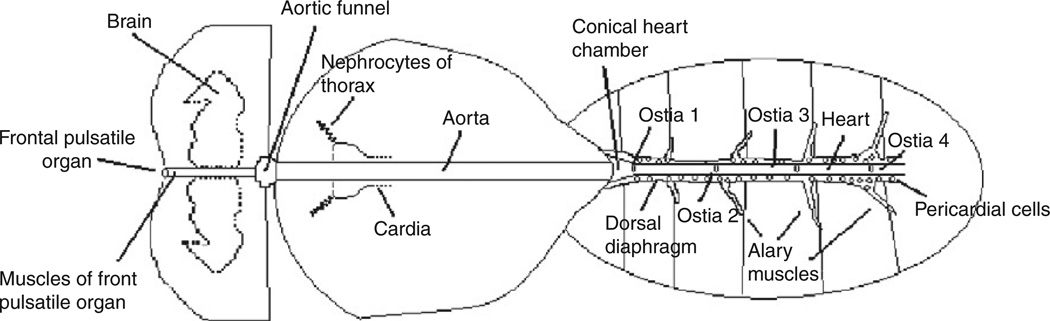
During pupal morphogenesis, the heart is one of a few structures that persist without being completely degraded and remodeled.119 However, a few modifications are made to the morphology of the heart before eclosion.81,82 The formation of the familiar head/thorax/abdomen insect body plan is overlaid with the heart in such a way that the heart proper is located in the abdomen of the adult. A conical chamber is formed near the thorax/abdomen boundary that serves to collect haemolymph for expulsion through the aorta, which proceeds through the thorax and sends haemolymph forward toward the head (Fig. 4).
Fig. 4.
Diagram of adult heart (adapted from Ref. 82).
Limited changes also occur to the structure of the heart itself. An additional layer of striated muscle is generated during pupation along the ventral surface of the heart, with striations that proceed in a longitudinal direction, rather than the transverse spiral “paper towel tube” shape of the fibers in the rest of the heart muscle. An additional pair of ostia are also generated during metamorphosis, such that the adult heart contains four pairs of ostia, as compared to the larval three.81,82 In addition, the character of these ostia is morphologically different. In the adult, they are no longer simple openings, but take on a more valve-like appearance.
Perhaps, the most important difference from the point of view of disease modeling is the introduction of direct innervations to the heart at adult stages, adding a layer of complexity to the regulation of heart rate and rhythm.121 Pairs of neurons in each segment innervate both the heart itself and the muscles attaching the heart to the body wall (alary muscles). Additional nerve projections known as Bipolar Neurons cluster toward the posterior of the heart where they serve as a point of release for Crustacean Cardioactive Peptide,121 and application of CCAP at these sites instigates anterograde contractions, suggesting that these neurons are important regulators of “forward” heartbeat.150 Meanwhile, bilateral, segmental innervations from glutaminergic neurons regulate the periodic reversal characteristic of insect heartbeat.150
Recently, another forward pair of ostia was discovered in two different species of Drosophilids using a recording technique from multiple sensor elements in sequence.151 These forward ostia are essential for retrograde heartbeat, as they collect and separate haemolymph supply from specialized thoracic spaces, the implications of which for circulation are just beginning to be understood.151 These separated flows of circulation may in the near future make it possible to use flies to model other circulatory disorders that had not previously been thought accessible using insect model systems.
Already, the fly system has been in use over the last few years as a model for various cardiac disorders, and offers several important advantages over vertebrate models. First, the rapidity and facility of the genetics available in the fly system are an obvious motivation to use the fly to examine complicated genetic interactions in multifactorial etiologies of disease. Second, the groundwork laid by the fly community and by the Drosophila genome project has made mutations, overexpression constructs, and inducible RNAi constructs available for the vast majority of the genome. Third, and perhaps most excitingly, the short lifespan and ability to monitor cardiac function longitudinally have made the fly system available as an important model for long-term studies, applicable either to progressive disease models, or even to changes to cardiac function brought about by the consequences of normal aging.7
In Section VIII.A, we will discuss the novel techniques developed in the last few years to make these studies possible, and then discuss the results and their application to vertebrate disease and aging.
A. Methods
An initial success in usage of adult flies to model cardiac functional disturbance was accomplished by examining maximal heart rate in cases of exogenous stress. Heart rate was increased either by exposing flies to increased temperature or to external currents, and the maximum tolerable increase in heart rate was measured at various ages.152 It was determined that the ability of the heart to tolerate externally induced increases in heart rate is an age-dependent phenomenon, since genetically identical flies at older ages entered cardiac arrest when exposed to similar conditions to those tolerated easily by younger flies.152
Further progress was made when edge-detection techniques were adapted from usage in larvae (see above) to become feasible in adults as well.123 This technique relies on pixel-tracing software to identify edges of the cardiac tube, identifiable by their differential contrast in high-resolution black and white video images. This technique allows long-term tracing of heart movements in unstressed and intact conditions. Importantly, this technique and others that do not rely on fluorescence offer the advantage of not requiring the introduction of additional stimuli that may affect endogenous cardiac regulation, such as ultraviolet light when detecting movement using Green Fluorescent Protein, for example.
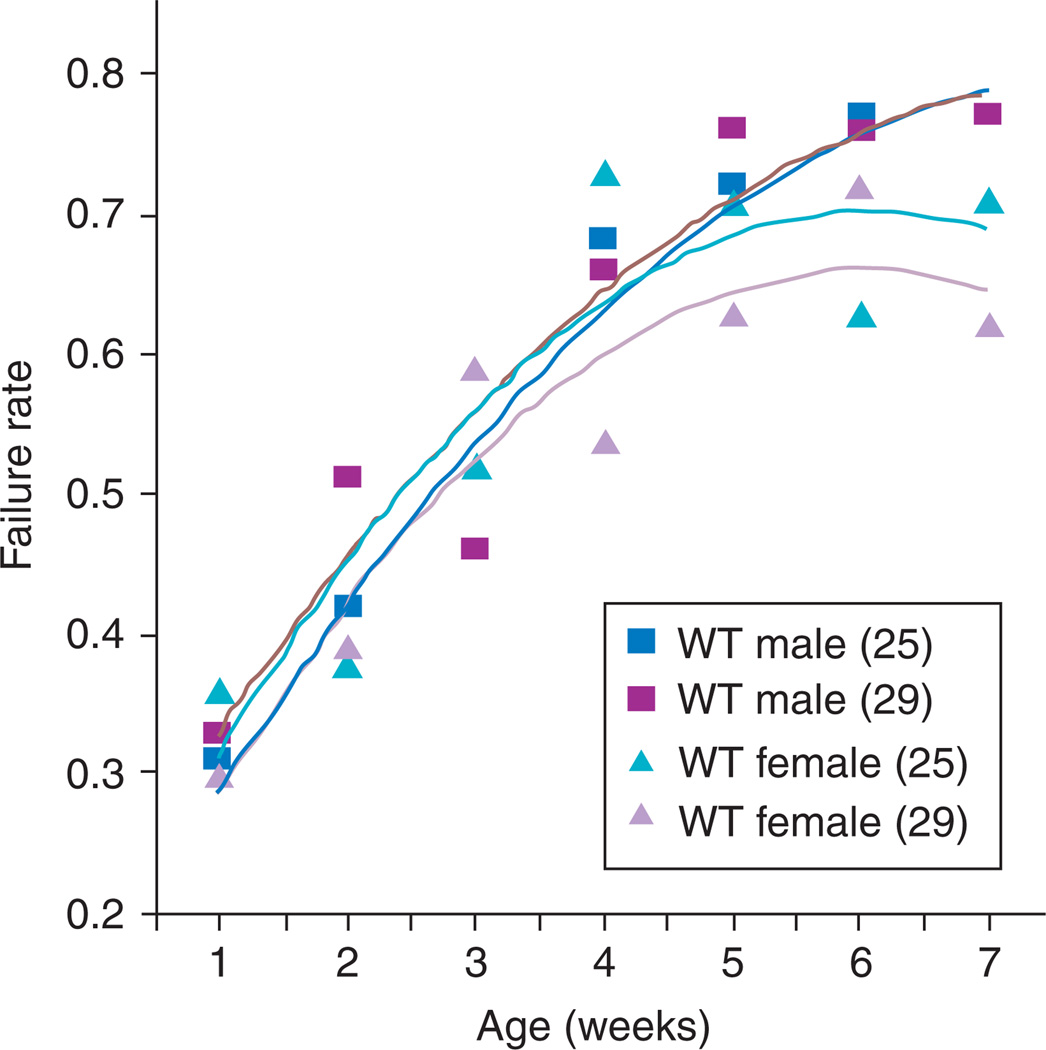
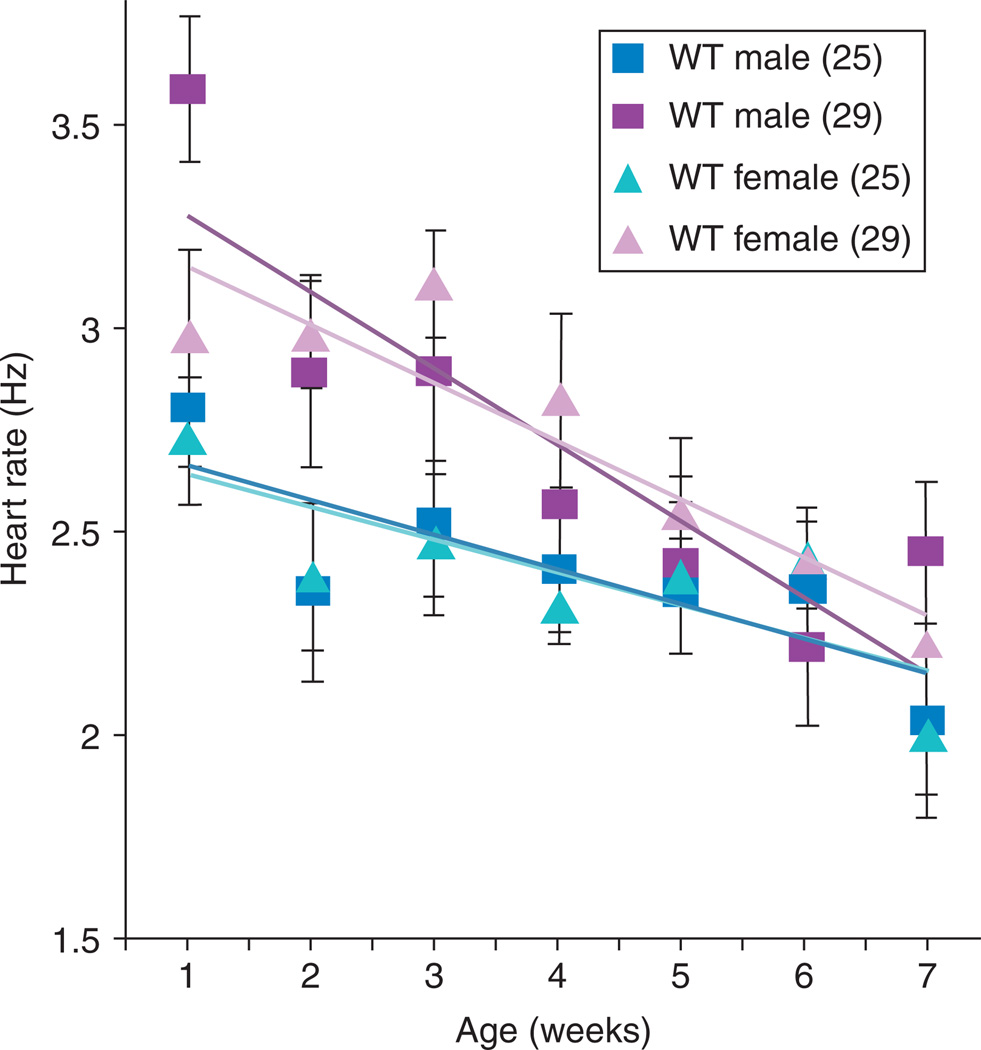
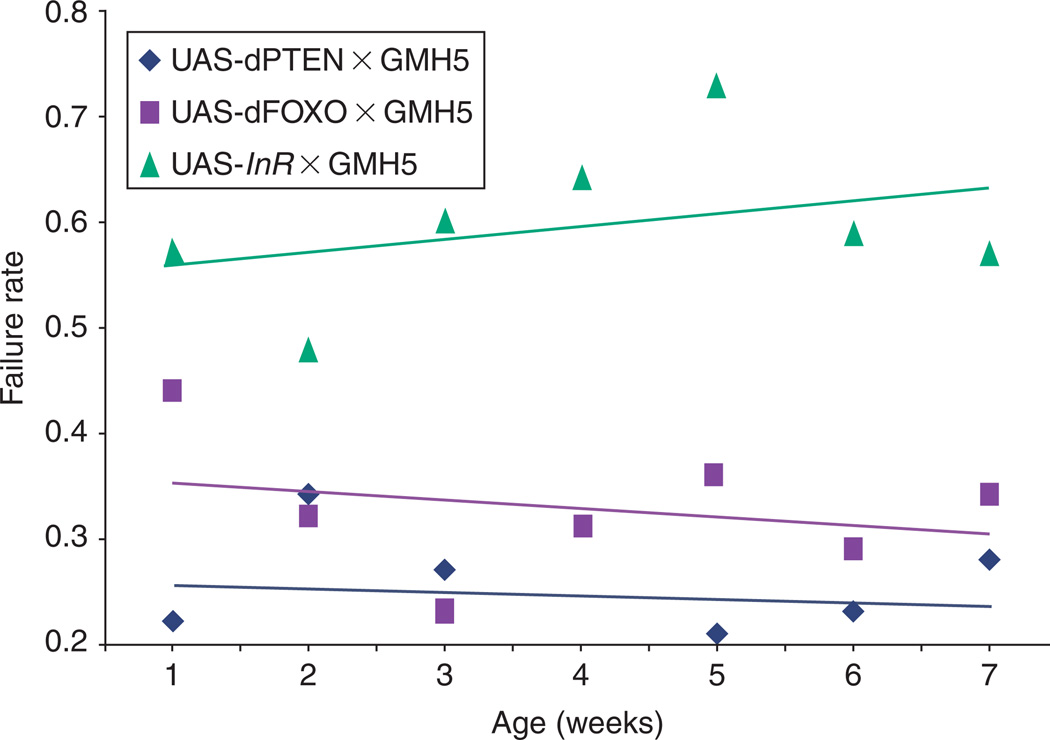
Refinement of stress testing followed rapidly with a stress test protocol that relied on a defined condition to test the response flies of various genotypes, ages, and environmental exposures.153 It offers the distinct advantage of being applicable to up to 10 flies at once, thus facilitating large study designs to take advantage of the large numbers common to fly studies, in comparison to vertebrates. Because this method does not depend on maximal tolerance of extreme stress, it is useful for picking up more subtle abnormalities and has been used effectively in screens to pick up mutations affecting short-term or long-term cardiac performance.123 By testing the percentage of populations that respond to low-level stress by entering fibrillations or arrest events, as well as the percentage of flies experiencing a cardiac event that successfully recover normal heartbeat regulation, this method serves as a useful marker for physiological age and health that can pick up not just direct abnormalities, but, importantly, population tendencies toward abnormalities (Fig. 5). In combination with genetics, this method has served as an important first step toward identifying new genotypes that can be used as models of disease154 or of aging.42
Fig. 5.
Heart failure as a function of age after external electrical pacing from outbred wild-type offspring (WT; yw × Canton S). This parameter is highly age-dependent, but is not affected by temperature or gender during the first 5 weeks of adult life.153
Further refinement to these techniques has involved partially dissected preps, bathed in physiological media that allows continued heart function for several hours.155 Algorithms have been developed to analyze video traces taken from such preps129 to extract several useful parameters, including fractional shortening, a two-dimensional measure that can be used proportionally to infer the volume expelled during contraction; arrhythmicity index, used to quantitate the frequency of arrhythmias; and period length, which is proportional to resting heart rate. The use of these measures in combination allows a significant degree of diagnostic precision, not just identifying cardiac abnormalities, but also classifying them into different subtypes of functional abnormalities.
Similar measures can also be achieved using video analysis of intact preps, although the use of intact preps involves some trade-offs in camera resolution as compared to dissected preps. In return, one gains the assurance that the animal is intact in its own physiological state rather than a mimicking media. Depending on experimental design, this trade-off may be worthwhile, in cases where longitudinal analysis of the same flies over time is critical156 or where retention of neuronal input is desired.
Another option for acquiring parametric measurements of live cardiac performance in intact animals is the use of OCT technology. In this technique, the fly is enclosed in a small chamber to prevent large movements, and the internal space of the heart tube is detected in real time as it shortens and expands through an ultrasound-like technology.157 This method combines the advantage of utilizing intact animals in their inherent physiological environment with some of the resolution advantages of the partial dissection/video method. The primary disadvantage of this powerful method is its requirement for specialized technology that is not readily available commercially.
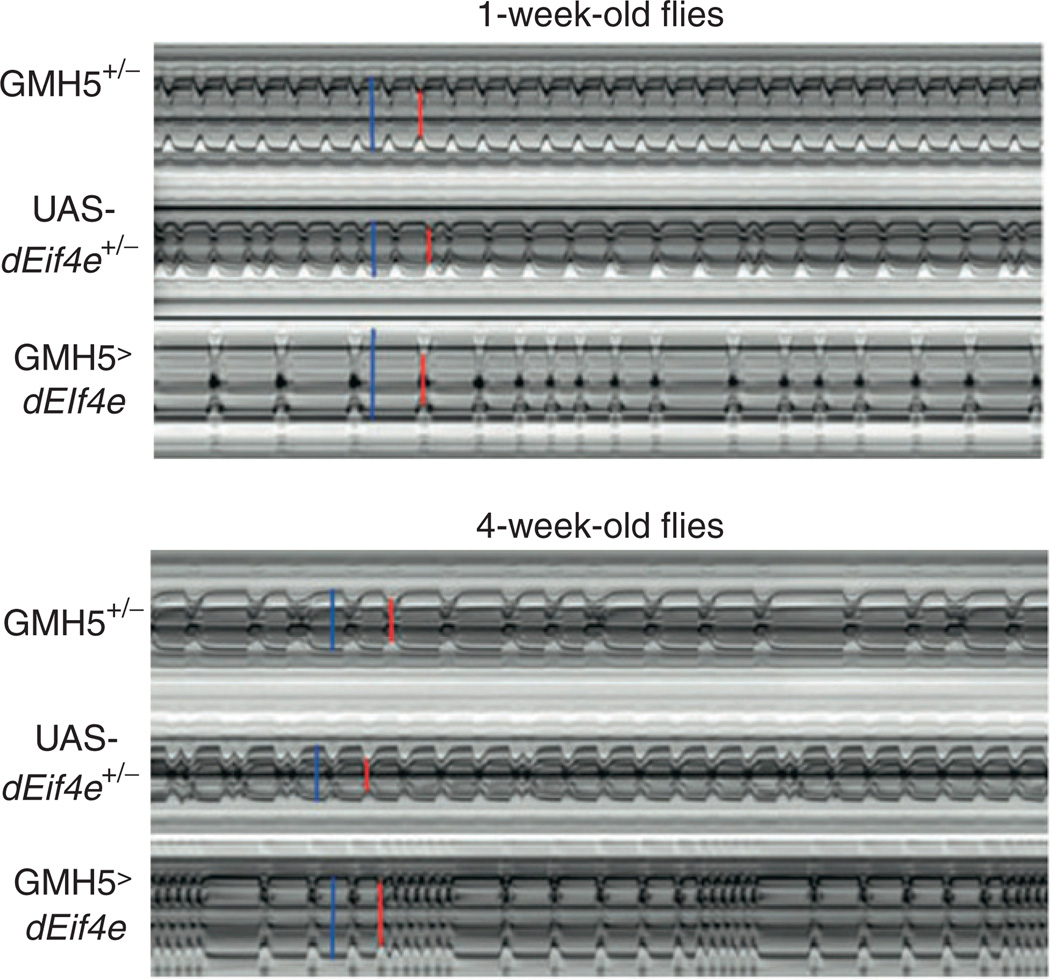
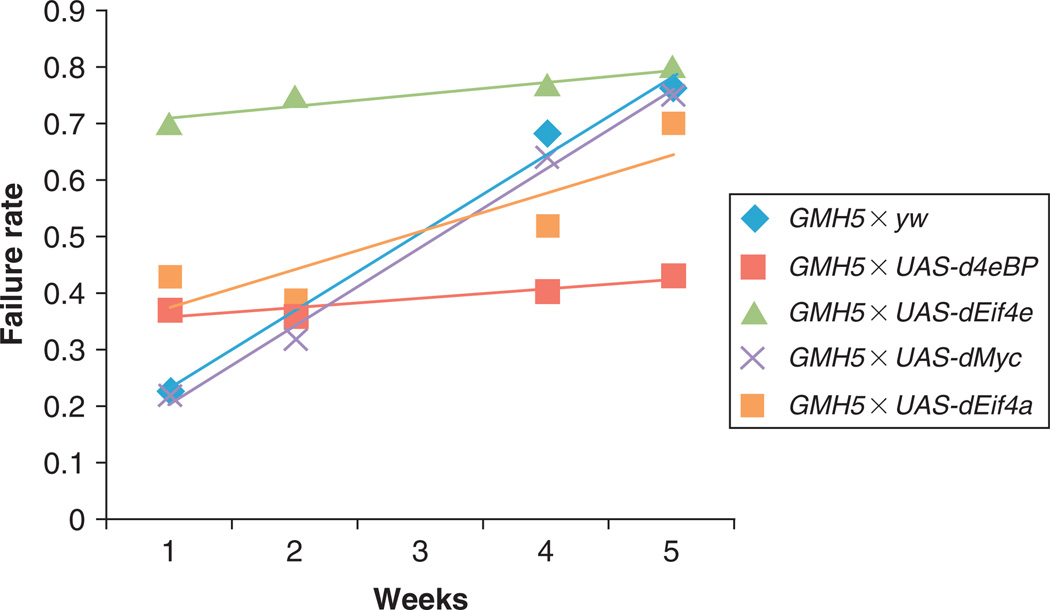
Either OCT or video-based traces can be utilized to generate M-mode displays (Fig. 6), which take a thin slice of an ongoing display of movement and then display repeated short intervals of that same slice across time. In effect, this creates a still-life visualization of the animated movement of that particular portion of the heart. This is valuable not just for visual purposes, but also can be used as input for analysis algorithms.155
Fig. 6.
Representative M-mode records showing the movement of the heart tube walls (y-axis) over time (x-axis). Blue bars indicate the diastolic diameter of each heart and the red bars indicate the systolic diameter. Records for 1-week-old flies (A) show predominately regular heart beat patterns as the GMH5 heterozygotes (as in Ref. 42), whereas most flies overexpressing UAS-dEif4e in the heart show arrhythmic heart beat patterns.55
While direct cardiac functional readouts have had a tremendous impact on the utility of the fly heart as a model, traditional genetic methods have also been effectively applied to generate tissue-specific data centered on the heart. For example, heart-specific microarrays have been generated,158 screens using inducible RNAi constructs driven in the heart have been conducted159 and genome-wide association studies have been conducted with cardiac performance as a quantitative output for QTL mapping.155
Excitingly, the fly heart has also been effective as a readout for testing human genes directly for effects on heart function. For example, genes from a human chromosomal region associated with congenital heart defects in Down’s Syndrome patients have been tested in all permutations of single and combined overexpression in the fly heart. This study has been successful in narrowing down the potential effectors of this phenotype to two genes that, when expressed in combination, generate cardiac defects. The same genes are now being expressed in mice to confirm the phenotypic results directly in a vertebrate model.160 Such studies are likely to become more common in the immediate future, as the power of fly genetics to test large numbers of factors in a short time will continue to make study designs possible that would be prohibitively expensive and time-consuming in vertebrate models. The information gained from bulk studies in the fly can then be used to intelligently design specific studies in vertebrate models limited to the best potential candidates.
In Section IX, we will describe in turn several recent studies that have effectively utilized the adult fly heart as a model for examining specific-genetic factors contributing to human disease.
IX. Single-Gene Disease Models
An effective entry point for modeling specific or general cardiac pathologies in flies is the candidate gene approach. A recent example of the effectiveness of such an approach is the recent work using the fly to model the cardiac dysfunction that occurs as a result of the progressive degeneration in muscular dystrophy patients.
The causal gene in multiple forms of the human disease muscular dystrophy has been identified as the structural protein Dystrophin.161 Dystrophin plays a dual role, being necessary to provide the structural framework for contractile activity, and also being involved in signaling events inside muscle cells through its interactions with other proteins in the Dystrophin/Dystroglycan complex.162,163
In both flies and vertebrates, the dystrophin(dys) gene is expressed in multiple isoforms, with different isoforms being expressed in different tissue-specific patterns. Two different splice variants of the fly dys gene are expressed in the adult myocardium, suggesting that Dys is likely to play an endogenous role in cardiac function in insects as well.
In mouse models of muscular dystrophy, deficiency of dys gene expression results in structural degeneration of the myocardium in a progressive fashion that is aggravated by age-related structural derangement.32 In flies mutant for the Drosophila homolog of dys, a similar structural degeneration of myocardial fiber arrays has been observed.33
Adult heart performance in dys mutant flies displays several abnormalities as a consequence of these structural defects, as measured by video tracking of the movements of dissected heart preparations.33 Such flies have an increased resting heart rate, which comes about due to a decrease in fractional shortening, defined as a decrease in heart wall diameter change during contraction, a measure that is proportional to cardiac output. Since dys mutants exhibit both wider diastolic diameter and reduced fractional shortening, they are said to have a dilated cardiomyopathy phenotype.33 This phenotype is a direct result of deficiency of Dys protein in cardiac tissue, since it can be rescued by cardiac-specific expression of the dys gene. Interestingly, the fly and mouse functional versions of these proteins are so similar that the fly phenotype can be rescued by cardiac expression of a truncated version of the homologous mouse gene.33
Another gene product that has been associated with muscular dystrophy in vertebrates is the Sarcoglycan protein, mutations in which are known to cause cardiomyopathy in vertebrates.34,35 Deletions of the homologous fly gene have been examined, and shown to cause a progressive impairment of locomotor ability along with reduced cardiac function.36 Cardiac phenotypes are qualitatively similar to those in dys mutants, with progressive structural derangement accompanied by increased diastolic diameter and reduced fractional shortening, characteristic of the dilated cardiomyopathy phenotype.36
These experiments, along with the others described below, serve well to illustrate several fundamental points. First, the fly model responds phenotypically to genetic interventions in ways that are recognizable and remarkably similar to vertebrates. Second, in some cases the genetic material involved is similar enough for cross-species experiments to produce useful data. Third, following upon the first two points, the essential similarity between vertebrate and insect models in isolated organ function makes the fly model a viable choice for structure–function analysis of vertebrate proteins. Indeed, the fly has already been effectively used to examine the role of various domains and splice variants of the Dys protein in skeletal muscle.164
The fly has also become a useful model for unraveling the effects of components of the contractile machinery in the myocardium. Due to genetic redundancy and multigene families in vertebrate lineages, the simpler fly model offers a desirable alternative for avoiding complications resulting from such redundancies. A recent example is found in the use of the fly system to study mutations in proteins from the myosin transducer complex.37 The molecular motor formed by a complex of myosin light and heavy chains is a conserved molecular regulator of contractile activity in both skeletal and cardiac muscles. Both reduction or stimulation of transducer activity can cause muscle pathology in vertebrates.38,39 In flies, there exists a single Mhc gene encoding all isoforms by means of differential splicing in different tissues.40,41
Using two existing point mutations in the Mhc gene, one that increases motor activity and one that inhibits motor activity, the effects of altering motor activity specifically in cardiac tissue were tested. As in vertebrates, either stimulation or inhibition of the myosin motor caused pathologies. Interestingly, the phenotypes generated were quite different, however.37 Flies with reduced myosin transducer activity become dilated at diastole, have reduced systolic shortening, and have an increased incidence of arrhythmias that grows progressively worse. This constellation of phenotypes is characteristic of the dilated cardiomyopathy phenotype (Table II). Meanwhile, flies with increased transducer activity display morphological constrictions in isolated areas of the heart tube where full relaxation does not occur. As these flies undergo aging, these constrictions appear in more areas of the heart in a progressive fashion. This phenotype is one of several indices that are collectively described as restrictive cardiomyopathy (Table II).
TABLE II.
Phenotypic Characterization of Cardiac Pathologies Elicited by Single-Gene Mutation in Drosophila
| Disease | Phenotypes |
|---|---|
| Dilated cardiomyopathy | Enlarged heart, abnormally functioning heart, reduced fractional shortening, enlarged tube diameters, impaired systolic function, increased diastolic diameter, increased systolic diameter, expanded heart period |
| Restrictive cardiomyopathy | Elevated arrhythmicity index, decreased resistance to external pacing stress, alterations to diastolic width and fractional shortening |
| Long QT syndrome | Increased risk of ventricular tachycardia and sudden death, sensitive to stress |
A similar approach was employed to test the fly homolog of the vertebrate protein Muscle LIM Protein (MLP). This protein has been found associated with the Z-discs at the boundaries of sarcomeres in cardiac muscle,165,166 and the collective assortment of proteins associated at the Z-disc has been demonstrated to be critical for cardiac contractile function.167 Mutations in several of these components, including MLP, lead to dilated cardiomyopathy.168
A fly homolog, mlp84B, is also expressed in sarcomeric Z-discs of the cardiac muscle in flies.169 RNAi knockdown of the fly gene did not cause overt structural defects, but did induce significant physiological deficiencies, including rhythmic defects and prolongation of the diastolic interval.169
Another grouping of conserved genetic factors associated with cardiac dysfunction in both vertebrates and insects is the group of genes encoding ion channels necessary for the generation or propagation of currents involved in electrical stimulation of cardiac contractions. Two recent studies have addressed specific single-gene mutations in flies affecting one such ion channel.
One such study focused on the fly homolog of the vertebrate KCNQ1 gene. This gene encodes a subunit of a potassium channel necessary for cardiac repolarization.43 Mutations in this gene have been associated in humans with a prolonged QT interval on electrocardiograms, a condition which, in turn, has been associated with Long QT Syndrome, a condition that causes increased risk of ventricular tachycardia and sudden death.44,45
Deletion mutants were generated for the fly homolog, also called KCNQ. Null mutants did not have overt structural abnormalities but, like animal models of Long QT syndrome,170 were extremely sensitive to stress, when induced by external pacing.42 These abnormalities could be rescued by cardiac-specific expression of a wild-type KCNQ transgene, indicating that the requirement for KCNQ protein is tissue autonomous in the myocardium.155 Using M-Mode video traces from semi-intact preparations, it was determined that KCNQ mutant hearts begin to exhibit substantial irregularities in heart rhythm early in life, in contrast to wild-type hearts, which are quite regular until age-related deterioration starts to induce spontaneous arrhythmias.155 Additionally, the period length of mutant heart beats rapidly degenerates to a much greater length, and hence, a much slower heart rate, than wild-type hearts of the same age. Again, as in vertebrates, electrical field potential recordings indicated a tendency toward failure to repolarize, suggesting that this is the underlying mechanism for the stress sensitivity and arrhythmias.155 Similar effects were reported for other conserved channel genes, including the homolog of the vertebrate HERG gene.
Another such study examined mutations in the dSUR gene, vertebrate homologs of which form essential subunits of the KATP channel. These channels are known to be important in adult heart function, particularly in protection from ischemic stress.171,172 Interestingly, cardiac expression of RNAi against dSUR rendered flies extremely susceptible to external pacing stress and to hypoxic stress.95 Interestingly, this gene has also served as a pioneering example in flies of reiterated usage of developmental transcription factors essential for cardiac specification to also regulate cardiac differentiation and adult function. The dSUR gene is coordinately regulated directly by the GATA family homolog Pnr and by the Nkx family homolog Tin.95
Several developmentally important cardiogenic factors have been implicated in adult cardiac pathologies, particularly in the induction of pathological hypertrophy.173 GATA4, for example, in addition to its association with several familial forms of congenital heart disease,174,175 also has been strongly associated with adult-onset pathological hypertrophy176 and is thought to contribute to hypertrophy by activating similar targets that are employed during initial cardiac growth during development.177
Using adult-specific expression drivers to drive RNAi constructs, the role of the Drosophila GATA family homolog pnr in maintenance of adult cardiac physiology was examined. Flies either expressing pnr RNAi, a dominant negative pnr construct, or carrying heterozygous genomic pnr mutations showed an enhanced sensitivity to external pacing stress, and these phenotypes could be rescued by cardiac expression of wild-type pnr.178 Flies heterozygous for pnr mutations exhibited a high-cardiac arrhythmia index and a high-period length (equivalent to low heart rate). Both of these phenotypes were also susceptible to rescue by expression of a pnr construct with a cardiac-specific driver.178 RT-PCR also revealed that expression of several structural components of muscle as well as essential ion channel subunits was downregulated in pnr heterozygotes.178
Heart-specific overexpression of the T-Box family homolog nmr was also able to rescue both pacing sensitivity and tendency to arrhythmia, suggesting that nmr is likely to be functionally downstream of pnr in this context.178 The Nmr protein had previously been shown to act synergistically with the Nkx2.5 homolog Tin to regulate adult function.179 Taken together, these results establish the fly as a model for conserved regulation of adult function by developmental genes. As such, the opportunity exists over the next few years to use the fly as a model to test alterations of such genes and their downstream targets, which are likely to involve similar targets to those in vertebrate pathological hypertrophy.
These studies are examples of how the fly heart can be used as a model system to unravel the etiology of diseases already known in humans, but difficult to test in vertebrate model systems. Such models, once developed, set the stage for testing of various interventions to rescue the disease phenotypes. Given the remarkable conservation of disease mechanisms, it seems likely that interventions identified in insect models will also find application in vertebrate models or human patients.
X. Disease Mechanisms
The usage of the fly model is not limited, of course, to forming models for the testing of interventions against known vertebrate diseases, however. The fly has also begun to emerge as a useful system for examining the role of various physiological interactions that are not yet well understood in vertebrates. A recent example is a study examining the role of the pericardial cells as a nonautonomous regulator of adult function. In vertebrates, the epicardial cells are known to be necessary for the proper maturation and adult function of the myocardium,180,181 but the mechanism by which epicardial cells accomplish this is not yet well understood.
The pericardial cells surrounding the fly myocardium may act in a homologous fashion to the vertebrate epicardium in some respects. For example, the transcription factor Evx2 is expressed in the mouse epicardium, and its homolog even-skipped (eve) is expressed in a key subset of the fly pericardial cells.46 If this expression is removed by genetic means, these cells become naïve and are deleted from the pericardial lineage, which results in several deleterious effects on both muscular mobility and myocardial performance, including reduced resting heart rate in both larvae and adults, as well as reduced resistance to pacing stress in adults.46 Taken together, these results suggest the possibility that cells marked by expression of Eve-family transcription factors may be essential to signal to associated myocardial cells to maintain adult heart function in both vertebrates and insects.
Recently, a fusion construct, in which the pericardial enhancer for eve was fused to a protein that represses eve expression, was used to examine more closely the consequences of deletion of Eve-positive pericardial cells on adult myocardial function. It was found that such hearts from flies with Eve-positive pericardial cells deleted have an elevated arrhythmicity index, decreased resistance to external pacing stress, and alterations to diastolic width and fractional shortening characteristic of a restrictive cardiomyopathy phenotype.
These phenotypes are consistent with a conserved role for vertebrate epicardium with insect pericardial cells, and further study in the fly model will be instrumental in unlocking the mechanism by which these interactions take place.
More evidence for a conserved regulation of adult cardiac function has been recently provided by the identification of the EGF Receptor signaling pathway as an important postdevelopmental regulator of cardiac function. A mutation in the rhomboid3 gene, which encodes a transmembrane protein necessary for the processing of a critical EGF-receptor ligand, was shown to cause a dilated cardiomyopathy phenotype.182 This effect was specific to maintenance of adult function, as no developmental abnormalities were detected. Cardiac expression of an activated version of the EGF Receptor rescued this phenotype, as did expression of preprocessed ligand.182 Furthermore, adult-specific expression of a dominant negative version of the EGF Receptor itself produced a progressive dilated cardiomyopathy phenotype as well. Taken together, these results argue strongly that active EGF Receptor is necessary to maintain postdevelopmental cardiac function in Drosophila.
This phenotypic role for EGF Receptor in the maintenance of postdevelopmental function appears to be conserved in vertebrates as well. This conservation has important consequences, particularly in the treatment of some forms of tumors. For example, inhibition of ErbB2, the closest human homolog of the Drosophila EGF Receptor, is a target of some chemotherapeutic treatments. Unfortunately, inhibition of ErbB2 has also been associated with dilated cardiomyopathy in humans.47–49
Another example of the usage of the fly model to shed light on mechanisms that are conceptually ambiguous in vertebrate comes from recent work on the role of oxidative stress in degenerative cardiac phenotypes resulting from disease or from normal aging. Oxidative stress has been proposed to be a causative agent in several cardiac disorders, including diabetic cardiomyopathy, where it may act indirectly through impairment of vascular function via induction of atherosclerosis or hypertension183,184 or directly in the myocardium, where increased levels of reactive oxygen species (ROS) have been associated with heart failure and ischemia/reperfusion injury.185,186 These effects are also exacerbated by a vicious cycle of coregulation with inflammatory cytokines.187 The inflammation associated transcription factor NF-kappa B has also been associated with arrhythmias, including atrial fibrillation.188 ROS levels in the myocardium itself have been shown to increase in response to inflammation and this process may be mediated by the proteins of the RAGE (receptor for advanced glycation end products) family, which is upregulated in response to hyperglycemia and triggers additional ROS producing activities.189
Evidence from a variety of vertebrate genetic models that increase ROS generation is in agreement that high levels of ROS are associated with pressure-induced cardiac hypertrophy190–193 and sensitivity to ischemic stress.194 Due to the cyclical nature of ROS as targets and regulators of numerous physiological states, it is difficult to resolve with certainty whether ROS are indeed causal factors in these pathologies or whether they are instead symptoms inevitably associated with dysfunctional cardiac tissue that exacerbate preexisting pathologies.
Two recent fly studies have used single-gene mutations to attempt to address these issues. The first of these used the fly homolog of the vertebrate Opa1 gene, whose product is expressed in the mitochondrial membrane in humans, and has been associated with several degenerative diseases of the eye.50 Flies heterozygous for a null mutation in the Drosophila homolog, dopa1, exhibit vision defects, indicating a high degree of functional conservation with the human gene.51 These flies also exhibit significant cardiac abnormalities, including a reduced resting heart rate, increased tendency toward arrhythmias, as well as decreases in fractional shortening and decreased resistance to external pacing stress.51 Significantly, however, these heart phenotypes do not appear to be directly a consequence of increased oxidative damage, because treating the same flies with antioxidants fails to rescue the cardiac phenotypes. This failure is not due to ineffectiveness of the antioxidant treatment, because the same treatment effectively rescues the eye phenotypes observed in the same model.195 Rather, the case is made that the eye phenotypes are a consequence of oxidative damage, but the cardiac phenotypes are related to deficiencies in mitochondrial respiratory function as a result of reduction in dOpa1 levels.
A common avenue for studying the effects of oxidative stress on specific functions is by examining the phenotypic effects of mutations or tissue-specific knockdowns of genes essential for scavenging ROS. A mouse knockout has been generated for superoxide dismutase 2 (SOD2),52 a gene that encodes a mitochondrially localized protein that scavenges superoxide produced during mitochondrial reactions and converts it to hydrogen peroxide, from which other proteins can convert it to water. Mice homozygous for the SOD2 knockout display serious cardiac defects, consistent with a diagnosis of dilated cardiomyopathy,52 in addition to other degenerative phenotypes and mitochondrial dysfunction.196 Mice heterozygous for this knockout are overtly normal but develop cancers at a high rate due to sensitivity to endogenous and exogenous oxidative stress.195,197 Conversely, treatment with an exogenous SOD2 mimetic can rescue some of these cardiac effects,198 while expression of an SOD2 transgene can alleviate effects of oxidative stress in cultured cardiomyocytes.193
Flies that carry a null mutation for Sod2 are able to reach adulthood, but have an extremely short lifespan, averaging less than 2 days.53 Since the lifespan is so short, flies were tracked using noninvasive video techniques throughout life to assess whether their progressive deterioration was qualitatively similar to the phenotypes seen in progressive human diseases or in aging.
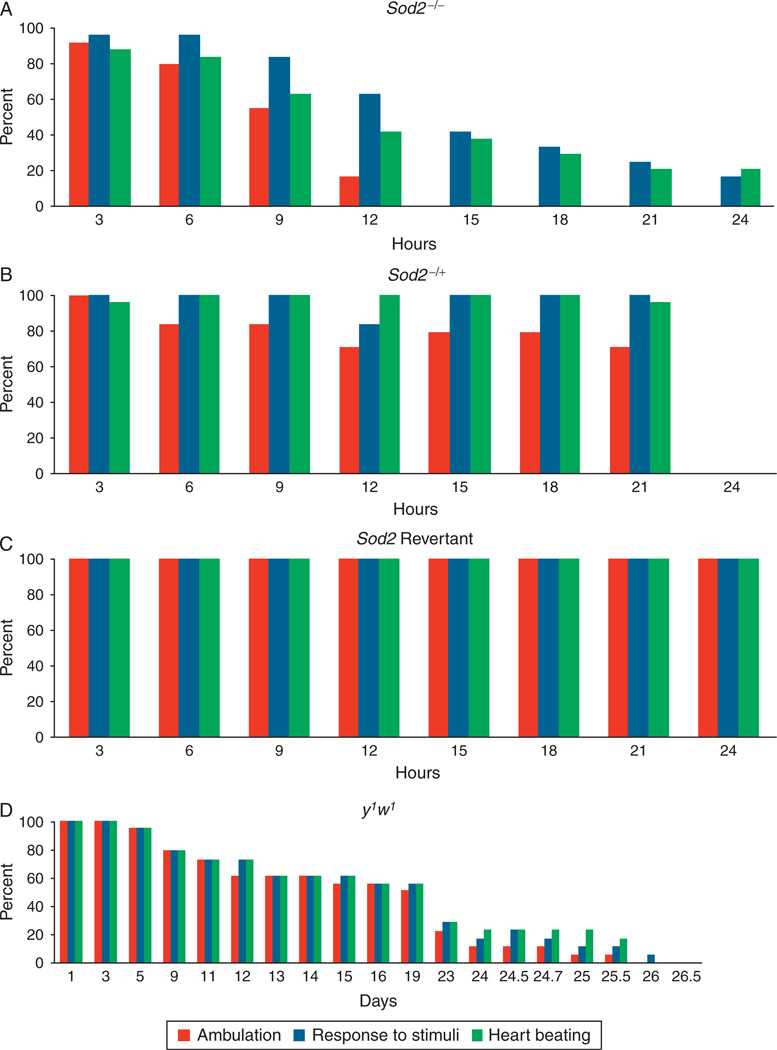
Sod2 mutant hearts have a dramatic reduction in net heart rate, due to frequent pauses, followed by episodic resumption of rapid, shallow heartbeats.156 During periods of beating, the heart rate is considerably faster than wild type, but with a reduced fractional shortening and presumably a lower contractile output as a result. Eventually, prior to the death of each individual animal, the heart ceases to beat at all, even in response to external stimuli. No evidence was seen for a progressive deterioration in high-oxidative stress conditions that resembled age-related or progressive disease-related pathology. Rather, there appeared to be a threshold effect, in which past a certain point of oxidative damage, the heart loses its ability to maintain effective function entirely, and up to that point, there is very little evidence of the progressive decline in resting function seen in aging wild-type flies (Fig. 7).42,153,156
Fig. 7.
Various measures of functionality decline at different rates in Sod2−/− flies. Percentage of flies observed to exhibit either spontaneous ambulation (red), response to gentle stimuli (blue), or spontaneous heartbeat (green) is charted for (A) Sod2−/−, (B) Sod2−/+, (C) Sod2 revertants or (D) y1w1. (A), (B) and (C) are shown over a 24-hour time period, whereas (D) is shown throughout a lifetime. In (D), day 1 in the graph represents day 1 of the experiment, but day 64 of the age of flies.156
Interestingly, when the same longitudinal tracking techniques were applied to wild-type flies at advanced ages, the same pattern was observed. Although there is a progressive decay in rhythmic control and ultrastructure over the lifespan of the animal (see aging section below), the chain of spontaneous functional failures that result in loss of effective cardiac performance do not actually begin until the last 36–48 h of life, and can, in fact, be used to predict which flies are in the process of dying 1–2 days prior to actual death156 (Fig. 7).
A distinct advantage of flies as a cardiac model is the ability of flies, as evident in the Sod2 mutant flies just described, to survive for relatively extended periods after extreme disruption of cardiac function. Since insects are not directly dependent on heart activity for oxygen, it is possible to observe the consequences of functional abnormalities that would result in sudden death in a vertebrate model. This advantage, in combination with the relative lack of redundancy already mentioned, has suggested the usage of flies to examine reiterative roles of gene products that are developmentally important to produce functional hearts and then are reused in temporally separable roles to maintain adult function.
XI. Unbiased Screens—Disease Phenotypes
The most historically important and obvious advantage of the Drosophila model system is the ability to do large-scale unbiased screens for a wide variety of phenotypes. Not only do the quick generation time and wide array of available techniques facilitate rapid identification of candidates, but the distance between primary screen candidates and specific retesting is also much shorter than in vertebrate model systems, allowing researchers to get from identification of loci to specific mechanisms within a reasonable time frame. The use of the fly model in this context has traditionally been associated with developmental biology, and indeed, most of the major developmental signaling pathways were first identified in this way. Only recently, as phenotypic characterization of physiological, rather than structural, abnormalities in flies has become more rapid and routine, has the concept of unbiased genetic screens become feasible for specific aspects of organ physiology, such as cardiac performance. Two recent examples demonstrate the inspiring possibilities now available for identifying novel regulators of adult cardiac function with the fly system.
One such screen has utilized the OCT technology recently developed to allow diagnosis of abnormalities in resting heart function in intact unanaesthetised animals (see Methods section above). Primary screening was done using a collection of deficiency stocks maintained at the public Drosophila stock center in Bloomington, Indiana. While these deficiencies are all homozygous lethal, they represent an excellent tool for identifying loci that are heteroinsufficient for physiological phenotypes. In this case, deficiencies on the second chromosome were screened for the dilated cardiomyopathy phenotype characteristic of adult flies with impaired adult functional regulation (Table II).
One gene identified in this screen, weary (wry), has already been further characterized and defined as a novel ligand for the conserved Notch receptor, based on cell aggregation assays.199 After identifying the importance of wry for maintenance of adult function, the same study went further to demonstrate that other known ligands for Notch, as well as the downstream transcription factor associated with Notch signaling, Supressor of Hairless, are also required autonomously in the heart for adult function.199 A role for Notch in the etiology of adult cardiac dysfunction has already been implicated in vertebrate studies.200 Now that the conservation of this importance has been extended to flies, the fly model system can be used to further dissect pathway components and mechanistic interactions in the heart in upcoming years.
A second example took a quite different approach, using a genome-wide collection of inducible RNAi constructs, also publicly available from the Vienna Drosophila RNAi Center. Each RNAi was driven specifically in the adult heart, and primary screening was done not for heart function directly, but indirectly by assessing lethality at an increased temperature. The rationale for this is that since heart rate is temperature-dependent in insect species, passage of adults through a period of high temperature is equivalent to a cardiac stress test. This approach proved fruitful as ~6% of the genome displayed evidence of inability to tolerate this stress, and these showed a significant enrichment for genes previously classified as being part of conserved functional classes.201
One pathway that generated multiple hits in this analysis was the CCR4-Not complex, previously identified in yeast202 as a chromatin regulator,203,204 but never before associated with cardiac function in any organism. Secondary screens confirmed that knockdown of the not3 gene in the myocardium by multiple methods generates flies with the classic dilated cardiomyopathy phenotype, including expanded heart period, reduced fractional shortening, and increased systolic diameter.201 These phenotypes may be a consequence of structural alterations, since disruption of myofiber arrangement was also observed. However, RT-PCR from hearts with knockdown of not3 revealed significant reduction in the gene expression from several genes previously identified to be critical for cardiac performance, including Serc2a, Mhc, and KCNQ,201 suggesting that the phenotype of not3 may be dependent on regulation of a cardiac gene program.
These observations were rapidly translated to vertebrate models by the construction of knockout mice for the mouse homolog of not3. Contractile abnormalities were observed in heterozygous knockout mice, and also in ex vivo explanted cardiomyocytes. Furthermore, hearts in heterozygous mice were extremely sensitive to cardiac stress, as assessed by transverse aortic constriction.201 In support of the idea that Not3 acts through influencing the chromatin state of a cardiac genetic program, these phenotypes were able to be rescued by HDAC inhibitors. Expanding the relevance of these results to humans, a recent genome-wide association study for Long QT Syndrome in humans also identified a member of the Not complex, supporting a conserved role in the human population.205
In Section XII, we will discuss the developing use of the fly as a model of long-term changes in cardiac function brought about by consequences of aging, or by changes in “lifestyle,” such as diet and exercise.
XII. Cardiac Aging
Invertebrate models have been instrumental in advancing the understanding of genetic regulation of lifespan. Both fly and worm models have been used extensively in screens and candidate approaches to identify single genes that are capable of extending lifespan when overexpressed, or when mutated.206,207 Several major mechanisms of lifespan extension have been shown to be conserved in vertebrate and invertebrate models, including dietary restriction,208 insulin/IGF signaling,209,210 and TOR Kinase signaling.211,212
Until recently, however, invertebrate models have not been suitable for investigations of detailed organ physiology during aging, primarily because of a lack of suitable measurement techniques that can be applied in large numbers or in longitudinal study designs. In recent years, functional aging has begun to be an increasingly fertile topic of study in flies and worms, however. Worms have been used as a model for deterioration of mobility and integumental integrity during aging,213 while flies have been used to analyze declines in functional mobility214 and immune response.215 Importantly, functional assays allow changes in lifespan associated with genetic modifications to be attributed causally to changes in actual function. Genetic alterations that improve lifespan may or may not also improve function,216 while interventions that improve function during aging may not always extend lifespan.217 Since extension of life without concomitant extension of function is of limited value, measurements of functional aging are likely to become an increasingly important function of invertebrate genetic model systems, such as worms and flies.
The development of increasingly powerful techniques for measurement of cardiac physiology in Drosophila in recent years has made cardiac aging an especially well-studied aspect of functional aging in flies. Given the extraordinary degree of conservation of functional genetics between fly and vertebrate hearts, as demonstrated by the multiple success stories in disease models discussed in the previous section, it seems likely that a similar degree of conservation will apply to mechanisms of cardiac age-related change as well.
Based on longitudinal human studies,218 and on a variety of studies in vertebrate animal models,218,219 conserved changes associated with aging cardiac function have been described. These fall into two categories, one being pathologies that are age-related simply because they are a result of accumulated damage that has had more time to accumulate in older individuals. These pathologies are often a result of prolonged exposure to various risk factors, such as high-fat diets, smoking, or family history, as well as environmental pathogens or irritants.220–223 However, it is generally accepted that even in healthy individuals, a gradual deterioration in cardiac function occurs as a function of normal aging.
For example, older humans exhibit an increase in diastolic end-filling, resulting in an increase of contraction duration.224,225 This increased contraction duration in turn leads to a reduction in resting heart rate, and an even more pronounced reduction in maximal heart rate during exercise. Reduction in maximal heart rate may also be a consequence of reduced number of functional atrial pacemaker cells, which contributes to loss of rhythmic homeostasis, as reflected in the age-related increase in atrial fibrillation events.226 Aging humans and rodents also exhibit decreased resistance to a variety of cardiac stresses, especially ischemic reperfusion stress.227
Work in isolated cardiomyocytes has demonstrated that cardiomyocytes from aging individuals also have reduced contractile strength,228 reflected in reduction of gene expression for components of the contractile machinery229 and a reduction in mitochondrial gene expression and mitochondrial enzymatic activity.230 Mitochondrial morphology is also altered in aged cardiomyocytes, as mitochondria from older cells appear swollen and fewer cristae.231 As a consequence of reduced mitochondrial efficiency, higher amounts of ROS are also produced in these cells, causing further damage to mitochondria.232,233 In addition, aging in humans has been associated with a progressive pathological hypertrophy, which may be driven in part by reiteration of a developmental program, since Nkx2.5 and GATA4 are reactivated during this process.229
Several aspects of cardiac function have been shown to decline progressively with age in a robust fashion detectable in widely divergent genetic backgrounds. Maximum heart rate declines steadily throughout life,152 while resting heart rate undergoes a steady decline between 1 and 5 weeks of age123,153 (Fig. 8). This reduction in beats per minute is largely a function of lengthening relaxation time, which gradually increases the period length of a contraction wave.155 In addition, a gradual loss of rhythmic regularity leads to a gradual increase in incidence of spontaneous tachycardia-like events occurring in older flies.155 Furthermore, a reduction in fractional shortening is evident during normal aging33 which produces phenotypes similar to disease models in which contractile machinery is impaired.36,37 This changing profile of performance is reminiscent of age-related dysfunctions in cardiac performance in the human population, who undergo increases in incidence of atrial fibrillation, reduction in maximal heart rate, and reduction in contractile output with age as well.7,218
Fig. 8.
Plot of heart rates from an outbred wild-type (WT; yw × Canton S) strain by age. Heart rate measurements were made at 25 and 29 °C for seven consecutive weeks. All flies were reared at the same temperature (25 °C). Heart rate measurements for both genders at each temperature are progressively lower with age (age main effect, F ratio = 54, P < 0.001).153
Thus, flies are an excellent system to test the long-term effects of genetic or environmental antiaging interventions on cardiac function. The first such experiments sought to test the effects on cardiac performance of the best known signaling pathway that regulates lifespan, the insulin/IGF signaling pathway.
Reduction in insulin/IGF signaling, in some contexts, extends lifespan in flies, worms, and mice.234–236 A series of alleles of the Drosophila Insulin Receptor (InR), some of which extend lifespan and some do not, were tested across ages for their effects on cardiac performance. Allelic combinations that did not extend lifespan also failed to extend cardiac performance, as measured by resistance to external pacing stress. By contrast, allelic combinations that did extend lifespan also delayed the age-related decrease in cardiac stress resistance.153 Similar results were observed when insulin/IGF signaling was reduced by other means, including mutations in the insulin receptor substrate homolog chico and ablation of cells responsible for secretion of insulin-like peptide.153 These results suggested that reduction of insulin/IGF signaling extends lifespan in part by extending functional characteristics of essential organ systems to more advanced ages.
Strikingly, it was also demonstrated that this function of insulin signaling acts autonomously in this context to regulate cardiac aging. Although blocking insulin signaling specifically in adipose tissue has been shown to extend lifespan in flies, mice, and worms,235,237–239 cardiac-specific reduction of insulin signaling was capable of impeding cardiac aging even in animals where insulin signaling was unaltered in any other tissue153 (Fig. 9). Taken together, these results support a model in which insulin/IGF signaling activity plays a conserved role in adipose tissue, where it regulates the production of secondary signals that regulate organ-specific aging. Importantly, one of those secondary signals is insulin itself, and blockage of this signal alone in cardiac tissue is sufficient to severely impede cardiac age-related dysfunction.
Fig. 9.
Stress-induced cardiac failure rate over 7 weeks in flies with heart-specific overexpression of InR, dPTEN, or dFOXO. UAS-InR × GMH5-Gal4 have a high failure rate at all ages (mean failure rate = 0.58). In contrast to heart-specific InR overexpression, both UAS-dPTEN × GMH5 and UAS-dFOXO × GMH5 progeny showed a low-failure rate at all ages (mean failure rate = 0.22 and 0.33, respectively), which does very with age (χ2 = 0.17, P = 0.7 and χ2 = 2.90, P = 0.9, respectively; n > 160 for each sample).153
Significantly, tissue-specific impedance of the aging process in the heart, despite its potent local effects, had no impact on overall lifespan,153 presumably due to normal age-related decline of other essential organ systems. This observation highlights a substantial advantage of the Drosophila system as a model of cardiac aging, since it allows age-related change in cardiac function to be studied separately from lifespan, an approach that would be impossible in a vertebrate model system, where acute dependence on cardiac function for immediate oxygen supply renders cardiac function inextricably tied to lifespan.
Another signaling pathway known to influence lifespan in multiple organisms is the TOR Kinase signaling pathway.211 TOR signaling interacts with, and experiences cross-talk with, insulin/IGF signaling at multiple points in the respective pathways, but nevertheless, produces phenotypes separable from those of the canonical insulin pathway.240 A heteroallelic combination of dTOR mutations is capable of extending lifespan in Drosophila.241 This same combination appears to alter the response to insulin-signaling downstream of the transcription factor dFoxo, as dTOR mutants block several effects of an activated dFoxo transgene.241 Both loss of function mutations in dTOR and cardiac expression of TOR antagonists are capable of providing protection against age-related decline in both resting heart rate and cardiac stress resistance.241
Further support for this idea has come from recent study of a negative feedback regulator of TOR signaling, Sestrin. Flies carrying mutations in the Drosophila homolog of Sestrin, dSesn, exhibit many phenotypes characteristic of hyperactive TOR signaling, including impaired heart performance that is qualitatively similar to that seen in flies of advanced ages. dSesn mutants displayed increased period length, and increased incidence of arrhythmias, disorganization of structural myofibrils, and dilation of the heart diameter during both systolic and diastolic phases.54 Slowing of heart rate and dilation of heart diameter could be rescued by feeding flies rapamycin, while rhythmic defects could be rescued by feeding of antioxidants.54 This seems to support a model in which TOR activity leads directly to increased period length and structural defects, but rhythmic defects are a secondary consequence of increased oxidative stress in flies where TOR is hyperactive.
Thus, two major nutrient responsive signaling pathways, TOR and insulin/IGF, act to regulate cardiac aging and lifespan. The possibility that they do so through a common regulatory target was tested using a candidate gene approach. Since both of these signaling pathways have a conserved role in regulating translation through control of either expression or activity of the conserved 4eBP protein,242–245 the role of 4eBP in controlling cardiac aging was tested directly. Cardiac-specific overexpression of d4eBP provided a similar degree of protection against age-related decline in rhythmicity and stress resistance to that provided by heart-specific impedance of TOR activity (Fig. 10).55 By contrast, heart-specific overexpression of the regulatory target of 4eBP, the eukaryotic initiation factor, deif4e, caused hearts at young ages to display impaired stress resistance in a profile characteristic of aging wild-type hearts (Fig. 10).55 Differential expression and/or activity of 4eBP seems to completely account for the tissue-autonomous effects of both insulin/IGF and TOR signaling, as a series of co-overexpressions consistently indicated that d4eBP and deif4e acted epistatically to both dFoxo and dTOR.55 This role in regulating cardiac aging is specific to d4eBP and is not general to all regulators of translation levels, as other regulators of translation were not able to generate such phenotypes.
Fig. 10.
Cardiac overexpression of d4eBP showed a significantly flattened slope of stress-induced failure. Indeed, the change in failure rate with age is no longer statistically significant (genotype-by-age, χ2 = 1, P = 0.4). Expression of dEif4e also flattened the slope throughout the time period, but showed a higher failure rate compared with controls at each time point (at week 1, χ2 = 22, P < 0.0001).55
Another major signaling molecule that interacts with both insulin and TOR signaling to regulate translational levels, for example, is S6 Kinase, which regulates translation in part by phosphorylating ribosomal S6 to regulate its activity.246 Hypomorphic mutations in the Drosophila S6K gene provided a potent extension of cardiac stress resistance to older ages, suggesting that this signaling molecule too plays an important role in the translation/cardiac aging axis. However, cardiac-specific expression of a dominant negative dS6K transgene had no effect on cardiac aging.55 Expression of dominant negative dS6K in the insulin producing cells (IPCs),247 however, greatly extended cardiac stress resistance in aging flies. This effect is likely to be mediated by the observed reduction in secretion of insulin-like peptides in these flies,55 since the cardiac aging profile of these flies is similar to flies in which the IPCs have been ablated.153
If TOR and insulin/IGF signaling regulate cardiac aging using a specific translational regulatory protein, 4eBP, as an effector, what are the likely targets for 4eBP that are of importance? A reasonable hypothesis is that genes regulating fatty-acid metabolism are likely to be crucial in this process. During vertebrate aging, both gene expression of genes encoding proteins involved in fatty-acid metabolism248 and fatty-acid substrate utilization are altered.249 Furthermore, both fatty-acid metabolism itself241 and gene expression of genes involved in lipid metabolism250 are altered as a consequence of dFoxo and dTOR activity. Thus, signaling pathways that modulate physiology in response to diet are likely to be of continuing importance for modeling not only effects of normal aging on cardiac physiology but also the effects of diets high in lipid on progressive cardiac disease states.
How does this compare with the effects of insulin/IGF axis on vertebrate cardiac aging? The role of insulin/IGF in vertebrates is more complicated than in flies, since the effects on aging related functional changes are superimposed on the potent effects of this pathway on growth and on repair mechanisms. The insulin receptor is required for postnatal cardiac growth in mice and also for hypertrophy in response to exercise.251–254 Insulin/IGF also plays a very different role in damaged hearts than in healthy aging hearts. For example, although chronic administration of IGF-1 in murine hearts induces pathological hypertrophy, IGF-1 administration to hearts following myocardial infarction improves survival of cardiomyocytes.255,256 Further complicating matters, IGF-1 exerts a protective effect on repair of damaged cardiac tissue, but these effects are dependent on which isoform is expressed in the heart, as some have tissue-specific and opposing effects.257
A significant advantage of the fly model in this context is that these effects can be removed from the equation, facilitating study of the direct effects of nutrient sensing pathways, such as insulin and TOR signaling, on cardiac functional aging, without complications from other effects of pathway manipulation. Since flies have an open circulatory system, indirect effects on the heart from alterations in vascular biology can be excluded. Since adult fly hearts are essentially postmitotic, effects derived from alterations in postnatal growth and repair can also be excluded.
In Section XIII, we will focus on recent work using the fly model to address genetic responses to “lifestyle choices” and their effects on cardiac physiology.
XIII. Diet and Exercise
Perhaps, the most important disease that can be modeled in flies is obesity. Obesity is arguably the most common contributing factor to cardiac disease worldwide,258,259 and extensive evidence from human studies and from vertebrate models indicates that exposure to high-fat diets and/or obesity induces a variety of pathological states that affect cardiac function, either directly or indirectly, including atherosclerosis,260,261 inflammation,262 hypertension,263,264 and dyslipidemia.265
Since the fly model system lacks a vasculature, it represents an excellent in vivo system for modeling direct effects of nutritional content and energy state on myocardial function, without complications from changes in vascular function. Fatty acids are the primary energy source for cardiomyocytes and during the aging process a decrease in fatty-acid usage has been observed,266 suggesting that ATP production may be compromised in aging hearts. This idea is consistent with age-related changes in myocardial expression of genes required for beta-oxidation.248,267
Recent work has also demonstrated that altering fatty-acid metabolism has potent effects on cardiac physiology in the fly. A dominant hypomorphic mutation in a gene encoding a conserved fatty-acid transporter, dFatp, has been examined for effects on a variety of stresses. Homologs of dFatp are necessary for long-chain fatty-acid uptake in a variety of species, from yeast to human.56,57 While null mutations for dFatp are lethal, hypomorphic mutations improve resistance to cold stress, heat stress, and starvation stress (Morley and Wessells, unpublished observation). Although dFatp hypomorphs display a dramatic increase in stored triglycerides and circulating fatty-acid levels, they nevertheless have an improved lifespan, not only on standard diets, but also on extreme high-fat diets, suggesting that they have an increased general resistance to lipotoxicity (Piazza and Wessells, unpublished observation).
The benefits of this mutation do not extend to cardiac function, however. Mutant flies have reduced cardiac contractility and reduced resistance to external pacing stress, consistent with the idea that they are compromised energetically. Defects in energetic function are tissue autonomous, since they can be reproduced by heart-specific expression of RNAi constructs against dFatp (Jennens and Wessells, unpublished observation) in otherwise wild-type flies. Overexpression of dFatp in the myocardium is also deleterious, producing phenotypes resembling lipotoxic cardiomyopathy. This is consistent with results in vertebrate models, where cardiac overexpression of the closest murine homolog of dFatp induces lipotoxicity and compromised cardiac function.268 Interestingly, the cardiac defects induced by hypomorphic mutation of dFatp are rescued by knocking down one copy of a gene required for triglyceride storage, suggesting that these phenotypes are not governed solely by the amount of transport activity available to the heart, but by the balance of transport activity available in comparison to the amount of circulating free fatty acids and energy requirements in times of stress. Excitingly, the detrimental cardiac phenotypes of reduced dFatp expression can also be rescued by inducing an endurance exercise program, which tends to also reduce stored triglycerides. These results highlight the value of the fruit fly model as a way for examining the impact of environmental factors on genetic diseases.
Indeed, upcoming years are likely to see a substantial increase in the usage of the fly system as a model for the complex interactions between diet, function, and aging. It is already becoming clear, as advances in assays for organ physiology in flies continue to become available, that lifespan extending mutations or interventions do not necessarily improve function concomitantly. Lifespan is a highly combinatorial readout for a wide ranging number of changes to the physiology of various organ systems. Mathematically speaking, lifespan extension may depend most directly on whether a particular intervention happens to improve aspects of physiology that are relevant to caged, laboratory lifespan. The effects on overall physiology or on wild-dwelling animals may in fact be quite different. In the previous example, as an illustration, reductions in overall fatty-acid intake and usage powerfully extend lifespan and resistance to external stresses, but the same animals are unlikely to survive for long in the wild, since they have especially low tolerance for temporary cardiac stresses such as are commonly experienced by wild animals, or city-dwelling humans, every day. The fly system, since it combines invertebrate genetic tools with functional assays for specific organs, such as the heart, offers an exceptional model system for scrutinizing details and trade-offs involved in various interventions that alter lifespan or cardiac performance.
Another development that will further this process in upcoming years is the development of flies as a model for exercise training. High levels of habitual exercise serve multiple modeling purposes. First, the ability of flies to undergo a training regimen is a useful assay to measure overall functional fitness of animals undergoing a genetic or environmental intervention. Second, flies undergoing training can be a useful model system to test the effects of varying exercise and diet levels on cardiac function across ages. Third, flies undergoing training can be a model to use in screens to identify genes that are necessary for the heart to experience functional improvement as a result of training.
Endurance exercise has long been known to produce improvements both to skeletal muscle function and to cardiac function in vertebrates. In addition to its preventive role, exercise training can even benefit patients already experiencing heart failure269,270 and protect against damage from ischemia and reperfusion.271 Improvements to vertebrate cardiac function following endurance training have been documented in both young and old animals.272,273 The Drosophila model, due to its relatively short lifespan, offers an excellent opportunity to study the effects of age across ages in long-term, longitudinal study designs.
A protocol for long-term endurance training has been devised for Drosophila. It relies on the instinct for negative geotaxis that causes flies to involuntarily run upwards when knocked to the bottom of a container. A machine, called the Power Tower, named after an amusement park ride with a similar motion, was built to take advantage of this instinct by repeatedly knocking flies down at regular intervals, inducing them to continuously run upwards as long as the machine is running217 (Fig. 11). A training paradigm was designed empirically that produces long-term improvements in mobility and cardiac stress resistance in multiple genotypes of wild-type flies.
Fig. 11.
Schematic representation of the Power Tower, a motor-operated enforced climbing apparatus that exercise-trains Drosophila. (A) As the rotation arm circles around in a clockwise direction, the lever pushes down and the two platforms of vial holders rise up. (B) As the rotating arm rolls-off, the lever lifts and the platform of vial holders drops down. The flies drop to the bottom of the vial, inducing their negative geotaxis instinct. The flies continue to repeat this process until the motor is shut-off. Exercise-training thus occurs through continuous climbing. (C) An image of the Power Tower.217
Flies that undergo endurance training gradually begin to show greater resistance to external pacing stress during and following training. Although no difference in rhythmicity was detected between cohorts, exercised groups displayed improvements in contractility and experienced a lower incidence of cardiac arrest in response to externally induced stress.217 In contrast to vertebrate studies, this improvement is only seen in flies that begin training at young ages. After a certain point of age-related decline is reached, flies are no longer able to execute the training program at the activity level necessary to induce benefits.
A hallmark of endurance training in humans and vertebrate models is a progressive alteration in mitochondrial number and efficiency.274,275 Benefits to cardiac function acquired during exercise training in flies do not seem to be dependent on alterations in mitochondrial number or efficiency, however. A fly line with an extraordinary degree of mitochondrial activity without increased generation of ROS276 displays dramatically improved mobility, with or without exercise, suggesting that improvements in mobility may be mediated in part by changes in mitochondrial activity in flies, as in vertebrates. However, these “mito-efficient” flies do not display a corresponding improvement in cardiac contractility or stress resistance, suggesting that the mechanisms by which endurance training improves cardiac function may be distinct from those by which it improves skeletal function.217
First steps have also been taken to begin to model the long-term effects of diet and exercise on cardiac function in combination. For example, different dietary constituents have been altered to form a matrix of varying dietary components. Flies were grown on each diet and resistance to external pacing stress tested throughout life as an indicator of cardiac functional health across ages on each diet (Morley and Wessells, unpublished). It was found that altering the level of carbohydrates in the diet had little, if any effect, on the normal age-related decline in cardiac stress resistance. However, reducing lipid content in the diet profoundly altered the aging profile of cardiac stress resistance. Reduction in yeast (source of both protein and lipid in the fly diet) has long been known to induce a lifespan extending dietary restriction process.235 Interestingly, the level of yeast reduction that produces lifespan extension has no effect on cardiac aging. However, if yeast is reduced still further, to a level that actually reduces lifespan, presumably through nutritional insufficiency, cardiac function is dramatically improved. At this reduced yeast level, cardiac stress resistance decreases much more slowly with age than in wild type (Morley and Wessells, unpublished), suggesting that cardiac function, especially under stress conditions, has quite different energy requirements from those influencing overall lifespan.
This difference becomes more pronounced when intensive exercise is introduced into the experimental design. Lifespan extension in response to diet has been shown to be more a function of the ratio of carbohydrate calories to protein and lipid calories than a function of actual caloric reduction.277 However, the ability to execute an endurance exercise program and derive benefits to cardiac function requires a much higher level of calories than the amount that is optimal for lifespan extension under inactive conditions. Furthermore, the amount of lipid contained in the diet is much more important to the ability to complete endurance exercise than the amount of carbohydrate.217
Significantly, there is also a qualitative difference between the effects of dietary restriction on cardiac performance and the effects of endurance exercise on cardiac performance. Dietary restriction benefits primarily late life control of rhythmicity, while endurance exercise benefits primarily late life retention of contractility and fractional shortening. This difference is reflected as well in the difference in resistance to external pacing between the two interventions, as dietary restricted flies have a reduced incidence of fibrillation events, while exercise trained flies have a reduced incidence of arrest events when paced (Morley and Wessells, unpublished observation217). Another important difference is that, at least in flies, dietary restriction does not improve measures of age-related functional decline such as mobility assays278 or cold tolerance,279 although it does extend lifespan.208 By contrast, endurance exercise does not increase maximal lifespan, but does improve several indices of cardiac and skeletal muscle function.217,280
Although endurance training in flies produces several physiological responses that resemble those observed in humans and vertebrate animal models, whether or not these effects occur through mechanisms that are genetically conserved is still an open question. A first step toward addressing this question has been to use a candidate gene approach to test whether genes known to be important in the vertebrate response to endurance exercise may also serve the same function in flies.
Perhaps, the best studied gene in the vertebrate exercise response is PGC1-α, a transcriptional cofactor that is strongly upregulated by exercise in both humans58 and in rats.59 Upregulation of PGC1-α causes an increase in mitochondrial biogenesis280 and an upregulation of fatty-acid oxidation.266
Overexpression of the fly homolog of PGC1-α, known as spargel, appears to provide some of the same benefits as an endurance training program. Flies overexpressing spargel in muscle experience an upregulation in global mitochondrial activity, an improvement in mobility, as measured by climbing assays, and an increase in oxidative stress resistance (Jennens and Wessells, unpublished observation).
Flies overexpressing spargel in the myocardium display a dramatic improvement in resistance to external pacing stress across ages. Interestingly, although spargel overexpression and endurance exercise provide a similar magnitude of pacing stress resistance, if spargel overexpressing flies are also subjected to endurance training, they receive a further increase in the magnitude of stress resistance (Jennens and Wessells, unpublished observation). This is consistent with a model in which spargel is involved in a conserved genetic response to endurance exercise that provides benefits to cardiac stress resistance, but that other, as yet unidentified genes are also necessary to provide the full potential benefits of this intervention. Future work using this model will detail further which aspects of cardiac function are altered by exercise or by spargel expression. The way is now open, as well, to utilize the traditional strength of the fly model system, which is to do unbiased screens to identify novel loci that are necessary or sufficient to induce cardioprotective effects of endurance exercise.
The use of adult flies as a model for functional aging, diet, and exercise is only in its infancy, as is the newly feasible use of flies as a model for adult cardiac physiology. It is to be expected, however, that many more researchers will begin to utilize this model system in these contexts. Eventually, the fly system may prove to be just as important a model for adult physiology and aging as it has been historically in the study of development.
XIV. Summary
The Drosophila model system has evolved into a powerful model for multiple aspects of cardiac function and disease. Using classical genetics, lineage tracing, and confocal microscopy, flies have been used to define conserved signaling pathways regulating the formation of the cardiac field, controlling migration of cardiac cells to the midline, governing repolarization of the cellular arrangement and fusion of opposing cells to form the heart tube, and instituting a cardiogenic network of genes that specify specific functional subsets of cardioblasts.
The larval stage of Drosophila development has been a fruitful model for isolating the effects of various genetic alterations on current formation and propagation of contractile waves, as well as the maintenance of rhythmic control. The development of isolated preps with beating hearts under physiological conditions has made possible the use of the larva as a model to test the effects of various biological and pharmacological compounds on cardiac rate, rhythm, and contractile force.
Novel methods for visualizing adult cardiac function and for measuring cardiac stress resistance in adults have led to further use for the fly model. Several fly lines have been created that genetically model specific human diseases that impair cardiac function. In some cases, these are genomic mutants, while in others, cardiac-specific gene alteration produces cardiac phenotypes in isolation from the rest of the body.
The ability to track cardiac function across ages in adult flies has led to the use of the fly as a model for cardiac aging. Several conserved genetic pathways have been identified that alter the rate of age-related change in cardiac performance in flies. The ease of maintenance and short lifespan of the fly have made it possible to begin using the fly to model the effects of alterations and diet and exercise level on cardiac function across ages. The intersection between diet, exercise, and aging is of critical importance to addressing the continued increase in heart disease linked to obesity and insulin resistance in the world’s population. As a model system, flies are well suited for dealing with multiple interventions simultaneously in large numbers. They are also well suited for tracking lifelong effects of such interventions. As such, we expect that flies will be used even more frequently in upcoming years as a valuable tool for understanding these interrelated problems.
References
- 1.Tao Y, Schulz RA. Heart development in Drosophila. Semin Cell Dev Biol. 2007;18:3–15. doi: 10.1016/j.semcdb.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Medioni C, Astier M, Zmojdzian M, Jagla K, Sémériva M. Genetic control of cell morphogenesis during Drosophila melanogaster cardiac tube formation. J Cell Biol. 2008;182:249–261. doi: 10.1083/jcb.200801100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodmer R. Heart development in Drosophila and its relationship to vertebrate systems. Trends Cardiovasc Med. 1995;5:21–28. doi: 10.1016/1050-1738(94)00032-Q. [DOI] [PubMed] [Google Scholar]
- 4.Garg V. Insights into the genetic basis of congenital heart disease. Cell Mol Life Sci. 2006;63:1141–1148. doi: 10.1007/s00018-005-5532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryantsev AL, Cripps RM. Cardiac gene regulatory networks in Drosophila. Biochim Biophys Acta. 2009;1789:343–353. doi: 10.1016/j.bbagrm.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodmer R, Wessells RJ, Johnson EC, Dowse H. Heart development and function. In: Gilbert LI, Iatrou K, Gills SS, editors. Comprehensive molecular insect science. Vol. 2. Amsterdam: Elsevier; 2005. pp. 199–250. Volumes 107. [Google Scholar]
- 7.Wessells RJ, Bodmer R. Cardiac aging. Semin Cell Dev Biol. 2007;18:111–116. doi: 10.1016/j.semcdb.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 8.Dunin-Borkowski OM, Brown NH, Bate M. Anterior–posterior subdivision and the diversification of the mesoderm in Drosophila. Development. 1995;121:4183–4193. doi: 10.1242/dev.121.12.4183. [DOI] [PubMed] [Google Scholar]
- 9.Lawrence PA, Bodmer R, Vincent JP. Segmental patterning of heart precursors in Drosophila. Development. 1995;121:4303–4308. doi: 10.1242/dev.121.12.4303. [DOI] [PubMed] [Google Scholar]
- 10.Wu X, Golden K, Bodmer R. Heart development in Drosophila requires the segment polarity gene wingless. Dev Biol. 1995;169:619–628. doi: 10.1006/dbio.1995.1174. [DOI] [PubMed] [Google Scholar]
- 11.Park M, Wu X, Golden K, Axelrod JD, Bodmer R. The wingless signaling pathway is directly involved in Drosophila heart development. Dev Biol. 1996;177:104–116. doi: 10.1006/dbio.1996.0149. [DOI] [PubMed] [Google Scholar]
- 12.Rugendorff A, Younossi-Hartenstein A, Hartenstein V. Embryonic origin and differentiation of the Drosophila heart. Rouxs Arch Dev Biol. 1994;203:266–280. doi: 10.1007/BF00360522. [DOI] [PubMed] [Google Scholar]
- 13.Frasch M. Induction of visceral and cardiac mesoderm by ectodermal Dpp in the early Drosophila embryo. Nature. 1995;374:464–467. doi: 10.1038/374464a0. [DOI] [PubMed] [Google Scholar]
- 14.Azpiazu N, Frasch M. Tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993;7:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- 15.Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- 16.Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, et al. Congenital heart disease caused by mutations in the transcription factor NKX2–5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- 17.Lee HH, Frasch M. Nuclear integration of positive Dpp signals, antagonistic Wg inputs and mesodermal competence factors during Drosophila visceral mesoderm induction. Development. 2005;132:1429–1442. doi: 10.1242/dev.01687. [DOI] [PubMed] [Google Scholar]
- 18.Reifers F, Walsh EC, Léger S, Stainier DY, Brand M. Induction and differentiation of the zebrafish heart require fibroblast growth factor 8 (fgf8/acerebellar) Development. 2000;127:225–235. doi: 10.1242/dev.127.2.225. [DOI] [PubMed] [Google Scholar]
- 19.Alsan BH, Schultheiss TM. Regulation of avian cardiogenesis by Fgf8 signaling. Development. 2002;129:1935–1943. doi: 10.1242/dev.129.8.1935. [DOI] [PubMed] [Google Scholar]
- 20.Marques SR, Lee Y, Poss KD, Yelon D. Reiterative roles for FGF signaling in the establishment of size and proportion of the zebrafish heart. Dev Biol. 2008;321:397–406. doi: 10.1016/j.ydbio.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shishido E, Ono N, Kojima T, Saigo K. Requirements of DFR1/heartless, a mesoderm-specific Drosophila FGF-receptor, for the formation of heart, visceral and somatic muscles, and ensheathing of longitudinal axon tracts in CNS. Development. 1997;124:2119–2128. doi: 10.1242/dev.124.11.2119. [DOI] [PubMed] [Google Scholar]
- 22.Gisselbrecht S, Skeath JB, Doe CQ, Michelson AM. Heartless encodes a fibroblast growth factor receptor (DFR1/DFGF-R2) involved in the directional migration of early mesodermal cells in the Drosophila embryo. Genes Dev. 1996;10:3003–3017. doi: 10.1101/gad.10.23.3003. [DOI] [PubMed] [Google Scholar]
- 23.Fossett N, Tevosian SG, Gajewski K, Zhang Q, Orkin SH, Schulz RA. The friend of GATA proteins U-shaped, FOG–1, and FOG–2 function as negative regulators of blood, heart, and eye development in Drosophila. Proc Natl Acad Sci USA. 2001;19:7342–7347. doi: 10.1073/pnas.131215798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klinedinst SL, Bodmer R. Gata factor Pannier is required to establish competence for heart progenitor formation. Development. 2003;130:3027–3038. doi: 10.1242/dev.00517. [DOI] [PubMed] [Google Scholar]
- 25.Lo PC, Skeath JB, Gajewski K, Schulz RA, Frasch M. Homeotic genes autonomously specify the anteroposterior subdivision of the Drosophila dorsal vessel into aorta and heart. Dev Biol. 2002;251:307–319. doi: 10.1006/dbio.2002.0839. [DOI] [PubMed] [Google Scholar]
- 26.Lo PC, Frasch M. A role for the COUP-TF-related gene seven-up in the diversification of cardioblast identities in the dorsal vessel of Drosophila. Mech Dev. 2001;104:49–60. doi: 10.1016/s0925-4773(01)00361-6. [DOI] [PubMed] [Google Scholar]
- 27.Perrin L, Monier B, Ponzielli R, Astier M, Semeriva M. Drosophila cardiac tube organogenesis requires multiple phases of Hox activity. Dev Biol. 2004;272:419–431. doi: 10.1016/j.ydbio.2004.04.036. [DOI] [PubMed] [Google Scholar]
- 28.Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 29.Zikova M, Da Ponte JP, Dastugue B, Jagla K. Patterning of the cardiac outflow region in Drosophila. Proc Natl Acad Sci USA. 2003;100:12189–12194. doi: 10.1073/pnas.2133156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schäfer K, Neuhaus P, Kruse J, Braun T. The homeobox gene Lbx1 specifies a subpopulation of cardiac neural crest necessary for normal heart development. Circ Res. 2003;92:73–80. doi: 10.1161/01.res.0000050587.76563.a5. [DOI] [PubMed] [Google Scholar]
- 31.Sanyal S, Jennings T, Dowse H, Ramaswami M. Conditional mutations in SERCA, the sarco-endoplasmic reticulum Ca2+ -ATPase, alter heart rate and rhythmicity in Drosophila. J Comp Physiol. 2006;176:252–263. doi: 10.1007/s00360-005-0046-7. [DOI] [PubMed] [Google Scholar]
- 32.Chamberlain JS, Metzger J, Reyes M, Townsend D, Faulkner JA. Dystrophin-deficient mdx mice display a reduced life span and are susceptible to spontaneous rhabdomyosarcoma. FASEB J. 2007;21:2195–2204. doi: 10.1096/fj.06-7353com. [DOI] [PubMed] [Google Scholar]
- 33.Taghli-Lamallem O, Akasaka T, Hogg G, Nudel U, Yaffe D, Chamberlain JS, et al. Dystrophin deficiency in Drosophila reduces lifespan and causes a dilated cardiomyopathy phenotype. Aging Cell. 2008;7:237–249. doi: 10.1111/j.1474-9726.2008.00367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu X, Wheeler MT, Hadhazy M, Lam MY, McNally EM. Cardiomyopathy is independent of skeletal muscle disease in muscular dystrophy. FASEB J. 2002;16:1096–1098. doi: 10.1096/fj.01-0954fje. [DOI] [PubMed] [Google Scholar]
- 35.Wheeler MT, Allikian MJ, Heydemann A, Hadhazy M, Zarnegar S, McNally EM. Smooth muscle cell-extrinsic vascular spasm arises from cardiomyocyte degeneration in sarcoglycan-deficient cardiomyopathy. J Clin Invest. 2004;113:668–675. doi: 10.1172/JCI20410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allikian MJ, Bhabha G, Dospoy P, Heydemann A, Ryder P, Earley JU, et al. Reduced life span with heart and muscle dysfunction in Drosophila sarcoglycan mutants. Hum Mol Genet. 2007;16:2933–2943. doi: 10.1093/hmg/ddm254. [DOI] [PubMed] [Google Scholar]
- 37.Cammarato A, Dambacher CM, Knowles AF, Kronert WA, Bodmer R, Ocorr K, et al. Myosin transducer mutations differentially affect motor function, myofibril structure, and the performance of skeletal and cardiac muscles. Mol Biol Cell. 2008;19:553–562. doi: 10.1091/mbc.E07-09-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geeves MA, Holmes KC. The molecular mechanism of muscle contraction. Adv Protein Chem. 2005;71:161–193. doi: 10.1016/S0065-3233(04)71005-0. [DOI] [PubMed] [Google Scholar]
- 39.Oldfors A. Hereditary myosin myopathies. Neuromuscul Disord. 2007;17:355–367. doi: 10.1016/j.nmd.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 40.Bernstein SI, Mogami K, Donady JJ, Emerson CP., Jr Drosophila muscle myosin heavy chain encoded by a single gene in a cluster of muscle mutations. Nature. 1983;302:393–397. doi: 10.1038/302393a0. [DOI] [PubMed] [Google Scholar]
- 41.Bernstein SI, Hansen CJ, Becker KD, Wassenberg DR, 2nd, Roche ES, Donady JJ, et al. Alternative RNA splicing generates transcripts encoding a thorax-specific isoform of Drosophila melanogaster myosin heavy chain. Mol Cell Biol. 1986;1986:2511–2519. doi: 10.1128/mcb.6.7.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ocorr K, Akasaka T, Bodmer R. Age-related cardiac disease model of Drosophila. Mech Ageing Dev. 2007;1:112–116. doi: 10.1016/j.mad.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanguinetti MC, Curran ME, Zou A, Shen J, Spector PS, Atkinson DL, et al. Coassembly of K (V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature. 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- 44.Priori SG, Napolitano C, Schwartz PJ, Grillo M, Bloise R, Ronchetti E, et al. Association of long QT syndrome loci and cardiac events among patients treated with beta-blockers. JAMA. 2004;292:1341–1344. doi: 10.1001/jama.292.11.1341. [DOI] [PubMed] [Google Scholar]
- 45.Roberts R. Genomics and cardiac arrhythmias. J Am Coll Cardiol. 2006;47:9–21. doi: 10.1016/j.jacc.2005.08.059. [DOI] [PubMed] [Google Scholar]
- 46.Fujioka M, Wessells RJ, Han Z, Liu J, Fitzgerald K, Yusibova GL, et al. Embryonic even skipped-dependent muscle and heart cell fates are required for normal adult activity, heart function, and lifespan. Circ Res. 2005;97:1108–1114. doi: 10.1161/01.RES.0000191546.08532.B2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crone SA, Zhao YY, Fan L, Gu Y, Minamisawa S, Liu Y, et al. ErbB2 is essential in the prevention of dilated cardiomyopathy. Nat Med. 2002;8:459–465. doi: 10.1038/nm0502-459. [DOI] [PubMed] [Google Scholar]
- 48.Lemmens K, Doggen K, De Keulenaer GW. Role of neuregulin-1/ErbB signaling in cardiovascular physiology and disease: implications for therapy of heart failure. Circulation. 2007;116:954–960. doi: 10.1161/CIRCULATIONAHA.107.690487. [DOI] [PubMed] [Google Scholar]
- 49.Ozcelik C, Erdmann B, Pilz B, Wettschureck N, Britsch S, Hübner N, et al. Conditional mutation of the ErbB2 (HER2) receptor in cardiomyocytes leads to dilated cardiomyopathy. Proc Natl Acad Sci USA. 2002;99:8880–8885. doi: 10.1073/pnas.122249299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carelli V, Ross-Cisneros FN, Sadun AA. Mitochondrial dysfunction as a cause of optic neuropathies. Prog Retin Eye Res. 2004;23:53–89. doi: 10.1016/j.preteyeres.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 51.Shahrestani P, Leung HT, Le PK, Pak WL, Tse S, Ocorr K, et al. Heterozygous mutation of Drosophila Opa1 causes the development of multiple organ abnormalities in an age-dependent and organ-specific manner. PLoS One. 2009;4:e6867. doi: 10.1371/journal.pone.0006867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y, Huang TT, Carlson EJ, Meloy S, Ursell PC, Olson JL, et al. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 53.Duttaroy A, Paul A, Kundu M, Belton A. A Sod2 null mutation confers severely reduced adult life span in Drosophila. Genetics. 2003;165:2295–2299. doi: 10.1093/genetics/165.4.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee JH, Budanov AV, Park EJ, Birse R, Kim TE, Perkins GA, et al. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science. 2010;327:1223–1228. doi: 10.1126/science.1182228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wessells R, Fitzgerald E, Piazza N, Ocorr K, Morley S, Davies C, et al. d4eBP acts downstream of both dTOR and dFoxo to modulate cardiac functional aging in Drosophila. Aging Cell. 2009;8:542–552. doi: 10.1111/j.1474-9726.2009.00504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Faergeman NJ, DiRusso CC, Elberger A, Knudsen J, Black PN. Disruption of the Saccharomyces cerevisiae homologue to the murine fatty acid transport protein impairs uptake and growth on long-chain fatty acids. J Biol Chem. 1997;272:8531–8538. doi: 10.1074/jbc.272.13.8531. [DOI] [PubMed] [Google Scholar]
- 57.Herrmann T, Buchkremer F, Gosch I, Hall AM, Berniohr DA, Stremmel W. Mouse fatty acid transport protein 4 (FAT4): characterization of the gene and functional assessment as a very long chain acyl-CoA synthetase. Gene. 2001;270:31–40. doi: 10.1016/s0378-1119(01)00489-9. [DOI] [PubMed] [Google Scholar]
- 58.Pilegaard H, Saltin B, Neufer PD. Exercise induces transient transcriptional activation of the PGC-1alpha gene in human skeletal muscle. J Physiol. 2003;546:851–858. doi: 10.1113/jphysiol.2002.034850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goto M, Terada S, Kato M, Katoh M, Yokozeki T, Tabata I, et al. cDNA cloning and mRNA analysis of PGC1 in epitrochlearis muscle in swimming-exercised rats. Biochem Biophys Res Commun. 2000;274:350–354. doi: 10.1006/bbrc.2000.3134. [DOI] [PubMed] [Google Scholar]
- 60.Lockwood WK, Bodmer R. The patterns of wingless, decapentaplegic, and tinman position the Drosophila heart. Mech Dev. 2002;114:13–26. doi: 10.1016/s0925-4773(02)00044-8. [DOI] [PubMed] [Google Scholar]
- 61.Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 62.Schultheiss TM, Burch JB, Lassar AB. A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev. 1997;15:451–462. doi: 10.1101/gad.11.4.451. [DOI] [PubMed] [Google Scholar]
- 63.Shi Y, Katsev S, Cai C, Evans S. BMP signaling is required for heart formation in vertebrates. Dev Biol. 2000;224:226–237. doi: 10.1006/dbio.2000.9802. [DOI] [PubMed] [Google Scholar]
- 64.Song L, Fässler R, Mishina Y, Jiao K, Baldwin HS. Essential functions of Alk3 during AV cushion morphogenesis in mouse embryonic hearts. Dev Biol. 2007;301:276–286. doi: 10.1016/j.ydbio.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 65.Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, et al. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2–5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 66.Lien CL, McAnally J, Richardson JA, Olson EN. Cardiac-specific activity of an Nkx2–5 enhancer requires an evolutionarily conserved Smad binding site. Dev Biol. 2002;244:257–266. doi: 10.1006/dbio.2002.0603. [DOI] [PubMed] [Google Scholar]
- 67.Tzahor E, Lassar AB. Wnt signals from the neural tube block ectopic cardiogenesis. Genes Dev. 2001;15:255–260. doi: 10.1101/gad.871501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marvin MJ, Di Rocco G, Gardiner A, Bush SM, Lassar AB. Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev. 2001;15:316–327. doi: 10.1101/gad.855501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schneider VA, Mercola M. Wnt antagonism initiates cardiogenesis in Xenopus laevis. Genes Dev. 2001;15:304–315. doi: 10.1101/gad.855601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Foley AC, Mercola M. Heart induction by Wnt antagonists depends on the homeodomain transcription factor Hex. Genes Dev. 2005;19:387–396. doi: 10.1101/gad.1279405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tzahor E. Wnt/beta-catenin signaling and cardiogenesis: timing does matter. Dev Cell. 2007;13:10–13. doi: 10.1016/j.devcel.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 72.Kwon C, Cordes KR, Srivastava D. Wnt/beta-catenin signaling acts at multiple developmental stages to promote mammalian cardiogenesis. Cell Cycle. 2008;7:3815–3818. doi: 10.4161/cc.7.24.7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jaspard B, Couffinhal T, Dufourcq P, Moreau C, Duplhàa C. Expression pattern of mouse sFRP-1 and mWnt-8 gene during heart morphogenesis. Mech Dev. 2000;90:263–267. doi: 10.1016/s0925-4773(99)00236-1. [DOI] [PubMed] [Google Scholar]
- 74.Garriock RJ, D’Agostino SL, Pilcher KC, Krieg PA. Wnt11-R, a protein closely related to mammalian Wnt11, is required for heart morphogenesis in Xenopus. Dev Biol. 2005;279:179–192. doi: 10.1016/j.ydbio.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 75.Ueno S, Weidinger G, Osugi T, Kohn AD, Golob JL, Pabon L, et al. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pandur P, Läsche M, Eisenberg LM, Kühl M. Wnt-11 activation of a non-canonical Wnt signalling pathway is required for cardiogenesis. Nature. 2002;418:636–641. doi: 10.1038/nature00921. [DOI] [PubMed] [Google Scholar]
- 77.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reim I, Frasch M. Genetic and genomic dissection of cardiogenesis in the Drosophila model. Pediatr Cardiol. 2010;31:325–334. doi: 10.1007/s00246-009-9612-1. [DOI] [PubMed] [Google Scholar]
- 79.Ponzielli R, Astier M, Chartier A, Gallet A, Thérond P, Sémériva M. Heart tube patterning in Drosophila requires integration of axial and segmental information provided by the Bithorax Complex genes and hedgehog signaling. Development. 2002;129:4509–4521. doi: 10.1242/dev.129.19.4509. [DOI] [PubMed] [Google Scholar]
- 80.Monier B, Astier M, Sémériva M, Perrin L. Steroid-dependent modification of Hox function drives myocyte reprogramming in the Drosophila heart. Development. 2005;132:5283–5293. doi: 10.1242/dev.02091. [DOI] [PubMed] [Google Scholar]
- 81.Molina MR, Cripps RM. Ostia, the inflow tracts of the Drosophila heart, develop from a genetically distinct subset of cardial cells. Mech Dev. 2001;109:51–59. doi: 10.1016/s0925-4773(01)00509-3. [DOI] [PubMed] [Google Scholar]
- 82.Curtis NJ, Ringo JM, Dowse HB. Morphology of the pupal heart, adult heart, and associated tissues in the fruit fly, Drosophila melanogaster. J Morphol. 1999;240:225–235. doi: 10.1002/(SICI)1097-4687(199906)240:3<225::AID-JMOR2>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 83.Zaffran S, Reim I, Qian L, Lo PC, Bodmer R, Frasch M. Cardioblast-intrinsic tinman activity controls proper diversification and differentiation of myocardial cells in Drosophila. Development. 2006;133:4073–4083. doi: 10.1242/dev.02586. [DOI] [PubMed] [Google Scholar]
- 84.Reim I, Mohler JP, Frasch M. Tbx20-related genes, mid and H15, are required for tinman expression, proper patterning, and normal differentiation of cardioblasts in Drosophila. Mech Dev. 2005;122:1056–1069. doi: 10.1016/j.mod.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 85.Miskolczi-McCallum CM, Scavetta RJ, Svendsen PC, Soanes KH, Brook WJ. The Drosophila melanogaster T-box genes midline and H15 are conserved regulators of heart development. Dev Biol. 2005;278:459–472. doi: 10.1016/j.ydbio.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 86.Qian L, Liu J, Bodmer R. Neuromancer Tbx20-related genes (H15/midline) promote cell fate specification and morphogenesis of the Drosophila heart. Dev Biol. 2005;279:509–524. doi: 10.1016/j.ydbio.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 87.Gajewski K, Fossett N, Molkentin JD, Schulz RA. The zinc finger proteins Pannier and GATA4 function as cardiogenic factors in Drosophila. Development. 1999;126:5679–5688. doi: 10.1242/dev.126.24.5679. [DOI] [PubMed] [Google Scholar]
- 88.Gajewski K, Zhang Q, Choi CY, Fossett N, Dang A, Kim YH, et al. Pannier is a transcriptional target and partner of tinman during Drosophila cardiogenesis. Dev Biol. 2001;233:425–436. doi: 10.1006/dbio.2001.0220. [DOI] [PubMed] [Google Scholar]
- 89.Alvarez AD, Shi W, Wilson BA, Skeath JB. Pannier and pointedP2 act sequentially to regulate Drosophila heart development. Development. 2003;130:3015–3026. doi: 10.1242/dev.00488. [DOI] [PubMed] [Google Scholar]
- 90.Klinedinst SL, Bodmer R. Gata factor Pannier is required to establish competence for heart progenitor formation. Development. 2003;130:3027–3038. doi: 10.1242/dev.00517. [DOI] [PubMed] [Google Scholar]
- 91.Reim I, Frasch M. The Dorsocross T-box genes are key components of the regulatory network controlling early cardiogenesis in Drosophila. Development. 2005;132:4911–4925. doi: 10.1242/dev.02077. [DOI] [PubMed] [Google Scholar]
- 92.Han Z, Yi P, Li X, Olson EN. Hand, an evolutionarily conserved bHLH transcription factor required for Drosophila cardiogenesis and hematopoiesis. Development. 2006;133:1175–1182. doi: 10.1242/dev.02285. [DOI] [PubMed] [Google Scholar]
- 93.Kremser T, Hasenpusch-Theil K, Wagner E, Buttgereit D, Renkawitz-Pohl R. Expression of the beta3 tubulin gene (beta Tub60D) in the visceral mesoderm of Drosophila is dependent on a complex enhancer that binds tinman and UBX. Mol Gen Genet. 1999;262:643–658. doi: 10.1007/s004380051127. [DOI] [PubMed] [Google Scholar]
- 94.Nasonkin I, Alikasifoglu A, Ambrose C, Cahill P, Cheng M, Sarniak A, et al. A novel sulfonylurea receptor family member expressed in the embryonic Drosophila dorsal vessel and tracheal system. J Biol Chem. 1999;274:29420–29425. doi: 10.1074/jbc.274.41.29420. [DOI] [PubMed] [Google Scholar]
- 95.Akasaka T, Klinedinst S, Ocorr K, Bustamante EL, Kim SK, Bodmer R. The ATP-sensitive potassium (KATP) channel-encoded dSUR gene is required for Drosophila heart function and is regulated by tinman. Proc Natl Acad Sci USA. 2006;103:11999–12004. doi: 10.1073/pnas.0603098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Han Z, Olson EN. Hand is a direct target of tinman and GATA factors during Drosophila cardiogenesis and hematopoiesis. Development. 2005;132:3525–3536. doi: 10.1242/dev.01899. [DOI] [PubMed] [Google Scholar]
- 97.Gajewski K, Kim Y, Lee YW, Olson EN, Schulz RA. D-mef2 is a target for tinman activation during Drosophila heart development. EMBO J. 1997;16:515–522. doi: 10.1093/emboj/16.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Goldmuntz E, Geiger E, Benson DW. NKX2.5 mutations in patients with tetralogy of fallot. Circulation. 2001;104:2565–2568. doi: 10.1161/hc4601.098427. [DOI] [PubMed] [Google Scholar]
- 99.Maitra M, Schluterman MK, Nichols HA, Richardson JA, Lo CW, Srivastava D, et al. Interaction of Gata4 and Gata6 with Tbx5 is critical for normal cardiac development. Dev Biol. 2009;326:368–377. doi: 10.1016/j.ydbio.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tu CT, Yang TC, Tsai HJ. Nkx2.7 and Nkx2.5 function redundantly and are required for cardiac morphogenesis of zebrafish embryos. PLoS One. 2009;4:e4249. doi: 10.1371/journal.pone.0004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Singh MK, Li Y, Cobb RM, Zhou D, Lu MM, Epstein JA, et al. Gata4 and Gata5 cooperatively regulate cardiac myocyte proliferation in mice. J Biol Chem. 2010;285:1765–1772. doi: 10.1074/jbc.M109.038539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- 103.Haag TA, Haag NP, Lekven AC, Hartenstein V. The role of cell adhesion molecules in Drosophila heart morphogenesis: faint sausage, shotgun/DE-cadherin, and laminin A are required for discrete stages in heart development. Dev Biol. 1999;208:56–59. doi: 10.1006/dbio.1998.9188. [DOI] [PubMed] [Google Scholar]
- 104.Wang J, Tao Y, Reim I, Gajewski K, Frasch M, Schulz RA. Expression, regulation, and requirement of the toll transmembrane protein during dorsal vessel formation in Drosophila melanogaster. Mol Cell Biol. 2005;25:4200–4210. doi: 10.1128/MCB.25.10.4200-4210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yarnitsky T, Volk T. Laminin is required for heart, somatic muscles, and gut development in the Drosophila embryo. Dev Biol. 1995;169:609–618. doi: 10.1006/dbio.1995.1173. [DOI] [PubMed] [Google Scholar]
- 106.Tepass U. Epithelial differentiation in Drosophila. Bioessays. 1997;19:673–682. doi: 10.1002/bies.950190807. [DOI] [PubMed] [Google Scholar]
- 107.Stark KA, Yee GH, Roote CE, Williams EL, Zusman S, Hynes RO. A novel alpha integrin subunit associates with betaPS and functions in tissue morphogenesis and movement during Drosophila development. Development. 1997;124:4583–4594. doi: 10.1242/dev.124.22.4583. [DOI] [PubMed] [Google Scholar]
- 108.Frémion F, Astier M, Zaffran S, Guiellèn A, Homburger V, Sémériva M. The heterotrimeric protein Go is required for the formation of heart epithelium in Drosophila. J Cell Biol. 1999;145:1063–1076. doi: 10.1083/jcb.145.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chartier A, Zaffran S, Astier M, Sémériva M, Gratecos D. Pericardin, a Drosophila type IV collagen-like protein is involved in the morphogenesis and maintenance of the heart epithelium during dorsal ectoderm closure. Development. 2002;129:3241–3253. doi: 10.1242/dev.129.13.3241. [DOI] [PubMed] [Google Scholar]
- 110.Qian L, Liu J, Bodmer R. Slit and Robo control cardiac cell polarity and morphogenesis. Curr Biol. 2005;15:2271–2278. doi: 10.1016/j.cub.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 111.Santiago-Martínez E, Soplop NH, Kramer SG. Lateral positioning at the dorsal midline: slit and roundabout receptors guide Drosophila heart cell migration. Proc Natl Acad Sci USA. 2006;103:12441–12446. doi: 10.1073/pnas.0605284103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Santiago-Martínez E, Soplop NH, Patel R, Kramer SG. Repulsion by Slit and Roundabout prevents Shotgun/E-cadherin-mediated cell adhesion during Drosophila heart tube lumen formation. J Cell Biol. 2008;182:241–248. doi: 10.1083/jcb.200804120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zmojdzian M, Da Ponte JP, Jagla K. Cellular components and signals required for the cardiac outflow tract assembly in Drosophila. Proc Natl Acad Sci USA. 2008;105:2475–2480. doi: 10.1073/pnas.0706402105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yi P, Han Z, Li X, Olson EN. The mevalonate pathway controls heart formation in Drosophila by isoprenylation of Ggamma1. Science. 2006;313:1301–1303. doi: 10.1126/science.1127704. [DOI] [PubMed] [Google Scholar]
- 115.Yi P, Johnson AN, Han Z, Wu J, Olson EN. Heterotrimeric G proteins regulate a noncanonical function of septate junction proteins to maintain cardiac integrity in Drosophila. Dev Cell. 2008;15:704–713. doi: 10.1016/j.devcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Horowitz A, Simons M. Branching morphogenesis. Circ Res. 2008;103:784–795. doi: 10.1161/CIRCRESAHA.108.181818. [DOI] [PubMed] [Google Scholar]
- 117.Chan PK, Lin CC, Cheng SH. Noninvasive technique for measurement of heartbeat regularity in zebrafish (Danio rerio) embryos. BMC Biotechnol. 2009;19:11. doi: 10.1186/1472-6750-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Miller TA. Comprehensive insect physiology, biochemistry and pharmacology. Vol 3, Integument, respiration and circulation. Oxford: Pergamon Press; 1985. Structure and physiology of the circulatory system; pp. 289–353. [Google Scholar]
- 119.Rizki TM. The circulatory system and associated cells and tissues. In: Ashbumer M, Wright TRF, editors. The genetics and biology of Drosophila. New York: Academic Press; 1978. pp. 397–452. [Google Scholar]
- 120.Siegmund T, Korge G. Innervation of the ring gland of Drosophila melanogaster. J Comp Neurol. 2001;431:481–491. doi: 10.1002/1096-9861(20010319)431:4<481::aid-cne1084>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 121.Dulcis D, Levine RB. Innervation of the heart of the adult fruit fly, Drosophila melanogaster. J Comp Neurol. 2003;465:560–578. doi: 10.1002/cne.10869. [DOI] [PubMed] [Google Scholar]
- 122.Dowse H, Ringo J, Power J, Johnson E, Kinney K, White L. A congenital heart defect in Drosophila caused by an action-potential mutation. J Neurogenet. 1995;10:153–168. doi: 10.3109/01677069509083461. [DOI] [PubMed] [Google Scholar]
- 123.Wessells RJ, Bodmer R. Screening assays for heart function mutants in Drosophila. Biotechniques. 2004;37:58–60. doi: 10.2144/04371ST01. 62, 64. [DOI] [PubMed] [Google Scholar]
- 124.Johnson E, Ringo J, Dowse H. Modulation of Drosophila heartbeat by neurotransmitters. J Comp Physiol. 1997;167:89–97. doi: 10.1007/s003600050051. [DOI] [PubMed] [Google Scholar]
- 125.Johnson E, Ringo J, Bray N, Dowse H. Genetic and pharmacological identification of ion channels central to the Drosophila cardiac pacemake. J Neurogenet. 1998;12:1–24. doi: 10.3109/01677069809108552. [DOI] [PubMed] [Google Scholar]
- 126.Papaefthmiou C, Theophilidis G. An in vitro method for recording the electrical activity of the isolated heart of the adult Drosophila melanogaster. In Vitro Cell Dev Biol Anim. 2001;37:445–449. doi: 10.1290/1071-2690(2001)037<0445:aivmfr>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 127.Cooper AS, Rymond KE, Ward MA, Bocook EL, Cooper RL. Monitoring heart function in larval Drosophila melanogaster for physiological studies. J Vis Exp. 2009;16:1596. doi: 10.3791/1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Vogler G, Ocorr K. Visualizing the beating heart in Drosophila. J Vis Exp. 2009;28:1425. doi: 10.3791/1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fink M, Callol-Massot C, Chu A, Ruiz-Lozano P, Izpisua Belmonte JC, Giles W, et al. A new method for detection and quantification of heartbeat parameters in Drosophila, zebrafish and embryonic mouse hearts. Biotechniques. 2009;46:101–113. doi: 10.2144/000113078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ocorr K, Fink M, Cammarato A, Bernstein S, Bodmer R. Semi-automated optical heartbeat analysis of small hearts. J Vis Exp. 2009;16:1435. doi: 10.3791/1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bradu A, Ma L, Bloor JW, Podoleanu A. Dual optical coherence tomography/fluorescence microscopy for monitoring Drosophila melanogaster larval heart. J Biophotonics. 2009;2:380–388. doi: 10.1002/jbio.200910021. [DOI] [PubMed] [Google Scholar]
- 132.Gu GG, Singh S. Pharmacological analysis of heartbeat in Drosophila. J Neurobiol. 1995;28:269–280. doi: 10.1002/neu.480280302. [DOI] [PubMed] [Google Scholar]
- 133.Sullivan KM, Scott K, Zuker CS, Rubin GM. The ryanodine receptor is essential for larval development in Drosophila melanogaster. Proc Natl Acad Sci USA. 2000;97:5942–5947. doi: 10.1073/pnas.110145997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Noyes BE, Katz FN, Schaffer MH. Identification and expression of the Drosophila adipokinetic hormone gene. Mol Cell Endocrinol. 1995;109:133–141. doi: 10.1016/0303-7207(95)03492-p. [DOI] [PubMed] [Google Scholar]
- 135.Zornik E, Paisley K, Nichols R. Neural transmitters and a peptide Drosophila heart rate. Peptides. 1999;20:45–51. doi: 10.1016/s0196-9781(98)00151-x. [DOI] [PubMed] [Google Scholar]
- 136.Dasari S, Cooper RL. Direct influence of serotonin on the larval heart of Drosophila melanogaster. J Comp Physiol. 2006;176:349–357. doi: 10.1007/s00360-005-0058-3. [DOI] [PubMed] [Google Scholar]
- 137.Johnson E, Ringo J, Dowse H. Native and heterologous neuropeptides are cardioactive in Drosophila melanogaster. J Insect Physiol. 2000;46:1229–1236. doi: 10.1016/s0022-1910(00)00043-3. [DOI] [PubMed] [Google Scholar]
- 138.Nichols R, Kaminski S, Wallin E, Zornik E. Regulating the activity of a cardioacceleratory peptide. Peptides. 1999;20:1153–1158. doi: 10.1016/s0196-9781(99)00118-7. [DOI] [PubMed] [Google Scholar]
- 139.Baker JD, McNabb SL, Truman JW. The hormonal coordination of behavior and physiology at adult ecdysis in Drosophila melanogaster. J Exp Biol. 1999;202:3037–3048. doi: 10.1242/jeb.202.21.3037. [DOI] [PubMed] [Google Scholar]
- 140.Gammie SC, Truman JW. Eclosion hormone provides a link between ecdysis-triggering hormone and crustacean cardioactive peptide in the neuroendocrine cascade that controls ecdysis behavior. J Exp Biol. 1999;202:343–352. doi: 10.1242/jeb.202.4.343. [DOI] [PubMed] [Google Scholar]
- 141.Meng X, Wahlström G, Immonen T, Kolmer M, Tirronen M, Predel R, et al. The Drosophila hugin gene codes for myostimulatory and ecdysis-modifying neuropeptides. Mech Dev. 2002;117:5–13. doi: 10.1016/s0925-4773(02)00175-2. [DOI] [PubMed] [Google Scholar]
- 142.Chen MS, Obar RA, Schroeder CC, Austin TW, Poodry CA, Wadsworth SC, et al. Multiple forms of dynamin are encoded by shibire, a Drosophila gene involved in endocytosis. Nature. 1991;351:583–586. doi: 10.1038/351583a0. [DOI] [PubMed] [Google Scholar]
- 143.Masur SK, Kim YT, Wu CF. Reversible inhibition of endocytosis in cultured neurons from the Drosophila temperature-sensitive mutant shibire. J Neurogenet. 1990;6:191–206. doi: 10.3109/01677069009107110. [DOI] [PubMed] [Google Scholar]
- 144.Johnson E, Ringo J, Dowse H. Dynamin, encoded by shibire, is central to cardiac function. J Exp Zool. 2001;289:81–89. doi: 10.1002/1097-010x(20010201)289:2<81::aid-jez1>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 145.Johnson E, Sherry T, Ringo J, Dowse H. Modulation of the cardiac pacemaker of Drosophila: cellular mechanisms. J Comp Physiol. 2002;172:227–236. doi: 10.1007/s00360-001-0246-8. [DOI] [PubMed] [Google Scholar]
- 146.Sokolowski MB. Foraging strategies of Drosophila melanogaster: a chromosomal anaylsis. Behav Genet. 1980;10:291–302. doi: 10.1007/BF01067774. [DOI] [PubMed] [Google Scholar]
- 147.de Belle JS, Hilliker AJ, Sokolowski MB. Genetic localization of foraging (for): a major gene for larval behavior in Drosophila melanogaster. Genetics. 1989;123:157–163. doi: 10.1093/genetics/123.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Price MD, Merte J, Nichols R, Koladich PM, Tobe SS, Bendena WG. Drosophila melanogaster flatline encodes a myotropin orthologue to Manduca sexta allatostatin. Peptides. 2002;23:787–794. doi: 10.1016/s0196-9781(01)00649-0. [DOI] [PubMed] [Google Scholar]
- 149.Lalevée N, Monier B, Sénatore S, Perrin L, Sémériva M. Control of cardiac rhythm by ORK1, a Drosophila two-pore domain potassium channel. Curr Biol. 2006;16:1502–1508. doi: 10.1016/j.cub.2006.05.064. [DOI] [PubMed] [Google Scholar]
- 150.Dulcis D, Levine RB. Glutamatergic innervation of the heart initiates retrograde contractions in adult Drosophila melanogaster. J Neurosci. 2005;25:271–280. doi: 10.1523/JNEUROSCI.2906-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wasserthal LT. Drosophila flies combine periodic heartbeat reversal with a circulation in the anterior body mediated by a newly discovered anterior pair of ostial valves and ‘venous’ channels. J Exp Biol. 2007;210:3707–3719. doi: 10.1242/jeb.007864. [DOI] [PubMed] [Google Scholar]
- 152.Paternostro G, Vignola C, Bartsch DU, Omens JH, McCulloch AD, Reed JC. Age-associated cardiac dysfunction in Drosophila melanogaster. Circ Res. 2001;88:1053–1058. doi: 10.1161/hh1001.090857. [DOI] [PubMed] [Google Scholar]
- 153.Wessells RJ, Fitzgerald E, Cypser JR, Tatar M, Bodmer R. Insulin regulation of heart function in aging. Nat Genet. 2004;36:1275–1281. doi: 10.1038/ng1476. [DOI] [PubMed] [Google Scholar]
- 154.Bier E, Bodmer R. Drosophila, an emerging model for cardiac disease. Gene. 2004;342:1–11. doi: 10.1016/j.gene.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 155.Ocorr K, Reeves NL, Wessells RJ, Fink M, Chen HS, Akasaka T, et al. KCNQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. Proc Natl Acad Sci USA. 2007;104:3943–3948. doi: 10.1073/pnas.0609278104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Piazza N, Hayes M, Martin I, Duttaroy A, Grotewiel M, Wessells R. Multiple measures of functionality exhibit progressive decline in a parallel, stochastic fashion in Drosophila Sod2 null mutants. Biogerontology. 2009;10:637–648. doi: 10.1007/s10522-008-9210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wolf MJ, Amrein H, Izatt JA, Choma MA, Reedy MC, Rockman HA. Drosophila as a model for the identification of genes causing adult human heart disease. Proc Natl Acad Sci USA. 2006;31:1394–1399. doi: 10.1073/pnas.0507359103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zeitouni B, Sénatore S, Séverac D, Aknin C, Sémvériva M, Perrin L. Signalling pathways involved in adult heart formation revealed by gene expression profiling in Drosophila. PLoS Genet. 2007;3:1907–1921. doi: 10.1371/journal.pgen.0030174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kim YO, Park SJ, Balaban RS, Nirenberg M, Kim Y. A functional genomic screen for cardiogenic genes using RNA interference in developing Drosophila embryos. Proc Natl Acad Sci USA. 2003;10:159–164. doi: 10.1073/pnas.0307205101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Grossman TR, Gamliel A, Wessells RJ, Taghli-Lamallem O, Jepsen K, Ocorr K, et al. DSCAM and COL6A2 cooperate to produce down syndrome congenital heart defects (personal communication) doi: 10.1371/journal.pgen.1002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Koenig M, Monaco AP, Kunkel LM. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 1988;53:219–228. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- 162.Petrof BJ, Shrager JB, Stedman HH, Kelly AM, Sweeney HL. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc Natl Acad Sci USA. 1993;90:3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Straub V, Campbell KP. Muscular dystrophies and the dystrophin-glycoprotein complex. Curr Opin Neurol. 1997;10:168–175. doi: 10.1097/00019052-199704000-00016. [DOI] [PubMed] [Google Scholar]
- 164.Shcherbata HR, Yatsenko AS, Patterson L, Sood VD, Nudel U, Yaffe D, et al. Dissecting muscle and neuronal disorders in a Drosophila model of muscular dystrophy. EMBO J. 2007;26:481–493. doi: 10.1038/sj.emboj.7601503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Geier C, Perrot A, Ozcelik C, Binner P, Counsell D, Hoffmann K, et al. Mutations in the human muscle LIM protein gene in families with hypertrophic cardiomyopathy. Circulation. 2003;107:1390–1395. doi: 10.1161/01.cir.0000056522.82563.5f. [DOI] [PubMed] [Google Scholar]
- 166.Mohapatra B, Jimenez S, Lin JH, Bowles KR, Coveler KJ, Marx JG, et al. Mutations in the muscle LIM protein and alpha-actinin-2 genes in dilated cardiomyopathy and endocardial fibroelastosis. Mol Genet Metab. 2003;80:207–215. doi: 10.1016/s1096-7192(03)00142-2. [DOI] [PubMed] [Google Scholar]
- 167.Gutstein DE, Morley GE, Tamaddon H, Vaidya D, Schneider MD, Chen J, et al. Conduction slowing and sudden arrhythmic death in mice with cardiac-restricted inactivation of connexin43. Circ Res. 2001;88:333–339. doi: 10.1161/01.res.88.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Knöll R, Hoshijima M, Hoffman HM, Person V, Lorenzen-Schmidt I, Bang ML, et al. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111:943–955. doi: 10.1016/s0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- 169.Mery A, Taghli-Lamallem O, Clark KA, Beckerle MC, Wu X, Ocorr K, et al. The Drosophila muscle LIM protein, Mlp84B, is essential for cardiac function. J Exp Biol. 2008;211:15–23. doi: 10.1242/jeb.012435. [DOI] [PubMed] [Google Scholar]
- 170.Robbins J. KCNQ potassium channels: physiology, pathophysiology, and pharmacology. Pharmacol Ther. 2001;90:1–19. doi: 10.1016/s0163-7258(01)00116-4. [DOI] [PubMed] [Google Scholar]
- 171.Suzuki M, Sasaki N, Miki T, Sakamoto N, Ohmoto-Sekine Y, Tamagawa M, et al. Role of sarcolemmal K(ATP) channels in cardioprotection against ischemia/reperfusion injury in mice. J Clin Invest. 2002;109:509–516. doi: 10.1172/JCI14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hanley PJ, Gopalan KV, Lareau RA, Srivastava DK, von Meitzer M, Daut J. Beta-oxidation of 5-hydroxydecanoate, a putative blocker of mitochondrial ATP-sensitive potassium channels. J Physiol. 2003;547:387–393. doi: 10.1113/jphysiol.2002.037044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Oka T, Xu J, Molkentin JD. Re-employment of developmental transcription factors in adult heart disease. Semin Cell Dev Biol. 2007;18:117–131. doi: 10.1016/j.semcdb.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pehlivan T, Pober BR, Brueckner M, Garrett S, Slaugh R, Van Rheeden R, et al. GATA4 haploinsufficiency in patients with interstitial deletion of chromosomes region 8p23.1 and congenital heart disease. Am J Med Genet. 1999;83:201–206. [PubMed] [Google Scholar]
- 175.Rajagopal SK, Ma Q, Obler D, Shen J, Manichaikul A, Tomita-Mitchell A, et al. Spectrum of heart disease associated with murine and human GATA4 mutation. J Mol Cell Cardiol. 2007;43:677–685. doi: 10.1016/j.yjmcc.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Liang Q, Wiese RJ, Bueno OF, Dai YS, Markham BE, Molkentin JD. The transcription factor GATA4 is activated by extracellular signal-regulated kinase 1- and 2-mediated phosphorylation of serine 105 in cardiomyocytes. Mol Cell Biol. 2001;21:7460–7469. doi: 10.1128/MCB.21.21.7460-7469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bisping E, Ikeda S, Kong SW, Tarnavski O, Bodyak N, McMullen JR, et al. Gata4 is required for maintenance of postnatal cardiac function and protection from pressure overload-induced heart failure. Proc Natl Acad Sci USA. 2006;103:14471–14476. doi: 10.1073/pnas.0602543103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Qian L, Bodmer R. Partial loss of GATA factor Pannier impairs adult heart function in Drosophila. Hum Mol Genet. 2009;18:3153–3163. doi: 10.1093/hmg/ddp254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Qian L, Mohapatra B, Akasaka T, Liu J, Ocorr K, Towbin JA, et al. Transcription factor neuromancer/TBX20 is required for cardiac function in Drosophila with implications for human heart disease. Proc Natl Acad Sci USA. 2008;105:19833–19838. doi: 10.1073/pnas.0808705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yu L, Lee T, Lin N, Wolf MJ. Affecting Rhomboid-3 function causes a dilated heart in adult Drosophila. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000969. e1000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Khullar M, Al-Shudiefat AA, Ludke A, Binepal G, Singal PK. Oxidative stress: a key contributor to diabetic cardiomyopathy. Can J Physiol Pharmacol. 2010;88:233–240. doi: 10.1139/Y10-016. [DOI] [PubMed] [Google Scholar]
- 184.Subramanian S, Kalyanaraman B, Migrino RQ. Mitochondrially targeted antioxidants for the treatment of cardiovascular diseases. Recent Pat Cardiovasc Drug Discov. 2010;5:54–65. doi: 10.2174/157489010790192601. [DOI] [PubMed] [Google Scholar]
- 185.Lakshmi SV, Padmaja G, Kuppusamy P, Kutala VK. Oxidative stress in cardiovascular disease. Indian J Biochem Biophys. 2009;46:421–440. [PubMed] [Google Scholar]
- 186.Misra MK, Sarwat M, Bhakuni P, Tuteja R, Tuteja N. Oxidative stress and ischemic myocardial syndromes. Med Sci Monit. 2009;15:RA209–RA219. [PubMed] [Google Scholar]
- 187.Khaper N, Bryan S, Dhingra S, Singal R, Bajaj A, Pathak CM, et al. Targeting the vicious inflammation-oxidative stress cycle for the management of heart function. Antioxid Redox Signal. 2010;13:1033–1049. doi: 10.1089/ars.2009.2930. [DOI] [PubMed] [Google Scholar]
- 188.Gao G, Dudley SC., Jr Redox regulation, NF-kappaB, and atrial fibrillation. Antioxid Redox Signal. 2009;11:2265–2277. doi: 10.1089/ars.2009.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yan SF, Ramasamy R, Schmidt AM. The RAGE axis: a fundamental mechanism signaling danger to the vulnerable vasculature. Circ Res. 2010;106:842–853. doi: 10.1161/CIRCRESAHA.109.212217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Aikawa R, Nagai T, Tanaka M, Zou Y, Ishihara T, Takano H, et al. Reactive oxygen species in mechanical stress-induced cardiac hypertrophy. Biochem Biophys Res Commun. 2001;289:901–907. doi: 10.1006/bbrc.2001.6068. [DOI] [PubMed] [Google Scholar]
- 191.Date T, Belanger AJ, Mochizuki S, Sullivan JA, Liu LX, Scaria A, et al. Adenovirus-mediated expression of p35 prevents hypoxia/reoxygenation injury by reducing reactive oxygen species and caspase activity. Cardiovasc Res. 2002;55:309–319. doi: 10.1016/s0008-6363(02)00412-1. [DOI] [PubMed] [Google Scholar]
- 192.Pimentel DR, Adachi T, Ido Y, Helbeck T, Jiang B, Lee Y, et al. Strain-stimulated hypertrophy in cardiac myocytes is mediated by reactive oxygen species-dependent Ras S-glutathiolation. J Mol Cell Cardiol. 2006;41:613–622. doi: 10.1016/j.yjmcc.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 193.Adiga IK, Nair RR. Multiple signaling pathways coordinately mediate reactive oxygen species dependent cardiomyocyte hypertrophy. Cell Biochem Funct. 2008;26:346–351. doi: 10.1002/cbf.1449. [DOI] [PubMed] [Google Scholar]
- 194.Asimakis GK, Lick S, Patterson C. Postischemic recovery of contractile function is impaired in SOD2(+/−) but not SOD1(+/−) mouse hearts. Circulation. 2002;26:981–986. doi: 10.1161/hc0802.104502. [DOI] [PubMed] [Google Scholar]
- 195.Van Remmen H, Ikeno Y, Hamilton M, Pahlavani M, Wolf N, Thorpe SR, et al. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol Genomics. 2003;16:29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- 196.Lebovitz RM, Zhang H, Vogel H, Cartwright J, Jr, Dionne L, Lu N, et al. Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice. Proc Natl Acad Sci USA. 1996;93:9782–9787. doi: 10.1073/pnas.93.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Van Remmen H, Williams MD, Guo Z, Estlack L, Yang H, Carlson EJ, et al. Knockout mice heterozygous for Sod2 show alterations in cardiac mitochondrial function and apoptosis. Am J Physiol Heart Circ Physiol. 2001;281:1422–1432. doi: 10.1152/ajpheart.2001.281.3.H1422. [DOI] [PubMed] [Google Scholar]
- 198.Ding G, Fu M, Qin Q, Lewis W, Kim HW, Fukai T, et al. Cardiac peroxisome proliferator-activated receptor gamma is essential in protecting cardiomyocytes from oxidative damage. Cardiovasc Res. 2007;76:269–279. doi: 10.1016/j.cardiores.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 199.Kim IM, Wolf MJ, Rockman HA. Gene deletion screen for cardiomyopathy in adult Drosophila identifies a new notch ligand. Circ Res. 2010;106:1233–1243. doi: 10.1161/CIRCRESAHA.109.213785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.High FA, Epstein JA. The multifaceted role of Notch in cardiac development and disease. Nat Rev Genet. 2008;9:49–61. doi: 10.1038/nrg2279. [DOI] [PubMed] [Google Scholar]
- 201.Neely GG, Kuba K, Cammarato A, Isobe K, Amann S, Zhang L, et al. A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell. 2010;141:142–153. doi: 10.1016/j.cell.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Denis CL. Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics. 1984;108:833–844. doi: 10.1093/genetics/108.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Laribee RN, Shibata Y, Mersman DP, Collins SR, Kemmeren P, Roguev A, et al. CCR4/NOT complex associates with the proteasome and regulates histone methylation. Proc Natl Acad Sci USA. 2007;104:5836–5841. doi: 10.1073/pnas.0607996104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Peng W, Togawa C, Zhang K, Kurdistani SK. Regulators of cellular levels of histone acetylation in Saccharomyces cerevisiae. Genetics. 2008;179:277–289. doi: 10.1534/genetics.107.085068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Newton-Cheh C, Eligelsheim M, Rice KM, de Bakker PI, Yin X, Estrada K, et al. Common variants at ten loci influence QT interval duration in the QTGEN study. Nat Genet. 2009;41:399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lepperdinger G, Berger P, Breitenbach M, Frohlich KU, Grillari J, Grubeck-Loebenstein B, et al. The use of genetically engineered model systems for research on human aging. Front Biosci. 2008;1:7022–7031. doi: 10.2741/3207. [DOI] [PubMed] [Google Scholar]
- 207.Partridge L. Some highlights of research on aging with invertebrates. Aging Cell. 2009;8:509–513. doi: 10.1111/j.1474-9726.2009.00498.x. [DOI] [PubMed] [Google Scholar]
- 208.Katewa SD, Kapahi P. Dietary restriction and aging, 2009. Aging Cell. 2010;9:105–112. doi: 10.1111/j.1474-9726.2010.00552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Broughton S, Partridge L. Insulin/IGF-like signalling, the central nervous system and aging. Biochem J. 2009;418:1–12. doi: 10.1042/BJ20082102. [DOI] [PubMed] [Google Scholar]
- 210.Narasimhan SD, Yen K, Tissenbaum HA. Converging pathways in lifespan regulation. Curr Biol. 2009;19:657–666. doi: 10.1016/j.cub.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Stanfel MN, Shamieh LS, Kaeberlein M, Kennedy BK. The TOR pathway comes of age. Biochim Biophys Acta. 2009;1790:1067–1074. doi: 10.1016/j.bbagen.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Sudarsanam S, Johnson DE. Functional consequences of mTOR inhibition. Curr Opin Drug Discov Dev. 2010;13:31–40. [PubMed] [Google Scholar]
- 213.Maricq AV, Peckol E, Driscoll M, Bargmann CI. Mechanosensory signaling in C. elegans mediated by the GLR-1 glutamate receptor. Nature. 1995;378:78–81. doi: 10.1038/378078a0. [DOI] [PubMed] [Google Scholar]
- 214.Grotewiel MS, Martin I, Bhandari P, Cook-Wiens E. Functional senescence in Drosophila melanogaster. Ageing Res Rev. 2005;4:372–397. doi: 10.1016/j.arr.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 215.Libert S, Chao Y, Zwiener J, Pletcher SD. Realized immune response in enhanced in long-lived puc and chico mutants but is unaffected by dietary restriction. Mol Immunol. 2008;45:810–817. doi: 10.1016/j.molimm.2007.06.353. [DOI] [PubMed] [Google Scholar]
- 216.Rhodenizer D, Martin I, Bhandari P, Pletcher SD, Grotewiel M. Genetic and environmental factors impact age-related impairment of negative geotaxis in Drosophila by altering age-dependent climbing speed. Exp Gerontol. 2008;43:739–748. doi: 10.1016/j.exger.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Piazza N, Gosangi B, Devilla S, Arking R, Wessells R. Exercise-training in young Drosophila melanogaster reduces age-related decline in mobility and cardiac performance. PLoS One. 2009;4:e5886. doi: 10.1371/journal.pone.0005886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Lakatta EG, Levy D. Arterial and cardiac aging: major shareholders in cardiovascular disease enterprises: Part II: the aging heart in health: links to heart disease. Circulation. 2003;107:346–354. doi: 10.1161/01.cir.0000048893.62841.f7. [DOI] [PubMed] [Google Scholar]
- 219.Dai DF, Wessells RJ, Bodmer R, Rabinovitch PS. Cardiac aging. In: Wolf N, editor. The comparative biology of aging. New York: Springer; 2009. pp. 259–286. [Google Scholar]
- 220.Yarnell J, Yu S, Patterson C, Cambien F, Arveiler D, Amouyel P, et al. Family history, longevity, and risk of coronary heart disease: the PRIME study. Int J Epidemiol. 2003;32:71–77. doi: 10.1093/ije/dyg038. [DOI] [PubMed] [Google Scholar]
- 221.Kratz M. Dietary cholesterol, atherosclerosis and coronary heart disease. Handb Exp Pharmacol. 2005;170:195–213. doi: 10.1007/3-540-27661-0_6. [DOI] [PubMed] [Google Scholar]
- 222.Kuller LH. Nutrition, lipids, and cardiovascular disease. Nutr Rev. 2006;64:15–26. doi: 10.1111/j.1753-4887.2006.tb00230.x. [DOI] [PubMed] [Google Scholar]
- 223.Menotti A, Lanti M, Maiani G, Kromhout D. Determinants of longevity and all-cause mortality among middle-aged men. Role of 48 personal characteristics in a 40-year follow-up of Italian Rural Areas in the Seven Countries Study. Aging Clin Exp Res. 2006;18:394–406. doi: 10.1007/BF03324836. [DOI] [PubMed] [Google Scholar]
- 224.Rodeheffer RJ, Gerstenblith G, Becker LC, Fleg JL, Weisfeldt ML, Lakatta EG. Exercise cardiac output is maintained with advancing age in healthy human subjects: cardiac dilation and increased stroke volume compensate for a diminished heart rate. Circulation. 1984;69:203–213. doi: 10.1161/01.cir.69.2.203. [DOI] [PubMed] [Google Scholar]
- 225.Lakatta EG, Gerstenblith G, Angell CS, Shock NW, Weisfeldt ML. Prolonged contraction duration in aged myocardium. J Clin Invest. 1975;55:61–68. doi: 10.1172/JCI107918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Cheitlin MD. Cardiovascular physiology-changes with aging. Am J Geriatr Cardiol. 2003;12:9–13. doi: 10.1111/j.1076-7460.2003.01751.x. [DOI] [PubMed] [Google Scholar]
- 227.Sadek HA, Nulton-Persson AC, Szweda PA, Szweda LI. Cardiac ischemia/reperfusion, aging, and redox-dependent alterations in mitochondrial function. Arch Biochem Biophys. 2003;420:201–208. doi: 10.1016/j.abb.2003.09.029. [DOI] [PubMed] [Google Scholar]
- 228.Chiamvimonvat N. Diastolic dysfunction and the aging heart. J Mol Cell Cardiol. 2002;34:607–610. doi: 10.1006/jmcc.2002.2010. [DOI] [PubMed] [Google Scholar]
- 229.Bodyak N, Kang PM, Hiromura M, Sulijoadikusumo I, Horikoshi N, Khrapko K, et al. Gene expression profiling of the aging mouse cardiac myocytes. Nucleic Acids Res. 2002;30:3788–3794. doi: 10.1093/nar/gkf497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Hudson EK, Tsuchiya N, Hansford RG. Age-associated changes in mitochondrial mRNA expression and translation in the Wistar rat heart. Mech Ageing Dev. 1998;103:179–193. doi: 10.1016/s0047-6374(98)00043-8. [DOI] [PubMed] [Google Scholar]
- 231.Coleman R, Silbermann M, Gershon D, Reznick AZ. Giant mitochondria in the myocardium of aging and endurance-trained mice. Gerontology. 1987;33:34–39. doi: 10.1159/000212851. [DOI] [PubMed] [Google Scholar]
- 232.Muscari C, Guarnieri C, Biagetti L, Finelli C, Caldarera CM. Influence of age on oxidative damage in mitochondria of ischemic and reperfused rat hearts. Cardioscience. 1990;1:275–278. [PubMed] [Google Scholar]
- 233.Sohal RS, Sohal BH. Hydrogen peroxide release by mitochondria increases during aging. Mech Ageing Dev. 1991;57:187–202. doi: 10.1016/0047-6374(91)90034-w. [DOI] [PubMed] [Google Scholar]
- 234.Richardson A, Liu F, Adamo ML, Van Remmen H, Nelson JF. The role of insulin and insulin-like growth factor-I in mammalian ageing. Best Pract Res Clin Endocrinol Metab. 2004;18:393–406. doi: 10.1016/j.beem.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 235.Partridge L, Piper MD, Mair W. Dietary restriction in Drosophila. Mech Ageing Dev. 2005;126:938–950. doi: 10.1016/j.mad.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 236.Walker G, Houthoofd K, Vanfleteren JR, Gems D. Dietary restriction in C. elegans: from rate-of-living effects to nutrient sensing pathways. Mech Ageing Dev. 2005;126:929–937. doi: 10.1016/j.mad.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 237.Tatar M. The neuroendocrine regulation of Drosophila aging. Exp Gerontol. 2004;39:1745–1750. doi: 10.1016/j.exger.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 238.Katic M, Kennedy AR, Leykin I, Norris A, McGettrick A, Gesta S, et al. Mitochondrial gene expression and increased oxidative metabolism: role in increased lifespan of fat-specific insulin receptor knock-out mice. Aging Cell. 2007;8:827–839. doi: 10.1111/j.1474-9726.2007.00346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 240.Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Luong N, Davies CR, Wessells RJ, Graham SM, King MT, Veech R, et al. Activated FOXO-mediated insulin resistance is blocked by reduction of TOR activity. Cell Metab. 2006;4:133–142. doi: 10.1016/j.cmet.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 242.Jünger MA, Rintelen F, Stocker H, Wasserman JD, Végh M, Radimerski T, et al. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J Biol. 2003;2:20. doi: 10.1186/1475-4924-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Puig O, Marr MT, Ruhf ML, Tjlan R. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 2003;17:2006–2020. doi: 10.1101/gad.1098703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 245.Teleman AA, Chen YW, Cohen SM. 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes Dev. 2005;19:1844–1848. doi: 10.1101/gad.341505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.McNeill H, Craig GM, Bateman JM. Regulation of neurogenesis and epidermal growth factor receptor signaling by the insulin receptor/target of rapamycin pathway in Drosophila. Genetics. 2008;170:843–853. doi: 10.1534/genetics.107.083097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- 248.Linford NJ, Beyer RP, Gollahon K, Krajcik RA, Malloy VL, Demas V, et al. Transcriptional response to aging and caloric restriction in heart and adipose tissue. Aging Cell. 2007;6:673–688. doi: 10.1111/j.1474-9726.2007.00319.x. [DOI] [PubMed] [Google Scholar]
- 249.Sample J, Cleland JG, Seymour AM. Metabolic remodeling in the aging heart. J Mol Cell Cardiol. 2006;40:56–63. doi: 10.1016/j.yjmcc.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 250.Vihervaara T, Puig O. dFOXO regulates transcription of a Drosophila acid lipase. J Mol Biol. 2008;376:1215–1223. doi: 10.1016/j.jmb.2007.12.042. [DOI] [PubMed] [Google Scholar]
- 251.Belke DD, Betuing S, Tuttle MJ, Graveleau C, Young ME, Pham M, et al. Insulin signaling coordinately regulates cardiac size, metabolism, and contractile protein isoform expression. J Clin Invest. 2002;109:629–639. doi: 10.1172/JCI13946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Shioi T, McMullen JR, Kang PM, Douglas PS, Obata T, Franke TF, et al. Akt/protein kinase B promotes organ growth in transgenic mice. Mol Cell Biol. 2002;22:2799–2809. doi: 10.1128/MCB.22.8.2799-2809.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.McMullen JR, Shioi T, Zhang L, Tarnavski O, Sherwood MC, Dorfman AL, et al. Deletion of ribosomal S6 kinases does not attenuate pathological, physiological, or insulin-like growth factor 1 receptor-phosphoinositide 3-kinase-induced cardiac hypertrophy. Mol Cell Biol. 2004;24:6231–6240. doi: 10.1128/MCB.24.14.6231-6240.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.DeBosch B, Treskov I, Lupu TS, Weinheimer C, Kovacs A, Courtois M, et al. Akt1 is required for physiological cardiac growth. Circulation. 2006;113:2097–2104. doi: 10.1161/CIRCULATIONAHA.105.595231. [DOI] [PubMed] [Google Scholar]
- 255.Li Q, Li B, Wang X, Leri A, Jana KP, Liu Y, et al. Overexpression of insulin-like growth factor-1 in mice protects from myocyte death after infarction, attenuating ventricular dilation, wall stress, and cardiac hypertrophy. J Clin Invest. 1997;15:1991–1999. doi: 10.1172/JCI119730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Delaughter MC, Taffet GE, Fiorotto ML, Entman ML, Schwartz RJ. Local insulin-like growth factor I expression induces physiologic, then pathologic, cardiac hypertrophy in transgenic mice. FASEB J. 1999;13:1923–1929. doi: 10.1096/fasebj.13.14.1923. [DOI] [PubMed] [Google Scholar]
- 257.Winn N, Paul A, Musaró A, Rosenthal N. Insulin-like growth factor isoforms in skeletal muscle aging, regeneration, and disease. Cold Spring Harb Symp Quant Biol. 2002;67:507–518. doi: 10.1101/sqb.2002.67.507. [DOI] [PubMed] [Google Scholar]
- 258.Higgins M, Kannel W, Garrison R, Pinsky J, Stokes J., 3rd Hazards of obesity—the Framingham experience. Acta Med Scand Suppl. 1988;723:23–36. doi: 10.1111/j.0954-6820.1987.tb05925.x. [DOI] [PubMed] [Google Scholar]
- 259.Kannel WB, Cupples LA, Ramaswami R, Stokes J, 3rd, Kreger BE, Higgins M. Regional obesity and risk of cardiovascular disease: the Framingham study. J Clin Epidemiol. 1991;44:183–190. doi: 10.1016/0895-4356(91)90265-b. [DOI] [PubMed] [Google Scholar]
- 260.McGill HC, Jr, McMahan CA, Herderick EE, Zieske AW, Malcom GT, Tracy RE, et al. Pathobiological Determinants of Atherosclerosis in Youth (PDAY) Research Group. Obesity accelerates the progression of coronary atherosclerosis in young men. Circulation. 2002;105:2712–2718. doi: 10.1161/01.cir.0000018121.67607.ce. [DOI] [PubMed] [Google Scholar]
- 261.King VL, Hatch NW, Chan HW, de Beer MC, de Beer FC, Tannock LR. A murine model of obesity with accelerated atherosclerosis. Obesity. 2010;18:35–41. doi: 10.1038/oby.2009.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Krauss RM, Winston M, Fletcher BJ, Grundy SM. Obesity: impact on cardiovascular disease. Circulation. 1998;98:1472–1476. [PubMed] [Google Scholar]
- 264.Narkiewicz K. Obesity and hypertension—the issue is more complex than we thought. Nephrol Dial Transplant. 2006;21:264–267. doi: 10.1093/ndt/gfi290. [DOI] [PubMed] [Google Scholar]
- 265.Howard BV, Ruotolo G, Robbins DC. Obesity and dyslipidemia. Endocrinol Metab Clin North Am. 2003;32:855–867. doi: 10.1016/s0889-8529(03)00073-2. [DOI] [PubMed] [Google Scholar]
- 266.Koves TR, Li P, An J, Akimoto T, Slentz D, Ilkayeva O, et al. Peroxisome proliferator-activated receptor-gamma co-activator 1alpha-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J Biol Chem. 2005;280:33588–33598. doi: 10.1074/jbc.M507621200. [DOI] [PubMed] [Google Scholar]
- 267.Dhahbi JM, Tsuchiya T, Kim HJ, Mote PL, Spindler SR. Gene expression and physiologic responses of the heart to the initiation and withdrawal of caloric restriction. J Gerontol A Biol Sci Med Sci. 2006;61:218–231. doi: 10.1093/gerona/61.3.218. [DOI] [PubMed] [Google Scholar]
- 268.Hsiu-Chiang C, Kovacs A, Blanton RM, Han X, Courtois M, Weinheimer CJ, et al. Transgenic expression of fatty acid transport protein 1 in the heart causes lipotoxic cardiomyopathy. Circ Res. 2005;96:225–233. doi: 10.1161/01.RES.0000154079.20681.B9. [DOI] [PubMed] [Google Scholar]
- 269.Kavanagh T, Myers MG, Baigrie RS, Mertens DJ, Sawyer P, Shephard RJ. Quality of life and cardiorespiratory function in chronic heart failure: effects of 12 months’ aerobic training. Heart. 1996;76:42–49. doi: 10.1136/hrt.76.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Erbs S, Höllriegel R, Linke A, Beck EB, Adams V, Gielen S, et al. Exercise training in patients with advanced chronic heart failure (NYHA IIIb) promotes restoration of peripheral vasomotor function, induction of endogenous regeneration, and improvement of left-ventricular function. Circ Heart Fail. 2010;3:486–494. doi: 10.1161/CIRCHEARTFAILURE.109.868992. [DOI] [PubMed] [Google Scholar]
- 271.Delbin MA, Antunes E, Zanesco A. Role of exercise training on pulmonary ischemia/reperfusion and inflammatory response. Rev Bras Cir Cardiovasc. 2009;24:552–561. doi: 10.1590/s0102-76382009000500017. [DOI] [PubMed] [Google Scholar]
- 272.Suvorava T, Lauer N, Kojda G. Physical inactivity causes endothelial dysfunction in healthy young mice. J Am Coll Cardiol. 2004;44:1320–1327. doi: 10.1016/j.jacc.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 273.Derumeaux G, Ichinose F, Raher MJ, Morgan JG, Coman T, Lee C, et al. Myocardial alterations in senescent mice and effect of exercise training: a strain rate imaging study. Circ Cardiovasc Imaging. 2008;1:227–234. doi: 10.1161/CIRCIMAGING.107.745919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Gollnick PD, Saltin B. Significance of skeletal muscle oxidative enzyme enhancement with endurance training. Clin Physiol. 1982;2:1–12. doi: 10.1111/j.1475-097x.1982.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 275.Davidson SR, Burnett M, Hoffman-Goetz L. Training effects in mice after long-term voluntary exercise. Med Sci Sports Exerc. 2006;38:250–255. doi: 10.1249/01.mss.0000183179.86594.4f. [DOI] [PubMed] [Google Scholar]
- 276.Soh JW, Hotic S, Arking R. Dietary restriction in Drosophila is dependent on mitochondrial efficiency and constrained by pre-existing extended longevity. Mech Ageing Dev. 2007;128:581–593. doi: 10.1016/j.mad.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 277.Skorupa DA, Dervisefendic A, Zwiener J, Pletcher SD. Dietary composition specifies consumption, obesity, and lifespan in Drosophila melanogaster. Aging Cell. 2008;7:478–490. doi: 10.1111/j.1474-9726.2008.00400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Bhandari P, Jones MA, Martin I, Grotewiel MS. Dietary restriction alters demographic but not behavioral aging in Drosophila. Aging Cell. 2007;6:631–637. doi: 10.1111/j.1474-9726.2007.00320.x. [DOI] [PubMed] [Google Scholar]
- 279.Burger JM, Promislow DE. Are functional and demographic senescence genetically independent? Exp Gerontol. 2006;41:1108–1116. doi: 10.1016/j.exger.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 280.Holloszy JO, Booth FW. Biochemical adaptations to endurance exercise in muscle. Annu Rev Physiol. 1976;38:273–291. doi: 10.1146/annurev.ph.38.030176.001421. [DOI] [PubMed] [Google Scholar]